Abstract
The synthesis of the triphosphates of 4′-thiouridine and 4′-thiocytidine, 4′-thioUTP (7; thioUTP) and 4′-thioCTP (8; thioCTP), and their utility for SELEX (systematic evolution of ligands by exponential enrichment) is described. The new nucleoside triphosphate (NTP) analogs 7 and 8 were prepared from appropriately protected 4′-thiouridine and -cytidine derivatives using the one-pot method reported by J. Ludwig and F. Eckstein [(1989) J. Org. Chem., 54, 631–635]. Because SELEX requires both in vitro transcription and reverse transcription, we examined the ability of 7 and 8 for SELEX by focusing on the two steps. Incorporation of 7 and 8 by T7 RNA polymerase to give 4′-thioRNA (thioRNA) proceeded well and was superior to those of the two sets of frequently used modified NTP analogs for SELEX (2′-NH2dUTP and 2′-NH2dCTP; 2′-FdUTP and 2′-FdCTP), when an adequate leader sequence of DNA template was selected. We revealed that a leader sequence of about +15 of DNA template is important for the effective incorporation of modified NTP analogs by T7 RNA polymerase. In addition, reverse transcription of the resulting thioRNA into the complementary DNA in the presence of 2′-deoxynucleoside triphosphates (dNTPs) also proceeded smoothly and precisely. The stability of the thioRNA toward RNase A was 50 times greater than that of the corresponding natural RNA. With these successful results in hand, we attempted the selection of thioRNA aptamers to human α-thrombin using thioUTP and thioCTP, and found a thioRNA aptamer with high binding affinity (Kd = 4.7 nM).
INTRODUCTION
SELEX (systematic evolution of ligands by exponential enrichment) is a biotechnological method used for isolating functional RNA molecules, such as ribozymes and aptamers from a random pool of RNA combinatorial libraries (1–3). This innovative technology has already been successfully employed to isolate a number of ribozymes and aptamers. Ribozymes, RNA molecules that catalyze not only biological processes, such as RNA cleavage and ligation but also organic reactions, including the Diels–Alder reaction and hydrolysis of esters, were isolated (4–6). Aptamers have also been obtained for a variety of target molecules, e.g. nucleotides, amino acids and proteins using SELEX (7–9). Since the resulting aptamers, such as antibodies, show a potent and selective binding affinity for the target molecule, they are expected to become not only useful biological tools, but also diagnostic and therapeutic agents (10,11). In addition, given the ease of chemical modification of aptamers consisting of nucleic acids rather than proteins, it is not surprising that applications such as biosensors and ligands in affinity probe capillary electrophoresis have also been reported (12,13). For these reasons, isolation of functional RNA molecules using SELEX has gained increasing attention. However, because it is generally acknowledged that RNA is a transient molecule in biological fluids, stabilization of functional RNA molecules toward nuclease digestion would be required for their versatile use and application.
Until now, three methods have been devised to improve the stability of RNA molecules obtained using SELEX. Nolte et al. (14) have reported a mirror-design RNA aptamer, which consists of selecting an RNA aptamer against the enantiomer of the target molecule, the mirror image of the target, by using standard SELEX. When the resulting RNA aptamer (d-RNA) was converted to l-RNA with the same sequence context, the l-RNA exhibited high binding affinity to the target molecule and high stability against nuclease digestion. Although this concept would appear to be an intriguing idea, the preparation of the phosphoramidite units of l-nucleosides is required. Furthermore, this strategy is limited to cases where an enantiomer of the target molecule is available. The second method for the stabilization of functional RNA molecules is a post-modification, which consists of a chemical modification of the RNA molecules by phosphoramidite units, such as 2′-O-methyl derivatives. However, this strategy causes structural changes of the RNA molecules and often results in loss of ribozyme and RNA aptamer activity (15). The third method uses SELEX to isolate the stable RNA molecules by employing modified nucleoside triphosphates, which arguably is a more reliable strategy. However, recognition of the nucleoside triphosphates by the T7 RNA polymerase is quite restrictive, and few of the modified triphosphates are considered good candidates for SELEX. Thus far, 2′-modified-2′-deoxynucleoside 5′-triphosphates (16), nucleoside 5′-(α-P-borano)triphosphates (17) and nucleoside 5′-(α-thio)triphosphates (18) are used as modified triphosphates for SELEX. Among them, 2′-amino- and 2′-fluoro-2′-deoxypyrimidine nucleoside triphosphates (2′-NH2dUTP and 2′-NH2dCTP; 2′-FdUTP and 2′-FdCTP) are frequently used compounds, and a number of nuclease-resistant RNAs, especially RNA aptamers, were isolated by SELEX using these two sets of 5′-triphosphates.
