Abstract
Nanoarchaeum equitans is a species of hyperthermophilic archaea with the smallest genome size. Its alanyl-tRNA synthetase genes are split into AlaRS-α and AlaRS-β, encoding the respective subunits. In the current report, we surveyed N. equitans AlaRS-dependent alanylation of RNA minihelices, composed only of the acceptor stem and the T-arm of tRNAAla. Combination of AlaRS-α and AlaRS-β showed a strong alanylation activity specific to a single G3:U70 base pair, known to mark a specific tRNA for charging with alanine. However, AlaRS-α alone had a weak but appreciable alanylation activity that was independent of the G3:U70 base pair. The shorter 16-mer RNA tetraloop substrate mimicking only the first four base pairs of the acceptor stem of tRNAAla, but with C3:G70 base pair, was also successfully aminoacylated by AlaRS-α. The end of the acceptor stem, including CCA-3ʹ terminus and the discriminator A73, was able to function as a minimal structure for the recognition by the enzyme. Our findings imply that aminoacylation by N. equitans AlaRS-α may represent a vestige of a primitive aminoacylation system, before the appearance of the G3:U70 pair as an identity element for alanine.
Electronic supplementary material
The online version of this article (10.1007/s00239-020-09945-1) contains supplementary material, which is available to authorized users.
Keywords: tRNA, Minihelix, Alanyl-tRNA synthetase, Nanoarchaeum equitans, G3:U70 independent aminoacylation, Evolution
Introduction
Aminoacyl-tRNA synthetases (aaRSs) play a central role in protein biosynthesis. Accurate primary protein sequences are assured by the proper attachment of cognate amino acids to their corresponding cognate tRNAs catalyzed by aaRSs (Schimmel 1987; Schimmel and Söll 1979). The aminoacylation reaction is generally performed by the activation of an amino acid (aminoacyl adenylate formation), followed by the transfer to a cognate tRNA. (Arginyl-, glutaminyl-, glutamyl-, and class I lysyl-tRNA synthetases require the presence of tRNA for amino acid activation (Schimmel and Söll 1979; Ibba et al. 1999).) To ensure accurate assignment, each aaRS recognizes specific regions of their corresponding tRNA. These identity elements have been extensively investigated. tRNAAla possesses a unique wobble base pair in the acceptor stem (G3:U70) and this position has been identified as a crucial identity element for recognition by alanyl-tRNA synthetase (AlaRS) (Hou and Schimmel 1988; McClain and Foss 1988). AlaRS is a class II aminoacyl-tRNA synthetase with characteristic antiparallel β-sheets and three conserved sequence motifs (Eriani et al. 1990; Ibba and Söll 2000). The crystal structures of AlaRS from the archaeon Archaeoglobus fulgidus, in complex with tRNAAla have been solved. These structures showed Asp450 and Asn359 in AlaRS (corresponding to Asp400 and Asn303 in Escherichia coli AlaRS, respectively) interacting with the amino group at position 2 of G3, and the carbonyl oxygen at position 4 of U70, respectively (Naganuma et al. 2014).
Nanoarchaeum equitans is a species of hyperthermophilic archaea with the smallest genome size (Huber et al. 2002). Some of tRNA- and protein-encoding genes of this archaeon, including the AlaRS gene, are split (Randau et al. 2005a, b; Waters et al. 2003). The genes for the two subunits of AlaRS are separated by half of the chromosome (Waters et al. 2003). Recently, in other members of the Nanoarchaeota phylum, i.e., Nanobsidianus stetteri (Nst1) (Wurch et al. 2016), Nanopusillus acidilobi (Wurch et al. 2016), and Nanoclepta minutus (St John et al. 2019), gene splitting of AlaRS has been estimated. In a previous study using the N. equitans AlaRS, a combination of both the subunits (α- and β-chains) of AlaRS was found to be active, but each individual subunit was not active when present alone (Waters et al. 2003). However, as the alanylation assay was performed using Methanococcus jannaschii tRNAAla, the target sequence of tRNA was not identical in a strict sense.
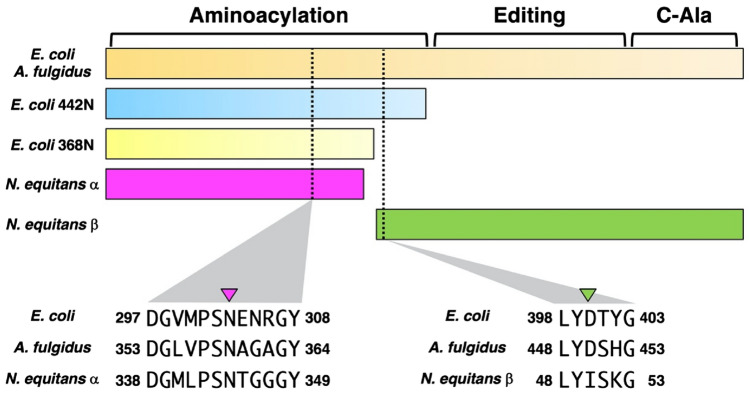
The multisequence alignment of N. equitans AlaRS-α with E. coli AlaRS shows that AlaRS-α almost entirely corresponds to E. coli AlaRS368N, a N-terminal fragment with 368 residues of E. coli AlaRS (Fig. 1 and Supplementary Material, Fig. S1). Previous studies have shown that the E. coli AlaRS368N cannot alanylate tRNAAla (Jasin et al. 1983; Regan et al. 1987; Chihade and Schimmel 1999), and indeed the minimal E. coli AlaRS variant with alanylation activity is the N-terminal 442-residue fragment (AlaRS442N) (Fig. 1 and Supplementary Material, Fig. S1) (Guo et al. 2009).
Fig. 1.
