Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV), also called human herpesvirus 8 (HHV-8), is the likely etiological agent of Kaposi's sarcoma and primary effusion lymphoma. Common to these malignancies is that tumor cells are latently infected with KSHV. Viral gene expression is limited to a few genes, one of which is the latency-associated nuclear antigen (LANA), the product of ORF73. Examination of the primary sequence of LANA reveals some structural features reminiscent of transcription factors, leading us to hypothesize that LANA may regulate viral and cellular transcription during latency. In reporter gene-based transient transfection assays, we found that LANA can have either positive or negative effects on gene expression. While expression of a reporter gene from several synthetic promoters was increased in the presence of LANA, expression from the human immunodeficiency virus (HIV) long terminal repeat (LTR)—and from NF-κB-dependent reporter genes—was reduced by LANA expression. In addition, the promoter of KSHV ORF73 itself is activated up to 5.5-fold by LANA. This autoregulation may be important in tumorigenesis, because two other genes (v-cyclin and v-FLIP) with likely roles in cell growth and survival are also controlled by this element. To identify cellular genes influenced by LANA, we employed cDNA array-based expression profiling. Six known genes (and nine expressed sequence tags) were found to be upregulated in LANA-expressing cell lines. One of these, Staf-50, is known to inhibit expression from the HIV LTR; most of the other known genes are interferon inducible, although the interferon genes themselves were not induced by LANA. These data demonstrate that LANA expression has effects on cellular and viral gene expression. We suggest that, whether direct or indirect in origin, these effects may play important roles in the pathobiology of KSHV infection.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), also called human herpesvirus 8 (HHV-8), is associated with KS and with two lymphoproliferative diseases: primary effusion lymphomas (PEL) and multicentric Castleman's disease (18). Common to these neoplasms is the fact that the majority of tumor cells are latently infected (5, 46). Viral gene expression in this stage is restricted to a small number of genes, one of which is the latency-associated nuclear antigen (LANA). This antigen was first identified by reactivity with sera from KS patients in immunofluorescence assays (IFA) on latently infected PEL cell lines (19, 25). Using Northern blot analysis and expression cloning, it was subsequently shown that LANA is encoded by ORF73 of KSHV (24, 26, 36). ORF73 encodes a protein of about 1,162 amino acids (aa) and is expressed from a singly spliced mRNA of 5.7 kb which also bears ORF72 and ORF71 coding sequences. In situ hybridization revealed that nearly all cells in the KS lesion express ORF73 mRNA (12), and the expression of LANA protein has been demonstrated by immunohistochemistry in all malignancies associated with KSHV (13).
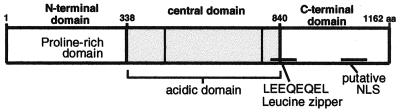
The identification of ORF73 as LANA opened the door for studies addressing the function of this protein during latency. Its nuclear localization and restriction to latency suggested that LANA was a formal analog of the Epstein-Barr virus (EBV) nuclear antigens (EBNAs). EBNAs play an important role in the pathogenesis of EBV, contributing directly to plasmid maintenance, as well as to host cell transformation (for reviews see references 28 and 35). However, comparison of the predicted primary structure of ORF73 did not reveal any amino acid homology to EBV, genes. Examination of the predicted amino acid sequence of LANA reveals several interesting features. ORF73 encodes a polypeptide with a predicted molecular mass of about 132 kDa. Sequence inspection suggests that the protein can be divided into three distinct domains (Fig. 1). The N-terminal 340 aa are extremely proline rich and contain several PXXP motifs, which are potential binding sites for SH3 domain-containing proteins (1). The central region contains three different highly repetitive blocks of acidic residues; similar domains often function in transcriptional activation in viral and cellular transcription factors (33, 42). The C-terminal domain contains a putative nuclear localization site, and partially overlapping with the central domain is a leucine zipper repeat motif, raising the possibility of homo- and hetero-oligomerization. Both the N- and C-terminal domains contain several potential phosphorylation sites.
FIG. 1.
Domain structure of LANA. The central domain has three families of simple repeats: EEDD, DEQQQ or DEEQQ, and LEEQEQEL. The length of the second repeat is variable between isolates (20, 24).
Many of these features are also found in proteins involved in regulation of gene expression and recall similar features of EBNA-1 and EBNA-2. EBNA-1 is the oriP binding protein required for plasmid maintenance during the latency of EBV. EBNA-1 also transactivates viral promoters while bound to oriP during latency (17, 34, 43). EBNA-2 is a transcriptional transactivator which regulates viral transcription during latency by up-regulating the oncogenic latency-associated membrane proteins (LMP1, LMP2A, and LMP2B). In addition, EBNA-2 regulates a variety of cellular target genes within latently infected cells. Most prominent is the strong activation of CD23; other EBNA-2 target genes have important roles in adhesion (ICAM-1) and the control of apoptosis (Bcl-2), to mention two examples. While EBNA-1 binds specific target sites on DNA, EBNA-2 activates promoters primarily through interaction with other transcription factors. In addition to EBNA-1 and EBNA-2, the EBNA-3 gene encodes three different proteins (two of which are important for transformation) generated by alternative splicing. EBV thus encodes an entire family of latency-associated nuclear proteins (for a review see reference 28 and references therein).
So far, LANA is the only latency-associated nuclear antigen identified for KSHV. We speculated that in view of its large size and modular architecture, LANA might have subsumed in one polypeptide many of the functions which in EBV are distributed among the several EBNAs. Indeed, it was recently shown that, like EBNA-1, LANA plays a role in plasmid maintenance (4, 8). Specifically, we hypothesized that (by analogy to EBNA-1 and EBNA-2) LANA may function as a transcriptional regulator and thereby modify viral and cellular gene expression during latency. Here we show, by using transient transfection assays in combination with cDNA array-based expression profiling, that LANA can both positively and negatively regulate viral and cellular gene expression, and we identify several targets of this regulation.
MATERIALS AND METHODS
Cell lines.
COS-7 and 293 cells were obtained from the American Type Culture Collection. Cell monolayers were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin at 37°C under a 5% CO2 atmosphere. BJAB cells (kindly provided by Elliot Kieff, Harvard University) were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 0.05 mM 2-mercaptoethanol, 1 mM sodium bicarbonate, 2 mM l-glutamine, penicillin, and streptomycin at 37°C under a 5% CO2 atmosphere.
Plasmids.
