Abstract
Most RNA-protein condensates are composed of heterogeneous immiscible phases. However, how this multiphase organization contributes to their biological functions remains largely unexplored. Drosophila germ granules, a class of RNA-protein condensates, are the site of mRNA storage and translational activation. Here, using super-resolution microscopy and single-molecule imaging approaches, we show that germ granules have a biphasic organization and that translation occurs in the outer phase and at the surface of the granules. The localization, directionality, and compaction of mRNAs within the granule depend on their translation status, translated mRNAs being enriched in the outer phase with their 5′end oriented towards the surface. Translation is strongly reduced when germ granule biphasic organization is lost. These findings reveal the intimate links between the architecture of RNA-protein condensates and the organization of their different functions, highlighting the functional compartmentalization of these condensates.
Subject terms: Super-resolution microscopy, Oogenesis, Translation
Biomolecular condensates are often composed of immiscible phases. Here, the authors reveal a functional biphasic organization of germ granules, where translational repression happens in the granule core and translation in the outer phase.
Introduction
Membraneless biomolecular condensates have recently emerged as fundamental in cell biology favoring the coordination and efficiency of biochemical reactions by concentrating substrates and enzymes in a common space. Their assembly depends on a demixing process that involves multivalent interactions between proteins and nucleic acids once their concentrations reach a specific threshold1. RNA-protein (RNP) condensates, also called RNA granules, exist in both the nucleus and cytoplasm and are linked to most aspects of RNA biology2,3. Yet, although the biophysical properties and assembly mechanisms of RNP condensates have been deeply investigated, the biological functions of most of them remain poorly understood4–6. Super-resolution microscopy approaches have revealed that many RNP condensates are not homogeneous but rather composed of multiple immiscible phases, reflected by a heterogeneous distribution of RNA and/or RNA binding proteins3,7. The best understood example of this phenomenon is the nucleolus in which three nested phases are linked to three different steps of ribosome biogenesis8–10. For other RNP condensates, the functional relevance of their higher-order organization remains elusive.
Germ granules are evolutionary conserved germline specific RNP condensates that instruct germ cell fate through mRNA regulation11,12. They are hubs for mRNA storage and translational control. Drosophila embryonic germ granules contain ≈200 different maternal mRNAs13,14 as well as four main proteins: the germ granule inducer Oskar (Osk), Vasa, an RNA helicase homolog of human DDX4, the scaffold protein Tudor (Tud) and Aubergine (Aub), an Argonaute protein of the PIWI clade15. Drosophila germ granules display both liquid-like and hydrogel-like properties, mRNA representing a stable component of the granule16,17. Multiple copies of the same mRNA molecules are grouped together in homotypic clusters18,19, although the function behind this organization is still unknown. In several species, germ granules have a multiphase architecture20,21, and understanding the relationships between this organization and germ granule functions is attracting a growing interest. In particular, although the function of germ granules in the storage of repressed mRNAs is well described, whether they are also sites for translational activation remains an open question. In C. elegans, translational activation of germ granule mRNAs coincides with their relocalization to the cytoplasm22,23. In Drosophila embryos, translation of germ granule mRNAs follows a tight temporal sequence13, implying that germ granules achieve temporal translational control by coordinating concomitant translational repression of some mRNAs and activation of others. We took advantage of this outstanding property to determine how these opposite functions are compartmentalized within germ granules and address the relationships between architecture and biological functions of RNP condensates.
Results
Drosophila germ granules have a biphasic organization
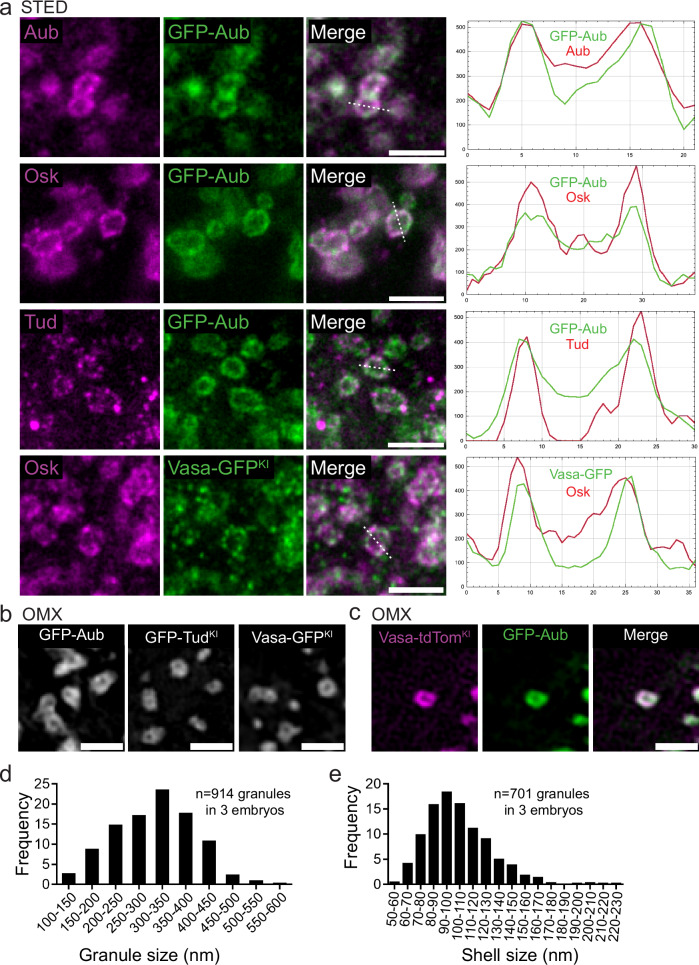
Due to the small size of germ granules, ranging from 200 to 500 nm24, we used super-resolution 2D Stimulated Emission Depletion (STED) microscopy to characterize the distribution of germ granule main protein components. We set up a method to slice and expose the posterior pole of embryos to the objective, in order to reduce the distance between the sample and the objective and get optimal resolution (Supplementary Fig. 1a–c and Supplementary Note). All four main components of germ granules were enriched in the outer most part of the granule that we termed outer phase or shell (Fig. 1a) and they colocalized as quantified using the Pearson Correlation Coefficient (PCC(Costes)) (Supplementary Fig. 1d). This organization was reminiscent of the germ granule structure observed previously by electron microscopy, showing an electron dense periphery and lucid center25,26. STED microscopy requires the use of an antibody to get a signal that stands the depletion. To rule out that the observed pattern was the consequence of poor antibody penetration within the granule, we used AiryScan and 3D-OMX microscopy to visualize the localization of GFP-tagged versions of Aub, Vasa, and Tud using GFP fluorescence. The same enrichment was observed in the shell (Fig. 1b, Supplementary Fig. 1c). Furthermore, GFP-Aub and Vasa-tdTomato visualized using 3D-OMX without antibody staining also colocalized in the shell of germ granules (Fig. 1c). GFP-Aub enrichment in the outer phase was visible in 3D (Supplementary Fig. 1e), consistent with previous observations based on electron microscopy of ultra-thin sections that led to propose a hollow sphere morphology of germ granules26. To further confirm that a substantial proportion of germ granules in early embryos had this hollow morphology, we visualized germ granules in two consecutive serial sections using transmission electron microscopy. This approach showed that of 16 germ granules present on both sections, 15 (94%) were visualized as hollow in at least one section, whereas of 11 granules present on a single section, only two had a hollow center (Supplementary Fig. 1f, g). These data revealed the importance of imaging through the granule volume when using ultra-thin sections (70 nm) to access granule morphology, as reported previously26. Of note, enrichment of germ granule major proteins in the outer phase was not described in previous studies based on structured illumination microscopy (SIM)17–19. We believe that this difference comes from the higher resolution (60 ± 12 nm, see “Methods”) reached using STED microscopy combined with our slicing method (Supplementary Fig. 1c), as compared to SIM (typically 100 nm). Further studies will be needed to fully resolve this discrepancy. Using STED acquisitions and Osk protein as a marker, we applied a Laplacian of Gaussian filter to define the edges of germ granule outer and inner phases (see “Methods”) and measured the size of germ granules and their outer phase. Germ granule diameters ranged from 110 to 570 nm with a mean of 308 ± 86 nm (Fig. 1d), and the thickness of the outer phase ranged from 54 to 224 nm with a mean of 104 ± 25 nm (Fig. 1e). We conclude that germ granules have a multiphase structure, with main protein components enriched in the outer phase.
Fig. 1. Drosophila germ granules have a biphasic organization.
a STED imaging of germ granule main components. Immunostaining of UASp-GFP-Aub; nos-Gal4 or vasa-GFPKI embryos with anti-Aub (magenta), anti-Osk (magenta), anti-Tud (magenta), and anti-GFP (green) to visualize GFP-Aub or Vasa-GFP. Fluorescence intensity (right) was measured along the path marked with a white dashed line and intensity profiles of each channel are shown on the graph. b 3D-OMX imaging of UASp-GFP-Aub; nos-Gal4 (left), GFP-TudKI (middle), Vasa-GFPKI (right) in which GFP was directly recorded without antibody staining. c 3D-OMX imaging of UASp-GFP-Aub/ vasa-tdTomKI; nos-Gal4/+ embryos in which GFP and tdTomato were directly recorded without antibody staining. d, e Measurement of germ granule size (d) and outer phase size (e) from wild-type embryos immunostained with Osk antibody and imaged using STED microscopy. Scale bars: 1 µm. Source data are provided as a Source Data file.
Aub is required for the biphasic architecture and maintenance of germ granules
Aub binds germ granule mRNAs through base-pairing with piRNAs and is a major actor of their recruitment to germ granules14,27,28. Through this function, Aub is expected to play an important role in maintaining the RNA content of germ granules. We thus addressed the contribution of mRNAs to germ granule biphasic organization by analyzing germ granule architecture in the absence of Aub. We used a specific genetic set up to visualize germ granules in aub mutant embryos. Indeed, due to early developmental defects during oogenesis in aub mutants, Osk protein is not produced at the posterior pole, and germ granules do not form in these mutants29. However, Osk does accumulate at the anterior pole of embryos following recruitment of osk mRNA to the anterior pole, even in the absence of Aub27,30. In this context, germ granule mRNAs localize very poorly to the anterior pole, consistent with the role of Aub in mRNA recruitment to germ granules27. Using STED imaging and Osk as a marker, we found that germ granules produced at the anterior pole when osk mRNA was delocalized had a similar biphasic organization as those present at the posterior pole of wild-type embryos (Supplementary Fig. 2a). In contrast, in aub mutant embryos, germ granule architecture was severely affected, with the disappearance of their biphasic organization and a diameter reduced to 158 ± 37 nm, instead of 324±66 nm in the aub+ background (Supplementary Fig. 2b, c). In line with Aub function in mRNA recruitment to germ granules, nos mRNA was not found in these germ granules using single molecule Fluorescent In Situ Hybridization (smFISH) and STED imaging (Supplementary Fig. 2d). In addition, these germ granules disappeared altogether in 79.3% of embryos (n = 53) after 2.5 h of development, while they were maintained in the aub+ background (100% of embryos, n = 50).
These data demonstrate the essential contribution of Aub to germ granule biphasic organization and maintenance, in part through its ability to recruit mRNAs.
nos mRNA translation occurs in the outer phase and at the immediate periphery of germ granules
To understand how germ granule mRNA translation integrates within germ granule architecture, we used the Suntag method that allows the visualization of ongoing translation31–35, to monitor nos mRNA translation. Briefly, the binding of single-chain antibodies fused to GFP (scFv-GFP) on an array of GCN4-based epitope (called Suntag) placed downstream of the start codon of the mRNA of interest allows the visualization of nascent peptides by creating bright GFP spots above background whose intensity reflects translation efficiency (Fig. 2a). Translating mRNAs are detected either by the MS2/MCP system36 or smFISH37. We inserted an array of 12 Suntag repeats after nos initiation codon (Supplementary Fig. 3a) in a genomic construct known to rescue nos RNA null mutant (nosBN) phenotypes38. We first recorded the efficiency of suntag-nos mRNA localization to the germ plasm, the cytoplasm containing germ granules at the posterior pole of embryos, by measuring suntag smFISH fluorescence intensity at the posterior pole and in whole embryos expressing suntag-nos and comparing this quantification to that of nos smFISH fluorescence intensity in wild-type embryos. While 3.8% of nos mRNA localized to the posterior pole in wild-type embryos as previously reported18,39, 2.15% of suntag-nos mRNA localized to the posterior pole (Supplementary Fig. 3b), indicating a reduced recruitment of the chimeric suntag-nos mRNA to the germ plasm. Nonetheless, the suntag-nos construct was able to rescue nosBN embryonic lethality (nosBN: 100% embryonic lethality, suntag-nos/+; nosBN: 48.7% embryonic lethality, n > 100 embryos), showing that the regulation of suntag-nos mRNA, including its level of localization and translational control allowed embryonic development.
