Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) is associated with three human tumors, Kaposi's sarcoma, primary effusion lymphoma (PEL), and multicentric Castleman's disease. KSHV encodes a number of homologs of cellular proteins involved in the cell cycle, signal transduction, and modulation of the host immune response. Of the virus complement of over 85 open reading frames (ORFs), the expression of only a minority has been characterized individually. We have constructed a nylon membrane-based DNA array which allows the expression of almost every ORF of KSHV to be measured simultaneously. A PEL-derived cell line, BC-3, was used to study the expression of KSHV during latency and after the induction of lytic replication. Cluster analysis, which arranges genes according to their expression profile, revealed a correlation between expression and assigned gene function that is consistent with the known stages of the herpesvirus life cycle. Furthermore, latent and lytic genes thought to be functionally related cluster into groups. The correlation between gene expression and function also infers possible roles for KSHV genes yet to be characterized.
KSHV (Kaposi's sarcoma-associated herpesvirus) is the eighth and most recently identified human herpesvirus (10). Many studies have causally linked KSHV to the pathogenesis of Kaposi's sarcoma (KS) (reviewed in reference 41). KSHV is also specifically associated with primary effusion lymphoma (PEL) (7) and a plasmablastic form of multicentric Castleman's disease (16, 42). KSHV is predicted to encode at least 85 open reading frames (ORFs) which include a number of homologs of cellular genes (30, 35). These cellular homologs may determine the pathogenicity of KSHV (32).
Like all herpesviruses, KSHV establishes a latent infection in cells, persisting as episomal DNA (13). During latency, viral gene expression is restricted to only a few genes (37, 43, 49). These latent genes are thought to maintain the viral episome (3), avoid antiviral host immune responses (15), and provide a growth advantage to infected cells (17, 20, 33). The full repertoire of viral gene expression occurs only during lytic replication, when virus progeny are produced and the host cell is destroyed (34). The study of KSHV has benefited from the establishment of cell lines from PEL (2, 8, 34). These cells harbor the virus in a latent form and can be induced to enter lytic replication by treatment with sodium butyrate or the phorbol ester 12-O-tetradecoylphorbol 13-acetate (TPA) (29, 34).
A number of studies have described KSHV gene expression in latency and during lytic replication (37, 44, 49, 50). KSHV transcripts have been categorized into three classes based on their expression in uninduced and lytically induced PEL (BC-1) cells: class I (constitutive), class II (present in uninduced cells but upregulated with TPA), and class III (only present after induction) (37). However, only one time point after lytic induction was studied, and large probes that were not specific for individual genes were used. The expression pattern of most KSHV genes therefore remains largely unknown.
DNA arrays provide a means to measure the expression of hundreds or thousands of genes simultaneously as well as allowing high-throughput characterization of samples (reviewed in reference 26). Comparison of mRNA populations of cells with a known phenotype under known conditions enables patterns of gene expression to be linked to the status of cell processes and events. The vast amount of data generated with these techniques have dictated the development of mathematical techniques to address the problem of analysis. Clustering algorithms that arrange data based on the coexpression of genes (18) are able to group together genes which have common roles in a cellular process or by the cell type in which they are expressed (1, 12). This method also accurately distinguishes samples from different sources solely by their patterns of gene expression (1). DNA arrays have recently been applied to studies of viral infection, including that of human cytomegalovirus (9, 21, 51). Herpesviruses make ideal candidates for DNA array technology, as every viral gene can be printed onto a single array. Therefore, the complete gene expression profile (transcriptome) of an entire genome can be elucidated from the results of one experiment.
Here, we report on the construction of a DNA array that allows the measurement of the expression level of almost every known KSHV ORF. We studied the expression of the viral genome during latency and during TPA-induced lytic replication in a PEL cell line and used cluster analysis to arrange genes according to their expression profile. This method groups together genes which may be involved in a common process and arranges genes in a temporal order consistent with the known stages of herpesvirus replication. This revealed that K10 has an expression profile similar to that of T0.7 RNA, a known latent transcript. The correlation between gene expression and function also suggests possible roles for genes that have yet to be characterized. We have made an initial demonstration of the feasibility of this approach by the discovery of a novel transcribed ORF (K10.7) with homology to known interferon regulatory factors (IRFs). Reverse transcription (RT)-PCR confirms array data suggesting that KSHV encodes four full-length IRF-related proteins.
MATERIALS AND METHODS
Array elements.
Approximately 350 bp of DNA sequence was amplified from the 5′ end of known KSHV ORFs. Similarly, primers were chosen to amplify sequences from cellular genes and nonhuman/nonviral sequences which were used as positive and negative hybridization controls. The genes chosen were those encoding ubiquitin (M26880), tyrosine 3-monooxygenase activation protein (NM_003404), pyrophosphate phosphoribosyltransferase (V00530), glyceraldehyde 3-phosphate dehydrogenase (X01677), α-tubulin (K00558), major histocompatibility complex (MHC) class I HLA-B (X75953), β-actin (X00351), highly basic protein (X56932), ribosomal protein S9 (U14971), luciferase (E15166), and tobacco mosaic virus 180-kDa protein (D78608). The primers used are listed at www.biochem.ucl.ac.uk/bsm/virus_database/KSHVarray.html. Each array element was cloned into either pBluescript (Stratagene) or pGEM-T Easy (Promega), and the sequence was verified (Beckman). Common vector primers were designed to amplify the cloned array elements. The PCR products were purified using a Qiagen 96-well PCR purification system and concentrated by ethanol precipitation. Finally, the DNA was resuspended in water at 400 ng/μl, and approximately 36 ng of DNA was spotted in duplicate on Hybond-N membrane (Amersham) using a high-density gridder (Eurogentec). The DNA on the arrays was denatured (in 0.66 M NaCl–0.5 M NaOH), neutralized (in 40 mM phosphate buffer, pH 7.3), and then UV cross-linked to the membrane.
Quality control of DNA deposition.
The quality of array element spotting was determined by the hybridization of 33P-labeled oligonucleotide probes specific for common primer sequences present at the ends of the PCR products. Oligonucleotide probes were end labeled with [γ-33P]ATP (ICN) using T4 polynucleotide kinase (Promega). Labeled probes were separated from unincorporated label by ethanol precipitation with lithium chloride. The arrays were preincubated for 30 min in ExpressHyb hybridization solution (Clontech) with denatured salmon sperm DNA (100 μg/ml; Roche) at 42°C. The labeled oligonucleotides were hybridized to the arrays for 18 h at 42°C in 5 ml of ExpressHyb with salmon sperm DNA (100 μg/ml). The arrays were washed twice at 50°C in 2× SSC (0.3 M NaCl plus 0.03 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS), with a final wash in 0.1× SSC–0.1% SDS. Bound probe was detected using a phosphor screen (Molecular Dynamics), and the signals were quantitated by phosphorimaging using ArrayVision software (Imaging Research). For quantitation, the local background was subtracted from each array element. Only complete, evenly spotted arrays were used in the subsequent experiments. The oligonucleotides were stripped from the arrays by the addition of boiling 0.5% SDS, which was allowed to cool to 22°C with agitation. Stripping efficiency was assessed by phosphorimaging. The Spearman rank order correlation of probe bound to each array element for each array compared to all others was found using the program Statistica (Statsoft).
