Abstract
The study of neurodegenerative diseases, particularly tauopathies like Pick’s disease (PiD) and Alzheimer’s disease (AD), offers insights into the underlying regulatory mechanisms. By investigating epigenomic variations in these conditions, we identified critical regulatory changes driving disease progression, revealing potential therapeutic targets. Our comparative analyses uncovered disease-enriched non-coding regions and genome-wide transcription factor (TF) binding differences, linking them to target genes. Notably, we identified a distal human-gained enhancer (HGE) associated with E3 ubiquitin ligase (UBE3A), highlighting disease-specific regulatory alterations. Additionally, fine-mapping of AD risk genes uncovered loci enriched in microglial enhancers and accessible in other cell types. Shared and distinct TF binding patterns were observed in neurons and glial cells across PiD and AD. We validated our findings using CRISPR to excise a predicted enhancer region in UBE3A and developed an interactive database (http://swaruplab.bio.uci.edu/scROAD) to visualize predicted single-cell TF occupancy and regulatory networks.
Introduction
Neurodegeneration is a key aspect of many neurological disorders, each with distinct molecular mechanisms and etiologies. Alzheimer’s disease (AD) is the most prevalent neurodegenerative disorder and is pathologically characterized by progressive accumulation of amyloid-beta plaques and neurofibrillary tangles (NFTs) of tau1. Alternatively, Pick’s disease (PiD) is a rare behavioral variant of frontotemporal dementia (FTD)2,3, which has a prevalence of 15 to 22 per 100,000 individuals and an incidence of 2.7 to 4.1 per 100,000 individuals per year4. PiD is characterized by the presence of pathological tau aggregates known as Pick bodies5. Abnormal tau aggregates like NFTs and Pick bodies alter cellular and molecular functions in the brain, but we currently do not understand the differences and similarities between these cellular changes across different tauopathies like AD and PiD6. The rare prevalence of PiD, combined with the challenges of clinical diagnosis, has hindered comprehensive research on this neurodegenerative condition, leaving its genetic and epigenetic etiology unclear. With these difficulties, comparative functional genomics analyses of different tauopathies may reveal shared and distinct molecular mechanisms underlying these disorders.
While recent genome-wide association studies (GWAS) and fine-mapping analyses have implicated numerous genetic loci in neurodegeneration7–12, much of the attention in this area is currently focused on AD over other disorders6, and the functional roles of these loci are often ambiguous since they frequently reside in non-coding regions13–15. The advent of single-cell epigenomics has allowed us to provide additional context for these genetic risk variants in specific cell types16, while single-cell transcriptomics has provided insights into the molecular states of NFT-bearing neurons and NFT susceptibility in AD17. While these technologies have broadened our understanding of altered cellular states and gene regulatory programs in AD16,18–26, much work remains to characterize these changes in other neurodegenerative disorders and to understand their shared and unique molecular signatures.
In this study, we employed single-nucleus assay for transposase-accessible chromatin using sequencing (snATAC-seq) to characterize the open chromatin landscape and single-nucleus RNA-seq (snRNA-seq) to profile the gene expression of the frontal cortex in Pick’s disease donors and cognitively normal controls. We performed parallel comparative analyses of PiD datasets with our previous AD datasets to facilitate our understanding of PiD. We leveraged cell-type-specific chromatin accessibility information to model the gene-regulatory landscape of PiD and AD, identifying sets of promoter-gene links for each disease in each cell type. We intersected these links with our internally conducted fine-mapping analyses, considering linkage disequilibrium (LD), at selected disease risk loci to nominate candidate cell types and genes associated with non-coding risk SNPs. Further, we modeled transcription factor (TF) binding activity in each cell type for disease and control to characterize regulatory networks and key gene-regulatory mechanisms mediated by enhancer-promoter links, allowing us to focus our attention directly on the regulators of these GWAS genes, differentially expressed genes and TFs. Furthermore, snRNA-seq of PiD donors corroborated some of our findings at the transcriptome level. To validate the robustness of our insights, we highlighted a previously unknown human-gained enhancer (HGE) in excitatory neurons regulating UBE3A, known for its role in regulating synaptic activity, that is altered in both PiD and AD. Using CRISPR-Cas9, we excised this HGE in induced pluripotent stem cell (iPSC) derived neurons, and we observed a subsequent downregulation of UBE3A using RNA-seq. Our data suggests both shared and distinct patterns of gene regulation in PiD and AD, particularly evident in the disease-enriched and specific TF activity. Furthermore, disruption in the imputed enhancer accessibility provides validation for the accurate identification of enhancer regions located more than 40kbp away from the UTR region of the disease-relevant gene.
Results
Single-nucleus ATAC and RNA profiling of the prefrontal cortex in PiD and AD.
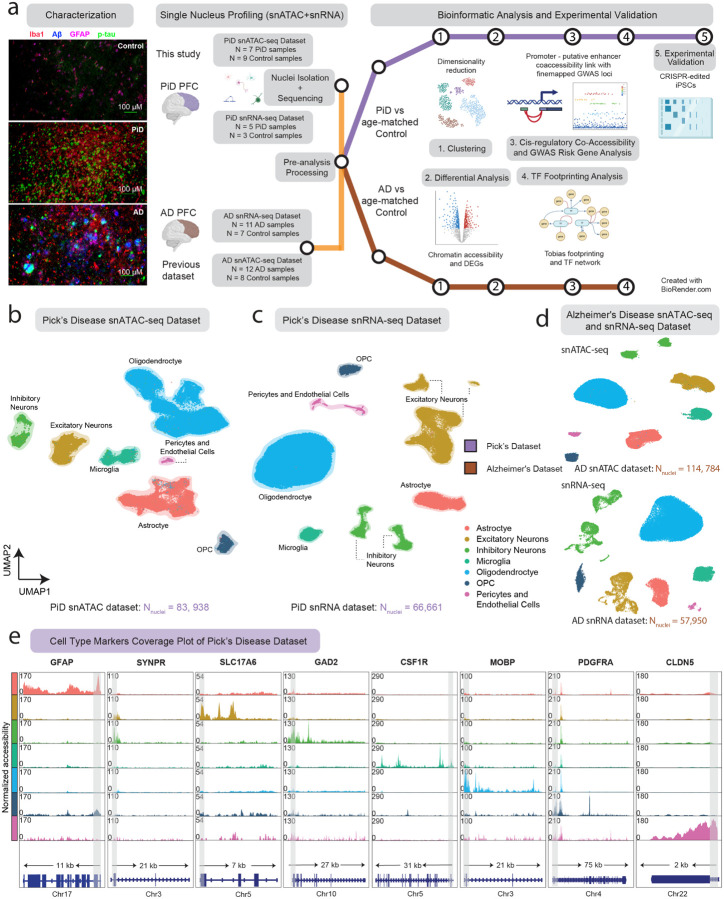
We applied snATAC-seq in frontal cortical tissue sections of PiD and cognitively normal control cases (10x Genomics; n = 7 PiD; n = 9 control), and snRNA-seq on the same PiD and control cases (Parse Bio; n = 5 PiD; n = 3 control). Notably, our study is the first to delineate the molecular landscape within frontal cortical regions of PiD at the single-cell level. We processed our single-nucleus data separately in PiD and our previously generated snATAC-seq of AD (10x Genomics; n = 12 late-stage AD; n = 8 control)16 and snRNA-seq (10x Genomics; n = 11 late-stage AD; n = 7 control)16,27 (Figure 1a). After quality control filtering, 83,938 snATAC-seq and 66,661 snRNA-seq profiles come from the newly generated PiD dataset (Figure 1b,c,d, S1a,b, Methods), and 114,784 nuclei originated from previously generated AD snATAC-seq and 57,950 nuclei were from AD snRNA-seq. In snATAC-seq, clustering analyses revealed seven major brain cell types in this dataset — excitatory neurons (EX), inhibitory neurons (INH), astrocytes (ASC), microglia (MG), oligodendrocytes (ODC), oligodendrocyte progenitor cells (OPC), and pericytes and endothelial cells (PER-END) — annotated based on chromatin accessibility at the promoter regions of known marker genes (Figure 1b,d,e). We performed label transfer using AD dataset16 as a reference and then confirmed the annotation of our excitatory and inhibitory neurons based on previously identified marker genes, SYNPR for both EX and INH neurons, SLC17A for EX, GAD2 for INH. Similarly, we sought to annotate our glial subpopulations, our astrocyte cluster based on GFAP promoter, which has been shown to increase in disease28; microglia cluster based on CSF1R promoter; oligodendrocyte cluster based on MOBP promoter; OPC cluster on PDGFRA promoter; and PER-END cluster on CLDN5 promoter (Figure 1e and S1f). Additionally, we further confirmed cell type identities by gene activity shown in the panel of canonical cell-type marker genes (Table S1)29. In the snRNA-seq dataset, we first clustered and identified seven major brain cell types in PiD using a panel of canonical cell-type marker genes (Figure 1c,d and S1d,e, Table S1, Methods). These robust cell type identifications laid a critical foundation for subsequent analyses, enabling us to explore cell type-specific alterations and molecular mechanisms underlying PiD pathogenesis with a high degree of confidence.
Figure 1. snATAC-seq for the study of cellular diversity in the PiD and AD brain.
a, Immunofluorescence characterization of PiD, AD and Control; and Schematic representation of the samples used in this study, sequencing experiments and downstream bioinformatic analyses, created with BioRender.com. Representative quadruple immunofluorescence images for Iba1 (red), GFAP (magenta), amyloid plaque (blue), and AT8/p-tau (green) from prefrontal cortex region of postmortem human brain tissues of age- and sex-matched control (n=3), AD (n=5) and Pick’s (n=5) cases. Images were captured using Nikon ECLIPSE Ti2 inverted microscope (20X). b,c, Uniform Manifold Approximation and Projection (UMAP) visualizations for single-nucleus ATAC-seq data (b) and single-nucleus RNA-seq data (c) from Pick’s disease and age-matched control. d, Uniform Manifold Approximation and Projection (UMAP) visualizations for single-nucleus ATAC-seq and RNA-seq data from Alzheimer’s disease and age-matched control. e, Coverage plots for canonical cell-type markers: GFAP (chr17:44905000–44916000) for astrocytes, SYNPR (chr3:63278010–63278510) for neurons, SLC17A6 (chr11:22338004–22345067) for excitatory neurons, GAD2 (chr10:26214210–26241766) for inhibitory neurons, CSF1R (chr5:150056500–150087500) for microglia, MOBP (chr3:39467000–39488000) for oligodendrocytes, PDGFRA (chr4:54224871–54300000) for pericytes and endothelial cells in PiD dataset. The grey bar within each box highlights the promoter regions.
Promoter-enhancer linkages enable better characterization of chromatin accessibility in PiD and AD.
From these single-nucleus ATACseq libraries, we compiled a combined set of 609,675 reproducible peaks29 (Table S2a). To identify cis-regulatory elements (CREs) with altered chromatin accessibility in disease, we systematically performed differential chromatin accessibility analyses in each cell type comparing PiD vs. controls and AD vs. controls, yielding a set of differentially accessible peaks (Table S2c). Our chromatin accessibility regions were broadly categorized by genomic features, including gene promoter, exonic, intronic, or distal regions, and we investigated these differential peaks in PiD and AD based on these categories (Figure 2a). The majority of the differential peaks in PiD (54%) and AD (53%) are located within intronic regions. Approximately 30% of the differential peaks in both PiD and AD are distal, while 9% and 10% correspond to promoters specifically in PiD and AD, respectively. Less than 10% of the identified differential peaks are exonic in both PiD and AD (Figure 2a and Table S2a). These percentages are more or less consistent with the peak type distribution of the entire peakset, where distal peaks comprise approximately 32%, promoter peaks make up 5%, intronic peaks constitute 55%, and exonic peaks represent 7% (Table S2a). Similar percentages were also observed in other published studies, reinforcing the robustness and consistency of our findings across different datasets26,30. Although EX was not the most sampled cell types in PiD, our differential analyses revealed that the most significant variance in the activity of CREs in the PiD dataset was observed in EX. Conversely, ASC exhibited the highest differential activity in the AD dataset (Figure S2a).
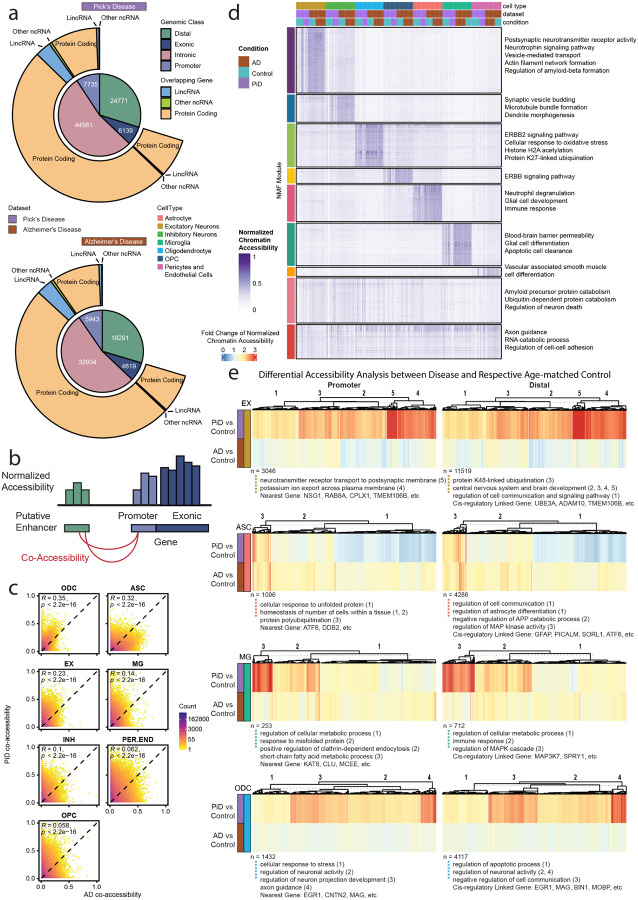
Figure 2. Open chromatin classification and epigenetically distinct cell types through putative promoter–enhancer links in the human PiD and AD prefrontal cortex.
a, Peaktype and biotype classification of differential open accessible peaks (P-value < 0.05). b, Schematics of promoter – putative enhancer linkage. c, Correlation heatmap of promoter – putative enhancer co-accessibility. d, NMF heatmap of putative enhancer scaled chromatin activity in PiD, AD and their matching controls. e, Heatmaps of fold changes (Disease vs. Control) on normalized chromatin accessibility of differential accessible promoters and distal in excitatory neurons, astrocytes, microglia and oligodendrocytes (FDR adjusted P-value < 0.05 and abs(log2FC) > 0.5), gene ontology acquired from GREAT and examples of promoters and distal regions’ cis-regulatory linked gene as in panel (b).
