Abstract
To better understand retroviral entry, we have characterized the interactions between subgroup A avian leukosis virus [ALV(A)] envelope glycoproteins and Tva, the receptor for ALV(A), that result in receptor interference. We have recently shown that soluble forms of the chicken and quail Tva receptor (sTva), expressed from genes delivered by retroviral vectors, block ALV(A) infection of cultured chicken cells (∼200-fold antiviral effect) and chickens (>98% of the birds were not infected). We hypothesized that inhibition of viral replication by sTva would select virus variants with mutations in the surface glycoprotein (SU) that altered the binding affinity of the subgroup A SU for the sTva protein and/or altered the normal receptor usage of the virus. Virus propagation in the presence of quail sTva-mIgG, the quail Tva extracellular region fused to the constant region of the mouse immunoglobulin G (IgG) protein, identified viruses with three mutations in the subgroup A hr1 region of SU, E149K, Y142N, and Y142N/E149K. These mutations reduced the binding affinity of the subgroup A envelope glycoproteins for quail sTva-mIgG (32-, 324-, and 4,739-fold, respectively) but did not alter their binding affinity for chicken sTva-mIgG. The ALV(A) mutants efficiently infected cells expressing the chicken Tva receptor but were 2-fold (E149K), 10-fold (Y142N), and 600-fold (Y142N/E149K) less efficient at infecting cells expressing the quail Tva receptor. These mutations identify key determinants of the interaction between the ALV(A) glycoproteins and the Tva receptor. We also conclude from these results that, at least for the wild-type and variant ALV(A)s tested, the receptor binding affinity was directly related to infection efficiency.
The avian leukosis-sarcoma viruses (ALV) are useful for studying early events in retrovirus infection because they use distinct cellular receptors to gain entry into cells. ALVs that infect chicken cells have been divided into six envelope subgroups, A through E and J [ALV(A) through ALV(E) and ALV(J)], based on host range, interference patterns, and cross-reactivity with neutralizing antibodies (65). Some members of these envelope subgroups have also been classified as either noncytopathic [ALV(A), ALV(C), and ALV(E)] or cytopathic [ALV(B) and ALV(D)]. Infection of primary avian fibroblasts by cytopathic ALVs usually causes a transient cytotoxicity that results in the death of 30 to 40% of the cells (66, 67). Replication of some subgroup ALV(B) and ALV(D) strains in DF-1 cells, a nontransformed cell line derived from line 0 chicken embryo fibroblasts (CEF), induces a transient cytotoxicity similar to the cytotoxicity observed in CEF (35, 56). Unexpectedly, some ALV(C) strains induce a transient cytotoxicity in DF-1 cells. DF-1 cells may be more sensitive to the cytotoxic effects of ALV(B), ALV(D), and ALV(C) than are CEF (35). However, not all ALV(B), ALV(D), and ALV(C) strains induce observable cytotoxicity. The degree of cytotoxicity in DF-1 cells and CEF appears to be related to the level of expression of the ALV envelope glycoproteins (56).
The susceptibility of chicken cells to infection by ALV(A) to ALV(E) strains is determined by three genetic loci designated tv-a, tv-b, and tv-c. The tv-a receptor controls susceptibility to ALV(A), the tv-c receptor controls susceptibility to ALV(C), and the tv-b receptor controls susceptibility to ALV(B), ALV(D), and ALV(E). Susceptibility or resistance to virus infection is conferred by alleles at these loci (65). Several cell surface proteins have been identified as ALV receptors. Tva, the receptor for ALV(A), contains sequences related to the ligand binding region of low-density lipoprotein receptors (LDLR) (7, 8, 69). TvbS3, a receptor for ALV(B) and ALV(D) (13, 60), TvbT, a receptor for ALV(E) (1), and TvbS1, a receptor for ALV(B), ALV(D), and ALV(E) (2), are all members of the tumor necrosis factor receptor family. The subgroup C receptor has not yet been identified.
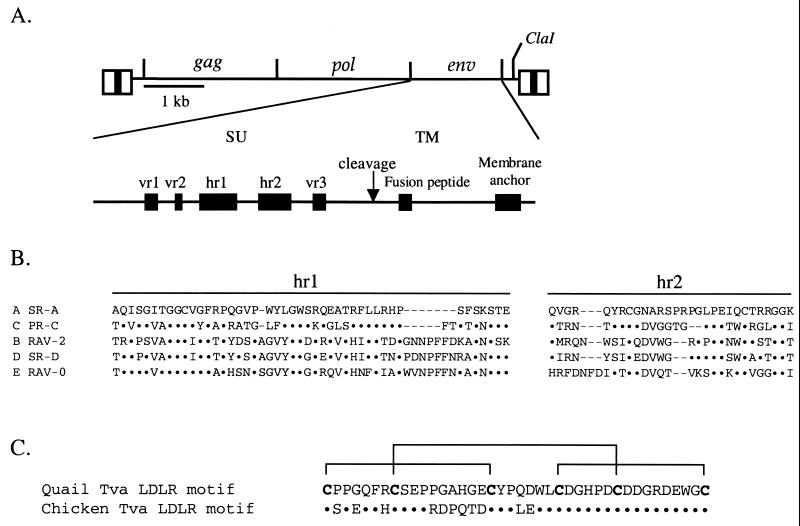
The ALV env gene (Fig. 1A) encodes a precursor polyprotein that assembles into a trimer in the endoplasmic reticulum (39). After transport to the Golgi, the polyproteins are glycosylated and subsequently cleaved to produce two glycoproteins: the surface glycoprotein (SU), which contains the major domains that interact with the host cell receptor, and the transmembrane glycoprotein (TM), which anchors SU to the cell membrane. The specific interaction of SU with its receptor is thought to trigger a conformational change in the envelope glycoprotein trimer necessary for the fusion peptide in TM to interact with the cell membrane and mediate the fusion of the viral and cellular membranes. The amino acid sequences of the ALV(A) to ALV(E) envelope glycoproteins are highly conserved except for five variable regions in SU (vr1, vr2, hr1, hr2, and vr3) (Fig. 1B) (11, 12, 22). It should be noted, however, that there is some variation in these five regions in individual ALV env subgroups (33).
FIG. 1.
(A) Schematic representations of the ALV-based RCASBP replication-competent retroviral vector and the major domains of the envelope glycoproteins. The five regions of amino acid sequence variation (vr1, vr2, hr1, hr2, and vr3) identified by comparing the sequences of the surface glycoproteins (SU) of ALV(A) to ALV(E) are also shown. (B) Comparison of the amino acid sequences of the two major SU variable domains, hr1 and hr2, of representative ALV(A) to ALV(E). The sequences were aligned with the ClustalW multiple-sequence alignment program of MacVector 6.5. Amino acids identical to SR-A are denoted by dots, and gaps in the alignment are denoted by dashes. (C) Comparison of the amino acid sequences of the quail and chicken Tva receptors homologous to the LDLR cysteine-rich ligand binding domain. Only the amino acid differences are shown for chicken Tva. The six cysteines are in bold, and the disulfide bonds are indicated by brackets.
Analysis of the different ALV env subgroups suggests that they evolved from a common ancestor, presumably because of host resistance (15). The results of several studies of recombinant env genes containing portions of the variable regions from ALV(B), ALV(C), or ALV(E), suggest that the principal binding interactions between the host receptor and the envelope glycoproteins occur in the two major variable regions, hr1 and hr2 (21, 63, 64). The vr3 region may play a role in receptor recognition but does not appear to participate in binding specificity; vr1 and vr2 do not appear to be essential for receptor specificity. A recent study demonstrated that basic residues in hr2 were important for efficient receptor interaction, although multiple basic residue mutations were required to significantly alter the binding of ALV(A) envelope glycoproteins to Tva and alter receptor-triggered envelope glycoprotein conformational changes (18, 52). Basic amino acids in ligands of the LDLR bind to the cysteine-rich domains of the receptor (50, 68).
It has been proposed that ALV variants with altered host range may have developed by both point mutations and exchange of the hr regions (15). A recent study by Taplitz and Coffin (61) tested this hypothesis by propagating an ALV(B) strain, td-Pr-RSV-B, on a mixture of permissive chicken (C/E) and nonpermissive quail (QT6/BD) cells. The selection for the ability to replicate in cells that lacked the ALV(B) receptor produced a variant virus with an extended host range. The variant virus, which contained two amino acid changes in the hr1 domain, was able to use both the chicken ALV(B) receptor and the quail ALV(E) receptor. In a different approach, Holmen and Federspiel (37) identified a variant ALV(A) virus with altered receptor usage by selecting for ALV(A) replication in chicken DF-1 cells expressing SU(A)-IgG, a soluble form of the ALV(A) SU domain fused to a rabbit immunoglobulin G (IgG) tag (72). The TF/SU(A)-19 cell line expressing SU(A)-rIgG was highly resistant to ALV(A) infection (∼185,000-fold) compared to DF-1 cells, and the antiviral effect was specific for ALV(A) strains, consistent with a receptor interference mechanism. A variant virus resistant to the SU(A)-rIgG antiviral effect was obtained and contained a 6-amino-acid deletion in the ALV(A) hr1 region. The variant virus could efficiently infect cells preinfected with another ALV(A) strain, indicating an altered receptor usage. However, the binding affinity of the variant virus envelope glycoproteins for Tva was unchanged. Interestingly, the deleted region in the ALV(A) variant overlaps the homologous hr1 region mutated in the ALV(B) variant identified by Taplitz and Coffin (61) and may indicate that this region of ALV SU hr1 plays a critical role in receptor specificity. These studies demonstrated that small changes within the retroviral env gene could alter receptor usage.