Cell culture.
The cell line BC-3 (2) was grown in RPMI 1640 medium (Gibco BRL) supplemented with 10% fetal calf serum, penicillin and streptomycin (100 U/μl), and ciprofloxacin (40 μg/ml). The culture was maintained between approximately 105 and 106 cells per ml. The herpesvirus-negative Burkitt's lymphoma cell line Ramos (24) was grown under the same conditions.
Induction of viral replication and RNA purification.
Before induction of virus replication, dead cells were removed from the cultures by density centrifugation through Lymphoprep (Nycomed). Cells were then resuspended in fresh medium, and viral replication was induced by the addition of TPA (Sigma) at 20 ng/μl. BC-3 cells were harvested after 0, 2, 4, 10, 24, 34, 48, and 72 h, pelleted, and washed once with phosphate-buffered saline. The cell pellet was frozen immediately at −80°C or lysed in RLT lysis buffer (Qiagen) before freezing. As a control, the same procedure was followed for BC-3 cells but without the addition of TPA. RNA was purified using Qiagen's RNeasy kit and quantitated by UV spectrophotometry. RNA quality was assessed by denaturing agarose gel electrophoresis. The RNA was DNase I treated (Promega) and repurified by phenol extraction and ethanol precipitation.
cDNA synthesis and array labeling.
Between 4 and 24 μg of total RNA was used in each RT reaction. The cDNA synthesis was primed using a pool of gene- and sense-strand-specific 3′ primers (0.2 μmol). [α-33P]dATP-labeled cDNA was prepared using a Strip-EZ RT kit (Ambion). cDNA was separated from unincorporated nucleotides using Microspin SR-400 columns (Amersham). The labeled probe was denatured by incubation in 0.1 M NaOH–0.1 mM EDTA at 68°C and then neutralized with 1 M NaH2PO4. Human Cot-1 DNA (Gibco BRL) was added to the probe to suppress cross-hybridization to repetitive DNA. The prehybridization and hybridization steps as described above except for a higher hybridization temperature of 64°C. Washes as described above were performed at 65°C. The signal was quantitated as before. Labeled cDNA probe was stripped from the arrays using the Strip-EZ system (Ambion), and the process was checked by phosphorimaging. Arrays were stored at 4°C on filter paper (Whatman) saturated with 0.1× SSC–0.1% SDS and were rinsed in 0.1× SSC–0.1% SDS at 68°C before being used again.
Data processing and cluster analysis.
Local background for each array element was subtracted, and the mean signal from the duplicate spots was calculated. This gives one data point for each viral gene and two for each cellular gene to assess hybridization consistency. The complete data set can be viewed at www.biochem.ucl.ac.uk/bsm/virus_database/KSHVarray.html. The mean uninduced expression values were calculated for each viral gene across all uninduced samples, and from this a median value was generated. This value was then doubled and used as the divisor to transform the data from absolute expression values to ratios. The data set was then converted to log base 2, and before the induced samples were clustered, genes were normalized across the experimental data set (the sum of the squares set to 1) to equalize the magnitude of the expression vectors (18). To control for differences in amounts of RNA and the age of isotope in each experiment, the arrays were mean centered using the expression of a subset of the cellular genes (those encoding glyceraldehyde 3-phosphate dehydrogenase, α-tubulin, and ribosomal protein S9) which were deemed nonchanging by their covariance across all arrays. The covarying housekeeping genes were determined by generating a Pearson correlation matrix for all the cellular genes using Statistica (Statsoft). The mean centered data set was then imported into the program Cluster (18), and the genes were ordered using a self-organizing map algorithm (12) (the number of nodes was set to √n). The genes and arrays were clustered by average linkage hierarchical clustering using the uncentered Pearson correlation as the similarity metric. To cluster the arrays, each gene was weighted based on the local density of row vectors in its vicinity. The results were visualized with the software TreeView (18). All viral genes were clustered except for ORFs 35, 50, 60, 68 and K15 exon 8 due to their cross-reaction with Ramos cell RNA and ORF 33, whose expression could not be detected.
RT-PCR.
RNA was prepared from BC-3 cells as for array analysis. cDNA was synthesized from 5 μg of total RNA primed with oligo(dT) primers (Stratagene), using SuperScript II (Gibco BRL) according to the manufacturer's instructions. Genomic DNA was purified from BC-3 cells using Qiagen's QIAamp DNA Mini kit. PCR was performed on 1/10 volume of cDNA using Taq polymerase (Roche) or the Expand Long Template PCR system (Roche). The primers used were as follows: for K9, ATGGACCCAGGCCAAAGACC and TTATTGCATGGCATCCCATAACG; for K10/10.1, ATGCCTAAAGCCGGTGGCTC and TCAATGTAGACTATCCCAAATGG; for K10.5/10.7, ATGGCGGGACGCAGGCTTAC and TTAGTCATCACATGTAACTGAACG; for K11/vIRF-2, ATGCCTCGCTACACGGAGTC and TTAGTCTCTGTGGTAAAATGGG; and for K10.1 splice (see Fig. 5D), GGACATTTGTCAAAGGAGCTA and CAAATGTGTCGCTGTACCGT. Splice sites were predicted and verified by software at the Berkeley Drosophila Genome Project (www.fruitfly.org/seq_tools/splice.html).
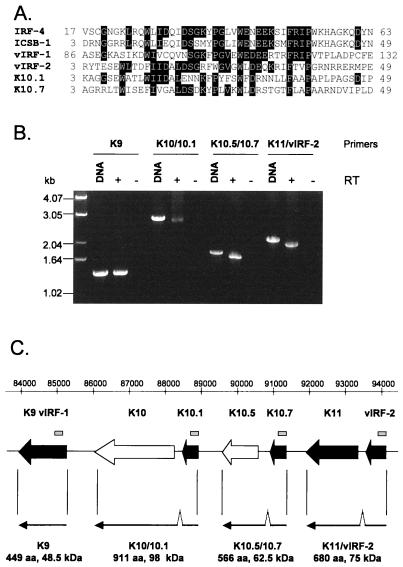
FIG. 5.
(A) Alignment of DNA binding domains of the human IRFs IRF-4 (GI:2497445) and ICSB-1 (GI:4504567) with those of vIRF-1 (GI:1718311), vIRF-2 (GI:3152729), K10.1, and a novel putative ORF, K10.7. The amino acid sequences for K10.1 and K10.7 were translated from the genomic sequence of KSHV (U75698). The sequence shown for each protein is bounded by the amino acid positions shown on either side. The alignment was performed with ClustalW (46). (B) RT-PCR products showing transcripts encoding vIRF-1 and the putative ORFs K10/10.1, K10.5/10.7, and K11/vIRF-2. The primers used are complementary to either end of the putative long ORFs (see Materials and Methods). RNA was extracted from BC-3 cells 24 h after the addition of TPA. RT-negative controls and positive controls from KSHV genomic DNA are included for each RT-PCR. The RT-PCR product sizes are predicted to be 1,350 (K9), 2,736 (K10/10.1), 1,701 (K10.5/10.7), and 2,043 (K11/vIRF-2) bp. (C) Positions (relative to genomic sequence U75698) and organization of the IRF-related genes of KSHV. The location of the novel putative ORF K10.7 is shown. Secondary lytic genes are shaded in black. The region of each ORF whose translated sequence is shown in panel A is indicated by the grey bars above. Sequenced transcripts shown in panel B are drawn below the corresponding ORFs. Each transcript encodes a predicted protein with full-length homology to known IRFs. The predicted size of the encoded protein is indicated below each transcript. Introns are located between 88343 and 88443 (K10/10.1), 90846 and 90939 (K10.5/10.7), and 93519 and 93639 (K11/vIRF-2). (D) Sequenced transcripts encoding K10/10.1 and K10. The positions of start and stop codons and introns are shown relative to U75698. RT-PCR products corresponding to the two transcripts are shown on the right. The primers used (see Materials and Methods) are labeled (small arrows). RNA templates was taken from BC-3 cells 0 (latent) and 24 (lytic) h after the addition of TPA. The PCR product from KSHV genomic DNA is shown for size comparison.