We next sought to provide functional context for non-coding distal regulatory elements with respect to cell type and disease status by performing cis co-accessibility analyses with Cicero31, revealing linkages between promoters and distal elements (Figure 2b, Methods). Using this information, we compared the co-accessibility strength of chromatin peak links from PiD and AD samples in the major cell lineages (Figure 2c). These analyses revealed relatively higher correlations between PiD and AD in ODCs (Pearson R=0.35) and ASCs (R=0.32), with weaker correlations in the other cell types, overall revealing a substantial amount of conserved epigenomic linkages across PiD and AD as well as those unique to each condition. We next applied non-negative matrix factorization (NMF) to pseudobulk chromatin accessibility profiles of all distal regulatory elements that were linked to gene promoter regions, and we identified matrix factors corresponding to epigenetic signatures of biological processes and specific cell states (Figure 2d). We grouped CREs into discrete epigenetic modules based on the matrix factor with the highest loading for each CRE, and we performed gene ontology analyses of the regulatory target genes of each of these modules, revealing the cell-type-function related pathways and processes regulated by these non-coding CREs, such as pathways associated with postsynaptic and synaptic activity in EX and INH, cell proliferation and migration-linked ERBB2 signaling pathway in ODC, and processes such as apoptotic cell clearance in MG, among others.
Using this cis-regulatory linkage approach coupled with differential analyses, we used heatmaps to depict the fold changes of normalized chromatin accessibility for differential accessible promoters, distal and intronic regions across cell types EX, ASC, MG, and ODC (Figure 2e and S2c). Additionally, we incorporated gene ontology information obtained from GREAT, presenting cluster numbers alongside specific gene names as examples below. Notably, by inspecting the distal, intronic, and promoter chromatin regions and their linked regulatory target genes, we identified changes containing AD and FTD genetic risk loci, including TMEM106B, ADAM10, SORL1, KAT8, CLU, BIN1 and genes involved in essential cellular activity, such as UBE3A. Moreover, while examining the absolute fold change of normalized chromatin accessibility, genes in EX in PiD exhibited much more robust changes than those in AD (Figure 2e). This potentially indicates that the neuronal changes are more pronounced in PiD, probably due to age-associated regional changes in frontal cortical regions2,3, and patients with frontotemporal lobar degeneration progress more rapidly than AD32.
Frontotemporal dementia and Alzheimer’s disease fine-mapping with cell-type-specific epigenomic annotations.
Given that the majority of variants reside in non-coding regions, around 80% of the deferentially accessible peaks in distal and intronic regions (Figure 2a), and the limited research on disease gene identification for PiD, we assert the importance of utilizing closely related FTD and AD GWAS data as reference points. Our analysis approach involves overlapping snATAC-seq accessible peaks with fine-mapped GWAS SNPs, enabling us to determine whether chromosomal regions surrounding these pathogenic-related SNPs exhibit accessibility in our dataset. However, it’s important to acknowledge the inherent limitations of our study, particularly the rarity of PiD and the consequent unavailability of specific PiD GWAS summary data with sufficient statistical power. This limitation restricts our analyses to leveraging existing knowledge and datasets to explore potential gene targets for PiD, rather than conducting direct PiD GWAS analyses. We conducted comprehensive fine-mapping, annotation, and cell-type-specific gene expression analyses, in addition to collecting publicly available predicted loss-of-function data33 (gnomAD v4.0 UCSC; See Methods). These efforts aimed to identify causal variants and explore the association of genetic variant-related genes with the risk of AD11 and FTD12 (Figure 3a). The fine-mapping analyses identified 72 lead GWAS risk SNPs with 113 credible sets and 716 SNPs in LD within the AD and FTD brain (posterior inclusion probability (PIP) > 0.95) overlapping with accessible peaks from the seven major cell types. Interestingly, we found that 36 out of 113 fine-mapped causal credible sets overlap with accessible peaks of a single or two cell type, and 16 out of 113 are present in all cell types (Figure 3b,c, Table S3a), suggesting that some disease risk variants are relevant to a particular cell type while others influence gene regulation across several cell types. To reinforce this notion, we integrated the snRNA-seq dataset from three previous studies of the AD cortex16,19,34 and plotted the expression of genes identified from GWAS summary statistics, where each gene was associated with the lead causal SNP, from its respective control group of distinct cell types. Additionally, we corroborated the association of the lead SNP and other fine-mapped SNPs in LD with its associated gene by cross-referencing cCREs and target genes, which you can access the data through our online interactive database, scROAD.
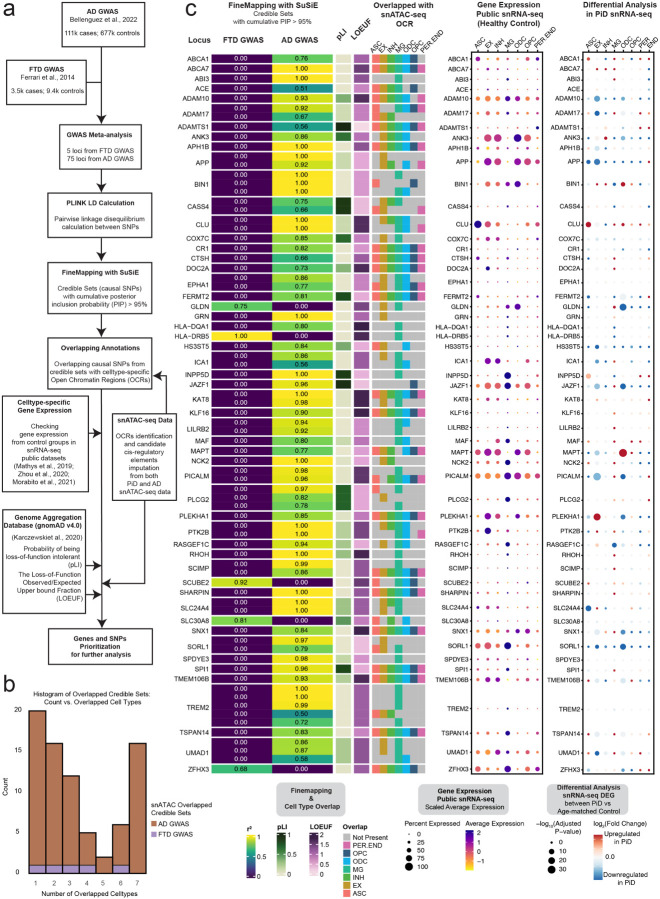
Figure 3. Cell-type-specific fine-mapped causal SNPs from FTD and AD GWAS risk locus.
a, The Schematic of analyses showing the summary of Frontotemporal Dementia (FTD) and Alzheimer’s Disease GWAS meta-analyses, fine-mapping, and other data processing steps to link causal SNPs to snATAC-seq accessible peaks in specific cell types. b, Histogram of Overlapped Credible Sets: Count vs. Overlapped Cell Types. It describes a histogram that displays the count of overlapped credible sets and their associated number of overlapped cell types. c, Fine-mapped SNPs from identified FTD and AD GWAS risk locus showing overlap with open chromatin regions from snATAC-seq and GWAS risk gene expression in major cell types. The fine-mapping column using SuSiE shows all of the snATAC-seq cell-type-specific open chromatin regions overlapped credible sets (PIP > 0.95). The closest gene to the credible set is indicated on the left. The r2 indicates the average correlation between the SNPs in the credible set. Both the probability of being loss-of-function intolerant (pLI) and loss-of-function observed/expected upper bound fraction (LOEUF) are from gnomAD33 (gnomAD v4.0 UCSC). In the pLI column, when the number closes to 1, it means that the gene cannot tolerate protein truncating variation. The LOEUF column indicates whether the GWAS risk gene is constrained or mutation intolerance when it is closer to 0. The overlapped snATAC-seq OCR column reflects the cell types of those causal SNPs from a credible set that are present or not. The middle dotplot shows the gene expression in each cell type from the control samples of three public snRNA-seq datasets16,19,34. The dotplot on the far right shows the snRNA-seq differentially expressed GWAS gene in each cell type between PiD and age-matched control samples. A complete set of the fine-mapped SNPs and credible sets with a PP > 0.95 shown for FTD and AD are available in Table S3. Data about finemapped SNPs with cCREs and their associated target genes can accessed through our online interactive database, scROAD.
For AD, our analyses revealed that more than half of the 113 fine-mapped signals overlap with accessible peaks found in microglia, a cell type of particular interest in AD research. Notably, these peaks encompass several classical genes that have been extensively studied in microglia, including ABCA1, ADAM10, ADAM17, BIN1, INPP5D, NCK2, PI-CALM, and TREM2 (Figure 3c). The enrichment of GWAS risk signals within microglia aligns with the established pathophysiological role of these cells, particularly their involvement in inflammation in AD35. AD risk variants at the INPP5D locus were found in accessible chromatin regions exclusively in microglia, and the INPP5D gene is expressed almost specifically in microglia as well (Figure 3c).
While previous studies have demonstrated enrichment of AD genetic risk SNPs specifically in microglia16, we note that these risk genes are expressed in several cell types. For example, the risk variants of ADAM10 overlap with accessible peaks from EX, INH, MG and ODC and its gene expression is present across all cell types. As the major constitutive of α-secretase, ADAM10 cleaves APP towards a non-amyloidogenic pathway, thereby preventing Aβ generation36.Furthermore, fine-mapping analyses revealed that BIN1 risk variants, a major risk factor for AD known to induce tau- and isoform-dependent neurotoxicity37,38, predominantly localize to accessible peaks associated with ASC, MG, ODC, and OPC. These findings give credence to previously reported disparate findings on the effects of BIN1 SNPs in microglia39 and oligodendrocytes16. Considering that a gene’s expression can be widespread across multiple cell types, it becomes crucial to exercise caution when analyzing the effects of variants, as these effects may vary significantly among different cell types. Similarly, GWAS variants in the TREM2 gene were identified within accessible peaks primarily associated with MG and EX. TREM2 plays a crucial role in various cellular processes, including cell proliferation, survival, phagocytosis, and regulation of inflammation40. Notably, its defensive response against AD pathology, coupled with its upregulation in reactive microglia surrounding amyloid plaques, has been consistently observed across multiple studies, both in mouse models and human samples35,41,42.
Complementing our analyses of AD risk loci, we also performed fine mapping analyses on GWAS risk loci for FTD, aiming to propose possible risk genes for FTD subtype PiD (Figure 3c and Table S3a,b), with five of them intersecting accessible chromatin regions in our snATAC-seq dataset. For example, one of the fine-mapped FTD risk loci, SLC30A8, encodes for a zinc transporter and is a susceptible GWAS locus for type 2 diabetes43. Strikingly, there is a notable increase in the prevalence of both type 2 diabetes and dementia in older adults44. We speculate that SLC30A8 could be an indirectly related risk locus for FTD. Moreover, among the identified FTD risk loci, GLDN stands out as another intriguing candidate. GLDN encodes for gliomedin, a crucial protein involved in the formation of the nodes of Ranvier45.These nodes are critical structures along the neural axons where action potentials are regenerated. Disruption of the nodes of Ranvier can result in the failure of the electrically resistive seal between the myelin and the axon, ultimately contributing to various neurological diseases46. Given the fundamental role of gliomedin in maintaining axonal integrity, investigating GLDN variants within specific cell types may provide valuable insights into their potential involvement in FTD pathogenesis, especially in our snRNA-seq differential analyses between PiD and age-matched control, has shown that GLDN is significantly downregulated. Besides GLDN, in our snRNA-seq analyses, some of the AD GWAS genes, such as ADAM10, ADAM17, BIN1, APP, CLU, JAZF1, MAPT, PI-CALM, PLEKHA1, SLC24A4, SORL1, and UMAD1, were also differentially expressed in PiD (Table S4a,b). While risk loci have been identified as significant in our GWAS studies and cis-regulatory-linked risk genes, the fact that AD GWAS genes show a highly significant overlap with differentially expressed genes in the PiD case (Fisher’s Exact Test: p-value < 2.2×10ˆ−16, Table S3c) suggests that these associations are not random. However, it remains crucial to determine how fine-mapped signals specifically relate to PiD. By integrating these genetic findings with our multi-omics data, we can gain deeper insights into the complex interplay between genetic risk factors and cellular processes contributing to PiD and AD pathology, particularly with regard to regulatory non-coding regions and gene expression in the corresponding cell types.
Excitatory neuron TF binding site occupancy and expression analyses reveals TF dysregulation in PiD and AD.
To further uncover gene regulatory mechanisms impacting the neurons and glial cells in PiD and AD, we sought to investigate co-accessible enhancer-promoter regions, where the genome-wide and gene-specific TF differential binding activities were observed. We want to further our understanding of FTD and AD genetic risk signals, especially their fine-mapped SNPs’ involvement in the putative regulatory functions. We performed chromatin cis co-accessibility analyses and the transcription factor occupancy prediction analyses on 609,675 cCREs (Table S2), and examined the disease-enriched signals in both PiD and AD. We implemented cis-regulatory co-accessibility31 and trans-regulatory occupancy prediction47 methods for each predominant cell type and divided each cell type into PiD, AD, and their corresponding control cells for the examination (Figure 4a).
