Abstract
Microarrays containing 1,126 nonredundant cDNAs selected from a chicken activated T-cell expressed sequence tag database (http://chickest.udel.edu) were used to examine changes in host cell gene expression that accompany infection of chicken embryo fibroblasts (CEF) with Marek's disease virus (MDV). Host genes that were reproducibly induced by infection of CEF with the oncogenic RB1B strain of MDV included macrophage inflammatory protein, interferon response factor 1, interferon-inducible protein, quiescence-specific protein, thymic shared antigen 1, major histocompatibility complex (MHC) class I, MHC class II, β2-microglobulin, clusterin, interleukin-13 receptor alpha chain, ovotransferrin, a serine/threonine kinase, and avian leukosis virus subgroup J glycoprotein.
Marek's disease (MD) is a lymphoproliferative disorder of chickens caused by a herpesvirus, Marek's disease virus (MDV) (5). MDV initially infects chickens via the respiratory tract and then causes early cytolytic infection in B cells followed by latent infection in T cells. In some infections, MDV transforms CD4+ T cells and results in the formation of massive lymphomas in a variety of tissues. MD has been controlled in the commercial poultry industry since 1970 by the use of live, nononcogenic vaccines; however, the mechanism of vaccine-induced immunity is not well understood. Vaccination efficiently prevents tumor formation but does not stop infection. As a result, variants of MDV with enhanced virulence have emerged throughout the second half of this century, and periodically new vaccines or vaccine regimens must be introduced to adequately control field challenges. Little is known about virus-host cell interactions that relate to MDV pathogenicity.
Microarrays have been widely applied to analyze gene expression changes on a genome wide scale. They have been used to assess differences in yeast transcription among strains in general (39), during growth under various conditions such as heat or cold shock (27), during growth using different carbon sources (27), and during aerobic versus anaerobic growth (11). High-density arrays have been used to identify yeast genes whose expression depends on transcriptional initiation factors (21), to profile gene expression changes that accompany activation of mouse T cells (41), and to explore and compare signal transduction pathways (14, 28). They have been used to identify human genes involved in the pathology of diseases such as rheumatoid arthritis and inflammatory bowel disease (17), to compare gene expression in cells expressing either a transformed or a nontransformed phenotype (12, 46), and to study hematopoietic differentiation (40). In combination with cluster analysis, microarrays have been used to assess variation in gene expression patterns of human cancers as a means to classify solid tumors (34). In plant genomics, microarrays have been exploited to examine differential gene expression for Arabidopsis (38). Microarray analysis is widely recognized as a key tool in drug discovery (16). With regard to virus infections, microarrays have been used to assess changes in host cell gene expression following human cytomegalovirus (HCMV) infection (49) and to characterize temporal classes of HCMV gene expression (7).
Microarrays.
Our microarray was designed to contain as many genes as possible with minimal redundancy. Sequences included were chosen from our poultry activated T-cell database (43). Cytokines, cytokine receptors, chemokines, and factors involved in apoptosis, transcription, and T-cell activation were among the arrayed cellular cDNAs. Inserts from selected clones were amplified using PCR and vector-specific primers. PCR products were examined by agarose gel electrophoresis for quality, yield, and concentration. Following alcohol precipitation, PCR products were resuspended in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and stored in 96-well format. Using a Flexys robot (Genomic Solutions, Ann Arbor, Mich.), samples were spotted (5 ng in 40 nl) onto 5- by 7-cm nylon membranes at a density of 2,418 spots per membrane. In addition to 26 positive and negative controls, each microarray contained 1,126 chicken activated T-cell cDNAs, 32 MDV sequences, and 51 herpesvirus of turkeys sequences, each spotted in duplicate. Once printed, membranes were treated using standard procedures for adhering DNA to nylon. Presence of DNA was confirmed by hybridization to a radiolabeled probe for the multiple cloning region of the vector. The sensitivity and linear range of array analysis was determined by hybridizing filters to increasing amounts of labeled RNA. The linear range covered at least 2 orders of magnitude, with inputs of 5 × 106 to 5 × 108 cpm [0.05 to 5 μg of poly(A) RNA].
