Abstract
How complex are the rules by which a protein’s sequence determines its function? High-order epistatic interactions among residues are thought to be pervasive, suggesting an idiosyncratic and unpredictable sequence-function relationship. But many prior studies may have overestimated epistasis, because they analyzed sequence-function relationships relative to a single reference sequence—which causes measurement noise and local idiosyncrasies to snowball into high-order epistasis—or they did not fully account for global nonlinearities. Here we present a reference-free method that jointly infers specific epistatic interactions and global nonlinearity using a bird’s-eye view of sequence space. This technique yields the simplest explanation of sequence-function relationships and is more robust than existing methods to measurement noise, missing data, and model misspecification. We reanalyze 20 experimental datasets and find that context-independent amino acid effects and pairwise interactions, along with a simple nonlinearity to account for limited dynamic range, explain a median of 96% of phenotypic variance and over 92% in every case. Only a tiny fraction of genotypes are strongly affected by higher-order epistasis. Sequence-function relationships are also sparse: a miniscule fraction of amino acids and interactions account for 90% of phenotypic variance. Sequence-function causality across these datasets is therefore simple, opening the way for tractable approaches to characterize proteins’ genetic architecture.
Subject terms: Epistasis, Protein function predictions, Statistical methods
Understanding protein sequence-function relationships is complicated by high order epistatic interactions among residues, although the extent of these interactions remains uncertain. Here, the authors present a reference-free method which suggests that sequence-function relationships are relatively simple, with little influence from high order epistatic interactions.
Introduction
If we had complete knowledge of a protein’s genetic architecture—the set of causal rules by which its sequence determines its function—we could predict and understand the functional and evolutionary consequences of any variant sequence. Whether such knowledge is possible in practice depends on the extent of epistatic interactions. If all residues in a protein acted independently, knowing the effects of point mutations on any genetic background would suffice to understand the functional contribution of every possible residue and predict the function of every possible sequence; moreover, any mutation’s evolutionary fate would be independent of the genetic context in which it may arise. A genetic architecture of such extreme simplicity could be reconstructed by moderate-throughput experiments. At the opposite extreme, pervasive high-order epistasis would cause a mutation’s effect to vary idiosyncratically across genetic backgrounds, and the evolutionary fate of any mutation would change unpredictably with each sequence substitution. Assessing the genetic architecture would require exhaustive characterization of all possible sequences.
High-throughput methods for characterizing large libraries of protein variants have made it possible to directly assess the complexity of sequence-function relationships. Studies to date disagree on the extent of epistasis within proteins. Some report extensive high-order interactions1–9, while others find that they account for less than 10% of variance in phenotype among sequences10–20. Even pairwise interactions are strong and widespread in some studies7,14,20–24 but weak and rare in others11,18,25,26. Some studies report a sparse genetic architecture in which a small fraction of possible amino acids and interactions dictate the function15,18, but others point to a more complex mapping in which many determinants of small effect contribute to function7,20,22,24.
These discrepancies may arise from the use of different methods to characterize epistasis. Two aspects of widely used approaches may lead to unnecessarily complex descriptions of genetic architecture. First, many studies have analyzed combinatorial mutagenesis data using a reference-based framework, which designates a single sequence as wild-type. If a mutation’s effect when introduced into a variant differs from its effect on the wild-type, the deviation is attributed to epistasis, even though this may reflect local idiosyncrasies in the wild-type architecture or propagation of error from measurement noise27. Second, many studies have not fully accounted for global nonlinearities in the relationship between sequence and function28. When this nonspecific epistasis is not incorporated, pervasive and complex amino acid interactions must be invoked to explain why every mutation’s effect varies across genetic backgrounds13,29,30.
Advances have been made in both areas of concern, but current methods have major limitations. Fourier analysis31,32, also known as simplex encoding33 or graph Fourier transform34, is reference-free: instead of focusing on the effects of states on a particular sequence, it captures their average effects across sequence space. But the application of Fourier analysis has been mostly limited to datasets that sample just two states per site, because the multi-state formalism is complicated and has no straightforward interpretation. For example, when all 20 amino acids are assessed, they must be recoded into 19 Fourier coefficients using Hadamard matrices or graph Fourier bases, and the resulting model terms do not correspond to any genetically or biochemically meaningful quantities. Another formalism, background-averaged analysis2,27,35,36, is a modified reference-based analysis in which the effects of mutations are averaged across all genetic backgrounds at other sites. It is less sensitive to idiosyncrasy around any particular sequence, but an arbitrary reference state is still chosen for each site. Implementing background-averaged analysis also requires large Hadamard matrices, and the multi-state formalism has only recently been derived36.
Existing methods to address nonspecific epistasis also have limitations. Sometimes the protein’s phenotype can be measured or transformed onto a scale that is expected to be less affected by nonspecific epistasis, such as thermodynamic free energy18,37,38. But protein phenotypes can scale nonadditively because of many causes, and the transformation required to remove nonspecific epistasis are seldom known in advance39. Even free energy must be measured using techniques that have limited dynamic range and thus entail nonlinearity. Several studies have addressed this issue by inferring a transformation that maximizes the fit of a first-order genetic model11,13,15,19,25,40,41, but many of these approaches rely on rigid convex or concave transformations that cannot incorporate common forms of nonlinearity, such as the bounding of measured phenotypes within lower and upper limits. Some studies employ flexible splines or neural networks11,25,40, but these approaches have not been widely adopted because they are cumbersome to implement and interpret.
Here we develop a simple and powerful reference-free framework that can be coupled with an effective model of nonspecific epistasis and applied to any number of states. We first explain our reference-free approach and show how it differs from existing frameworks. We then systematically reanalyze available combinatorial mutagenesis datasets to assess the complexity of sequence-function relationship. Finally, we explore strategies to infer the genetic architecture when only a small fraction of possible sequences can be experimentally characterized.
Results
We have several goals in dissecting a protein’s genetic architecture. First, we would like to know how sequence determines function across the space of all possible variants, including the effects and interactions of each amino acid and any systematic nonlinearity in sequence-function map. Second, we would like to use these fine-scale causal rules for macroscopic descriptions of the genetic architecture, such as the overall importance of effects at each epistatic order or of sequence variation at each site or set of sites. Third, knowing the rules of genetic architecture inferred from a sample of genotypes could allow us to predict the function of uncharacterized variants. Finally, once the rules of genetic architecture are known, they can be interpreted in biochemical and structural terms to understand the physical mechanisms by which sequence shapes function. They also explain why protein phenotypes are distributed as they are across sequence space, which shapes the trajectory and outcome of evolution. In these ways, analyzing genetic architecture allows us to deepen our understanding of how and why a protein works as it does.
To achieve these ends, an ideal method of analysis would meet three criteria: (1) the structure of the model yields a transparently interpretable description of the causal rules by which sequence determines function; (2) the model’s terms can be accurately estimated from real datasets, which usually contain experimental noise and are missing measurement for some variants; and (3) the model decomposes the genotype-phenotype relationship parsimoniously, explaining the observed data while minimizing gratuitous complexity.
Reference-free analysis of genetic architecture
We designed reference-free analysis (RFA) to achieve these goals. It uses Fisher’s statistical formalism for decomposing genetic architecture42—and for analyzing interaction effects in factorial designs more generally—and applies it to protein sequence space.
RFA takes a bird’s-eye view of genetic architecture. The causal factors are sequence states rather than mutations, and their effects on the phenotype are defined relative to the global average of all variants (Fig. 1a). The formalism is simple and interpretable. The zero-order term, which affects all genotypes, is the mean phenotype across sequence space. The first-order effect of a state at a site is its context-independent effect on the phenotype, calculated as the difference between the mean phenotype of all sequences containing that state and the global mean. The epistatic effect of a combination of states is the difference between the mean phenotype of all sequences containing the combination and that expected given the lower-order effects. The phenotype of any genotype is simply the sum of the effects of the genetic states in its sequence (Fig. 1b).
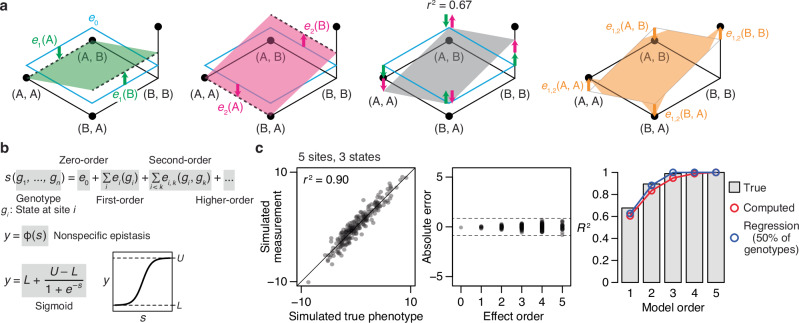
Fig. 1. Reference-free analysis (RFA) of genetic architecture.
a Illustration of RFA on a 2-site, 2-state genetic architecture. The four possible genotypes are arranged on a plane with their phenotype indicated by elevation. (First panel) The zero-order effect (e0) is the mean phenotype of all genotypes, marked by the clear plane with cyan edges. The first-order effect of state A or B at site 1 [e1(A) or e1(B), green arrows] measures how the mean phenotype of all genotypes containing the state (dashed line) differs from the global mean. The green plane predicts the phenotype based on the state at site 1. (Second panel) First-order effects at site 2 are defined similarly and shown in pink. (Third panel) The first-order model predicts phenotypes by summing the first-order effects of all genetic states plus the global mean, shown as the gray plane tilted in both dimensions; the fraction of phenotypic variance explained is shown. (Fourth panel) The pairwise interaction between states A and B at sites 1 and 2 [e1, 2(A, B)] measures how the mean phenotype of all genotypes containing the two states [here just one genotype (A, B)] differs from the first-order prediction. b We implement RFA with a nonlinear link function to incorporate nonspecific epistasis. Each variant’s genetic score (s) is the sum of the effects of its genetic states. The link function transforms s of each variant into its phenotype, y. Although the link function can take any form, here we use a simple sigmoid defined by two parameters representing the upper and lower bounds of the measurable phenotype. c (Left) A 5-site, 3-state genetic architecture was simulated by drawing reference-based effects from the standard normal distribution (but setting all fifth-order effects to zero); a small amount of simulated noise was added to the phenotypes. (Middle) Absolute error of RFA terms computed from the simulated measurements. Dashed lines, mean absolute error of individual phenotypes. Supplementary Fig. 1 shows the individual inferred terms. (Right) The fraction of phenotypic variance explained by the true, directly computed, and regression-estimated RFA terms. Supplementary Section 1.1 analyzes additional simulated genetic architectures.