KSHV DNA was derived from a KSHV λ library derived from a KS lesion (47) unless otherwise indicated. The effector plasmid pcDNA3/73, which we had generated during our initial studies on the identification of LANA (25), contains viral sequences from nucleotide (nt) 127394 to nt 123663 (all nucleotide numbering refers to GenBank sequence no. U75698 [41]). The ORF73 fragment was cloned from a λ clone as a KpnI/NheI fragment into the KpnI/XbaI sites of the pcDNA3 polylinker (Invitrogen). PCEP4/73 was generated similarly by cloning this fragment into the KpnI/NheI sites of the pCEP4 (Invitrogen) polylinker. In IFA, COS-7 and 293 cells transfected with pcDNA3/73 produce the characteristic nuclear punctate pattern. By Western blot analysis we detected a doublet running at about 200 and 220 kDa, which is identical to the pattern observed in BCBL-1 cells (25). The unusually slow mobility of this protein (computed molecular mass, 132 kDa) was also observed by other investigators and is believed to be, in part, due to its central acidic region and possible posttranslational modifications (24, 36; A. Polson and D. Ganem, unpublished data). The synthetic promoter vectors were a gift from J. Alwine, University of Pennsylvania (29). The human immunodeficiency virus (HIV) long terminal repeat (LTR) reporter construct (pC15CAT) was obtained from the National Institutes of Health (NIH) AIDS Research and Reference Program. The HIV LTR luciferase reporter was generated by ligating a HindIII fragment from pC15CAT into pGL3/Basic (Promega). PDD83 contains the LANA promoter harboring KSHV sequences from nt 128159 to nt 127607 cloned into pGL3/Basic (Promega) as previously described (12). The NF-κB luciferase reporters contained dimerized NF-κB consensus sites from the major histocompatibility complex class I (MHC-I) promoter and the HIV promoter upstream of a simian virus 40 (SV40) promoter without enhancer.
Transient transfection assays.
COS-7 and 293 cells were plated at 4 × 105 cells per well in 6-well plates 8 to 12 h prior to transfection. DNA for reporter and effector plasmids was transfected using Fugene (Gibco-BRL) according to the manufacturer's instructions. To monitor transfection efficiency, we transfected pcDNA3/lacZ into parallel wells and stained fixed cells for β-galactosidase activity. Transfection efficiencies were generally between 25 and 35%.
We did not use internal standards in each transfection for normalization because the presence of LANA influenced a wide range of reporters in these experiments. An SV40 promoter as well as a cytomegalovirus (CMV) promoter driving β-galactosidase was activated by LANA at low concentrations but was inhibited at higher concentrations. Additionally, we tested pGKβ-gal (a gift from J. Jung, Harvard University), driving β-galactosidase expression from a phosphoglucokinase promoter, a housekeeping gene, as an internal standard. This reporter was consistently down-regulated between two- and fivefold by the presence of different concentrations of LANA expression plasmid—even in experiments where the LANA promoter (pDD83) was up-regulated by the same construct (see Fig. 3). We therefore performed Bradford assays on all lysates and normalized all relative light unit (RLU) values to the protein concentrations as previously reported for other proteins which have the ability to affect a wide range of different reporters in transient transfection assays (40a).
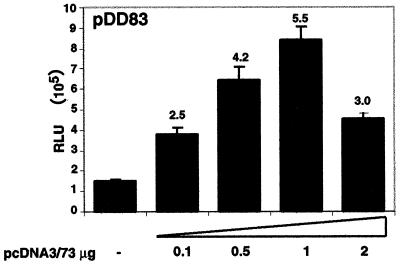
FIG. 3.
LANA up-regulates its own promoter. Each transfection contained 0.1 μg of pDD83 as the reporter either alone or with varying amounts of the LANA expression vector (pcDNA3/73). The total DNA amounts were kept identical by adding pBSII as filler DNA, and cell extracts were prepared 48 h after transfection. Bars represent means ± standard deviations from two experiments carried out in duplicate. Numbers above bars show relative activities based on mean values. All relative light unit values are normalized to protein concentrations.
For chloramphenicol acetyltransferase (CAT) assays, cells were harvested 48 h after transfection and CAT activity was determined as previously described (39). Briefly, cells were washed with phosphate-buffered saline (PBS) and scraped into PBS. After centrifugation cells were resuspended in 100 μl of 0.25 M Tris-HCl and lysed by three freeze-thaw cycles. Debris was spun down, and cell extracts (30 μl) were assayed for CAT activity in a reaction mixture containing 14C-labeled chloramphenicol and n-butyryl coenzyme A (CoA) in 0.25 M Tris-HCl. Acetylated chloramphenicol was collected by organic extraction using hexene-xylene (2:1) and transferred into scintillation fluid; then counts per minute were measured in a Beckman scintillation counter. Luciferase assays were performed as recommended by the manufacturer's manual. Briefly, 48 h after transfection, cells were washed with PBS and lysed using 200 μl of lysis buffer (Promega). After centrifugation of cell debris, 10 μl of cell extracts was used to determine relative light units in a Monolight 2010 luminometer. Protein content in lysates was measured by Bradford assays as recommended by the supplier (Bio-rad), and all relative light unit values were normalized to protein concentrations.
Expression profiling.
One microgram of mRNA was used for synthesis of the fluorescently labeled cDNA probes for hybridization to the microarrays using the protocol described previously (11). cDNA probes from BJAB/ORF73 and BJAB/pCEP4 cells were synthesized in the presence of Cy3 and Cy5 fluorescently labeled deoxynucleoside triphosphates (dNTPs). Ten micrograms of yeast tRNA, 10 μg of polydeoxyadenylic acid, and 20 μg of human CoT1 DNA (Gibco-BRL) were added to the mixture of labeled probes in a solution containing 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.3% sodium dodecyl sulfate (SDS) and allowed to prehybridize at room temperature for 30 min before the probe was added to the surface of the microarray. Hybridizations, washes, and fluorescent scans were performed as described previously (11, 14). All measurements were stored in a computer database for analysis and interpretation.
Northern blot analysis and reverse transcription-PCR (RT-PCR).