Cultures of secondary chicken embryo fibroblasts (CEF; 1.2 × 107 cells per 75-cm2 tissue culture flask) were mock infected or infected with cell-associated RB1B (105 PFU per flask) at passage level 18 and incubated for either 48 h or 96 h. Total RNA was purified using guanidinium isothiocyanate followed by centrifugation through cesium chloride. Poly(A)+ RNA was purified using a PolyATtract mRNA isolation system (Promega, Madison, Wis.). Poly(A)+ mRNA (0.5 to 1 μg) was heat denatured and annealed to oligo(dT) (1 μg). Thereafter, the RNA was incubated with 3 mM dithiothreitol, 0.8 mM dATP, dGTP, or dTTP, 40 U of RNasin, 100 μCi of [α-32P]dCTP, and 400 U of Superscript II reverse transcriptase (Life Technologies, Gaithersburg, Md.) in a total volume of 30 μl of the manufacturer's recommended buffer at 42°C for 1 h. Labeled cDNA was purified using a ProbeQuant G-50 Micro Column (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). cDNAs were labeled to a specific activity of about 108 cpm/μg of poly(A)+ RNA.
Hybridization of cDNAs to membrane arrays was similar to protocols routinely used for DNA hybridizations except that filters were prehybridized for 1 h at 48°C in 6 ml of hybridization solution containing sheared, denatured chicken genomic DNA (50 μg/ml) and salmon sperm DNA (100 μg/ml). For additional blocking, 50 μg of chicken genomic DNA was added to the entire probe reaction prior to denaturation for 2 min at 98°C. Hybridization took place overnight at 48°C. Following hybridization, filters were washed to high stringency and exposed to phosphorimager screens, which were scanned using a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) TIFF files were imported to a Sun workstation (Sun Microsystems, Inc., Palo Alto, Calif.) for analysis using Visage high-density grid analysis software (Genomic Solutions). Spreadsheets were merged with Excel files containing addresses and identities of each spot. Excel spreadsheets used in this study are accessible at the University of Delaware chicken expressed sequence tag website (http://chickest.udel.edu).
Induced host gene expression.
Differences in host gene expression following infection of CEF with MDV were easily detected. Signals on arrays hybridized with cDNA from MDV-infected CEF were considered further if they were present in duplicate and differed 2-fold or more from signals on arrays hybridized with cDNA from uninfected CEF. Normalization among arrays was done using global normalization (http: //www.geneindex.org), a procedure in which the output from each array is multiplied by a normalization factor such that the average signal intensities of all arrays are equivalent.
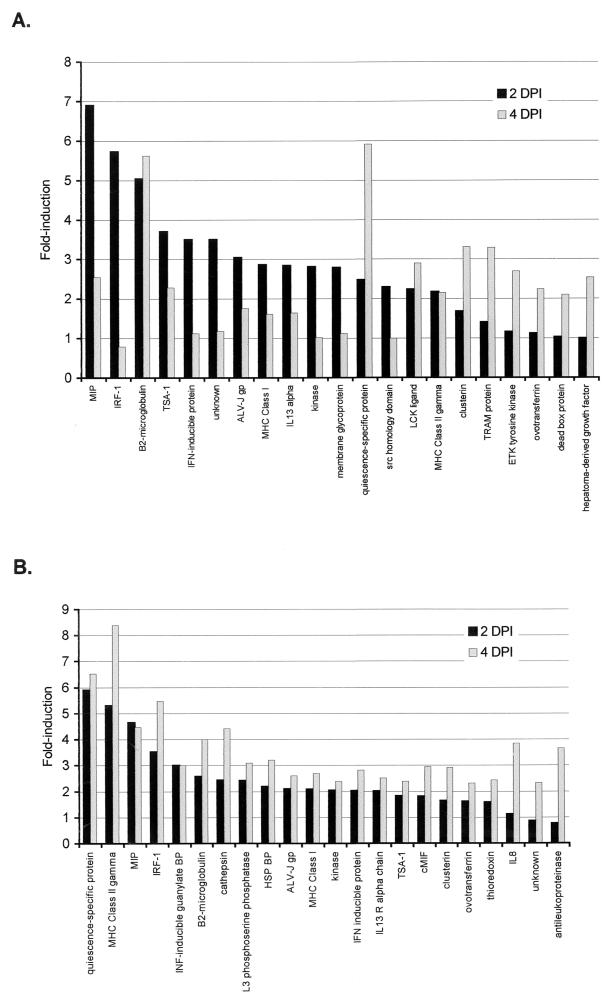
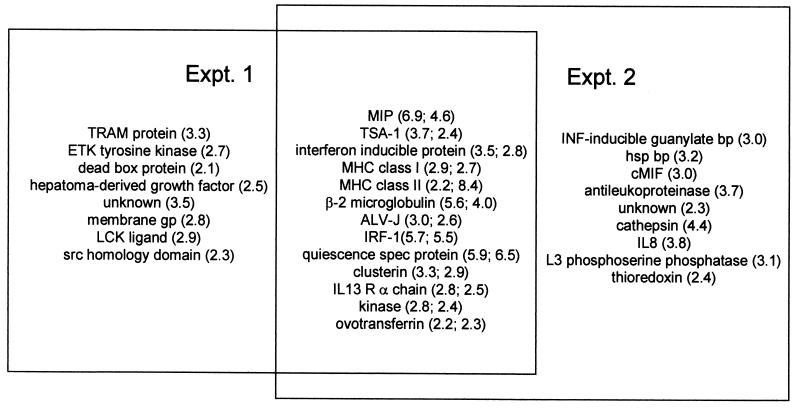
All host genes showing twofold or greater induction above the uninfected cell background at either 2 or 4 days postinfection (dpi) in either of two replicate experiments are shown (Fig. 1). Twenty-one host genes in one experiment and 22 in the other appeared induced at either 2 or 4 dpi, the overlap between the two replicate experiments being 13 genes (Fig. 2). We have elected to maintain the cutoff for further consideration in both experiments at twofold, a value of minimum stringency that may allow identification of some false positives. All of these data can be accessed, downloaded, and subsequently manipulated from our website (http://chickest.udel.edu) for