Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8, is associated with three proliferative diseases ranging from viral cytokine-induced hyperplasia to monoclonal neoplasia: multicentric Castleman's disease (CD), Kaposi's sarcoma (KS), and primary effusion lymphoma (PEL). Here we report a new latency-associated 1,704-bp KSHV spliced gene belonging to a cluster of KSHV sequences having homology to the interferon regulatory factor (IRF) family of transcription factors. ORFK10.5 encodes a protein, latency-associated nuclear antigen 2 (LANA2), which is expressed in KSHV-infected hematopoietic tissues, including PEL and CD but not KS lesions. LANA2 is abundantly expressed in the nuclei of cultured KSHV-infected B cells. Transcription of K10.5 in PEL cell cultures is not inhibited by DNA polymerase inhibitors nor significantly induced by phorbol ester treatment. Unlike LANA1, LANA2 does not elicit a serologic response from patients with KS, PEL, or CD as measured by Western blot hybridization. Both KSHV vIRF1 (ORFK9) and LANA2 (ORFK10.5) appear to have arisen through gene duplication of a captured cellular IRF gene. LANA2 is a potent inhibitor of p53-induced transcription in reporter assays. LANA2 antagonizes apoptosis due to p53 overexpression in p53-null SAOS-2 cells and apoptosis due to doxorubicin treatment of wild-type p53 U2OS cells. While LANA2 specifically interacts with amino acids 290 to 393 of p53 in glutathione S-transferase pull-down assays, we were unable to demonstrate LANA2-p53 interaction in vivo by immunoprecipitation. These findings show that KSHV has tissue-specific latent gene expression programs and identify a new latent protein which may contribute to KSHV tumorigenesis in hematopoietic tissues via p53 inhibition.
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV8), is the most recently described DNA tumor virus. It is the infectious trigger for Kaposi's sarcoma (KS), body cavity-based primary effusion lymphomas (PEL), and some subtypes of multicentric Castleman's disease (CD) (for review, see reference 37). KSHV-related CD is a polyclonal B-cell hyperplasia that is presumably driven by KSHV vIL-6 secretion as well as other viral proteins. In contrast, PEL are B-cell lymphomas that generally have a monoclonal origin as determined by immunoglobulin gene rearrangement and viral terminal repeat analyses (7, 20, 36). Terminal repeat analyses by Judde and colleagues (20) have also demonstrated that KS tumors can have an oligo- or monoclonal pattern and may evolve from a polyclonal hyperplasia into a monoclonal tumor. Thus, KSHV may contribute to cell proliferation through secretion of viral cytokines and induction of cellular cytokines, as in the case of CD, as well as through the expression of transforming viral oncogenes, particularly in the case of PEL.
The KSHV genome has significant sequence homology to all classes of herpesviruses but is unique among the human herpesviruses in encoding an extensive number of regulatory genes which have been pirated from the host genome during its evolution (30, 36). While a number of these genes have homology to known cellular oncogenes or transform rodent cell lines in vitro (2, 14, 26), only a small number of KSHV genes are routinely found to be expressed in tumor tissues. vBCL-2, vIRF1, vGPCR, and K01 are examples of KSHV proteins which might contribute to cell transformation in vitro but are not appreciably expressed in most KSHV-infected KS or PEL tumors (21, 24, 32, 38).
KSHV-infected PEL cell lines constitutively express three viral genes, vFLIP (open reading frame K13 [ORFK13]), vCYC (ORF72), and the latency-associated nuclear antigen 1, LANA1 (ORF73), which are not inducible by tetradecanoyl phorbol acetate (TPA) or inhibited by phosphonoformic acid (PFA) and thus are unambiguously designated as latent or class I genes. These three proteins are transcribed on the major polycistronic latent transcripts, LT1 and LT2 (10, 39, 42). In vitro studies demonstrate that the viral cyclin associates with cyclin-dependent kinase 4 and 6 and phosphorylates pRB (8, 16, 28). LANA1 is believed to bind to the origin of replication to tether the viral genome to host chromatin during mitosis, effecting equal segregation of viral genome during division (3). LANA1 also binds to p53 and inhibits p53-mediated transcriptional activity and apoptosis (13). vCYC overexpression induces apoptosis (31), and it is at least theoretically possible that this may be inhibited in situ by the antiapoptotic activities of other latency expressed proteins, such as vFLIP and LANA1.
Viral protein expression is highly restricted in KS and PEL tumors. Presently, only LANA1 protein has been shown by immunohistochemistry to be expressed in situ in all cells infected by KSHV (11, 22, 32). Viral cyclin and ORFK12 transcripts have been identified by in situ hybridization in all KSHV-infected cells (9, 34); however, protein localization has yet to be performed. No other viral proteins examined thus far, including vIL-6 (K2), minor capsid protein (ORF26), K8, K8.1, vIRF1 (K9), K10, K11, PF-8 (ORF59), and ORF65, have a similar in situ constitutive pattern of expression (21, 32).
KSHV gene expression studies remain controversial. Since PEL cell lines can be manipulated into lytic replication by TPA and butyrate, studies on cultured cell lines have been used to classify KSHV genes into mutually exclusive latent and lytic classes based on transcription kinetics (40). Frequently, KSHV expression patterns from cultured cell studies are assumed to be similar in tumor tissues in situ without direct evidence. However, a number of KSHV genes are expressed at low levels in resting PEL cell lines but are induced to high expression levels during TPA treatment and thus have properties of both latent and lytic genes (analogous to the Epstein-Barr virus [EBV] LMP1 expression pattern). This pattern of gene expression has been referred to as class II expression (37). Recent studies demonstrate that results from expression studies in tissue culture cannot be uniformly applied to human tumor tissues, in part because KSHV may have tissue-specific gene expression patterns. vIL-6, for example, behaves as a class II protein in tissue culture cell lines and is expressed in hematopoietic-derived cells but generally not in KS lesions (29). Thus, determining precisely which viral genes are likely to play a role in KSHV-related pathogenesis requires direct tissue examination of each tumor type. Discovery of additional genes that are constitutively expressed in KSHV-induced disorders is particularly important since these genes are likely to play a role in cell growth dysregulation.