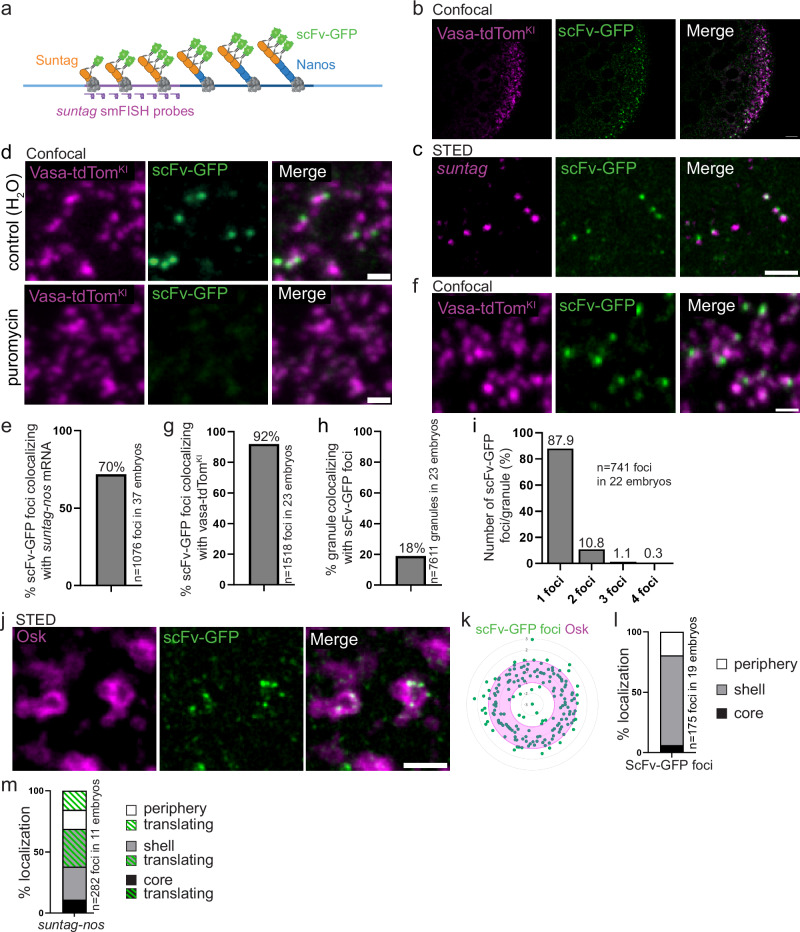
Fig. 2. Visualization of nos mRNA translation at germ granules using Suntag.
a Principle of the Suntag technique to visualize nos mRNA translation. Blue lines: nos mRNA (light blue: UTRs, dark blue: CDS). Purple line: Suntag array. The polysomes are in grey. The Suntag and Nos nascent peptides are in orange and blue, respectively. The scFv-GFP antibody (green) binds the Suntag peptide. suntag smFISH probes are in purple. Created on Biorender.com. b Confocal images of vasa-tdTomKI/nos-suntag-nos; nos-scFv-GFP/+ embryos. Vasa-tdTomato (magenta) and scFv-GFP (green) fluorescence were directly recorded without antibody staining. c STED images of immuno-smFISH of nos-suntag-nos/+; nos-scFv-GFP/+ embryos with anti-GFP nanobody (green) to detect scFv-GFP and suntag smFISH probes (magenta). d Fluorescent confocal images of vasa-tdTomKI/nos-suntag-nos; nos-scFv-GFP/+ permeabilized embryos without (top) and with puromycin treatment (bottom). Vasa-tdTomato (magenta) and scFv-GFP (green) fluorescence were directly recorded. e Percentage of scFv-GFP foci colocalizing with suntag-nos mRNA from images as in (c). f Close-up view of fluorescent confocal images of vasa-tdTomKI/nos-suntag-nos; nos-scFv-GFP/+ embryos showing scFv foci with germ granules. Vasa-tdTomato (magenta) and scFv-GFP (green) fluorescence were directly recorded. g Percentage of scFv-GFP foci colocalizing with germ granules marked with Vasa-tdTom from images as in (f). h Percentage of germ granules marked with Vasa-tdTom colocalizing with scFv-GFP foci, i.e., undergoing translation, from images as in (f). i Quantification of the number of scFv-GFP foci per granule from images as in (f). j STED imaging of nos-suntag-nos/+; nos-scFv-GFP/+ embryos immunostained with anti-Osk antibody (magenta) and anti-GFP nanobody (green) to reveal scFv-GFP. k, l Quantification of svFv-GFP foci localization within the germ granule biphasic structure from STED images as in (j). Radar plot of the relative distance of scFv-GFP foci (green dots) within Osk immunostaining (magenta). The granule shell is in pink (k). Percentage of scFv-GFP foci localized in the core (black), in and at the surface of the shell (grey), and at the immediate granule periphery (white) (l). m Percentage of suntag-nos mRNA foci localized in the core (black), in and at the surface of the shell (grey), and at the immediate granule periphery (white). The green dashed part in each category represents the proportion of suntag-nos foci undergoing translation (i.e., colocalizing with scFv-GFP) from images as in Supplementary Fig. 3e, f. Scale bars: 5 µm in (b), 1 µm in (c–e, j). Source data are provided as a Source Data file. a Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Using confocal microscopy, we found that suntag-nos embryos containing maternally-provided scFv-GFP (nos-scFv-GFP40) showed bright GFP foci at the posterior pole as expected for Nos protein production site (Fig. 2b). To determine whether these GFP foci reflected suntag-nos mRNA translation, first, we analyzed their colocalization with cognate mRNA using smFISH probes directed against the suntag sequence, and STED super-resolution imaging (Fig. 2c). We quantified that 70% of scFv-GFP foci colocalized with suntag mRNA (Fig. 2e), revealing translating mRNAs. Second, we showed that these scFv-GFP foci disappeared upon treatment of embryos with puromycin that releases translating ribosomes31 (Fig. 2d), confirming that scFv-GFP foci corresponded to translation sites or accumulation of freshly produced Suntag-Nos protein. We next addressed the localization of scFv-GFP foci in relation with germ granules marked with Vasa-tdTomato. Confocal imaging revealed that 92% of scFv-GFP foci colocalized with or overlapped germ granules (Fig. 2f, g). This suggested that suntag-nos mRNA translation occurred exclusively at germ granules and not in the intergranular space. To confirm this result, we performed immuno-smFISH to visualize suntag-nos mRNA, scFv-GFP foci, and germ granules marked with Osk (Supplementary Fig. 3c). Quantification of confocal images (see ”Methods”) showed that all (99.85%) translation events were restricted to the germ plasm and adjacent or colocalizing with germ granules (Supplementary Fig. 3c). Therefore suntag-nos mRNA translation occurred specifically at germ granules.
We quantified that 18% of germ granules marked with Vasa-tdTomato colocalized with scFv-GFP foci, and were thus translating suntag-nos mRNA (Fig. 2f, h). Most translating germ granules (87.9%) had a single scFv-GFP foci, but up to four scFv-GFP foci could be visualized per granule (Fig. 2i). However, it should be noted that these numbers would be expected to be higher for endogenous nos mRNA translation as a lower level of suntag-nos localized to the germ plasm (Supplementary Fig. 3b), and we quantified using FISH-quant41 a lower number of suntag-nos mRNA molecules per cluster compared to that of endogenous nos mRNA42 (Supplementary Fig. 3d).
We next used STED super-resolution microscopy to record the localization of translation events within germ granules (Fig. 2j). We found that most scFv-GFP foci localized in the outer phase and at the granule immediate periphery (74.3% and 19.4%, respectively), with only 6.3% localizing in the inner phase of the granule (Fig. 2j-l). We performed immuno-smFISH to visualize scFv-GFP, suntag-nos mRNA, and germ granule marked by Osk, using STED microscopy. We observed that suntag-nos mRNA and scFv-GFP translation foci were not organized in a specific order within the outer phase, scFv-GFP foci appearing either at the same level or in a more internal or more external position than suntag-nos mRNA within the outer phase (Supplementary Fig. 3e). Importantly, when suntag-nos mRNA was detected in the inner phase or core of the granule, it was not associated with scFv-GFP foci (Supplementary Fig. 3f, 100% n = 31 granules), indicating a lack of translation. Using this triple staining and STED microscopy, we recorded the localization of translating (associated with scFv-GFP foci) and non-translating suntag-nos mRNA within germ granules and found that among the 11% of suntag-nos mRNA localizing in the core, none were translating, whereas among the 89% of suntag-nos mRNA localizing in the outer phase and periphery, about half (52%) were translating (Fig. 2m). To confirm that this localization of scFv-GFP foci did not reflect reduced scFv-GFP availability in embryos, or its inability to reach the core of germ granules, we visualized suntag-nos translation using direct Suntag immunostaining with anti-GCN4 antibody. This approach revealed the same localization of translation mostly in the outer phase and periphery of germ granules (94.4 %) (Supplementary Fig. 3g, h). Furthermore, we designed a construct in which the 12 Suntag repeats were fused to MCP in order to recruit the MCP-Suntag fusion protein to germ granules using nos-MS2 (MS2 inserted into nos 3′UTR) (Supplementary Fig. 3i). When MCP-Suntag was visualized with scFv-GFP, we found a high colocalization (93%) with nos-MS2 (Supplementary Fig. 3j, k). Importantly, co-staining of scFv-GFP with Osk to visualize germ granules showed that 31.7% of scFv-GFP foci localized to the core of the granules, demonstrating its capability to access the core (Supplementary Fig. 3l, m).
We conclude that mRNA translation is compartmentalized within germ granules, occurring in the outer phase and at the immediate periphery of the granules. In addition, mRNAs localized in the core of the granules are translationally repressed.