Initial characterization of the extracellular domain of the quail Tva receptor showed that the principal domains of Tva required for efficient binding to the ALV(A) envelope glycoproteins and infection of cells expressing Tva are contained within the extracellular region of Tva homologous to the LDLR cysteine-rich ligand binding domain (51, 71). Further characterization of the quail Tva receptor revealed that the carboxy-terminal half of the LDLR motif was required for receptor function (53, 54, 72). The overall structure of the Tva cysteine-rich region (amino acids 11 to 50 in quail Tva) is formed by three disulfide bonds (C1-C3, C2-C5, and C4-C6) between the six cysteine residues (Cys-11, Cys-18, Cys-28, Cys-35, Cys-41, and Cys-50) (Fig. 1C) (9). Three amino acid residues (Asp-46, Glu-47, and Trp-48) (54, 72) contained in the protein loop formed by the C4-C6 disulfide bond, as well as several other residues in the carboxy-terminal half of this region (Leu-34, His-38, and Gly-49) (53), have been identified as critical for efficient interaction between the quail Tva receptor and ALV(A) in a variety of assays. The chicken Tva receptor has not been extensively characterized since the complete coding region of the gene has not been identified (8, 69). However, the extracellular domain of the chicken Tva receptor was identified and shown to correspond to amino acids 7 to 83 of the quail Tva receptor. The carboxy-terminal halves of the cysteine-rich domains of the two Tva receptors are identical; however, the N-terminal regions are only 50% identical (Fig. 1C).
Soluble forms of viral receptors are effective reagents for characterizing viral envelope glycoprotein-receptor interactions. Soluble forms of the quail Tva receptor are capable of inducing conformational changes in subgroup A envelope glycoproteins that may be similar to receptor-triggered events required for membrane fusion (19, 31, 32, 34). A soluble receptor chimera consisting of the leader and first 6 amino acids of the quail Tva receptor fused to the extracellular region of the chicken Tva receptor efficiently and specifically blocked infection of ALV(A) (16). Similarly, a soluble form of the 83-amino-acid extracellular domain of the quail Tva receptor, produced and purified with a baculovirus expression system, efficiently bound ALV(A) envelope glycoproteins and specifically blocked ALV(A) infection (5). We recently showed that soluble forms of the chicken and quail Tva receptors, expressed from genes delivered by retroviral vectors, block ALV(A) infection of cultured chicken cells (∼200-fold antiviral effect) and chickens (>98% of the birds were not infected) (38). The antiviral effect was specific for ALV(A), consistent with a receptor interference mechanism.
In this study, we tested whether the antiviral effect of soluble quail Tva could select for resistant viruses. We hypothesized that virus variants with mutations in SU that reduced the affinity for the soluble quail Tva protein and/or had altered receptor usage would be selected and that these mutations would identify specific amino acids in subgroup A envelope glycoproteins critical for productive interaction with the Tva receptor. A pool of viruses resistant to soluble quail Tva receptor interference was selected in the chicken DF-1 cell line expressing quail sTva-mIgG, the extracellular domain of quail Tva fused to a mouse IgG tag. ALV(A) variants were obtained with mutations in the subgroup A hr1 region that were capable of efficiently infecting cells expressing the chicken but not the quail Tva receptor. The mutations significantly reduced the binding affinity of the mutant subgroup A envelope glycoproteins for quail sTva-mIgG without affecting the affinity for chicken sTva-mIgG.
MATERIALS AND METHODS
Virus sequence alignment.
The deduced amino acid sequences of the SU regions of ALV(A) to ALV(E) were compared using the ClustalW multiple-sequence alignment program of MacVector 6.5 (Oxford Molecular Ltd., Oxford, England). The Schmidt-Ruppin subgroup A strain of Rous sarcoma virus (SR-A; GenBank accession no. M14901), Prague subgroup C strain of Rous sarcoma virus (PR-C; GenBank accession no. J02342), Rous-associated virus type 2 (RAV-2; GenBank accession no. M14902), Schmidt-Ruppin subgroup D strain of Rous sarcoma virus (SR-D; GenBank accession no. D10652), and Rous-associated virus type 0 (RAV-0; GenBank accession no. M12171) were used in the sequence alignments.
Vector constructions.
The construction of the quail stva-mIgG gene in the CLA12NCO adapter plasmid and the RCASBP(C)stva-mIgG retrovirus has been described previously (38). The stva-mIgG gene encodes the signal peptide and 83-amino-acid extracellular domain of the quail pg950 Tva cDNA, fused to the constant region of the mouse IgG heavy chain. The stva-mIgG gene was isolated as a ClaI fragment from the CLA12NCO/stva-mIgG adapter plasmid and subcloned into the TFANEO expression plasmid (TFANEO/stva-mIgG). The expression cassette of TFANEO consists of two long terminal repeats derived from the RCAS vector that provide strong promoter, enhancer, and polyadenylation sites flanking a unique ClaI insertion site (26, 27). The TFANEO plasmid also contains a neo resistance gene expressed under the control of the chicken β-actin promoter and an ampicillin resistance gene for selection in Escherichia coli.
A soluble form of the chicken Tva receptor, encoding the leader and first six amino acids of quail Tva fused to the extracellular region of chicken Tva (residues 7 to 83), was described previously (16). The chicken stva-mIgG (ckstva-mIgG) gene was constructed by replacing the region encoding the quail sTva domain with the region encoding the chicken sTva domain, isolated as a NcoI-EagI fragment from pLC126, into the NcoI and EagI sites of CLA12NCO/stva-mIgG (38). The ckstva-mIgG gene cassette was isolated as a ClaI fragment and subcloned into the ClaI site of RCASBP(C).
The construction of the RCASBP(A)AP retroviral vector, the ALV-based replication-competent RCASBP vector with a subgroup A env gene and the heat-stable human placental alkaline phosphatase (AP) gene, has been described previously (27–29). The SalI sites flanking the AP gene were made blunt with the Klenow fragment of DNA polymerase type I (New England Biolabs, Beverly, Mass.), and the modified fragment was cloned into the SalI site (also made blunt) of the CLA12 adapter plasmid. The APSal− gene was subcloned into the RCASBP(A) vector as a ClaI fragment to produce RCASBP(A)APSal−. The mutant SU regions were isolated as Asp718-SalI fragments from the pBluescript KS clones, clone 17 (Y142N), clone 13 (E149K), or clone 22 (Y142N/E149K) (see Fig. 3), and cloned into the unique Asp718 and SalI sites of the RCASBP(A)APSal− vector. The mutation(s) in the env gene of the recombinant RCASBP(A)APSal− clones was verified by nucleotide sequence analysis.
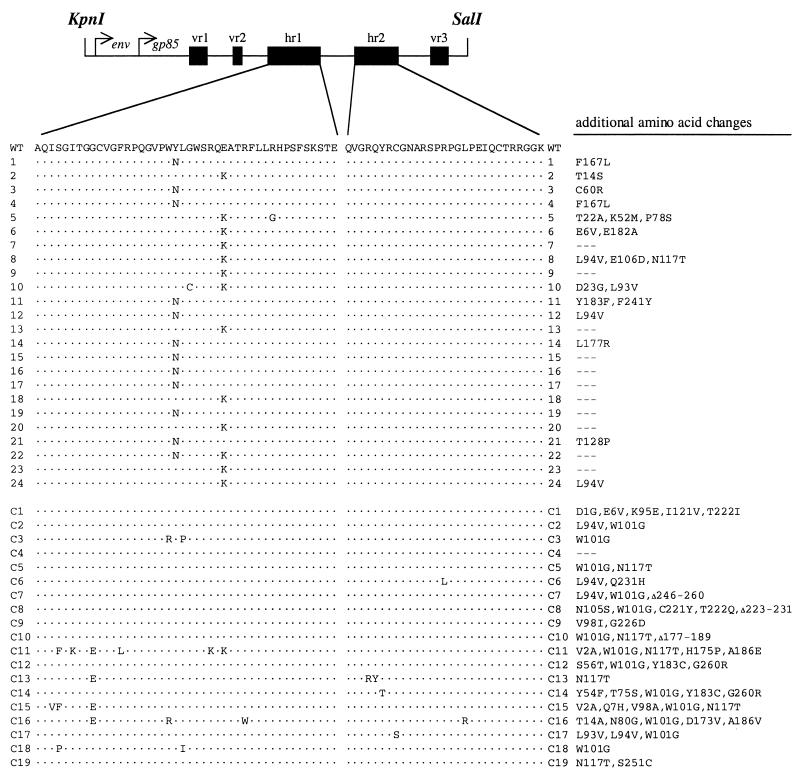
FIG. 3.
Identification of ALV(A) variants with env mutations. The SU regions of the env genes of ALV proviruses in genomic DNA isolated from each infected culture were amplified by PCR and cloned as KpnI-SalI fragments, and the nucleotide sequences were determined. A schematic of the cloned region is included at the top of the figure. The amino acids are numbered from the start of the mature SU glycoprotein (gp85). The amino acids contained in the five variable regions are vr1, 64 to 75; vr2, 100 to 105; hr1, 122 to 165; hr2, 199 to 227; and vr3, 261 to 269. The deduced amino acid sequences of the hr1 and hr2 regions of SU cloned from the sTva-mIgG-selected viral population (clones 1 to 24) and the control (unselected) viral population (clones C1 to C19) are shown. Clones 1 to 12 and C1 to C9 were amplified by Taq DNA polymerase, whereas clones 13 to 24 and C10 to C19 were amplified by Vent DNA polymerase. Only the differences in amino acid sequence compared to SR-A SU (WT) are shown. Additional amino acid changes in this region of SU are listed for each clone.
