Abstract
γ‐Hydroxybutyric acid (GHB) analogs are small molecules that bind competitively to a specific cavity in the oligomeric CaMKIIα hub domain. Binding affects conformation and stability of the hub domain, which may explain the neuroprotective action of some of these compounds. Here, we describe molecular details of interaction of the larger‐type GHB analog 2‐(6‐(4‐chlorophenyl)imidazo[1,2‐b]pyridazine‐2‐yl)acetic acid (PIPA). Like smaller‐type analogs, PIPA binding to the CaMKIIα hub domain promoted thermal stability. PIPA additionally modulated CaMKIIα activity under sub‐maximal CaM concentrations and ultimately led to reduced substrate phosphorylation. A high‐resolution X‐ray crystal structure of a stabilized CaMKIIα (6x mutant) hub construct revealed details of the binding mode of PIPA, which involved outward placement of tryptophan 403 (Trp403), a central residue in a flexible loop close to the upper hub cavity. Small‐angle X‐ray scattering (SAXS) solution structures and mass photometry of the CaMKIIα wild‐type hub domain in the presence of PIPA revealed a high degree of ordered self‐association (stacks of CaMKIIα hub domains). This stacking neither occurred with the smaller compound 3‐hydroxycyclopent‐1‐enecarboxylic acid (HOCPCA), nor when Trp403 was replaced with leucine (W403L). Additionally, CaMKIIα W403L hub was stabilized to a larger extent by PIPA compared to CaMKIIα hub wild type, indicating that loop flexibility is important for holoenzyme stability. Thus, we propose that ligand‐induced outward placement of Trp403 by PIPA, which promotes an unforeseen mechanism of hub domain stacking, may be involved in the observed reduction in CaMKIIα kinase activity. Altogether, this sheds new light on allosteric regulation of CaMKIIα activity via the hub domain.
Keywords: allosteric regulation, CaMKIIα, hub domain, ligands, self‐association, small‐angle X‐ray scattering, X‐ray crystallography
1. INTRODUCTION
Recently, the calcium/calmodulin‐dependent protein kinase II subtype alpha (CaMKIIα) was discovered as the long‐sought‐after high‐affinity neuronal target for γ‐hydroxybutyric acid (GHB; Figure 1a) (Bay et al., 2014; Leurs et al., 2021). This was surprising since GHB is a natural metabolite of γ‐aminobutyric acid (GABA) involved in inhibitory neurotransmission. GHB is a well‐characterized agonist for the inhibitory GABAB receptors displaying diverse functional properties yet binds with low millimolar affinity to this target (Kaupmann et al., 2003). On the other hand, GHB and specific GHB analogues bind with high nanomolar affinity to CaMKIIα and a complete lack of binding of GHB ligands to CaMKIIα knock‐out mice was observed (Leurs et al., 2021). Intriguingly, GHB and synthetic small‐molecule analogs were found to bind to a pocket in the hub domain of the multimeric CaMKIIα. This offers an entirely novel pharmacological approach to CaMKIIα regulation where oligomeric stabilization has been proposed as the mechanistic basis for the neuroprotective action of such analogs (Griem‐Krey et al., 2022, 2023; Leurs et al., 2021). CaMKIIα holds a key role in synaptic plasticity, learning, and memory (Bayer & Schulman, 2019), and mutations in human CaMKII genes have been uncovered to cause intellectual disability (Chia et al., 2018; Kury et al., 2017; Mohanan et al., 2022; Proietti Onori et al., 2018; Rigter et al., 2024; Stephenson et al., 2017). Furthermore, after ischemic brain injury, aberrant calcium (Ca2+) levels trigger cell death involving excessive CaMKIIα activity and peptide inhibitors have been found to be neuroprotective post‐injury, especially by inhibiting the autonomous CaMKIIα activity (Coultrap et al., 2011; Vest et al., 2010). By contrast, GHB analogs act via a direct stabilization of the hub domain and are not CaMKIIα inhibitors per se. Instead, they promote neuroprotection by yet undisclosed molecular mechanisms, which appear to centrally involve the hub domain.
FIGURE 1.
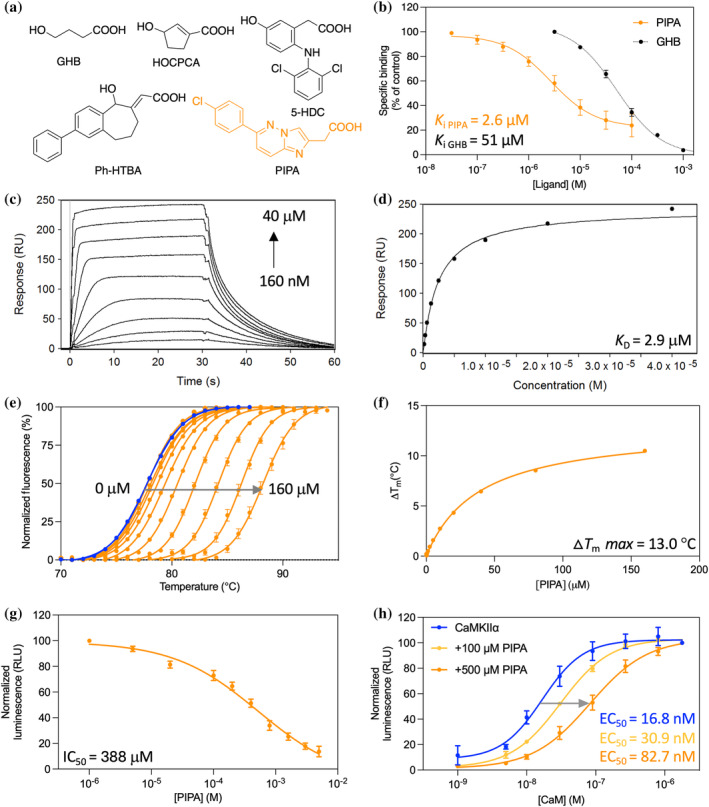
PIPA binds to CaMKIIα WT hub and inhibits substrate phosphorylation. (a) Chemical structures of γ‐hydroxybutyric acid (GHB), 3‐hydroxycyclopenten‐1‐enecarboxylic acid (HOCPCA), 5‐hydroxydiclofenac (5‐HDC), (E)‐2‐(5‐hydroxy‐2‐phenyl‐5,7,8,9‐tetrahydro‐6H‐benzo[7]annulen‐6‐ylidene)acetic acid (Ph‐HTBA), and 2‐(6‐(4‐chlorophenyl)imidazo[1,2‐b]pyridazine‐2‐yl)acetic acid (PIPA). (b) Concentration‐dependent inhibition of [3H]HOCPCA binding by PIPA in whole‐cell homogenates from HEK293T cells transfected with CaMKIIα (pooled data, n = 4; GHB for comparison; Leurs et al., 2021). (c) Concentration‐dependent binding of PIPA to immobilized CaMKIIα WT hub measured by SPR, and (d) associated Langmuir‐binding isotherm (representative data of n = 3). (e) Right‐shifted TSA melting curves of CaMKIIα WT hub upon binding of PIPA (representative data of n = 3). (f) Saturation isotherm (representative). (g) Concentration‐dependent inhibition of CaMKIIα syntide‐2 by PIPA using sub‐maximal (30 nM) CaM concentration in the luminescence‐based ADP‐Glo kinase assay (pooled data, n = 4, mean ± SEM). (h) Right‐shifted CaM curve of CaMKIIα syntide‐2 phosphorylation in the presence of PIPA (pooled data, n = 3). For clarity, only mean values without variation are given; all mean values can be found in the respective results section.
The CaMKIIα holoenzyme is multimeric, where subunits are organized into dodecamers and tetradecamers (12 and 14 subunits). Each subunit consists of an N‐terminal kinase domain, a regulatory segment, a linker region, and a C‐terminal hub domain. The latter governs the oligomerization of individual subunits into functional holoenzymes (Rosenberg et al., 2006) and is also emerging as regulator of kinase activity (Sloutsky et al., 2020). Although debated in the field (Lučić et al., 2023), hub domain dynamics have also been suggested to play a role in so‐called activation‐triggered subunit exchange, which can lead to the activation of neighboring holoenzymes (Lee et al., 2022; Stratton et al., 2014).
We recently revealed structural details of the small‐molecule binding pocket of CaMKIIα hub from a 2.2 Å resolution co‐crystal structure of the tetradecamer‐stabilized human CaMKIIα hub mutant (CaMKIIα 6x hub; McSpadden et al., 2019) with the GHB analog 5‐hydroxydiclofenac (5‐HDC; Figure 1a) (Leurs et al., 2021). 5‐HDC was found bound to 12 out of 14 subunits of the tetradecameric CaMKIIα hub and to interact specifically with the positively charged residues Arg433, Arg453, and Arg469 as well as His395 that can be either neutral or positively charged. In addition, intrinsic tryptophan fluorescence (ITF) measurements with purified CaMKIIα hub domain demonstrated that binding of 5‐HDC displaced a flexible loop located at the edge of the binding pocket, which contains the central residue tryptophan 403 (Trp403).