Schematic diagram of E. coli, A. fulgidus, and N. equitans AlaRS (α and β) sequences. The Asn (N) and Asp (D) residues involved in G3:U70 recognition in E. coli and A. fulgidus AlaRS are indicated with the amino acid context shown (multisequence alignment of these AlaRSs are given in Supplementary Material, Fig. S1)
Minihelices (the coaxial stack of the acceptor stem on the T-arm of the tRNA) have been shown to function as substrates of many aaRSs (Francklyn and Schimmel 1989; Frugier et al. 1994; Martinis and Schimmel 1997; Musier-Forsyth and Schimmel 1999). Similarly, oligonucleotide substrates based on the acceptor stem of tRNAAla (microhelix) (Francklyn et al. 1992), or a shortened acceptor stem containing a G3:U70 base pair (Shi et al. 1992), are also aminoacylated by AlaRS. Evidence suggests that the top half of the L-shaped tRNA structure may have appeared in evolution prior to the addition of the other half of the structure, which includes the anticodon (Schimmel et al. 1993; Schimmel and Ribas de Pouplana 1995; Tamura 2015). The split forms of the N. equitans AlaRS may therefore be connected with the evolutionary history of aaRS enzyme activity.
In this report, we show a very important fact that might be closely related to the evolution of aminoacyl-tRNA synthetases: N. equitans AlaRS-α shows the G3:U70 base pair-independent aminoacylation of RNA minihelices with alanine. Our data support the existence of a simplified aminoacylation of tRNAAla by AlaRS early in the evolutionary process, prior to the appearance of the G3:U70 base pair.
Materials and Methods
Plasmid Construction
The coding sequences for AlaRS-α (NEQ547) and AlaRS-β (NEQ211) of N. equitans were PCR-amplified from genomic DNA of N. equitans (a gift from Dr. Harald Huber, Universität Regensburg, Germany) using the pairs of primers 5ʹ-CAAAAATTCATATGGTTAAAG-3ʹ and 5ʹ-GATAACATGGATCCCGCAAAAAG-3ʹ for AlaRS-α, and 5ʹ-GTTTTTAGATTCATATGATACC-3ʹ and 5ʹ-CAATTGTGGATCCTTTTATAACAAAC-3ʹ for AlaRS-β. These primers were synthesized by Eurofins Genomics K.K. (Tokyo, Japan). After digestion by NdeI and BamHI, the fragments were first incorporated into pET11a (Novagen, Madison, WI), and then the NdeI- and BlpI-digested fragments were finally transferred into pET15b (Novagen, Madison, WI), to facilitate the purification of the expressed proteins using a 6 × His encoding tag at the N-terminus. The DNA sequences were confirmed by Eurofins Genomics K.K. (Tokyo, Japan).
Expression and Purification of N. equitans AlaRS-α and AlaRS-β
Escherichia coli BL21-Codon Plus(DE3)-RIL (Stratagene, La Jolla, CA) containing the N. equitans AlaRS-α or AlaRS-β were grown and induced with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG), respectively. Cultures were harvested and cells were suspended in lysis buffer (20 mM Tris–HCl (pH 8.0), 300 mM NaCl, 10 mM imidazole), followed by the disruption of the cells by sonication on ice. The supernatant was collected after centrifugation and heated to 80 °C for 30 min and re-centrifuged. The supernatant was charged onto a Ni–NTA agarose (QIAGEN, Valencia, CA, USA) column equilibrated with lysis buffer and washed with wash buffer (20 mM Tris–HCl (pH 8.0), 300 mM NaCl, and 20 mM imidazole), and His-tagged AlaRS was eluted with elution buffer (20 mM Tris–HCl (pH 8.0), 300 mM NaCl, and 250 mM imidazole). Fractions containing a homogeneous protein enzyme were pooled and dialyzed twice in 1 L of dialysis buffer [40 mM Tris–HCl (pH 8.0), 200 mM NaCl, 0.2 mM EDTA] followed by concentration by Amicon Ultra Ultracel-30K (Merck Millipore, Billerica, MA, USA). Finally, the enzymes were stored in 50% glycerol.
Preparation of RNA Substrates
Chemically synthesized and/or PCR-amplified DNAs carrying the T7 promoter and the sequences corresponding to N. equitans tRNAAla, or minihelixAla and its mutants were used for RNA transcription. The in vitro transcripts were prepared in a reaction mixture containing 40 mM Tris–HCl (pH 8.0), 8 mM MgCl2, 2 mM spermidine, 1 mM dithiothreitol, 4 mM each NTP, 40 mM 5ʹGMP, template DNA (~ 0.2 mg/ml) using T7 RNA polymerase (Sampson and Uhlenbeck 1988; Hamachi et al. 2013). The transcripts were purified by denaturing 12% polyacrylamide gel electrophoresis. The concentrations of purified obtained RNA were determined from the UV absorbance at a wavelength of 260 nm using Implen NanoPhotometer (München, Germany). RNA tetraloop substrate was synthesized by Eurofins Genomics K.K. (Tokyo, Japan). Periodate oxidation of the 3ʹ-terminal adenosine residue of minihelixAla for generating 2ʹ,3ʹ-dialdehydes was performed using the method reported by Watanabe and coworkers (Kurata et al. 2003). Briefly, the reaction was performed for 60 min at 0 °C in the dark containing 10 mM NaIO4 and the total RNA was recovered by ethanol precipitation.