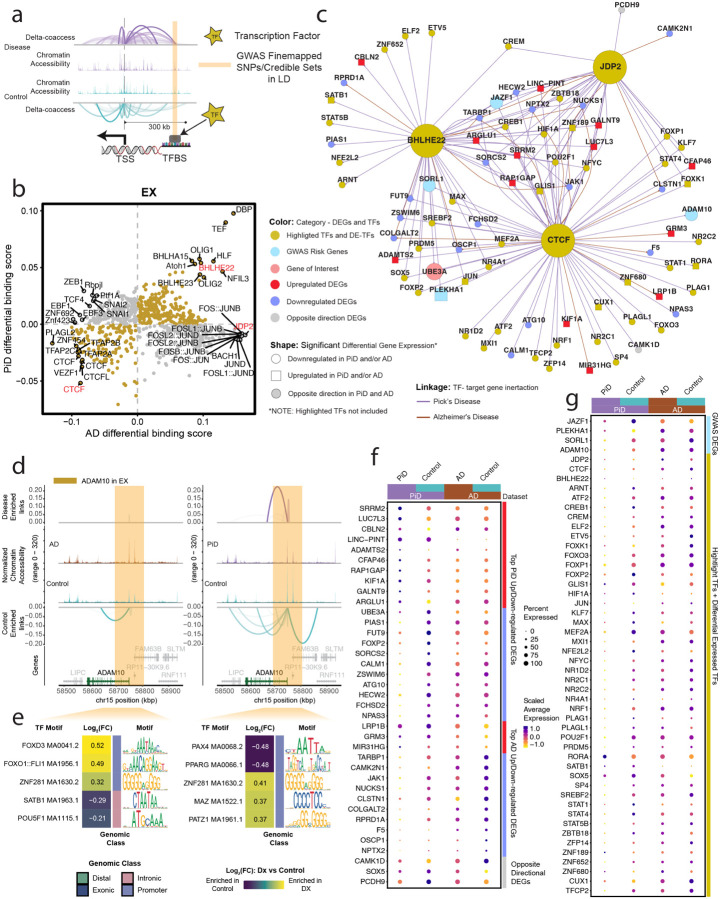
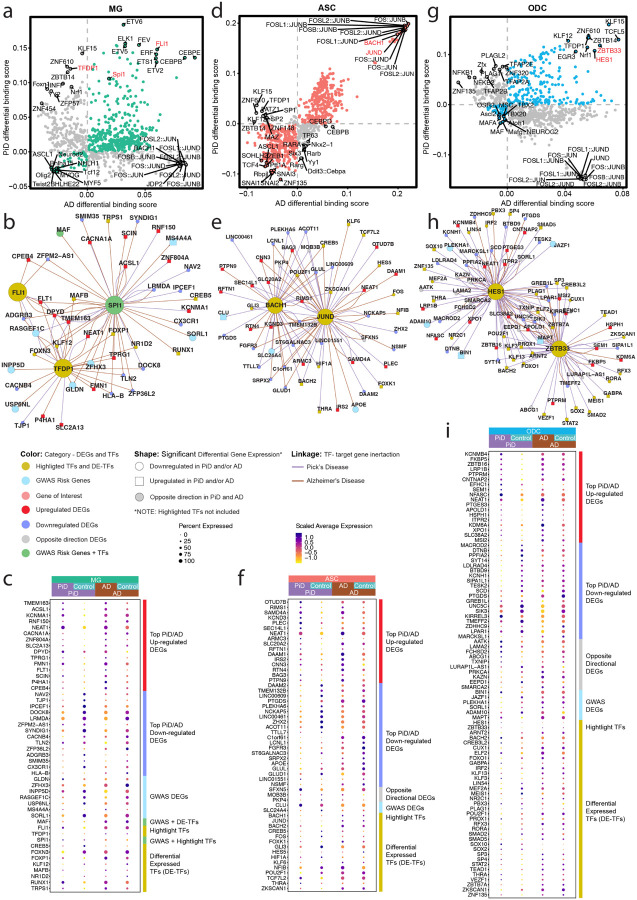
Figure 4. Excitatory neuronal related transcription factor dysregulation and gene expression changes associated with PiD and AD pathology.
a, The schematic diagram for the co-accessible mapping between putative enhancer and promoter for the target gene as well as the TF binding activity at its local regions. b, Genome-wide Tn5 bias-subtracted TF differential footprinting binding score of PiD and AD in EX compared to the corresponding controls. c, TF regulatory networks showing the predicted candidate target genes for the following TFs: CTCF, BHLHE22, and JDP2 in EX. Highlighted transcription factors and other differential expressed TFs are shown in yellow. Upregulated differential genes are shown in red and square. Downregulated differential genes are shown in blue and in a circle. The gene of interest is downregulated, shown in pink and in a circle. The differential expressed GWAS risk genes are displayed in bright blue. Edges representing the linkage of TF - target gene regulation are shown in purple for PiD and sienna for AD. d, delta co-accessibility of ADAM10 and its open chromatin regions in EX for both AD and PiD with their corresponding controls. Highlighted regions in dark yellow represent SuSiE fine-mapped all SNPs (Figure 3c) closed to the target gene in 4d. e, fold changes of TFs binding in the SuSiE fine-mapped regions for both AD and PiD. f, Dotplot of the differentially expressed gene in PiD and AD versus their respective controls. g, Dotplot of differentially expressed GWAS risk genes and TFs in PiD and AD versus their respective controls.
Our integrated cis- and trans-regulatory analyses approach allows us to explore disease-enriched enhancer-promoter links, TF differential binding activity, and motif binding site disruption in neurons. Genome-wide transcription factors’ differential binding scores were calculated in PiD and AD with their matching controls in neurons (Figure 4b). We identified transcription factors BHLHE22, a TF previously indicated to play a key role in neural cell fate48, along with other TFs (p-value < 0.05), exhibit shared and enhanced binding activity in both PiD and AD compared with their respective controls. JDP2, a TF involved in apoptosis49, along with other TFs, demonstrates increased binding activity only in AD. CTCF, a transcriptional regulator that acts on enhancers, promoters, and gene bodies50, together with other TFs in the lower left quadrant, displays decreased binding activity in both PiD and AD compared to their controls. We aimed to further contextualize the varying binding activity for highlighted transcription factors as an example to investigate the binding activity at the enhancer regions of their specific target genes in neurons (Figure 4c–e). To accomplish this, a gene regulatory network for the transcription factors BHLHE22, CTCF, and JDP2 was established in both PiD and AD datasets for excitatory neurons (Figure 4b,c, and S4). Several genes implicated in AD GWAS, including JAZF1, SORL1, PLEKHA1, and ADAM10, exhibited differential expression in EX in individuals with PiD, providing possible insights into shared molecular mechanisms between PiD and AD, suggesting potential convergent pathways underlying neurodegeneration in these conditions (Figure 4c). The differentially expressed TFs and genes we identified, positioned in the center of the network, are under the regulation of all three highlighted factors: CTCF, JDP2, and BHLHE22. Those regulated by two or a single TF are depicted on the outer ring of the network. We stress that these findings merely represent a simplified depiction of a highly complex regulatory network. Gene targets within this network are acknowledged to be subject to regulation, but it’s important to note that the highlighted transcription factors do not solely govern their regulation.
To complement our analyses of TF trans-regulatory network in neurons, we aimed to discern cis-regulatory elements and DNA-binding motifs that are enriched in either disease or control conditions, particularly within regions containing fine-mapped SNPs. Through the integration of the co-accessibility map with chromatin accessibility signals and GWAS statistics across the genomic axis, we elucidated potential disruptions in cis-regulatory relationships caused by causal disease variants in a GWAS gene, ADAM10, which is also differentially expressed genes (Figure 4d,e). Additionally, we conducted analyses of the sequences to identify motifs that are disrupted in comparison to control conditions. This procedure was executed with the aim of assessing disease or control gene local enhancer accessibility and predicting potential disruptions in TF binding.
We found alterations in the cis-regulatory mechanisms of ADAM10 in AD, a prominent anti-amyloidogenic candidate gene in AD pathology36 (Figure 4d). This change was identified in proximity to the fine-mapped lead ADAM10 SNP, rs602602, and in its strong LD block, potentially disrupting the DNA-binding motif. Consequently, these disruptions may result in diminished transcription factor (TF) binding activity in disease compared to their corresponding control group. We further investigated the gene locus TF binding activity in those highlighted fine-mapped accessible regions. We selected five TFs from the top-ranked TFs based on the average log2(fold change) of the TF binding score. For example, we found forkhead box O1 (FOXO1), SATB homeobox 1 (SATB1), POU class 5 homeobox 1 (POU5F1), Paired box 4 (PAX4), and peroxisome proliferator-activated receptor (PPAR) transcription factors enriched in highlighted regions identified for ADAM10 in EX (Figure 4e). Previous studies have investigated the potential roles of FOXO1, SATB1, and POU5F1 in the development of AD51–53. Notably, FOXO TF families were indicated as mediators of stress adaptation, which promotes the resilience of cells as a key regulator in other pathways, such as metabolism, cell cycle, and redox regulation54. The transcription factor PAX4 has been investigated in the contexts of both AD and Type 2 Diabetes (T2D), and is known to function as a key link in the common pathways of both diseases55.
To thoroughly examine the differences in gene expression in EX between disease and control groups, we arranged and compared all selected differentially expressed genes (DEGs) and transcription factors (TFs) side by side for PiD and AD (Figure 4f,g). Not only does the fold change in gene expression indicate the robustness of biological changes between diseases, but it is also important to highlight certain genes and TFs, particularly regarding their roles in disease development. Among those top-selected genes, CALM1 has been linked to the progression from mild cognitive impairment (MCI) to AD through involvement in the neurotrophin signaling pathway, which contributes to neuronal development, survival, and plasticity56. Additionally, CALM1 participates in dysregulated ligand-receptor (LR) interactions57. So, by exhibiting a significant downregulation in both PiD (FDR-adjusted p-value = 5.57×10ˆ−6, Table S4a) and AD (FDR-adjusted p-value = 0.011, Table S4b) samples, it may indicate a potential common role in the pathogenesis of both diseases. Similarly, TARBP1 showed a notable decrease in both PiD and AD (Figure 4c,f). TARBP1 encodes the TAR RNA binding protein 1 (TRBP), which participates as a methyltransferase enzyme in post-transcriptional gene regulation through its involvement in RNA processing pathways, and it is associated with inattention symptoms58. Moreover, as we explore further genes implicated in neurodegenerative processes, another significant player is UBE3A, for which we have already identified the regulation of its distal enhancer (Figure 2e). But here, we found a significant decrease of UBE3A, regulated by CTCF and BHLHE22, in EX in PiD (FDR-adjusted p-value = 1.04 ×10ˆ−14) (Figure 4c,f). We conducted a detailed examination of the alterations in TFs’ expression levels between diseased and control states. Our analyses revealed a pervasive downregulation of TF expression across PiD samples, when compared to the changes observed between Alzheimer’s disease (AD) and its respective control group (Figure 4g). Among those TFs with differentially expressed levels, we observed RORA, a transcription factor that plays an essential role in energy and lipid metabolism59, is significantly upregulated in both PiD (FDR-adjusted p-value = 1.14 ×10ˆ−20, Table S4a) and AD (FDR-adjusted p-value = 0.002, Table S4b). Aberrant energy metabolism is the critical factor for cell integrity maintenance and neurodegeneration. Another notable differentially expressed TF, STAT1, demonstrated a different expression pattern across PiD (FDR-adjusted p-value = 4.61 ×10ˆ−14, Table S4a) and AD (not significant), implying its distinct involvement in the regulatory mechanisms underlying different neurodegenerative disorders or different stages of disorders. Prior research has indicated that decreased STAT1 expression correlates with a higher risk of conversion to MCI and can be considered as a preclinical indication of AD development60.The preceding analyses and these data provide a likely genetic mechanism for two distinct dementias, based on differential TF binding activity on the enhancer or promoter regions of its target gene, coupled with analyses shown on gene expression.
Utilizing TF binding occupancy to identify the shared and distinct glial responses in PiD and AD.
We investigated the regulatory role of several TFs in glial cells in PiD and AD. Given the importance of TFs in modulating gene expression, we focused on identifying the top differential binding TFs, distinguishing those specific to PiD and those shared with AD. Among the selected TFs, we explored the regulatory effects of microglial TF SPI1, a well-known AD GWAS risk gene11, friend leukemia integration 1 (FLI1), and Transcription Factor Dp-1 (TFDP1) (Figure 5a, S5a, and Table S4ab), to shed light on the potential roles of these TFs in the pathogenesis of PiD and AD. In our snATAC-seq analyses of microglial cells, we observed increased differential binding activities of FLI1 and SPI1 in both PiD and AD. SPI1 is known to be associated with the normal development of microglial cells in the brain61, and Ets-related transcription factor FLI1 has been established as a regulator of gene activity during cellular differentiation62. However, TFDP1, a potential global modulator of chromatin accessibility by controlling histone transcription63, shows contrasting differential binding activities when comparing PiD with AD (Figure 5a and S5a), suggesting potential discrepancy in genome-wide TFDP1 TF binding action between diseases. Among the top-selected targets, we observed a significant downregulation of MAF in AD (FDR-adjusted p-value = 2.28×10ˆ−8, Table S4b), a gene identified as an AD GWAS risk gene and a differentially expressed TF11, regulated by SPI1. Interestingly, we did not observe any significant difference in MAF expression in PiD (Figure 5b,c). Additionally, we detected another AD DEG, CX3CR1 (FDR-adjusted p-value = 1.53×10ˆ−25, Table S4b), also regulated by SPI1, but not significantly dysregulated in PiD. CX3CR1 has been implicated in both neuroprotective and detrimental effects by regulating inflammation in neurological disorders64. Furthermore, our analyses revealed the regulation of several other GWAS risk genes, including GLDN, ZFHX3, USP6NL, SORL1, MS4A4A, INPP5D, and RASGEF1C11, by TFDP1, FLI1, and SPI1.