The GHB analogs 3‐hydroxycyclopent‐1‐enecarboxylic acid (HOCPCA; Figure 1a) and (E)‐2‐(5‐hydroxy‐2‐phenyl‐5,7,8,9‐tetrahydro‐6H‐benzo[7]annulen‐6‐ylidene)acetic acid (Ph‐HTBA; Figure 1a) bind to the same site and increase oligomeric thermal stability (Griem‐Krey et al., 2022; Leurs et al., 2021). Previous work also demonstrated a pronounced CaMKIIα selectivity since binding of tritiated HOCPCA was not observed at CaMKIIβ, γ, and δ variants (Leurs et al., 2021). Especially, the affected oligomerization has been suggested to explain their neuroprotection (Griem‐Krey et al., 2022; Tian et al., 2022). Recently, Ph‐HTBA (Tian et al., 2022), but not the smaller‐size compound HOCPCA (Leurs et al., 2021), was found to cause an outward flip of Trp403 in the flexible hub domain loop, and to reduce Ca2+‐stimulated CaMKIIα Thr286 autophosphorylation in primary cortical neurons and substrate phosphorylation of recombinant CaMKIIα holoenzyme (Griem‐Krey et al., 2022). Consequently, we hypothesize that distinct ligand‐induced molecular interactions involving conformational changes of Trp403 and its CaMKIIα hub‐associated loop contribute further to the function of CaMKIIα hub ligands. In this study, we provide evidence for this hypothesis using model compounds binding competitively with GHB: the larger‐type GHB analog 2‐(6‐(4‐chlorophenyl)imidazo[1,2‐b]pyridazine‐2‐yl)acetic acid (PIPA), and the smaller size GHB analogs, acetate (Wellendorph et al., 2009) and HOCPCA (Leurs et al., 2021) (Figure 1a). PIPA was previously reported as a high‐affinity GHB‐site competitive ligand (K i value of 0.22 μM in rat native synaptic membranes; Krall et al., 2019). Using X‐ray crystallography, biophysical and biochemical approaches, we show that both PIPA and acetate bind to the CaMKIIα hub but produce distinct structural and functional outcomes. In particular, we confirm effects of ligand size on hub domain loop movement and allosteric control of CaMKIIα function. Furthermore, we identify PIPA as a CaMKIIα hub ligand for studying aspects of CaMKIIα hub where Trp403 is restricted in the outward‐flipped position. Explicitly, we pinpoint Trp403—unique to CaMKIIα—as a key molecular determinant.
2. RESULTS
2.1. PIPA binds directly to the CaMKIIα hub domain
PIPA has previously been shown to bind to the native GHB high‐affinity site in rat cortical membranes (K i = 0.22 μM) (Krall et al., 2019). Here, we confirm binding of PIPA to recombinant CaMKIIα holoenzyme, albeit with micromolar affinity. This was shown by concentration‐dependent inhibition of [3H]HOCPCA binding by PIPA to whole‐cell homogenates of HEK293T cells transiently overexpressing homomeric CaMKIIα holoenzyme (K i = 2.6 μM; pK i = 5.65 ± 0.15) (Figure 1b). To confirm binding of PIPA directly to the CaMKIIα hub domain, surface plasmon resonance (SPR) binding studies were performed, revealing binding of PIPA to immobilized, purified CaMKIIα wild‐type (WT) hub with a mean K D value of 2.9 ± 0.6 μM (Figure 1c,d). Similar binding affinities were found for PIPA to the immobilized, purified CaMKIIα 6x hub construct and the CaMKIIα holoenzyme (Figure S1). Thus, although the affinity of PIPA was found to be slightly lower at recombinant CaMKIIα compared to native conditions, these experiments show that PIPA binds directly to the isolated CaMKIIα hub domain.
2.2. Thermal stabilization of the CaMKIIα hub domain by PIPA
Increased thermal stability of the purified CaMKIIα WT hub has been shown as a characteristic upon binding of GHB and several other known GHB analogs (Leurs et al., 2021; Tian et al., 2022). By using the described thermal shift assay (TSA) (Leurs et al., 2021), the thermal melting point (T m) for the CaMKIIα WT hub was found to be 77.4 ± 0.2°C (Figure 1e). Upon PIPA binding, a concentration‐dependent increase in CaMKIIα WT hub stability was observed (∆T m = 13.04 ± 0.09°C) (Figure 1e,f). However, the maximum ∆T m for the CaMKIIα WT hub upon PIPA binding was 10.7°C less compared to the analog 5‐HDC, which had a maximum ∆T m of 23.7 ± 0.2°C (Figure S2A–C). Contrary, binding of the smaller‐size compound HOCPCA induced a similar extent of hub stabilization as PIPA, with a maximum ∆T m of 11.5 ± 0.6°C (Figure S2D–F).
2.3. PIPA reduces CaMKIIα‐mediated substrate phosphorylation
The larger‐type GHB analog Ph‐HTBA was previously found to reduce substrate phosphorylation catalyzed by the CaMKIIα holoenzyme (Griem‐Krey et al., 2022; Tian et al., 2022). Specifically, Ph‐HTBA reduced CaMKIIα‐dependent syntide‐2 phosphorylation in the ADP‐Glo kinase assay using purified CaMKIIα holoenzyme and under sub‐maximal calmodulin (CaM) concentration (30 nM) (Griem‐Krey et al., 2022). Similarly, in the present study, we observed that PIPA reduced syntide‐2 phosphorylation (IC50 = 388.6 μM; pIC50 = 3.44 ± 0.09) (Figure 1g). A reduced CaM sensitivity was observed with increasing PIPA concentration, with EC50 of CaM of 30.9 μM using 100 μM PIPA and 82.7 μM using 500 μM PIPA (Figure 1h). We also tested the smaller‐size compounds HOCPCA and acetate, using corresponding sub‐maximal (30 nM) CaM concentrations. Under this condition, neither of the compounds affected substrate phosphorylation, even at concentrations up to 5–10 mM (Figure S3). Of note, in previous studies, all GHB analogs were found inactive in the ADP‐Glo kinase assay when performed at maximal CaM concentrations (1781 nM) (Leurs et al., 2021).
2.4. PIPA flips and restricts Trp403 positioning
Using the recombinantly expressed and purified tetradecamer‐stabilized CaMKIIα 6x hub (McSpadden et al., 2019), also previously successfully used for crystallization with the ligand 5‐HDC (Leurs et al., 2021), we obtained a co‐crystal structure of the CaMKIIα hub oligomer bound to PIPA and acetate determined at 2.1 Å resolution (Figure 2, PDB entry 9EOY and Table S1). In this structure, PIPA occupied two out of seven subunits in the equatorial plane, while the rest of the subunits were occupied by the low‐affinity (K i >100 μM) acetate (Wellendorph et al., 2009) (from the buffer) (Figure 2a,b). In one of the subunits, PIPA bound at the conventional binding site, whereas in the other subunit, it was located in the vicinity of the conventional binding site and with acetate and a polyethylene (PEG) moiety observed in the conventional binding site. Intriguingly, PIPA and acetate differentially directed movements of Trp403 in the loop at the edge of the binding pocket.
FIGURE 2.
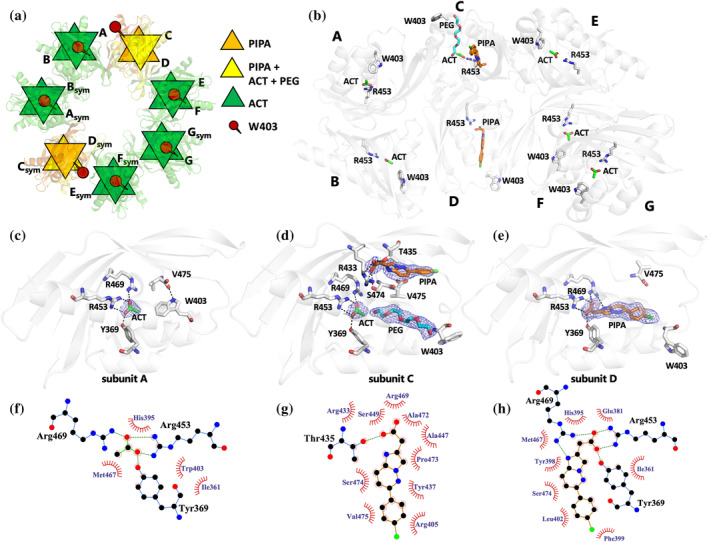
PIPA bound to CaMKIIα 6x hub promotes Trp403 flip while acetate does not. (a) X‐ray crystal structure of tetradecameric CaMKIIα 6x hub (PDB entry 9EOY) in schematic representation, with two subunits bound to PIPA (orange), 10 subunits to acetate (ACT) only (green), and two to ACT + PIPA (yellow). Trp403 is shown in red. (b) The structure of CaMKIIα 6x hub with seven subunits A–G in the asymmetric unit of the crystal, forming a tetradecamer with symmetry‐related molecules, is shown in white cartoon representation with PIPA (orange carbon atoms), ACT (green), PEG (cyan), Arg453 (white), and Trp403 (white) shown in sticks representation. (c) Zoom‐in on binding mode and interactions of acetate in subunit A. Salt bridges and hydrogen bonds between acetate and corresponding amino‐acid residues are shown as black dashed lines and with compounds and residues as sticks representation. In addition, a hydrogen bond between the C‐terminal carboxylate group of Val475 and Trp403 is shown. The 2F o − F c electron densities for the ligands are contoured at 1.0 sigma and carved at 1.6 Å. (d) Alternative binding mode and interactions of PIPA in subunit C. Hydrogen‐bonding interactions between PIPA, ACT, and corresponding amino‐acid residues are shown as black dashed lines and with compounds and residues as sticks representation. The 2F o − F c electron densities for PIPA (carved at 1.6 Å), ACT (carved at 1.75 Å), and PEG (carved at 2 Å) have been contoured at 0.5 sigma. In addition, Trp403 and Val475 are shown as sticks representation. (e) Binding mode and interactions of PIPA in subunit D. Salt bridges and hydrogen bonds between PIPA and corresponding amino‐acid residues (as sticks representation) are shown as black dashed lines. In addition, Trp403 and Val475 are shown as sticks representation. The 2F o − F c electron density for PIPA has been contoured at 0.5 sigma and carved at 1.8 Å. (f–h) Binding site interactions between CaMKIIα 6x hub and acetate (f) and PIPA (g, h) analyzed using the LigPlot+ software (Laskowski & Swindells, 2011). Hydrogen bonds and hydrophobic interactions between amino‐acid residues and ligands are depicted as green dotted lines and red semicircles, respectively.