This way of dissecting the sequence-function relationship gives RFA several desirable properties. First, RFA offers a maximally efficient description of the global sequence-function relationship. An RFA model truncated at any epistatic order captures the maximum amount of phenotypic variance that can be captured by any linear model of the same order (Supplementary Section 2.6). Consider all zero-order models, which predict the phenotype of every sequence by a single number. The RFA zero-order term is the mean phenotype of all sequences and is therefore the best predictor in the sense of minimizing the total squared error. The first-order RFA model predicts each variant’s phenotype as the sum of the first-order effects of its constituent states and the global mean. This predictor again achieves the minimum total squared error among all possible first-order models and therefore explains the maximum possible amount of phenotypic variance. This property continues as the model order increases. To the greatest extent possible, RFA explains the sequence-function relationship by low-order causal factors, which are relatively few in number and apply most broadly, rather than by high-order factors, which at the limit explain every single data point as the result of a unique set of idiosyncratic causes.
Second, RFA is robust to measurement noise, because its terms are defined using average phenotypes over sets of genotypes. To illustrate this property, we simulated a genetic architecture in which phenotypic measurements are determined by up to fourth-order effects plus a moderate amount of measurement noise (Fig. 1c). The RFA terms computed from the simulated measurements accurately estimate the true effects; errors in the estimated terms are smaller than the noise in the individual phenotypic measurements, even for the highest-order terms. The fraction of phenotypic variance explained by the computed terms is also accurate.
Third, when data are partially sampled, RFA models can be accurately estimated by least-squares regression. When 50% of genotypes are missing from the simulated example, the estimated terms of the model and the variance partition are highly accurate (Fig. 1c, Supplementary Fig. 1). RFA can be accurately estimated by regression because its true terms minimize the sum of squared error across all genotypes, so least-squares estimates converge on the true values as long as noise and sampling are unbiased. Truncated models can be estimated accurately because the patterns of variation produced by the unmodeled higher-order interactions appear as noise around lower-order predictions, so they cannot be absorbed by the model (Supplementary Section 2.9).
Shortcomings of reference-based analysis
Reference-based analysis (RBA) is less suited in both theory and practice for analyzing a protein’s global genetic architecture. The causal genetic factors in RBA are not amino acid states but mutations when introduced into a designated wild-type reference sequence (Fig. 2a). Each first-order effect is defined as the difference in phenotype between the one variant that contains that single mutation and the wild-type. Each second-order interaction effect is the difference between the phenotype of the one double mutant and that expected from the sum of the first-order effects. This structure continues for higher-order mutants, invoking interactions whenever one variant’s phenotype deviates from the sum of lower-order effects.
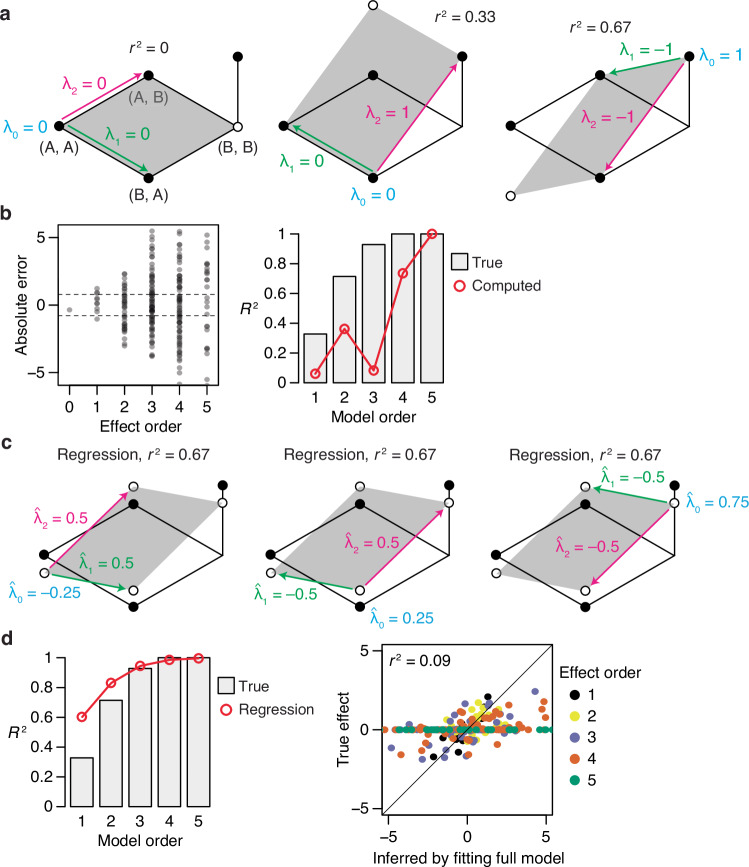
Fig. 2. Reference-based analysis (RBA) is unsuitable for inferring global genetic architecture.
a The apparent complexity of genetic architecture varies depending on the choice of wild-type genotype. A first-order RBA model is directly fitted to the genetic architecture in Fig. 1a, with (A, A), (B, A), or (B, B) as wild-type. λ0, wild-type phenotype; λ1, λ2, first-order effects of mutations at sites 1 and 2; empty circle, predicted phenotype of the double mutant; r2, fraction of phenotypic variance explained by the model. b (Left) Absolute error of RBA terms computed from the simulated measurements with a small amount of noise in Fig. 1c. Dashed lines, mean absolute error of individual phenotypes. (Right) Fraction of phenotypic variance explained by the true and computed RBA terms. c Regression yields incorrect estimates of RBA terms and overestimates the fraction of phenotypic variance they explain. The first panel fits the first-order RBA model defined with respect to (A, A) by regression; the estimated terms, predicted phenotype for each genotype, and r2 are shown. The next two panels repeat the analysis by choosing (B, A) or (B, B) as wild-type. d (Left) Fraction of phenotypic variance explained by the true and regression-estimated RBA terms for the simulated measurements in Fig. 1c. (Right) RBA terms estimated by fitting the full model, compared with their true values.
RBA is useful in principle if one is interested in the effects and interactions of mutations when introduced into a particular sequence of interest43,44. Its structure is not suited, however, for understanding how sequence determines function across the space of possible variants. First, the wild-type-centric view means that the genetic architecture varies depending on the choice of wild-type genotype; in the example of Fig. 2a, first-order effects may make zero contribution to phenotypic variance or explain most of it, depending on the reference sequence chosen, and the pairwise interaction switches in both magnitude and sign. Second, the RBA formalism implies that proteins containing wild-type residues are unaffected by any of those states. The wild-type protein has no genetic determinants whatsoever because it contains no mutations. A point mutant is subject to the first-order effect of one mutation but is by definition unaffected by epistasis. A double mutant is shaped by one pairwise interaction but no higher-order interactions, and so on. In reality, these proteins have a genetic architecture just as interesting and complex as those of sequences distant from the wild-type. Finally, RBA efficiently explains phenotypic variation in the neighborhood of the reference sequence but produces a less parsimonious description of a protein’s genetic architecture over sequence space as a whole. In the absence of noise, the zero-order RBA term predicts the wild-type sequence with perfect accuracy but is less accurate across all sequences than the global mean. The first-order RBA terms perfectly predict the point mutants, and the second-order terms exactly predict the double mutants, but across the vast number of other sequences these terms are less accurate predictors and thus leave more variation to be explained by higher-order terms. RBA thus infers a genetic architecture that is more complicated and idiosyncratic than is necessary to explain the distribution of phenotype across sequence space.
A second concern is that in practice, the RBA model cannot be accurately estimated from noisy and partially sampled datasets, either by exact computation or by regression. Exact RBA is hypersensitive to measurement noise: each term is calculated as a chain of sums and subtractions of phenotypic measurements, so the noise of each measurement propagates when estimating high-order terms. This phenomenon is illustrated in Fig. 2b: using the same simulated measurements in Fig. 1c, the calculated RBA terms are dramatically incorrect, with errors larger than that of the individual measurements and snowball as the order increases. When the computed terms at each order are used to predict the phenotype, high-order epistasis appears to be far more important than it actually is under the true RBA architecture (Fig. 2b). Exact computation of RBA is also incompatible with missing data: if a variant is unmeasured, it becomes impossible to compute the effect of the mutation and all the interactions that involve it.
To cope with this limitation of exact estimation of RBA models, an alternative approach has been to use least-squares regression: a series of truncated RBA models are fit to the data to estimate the variance explained by the model at each order, and the complete RBA model is then used to estimate the individual effects7,19,20. This procedure yields biased estimates that oversimplify the genetic architecture under the true RBA model. Consider the simple example of Fig. 2c, setting (A, A) as the reference genotype. In the true RBA model, first-order terms explain no variance, all of which is caused by the pairwise interaction; when fit by regression, however, 67% of variance is explained at the first order, leaving only 33% attributable to the interaction. The estimated terms of the truncated first-order model are also inflated in magnitude. Another anomaly is that the fraction of first-order variance estimated by regression is the same irrespective of the reference genotype chosen, whereas under the true RBA architecture this quantity depends strongly on the reference.