For these reasons, discovery of a KSHV gene having a tissue-specific expression profile is important, particularly if the encoded protein is functionally capable of contributing to cell proliferation. In this paper we describe a new KSHV gene (K10.5) expressed in KSHV-infected hematopoietic tissues. This gene is located in a region containing a cluster of viral sequences with limited homology to the interferon regulatory factor (IRF) family of proteins (36). vIRF1 is encoded by ORF K9 and inhibits interferon-induced transcription and fully transforms NIH 3T3 cells (12, 14, 27, 44). vIRF1 binds to histone acetyltransferase transcriptional coadaptors (5, 19) and induces cell transformation by activating the cMYC oncogene through an interferon-stimulated response element called the PRF element (19). Based on these findings and the fact that other tumor viruses target the same tumor suppressor pathways as KSHV, Jayachandra et al. found that both EBV (or HHV4) EBNA2 and adenovirus E1A proteins also activate cMYC but use differing sets of coadaptors from those used by vIRF1 (19). vIRF1 additionally inhibits p53- and Fas-induced apoptosis (5; S. Jayachandra, P. S. Moore, and Y. Chang, unpublished observations). vIRF1, however, is not generally expressed in PEL or KS and is therefore unlikely to contribute to these diseases, although it may be important in the pathogenesis of CD (21, 32). Another IRF-like KSHV ORF encoding vIRF2 and having NF-κB-inhibitory activity has been described (6). We show here that LANA2 is a B-cell-specific factor that antagonizes p53 tumor suppressor functions and is expressed during latency.
MATERIALS AND METHODS
Cell cultures.
BC-1, BCP-1, BCBL-1, BJAB, Ramos, and P3HR1 (obtained from the American Type Culture Collection [ATCC]) cells were maintained in RPMI 1640 (Gibco BRL, Gaithersburg, Md.) supplemented with 10 to 20% fetal bovine serum (Gibco BRL). SAOS-2, U2OS, and COS7 cells (obtained from ATCC) and IRF1/2 (−/−) cells (a gift from T. Taniguchi [43]) were maintained in Dulbecco modified Eagle medium (Gibco BRL) with 10% fetal bovine serum. Induction of viral lytic replication and gene transcription were performed by treatment of cells with 20 ng of TPA (Sigma Chemical Co., St. Louis, Mo.)/ml. Cells were harvested 48 h after treatment. To inhibit viral DNA replication, PFA (Sigma) was added at a concentration of 0.5 mM either alone or in the presence of 20 ng of TPA/ml for 48 h.
Northern analysis.
Total RNA was extracted by the RNAzol method (TelTest, Friendswood, Tex.) followed by mRNA selection using a PolyATract mRNA isolation kit (Promega, Madison, Wis.). Five hundred nanograms of the poly(A)-selected mRNA was loaded per lane on formaldehyde 1% agarose gel and transferred onto nylon membranes (GeneScreen; NEN Research Products, Boston, Mass.). The V1 probe consists of the entire K9 ORF (14). The V1 probe as well as V2, V3, and V4 probes derived from PCR products (see Fig. 2) (V2F [5′-GGGAATTCGATGCCTAAAGCCGGTGGC-3′], V2R [5′-TGCGGCCGCTCAAACCTCACACCCCCT-3′], V3F [5′-GGGAATTCGATGTACCACGTGGGACAG-3′], V3R [5′-TGCGGCCGCTTAGTCATCACATGTAAC-3′], V4F [5′-GGGAATTCGATGCCTCGCTACACGGAG-3′], and V4R [5′-GGGAATTCGCTACCTCTGGGCTTTTTT-3′]) were labeled by random priming using synthetic hexanucleotide primers (RediPrime DNA labeling system; Amersham International, Amersham, England) and [32P]dCTP. Hybridization was performed in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–50% formamide–5× Denhardt's solution–2% sodium dodecyl sulfate (SDS)–10% dextran sulfate–100 μg of denatured sheared salmon sperm DNA per ml at 42°C. A β-actin probe was used to standardize the amount of RNA loaded.
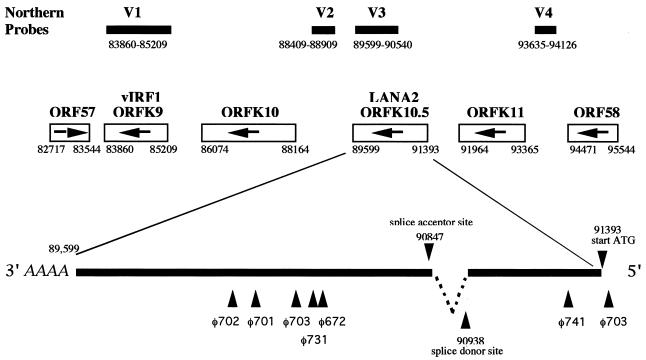
FIG. 2.
Transcript map of LANA2 in KSHV genomic environs showing V1 (vIRF1), V2, V3, and V4 probes used for Northern blot hybridization. The V3 probe was used to screen a TPA-induced BC-1 cDNA library. Six phages (φ672, φ701, φ702, φ703, φ731, and φ741) were isolated containing inserts of various sizes. One full-length, 1,735-bp cDNA starting at nt 91425 and terminating at nt 89599 was identified and sequenced from phage φ703. This cDNA contained a start ATG at position 91393 and a splice donor/acceptor site corresponding to nt 90938/90837.
cDNA library screening.
cDNA phage libraries of TPA-stimulated BC-1 cells were constructed and amplified in the ZAP Express vector according to the manufacturer's protocol (Stratagene, La Jolla, Calif.). Plaques (3.2 × 105) were screened following the manufacturer's suggested protocols, with the V3 probe made from the V3F and V3R PCR primers (used in Northern analysis, see above).
Plasmids.
pcDNA.LANA2 was obtained by excising the full-length LANA2 insert with EcoRI and NotI digestion from phagemid pBK-CMV-LANA2 (φ703 screened from the cDNA library). This insert was then cloned in frame into EcoRI/NotI-prepared pcDNAHis3.1B vector (Invitrogen, Carlsbad, Calif.). pMET7.LANA2 was constructed by digesting pBK-CMV-LANA2 with PstI and XbaI. The isolated insert was then cloned into the PstI/XbaI-prepared pMET7 mammalian expression vector (41). The fidelity of all cloning junctions was verified on an ABI 377 Sequenator (Applied Biosystems Inc., Foster City, Calif.). pG13-Luc, a reporter plasmid containing 13 tandem p53 response elements derived from the p21 promoter, was a gift from W. El-Deiry and B. Volgelstein (4). pGL-3 control (Promega) was used as a control vector for luciferase transient transfection assays. The glutathione S-transferase (GST)-p53 (full-length) plasmid and the C-terminal fragment of p53 (GST-p53 [290 to 393]) plasmid were a gift from W. Gu (17). DNA sequences corresponding to amino acids (aa) 1 to 100 and 100 to 290 of human p53 were amplified by PCR and subcloned into pGEX-KG (18) to generate the protein expression plasmids GST-p53 (1 to 100) and GST-p53 (100 to 290). The pcDNA.p53 expression plasmid was a gift of R. T. Hay (35). pEGFP-F* (gift of W. Jiang) expresses green fluorescent protein (GFP) and was used as a marker for pcDNA.LANA2 and/or pcDNA.p53 transfection to gate fluorescent cells by a fluorescence-activated cell sorter (FACS). The plasmids containing the Gal4 binding domain (Gal4 BD), pAS2-1, and the Gal4 activation domain (Gal4 AD), pGAD424, as well as the plasmids containing the DNA BD-murine p53 fusion protein PVA3 and the DNA AD-murine p53 fusion protein pGADp53 and control plasmids pCL1, PLAM5′, pGBT9, and pTD1 were obtained from Clontech Laboratories (Palo Alto, Calif.).