The key molecular interactions of PIPA and acetate are illustrated in Figure 2. Acetate binds into the hub cavity of CaMKIIα 6x hub with the carboxylate moiety forming salt bridges with Arg453 and Arg469, and a charge‐assisted hydrogen bond with Tyr369 (Figure 2c,f, subunit A). The binding of the smaller‐size acetate allows Trp403 to adopt an inward‐flipped conformation and the C‐terminal carboxylate group of Val475 to form a hydrogen bond to the indole nitrogen of Trp403 (Figure 2a–c). In subunit C, PIPA occupies the binding pocket together with acetate and a PEG molecule from the crystallization buffer. Here, PIPA binds at a site (site 1) in the vicinity of the conventional binding site (site 2), with the carboxylate group of PIPA forming hydrogen‐bonding interactions with the hydroxyl groups of Ser474 and Thr435 and a potential salt bridge with Arg433 (Figure 2d,g). The PEG molecule binds at the conventional site near acetate (Figure 2d). In subunit C, Trp403 adopts an outward‐facing conformation (Figure 2a,b,d). In subunit D, PIPA bound to the conventional binding site (site 2) as previously observed for 5‐HDC, with the carboxylate group of PIPA forming a salt bridge with Arg453 and Arg469, and a charge‐assisted hydrogen bond to Tyr369 (Figure 2e,h), as also observed for acetate. In addition, a potential hydrogen bond is formed between the heteroaromatic bicyclic ring system of PIPA and Arg469. The binding of PIPA leads Trp403 to flip outward in a similar fashion as observed in subunit C (Figure 2a,b,d,e). Overall, the movement of the Trp403‐containing loop is the only major structural difference observed in subunits occupied by PIPA compared to acetate (Figure S4A).
2.5. CaMKIIα WT hub self‐association is induced by ligand binding
Dissociation of subunits from the dodecameric or tetradecameric CaMKIIα holoenzyme was previously found to be extremely slow with a K D estimated at 10–20 nM (Torres‐Ocampo et al., 2020). As the CaMKIIα holoenzyme concentration in dendritic spines is estimated to be 100 μM (Otmakhov & Lisman, 2012), it is therefore likely that these dodecameric or tetradecameric oligomers represent functionally relevant native structures. As the GHB analogs induce large stabilizing effects by binding to the isolated CaMKIIα WT hub domain, we hypothesized these analogs to change the proportion of CaMKIIα hub dodecamers and tetradecamers. Therefore, the oligomeric structures of the purified CaMKIIα WT hub and the tetradecamer‐stabilized CaMKIIα 6x hub mutant were investigated using small‐angle X‐ray scattering (SAXS), to elucidate the solution state and potential oligomeric changes upon ligand binding. Furthermore, as Trp403, based on the X‐ray crystallography data, seemed to play a central role in the binding of PIPA, we also investigated the ligand interaction with the purified CaMKIIα W403L hub mutant.
The SAXS data of neither CaMKIIα WT hub nor CaMKIIα 6x hub domains revealed any concentration‐dependent effects when investigated without ligand (Figure S5A–D). Distinct differences between the CaMKIIα WT hub and the tetradecamer‐stabilized CaMKIIα 6x hub were observed, while the CaMKIIα W403L hub resembled the CaMKIIα WT hub (Figure 3a). Based on the primary analyses (Figure S5E,F) and knowledge from the previously determined crystal structures, we tested the respective fits of dodecamers and tetradecamers, as well as mixtures of the two, to the data. The best fits were obtained by the use of single oligomers, i.e., tetradecamers for the CaMKIIα 6x hub and dodecamers for the CaMKIIα WT hub (Figure 3b) as also seen in the respective crystal structures (PDB entries 6OF8 and 5IG3) (Bhattacharyya et al., 2016; McSpadden et al., 2019). The dodecamer fit to the CaMKIIα WT hub is not perfect, and we cannot exclude, for example, minor fractions of tetradecamers or other aggregates (observed upon storage); however, the fit was not improved by inclusion of tetradecamers. Although some reports on the isolated CaMKIIα WT hub have suggested a 1:1 distribution of dodecamers and tetradecamers (Bhattacharyya et al., 2016; McSpadden et al., 2019), our data suggest dodecamers as the absolute prominent species for the CaMKIIα WT hub, consistent with observations on the CaMKIIα holoenzyme (Myers et al., 2017).
FIGURE 3.
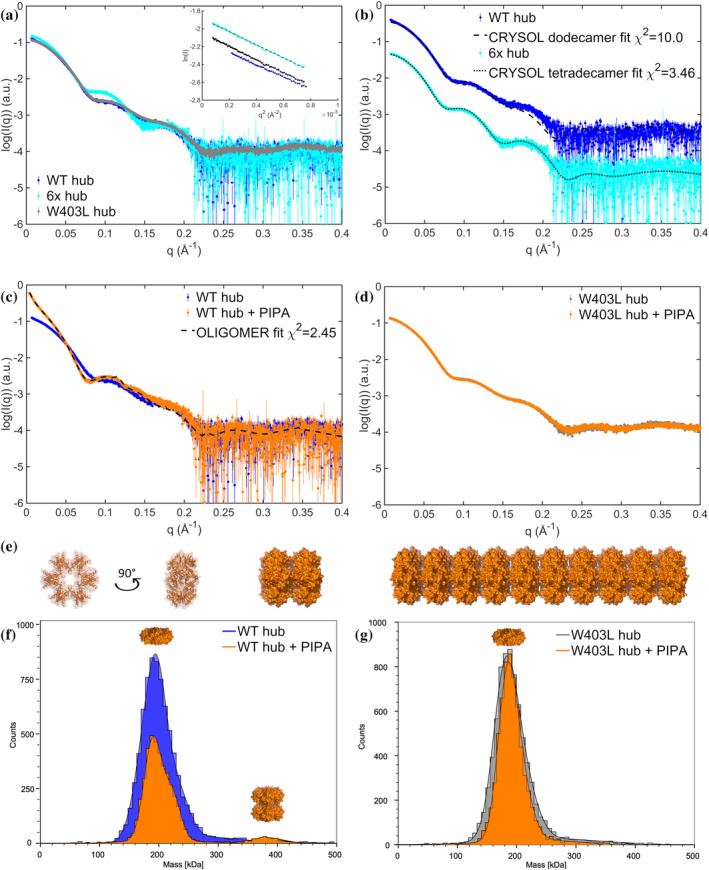
Solution structures of CaMKIIα WT hub reveals PIPA‐induced structured self‐association. (a) SAXS scattering curves for the CaMKIIα WT, 6x, and W403L hub domains, with corresponding Guinier fits (insert). Full concentration series of CaMKIIα WT hub and CaMKIIα 6x hub are included in Figure S5A–D. (b) Comparison of CaMKIIα WT hub and CaMKIIα 6x hub curves to atomic models of the dodecamer (PDB entry 5IG3; Bhattacharyya et al., 2016) and tetradecamer (PDB entry 6OF8; McSpadden et al., 2019), respectively. The scattering curves have been translated for clarity. (c) SAXS scattering curves of CaMKIIα WT hub in the presence and absence of PIPA, including oligomer fit using model of self‐associated dodecamers. (d) SAXS scattering curves of CaMKIIα W403L hub in the presence and absence of PIPA. (e) Examples of the stacked self‐associated dodecamers used to model CaMKIIα WT hub with PIPA data. A single dodecamer is shown in two orientations. In addition, a dimer of dodecamers and a stacked model including 10 dodecamers are illustrated. Models and figure were created using PyMOL. (f) Representative MP histograms showing CaMKIIα WT hub diluted to 500 nM with and without the addition of PIPA. (g) Representative MP histograms showing W403L CaMKIIα hub diluted to 500 nM, with and without PIPA. Each count indicates a single molecule. Additional MP data are shown in Figure S7.