Cell culture and virus propagation.
DF-1 cells (35, 56), QT6 cells (48) and CEF derived from 10-day, line 0 embryos (C/E) (3) were grown in Dulbecco's modified Eagle's medium (GIBCO/BRL) supplemented with 10% fetal bovine serum (GIBCO/BRL), 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Quality Biological, Inc., Gaithersburg, Md.) at 39°C and 5% CO2. The generation of the 3T3pg950 cell line, an NIH 3T3 cell line stably transfected with the quail pg950 Tva receptor cDNA, was previously described (31). The 3T3pg950 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% calf serum (CS) (GIBCO/BRL), 250 μg of G418 per ml (GIBCO/BRL), 100 U of penicillin per ml, and 100 μg of streptomycin per ml at 37°C and 5% CO2. The cultures were passaged 1:3 or 1:6 when confluent.
Virus propagation was initiated either by transfection of plasmid DNA that contained the retroviral vector in proviral form (27) or by direct infection. In standard transfections, 5 μg of purified plasmid DNA was introduced into DF-1 cells by the calcium phosphate precipitation method (43). Viral spread was monitored by assaying culture supernatants for ALV capsid protein (CA) by enzyme-linked immunosorbent assay (ELISA) (59). Virus stocks were generated from cell supernatants. The supernatants were cleared of cellular debris by centrifugation at 2,000 × g for 10 min at 4°C and stored in aliquots at −80°C. DF-1 cells transfected with TFANEO or TFANEO/stva-mIgG plasmid DNA were grown in 500 μg of G418 per ml to select for neomycin-resistant cells. Clones were isolated using cloning cylinders (Bellco Glass Inc., Vineland, N.J.), expanded, and maintained with standard medium supplemented with 250 μg of G418 per ml.
DF-1 cell cultures chronically infected with RCASBP(A), RCASBP(B), RCASBP(C), or HPRS-103 were produced. The RCASBP viruses with subgroup A, B, and C env genes have been described previously (27). HPRS-103 (GenBank accession no. Z46390) is an ALV strain with a subgroup J env gene (4) and was obtained from Michael A. Skinner (Institute for Animal Health, Compton, Berks., United Kingdom).
ELISA.
The ALV CA protein was detected in culture supernatants by ELISA as previously described (59). The levels of sTva-mIgG and cksTva-mIgG were determined in culture supernatants by ELISA for the mouse IgG tag as previously described (38). The linear range for a standard experiment was between 0.5 and 50 ng of ImmunoPure mouse IgG Fc fragment per ml.
Cloning and nucleotide sequence analysis of integrated viral DNA.
DNA was isolated from cells in culture using the QIAamp tissue kit (Qiagen). The SU region of the env gene was amplified by PCR using either Taq DNA polymerase (Promega, Madison, Wis.) or Vent DNA polymerase (New England Biolabs) with the primers 5′-GGGACGAGGTTATGCCGCTG-3′ (∼50 bp upstream of the Asp718 site) and 5′-ATGAGGAAAATTGCGGGTGG-3′ (downstream of the SalI site). Each Taq PCR mixture contained 1.25 μl of 10× PCR buffer (final concentrations, 50 mM Tris-Cl [pH 8.3], 50 mM KCl, 7 mM MgCl2, and 1.1 mM β-mercaptoethanol), 1.25 μl of 1.7-mg/ml bovine serum albumin, 0.5 μl of each deoxynucleoside triphosphate at 25 mM, 0.5 μl of each primer (absorbance at 260 nm = 5), 6.0 μl of H2O, and 1.0 μl of DNA (genomic DNA, ∼100 ng/μl; plasmid DNA, ∼2 ng/μl). The reaction mixtures were heated to 90°C for 1 min, and the reactions were initiated by the addition of 1.5 μl of Taq DNA polymerase diluted 1:10 (vol/vol) (0.75 U). Thirty cycles of PCR were carried out as follows: 90°C for 40 s and then 59°C for 80 s. Each Vent PCR mixture contained 1.25 μl of 10× Vent Pol buffer and 0.125 μl of 100× acetylated bovine serum albumin (supplied by the manufacturer), 1.0 μl of each deoxynucleoside triphosphate at 25 mM, 0.5 μl of each primer (absorbance at 260 nm = 5), 0.625 μl of 0.1 M MgSO4, 6.0 μl of H2O, 1.0 μl of 50% dimethyl sulfoxide, and 1.0 μl of DNA (genomic DNA, ∼100 ng/μl; plasmid DNA, ∼2 ng/μl). The reaction mixtures were heated to 90°C for 1 min, and the reactions were initiated by the addition of 0.5 μl of Vent DNA polymerase diluted 1:2 (vol/vol) (0.5 U). Thirty cycles of PCR were carried out as follows: 90°C for 40 s and then 59°C for 80 s.
The amplified products were separated by agarose gel electrophoresis, and the 1.1-kb product was purified and digested with Asp718 and SalI. The digested product was cloned into pBluescript KS (Stratagene, La Jolla, Calif.) digested with Asp718 and SalI. The nucleotide sequences of the SU regions were determined by the Mayo Clinic Molecular Biology Core on an ABI PRISM 377 DNA sequencer (with XL upgrade) with a ABI PRISM dRhodamine Terminator cycle-sequencing ready reaction kit and AmpliTaq DNA polymerase (PE Applied Biosystems, Foster City, Calif.).
ALV AP challenge assay.
In a direct AP challenge assay, DF-1 cell cultures (∼30% confluent) were incubated with 10-fold serial dilutions of the RCASBP/AP virus stocks for 36 to 48 h at 39°C. In a preadsorption AP challenge assay, the 10-fold viral serial dilutions were first mixed for 3 h at 4°C with 2 ml of supernatant containing quail sTva-mIgG and then assayed as above. The assay for AP activity was described previously (38).
Fluorescence-activated cell sorting (FACS) analysis of envelope glycoprotein binding to sTva-mIgG.
Uninfected DF-1 cells or DF-1 cells infected with either wild-type or mutant RCASBP(A)AP virus were removed from culture with Trypsin de Larco (Quality Biological, Inc.) and washed with Dulbecco's phosphate-buffered saline (PBS). The cells were fixed with 4% paraformaldehyde in PBS at room temperature for 15 min and then washed with PBS. Approximately 106 cells in PBS supplemented with 1% calf serum (PBS-CS) were incubated on ice for 30 min with supernatant containing either chicken or quail sTva-mIgG. The cells were then washed with PBS-CS and incubated with 5 μl of goat anti-mouse IgG (heavy plus light chains) linked to phycoerythrin (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) in PBS-CS (1-ml total volume) on ice for 30 min. The cell–sTva-mIgG–Ig-phycoerythrin complexes were washed with PBS-CS, resuspended in 0.5 ml of PBS-CS, and analyzed with a Becton Dickinson FACSCalibur using CELLQuest 3.1 software.
Apparent dissociation constant (Kd) calculations.
The maximum possible bound fluorescence for each data set obtained from the FACS binding assays was estimated by fitting the data via a nonlinear least-squares method to a log logistic growth curve function: f(y)=M/[1 + e−r(log x − logKd)], where y is the mean fluorescence, M is the maximum fluorescence, r is the rate, x is the concentration of sTva-mIgG, and Kd is the dissociation constant, defined as the concentration of sTva-mIgG at half-maximal binding. The estimated maximum fluorescence was used to convert the FACS fluorescence bound to a percentage of the maximum values and plotted against the sTva-mIgG concentration. The statistical significance of the estimated Kd values was determined using multiple t tests and the Bonferonni correction for multiple testing, where the P values must be smaller than 0.003 to be considered significant at a 95% confidence level.
RESULTS
Generation of DF-1 cell lines expressing quail sTva-mIgG.
Clonal cell lines expressing quail sTva-mIgG were produced by calcium phosphate transfection of the expression plasmid TFANEO/stva-mIgG in DF-1 cells. DF-1 cells are derived from line 0 (C/E) CEF that do not contain endogenous sequences closely related to the ALV-based vectors (3), eliminating possible recombinants between the vector and endogenous viruses. The cell lines were challenged with RCASBP(A)AP, and the level of resistance to ALV(A) infection was between 40- and 250-fold. The TF/sTva-4 cell line had the highest level of resistance to infection by RCASBP(A)AP in a direct AP challenge assay. Preabsorption of RCASBP(A)AP with supernatant from a confluent TF/sTva-4 culture increased the antiviral effect to ∼15,600-fold (Table 1). The TF/sTva-4 cell line expressed relatively high levels of quail sTva-mIgG in the supernatant: 4.6 ± 0.2 μg/ml measured by ELISA for mouse IgG. The level of sTva-mIgG expressed by the TF/sTvaI4 culture did not vary significantly over a 50-day period.
TABLE 1.
Relative resistance of TF/sTva-4 to infection by wild-type and mutant RCASBP(A)AP viruses
| Virus | Titer (mean ± SD) on:
|
Resistanceb | |
|---|---|---|---|
| DF-1 cells | TF/sTva-4 cellsa | ||
| Wild type | (5.3 ± 1.9) × 106 | (3.4 ± 1.2) × 102 | 15,590 |
| E149K | (1.3 ± 0.1) × 107 | (5.6 ± 1.6) × 103 | 2,320 (7) |
| Y142N | (7.7 ± 1.1) × 106 | (5.4 ± 1.2) × 104 | 140 (111) |
| Y142N/E149K | (1.2 ± 0.2) × 107 | (5.8 ± 1.5) × 105 | 21 (742) |
Virions were preabsorbed with supernatant from a confluent TF/sTva-4 culture containing sTva-mIgG protein prior to the assay.