On the other hand, 4′-thioribonucleoside is a nucleoside analog possessing a sulfur atom instead of the furanose ring oxygen. We have already reported an effective and stereoselective synthesis of 4′-thioribonucleosides (19–21). Furthermore, it was revealed that RNAs consisting of 4′-thioribonucleosides exhibit an enhancement of thermal stability and nuclease resistance (22,23). Because the RNAs consisting of 4′-thioribonucleosides possess 2′-OH groups, a distinctive characteristic of RNA, which would contribute to the catalytic activity and diversity of the tertiary structure and of the interaction with the target molecules, we were prompted to prepare 4′-thioribonucleoside 5′-triphosphates and investigate whether the 5′-triphosphates are applicable to the SELEX process. Although the synthesis of 5-ethyl-2′-deoxy-4′-thiouridine 5′-triphosphate and its substrate properties for DNA synthesizing enzymes, such as DNA polymerase α and reverse transcriptases, have been reported previously (24), there is no literature precedent for a series of 4′-thioribonucleosides. Here, we describe the synthesis of the new nucleoside triphosphates, 4′-thioUTP (7; thioUTP) and 4′-thioCTP (8; thioCTP) and their utility for SELEX. The 5′-triphosphates obtained were effectively incorporated into RNA by T7 RNA polymerase to give 4′-thioRNA (thioRNA; in this paper, thioRNA indicates an RNA molecule containing 4′-thiouridine and 4′-thiocytidine units. Similarly, aminoRNA and fluoroRNA indicate RNA molecules containing 2′-amino- and 2′-fluoro-2′-deoxypyrimidine nucleoside units.). The resulting thioRNA was then converted into complementary DNA without misincorporation by reverse transcriptase in the presence of dNTPs. Selection of thioRNA aptamers to human α-thrombin is also described.
MATERIALS AND METHODS
General methods and materials
Physical data were measured as follows: 1H, 13C and 31P NMR spectra were recorded on JEOL-EX270 or JEOL-AL400 instruments in CDCl3 or D2O as the solvent with tetramethylsilane (1H and 13C) or H3PO4 (31P) as an internal standard. Chemical shifts are reported in p.p.m. (δ), and signals are expressed as s (singlet), d (doublet), t (triplet), m (multiplet) or br (broad). Mass spectra were measured on JEOL JMS-D300 spectrometer. The analytical data of the synthetic compounds were presented in the Supplementary Material. TLC was carried out on Merck Kieselgel F254 precoated plates. Silica gel used for column chromatography was Merck silica gel 5715. T7 RNA polymerase and TaKaRa Ex Taq™ were purchased from TaKaRa. Superscript™ II (RNase H−) Reverse Transcriptase and RNaseOUT™ were from Invitrogen. RNase A was from NIPPON GENE. Nitrocellulose filter (HAWP filter, 0.45 μm) was from Millipore. Human α-thrombin was from Enzyme Research Laboratories. pGEM-T Easy Vector System was from Promega. BigDye Terminator v3.1 Cycle Sequence Kit was from Applied Biosystems. [γ-32P]GTP was purchased from PerkinElmer. Unlabeled NTPs were from Amersham. Modified NTPs were from TriLink (2′-NH2dUTP and 2′-NH2dCTP) and EPICENTRE (2′-FdUTP and 2′-FdCTP). Radioactive densities of the gel were visualized using a Bio-imaging analyzer (Bas 2500, Fuji). DNA was sequenced using a BigDye Terminator v3.1 Cycle Sequence Kit (Applied Biosystems) on a 377 DNA sequencer (Applied Biosystems). DNA templates for transcription and DNA-G4 were purchased from Sigma-Genosys, and RNA-24 was from Hokkaido System Science.
1-[5-O-(4-Methoxytrityl)-2,3-di-O-acetyl-4-thio-β-D-ribofuranosyl]uracil (3)
To a solution of 1 (131 mg, 0.5 mmol) in pyridine (4 ml) was added 4-methoxytrityl chloride (232 mg, 0.75 mmol), and the reaction mixture was stirred at room temperature. After 14 h, acetic anhydride (188 μl, 2.0 mmol) and 4-(dimethylamino)pyridine (DMAP) (5 mg, 0.05 mmol) was added to the reaction mixture and being stirred for additional 2 h at the same temperature. The reaction was quenched by the addition of methanol, and the solvent was removed in vacuo. The residue was dissolved in AcOEt, and the organic solution was washed with H2O, saturated aqueous NaHCO3 and brine. The separated organic layer was dried (Na2SO4) and concentrated in vacuo. The residue was purified by a silica gel column, eluted with hexane/AcOEt (2:1–1:1) to give 3 (214 mg, 70%), as a colorless glass.
1-(2,3-Di-O-acetyl-4-thio-β-D-ribofuranosyl)uracil (5)
A solution of 3 (199 mg, 0.32 mmol) in 80% aqueous acetic acid (3 ml) was stirred for 11 h at room temperature. The reaction mixture was partitioned between CHCl3 and saturated aqueous NaHCO3. The organic layer was washed with saturated aqueous NaHCO3 and brine. The separated organic layer was dried (Na2SO4) and concentrated in vacuo. The residue was purified by a silica gel column, eluted with hexane/AcOEt (2:1–0:1) to give 5 (93 mg, 84%), as a white foam.
4′-Thiouridine 5′-triphosphate (7)
To a solution of 5 (89 mg, 0.26 mmol) in pyridine–DMF (260–780 μl) was added a freshly prepared 1 M solution of 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one in dioxane (290 μl, 0.29 mmol), and the mixture was stirred at room temperature. After 10 min, a 0.5 M solution of bis(tri-n-butylammonium)pyrophosphate in DMF (780 μl, 0.39 mmol) and tri-n-butylamine (278 μl, 1.17 mmol) was quickly added. The reaction mixture was stirred for an additional 10 min, and a solution of 1% iodine in pyridine/H2O (98/2, v/v) (5 ml) was added. After 30 min, the excess iodine was destroyed by adding a few drops of a 5% aqueous NaHSO3 followed by ammonium hydroxide (28%, 8 ml). After 3.5 h, the solvent was removed in vacuo, and the residue was dissolved in H2O (300 ml). The solution was applied to a DEAE Sephadex column, which was eluted with a linear gradient of 1 liter each of H2O and 1 M TEAB buffer. The fractions containing the desired triphosphate were collected, concentrated and coevaporated with H2O. The residue was dissolved in H2O (5 ml), and the solution was passed through ion-exchange resins DIAION PK212 (H+ form) and DIAION WK 20 (Na+ form) to give 7 as a trisodium salt (80 mg, 55%).