RESULTS
A DNA array for KSHV.
To fully represent the coding potential of the KSHV genome on the array, we designed a set of PCR primers to amplify sequences from 88 known ORFs and some KSHV gene-specific exons. Cloned array elements were checked for the possibility of cross-hybridization both against each other and against host genes by searching GenBank using pairwise sequence analysis. No array elements were found to have significant homology to sequences other than those they represent. The arrays consist of a total of 288 separate elements spotted onto nylon membranes. Every KSHV PCR product was spotted in duplicate, and every cellular PCR product was spotted in quadruplicate. The cellular genes were split between different parts of the array to assess the consistency of hybridization.
The spotting of every element on each array was checked by hybridization of 33P-labeled oligonucleotides complementary to the common PCR primers. Quantitation of these signals showed the spotting to be very consistent. Within each array, the average variation in the amount of bound probe detected between duplicate spots was measured at ±5%. Patterns of spotting between arrays were also found to be almost identical; the Pearson correlation coefficient of bound probe to array elements between different arrays was 0.93 or above (data not shown).
The arrays were used to examine virus gene expression during latency and lytic viral replication. Total RNA was extracted from BC-3 cells at different time points (0 to 72 h) with or without the addition of TPA, which induces lytic replication. To confirm the reproducibility of the arrays, we performed duplicate experiments representing 0, 24, 34, 48, and 72 h after induction. To control for the sensitivity of the arrays, luciferase RNA was added to the samples before labeling. This showed that RNA present at 1:100,000 (wt/wt), approximately equivalent to 10 copies per cell, could be detected at 10-fold above background. This sensitivity compares well with the known sensitivity of glass microarrays (40, 51). Tobacco mosaic virus elements acted as negative controls and on average gave a signal only 1.3-fold above background (standard deviation of 0.3). To further test the specificity of the arrays, we extracted RNA from Ramos cells, a KSHV-negative B-cell line, and hybridized it to an array. Almost all of the virus genes appeared negative, showing that the vast majority of the probes are specific for KSHV RNA. However, five elements, corresponding to ORFs 35, 50, 60, and 68 and K15 exon 8 exhibited cross-hybridization with Ramos cellular RNAs and therefore were not included in further analyses.
Gene expression analysis by KSHV arrays.
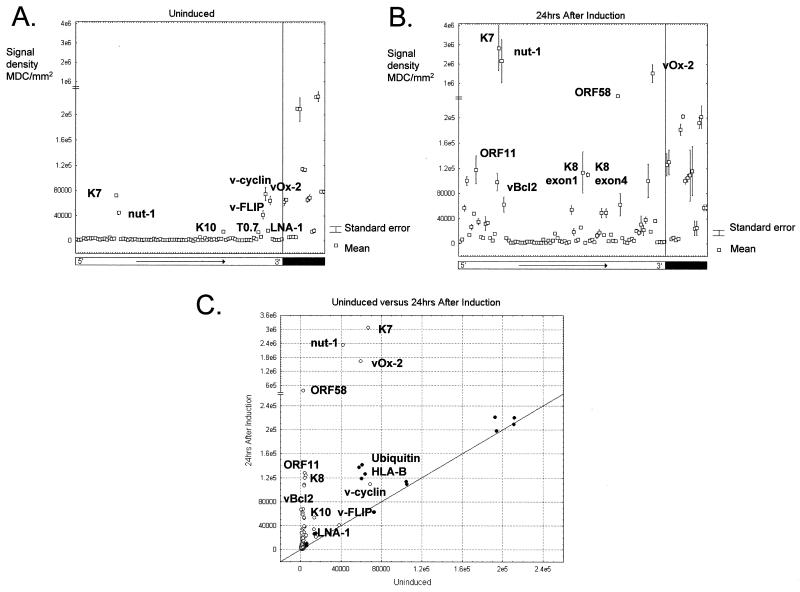
Examples of results from the arrays are shown in Fig. 1 and represent genes expressed during latency and 24 h after the induction of lytic replication with TPA. Both conditions are represented by the results from two independent experiments. The signal intensities are identical between the duplicate sets of cellular genes on the individual arrays, suggesting consistent hybridization results. Gene expression values (means and standard errors from two samples) for uninduced cells show that the genes fall into two main classes at these time points (Fig. 1A). The signal from the majority of array elements forms a baseline, while the expression of a few genes (those encoding v-FLIP [ORF 71], v-cyclin [ORF 72], LNA-1 [ORF 73], K7, T1.1 [nut-1], T0.7, K10, and vOx-2 [K14]) is detected at significantly higher levels. The small standard errors indicate that duplicate samples give identical gene expression patterns. Even genes expressed at low levels, such as the LNA-1 gene, are consistently and significantly detected above the baseline, as shown by the absence of significant error bars. Therefore, the arrays are accurate and highly reproducible.
FIG. 1.
(A) Quantitation of results from two independent uninduced samples. The data from the two arrays were normalized using a subset of the cellular genes. The signal variation between the two samples is shown by the error bar for each element which represents the mean value. KSHV genes are indicated by the open bar and are ordered colinearly with the genome from 5′ to 3′ as indicated. Cellular genes are indicated by a solid bar and are separated from viral genes by a vertical line. The signal from the majority of array elements forms a baseline, while the expression of a few genes is detected at levels significantly above this (labeled). MDC, Molecular Dynamics counts. (B) Quantitation of the results for duplicate samples taken at 24 h after TPA induction. The chart is in the same format as panel A. Array elements showing strong signals are labeled. (C) Scatter plot comparing the results from panels A and B. The values plotted represent the means from duplicate experiments. KSHV genes are indicated by open circles; cellular genes are indicated by filled circles. Identities of the points representing those labeled in panels A and B are shown.
The array data demonstrate that KSHV is under tight transcriptional control in BC-3 cells and remains in a latent state in the vast majority of cells (37, 50). The induction of lytic replication leads to a marked difference in the hybridization pattern on the array (Fig. 1B and C). This reflects the increase in the transcriptional activity of the viral genome during lytic replication (34, 37). The increase in transcription varies between regions of the genome and individual genes. Genes markedly upregulated within 24 h include those encoding nut-1, vOx-2, K8, and v-Bcl-2, in agreement with previously published data (37, 38, 44, 50). Other genes for which expression has not previously been analyzed, for example, ORFs 11 and 58, are upregulated to similar extents. TPA induction of viral replication also increases the expression of two of the cellular genes, encoding ubiquitin and the MHC class I antigen HLA-B. Interestingly, the expression of HLA-E was found to be induced by lytic replication of another human herpesvirus, cytomegalovirus (51).