individuals wishing to analyze them more stringently or using customized software tools. Table 1 indicates the relative induction of each species at both 2 and 4 dpi. In some cases, the patterns of expression were similar in the two experiments; i.e., genes that were expressed more heavily at 2 dpi than at 4 dpi showed the same trend in both trials. In other cases, trends for the two experiments were not the same. The species that were induced most strongly (fivefold or greater in both replicates at either time point) in these experiments were macrophage inflammatory protein (MIP), quiescence-specific protein, and interferon (IFN) response factor 1 (IRF-1). Species that were moderately (fivefold or greater in one experiment and twofold or greater in the other) or somewhat (threefold or greater in one experiment and twofold or greater in the other) induced included β2-microglobulin, major histocompatibility complex (MHC) class II, thymic shared antigen 1 (TSA-1; also known as stem cell antigen 2), IFN-inducible protein, avian leukosis virus (ALV) subgroup J (ALV-J) envelope glycoprotein, and clusterin. Weakly (approximately twofold in both experiments) induced species included interleukin-13 (IL-13) receptor alpha chain (IL-13Rα), MHC class I, a serine/threonine kinase, and ovotransferrin.
FIG. 1.
Induction of host gene expression following infection of CEF with MDV. For experiment 1 (A) and 2 (B), data are presented as fold induction above the background microarray, which was hybridized with cDNA from uninfected CEF. All samples that were twofold or greater above the background in duplicate spots on either the 2-dpi microarray, the 4-dpi microarray, or both microarrays are shown. Each bar represents the average of duplicate spots on the microarrays. LCK, p56lck; TRAM, translocating chain-associating membrane; ETK, epithelial and endothelial tyrosine kinase; BP, binding protein; HSP, heat shock protein; cMIF, macrophage migration inhibitory factor.
FIG. 2.
Venn diagram showing relationship between induced genes in experiments 1 and 2. Numbers in parentheses indicate maximum fold induction seen in experiments 1 and 2, respectively. Abbreviations are as in Fig. 1.
TABLE 1.
Classification of changes in CEF gene expression following infection with the very virulent RB1B strain of MDV
| Category | Gene | Fold induction in MDV-infected CEFa
|
|||
|---|---|---|---|---|---|
| Expt 1
|
Expt 2
|
||||
| 2 dpi | 4 dpi | 2 dpi | 4 dpi | ||
| Strongly induced (5-fold in both expts) | MIP | 6.9 | 2.5 | 4.6 | 4.5 |
| Quiescence-specific protein | 2.5 | 5.9 | 5.9 | 6.5 | |
| IRF-1 | 5.7 | 0.79 | 3.5 | 5.5 | |
| Moderately induced (5-fold in 1 expt; 2-fold in the other) | β2-Microglobulin | 5.0 | 5.6 | 2.3 | 4.0 |
| MHC class II | 2.2 | 2.2 | 5.3 | 8.4 | |
| Somewhat induced (3-fold in 1 expt; 2-fold in the other) | TSA-1 | 3.7 | 2.3 | 1.8 | 2.4 |
| IFN-inducible protein | 3.5 | 1.1 | 2.0 | 2.8 | |
| ALV-J | 3.0 | 1.8 | 2.1 | 2.6 | |
| Clusterin | 1.7 | 3.3 | 1.6 | 2.9 | |
| Weakly induced (2-fold in both expts) | IL-13Rα | 2.8 | 1.7 | 2.0 | 2.5 |
| MHC class I | 2.9 | 1.6 | 2.1 | 2.7 | |
| Kinase | 2.8 | 1.0 | 2.0 | 2.4 | |
| Ovotransferrin | 1.1 | 2.2 | 1.6 | 2.3 | |
Each value is the average of duplicate samples and is the value obtained using MDV-infected CEF RNA for hybridization divided by the value obtained using uninfected CEF RNA for hybridization.