Reporter assays.
SAOS-2 or U2OS cells were seeded at a density of 5 × 104 cells per plate in six-well plates 1 day before transfection. Transient transfections with plasmid DNA were performed using Cell Phect (Pharmacia Biotech, Piscataway, N.J.). In all experiments, total amounts of transfected DNA were equalized between wells using empty pcDNA3.1HisC (Invitrogen). Cells were harvested and lysed, and luciferase activity was measured by using standard protocols after 48 h. pcDNAHis3.1LacZ (Invitrogen) was used to normalize luciferase activity to transfection efficiency. In this way, reporter expression levels were normalized to the amount of transfected plasmid for each experimental condition. Each measurement was performed in triplicate, with experiments independently replicated at least three times. p53-null SAOS-2 cells were cotransfected with 2 μg of pG13-Luc in the presence or absence of 0.5 μg of pcDNA.p53 with or without pcDNA.LANA2 (0.5 to 1 μg). U2OS cells were cotransfected with 2 μg of pG13-Luc in the presence or absence of pcDNA.LANA2 (0.5 to 1 μg/well) and were treated with 0.4 μM doxorubicin (Sigma) 18 h posttransfection.
FACS analysis.
SAOS-2 cells (106) were transfected (Cell Phect) with 1 μg of the GFP-expressing plasmid, pEGFP-F*, in the presence of pcDNA.p53 (4.5 μg) and/or pcDNA.LANA2 (4.5 μg) or the empty expression vector. U2OS cells were transfected with 1 μg of pEGFP-F* in the presence or absence of the expression vector pcDNA.LANA2 (4.5 to 9 μg) and were treated with doxorubicin 18 h posttransfection. Cells were washed 48 h after transfection in phosphate-buffered saline (PBS) and were fixed at 4°C in 80% ethanol in PBS for 1 h. Cells were then washed three times with PBS and incubated for 30 min at 37°C in 0.1% Triton X-100–0.1% trisodium citrate–0.5 μg of RNase A/ml–50 μg of propidium iodide/ml. The DNA content of cells gated for GFP expression was then analyzed using a FACScan flow cytometer.
Activation of caspase 8.
Caspase 8 activation was determined using the synthetic oligopeptide substrate Ac-LETD-AFC from Bio-Rad Laboratories (Hercules, Calif.) as described by the manufacturer, and the samples were read on a Bio-Rad VersaFluor fluorometer.
GST pull-down assay.
GST in vitro binding assays were performed using in vitro translated [35S]methionine-labeled LANA2 incubated with p53 GST fusion proteins (GST-p53 [full length], GST-p53 (1 to 100), GST-p53 (100 to 290), GST-p53 (290 to 393), and GST alone. In vitro translated [35S]methionine-labeled p53 was incubated with GST-LANA2 and GST alone.
Coimmunoprecipitation.
LANA2 (20 μg of pcDNA.LANA2) and p53 (20 μg of pcDNA.p53) were expressed in SAOS-2 cells by cotransfection and were immunoprecipitated with anti-LANA2 CM-8B6 or CM-10A2 antibodies or with D0-1 (Santa Cruz Biotech, Santa Cruz, Calif.), Pab 1801 (Santa Cruz), and Ab-1 (Oncogene, Cambridge, Mass.) anti-p53 antibodies. Protein complexes were resolved by SDS–10% polyacrylamide gel electrophoresis (PAGE) and transferred onto nitrocellulose membrane. LANA2 was detected using CM-8B6, and CM-10A2, and p53 was detected using D0-1, Pab 1801, and Ab-1 by immunoblotting and enhanced chemiluminescence (ECL; Amersham).
Immunohistochemistry of KSHV-infected tissues and controls.
Glass slides were obtained with the Cytospin 3 apparatus (Shandon Lipshaw, Pittsburgh, Pa.) using 25,000 washed cells per spot. These cytospins were air dried overnight, fixed in acetone for 4 min at room temperature, air dried for 30 min, and processed for immunohistochemistry. Ten KS skin lesions, five lymph nodes from patients with CD, and biopsies from two cases of PEL were investigated for protein expression of LANA2 by immunohistochemistry. One CD lymph node also contained KS. Control tissues were tonsil biopsies from KSHV-negative children. Mouse monoclonal antibody clone CM-10A2 was made against bacterially produced GST-LANA2 and was confirmed to be specific to the 80-kDa LANA2 protein on Western blot hybridization of KSHV-infected cell lysates (BC-1, BCBL-1, BCP-1) compared to KSHV-uninfected cell lysates (BJAB, P3HR1, Ramos). CM-10A2 was nonreactive to GST protein by both enzyme-linked immunosorbent assay and Western blot hybridization. The rabbit polyclonal antibody against LANA1, R UK163, was the kind gift of B. Chandran. Microwave EDTA pretreatment was required for antigen retrieval. Antibody binding was revealed using peroxidase-labeled goat anti-mouse antisera (DAKO, Glostrupp, Denmark) followed by tyramide amplification (DuPont/NEN, Boston, Mass.). Reactions were developed using diaminobenzidine (DAB; Sigma) or amino ethyl carbazole (AEC; DAKO) as chromogenic substrates, and sections were counterstained with hematoxylin. Antibodies to KSHV vIL-6 (cytoplasmic staining) and PF-8 (perinuclear staining) were used for comparative antibody controls for tissue staining (32). For fluorescence double-immunostaining with LANA1 and LANA2, fluorescein isothiocyanate-conjugated goat anti-mouse was used in combination with goat anti-rabbit antisera (Southern Biotechnology) followed by avidin Texas red (Vector Laboratories, Burlingame, Calif.).
Serologic analysis.