Measurement noise further undermines the accuracy of RBA-regression. For the simulation with mild noise (Fig. 1c), the terms estimated by RBA-regression using each truncated model deviate substantially from the true RBA terms (Supplementary Fig. 1), and the variance partition implies a genetic architecture far simpler than the true RBA architecture (Fig. 2d). When the complete model is finally fit, the estimated terms deviate wildly from the true terms, with particularly large errors for the high-order interactions. If taken at face value, these two observations would suggest the anomalous conclusion that high-order interactions are widespread and very large but somehow contribute negligibly to phenotypic variance.
The bias of RBA-regression has been previously established29, and it exemplifies a general bias that arises whenever regression is used to fit uncentered interaction models in which variables are correlated across orders45. RBA-regression oversimplifies the RBA variance partition because regression finds the parameter values that minimize the sum of squared error between observed and model-predicted phenotypes across all variants. The true RBA terms are defined only by the phenotypes of mutants at that order and below; they do not minimize the squared error for higher-order variants if those variants are affected by noise or higher-order interactions. Regression therefore optimizes the low-order terms to fit the variation caused by effects that are excluded from the truncated model; the resulting estimates of low-order terms are incorrect and the fraction of variance they explain is overestimated. When the complete model is used for the final coefficient estimates, using regression is identical to exact computation, so measurement noise propagates into wildly inflated epistatic terms. Because RBA-regression produces biased and anomalous results, we do not explore the behavior of this method further.
RFA is more interpretable and robust than other global formalisms
Like RFA, Fourier analysis (FA) and background-averaging (BA) take a bird’s-eye view of genetic architecture, but RFA has a more interpretable structure and is more robust to missing data. In RFA, each model term directly expresses the global phenotypic effect of an amino acid or combination, and a variant’s phenotype is the sum of the effects of only the states in its sequence (Fig. 3a). In FA, each sequence state is recoded as a series of (1, –1) coordinates over (q – 1) Fourier dimensions, where q is the number of states (Supplementary Section 1.2). Each first-order Fourier term represents the effect of having a positive or negative coordinate along one of these dimensions. If more than two amino acid states are present, Fourier terms have no straightforward genetic or biochemical meaning. With 20 states, for example, the effect of each amino acid at a site is a uniquely signed sum over 19 first-order Fourier terms, each pairwise amino acid interaction is a signed sum over 361 second-order Fourier terms, and so on. The phenotype of any variant is therefore a sum over every term in the entire model (Fig. 3a). This complex mapping makes it difficult to understand how a variant’s phenotype arises from its sequence.
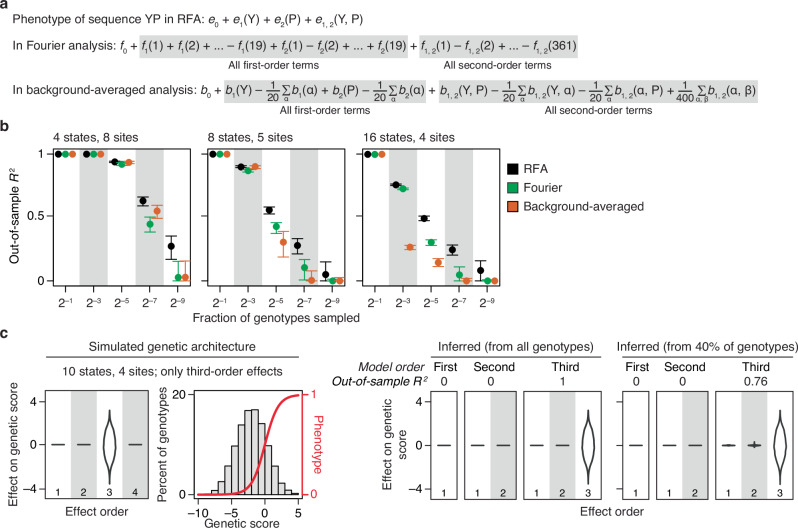
Fig. 3. RFA is more interpretable and robust than Fourier analysis (FA) and background-averaged analysis (BA).
a Mapping of model terms to phenotype in RFA, FA, and BA shown for a two-amino-acid sequence YP. fi(k) and fi, j(k), k-th first- and second-order FA terms for site i and site-pair (i, j). bi(α) and bi, j(α, β), first- and second-order BA terms for amino acids α and β; the summations shown are over all 19 mutant amino acids. b RFA is more robust to missing genotypes than FA and BA. Genetic architectures with the indicated number of states and sites were simulated by drawing up to third-order reference-based effects from the standard normal distribution. Third-order models were fit to a varying fraction of genotypes and evaluated by predicting the phenotypes of the remaining genotypes. The mean and 95% interval of out-of-sample R2 across 200 trials are shown. c RFA with nonspecific epistasis does not misinterpret high-order epistasis as clusters of low-order interactions. Phenotypes were simulated with only third-order determinants (distribution shown in the first panel) under a sigmoid link function (second panel). First-, second-, and third-order reference-free effects were inferred from all genotypes or a random 40%. The distribution of inferred effects and the variance they explain are shown in the right panels.
In BA, each term is defined as the average effect of a state (or combination) relative to some arbitrarily chosen reference state (typically the first “letter” in the alphabet of sequence states), and the phenotype is a weighted sum over all terms in the entire model, including the coefficients for states not in the genotype of interest (Fig. 3a). As in FA, the effects of each amino acid or combination can be derived from the model terms only via an elaborate set of equations when more than two amino acids per site are considered (Supplementary Section 1.3).
FA and BA models can be estimated by regression, but RFA is more robust to partial sampling. We simulated genetic architectures of varying shape and removed a variable fraction of genotype measurements; we then fit the three models to the remaining sequences by regression and predicted the phenotypes of the excluded genotypes using the estimated models (Fig. 3b). When there are only four states per site, all models have high predictive accuracy, which declines only when the fraction of sampled sequences drops below 1%, at which point RFA is slightly more accurate. When there are 16 states, however, RFA is much more robust than BA, the accuracy of which degrades rapidly as sample size shrinks; it is also more robust than FA, but to a smaller extent. RFA is more robust to missing genotypes because the phenotype of each unsampled variant is predicted as the sum of only the terms for its genetic states; FA and BA predict the phenotype as a weighted sum of all model terms, so the error associated with every model term propagates to all genotypes. This difference is exacerbated as more states are considered, because the total number of terms increases exponentially with the number of states.
Incorporating nonspecific epistasis
Nonspecific epistasis can be incorporated into RFA by using a generalized linear model in which the phenotype of a variant is a nonlinear transformation of the effects of its genetic states25 (Fig. 1b). The total effect of a variant’s genetic states is its genetic score, and its phenotype is a nonlinear transformation of the score. The parameters of the link function from score to phenotype can be inferred by regression in a joint fitting procedure along with the specific RFA genetic effects.
We explore using a sigmoid link function to incorporate nonspecific epistasis (Fig. 1b). We reasoned that most DMS datasets are likely to involve a limited dynamic range, and the sigmoid function can account for the diminishing effects of amino acid states in variants that are near the minimum or maximum of this range. The sigmoid also contains only two free parameters, which facilitates estimation and interpretation. Although the mechanisms and precise forms of nonlinearity are likely to be complex and vary among datasets, we explore here whether this simple and common form of nonspecific epistasis might be an important factor in protein genetic architecture.
We used simulations to determine whether regression can be used to accurately estimate the RFA model coupled with sigmoid nonspecific epistasis. We were particularly interested in whether this procedure might oversimplify the genetic architecture by misinterpreting true high-order interactions as nonspecific epistasis or as clusters of low-order interactions. We first simulated phenotypes under a genetic architecture that contains only third-order effects plus nonspecific epistasis and then fitted RFA models (with the sigmoid link) truncated at various orders (Fig. 3c). The first- and second-order truncated models correctly explain no phenotypic variance and detect no first- or second-order effects. When the third-order model is used, all variance is correctly attributed to third-order effects. Similar results hold when variants are only partially sampled.
We next explored whether including the link function might absorb specific epistasis when the true phenotypes are unaffected by global nonlinearity. We simulated measurements with specific epistasis derived from a real DMS dataset but imposed no nonspecific epistasis; we then fitted the RFA model with and without the sigmoid link function to these data (Supplementary Fig. 2). We found that variance partition across orders is estimated accurately, and the link function has no effect on these inferences. The minimum and maximum of the sigmoid function are estimated to be well beyond the range of phenotypic prediction, so the transformation has no effect.
Taken together, these data indicate that the impact of limited dynamic range on genetic architecture can be effectively inferred by coupling RFA with a sigmoid link function. Under the realistic conditions we examined, this procedure does not artifactually absorb specific epistatic interactions or underestimate the true complexity of genetic architecture when nonspecific epistasis is weak or absent.