Figure 5. Glial changes in transcription factor dysregulation and gene expression in PiD and AD progression.
a, d, g, Genome-wide Tn5 bias-subtracted TF differential footprinting binding score of PiD and AD in MG (a), ASC (d), and ODC (g) compared to their corresponding controls. b, e, h, TF regulatory networks showing the predicted candidate target genes for MG (b), ASC (e), and ODC (h). Highlighted transcription factors and other differential expressed TFs are shown in yellow. Upregulated differential genes are shown in red and square. Downregulated differential genes are shown in blue and in a circle. The gene of interest is downregulated, shown in pink and in a circle. The differential expressed GWAS risk genes are displayed in bright blue. Edges representing the linkage of TF - target gene regulation are shown in purple for PiD and sienna for AD. c, f, i, Dotplot of the differentially expressed gene, differentially expressed GWAS risk genes, and TFs in PiD and AD versus their respective controls.
In astrocytes, we observed a consistent trend among most TFs, where the majority displayed either increased or decreased binding scores in both PiD and AD. Notably, a subgroup of TFs from the activating protein-1 (AP-1) family, namely JUND, JUNB, and FOS, exhibited significant enrichment in both PiD and AD (Figure 5d and S5b). For instance, JUND from the AP-1 TF family, known for its strong correlations with pTau and amyloid-beta65, demonstrated similar patterns. Additionally, BACH1, primarily recognized as a transcriptional suppressor66, showed a positive correlation with both PiD and AD. These findings suggest some potential convergence of top-selected TFs’ activity in astrocytes across PiD and AD pathologies. Specifically, JUND’s inferred role in astrocyte APOE expression, which is shown to be downregulated in AD (FDR-adjusted p-value = 3.14×10ˆ−4, Table S4b) but not statistically significant in PiD (Figure 5e,f), underscores its involvement in AD-related processes. At the same time, we identified hypoxia-inducible factor-1 alpha (HIF1A), regulated by both JUND and BACH1, as downregulated in PiD (FDR-adjusted p-value = 4.35×10ˆ−6, Table S4a) but not significant in AD, which may align with previous reports suggesting that the loss of HIF1A within astrocytes protects neurons from cell death67. Our observations underscore potential regulatory changes in astrocytes, characterized by the regulatory activation mediated by AP-1 family TFs and the transcriptional suppression facilitated by BACH1. Furthermore, the dysregulation of APOE expression and HIF1A levels in astrocytes emphasizes the intricate interplay between transcriptional regulation and disease progression in these neurodegenerative disorders.
In oligodendrocytes, we observed a predominant trend where the majority of TFs exhibited either increased binding activity in both PiD and AD or unique patterns specific to each disease state (Figure 5g and S5c). Noteworthy among these are the transcriptional suppressors HES1 and ZBTB3368,69, which displayed enriched differential binding scores in both PiD and AD. Moreover, our analyses revealed that these two transcriptional repressors were not only associated with the downregulation of ADAM10, PLEKHA1, and JAZF1, among other downregulated DEGs, but also with the upregulation of BIN1 and MAPT, along with other upregulated DEGs (Figure 5h). This suggests the intricate and multifaceted nature of the transcriptional processes, which may be relevant to both PiD and AD, or specific to one of these conditions, indicating shared or condition-specific regulatory mechanisms. Furthermore, MAPT, a gene encoding tau protein to keep the function of microtubules and axonal transport, which ZBTB33 also regulates, is differentially expressed in both PiD and AD. Additionally, the downregulation of FOXO1, known to protect against age-progressive axonal degeneration70, further underscores the intricate interplay between transcriptional regulation and neurodegenerative processes in oligodendrocytes.
Cis-regulatory linked HGE impacts the expression of a target gene in disease synaptic pathology.
We have elucidated the shared and distinct changes in the pathways between these two frontal cortical degenerative diseases related to the prominent features, glial activation, neuroinflammation, synaptic dysfunction, and synapse loss of AD and related dementia71,72. Building upon these findings, we reasoned that these data further provide a unique opportunity to identify human-specific regulatory elements responsible for maintaining the integrity of human cortical neurons and driving cortical neurogenesis.
In order to uncover regulatory elements unique to humans that drive cortical neurogenesis, we further delved into a previously compiled highly expressed gene list that shows increased activity specifically in the developing human brain, when comparing gene expression between mice, macaques, and humans73. Through overlapping human-gained enhancer (HGE) with snATAC-seq peaks from PiD and AD, we identified an enhancer element that is both a differentially accessible peak in PiD and an HGE. Using chromatin co-accessibility analyses, we bioinformatically linked this differential accessible enhancer to UBE3A, even though it is located more than 40Kbp away from its UTRs and around 80kbp away from its coding region (Figure 6a,b). As a gene implicated in neuronal activity, UBE3A codes for a protein that plays a critical role in neuronal functioning, regulating proliferation and apoptosis74. UBE3A loss of function mutation has been observed in individuals with Angelman Syndrome, while autism-linked UBE3A gain of function mutation was recently reported in a mouse model showing neurobehavioral deficits75,76. The cis-regulatory identified distal enhancers and HGE of UBE3A in neurons are more accessible in PiD (FDR-adjusted p-value = 4.40 ×10ˆ−5) (Figure 2e and 6a,b).
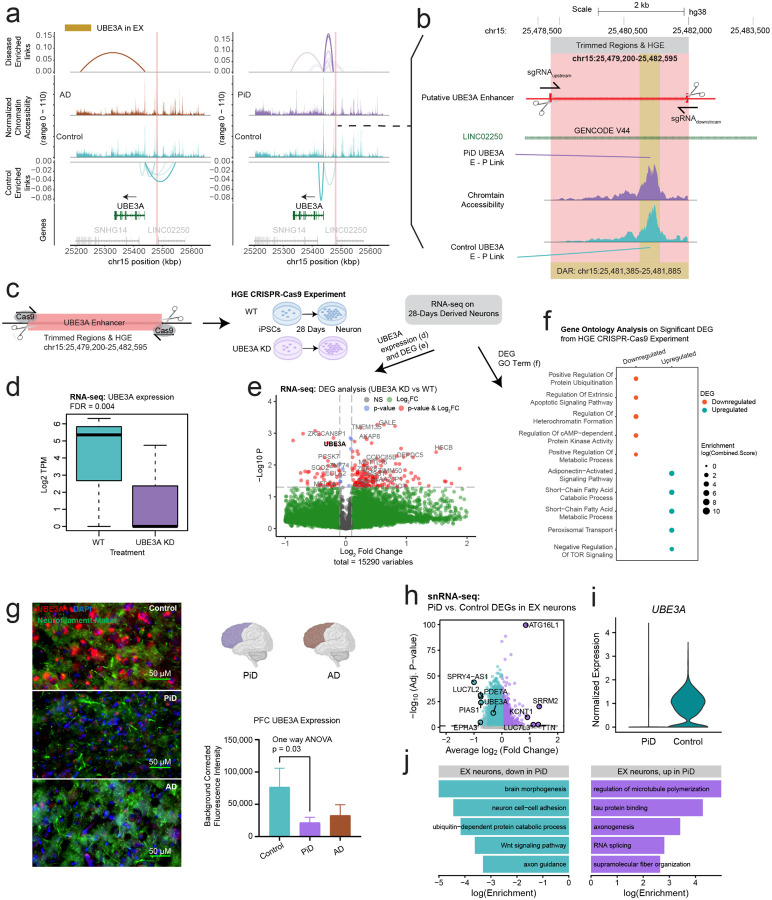
Figure 6. Mapping DREs involved in synaptic function to their target genes.
a, Delta co-accessibility of UBE3A and its open chromatin regions in EX for both AD and PiD with their corresponding controls. Highlighted regions in salmon represent CRISPR-edited enhancer regions to the UBE3A. b, Enlarged CRISPR-edited enhancer regions of UBE3A in salmon and differentially accessible peaks in yellow overlap with intronic regions of long intergenic non-protein coding RNA 22 (LINC02250). c, Experimental design for HGE CRISPR-Cas9 for UBE3A, RNA-seq performed on iPSC-derived neurons after 28 days of development. d, UBE3A expression from RNA-seq. e, Volcano plot of DEGs from RNA-seq (UBE3AKD vs WT). e, Gene ontology of upregulated and downregulated DEGs from RNA-seq (UBE3AKD vs WT). g, Representative triple immunofluorescence images for UBE3A (red), neurofilament marker (green), and DAPI (blue) from postmortem human brain tissue (PFC) of control (n=3), AD (n=5) and Pick’s (n=5) cases. 60 X Images were captured using Nikon ECLIPSE Ti2 inverted microscope. h, snRNA-seq DEG analyses in EX. i, UBE3A expression from snRNA-seq in EX. j, Gene ontology of upregulated and downregulated DEGs from snRNA-seq (PiD vs Control).
We hypothesized that the active HGE would likely enhance the expression of UBE3A or mitigate suppressive effects leading to its downregulation. Conversely, the elimination of this active HGE would presumably result in reduced levels of UBE3A. To validate whether this imputed enhancer is indeed the putative enhancer of UBE3A, we conducted CRISPR-edited experiments in iPSCs, wherein we targeted and trimmed the HGE region (chr15:25,479,200–25,482,595) (Figure 6b,c). In theory, if the predicted enhancer does regulate gene activity, removing it would interfere with its control mechanisms, resulting in reduced activity of the target gene. This approach has previously been employed to identify enhancers that regulate neocortical development77. Following RNA-seq analyses after 28 days in derived neurons, perturbation of UBE3A expression led not only to its downregulation (Figure 6d,e), but also to differential expression in other genes (Figure 6e). Gene ontology analyses of the RNA-seq data from iPSCs revealed that downregulation of UBE3A was associated with processes including downregulation of protein ubiquitination, apoptosis, heterochromatin organization, cAMP-dependent protein kinase activity, and disruptions in various metabolic processes (Figure 6f).
Given the intricate nature of human tissue, particularly in the context of disease conditions, our subsequent analyses noted an enriched activity of chromatin accessibility (average log2FC > 0, Table S2) for all distal peaks associated with UBE3A in the EX. Despite this, we observed a decrease in the proteomic and transcriptomic levels of UBE3A. In our immunofluorescence staining of UBE3A, we noted a statistically significant decrease in UBE3A levels in human PFC (Figure 6g). Furthermore, our analyses of DEGs also revealed UBE3A as one of the downregulated genes (Figure 6h,i). In our gene ontology analyses, we found that the downregulated genes were involved in various processes related to neuronal integrity, brain morphogenesis, neuron cell-cell adhesion, axon guidance, cell fate determination via the Wnt signaling pathway, and UBE3A-related ubiquitin-dependent protein catabolic processes. Conversely, among the upregulated terms, we observed enrichment in processes related to microtubule organization and tau protein regulation (Figure 6j). This discrepancy may be attributed to the chromatin region becoming more accessible as a compensatory mechanism to counteract the downregulation of its target genes (Figure S2d), highlighting the need for further investigation into the regulatory processes underlying these observations.
Discussion
Single-cell sequencing has been used to characterize the cell type and cell state-specific changes in Alzheimer’s disease pathology extensively. However, comparatively, other tauopathies, including Pick’s disease, have been understudied. In this study, we generated single-nucleus epigenomic and transcriptomic data from postmortem human brain tissue samples of Pick’s disease and cognitively normal controls. By integrating the analyses on cis- and trans-regulatory mechanisms with gene expression data, our approach at the single-cell resolution enabled us to investigate the cellular diversity of the human PFC to compare shared and distinct regulatory mechanisms between these two tauopathies in excitatory neurons, astrocytes, microglia, and oligodendrocytes, and pinpoint the cell-type specific disease-associated alterations. Meta-analyses in genome-wide association studies, supplemented with the assistance of snATAC and snRNA data, utilized AD and FTD GWAS genes and revealed putative and dysregulated risk genes for PiD. Systematic analyses of alternation in TF binding activity on promoter-enhancer links in both a genome-wide scale and gene-local region in PiD and AD revealed distinct and shared TF-regulatory networks from neurons and glial cells. Our single-nucleus data and customized approach to investigating cis- and trans-regulatory mechanisms altered in PiD and AD pathology led to the creation of an online interactive database, scROAD, which researchers are free to explore. We additionally generated RNA-seq data from iPSC-derived neurons in the CRISPR-Cas9 experiment, allowing us to validate imputed promoter-enhancer regulatory linkage from possible target genes involved in disease progression.
Although the precise molecular mechanisms driving PiD pathology remain elusive, our study provides novel insights into the intricate landscape of gene regulation in PiD, particularly the challenges in interpreting distal regulatory elements. Our differential analyses highlight the utility of our identified promoter-enhancer links in elucidating regulatory mechanisms, and revealed widespread chromatin accessibility and gene expression changes linked to PiD and AD pathology across major cell types. Some of these changes, including the increased chromatin accessibility and dysregulated gene expression involved in synaptic signaling, apoptotic process, regulation of neuronal activity, cellular response to stress, and cell communication, may represent an attempt of neurons and oligodendrocytes interaction to reestablish homeostasis through necessary attempts to different genes. Some promoter-enhancer connections facilitated increased chromatin accessibility, potentially serving as a compensatory mechanism to mitigate the dysregulation of target genes. Other alterations, including positive regulation of endocytosis, genes responsible for cellular metabolic process, and genes encoding cellular response to unfolded/misfolded protein in astrocytes and microglia, may contribute to glial cell differentiation or immune activation in PiD and AD. Disruptions in the metabolic processes and cellular stress response compromise the balance in the cellular microenvironment and consequently contribute to the progression of PiD and AD.