Inconsistency among array experiments makes replication of results essential for studies of MDV. This inconsistency is not unexpected given the complexities and limitations of the MDV system. First, there are no cell lines for propagation of MDV, and the virus is generally grown in primary or secondary CEF. Therefore, it is not possible to synchronize infections to the degree that would be easily achieved with other viruses. We chose 2 and 4 dpi as the times to sample, as these represent a relatively early point in the infection before cytopathic effects are apparent and a relatively late time in the infection when plaques are visible. Second, MDV is a cell-associated virus and therefore cannot be used at high multiplicities of infection. We infect as heavily as possible (105 infected cells/1.2 × 107 uninfected CEF), but only a fraction of the cells plated eventually become infected. This means that a large portion of the mRNA being purified at the time of harvest is from uninfected cells, which probably obscures the true magnitude of any differences observed. One must keep in mind that differences seen are the result of mRNA levels both in infected cells and in uninfected cells also present in the culture.
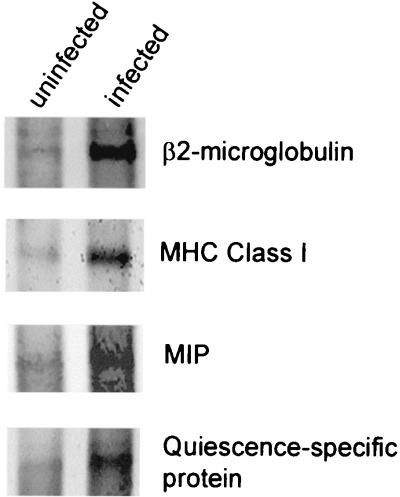
Northern hybridizations were used to confirm gene expression changes first observed on microarrays for selected genes (Fig. 3). Poly(A)+ RNA (1 μg) purified from uninfected CEF or RB1B-infected CEF was electrophoresed, blotted, and probed using riboprobes transcribed by T7 RNA polymerase off of NotI-linearized cDNA clones. mRNA levels were elevated at 1 dpi for MHC class I and at 2 dpi for MIP, quiescence-specific protein, and β2-microglobulin. The RNA preparations used for Northern analysis were different from those used for microarrays and therefore reflect variation in the timing of the infection at harvest. Nevertheless, induced levels of these species were apparent qualitatively.
FIG. 3.
Northern analysis of poly(A)+ RNA purified from CEF that were either uninfected or infected with RB1B. Probes (shown at the right) consisted of riboprobes transcribed by T7 RNA polymerase off of NotI-linearized cDNA clones. RNA was harvested at 1 dpi for MHC class I and at 2 dpi for all other samples.
Host genes reproducibly induced upon infection of CEF with MDV can be grouped into those involved in inflammation and cellular stress (MIP and clusterin), cell growth and differentiation (quiescence-specific protein, TSA-1, and IL-13R), antigen presentation (MHC class I, MHC class II, and β2-microglobulin), and IFN responses (IRF-1 and IFN-inducible protein). Species that did not fall obviously into these classes were ALV-J, a serine/threonine kinase, and ovotransferrin.
MIP induction in these and other experiments was strong and striking, a result that is not surprising given that MDV infection is expected to induce an inflammatory response. MIP-1α is a small, inducible cytokine that belongs to the C-C chemokine subfamily (9). MIP-1α has proinflammatory activities and also inhibits growth of hematopoietic stem cells (9). Null mice that lack MIP-1α are hematopoietically normal but resist coxsackievirus-induced myocarditis and show decreased pneumonitis and delayed viral clearance following exposure to influenza virus (10). Thus, MIP-1α appears to mediate inflammatory responses during viral infection.