N4-Benzoyl-1-[5-O-(4-methoxytrityl)-2,3-di-O-acetyl-4-thio-β-D-ribofuranosyl]cytosine (4)
In a similar manner as described for 3, 2 (25) (302 mg, 0.83 mmol) was treated with 4-methoxytrityl chloride, followed by acetic anhydride to give 4 (579 mg, 97%), as a white foam.
N4-Benzoyl-1-(2,3-di-O-acetyl-4-thio-β-D-ribofuranosyl)cytosine (6)
In a similar manner as described for 5, 4 (420 mg, 0.58 mmol) was treated with 80% aqueous acetic acid to give 6 (239 mg, 92%), as a white foam.
4′-Thiocytidine 5′-triphosphate (8)
In a similar manner as described for 7, 6 (71 mg, 0.16 mmol) was successively treated with 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one, bis(tri-n-butylammonium)pyrophosphate, tri-n-butylamine, and then with ammonium hydroxide to give 8 as a trisodium salt (68 mg, 75%).
General procedure for in vitro transcription by T7 RNA polymerase
The in vitro transcription was performed with 40 pmol of template DNA in 20 μl of transcription buffer. The mixture contained either 0.4 mM of natural NTP(s) and/or modified NTP(s), 100 U of T7 RNA polymerase in a 40 mM Tris–HCl buffer (pH 8.0) containing 8 mM MgCl2 and 2 mM spermidine, and 5 mM DTT. Reaction mixtures were incubated at 37°C. At appropriate time points, 2 μl aliquots of the reaction mixture were added to 4 μl of loading buffer (50 mM EDTA, 10 M urea, 0.1% bromophenol blue and 0.1% xylene cyanol). The mixtures were then analyzed by electrophoresis on 20% polyacrylamide gel containing 7 M urea. The precursor [γ-32P]GTP (10 μCi) was used when labeled transcripts were needed.
Reverse transcription by Superscript™ II reverse transcriptase
Reverse transcription was carried out using RNA59 or thioRNA59 as templates. A 16mer of oligodeoxynucleotide labeled with 32P at the 5′ end (5′-pCTGGTACCGTCACCTC-3′) was used as a primer. Reverse transcription was performed with 20 pmol each of the template RNA and the primer containing 2 pmol of the labeled substrate in 20 μl of transcription buffer. The mixture contained 0.5 mM of four dNTPs, 200 U of Superscript™ II (RNase H−) in a 50 mM Tris–HCl buffer (pH 8.3) containing 75 mM KCl and 3 mM MgCl2, 40 U of RNaseOUT™ and 10 mM DTT. Reaction mixtures were incubated at 42°C. After 50 min, 2 μl aliquots of the reaction mixture were added to 4 μl of the loading buffer (50 mM EDTA, 10 M urea, 0.1% bromophenol blue and 0.1% xylene cyanol). The mixtures were then analyzed by electrophoresis on 12% polyacrylamide gel containing 7 M urea.
Sequencing
The complementary DNA was amplified with PCR using primers (5′-CTGGTACCGTCACCTC-3′ and 5′-GGGAGAAGGGTATCCGGA-3′) and TaKaRa Ex Taq™. The PCR product was cloned using a pGEM-T Easy Vector System. The resulting plasmid DNA was isolated by the alkaline-lysis method and sequenced.
Stability toward RNase A
To test for RNase A resistance, a mixture of 5 pmol of RNA59 (or thioRNA59 and aminoRNA59) labeled with 32P at the 5′ end and the corresponding unlabeled RNA (400 pmol) was used. The RNA sample was incubated with RNase A (2 ng) in a buffer containing 10 mM Tris–HCl (pH 7.4), 0.3 M NaCl and 5 mM EDTA (total 40 μl) at 37°C. At appropriate time points, 2 μl aliquots of the reaction mixture were added to 4 μl of loading buffer (50 mM EDTA, 10 M urea, 0.1% bromophenol blue and 0.1% xylene cyanol). The mixtures were then analyzed by electrophoresis on 16% polyacrylamide gel containing 7 M urea.