Translational repressors are not present within germ granules
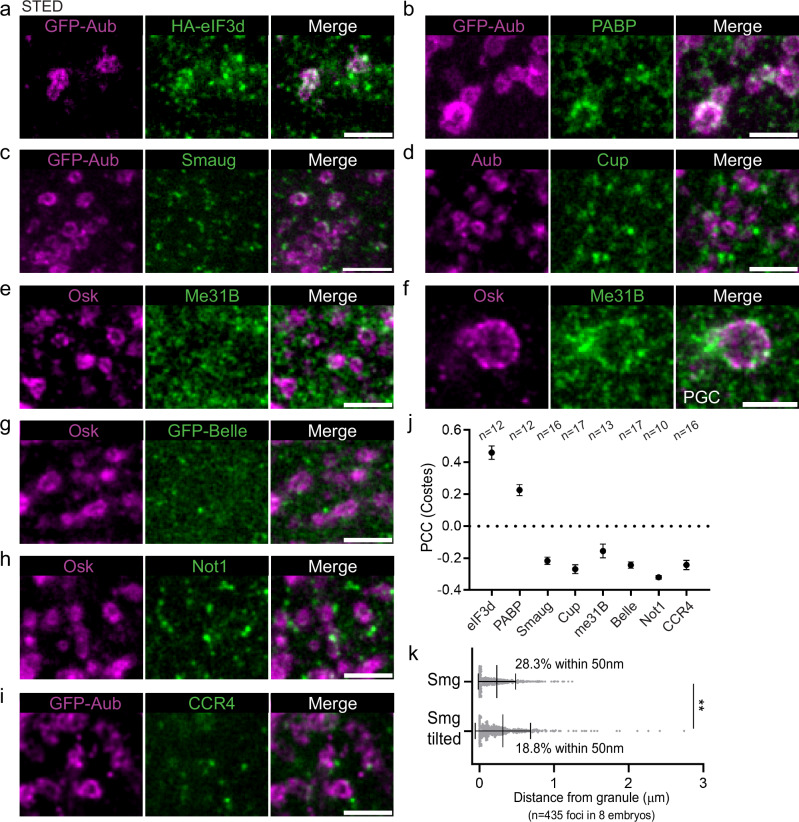
The regionalization of suntag-nos mRNA translation within germ granules implied that translational regulators should be heterogeneously distributed within germ granules. We previously reported a role of Aub in nos mRNA translation activation through interaction with the translation initiation factors eIF3d and PABP that localized at the edge of germ granules43. Using STED imaging, we found that eIF3d and PABP accumulated in the outer phase and at the surface of germ granules, but not in the inner phase (Fig. 3a, b), and they colocalized with GFP-Aub using PCC(Costes) (Fig. 3j). We next investigated the localization of translational repressors known to regulate nos mRNA in early embryos. Whereas nos mRNA translation is activated in the germ plasm, it is repressed in the somatic part of early embryos through deadenylation by the CCR4-NOT deadenylation complex, and by a repressor complex containing Smaug, Cup, Trailer Hitch, and the RNA helicases Me31B -a DDX6 homolog- and Belle -a DDX3 homolog-44. We analyzed the localization of Smaug, Cup, Me31B and Belle in the germ plasm using STED microscopy and did not find colocalization of any of these proteins with the germ granule markers Osk or Aub using PCC(Costes) (Fig. 3c–e, g, j). In particular, these translational repressors never accumulated in the core of germ granules where mRNAs are not translated. However, we could observe Smaug, Cup, and Me31B foci at the surface of germ granules (Fig. 3c–e). Quantification of the distance between Smaug foci and germ granules (see “Methods”), revealed a significant localization of Smaug in germ granules and close proximity, with 28.3% of Smaug foci within 50 nm of germ granules (Fig. 3k). Smaug is known to interact with Osk in the germ plasm, preventing Smaug binding to nos mRNA and relieving translational repression45,46. Given Smaug foci localization at the surface of germ granules (Fig. 3c, k), this might be the site where the Smaug-Osk interaction takes place. Me31B was previously reported to be a component of germ granules47. Although Me31B was not found within germ granules in the germ plasm, it accumulated in the outer phase of germ granules at a later stage when germ granules enlarge by fusion in primordial germ cells (Fig. 3f), consistent with a recent study describing Me31B enrichment in germ granules at this stage48. We then addressed the relationships of the deadenylation machinery with germ granules by recording the localization of two subunits of the deadenylation complex, Not1, and CCR4, in the germ plasm. None of the subunits showed association with germ granules marked with Osk or Aub (Fig. 3h–j).
Fig. 3. Localization of translation initiation factors and translational repressors in the germ plasm.
a, b Localization of translation initiation factors. STED imaging of immunostaining of UASp-GFP-Aub/UASp-HA-eIF3d; nos-Gal4/+ (a) and UASp-GFP-Aub; nos-Gal4 (b) embryos with anti-GFP (magenta) to detect Aub, anti-HA (green) to detect eIF3d (a) and anti-PABP (green) (b). c–i Localization of translational repressors. STED imaging of immunostaining of UASp-GFP-Aub; nos-Gal4, GFP-belle, and wild-type embryos with antibodies to reveal the indicated proteins. Germ granule markers are in magenta and translational repressors are in green. A larger germ granule exemplifying those present at later stages in primordial germ cells (PGC) is shown in (f). j Quantification of colocalization of GFP-Aub or Osk with the indicated proteins using PCC(Costes) from images as in (a–e and g–i). Black circles represent the mean and error bars represent SEM. The number of embryos is indicated (n). k Quantification of the distance between Smaug (Smg) foci and the edge of germ granules marked with GFP-Aub from images as in (c). Vertical bars represent the mean and SD. **p < 0.01 using χ2 test. p = 0.0033. Scale bars: 1 µm. Source data are provided as a Source Data file.
The lack of translational repressors inside germ granules suggests that mRNAs accumulating in the core of germ granules may be repressed due to their high mRNA compaction and/or partitioning of translational activators into the shell.
mRNA localization within the germ granule biphasic architecture depends on their translation status
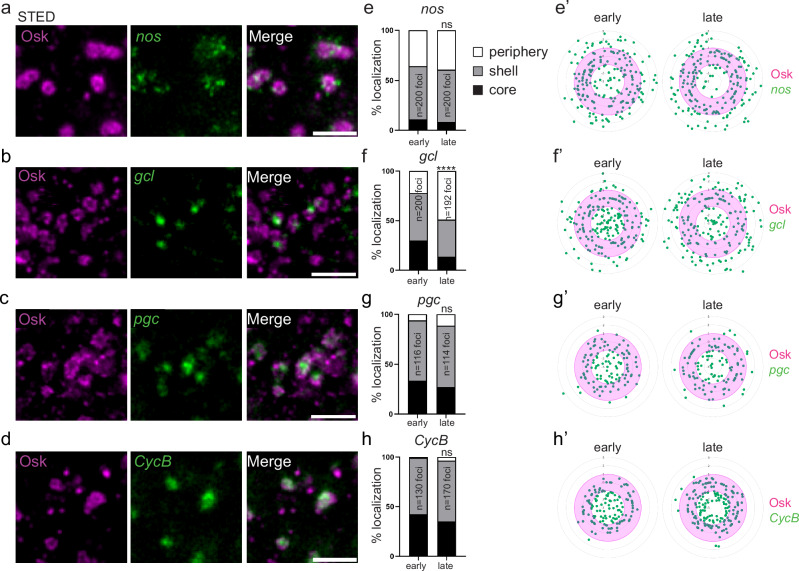
Translation of the germ granule mRNAs, nos, germ cell less (gcl), polar granule component (pgc) and Cyclin B (CycB) is sequential13: nos translation is already active at the onset of embryogenesis; gcl translation starts soon after, Gcl protein being visible 20–30 min after embryo deposition at 25 °C13,49; pgc translation takes place after primordial germ cell formation, 90 min after embryo deposition13; and CycB translation starts after primordial germ cell incorporation into the gonads, at 12–13 h of embryogenesis50. To understand how these mRNAs integrate within the biphasic organization of germ granules in relation with their translation status, we analyzed their localization using immuno-smFISH with anti-Osk immunostaining to visualize germ granules, combined with STED microscopy. We found that gcl, pgc, and CycB mRNAs formed one to two foci within germ granules, consistent with their assembly in homotypic clusters18,19 and that nos mRNA assembled in multiple foci, as previously reported19, within and at the immediate periphery of germ granules (Fig. 4a–d). We used the number of nuclei to carefully stage embryos and classify them into early (≤2 nuclei, age 0–20 min) and late (>2 nuclei and before primordial germ cell formation, age 20–90 min) stages. The localization of mRNA foci within germ granules was recorded in early and late-stage embryos. Each mRNA focus was categorized as localized in the core, the shell, or at the immediate periphery of germ granules. The localization of nos mRNA whose translation was active in both early and late stage embryos did not significantly change between both time points (Fig. 4e, e’). Similarly, the localization of pgc and CycB mRNAs whose translation started beyond the developmental period analyzed did not change between both stages (Fig. 4g–h’). In contrast, gcl mRNA localization changed after the onset of its translation (Fig. 4f, f’). While in early-stage embryos, 30% of gcl mRNA foci localized in the core of germ granules, this percentage decreased to 13.5% in late stage embryos. Therefore, upon translation, gcl mRNA relocalized from the core to the periphery of germ granules, showing that the distribution of mRNAs within germ granules correlated with their translation status.
Fig. 4. mRNA localization within germ granules.
a–d STED images of immuno-smFISH of wild-type embryos (20–120 min) with anti-Osk (magenta) and smFISH probes (green) against nos (a), gcl (b), pgc (c), and CycB (d) mRNAs. e–h Percentage of localization of nos, gcl, pgc, and CycB mRNA foci in early (0–20 min) and late (20–90 min) wild-type embryos, in the core (black), in and at the surface of the shell (grey) and at the immediate periphery (white) of germ granules from images as in (a–d). ns: non-significant, ****p < 0.0001 using the χ2 test. p = 0.61 in (e), 1.49 × 10−18 in (f), 0.26 in (g) and 0.17 in (h). e’–h’, Radar plots of the relative localization of nos, gcl, pgc, and CycB mRNAs (green dots) within Osk immunostaining (magenta) in early (0–20 min) and late (20–90 min) embryos from images as in (a–d). The granule shell is in pink. Scale bars: 1 µm. Source data are provided as a Source Data file.
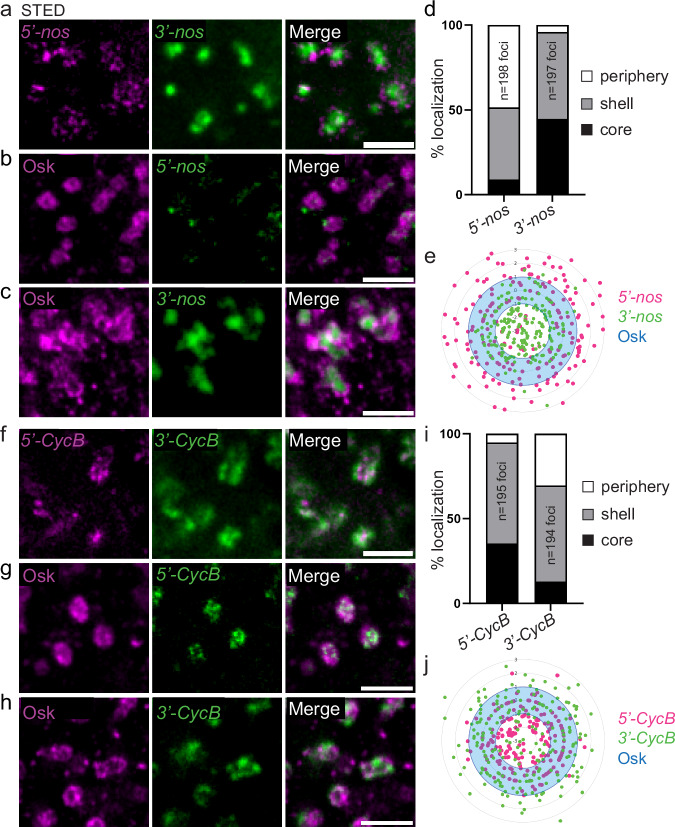
To address mRNA orientation in relation with their translation within germ granules, we next designed smFISH probes directed against the 5′end and 3′end of nos (5′-nos and 3′-nos) as an example of translated mRNA, and CycB (5′-CycB and 3′-CycB) as an example of repressed mRNA (Supplementary Fig. 4a), and performed double smFISH or immuno-smFISH with anti-Osk antibody of embryos. Notably, nos mRNA 3′ends assembled in a single cluster per granule, surrounded by many smaller foci corresponding to nos 5′ends (Fig. 5a). The specificity of nos smFISH probes was confirmed by the lack of signal obtained with these probes in nosBN mutant embryos (Supplementary Fig. 4b). In addition, we swapped the fluorophores between 5′-nos and 3′-nos probes and confirmed nos orientation with this new set of probes (Supplementary Fig. 4c). Using the new 3′-nos probe that provides a more defined signal, we could identify discrete central nos mRNA 3′ends (Supplementary Fig. 4c), and the same number of 5′-nos and 3′-nos foci per granule (Supplementary Fig. 4d). With regards to germ granule core/shell organization defined by Osk staining, nos 5′ends were predominantly localized in the shell and at the immediate periphery of germ granules, whereas nos 3′ends localized within the core and shell of germ granules (Fig. 5b–e). Interestingly, CycB mRNA showed the reverse orientation. CycB 5′ends were localized more internally than their 3′ends (Fig. 5f), and when compared to germ granule organization, CycB 5′ends localized within the core and shell of germ granules, whereas its 3′ends localized more externally in the shell and at the periphery of the granules (Fig. 5g-j).