Simplicity of protein sequence-function relationships
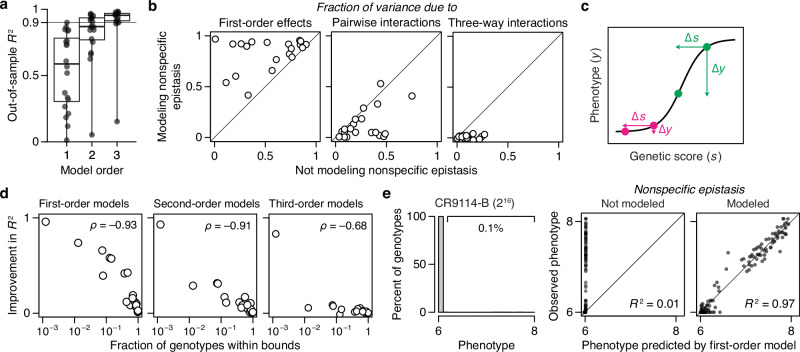
To understand the genetic architecture of real proteins, we performed RFA on 20 combinatorial mutagenesis datasets available for antibodies, enzymes, fluorescent proteins, transcription factors, viral surface proteins, and toxin-antitoxin pairs (Table 1). We considered only datasets with precise measurement (r2 > 0.9 among replicates) and sampling of at least 40% of possible variants. We focused primarily on large libraries but included three small ones in which high-order epistasis has been reported. The datasets range in size from 32 to 160,000 possible genotypes, with the number of variable sites ranging from 3 to 16 and the number of sampled states per site from 2 to 20. To assess the complexity of each dataset, we fitted a series of truncated reference-free models of increasing order, each time using the sigmoid link function to incorporate nonspecific epistasis and L1 regularization to reduce overfitting; we then used cross-validation to estimate the fraction of phenotypic variance explained at each order as the out-of-sample R2, which measures how well a model inferred from a random subset of data can predict the phenotypes of unsampled variants.
Table 1.
Combinatorial mutagenesis datasets analyzed in this study
| Protein | Genotype space | Phenotypes |
|---|---|---|
| Methyl-parathion hydrolase49 | 25 (32) | Catalytic activity |
| β-lactamase51 | 25 (32) | Antibiotics resistance (minimum antibiotics conc. inhibiting growth) |
| Dihydrofolate reductase3 | 3 × 24 (48) | Antibiotics resistance (antibiotics conc. reducing growth rate by 75%) |
| Influenza A H3N2 hemagglutinin41 | 22 × 32 × 42 (576) | Viral replication fitness |
| Antibody CR626119 | 211 (2,048) | Affinity for influenza hemagglutinin strain H1 or H9 |
| Bacterial antitoxin ParD352 | 203 (8000) | Fitness conferred by binding to toxin ParE3 or ParE2 |
| Aequorea victoria GFP (avGFP)15 | 213 (8192) | Fluorescence |
| Bacterial antitoxin ParD353 | 13 × 12 × 10 × 6 (9360) | Fitness conferred by binding to toxin ParE3 or ParE2 |
| SARS-CoV-2 spike protein7 | 215 (32,768) | Affinity for human ACE2 |
| Antibody CH6520 | 216 (65,536) | Affinity for influenza hemagglutinin strain MA90, MA90-G189E, or SI06 |
| Antibody CR911419 | 216 (65,536) | Affinity for influenza hemagglutinin strain B, H1, or H3 |
| Transcription factor ParB50 | 204 (160,000) | Fitness conferred by transcription |
| Protein G B1 domain (GB1)12 | 204 (160,000) | Binding enrichment for IgG-Fc |
Across all proteins examined, most phenotypic variance is explained by first-order effects of amino acids and virtually all of the remainder by pairwise interactions. The first-order model achieves a median R2 of 0.91 across the 20 datasets—with a maximum of 0.97 and greater than 0.75 in all but four cases (Fig. 4a). When pairwise interactions are included, virtually all genetic variance is explained, with a median out-of-sample R2 of 0.96 and a minimum of 0.92 across the datasets. There is no relationship between the fraction of variance explained at low orders and the number of sites or states assayed (Supplementary Fig. 3).
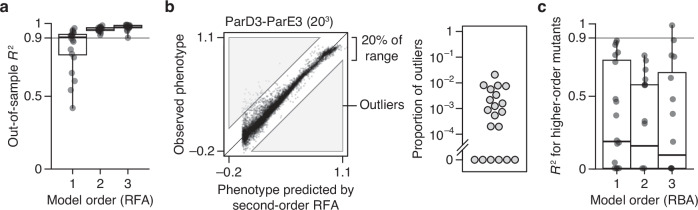
Fig. 4. Simplicity of protein sequence-function relationships.
a RFA of 20 combinatorial mutagenesis datasets (Table 1). First-, second-, and third-order models with the sigmoid link function were evaluated by cross-validation—by inferring the model from a subset of data and predicting the rest of data. Each dot shows the mean out-of-sample R2 for one dataset; boxplots show the median, interquartile range, and total range across the datasets. Supplementary Fig. 3 shows the out-of-sample R2 for individual datasets. b Variants possibly affected by strong high-order epistasis were identified as outliers in the second-order model (residual > 20% of the phenotype range). (Left) Outliers in the ParD3-ParE3 (203) dataset. Each point is a variant, plotted by its observed and predicted phenotype. (Right) Proportion of outliers in each dataset. c RBA of the 20 datasets. Each truncated model was fit to the phenotype of all mutants up to the specified model order using as a reference the genotype designated as wild-type in the original publication; nonspecific epistasis was accounted for as in (a). Accuracy of prediction of higher-order mutants by each model is shown as R2, with negative values plotted as zero. Only higher-order mutants for which all the relevant lower-order effects can be computed were predicted. Supplementary Fig. 6 shows the R2 for individual datasets.
Incorporating third-order terms confers only a marginal or no improvement in fit (median change in out-of-sample R2 of 0.02, maximum 0.04). The very small fraction of phenotypic variance unexplained by the third-order model represents some combination of fourth- and higher-order epistasis, measurement noise, and limitations in the sigmoid link function to accurately capture nonspecific epistasis. The inferred simplicity of the architecture is not attributable to the use of regularization (Supplementary Fig. 4). The estimated third-order effects are generally of small magnitude, and by nature each one affects fewer genotypes than the low-order effects, explaining why together they have a small impact on genetic variation (Supplementary Fig. 5).
Although high-order epistasis is negligible across sequence space as a whole, there could still be a subset of genotypes shaped by strong high-order epistasis. To address this possibility, we analyzed the residuals of the second-order model, which represent the sum of all higher-order interactions and measurement noise. Genotypes with a residual greater than 20% of the phenotype range were considered candidates for strong higher-order epistasis, although erratic measurement noise cannot be excluded. The proportion of such genotypes is zero in six datasets and between 0.02 and 2% in the others (Fig. 4b). Only a tiny fraction of genotypes is therefore potentially affected by strong high-order epistasis.
These analyses show that the genetic architecture of proteins is simple: knowing just the first-order effects and pairwise interactions, coupled with a simple model of nonspecific epistasis, is sufficient to accurately predict and explain phenotypes across the entire ensemble of sequences. Higher-order interactions are not completely absent, but they are weak and limited to a very small fraction of genotypes.
We also examined the 20 datasets using RBA. We exactly computed the first-, second-, and third-order RBA models, using the sigmoid link function with parameters that maximize predictive accuracy for all genotypes. We then used each fitted model to predict the phenotypes of the higher-order mutants not used to compute the model. The median R2 across datasets is less than 0.2 for all three model orders; the vast majority of phenotypic variation is thus left to be explained by higher-order epistasis (Fig. 4c). The RBA formalism therefore leads to a complex and idiosyncratic description of the genetic architecture of these proteins.
Phenotype bounding is the major cause of nonspecific epistasis
To understand the impact of incorporating nonspecific epistasis, we compared RFA of the empirical datasets when estimated with and without the sigmoid link function. We found that incorporating nonspecific epistasis dramatically improves phenotype prediction and reduces the variance attributed to epistasis (Fig. 5a, b). Using the sigmoid link raises the median out-of-sample R2 of first-order models from 0.59 to 0.92, reducing the variance attributable to specific epistasis by a factor of 5. For second-order models, it improves the median R2 from 0.87 to 0.96, reducing the variance explained by higher-order epistasis by a factor of 3. For third-order models, incorporating nonspecific epistasis increases the median R2 from 0.95 to 0.98.
Fig. 5. The primary cause of nonspecific epistasis is phenotype bounding.
a RFA of the 20 datasets without incorporating nonspecific epistasis, shown as in Fig. 4a. b Incorporating nonspecific epistasis reduces the fraction of phenotypic variance attributed to pairwise and higher-order interactions. Each dot shows the variance component attributed to each model order for one dataset when computed with or without incorporating nonspecific epistasis. c Nonspecific epistasis causes the phenotypic effect of a mutation (∆y) to vary among genetic backgrounds (magenta versus green) even when the effect on genetic score (∆s) is constant. Phenotype bounding is a particularly strong form of nonspecific epistasis that causes mutations to appear nearly neutral on backgrounds close to the bounds but not on others. d The extent to which the sigmoid link function improves the model fit (the difference between out-of-sample R2 in (a) and that in Fig. 4a) is proportional to the fraction of genotypes at the phenotype bounds. e In an experimental dataset in which only 0.1% of genotypes are within the bounds, incorporating nonspecific epistasis improves the fraction of variance explained by first-order effects from 0.01 to 0.97.
The dramatic improvement in fit conferred by the simple sigmoid function suggests that phenotype bounds—biological or technical limits on the dynamic range over which genetic states have measurable effects on function—are the major cause of nonspecific epistasis in these datasets (Fig. 5c). Corroborating this conclusion, the degree to which the link function improves the R2 is tightly correlated with the fraction of genotypes at the phenotype bounds (Fig. 5d). In the CR9114-B dataset, for example, 99.9% of genotypes are at the lower bound, and incorporating nonspecific epistasis improves the first-order variance explained from 1% to 97% (Fig. 5e). Conversely, in the CH65-MA90 dataset, virtually all genotypes are within the dynamic range, and using the sigmoid link function has little effect on the variance partition.