COS7 cells were plated at a density of 106 cells per 90-mm plate 1 day prior to transfection. Transfections were performed (Cell Phect) using 6 μg of pMET7.LANA2 or pMET7 empty vector as control. Cells were harvested 48 h posttransfection, placed in 500 μl of lysis buffer, incubated on ice for 20 min, centrifuged, and resuspended in 200 μl of nuclear extraction buffer (20 mM HEPES [pH 7.9], 0.4 M NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium vanadate, 1-μg/ml (each) aprotinin, leupeptin, and pepstatin). Protein (15 μg) was loaded into a single-well comb slot for Western blot analysis by SDS–12.5% PAGE. After transferring the protein onto nitrocellulose membrane, strips were cut and incubated in CM-10A2 primary antibody or patient serum overnight. After washing, strips were incubated for 1 h in anti-mouse or anti-human immunoglobulin G alkaline phosphatase-conjugated secondary antibody (1:3,500 dilution; Sigma).
Yeast two-hybrid assay.
LANA2 was fused either to Gal4 AD in the plasmid pGAD424 or to Gal4 DNA BD in the plasmid pAS2-1. The plasmids containing the murine p53 fused to Gal4 AD or Gal4 BD were provided by Clontech. The yeast strain Y-190 was used for this two-hybrid assay. Plasmids are introduced into Y-190 by the standard lithium acetate transformation method. To test for potential protein-protein interaction, transformants were screened for growth in medium lacking histidine but in the presence of 15 mM 3-aminotriazol (His + phenotype) or assayed for β-galactosidase activity (blue phenotype) in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
Nucleotide sequence accession number.
The sequence of the new latency-associated 1,704-bp KSHV spliced gene reported here has been submitted to GenBank and assigned accession number AY008303.
RESULTS
Identification of the K10.5 (LANA2) transcript.
ORFK10.5 was originally described as a sequence feature rather than an open reading frame in Russo et al.'s conservative annotation of the BC-1 genome (36). This gene was one of four KSHV sequences showing limited homology to cellular IRFs. Neipel and colleagues subsequently annotated a theoretical ORFK10.1 based on their sequencing of the KSHV genome from a KS lesion (30). We therefore sought to directly determine whether the four IRF-like motifs, including the putative K10.1 gene, in the KSHV genomic sequence represent expressed gene products using TPA-induced and uninduced BC-1 cell mRNAs.
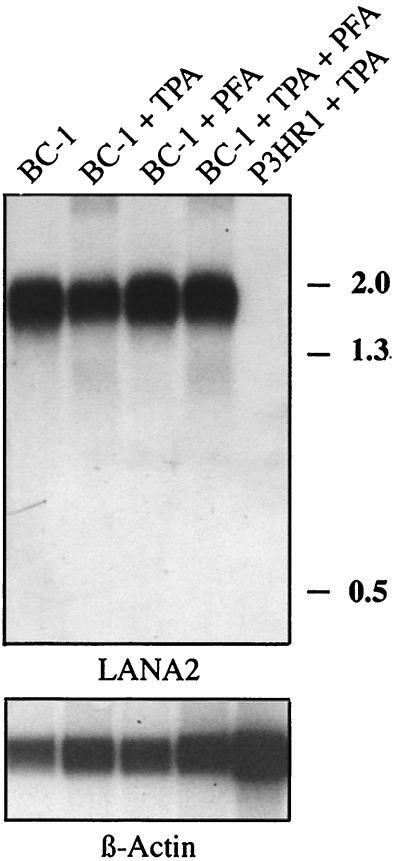
Four probes spanning nucleotide (nt) 83860 to 85209 (V1 probe), nt 88409 to 88909 (V2 probe), nt 89599 to 90540 (V3 probe), and nt 93635 to 94126 (V4 probe) were generated by PCR (see Fig. 2). The V1 probe corresponds to the ORFK9 region encoding vIRF, and expression patterns for this probe matched those previously described (27, 29, 37) in that the 1.5-kb mRNA is weakly detected in unstimulated BC-1 cells and induced to high levels of expression after TPA treatment. While probes V2 and V4 (corresponding to the vIRF2 protein gene 6) did not hybridize to detectable transcripts in unstimulated or stimulated BC-1 cells, the V3 probe corresponding to ORFK10.5 hybridized to a 1.8-kb transcript that was absent from the KSHV negative control cell line, P3HR1 (Fig. 1). Expression of the K10.5 transcript is not affected by TPA stimulation or PFA inhibition, thereby qualifying it as a latent transcript in BC-1 cells. Similar results were also obtained using BCBL-1 cells (data not shown).
FIG. 1.
Northern hybridization of BC-1 mRNA with an ORFK10.5 probe. Probe hybridization for mRNA from uninduced BC-1 cells (lane 1), BC-1 cells treated with 20 ng of TPA/ml for 48 h (lane 2), BC-1 cells treated with 0.5 mM PFA (lane 3), BC-1 cells treated with 20 ng of TPA/ml and 0.5 mM PFA (lane 4), and KSHV-negative, EBV-infected P3HR1 cells treated with 20 ng of TPA/ml (lane 5) are shown. The probe hybridizes to a 1.8-kb band which is not induced by TPA nor inhibited by PFA treatment, consistent with a latent pattern of viral gene transcription. The same blot is stripped and reprobed with β-actin as a control for loading.
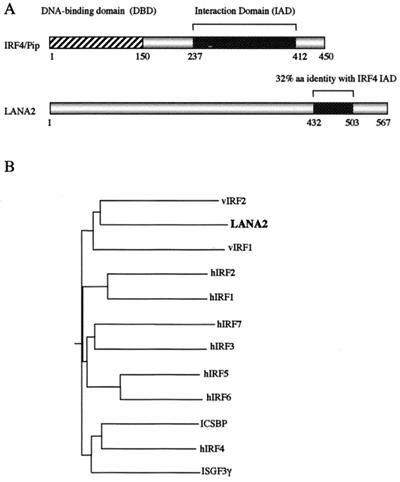
Since the transcript size identified by the V3 probe is incompatible with the predicted transcript for putative ORFK10.5, we screened a cDNA library made from TPA-stimulated BC-1 cells to identify spliced transcripts. Of 3.2 × 105 plaques screened, six positive phages were found with inserts ranging between 503 and 1,735 bp in length (Fig. 2). The clones were sequenced and one (φ703) contained the full-length cDNA transcript beginning 31 bp upstream from a putative start ATG (nt 91393) and having a stop codon at nt 89599. All six cDNAs have a 3′ termination coordinate at nt 89599. Conserved splice-donor sites (nt 90938 and 90847) are present in the φ703 insert, but only one of the five other phage inserts extended through the 5′ splice junction. Splicing results in a 1,704-bp full-length transcript for the newly annotated gene, which is designated ORFK10.5 to distinguish it from the unspliced 3′ exon previously designated K10.1 (Fig. 2). This ORF is composed of a novel 455-bp 5′ exon that is joined to the 1,339-bp 3′ exon previously annotated as ORFK10.1 from genome sequence analysis (30). Based on its constitutive expression in BC-1 cells and its nuclear localization (see below), we refer to the protein encoded by ORFK10.5 as latency-associated nuclear antigen 2 (LANA2). LANA2 has low overall homology to members of the IRF family. Members of the IRF family of proteins have at least two common functional domains: an amino-terminal DNA BD and a carboxyl-terminal activation domain. LANA2 does not have conserved tryptophans in its amino terminus, which are required for DNA binding by IRF members, but has 32% amino acid identity over a 71-bp region corresponding to the IRF4 interaction domain (Fig. 3A). Comparative phylogenetic analysis shows that the KSHV proteins vIRF1, vIRF2, and LANA2 have a common branch point and appear to have arisen through gene duplication of a captured ancestral IRF-like cellular gene (Fig. 3B). A previous study from our laboratory surveying transcription of the KSHV genome in BC-1 cells (37) failed to detect this transcript, possibly due to the use of large probes covering this region, which resulted in a low signal intensity on Northern blotting.