Fig. 5. mRNA orientation within germ granules.
a STED images of smFISH of wild-type embryos with nos 5′end (5′-nos, magenta) and 3′end (3′-nos, green) probes. b, c STED images of immuno-smFISH with anti-Osk antibody (magenta) as a marker of germ granules and nos 5′end (b) or 3′end (c) probes (green). d Percentage of nos 5′end and 3′end foci localized in the core (black), in and at the surface of the shell (grey), and at the immediate periphery (white) of germ granules from images as in (b, c). e Radar plot of the relative localization of nos 5′end (magenta dots) and 3′end (green dots) within Osk immunostaining (blue) in wild-type embryos from images as in (b, c). The granule shell is in blue. f STED images of smFISH of wild-type embryos with CycB 5′end (5′-CycB, magenta) and 3′end (3′-CycB, green) probes. g, h, STED images of immuno-smFISH with anti-Osk antibody (magenta) as a marker of germ granules and CycB 5′end (g) or 3′end (h) probes (green). i Percentage of CycB 5′end and 3′end foci localized in the core (black), in and at the surface of the shell (grey), and at the immediate periphery (white) of germ granules from images as in (g, h). j Radar plot of the relative localization of CycB 5′end (magenta dots) and 3′end (green dots) within Osk immunostaining (blue) in wild-type embryos from images as in (g, h). The granule shell is in blue. Scale bars: 1 µm. Source data are provided as a Source Data file.
Together, these results show mRNAs translocate from the core towards the shell and periphery of germ granules upon translation and that they have a specific orientation within the granules, 5′ends of translated mRNAs being localized externally.
Reducing translation increases mRNA compaction in germ granules
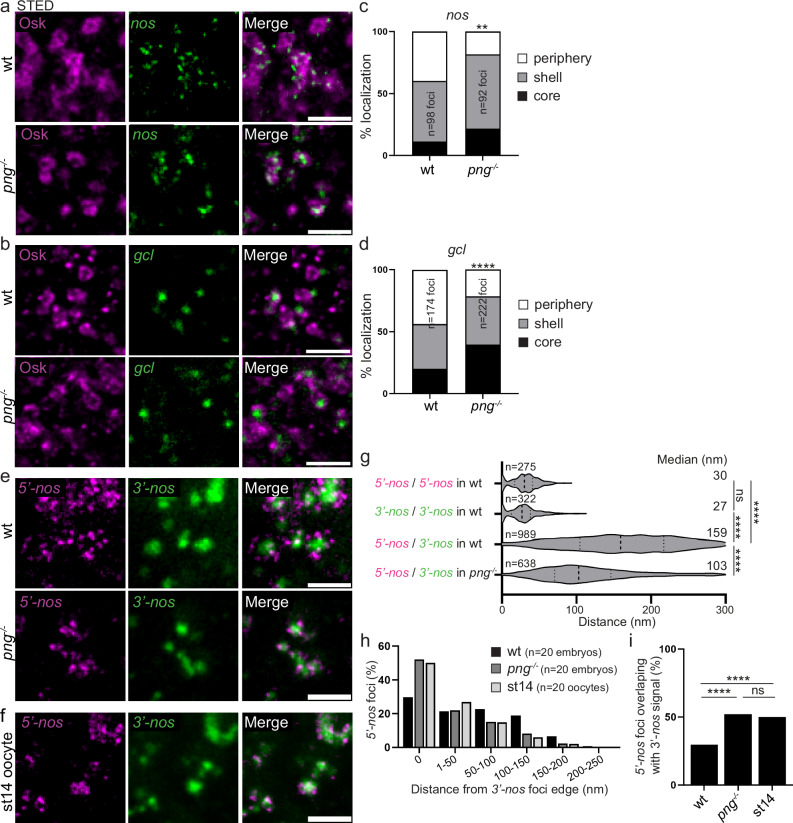
Egg activation that occurs upon egg laying triggers a massive remodeling of the maternal transcriptome landscape, involving drastic changes in mRNA stability and translation efficiency51,52. The Pan gu (Png) kinase has a critical role in promoting translation at egg activation through the phosphorylation of several translational repressors53. We addressed whether nos mRNA translation at germ granules would be affected in png mutant embryos, making them an excellent genetic system to analyze the links between mRNA translation and localization within germ granules, if it was the case. Using immuno-smFISH and confocal imaging to visualize suntag-nos mRNA and scFv-GFP foci, we found a significant decrease in the number of suntag-nos mRNA clusters colocalizing with scFv-GFP, i.e., undergoing translation, in png mutant compared to control embryos (Supplementary Fig. 5a, b), whereas the number of suntag-nos mRNA molecules per cluster was not reduced (Supplementary Fig. 5c). Therefore, suntag-nos mRNA translation was reduced in png mutant embryos. We analyzed the distribution of nos and gcl mRNAs within germ granules in png mutant embryos using immuno-smFISH with anti-Osk antibody and STED imaging. Importantly, both nos and gcl mRNA localization were affected in png mutant embryos where the percentage of mRNA foci localized in the core of germ granules increased at the expense of that at the periphery (Fig. 6a–d). In contrast, the localization within germ granules of CycB mRNA that was not translated at this stage, did not change in png mutant embryos (Supplementary Fig. 6a, b). Thus, decreasing translation led to a redistribution of translating mRNAs towards the inside of germ granules.
Fig. 6. mRNA localization and compaction within germ granules depend on their translation.
a, b STED imaging of immuno-smFISH of wild-type (top) and png1058 (bottom) embryos with anti-Osk antibody (magenta) as a marker of germ granule and smFISH probes (green) against nos (a) and gcl (b) mRNAs. c, d Percentage of localization of nos (c) and gcl (d) mRNA foci in wild-type and png1058 mutant embryos, in the core (black), in and at the surface of the shell (grey), and at the immediate periphery (white) of germ granules from images as in (a, b). **p < 0.01, ****p < 0.0001 using the χ2 test. p = 0.0031 in (c) and 1.75 × 10−22 in (d). e STED images of smFISH against nos 5′end (5′-nos, magenta) and 3′end (3′-nos, green) in wild-type and png1058 mutant embryos. f STED images of smFISH against nos 5′end (5′-nos, magenta) and 3′end (3′-nos, green) in wild-type stage 14 oocytes. g Violin plots showing distance distribution of colocalizing foci (5′-nos to 5′-nos and 3′-nos to 3′-nos) and 5′end to 3′end distances for nos mRNAs at germ granules in wild-type and png1058 mutant embryos, from STED images as in (e). Dashed lines inside the violin plots show first quartile, median, and third quartile. Median distances are indicated on the right. ns: non-significant, ****p < 0.0001 using unpaired two-tailed Student’s t-test. p = 0.2 between 5′-nos/5′-nos and 3′-nos/3′-nos, p = 3.5 × 10−191 between 5′-nos/5′-nos and 5′-nos/3′-nos in wt, p = 1.3 × 10−218 between 3′-nos/3′-nos and 5′-nos/3′-nos in wt and p = 4.5 × 10−58 between 5′-nos/3′-nos in wt and png−/−. The number of measured distances is indicated (n). h Measurement of the distance between nos 5′end foci and the edge of nos 3′end foci in wild-type embryos, png1058 embryos, and wild-type stage 14 oocytes (st14), from images as in (e, f). The histogram shows the percentage of nos 5′end foci in each distance class. i Percentage of nos 5′end foci overlaps with nos 3′end foci in wild-type embryos, png1058 embryos and wild-type stage 14 oocytes (st14). ns: non-significant, ****p < 0.0001 using the χ2 test. p = 2.64 × 10−129 between wt and png−/−, p = 1.79 × 10−167 between wt embryos and stage 14 oocytes, and p = 0.15 between png−/− embryos and stage 14 oocytes. Scale bars: 1 µm. Source data are provided as a Source Data file.
We next analyzed the orientation and conformation of nos mRNA within germ granules upon reduced translation using double smFISH with nos 5′end and 3′end specific probes and STED imaging. The 5′−3′ orientation of nos mRNA within germ granules did not change in png mutant embryos, nos 5′end remaining localized externally to nos 3′end (Fig. 6e). However, both ends appeared closer to each other than in wild-type embryos. To measure the distance between nos 5′end and 3′end, we first determined the colocalization precision in our experimental conditions. We performed smFISH with the same nos probe labeled with two different fluorophores; we identified the centers of each signal using a 2D Gaussian fitting and measured the distances between colocalizing signals (see “Methods”)54. We recorded the median distances to be 30 nm and 27 nm for the 5′-nos and 3′-nos probes, respectively, indicating that we can resolve mRNA 5′end and 3′end when they are distant by more than 30 nm (Fig. 6g). Using this approach, we found that the median distance between nos 5′end and 3′end in wild-type germ granules was 159 nm, whereas it was reduced to 103 nm in png mutant embryos (Fig. 6g). We then recorded the range of distances between nos 5′end and 3′end by measuring the distance between the center nos 5′end signal and the edge of nos 3′end signal (see “Methods” and Supplementary Fig. 7) in wild-type and png mutant embryos. We observed that the proportion of nos 5′end overlapping with nos 3′end (distance = 0) increased in png mutant compared to wild-type embryos (Fig. 6h, i), at the expense of nos 5′end separated from nos 3′end by more than 50 nm. A similar increase in mRNA compaction was not observed with CycB mRNA in pgn mutant embryos (Supplementary Fig. 6c–e). Furthermore, the distance between 5′end and 3′end for CycB was closer than that for nos mRNA in wild-type embryos (compare Fig. 6h, i and Supplementary Fig. 6d, e).
In addition to png mutant embryos, we took advantage of another context where translation is reduced. A previous study based on GFP-Nos fusion protein reported that nos mRNA translation started at the posterior pole during oogenesis, in stage 13 oocytes55. However, using suntag-nos mRNA, we found that translation did not occur in stage 14 oocytes (Supplementary Fig. 8), suggesting that nos translation at germ granules was initiated or strongly increased following egg activation. We found that the distance between nos 5′end and 3′end was closer in stage 14 oocytes than in embryos, and similar to that in png mutant embryos (Fig. 6f–i).
These results strengthen the conclusion that mRNAs move towards the outer phase and periphery of germ granules when they are translated. They also reveal that mRNA conformation depends on their translational status, showing higher compaction when they are not translated and decompaction upon translation.