Whereas neither 5‐HDC nor HOCPCA had any observable influence on the oligomeric state of the CaMKIIα WT hub construct (Figure S5G–I), PIPA induced self‐association of the CaMKIIα WT hub (Figure 3c). A varying degree of self‐association was observed in different preparations of CaMKIIα WT hub with PIPA, depending on the protein concentration upon buffer exchange and the storage time. The scattering data from the CaMKIIα WT hub with PIPA are characterized by the presence of rod‐like structures as revealed by the pair‐distance distribution function (Figure S5J). In addition, we observe an indication of a Bragg peak (at q = 0.115 Å−1, corresponding to a repeated real space distance of 54.5 Å) in one curve from data of the sample stored overnight. Examining previous crystal structures of the CaMKIIα hub domains, crystal packing patterns were observed with either the tetradecameric hub surrounded by tetradecamers positioned perpendicularly (face‐to‐edge) (e.g., as in PDB entry 5IG3) or stacks (as in PDB entry 6OF8). A self‐association of the oligomers of CaMKIIα WT hub with PIPA through stacking (Figure 3e) is consistent with the rod‐like structure and repeated distance of 54.5 Å between the dodecamers, i.e., the height of the dodecamers is consistent with the Bragg peak observation. Based on this observation, a range of stacked dodecamer models were created. Using combinations of these models, reflecting the degree of self‐association, we could fit the scattering data of the CaMKIIα WT hub with PIPA (Figure 3c, Figure S6, and Table S2).
Despite differences in sample preparation, storage time, and data collection, convincingly, these stacked models could fit the different data measured with volume fractions of the different components consistent with the variation in concentration of the protein during sample preparation (Figure S6 and Table S2). In the lowest concentration sample, we observed mainly dodecamers (~82%), a fraction of paired dodecamers (~10%), and only minor fractions of the larger stacks. In the more concentrated samples, these volume fractions are shifted toward more paired dodecamers and larger stacks (Table S2). The most concentrated sample of CaMKIIα WT hub with PIPA aggregated immediately upon buffer exchange. SAXS data were in that case collected on the soluble fraction after removal of the non‐soluble aggregates by centrifugation (Figure 3c). This soluble fraction was fitted by a combination of dodecamers (~37%), dimers of dodecamers (~15%), and significant fractions of larger multimers (using models composed of 3–40 dodecamers; examples illustrated in Figure 3e). Although the finite size of these stacks may exceed the resolution limit of the data and the resulting fits do not exclude a broader variation in the lengths of the stacks, the trend from these fits consistently shows that PIPA under these experimental conditions induces formation of self‐associated stacks. The self‐association induced by PIPA was observed for the CaMKIIα WT hub domain in the concentration range examined (1.2–3.9 mg/mL), while no self‐association was detected for the CaMKIIα 6x hub and CaMKIIα W403L hub constructs with PIPA despite investigating higher protein concentrations (6.7 and 6.8 mg/mL, respectively) (Figure 3d and Figure S5H).
To validate the occurrence of PIPA‐induced CaMKIIα WT hub self‐association at lower protein concentrations (100–500 nM (1.6–7.7 μg/mL)), mass photometry studies were performed. In the absence of PIPA, purified CaMKIIα WT hub and CaMKIIα W403L hub both adopted a structure with molecular weight (MW) around 188–198 kDa, compatible with dodecamers (Figure 3f,g and Figure S7). However, CaMKIIα WT hub with PIPA displayed a shoulder on the main peak around 220 kDa as well as a much larger species at 372 kDa (corresponding to two dodecamers) (Figure 3f and Figure S7). CaMKIIα W403L hub did not display any change in MW upon PIPA addition, indicating that there was no significant change in its oligomerization state. Based on SAXS and mass photometry data, we conclude that PIPA introduces ordered self‐association of CaMKIIα WT hub and that this self‐association involves Trp403 as mutation of this residue prevents formation of larger species.
2.6. The W403L mutation increases PIPA‐induced hub stability
As conformational change of the CaMKIIα‐specific residue Trp403 at the upper edge of the binding pocket has previously been demonstrated for the compound 5‐HDC using ITF measurements (Leurs et al., 2021), this was also probed for PIPA. A concentration‐dependent quenching of a fluorescence signal of purified CaMKIIα 6x hub in the presence of PIPA was observed (IC50 = 5.1 μM; pIC50 = 5.30 ± 0.06) (Figure 4a,b). These data support that PIPA reaches into the hydrophobic space in the upper CaMKIIα hub cavity, as observed in the X‐ray structure of CaMKIIα 6x hub, and leads to an outward placement of Trp403.
FIGURE 4.
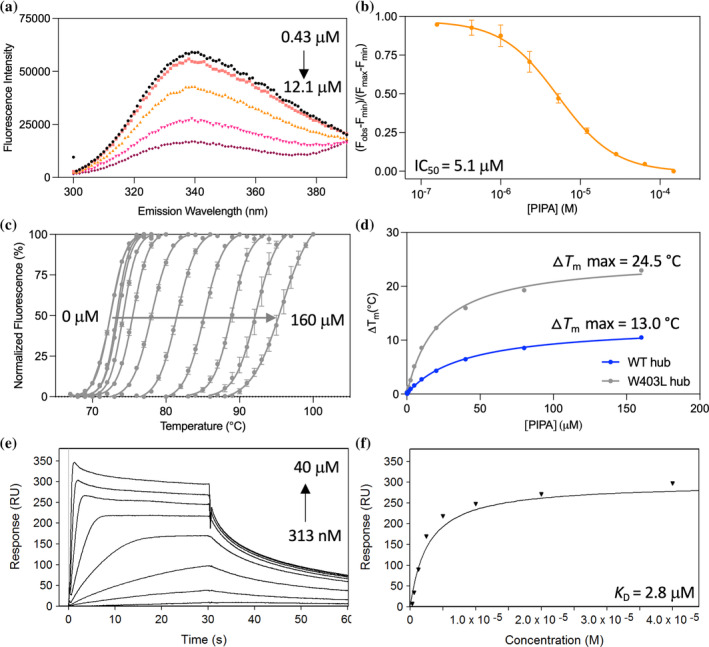
CaMKIIα hub mutation confirms a central role for Trp403 in PIPA binding. (a) Quenching of intrinsic tryptophan fluorescence caused by Trp403 flip in the CaMKIIα 6x hub with increasing concentrations of PIPA: 0.43 μM (black), 1 μM (red), 2.3 μM (orange), 5.3 μM (pink), 12.1 μM (purple), and (b) resulting normalized inhibition curve (pooled data, n = 3). (c) Right‐shifted TSA melting curves of CaMKIIα W403L hub upon binding of PIPA (representative data). (d) Thermal melting point (T m) of CaMKIIα WT hub (blue) and CaMKIIα W403L hub (gray) plotted against increasing concentrations of PIPA (representative data of n = 3). (e) Concentration‐dependent binding of PIPA to immobilized CaMKIIα W403L hub measured by SPR (representative data), and (f) associated Langmuir‐binding isotherm (representative data of n = 2).
To investigate impact of the ligand‐induced Trp403 conformational restriction on thermal stability, the purified CaMKIIα W403L hub was evaluated in the TSA in the presence of ligands. In the absence of ligand, the T m for the CaMKIIα W403L hub was found to be 73.0 ± 0.4°C, compared to 77.4 ± 0.2°C for the CaMKIIα WT hub. Thus, replacing Trp403 with a leucine slightly reduced the thermal stability of the hub. However, in the presence of PIPA, the maximum ∆T m for the CaMKIIα W403L hub was found increased with 24.5 ± 0.4°C (Figure 4c), compared to only 13.04 ± 0.09°C for the CaMKIIα WT hub (Figure 4d). In contrast, the maximum ∆T m for the CaMKIIα W403L hub in the presence of 5‐HDC and HOCPCA was only slightly increased relative to the CaMKIIα WT hub with these ligands (from 11.5 ± 0.6°C to 15.2 ± 1.0°C with HOCPCA and from 23.7 ± 0.4°C to 28.21 ± 0.03 with 5‐HDC) (Figure S2A–F).
To explore potential changes in the binding affinity of PIPA to the purified CaMKIIα W403L hub domain compared to CaMKIIα WT hub, SPR binding studies were performed, revealing unchanged binding between PIPA and immobilized CaMKIIα W403L hub (K D of 2.80 ± 0.13 μM) (Figure 4e,f) and CaMKIIα WT hub.
3. DISCUSSION
The CaMKIIα hub domain has previously been proposed to be involved in allosteric regulation of the kinase activity by direct interaction with the kinase domain of the CaMKIIα holoenzyme (Sloutsky et al., 2020). In this study, the GHB analog PIPA was found to bind to the CaMKIIα hub domain leading to a Trp403 outward‐flip and to modulate kinase activity.
Based on the X‐ray crystal structure, it is tempting to suggest that PIPA binds to the CaMKIIα 6x hub in a two‐step process as illustrated in Figure 5. First, PIPA enters the binding site via the accessible intermediate site 1 (represented by subunit A, Figure 2c) and causes Trp403 and the connecting loop to flip outwards. Then, PIPA moves into the more buried site 2 (represented by subunit D, Figure 2e) where it binds to Arg453 and restricts the Trp403 in the outward‐flipped position. We further observed that the much smaller compound acetate binds to the same pocket and binding site residues as PIPA. However, with acetate bound, Trp403 is pointing inward as also observed for CaMKIIα 6x hub without ligand (PDB entry 6OF8). Thus, the similarly small compounds GHB and HOCPCA binding within the same CaMKIIα hub pocket may likewise be predicted to allow an inward positioning of Trp403, possibly stabilizing a slightly different conformational state of the CaMKIIα hub domain.
FIGURE 5.