Northern blotting and hybridization have been described previously (16). Total cellular RNA was isolated with RNAzol (Tel-Test Inc., Friendswood, Tex.) and poly(A) enriched using Oligotex beads (Oiagen) as recommended by the supplier. RNA was quantified, electrophoretically separated on denaturing agarose gels, blotted to Hybond membranes (Amersham), hybridized to specific probes overnight at 65°C in Church buffer (5) (1% [wt/vol] bovine serum albumin [BSA], 1 mM EDTA, 0.5 M NaHPO4 [pH 7.2], 7% [wt/vol] SDS), washed in a solution of 40 mM NaHPO4 (pH 7.2), 0.1% SDS, and 1 mM EDTA, and exposed to film for 48 h. Probes were randomly labeled with [32P]dCTP by using the Redivue random priming kit (Amersham).
RT-PCR assays.
One microgram of total RNA was reverse transcribed by using 200 U of Moloney murine leukemia virus reverse transcriptase (Gibco BRL) in a total volume of 20 μl containing 125 μM dATP, dGTP, and dTTP, 20 U of RNasin (Promega), and 120 pmol of random hexanucleotide primers (Boehringer Mannheim). After incubation at 42°C for 60 min, the reaction was stopped by heating to 95°C for 5 min. Five microliters of this cDNA pool was amplified in 50 μl of a PCR mix containing 10× PCR buffer and 100 pmol of each primer, and 5 U of Taq polymerase (Perkin-Elmer) was added. Each reaction mixture was overlaid with 50 μl of mineral oil prior to amplification for 30 cycles (30 s at 94°C, 1 min at 58°C, and 1 min 30 s at 72°C). To perform semi- quantitative RT-PCR, we serially diluted (in threefold steps) cDNA pools generated from BJAB/pCEP4 and BJAB/LANA cells and performed PCR amplification for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and STAT1 as described above. Amplification products were electrophoresed in 1.5% agarose gels. Primers for STAT1 amplification were STAT1/F (CGGTTGAACCCTACACGAAG) and STAT1/R (CAAGTTCCATTGGCTCTGGT), and primers for the amplification of GAPDH were GAPDH/F (CGACCACTTTGTCAAGCTCA) and GAPDH/R (AGGGGAGATTCAGTGTGGTG).
IFA.
Cells were washed twice in PBS and resuspended at a density of 5 × 106 cells/ml in PBS. Ten microliters of the cells was added to Teflon-coated 32-well IFA slides, and cells were allowed to settle for 30 min. After careful aspiration, slides were transferred into precooled methanol-acetone (1:1) for 10 min. Slides were dried and rehydrated with PBS containing 3% BSA. For blocking, cells were incubated with PBS containing 3% BSA and 1% glycine for 30 min. To detect LANA, we used a polyclonal rabbit antibody raised against a synthetic peptide from the acidic domain of LANA which is highly specific (Polson and Ganem, unpublished). The primary antibody was diluted 1:400 in blocking buffer, and cells were incubated for 1 h, followed by two washes with PBS–4% Tween 20 for 30 min. The secondary antibody was a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit whole antibody used at a dilution of 1:300. Incubation was carried out for 1 h, followed by washing as described above. Slides were mounted and microscopy was performed using a Zeiss IFA microscope.
RESULTS
LANA can function as a transcriptional modulator.
To test our hypothesis we first performed transient transfection assays using a LANA expression vector and asked whether LANA augments transcription from a set of reporter plasmids.
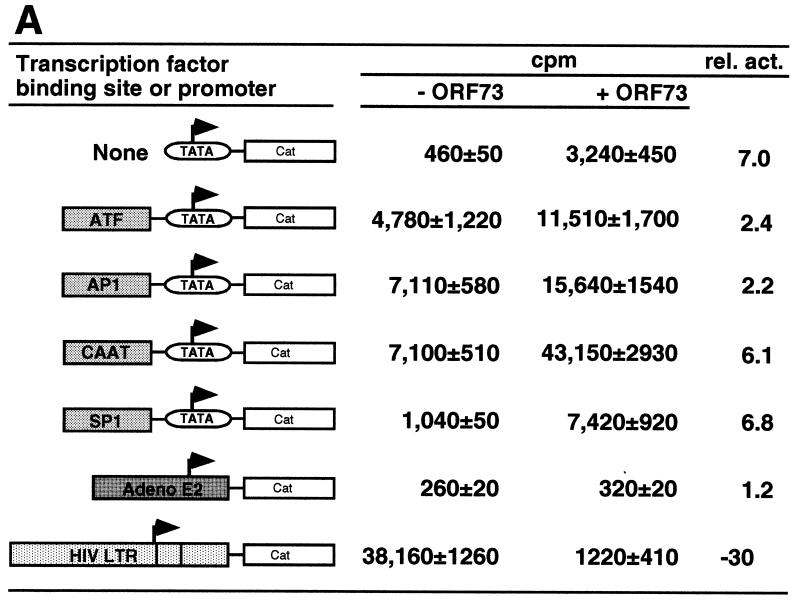
The effector plasmid pcDNA3/73 contains viral sequences from nt 127394 to nt 123663 (all nucleotide numbering refers to sequence U75698 [40]) and was previously shown to express LANA (24). As reporter plasmids we used a series of synthetic promoter constructs each containing a single transcription factor binding site linked in cis to the minimal TATA box from the SV40 early promoter; these promoters drive expression of the CAT reporter gene. COS-7 cells were transiently transfected by lipofection with 1 μg of reporter plasmid and 2 μg of either pcDNA3/73 or empty vector (pcDNA3). After incubation for 48 h, cell extracts were prepared and analyzed for the amount of CAT activity as previously described (39). As shown in Fig. 2A, the presence of LANA transactivates most of the constructs between two- and sixfold. A construct containing only the basic TATA box was significantly activated by the presence of LANA (Fig. 2A), suggesting that, directly or indirectly, LANA can affect the basal transcription machinery. Constructs bearing additional upstream activating sequences (UAS) were similarly up-regulated, although we cannot determine if this effect is due to the action of LANA at the TATA element or the UAS. Using a second set of reporter constructs containing identical transcription factor binding sites tethered to a different TATA box (HSP70 versus SV40 early), we also found a similar pattern of transactivation in these assays, including an activation of the HSP70 TATA element alone up to sevenfold (data not shown). These results demonstrate that LANA can function, directly or indirectly, to regulate transcription.
FIG. 2.