Aminoacylation Assay
The aminoacylation reaction was performed at 60 °C in a reaction mixture containing 50 mM HEPES–NaOH (pH 7.4), 10 mM MgCl2, 30 mM KCl, 2 mM dithiothreitol, 2 mM ATP and 10 μM l-[U–14C]alanine (132.0 mCi/mmol) (Moravek, Inc., Brea, CA, USA), with 15 μM RNA transcripts and either with [1] 0.5 μM AlaRS-α and 0.5 μM AlaRS-β, or [2] 0.5 μM AlaRS-α, or [3] 0.5 μM AlaRS-β, or [4] 5 μM AlaRS-α, or [5] 5 μM AlaRS-β. Aliquots were removed at specified time points, spotted onto trichloroacetic acid-soaked filter pads, washed with cold 5% trichloroacetic acid, and measured by scintillation counting (Schreier and Schimmel 1972). Kinetic parameters were determined by a Lineweaver–Burk plot of 1/v against 1/[S] ([S] is the RNA concentration and v is the observed initial velocity of alanylation) using various concentrations of RNA and proper concentrations of the enzymes that produce linear initial velocities (Table 1 and Supplementary Material, Fig. S2).
Table 1.
Kinetic parameters for aminoacylation of tRNAAla and minihelixAla by N. equitans AlaRS
| AlaRS-α + AlaRS-β | kcat (s−1) | Km (μM) |
kcat/Km (μM−1 s−1) |
kcat/Km (relative) |
|---|---|---|---|---|
| tRNAAla | 3.0 ± 0.36 | 1.6 ± 0.067 | 1.9 ± 0.31 | 1 |
| MinihelixAla | 1.9 ± 0.46 | 2.1 ± 0.42 | 0.9 ± 0.44 | 0.47 |
| AlaRS-α | kcat (s−1) | Km (μM) |
kcat/Km (μM−1 s−1) |
kcat/Km (relative) |
|---|---|---|---|---|
| tRNAAla | 0.021 ± 0.0021 | 7.7 ± 0.93 | 0.0027 ± 0.00059 | 1 |
| MinihelixAla | 0.028 ± 0.00050 | 23 ± 1.6 | 0.0012 ± 0.00030 | 0.44 |
The data represent the mean of triplicate experiments ± SEM
Results
Alanylation Activity of N. equitans AlaRS Toward tRNAAla and MinihelixAla
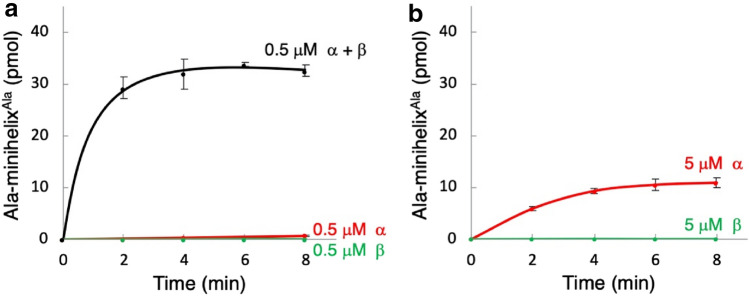
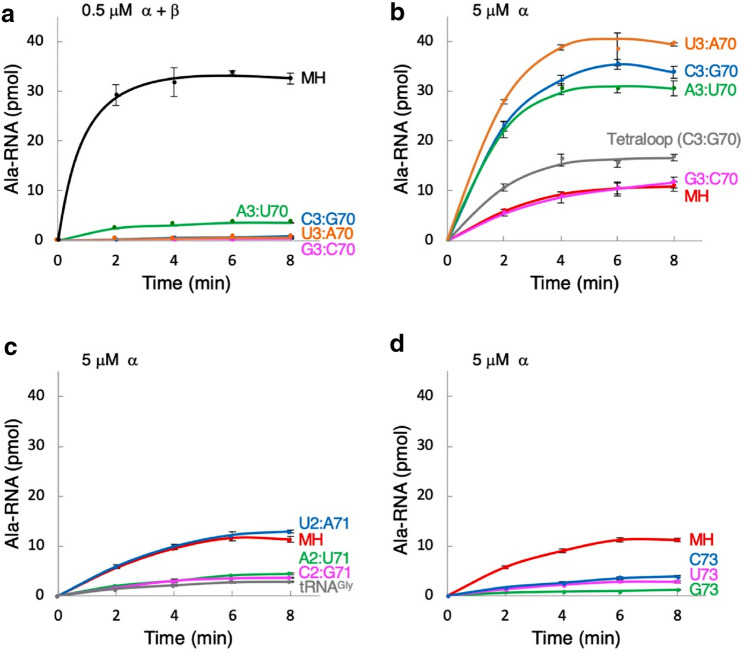
Although N. equitans grows at 80 °C (Huber et al. 2002), alanylation assays were performed at 60 °C to minimize evaporation of the reaction solution during the measurement. We first performed the alanylation assays using a combination of enzymes composed of [1] both AlaRS-α and AlaRS-β, or [2] AlaRS-α alone, or [3] AlaRS-β alone on the minihelixAla, which is made up of the acceptor stem on the T-arm of tRNAAla (Fig. 2a). At a concentration of 0.5 μM enzyme, minihelixAla was aminoacylated with the combination of enzymes containing AlaRS-α and AlaRS-β, but not with either AlaRS-α alone or with AlaRS-β alone (Fig. 3a). However, an increase in the concentration of AlaRS-α to 5 μM resulted in an apparent alanylation activity, whereas 5 μM AlaRS-β did not show any measurable activity (Fig. 3b). A combination of AlaRS-α and AlaRS-β was also active on tRNAAla and the kinetic parameters of aminoacylation for both on tRNAAla and the minihelixAla substrates were similar (Table 1). In the case of using AlaRS-α alone, the kinetic parameters of aminoacylation of minihelixAla was also nearly comparable to those of tRNAAla (Table 1).