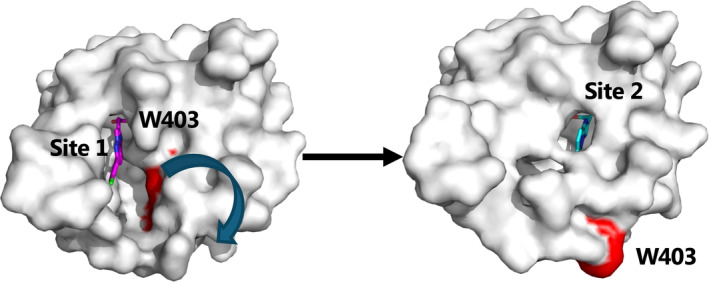
Model for sequential two‐step process of PIPA binding to CaMKIIα 6x hub (PDB entry 9EOY). Surface representation of subunit A, where Trp403 (marked in red) is flipped inward as shown to the left. PIPA (magenta sticks) has been modeled into site 1 to illustrate entrance of PIPA into the CaMKIIα 6x hub. Subunit D is shown to the right where PIPA (cyan sticks) binds at the conventional binding site 2, where Trp403 is flipped outward as shown to the right.
No Trp403 flip was previously observed for HOCPCA based on ITF measurements, and no kinase inhibition could be demonstrated (Leurs et al., 2021). Thus, it appears that there is a correlation between the extent of the displacement of the flexible loop containing Trp403 and reduced substrate phosphorylation. In other words, smaller differences in the binding mode of these compounds seem to dictate functional outcomes. Such differences seem linked to the molecular size of the ligands where PIPA, because of a larger size, sterically prevents an inward‐posing of the loop containing Trp403 and restricts it in an outward‐flipped conformation. When comparing the CaMKIIα 6x hub subunit binding of PIPA (subunit D) and acetate (subunit E), respectively, especially the two residues Trp403 and Ser404 in the acetate bound conformation of the CaMKIIα 6x hub would severely clash with PIPA (Figure S4B). Potential clashes with His395 and Ser474 might be relieved by smaller side‐chain conformational changes. Therefore, PIPA requires an outward‐flipped conformation to bind.
We further revealed that PIPA stabilizes the CaMKIIα WT hub to a lesser extent than 5‐HDC, shown by a 10.7°C lower maximum ∆T m in the TSA. However, upon binding to the CaMKIIα W403L hub, both compounds were equally effective at stabilizing the hub domain. Overall, this suggests that an outward‐flipped Trp403, provoked by PIPA, results in lesser stabilization of the CaMKIIα hub‐PIPA complex compared to when Trp403 is not present (CaMKIIα W403L hub). Moreover, it suggests that the Trp403 is not similarly restricted when binding to 5‐HDC, although previously shown to cause a tryptophan flip (Leurs et al., 2021). This was also supported when comparing the structure of CaMKIIα 6x hub with PIPA and the previously published structure with 5‐HDC (Leurs et al., 2021), which revealed subtle differences in the extent of Trp403 flip induced by the two ligands (Figure S8). In the structure with 5‐HDC, more space is observed, potentially allowing for Trp403 movement, while the steric bulk of PIPA appears to push and restrict the Trp403 loop into an even more outward pose than 5‐HDC. Overall, the data suggest that conformational changes of Trp403 and its connecting loop are a determinant for CaMKIIα hub stability.
An alternative explanation to the lesser stabilization of the CaMKIIα hub‐PIPA complex compared to with 5‐HDC could be a substoichiometric binding of PIPA to the CaMKIIα hub. In the X‐ray crystal structure, under the specific crystallization conditions, PIPA was bound in four out of the 14 CaMKIIα 6x hub subunits, while the remaining subunits were occupied by acetate. The presence of acetate in the crystallization buffer likely competes with PIPA for binding, stabilizing favorable conformations, which could explain the lower number of subunits occupied by PIPA in the crystal structure. However, we cannot exclude that the stoichiometry of PIPA binding could be different under other conditions. In the previously reported structure of CaMKIIα 6x hub with the GHB analog 5‐HDC (Leurs et al., 2021), the ligand occupied 12 out of 14 subunits, which confirms that the hub pocket is available for binding of several ligand molecules at the same time.
As the GHB analogs induce large stabilizing effects by binding to the CaMKIIα hub domain, we hypothesized that this stabilization might lead to changes in the proportional distribution of the different CaMKIIα hub oligomers. Importantly, the SAXS data in this study did not suggest any change in the proportions of dodecameric and tetradecameric forms of the isolated CaMKIIα WT hub upon ligand binding. However, this does not exclude the possibility that these ligands bind and stabilize the existing complexes of the hub domains, and thus prevent any natural dynamic exchange between these in the holoenzyme. The so‐called activation‐triggered subunit exchange has previously been reported to require CaMKIIα holoenzyme activation and was not observed when CaMKIIα was lacking the autoinhibitory segment and the linker (Bhattacharyya et al., 2016; Karandur et al., 2020; Stratton et al., 2014). Thus, further studies exploring the effect of the GHB analogs on the proportional distribution of the different CaMKIIα holoenzyme oligomers would be important. CaMKIIα hub stabilization in vivo may, however, involve other effects of CaMKIIα, for example, an altered subcellular localization of CaMKIIα as seen after ischemic stroke (Griem‐Krey et al., 2023). Furthermore, it would be interesting to also consider whether hub ligands (e.g., endogenous GHB) could affect intrinsic hub lability (Chien et al., 2024) and activity‐independent (structural) LTP induction (Tullis et al., 2023).
The SAXS data revealed another unexpected effect of PIPA binding. For the CaMKIIα WT hub, PIPA induced highly structured self‐association (dodecamer stacking as illustrated in Figure 3e). This effect was, however, not observed for the CaMKIIα WT hub alone nor for the CaMKIIα W403L hub. Furthermore, self‐association of the CaMKIIα WT hub was not observed in the presence of HOCPCA or 5‐HDC. Overall, this infers a probe‐dependent effect on stacking involving the altered peripheral surface of the CaMKIIα WT hub oligomers when the Trp403‐containing loop is restricted in an outward‐flipped conformation by the bulky PIPA. A limitation in the SAXS study is the high‐protein concentrations required for the measurements. Importantly, mass photometry data also showed the appearance of PIPA‐induced self‐association down to 100 nM of CaMKIIα WT hub. This observation at lower CaMKIIα WT hub concentration further indicates that this structural effect could be physiologically relevant. Thus, it is tempting to speculate that the binding of PIPA promotes self‐association of CaMKIIα WT hub by restricting Trp403 in the outward flip. We did not observe PIPA‐induced stacking of CaMKIIα 6x hub under similar conditions as for CaMKIIα WT hub. This indicated that additional factors than Trp403 may influence the stacking, including the oligomeric state and the multiple mutations in CaMKIIα 6x hub.
Although being different (face‐to‐edge; Figure S9) than the pattern of self‐association observed in the CaMKIIα WT hub by SAXS (stacking; Figure 3e), it is apparent that Trp403 plays an important role in self‐association per se in the X‐ray crystal packing of the CaMKIIα 6x hub with PIPA, as in each interface an outward‐flipped Trp403 in one tetradecamer participates in π–π interaction to another outward‐flipped Trp403 from a neighboring tetradecamer (Figure S9). Investigating the previously published X‐ray crystal structure of the CaMKIIα 6x hub without ligand (PDB entry 6OF8), a similar stacking pattern as observed in SAXS for CaMKIIα WT hub with PIPA could be observed in the structure with a repetitive distance of 56.2 Å (McSpadden et al., 2019). As illustrated in Figure S10, the close positioning of Trp403 between the connecting oligomers in the stacked packing further supports that the Trp403 residues are available for π–π interaction between them when flipped outward. This could suggest self‐association as a general feature of the CaMKIIα hub domain, possibly depending on loop flexibility and the proportion of outward‐flipped Trp403 available for π–π‐stacking.
Although binding of PIPA to the CaMKIIα holoenzyme ultimately inhibited substrate phosphorylation in the ADP‐Glo kinase assay, it remains to be shown whether the observed self‐association also happens to the CaMKIIα holoenzyme in the presence of PIPA and how such potential stacking might affect holoenzyme activity under different physiological or pathophysiological conditions. Interestingly, self‐association has previously been observed for the CaMKIIα holoenzyme (Barcomb et al., 2015; Hudmon et al., 1996, 2005; Petersen et al., 2003) and suggested as an inactivation mechanism of CaMKIIα following ischemia (Barcomb et al., 2015; Hudmon et al., 1996, 2005), which supports self‐association as a potential relevant regulatory or protective mechanism. Importantly, similar stacking pattern, as here observed for the isolated hub by SAXS, has previously been observed for the holoenzyme (Petersen et al., 2003).
The potential allosteric effect of the CaMKIIα hub on the kinase activity was previously proposed to involve specifically the loop containing Trp403 (W403‐P409) (Sloutsky et al., 2020). In line with this, by binding to the hub domain, PIPA may modulate the activity of the CaMKIIα holoenzyme by leading to an outward‐flipped conformation of Trp403 and thereby interfere with docking of the kinase domain onto the hub domain. Furthermore, the reduced CaM sensitivity observed upon increasing PIPA concentrations suggests interference with the activation of CaMKIIα by Ca2+/CaM and ultimately inhibition of substrate phosphorylation. One explanation for the high concentrations of PIPA needed could be a reduced ligand access to the hub cavity in the presence of CaM. In addition, we cannot exclude that PIPA stoichiometry could also play a role. However, the exact structural mechanism for how PIPA modulates CaMKIIα activity is still unclear. Due to the requirement of sub‐maximal CaM concentrations to observe an effect of PIPA, and the apparent high micromolar potency of PIPA to achieve kinase modulation compared to the high nanomolar hub affinity, it cannot be excluded that PIPA interacts with other sites at CaMKIIα or with CaM (awaits further studies). However, SPR data on the CaMKIIα holoenzyme and CaMKIIα WT hub showed comparable binding affinity to PIPA and therefore suggests specific interaction of PIPA with the hub domain. As such, PIPA together with Ph‐HTBA (Griem‐Krey et al., 2022) represents first‐in‐class allosteric modulators of CaMKIIα enzymatic activity acting through binding to the CaMKIIα hub domain.