Although the causes of nonspecific epistasis are likely to be complex and vary among datasets, these results indicate that the simple sigmoid link function effectively captures its most salient features and allows the specific genetic architecture to be described economically.
Sparsity of protein sequence-function relationships
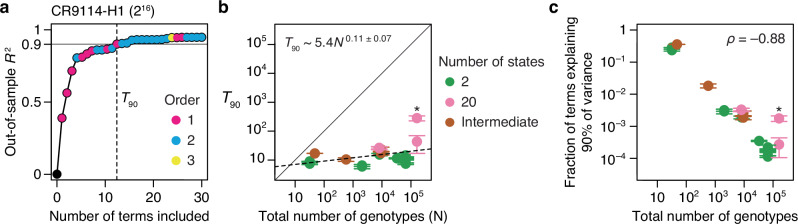
We next asked whether protein function is determined by many genetic states and interactions of small effect or by a few determinants of large effect. For each dataset, we estimated the minimal number of reference-free terms required to predict the phenotype with 90% accuracy (T90): we ranked the terms in the fitted third-order model by their contribution to variance, constructed increasingly complex models by sequentially including each term, and estimated the accuracy of each model by cross-validation (Fig. 6a).
Fig. 6. Sparsity of protein sequence-function relationships.
a Measuring the sparsity of genetic architecture, illustrated using the CR9114-H1 dataset. RFA terms up to third order were estimated and ranked by the fraction of variance they explain (calculated using the simple method for computing the variance contribution of each term described in Methods). Models of increasing complexity were constructed by sequentially including each term, and each model was evaluated by cross-validation. Each dot represents a model, colored by the order of the last term added. Vertical line marks T90, the minimal number of terms required for an out-of-sample R2 of 0.9. b T90 as a function of the total number of genotypes. Dotted line, best-fit power function. Asterisk, GB1 dataset. Each T90 was estimated in two ways: as the number of terms required to reach R2 of 0.9 (upper error bar)—an overestimate because measurement noise prevents any model from attaining an out-of-sample R2 of 1—and as the number of terms required for an R2 equal to 90% of that of the full third-order model (lower error bar); circles show the average of the two estimates. c Fraction of all terms required to explain 90% of phenotypic variance shown against the total number of genotypes. Asterisk, GB1 dataset. Error bars show the possible maximum and minimum computed as in (b).
The genetic architecture of proteins is very sparse (Fig. 6b). Out of up to 160,000 possible terms in each model, T90 ranges from just 6 to 44 across all datasets except for GB1, in which the mutated sites were specifically chosen to be enriched for epistatic interactions12. As the total number of possible genotypes (N) increases, T90 increases very slowly, so that the fraction of all terms required for an R2 of 0.9 declines almost linearly (Fig. 6c). These relationships hold irrespective of the number of states per variable site.
Our findings suggest that even a very large genetic architecture should be describable with a compact set of terms. For example, the relationship between T90 and N predicts that a very large genetic architecture—two states at 100 variable sites, ~1030 possible genotypes and model terms—could be described with 90% accuracy by a model with just ~10,000 key terms.
Inferring genetic architecture by sparse sampling
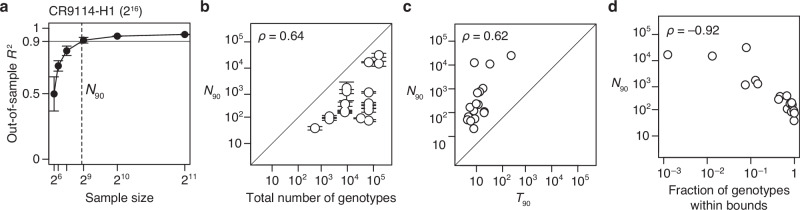
Although a protein’s genetic architecture is defined by relatively few causal factors, identifying them could be challenging. Comprehensive experimental characterization is impractical for sequence spaces much larger than those we have analyzed, so a critical question is whether the important terms can be inferred from a small sample of genotypes by sparse learning methods15. To address this possibility, we sampled a variable number of genotypes from the datasets, fitted RFA models using regression with L1 regularization, predicted phenotypes of the unsampled genotypes, and determined N90, the minimum sample size required for R2 of 0.9 (Fig. 7a).
Fig. 7. Inferring genetic architecture by random sampling.
a Illustration using the CR9114-H1 dataset. Up to third-order RFA terms were inferred from a varying number of genotypes randomly sampled from the experimental results, and the estimated models were then evaluated by predicting the phenotypes of all unsampled genotypes. For each sample size, the mean and standard deviation of out-of-sample R2 across 10 trials are shown. Dashed line marks N90, the minimal sample size required for a mean out-of-sample R2 ≥0.9. b N90 as a function of the total number of genotypes. Error bars were computed as in Fig. 6b. The three datasets with 48 or fewer genotypes are not shown. c N90 as a function of T90, the minimal number of terms required to explain 90% of phenotypic variance (Fig. 6). d N90 as a function of the fraction of genotypes within phenotype bounds.
We found that genetic architecture of proteins cannot be efficiently inferred from sparse random samples (Fig. 7b). Excluding the three small datasets, N90 ranges from 0.2 to 25% of the total number of genotypes, with a median of 5%. Even the lowest end of this range does not bode well for inferring the architecture of large sequence spaces with many states at many variable sites.
We evaluated several factors that might determine the necessary sample size. First, we found that large sequence spaces require larger samples: N90 increases with the total number of genotypes, although there is a considerable scatter in this relationship (Fig. 7b). Second, the complexity of the genetic architecture is not a major factor: N90 depends only weakly on T90 (Fig. 7c). Finally, we found that the fraction of genotypes within the dynamic range of measurement is a critical factor: N90 increases sharply with the degree of phenotype bounding (Fig. 7d). An extreme case is the CR9114-B dataset (65,536 genotypes), where just 10 first-order effects account for 90% of phenotypic variance but approximately 16,000 genotypes are needed to identify them. This is because 99.9% of genotypes are at the lower bound, providing little quantitative information on genetic effects. By contrast, the CH65-MA90 dataset consists of the same number of genotypes, but the genetic architecture can be inferred from just 99 random genotypes because there is virtually no phenotype bounding.
We conclude that despite the global simplicity of proteins’ genetic architecture, the important causal factors cannot be efficiently identified by sparse random sampling. A critical step is therefore to develop a sampling strategy that can efficiently identify the key first-order effects and pairwise interactions that define a genetic architecture.
Understanding genetic architecture
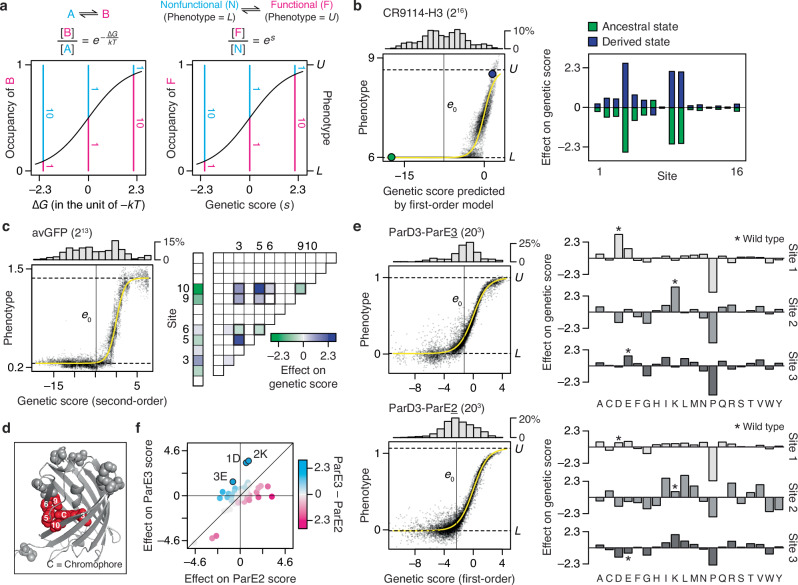
A benefit of coupling RFA with the sigmoid link function is that the genetic effects are expressed in a unit that is intelligible through a simple biophysical analogy, and they become comparable across datasets, even when different phenotypes are measured. The sigmoid model describes the phenotype of a variant as an equilibrium between two thermodynamic states: the functional state, whose phenotype is U, and the nonfunctional state, whose phenotype is L (Fig. 8a). A variant’s phenotype, lying between U and L, reflects the relative occupancy of the functional to nonfunctional state, which is determined by its genetic score (s) as es. The genetic score takes the role of the Gibbs free energy difference between the two states (∆G) expressed in the unit of –kT (the product of Boltzmann constant and absolute temperature). If a variant’s genetic score is 0, the two states are equally populated and its phenotype is midway between U and L. A sequence state or combination that increases the genetic score by 2.3 always causes a ten-fold increase in the relative occupancy of the functional state, corresponding to an apparent ∆∆G of –1.4 kcal/mol at 37 °C. This relationship holds across proteins, functions, and experimental systems.