While the causative molecular mechanisms of PiD remain unknown, our work offers new insights that assist in unraveling the nature of gene regulation in PiD, especially regarding genomic loci with well-described heritable disease risk. We capitalized on the AD and FTD GWAS data to identify genes associated with phenotypic variability between PiD and AD because of similar pathological and clinical traits, such as tauopathies and cognitive decline. GWASs have been widely used to enhance our understanding of polygenic human traits and to reveal clinically relevant risk variants for neurodegeneration. Notably, we identified genetic risk variants that overlapped with specific cell types to narrow down the potential non-coding variants underlying disease susceptibility. Furthermore, our analysis revealed that AD GWAS genes showed a highly significant overlap with differentially expressed genes in PiD cases, suggesting that these associations are not random. This highlights the potential convergent regulatory mechanisms that may be shared between PiD and AD, despite the distinct clinical manifestations. Although this method has enabled the investigation of cell-type-specific disease-associated regulatory mechanisms, key limitations of the snATAC-seq assay without variant calling in PiD samples leave the opportunity for future studies and improvements.
Cell demise constitutes a defining characteristic of neurodegenerative ailments, including Pick’s and Alzheimer’s disease. Significantly more pronounced alterations in chromatin accessibility and gene expression were observed in excitatory neurons and oligodendrocytes in PiD compared to AD. In agreement with a previously observed association between rapid progression and early disease onset in PiD than AD3,32, we found an elevation in the fold change in chromatic accessibility of dysregulation among genes and TFs, especially in excitatory neurons. Additionally, in excitatory neurons from PiD, we identified a possible compensatory mechanism that downregulated genes strongly associated with increased chromatin-accessible regions for the same genes through cis-regulated promoter-enhancer links, including genes responsible for neuronal activity and signaling, for example, UBE3A. A major contribution of our study lies in the identification of cell-type-specific enhancer-promoter pairs, potentially facilitating gene-regulatory alterations in PiD and AD, along with the TFs likely to bind to these regulatory elements within the respective cell types. Our investigation into cis-regulatory elements and DNA-binding motifs, particularly in regions harboring fine-mapped SNPs, has uncovered potential disruptions in regulatory relationships, exemplified by the anti-amyloidogenic gene ADAM10. These disruptions, proximal to disease-associated SNPs, may lead to diminished TF binding activity and subsequent dysregulation of target gene expression. Furthermore, our analyses utilized the gene-specific-enhancer-binding TFs’ information to construct a TF regulatory network in neurons and demonstrated alterations in PiD and AD. We also provide insights into the regulatory landscape of TFs in glial cells across PiD and AD. We identified differential binding activities of TFs, such as SPI1, known as a significant AD GWAS risk gene in microglia and associated with its development, JUND in astrocytes, known for its strong correlations with pTau and amyloid-beta, and transcriptional suppressors HES1 and ZBTB33 in oligodendrocytes, shedding light on their potential roles in disease pathogenesis. Moreover, the downstream dysregulation of TFs and genes associated with the highlighted TFs, including CX3CR1, MAPT, and FOXO1, emphasizes the intricate regulatory mechanisms implicated in neurodegenerative processes, with some alterations shared between PiD and AD, while others are uniquely observed in either condition.
The identification of functional regulatory elements in human excitatory neurons and the validation of their functions in iPSC-derived neurons enhance our understanding of epigenomic discovery. Leveraging these findings, we identified human-specific regulatory elements crucial for maintaining the integrity of cortical neurons in a neurodegenerative disorder, providing valuable annotations. Subsequent CRISPR-edited experiments in iPSCs confirmed the regulatory role of a putative enhancer in UBE3A expression. Furthermore, our observations of enriched chromatin accessibility near UBE3A in excitatory neurons, despite decreased UBE3A levels in snRNA-seq, suggest compensatory mechanisms in response to its downregulation.
Overall, our findings provide valuable insights into the regulatory landscape of PiD and AD, emphasizing the importance of integrated genomic approaches for elucidating the molecular underpinnings of neurodegenerative disorders. These findings underscore the intricate interplay between transcriptional regulation and disease progression in PiD and AD, highlighting the importance of understanding these mechanisms for developing effective therapeutic strategies.
Methods
Postmortem human brain tissue.
Human postmortem frontal cortex brain samples were obtained from UCI MIND’s Alzheimer’s Disease Research Center (ADRC), Harvard and Mt.Sinai tissue repositories. All participants, or participants’ legal representatives, provided written informed consent for the study. 50 mg tissue from each sample (n= 9 control brain and n= 7 Pick’s brain) was dissected and aliquoted into a 1.5 ml tube inside a prechilled tissue dissection box as described previously78. Samples were also selected based upon several covariates, including age, sex, postmortem interval (PMI), and disease comorbidity. Sample information is available in Table S1.
Immunofluorescence.
PFA fixed human postmortem brain tissues (PFC region) were sectioned at 30μm using a cryotome (Leica SM2010R). Sections were then rehydrated and washed in 1X sterile PBS and permeabilized using 1X sodium citrate buffer pH 6.0 (heated at 95°C for 10 mins). After blocking with 3% BSA solution or serum, sections were incubated with diluted primary antibodies (as per manufactures recommendation) at 4°C overnight (IBA1 antibody; Cat #NC9288364; 1:1000; Fisher Scientific, GFAP Polyclonal Antibody; Cat #PA3–16727; 1:500; ThermoFisher, p-tau (AT8) Cat #MN1020; 1:250; ThermoFisher; UBE3A; Cat #10344–1-AP; 1:1000; Proteintech, Anti neurofilament protein; Cat #837904; 1:1000; Biolegend). Secondary antibodies were selected and diluted according to the manufacturer’s instruction and incubated for 1.5–2 hrs. Sections were then washed (3X with PBS), mounted and cover slipped using anti fade mounting media. Slides were imaged (20X/40X/60X) using Nikon ECLIPSE Ti2 inverted microscope. Images from 3 randomly selected areas of each slice were used for analyses.
snATAC-seq tissue processing and nuclei isolation.
Frozen brain tissue pieces were placed in 500 μL chilled 0.1× Lysis Buffer (1X lysis buffer diluted with lysis dilution buffer; please refer to snATAC-seq protocol78 for more details) and immediately homogenized 15 times using a pellet pestle (Fisherbrand™ Pellet Pestle™ Cordless Motor with RNase-Free Disposable Pellet Pestles, Cat#12–141-364). The homogenized tissues were then incubated for 15 mins followed by addition of 500 μL of chilled Wash Buffer and filtration through a 70 μm Cell Strainer (Miltenyi Biotech). In the next step, a sucrose gradient (Nuclei PURE Prep Nuclei Isolation Kit, Cat #NUC201–1KT, Sigma) was prepared and nuclei were spun at 13,000 g for 45 minutes at 4°C. After centrifugation, the debris and myelin from the top of the sucrose gradient were removed. Nuclei were resuspended, washed, filtered (through a 40 μm cell strainer), counted (using a cell counter), and then incubated in a Transposition Mix.
snATAC-seq library preparation and sequencing.
Transposed nuclei were loaded on 10X Genomics Next GEM Chip H (10x Genomics) to generate single-cell GEMS. GEMs were then transferred, incubated, and cleaned for further processing. Single nuclei ATAC-seq libraries were prepared using the Chromium Single Cell ATAC v2 (10x Genomics) reagents kit as per the manufacturer’s instructions. Library size distribution and average fragment length, of each library, were assessed with Agilent TapeStation High Sensitivity D5000 ScreenTapes and the concentrations were quantitated using Qubit Fluorometer. Libraries were sequenced on a NovaSeq 6000 (Illumina) in paired-end mode (read1N: 50 cycles, index i7: 8, index i5:16 cycles, read 2N:50 cycles) to generate approximately 500 M reads per sample.
snRNA-seq library preparation and sequencing.
45–50 mg of fresh frozen brain tissue (PFC) was homogenized in EZ Lysis buffer (Cat #NUC101–1KT, Sigma-Aldrich) and incubated for 10 min on ice before being passed through a 70μm filter. The new tube with filtered homogenate was then centrifuged at 500 g for 5 min at 4°C and resuspended in additional 1ml of lysis buffer. After another centrifugation samples were incubated in Nuclei Wash and Resuspension buffer (1xPBS, 1% BSA, 0.2U/μl RNase inhibitor) for 5 min. To remove myelin contaminants and debris, we prepared sucrose gradients and centrifuged the tubes at 13,000 g for 45 min at 4°C. Next, a debris removal solution (Cat #130–109–398, Miltenyi Biotec) was added to the nuclei suspension (and centrifuged at 3,000 g for 10 mins at 4°C) for a second round of cleanup. Debris free clean nuclei suspension was then diluted in nuclei buffer (with BSA and RNase) before processing with the Nuclei Fixation Kit (Parse Biosciences). After fixation and permeabilization, nuclei were cryopreserved with DMSO until day of library preparation. Libraries were prepared using using EVERCODE™ WT V3 kit (Parse Bioscience) and quantified using Qubit dsDNA HS assay kit (Cat #Q32851, Invitrogen). D5000 HS kit (Cat #5067–5592, Cat #5067–5593; Agilent) was used for measuring the average fragment length of each library. Libraries were sequenced using Illumina Novaseq 6000 S4 platform (paired-end sequencing) for a sequencing depth of 50,000 read pairs/nuclei.
Cortical Neuron Pellet Generation.
Cortical neurons were generated as previously described79 with some modifications. Induced pluripotent cell lines were maintained in mTeSR Plus medium (Stem Cell Technolgies Cat #100–0276) on GelTrex basement membrane (ThremoFisher Cat #A1413302) and passaged using ReLeSR (Stem Cell Technologies Cat #100–0484) at 80% confluence in the presence of CEPT (Chroman1-Tocris Cat #7163, Emricasan-Seleck Chemicals Cat #S7775, Polyamine supplement-Sigma Cat #P8483, Trans-ISRIB-R&D Systems-528480). UBE3A mutant and parental lines were transfected via Nucleofection (LONZA Cat #VPH-5022) of the PB-TO-hNGN2 (Addgene Cat #172115*) plasmid and purified in the presence of 200 ng/mL Puromycin (Invivogen ant-pr-1) until the majority of cells showed plasmid expression as determined by BFP expression. Once a high BFP expression had been established, iPSCs dissociated to single cell with accutase (ThermoFisher Cat #NC9464543) and seeded at 1×106 cells per GelTrex coated 6 well in Induction media: Knockout DMEM/F12 (ThermoFisher); N2 supplement 100X (ThermoFisher); non-essential amino acids 100X (ThermoFisher), and supplemented with Doxycycline at a final concentration of 1μM (Sigma) and CEPT. The medium was changed every day. After 3 days, Uridine (U) and Fluorodeoxyuridine (FdU) were both added at 1mM (Sigma Cat #3750, Sigma Cat #0503). On day 4, the induced cells were passaged as single cells with Accutase and seeded at 2×106 cells per Poly-D-Lysine coated 6 wells (Sigma Cat #P6407) in Cortical Neuron Culture Medium 1 (CM!): 1:1 Knockout DMEM/F12: BrainPhys neuronal medium without Phenol-Red (STEMCELL Technologies); B27 supplement, 50X (ThermoFisher); BDNF (10 μg/ml, STEMCELL Technolgies) in PBS containing 0.1% BSA (ThermoFisher); NT-3 (10 μg/ml, Preprotech) in PBS containing 0.1% BSA, GDNF (10 μg/ml, STEM-CELL Technologies) in PBS containing 0.1% BSA; laminin final con. 1 μg/ml (ThermoFisher), Doxycycline (1uM), U (1uM), and FdU (1uM). Cells were maintained an additional 24 days with half media changes every 3–4 days first with CM1 (day 7), then Cortical Neuron Culture Medium 2 (CM2) starting at day 10. CM2: BrainPhys neuronal medium without Phenol-Red (STEMCELL Technologies); B27 supplement, 50X (ThermoFisher); BDNF (10 μg/ml, STEMCELL Technolgies) in PBS containing 0.1% BSA (ThermoFisher); NT-3 (10 μg/ml, Preprotech) in PBS containing 0.1% BSA, GDNF (10 μg/ml, STEMCELL Technologies) in PBS containing 0.1% BSA; laminin final con. 1 μg/ml (ThermoFisher), Doxycycline (1uM), U (1uM), and FdU (1uM). Three successive passages of each cell line were differentiated in parallel with pellets collected and flash froze for RNAseq at D0, D4, and D28 along with PFA fixed coverslips.
Processing snATAC-seq data.
We used Cellranger-atac count (v 2.0.0) to map raw snATAC-seq reads to the GRCh38 reference genome (downloaded from the 10X Genomics website) in each sample, quantifying chromatin accessibility for each cell barcode. First, we used the ArchR function createArrowFiles to format the output of Cellranger-atac, removing barcodes with transcription start site (TSS) enrichment less than 4 and fewer than 1000 fragments. This function also yields a barcodes-by-genomic-bins “tile matrix” and a “gene score matrix“ which aggregates chromatin accessibility information proximal to each gene. We next used the R package ArchRtoSignac to convert our dataset from ArchR to Signac format to proceed with downstream analyses in Signac. We next performed analyses of our new snATAC-seq samples from PiD donors and cognitively normal controls with our previous snATAC-seq dataset of AD donors and controls as the reference dataset. We created a merged object of the PiD and AD snATAC-seq datasets, and then created an integrated dimensionally-reduced representation of them using the Seurat function FindIntegrationAnchors, using reciprocal latent semantic indexing (RLSI) as the dimensionality reduction method. Using this anchor set, we performed transfer learning to predict the cell type identity of the nuclei in the PiD dataset based on the annotations in the AD dataset with the Seurat function FindTransferAnchors. This transfer learning analysis provides a probability for each nucleus in the PiD dataset for its cell-type assignment, where some nuclei are confidently mapped to a single cell type while others are more ambiguously mapped to multiple cell types. We next filtered the PiD dataset by nuclei with a max prediction probability greater than or equal to 0.95 to retain nuclei with highly confident mappings across the datasets. We performed a final integrated analysis using LSI dimensionality reduction and Harmony on the basis of the biological sample.
Processing snRNA-seq data.