The resistance of TF/sTva-4 cells to virus infection was determined by dividing the mean titer obtained in the control DF-1 cells by the mean titer obtained for each virus in TF/sTva-4 cells. The fold difference in the resistance of the mutant viruses compared to wild-type virus is given in parentheses.
Selection of RCASBP(A) variants resistant to the quail sTva-mIgG antiviral effect.
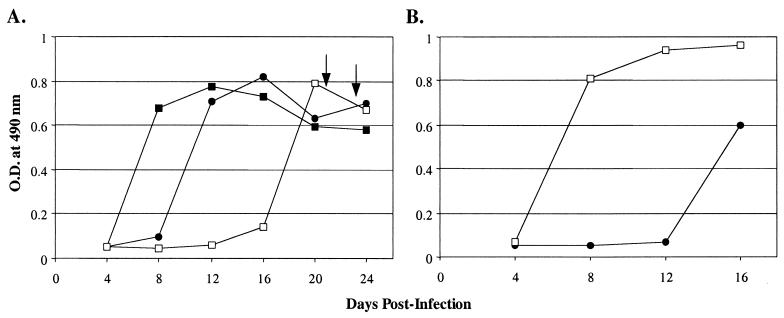
To expose the TF/sTva-4 cell line to a high level of virus yet infect a small number of cells, TF/sTva-4 cells were challenged with ∼5 × 106 infection-forming units (IFU) of RCASBP(A)AP preadsorbed with supernatant containing quail sTva-mIgG. Two days after virus challenge, the culture was split among three plates. On day 4 postchallenge, one of the three plates contained 115 infected foci as determined by the AP assay. As a control, a cell line transfected with TFANEO alone was challenged with ∼1 and ∼10 IFU of RCASBP(A)AP and passaged as above, which resulted in one plate of each culture with 15 and 256 infected AP-positive foci 4 days postchallenge. These controls allowed us to monitor the rate of viral spread from approximately the same number of infected cells in the absence of sTva-mIgG. The infected cell lines were passaged, and the supernatants were assayed for the ALV CA protein by ELISA to monitor for virus production. In the control cultures, ALV CA protein levels rose rapidly, reaching peaks 8 to 12 days postinfection (Fig. 2A). ALV CA protein was not observed in the TF/sTva-4 culture until 16 days postinfection, reaching a peak on day 20 (Fig. 2A). There was a transient period of cytotoxicity in the infected TF/sTva-4 culture 16 days postinfection. The observed cytotoxicity was similar to the transient cytotoxicity observed in DF-1 cultures infected with some ALV(B), ALV(D), and ALV(C) strains (35, 56). Since wild-type ALV(A) strains do not induce cytotoxicity and the cultures were negative for other ALV envelope subgroups as assayed by PCR with subgroup-specific primers (data not shown), the infected TF/sTva-4 culture may be producing a mutant virus pool containing ALV(A) variants which interact with an alternate receptor, resulting in cytotoxicity.
FIG. 2.
Selection of ALV(A) variants resistant to quail sTva-mIgG receptor interference. Viral growth was monitored by ELISA for the ALV CA protein. (A) TF/sTva-4 cells, a cell line derived from DF-1 cells transfected with the TFANEO/stva-mIgG plasmid and expressing quail sTva-mIgG protein, were challenged with ∼5 × 106 IFU of RCASBP(A)AP preadsorbed with supernatant containing quail sTva-mIgG protein prior to infection (□). As controls, the TFANEO-alone DF-1 cell line was challenged with ∼1 IFU (●) or ∼10 IFU (■) of RCASBP(A)AP. The arrows indicate the 3-day period of transient cytotoxicity observed in the challenged TF/sTva-4 culture. (B) Uninfected TF/sTva-4 cells were infected with 0.1 ml of day 24 supernatant from the challenged TF/sTva-4 culture (□) and 0.1 ml of supernatant from a TFANEO-alone culture (●) (see panel A). O.D., optical density.
To determine if the viral population produced by the TF/sTva-4 culture now had a replication advantage in TF/sTva-4 cells compared to wild-type RCASBP(A)AP, uninfected TF/sTva-4 cells were challenged with the virus pool from the infected TF/sTva-4 culture or wild-type RCASBP(A)AP. In this assay, 0.1 ml of day 24 supernatant was preadsorbed with sTva-mIgG prior to infection. The cultures were passaged, and the supernatants were monitored by ELISA for virus production. The virus pool produced by the TF/sTva-4 culture (Fig. 2A) had a significant replication advantage over wild-type RCASBP(A)AP in TF/sTva-4 cells (Fig. 2B).
The selected viruses contain mutations in the hr1 region of SU.
The ∼1.1-kb SU coding regions of integrated proviruses were amplified by PCR from genomic DNA isolated from the day 16 TF/sTva-4 culture (Fig. 2B) and from a control TFANEO culture infected with wild-type RCASBP(A)AP (Fig. 2A). The SU regions were amplified in two different reactions, one using Taq DNA polymerase and one using Vent DNA polymerase, to control for any changes that might have been introduced by PCR. The amplified products were cloned, and the nucleotide sequence of SU was compiled from 19 control clones and 24 clones isolated from TF/sTva-4 culture DNA. The deduced amino acid sequence of each clone was compared to the sequence of the SU region of the parental virus, RCASBP(A). All of the SU coding regions cloned from the TF/sTva-4 culture contained changes in the hr1 region (Fig. 3). There was either a tyrosine (TAT)-to-asparagine (AAT) mutation at codon 142 (Y142N) (11 clones) or a glutamic acid (GAG)-to-lysine (AAG) mutation at codon 149 (E149K) (12 clones), or both mutations (Y142N/E149K) (1 clone). Several clones contained additional amino acid changes in SU compared to RCASBP(A). The amino acid sequences of the SU regions cloned from the control “unselected” viral population in the TFANEO culture contained significantly more differences (average of four changes per clone) than did the sequences in the TF/sTva-4-selected clones (average of two changes per clone) (Fig. 3). Only one amino acid change (W101G) was present in a significant proportion of the controls (13 of 19).
RCASBP(A)AP molecular clones containing the putative mutations display a resistant phenotype.
To determine if the Y142N, E149K, and Y142N/E149K mutations provided an advantage for ALV(A) replication in cells expressing quail sTva-mIgG, we replaced the SU fragment in the wild-type RCASBP(A)AP molecular clone with the mutant SU fragments containing either the Y142N or E149K mutation or both (Y142N/E149K) mutations. Wild-type and mutant RCASBP(A)AP plasmid DNAs were transfected into both DF-1 and TF/sTva-4 cells. The culture supernatants were assayed on DF-1 cells for infectious virus. In DF-1 cells, the wild-type RCASBP(A)AP virus and the three mutant viruses produced similar titers of infectious virus (Fig. 4). To determine if the infected DF-1 cultures produced the expected mutant viruses or revertants, the SU regions in day 16 genomic DNA of each culture were amplified by PCR and cloned, and the nucleotide sequences of 10 clones were determined as described above. All of the clones analyzed contained the expected hr1 mutation(s) with no additional second-site mutations (data not shown). In TF/sTva-4 cells, the Y142N and Y142N/E149K viruses replicated to high titers, as in DF-1 cells (Fig. 4). However, the E149K virus produced a lower level of infectious virus in TF/sTva-4 cells than in DF-1 cells (Fig. 4), and there was a prolonged period of cytotoxicity (13 days), after which the cells recovered (data not shown). The cytotoxicity phenotype observed during the selection of variant ALV(A) strains (Fig. 2A) appears to correlate with the E149K mutation, but only when the virus is produced in the presence of quail sTva-mIgG in TF/sTva-4 cells.
FIG. 4.
Analysis of the receptor interference patterns of the mutant and wild-type RCASBP(A)AP viruses produced in DF-1 cells 12 days posttransfection (A) and in TF/sTva-4 cells for the Y142N and Y142N/E149K mutants 12 days posttransfection, and the E149K mutant 24 days posttransfection (B). Uninfected DF-1 cells (DF-1) and DF-1 cells chronically infected with RCASBP(A) (A), RCASBP(B) (B), RCASBP(C) (C), or subgroup J HPRS-103 (J) viruses were infected with 10-fold serial dilutions of the culture supernatants, and the titer was determined by the AP assay. The results shown are an average of three different experiments. Error bars show standard deviations.
The hr1 mutations reduce the antiviral effect of quail sTva-mIgG.
The antiviral effect of quail sTva-mIgG on wild-type and mutant virions was determined as one measure of the interaction of the wild-type and mutant envelope glycoproteins with the quail Tva receptor. Virions collected from the DF-1 cultures infected with Y142N, E149K, Y142N/E149K, or wild-type RCASBP(A)AP were preadsorbed with quail sTva-mIgG, and the infectious-virus titer was determined on uninfected TF/sTva-4 cells. Treatment of wild-type RCASBP(A)AP virus with quail sTva-mIgG in this assay reduced the titer ∼15,600-fold compared to that of untreated virus determined on DF-1 cells (Table 1). However, preadsorption of the mutant RCASBP(A)AP viruses with quail sTva-mIgG resulted in significantly lower antiviral effects: ∼2,300-fold for E149K, ∼140-fold for Y142N, and ∼20-fold for Y142N/E149K. Since we believe that the antiviral effect of sTva-mIgG depends on competitive binding to the envelope glycoproteins on ALV(A) virions, the results of this assay suggest that the antiviral effect of quail sTva-mIgG selected envelope glycoproteins with a lower binding affinity for quail sTva-mIgG.
Analysis of receptor interference patterns.