KSHV gene expression during latency.
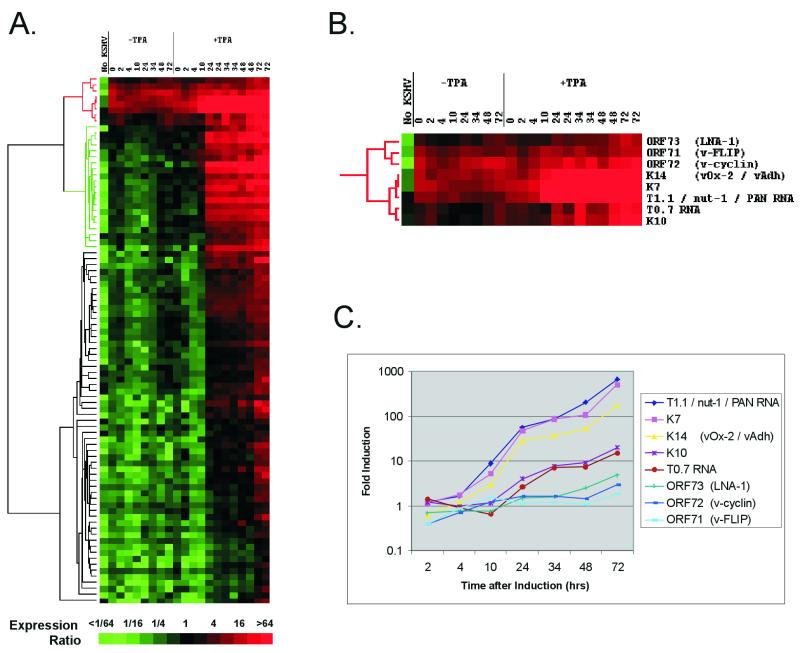
Although KSHV is under tight transcriptional control in uninduced BC-3 cells, lytic replication can clearly be induced with TPA, leading to differential KSHV gene expression (Fig. 1C). Lytic replication also occurs spontaneously in around 1% of uninduced BC-3 cells (52). To order and compare those genes that were being expressed in latent and lytic cells, we assembled all data from both uninduced and induced cells at all time points and clustered the viral genes using the software Cluster (18). The hierarchical clustering algorithm groups genes based on how their expression varies over all samples. A dendrogram is generated with genes sharing similar patterns of expression situated on the same branch. The branch lengths represent the degree of similarity between the genes. The output of this algorithm as applied to our data is shown in Fig. 2A. Shades of red signify detection of expression at levels sufficiently above the baseline (as defined by the median expression level [Materials and Methods]) to be judged significant. All genes are colored green or black in Ramos cells, a B-cell line negative for KSHV, as are most genes in all uninduced samples, indicating that these genes are not detectable (not expressed) in these samples. The small number of genes expressed in every uninduced sample (red) correspond exactly to those shown to be above the baseline signal (Fig. 1A). After hierarchical clustering, these genes also form their own branch of the tree, corresponding to genes for which expression can consistently be detected in uninduced cells. The branch splits into two subbranches, one corresponding to v-FLIP, v-cyclin, and LNA-1 (ORFs 71, 72, and 73, respectively) and the other containing vOx-2 (K14), K7, nut-1, T0.7, and K10 (Fig. 2B). Previous work has shown that transcripts encoding v-FLIP, v-cyclin, and LNA-1 are present during latent infection of PEL cell lines and show a slight increase upon the induction of lytic replication (14, 45). This branch thus represents latent (class I) transcripts. All three genes are transcribed together from the same initiation site on two overlapping transcripts (14, 39, 45). Their clustering together due to their similar expression profiles therefore gives us further confidence that the arrays are able to represent accurately biological information.
FIG. 2.
(A) Hierarchical clustering of gene expression data. Each row represents a separate amplicon on the array; each column represents the results from one array, hybridized with the sample detailed above. Columns 2 to 8 (−TPA) show untreated cells at successive time points shown in hours; lanes 9 to 21 (+TPA) show time points after the induction of lytic replication with TPA. Two independent experiments were conducted for the 24-, 34-, 48-, and 72-h time points for TPA induction. Column 1 (No KSHV) shows the array results from Ramos cell RNA, a KSHV-negative B-cell line. Levels of expression are relative to double the median level of expression for all genes averaged over all uninduced samples. The magnitude of this ratio is color coded according to the scale; shades of red signify detectable expression, and shades of green illustrate expression below the baseline level. The dendrogram on the left represents similarities of patterns of gene expression. The branch colored green is discussed in the text. (B) Expanded view of the uppermost cluster (red in panel A), which represents genes whose expression is detectable in uninduced cells. (C) Fold increase in expression at each time point (relative to time zero) after the induction of lytic replication with TPA. The genes form the cluster shown in panel B. The values are relative to the mean of the two time zero samples, with the values for 24, 34, 48, and 72 h being the averages of two experiments.
The expression of all genes in the second subbranch increases significantly upon the induction of lytic replication. These results have previously been shown by Northern blotting in the cell line BC-1 (37). These genes could either be expressed in latent cells but increase upon lytic induction or be expressed at a high level in the small percentage of spontaneously lytic cells present in uninduced cultures which form an increasing proportion of the total population during induction. To distinguish between these two possibilities, we determined the fold induction over time for each gene (as opposed to expression relative to the median expression level) (Fig. 2C). v-FLIP, v-cyclin, and LNA-1 show an increase of less than twofold by 24 h (also shown in Fig. 1C) and up to fourfold over 72 h, consistent with previous data (14, 45). Although these increases are significant, they are below that for almost every other gene on the array (data not shown).
The expression of K7, nut-1 (T1.1), and vOx-2 increases over 100-fold from uninduced cells, and the genes form a further subbranch from the v-FLIP/v-cyclin/LNA-1 branch (Fig. 2B and C). The nut-1 signal increases up to a maximum of over 640-fold at 72 h postinduction. This is well above previous Northern blot analysis, which estimated an increase of 20- to 30-fold (48) or 48-fold (44). Results of experiments in which known amounts of luciferase mRNA were added to the labeling reaction suggest that the expression level of nut-1 reaches at least 50,000 copies per cell, in close agreement with previous studies which estimated that there were approximately 25,000 copies per cell in transfected cell lines (48) and at least 10,000 copies per positive cell in a KS biopsy (43). This large amount of RNA in each cell would account for its detection in the few spontaneously lytic cells that exist in an otherwise uninduced cell population (50,000 copies in 1% of cells is the equivalent of 500 copies per cell). vOx-2 also shows a large (168-fold) increase in signal during lytic induction. This, and its clustering with nut-1 and K7, leads us to believe that the positive signal for vOx-2 during latency is again from a minority of spontaneously lytic cells.