SELEX protocols
SELEX was carried out essentially according to reported methods (26). Single-stranded DNA (ssDNA) templates with a 30 nt variable region (5′-GGGAGAAGGGAAGTAACAGG-N30-GTGAGAAGAGGTGACGGTACCAG-3′; 73mer), a forward primer (5′-GCTCTAGATAATACGACTCACTATAGGGAGAAGGGAAGTAACAGG-3′; 45mer) and a reverse primer (5′-CTGGTACCGTCACCTCTTCTCAC-3′; 23mer) were prepared. The ssDNA templates were converted to double-stranded DNA (dsDNA) by PCR with the forward and reverse primers. The resulting DNA templates were transcribed using T7 RNA polymerase in the presence of thioUTP and thioCTP, together with unmodified ATP and GTP. The transcribed thioRNA library was purified on 12% denaturing polyacrylamide gel. The resulting thioRNA library was denatured in annealing buffer (50 mM Tris–HCl, pH 7.4, 100 mM NaCl and 1 mM MgCl2) at 90°C for 3 min and allowed to cool at 23°C for 10 min before each round of selection. In addition, the thioRNA library was counterselected by passing through a nitrocellulose filter to remove filter-binding species. For the first round of selection, human α-thrombin was mixed with an equimolar amount of the thioRNA library (1000 nM:1000 nM) in the selection buffer (50 mM Tris–HCl, pH 7.4, 100 mM NaCl, 1 mM MgCl2 and 1 mM DTT). After incubation at 25°C for 10 min, the mixture was separated by filtration through a nitrocellulose filter to collect human α-thrombin with the thioRNAs and washed with 1 ml of the selection buffer. The thioRNAs were eluted from the filter by phenol (400 μl) and freshly prepared 7 M urea (200 μl). Ethanol precipitation with 20 μg of glycogen was carried out and the thioRNAs recovered were annealed to reverse primer, and reverse transcribed by Superscript™ II (RNase H−) at 42°C for 50 min to give the cDNAs. Following PCR amplification, the resulting dsDNA templates were transcribed in vitro to give the thioRNA library for the next round of selection. The concentration of the thioRNA library was fixed to 1000 nM in each selection, while that of human α-thrombin in the selection buffer was decreased gradually in successive rounds from 1000 to 10 nM. The selection process was carried out at 25°C for rounds 1–7 and at 37°C for rounds 8–10 to progressively increase the selective pressure. After 10 rounds of selection, the selected RNA pool was reverse transcribed and PCR amplified. The resulting DNAs were cloned and sequenced as described above. The secondary structure predictions for the selected aptamers were made using the Zuker RNA mfold computer algorithm (27).
Chemical synthesis of thioRNA aptamers
The predicted thioRNA aptamers, CI-2-23 and CII-1-37, were synthesized on an Applied Biosystem 3400 DNA synthesizer using 4′-thiouridine and -cytidine phosphoramidite units, and commercially available phosphoramidite units of purine nucleosides. The synthesis and purification of the thioRNAs were carried out according to our previous methods (23). The structure of each thioRNA was confirmed by the measurement of matrix-assisted laser desorption ionization time-of-flight/mass spectrometry on a Voyager-DE pro. CI-2-23: calculated mass, C223H273N97O150P22S7 7613.9 (M–H); observed mass, 7619.9. CII-1-37: calculated mass, C354H434N142O244P36S16 12 203.2 (M–H); observed mass, 12 205.9.
Filter-binding assay and determination of dissociation constant
The equilibrium dissociation constants (Kds) for the selected aptamers were determined by using a constant amount of 5′ end labeled oligonucleotides (0.1 nM) in the selection buffer with increasing concentrations of human α-thrombin (0.1 to 111 or 333 nM). After incubation at 37°C for 5 min, the mixture was passed through a nitrocellulose filter, and the filter was immediately washed with 1 ml of selection buffer. Radioactivity retained on the filter was quantified using a Bio-imaging analyzer and evaluated by calculating the percentage of the input oligonucleotides retained on each nitrocellulose filter in complexes with protein. The data points were fitted to a Scatchard plot to determine the equilibrium dissociation constant by PRISM.
RESULTS AND DISCUSSION
Synthesis of thioUTP and thioCTP
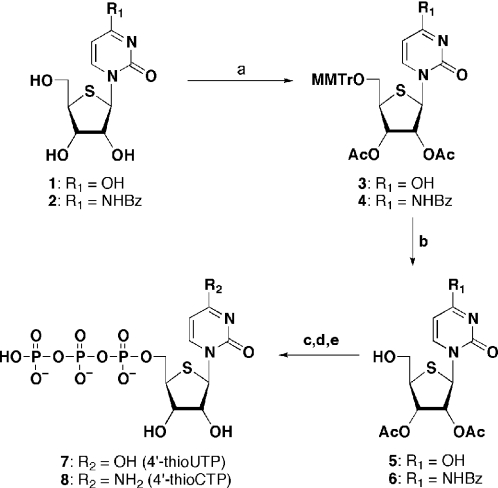
As mentioned in the introduction, one example of the 5′-triphosphate synthesis of 4′-thionucleoside derivative has been reported by Alexandrova et al. (24). Because the initial monophosphorylation of 4′-thionucleoside derivative by the standard method [POCl3, (MeO)3P=O] did not take place because of the formation of bicyclic episulfonium intermediate (28), the desired triphosphate was prepared in a lengthy route with low yield. Therefore, we sought a more practical method. Ludwig and Eckstein (29) reported an efficient one-pot synthesis of nucleoside 5′-triphosphate starting with an appropriately protected nucleoside using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. This method was examined for the synthesis of 7 and 8 (Scheme 1). 4′-Thiouridine (1) was first converted to protected derivative 3 by treatment with 4-methoxytrityl chloride (MMTrCl), followed by acetic anhydride in pyridine. The MMTr group of 3 was then removed by treatment with 80% aqueous acetic acid to give 5, a substrate for subsequent phosphitylation reaction. In the same manner, N4-benzoyl-4′-thiocytidine (2) (25) was converted into 2′,3′-di-O-acetyl derivative 6. Because the one-pot 5′-triphosphate synthesis requires oxidation step of the cyclotriphosphite intermediate, 5 was treated under the oxidative conditions (1% iodine in pyridine–H2O) before the 5′-triphosphate synthesis. Consequently, the sulfur atom of 5 was resistant under the oxidative conditions (data not shown). Thus, 5 was treated with 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one, followed by bis(tri-n-butylammonium)pyrophosphate to give the cyclotriphosphite intermediate. The resulting intermediate was then oxidized by 1% iodine in pyridine–H2O, and successive treatment with NH4OH to give the desired thioUTP (7) in 55% yield after a DEAE Sephadex column chromatographic purification. In the same manner, 8 was also prepared from 6 in 75% yield by the one-pot reaction.