To determine if the envelope glycoprotein mutations altered the receptor usage of the recombinant viruses, we performed receptor interference assays. Uninfected DF-1 cells and DF-1 cells chronically infected with subgroup A [RCASBP(A)], subgroup B [RCASBP(B)], subgroup C [RCASBP(C)] or subgroup J (HPRS-103) ALVs were used to determine the titers of the Y142N, E149K, Y142N/E149K, or wild-type RCASBP(A)AP virus stocks produced in DF-1 cells by the AP assay. The susceptibility of this panel of cells to a wild-type ALV(A) strain was demonstrated by RCASBP(A)AP infection (Fig. 4, WT). Uninfected DF-1 cells and DF-1 cells infected with ALV(B), ALV(C), or ALV(J) strains were efficiently infected by RCASBP(A)AP. However, preinfection with ALV(A) inhibited RCASBP(A)AP superinfection by 5 log units (Fig. 4A), and superinfection of ALV(B)-infected cells with RCASBP(B)AP or ALV(C)-infected cells with RCASBP(C)AP produced a 4-log-unit decrease in titer (data not shown). The three mutant RCASBP(A)AP viruses propagated in DF-1 cells had receptor interference patterns similar to that of wild-type RCASBP(A)AP (Fig. 4A). The only difference that may be significant was that both the Y142N and Y142N/E149K viruses were somewhat less efficient at infecting cells infected with ALV(B) or ALV(C) than were wild-type and E149K viruses.
A very different pattern of receptor interference was produced when the mutant viruses were propagated in TF/sTva-4 cells expressing quail sTva-mIgG. All three mutant viruses had different receptor interference profiles compared to virus produced in DF-1 cells (Fig. 4B). The E149K virus-infected TF/sTva-4 culture produced virus that infected the ALV(A)-infected DF-1 cells relatively efficiently (within 10-fold of the titer on DF-1 cells) and was ∼20-fold less efficient at infecting ALV(B)- or ALV(C)-infected DF-1 cells (Fig. 4B). As described above, the E149K virus grew to a much lower titer in TF/sTva-4 cells than did the other mutant RCASBP(A)AP viruses, and it induced a prolonged period of cytotoxicity (data not shown). The Y142N virus-infected TF/sTva-4 culture produced virus capable of infecting ALV(A)-infected DF-1 cells ∼200-fold more efficiently than did Y142N or wild-type RCASBP(A)AP propagated in DF-1 cells (Fig. 4). The Y142N/E149K RCASBP(A)AP virus-infected TF/sTva-4 culture produced virus capable of infecting ALV(A)-infected DF-1 cells ca. fivefold more efficiently than did Y142N/E149K or wild-type RCASBP(A)AP propagated in DF-1 cells (Fig. 4). These results indicate that the mutations in the ALV(A) envelope glycoproteins may alter the receptor usage of the mutant viruses in the presence of quail sTva-mIgG.
The mutant viruses efficiently infect cells expressing the chicken but not the quail Tva receptor.
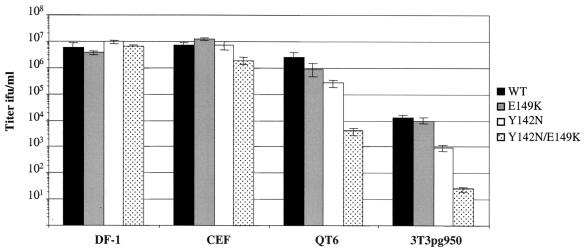
The inability of the mutant RCASBP(A)AP viruses propagated in DF-1 cells to efficiently infect ALV(A)-infected DF-1 cells (Fig. 4A), even though the mutants were resistant to the antiviral effects of quail sTva-mIgG (Table 1), presented a conundrum. One possible explanation may be that virus replication in chicken DF-1 cells and in the presence of quail sTva-mIgG selected envelope glycoprotein mutants which had lower affinities for the quail Tva receptor but retained high binding affinities for the chicken Tva receptor. As an initial test of this hypothesis, the ability of mutant and wild-type RCASBP(A)AP viruses to infect cells expressing the chicken Tva receptor (chicken DF-1 cells [C/E] and line 0 CEF [C/E]), and cells expressing the quail Tva receptor (quail QT6 cells [QT6/BD] and mouse NIH 3T3 cells that express the quail pg950 Tva cDNA [3T3pg950]), was determined. NIH 3T3 cells are not susceptible to efficient infection by ALV because they lack functional receptors. As shown above, the three mutant viruses infected DF-1 cells as efficiently as the wild-type RCASBP(A)AP did, as demonstrated by the AP assay (Fig. 5). Two of the mutant viruses also infected CEF as efficiently as the wild type did. However, the Y142N/E149K virus was ca. twofold less efficient at infecting CEF than the wild-type virus was. Overall, the envelope glycoprotein mutations had little or no effect on the efficiency of the mutant viruses to infect cells expressing the chicken Tva receptor.
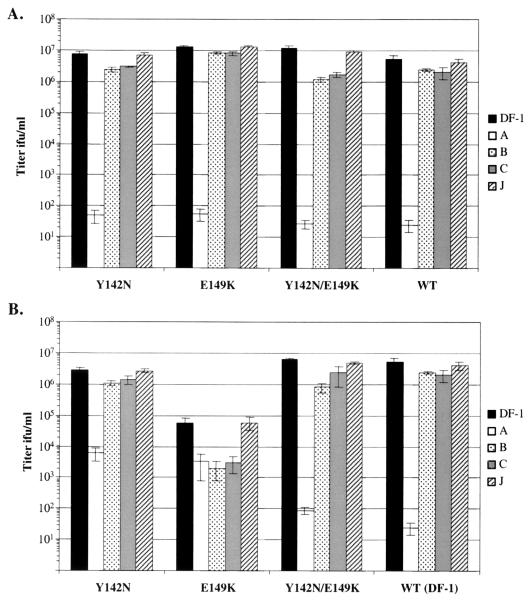
FIG. 5.
Analysis of the ability of the mutant viruses to use different Tva receptors. Cell lines expressing the chicken Tva receptor, chicken DF-1 cells (DF-1) and line 0 chicken embryo fibroblasts (CEF), or the quail Tva receptor, quail QT6 cells (QT6) and NIH 3T3 cells expressing the quail pg950 Tva (3T3pg950), were infected with 10-fold serial dilutions of Y142N, E149K, Y142N/E149K, or wild-type RCASBP(A)AP DF-1 virus stocks, and the viral titer was determined by the AP assay. The results shown are an average of three different experiments. Error bars show standard deviations.
In contrast, the ability of the mutant viruses to infect cells expressing the quail Tva receptor was reduced and directly related to the susceptibility of the viruses to the antiviral effect of quail sTva-mIgG. The Y142N/E149K virus, which was the least susceptible to sTva-mIgG neutralization, was least efficient at infecting QT6 and 3T3pg950 cells, producing titers ∼600-fold lower than those of the wild-type RCASBP(A)AP (Fig. 5). The Y142N virus, which was more susceptible to sTva-mIgG neutralization, infected QT6 and 3T3pg950 cells at ∼10-fold-lower levels than wild-type virus did. The E149K virus, which was susceptible to sTva-mIgG neutralization at a level similar to wild-type virus, infected QT6 and 3T3pg950 cells at near wild-type levels (ca. twofold lower). While the 3T3pg950 cells were susceptible to ALV(A) infection, the efficiency of wild-type RCASBP(A)AP infection was 200- to 500-fold lower in 3T3pg950 cells than in avian cells (Fig. 5). In a study where mouse embryonic stem cells were infected by a similar ALV-based retroviral vector containing a selectable marker, only one-third of the infected stem cells could be selected with the appropriate selective agent (70). Therefore, it seems likely that the low wild-type RCASBP(A)AP titer obtained on 3T3pg950 cells was due, at least in part, to infected cells producing undetectable levels of AP.
Analysis of the binding affinities of the mutant envelope glycoproteins for Tva.
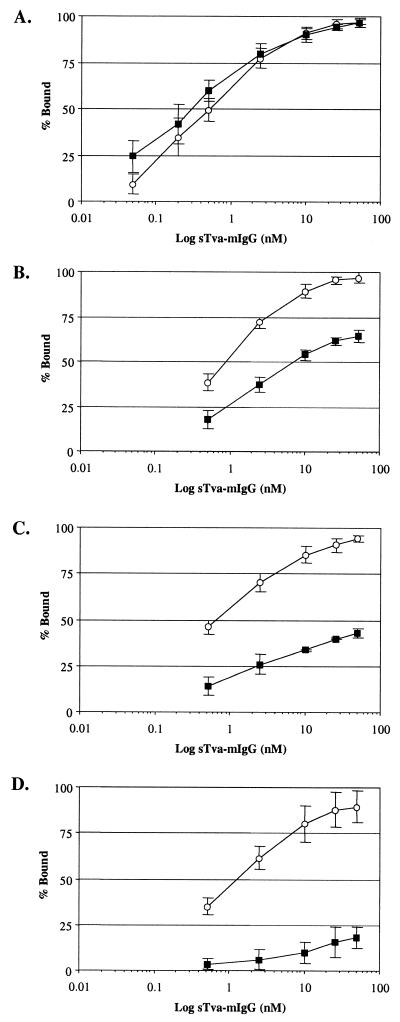
To determine if the hr1 mutations altered the binding affinities of the mutant envelope glycoproteins for the Tva receptors, the Y142N, E149K, Y142N/E149K, and wild-type RCASBP(A)AP envelope glycoproteins expressed on DF-1 cells were assayed for binding to chicken and quail sTva-mIgG by FACS. The sTva-mIgG proteins were produced by DF-1 cells chronically infected with either the RCASBP(C)stva-mIgG virus or the RCASBP(C)ckstva-mIgG virus. The level of sTva-mIgG in the culture supernatants was quantitated by ELISA for the mouse IgG tag. Uninfected DF-1 cells and DF-1 cells expressing wild-type RCASBP(A)AP envelope glycoproteins were incubated with different amounts of supernatant containing either chicken or quail sTva-mIgG (0.05 to 51 nM). The sTva-mIgG proteins bound to the cells were incubated with an antimouse antibody conjugated to phycoerythrin, and the fluorescence was quantitated by FACS. The results were corrected for nonspecific binding of sTva-mIgG and the secondary antibody by subtracting the level of fluorescence obtained from uninfected DF-1 cells. Both the chicken and quail sTva-mIgG proteins bound to wild-type subgroup A envelope glycoproteins in a concentration-dependent manner, reaching near-saturable levels of binding at 25 to 50 nM. The maximum mean fluorescence was estimated for each data set by fitting the data via a nonlinear least-squares method to a log logistic growth curve function (see Materials and Methods). The data from four experiments were plotted as the percentage of the maximum mean fluorescence bound against the log of the sTva-mIgG concentration (Fig. 6A). From these data, the binding affinities of the wild-type ALV(A) envelope glycoproteins for the chicken and quail sTva-mIgG proteins were estimated to have similar apparent Kd values, 0.51 and 0.31 nM, respectively (Table 2). The Kd values obtained are an approximation of the equilibrium dissociation constant, since equilibrium was not maintained during the wash and detection steps of the FACS binding assay.