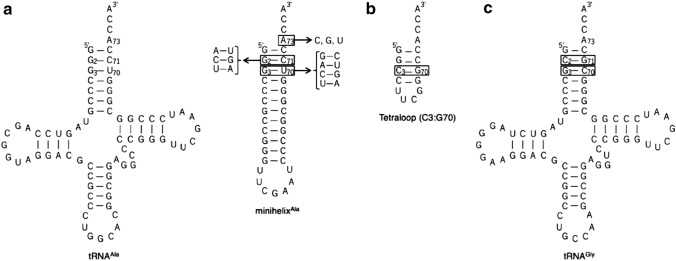
Fig. 2.
a Sequence of N. equitans tRNAAla (Left) and minihelixAla (Right). Numbering is based on that for wild type tRNAAla. MinihelixAla comprises the acceptor stem and the T-arm of the tRNAAla. Arrows indicate the substitutions performed in this study. b Sequence of 16-mer RNA tetraloop substrate including only the first four base pairs of the acceptor stem of tRNAAla, but with C3:G70 base pair. c Sequence of N. equitans tRNAGly
Fig. 3.
a Alanylation of 15 μM minihelixAla by the mixture of 0.5 μM AlaRS-α and AlaRS-β, by 0.5 μM AlaRS-α, and by 0.5 μM AlaRS-β. b Alanylation of 15 μM of minihelixAla by 5 μM AlaRS-α and by 5 μM AlaRS-β. Error bars represent the SEM of triplicate experiments
Alanylation Activity of AlaRS on MinihelixAla Mutants
The unexpected new result that AlaRS-α alone caused the alanylation activity led us to survey the role of G3:U70 of minihelices in aminoacylation by AlaRS-α as well as by the wild-type system composed of both AlaRS-α and AlaRS-β. We created four mutants with Watson–Crick base pairing at position 3–70 (A3:U70, G3:C70, C3:G70, and U3:A70), and assayed their alanylation activities. All G3:U70 mutants tested decreased the aminoacylation activity of 15 μM substrate using 0.5 μM of both AlaRS-α and AlaRS-β, (Fig. 4a). This is consistent with previous data showing that the G3:U70 base pair of tRNAAla is a major identity element by AlaRS (Hou and Schimmel 1988; McClain and Foss 1988). However, using 5 μM AlaRS-α, G3:C70 mutation had little effect on alanylation compared to the wild-type minihelixAla (Fig. 4b). Interestingly, the three remaining mutants (A3:U70, C3:G70, and U3:A70) showed higher level of activities compared with the wild-type minihelixAla (Fig. 4b). We also investigated the alanylation activity using an RNA tetraloop substrate. The tetraloop substrate was synthesized on the basis of the UUCG tetraloop (Tuerk et al. 1988; Woese et al. 1990) and the first four base pairs of the acceptor stem of tRNAAla (Shi et al. 1992), but the third base pair (G3:U70) was replaced with C3:G70 (Fig. 2b). AlaRS-α aminoacylated the RNA tetraloop substrate (Fig. 4b). Of the three mutants with Watson–Crick base pairing at position 2–71 (A2:U71, C2:G71, and U2:A71), only U2:A71 mutant showed wild-type levels of alanylation (Fig. 4c). A2:U71 and C2:G71 showed reduced activities (Fig. 4c). Finally, we examined the effect of the discriminator A73 substitution on alanylation of minihelixAla. Substitution of A73 with any other nucleotides caused a significant decrease in alanylation activity (Fig. 4d).
Fig. 4.
a Alanylation of wild-type minihelixAla (MH) and its mutants at positions 3–70 by the mixture of 0.5 μM AlaRS-α and AlaRS-β. b Alanylation of MH, its mutants at positions 3–70, and the RNA tetraloop substrate shown in Fig. 2b by 5 μM AlaRS-α. c Alanylation of MH, its mutants at positions 2–71, and tRNAGly by 5 μM AlaRS-α. d Alanylation of MH and its mutants at position 73 by 5 μM AlaRS-α. The concentration of RNA substrates was 15 μM. Error bars represent the SEM of triplicate experiments
Confirmation for Excluding Artifacts in the Aminoacylation by N. equitans AlaRS-α
In order to confirm that other factors did not contribute to the observed charging, we further monitored the reaction. To confirm that aminoacylation occurred at the 3′-end, the minihelixAla was pre-treated with NaIO4 to oxidize cis-hydroxyl groups (Kurata et al. 2003). No aminoacylation was detected in the reaction using N. equitans AlaRS-α (Fig. 5). In addition, the controls without any addition of enzyme or RNA did not cause alanylation (Fig. 5). Then, we studied whether AlaRS-α could alanylate tRNAGly to provide evidence that alanylation by AlaRS-α was not dependent on G3:U70 at the tRNA level. N. equitans tRNAGly has a sequence similarity with the tRNAAla despite the presence of C2:G71 and G3:C70 (Fig. 2c). Previously, we solved the X-ray crystallographic structure of N. equitans glycyl-tRNA synthetase (GlyRS), and by performing mutational analysis of the discriminator base A73 and the second base-pair (C2:G71) present on the acceptor stem of tRNAGly, we elucidated the importance of these positions as key identity elements for the recognition by GlyRS (Fujisawa et al. 2019). tRNAGly was alanylated by N. equitans AlaRS-α to a level comparable to that of the minihelixAla mutant with C2:G71 (Fig. 4c).
Fig. 5.