Scheme 1.
(a) MMTrCl, pyridine then Ac2O, DMAP; (b) 80% aq. AcOH; (c) 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one, pyridine–DMF; (d) bis(tri-n-butylammonium)pyrophosphate in DMF, Bu3N; (e) 1% iodine in pyridine–H2O then NH4OH.
Transcription by T7 RNA polymerase in the presence of thioUTP and thioCTP
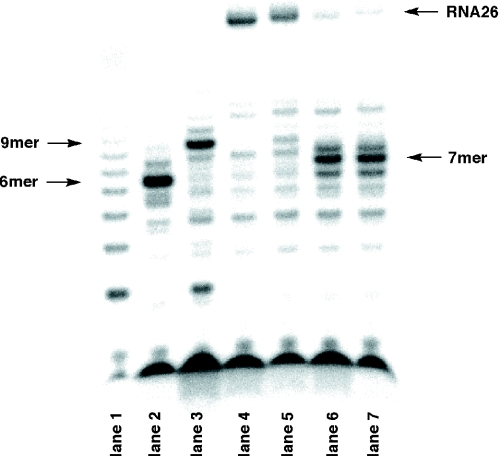
To conduct the SELEX strategy, it is necessary to determine whether in vitro transcription by T7 RNA polymerase in the presence of modified NTP analogs and reverse transcription of the resulting RNA containing modified nucleoside units to the complementary DNA proceed effectively and precisely. With the desired 7 and 8 in hand, the in vitro transcription by T7 RNA polymerase was first examined using synthetic short DNA templates, including the T7 promoter sequence. The in vitro transcription experiments were performed at 37°C for 3 h in the presence of appropriate unmodified and modified NTPs (0.4 mM each), including [γ-32P]GTP using template1 (43mer) to form 26mer RNA, RNA26 (Figure 1). PAGE analysis of the experiments is shown in Figure 2. In lane 1 (+GTP), formation of the G ladder was observed (30). In lanes 2 (+GTP and ATP) and 3 (+GTP, ATP and CTP), the transcriptions were stopped at the corresponding positions to form the 6mer and 9mer RNAs, respectively. In the presence of all NTPs (lane 4), a significant amount of RNA26 was observed along with some abortive sequences. When UTP was replaced by thioUTP (lane 5), the transcription proceeded well and the full-length RNA was also formed under the same conditions. In contrast, the transcription efficiencies were drastically decreased and a significant amount of abortive RNAs (mainly 7mer RNA) was observed when CTP was replaced by thioCTP (lanes 6 and 7). In the presence of thioUTP, the full-length RNA was formed with 70% of the efficiency relative to that of the unmodified NTPs (lane 4 versus lane 5). In contrast, the efficiency of thioCTP was only 14% (lane 4 versus lane 6). In a combination of thioUTP and thioCTP, the efficiency to give thioRNA26 was decreased to 11% (lane 4 versus lane 7). Although we examined the transcription using 2- and 10-fold concentrations of thioCTP, the efficiency did not improve significantly (16 and 22%, respectively). One reason for the low thioCTP incorporation compared with that of thioUTP is perhaps due to the RNA sequence formed. Thus, in this particular RNA sequence, the first position of a uridine (U) residue is at the +10 position, while that of cytidine (C) is at the +7 position. In addition, consecutive introduction of the C residue is required (see the sequence of RNA26). In the transcription using the T7 RNA polymerase, it is known that quenching the chain elongation to form abortive RNA products often takes place within about the first 10 nt incorporated (termed the initiation phase) (31,32). This abortive formation is especially enhanced when modified NTP(s) are incorporated within the first 10 nt (33).
Figure 1.
Sequences of DNA templates and the resulting RNAs by in vitro transcription. The italic regions were T7 promoter sequence.
Figure 2.
PAGE analysis of in vitro transcription by T7 RNA polymerase using template1. In vitro transcription was carried out as described in Materials and Methods in the presence of GTP (lane 1); GTP and ATP (lane 2); GTP, ATP and CTP (lane 3); four NTPs (lane 4); GTP, ATP and CTP plus thioUTP (lane 5); GTP, ATP and UTP plus thioCTP (lane 6); and GTP and ATP plus thioUTP and thioCTP (lane 7).