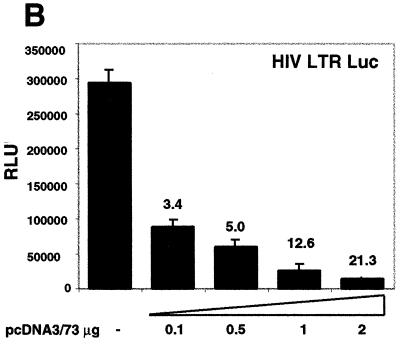
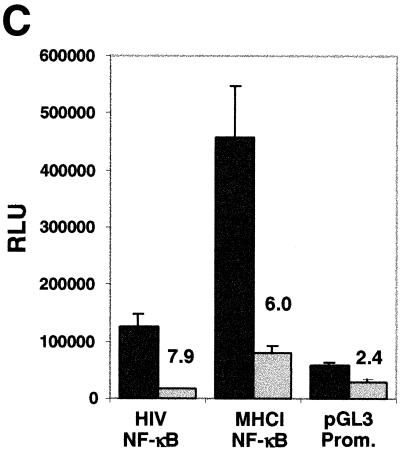
LANA can augment or repress transcription. All transient transfection assays were carried out in COS-7 cells. (A) One microgram of each reporter construct was cotransfected with 2 μg of pcDNA3/ORF73 as effector. Values are mean counts per minute ± standard deviations and relative activities based on means from two experiments, each carried out in duplicate. (B) Each transfection mixture contained 1 μg of the HIV LTR reporter construct (pHIV/LTR/Luc) and the indicated amounts of pcDNA3/ORF73. The total DNA amounts in the transfections were kept identical by adding pBSII as filler DNA. (C) NF-κB-dependent transcription is down-regulated by LANA. One microgram of NF-κB–luciferase reporter constructs was cotransfected with 2 μg of pcDNA3/ORF73 as effector. As controls we transfected pGL3/promoter. Bars represent means ± standard deviations from two experiments carried out in duplicate. Numbers above bars show relative activities based on mean values. All relative light unit values are normalized to protein concentrations determined by Bradford assays.
In addition to these artificial promoter constructs, we tested two more complex viral promoters: the adenovirus E2 promoter and the HIV LTR. The adenovirus E2 promoter construct was not up-regulated by ORF73 (Fig. 2A), indicating that not all promoters are subject to LANA-mediated regulation. Surprisingly, however, basal transcription from the HIV LTR was dramatically repressed (30-fold) by the presence of LANA (Fig. 2A). To confirm this observation, we tested the HIV LTR in a reporter driving the luciferase gene and cotransfected a constant amount of the HIV reporter construct together with increasing amounts of the LANA expression construct pcDNA3/73. As little as 0.5 μg of LANA expression vector led to a fivefold inhibition compared to transfection with the control vector. Increasing the amount of the LANA expression construct pcDNA3/73 progressively increased the inhibition of this promoter up to 21-fold (Fig. 2B). Taken together, these data demonstrate that LANA can augment transcription from some promoters while it can function to antagonize gene expression from others.
The HIV LTR promoter is controlled by a complex enhancer containing several transcription factor binding sites (41). In activated T cells, the basal level of HIV transcription is mainly regulated by a tandem element which contains two NF-κB binding sites. The inhibitory effect of alpha interferon (IFN-α) on HIV transcription is also thought to be mediated through these NF-κB sites (32). We therefore asked whether the inhibitory effect of LANA on the basal activity of the HIV LTR might also be NF-κB mediated, at least in part. Accordingly, we measured gene expression from an NF-κB reporter in the presence and absence of LANA. In addition to the NF-κB consensus sequence found in the HIV LTR, we also tested a construct containing an NF-κB site from the MHC-I promoter. Transcription from both luciferase reporter constructs was inhibited 7.9- and 6-fold by the presence of LANA; in contrast, a reporter containing a basic SV40 promoter was inhibited only 2.4-fold under these conditions (Fig. 2C). The magnitude of this reduction is less than that observed for the intact HIV LTR, suggesting that other factors may also play a role in the down-regulation of the latter; evidence consistent with this inference will be presented below.
We have previously identified and mapped the KSHV ORF73 promoter, which drives the expression of v-cyclin (ORF72) and v-FLIP (ORF71) in addition to LANA (12). As we have reported earlier, the LANA promoter is active in 293 cells, which are semipermissive for KSHV infection (15, 37). To investigate whether LANA can regulate its own synthesis, we cotransfected a luciferase reporter construct driven by the LANA promoter with increasing amounts of pcDNA3/73 into 293 cells. Expression of LANA transactivates this promoter consistently up to 5.5-fold in a dose-dependent manner (high levels of LANA expression vector in the transfection depressed activation of this promoter) (Fig. 3). Since the ORF73 promoter also governs the expression of v-cyclin and v-FLIP, these important gene products are presumably also subject to regulation by LANA (30). These results thus identify the first KSHV genes whose expression is in part regulated by LANA.
LANA activates cellular genes.
Since LANA is highly expressed in the tumor cells of KS and Castleman's disease (12, 13), we asked whether the expression of host genes could be altered by the presence of LANA. To probe for putative cellular target genes, we performed expression profiling experiments utilizing cDNA microarrays, which allow analysis of differences in expression of several thousand cellular genes in a single experiment (6). To detect expression differences in two different tissues or cell lines, mRNA from both samples is reverse transcribed into cDNA in the presence of two different fluorescent dyes. Equal amounts of both probes are mixed and hybridized to the cDNA array. After washing, the hybridized array is scanned for fluorescence intensities in each spot by using a confocal laser array scanner. Because KSHV establishes latency in CD19-positive B cells in infected individuals, we decided to use a chip containing approximately 6,000 genes and/or expressed sequence tags (ESTs) which had previously been shown to be expressed in lymphoid cell lines or tissues (2).