Altogether, the presented data shed new light on allosteric regulation of CaMKIIα activity via the hub domain, where positioning of Trp403 in the flexible hub loop is central. Furthermore, mechanistic insight into this regulation is warranted in future experiments.
4. MATERIALS AND METHODS
Further details to methods are provided in Supporting Information.
4.1. Cell culturing and transfection of CaMKIIα in HEK293T cells
Human embryonic kidney (HEK) 293T (#CRL‐3216, ATCC) cells were cultured as previously described (Leurs et al., 2021).
4.2. [ 3H]HOCPCA binding assay to whole cell homogenates
This assay was performed as previously described (Leurs et al., 2021). Experiments were performed in technical triplicates and curves generated from pooled data from four individual experiments. K i values were calculated from IC50 values using the Cheng–Prusoff equation and previously determined K D value (Leurs et al., 2021). Mean pK i values are reported as mean ± SEM.
4.3. Expression and purification of CaMKIIα hub domains
The human CaMKIIα WT hub (UniProtKB Q9UQM7, residues 345–475), CaMKIIα W403L hub, and CaMKIIα 6x hub (containing the six mutations Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met) were expressed and purified as previously described (Hoelz et al., 2003; Leurs et al., 2021; McSpadden et al., 2019).
4.4. Surface plasmon resonance
SPR measurements were performed at 25°C using a Pioneer FE instrument (Sartorius). Recombinant purified CaMKIIα WT hub, CaMKIIα 6x hub, CaMKIIα W403L hub, and CaMKIIα holoenzyme (#02‐109, Carna Biosciences) were immobilized by amine coupling on to a biosensor using 20 mM NaAc, pH 5 buffer. PIPA was injected in twofold dilution series over the immobilized CaMKIIα hub constructs using a MES running buffer (20 mM MES, 150 mM NaCl, 1 mM DTT, pH 6) and CaMKIIα holoenzyme using an HBS‐P running buffer (10 mM Hepes, 150 mM NaCl, 0.005% Tween, 1 mM DTT, pH 7.4). The data were analyzed using Qdat Data Analysis Tool version 2.6.3.0 (Sartorius). Sensorgrams were corrected for buffer bulk effects and unspecific binding of the samples to the chip matrix by blank and reference surface subtraction (activated flow cell channel by injection of EDC/NHS and inactivated by injection of ethanolamine). The dissociation constants (K D) were estimated by plotting responses at equilibrium (R eq) against the injected concentration and curve‐fitted to a Langmuir (1:1) binding isotherm. The calculated K D value is mean ± SEM of three individual experiments.
4.5. Thermal shift assay
T m of CaMKIIα WT hub and CaMKIIα W403L hub were determined with and without 0.3–160 μM PIPA by differential scanning fluorometry (DSF), as previously described (Leurs et al., 2021). Data analysis was performed in GraphPad Prism (v. 10). Data (mean ∆T m max values ± SEM) were obtained from three independent experiments performed in technical triplicates.
4.6. ADP‐Glo kinase assay
CaMKIIα kinase activity was assessed using the ADP‐Glo Kinase Assay kit (#V9101, Promega) with purified CaMKIIα holoenzyme (#PR4586C, Thermo Fisher). The kinase detection reagent and the kinase reaction buffer (40 mM Tris, 0.5 mM CaCl2, 20 mM MgCl2, 0.1 mg/mL BSA, 50 μM DTT, pH 7.5) were prepared according to the manufacturer. All experiments were performed with 20 μL in 384‐well white polypropylene plates (#784075, Greiner). The kinase reaction was performed with a final concentration of 3 ng CaMKIIα, 25 μM ATP, 50 μM syntide‐2, and varying compound and CaM concentrations. CaM curves with 1–1781 nM CaM were obtained in the absence or presence of 100 μM or 500 μM compound. Compound inhibition curves were generated with 1–5000 μM compound and submaximal concentration of CaM (30 nM) in line with a recently published CaMKIIα ligand, Ph‐HTBA (Griem‐Krey et al., 2022). Likewise, the chosen ATP concentration was based on previous studies (Griem‐Krey et al., 2022; Leurs et al., 2021) and reduces the risk of limiting potency of ATP‐competing ligands. The kinase reactions were carried out for 55 min at 37°C. Hereafter, excess ATP was depleted by incubation with 5 μL ADP‐Glo Reagent for 40 min at room temperature. Finally, 10 μL Kinase Detection Reagent was added to each well to convert ADP to ATP and measured after 30 min incubation at room temperature. Luminescence was measured on a LUMIstar Omega plate reader, and data analysis and curve fitting were performed using GraphPad (v. 9). IC50 and EC50 values were determined using ‘log(inhibitor) vs. response with variable slope’. Individual experiments were performed in technical triplicates, and all curves are pooled data (mean ± SEM) of three independent experiments.
4.7. Co‐crystallization of PIPA with CaMKIIα 6x hub
Crystals were grown via sitting drop vapor diffusion at 20°C with reservoir solution containing 11% w/v PEG3350, 300 mM potassium acetate, pH 8.0 as previously established for CaMKIIα hub crystals (Bhattacharyya et al., 2016; McSpadden et al., 2019). PIPA was co‐crystallized by preincubating the protein (16 mg/mL) with 2.65 mM PIPA for 1 h at 20°C. 1.5 μL sitting drops were dispensed by adding 500 nL of protein stock to 1000 nL of reservoir solution. Drops were equilibrated against 50 μL of reservoir solution. Crystals appeared in 2 weeks and were cryoprotected (11% PEG, 300 mM potassium acetate pH 8.0, 25% glycerol, and 2.65 mM PIPA) prior to flash cooling in liquid nitrogen for data collection. For statistics on X‐ray diffraction data, see Table S1.
4.8. X‐ray structure determination
X‐ray diffraction data were collected at the Advanced Light Source (ALS) beamline 8.2.2 at wavelength 1.0000 Å and temperature of 100 K. Data were processed with XDS (Kabsch, 2010), scaled, and merged with Aimless (Evans & Murshudov, 2013) in the CCP4 suite; Agirre et al., 2023). The space group was C2221 with cell dimensions a = 103.05 Å, b = 182.92 Å, c = 107.76 Å, α = β = γ = 90°. Initial phases were obtained by molecular replacement using Phenix Phaser‐MR (McCoy et al., 2007) and the structure of CaMKIIα 6x hub with 5‐HDC (PDB entry 7REC; Leurs et al., 2021) as search model. AutoBuild was used for model building, refinement, and density modification. Further refinement of the structure was performed using Phenix (version 13) (Liebschner et al., 2019), and Coot (Emsley et al., 2010) was used for manual model building. The CIF dictionary file for PIPA was generated using Phenix eLBOW (Moriarty et al., 2009). The 2F o − F c electron density for PIPA, acetate, and PEG is shown in Figure 2. Statistics for structure refinement are available in Table S1. PyMOL (The PyMOL Molecular Graphics System, Version 2.5.2 Schrödinger, LLC) was used to prepare all structure figures. 2D ligplots were generated using the LigPlot+ software (Laskowski & Swindells, 2011).
4.9. Small‐angle X‐ray scattering
For the SAXS studies, purified CaMKIIα hub proteins were buffer exchanged into a MES buffer (20 mM MES, 150 mM NaCl, 1 mM DTT, pH 6.0) without or with ligands (PIPA 200 μM, HOCPCA 1000 μM, 5‐HDC 100 μM) using Zeba™ Spin Desalting Columns (Thermo Fisher Scientific, 0.5 mL 7K MWCO). Dilution series were made of CaMKIIα WT hub, CaMKIIα 6x hub, and CaMKIIα W403L hub in concentrations ranging from 1 to 7 mg/mL. Several preparations were made of CaMKIIα WT hub with PIPA at different protein concentrations prior to buffer exchange. During the preparation of CaMKIIα WT hub with PIPA (sample iv), significant aggregation was visible immediately upon buffer exchange; thus, the sample was centrifuged (2 min, 4°C, 10,000 g ) and SAXS data were measured only on the soluble fraction (supernatant).
SAXS data were collected at the CPHSAXS facility (University of Copenhagen, Denmark) and at the P12 beamline operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany) (Blanchet et al., 2015). Data from the CPHSAXS facility were collected on a BioXolver L (Xenocs) using metal jet source (Excillum) equipped with a Pilatus3 R 300K detector (Dectris). Samples were automatically loaded using the BioCUBE sample handling robot from a 96‐well tray. Initial data processing was done using BioXTAS RAW (Hopkins et al., 2017). Data from P12 were collected on a Pilatus6M detector (Dectris). Samples were loaded automatically using the ARINAX BioSAXS sample changer and sample flow during exposure. Data reduction was done using the SASflow pipeline (Franke et al., 2012).
Scattering intensities were measured at room temperature (20–22°C) as a function of the momentum transfer q = (4πsinθ)/λ with 2θ being the scattering angle and λ the X‐ray wavelength. Details of the individual measurements are listed in Table S3.