With regard to herpesvirus infections, MIP-1α plays a key role in the development of herpetic stromal keratitis, a blinding inflammatory condition that develops in mice infected with replication-competent herpes simplex virus (42, 48). The Kaposi's sarcoma-associated herpesvirus genome encodes MIP-related chemokine homologs, namely, vMIP-I, vMIP-II, and vMIP-III (3, 13). vMIP-I may affect the Th1/Th2 balance during host immune responses by antagonizing C-C chemokine receptor 8 (CCR8), a receptor expressed on Th2 T cells. vMIP-II has been shown to activate and attract human eosinophils via CCR3. vMIP-III has not been functionally characterized. It is possible that MIP induction following MDV infection is important for T-cell attraction and infiltration at the site of infection. Calnek (4) has pointed out that in the case of MD, local T-cell immune responses may contribute significantly to disease, as they provide targets for latent infection and subsequent transformation.
Another induced species likely to be related to inflammation and/or cellular damage is clusterin (29). Clusterin is a conserved glycoprotein, also known as apolipoprotein J, whose expression is increased in many cell types in response to stress. Clusterin has been reported to have chaperonin-like activity (22) and anti-inflammatory activity (32), and it is expressed during tissue differentiation and remodeling involving apoptosis (26, 29).
Quiescence-specific protein is a 20-kDa protein reported to be present in contact-inhibited CEF (2, 30). Factors that induce quiescence, such as serum starvation and hydroxyurea treatment, induce this protein, whereas mitogen treatment reduces its levels. Transient expression assays have indicated that regulation of quiescence-specific protein is, at least in part, at the transcriptional level. Induction of quiescence-specific gene expression following MDV infection suggests that viral infection inhibits cellular proliferation. This is consistent with the idea that upon infection, a herpesvirus poises cells to accumulate factors needed for DNA synthesis and simultaneously inhibits cell cycle progression such that the virus can exploit the replication-ready environment for its own benefit.
TSA-1 is a developmentally regulated glycosylphosphatidylinositol-anchored differentiation antigen that is identical to stem cell antigen 2 (33, 37). It plays a key role in T-cell differentiation by participating in T-cell receptor/CD3-mediated apoptosis of immature thymocytes (33). It also functions in T-cell activation via the T-cell receptor signaling pathway (37). Our results unexpectedly indicate that chicken TSA-1 is expressed in MDV-infected fibroblasts. Since MDV latently infects and can transform T cells in vivo, the finding that infection induces a factor important for T-cell activation and differentiation is provocative.
Human IL-13 is known to stimulate proliferation, differentiation, and effector functions of B lymphocytes and macrophages (47). IL-13 is closely related to IL-4 (8). Receptors for these human interleukins have been characterized and compared (6). The IL-13 receptor has not been previously described for chickens.
Elevation of MHC class I, MHC class II, and β2-microglobulin mRNA levels was unexpected since many viruses, including herpesviruses, have been reported to down-regulate cell surface expression of species involved in antigen presentation, particularly MHC class I. For example, herpes simplex virus ICP47 associates with transporter for antigen processing (TAP) in a species-specific manner and blocks peptide binding and subsequent translocation of antigenic peptides across the endoplasmic reticulum (15, 19, 31, 45). Bovine herpesvirus 1 inhibits MHC class I cell surface expression by down-regulating TAP activity in bovine epithelial cells, although viral gene products involved are not known (20). HCMV uses several mechanisms to interfere with antigen presentation. TAP activity continuously declines during HCMV infection of fibroblasts (18). HCMV US3 binds β2-microglobulin-associated class I heavy chains, making them susceptible to destabilization mediated by both HCMV US2 and US11 gene products (25). Likewise, murine cytomegalovirus m06 binds β2-microglobulin-associated class I molecules and redirects them for endocytosis (36). HCMV US2 mediates degradation of two components of the MHC class II antigen presentation pathway, namely, DRα and DMα (44).