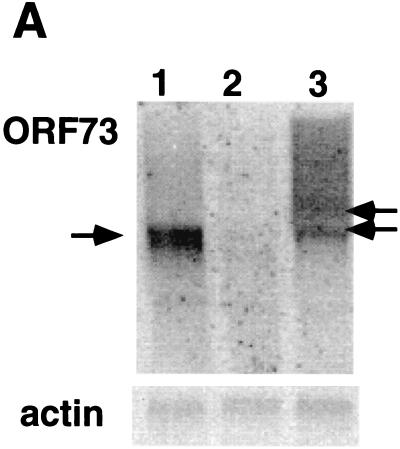
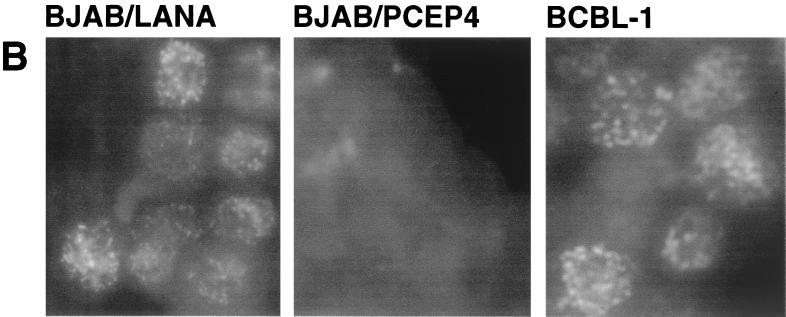
To identify candidate cellular target genes for LANA, we generated a LANA-expressing B-cell line using BJAB cells. BJAB is an EBV-negative Burkitt's lymphoma cell line. We constructed a LANA expression vector using the pCEP4 vector (Invitrogen), which bears the oriP sequence and the entire EBNA-1 open reading frame (ORF), allowing for stable plasmid maintenance without integration into the host genome—exactly analogous to the state of the KSHV genome in latency (38). We are aware that EBNA-1 does have transcription factor activity and that therefore our approach could potentially miss target genes which are equally responsive to both proteins. We transfected pCEP4/73 or the control pCEP4 vector into BJAB cells and selected with hygromycin B for 4 weeks. After selection, total RNA was extracted and analyzed on a Northern blot using an ORF73-specific probe. As expected, in cells transfected with pCEP4/73 (BJAB/LANA), a discrete band of about 4 kb is detectable (Fig. 4A, lane 1); no signal can be detected in cells transfected with vector only (lane 2). To compare the levels, of expression, Fig. 4A, lane 3, shows the expression of ORF73 in latently infected BCBL-1 cells. Even though the size of ORF73 transcripts is different in BCBL-1 cells, where several transcripts are expressed from this promoter (12), this Northern blot indicates that ORF73 is not dramatically overexpressed in BJAB/73 cells. (Control experiments with actin probes confirmed equal loading of RNA in the lanes of this blot.) In addition, we analyzed BJAB/LANA cells for the expression of LANA by IFA using a polyclonal antibody raised against LANA (Polson and Ganem, unpublished). The characteristic nuclear speckled IFA pattern is detected in BCBL-1 cells; no signal can be detected in BJAB cells transfected with empty vector. BJAB/LANA cells show a bright, nuclearly localized speckled IFA pattern, although we note that the number of speckles as well as their size and intensity differ between BJAB/LANA and BCBL-1 cells (Fig. 4B). However, this result clearly demonstrates that BJAB cells transfected with pCEP4/73 express LANA in their nuclei.
FIG. 4.
BJAB/73 cells express ORF73-specific mRNA and the LANA protein. (A) Northern blot analysis of RNA extracted from BJAB cells transfected with pCEP4/73 (lane 1) or pCEP4 (lane 2) and from BCBL-1 cells (lane 3). BJAB cells were maintained in selection medium containing hygromycin B for 4 weeks prior to the analysis. Each lane contains 10 μg of total RNA. A SmaI/BamHI fragment containing the N-terminal domain of LANA was used to prepare a radioactively labeled probe. We have previously shown that BCBL-1 cells express two transcripts from this region (indicated by arrows) (24). (B) Immunofluorescence patterns of BJAB cells transfected with pCEP4/73 or pCEP4 and of BCBL-1 cells. Cells were reacted with a 1:400 dilution of a LANA-specific polyclonal rabbit antiserum.
After poly(A) selection, equal amounts of mRNA from BJAB/LANA and BJAB/pCEP4 cells were used to produce cDNA probes by reverse transcription in the presence of Cy3 or Cy5 fluorescent-tagged dNTPs. Both probes were then hybridized to the array described above and analyzed by confocal laser scanning measuring the fluorescence intensity for each spot. The results can also be visualized by representing Cy3 fluorescence as green and Cy5 fluorescence as red. Merging both images gives an estimate of the relative abundance of transcripts from each gene in both samples. Genes whose transcripts are more abundant in the sample labeled “red” are represented by red spots in the array, genes whose transcripts are more abundant in the sample labeled “green” are represented by green spots, and genes whose expression is identical in the two samples are recognized by yellow spots.
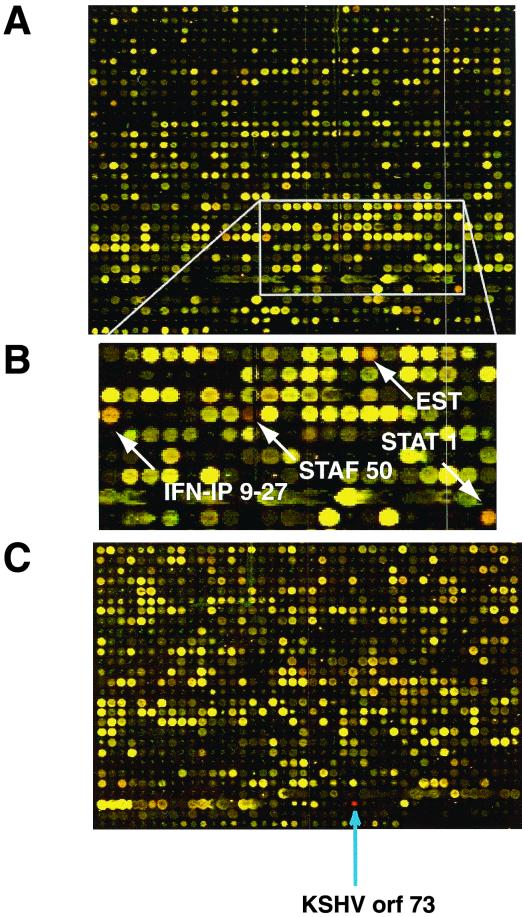
Figure 5 shows two different regions, containing approximately 2,200 spots of the merged and colored fluorescence image of the array after hybridization. The signal from mRNA extracted from LANA-expressing cells is shown in red. As a positive control, a PCR-amplified DNA fragment representing ORF73, encoding LANA, was imprinted on the array. As expected, this ORF73 spot, which is present only in cDNA from BJAB/LANA cells but not in control cells transfected with pCEP4, is bright red (Fig. 5C). Fluorescence measurements were statistically analyzed by a software program, which after several corrections and normalizations calculates a fold induction or inhibition for each gene on the array. Using this technique, every reading above twofold is scored as significant, as previously described (14, 23). It should be noted that a twofold induction or repression by this technique may not correspond quantitatively to the same induction ratio determined by Northern blot analysis. The dynamic range of the expression ratios measured by this technique is often somewhat compressed compared to that obtained by Northern analysis (14, 22). This is reflected by the computed induction ratio of 17 for ORF73, an RNA which is absent in one of the two samples and for which the “true” induction ratio is therefore infinite.