Fig. 8. Understanding genetic architecture.
a Interpreting genetic score (s) as free energy difference (∆G). (Left) Relative occupancy of two thermodynamic states as a function of their ∆G. k, Boltzmann constant; T, absolute temperature. (Right) The sigmoid link function can be interpreted as describing an equilibrium between two states—the “functional” state, which produces a phenotype of U, and the “nonfunctional” state, which produces a phenotype of L. Their relative occupancy (pink versus blue lines) equals es, allowing s to be interpreted as ∆G in the unit of –kT. b CR9114-H3 dataset, which measures the affinity of all combinations of ancestral and derived amino acids at 16 sites in an antibody towards a hemagglutinin. (Left) First-order RFA. Each dot is a genotype, plotted by its estimated genetic score and measured phenotype. Histogram, distribution of genetic score; yellow curve, inferred sigmoid link; horizontal lines, inferred phenotype bounds; vertical line, mean genetic score; green and purple dots, ancestral and derived genotypes. (Right) First-order effects of amino acids at each site. c avGFP dataset, which measures the fluorescence of all combinations of pairs of amino acids at 13 sites. (Left) Second-order RFA. (Right) First-order effects and pairwise interactions, which account for 57 and 38% of phenotypic variance, respectively. Values are shown for one of the two of amino acids at each site. The ten pairwise interactions possible among sites 3, 5, 6, 9, and 10 are outlined. d Crystal structure of avGFP (PDB ID: 3e5w). The 13 mutated sites are shown in spheres, and the chromophore and its five surrounding sites are colored in red. e ParD3-ParE3 and ParD3-ParE2 (203) datasets, which measure the binding of all possible variants of ParD3 at three sites to ParE3, the cognate substrate, and ParE2, a noncognate substrate. (Left) First-order RFA. (Right) First-order effects at each site. Asterisk, wild-type amino acid. f Comparing the effect of each amino acid on ParE3 versus ParE2 binding. Wild-type amino acids are marked.
We applied this framework to understand the genetic architecture of several example proteins. The CR9114-H3 dataset (Fig. 8b) consists of affinity measurements for binding of 216 antibody variants (all possible combinations of ancestral and derived amino acids at 16 sites that evolved during affinity maturation) to an influenza hemagglutinin. The vast majority of variants are at the lower bound of detectable binding, so the average genetic score is –7.8, corresponding to just 0.04% occupancy of the bound state, or ∆Gapp = 5.6 kcal/mol. The best variant has a score of just 2.6, corresponding to 93% occupancy and ∆Gapp = –1.9 kcal/mol. There is virtually no specific epistasis in this genetic architecture (Supplementary Fig. 3). First-order effects at three key sites mostly determine the phenotype: each favorable state increases the genetic score by 2.1 to 2.6 (∆∆Gapp < –2 kcal/mol); together, these states increase the relative occupancy by almost three orders of magnitude compared with the global average but still yield absolute occupancy of the bound state of just 36%. Five other sites make modest contributions, each changing the genetic score by ~0.5 and shifting the relative occupancy by ~1.3 fold. The remaining eight have even smaller effects. A variant must therefore have all three large-effect favorable states to achieve measurable binding, and the particular combination of states at the other sites modulates the affinity.
Specific pairwise interactions are important in the avGFP dataset (Fig. 8c), accounting for 38% of variance in fluorescence measurements. There are many functional variants in this library, including a large number at the measurement maximum, so the average variant has a genetic score of –1 with the occupancy of the fluorescent state at 20%. First- and second-order effects involving just five of 13 variable sites account for 86% of variance. These sites, which tightly surround the chromophore in the crystal structure (Fig. 8d), engage in a dense epistatic network in which nine of the ten possible pairwise interactions are nonzero. Only four of these interactions alter the genetic score by more than 1, but their total impact is substantial, conferring an increase in genetic score by 7.8 and relative occupancy by 2400-fold (∆∆Gapp = –5.6 kcal/mol) when all are in the most favorable combination. Not all of these are necessary to achieve high fluorescence, however: because the global average has measurable fluorescence, one or more favorable states can be removed while leaving the other interactions intact.
RFA terms can also be used to understand the determinants of functional specificity in multistate sequence space and when multiple functions are measured. The ParD3 library (all combinations of 20 states at 3 sites in the binding interface) was assayed separately for binding its cognate ligand ParE3 and the noncognate ligand ParE2. Effects on specificity can be quantified as the difference between a state’s effects on the genetic score with the two ligands. The average variant displays a weak but measurable binding to both ligands, with a preference for ParE3 over ParE2 by a genetic score of ~ 1 (difference in relative occupancy of 2.5-fold). For both ligands, first-order effects account for the vast majority of variance in binding (Fig. 8e). There are only eight amino acid states that can change the genetic score in favor of one ligand over the other by more than 1.6, each equivalent to more than 5-fold difference in occupancy (Fig. 8f). The three strongest of these each favor ParE3 by scores of 2.2 to 2.8 (~10-fold preference in occupancy, ∆∆Gapp ~ 2 kcal/mol). Two of these change specificity by increasing affinity for both ligands but more strongly enhancing ParE3 binding, and the third has opposite effects on the two ligands. The wild-type protein in this case possesses these three specificity-optimal states.
Discussion
Our finding that first-order effects and pairwise interactions account for virtually all genetic variation within proteins contrasts with several reports of extensive high-order epistasis1–8. Use of reference-based analysis and incomplete accounting of nonspecific epistasis have led prior studies to invoke more high-order epistasis than is necessary to explain the data.
We expect our finding to be general across proteins and biochemical phenotypes, but the available datasets have some important limitations. The datasets we analyzed comprise proteins with diverse structures and functions. It is unlikely that the particular sites varied in the datasets biased the architectures towards simplicity. In most cases, the sites were chosen because of prior structural evidence that they are functionally important or they vary between functional homologs. The sites are dispersed across the structure in some datasets but clustered in others, so our results are unlikely to be the consequence of spatially biased sampling. A limitation is that each dataset assessed a single phenotype, so the genetic architecture of functional specificity could be more complex; however, a recent study using a similar approach as ours found that high-order interactions within a transcription factor are relatively unimportant for determining its DNA binding specificity46. Allosteric phenotypes, in which multiple functions within a protein modulate each other across a protein’s structure, may have more complex genetic architectures. The relative simplicity of global genetic architecture does not necessarily imply that epistasis does not affect evolutionary processes; a moderate degree of pairwise epistasis could be sufficient to introduce substantial contingency into protein sequence evolution46,47.
The lack of high-order epistasis within proteins may seem surprising from a structural perspective, because proteins often contain clusters of three or more residues that contact each other directly. Our results indicate that the phenotypic variation encoded by these physical clusters can largely be explained as the sum of the their pairwise interactions. But any pairwise coupling depends on the fold of the protein, which in turn depends on states at other sites. A mutation that changes the conformation should alter pairwise couplings and induce high-order epistasis. In the datasets we examined, such conformational epistasis seems rare or inconsequential. A possible explanation is that these datasets held most sites in the protein constant and therefore presumably maintained the overall conformation (or caused it to unfold entirely). High-order interactions that specify a protein’s fold might be revealed in a library large enough to contain variants with multiple folds, or if phenotypes involving multiple conformations within a single fold were measured. Direct insight into the physical reasons why genetic architecture is so simple in the protein datasets we examined will require contrasting them to proteins that manifest more high-order epistasis, but those in the latter category have not yet been found.
The effectiveness of the sigmoid link to capture nonspecific epistasis may seem surprising, because nonlinearities in sequence-function relationships can arise from complex biological and technical causes that vary among proteins, phenotypes, and assays. Our results suggest that bounds on the range over which a phenotype can be produced and measured are the major cause of nonspecific epistasis in these datasets. Irrespective of the underlying causes, incorporating this nonlinearity using a simple sigmoid with RFA yields a parsimonious and efficient description of a protein’s genetic architecture. It is possible that other link functions could offer superior accuracy for some proteins; further research is warranted to examine their performance under a variety of conditions.
Our finding that RFA outperforms RBA in providing a compact and accurate description of the global sequence-function relationship does not mean that RBA is never useful. RBA is appropriate in principle if the object of interest is interactions among a few mutations in the background of a particular wild-type or ancestral protein. In such cases, exact RBA should be used with caution because of its tendency to infer interactions from measurement noise and local idiosyncrasies and its limitations when data are incomplete. Regression should not be used to fit RBA models because of bias in the variance partition and propagating error in the estimated coefficients.
For scientists who would like to understand how proteins work, our findings are reassuring, but they also clarify a challenge. Proteins’ genetic architecture is intelligible: a small fraction of low-order model terms explains most functional variation. It is therefore unnecessary to exhaustively characterize complete combinatorial libraries or estimate high-order models, which would quickly become intractable as the number of sites or states increases. But random sampling from combinatorial libraries cannot efficiently identify the important genetic determinants if the sequence space is very large and most random sequences are nonfunctional. Analyzing the effects of low-order combinations of mutations on a single functional protein would not work either, because this approach would be subject to the same kind of errors and idiosyncrasies that plague RBA. An effective strategy may be to perform single- and double-mutant scans using as starting points a diverse set of functional proteins, such as distantly related homologs48, while also improving the dynamic range of measurement. Future research is warranted to define how distant from each other such proteins must be. The potential of this strategy to efficiently learn the rules of sequence-function relationships has not been previously considered, perhaps because the genetic architecture of proteins was thought to be much more complex than it is.
Methods
Reference-free analysis (RFA)
Here we define RFA and summarize its key properties. Proofs for the properties and detailed comparisons with other formalisms are in Supplementary Information. Scripts and tutorials for performing RFA are on GitHub (github.com/JoeThorntonLab/RFA).
Consider a genotype space defined by q states across n sites. Let g denote a genotype, y(g) its phenotype, and G the set of all qn possible genotypes. RFA decomposes the phenotype into the contribution of individual states and their interactions relative to the global mean phenotype, which is denoted
where the brackets indicate averaging y over G. The first-order effect of state s in site i is the difference between the mean phenotype of the subset of genotypes sharing that state (denoted ) and the global mean:
The pairwise interaction between states s1 and s2 in sites i1 and i2 is the difference between the mean phenotype of the subset of genotypes sharing that state-pair () and the global mean after accounting for the first-order effects:
Likewise, a higher-order effect is the difference between the mean phenotype of a subset of genotypes sharing a set of states and the global mean after accounting for the relevant lower-order effects.