Translation is perturbed in a tud mutant that lost germ granule biphasic organization
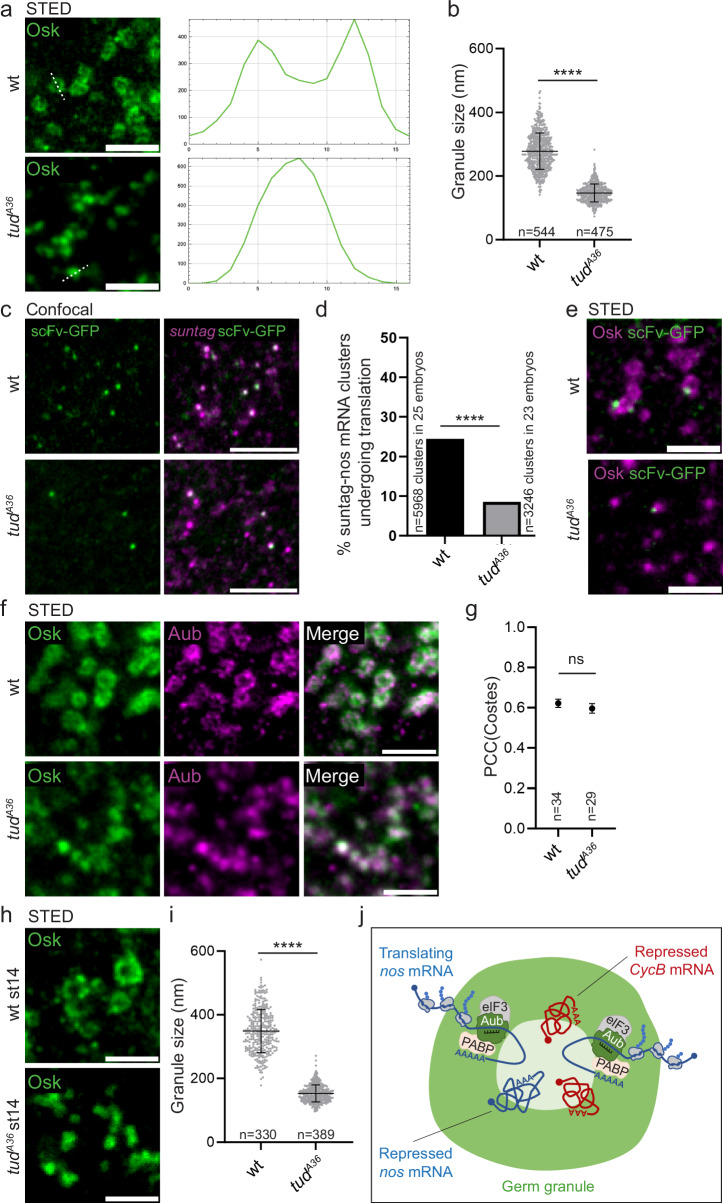
We sought to perturb germ granule architecture in order to address its contribution to germ granule functions. We took advantage of a single point mutant of tud in the first Tudor domain, tudA36 (Supplementary Fig. 9a), in which germ granules were described as rods instead of hollow spheres by electron microscopy26. Tud protein contains 11 Tudor domains. These domains are known to mediate interactions with other proteins, in particular through dimethylated arginines. Tudor domains in Tud have been proposed to serve as docking platforms for germ granule assembly26, and indeed the multiple Tudor domains should generate multivalent interactions known to be instrumental in the formation of condensates by phase separation56. Aub undergoes symmetric arginine dimethylation and Tud recruits Aub to germ granules through interaction between several Tudor domains and Aub dimethylated arginines57–60. Interestingly, in tudA36 mutant embryos, although germ granule mRNAs were shown to localize to the posterior pole, the formation of germ cells was strongly affected26, suggesting a defect in mRNA translation at germ granules. Analysis of germ granules in tudA36 embryos, using Osk immunostaining and STED imaging revealed a drastic alteration of germ granule organization. The biphasic structure was lost (Fig. 7a) and the size of germ granule was reduced to an average of 147 ± 28 nm instead of 279 ± 57 nm in wild-type embryos (Fig. 7b). Investigating suntag-nos translation in tudA36 mutant embryos using immuno-smFISH and confocal imaging to record suntag-nos mRNA clusters and scFv-GFP foci, we found a strong decrease in translation (Fig. 7c). The percentage of translating suntag-nos mRNA clusters, i.e., colocalizing with scFv-GFP foci, decreased from 24.4% in wild-type embryos to 8.5% in tudA36 mutant embryos (Fig. 7d), whereas suntag-nos mRNA localization to germ granules was only slightly reduced (mean of 2.4 suntag-nos molecules per cluster in wild-type germ granules versus 2.1 in tudA36) (Supplementary Fig. 9b). We analyzed the localization of suntag-nos translation within germ granules using immunostaining to reveal scFv-GFP foci and Osk as a germ granule marker, visualized with STED microscopy. We confirmed a lower number of scFv-GFP foci in germ granules in tudA36 embryos and these foci localized mostly at the periphery of germ granules or close to their surface (Fig. 7e, Supplementary Fig. 9c), indicating that the structure of tudA36 germ granules was not compatible with translation inside the granules.
Fig. 7. nos mRNA translation depends on germ granule biphasic architecture.
a STED images of wild-type and tudA36/Df(2 R)PurP133 (tudA36) embryos immunostained with anti-Osk antibody. Fluorescence intensity was recorded along the white dotted line (right). b Measurement of germ granule size in wild-type and tudA36/Df(2 R)PurP133 (tudA36) embryos from images as in (a). Horizontal bars represent the mean and SD. ****p < 0.0001 using unpaired two-tailed Student’s t-test. p = 2.4 × 10−248. c Immuno-smFISH of nos-suntag-nos/+; nos-scFv-GFP/+ (wild-type) and nos-suntag-nos tudA36/Df(2 R)PurP133; nos-scFv-GFP/+ (tudA36) embryos with anti-GFP nanobody (green) to reveal scFv-GFP and suntag smFISH probe (magenta). d Percentage of suntag-nos mRNA clusters colocalizing with scFv-GFP foci, i.e., undergoing translation, in nos-suntag-nos/+; nos-scFv-GFP/+ (wild-type) and nos-suntag-nos tudA36/Df(2 R)PurP133; nos-scFv-GFP/+ (tudA36) embryos from images as in (c). ****p < 0.0001 using χ2 test. p = 2 × 10−77. e STED images of immunostaining of nos-suntag-nos/+; nos-scFv-GFP/+ (wild-type) and nos-suntag-nos tudA36/Df(2 R)PurP133; nos-scFv-GFP/+ (tudA36) embryos with anti-Osk antibody (magenta) and anti-GFP nanobody (green) to reveal scFv-GFP. f STED images of immunostaining of wild-type and tudA36/Df(2 R)PurP133 (tudA36) embryos with anti-Osk (green) and anti-Aub (magenta) antibodies. g Quantification of colocalization between Osk and Aub using PCC(Costes). Black circles represent the mean and error bars represent SEM. The number of embryos is indicated (n). ns: non-significant using unpaired two-tailed Student’s t-test. p = 0.42. h STED images of immunostaining of wild-type and tudA36/Df(2 R)PurP133 (tudA36) stage 14 oocytes with anti-Osk antibody. i Measurement of germ granule size in wild-type and tudA36/Df(2 R)PurP133 (tudA36) stage 14 oocytes from images as in (h). Horizontal bars represent the mean and SD. ****p < 0.0001 using unpaired two-tailed Student’s t-test. p = 6.2 × 10−209. j Model of functional compartmentalization of germ granules. The scheme represents a biphasic germ granule with main protein components accumulating in the outer phase (green). Repressed mRNAs in a condensed state accumulate towards the granule core. Translation takes place in the outer phase and at the periphery of the granule. Translational activators (eIF3 and PABP) are found in the outer phase. Translated mRNAs are less condensed and anchored to the granule core from their 3′region, whereas their 5′end is oriented toward the outer phase and periphery of the granule. mRNAs translocate from the core to the outer phase during translation. Scale bars: 1 µm. Source data are provided as a Source Data file.
Aub activates nos mRNA translation initiation43 and Vasa was also reported to play a role in activating translation61. We, therefore, asked whether Aub or Vasa recruitment to germ granules was impaired in tudA36 embryos, which might contribute to reduced suntag-nos mRNA translation. Immunostaining to examine the posterior recruitment of Aub and Vasa together with Osk in tudA36 mutant embryos showed that their recruitment was not affected as quantified using confocal imaging and measuring immunostaining intensity relative to that of Osk (Supplementary Fig. 9d–g). In addition colocalization of Aub with Osk analyzed using STED microscopy showed that Aub was recruited to germ granules in tudA36 embryos, with a colocalization with Osk similar to that in wild-type embryos (Fig. 7f, g). Germ granule altered architecture in tudA36 mutant might be the consequence rather than the cause of reduced translation levels. To address this question, we analyzed germ granule organization in tudA36 stage 14 oocytes in which translation has not started yet or was occurring at very low levels. We found that the defects in germ granule architecture in stage 14 oocytes were similar to that recorded in tudA36 embryos (Fig. 7h, i), indicating that altered granule organization preceded the burst of translation that took place at egg activation.
We conclude that translation is strongly reduced in tudA36 mutant embryos due to the defective architecture of germ granules, although we cannot totally exclude the contribution of another Tud function. These results reveal tight links between germ granule core/shell organization and mRNA translation.
Discussion
In this manuscript, we use Drosophila germ granules as a model system to address the relationships between the multi-layered organization of biomolecular condensates and their functions. Drosophila germ granules ensure two opposite functions: storage of translationally repressed mRNAs and sequential translational activation of the same mRNAs. Using super-resolution STED microscopy, we find that these germ granules have a biphasic organization with an accumulation of protein components in their outer phase. Analyses of mRNA localization within germ granules combined with the suntag approach to record ongoing translation reveal that the storage of translationally repressed mRNAs takes place in the core of germ granules, whereas translation occurs in the shell and at the periphery of the granules. Therefore, the granule shell and periphery are permissive for translation, while the core is not. Using 5′end and 3′end probes of the same mRNAs, we show that mRNA orientation within germ granules reflects their translation status, 5′ends of translated mRNAs pointing towards the surface and periphery of germ granules. Moreover, mRNA compaction correlates with their translation levels, reduced translation leading to higher mRNA compaction, as it was described recently in stress granules54. Finally, taking advantage of a mutant in which the biphasic architecture of germ granules is lost, we show that translation is linked to their core/shell organization. These results demonstrate the functional compartmentalization of germ granules, a process that is central for germ cell specification and development.
We propose a model in which repressed mRNAs are stored in a compacted state in the core of germ granules and translocate to the outer phase for translation (Fig. 7j). To our surprise, we did not identify translational repressors accumulating in the core of germ granules. Together with the information that untranslated mRNAs have their 5′- and 3′regions closer than translated mRNAs, and are thus more compacted, these data suggest that the lack of translation in the core of germ granules would involve a mechanism based on mRNA compaction and compartmentalization away from the translation machinery, independent of specific translational repressors. Nonetheless, several lines of evidence point to a second mechanism involving translational repressors. First, the 3′end of the repressed mRNA, CycB is oriented towards the shell and periphery of germ granules, and we also find the association of Smaug translational repressor with the surface and periphery of the granules, suggesting that Smaug might bind repressed mRNA 3′UTRs in this location. Smaug interaction with Osk in this external part of germ granules would relieve translational repression45,46. Second, suntag-nos translation at germ granules is reduced in png mutant embryos and Png kinase is known to relieve translational repression through phosphorylation of translational repressors53. These data suggest the implication of translational repressors in translational regulation at germ granules. Third, consistent with this, translational activation of germ granule mRNAs is sequential, again suggesting the role of specific repressors, or combinations of repressors whose effect would be relieved sequentially to achieve timely translation of specific mRNAs.
A major information from this study is that Drosophila germ granules have a function in translation. These granules are not incidental condensates regarding the translation of germ granule mRNAs4 as translation of these mRNAs does not take place anywhere else in the embryo. Therefore, germ granules have an essential function in localized translation of specific mRNAs, which is required for germ cell development. Another key finding is the fact that translation takes place in a defined region of germ granules, the outer phase, indicating that the biophysical properties of this outer phase allow translation. Quantifications of nos mRNA levels in germ granules have shown that these levels do not decrease with time, up to the formation of primordial germ cells62. This indicates that nos mRNAs might be translated several times at germ granules without decay. These consecutive rounds of translation would be possible due to the capacity of the outer phase to allow translation, bypassing the necessity for mRNAs to leave germ granules to be translated, as it is the case in C. elegans22,23. Indeed, we found that translation initiation factors concentrate in this phase of germ granules. Thus, in addition to a role in localizing translation within the embryo, germ granules might be involved in increasing translation efficiency through the outer phase properties that would allow consecutive rounds of mRNA translation in the absence of decay.
RNA granules have a recognized role in mRNA storage (e.g., P bodies and stress granules), but their function in translational activation has remained more elusive and is currently an emerging question. A recent study showed that translation can occur in stress granules, although these granules are mostly composed of untranslated mRNAs63. In addition, translating mRNAs were also found capable of transiently docking to the surface of stress granules64. More recently, another study revealed that the formation of RNA granules driven by the FXR1 protein led to the translation of mRNA targets through translation initiation factor recruitment, thus identifying the first RNA condensate specialized in translation activation65. A recent analysis of zebrafish embryo germ granules reported that mRNA translation required their translocation to the granule periphery66. Finally, while this manuscript was under review, another study reporting translation at the surface of Drosophila germ granules was published67. These data align with previous electron microscopy analyses showing polysomes emerging from the surface of germ granules25. Here, we found that translation occurs in a specific phase of germ granules, the outer phase, highlighting the functional relevance of RNA granule higher-order organization. Whether such a functional organization is a conserved feature of germ granules remains an open question. In favor of this possibility, it was shown that during mouse spermatogenesis, translational activation coincided with the recruitment of eIF3f by Miwi (the mouse homolog of Aub) to the edge of the chromatoid body68. Understanding how translation is linked to the formation and biophysical properties of a specific phase of germ granules represents an interesting challenge for future studies.