Images were radially averaged and overlap of the individual frames checked before averaging. Corresponding buffer measurements were subtracted and, when relevant, the data from different configurations were merged and scaled by concentration to the final data files. Primary data analysis, Guinier analyses, indirect Fourier transformation, and MW estimation were done using PRIMUS (Manalastas‐Cantos et al., 2021). CRYSOL (Franke et al., 2017) was used for the evaluation of single structures against the experimental data, and for mixtures, OLIGOMER (Konarev et al., 2003) was used to determine best fit and corresponding volume fractions.
The dodecamer (PDB entry 5IG3) (Bhattacharyya et al., 2016) and tetradecamer (PDB entry 6OF8) (McSpadden et al., 2019) high‐resolution models were used for comparison and analysis. In addition, models of stacked dodecamers and tetradecamer were made by simple translation of the individual structures using PyMOL (The PyMOL Molecular Graphics System, Version 2.5.2 Schrödinger, LLC).
4.10. Mass photometry
Mass photometry experiments were performed on a Refeyn One MP. Standard calibration was performed using 2.25 nM apoferritin, 15 nM bovine serum albumin, and 15 nM thioglycolic acid with known molecular weights: 440 kDa, 66.5 kDa, and 92.11 Da, respectively. 3 μL of standard protein was diluted with 17 μL of filtered buffer (25 mM Tris–HCL, 150 mM KCl, pH 8.0) and measured for 60 sec. For experiments without PIPA, purified CaMKIIα WT hub and CaMKIIα W403L hub were diluted to final concentrations of 500, 200, and 100 nM in MES buffer (20 mM MES, 150 mM NaCl, 1 mM DTT, pH 6.0) and measurements were made for 60 s. For experiments with PIPA added, 1.5 mg/mL purified CaMKIIα WT hub and CaMKIIα W403L hub were incubated with 200 μM PIPA for 90 min and desalted into MES buffer + PIPA (ZebaTM spin, 7K MWCO). The eluant was diluted to 500, 200, and 100 nM for each measurement. Buffer alone was measured before each measurement, and histograms were generated using the Refeyn's DiscoverMP software.
4.11. Intrinsic tryptophan fluorescence assay
ITF measurements targeting the Trp403 were recorded at 25°C on a Safire2 plate reader (Tecan) and performed as previously described (Leurs et al., 2021). PIPA showed absorbance for the higher concentrations and was corrected for inner filter effect with a factor between 0.99 and 1.4 calculated from:
The fluorescence intensities were normalized according to:
F obs is the observed fluorescence intensity and F b is the background fluorescence for compound in buffer alone. F max is the fluorescence intensity of CaMKIIα hub alone without compound, and F min is the fluorescence intensity when plateau is reached at high compound concentrations in the presence of the CaMKIIα hub domain. Since PIPA did not reach a plateau at high compound concentrations, F min was set to the fluorescence intensity of buffer for all compounds tested. Fluorescence intensities usually spanned from 3000 to 40,000 for PIPA, while the fluorescence intensity for buffer was around 1000. Non‐linear regression was used for curve‐fitting using the equation for “log(inhibitor) vs. response with variable slope” to determine IC50 values (GraphPad Prism, v. 8).
AUTHOR CONTRIBUTIONS
Dilip Narayanan: Methodology; investigation; formal analysis; validation; writing – original draft. Anne Sofie G. Larsen: Investigation; formal analysis. Stine Juul Gauger: Methodology; investigation; formal analysis; validation; writing – review and editing. Ruth Adafia: Investigation; formal analysis. Rikke Bartschick Hammershøi: Investigation; formal analysis. Louise Hamborg: Investigation; formal analysis. Jesper Bruus‐Jensen: Investigation; formal analysis. Nane Griem‐Krey: Investigation; formal analysis. Christine L. Gee: Methodology; investigation; formal analysis. Bente Frølund: Validation; supervision; writing – review and editing. Margaret M. Stratton: Methodology; investigation; formal analysis; validation; writing – review and editing; supervision. John Kuriyan: Supervision. Jette Sandholm Kastrup: Formal analysis; methodology; investigation; validation; supervision; writing – review and editing. Annette E. Langkilde: Methodology; validation; investigation; writing – review and editing; formal analysis. Petrine Wellendorph: Conceptualization; methodology; formal analysis; validation; supervision; writing – review and editing. Sara M. Ø. Solbak: Conceptualization; investigation; writing – original draft; writing – review and editing; validation; methodology; formal analysis; project administration; supervision.
CONFLICT OF INTEREST STATEMENT
The University of Copenhagen has licensed patent rights (EP 3952873) of PIPA and analogs to Ceremedy Ltd. of which P.W. and B.F. are co‐founders, and N.G.‐K. is an employee. All remaining authors declare no conflict of interest.
Supporting information
Data S1.
ACKNOWLEDGMENTS
This work was supported through financial support from the following foundations and grants: The Lundbeck Foundation (R277‐2018‐260 to P.W.), The Novo Nordisk Foundation (NNF17OC0028664 and NNF21OC0067835 to P.W.), the Independent Research Fund Denmark (1026‐00335B to P.W.), and the Lundbeck Foundation pregraduate scholarships in pharmaceutical neuroscience under the auspieces of the Drug Research Academy (to A.S.G.L. and R.B.H.). We acknowledge the University of Copenhagen Small‐angle X‐ray facility, CPHSAXS, funded by the Novo Nordisk Foundation (grant no. NNF19OC0055857) and assistance from Pernille S. Tuelung. https://drug.ku.dk/core-facilities/cphsaxs/. The synchrotron SAXS data were collected at beamline P12 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). We thank Cy Jeffries for the assistance in using the beamline as well as financial support through DANSCATT (funded by the Danish Agency for Science, Technology, and Innovation). X‐ray diffraction data were collected at the Advanced Light Source beamline 8.2.2. The Berkeley Center for Structural Biology is supported in part by the Howard Hughes Medical Institute. The Advanced Light Source is a Department of Energy Office of Science User Facility under Contract No. DE‐AC02‐05CH11231. The ALS‐ENABLE beamlines are supported in part by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169.
Narayanan D, Larsen ASG, Gauger SJ, Adafia R, Hammershøi RB, Hamborg L, et al. Ligand‐induced CaMKIIα hub Trp403 flip, hub domain stacking, and modulation of kinase activity. Protein Science. 2024;33(10):e5152. 10.1002/pro.5152
Review Editor: Aitziber L. Cortajarena
Contributor Information
Jette Sandholm Kastrup, Email: jsk@sund.ku.dk.
Annette E. Langkilde, Email: annette.langkilde@sund.ku.dk.
Petrine Wellendorph, Email: pw@sund.ku.dk.
Sara M. Ø. Solbak, Email: sara.solbak@sund.ku.dk.
DATA AVAILABILITY STATEMENT
The structure coordinates and corresponding structure factor file of the CaMKIIα 6x hub in complex with PIPA, acetate, and PEG have been deposited in the Protein Data Bank under the accession code 9EOY. SAXS data and selected models are deposited in SASBDB (Kikhney et al., 2020) under accession codes SASDUQ4, SASDUR4, SASDUS4, SASDUT4, SASDUU4, SASDUV4, SASDUW4, SASDUX4, SASDUY4, SASDUZ4, SASDU25, SASDU35, SASDU45, SASDU55, SASDU65, SASDU75.