FIG. 3.
(A) Comparison of motif domains between IRF4/Pip and LANA2. IRF4 encodes a 450-aa protein with an N-terminal DNA-binding domain (DBD) defined by five tryptophan residues. This characteristic is not found in LANA2 (567 aa); however, a 213-bp region of LANA2, between aa 432 and 503, shows 32% amino acid identity with the C-terminal interaction domain (IAD) of IRF4. (B) Phylogenetic tree for KSHV and human IRF proteins. LANA2 is most closely related to vIRF1 and vIRF2, suggesting a common origin from an ancestral IRF-like gene. Amino acid sequences were aligned using ClustalW, and the phylogenetic tree was generated using the Bootstrap NJ tree 1000 program. Protein peptide sources (GenBank accession numbers) are as follows: hIRF1 [87992], hIRF2 [539621], hIRF3 [4504725], hIRF4 [2497445], hIRF5 [4504727], hIRF6 [3122293], hIRF7 [4809288], ICSBP(hIRF8) [6016308], ISGF3γ [266392], KSHV vIRF1 [4929348], KSHV vIRF2 [3152728], and KSHV LANA2 [AY008303]. A phylogenetic tree comparing the IRF-like proteins from the RRV26-95 isolate and the KSHV IRF-like proteins has been published by Alexander et al (1).
LANA2 expression in vitro and in vivo.
Immunostaining using CM-10A2 mouse monoclonal antibody against LANA2 on KSHV-infected cell lines (BC-1, BCBL-1, BCP-1) shows a fine granular nuclear pattern in all preparations (Fig. 4). This is similar to the subnuclear distribution of LANA1 (ORF73) (11, 15, 23, 33). Double staining for LANA1 and LANA2 shows that the two proteins can colocalize to some degree but that LANA2 has a much more diffuse pattern (Fig. 5). In mitotic cells, in which LANA1 bridges viral and cellular chromosomes to allow equal viral episome segregation, LANA1 aggregates with the mitotic spindle (3). LANA2, however, is excluded from these LANA1-containing mitotic figures, suggesting that LANA2, unlike LANA1, does not play an important role in episome segregation during mitosis (Fig. 5).
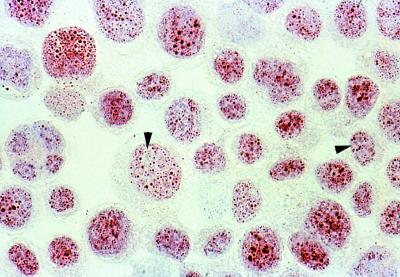
FIG. 4.
Cytospin preparation of TPA-stimulated BCBL-1 cells immunostained with CM-10A2 mouse monoclonal antibody against LANA2. LANA2 demonstrates a finely speckled nuclear pattern exclusive of nucleolar zones in essentially all BCBL-1 cells (magnification, ×60 hematoxylin counterstain).
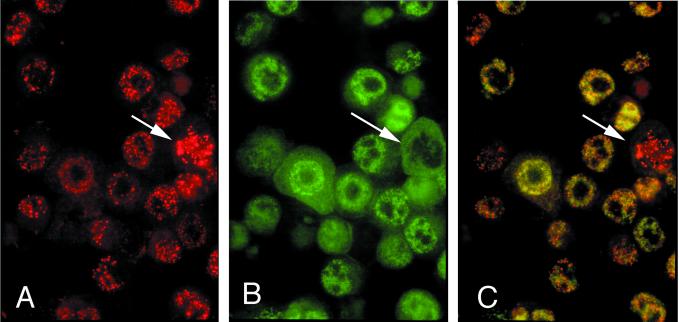
FIG. 5.
Immunofluorescence double colocalization of LANA1 and LANA2 in KSHV-infected BCBL-1 cells. (A) LANA1 protein (red) in a coarsely speckled nuclear distribution. (B) Diffuse, finely speckled nuclear pattern of LANA2 protein (green). (C) Double filter; colocalization of LANA1 and LANA2. (A, B, C) Although some subnuclear regions show the distinct dispersal of the two proteins exclusive of each other, yellow nuclear staining is also evident in other areas, possibly representing colocalization of a subfraction of LANA1 and LANA2. Cells undergoing mitosis (arrow) appear to express only LANA1 exclusive of LANA2 (C) (magnification, ×100; Texas red and fluorescein isothiocyanate).
Previous studies demonstrate that some genes (e.g., ORFK9) become dysregulated in PEL tissue culture and are expressed in established in vitro cell lines but not parental PEL tumors. Other proteins, such as vIL-6, are expressed only in situ in a minority of PEL tumor cells (32). LANA2, in contrast, is expressed in virtually all KSHV-infected cells in PEL and in the majority of the KSHV-infected cells in Castleman's disease tumors (Fig. 6D and E and 7B). LANA2 is not appreciably expressed in KS spindle cells taken from skin biopsies (Fig. 6F). This is most clearly seen in Fig. 7 in a lymph node containing both KS (endothelial cell origin) and CD (B-cell origin) tumors. LANA2 expression in this lymph node occurs exclusively in the CD tumor cells but not in KS spindle cells (Fig. 7B).
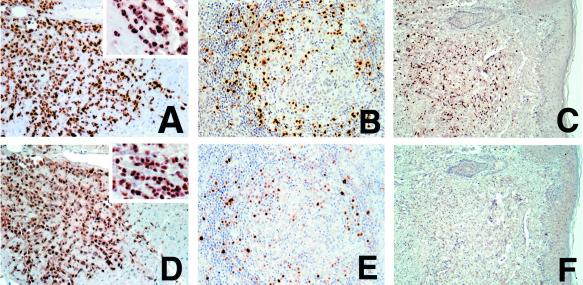
FIG. 6.