FIG. 6.
Binding of chicken and quail sTva-mIgG to wild-type and mutant ALV(A) glycoproteins. Uninfected DF-1 cells and DF-1 cells infected with wild-type RCASBP(A)AP virus (A) or the E149K (B), Y142N (C), or Y142N/E149K (D) mutant viruses (Fig. 4) were fixed with paraformaldehyde, incubated with different amounts of the chicken (○) or quail (■) sTva-mIgG protein, and the envelope glyco- protein–sTva-mIgG complexes were bound to goat anti-mouse Ig antibody linked to phycoerythrin. The levels of phycoerythrin bound to the cells were measured by FACS, and the maximum fluorescence was estimated (see Materials and Methods). The data were plotted as percent maximum fluorescence bound against sTva-mIgG concentration. The values shown are an average of four different experiments. Error bars show standard error.
TABLE 2.
Estimated binding affinities of wild-type and mutant ALV(A) glycoproteins for soluble forms of the chicken and quail Tva receptors
| Envelope glycoprotein | Apparent Kd (nM)a
|
|
|---|---|---|
| Chicken sTva-mIgG | Quail sTva-mIgG | |
| Wild type | 0.51 ± 0.18cd | 0.31 ± 0.15ce |
| E149K | 0.86 ± 0.10df | 9.68 ± 2.94ef |
| Y142N | 0.67 ± 0.21dg | 100.4 ± 11.7beg |
| Y142N/E149K | 1.16 ± 0.13dh | 1469 ± 768beh |
The apparent Kd values were estimated by fitting the data (see Fig. 6) via the nonlinear least-squares method to a log logistic growth curve function as described in Materials and Methods. Each result is the mean and standard error of four experiments.
The apparent Kd was estimated by extrapolating the data in Fig. 6 to 50% maximum sTva-mIgG bound.
These results are not statistically different (P > 0.1).
These results are not statistically different (P > 0.02).
Statistical difference of P < 0.0002.
Statistical difference of P < 0.0001.
Statistical difference of P < 0.0001.
Statistical difference of P < 0.0001.
The binding affinities of the E149K, Y142N, and Y142N/E149K envelope glycoproteins for the chicken and quail sTva-mIgG proteins were estimated in the same way. However, in contrast to the wild-type envelope glycoproteins having similar binding affinities for both sTva-mIgG proteins, the binding affinities of all three mutant glycoproteins for quail sTva-mIgG were significantly reduced, while near-wild-type binding affinities for chicken sTva-mIgG were retained. Since the mutant glycoproteins bound the chicken sTva-mIgG with high affinity, the maximum mean fluorescence was calculated using the data from the chicken sTva-mIgG binding assays for each mutant envelope glycoprotein. The data from both the chicken and quail sTva-mIgG binding assays were plotted as the percentage of the chicken sTva-mIgG maximum mean fluorescence bound against the log of the sTva-mIgG concentration (Fig. 6B to D). The binding affinities of the mutant glycoproteins for chicken sTva-mIgG were estimated to have apparent Kd values similar to those of wild-type ALV(A) envelope glycoproteins (Table 2). However, the mutant glycoproteins bound quail sTva-mIgG at significantly lower affinities than did the wild-type ALV(A) glycoproteins. The binding affinity of the mutant glycoproteins for quail sTva-mIgG was directly related to the ability of the mutant viruses to infect cells expressing the quail Tva receptor; i.e., the Y142N/E149K envelope glycoproteins had the lowest binding affinity for quail sTva-mIgG and the Y142N/E149K virus was the least efficient at infecting QT6 and 3T3pg950 cells.
DISCUSSION
As we had hypothesized, sTva-mIgG receptor interference selected SU mutations that significantly reduced the binding affinity of the envelope glycoproteins for sTva-mIgG. We did not expect that all three mutant viruses produced in DF-1 cells would be efficiently blocked from infecting DF-1 cells preinfected with ALV(A) strains at levels similar to those of wild-type virus (Fig. 4A). We believe that selecting for viruses able to escape from receptor interference produced by quail sTva-mIgG in DF-1 cells which express the chicken Tva receptor identified mutants that selectively recognize chicken Tva in preference to quail Tva. We show that the mutant envelope glycoproteins retain near-wild-type binding affinities for the soluble chicken Tva receptor but have significantly lower binding affinities for the soluble quail Tva receptor (Fig. 6; Table 2). The lower binding affinities of the mutant glycoproteins for quail sTva-mIgG most probably explain the resistance of the mutant virus to neutralization by quail sTva-mIgG (Table 1) and the reduced infection efficiency of the mutant viruses for cells expressing the quail Tva receptor (Fig. 5). The region of quail Tva homologous to the LDLR ligand binding domain, principally the carboxy-terminal half of the region, is thought to contain the major sites of interaction with ALV(A) envelope glycoproteins. The chicken and quail Tva receptors are identical in the carboxy-terminal half of the 40-amino-acid cysteine-rich region of the LDLR motif; however, they are only ∼50% identical in the amino-terminal half of this region (Fig. 1C) (8). These results indicate that the mutations identify specific amino acids in ALV(A) SU important for the interaction of SU with the nonconserved amino-terminal region of the Tva receptor.
Soluble forms of the Tva receptor specifically block ALV(A) infection (5, 38, 72) and induce conformational changes in the envelope glycoproteins similar to events thought to be required for membrane fusion (19, 31, 32, 34). In this study, the binding affinities of both chicken and quail sTva-mIgG produced in chicken cells, for ALV(A) envelope glycoproteins expressed on the surface of infected chicken cells, were estimated to have apparent Kd values of 0.51 and 0.31 nM, respectively (Table 2). Since the sTva-mIgG proteins were most probably dimers due to the IgG tag and the Tva receptor is thought to be a monomer, our estimate of the binding affinity of ALV(A) glycoprotein for sTva-mIgG may be lower than the actual binding affinity for membrane-bound Tva. Two other studies have estimated the apparent dissociation constant of quail Tva for ALV(A) envelope glycoproteins by using different assays. One study used a FACS binding assay to quantitate soluble ALV(A) SU immunoadhesin produced in human 293 cells, bound to 293 cells transiently expressing quail Tva (Kd = 1.5 nM) (72). The other study used an ELISA-based binding assay to quantitate purified quail sTva expressed from insect cells, bound to an immobilized epitope-tagged ALV(A) envelope glycoprotein produced in 293 cells (Kd = 0.3 nM) (5). In none of the three studies was an equilibrium maintained in the binding assays, and therefore the apparent Kd values are only approximations of the actual dissociation constant. However, despite the differences in the three assays, all three studies estimated very similar apparent Kd values for the binding affinity of the ALV(A) envelope glycoprotein-quail Tva interaction.
Soluble forms of a variety of viral receptors are effective at inhibiting viral entry (14, 20, 30, 40, 41, 46, 49, 55, 58, 62). Several studies have identified and characterized mutant viruses that are resistant to the antiviral effect of the soluble receptor (42, 44, 47, 55). Invariably, the resistant viruses have a reduced binding affinity for the soluble receptor but may or may not have a reduced affinity for the membrane-bound receptor compared to wild-type virions. Poliovirus variants resistant to soluble poliovirus receptor showed reduced binding to soluble and membrane-bound receptors, but the reduced binding did not alter poliovirus replication (42). Mouse hepatitis virus variants resistant to soluble mouse hepatitis virus receptor showed reduced binding to only the soluble receptor but showed near-wild-type binding to membrane-bound receptors (55). Several human immunodeficiency virus (HIV) variants resistant to neutralization by soluble forms of the primary receptor CD4 have been isolated and exhibit lower binding affinities for soluble and membrane-bound CD4 (44, 47). However, in addition to the lower receptor binding affinity for CD4, postbinding changes may be necessary to explain the resistant phenotype of the HIV variants. For example, since HIV also requires a coreceptor for efficient entry into cells (39) and since there are HIV strains that do not require CD4 for entry (23–25, 36, 45), the mutations in the soluble CD4-resistant HIV envelope glycoproteins may increase the ability of the virus to use the coreceptor alone for virus entry. In this study, we show that ALV(A) variants could be selected with a wide range of binding affinities for quail sTva-mIgG (Table 2), which directly correlated with the ability of the viruses to infect cells expressing quail Tva (Fig. 5). However, even though the binding affinity of the Y142N/E149K envelope glycoproteins for quail sTva-mIgG was >4,000-fold lower than that of the wild-type glycoproteins, the mutant virus could still infect cells expressing the quail Tva receptor, albeit inefficiently [600-fold lower than wild-type ALV(A)]. The lower binding affinity of the virus for the receptor may result in an inefficient conformational change in the envelope glycoproteins and/or poor recruitment of additional viral glycoprotein-Tva interactions necessary for efficient fusion of the viral and cellular membranes.