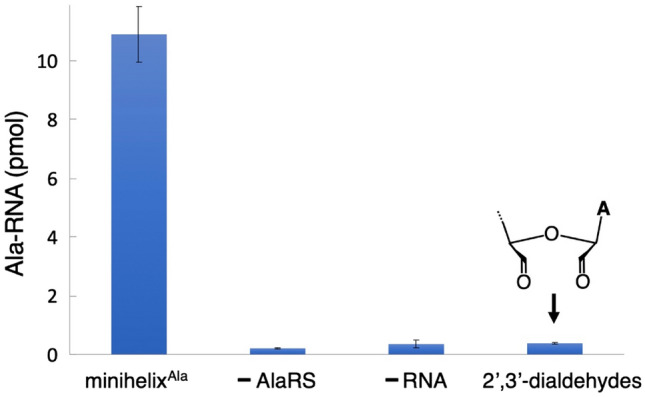
A comparative study of the aminoacylation reaction. From left to right: minihelixAla with AlaRS-α (minihelixAla), minihelixAla without AlaRS-α (-AlaRS), no minihelixAla with AlaRS-α (-RNA), and periodate-oxidized minihelixAla with AlaRS-α (2ʹ,3ʹ-dialdehydes). The scintillation count values after 8 min were used for comparison purposes. All reactions were performed with/without 15 μM of RNA substrates and 5 μM AlaRS-α. Error bars represent the SEM of triplicate experiments
Discussion
The unique G3:U70 base pair in the acceptor stem of tRNAAla has been shown to be a critical recognition site by AlaRS (Hou and Schimmel 1988; McClain and Foss 1988). Although many aaRSs specifically recognize the anticodons of the corresponding cognate tRNAs, the primordial genetic code is likely to have resided in the minihelix region as the modern tRNA-aaRS system is thought to have evolved from a primordial tRNA-aaRS system (Schimmel et al. 1993; Schimmel and Ribas de Pouplana 1995; Tamura 2015). Although we observed a higher aminoacylation activity in the condition involving a combination of AlaRS-α and AlaRS-β than that observed in a condition with AlaRS-α alone, our data supported two proposed features of the evolution of AlaRS: [1] N. equitans AlaRS behaves in a G3:U70 dependent manner when both AlaRS-α and AlaRS-β are present, and [2] N. equitans AlaRS-α alone can alanylate tRNAAla and minihelixAla, but it behaves in a G3:U70 independent manner. In this context, the G3:U70 may be a late-coming “operational RNA code” (Schimmel et al. 1993; Schimmel and Ribas de Pouplana 1995; Carter and Wills 2018), relevant to later alanylation systems incorporating further specificity through evolution of the AlaRS-β subunit.
N. equitans is a species of hyperthermophilic archaea that grows only in co-culture with another archaeon, Ignicoccus sp. strain KIN4/I (Huber et al. 2002). The existence of both genes of AlaRS-α and AlaRS-β in N. equitans suggests that both AlaRS-α and AlaRS-β function cooperatively in the organism. However, the behavior of AlaRS-α alone may inform our understanding of early evolution. Although N. equitans AlaRS-α is homologous to the E. coli AlaRS368N (Fig. 1 and Supplementary Material, Fig. S1), the former can alanylate tRNAAla, but the latter enzyme cannot alanylate (Jasin et al. 1983; Regan et al. 1987; Chihade and Schimmel 1999). In E. coli AlaRS, the region consisting of the residues 369 to 442 is also necessary to perform this reaction (Guo et al. 2009). Especially, a crucial residue for the recognition of G3 in E. coli AlaRS is located at 400th position (Asp400), that has been confirmed by an analysis of the crustal structure of A. fulgidus AlaRS (Naganuma et al. 2014).
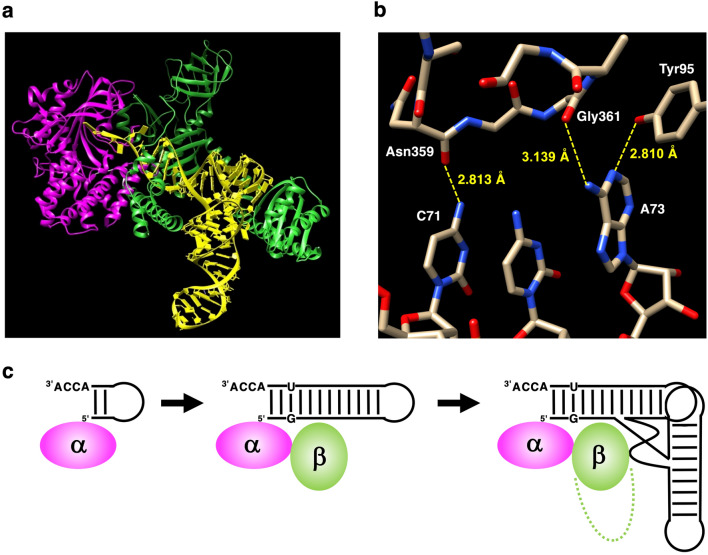
Further analysis of the sequence alignment in combination with the crystal structures of A. fulgidus AlaRS complexed with tRNAAla (Fig. 6a), suggests that AlaRS residues capable of hydrogen bond-formation with the tRNA are conserved. Asn359 in A. fulgidus recognizes the carbonyl oxygen at position 4 of U70 and this Asn is also conserved in N. equitans AlaRS-α (Fig. 1 and Supplementary Material, Fig. S1). The sequence LYISKG (LYDSHG in A. fulgidus) present in N. equitans AlaRS-β is likely important for the recognition of G3 (Fig. 1 and Supplementary Material, Fig. S1) (Naganuma et al. 2014). However, the residue corresponding to Asp450 of A. fulgidus AlaRS is Ile in N. equitans AlaRS-β (Fig. 1 and Supplementary Material, Fig. S1), and the Ile cannot form hydrogen bond seen in the crystal structure of A. fulgidus AlaRS and tRNAAla (Naganuma et al. 2014). Indeed, the “Asp-plus-Asn” architecture of AlaRSs may select G:U in different unconventional ways depending on the three domains of life (Chong et al. 2018).