We used split-pipe ParseBio pipeline (v 1.0.3) to map snRNA-seq reads to the GRCh38 reference transcriptome (downloaded from the Ensembl website) in each sample, quantifying unique molecular identifiers (UMI) for each cell barcode. Next, we accounted for potential ambient RNA contamination by applying Cellbender remove-background (v 0.2.0) to model the ambient signal and remove it from the UMI counts matrix for each sample. We then identified barcodes mapping to multiple nuclei (multiplets) by applying Scrublet (v 0.2.3) with default settings to each sample. We applied an initial quality control (QC) filter to remove barcodes with fewer than 250 UMI. Further, we applied sample-specific filters to remove barcodes in the top 5% of UMI, percentage of mitochondrial reads, and multiplet score within each sample. We finally applied a dataset-wide cutoff to remove barcodes with greater than 20,000 UMI, greater than 0.2 multiplet score, and greater than 5% mitochondrial reads, resulting in 68,999 barcodes for clustering analysis. We next performed clustering analysis with Scanpy with the following steps. First, we normalized gene expression for each cell by the total UMI counts in all genes and log transform using sc.pp.normalize_total and sc.pp.log1p. Second, we performed feature selection using sc.pp.highly_-variable_genes using the “Seurat_v3” option for the feature selection method, retaining 3,000 genes for downstream analyses. Third, we scaled the normalized expression matrix for these 3,000 genes to unit variance and centered at zero mean using the sc.pp.scale function. Fourth, we performed linear dimensionality reduction with principal component analysis (PCA) using the sc.tl.pca function, which we then corrected on the basis of sample of origin using Harmony. Fifth, we constructed a cell neighborhood graph using the top 30 harmonized PCs using sc.pp.neighbors function. We visualized this cell neighborhood graph using UMAP with the function sc.tl.umap. We performed an initial round of Leiden clustering with a high resolution parameter (resolution=3) to reveal additional clusters of low-quality cells which may have escaped our previous QC filtering, and to annotate major cell types based on a panel of canonical marker genes. After removing two low-quality clusters, we split apart the dataset by major cell lineages (excitatory neurons, inhibitory neurons, oligodendrocytes, and astrocytes) to perform subclustering analyses, yielding our final clustered and processed snRNA-seq dataset.
Differential accessible open chromatin analyses.
We systematically performed the analyses of differential open chromatin accessibility across each cellular type. This involved contrasting the disease states with their respective control conditions. For all the differential analyses employed, differentially accessible peak scrutiny was facilitated by implementing logistic regression (test.use=‘LR’) to draw comparisons between cellular groupings. Logistics regression was utilized based on the accessibility interface of a specified open chromatin region (OCR) within varying groups of the selected cell type. This is a protocol recommended by the Signac package (v1.9.0)81. The differential analyses were executed in Signac by deploying the same FindMarkers function found in Seurat (v4.3.0). The accessible peaks that exhibited an adjusted P-value (corrected by Bonferroni method) of less than 0.05, accompanied by a minimum cellular fraction (min.pct > 0.05) in either of the two groups, were categorized as differentially accessible peak between the cellular groupings. We ran a comparative analysis of chromatin accessibility between the two diagnosis groups, specifically Pick’s disease (PiD) and Alzheimer’s disease (AD), and their age-appropriate cognitively normal counterparts. This was conducted within the human single nucleus ATAC-seq dataset. The differential accessibility findings were visualized using a Complexheatmap82, divided by diagnosis comparison and hierarchically aggregated based on the avg_log2FC of differentially accessible peaks. This enabled us to focus on changes specific to each cellular type within each genomic classification. Finally, to single out the biological pathways and processes exhibiting a significant presence within our promoter differentially accessible peak sets or promoters of cis-regulatory-associated differentially accessible peaks present in distal and intronic regions, we invoked the support of the GREAT R package (v2.0.2)83,84.
Differential gene expression analyses.
We identified unbiased marker genes in each of our snRNA-seq clusters by a one-versus-all differential gene expression test using the Seurat function FindAllMarkers with MAST as our differential expression model. We used sequencing biological sample and total number of UMIs per cell as model covariates. We performed differential expression analyses to compare gene expression signatures in cells from PiD and control samples in each of our major cell types (excitatory neurons, inhibitory neurons, oligodendrocytes, OPCs, astrocytes, pericytes, endothelial cells, and microglia). Similar to our cluster marker gene test, we used MAST as our differential expression module with biological sample, sex, and number of UMI as model covariates. We used the R package enrichR to perform pathway enrichment analyses for the DEGs in our excitatory neuron population.
Statistical fine-mapping of candidate causal variants residue within cell-type specific accessible peaks from the snATAC-seq data.
We sourced comprehensive genome-wide association studies (GWAS) pertinent to Alzheimer’s Disease (AD)11 and frontotemporal degeneration (FTD)12. The summary data pertaining to the AD GWAS was procured from the European Bioinformatics Institute GWAS Catalog (accession number: GCST90027158), whilst the FTD GWAS summary data was retrieved from the International Frontotemporal Dementia Genetics Consortium. To streamline the output files of the GWAS summary statistics from each dataset, we employed a uniformly designed pipeline, MungeSumstats85. The application of this tool was governed by parameters that have been specified comprehensively in our GitHub repository. To further elucidate the role of single nucleotide polymorphisms (SNPs) pertaining to AD, we fine-mapped these SNPs within a 1-Mb window of the lead variants of AD risk loci that had been unearthed in the initial GWAS investigation11. In addition to the AD SNPs, the detection of lead SNPs associated with FTD12 required the identification of specific genetic markers encased within a 1-Mb spectrum present on all chromosomes. The selection criteria for these markers were established based on the statistical significance of their corresponding P-values. To accommodate all SNPs within the linkage disequilibrium (LD) block, we estimated pairwise LD between SNPs within the 1-Mb window of the GWAS lead variant. This estimation was performed using PLINK (v1.9 and v2.0)86. Once the lead SNPs from the FTD and AD GWAS had been secured, the identified data was customized according to the corresponding 1-MB range LD matrix, within the sparse multiple regression model. This model was then implemented in the fine-mapping instrument, Sum of Single Effects (SuSiE)87,88. We managed to acquire a number of credible sets (CSs) for identified FTD and AD GWAS risk loci with high probability (a posterior inclusion probability: PIP > 0.95). In order to prioritize these credible sets, we aligned SNP locations with our snATAC-seq open chromatin regions. The fine-mapped casual SNPs within the identified cell types were assessed for credibility by cross-referencing the GWAS risk genes’ expression level across all cell types using control data from published resources16,19,34. The final step included checking two scores - the probability of being loss-of-function intolerant (pLI) and the loss-of-function observed/expected upper-bound fraction (LOEUF) for the prioritized GWAS risk loci. These scores reflect the integrity of a gene or transcript in tolerating protein truncating variation33.
Reprocessing publicly available single-nucleus gene expression datasets.
We obtained the sequence data from three peer-reviewed single-nucleus RNA sequencing (snRNA-seq) studies related to Alzheimer’s Disease (AD)16,19,34. The datasets represented in the works of Mathys et al. (2019), and Zhou et al. (2020) were accessed via the Synapse platform (referenced under syn18485175 and syn21670836, respectively). In the context of the dataset for the Morabito et al. (2021) study, which was formulated by our research team, a download was not necessitated, but you can access it under syn22079621. Although the pipeline was largely consistent, there were minor deviations in terms of parameter adjustments as per the individual requirements of each dataset. A comprehensive delineation of these nuanced changes is documented in our GitHub repository.
Finding co-accessible peaks with Cicero to establish putative enhancer-promoter linkage.
We initiated the conversion of the SeuratObject into the CellDataSet framework utilizing the as.cell_data_set function offered within the SeuratWrappers toolkit (v0.3.0). This was subsequently transformed into a Cicero object through the application of the make_cicero_cds function taken from the Cicero package (v1.3.4.11). The run_cicero function, a significant component of the Cicero suite, was then employed to calibrate the co-accessibility of open chromatin peaks across the genome for each cell type. The predominant objective here was to predict cis-regulatory interactions within a genomic window of 300,000 base pairs. The construction of a linkage co-accessibility score for each associating pair of accessible peaks was completed using a graphical LASSO regression model. This package and approach were based on techniques detailed in the Cicero method31. The understanding being that an increased co-accessibility score denoted a stronger bond between an OCR pair and hence, greater confidence could be assigned to this pairing within a given dataset. Within the total ensemble of OCR pairs, we prioritized our examination on pairs identified as enhancer-promoter. The rationale for this selective focus stemmed from the potential for the enhancer-enhancer pair’s co-accessibility score to originate from inherent enhancer-enhancer interactions. This in turn could lead to a perceivable reduction in the co-accessibility scoring for the enhancer-promoter pair. Lastly, a comparative study was undertaken to calculate the delta co-accessibility score within identical OCR pairs. In this step, diseased states were compared with their corresponding control settings. The purpose of this comparison was to highlight any enhanced enhancer-promoter linkages that could potentially be contributing to the advancement of the disease.
Transcription factor Occupancy prediction on snATAC-seq chromatin accessibility.
TOBIAS47 stands as a robust, precise, and rapid footprinting framework, facilitating a comprehensive exploration of TF binding occupancy for numerous TFs concurrently on a genome-wide scale as well as at the gene local region. We want to use this ATAC-seq analysis toolkit to investigate the kinetics of transcription factor (TF) binding in PiD, AD and their distinctions compared to respective control conditions, we turn to TOBIAS for its capabilities as the ATAC-seq TF footprinting analyses toolkit. Our initial steps involved the extraction and categorization of cell barcodes based on both cell type and diagnosis. Subsequently, we compiled distinct .bam files for each condition, serving as the requisite input format for the TOBIAS ATACorrect step. This particular tool within TOBIAS corrects the inherent insertion bias of Tn5 transposition. Following this correction process, the central task in footprinting commenced with the identification of protein binding regions across the entire genome. Utilizing single-base pair cutsite tracks generated by ATACorrect, TOBIAS FootprintScores was employed to compute a continuous footprinting score across these regions. This approach enhances the prediction of binding for transcription factors even with lower footprintability, characterized by weaker footprints. Subsequently, the footprints were plotted using function PlotAggregate to visualize and compare the aggregated signals across the specified conditions. This step serves to provide a tangible representation of TF binding occupancy and facilitates comparative analyses of these changes under different diagnosis conditions in each cell type. We developed a wrapper package, scCis-TF, designed to streamline information extraction to integrate data from Signac into Cicero and TOBIAS and perform analysis.
Transcription factor Regulatory Network Construction.
To construct a comprehensive TF regulatory network, we integrated insights from Cicero and TOBIAS. First, leveraging Cicero, we focused on the predicted cis-regulatory interactions within a 300,000 base pair window, and grouped them based on the genomic class around its target gene identified by accessible promoter peaks. So by prioritizing the examination of enhancer-promoter pairs within the ensemble of OCR pairs, we discerned potential interactions crucial for regulatory difference. Simultaneously, using TOBIAS, we explored TF binding activity in the selected gene local region by applying ATAC-seq footprinting analyses to identify protein binding regions across the genome. Following that, we utilized knowledge of accessible peaks’ cis-regulatory activity and gene local region TF binding activity to construct a TF regulatory network for selected target genes using R package igraph (v2.0.1.9005). This approach combines co-accessibility from Cicero and footprinting from TOBIAS, providing a nuanced perspective on the regulatory landscape. The resultant TF regulatory network offers a multifaceted depiction of the interplay between TFs, enhancers, and promoters, enhancing our ability to decipher the intricacies of gene regulation in the context of PiD and AD.
Supplementary Material
ACKNOWLEDGEMENTS
Funding for this work was provided by National Institutes on Aging, Neurological Disorders and Stroke, and Drug Abuse grants 1RF1AG071683, P01NS084974-06A1, 1U01DA053826, U54 AG054349-06 (MODEL-AD), and 3U19AG068054-02S, Adelson Medical Research Foundation funds to V.S, T32AG000096-38 to E.M., and the National Institute on Aging predoctoral fellowship 1F31AG076308-01 to S.M. We thank the International FTD-Genetics Consortium (IFGC) for summary data. We thank the UCI MIND’s ADRC and NIH NeuroBioBank for providing tissue. We thank the UCI Stem Cell Center and CRISPR Core for providing iPSC stem cells for CRISPR validation. We thank the UCI Genomics Research and Technology Hub for providing their facilities and sequencing our single-nucleus RNA-seq and ATAC-seq libraries. This work utilized the infrastructure for high-performance and high-throughput computing, research data storage and analyses, and scientific software tool integration built, operated, and updated by the Research Cyberinfrastructure Center (RCIC) at the University of California, Irvine (UCI).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing interests.
CODE AVAILABILITY
The data analyses code used for this study is available on GitHub at snMultiome_-PiDnAD. Single-cell Regulatory Occupancy Archive in Dementia scROAD. Code for the wrapper package designed to streamline information extraction to integrate data from Signac into Cicero and TOBIAS and perform analysis scCis-TF
DATA AVAILABILITY
All raw and processed single-nucleus ATAC, single-nucleus RNA and bulk RNA sequencing data have been deposited into the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database and will be made publicly available upon publication. All data in this manuscript have been deposited in the NIH GEO database. Data will be made available upon the acceptance of the paper.