Immunolocalization of LANA1 compared with LANA2 in KSHV-infected disorders. Shown are LANA1 immunolocalization in a pericardial PEL-infiltrating cardiac muscle (A), a germinal center from a lymph node with multicentric Castleman's disease (B), and a cutaneous KS lesion biopsy (C). Adjacent sections of the same tissues are immunostained for LANA2 (D, E, and F). All tumor cells in PEL express both LANA1 (A) and LANA2 (D), and the majority of the KSHV-infected mantle zone lymphocytes in CD express both LANA1 (B) and LANA2 (E). However, while the majority of KS spindle cells express LANA1 (C), none express LANA2 (F).
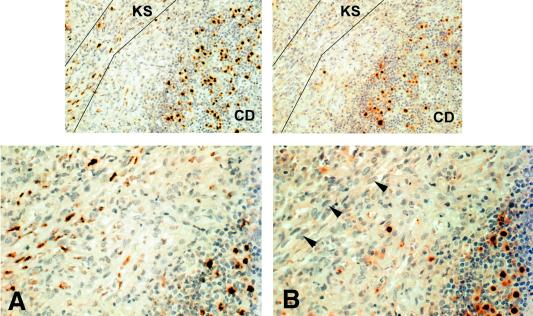
FIG. 7.
Immunolocalization of LANA1 (A) compared with LANA2 (B) in a lymph node with CD as well as KS. While the KS spindle cells (area within guide lines) and some of the mantle zone lymphocytes show strong nuclear positivity to LANA1, the adjacent section immunostained with LANA2 only shows this protein expressed in the lymphocyte subpopulation of KSHV-infected cells in the mantle. The CD serves as an internal positive control for the negative LANA2 immunostaining of KS spindle cells.
Lack of seroreactivity to LANA2 in serum from KSHV-infected patients.
Unlike LANA1, LANA2 is unlikely to be a useful Western blot antigen for detecting KSHV antibodies. LANA2 expressed in COS7 cells failed to react on Western blotting with serum from patients with various KSHV-related disorders. None of 14 sera from individuals with AIDS-KS (n = 4), classical KS (n = 4), KSHV-seropositive Castleman's disease (n = 4), or PEL (n = 2) showed serologic reactivity to LANA2 (Table 1). Negative control sera from four blood donors (seronegative for ORF65 and LANA1 antigens) were also nonreactive whereas the supernatants from two mouse monoclonal LANA2 hybridoma clones (CM-10A2 and CM-8B6) were positive. We cannot exclude the possibility that other antigen formats (e.g., enzyme-linked immunoassay) might reveal a useful pattern for LANA2 seroreactivity.
TABLE 1.
Negative seroreactivity to LANA2 in patients with KS, PEL, and MCDa
| Donor category | No. of sera | Result (no. seroreactive/total no.) from:
|
||
|---|---|---|---|---|
| Western blot with ORF65 | IFA with LANA1 | Western blot with LANA2 | ||
| AIDS-KS | 4 | 4/4 | 4/4 | 0/4 |
| Classical KS | 4 | 4/4 | 4/4 | 0/4 |
| MCD | 4 | 4/4 | 4/4 | 0/4 |
| PEL | 2 | 2/2 | 2/2 | 0/2 |
| Blood donors | 4 | 0/4 | 0/4 | 0/4 |
| LANA2 hybridoma clonesb | 2 | ND | ND | 2/2 |
MCD, multicentric Castleman's disease; IFA, immunofluorescence assay; ND, not done.
Clones CM-10A2 and CM-8B6.
LANA2 inhibits p53 transactivation.
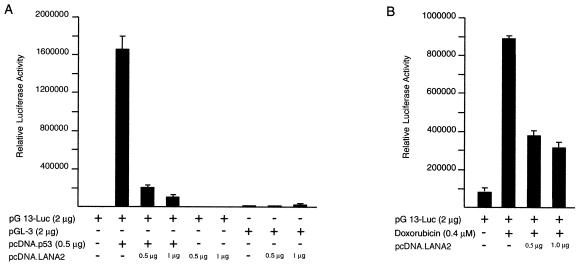
Since LANA1 inhibits p53-mediated transcription and apoptosis (13), we examined the effects of LANA2 on p53 function using the pG13-Luc promoter reporter (containing 13 copies of the p53 response element) transiently transfected into SAOS-2 (p53 null) osteosarcoma cells. Transient expression of 0.5 μg of p53 plasmid in SAOS-2 cells resulted in an 800-fold activation of the pG13-Luc reporter, which was inhibited by 87% on cotransfection of 0.5 μg of pcDNA.LANA2 expression plasmid. This transcriptional repression was specific since the pGL-3 control promoter activity (Fig. 8A), as well as the activity of a Gal4 reporter plasmid (not shown), were unaffected by pcDNA.LANA2 cotransfection. This effect is not due to squelching since no transcriptional activation was seen at low levels of LANA2 expression and increasing amounts of pcDNA.LANA2 resulted in a monotonic repression of p53 activity on the pG13 reporter.
FIG. 8.
Inhibition of p53 transcriptional activity by LANA2. A representative luciferase assay shows inhibition of reporter gene expression by transient transfection of pcDNA.LANA2. (A) SAOS-2 cells were transfected with 2 μg of plasmid pG13-Luc reporter plasmid together with 0.0 or 0.5 μg of pcDNA.p53 and 0.5 or 1 μg of pcDNA.LANA2, as indicated. For control, SAOS-2 cells were transfected with the reporter plasmid pGL3-control and 0.0, 0.5, or 1 μg of pcDNA.LANA2. (B) U2OS cells were transfected with 2 μg of plasmid pG13-Luc reporter plasmid with or without 0.5 or 1 μg of pcDNA.LANA2 and were treated with 0.4 μM doxorubicin.
To determine if the same effect is present during endogenous p53 activation, these experiments were repeated in U2OS cells (wild type for p53) with and without treatment with 0.4 μM doxorubicin, a chemotherapeutic agent which induces p53-mediated apoptosis. Doxorubicin treatment resulted in 13-fold activation of the pG13-Luc reporter, and this effect was inhibited 57% by 0.5 μg of pcDNA.LANA2 transfection (Fig. 8B).
LANA2 protein-protein interactions.