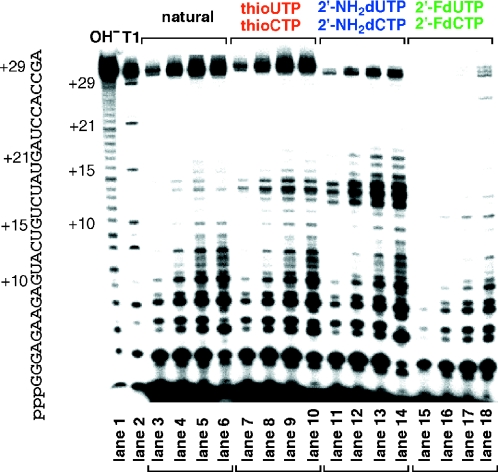
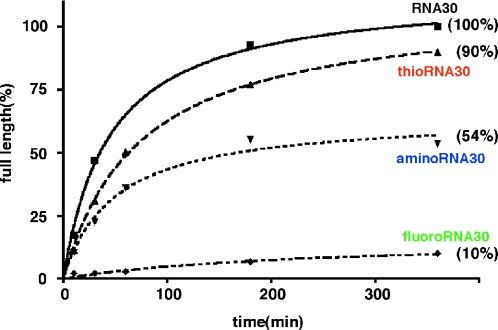
With these considerations in mind, we decided to examine the in vitro transcription using the new DNA template2 to form the 30mer RNA, RNA30 (Figure 1). In this case, the initial position of the U residue incorporation is +11 and that of C residue is at the +13 position. When the transcription was carried out using template2, the desired full-length RNA was obtained with high efficiency in the presence of not only thioUTP but also thioCTP and a combination of thioUTP and thioCTP (data not shown). Because the in vitro transcription using thioUTP and thioCTP proceeded well, the transcription efficiency of these triphosphates was then compared with those of 2′-NH2dUTP and 2′-NH2dCTP, and 2′-FdUTP and 2′-FdCTP, which are generally used for SELEX. The transcription by T7 RNA polymerase was performed at 37°C in the presence of ATP, GTP and modified pyrimidine nucleoside triphosphates (0.4 mM each), and the transcription products at various time periods were detected using PAGE analysis. As shown in Figure 3, a significant amount of the complete transcription product thioRNA30 was observed under the conditions in which UTP and CTP were replaced by thioUTP and thioCTP, and the efficiency was almost equal to that with the natural NTPs (lanes 3–6 versus lanes 7–10). In contrast, the transcription efficiencies were decreased when 2′-NH2dUTP and 2′-NH2dCTP, and 2′-FdUTP and 2′-FdCTP were used (lanes 11–14 and 15–18). The time course of formation of the complete transcription products (0–6 h) under various conditions is shown in Figure 4. With thioUTP and thioCTP as substrates, the amount of thioRNA30 was 90% of that with the natural NTPs as substrates, whereas with 2′-NH2dUTP and 2′-NH2dCTP it was 54%, to give aminoRNA30. When 2′-FdUTP and 2′-FdCTP were used, the relative efficiency decreased to only 10% giving fluoroRNA30. The DuraScribe™ T7 RNA polymerase, a mutant polymerase for effective incorporation of 2′-FdUTP and 2′-FdCTP, was also examined. The relative efficiency increased to ∼20% giving fluoroRNA30. In contrast, the incorporation of thioUTP and thioCTP using this mutant polymerase was ineffective and the relative efficiency was decreased to <10% (data not shown).
Figure 3.
PAGE analysis of in vitro transcription by T7 RNA polymerase using template2. In vitro transcription was carried out as described in Materials and Methods in the presence of four NTPs (lanes 3–6, for 10, 30, 60 and 180 min, respectively); GTP and ATP plus thioUTP and thioCTP (lanes 7–10, for 10, 30, 60 and 180 min, respectively); GTP and ATP plus 2′-NH2dUTP and 2′-NH2dCTP (lanes 11–14, for 10, 30, 60 and 180 min, respectively), and GTP and ATP plus 2′-FdUTP and 2′-FdCTP (lanes 15–18, for 10, 30, 60 and 180 min, respectively). Lanes 1 and 2 are sequence marker obtained by treatment of RNA30 with 50 mM aq. Na2CO3 (pH 9.0) and RNase T1, respectively.
Figure 4.
Time course of transcription efficiency. The amount of full-length RNAs is presented in % compared with the amount of RNA30 (100%) after 360 min.
As mentioned above, formation of the abortive RNA products predominantly occurs in the initiation phase because of an unstable ternary complex of the T7 RNA polymerase, the template DNA and shorter RNA fragments. After synthesis of an RNA longer than 10 nt, the polymerase reaction enters the elongation phase to give full-length RNA without dissociation of the ternary complex. In our reaction system using template2, formation of the abortive RNA products was observed not only under conditions using the modified NTPs (lanes 7–10 and 11–14) but also under those using the natural NTPs (lanes 3–6) to a similar extent. Drastic differences of transcription were observed at the positions around +11 to +15, the presumed positions of the transition from the initiation phase to the elongation phase. Under conditions using the natural NTPs, the transcription proceeded smoothly until termination, although a trace amount of the abortive products between +11 and +15 was observed. In contrast, formation of the abortive products between +11 and +15 was critical under the conditions using modified NTPs, especially those using 2′-NH2dUTP and 2′-NH2dCTP. In the RNA product formed by the transcription, consecutive introduction of pyrimidine residues is required at the positions +13 and +14, and this would presumably be responsible for the low efficiency of the transcription using 2′-NH2dUTP and 2′-NH2dCTP. Under conditions using thioUTP and thioCTP, the proportion of the abortive RNA products between +11 and +15 was smaller, thus providing a higher amount of thioRNA30 than aminoRNA30. Although there are three consecutive positions where pyrimidine residues can be introduced at +16 to +18 and +23 to +25 into the RNA product, formation of the abortive RNA products corresponding to these positions is negligible. From these results, it was revealed that thioUTP and thioCTP are better substrates of the T7 RNA polymerase than the 2′-amino- and 2′-fluoro-2′-deoxypyrimidine nucleoside triphosphates. In addition, our results showed that a leader sequence of about +15 of the resulting RNA is critical for effective incorporation of the modified pyrimidine nucleoside triphosphates for in vitro transcription.