Bibliography
- 1.Holtzman D. M., Morris J. C., and Goate A. M.. Alzheimer’s disease: the challenge of the second century. Sci Transl Med, 3(77):77sr1, 2011. ISSN 1946–6234 (Print) 1946–6234. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rascovsky K., Hodges J. R., Knopman D., Mendez M. F., Kramer J. H., Neuhaus J., van Swieten J. C., Seelaar H., Dopper E. G., Onyike C. U., et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain, 134:2456–77, 2011. ISSN 0006–8950 (Print) 0006–8950. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irwin D. J., Brettschneider J., McMillan C. T., Cooper F., Olm C., Arnold S. E., Van Deerlin V. M., Seeley W. W., Miller B. L., Lee E. B., et al. Deep clinical and neuropathological phenotyping of pick disease. Ann Neurol, 79(2):272–87, 2016. ISSN 0364–5134 (Print) 0364–5134. doi: 10.1002/ana.24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onyike Chiadi U. and Diehl-Schmid Janine. The epidemiology of frontotemporal dementia. International review of psychiatry (Abingdon, England), 25(2):130–137, 2013. ISSN 0954–0261. doi: 10.3109/09540261.2013.776523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickson D. W., Kouri N., Murray M. E., and Josephs K. A.. Neuropathology of frontotemporal lobar degeneration-tau (FTLD-tau). J Mol Neurosci, 45(3):384–9, 2011. ISSN 0895–8696 (Print) 0895–8696. doi: 10.1007/s12031-011-9589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyoshi E., Morabito S., and Swarup V.. Systems biology approaches to unravel the molecular and genetic architecture of Alzheimer’s disease and related tauopathies. Neurobiology of Disease, page 105530, 2021. ISSN 0969–9961. doi: 10.1016/j.nbd.2021.105530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert J. C., Ibrahim-Verbaas C. A., Harold D., Naj A. C., Sims R., Bellenguez C., DeStafano A. L., Bis J. C., Beecham G. W., Grenier-Boley B., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for alzheimer’s disease. Nat Genet, 45(12): 1452–8, 2013. ISSN 1061–4036 (Print) 1061–4036. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen I. E., Savage J. E., Watanabe K., Bryois J., Williams D. M., Steinberg S., Sealock J., Karlsson I. K., Hägg S., Athanasiu L., et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing alzheimer’s disease risk. Nat Genet, 51(3):404–413, 2019. ISSN 1061–4036 (Print) 1061–4036. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkle B. W., Grenier-Boley B., Sims R., Bis J. C., Damotte V., Naj A. C., Boland A., Vronskaya M., van der Lee S. J., Amlie-Wolf A., et al. Genetic meta-analysis of diagnosed alzheimer’s disease identifies new risk loci and implicates aβ, tau, immunity and lipid processing. Nat Genet, 51(3):414–430, 2019. ISSN 1061–4036 (Print) 1061–4036. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartzentruber J., Cooper S., Liu J. Z., Barrio-Hernandez I., Bello E., Kumasaka N., Young A. M. H., Franklin R. J. M., Johnson T., Estrada K., et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new alzheimer’s disease risk genes. Nat Genet, 53(3):392–402, 2021. ISSN 1061–4036 (Print) 1061–4036. doi: 10.1038/s41588-020-00776-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellenguez C., Küçükali F., Jansen I. E., Kleineidam L., Moreno-Grau S., Amin N., Naj A. C., Campos-Martin R., Grenier-Boley B., Andrade V., et al. New insights into the genetic etiology of alzheimer’s disease and related dementias. Nat Genet, 54(4):412–436, 2022. ISSN 1061–4036 (Print) 1061–4036. doi: 10.1038/s41588-022-01024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari R., Hernandez D. G., Nalls M. A., Rohrer J. D., Ramasamy A., Kwok J. B., Dobson-Stone C., Brooks W. S., Schofield P. R., Halliday G. M., et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol, 13(7):686–99, 2014. ISSN 1474–4422 (Print) 1474–4422. doi: 10.1016/s1474-4422(14)70065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visscher P. M., Wray N. R., Zhang Q., Sklar P., McCarthy M. I., Brown M. A., and Yang J.. 10 years of GWAS discovery: Biology, function, and translation. Am J Hum Genet, 101(1): 5–22, 2017. ISSN 0002–9297 (Print) 0002–9297. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boix C. A., James B. T., Park Y. P., Meuleman W., and Kellis M.. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature, 590(7845):300–307, 2021. ISSN 0028–0836. doi: 10.1038/s41586-020-03145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaulton K. J., Preissl S., and Ren B.. Interpreting non-coding disease-associated human variants using single-cell epigenomics. Nat Rev Genet, 24(8):516–534, 2023. ISSN 1471–0056. doi: 10.1038/s41576-023-00598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morabito S., Miyoshi E., Michael N., Shahin S., Martini A. C., Head E., Silva J., Leavy K., Perez-Rosendahl M., and Swarup V.. Single-nucleus chromatin accessibility and transcriptomic characterization of alzheimer’s disease. Nat Genet, 53(8):1143–1155, 2021. ISSN 1061–4036 (Print) 1061–4036. doi: 10.1038/s41588-021-00894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otero-Garcia M., Mahajani S. U., Wakhloo D., Tang W., Xue Y. Q., Morabito S., Pan J., Oberhauser J., Madira A. E., Shakouri T., et al. Molecular signatures underlying neurofibrillary tangle susceptibility in Alzheimer’s disease. Neuron, 2022. ISSN 0896–6273. doi: 10.1016/j.neuron.2022.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grubman A., Chew G., Ouyang J. F., Sun G., Choo X. Y., McLean C., Simmons R. K., Buckberry S., Vargas-Landin D. B., Poppe D., et al. A single-cell atlas of entorhinal cortex from individuals with alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat Neurosci, 22(12):2087–2097, 2019. ISSN 1097–6256. doi: 10.1038/s41593-019-0539-4. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y., Song W. M., Andhey P. S., Swain A., Levy T., Miller K. R., Poliani P. L., Cominelli M., Grover S., Gilfillan S., et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in alzheimer’s disease. Nat Med, 26(1):131–142, 2020. ISSN 1078–8956 (Print) 1078–8956. doi: 10.1038/s41591-019-0695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau S. F., Cao H., Fu A. K. Y., and Ip N. Y.. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in alzheimer’s disease. Proc Natl Acad Sci U S A, 117(41):25800–25809, 2020. ISSN 0027–8424 (Print) 0027–8424. doi: 10.1073/pnas.2008762117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leng K., Li E., Eser R., Piergies A., Sit R., Tan M., Neff N., Li S. H., Rodriguez R. D., Suemoto C. K., et al. Molecular characterization of selectively vulnerable neurons in alzheimer’s disease. Nat Neurosci, 24(2):276–287, 2021. ISSN 1097–6256 (Print) 1097–6256. doi: 10.1038/s41593-020-00764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang A. C., Vest R. T., Kern F., Lee D. P., Agam M., Maat C. A., Losada P. M., Chen M. B., Schaum N., Khoury N., et al. A human brain vascular atlas reveals diverse mediators of alzheimer’s risk. Nature, 603(7903):885–892, 2022. ISSN 0028–0836 (Print) 0028–0836. doi: 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathys H., Peng Z., Boix C. A., Victor M. B., Leary N., Babu S., Abdelhady G., Jiang X., Ng A. P., Ghafari K., et al. Single-cell atlas reveals correlates of high cognitive function, dementia, and resilience to alzheimer’s disease pathology. Cell, 186(20):4365–4385.e27, 2023. ISSN 0092–8674. doi: 10.1016/j.cell.2023.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gazestani V., Kamath T., Nadaf N. M., Dougalis A., Burris S. J., Rooney B., Junkkari A., Vanderburg C., Pelkonen A., Gomez-Budia M., et al. Early alzheimer’s disease pathology in human cortex involves transient cell states. Cell, 186(20):4438–4453.e23, 2023. ISSN 0092–8674. doi: 10.1016/j.cell.2023.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun N., Victor M. B., Park Y. P., Xiong X., Scannail A. N., Leary N., Prosper S., Viswanathan S., Luna X., Boix C. A., et al. Human microglial state dynamics in alzheimer’s disease progression. Cell, 186(20):4386–4403.e29, 2023. ISSN 0092–8674. doi: 10.1016/j.cell.2023.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong X., James B. T., Boix C. A., Park Y. P., Galani K., Victor M. B., Sun N., Hou L., Ho L. L., Mantero J., et al. Epigenomic dissection of alzheimer’s disease pinpoints causal variants and reveals epigenome erosion. Cell, 186(20):4422–4437.e21, 2023. ISSN 0092–8674. doi: 10.1016/j.cell.2023.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morabito S., Reese F., Rahimzadeh N., Miyoshi E., and Swarup V.. hdWGCNA identifies co-expression networks in high-dimensional transcriptomics data. Cell Reports Methods, 3(6):100498, 2023. ISSN 2667–2375. doi: 10.1016/j.crmeth.2023.100498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habib N., McCabe C., Medina S., Varshavsky M., Kitsberg D., Dvir-Szternfeld R., Green G., Dionne D., Nguyen L., Marshall J. L., et al. Disease-associated astrocytes in alzheimer’s disease and aging. Nat Neurosci, 23(6):701–706, 2020. ISSN 1097–6256 (Print) 1097–6256. doi: 10.1038/s41593-020-0624-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granja J. M., Corces M. R., Pierce S. E., Bagdatli S. T., Choudhry H., Chang H. Y., and Greenleaf W. J.. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat Genet, 53(3):403–411, 2021. ISSN 1061–4036 (Print) 1061–4036. doi: 10.1038/s41588-021-00790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarropoulos I., Sepp M., Frömel R., Leiss K., Trost N., Leushkin E., Okonechnikov K., Joshi P., Giere P., Kutscher L. M., et al. Developmental and evolutionary dynamics of cis-regulatory elements in mouse cerebellar cells. Science, 373(6558):eabg4696, 2021. doi: 10.1126/science.abg4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pliner H. A., Packer J. S., McFaline-Figueroa J. L., Cusanovich D. A., Daza R. M., Aghamirzaie D., Srivatsan S., Qiu X., Jackson D., Minkina A., et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Molecular Cell, 71(5):858–871.e8, 2018. ISSN 1097–2765. doi: 10.1016/j.molcel.2018.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberson E. D., Hesse J. H., Rose K. D., Slama H., Johnson J. K., Yaffe K., Forman M. S., Miller C. A., Trojanowski J. Q., Kramer J. H., and Miller B. L.. Frontotemporal dementia progresses to death faster than alzheimer disease. Neurology, 65(5):719–725, 2005. doi: 10.1212/01.wnl.0000173837.82820.9f. [DOI] [PubMed] [Google Scholar]
- 33.Karczewski K. J., Francioli L. C., Tiao G., Cummings B. B., Alföldi J., Wang Q., Collins R. L., Laricchia K. M., Ganna A., Birnbaum D. P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581(7809):434–443, 2020. ISSN 1476–4687. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J. Z., Menon M., He L., Abdurrob F., Jiang X., et al. Single-cell transcriptomic analysis of alzheimer’s disease. Nature, 570(7761):332–337, 2019. ISSN 0028–0836 (Print) 0028–0836. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leng F. and Edison P.. Neuroinflammation and microglial activation in alzheimer disease: where do we go from here? Nature Reviews Neurology, 17(3):157–172, 2021. ISSN 1759–4766. doi: 10.1038/s41582-020-00435-y. [DOI] [PubMed] [Google Scholar]
- 36.Kuhn P., Wang H., Dislich B., Colombo A., Zeitschel U., Ellwart J. W., Kremmer E., Roßner S., and Lichtenthaler S. F.. ADAM10 is the physiologically relevant, constitutive α-secretase of the amyloid precursor protein in primary neurons. The EMBO Journal, 29(17):3020–3032, 2010. ISSN 0261–4189. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voskobiynyk Y., Roth J. R., Cochran J. N., Rush T., Carullo N. V., Mesina J. S., Waqas M., Vollmer R. M., Day J. J., McMahon L.L., and Roberson E.D.. Alzheimer’s disease risk gene BIN1 induces tau-dependent network hyperexcitability. eLife, 9:e57354, 2020. ISSN 2050–084X. doi: 10.7554/eLife.57354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert E., Saha O., Soares Landeira B., Melo de Farias A. R., Hermant X., Carrier A., Pelletier A., Gadaut J., Davoine L., Dupont C., et al. The alzheimer susceptibility gene BIN1 induces isoform-dependent neurotoxicity through early endosome defects. Acta Neuropathologica Communications, 10(1):4, 2022. ISSN 2051–5960. doi: 10.1186/s40478-021-01285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nott A., Holtman I. R., Coufal N. G., Schlachetzki J. C. M., Yu M., Hu R., Han C. Z., Pena M., Xiao J., Wu Y., et al. Brain cell type–specific enhancer–promoter interactome maps and disease-risk association. Science, 366(6469):1134–1139, 2019. doi: 10.1126/science.aay0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colonna M. and Wang Y.. TREM2 variants: new keys to decipher alzheimer disease pathogenesis. Nature Reviews. Neuroscience, 17(4):201–207, 2016. ISSN 1471–0048. doi: 10.1038/nrn.2016.7. [DOI] [PubMed] [Google Scholar]