Recent microscopy results suggest that MDV MHC class I expression is actually down-regulated within individual infected CEF but consistently up-regulated in neighboring cells present in the culture (J. Kent, E. L. Bernberg, and R. W. Morgan, unpublished data). Thus, our microarray results reflected transcription changes occurring in the entire culture, one that contained a majority of uninfected cells. Up-regulation in neighboring cells may be IFN mediated. Indeed, a chicken fibroblast cell line, C32, stably transfected to constitutively overexpress IFN-1 showed enhanced MHC class I surface antigen expression (50). Furthermore, elevation of IRF-1 (24) mRNA was consistently seen in the microarray analyses. IRF-1 is a highly conserved transcription factor that mediates responses to viral infections and IFNs. In CEF, IRF-1 is strongly induced by IFN treatment.
Three other consistently induced species that are less well understood are ALV-J, a serine/threonine kinase, and ovotransferrin. It is likely that ALV-J-specific hybridization in these experiments was due to gene expression from endogenous retroviruses related to ALV-J. ALV-J is currently a problematic chicken virus present in flocks worldwide. Serotype 2 MDV is known to augment ALV-induced lymphoid leukosis in some genetic lines of chickens (1). In addition, serotype 2 MDV has been reported to enhance ALV gene expression and protein accumulation in coinfected cell cultures (35). Ovotransferrin (23) is a key iron delivery and iron-scavenging protein. The chicken serine/threonine protein kinase represents a novel homolog, poorly understood at this time.
In summary, we have used a microarray containing more than 1,000 selected, nonredundant chicken cDNAs to learn that MDV infection of CEF results in reproducible elevation of steady-state levels of certain cellular mRNAs. We have learned that genes involved in virus-induced inflammation and IFN responses appear consistently induced. Even in CEF, MDV infection appears to induce expression of TSA-1, a gene important for T-cell differentiation and activation. These results provide a powerful platform for additional studies aimed at understanding the biology of MDV. For example, similar studies using oncogenic strains with different pathogenicities, nononcogenic and vaccine strains of MDV, chicken tissues following in vivo infections of susceptible and resistant lines of chickens, lymphoblastoid cell lines, and tumors promise to be revealing and are in progress.
REFERENCES
- 1.Bacon L D, Witter R L, Fadly A M. Augmentation of retrovirus-induced lymphoid leukosis by Marek's disease herpesviruses in white leghorn chickens. J Virol. 1989;63:504–512. doi: 10.1128/jvi.63.2.504-512.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedard P A, Yannoni Y, Simmons D L, Erikson R L. Rapid repression of quiescence-specific gene expression by epidermal growth factor, insulin, and pp60v-src. Mol Cell Biol. 1989;9:1371–1375. doi: 10.1128/mcb.9.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 4.Calnek B W. Marek's disease—a model for herpesvirus oncology. Crit Rev Microbiol. 1986;12:293–320. doi: 10.3109/10408418509104432. [DOI] [PubMed] [Google Scholar]
- 5.Calnek B W, Witter R L. Marek's disease. In: Calnek B W, editor. Diseases of poultry. 10th ed. Ames, Iowa: Iowa State University Press; 1997. pp. 369–413. [Google Scholar]
- 6.Caput D, Laurent P, Kaghad M, Lelias J M, Lefort S, Vita N, Ferrara P. Cloning and characterization of a specific interleukin (IL)-13 binding protein structurally related to the IL-5 receptor alpha chain. J Biol Chem. 1996;271:16921–16926. doi: 10.1074/jbc.271.28.16921. [DOI] [PubMed] [Google Scholar]
- 7.Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan J, Bittner A, Frueh K, Jackson M, Peterson P, Erlander M, Ghazal P. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73:5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomarat P, Banchereau J. Interleukin-4 and interleukin-13: their similarities and discrepancies. Int Rev Immunol. 1998;17:1–52. doi: 10.3109/08830189809084486. [DOI] [PubMed] [Google Scholar]
- 9.Cook D N. The role of MIP-1 alpha in inflammation and hematopoiesis. J Leukoc Biol. 1996;59:61–66. doi: 10.1002/jlb.59.1.61. [DOI] [PubMed] [Google Scholar]