Reverse transcription and characterization of the resulting complementary DNA
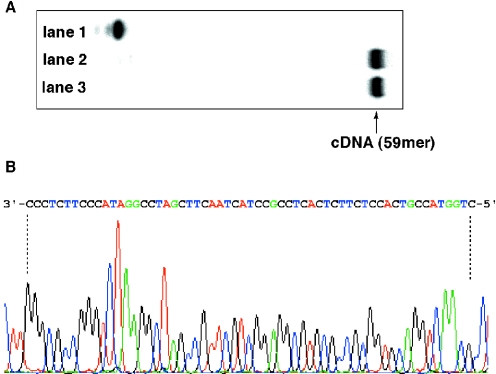
As described above, in vitro transcription using thioUTP and thioCTP afforded thioRNA efficiently. Accordingly, we next examined whether the resulting thioRNA acted as a template for reverse transcription in the presence of natural dNTPs to give complementary DNA. To test this function, RNA59 and thioRNA59, longer sequences for the reverse transcription, were prepared by in vitro transcription using template3 (Figure 1). The transcription efficiencies were almost equal for both RNA products (data not shown). After annealing RNA59 or thioRNA59 with the labeled DNA primer (5′-pCTGGTACCGTCACCTC-3′; 16mer), the reverse transcription using Superscript™ II (RNase H−) was carried out at 42°C for 1 h in the presence of dNTPs (0.5 mM each). As shown in Figure 5A, the reverse transcription of thioRNA59 proceeded smoothly and effectively, as that of RNA59, to give the 59mer of the complementary DNA (lanes 2 and 3). To check the accuracy of the reverse transcription, the resulting complementary DNA from thioRNA59 was amplified by PCR and then cloned into a pGEM-T Easy Vector. Plasmids containing the 59mer region were sequenced and compared with the sequence of template3. The results show that in vitro transcription using thioUTP and thioCTP, and the reverse transcription of the resulting thioRNA as template proceeded with high fidelity (Figure 5B).
Figure 5.
(A) PAGE analysis of reverse transcription by Superscript™ II (RNase H−). Reverse transcription was carried out as described in Materials and Methods in the presence of labeled primer only (lane 1); labeled primer and RNA59 as a template (lane 2); labeled primer and thioRNA59 as a template (lane 3). (B) Electropherogram of DNA-sequencing fragments.
Stability of thioRNA toward RNase A
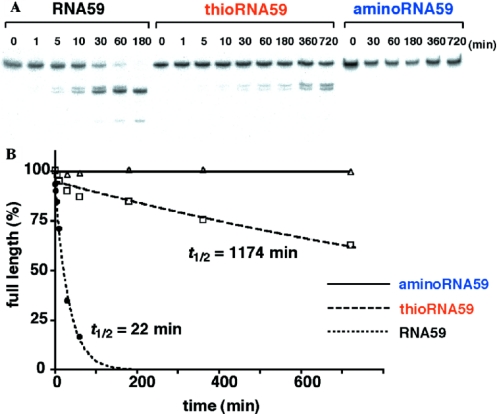
We have already reported high stability of fully modified thioRNA against nuclease digestion (23). The thioRNA prepared in this study is not fully modified, but rather partially modified by the 4′-thiouridine and 4′-thiocytidine units. Therefore, the stability of this partially modified thioRNA toward RNase A, the pyrimidine-specific predominant nuclease in human serum (34), was examined. The stability was tested with labeled thioRNA59, and compared with that of the labeled RNA59 and aminoRNA59. The results of enzymatic degradation by RNase A are shown in Figure 6. As shown in Figure 6A, thioRNA59 was resistant to RNase A digestion. More than 60% of full-length RNA was still intact after 12 h, while under the same conditions RNA59 was completely consumed within 3 h. The half-lives (t1/2 s) were estimated based on the ratio of full-length RNA at each time point, and the t1/2 of thioRNA59 was calculated to be 1174 min, which was 50 times greater than that of RNA59 (Figure 6B). Under the same conditions, aminoRNA59, which lacks the critical 2′-OH groups for RNA cleavage, was completely intact. Although thioRNA59 was less stable than aminoRNA59, the higher stability of thioRNA relative to natural RNA is worth noting.
Figure 6.
Stabilities of RNA59, thioRNA59 and aminoRNA59 toward RNase A. (A) PAGE analysis of labeled RNAs incubated at 37°C in the presence of RNase A. (B) Time course of degradation of RNAs by RNase A.