While the primary mechanism of antiviral resistance of the mutant viruses propagated in DF-1 cells appears to be a change in the binding affinity of the mutant glycoproteins for quail sTva-mIgG, the picture appears more complex when the mutant viruses are propagated in cells expressing quail sTva-mIgG. All three mutant viruses had an altered pattern of receptor usage when propagated in TF/sTva-4 cells that express quail sTva-mIgG than when propagated in DF-1 cells (Fig. 4), and the degree of altered receptor usage correlated with quail sTva-mIgG binding affinity (Table 2). We hypothesize that in virions produced by TF/sTva-4 cells the mutant envelope glycoproteins are complexed with quail sTva-mIgG that effectively blocks access to membrane-bound Tva on the target cell, thereby requiring the use of an alternate receptor for entry. For example, the E149K envelope glycoprotein binds sTva-mIgG relatively well, and virions carrying this mutation show the greatest alteration in receptor usage. The E149K virus also induced a period of cytotoxicity when propagated in TF/sTva-4 cells expressing quail sTva-mIgG, similar to that seen with some ALV(B), ALV(D), and ALV(C) strains in DF-1 cells but not with ALV(A) strains (35, 56). One possible explanation for the alteration in receptor usage is that unique complexes of the mutant envelope glycoproteins and quail sTva-mIgG are formed in the TF/sTva-4 cells as the proteins are processed in the endoplasmic reticulum and Golgi. Virions produced with envelope glycoproteins from these complexes may have lost receptor specificity due to envelope glycoprotein preactivation (e.g., they may have undergone a partial conformational change) and may be capable of infecting cells by interacting with a variety of cell surface proteins. Several other studies demonstrated that prebinding soluble forms of Tva to ALV(A) virions produced activated viruses capable of infecting Tva-negative cells (10, 17, 60). Viral envelope preactivation has also been proposed for the observed enhancement of infection of CD4-negative cells by some HIV and simian immunodeficiency virus variants preadsorbed with soluble CD4 (6, 57).
A variety of selective forces, including development of resistance to viral entry by the host, are probably responsible for retroviruses evolving to use different receptors. The highly conserved ALV envelope glycoprotein subgroups are presumably a result of these selective forces. We believe that our model system can mimic the adaptation of ALV to blocks in viral entry provided by the selective pressure of receptor interference, either with soluble forms of the Tva receptor (this report) or with SU (37). Using this system, key interaction determinants of both Tva binding affinity and receptor specificity have been identified in the hr1 region of ALV(A) SU. The selection of variant ALV(A) strains with quail sTva-mIgG in chicken cells further restricted the receptor use of the mutant ALV(A) strains to the chicken Tva receptor. This result highlights the importance of differences in homologous receptor proteins on virus evolution and adaptation between species. We also conclude from these results that at least for the wild-type and variant ALV(A) strains tested, the receptor binding affinity was directly related to infection efficiency.
ACKNOWLEDGMENTS
We thank Lynn Connolly and John A. T. Young for providing the pLC126 plasmid, Douglas Foster for the DF-1 cell line, Michael Skinner for the pHPRS-103 virus, Judith White for the 3T3pg950 cell line, and Stephen Hughes, Stephen Russell, Roberto Cattaneo, and the members of the Federspiel laboratory for helpful discussions and critical reading of the manuscript. We especially thank V. Shane Pankratz (Department of Health Sciences Research Section of Biostatistics, Mayo Clinic) for the statistical analysis of the data and Matthew VanBrocklin for constructing the ckstva-mIgG constructs.
This work was supported in part by the USDA NRI Competitive Grants Program (98-35204-6392), the Siebens Foundation under the Harold W. Siebens Research Scholar Program, and the Mayo Foundation (M.J.F.) and by a Mayo Graduate School and National Cancer Institute predoctoral training grant (T32CA75926) (S.L.H.).
REFERENCES
- 1.Adkins H B, Brojatsch J, Naughton J, Rolls M M, Pesola J M, Young J A T. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc Natl Acad Sci USA. 1997;94:11617–11622. doi: 10.1073/pnas.94.21.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins H B, Brojatsch J, Young J A T. Identification and characterization of a shared TNFR-related receptor for subgroup B, D and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J Virol. 2000;74:3572–3578. doi: 10.1128/jvi.74.8.3572-3578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astrin S M, Buss E G, Hayward W S. Endogenous viral genes are non-essential in the chicken. Nature. 1979;282:339–341. doi: 10.1038/282339a0. [DOI] [PubMed] [Google Scholar]
- 4.Bai J, Payne L N, Skinner M A. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J Virol. 1995;69:779–784. doi: 10.1128/jvi.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balliet J W, Berson J, D'Cruz C M, Huang J, Crane J, Gilbert J M, Bates P. Production and characterization of a soluble, active form of Tva, the subgroup A avian sarcoma and leukosis virus receptor. J Virol. 1999;73:3054–3061. doi: 10.1128/jvi.73.4.3054-3061.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandres J C, Wang Q F, O'Leary J, Baleaux F, Amara A, Hoxie J A, Zolla-Pazner S, Gorny M K. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J Virol. 1998;72:2500–2504. doi: 10.1128/jvi.72.3.2500-2504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bates P, Rong L, Varmus H E, Young J A T, Crittenden L B. Genetic mapping of the cloned subgroup A avian sarcoma and leukosis virus receptor gene to the TVA locus. J Virol. 1998;72:2505–2508. doi: 10.1128/jvi.72.3.2505-2508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates P, Young J A T, Varmus H E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 9.Belanger C, Zingler K, Young J A. Importance of cysteines in the LDLR-related domain of the subgroup A avian leukosis and sarcoma virus receptor for viral entry. J Virol. 1995;69:1019–1024. doi: 10.1128/jvi.69.2.1019-1024.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boerger A L, Snitkovsky S, Young J A T. Retroviral vectors preloaded with a receptor-ligand bridge protein are targeted to specific cell types. Proc Natl Acad Sci USA. 1999;96:9867–9872. doi: 10.1073/pnas.96.17.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bova C A, Manfredi J P, Swanstrom R. Env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology. 1986;152:343–354. doi: 10.1016/0042-6822(86)90137-6. [DOI] [PubMed] [Google Scholar]
- 12.Bova C A, Olsen J C, Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brojatsch J, Naughton J, Rolls M M, Zingler K, Young J A T. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 14.Clapham P R, Weber J N, Whitby D, McIntosh K, Dalgleish A G, Maddon P J, Deen K C, Sweet R W, Weiss R A. Soluble CD4 blocks the infectivity of diverse strains of HIV and SIV for T cells and monocytes but not for brain and muscle cells. Nature. 1989;337:368–370. doi: 10.1038/337368a0. [DOI] [PubMed] [Google Scholar]
- 15.Coffin J M. Genetic diversity and evolution of retroviruses. Curr Top Microbiol Immunol. 1992;176:143–164. doi: 10.1007/978-3-642-77011-1_10. [DOI] [PubMed] [Google Scholar]
- 16.Connolly L, Zingler K, Young J A T. A soluble form of a receptor for subgroup A avian leukosis and sarcoma viruses (ALSV-A) blocks infection and binds directly to ALSV-A. J Virol. 1994;68:2760–2764. doi: 10.1128/jvi.68.4.2760-2764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damico R, Bates P. Soluble receptor-induced retroviral infection of receptor-deficient cells. J Virol. 2000;74:6469–6475. doi: 10.1128/jvi.74.14.6469-6475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damico R, Rong L, Bates P. Substitutions in the receptor-binding domain of the avian sarcoma and leukosis virus envelope uncouple receptor-triggered structural rearrangements in the surface and transmembrane subunits. J Virol. 1999;73:3087–3094. doi: 10.1128/jvi.73.4.3087-3094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damico R L, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deen K C, McDougal J S, Inacker R, Folena-Wasserman G, Arthos J, Rosenberg J, Maddon P J, Axel R, Sweet R W. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature. 1988;331:82–84. doi: 10.1038/331082a0. [DOI] [PubMed] [Google Scholar]
- 21.Dorner A J, Coffin J M. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell. 1986;45:365–374. doi: 10.1016/0092-8674(86)90322-3. [DOI] [PubMed] [Google Scholar]
- 22.Dorner A J, Stoye J P, Coffin J M. Molecular basis of host range variation in avian retroviruses. J Virol. 1985;53:32–39. doi: 10.1128/jvi.53.1.32-39.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edinger A L, Mankowski J L, Doranz B J, Marguilies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent SIV strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endres M J, Clapham P R, Marsh M, Ahija M, Turner J D, McNight A, Thomas J F, Stoebenau-Haggerty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 26.Federspiel M J, Crittenden L B, Hughes S H. Expression of avian reticuloendotheliosis virus envelope confers host resistance. Virology. 1989;173:167–177. doi: 10.1016/0042-6822(89)90232-8. [DOI] [PubMed] [Google Scholar]
- 27.Federspiel M J, Hughes S H. Retroviral gene delivery. Methods Cell Biol. 1997;52:179–214. [PubMed] [Google Scholar]