- 10.Cook D N, Beck M A, Coffman T M, Kirby S L, Sheridan J F, Pragnell I B, Smithies O. Requirement of MIP-1 alpha for an inflammatory response to viral infection. Science. 1995;269:1583–1585. doi: 10.1126/science.7667639. [DOI] [PubMed] [Google Scholar]
- 11.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 12.DeRisi J L, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Use of a cDNA microarray to analyze gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 13.Endres M J, Garlisi C G, Xiao H, Shan L, Hedrick J A. The Kaposi's sarcoma-related herpesvirus (KSHV)-encoded chemokine vMIP-I is a specific agonist for the CC chemokine receptor (CCR)8. J Exp Med. 1999;189:1993–1998. doi: 10.1084/jem.189.12.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fambrough D, McClure D, Kazlauskas A, Lander E S. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 15.Fruh K, Ahn K, Djaballah H, Sempe P, Van Endert P M, Tampe R, Peterson P A, Yang Y. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 16.Gray N S, Wodicka L, Thunnissen A-M, Norman T C, Kwon S, Espinoza F H, Morgan D O, Barnes G, LeClerc S, Meijer L, et al. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 17.Heller R A, Schena M, Chai A, Shalon D, Bedoin T, Gilmore J, Woolley D E, Davis R W. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci USA. 1997;94:2150–2155. doi: 10.1073/pnas.94.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hengel H, Flohr T, Hammerling G J, Koszinowski U H, Momburg F. Human cytomegalovirus inhibits peptide translocation into the endoplasmic reticulum for MHC class I assembly. J Gen Virol. 1996;77:2287–2296. doi: 10.1099/0022-1317-77-9-2287. [DOI] [PubMed] [Google Scholar]
- 19.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 20.Hinkley S, Hill A B, Srikumaran S. Bovine herpesvirus-1 infection affects the peptide transport activity in bovine cells. Virus Res. 1998;53:91–96. doi: 10.1016/s0168-1702(97)00128-7. [DOI] [PubMed] [Google Scholar]
- 21.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 22.Humphreys D T, Carver J A, Easterbrook-Smith S B, Wilson M R. Clusterin has chaperone-like activity similar to that of small heat shock proteins J. Biol Chem. 1999;274:6875–6881. doi: 10.1074/jbc.274.11.6875. [DOI] [PubMed] [Google Scholar]
- 23.Jeltsch J, Chambon P. The complete nucleotide sequence of chicken ovotransferrin mRNA. Eur J Biochem. 1982;122:291–295. doi: 10.1111/j.1432-1033.1982.tb05879.x. [DOI] [PubMed] [Google Scholar]
- 24.Jingwirth C, Rebbert M, Ozato K, Degen H J, Schultz U, Dawid I B. Chicken interferon consensus sequence-binding protein (ICSBP) and interferon regulatory factor (IRF) 1 genes reveal evolutionary conservation in the IRF gene family. Proc Natl Acad Sci USA. 1995;92:3105–3109. doi: 10.1073/pnas.92.8.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones T R, Sun L. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J Virol. 1997;71:2970–2979. doi: 10.1128/jvi.71.4.2970-2979.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klock G, Storch S, Rickert J, Gutacker C, Koch-Brandt C. Differential regulation of the clusterin gene by Ha-ras and c-myc oncogenes and during apoptosis. J Cell Physiol. 1998;177:593–605. doi: 10.1002/(SICI)1097-4652(199812)177:4<593::AID-JCP10>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 27.Lashkari D A, DeRisi J L, McCusker J H, Namath A F, Gentile C, Hwang S Y, Brown P O, Davis R W. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madhani H D, Galitski T, Lander E S, Fink G R. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci USA. 1999;96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahon M G, Lindstedt K A, Hermann M, Nimpf J, Schneider W J. Multiple involvement of clusterin in chicken ovarian follicle development. Binding to two oocyte-specific members of the low density lipoprotein receptor gene family. J Biol Chem. 1999;274:4036–4044. doi: 10.1074/jbc.274.7.4036. [DOI] [PubMed] [Google Scholar]
- 30.Mao P L, Beauchemin M, Bedard P A. Quiescence-dependent activation of the p20K promoter in growth-arrested chicken embryo fibroblasts. J Biol Chem. 1993;268:8131–8139. [PubMed] [Google Scholar]
- 31.Neumann L, Kraas W, Uebel S, Jung G, Tampe R. The active domain of the herpes simplex virus protein ICP47: a potent inhibitor of the transporter associated with antigen processing. J Mol Biol. 1997;272:484–492. doi: 10.1006/jmbi.1997.1282. [DOI] [PubMed] [Google Scholar]