FIG. 5.
LANA can activate cellular genes. Shown are two sections of the microarray after hybridization to cDNA prepared from LANA-expressing and non-LANA-expressing cells. LANA-expressing cells are labeled in red, and non-LANA-expressing cells are labeled in green. (A) Larger area of the chip showing a representative result of this hybridization experiment. (B) Enlargement of an area of panel A where genes apparently induced by LANA expression (red spots) are indicated by arrows. (C) An area of the chip containing the positive control ORF73, whose transcripts are present only in LANA-expressing cells; therefore, this spot is bright red.
Using these criteria, we found 15 mRNAs (6 known human genes and 9 ESTs) induced between 2- and 3.3-fold in LANA-expressing B cells (Table 1). Five of the six known human genes were known to be IFN-stimulated genes (ISGs). Signals of three of these genes are indicated in Fig. 5B. However, none of the IFN genes themselves were induced in LANA-expressing cells (Table 1).
TABLE 1.
Genes or ESTs up-regulated in the presence of LANA in BJAB cells
| Gene | Induction ratio |
|---|---|
| KSHV ORF73 (positive control) | 17.30 |
| EST (example)a | 3.30 |
| Staf-50 (IFN induced) | 3.20 |
| Evi5 | 2.99 |
| STAT1 (ISGF-3 β-subunit) | 2.71 |
| IFI 6-16 (IFN induced) | 2.46 |
| MxA (IFN induced) | 2.43 |
| IFI 9-27 (IFN induced) | 2.08 |
| IFN-α | 1.18 |
| IFN-β1 | 0.78 |
| IFN-γ | 0.78 |
In addition, eight ESTs showed induction levels ranging between 2.0 and 2.5.
While the cellular functions of the IFI 9-27 and IFI 6-16 proteins are not very well characterized, MxA, STAT1, and, to a lesser extent, Staf-50 have been extensively studied. MxA is a GTP-binding IFN-inducible protein with antiviral activity against influenza virus and a variety of other RNA viruses through inhibition of the viral RNA-dependent RNA polymerases (21). STAT1 (signal tranducers and activators of transcription) encodes a β-subunit of ISGF-3 (ISG factor 3), a transcription factor which upon phosphorylation in the cytoplasm gets translocated into the nucleus, where it can bind upstream of ISGs and activate or repress gene expression (for a review see reference 9).
A second gene involved in transcriptional control is Staf-50 (stimulated trans-acting factor of 50 kDa). Staf-50 belongs to the Ring finger family, many members of which are cellular and viral zinc finger motif-containing DNA binding proteins involved in gene regulation, DNA recombination, and DNA repair. Surprisingly, Staf-50 was shown to be a potent inhibitor of expression from the HIV LTR (45). The result that Staf-50 was up-regulated in LANA-expressing cells suggested to us the possibility that the inhibitory effect of LANA on the HIV LTR might be indirect, and might be mediated in part by activation of a transcriptional repressor molecule (Staf-50). Importantly, other experiments (see Fig. 2C) suggest that additional factors involved in LTR-promoted gene expression are also regulated by LANA, so that multiple mechanisms may contribute to this effect.
Also on our list of induced genes (Table 1) is Evi5. Human Evi5, originally described as a locus for increased frequency of murine retroviral integration events, is associated with human cancers, often in combination with chromosomal translocations. Evi5 is a homolog of the murine Tre-2 oncogene, which shows sequence homology to cell cycle regulators (27). At this point we have not further characterized any of the nine induced ESTs.
Confirmation by Northern blot analysis.
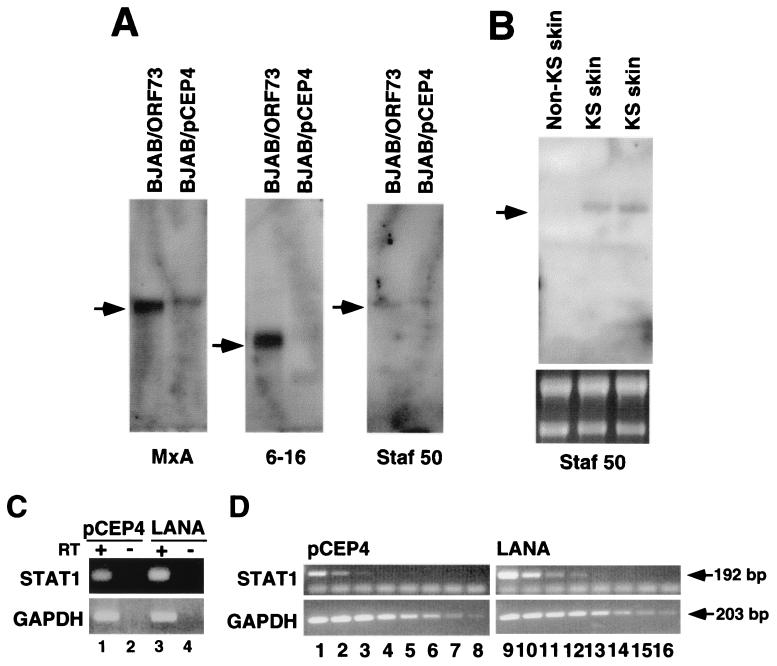
To confirm the observed up-regulated genes, we performed Northern blot analysis on RNA extracted from BJAB/LANA cells and from control cells (BJAB/pCEP4). MxA mRNA was detectable at extremely low levels in control cells but was clearly induced in BJAB/73 cells. IFI 6-16 and Staf-50 were detectable only in cells expressing LANA. However, the expression level of Staf-50 was close to the detection level of our Northern blot assays (Fig. 6A). This low level of expression is in agreement with recently published data on the induction of Staf-50 by IFN in HT1080 cells (human fibrosarcoma) (10). We were not able to detect mRNA for STAT1, IFI 9-27, or the Evi5 gene by Northern blot analysis. However, we detected STAT1 mRNA expression in BJAB/LANA cells by RT-PCR assays. Total RNAs from LANA-expressing and control cells were reverse transcribed, and the resulting cDNAs were amplified with STAT1-specific primers. As shown in Fig. 6C, STAT1 mRNA expression is slightly higher in LANA-expressing cells (lanes 1 and 3). To show that we indeed amplified RNA, we carried out control experiments with no reverse transcriptase, which did not lead to any amplification (Fig. 6C, lanes 2 and 4). We also amplified GAPDH to control for equal RNA amounts in the reaction mixture (Fig. 6C). To determine differences in STAT1 expression in a more quantitative analysis, we performed dilution PCR assays. cDNA pools from LANA-expressing and control cells were diluted in threefold steps and analyzed for the presence of STAT1 and GAPDH by PCR. A GAPDH-specific band is detectable throughout the entire dilution series in both BJAB/pCEP4 cells (Fig. 6 D, lanes 1 to 8) and BJAB/LANA cells (lanes 9 to 16), demonstrating equal amounts of input RNA. In contrast, in BJAB/pCEP4 cells a STAT1-specific band can be detected only in the first three lanes, while in BJAB/LANA cells this signal can be detected up to lane 4, confirming the induction of this gene (Fig. 6D).