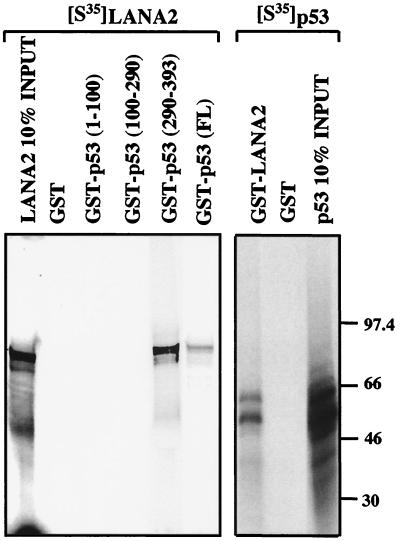
To determine if inhibition of p53 transactivation is due to direct interaction with p53 protein, we performed full-length and truncated GST-p53 pull-down assays using in vitro translated [35S]methionine-labeled LANA2. As shown in Fig. 9, GST-p53 fusion protein precipitates LANA2 in vitro whereas no interaction is seen with GST protein alone. LANA2 interaction is localized to the region of p53 comprising aa 290 to 393, and no interaction occurs with the truncated p53 constructs containing aa 1 to 100 or 100 to 290. In the reverse pull-down experiments, GST-LANA2 but not GST alone showed specific interaction with in vitro-translated full-length p53.
FIG. 9.
In vitro GST pull-down assays using [35S]methionine-labeled LANA2 or p53. LANA2 interacts with full-length p53 protein as well as the p53 region between aa 290 and 393.
In vivo coimmunoprecipitation experiments, however, failed to demonstrate direct interaction between LANA2 and p53 (data not shown). In experiments using naturally abundant p53 from BCBL-1 cells or SAOS-2 cells in which p53 protein was overexpressed, no coimmunoprecipitation was detected for LANA2 and p53 using either LANA2 (CM-10A2 and CM-8B6) or p53 (D0-1, Pab 1801, Ab-1) monoclonal antibodies. In part these experiments were inconclusive since we noted an unusual phenomenon in that D0-1 (Santa Cruz), Pab 1801 (Santa Cruz) and Ab-1 (Oncogene) antibodies directed against p53 directly cross-reacted with LANA2. This was confirmed by direct Western blotting with these antibodies and the bacteria-derived GST-LANA protein in the absence of p53. We thus cannot exclude artifactual p53-LANA2 interactions in the GST pull-down assays or that antibody binding occurs at the LANA2-p53 interaction site(s) which interferes with the immunoprecipitation reaction, since the binding was done under native conditions. Yeast two-hybrid assays between LANA2 and full-length p53 failed to clarify whether direct protein-protein interactions occur in vivo (data not shown). LANA2 cloned into the Gal4 BD cassette is toxic to the yeast and could not be evaluated. LANA2 cloned into the Gal4 AD cassette and p53 into the Gal4 BD cassette, however, showed no interaction by the β-galactosidase assay.
LANA2 inhibits p53-mediated apoptosis.
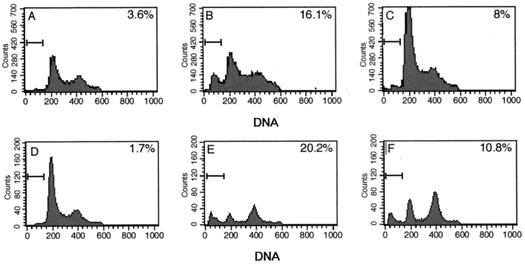
SAOS-2 cells are null for pRB as well as p53, and overexpression of wild-type p53 in SAOS-2 cells results in apoptosis, as indicated by the subdiploid fraction (20%) of cells staining with propidium iodide in a cell-sorting profile (Fig. 10). In this experiment, cells were cotransfected with p53 and GFP expression plasmids, and DNA content analysis was performed only on cells gated for GFP. When LANA2 was expressed together with p53 in SAOS-2 cells (Fig. 10C), a marked diminution in subdiploid cells (from 20 to 10.8%) occurred, indicating a specific inhibition of p53-mediated apoptosis and genomic fragmentation. Similar results were obtained for U2OS cells, which have wild-type p53, treated with 0.4 μM doxorubicin for 30 h, indicating that LANA2 can inhibit activation of endogenous p53 resulting from doxorubicin treatment (Fig. 10F). This was confirmed by caspase 8 activation fluorometric assays. Doxorubicin-treated U2OS cells transfected with pcDNA.LANA2 showed lower levels of caspase 8 activation than doxoribicin-treated U2OS cells transfected with pcDNA empty vector control (data not shown).
FIG. 10.
LANA2 inhibits p53-induced apoptosis. SAOS-2 cells were transfected with pEGFP-F* and the empty expression vector pCDNA3.1 (A), pCDNA.p53 (B), or pCDNA.p53 and pCDNA.LANA2 (C). Total DNA in all transfections was normalized using the empty expression vector. After 48 h, cells were fixed and stained with propidium iodide. The cellular DNA content was analyzed by flow cytometry. U2OS cells were transfected with pEGFP-F* and the empty expression vector pcDNA (D and E) or pcDNA.LANA2 (F). Eighteen hours after transfection, cells were treated with doxorubicin (0.4 μM) (E and F), and the cells were processed for DNA content analysis 30 h posttreatment. Numbers indicate the percentage of cells in the sub-G1 phase of the cell cycle.
DISCUSSION
LANA2 is one of the few KSHV proteins which has been found to be expressed in PEL and CD cells in vivo (11, 22, 32). However, unlike LANA1, LANA2 is not expressed in the vast majority of KS spindle cells. These findings reinforce the concept that KSHV is capable of multiple latency expression programs, and genes that are expressed in some tissues or cell lines may be silenced in others. LANA2 differs from vIL-6, another KSHV protein whose protein expression is also limited to B cells, in that vIL6 is expressed in a minority population of PEL tumor cells. Since vIL-6 is a secreted cytokine, limited expression of vIL-6 may nonetheless contribute to the pathogenesis of PEL tumors. In contrast, LANA2 expression is uniformly present in PEL tumor cells, indicating that it too may have a critical role in maintaining the PEL tumor cell phenotype. These patterns of expression could be expected if the vIL-6 promoter is activated by cytokine signaling pathways that are dependent on the local cellular milieu (J. Osborne, Y. Chang, and P. S. Moore, unpublished observation), whereas the LANA2 promoter is activated by B-cell transcription factors.