- 32.Newkirk M M, Apostolakos P, Neville C, Fortin P R. Systemic lupus erythematosus, a disease associated with low levels of clusterin/apoJ, an antiinflammatory protein. J Rheumatol. 1999;26:597–603. [PubMed] [Google Scholar]
- 33.Noda S, Kosugi A, Saitoh S, Narumiya S, Hamaoka T. Protection from anti-TCR/CD3-induced apoptosis in immature thymocytes by a signal through thymic shared antigen-1/stem cell antigen-2. J Exp Med. 1996;183:2355–2360. doi: 10.1084/jem.183.5.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perou C M, Jeffrey S S, Van de Rijn M, Rees C A, Eisen M B, Ross D T, Pergamenschikov A, Williams C F, Zhu S X, Lee J C F, Lashkari D, Shalon D, Brown P O, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulaski J T, Tieber V L, Coussens P M. Marek's disease virus-mediated enhancement of avian leukosis virus gene expression and virus production. Virology. 1992;186:113–121. doi: 10.1016/0042-6822(92)90065-w. [DOI] [PubMed] [Google Scholar]
- 36.Reusch U, Muranyi W, Lucin P, Burgert H G, Hengel H, Koszinowski U H. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 1999;15:1081–1091. doi: 10.1093/emboj/18.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitoh S, Kosugi A, Noda S, Yamamoto N, Ogata M, Minami Y, Miyake K, Hamaoka T. Modulation of TCR-mediated signaling pathway by thymic shared antigen-1 (TSA-1)/stem cell antigen-1 (Sca-2) J Immunol. 1995;155:5574–5581. [PubMed] [Google Scholar]
- 38.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 39.Shalon D, Smith S, Brown P O. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 40.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander E S, Golub T R. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teague T K, Hildeman D, Kedl R M, Mitchell T, Rees W, Schaefer B C, Bender J, Kappler J, Marrack P. Activation changes the spectrum but not the diversity of genes expressed by T cells. Proc Natl Acad Sci USA. 1999;96:12691–12696. doi: 10.1073/pnas.96.22.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas J, Kanangat S, Rouse B T. Herpes simplex virus replication-induced expression of chemokines and proinflammatory cytokines in the eye: implications in herpetic stromal keratitis. J Interferon Cytokine Res. 1998;18:681–690. doi: 10.1089/jir.1998.18.681. [DOI] [PubMed] [Google Scholar]
- 43.Tirunagaru V G, Sofer L, Cui J, Burnside J. An expressed sequence tag database of T-cell-enriched activated chicken splenocytes: sequence analysis of 5251 clones. Genomics. 2000;66:144–151. doi: 10.1006/geno.2000.6189. [DOI] [PubMed] [Google Scholar]
- 44.Tomazin R, Boname J, Hegde N R, Lewinsohn D M, Altschuler Y, Jones T R, Cresswell P, Nelson J A, Riddell S R, Johnson D C. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat Med. 1999;5:1039–1043. doi: 10.1038/12478. [DOI] [PubMed] [Google Scholar]
- 45.Tomazin R, Van Schoot N E, Goldsmith K, Jugovic P, Sempe P, Fruh K, Johnson D C. Herpes simplex virus type 2 ICP47 inhibits human TAP but not mouse TAP. J Virol. 1998;72:2560–2563. doi: 10.1128/jvi.72.3.2560-2563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trent J M, Bittner M, Zhang J, Wiltshire R, Ray M, Su Y, Garcia E, Meltzer P, DeRisi J, Penland L, Brown P. Use of microgenomic technology for analysis of alterations in DNA copy number and gene expression in malignant melanoma. Clin Exp Immunol. 1997;107(Suppl. 1):33–40. [PubMed] [Google Scholar]
- 47.Trigona W L, Hirano A, Brown W C, Estes D M. Immunoregulatory roles of interleukin-13 in cattle. J Interferon Cytokine Res. 1999;19:1317–1324. doi: 10.1089/107999099312993. [DOI] [PubMed] [Google Scholar]
- 48.Tumpey T M, Cheng H, Cook D N, Smithies O, Oakes J E, Lausch R N. Absence of macrophage inflammatory protein 1 alpha prevents the development of blinding herpes stromal keratitis. J Virol. 1998;72:3705–3710. doi: 10.1128/jvi.72.5.3705-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu H, Cong J-P, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoeller B, Popp M, Walter A, Redmann-Muller I, Lodemann E, Jungwirth C. Overexpression of chicken interferon regulatory factor-1 (CH-IRF-1) induces constitutive expression of MHC class I antigens but does not confer virus resistance to a permanent chicken fibroblast cell line. Gene. 1998;222:269–278. doi: 10.1016/s0378-1119(98)00504-6. [DOI] [PubMed] [Google Scholar]