Fig. 6.
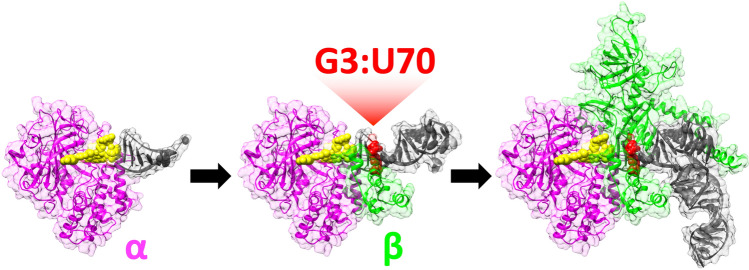
a Tertiary structure of A. fulgidus AlaRS complexed with tRNA (PDB ID: 3WQY). The corresponding regions to N. equitans AlaRS-α and AlaRS-β are colored in magenta and green, respectively, based on the multisequence alignment (Supplementary Material, Fig. S1). tRNA is shown in yellow. b Detailed stick representation of interactions between A. fulgidus AlaRS and C71 and A73 of tRNAAla. Distances within 3.5 Å between the amino acid residues of A. fulgidus AlaRS and any atoms of C71 and A73 are shown. c Schematic simplified representation of the proposed evolutionary processes of AlaRS-tRNAAla system. Before the appearance of the specific G3:U70, minimalist RNA and protein structures and their interactions resulted in requisite aminoacylation activities (Color figure online)
In the A. fulgidus AlaRS-tRNAAla (A3:U70) complex, the shifted A3 towards the major groove side finally causes the CCA-3′ region to be folded back into the non-reactive route (Naganuma et al. 2014). By the addition of β-chain to the α-chain of N. equitans AlaRS, it is possible that this selection may have occurred in a similar manner. However, the G3:U70 base pair-independent aminoacylation caused by AlaRS-α alone could have been performed by a different mechanism. The increase in alanylation activity in the case of three minihelixAla mutants with Watson–Crick base pairing at position 3–70 (A3:U70, C3:G70, and U3:A70) may suggest that the stability of the acceptor stem affected the alanylation potential or that this base pair serves additional unknown roles in the process. Subtle changes could induce a structural effect on the acceptor stem and affect binding of minihelixAla to the active site of AlaRS-α. Our data using the RNA tetraloop substrate with a C3:G70 base pair (Fig. 2b and Fig. 3b) are quite noteworthy, which strengthens the assertion that the G3:U70 base pair is not important for specific aminoacylation by N. equitans AlaRS-α. The thermal melting temperatures of these RNA tetraloop substrates were extensively investigated and they ranged over 70 °C when the discriminator was A (Shi et al. 1992). This could be a possible reason why the RNA tetraloop consisting of a C3:G70 base pair worked as a good substrate with N. equitans AlaRS-α at 60 °C (Fig. 2b and Fig. 3b).
In the complex structure of A. fulgidus AlaRS-tRNAAla (Naganuma et al. 2014), the distance between the amino nitrogen at position 6 of A73 and the carbonyl oxygen of Gly361 main chain is 3.14 Å, which may contribute the formation of a hydrogen bond between them (Fig. 6b). In addition, the distance between the nitrogen at position 1 of A73 and the hydroxyl oxygen of Tyr95 is 2.81 Å (Fig. 6b). In our present experiment using N. equitans AlaRS-α and minihelixAla system, the significant decrease in alanylation activity was detected by substitution of A73 with any other nucleotides (Fig. 4d). Although the electron donor–acceptor relationship of these two positions of A73 is the same as that of C73 (positions 4 and 3, respectively), productive interactions may be disrupted by the A73 to C73 mutation due to differences from the glycosyl bond. In the case of the 2–71 mutants, the U2:A71 mutant retained almost the same activity as that of the wild-type minihelixAla (with G2:C71) (Fig. 4c). In the G2:C71 base pair, the amino group at position 4 of C71 hydrogen-bonds with the carbonyl group at position 6 of G2 in the major groove. In the A. fulgidus AlaRS-tRNAAla complex (Naganuma et al. 2014), the distance between the amino nitrogen and the carbonyl oxygen of Asn359 main chain is 2.81 Å (Fig. 6b), which allows formation of a hydrogen bond between the amino hydrogen and the carbonyl oxygen. The electron donor–acceptor relationship of the amino group at position 4 of C71 is the same as that of the amino group at position 6 of A71. Therefore, the interaction of the amino hydrogen and the carbonyl oxygen may be retained on G2:C71 to U2:A71 substitution (Fig. 6b).