Methods
Drosophila lines
w1118 was used as wild-type (wt). Mutant alleles and transgenic lines were: UASp-GFP-Aub69, nos-Gal4-VP1670, UASp-HA-eIF3d43, UASp-osk-bcd3′UTR (short osk)71, Vasa-GFPKI, GFP-TudKI and vasa-tdTomKI72, nos-scFv-GFP40, yw nos-MS2-5 nos-MS2-2573, nosBN/TM3sb74, belCC0086944,75, png1058/FM6a76 and tudA36/Cyo26, Df(2 R)PurP133/CyO. The nos-suntag-nos and nos-MCP-suntag lines were generated in this study by PhiC31 recombination into the attP40 and attP9A sites, respectively. To record translation, females from the nos-scFv-GFP line were crossed with males of the nos-suntag-nos line. The genotypes of embryos (aged 0–2 h or less) indicated throughout were the genotypes of mothers. Females of the indicated genotypes were crossed with wt males.
Cloning and recombineering
To produce the nos-suntag-nos transgene, the sequence coding for 12 Suntag repeats was amplified by PCR from a clone provided by Bertrand34. The PCR fragment was cloned after the nos start codon into the pBSKS-R5561 plasmid that contains a 5.7 kb nos genomic fragment (a gift from Wharton77), using NEBuilder HiFi DNA Assembly (NEB). The nos genomic fragment containing suntag sequence was digested by EcoRI and NotI and cloned into the pattB vector (DGRC #1420) digested by EcoRI and NotI. The resulting plasmid was validated by sequencing and sent for injection (BestGene Inc) to be inserted into the Drosophila genome by PhiC31 recombination at the attP40 site. To produce the nos-MCP-suntag transgene the sequence coding for 12 Suntag repeats was amplified by PCR. The PCR fragment was cloned using NEBuilder HiFi DNA Assembly (NEB) in frame with MCP coding sequence into the pNosPE_MCP-TagRFPT-NLS plasmid that contains a nos promoter, nos 5′UTR, MCP coding sequence in frame with RFPt coding sequence (a gift from Dufourtt40) digested by HindIII and BamHI to remove the RFPt sequence. The resulting plasmid was validated by sequencing and sent for injection (BestGene Inc) to be inserted into the Drosophila genome by PhiC31 recombination at the attP9A site.
Immunostaining
0–2 h-embryos were collected in a basket from plates, washed in tap water and dechorionated using commercial bleach for 2 min, and rinsed. Embryos were then fixed at the interface of a 1:1 solution of formaldehyde 36%/heptane for 5 min, followed by 100% methanol devitellinization. Embryos were progressively rehydrated in 75%, 50%, and 25% methanol diluted in PBS 0.1% Tween, blocked in 1% BSA for 1 h for confocal imaging or 10% BSA for 3 h for STED imaging, and incubated overnight with primary antibodies. Secondary antibody incubation, after washes in PBS 0.1% Tween, was performed for 1 h at room temperature. Embryos were mounted in Vectashield (Vector Laboratories) for confocal imaging or Abberior liquid anti-fade (Abberior) for super-resolution imaging (STED, OMX and Airyscan). For super-resolution imaging the posterior pole of embryos was sliced using a thin needle in mounting medium and mounted posterior side up (Supplementary Fig. 1a). For stage 14 oocytes, ovaries were dissected from well fed females in Schneider medium and fixed in a chelating solution (1:1 heptane/PBS 1X, 50 mM EGTA pH8, 10.24% formaldehyde) to prevent egg activation, for 1.5 h at room temperature. Then ovaries were rinsed three times in PBS 0.1% Tween, transferred to a dissection dish where oocytes were dissociated from the rest of the ovary by multiple pipetting. Oocytes were put on a frosted slide and rolled with another frosted slide to remove chorion and vitelline membrane. Oocytes were transferred in a tube and processed for immunostaining as described for early embryos. Primary antibodies used were: mouse anti-Aub (1/1000, clone 4D10, a gift from M. Siomi,78), rabbit anti-CCR4 (1/200)79, rabbit anti-Cup (1/1000, a gift from R. Wharton,80), rabbit anti-Me31B (1/2000, a gift from A. Nakamura,81), rabbit anti-Nos (1/1000, a gift from A. Nakamura), mouse anti-Not1 (1/100, clone 2G5,79), rabbit anti-Osk (1/1000, this study), rabbit anti-PABP (1/500, a gift from A. Vincent), rabbit anti-Smg (1/1000,82), rabbit anti-Tud (1/500, a gift from P. Lasko,83), mouse anti-HA (1/2000, ascites produced from clone 12CA5), rat anti-Vasa (1/50, Developmental Studies Hybridoma Bank), rabbit anti-GFP (1/1000, Invitrogen), mouse anti-GCN4 (1/1000, clone C11L34, Bio-Techne) and mouse anti-GFP (1/1000, Roche). Secondary antibodies used were: FluoTag®-X4 anti-GFP nanobodies StarRed or Atto488 (1/500, NanoTag Biotechnologies), goat anti-rabbit IgG Alexa-488 (1/800, Invitrogen), donkey anti-rabbit IgG Cy5 (1/1000, Jackson ImmunoResearch), goat anti-mouse IgG StarRed (1/1000, Abberior), goat anti-rabbit IgG StarRed (1/1000, Abberior), and goat anti-rabbit IgG Star580 (1/1000, Abberior).
smFISH and Immuno-smFISH
smFISH on 0–2 h-embryos were performed as previously described43 with smFISH probes from Stellaris. For immuno-smFISH, rehydrated embryos were post-fixed in 4% formaldehyde for 20 min, rinsed three times for 10 min in PBS 0.1% Tween, and processed for smFISH followed by immunostaining. smFISH on stage 14 oocytes were performed as previously described84 using 10% formamide. Probe sequences for nos, gcl, pgc, and CycB are listed in ref. 18; the MS2 probe is described in ref. 85. Probe sequences to detect suntag, nos 5′end, nos 3′end, CycB 5′end and CycB 3′end are listed in Supplementary Tables 1–5. Probes were coupled to Cal Fluor 590 (nos, gcl, pgc, CycB, suntag, 5′-nos, 3′-nos, 3′-CycB, and MS2) or Quasar 670 (nos, 5′-nos, 3′-nos, and 5′-CycB).
Puromycin treatment
To treat embryos with puromycin, we adapted the permeabilization protocol previously described86. Dechorionized embryos were put in a 1:1 solution of DL-limonene (Sigma)/heptane with a 100 µl drop of PBS 1X containing 2 mg/ml puromycin (Sigma). Puromycin was initially diluted in water at 50 mg/ml. For control embryos, the puromycin solution was replaced by water alone. Embryos were shaken at maximum speed on a rotating plate for 40 min, rinsed in heptane, and processed for immunostaining.
Generation of anti-Osk antibody
The open reading frame of osk (short isoform) without the start codon was cloned after the GST sequence into the pGEX-4T-1 vector to express a GST-Osk fusion protein in BL21 E. coli bacteria. The GST-Osk fusion protein was isolated on an acrylamide gel, purified using columns (Amicon), and injected into rabbits by Agro-Bio. The polyclonal rabbit antibody against this fusion protein was validated using immunostaining in ovaries and embryos.
Fluorescence microscopy
Confocal microscopy was performed using a Leica SP8 confocal scanning microscope with objectives 20X Plan Apochromat 0.75 NA Imm Corr for whole embryos (Supplementary Fig. 2a, b and 3b) or 63X Plan Apochromat 1.4 NA oil DIC for other confocal images. STED microscopy was performed using an Abberior STED super-resolution microscope controlled by Imspector software (Abberior Instruments) using a 100X Plan SuperApochromat 1.4 Oil objective. Excitation and depletion laser powers were adjusted according to the strength of the signal to optimize the resolution without bleaching the sample as follows. Excitation laser powers were set to collect, in confocal mode, approximately 200 grey levels for granule staining and 100 grey levels for other stainings. The optimal STED laser power was defined as the value giving the best resolution, and at the point where resolution was not improved while signal-to-noise ratio was decreasing. Resolution scales with the square root of the applied intensity and increasing laser power results in a marginal benefit in resolution when the optimal resolution is reached87. Pixel size was set to 30 nm. Resolution in our imaging conditions was 60 ± 12 nm, assessed by measuring the full width at half maximum of the PSF (Point Spread Function) of the smallest structure imaged in our samples (5′-nos signal in the soma). No pixel shift correction was applied as red and far-red signals were obtained from the same depletion laser aligned on the excitation lasers and photons collected followed the same optic path before spectral detection. In addition, SuperApochromat objective corrects the chromatic and spherical aberrations. Laser alignments were checked monthly and alignment measurements on tretraspeck 100 nm beads showed a shift of less than half a pixel. For OMX microscopy, 3D-SIM acquisitions were performed on a Deltavision OMX V4 and reconstructed using SoftWoRx software. The OMX used was equipped with a 100X Plan Apochromat NA1.4 Oil PSF graded objective and EMCCD cameras. For two color imaging in OMX, both channels were acquired sequentially but in immediate temporal proximity to minimize drift and crosstalk. In addition, multiple channels were realigned following manufacturer’s procedures and calibration using the provided target slide. OMX image quality was assessed using SIMcheck, a FIJI plugin that assesses SIM reconstructed image quality through metric evaluation on the raw images, as previously described88. All images conformed to the requirements for intensity profile, modulation contrast to noise ratio and minimum to maximum intensity ratio. Airyscan microscopy was performed using confocal Zeiss LSM980 AiryScan II 8Y run by Zeiss Zen Blue software using a 63X Plan Apo oil 1.4NA objective.
Transmission electron microscopy (TEM)
For TEM, 0–1 h-embryos were collected in a basket from plates, washed in tap water and dechorionated using commercial bleach for 2 min and rinsed. Embryos were then fixed in heptane saturated with 12.5% glutaraldehyde in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 4 mM MgSO4·7 H20; EM grade) for 20 min at room temperature. The vitelline membrane was manually removed and the posterior poles were dissected and fixed in 2.5% glutaraldehyde in PHEM buffer overnight at 4 °C. Embryo posterior poles were post-fixed in a 0.5% osmic acid + 0.8% potassium Hexacyanoferrate trihydrate for 2 h at dark and room temperature. After two washes in PHEM buffer, they were dehydrated in a graded series of ethanol solutions (30–100%). The embryo posteriors were embedded in EmBed 812 using an Automated Microwave Tissue Processor for Electronic Microscopy (Leica EM AMW). Ultra-thin sections of 70 nm were cut (Leica-Reichert Ultracut E), mounted on formvar-coated slotted copper grids, and counterstained with uranyl acetate 1.5% in 70% ethanol and lead citrate. Stained grids were observed using a Tecnai F20 transmission electron microscope at 120KV, at the Electronic Microscopy facilities (Institut des Neurosciences de Montpellier).
Image analyses and quantifications
Before image analysis, all images were filtered with a Gaussian blur to reduce the noise and smooth the image. All images presented in the figures were filtered with Gaussian blur. No deconvolution was applied.
Quantification of mRNA molecule numbers
mRNA content in germ granules was quantified using FISH-quant34,41. Briefly, the mean intensity of single mRNA molecule was calculated from the signal of single particles in the soma as they mostly correspond to single mRNA molecules (83% of single particles for nos mRNA42). To determine the number of mRNA molecules present in each mRNA cluster at the posterior pole, the integrated intensity of each cluster was divided by the integrated intensity of the mean single mRNA molecules.
Quantification of germ granule size and shell size
On STED acquisitions of Osk or Aub immunostaining, we applied a Laplacian Gaussian filter to better define the signal edges. A line was drawn through the diameter of each granule and intensities along the line were measured using FIJI plot profile, giving two peaks that reflected the donut-like shape of germ granules (Fig. 1a). To determine the limit of the signal, we measured the full width at half maximum of the PSF for each peak and between the most external part of each peak, allowing the measurement of the granule diameters and shell thickness. From this, we defined the core and shell of germ granules. The surface of germ granules corresponded to their external edge, and the periphery corresponded to the cytoplasm surrounding the granule up to 200 nm. This value was chosen as we never observed RNAs or scFv-GFP foci with distances from the granule surface larger than this.