REFERENCES
- Agirre J, Atanasova M, Bagdonas H, Ballard CB, Basle A, Beilsten‐Edmands J, et al. The CCP4 suite: integrative software for macromolecular crystallography. Acta Crystallogr D Struct Biol. 2023;79:449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcomb K, Goodell DJ, Arnold DB, Bayer KU. Live imaging of endogenous Ca(2+)/calmodulin‐dependent protein kinase II in neurons reveals that ischemia‐related aggregation does not require kinase activity. J Neurochem. 2015;135:666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay T, Eghorn LF, Klein AB, Wellendorph P. GHB receptor targets in the CNS: focus on high‐affinity binding sites. Biochem Pharmacol. 2014;87:220–228. [DOI] [PubMed] [Google Scholar]
- Bayer KU, Schulman H. CaM kinase: still inspiring at 40. Neuron. 2019;103:380–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M, Stratton MM, Going CC, McSpadden ED, Huang Y, Susa AC, et al. Molecular mechanism of activation‐triggered subunit exchange in Ca(2+)/calmodulin‐dependent protein kinase II. elife. 2016;5:e13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet CE, Spilotros A, Schwemmer F, Graewert MA, Kikhney A, Jeffries CM, et al. Versatile sample environments and automation for biological solution X‐ray scattering experiments at the P12 beamline (PETRA III, DESY). J Appl Crystallogr. 2015;48:431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PH, Zhong FL, Niwa S, Bonnard C, Utami KH, Zeng R, et al. A homozygous loss‐of‐function CAMK2A mutation causes growth delay, frequent seizures and severe intellectual disability. elife. 2018;7:e32451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Puhl H, Vogel SS, Molloy JE, Chiu W, Khan S. Hub stability in the calcium calmodulin‐dependent protein kinase II. Commun Biol. 2024;7:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coultrap SJ, Vest RS, Ashpole NM, Hudmon A, Bayer KU. CaMKII in cerebral ischemia. Acta Pharmacol Sin. 2011;32:861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Kikhney AG, Svergun DI. Automated acquisition and analysis of small angle X‐ray scattering data. Nucl Instrum Methods Phys Res, Sect A. 2012;689:52–59. [Google Scholar]
- Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, et al. ATSAS 2.8: a comprehensive data analysis suite for small‐angle scattering from macromolecular solutions. J Appl Crystallogr. 2017;50:1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griem‐Krey N, Gauger SJ, Gowing EK, Thiesen L, Frølund B, Clarkson AN, et al. The CaMKIIα hub ligand Ph‐HTBA promotes neuroprotection after focal ischemic stroke by a distinct molecular interaction. Biomed Pharmacother. 2022;156:113895. [DOI] [PubMed] [Google Scholar]
- Griem‐Krey N, Klein AB, Clausen BH, Namini MR, Nielsen PV, Bhuiyan M, et al. The GHB analogue HOCPCA improves deficits in cognition and sensorimotor function after MCAO via CaMKIIα. J Cereb Blood Flow Metab. 2023;43:1419–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A, Nairn AC, Kuriyan J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin‐dependent kinase II. Mol Cell. 2003;11:1241–1251. [DOI] [PubMed] [Google Scholar]
- Hopkins JB, Gillilan RE, Skou S. BioXTAS RAW: improvements to a free open‐source program for small‐angle X‐ray scattering data reduction and analysis. J Appl Crystallogr. 2017;50:1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudmon A, Aronowski J, Kolb SJ, Waxham MN. Inactivation and self‐association of Ca2+/calmodulin‐dependent protein kinase II during autophosphorylation. J Biol Chem. 1996;271:8800–8808. [DOI] [PubMed] [Google Scholar]
- Hudmon A, LeBel E, Roy H, Sik A, Schulman H, Waxham MN, et al. A mechanism for Ca2+/calmodulin‐dependent protein kinase II clustering at synaptic and nonsynaptic sites based on self‐association. J Neurosci. 2005;25:6971–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandur D, Bhattacharyya M, Xia Z, Lee YK, Muratcioglu S, McAffee D, et al. Breakage of the oligomeric CaMKII hub by the regulatory segment of the kinase. elife. 2020;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupmann K, Cryan JF, Wellendorph P, Mombereau C, Sansig G, Klebs K, et al. Specific gamma‐hydroxybutyrate‐binding sites but loss of pharmacological effects of gamma‐hydroxybutyrate in GABA(B)(1)‐deficient mice. Eur J Neurosci. 2003;18:2722–2730. [DOI] [PubMed] [Google Scholar]
- Kikhney AG, Borges CR, Molodenskiy DS, Jeffries CM, Svergun DI. SASBDB: towards an automatically curated and validated repository for biological scattering data. Protein Sci. 2020;29:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a windows PC‐based system for small‐angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Krall J, Bavo F, Falk‐Petersen CB, Jensen CH, Nielsen JO, Tian Y, et al. Discovery of 2‐(Imidazo[1,2‐b]pyridazin‐2‐yl)acetic acid as a new class of ligands selective for the gamma‐hydroxybutyric acid (GHB) high‐affinity binding sites. J Med Chem. 2019;62:2798–2813. [DOI] [PubMed] [Google Scholar]
- Kury S, van Woerden GM, Besnard T, Proietti Onori M, Latypova X, Towne MC, et al. De novo mutations in protein kinase genes CAMK2A and CAMK2B cause intellectual disability. Am J Hum Genet. 2017;101:768–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Swindells MB. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. [DOI] [PubMed] [Google Scholar]
- Lee J, Chen X, Nicoll RA. Synaptic memory survives molecular turnover. Proc Natl Acad Sci USA. 2022;119:e2211572119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leurs U, Klein AB, McSpadden ED, Griem‐Krey N, Solbak SMØ, Houlton J, et al. GHB analogs confer neuroprotection through specific interaction with the CaMKIIα hub domain. Proc Natl Acad Sci USA. 2021;118:e2108079118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, et al. Macromolecular structure determination using X‐rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr D Struct Biol. 2019;75:861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lučić I, Héluin L, Jiang P‐L, Castro Scalise AG, Wang C, Franz A, et al. CaMKII autophosphorylation can occur between holoenzymes without subunit exchange. elife. 2023;12:e86090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manalastas‐Cantos K, Konarev PV, Hajizadeh NR, Kikhney AG, Petoukhov MV, Molodenskiy DS, et al. ATSAS 3.0: expanded functionality and new tools for small‐angle scattering data analysis. J Appl Crystallogr. 2021;54:343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSpadden ED, Xia Z, Chi CC, Susa AC, Shah NH, Gee CL, et al. Variation in assembly stoichiometry in non‐metazoan homologs of the hub domain of Ca(2+)/calmodulin‐dependent protein kinase II. Protein Sci. 2019;28:1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanan AG, Gunasekaran S, Jacob RS, Omkumar RV. Role of Ca(2+)/calmodulin‐dependent protein kinase type II in mediating function and dysfunction at glutamatergic synapses. Front Mol Neurosci. 2022;15:855752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty NW, Grosse‐Kunstleve RW, Adams PD. Electronic ligand builder and optimization workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JB, Zaegel V, Coultrap SJ, Miller AP, Bayer KU, Reichow SL. The CaMKII holoenzyme structure in activation‐competent conformations. Nat Commun. 2017;8:15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otmakhov N, Lisman J. Measuring CaMKII concentration in dendritic spines. J Neurosci Methods. 2012;203:106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JD, Chen X, Vinade L, Dosemeci A, Lisman JE, Reese TS. Distribution of postsynaptic density (PSD)‐95 and Ca2+/calmodulin‐dependent protein kinase II at the PSD. J Neurosci. 2003;23:11270–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti Onori M, Koopal B, Everman DB, Worthington JD, Jones JR, Ploeg MA, et al. The intellectual disability‐associated CAMK2G p.Arg292Pro mutation acts as a pathogenic gain‐of‐function. Hum Mutat. 2018;39:2008–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigter PMF, de Konink C, Dunn MJ, Proietti Onori M, Humberson JB, Thomas M, et al. Role of CAMK2D in neurodevelopment and associated conditions. Am J Hum Genet. 2024;111:364–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg OS, Deindl S, Comolli LR, Hoelz A, Downing KH, Nairn AC, et al. Oligomerization states of the association domain and the holoenyzme of Ca2+/CaM kinase II. FEBS J. 2006;273:682–694. [DOI] [PubMed] [Google Scholar]
- Sloutsky R, Dziedzic N, Dunn MJ, Bates RM, Torres‐Ocampo AP, Boopathy S, et al. Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub‐dependent regulation. Sci Signal. 2020;13:eaaz0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson JR, Wang X, Perfitt TL, Parrish WP, Shonesy BC, Marks CR, et al. A novel human CAMK2A mutation disrupts dendritic morphology and synaptic transmission, and causes ASD‐related behaviors. J Neurosci. 2017;37:2216–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton M, Lee IH, Bhattacharyya M, Christensen SM, Chao LH, Schulman H, et al. Activation‐triggered subunit exchange between CaMKII holoenzymes facilitates the spread of kinase activity. elife. 2014;3:e01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Shehata MA, Gauger SJ, Ng CKL, Solbak S, Thiesen L, et al. Discovery and optimization of 5‐hydroxy‐diclofenac toward a new class of ligands with nanomolar affinity for the CaMKIIα hub domain. J Med Chem. 2022;65:6656–6676. [DOI] [PubMed] [Google Scholar]
- Tian Y, Shehata MA, Gauger SJ, Veronesi C, Hamborg L, Thiesen L, et al. Exploring the NCS‐382 scaffold for CaMKIIα modulation: synthesis, biochemical pharmacology, and biophysical characterization of Ph‐HTBA as a novel high‐affinity brain‐penetrant stabilizer of the CaMKIIα hub domain. J Med Chem. 2022;65:15066–15084. [DOI] [PubMed] [Google Scholar]
- Torres‐Ocampo AP, Ozden C, Hommer A, Gardella A, Lapinskas E, Samkutty A, et al. Characterization of CaMKIIα holoenzyme stability. Protein Sci. 2020;29:1524–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullis JE, Larsen ME, Rumian NL, Freund RK, Boxer EE, Brown CN, et al. LTP induction by structural rather than enzymatic functions of CaMKII. Nature. 2023;621:146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vest RS, O'Leary H, Coultrap SJ, Kindy MS, Bayer KU. Effective post‐insult neuroprotection by a novel Ca(2+)/calmodulin‐dependent protein kinase II (CaMKII) inhibitor. J Biol Chem. 2010;285:20675–20682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellendorph P, Høg S, Skonberg C, Bräuner‐Osborne H. Phenylacetic acids and the structurally related non‐steroidal anti‐inflammatory drug diclofenac bind to specific γ‐hydroxybutyric acid sites in rat brain. Fundam Clin Pharmacol. 2009;23:207–213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1.
Data Availability Statement
The structure coordinates and corresponding structure factor file of the CaMKIIα 6x hub in complex with PIPA, acetate, and PEG have been deposited in the Protein Data Bank under the accession code 9EOY. SAXS data and selected models are deposited in SASBDB (Kikhney et al., 2020) under accession codes SASDUQ4, SASDUR4, SASDUS4, SASDUT4, SASDUU4, SASDUV4, SASDUW4, SASDUX4, SASDUY4, SASDUZ4, SASDU25, SASDU35, SASDU45, SASDU55, SASDU65, SASDU75.