In summary, our data support a model where N. equitans AlaRS-α alone interacts with the end of the acceptor stem and the ACCA-3ʹ of tRNAAla, but not the G3:U70 base pair (Fig. 6c). This is contrast to prior knowledge regarding tRNAAla and AlaRS system. Although a specific mechanism of the G3:U70 pair recognition by the protein enzyme is not conserved, the G3:U70 pair as an identity element for alanine is conserved. This suggests that the G3:U70 pair appeared early during genetic code evolution and used as a “second genetic code” (Chong et al. 2018). However, before the appearance of the specific G3:U70, minimalist RNA–protein interactions may have resulted in requisite aminoacylation activities. The activity of AlaRS-α shown in the present study may reflect a vestige of such a primordial aminoacylation of an RNA comprising a small number of nucleotides, including the NCCA-3ʹ (Fig. 6c). In related to this hypothesis, it is suggested that the G3:U70 base pair recognition is actually a later addition to the original operational code (Carter and Wills 2018), as it is likely implemented by segments of the enzyme that were not accessible to the earliest ancestral aaRS forms (Carter 2014). Furthermore, the genomic tag model proposes that ancient linear RNA genomes possessed tRNA-like structures with a 3ʹ-terminal CCA (Weiner and Maizels 1987). A hairpin RNA with NCCA-3ʹ has also been proposed as the origin of homochiral aminoacylation in the RNA world (Tamura and Schimmel 2004, 2006; Ando et al. 2018; Tamura 2019). Recently, our in vitro selection study also isolated RNA aptamers composed of 21 nucleotides which bind to a tRNA-binding protein (Trbp) from an extremophile archaeon Aeropyrum pernix, and we noticed that the 3′-terminal single stranded CA nucleotides were essential for this binding (Ohmori et al. 2020). The C-terminal domain of methionyl-tRNA synthetase (MetRS-C) from N. equitans is homologous to Trbp and the binding of the split 3′-half tRNA with CCA to MetRS-C was stronger than that of the 5′-half tRNA (Suzuki et al. 2017). Extensive investigations using the mutants of N. equitans AlaRS-α and structural analysis will be our focus for future study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Harald Huber (Universität Regensburg, Germany) for kindly providing genomic DNA of N. equitans.
Funding
Grants-in Aid for Scientific Research from The Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (Grant No. 17K19210 to KT).
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
The original online version of this article was revised due to a retrospective Open Access order.
The original online version of this article was revised: ‘G73’ in the Figure 6b has been corrected as ‘A73’.
Misa Arutaki and Ryodai Kurihara have contributed equally to this work.
Change history
9/12/2024
A Correction to this paper has been published: 10.1007/s00239-024-10203-x
Change history
11/25/2020
A Correction to this paper has been published: 10.1007/s00239-020-09975-9
References
- Ando T, Takahashi S, Tamura K (2018) Principles of chemical geometry underlying chiral selectivity in RNA minihelix aminoacylation. Nucleic Acids Res 46:11144–11152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW Jr (2014) Urzymology: experimental access to a key transition in the appearance of enzymes. J Biol Chem 289:30213–30220 10.1074/jbc.R114.567495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW Jr, Wills PR (2018) Hierarchical groove discrimination by class I and II aminoacyl-tRNA synthetases reveals a palimpsest of the operational RNA code in the tRNA acceptor-stem bases. Nucleic Acids Res 46:9667–9683 10.1093/nar/gky600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihade JW, Schimmel P (1999) Assembly of a catalytic unit for RNA microhelix aminoacylation using nonspecific RNA binding domains. Proc Natl Acad Sci USA 96:12316–12321 10.1073/pnas.96.22.12316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong YE, Guo M, Yang XL, Kuhle B, Naganuma M, Sekine SI, Yokoyama S, Schimmel P (2018) Distinct ways of G: U recognition by conserved tRNA binding motifs. Proc Natl Acad Sci USA 115:7527–7532 10.1073/pnas.1807109115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G, Delarue M, Poch O, Gangloff J, Moras D (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347:203–206 10.1038/347203a0 [DOI] [PubMed] [Google Scholar]
- Francklyn C, Schimmel P (1989) Aminoacylation of RNA minihelices with alanine. Nature 337:478–481 10.1038/337478a0 [DOI] [PubMed] [Google Scholar]
- Francklyn C, Shi JP, Schimmel P (1992) Overlapping nucleotide determinants for specific aminoacylation of RNA microhelices. Science 255:1121–1125 10.1126/science.1546312 [DOI] [PubMed] [Google Scholar]
- Frugier M, Florentz C, Giegé R (1994) Efficient aminoacylation of resected RNA helices by class II aspartyl-tRNA synthetase dependent on a single nucleotide. EMBO J 13:2219–2226 10.1002/j.1460-2075.1994.tb06499.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa A, Toki R, Miyake H, Shoji T, Doi H, Hayashi H, Hanabusa R, Mutsuro-Aoki H, Umehara T, Ando T, Noguchi H, Voet A, Park SY, Tamura K (2019) Glycyl-tRNA synthetase from Nanoarchaeum equitans: the first crystal structure of archaeal GlyRS and analysis of its tRNA glycylation. Biochem Biophys Res Commun 511:228–233 10.1016/j.bbrc.2019.01.142 [DOI] [PubMed] [Google Scholar]
- Guo M, Chong YE, Beebe K, Shapiro R, Yang XL, Schimmel P (2009) The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science 325:744–747 10.1126/science.1174343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamachi K, Hayashi H, Shimamura M, Yamaji Y, Kaneko A, Fujisawa A, Umehara T, Tamura K (2013) Glycols modulate terminator stem stability and ligand-dependency of a glycine riboswitch. BioSystems 113:59–65 10.1016/j.biosystems.2013.05.004 [DOI] [PubMed] [Google Scholar]
- Hou YM, Schimmel P (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333:140–145 10.1038/333140a0 [DOI] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO (2002) A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63–67 10.1038/417063a [DOI] [PubMed] [Google Scholar]