Quantification of the localization of mRNAs or scFv-GFP foci within the germ granule biphasic structure
To analyze the localization of mRNA foci (visualized either with the full-length, the 5′end, or the 3′end probes) or scFv-GFP foci within germ granule biphasic organization, we first applied a Laplacian Gaussian filter on the channel showing the germ granule staining to define the signal edges and thus the compartments (core, shell, periphery). Then mRNA coordinates were determined using RS-FISH ImageJ plugin89. A line was drawn across the granule and mRNA foci and pixel intensities along the line were recorded on Fiji Plot profile. The distance between the peak of the shell and the peak of the mRNA focus was measured on the plot. If the mRNA focus was located toward the external part of the granule, the measure was given a positive value. If the mRNA focus was toward the internal part of the granule, it was given a negative value. As the size of the shell is not identical between granules, the shell size was also measured for each measurement of the distance between the shell peak and mRNA focus, allowing us to determine the position of the mRNA focus within the granule using the following ratio:
distance between the peak of mRNA focus and the peak of the shell / (shell size/2).
If the ratio was > 1, the RNA focus was classified as “periphery”. If the ratio was ≤ −1, the RNA focus was classified as “core”. If the ratio was > −1 and ≤ 1, the RNA focus was classified as “shell”. Quantifications of these ratios were shown as graphs, and the localization of mRNA foci were represented on hypothetic granules using radar plots. The same method was applied to quantify the position of scFv-GFP foci within germ granules.
Analysis and quantification of colocalization of scFv-GFP foci with suntag mRNA clusters and/or germ granules
On confocal images, germ granules appear as full dots or foci. To measure the association of scFv-GFP foci with germ granules or suntag mRNA clusters, we used FIJI plugin ComDet v.0.5.5 (https://github.com/UU-cellbiology/ComDet) to define the different foci and measure their colocalization. To visualize the colocalization of scFv-GFP foci with suntag mRNA clusters at germ granules (i.e., translation at germ granules, Supplementary Fig. 3c), we segmented the signals from suntag smFISH and scFv-GFP and created a mask that showed their colocalization (translation foci) using FIJI Image Calculator function. We then overlaid this mask on the channel showing germ granules visualized with anti-Osk immunostaining and quantified the colocalization of translation foci with germ granules.
Colocalization precision
To determine the colocalization precision of smFISH probes under our imaging conditions, distances between signals obtained with the same probe labeled with two different fluorophores were measured. Embryos were hybridized with a mix of oligos labeled with Quasar 670 and the same oligos labeled with Cal Fluor Red 590. This was performed for the 5′-nos and 3′-nos probes. The center of each signal was determined by 2D Gaussian fitting using RS-FISH plugin89, and the distance between the signal centers from the two channels was measured using a FIJI plugin developed previously90. For measurements of 5′-nos to 3′-nos distances in wild-type and png mutant embryos, 300 nm was chosen as cutoff to avoid assigning signals from neighboring granules. This value was chosen from observations that 5′end to 3′end distances rarely extended beyond this cutoff and those that did corresponded to wrongful assignments.
Distance and colocalization between mRNA 5′end and 3′end
To measure the distance between nos mRNA 5′end and 3′end probes, the 3′end signal was segmented and defined as ROI. 5′end foci were identified using FIJI plugin ComDet v.0.5.5 (https://github.com/UU-cellbiology/ComDet) and their center was mapped by multipoint selection and added as ROI. The minimal distance between the edge of the 3′end ROI and the center of the 5′end foci was calculated using a FIJI plugin developed previously90. Overlap corresponds to a distance of 0.
Distance between Smaug foci and germ granules
To analyze the distance between Smaug foci and germ granules, Smaug signals above threshold were identified using the Fiji Find Maxima function. The signal for the granule corresponding to anti-Osk immunostaining was segmented and defined as ROI. The minimal distance between the edge of the granule ROI and the center of Smaug foci was calculated using a FIJI plugin developed previously90. As a control (Smg tilted), Smaug channel was horizontally and vertically rotated and the same measurement was performed.
Protein colocalization within germ granules
For images acquired with STED imaging, the degree of colocalization between signals corresponding to two immunostaining was quantified using PCC(Costes). The PCC method determines a threshold based on the mean fluorescence intensity for each signal. Then, the method analyzes each pixel and evaluates if signals are above or below their respective threshold, increasing PCC value if both signals go to the same direction, decreasing its value if they go to opposite directions. PCC ranges from 1 that indicates perfect colocalization, to −1 that indicates exclusion. To validate the significance of this colocalization, the PCC(Costes) value was calculated for each image. Briefly, images were randomized by shuffling pixels, the PCC value was calculated and compared to the original image. This process was repeated 200 times to evaluate the significance of the original picture. Costes p-value ranges from 0 to 1, where 0 indicates random colocalization and 1 indicates significant colocalization.
Quantification of germ granule main component levels
To quantify the level of Aub and Vasa in tudA36 mutants, ROIs delimiting the germ plasm on maximum Z projection of 40 planes from confocal images were defined. The fluorescence-integrated density of Aub or Vasa was measured and normalized to Osk fluorescence-integrated density levels.
Statistics and reproducibility
Statistical tests were performed using GraphPad. For each figure, the tests are indicated in the figure legend. Each experiment was repeated two to thirteen times (see detail below); quantifications were at least from two independent replicates. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The Investigators were not blinded to allocation during experiments and outcome assessment. Randomization was used in Figs. 3j, 7g and Supplementary Fig. 1d, and performed as explained above in the Image analyses and quantifications section. The number of times that experiments were repeated with similar results are as follows. Figure 1a: 2, 5, 3 and 4; Fig. 1b, c: 2; Fig. 2b, e: 13; Fig. 2d: 4; Fig. 2j: 5; Fig. 3a, d, e, f, h: 2; Fig. 3b, c, g, i: 3; Fig. 4a, b: 3; Fig. 4c, d: 2; Fig. 5a: 13; Fig. 5b: 6; Fig. 5c, f: 5; Fig. 5g, h: 3; Fig. 6a: 6; Fig. 6b: 4; Fig. 6e: 2; Fig. 6f: 4; Fig. 7a, f: 3; Fig. 7c: 6; Fig. 7e: 2; Fig. 7h: 3 and 4; Supplementary Fig. 1b: 4; Supplementary Fig. 1c, e, f: 2; Supplementary Fig. 2a, b: 3; Supplementary Fig. 2d: 2; Supplementary Fig. 3b, e, f, g, j: 2; Supplementary Fig. 3c, l: 4; Supplementary Fig. 4b: 2; Supplementary Fig. 4c, d: 3; Supplementary Fig. 5a: 2; Supplementary Fig. 6a, b: 2; Supplementary Fig. 8: 3; Supplementary Fig. 9d, f: 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We are grateful to A. Arkov, E. R. Gavis, M. Lagha, R. Lehmann and the Bloomington Drosophila Stock Center for providing Drosophila stocks. We thank E. Izaurralde, P. Lasko, A. Nakamura, M. Siomi, A. Vincent, and R. Wharton for the gifts of antibodies. We thank C. Jahan for producing the anti-Osk antibody. We are very grateful to A. Hubstenberger for helpful comments on the manuscript and to E. Bertrand and F. Slimani for their help with image quantification. We thank the MRI-IGH imaging facility, in particular M. P. Blanchard for her help with STED microscopy and J. M. Langerak for OMX imaging. This work was supported by the CNRS-University of Montpellier UMR9002, ANR (ANR-19-CE12-0031, ANR-21-CE12-0035-01), MSDAVENIR and FRM (Equipe FRM EQU202303016322). A. R. held a salary from ANR, Fondation ARC, and CNRS, A. H. held a PhD fellowship from the French Ministry and C. G. held a salary from ANR and MSDAVENIR.
Author contributions
Conceptualization: A.R., M.S. Methodology: A.H., A.R., M.S. Investigation: A.H., A.R., C.G. Visualization: A.H., A.R., C.G. Funding acquisition: M.S. Project administration: M.S. Supervision: M.S. Writing—original draft: A.R., M.S. Writing—review & editing: A.R., M.S.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data are available in the main text or Supplementary Information. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Ethics approval
We support inclusive, diverse, and ethical conduct in research.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Anne Ramat, Ali Haidar.
Contributor Information
Anne Ramat, Email: Anne.Ramat@igh.cnrs.fr.
Martine Simonelig, Email: Martine.Simonelig@igh.cnrs.fr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52346-x.
References
- 1.Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol.18, 285–298 (2017). 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol.28, 420–435 (2018). 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirose, T., Ninomiya, K., Nakagawa, S. & Yamazaki, T. A guide to membraneless organelles and their various roles in gene regulation. Nat. Rev. Mol. Cell Biol.24, 288–304 (2023). 10.1038/s41580-022-00558-8 [DOI] [PubMed] [Google Scholar]
- 4.Putnam, A., Thomas, L. & Seydoux, G. RNA granules: functional compartments or incidental condensates? Genes Dev.37, 354–376 (2023). 10.1101/gad.350518.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittag, T. & Pappu, R. V. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell82, 2201–2214 (2022). 10.1016/j.molcel.2022.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ripin, N. & Parker, R. Formation, function, and pathology of RNP granules. Cell186, 4737–4756 (2023). 10.1016/j.cell.2023.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fare, C. M., Villani, A., Drake, L. E. & Shorter, J. Higher-order organization of biomolecular condensates. Open Biol.11, 210137 (2021). 10.1098/rsob.210137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol.22, 165–182 (2021). 10.1038/s41580-020-0272-6 [DOI] [PubMed] [Google Scholar]
- 9.Riback, J. A. et al. Composition-dependent thermodynamics of intracellular phase separation. Nature581, 209–214 (2020). 10.1038/s41586-020-2256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell165, 1686–1697 (2016). 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodson, A. E. & Kennedy, S. Phase separation in germ cells and development. Dev. Cell55, 4–17 (2020). 10.1016/j.devcel.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voronina, E., Seydoux, G., Sassone-Corsi, P. & Nagamori, I. RNA granules in germ cells. Cold Spring Harb. Perspect. Biol.3, a002774 (2011). 10.1101/cshperspect.a002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rangan, P. et al. Temporal and spatial control of germ-plasm RNAs. Curr. Biol.19, 72–77 (2009). 10.1016/j.cub.2008.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barckmann, B. et al. Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep.12, 1205–1216 (2015). 10.1016/j.celrep.2015.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann, R. Germ plasm biogenesis—an Oskar-centric perspective. Curr. Top. Dev. Biol.116, 679–707 (2016). 10.1016/bs.ctdb.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kistler, K. E. et al. Phase transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells. eLife7, e37949 (2018). 10.7554/eLife.37949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trcek, T. et al. Sequence-independent self-assembly of germ granule mRNAs into homotypic clusters. Mol. Cell78, 941–950.e12 (2020). 10.1016/j.molcel.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trcek, T. et al. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun.6, 7962 (2015). 10.1038/ncomms8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niepielko, M. G., Eagle, W. V. I. & Gavis, E. R. Stochastic seeding coupled with mRNA self-recruitment generates heterogeneous Drosophila germ granules. Curr. Biol.28, 1872–1881.e3 (2018). 10.1016/j.cub.2018.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putnam, A., Cassani, M., Smith, J. & Seydoux, G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol.26, 220–226 (2019). 10.1038/s41594-019-0193-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roovers, E. F. et al. Tdrd6a regulates the aggregation of Buc into functional subcellular compartments that drive germ cell specification. Dev. Cell46, 285–301.e9 (2018). 10.1016/j.devcel.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, C.-Y. S. et al. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. eLife9, e52896 (2020). 10.7554/eLife.52896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cassani, M. & Seydoux, G. Specialized germline P-bodies are required to specify germ cell fate in Caenorhabditis elegans embryos. Development149, dev200920 (2022). 10.1242/dev.200920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahowald, A. P. Fine structure of pole cells and polar granules in Drosophila melanogaster. J. Exp. Zool.151, 201–215 (1962). 10.1002/jez.1401510302 [DOI] [Google Scholar]