The patterns of K10 and T0.7 expression indicate that these may be expressed as latent transcripts that are upregulated with TPA (19- and 15-fold, respectively). T0.7 RNA is expressed in the majority of KS spindle cells (43, 49) and in uninduced PEL cell lines (34). A longer but overlapping transcript is detectable with a T0.7-specific probe in uninduced BC-3 cells and has been shown to be upregulated by TPA (36). Our results are in agreement with these data. K10 clusters with T0.7 (Fig. 2B) and is induced to a similar extent (Fig. 2C). Although K10 was previously described as a class III transcript, it could also be detected in uninduced BC-1 cells (37).
A number of other genes have been described as class II (37). These all have low signals on arrays hybridized with uninduced samples, and cluster analysis places most of them in the adjacent major branch (dendogram colored green in Fig. 2A). Genes in this branch can be detected only at low levels in uninduced cells and are the first to be expressed at high levels after lytic induction. Therefore, it is likely that these genes were originally classified as class II due to their accumulation in the minority of lytic cells present in uninduced cultures (37). Taken together, these data suggest that care must be taken in assigning KSHV gene expression classes, as genes that are expressed abundantly in the lytic cycle (K7, nut1, and vOx-2) will be detected in a latent cell culture where a small fraction of cells enter the lytic cycle spontaneously.
KSHV gene expression during lytic replication: genes sharing similar functions are coordinately expressed.
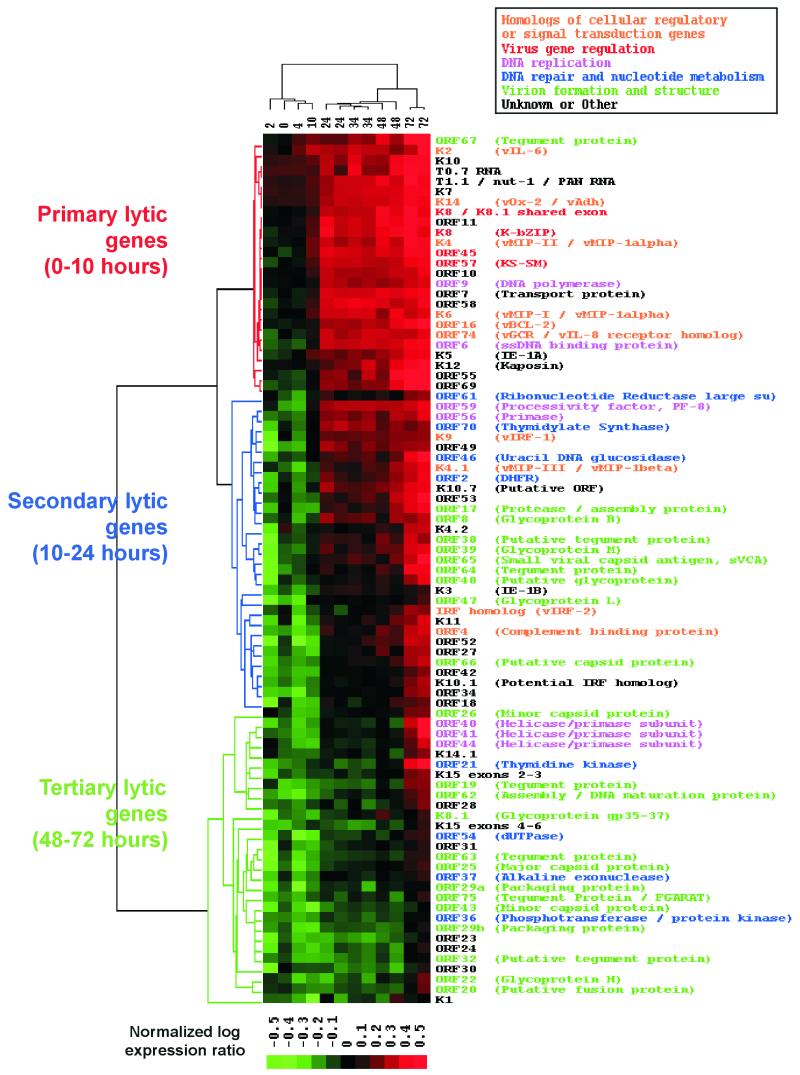
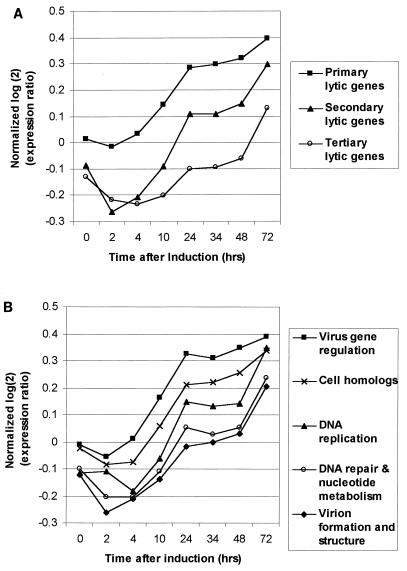
Most of the KSHV genes are not detectable in latent BC-3 cells. However, their expression increases over time after the induction of lytic replication (Fig. 2A). Different genes reach significant levels of expression at different time points, and cluster analysis arranges them into three main groups. To determine whether the timing of gene expression correlated with proposed gene function, we first ordered the data from the TPA induction time course with a self-organizing map (12) and then grouped the lytic genes by hierarchical clustering (Fig. 3). The order of the genes reflects the relative time when expression is first detected, from earliest to latest. Hierarchical clustering resulted in three main branches comprising genes with similar patterns of expression; we have named these classes primary lytic genes, secondary lytic genes, and tertiary lytic genes. The mean expression pattern of genes in each of these classes was determined, illustrating the time at which each group of genes was detected at levels greater than double the uninduced median gene expression level (Fig. 4A). Although expression of the tertiary lytic genes was not significantly above the uninduced median expression level until after 48 h, their level of expression starts to increase prior to this (Fig. 4A). Therefore, the relative times at which gene expression is detected by any method is intrinsically linked to the sensitivity of the detection system used. In addition, to perform extensive cross-comparisons, all genes should be measured by the same method at the same time.
FIG. 3.
Hierarchical clustering of genes and samples after the induction of lytic replication with TPA. The genes are ordered using a self-organizing map algorithm (18). The normalized log expression ratio is color coded according to the scale at the bottom. ORFs and corresponding gene names are listed on the right and color coded according to putative function shown by the key above. The dendrogram on the left represents relatedness of the patterns of gene expression. The three major branches are color coded according to the class of genes they represent and the times at which expression is first detected: primary lytic genes (0 to 10 h), secondary lytic genes (10 to 24 h), and tertiary lytic genes (48 to 72 h). Each column represents a sample taken at different times in hours after TPA induction (labeled above). The dendrogram at the top relates the samples according to the pattern of gene expression.
FIG. 4.
(A) Mean patterns of expression of the genes contained in each major branch of the tree shown in Fig. 4. The data shown are identical to those used in the cluster analysis. Values above the line y = 0 are red in Fig. 4; values below are green. The values for 24, 34, 48, and 72 h represent the averages of two experiments. (B) Mean patterns of expression of genes grouped by putative function (Fig. 3). The initial time when significant expression is detected is extrapolated from the point where the lines cross y = 0 and thus become significantly above the baseline expression in uninduced cells. The values for 24, 34, 48, and 72 h represent the averages of two experiments.