RFA predicts the phenotype by summing the effects of all states in the genotype. For a genotype with state gi in site i, the predicted phenotype under RFA of order k is
The overall accuracy of this prediction can be quantified by the sum of squared errors
Among all linear models of the same order, including reference-based models under any choice of wild-type genotype, RFA minimizes εG for any k for any set of sequence-function associations. For example, when k is zero (all phenotypes predicted by a single number), εG is minimized by the global mean phenotype, which is the RFA zero-order term. By minimizing εG, RFA explains the maximum fraction of phenotypic variance that can be explained by any linear model of the same order. Fourier and background-averaged analyses share this property.
RFA facilitates the analysis of genetic architecture by partitioning the phenotypic variance into components attributable to each state and interaction:
where e denotes any nonzero-order effect and O(e) its order. Note that an effect of order k is involved in the phenotype of one in qk genotypes. The amount of phenotypic variance attributable to an effect is therefore the square of its magnitude normalized by the fraction of genotypes involving that effect.
Applying RFA on noisy and incomplete data
When individual phenotypes are subject to measurement noise of variance ω, a reference-free effect of order k computed from them has a variance
This is always smaller than ω and typically miniscule for low-order effects. The extensive averaging of phenotypic measurements in the computation of reference-free effects confers robustness to measurement noise.
When some genotypes are missing from data, reference-free effects can be inferred by regression. To infer effects of order up to k, we model
where the residual ε(g) is the sum of all higher-order effects and measurement noise. Let G* be the set of sampled genotypes. The regression estimates are obtained by minimizing the sum of squared errors across G*,
Because reference-free effects minimize the sum of squared errors across genotype space, the regression estimates converge to the true effects as more genotypes are sampled. The estimates are unbiased as long as genotypes are randomly sampled, because the unmodeled higher-order effects appear as noise to any lower-order model and therefore do not bias the regression.
Nonspecific epistasis
We account for nonspecific epistasis by assuming that the effects of sequence states are transformed by a nonlinear link function into the observed phenotype. We modeled the link function as a simple sigmoid, which is defined by two parameters corresponding to the lower (L) and upper (U) bound of phenotype:
where s(g) is the genetic score—the sum of the reference-free effects of all states in the genotype g. The sigmoid link allows the genetic score to be interpreted in the free-energy scale, but any link function able to model phenotype bounds could be used, with the exact curvature between the bounds reflecting the properties of the particular dataset. To keep the unit of genetic score identical to that of phenotypic measurement, a bounded identity function can be used.
Implementation
We inferred the link function and reference-free effects jointly by regression. The joint inference25 is desirable over a widely used two-step approach, which infers the link function first and applies its inverse transformation on the observed phenotype to compute the effects of sequence states13. The two-step approach infers the link function by fitting a first-order model under the assumption of no nonspecific epistasis and by identifying any systematic nonlinearity between the observed and predicted phenotype. Because the first-order model is fit under the incorrect assumption that nonspecific epistasis is absent, this approach cannot uncover the true link function. Furthermore, the inverse transformation can dramatically amplify measurement noise for genotypes near the phenotype bounds.
The joint regression was performed with L1 regularization to reduce overfitting. The optimal regularization strength was determined by maximizing the out-of-sample R2 in cross-validation. Except for four datasets, cross-validation was performed by randomly partitioning the genotypes into training and test sets. For the three datasets with 48 or fewer genotypes and the CR9114-B dataset where only 81 genotypes are above the lower phenotype bound, cross-validation was performed by leaving out each measurement replicate in turn. The R package lbfgs was used for numerical optimization. To estimate variance explained using truncated models, we used ten-fold cross-validation, which may slightly underestimate accuracy, but this bias is expected to be weak because RFA uses many genotypes to estimate each model term at low orders.
For datasets that sample only two amino acids per site, we estimated RFA terms by first performing Fourier analysis and then computing the RFA terms from the Fourier coefficients. In a binary state space, there are fewer Fourier coefficients to model than there are RFA terms, and the two sets of terms are easily interconvertible (Supplementary Section 1.2). The best-fit Fourier coefficients and link function were determined by cross-validation as described above.
For incorporating nonspecific epistasis into reference-based analysis (RBA), the regression approach should not be used, because regression misestimates RBA terms (Fig. 2). For each candidate set of link function parameters, RBA terms were computed to recapitulate the observed phenotype for mutants up to the model order. For example, the first-order model was constrained to be exact for the wild-type and its point mutants, consistent with the definition of first-order RBA. The effects and the link function were then used to predict the phenotypes of higher-order mutants, and this procedure was repeated for other parameter values to identify the link function that maximizes the R2 for higher-order mutants.
Background-averaged analysis was originally developed only for binary state space2,27. We extended the recursive matrix formalism to multiple states and implemented it in a custom R script. The same multi-state formalism was recently independently derived36.
Combinatorial mutagenesis datasets
We systematically mined the literature for mutagenesis experiments with a combinatorially complete design. Among the many datasets comprising fewer than 100 genotypes, we chose three datasets where high-order epistasis has been reported. Any larger dataset in which precise measurement (r2 > 0.9 between replicates) is available for at least 40% of possible genotypes was included for analysis. Several datasets were edited as described below.
The methyl-parathion hydrolase activity49 was measured in the presence of seven different metal cofactors. In every case, the second-order RFA with the sigmoid link function explains more than 90% of phenotypic variance. Only the Ni2+ dataset, in which epistasis accounts for the greatest fraction of phenotypic variance, is presented here.
The original dihydrofolate reductase dataset3 includes a noncoding mutation for a total of 96 variants. We only analyzed the 48 protein variants fixed for the mutant state in the noncoding site. IC75—the antibiotics concentration that reduces the growth rate by 75%—was originally reported in logarithmic scale, set arbitrarily as –2 when the variant is unviable at any concentration. We reverted the logarithm, making IC75 equal to 0 when the variant is unviable.
The influenza A H3N2 hemagglutinin dataset41 characterized an identical set of genetic variants in six different genetic backgrounds. We analyzed only the genetic background for which the measurement is most precise (Bei89).
In the avGFP dataset15, fluorescence is systematically higher in the second measurement replicate by a factor of 1.31. This difference was normalized when combining the two replicates.
The ParB study50 measures how the transcription factor ParB binds to two DNA motifs, parS and NBS. Because measurement r2 is less than 0.9 for the NBS dataset, only the parS dataset was analyzed. The absolute fitness of each variant was inferred by comparing the read count before and after the bulk competition assay. Variants with the pre-competition read count fewer than 15 were excluded, resulting in 42.2% coverage of the 160,000 possible genotypes—down from 97.0% in the original study.
The extent of measurement noise in the protein G B1 domain dataset12 could not be directly determined because measurement was not replicated, but comparison to an independent dataset for a subset of variants showed that r2 is greater than 0.9. Variants with a pre-competition read count fewer than 100 were excluded, resulting in 68.6% coverage of the 160,000 possible genotypes—down from 93.4% in the original study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank members of the Thornton Laboratory and R. Ranganathan for discussion, and the University of Chicago Research Computing Center for high-performance computing. This work was supported by the National Institutes of Health grants R35GM145336 (J.W.T), R01GM131128 (J.W.T.), R01GM121931 (J.W.T.), and F32GM122251 (B.P.H.M.) and Samsung Scholarship (Y.P.).
Author contributions
Y.P., B.P.H.M., and J.W.T. designed research; Y.P. developed methods and analyzed data; Y.P. and J.W.T. wrote the paper with input from B.P.H.M.
Peer review
Peer review information
Nature Communications thanks Willow Coyote-Maestas, Juannan Zhou, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All sequence-function data were gathered from published studies (Table 1) and are available on GitHub (https://github.com/whatdoidohaha/RFA) and Zenodo (10.5281/zenodo.8307147).
Code availability
All scripts used for data analysis as well as tutorial scripts for performing reference-free analysis are available on GitHub (https://github.com/JoeThorntonLab/RFA) and Zenodo (10.5281/zenodo.8307147).
Competing interests
The authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51895-5.