FIG. 6.
Confirmation of up-regulation of cellular genes by the presence of LANA by Northern blot analysis and RT-PCR. (A) Each lane contains poly(A)-enriched mRNA from 50 μg of total RNA based on loading quantities shown in Fig. 4A. Probes were prepared from cDNA clones with accession number AA419365 for Staf-50, W49674 for IFI 6-16, and AA53117 for MxA. (B) Detection of Staf-50 expression in two KS skin lesions. Again, poly(A)-enriched mRNA extracted from 50 μg of total RNA was loaded in each lane. The lower panel shows 5 μg of total RNA prior to poly(A) selection. Arrows indicate mRNA species detected. (C) RT-PCR analysis of STAT1-specific mRNA in BJAB/LANA cells (lane 3) and BJAB/pCEP cells (lane 1). Lanes 2 and 4 show control reactions where reverse transcriptase was omitted. The lower panel shows the amplification of GAPDH as a control for RNA input. (D) Semiquantitative RT-PCR analysis of STAT1 mRNA in BJAB/LANA and BJAB/pCEP4 cells. cDNA pools were serially diluted in threefold steps. PCR amplification products from BJAB/pCEP4 cells are shown in lanes 1 to 8, and those from BJAB/LANA cells are shown in lanes 9 to 16. The lower panel shows GAPDH as a control for RNA input. Arrows indicate amplification products and their sizes.
Detection of Staf-50 mRNA in KS tumors.
To validate that the up-regulation of Staf-50 in LANA-expressing cells in culture reflects events in vivo, we asked whether Staf-50 is expressed in primary dermal KS lesions. We analyzed mRNA extracted from KS lesions of two patients and, as a control, from a healthy skin necropsy specimen. A specific band for Staf-50 was detectable in both skin lesions but not in healthy skin (Fig. 6B). These data suggest that Staf-50 is indeed up-regulated in KS tissues.
DISCUSSION
By performing transient transcription assays, we have shown that LANA expression can up-regulate its own promoter as well as synthetic promoter constructs as much as sixfold (Fig. 2). In contrast, the basal transcriptional activity of the HIV LTR was drastically repressed in the presence of LANA, in a dose-dependent fashion. At least two mechanisms can be invoked to explain the suppression of LTR-based transcription. First, LANA expression had a negative effect on NF-κB-dependent transcription, which is known to be important for LTR function; second, LANA induces Staf-50, a known inhibitor of LTR-driven expression. We do not know which of these mechanisms predominates, nor can we exclude additional contributions from as yet unexplained mechanisms. However, these initial experiments clearly demonstrate that LANA can modulate transcription both positively and negatively.
It was recently shown that LANA can interact with both RING3 (a homolog of the Drosophila female sterile homeotic (fsh) gene product) and the tumor suppressor p53. Fsh belongs to a class of proteins implicated in chromatin structure and transcriptional regulation (31). p53 is a known transcriptional activator, and binding of LANA to p53 impairs its transactivation activity (16). Both of these interactions are consistent with a role for LANA in the regulation of gene expression. Recently, it was demonstrated that LANA is required for maintenance of the episomal viral DNA in dividing cells, presumably by tethering episomal KSHV DNA to mitotic chromosomes (4). In agreement with this finding, it was demonstrated that LANA binds in an in vitro assay to a putative oriP at the left side of the genome overlapping with the terminal repeats (8). Taken together with our data, these findings indicate that LANA, like EBNA-1, has at least two activities: transcriptional regulation and episome maintenance.
In cells latently infected by EBV, EBNA-1 is also subject to autoregulation at the transcriptional level (3, 43). Our observation that the ORF73 promoter can be up-regulated by LANA coexpression suggests the existence of LANA autoregulation in KSHV, although we do not know if this autoregulation is based on direct binding of LANA to DNA targets in its own promoter. It should be noted that the basal activity of the LANA promoter in 293 cells is very high; accordingly, autoregulation by LANA might be more dramatic in cells in which this promoter is less active. For example, it has been shown that the LANA promoter is much weaker in cells of endothelial origin—a major target for KSHV infection in KS pathogenesis (44). Experiments to study the autoregulation of LANA in endothelial and lymphoid cell lines are currently in progress. However, our data provide the first identification of a LANA-responsive KSHV promoter. Moreover, this regulation by LANA is also expected to augment the expression of ORF72 (encoding a viral cyclin D homolog) and ORF71 (which encodes a putative FLICE-inhibitory protein, v-FLIP). Since both of these genes have potential roles in the deregulation of cellular growth and survival, this regulation may play an important role in driving proliferation in KSHV-linked disorders (12, 30).
To identify putative cellular target genes whose expression is altered by the presence of LANA, we first generated cells constitutively expressing LANA and then analyzed these cells by expression profiling using cDNA microarrays. This technique can be used not only to study very complex changes in gene expression patterns but also for the identification of genes which are changed in response to more subtle differences. For example, 570 of 8,600 genes changed their expression in human fibroblasts after treatment with serum (23). In contrast, using this technology, only a few genes have been identified as major targets for BRCA1, a gene often mutated in breast cancer, which was long thought to function as a transcription factor due to its primary structure (22). Gene expression profiling has also been used to study viral gene expression patterns following infection by large DNA viruses such as human CMV (7). Our approach was to use microarray technology to examine the effect of a single KSHV gene product on cellular gene expression. Using this technique, we identified 15 genes or ESTs that are expressed at significantly higher levels in the presence of LANA (Fig. 5 and Table 1). Interestingly, five of these genes are known to be IFN-responsive genes, with two of them being transcriptional regulators (Staf-50 and STAT1). The up-regulation of STAT1 may play a role in the up-regulation of other IFN-inducible genes. We do not yet know the biological role of the induction of these genes. Conceivably, they may play a role in the suppression of lytic induction and the maintenance of viral latency, but many other models are possible.