We also used hierarchical clustering to group the samples on the basis of similarities between patterns of gene expression. Samples representing repeated conditions are clustered in immediately adjacent columns, indicating that these time points have individually distinct patterns of expression (1). This also shows that the effects of experimental noise or artifact are minimal. Expression of the primary lytic genes becomes detectable within the first 10 h after the induction of lytic replication. The branching of the sample dendrogram shows that the largest change in gene expression occurs between 10 and 24 h after the addition of TPA, corresponding to the time of expression of the secondary lytic genes. A second significant change occurs between 48 and 72 h and corresponds to expression of the tertiary lytic genes.
To determine correlation between gene expression and function, we assigned functional annotations to genes, where known, from GenBank records. In addition, by comparison with orthologous sequences in other herpesvirus genomes, we were able to assign further putative KSHV gene functions. ORFs were broadly assigned to five functional groups: homologs of cellular regulatory or signal transduction genes, virus gene regulation, DNA replication, DNA repair and nucleotide metabolism, and virion formation and structure. Genes that could not be assigned to any of these groups were designated unknown/other (Fig. 3). Cluster analysis shows that genes belonging to the same functional group tend to have similar expression profiles. For example, viral regulatory genes are all primary lytic genes, whereas genes involved in virion formation are almost exclusively secondary and tertiary lytic genes. Also, the relative time at which the expression of different genes is detected correlates with the stage of the life cycle in which that gene is thought to act. This is known for other herpesviruses and can be seen more clearly for KSHV in Fig. 4B, which shows the mean expression profile for genes in each functional group. Genes thought to be involved in virus gene regulation are the first to be detected (at 4 h postinduction), consistent with their presumed role in activating lytic gene expression. Genes involved in DNA replication can be detected at significant levels by 14 h, followed by those presumed to function in DNA repair and nucleotide metabolism at 20 h. Genes encoding proteins that form part of the virus particle (tegument, capsid, and envelope glycoprotein) and those involved in virus assembly are not expressed until later in the virus life cycle (average of 34 h). Viral homologs of cellular genes involved in regulation or signal transduction are expressed after the viral regulatory genes but before those involved in DNA replication. This time of expression is consistent with the presumed role of these genes to overcome host responses to viral infection (31, 32), thus allowing replication to proceed. Thus, this type of classification, based on gene expression and function, provides insight into the KSHV lytic replication cycle.
Array analysis can predict the organization and function of viral genes.
It has previously been noted that the coexpression of uncharacterized genes with those of known function may provide a means of gaining clues to the functions of these genes (18). In addition to containing clones of previously identified ORFs, the array also includes a number of elements that correspond to presumed noncoding intergenic regions (data not shown). Preliminary data suggest that some of these may be transcribed. One such clone corresponds to a sequence located between K10.5 and K11 (91091 to 91380 in U75698) in block g (35). This region of the genome encodes two ORFs, vIRF-1 (ORF K9) (5, 32, 35) and vIRF-2 (6), which have both sequence and functional homology to cellular IRFs. Block g also contains a further two putative ORFs which have homology to IRFs (35), named K10.1 and K10.5 (GenBank accession no. U93872, submitted by F. Neipel et al. [6]). The ORF that we refer to as K10.5 has also been named K10.1 (32). K10, K10.1, K10.5, K11, and vIRF-2 proteins all share homology with vIRF-1 by pairwise sequence analysis. The vIRF-1, vIRF-2, K10.1, and K11 genes are all classified as secondary lytic genes by cluster analysis (Fig. 4). The array element corresponding to the region between K10.5 and K11 is also clustered in this branch [labeled K10.7 (Putative ORF) in Fig. 3]. This information, along with its genomic location, suggested that this clone might represent a novel ORF with homology to the known vIRFs. ORF analysis showed that the array element formed part of a novel ORF (named K10.7) that corresponds to nucleotides 91394 to 90936 in KSHV genomic sequence U75698. Multiple sequence alignment shows that this ORF shares homology with vIRF-1, vIRF-2, K10.1, and two human IRFs in part of the N-terminal DNA binding domain (6) (Fig. 5A).
The adjacent clustering of K11 and vIRF-2 on the array (Fig. 3) suggests that these two ORFs may be cotranscribed. K10, K10.5, and K11 share homology with vIRF-1 and human IRFs in the C-terminal region (data not shown). We therefore tested whether K10 and K10.1, K10.5 and K10.7, and K11 and vIRF-2 were spliced together in pairs to give three longer ORFs encoding putative proteins with homology to both the C termini and N-terminal DNA binding domains of known IRFs. RT-PCR across the predicted long ORFs showed that the transcripts were indeed spliced (Fig. 5B). Sequencing showed that the removal of an intron generated a complete ORF across each transcript (Fig. 5C). These transcripts encode for three putative IRF-related proteins, K10/10.1, K10.5/10.7, and K11/vIRF-2, with predicted molecular masses of 98, 62.5, and 75 kDa, respectively. Therefore, together with vIRF-1, KSHV encodes four proteins with homology to full-length IRFs.
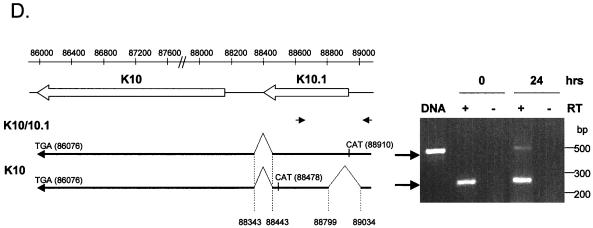
The array results suggest that K10.1 is not spliced to K10 during latency. To test this hypothesis, we searched for alternative splice sites around K10 and K10.1 and found a putative donor site upstream of K10.1 at 89034. RT-PCR with a primer complementary to sequence immediately upstream of this site showed that the first 112 bp of K10.1 are spliced out in latent cells (Fig. 5D). This splice removes an intron complementary to the K10.1 probe on the array. This transcript could not be detected by PCR of the entire K10/10.1 ORF (Fig. 5B) because the 5′ end of K10.1 lies within this intron. This alternatively spliced transcript encodes K10, a putative protein of 767 amino acids (82 kDa) which is missing the DNA binding domain encoded by K10.1. Transcripts containing this intron and therefore encoding the putative DNA binding domain can be detected only in cells undergoing lytic replication by both RT-PCR and array analysis (Fig. 3 and 5D).
DISCUSSION
We have described the creation of a nylon membrane-based array for the study of KSHV that has yielded highly consistent and reproducible results. We have analyzed KSHV gene expression during latency and lytic replication. The arrays are able to detect the small number of transcripts that were previously shown to be expressed during latency. The ordering of the array data by cluster analysis separates latent and lytic genes and generates a temporal program of lytic gene expression. The division of the lytic genes into three main classes (primary, secondary, and tertiary) gives a simple means of assessing when the expression of these genes becomes detectable.
Analysis of the relative levels of gene expression and predicted functions of the KSHV genes provides detailed insight into the biology of this herpesvirus. The array results confirm previous Northern blot analysis that the v-FLIP, v-cyclin, and LNA-1 genes are latent genes only marginally upregulated during lytic infection and that the T0.7 transcript is also present during latency (34, 36, 45). Cluster analysis and RT-PCR suggest that K10 may be a latent gene that is induced after the induction of lytic replication. Array data and RT-PCR show that K10 is alternatively spliced to give two forms, K10 and K10/10.1. Only K10/10.1, which is a lytic transcript, encodes a protein with a putative DNA binding domain. A protein of 100 kDa, closely matching the predicted size of K10/10.1, was previously found to be induced after TPA treatment of PEL cell lines (23). Further work is needed to confirm whether K10 protein is present in latent cells and whether the gene is alternatively spliced during lytic replication.