References
- 1.Sadovsky, E. & Yifrach, O. Principles underlying energetic coupling along an allosteric communication trajectory of a voltage-activated K+ channel. Proc. Natl Acad. Sci. USA104, 19813–19818 (2007). 10.1073/pnas.0708120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinreich, D. M., Lan, Y., Wylie, C. S. & Heckendorn, R. B. Should evolutionary geneticists worry about higher-order epistasis? Curr. Opin. Genet Dev.23, 700–707 (2013). 10.1016/j.gde.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer, A. C. et al. Delayed commitment to evolutionary fate in antibiotic resistance fitness landscapes. Nat. Commun.6, 7385 (2015). 10.1038/ncomms8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sailer, Z. R. & Harms, M. J. Molecular ensembles make evolution unpredictable. Proc. Natl Acad. Sci. USA114, 11938–11943 (2017). 10.1073/pnas.1711927114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerrero, R. F., Scarpino, S. V., Rodrigues, J. V., Hartl, D. L. & Ogbunugafor, C. B. Proteostasis environment shapes higher-order epistasis operating on antibiotic resistance. Genetics212, 565–575 (2019). 10.1534/genetics.119.302138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozovsky, E. R., Daniels, R. F., Heffernan, G. D., Jacobus, D. P. & Hartl, D. L. Relevance of higher-order epistasis in drug resistance. Mol. Biol. Evol.38, 142–151 (2021). 10.1093/molbev/msaa196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moulana, A. et al. Compensatory epistasis maintains ACE2 affinity in SARS-CoV-2 Omicron BA.1. Nat. Commun.13, 7011 (2022). 10.1038/s41467-022-34506-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buda, K., Miton, C. M. & Tokuriki, N. Pervasive epistasis exposes intramolecular networks in adaptive enzyme evolution. Nat. Commun.14, 8508 (2023). 10.1038/s41467-023-44333-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou, J. et al. Higher-order epistasis and phenotypic prediction. Proc. Natl Acad. Sci. USA119, e2204233119 (2022). 10.1073/pnas.2204233119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J. & Stites, W. E. Higher-order packing interactions in triple and quadruple mutants of staphylococcal nuclease. Biochemistry40, 14012–14019 (2001). 10.1021/bi011269d [DOI] [PubMed] [Google Scholar]
- 11.Sarkisyan, K. S. et al. Local fitness landscape of the green fluorescent protein. Nature533, 397–401 (2016). 10.1038/nature17995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu, N. C., Dai, L., Olson, C. A., Lloyd-Smith, J. O. & Sun, R. Adaptation in protein fitness landscapes is facilitated by indirect paths. eLife5, e16965 (2016). 10.7554/eLife.16965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sailer, Z. R. & Harms, M. J. Detecting high-order epistasis in nonlinear genotype-phenotype maps. Genetics205, 1079–1088 (2017). 10.1534/genetics.116.195214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams, R. M., Kinney, J. B., Walczak, A. M. & Mora, T. Epistasis in a fitness landscape defined by antibody-antigen binding free energy. Cell Syst.8, 86–93.e3 (2019). 10.1016/j.cels.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poelwijk, F. J., Socolich, M. & Ranganathan, R. Learning the pattern of epistasis linking genotype and phenotype in a protein. Nat. Commun.10, 4213 (2019). 10.1038/s41467-019-12130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamer, Y. T. et al. High-order epistasis in catalytic power of dihydrofolate reductase gives rise to a rugged fitness landscape in the presence of trimethoprim selection. Mol. Biol. Evol.36, 1533–1550 (2019). 10.1093/molbev/msz086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, G. et al. Higher-order epistasis shapes the fitness landscape of a xenobiotic-degrading enzyme. Nat. Chem. Biol.15, 1120–1128 (2019). 10.1038/s41589-019-0386-3 [DOI] [PubMed] [Google Scholar]
- 18.Ballal, A. et al. Sparse epistatic patterns in the evolution of terpene synthases. Mol. Biol. Evol.37, 1907–1924 (2020). 10.1093/molbev/msaa052 [DOI] [PubMed] [Google Scholar]
- 19.Phillips, A. M. et al. Binding affinity landscapes constrain the evolution of broadly neutralizing anti-influenza antibodies. eLife10, e71393 (2021). 10.7554/eLife.71393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips, A. M. et al. Hierarchical sequence-affinity landscapes shape the evolution of breadth in an anti-influenza receptor binding site antibody. eLife12, e83628 (2023). 10.7554/eLife.83628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinkley, T. et al. A systems analysis of mutational effects in HIV-1 protease and reverse transcriptase. Nat. Genet43, 487–489 (2011). 10.1038/ng.795 [DOI] [PubMed] [Google Scholar]
- 22.Olson, C. A., Wu, N. C. & Sun, R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr. Biol.24, 2643–2651 (2014). 10.1016/j.cub.2014.09.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podgornaia, A. I. & Laub, M. T. Pervasive degeneracy and epistasis in a protein-protein interface. Science347, 673–677 (2015). 10.1126/science.1257360 [DOI] [PubMed] [Google Scholar]
- 24.Diss, G. & Lehner, B. The genetic landscape of a physical interaction. eLife7, e32472 (2018). 10.7554/eLife.32472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski, J., McCandlish, D. M. & Plotkin, J. B. Inferring the shape of global epistasis. Proc. Natl Acad. Sci. USA115, E7550–E7558 (2018). 10.1073/pnas.1804015115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding, D. et al. Protein design using structure-based residue preferences. Nat. Commun.15, 1639 (2024). 10.1038/s41467-024-45621-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poelwijk, F. J., Krishna, V. & Ranganathan, R. The context-dependence of mutations: a linkage of formalisms. PLoS Comput. Biol.12, e1004771 (2016). 10.1371/journal.pcbi.1004771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domingo, J., Baeza-Centurion, P. & Lehner, B. The causes and consequences of genetic interactions (epistasis). Annu Rev. Genom. Hum. G20, 433–460 (2019). 10.1146/annurev-genom-083118-014857 [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski, J. & Plotkin, J. B. Inferring fitness landscapes by regression produces biased estimates of epistasis. Proc. Natl Acad. Sci. USA111, E2301–E2309 (2014). 10.1073/pnas.1400849111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starr, T. N. & Thornton, J. W. Epistasis in protein evolution. Protein Sci.25, 1204–1218 (2016). 10.1002/pro.2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinberger, E. D. Fourier and taylor series on fitness landscapes. Biol. Cyber.65, 321–330 (1991). 10.1007/BF00216965 [DOI] [Google Scholar]
- 32.Stadler, P. F. Landscapes and their correlation functions. J. Math. Chem.20, 1–45 (1996). 10.1007/BF01165154 [DOI] [Google Scholar]
- 33.Stormo, G. D. Maximally efficient modeling of DNA sequence motifs at all levels of complexity. Genetics187, 1219–1224 (2011). 10.1534/genetics.110.126052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brookes, D. H., Aghazadeh, A. & Listgarten, J. On the sparsity of fitness functions and implications for learning. Proc. Natl Acad. Sci. USA119, e2109649118 (2022). 10.1073/pnas.2109649118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinreich, D. M., Lan, Y., Jaffe, J. & Heckendorn, R. B. The influence of higher-order epistasis on biological fitness landscape topography. J. Stat. Phys.172, 208–225 (2018). 10.1007/s10955-018-1975-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faure, A. J., Lehner, B., Miró Pina, V., Serrano Colome, C. & Weghorn, D. An extension of the Walsh-Hadamard transform to calculate and model epistasis in genetic landscapes of arbitrary shape and complexity. bioRxiv10.1101/2023.03.06.531391 (2023). [DOI] [PMC free article] [PubMed]
- 37.Anderson, D. W., McKeown, A. N. & Thornton, J. W. Intermolecular epistasis shaped the function and evolution of an ancient transcription factor and its DNA binding sites. eLife4, e07864 (2015). 10.7554/eLife.07864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell182, 1295–1310.e20 (2020). 10.1016/j.cell.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Domingo, J., Diss, G. & Lehner, B. Pairwise and higher-order genetic interactions during the evolution of a tRNA. Nature558, 117–121 (2018). 10.1038/s41586-018-0170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pokusaeva, V. O. et al. An experimental assay of the interactions of amino acids from orthologous sequences shaping a complex fitness landscape. PLoS Genet15, e1008079 (2019). 10.1371/journal.pgen.1008079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, N. C. et al. Major antigenic site B of human influenza H3N2 viruses has an evolving local fitness landscape. Nat. Commun.11, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynch, M. & Walsh, B. Genetics and Analysis of Quantitative Traits Vol. 980 (OUP USA, 1998).
- 43.Horovitz, A. & Fersht, A. R. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J. Mol. Biol.214, 613–617 (1990). 10.1016/0022-2836(90)90275-Q [DOI] [PubMed] [Google Scholar]
- 44.Kondrashov, A. S., Sunyaev, S., Kondrashov, F. A. & Dobzhansky Muller incompatibilities in protein evolution. Proc. Natl Acad. Sci. USA99, 14878–14883 (2002). 10.1073/pnas.232565499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Afshartous, D. & Preston, R. A. Key results of interaction models with centering. J. Stat. Edu.10.1080/10691898.2011.11889620 (2011).
- 46.Metzger, B. P. H., Park, Y., Starr, T. N. & Thornton, J. W. Epistasis facilitates functional evolution in an ancient transcription factor. eLife12, RP88737 (2023). 10.7554/eLife.88737.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, Y., Metzger, B. P. H. & Thornton, J. W. Epistatic drift causes gradual decay of predictability in protein evolution. Science376, 823–830 (2022). 10.1126/science.abn6895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faure, A. J. et al. Mapping the energetic and allosteric landscapes of protein binding domains. Nature604, 175–183 (2022). 10.1038/s41586-022-04586-4 [DOI] [PubMed] [Google Scholar]
- 49.Anderson, D. W., Baier, F., Yang, G. & Tokuriki, N. The adaptive landscape of a metallo-enzyme is shaped by environment-dependent epistasis. Nat. Commun.12, 3867 (2021). 10.1038/s41467-021-23943-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jalal, A. S. B. et al. Diversification of DNA-binding specificity by permissive and specificity-switching mutations in the ParB/Noc protein family. Cell Rep.32, 107928 (2020). 10.1016/j.celrep.2020.107928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinreich, D. M., Delaney, N. F., Depristo, M. A. & Hartl, D. L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science312, 111–114 (2006). 10.1126/science.1123539 [DOI] [PubMed] [Google Scholar]
- 52.Lite, T. V. et al. Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library. eLife9, e60924 (2020). 10.7554/eLife.60924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aakre, C. D. et al. Evolving new protein-protein interaction specificity through promiscuous intermediates. Cell163, 594–606 (2015). 10.1016/j.cell.2015.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence-function data were gathered from published studies (Table 1) and are available on GitHub (https://github.com/whatdoidohaha/RFA) and Zenodo (10.5281/zenodo.8307147).
All scripts used for data analysis as well as tutorial scripts for performing reference-free analysis are available on GitHub (https://github.com/JoeThorntonLab/RFA) and Zenodo (10.5281/zenodo.8307147).