- 41.Tran K. M., Kawauchi S., Kramár E. A., Rezaie N., Liang H. Y., Sakr J. S., Gomez-Arboledas A., Arreola M. A., Cunha C., Phan J., et al. A trem2r47h mouse model without cryptic splicing drives age- and disease-dependent tissue damage and synaptic loss in response to plaques. Molecular Neurodegeneration, 18(1):12, 2023. ISSN 1750–1326. doi: 10.1186/s13024-023-00598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyoshi E., Morabito S., Henningfield C. M., Rahimzadeh N., Kiani Shabestari S., Das S., Michael N., Reese F., Shi Z., Cao Z., et al. Spatial and single-nucleus transcriptomic analysis of genetic and sporadic forms of alzheimer’s disease. bioRxiv: The Preprint Server for Biology, page 2023.07.24.550282, 2023. doi: 10.1101/2023.07.24.550282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flannick J., Thorleifsson G., Beer N. L., Jacobs S. B., Grarup N., Burtt N. P., Mahajan A., Fuchsberger C., Atzmon G., Benediktsson R., et al. Loss-of-function mutations in SLC30a8 protect against type 2 diabetes. Nature genetics, 46(4):357–363, 2014. ISSN 1061–4036. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cholerton L. D., nd Baker B., Montine T. J., and Craft S.. Type 2 diabetes, cognition, and dementia in older adults: Toward a precision health approach. Diabetes Spectrum: A Publication of the American Diabetes Association, 29(4):210–219, 2016. ISSN 1040–9165. doi: 10.2337/ds16-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maluenda J., Manso C., Quevarec L., Vivanti A., Marguet F., Gonzales M., Guimiot F., Petit F., Toutain A., Whalen S., et al. Mutations in GLDN, encoding gliomedin, a critical component of the nodes of ranvier, are responsible for lethal arthrogryposis. American Journal of Human Genetics, 99(4):928–933, 2016. ISSN 0002–9297. doi: 10.1016/j.ajhg.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arancibia-Carcamo I. L. and Attwell D.. The node of ranvier in CNS pathology. Acta Neuropathologica, 128(2):161–175, 2014. ISSN 0001–6322. doi: 10.1007/s00401-014-1305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bentsen M., Goymann P., Schultheis H., Klee K., Petrova A., Wiegandt R., Fust A., Preussner J., Kuenne C., Braun T., et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun, 11(1):4267, 2020. ISSN 2041–1723. doi: 10.1038/s41467-020-18035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dennis D. J., Han S., and Schuurmans C.. bHLH transcription factors in neural development, disease, and reprogramming. Brain Research, 1705:48–65, 2019. ISSN 0006–8993. doi: 10.1016/j.brainres.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 49.Ku C. C., Wuputra K., Kato K., Lin W. H., Pan J. B., Tsai S. C., Kuo C. J., Lee K. H., Lee Y. L., Lin Y. C., et al. Jdp2-deficient granule cell progenitors in the cerebellum are resistant to ROS-mediated apoptosis through xCT/slc7a11 activation. Scientific Reports, 10:4933, 2020. ISSN 2045–2322. doi: 10.1038/s41598-020-61692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holwerda S. J. B. and de Laat W.. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philosophical Transactions of the Royal Society B: Biological Sciences, 368(1620):20120369, 2013. ISSN 0962–8436. doi: 10.1098/rstb.2012.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L., Bai J., Liu F., Xu Y., Zhao M., Zhao C., and Zhou Z.. Cross-talking pathways of forkhead box o1 (FOXO1) are involved in the pathogenesis of alzheimer’s disease and huntington’s disease. Oxidative Medicine and Cellular Longevity, 2022:7619255, 2022. ISSN 1942–0900. doi: 10.1155/2022/7619255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cancio-Bello A. and Saez-Atienzar S.. SATB1 is a dopaminergic neuron specific regulator of cellular senescence. Movement disorders : official journal of the Movement Disorder Society, 35(2):235, 2020. ISSN 0885–3185. doi: 10.1002/mds.27978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hossini A. M., Megges M., Prigione A., Lichtner B., Toliat M. R., Wruck W., Schröter F., Nuernberg P., Kroll H., Makrantonaki E., et al. Induced pluripotent stem cell-derived neuronal cells from a sporadic alzheimer’s disease donor as a model for investigating AD-associated gene regulatory networks. BMC Genomics, 16(1):84, 2015. ISSN 1471–2164. doi: 10.1186/s12864-015-1262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez-Colman M. J., Dansen T. B., and Burgering B. M. T.. FOXO transcription factors as mediators of stress adaptation. Nature Reviews Molecular Cell Biology, 25(1):46–64, 2024. ISSN 1471–0080. doi: 10.1038/s41580-023-00649-0. [DOI] [PubMed] [Google Scholar]
- 55.Majumder P., Chanda K., Das D., Singh B. K., Chakrabarti P., Jana N. R., and Mukhopadhyay D.. A nexus of miR-1271, PAX4 and ALK/RYK influences the cytoskeletal architectures in alzheimer’s disease and type 2 diabetes. The Biochemical Journal, 478(17):3297–3317, 2021-09-17. ISSN 1470–8728. doi: 10.1042/BCJ20210175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H., Liu H., Lutz M. W., and Luo S.. Novel genetic variants in TP37, PIK3r1, CALM1, and PLCG2 of the neurotrophin signaling pathway are associated with the progression from mild cognitive impairment to alzheimer’s disease. Journal of Alzheimer’s disease : JAD, 91(3): 977–987, 2023. ISSN 1387–2877. doi: 10.3233/JAD-220680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu A., Fernandes B. S., Citu C., and Zhao Z.. Unraveling the intercellular communication disruption and key pathways in alzheimer’s disease: an integrative study of single-nucleus transcriptomes and genetic association. Alzheimer’s Research & Therapy, 16(1):3, 2024. ISSN 1758–9193. doi: 10.1186/s13195-023-01372-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiß A. L., Meijer M., Budeus B., Pauper M., Hakobjan M., Groothuismink J., Shi Y., Neveling K., Buitelaar J. K., Hoogman M., et al. DNA methylation associated with persistent ADHD suggests TARBP1 as novel candidate. Neuropharmacology, 184:108370, 2021. ISSN 0028–3908. doi: 10.1016/j.neuropharm.2020.108370. [DOI] [PubMed] [Google Scholar]
- 59.Dharshini S. A. P., Taguchi Y. H., and Gromiha M. M.. Investigating the energy crisis in alzheimer disease using transcriptome study. Scientific Reports, 9(1):18509, 2019. ISSN 2045–2322. doi: 10.1038/s41598-019-54782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song L., Chen J., Lo Chun-Yi Z., Guo Q., Feng J., and Zhao X.. Impaired type i interferon signaling activity implicated in the peripheral blood transcriptome of preclinical alzheimer’s disease. eBioMedicine, 82:104175, 2022. ISSN 2352–3964. doi: 10.1016/j.ebiom.2022.104175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith A. M., Gibbons H. M., Oldfield R. L., Bergin P. M., Mee E. W., Faull R. L., and Dragunow M.. The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia, 61(6):929–942, 2013. ISSN 1098–1136. doi: 10.1002/glia.22486. [DOI] [PubMed] [Google Scholar]
- 62.Tamir A., Howard J., Higgins R. R., Li Y. J., Berger L., Zacksenhaus E., Reis M., and Ben-David Y.. Fli-1, an ets-related transcription factor, regulates erythropoietin-induced erythroid proliferation and differentiation: Evidence for direct transcriptional repression of the rb gene during differentiation. Molecular and Cellular Biology, 19(6):4452–4464, 1999. ISSN 0270–7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishii S., Kakizuka T., Park S. J., Tagawa A., Sanbo C., Tanabe H., Ohkawa Y., Nakanishi M., Nakai K., and Miyanari Y.. Genome-wide ATAC-see screening identifies TFDP1 as a modulator of global chromatin accessibility. Nature Genetics, 56(3):473–482, 2024. ISSN 1546–1718. doi: 10.1038/s41588-024-01658-1. [DOI] [PubMed] [Google Scholar]
- 64.Pawelec P., Ziemka-Nalecz M., Sypecka J., and Zalewska T.. The impact of the CX3cl1/CX3cr1 axis in neurological disorders. Cells, 9(10):2277, 2020. ISSN 2073–4409. doi: 10.3390/cells9102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith A. M., Davey K., Tsartsalis S., Khozoie C., Fancy N., Tang S. S., Liaptsi E., Weinert M., McGarry A., Muirhead R. C. J., et al. Diverse human astrocyte and microglial transcriptional responses to alzheimer’s pathology. Acta Neuropathologica, 143(1):75–91, 2022. ISSN 0001–6322. doi: 10.1007/s00401-021-02372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X., Guo J., Wei X., Niu C., Jia M., Li Q., and Meng D.. Bach1: Function, regulation, and involvement in disease. Oxidative Medicine and Cellular Longevity, 2018:1347969, 2018. ISSN 1942–0900. doi: 10.1155/2018/1347969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vangeison G., Carr D., Federoff H. J., and Rempe D. A.. The good, the bad, and the cell type-specific roles of hypoxia inducible factor-1α in neurons and astrocytes. Journal of Neuroscience, 28(8):1988–1993, 2008. ISSN 0270–6474, 1529–2401. doi: 10.1523/JNEUROSCI.5323-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kageyama R., Ohtsuka T., and Kobayashi T.. The hes gene family: repressors and oscillators that orchestrate embryogenesis. Development, 134(7):1243–1251, 2007. ISSN 0950–1991. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 69.Kulikov Al. V., Korostina V. S., Kulikova E. A., Fursenko D. V., Akulov An. E., Moshkin M. P., and Prokhortchouk E. B.. Knockout zbtb33 gene results in an increased locomotion, exploration and pre-pulse inhibition in mice. Behavioural Brain Research, 297:76–83, 2016. ISSN 1872–7549. doi: 10.1016/j.bbr.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 70.Hwang I., Oh H., Santo E., Kim D. Y., Chen J. W., Bronson R. T., Locasale J. W., Na Y., Lee J., Reed S., et al. FOXO protects against age-progressive axonal degeneration. Aging Cell, 17(1):e12701, 2018. ISSN 1474–9726. doi: 10.1111/acel.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Merluzzi A. P., Carlsson C. M., Johnson S. C., Schindler S. E., Asthana S., Blennow K., Zetterberg H., and Bendlin B. B.. Neurodegeneration, synaptic dysfunction, and gliosis are phenotypic of alzheimer dementia. Neurology, 91(5):e436–e443, 2018. ISSN 0028–3878. doi: 10.1212/WNL.0000000000005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nordengen K., Kirsebom B. E., Henjum K., Selnes P., Gísladóttir B., Wettergreen M., Torsetnes S. B., Grøntvedt G. R., Waterloo K. K., Aarsland D., et al. Glial activation and inflammation along the alzheimer’s disease continuum. Journal of Neuroinflammation, 16(1):46, 2019. ISSN 1742–2094. doi: 10.1186/s12974-019-1399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reilly S. K., Yin J., Ayoub A. E., Emera D., Leng J., Cotney J., Sarro R., Rakic P., and Noonan J. P.. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science, 347(6226):1155–1159, 2015. doi: 10.1126/science.1260943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simchi L., Panov J., Morsy O., Feuermann Y., and Kaphzan H.. Novel insights into the role of UBE3a in regulating apoptosis and proliferation. Journal of Clinical Medicine, 9(5):1573, 2020. ISSN 2077–0383. doi: 10.3390/jcm9051573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kishino T., Lalande M., and Wagstaff J.. UBE3a/e6-AP mutations cause angelman syndrome. Nature Genetics, 15(1):70–73, 1997. ISSN 1546–1718. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 76.Xing L., Simon J. M., Ptacek T. S., Yi J. J., Loo L., Mao H., Wolter J. M., Mc-Coy E. S., Paranjape S. R., Taylor-Blake B., and Zylka M. J.. Autism-linked UBE3a gain-of-function mutation causes interneuron and behavioral phenotypes when inherited maternally or paternally in mice. Cell Reports, 42(7):112706, 2023. ISSN 22111247. doi: 10.1016/j.celrep.2023.112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de la Torre-Ubieta L., Stein J. L., Won H., Opland C. K., Liang D., Lu D., and Geschwind D. H.. The dynamic landscape of open chromatin during human cortical neurogenesis. Cell, 172(1):289–304.e18, 2018. ISSN 0092–8674, 1097–4172. doi: 10.1016/j.cell.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi Z., Das S., Morabito S., Miyoshi E., and Swarup V.. Protocol for single-nucleus ATAC sequencing and bioinformatic analysis in frozen human brain tissue. STAR Protocols, 3(3): 101491, 2022. ISSN 2666–1667. doi: 10.1016/j.xpro.2022.101491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pantazis C. B., Yang A., Lara E., McDonough J. A., Blauwendraat C., Peng L., Oguro H., Kanaujiya J., Zou J., Sebesta D., et al. A reference human induced pluripotent stem cell line for large-scale collaborative studies. Cell stem cell, 29(12):1685–1702.e22, 2022. ISSN 1934–5909. doi: 10.1016/j.stem.2022.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y., Tristan C. A., Chen L., Jovanovic V. M., Malley C., Chu P. H., Ryu S., Deng T., Ormanoglu P., Tao D., et al. A versatile polypharmacology platform promotes cytoprotection and viability of human pluripotent and differentiated cells. Nature Methods, 18(5):528–541, 2021. ISSN 1548–7105. doi: 10.1038/s41592-021-01126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stuart T., Srivastava A., Madad S., Lareau C. A., and Satija R.. Single-cell chromatin state analysis with signac. Nature Methods, 18(11):1333–1341, 2021. ISSN 1548–7105. doi: 10.1038/s41592-021-01282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gu Z., Eils R., and Schlesner M.. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics, 32(18):2847–2849, 2016. ISSN 1367–4803. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 83.McLean C. Y., Bristor D., Hiller M., Clarke S. L., T Schaar B., Lowe C. B., Wenger A. M., and Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nature biotechnology, 28(5):495–501, 2010. ISSN 1087–0156. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gu Z. and Hübschmann D.. rGREAT: an r/bioconductor package for functional enrichment on genomic regions. Bioinformatics, 39(1):btac745, 2023. ISSN 1367–4811. doi: 10.1093/bioinformatics/btac745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Skene N. G., Bryois J., Bakken T. E., Breen G., Crowley J. J., Gaspar H. A., Giusti-Rodriguez P., Hodge R. D., Miller J. A., Muñoz-Manchado A. B., et al. Genetic identification of brain cell types underlying schizophrenia. Nature Genetics, 50(6):825–833, 2018. ISSN 1546–1718. doi: 10.1038/s41588-018-0129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., Bender D., Maller J., Sklar P., de Bakker P. I. W., Daly M. J., and Sham P. C.. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics, 81(3):559–575, 2007. ISSN 0002–9297. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang G., Sarkar A., Carbonetto P., and Stephens M.. A simple new approach to variable selection in regression, with application to genetic fine mapping. Journal of the Royal Statistical Society Series B: Statistical Methodology, 82(5):1273–1300, 2020. ISSN 1369–7412. doi: 10.1111/rssb.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zou Y., Carbonetto P., Wang G., and Stephens M.. Fine-mapping from summary data with the “sum of single effects” model. PLOS Genetics, 18(7):e1010299, 2022. ISSN 1553–7404. doi: 10.1371/journal.pgen.1010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed single-nucleus ATAC, single-nucleus RNA and bulk RNA sequencing data have been deposited into the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database and will be made publicly available upon publication. All data in this manuscript have been deposited in the NIH GEO database. Data will be made available upon the acceptance of the paper.