The mechanism by which LANA modulates gene expression from its targets remains a subject for further investigation. In our microarray experiments we chose to express LANA constitutively in stable cell lines, so as to mimic the situation that obtains in latent infection in vivo. While the up-regulation we observed in this system could be due to the direct action of LANA on its targets, it is equally possible that these genes are induced by other regulatory molecules that are themselves controlled by LANA. The binding of p53 by LANA (16) would be one example of such a mechanism; our experiments similarly show that LANA expression can affect NF-κB-dependent transcription (Fig 2C) and can up-regulate known transcription factors STAT1 and Staf-50. We are currently developing cell lines in which LANA expression is inducible in order to examine gene regulation by LANA more directly. But whether activation of these targets is direct or indirect, the target genes identified herein are likely to be in latently infected cells—an inference we have directly confirmed for Staf-50 in KS tumors. A better understanding of the mechanisms by which LANA modifies cellular and viral gene expression is likely to be important for deciphering the role(s) of this protein in the pathobiology of KSHV infection.
ACKNOWLEDGMENTS
We thank Andy Polson for providing the LANA antibody and Karen E. Tucker for help with microscopy and digital imaging.
R.R. is a fellow of the Leukemia Society of America and a Mount Sinai Healthcare foundation scholar. This work was supported by the Howard Hughes Medical Institute (HHMI) and grants from the NIH to D.G. (CA73506-04) and R.R. (CA CA88763-01). D.G. is an investigator and P.O.B. is an associate investigator of the HHMI.
REFERENCES
- 1.Alexandropoulos K, Cheng G, Baltimore D. Proline-rich sequences that bind to Src homology 3 domains with individual specificities. Proc Natl Acad Sci USA. 1995;92:3110–3114. doi: 10.1073/pnas.92.8.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alizadeh A, Eisen M, Botstein D, Brown P O, Staudt L M. Probing lymphocyte biology by genomic-scale gene expression analysis. J Clin Immunol. 1998;18:373–379. doi: 10.1023/a:1023293621057. [DOI] [PubMed] [Google Scholar]
- 3.Ambinder R F, Mullen M A, Chang Y N, Hayward G S, Hayward S D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 6.Brown P O, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21(Suppl. 1):33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 7.Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan J S, Bittner A, Frueh K, Jackson M R, Peterson P A, Erlander M G, Ghazal P. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73:5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotter M A, II, Robertson E S. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 9.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 10.Der S D, Zhou A, Williams B R, Silverman R H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande J P, Weiss R A, Alitalo K, Boshoff C. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci USA. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen M B, Brown P O. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 15.Foreman K E, Friborg J, Jr, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 16.Friborg J, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 2000;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 17.Gahn T A, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 19.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 20.Gao S J, Zhang Y J, Deng J H, Rabkin C S, Flore O, Jenson H B. Molecular polymorphism of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent nuclear antigen: evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J Infect Dis. 1999;180:1466–1476. doi: 10.1086/315098. [DOI] [PubMed] [Google Scholar]
- 21.Haller O, Frese M, Kochs G. Mx proteins: mediators of innate resistance to RNA viruses. Rev Sci Tech. 1998;17:220–230. doi: 10.20506/rst.17.1.1084. [DOI] [PubMed] [Google Scholar]
- 22.Harkin D P, Bean J M, Miklos D, Song Y H, Truong V B, Englert C, Christians F C, Ellisen L W, Maheswaran S, Oliner J D, Haber D A. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 23.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, Lashkari D, Shalon D, Botstein D, Brown P O. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 24.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. . (Erratum, 2:1041.) [DOI] [PubMed] [Google Scholar]
- 26.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 27.Liao X, Du Y, Morse III H C, Jenkins N A, Copeland N G. Proviral integrations at the Evi5 locus disrupt a novel 90-kDa protein with homology to the Tre2 oncogene and cell-cycle regulatory proteins. Oncogene. 1997;14:1023–1029. doi: 10.1038/sj.onc.1200929. [DOI] [PubMed] [Google Scholar]
- 28.Liebowitz D, Kieff E. Epstein-Barr virus. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 107–172. [Google Scholar]
- 29.Lukac D M, Manuppello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 31.Platt G M, Simpson G R, Mittnacht S, Schulz T F. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popik W, Pitha P M. Transcriptional activation of the tat-defective human immunodeficiency virus type-1 provirus: effect of interferon. Virology. 1992;189:435–447. doi: 10.1016/0042-6822(92)90567-9. [DOI] [PubMed] [Google Scholar]
- 33.Ptashne M. A genetic switch. 2nd ed. Cambridge, Mass: Cell Press & Blackwell Scientific Publications; 1992. [Google Scholar]
- 34.Puglielli M T, Desai N, Speck S H. Regulation of EBNA gene transcription in lymphoblastoid cell lines: characterization of sequences downstream of BCR2 (Cp) J Virol. 1997;71:120–128. doi: 10.1128/jvi.71.1.120-128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raab-Traub N. Pathogenesis of Epstein-Barr virus and its associated malignancies. Virology. 1996;7:315–323. [Google Scholar]
- 36.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renne R, Mergia A, Renshaw-Gegg L W, Neumann-Haefelin D, Luciw P A. Regulatory elements in the long terminal repeat (LTR) of simian foamy virus type 3 (SFV-3) Virology. 1993;192:365–369. doi: 10.1006/viro.1993.1045. [DOI] [PubMed] [Google Scholar]
- 40.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Shen Y, Shenk T. Relief of p53-mediated transcriptional repression by the adenovirus E1B 19-kDa protein or the cellular Bcl-2 protein. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheridan P L, Sheline C T, Milocco L H, Jones K A. Tat and the HIV-1 promoter: a model for RNA-mediated regulation of transcription. Semin Virol. 1993;4:69–80. [Google Scholar]
- 42.Struhl K. Yeast transcriptional regulatory mechanisms. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 43.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 45.Tissot C, Mechti N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J Biol Chem. 1995;270:14891–14898. doi: 10.1074/jbc.270.25.14891. [DOI] [PubMed] [Google Scholar]
- 46.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]