- Ibba M, Losey HC, Kawarabayasi Y, Kikuchi H, Bunjun S, Söll D (1999) Substrate recognition by class I lysyl-tRNA synthetases: a molecular basis for gene displacement. Proc Natl Acad Sci USA 96:418–423 10.1073/pnas.96.2.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Söll D (2000) Aminoacyl-tRNA synthesis. Annu Rev Biochem 69:617–650 10.1146/annurev.biochem.69.1.617 [DOI] [PubMed] [Google Scholar]
- Jasin M, Regan L, Schimmel P (1983) Modular arrangement of functional domains along the sequence of an aminoacyl tRNA synthetase. Nature 306:441–447 10.1038/306441a0 [DOI] [PubMed] [Google Scholar]
- Kurata S, Ohtsuki T, Suzuki T, Watanabe K (2003) Quick two-step RNA ligation employing periodate oxidation. Nucleic Acids Res 31:e145 10.1093/nar/gng145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis SA, Schimmel P (1997) Small RNA oligonucleotide substrates for specific aminoacylations. tRNA: Structure, Biosynthesis, and Function. ASM Press, Washington DC, pp 349–370 [Google Scholar]
- McClain WH, Foss K (1988) Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science 240:793–796 10.1126/science.2452483 [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K, Schimmel P (1999) Atomic determinants for aminoacylation of RNA minihelices and relationship to genetic code. Acc Chem Res 32:368–375 10.1021/ar970148w [DOI] [Google Scholar]
- Naganuma M, Sekine S, Chong YE, Guo M, Yang XL, Gamper H, Hou YM, Schimmel P, Yokoyama S (2014) The selective tRNA aminoacylation mechanism based on a single G•U pair. Nature 510:507–511 10.1038/nature13440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori S, Wani M, Kitabatake S, Nakatsugawa Y, Ando T, Umehara T, Tamura K (2020) RNA aptamers for a tRNA-binding protein from Aeropyrum pernix with homologous counterparts distributed throughout evolution. Life 10:11 10.3390/life10020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau L, Calvin K, Hall M, Yuan J, Podar M, Li H, Söll D (2005a) The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge-helix-bulge motifs of joined tRNA halves. Proc Natl Acad Sci USA 102:17934–17939 10.1073/pnas.0509197102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau L, Münch R, Hohn MJ, Jahn D, Söll D (2005b) Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature 433:537–541 10.1038/nature03233 [DOI] [PubMed] [Google Scholar]
- Regan L, Bowie J, Schimmel P (1987) Polypeptide sequences essential for RNA recognition by an enzyme. Science 235:1651–1653 10.1126/science.2435005 [DOI] [PubMed] [Google Scholar]
- Sampson JR, Uhlenbeck OC (1988) Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci USA 85:1033–1037 10.1073/pnas.85.4.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P (1987) Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem 56:125–158 10.1146/annurev.bi.56.070187.001013 [DOI] [PubMed] [Google Scholar]
- Schimmel P, Giegé R, Moras D, Yokoyama S (1993) An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci USA 90:8763–8768 10.1073/pnas.90.19.8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P, Ribas de Pouplana L (1995) Transfer RNA: from minihelix to genetic code. Cell 81:983–986 10.1016/S0092-8674(05)80002-9 [DOI] [PubMed] [Google Scholar]
- Schimmel PR, Söll D (1979) Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem 48:601–648 10.1146/annurev.bi.48.070179.003125 [DOI] [PubMed] [Google Scholar]
- Schreier AA, Schimmel PR (1972) Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry 11:1582–1589 10.1021/bi00759a006 [DOI] [PubMed] [Google Scholar]
- Shi JP, Martinis SA, Schimmel P (1992) RNA tetraloops as minimalist substrates for aminoacylation. Biochemistry 31:4931–4936 10.1021/bi00136a002 [DOI] [PubMed] [Google Scholar]
- St John E, Liu Y, Podar M, Stott MB, Meneghin J, Chen Z, Lagutin K, Mitchell K, Reysenbach AL (2019) A new symbiotic nanoarchaeote (Candidatus Nanoclepta minutus) and its host (Zestosphaera tikiterensis gen. nov., sp. nov.) from a New Zealand hot spring. Syst Appl Microbiol 42:94–106 10.1016/j.syapm.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kaneko A, Yamamoto T, Nambo M, Hirasawa I, Umehara T, Yoshida H, Park SY, Tamura K (2017) Binding properties of split tRNA to the C-terminal domain of methionyl-tRNA synthetase of Nanoarchaeum equitans. J Mol Evol 84:267–278 10.1007/s00239-017-9796-6 [DOI] [PubMed] [Google Scholar]
- Tamura K (2015) Origins and early evolution of the tRNA molecule. Life 5:1687–1699 10.3390/life5041687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K (2019) Perspectives on the origin of biological homochirality on Earth. J Mol Evol 87:143–146 10.1007/s00239-019-09897-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Schimmel P (2004) Chiral-selective aminoacylation of an RNA minihelix. Science 305:1253 10.1126/science.1099141 [DOI] [PubMed] [Google Scholar]
- Tamura K, Schimmel PR (2006) Chiral-selective aminoacylation of an RNA minihelix: mechanistic features and chiral suppression. Proc Natl Acad Sci USA 103:13750–13752 10.1073/pnas.0606070103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gauss P, Thermes C, Groebe DR, Gayle M, Guild N, Stormo G, d’Aubenton-Carafa Y, Uhlenbeck OC, Tinoco I Jr (1988) CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci USA 85:1364–1368 10.1073/pnas.85.5.1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M, Beeson KY, Bibbs L, Bolanos R, Keller M, Kretz K, Lin X, Mathur E, Ni J, Podar M, Richardson T, Sutton GG, Simon M, Söll D, Stetter KO, Short JM, Noordewier M (2003) The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci USA 100:12984–12988 10.1073/pnas.1735403100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM, Maizels N (1987) tRNA-like structures tag the 3ʹ ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc Natl Acad Sci USA 84:7383–7387 10.1073/pnas.84.21.7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Winker S, Gutell RR (1990) Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci USA 87:8467–8471 10.1073/pnas.87.21.8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurch L, Giannone RJ, Belisle BS, Swift C, Utturkar S, Hettich RL, Reysenbach AL, Podar M (2016) Genomics-informed isolation and characterization of a symbiotic Nanoarchaeota system from a terrestrial geothermal environment. Nat Commun 7:12115 10.1038/ncomms12115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.