- 28.Fekete D M, Cepko C L. Retroviral infection coupled with tissue transplantation limits gene transfer in the chicken embryo. Proc Natl Acad Sci USA. 1993;90:2350–2354. doi: 10.1073/pnas.90.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fields-Berry S C, Halliday A L, Cepko C L. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc Natl Acad Sci USA. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher R A, Bertonis J M, Meier W, Johnson V A, Costopoulos D S, Liu T, Tizard R, Walker B D, Hirsch M S, Schooley R T, et al. HIV infection is blocked in vitro by recombinant soluble CD4. Nature. 1988;331:76–78. doi: 10.1038/331076a0. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert J M, Bates P, Varmus H E, White J M. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J Virol. 1994;68:5623–5628. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert J M, Hernandez L D, Balliet J W, Bates P, White J M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hara H, Tanaka A, Kaji A. Presence of a hypervariable region within the hr2 domain of the host range determining sequences of the envelope glycoprotein gp85 (SU) of subgroup-A avian sarcoma-leukosis viruses. Virus Genes. 1996;12:37–46. doi: 10.1007/BF00369999. [DOI] [PubMed] [Google Scholar]
- 34.Hernandez L D, Peters R J, Delos S E, Young J A T, Agard D A, White J M. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Himly M, Foster D N, Bottoli I, Iacovoni J S, Vogt P K. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmen S L, Federspiel M J. Selection of a subgroup A avian leukosis virus [ALV(A)] envelope resistant to soluble ALV(A) surface glycoprotein. Virology. 2000;273:364–373. doi: 10.1006/viro.2000.0424. [DOI] [PubMed] [Google Scholar]
- 38.Holmen S L, Salter D W, Payne W S, Dodgson J B, Hughes S H, Federspiel M J. Soluble forms of the subgroup A avian leukosis virus [ALV(A)] receptor Tva significantly inhibit ALV(A) infection in vitro and in vivo. J Virol. 1999;73:10051–10060. doi: 10.1128/jvi.73.12.10051-10060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–120. [PubMed] [Google Scholar]
- 40.Hussey R E, Richardson N E, Kowalski M, Brown N R, Chang H C, Siliciano R F, Dorfman T, Walker B, Sodroski J, Reinherz E L. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature. 1988;331:78–81. doi: 10.1038/331078a0. [DOI] [PubMed] [Google Scholar]
- 41.Kaplan G, Freistadt M S, Racaniello V R. Neutralization of poliovirus by cell receptors expressed in insect cells. J Virol. 1990;64:4697–4702. doi: 10.1128/jvi.64.10.4697-4702.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplan G, Peters D, Racaniello V R. Poliovirus mutants resistant to neutralization with soluble cell receptors. Science. 1990;250:1596–1599. doi: 10.1126/science.2177226. [DOI] [PubMed] [Google Scholar]
- 43.Kingston R E, Chen C A, Okayama H. Introduction of DNA into eukaryotic cells. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1989. pp. 911–919. [Google Scholar]
- 44.Klasse P J, McKeating J A. Soluble CD4 and CD4 immunoglobulin-selected HIV-1 variants: a phenotypic characterization. AIDS Res Hum Retroviruses. 1993;9:595–604. doi: 10.1089/aid.1993.9.595. [DOI] [PubMed] [Google Scholar]
- 45.LaBranche C C, Hoffman T L, Romano J, Haggarty B S, Edwards T G, Matthews T J, Doms R W, Hoxie J A. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J Virol. 1999;73:10310–10319. doi: 10.1128/jvi.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marlin S D, Staunton D E, Springer T A, Stratowa C, Sommergruber W, Merluzzi V J. A soluble form of intercellular adhesion molecule-1 inhibits rhinovirus infection. Nature. 1990;344:70–72. doi: 10.1038/344070a0. [DOI] [PubMed] [Google Scholar]
- 47.McKeating J, Balfe P, Clapham P, Weiss R A. Recombinant CD4-selected human immunodeficiency virus type 1 variants reduced gp120 affinity for CD4 and increased cell fusion capacity. J Virol. 1991;65:4777–4785. doi: 10.1128/jvi.65.9.4777-4785.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moscovici C, Moscovici M, Jimenez H, Lai M M C, Hayman M J, Vogt P K. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell. 1977;11:95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- 49.Nemerow G R, Mullen J J, Dickson P W, Cooper N R. Soluble recombinant CR2 (CD21) inhibits Epstein-Barr virus infection. J Virol. 1990;64:1348–1352. doi: 10.1128/jvi.64.3.1348-1352.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielson K L, Holtet T L, Etzerodt M, Moestrup S K, Gliemann J, Sottrup-Jensen L, Thogersen H C. Identification of residues in alpha-macroglobulins important for binding to the α2-macroglobulin receptor/low density lipoprotein receptor-related protein. J Biol Chem. 1996;271:12909–12912. doi: 10.1074/jbc.271.22.12909. [DOI] [PubMed] [Google Scholar]
- 51.Rong L, Bates P. Analysis of the subgroup A avian sarcoma and leukosis virus receptor: the 40-residue, cysteine-rich, low-density lipoprotein receptor repeat motif of Tva is sufficient to mediate viral entry. J Virol. 1995;69:4847–4853. doi: 10.1128/jvi.69.8.4847-4853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rong L, Edinger A, Bates P. Role of basic residues in the subgroup-determining region of the subgroup A avian sarcoma and leukosis virus envelope in receptor binding and infection. J Virol. 1997;71:3458–3465. doi: 10.1128/jvi.71.5.3458-3465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rong L, Gendron K, Bates P. Conversion of a human low-density lipoprotein receptor ligand-binding repeat to a virus receptor: identification of residues important for ligand specificity. Proc Natl Acad Sci USA. 1998;95:8467–8472. doi: 10.1073/pnas.95.15.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rong L, Gendron K, Strohl B, Shenoy R, Wool-Lewis R J, Bates P. Characterization of determinants for envelope binding and infection in Tva, the subgroup A avian sarcoma and leukosis virus receptor. J Virol. 1998;72:4552–4559. doi: 10.1128/jvi.72.6.4552-4559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saeki K, Ohtsuka N, Taguchi F. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J Virol. 1997;71:9024–9031. doi: 10.1128/jvi.71.12.9024-9031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaefer-Klein J, Givol I, Barsov E V, Whitcomb J M, VanBrocklin M, Foster D N, Federspiel M J, Hughes S H. The EV-0-derived cell line DF-1 supports efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 57.Schenten D, Marcon L, Karlsson G B, Parolin C, Kodama T, Gerard N, Sodroski J. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J Virol. 1999;73:5373–5380. doi: 10.1128/jvi.73.7.5373-5380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith D H, Byrn R A, Marsters S A, Gregory T, Groopman J E, Capon D J. Blocking of HIV infectivity by a soluble, secreted form of the CD4 antigen. Science. 1987;238:1704–1707. doi: 10.1126/science.3500514. [DOI] [PubMed] [Google Scholar]
- 59.Smith E J, Fadly A M, Okazaki W. An enzyme-linked immunoabsorbant assay for detecting avian leukosis sarcoma viruses. Avian Dis. 1979;23:698–707. [PubMed] [Google Scholar]
- 60.Snitkovsky S, Young J A T. Cell-specific viral targeting mediated by a soluble retroviral receptor-ligand fusion protein. J Virol. 1998;95:7063–7068. doi: 10.1073/pnas.95.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taplitz R A, Coffin J M. Selection of an avian retrovirus mutant with extended receptor usage. J Virol. 1997;71:7814–7819. doi: 10.1128/jvi.71.10.7814-7819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traunecker A, Luke W, Karjalainen K. Soluble CD4 molecules neutralize human immunodeficiency virus type 1. Nature. 1988;331:84–86. doi: 10.1038/331084a0. [DOI] [PubMed] [Google Scholar]
- 63.Tsichlis P N, Coffin J M. Recombinants between endogenous and exogenous avian tumor viruses: role of the C region and other portions of the genome in the control of replication and transformation. J Virol. 1980;33:238–249. doi: 10.1128/jvi.33.1.238-249.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsichlis P N, Conklin K F, Coffin J M. Mutant and recombinant avian retroviruses with extended host range. Proc Natl Acad Sci USA. 1980;77:536–540. doi: 10.1073/pnas.77.1.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiss R A. Cellular receptors and viral glycoproteins involved in retrovirus entry. In: Levy J A, editor. The retroviruses. Vol. 2. New York, N.Y: Plenum Press; 1992. pp. 1–108. [Google Scholar]
- 66.Weller S K, Joy A E, Temin H M. Correlation between cell killing and massive second-round superinfection by members of some subgroups of avian leukosis viruses. J Virol. 1980;33:494–506. doi: 10.1128/jvi.33.1.494-506.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weller S K, Temin H M. Cell killing by avian leukosis viruses. J Virol. 1981;39:713–721. doi: 10.1128/jvi.39.3.713-721.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson C, Wardell M R, Weisgraber K H, Mahley R W, Agard D A. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 69.Young J A T, Bates P, Varmus H E. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng X H, Hughes S H. An avian sarcoma/leukosis virus-based gene trap vector for mammalian cells. J Virol. 1999;73:6946–6952. doi: 10.1128/jvi.73.8.6946-6952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zingler K, Belanger C, Peters R, Agard D, Young J A T. Identification and characterization of the viral interaction determinant of the subgroup A avian leukosis virus receptor. J Virol. 1995;69:4261–4266. doi: 10.1128/jvi.69.7.4261-4266.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zingler K, Young J A T. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J Virol. 1996;70:7510–7516. doi: 10.1128/jvi.70.11.7510-7516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]