The expression patterns of the lytic genes also correlates well with previously published Northern blot analyses. The primary lytic genes encoding vIL-6, K12, K5, nut-1, K8, and K4 have previously been shown to be expressed between 8 and 13 h after induction (44). ORF65 (25, 44) and ORF17 (47), which encode the small viral capsid antigen and the KSHV protease/assembly protein, respectively, are expressed late during lytic replication and are both classified as secondary lytic genes on the array. Similarly, most of the secondary lytic genes and all of the tertiary lytic genes have been classified as class III transcripts (37).
The expression of most of the KSHV genes has not been analyzed before. The functions of only a few genes have been shown experimentally, but putative functions have been assigned to most of the genes based on their homology to previously characterized herpesviruses (35). Our analysis shows that the relative time at which the expression of different genes is detected correlates well with the stage of the virus life cycle in which that gene is thought to act. For example, genes which encode proteins thought to be involved in virus gene regulation are all expressed early and precede those that form the virus particle or which are involved in its assembly (Fig. 3 and 4B). The one exception to this is ORF 67, which is already detectable by 4 h postinduction. This gene is thought to encode a tegument protein due to its homology to the Epstein-Barr virus gene BFRF1 (35), which is also expressed early in lytic replication (19). The expression of ORF 67 as a primary lytic gene suggests functions consistent with those of the other genes expressed at this time, namely, cell regulation, signal transduction, and virus gene regulation.
Analysis of functional annotation in the context of gene expression also shows differences in temporal expression of certain functional classes. Genes encoding products involved in DNA replication such as DNA polymerase (ORF 9), PF-8 processivity factor (ORF 59), single-stranded DNA binding protein (ORF 6), and the viral primase (ORF 56) are detected early and show similar patterns of expression (Fig. 3, pink text). ORFs 40, 41, and 44 are also thought to be involved in DNA replication but are not detected until later during the life cycle (Fig. 3, pink text). It is possible that these proteins are not required for DNA replication from latent episomes but are packaged by the virus and act to replicate viral DNA after infection. Similarly, genes involved in DNA repair and nucleotide metabolism are also split into two groups (Fig. 3, blue text); ORFs 2, 46, 61, and 70 are expressed quite early with genes such as the primase and processivity factor genes, while ORFs 21, 36, 37, and 54 are expressed later with structural genes. Therefore, the requirement for genes with functions in this class may be split between initial DNA replication and subsequent DNA maturation and packaging. Such data are consistent with the alkaline exonuclease of herpes simplex virus type 1, which is dispensable for DNA replication but required for DNA maturation (28).
Viral homologs of cellular regulatory or signal transduction proteins are primary or secondary lytic genes (Fig. 3, orange text). Of these, the secondary lytic genes (encoding vMIP-III, vIRFs, and complement binding protein) are all thought to interact with the host immune system. Of particular interest is the relationship between expression of the three macrophage inflammatory protein homologs vMIP-I, -II, and -III. vMIP-I and -II are closely related and are thought to have evolved by gene duplication within the virus genome (4). vMIP-III is more distantly related and probably derived independently from another member of the CC chemokine family (32, 35). vMIP-I and -II are both primary lytic genes, whereas vMIP-III is in the secondary lytic gene cluster. Thus, the array data suggest that these genes are also differentially regulated at the level of transcription and may have distinct functions in the virus life cycle.
The expression profiles of the many uncharacterized KSHV genes provide clues to their function by virtue of their association with previously characterized genes. The discovery of a novel ORF, K10.7, by this approach coupled with other techniques shows that KSHV encodes four full-length IRF-related proteins. K10.5/10.7 has also recently been identified as a novel lytic gene in another study, where it was named vIRF-3 (27). Therefore, this kind of analysis should lead to further insights into KSHV gene function.
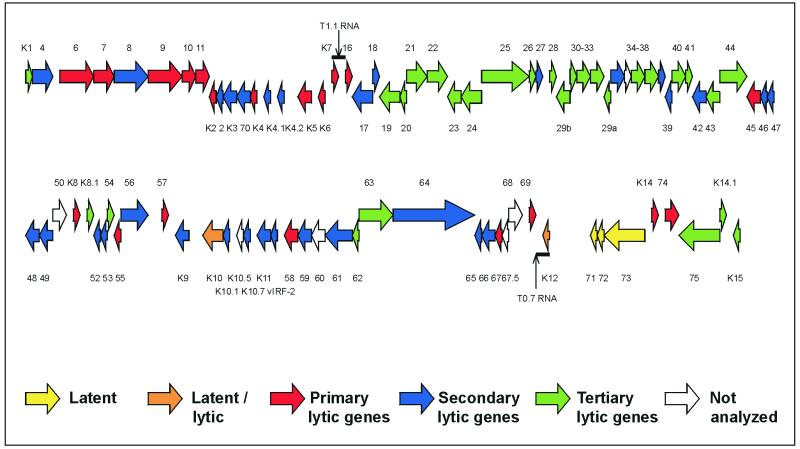
Many elements from adjacent genes on the genome also appear to be clustered near one another on the array. This may represent overlapping transcriptional units where one ORF forms the 3′ untranslated region of the transcript from another upstream ORF. An example of such an event is the extension of K15 transcripts through K14.1 and ORF75 (22). The arrays are unable to completely discriminate against presumably noncoding transcripts such as these. We have tried to minimize their detection by priming cDNA synthesis with sense-strand-specific primers. However, this does not completely prevent the K14.1 probe from detecting K15 transcripts primed by the ORF75 primer. The correlation between genomic location and expression could also be due to probes detecting spliced or polycistronic transcripts or due to coregulation of genes by common or related promoters. An example of a polycistronic transcript whose component genes cluster in terms of expression are the latent v-FLIP, v-cyclin, and LNA-1 genes. Examples of spliced ORFs sharing expression profiles are the different exons of K15, ORFs 29a and 29b, and, as shown here, K11 and vIRF-2. Figure 6 shows the locations of genes belonging to each expression class. The map indicates other areas of the KSHV genome where more detailed transcript mapping would help discriminate between noncoding transcriptional read-through events, polycistronic transcripts, splicing, and shared promoters.
FIG. 6.
Map of the KSHV genome (updated from reference 35). Each ORF is color coded according to its expression pattern: latent (class I), latent/lytic (induced by TPA), primary lytic genes, secondary lytic genes, and tertiary lytic genes. ORFs 35, 50, 60, 67.5, and 68 and K10.5 were not analyzed due to cross-hybridization or missing probes.