KSHV is a gammaherpesvirus which, like EBV, has part of its natural lifecycle in CD19+ B lymphocytes. It is apparent that a portion of the KSHV genome is devoted to maintenance of the virus in the B-cell environment. B cells, for example, respond to antigen by activating immunoreceptor signaling pathways to achieve rapid clonal expansion. Under normal circumstances, induction of cell death by apoptosis occurs after B-cell expansion to prevent lymphocytic hyperplasia (25). The ability of LANA2 to prevent p53-mediated B-cell apoptosis would be an apparent benefit in maintaining an expanded population of infected cells or in preventing p53 pathway activation as part of a cellular antiviral response. While our in vitro studies suggest that LANA2 inhibition of p53 activity is through direct protein-protein interaction, caution is necessary in interpreting these results since they were not confirmable through in vivo interaction assays. The p53 region binding LANA2 (aa 290 to 393) in GST pull-down assays includes the p53 tetramerization and regulatory domains, as well as residues acetylated by p300 (17), suggesting a plausible mechanism.
The reasons why KSHV possesses two latency-expressed viral proteins, LANA1 and LANA2, to target the same p53 tumor suppressor protein are unclear. LANA1 is constitutively expressed in both KS lesions and KSHV-infected hematopoietic tissues and therefore appears to have a broader functional spectrum than LANA2. It is important to note that our LANA2 experiments showing functional p53 inhibition were performed in osteosarcoma cell lines and so, at least under the conditions of our assays, LANA2 inhibition of p53 is not unique to B cell lines.
Regardless of the mechanism for p53 inhibition, LANA2 is a likely candidate protein involved in cell proliferation in hematopoietic tissues. Inhibition of p53-induced apoptosis may contribute to B-cell hyperplasia in Castleman's disease and to cell transformation in PEL cells. Although KSHV vCYC is constitutively expressed on LT1 and LT2 in all infected cell lines, stable expression of this cyclin homolog has been difficult to achieve in vitro since it induces apoptosis (31). Direct inhibition of both pRB and p53 signaling pathways by vCYC together with LANA1 and LANA2 could theoretically contribute to proliferative and/or neoplastic expansion of infected B cells.
ACKNOWLEDGMENTS
We thank Evelyn Hale and Marie Weiss for excellent technical help. We thank the scientists who have kindly provided reagents used in the study and who are individually listed in Materials and Methods. We thank Mary Ann Accavitti and the UAB hybridoma core facility for assistance with the production of anti-LANA2 monoclonal antibodies.
This work was supported by National Cancer Institute grant R01 CA67391.
ADDENDUM
We are aware that another group, Mori and colleagues, has independently characterized LANA2 as a novel KSHV latency protein (submitted for publication). While our manuscript was under revision, Lubyova and Pitha (J. Virol. 74:8194–8201, 2000) published a description of the K10.5 gene product (called “vIRF-3”) but found that it is an inducible gene using reverse transcription-PCR, in contrast to our data and that of Mori et al.
REFERENCES
- 1.Alexander L, Denekamp L, Knapp A, Auerbach M R, Damania B, Desrosiers R C. The primary sequence of rhesus monkey rhadinovirus isolate 26–95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol. 2000;74:3388–3398. doi: 10.1128/jvi.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bais C, Santomasso B, Coso O, Arvanitakis L, Geras Raaka E, Gutkind J S, Asch A S, Cesarman E, Gerhengorn M C, Mesri E A. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 4.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 5.Burysek L, Yeow W S, Lubyova B, Kellum M, Schafer S L, Huang Y Q, Pitha P M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burysek L, Yeow W S, Pitha P M. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2) J Hum Virol. 1999;2:19–32. [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences are present in AIDS-related body cavity based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden-Kent D, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 9.Davis M A, Sturzl M A, Blasig C, Schreier A, Guo H G, Reitz M, Opalenik S R, Browning P J. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1868–1874. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- 10.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande J P, Weiss R A, Alitalo K, Boshoff C. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci USA. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flowers C, Flowers S, Nabel G. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor confers resistance to the antiproliferative effect of interferon-alpha. Mol Med. 1998;4:402–412. [PMC free article] [PubMed] [Google Scholar]
- 13.Friborg J, Jr, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 14.Gao S-J, Boshoff C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (vIRF) is an oncogene that inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1986. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 15.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians, and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 16.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 18.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 19.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Judde J G, Lacoste V, Briere J, Kassa-Kelembho E, Clyti E, Couppie P, Buchrieser C, Tulliez M, Morvan J, Gessain A. Monoclonality or oligoclonality of human herpesvirus 8 terminal repeat sequences in Kaposi's sarcoma and other diseases. J Natl Cancer Inst. 2000;92:729–736. doi: 10.1093/jnci/92.9.729. [DOI] [PubMed] [Google Scholar]
- 21.Katano H, Sato Y, Kurata T, Mori S, Sata T. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease. Virology. 2000;269:335–344. doi: 10.1006/viro.2000.0196. [DOI] [PubMed] [Google Scholar]
- 22.Katano H, Sato Y, Kurata T, Mori S, Sata T. High expression of HHV-8-encoded ORF73 protein in spindle-shaped cells of Kaposi's sarcoma. Am J Pathol. 1999;155:47–52. doi: 10.1016/S0002-9440(10)65097-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 24.Kirshner J R, Staskus K, Haase A, Lagunoff M, Ganem D. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J Virol. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein G. Epstein-Barr virus strategy in normal and neoplastic B cells. Cell. 1994;77:791–793. doi: 10.1016/0092-8674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 26.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A, Desrosiers R C, Jung J U. Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpesvirus. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung J U. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72:5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 30.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ojala P M, Tiainen M, Salven P, Veikkola T, Castanos-Velez E, Sarid R, Biberfeld P, Makela T P. Kaposi's sarcoma-associated herpesvirus-encoded v-cyclin triggers apoptosis in cells with high levels of cyclin-dependent kinase 6. Cancer Res. 1999;59:4984–4989. [PubMed] [Google Scholar]
- 32.Parravicini C, Chandran B, Corbellino M, Berti E, Paulli M, Moore P S, Chang Y. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed J A, Nador R G, Spaulding D, Tani Y, Cesarman E, Knowles D M. Demonstration of Kaposi's sarcoma-associated herpes virus cyclin D homolog in cutaneous Kaposi's sarcoma by colorimetric in situ hybridization using a catalyzed signal amplification system. Blood. 1998;91:3825–3832. [PubMed] [Google Scholar]
- 35.Rodriguez M S, Desterro J M P, Lain S, Midgley C A, Lane D P, Hay R T. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc. Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 39.Sarid R, Wiezorek J S, Moore P S, Chang Y. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and R-U5 segment of the human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, Lamphier M S, Aizawa S, Mak T W, Taniguchi T. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell. 1994;77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 44.Zimring J C, Goodbourn S, Offermann M K. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]