- 25.Mahowald, A. P. Polar granules of Drosophila. II. Ultrastructural changes during early embryogenesis. J. Exp. Zool.167, 237–261 (1968). 10.1002/jez.1401670211 [DOI] [PubMed] [Google Scholar]
- 26.Arkov, A. L., Wang, J.-Y. S., Ramos, A. & Lehmann, R. The role of Tudor domains in germline development and polar granule architecture. Development133, 4053–4062 (2006). 10.1242/dev.02572 [DOI] [PubMed] [Google Scholar]
- 27.Dufourt, J. et al. piRNAs and Aubergine cooperate with Wispy poly(A) polymerase to stabilize mRNAs in the germ plasm. Nat. Commun.8, 1305 (2017). 10.1038/s41467-017-01431-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vourekas, A., Alexiou, P., Vrettos, N., Maragkakis, M. & Mourelatos, Z. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature531, 390–394 (2016). 10.1038/nature17150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klattenhoff, C. et al. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell12, 45–55 (2007). 10.1016/j.devcel.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 30.Wilson, J. E., Connell, J. E. & Macdonald, P. M. aubergine enhances oskar translation in the Drosophila ovary. Development122, 1631–1639 (1996). 10.1242/dev.122.5.1631 [DOI] [PubMed] [Google Scholar]
- 31.Yan, X., Hoek, T. A., Vale, R. D. & Tanenbaum, M. E. Dynamics of translation of single mRNA molecules in vivo. Cell165, 976–989 (2016). 10.1016/j.cell.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, C., Han, B., Zhou, R. & Zhuang, X. Real-time imaging of translation on single mRNA transcripts in live cells. Cell165, 990–1001 (2016). 10.1016/j.cell.2016.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morisaki, T. et al. Real-time quantification of single RNA translation dynamics in living cells. Science352, 1425–1429 (2016). 10.1126/science.aaf0899 [DOI] [PubMed] [Google Scholar]
- 34.Pichon, X. et al. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol.214, 769–781 (2016). 10.1083/jcb.201605024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, B., Eliscovich, C., Yoon, Y. J. & Singer, R. H. Translation dynamics of single mRNAs in live cells and neurons. Science352, 1430–1435 (2016). 10.1126/science.aaf1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertrand, E. et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell2, 437–445 (1998). 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- 37.Raj, A., Van Den Bogaard, P., Rifkin, S. A., Van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods5, 877–879 (2008). 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahanukar, A. & Wharton, R. P. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev.10, 2610–2621 (1996). 10.1101/gad.10.20.2610 [DOI] [PubMed] [Google Scholar]
- 39.Bergsten, S. E. & Gavis, E. R. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development126, 659–669 (1999). 10.1242/dev.126.4.659 [DOI] [PubMed] [Google Scholar]
- 40.Dufourt, J. et al. Imaging translation dynamics in live embryos reveals spatial heterogeneities. Science372, 840–844 (2021). 10.1126/science.abc3483 [DOI] [PubMed] [Google Scholar]
- 41.Mueller, F. et al. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods10, 277–278 (2013). 10.1038/nmeth.2406 [DOI] [PubMed] [Google Scholar]
- 42.Little, S. C., Sinsimer, K. S., Lee, J. J., Wieschaus, E. F. & Gavis, E. R. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol.17, 558–568 (2015). 10.1038/ncb3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramat, A. et al. The PIWI protein Aubergine recruits eIF3 to activate translation in the germ plasm. Cell Res. 30, 421–435 (2020). 10.1038/s41422-020-0294-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Götze, M. et al. Translational repression of the Drosophila nanos mRNA involves the RNA helicase Belle and RNA coating by Me31B and Trailer hitch. RNA23, 1552–1568 (2017). 10.1261/rna.062208.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaessinger, S., Busseau, I. & Simonelig, M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development133, 4573–4583 (2006). 10.1242/dev.02649 [DOI] [PubMed] [Google Scholar]
- 46.Kubíková, J., Ubartaitė, G., Metz, J. & Jeske, M. Structural basis for binding of Drosophila Smaug to the GPCR Smoothened and to the germline inducer Oskar. Proc. Natl Acad. Sci. Usa.120, e2304385120 (2023). 10.1073/pnas.2304385120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomson, T., Liu, N., Arkov, A., Lehmann, R. & Lasko, P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev.125, 865–873 (2008). 10.1016/j.mod.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hakes, A. C. & Gavis, E. R. Plasticity of Drosophila germ granules during germ cell development. PLoS Biol.21, e3002069 (2023). 10.1371/journal.pbio.3002069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jongens, T. A., Hay, B., Jan, L. Y. & Jan, Y. N. The germ cell-less gene product: a posteriorly localized component necessary for germ cell development in Drosophila. Cell70, 569–584 (1992). 10.1016/0092-8674(92)90427-E [DOI] [PubMed] [Google Scholar]
- 50.Dalby, B. & Glover, D. M. Discrete sequence elements control posterior pole accumulation and translational repression of maternal cyclin B RNA in Drosophila. EMBO J.12, 1219–1227 (1993). 10.1002/j.1460-2075.1993.tb05763.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eichhorn, S. W. et al. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife5, e16955 (2016). 10.7554/eLife.16955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kronja, I. et al. Widespread changes in the posttranscriptional landscape at the drosophila oocyte-to-embryo transition. Cell Rep.7, 1495–1508 (2014). 10.1016/j.celrep.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hara, M. et al. Identification of PNG kinase substrates uncovers interactions with the translational repressor TRAL in the oocyte-to-embryo transition. eLife7, e33150 (2018). 10.7554/eLife.33150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adivarahan, S. et al. Spatial organization of single mRNPs at different stages of the gene expression pathway. Mol. Cell72, 727–738.e5 (2018). 10.1016/j.molcel.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forrest, K. M., Clark, I. E., Jain, R. A. & Gavis, E. R. Temporal complexity within a translational control element in the nanos mRNA. Development131, 5849–5857 (2004). 10.1242/dev.01460 [DOI] [PubMed] [Google Scholar]
- 56.Bracha, D. et al. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell175, 1467–1480.e13 (2018). 10.1016/j.cell.2018.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirino, Y. et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol.11, 652–658 (2009). 10.1038/ncb1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishida, K. M. et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J.28, 3820–3831 (2009). 10.1038/emboj.2009.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirino, Y. et al. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA16, 70–78 (2010). 10.1261/rna.1869710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vo, H. D. L. et al. Protein components of ribonucleoprotein granules from Drosophila germ cells oligomerize and show distinct spatial organization during germline development. Sci. Rep.9, 19190 (2019). 10.1038/s41598-019-55747-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnstone, O. & Lasko, P. Interaction with eIF5B is essential for Vasa function during development. Development131, 4167–4178 (2004). 10.1242/dev.01286 [DOI] [PubMed] [Google Scholar]
- 62.Eichler, C. E., Hakes, A. C., Hull, B. & Gavis, E. R. Compartmentalized oskar degradation in the germ plasm safeguards germline development. eLife9, e49988 (2020). 10.7554/eLife.49988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mateju, D. et al. Single-molecule imaging reveals translation of mRNAs Localized to Stress Granules. Cell183, 1801–1812.e13 (2020). 10.1016/j.cell.2020.11.010 [DOI] [PubMed] [Google Scholar]
- 64.Moon, S. L. et al. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol.21, 162–168 (2019). 10.1038/s41556-018-0263-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang, J.-Y. et al. LLPS of FXR1 drives spermiogenesis by activating translation of stored mRNAs. Science377, eabj6647 (2022). 10.1126/science.abj6647 [DOI] [PubMed] [Google Scholar]
- 66.Westerich, K. J. et al. Spatial organization and function of RNA molecules within phase-separated condensates in zebrafish are controlled by Dnd1. Dev. Cell58, 1578–1592.e5 (2023). 10.1016/j.devcel.2023.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen, R., Stainier, W., Dufourt, J., Lagha, M. & Lehmann, R. Direct observation of translational activation by a ribonucleoprotein granule. Nat. Cell Biol.10.1038/s41556-024-01452-5 (2024). [DOI] [PMC free article] [PubMed]
- 68.Dai, P. et al. A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell179, 1566–1581.e16 (2019). 10.1016/j.cell.2019.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harris, A. N. & Macdonald, P. M. aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development128, 2823–2832 (2001). 10.1242/dev.128.14.2823 [DOI] [PubMed] [Google Scholar]
- 70.Rørth, P. Gal4 in the Drosophila female germline. Mech. Dev.78, 113–118 (1998). 10.1016/S0925-4773(98)00157-9 [DOI] [PubMed] [Google Scholar]
- 71.Hurd, T. R. et al. Long Oskar controls mitochondrial inheritance in Drosophila melanogaster. Dev. Cell39, 560–571 (2016). 10.1016/j.devcel.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kina, H., Yoshitani, T., Hanyu‐Nakamura, K. & Nakamura, A. Rapid and efficient generation of GFP ‐knocked‐in Drosophila by the CRISPR ‐Cas9‐mediated genome editing. Dev. Growth Differ61, 265–275 (2019). 10.1111/dgd.12607 [DOI] [PubMed] [Google Scholar]
- 73.Forrest, K. M. & Gavis, E. R. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol.13, 1159–1168 (2003). 10.1016/S0960-9822(03)00451-2 [DOI] [PubMed] [Google Scholar]
- 74.Murata, Y. & Wharton, R. P. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in drosophila embryos. Cell80, 747–756 (1995). 10.1016/0092-8674(95)90353-4 [DOI] [PubMed] [Google Scholar]
- 75.Buszczak, M. et al. The Carnegie protein trap library: a versatile tool for drosophila developmental studies. Genetics175, 1505–1531 (2007). 10.1534/genetics.106.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shamanski, F. L. & Orr-Weaver, T. L. The Drosophila plutonium and pan gu genes regulate entry into S phase at fertilization. Cell66, 1289–1300 (1991). 10.1016/0092-8674(91)90050-9 [DOI] [PubMed] [Google Scholar]
- 77.Rouget, C. et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature467, 1128–1132 (2010). 10.1038/nature09465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gunawardane, L. S. et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science315, 1587–1590 (2007). 10.1126/science.1140494 [DOI] [PubMed] [Google Scholar]
- 79.Temme, C., Zaessinger, S., Meyer, S., Simonelig, M. & Wahle, E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J.23, 2862–2871 (2004). 10.1038/sj.emboj.7600273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verrotti, A. C. & Wharton, R. P. Nanos interacts with Cup in the female germline of Drosophila. Development127, 5225–5232 (2000). 10.1242/dev.127.23.5225 [DOI] [PubMed] [Google Scholar]
- 81.Nakamura, A., Amikura, R., Hanyu, K. & Kobayashi, S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development128, 3233–3242 (2001). 10.1242/dev.128.17.3233 [DOI] [PubMed] [Google Scholar]
- 82.Chartier, A. et al. Mitochondrial dysfunction reveals the role of mRNA Poly(A) tail regulation in oculopharyngeal muscular dystrophy pathogenesis. PLoS Genet11, e1005092 (2015). 10.1371/journal.pgen.1005092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomson, T. & Lasko, P. Drosophila Tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis40, 164–170 (2004). 10.1002/gene.20079 [DOI] [PubMed] [Google Scholar]
- 84.Abbaszadeh, E. K. & Gavis, E. R. Fixed and live visualization of RNAs in Drosophila oocytes and embryos. Methods98, 34–41 (2016). 10.1016/j.ymeth.2016.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmidt, U. et al. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J. Cell Biol.193, 819–829 (2011). 10.1083/jcb.201009012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schulman, V. K., Folker, E. S. & Baylies, M. K. A method for reversible drug delivery to internal tissues of Drosophila embryos. Fly7, 193–203 (2013). 10.4161/fly.25438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jahr, W., Velicky, P. & Danzl, J. G. Strategies to maximize performance in STimulated Emission Depletion (STED) nanoscopy of biological specimens. Methods174, 27–41 (2020). 10.1016/j.ymeth.2019.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ball, G. et al. SIMcheck: a toolbox for successful super-resolution structured illumination microscopy. Sci. Rep.5, 15915 (2015). 10.1038/srep15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bahry, E. et al. RS-FISH: precise, interactive, fast, and scalable FISH spot detection. Nat. Methods19, 1563–1567 (2022). 10.1038/s41592-022-01669-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sharma, V. P. et al. Live tumor imaging shows macrophage induction and TMEM-mediated enrichment of cancer stem cells during metastatic dissemination. Nat. Commun.12, 7300 (2021). 10.1038/s41467-021-27308-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or Supplementary Information. Source data are provided with this paper.