There are some discrepancies between our results and those of others. We were unable to confirm detection of K15 expression in latent cells (11, 22), possibly because of differences in the cell lines used. However, both studies detected different banding patterns on Northern blots depending on the K15 probe used, which may be due to cross-hybridization with an unknown RNA (11). This explanation is consistent with the array results showing cross-hybridization of K15 exon 8 with RNA from Ramos cells, which occurred despite the use of stringent hybridization conditions (64°C). A decrease in the abundance of early gene transcripts 48 h after induction has also been reported (44). However, our results indicate that these transcripts remain constant or continue to accumulate during lytic replication. RNA loading controls suggest that the apparent decrease in the expression of KSHV genes may be due to a progressive reduction in the total amount of RNA present after induction (44). Consistent with this, we found that late time points after induction showed a decrease in the amount of total RNA extracted from equal amounts of cells and an additional decrease in the absolute amount of all cellular mRNAs hybridizing to the arrays. This is most likely due to the proapoptotic effect of TPA (53) and virus-induced shutdown of cellular genes (38). To maintain similar levels of hybridization on the arrays, we used greater amounts of total RNA in the cDNA synthesis reaction and discarded data from any array where the housekeeping gene signal was more than threefold different from the mean level.
The KSHV array provides a rapid, accurate, and reproducible method to analyze the global gene expression that constitutes the virus transcriptome. Such arrays provide a means of determining the KSHV transcriptome in the three tumor types that result from KSHV infection. The combination of viral and cellular genes in a single array will also lead to a greater understanding of the host-pathogen interactions that occur during KSHV infection. The use of post-genomic research methods and detailed bioinformatics analysis of such results will expand our understanding of virus-induced diseases.
ACKNOWLEDGMENTS
We gratefully acknowledge the support and advice of Robin Weiss. We also acknowledge Pieter Goedhart and Driss Talibi at Eurogentec for gridding of the KSHV array.
This work was funded by the Medical Research Council (MRC) (R.G.J. and P.K.), the Biotechnology and Biological Sciences Research Council (BBSRC) (M.M.A.), and the Cancer Research Campaign (CRC) (C.B.). R.G.J. is enrolled in the Graduate Program of the MRC Laboratory for Molecular Cell Biology.
REFERENCES
- 1.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, Powell J I, Yang L, Marti G E, Moore T, Hudson J, Jr, Lu L, Lewis D B, Tibshirani R, Sherlock G, Chan W C, Greiner T C, Weisenburger D D, Armitage J O, Warnke R, Staudt L M, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 3.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 5.Burysek L, Yeow W S, Lubyova B, Kellum M, Schafer S L, Huang Y Q, Pitha P M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burysek L, Yeow W S, Pitha P M. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2) J Hum Virol. 1999;2:19–32. [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 9.Chambers J, Angulo A, Amaratunga D, Guo H, Jiang Y, Wan J S, Bittner A, Frueh K, Jackson M R, Peterson P A, Erlander M G, Ghazal P. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 1999;73:5757–5766. doi: 10.1128/jvi.73.7.5757-5766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 11.Choi J K, Lee B S, Shim S N, Li M, Jung J U. Identification of the novel K15 gene at the rightmost end of the Kaposi's sarcoma-associated herpesvirus genome. J Virol. 2000;74:436–446. [PMC free article] [PubMed] [Google Scholar]
- 12.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 13.Decker L L, Shankar P, Khan G, Freeman R B, Dezube B J, Lieberman J, Thorley-Lawson D A. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J Exp Med. 1996;184:283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djerbi M, Screpanti V, Catrina A I, Bogen B, Biberfeld P, Grandien A. The inhibitor of death receptor signaling, FLICE-inhibitory protein defines a new class of tumor progression factors. J Exp Med. 1999;190:1025–1032. doi: 10.1084/jem.190.7.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupin N, Diss T L, Kellam P, Tulliez M, Du M Q, Sicard D, Weiss R A, Isaacson P G, Boshoff C. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood. 2000;95:1406–1412. [PubMed] [Google Scholar]
- 17.Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande J P, Weiss R A, Alitalo K, Boshoff C. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci USA. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen M B, Spellman P T, Brown P O, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farina A, Santarelli R, Gonnella R, Bei R, Muraro R, Cardinali G, Uccini S, Ragona G, Frati L, Faggioni A, Angeloni A. The BFRF1 gene of Epstein-Barr virus encodes a novel protein. J Virol. 2000;74:3235–3244. doi: 10.1128/jvi.74.7.3235-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friborg J, Jr, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 21.Geiss G K, Bumgarner R E, An M C, Agy M B, van 't Wout A B, Hammersmark E, Carter V S, Upchurch D, Mullins J I, Katze M G. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology. 2000;266:8–16. doi: 10.1006/viro.1999.0044. [DOI] [PubMed] [Google Scholar]
- 22.Glenn M, Rainbow L, Aurad F, Davison A, Schulz T F. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J Virol. 1999;73:6953–6963. doi: 10.1128/jvi.73.8.6953-6963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katano H, Sato Y, Kurata T, Mori S, Sata T. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease. Virology. 2000;269:335–344. doi: 10.1006/viro.2000.0196. [DOI] [PubMed] [Google Scholar]
- 24.Klein G, Giovanella B, Westman A, Stehlin J S, Mumford D. An EBV-genome-negative cell line established from an American Burkitt lymphoma; receptor characteristics. EBV infectibility and permanent conversion into EBV-positive sublines by in vitro infection. Intervirology. 1975;5:319–334. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- 25.Lin S F, Sun R, Heston L, Gradoville L, Shedd D, Haglund K, Rigsby M, Miller G. Identification, expression, and immunogenicity of Kaposi's sarcoma-associated herpesvirus-encoded small viral capsid antigen. J Virol. 1997;71:3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lockhart D J, Winzeler E A. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 27.Lubyova B, Pitha P M. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J Virol. 2000;74:8194–8201. doi: 10.1128/jvi.74.17.8194-8201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 31.Moore P S, Chang Y. Antiviral activity of tumor-suppressor pathways: clues from molecular piracy by KSHV. Trends Genet. 1998;14:144–150. doi: 10.1016/s0168-9525(98)01408-5. [DOI] [PubMed] [Google Scholar]
- 32.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radkov S A, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and together with the oncogene hras transforming primary rat cells. Nat Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 34.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 35.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadler R, Wu L, Forghani B, Renne R, Zhong W, Herndier B, Ganem D. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5722–5730. doi: 10.1128/jvi.73.7.5722-5730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 39.Sarid R, Wiezorek J S, Moore P S, Chang Y. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schena M, Shalon D, Heller R, Chai A, Brown P O, Davis R W. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharp T V, Boshoff C. Kaposi's sarcoma-associated herpesvirus: from cell biology to pathogenesis. IUBMB Life. 2000;49:97–104. doi: 10.1080/15216540050022395. [DOI] [PubMed] [Google Scholar]
- 42.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 43.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 46.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unal A, Pray T R, Lagunoff M, Pennington M W, Ganem D, Craik C S. The protease and the assembly protein of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:7030–7038. doi: 10.1128/jvi.71.9.7030-7038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong W, Ganem D. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:1207–1212. doi: 10.1128/jvi.71.2.1207-1212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu H, Cong J P, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoeteweij J P, Eyes S T, Orenstein J M, Kawamura T, Wu L, Chandran B, Forghani B, Blauvelt A. Identification and rapid quantification of early- and late-lytic human herpesvirus 8 infection in single cells by flow cytometric analysis: characterization of antiherpesvirus agents. J Virol. 1999;73:5894–5902. doi: 10.1128/jvi.73.7.5894-5902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]