Selection of thioRNA aptamers to human α-thrombin using thioUTP and thioCTP
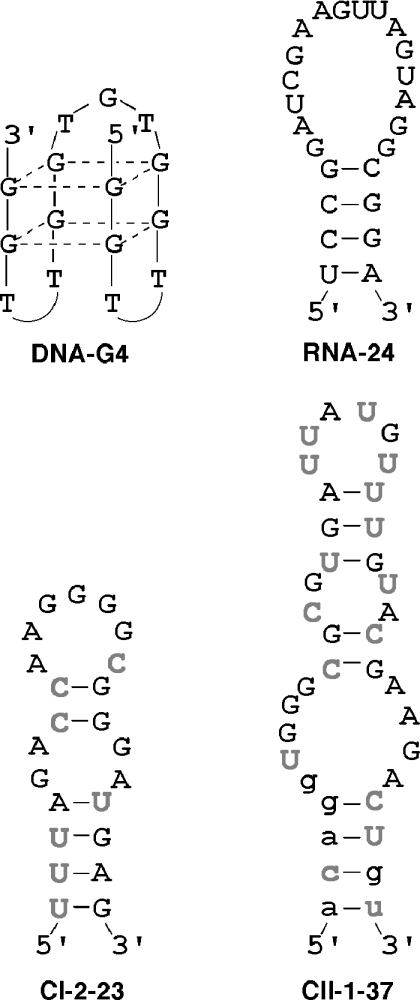
As described above, we showed that thioUTP and thioCTP can be used for the process of SELEX. Accordingly, selection of thioRNA aptamers using thioUTP and thioCTP was next examined. As a target protein for SELEX, human α-thrombin was chosen because the RNA and the DNA aptamers have been isolated and their structures and functions have been thoroughly investigated (26,35). The ssDNA templates comprising a 30 nt in randomized positions flanked by constant region were prepared. The constant regions included a sequence of 5′-GGGAGAAGGG-3′ right after a T7 promoter for the efficient transcription (see results shown in Figure 3) and targets for PCR primers and cloning sites. After amplification by PCR, the dsDNA templates were transcribed by T7 RNA polymerase in the presence of thioUTP, thioCTP, ATP and GTP. The resulting 1014 thioRNA library was used for the first round of the selection, and the selection processes were repeated under various conditions (see Materials and Methods for detail). As there was no further improvement in the binding affinity between the 8th and 10th rounds, the thioRNA library from the 10th round was reverse transcribed, amplified and cloned. Three major classes, i.e. classes I, II and III, emerged from analysis of the 24 clones (Figure 7). In each class, the most abundant sequences represented 6 of the 24 sequences. Affinities of individual full-length thioRNAs consisting of a 30 nt in randomized positions and the constant region were tested by nitrocellurose filter-binding assay. The results showed that CI-2 from class I and CII-1 from class II had the highest binding affinity to human α-thrombin. In contrast, all thioRNAs of class III showed the filter-binding affinity along with the affinity to human α-thrombin (data not shown). Therefore, the secondary structure predictions for CI-2 and CII-1 were made using the Zuker RNA mfold computer algorithm (27). For CI-2, a 23mer of the stem–bulge–loop structure CI-2-23 was predicted, while the longer, similar stem-bulge-loop structure CII-1-37 was predicted for CII-1 (Figure 8). The resulting CI-2-23 consisted of a part of the randomized region, and 7 of the 23 nt length came from thioUTP and thioCTP (gray letters). In contrast, the CII-1-37 contained the constant sequence (lower case) on its stem region, and 16 of the 37 nt length came from thioUTP and thioCTP (gray letters). The predicted thioRNAs were chemically synthesized according to our previous method (23). Because the predicted structures were different from the reported RNA aptamers to human α-thrombin, composed of stem–loop structures, the binding assay of CI-2-23 and CII-1-37 with the protein was examined along with RNA-24, which had previously been selected as the high-affinity RNA aptamer. In addition, the DNA aptamer DNA-G4 (35) to human α-thrombin was also prepared for comparison.
Figure 7.
Sequences identified in the 10th round thioRNA library derived from the affinity selection performed on human α-thrombin. The frequency of clones carrying the same sequence is indicated in parenthesis. The conserved sequences in each individual class are indicated by back ground colors. The sequences common to all ligands are shown in lower case letters and the sequences of the variable region in upper case letters.
Figure 8.
The secondary structures of DNA-G4, RNA-24 and thioRNA aptamers, CI-2-23 and CII-1-37.
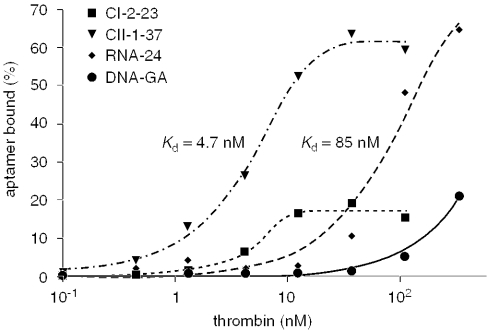
The binding assay of each aptamer was carried out by the filter-binding assay. The experiment performed with a constant concentration of each aptamer (0.5 nM) and the concentration of human α-thrombin was titrated from 0.1 to 111 or 333 nM. As shown in Figure 9, the aptamer possessing the highest binding affinity was CII-1-37. When the molar ratio of human α-thrombin and CII-1-37 was 222:1 (111 nM:0.5 nM), ∼60% maximal binding activity was observed, and the Kd value of CII-1-37 was estimated as 4.7 nM. The binding activity of the other thioRNA aptamer CI-2-23 was lower. Since the Kd value of RNA-24, prepared for comparison, was estimated to be 85 nM in our hands, we succeeded in isolating a high-affinity thioRNA aptamer to human α-thrombin using thioUTP and thioCTP. Although further analyses of the resulting thioRNA aptamers for their structures and functions may be required, our results demonstrated that thioUTP (7) and thioCTP (8) should be considered new NTP analogs intended for the development of nuclease-resistant functional RNA molecules by SELEX.
Figure 9.
The binding of DNA-G4, RNA-24, Cl-2-23 and CII-1-37 to varying concentration of human α-thrombin was determined by nitrocellulose filter partitioning as described in Materials and Methods.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors would like to thank Ms Y. Misawa (Hokkaido University) for technical assistance and Ms S. Oka (Center for Instrumental Analysis, Hokkaido University) for technical assistance with the manuscript. This paper constitutes part 235 of Nucleosides and Nucleotides [for part 234 in this series, see (36)]. This work was supported in part by Grant-in-Aid for Scientific Research from the Japan Society for Promotion of Science (No. 15510169 to N.M. and No. 15209003 to A.M.). Funding to pay the Open Access publication charges for this article was provided by the Japan Society for Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Robertson D.L., Joyce G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 3.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Breaker R.R. In vitro selection of catalytic polynucleotides. Chem. Rev. 1997;97:371–390. doi: 10.1021/cr960008k. [DOI] [PubMed] [Google Scholar]
- 5.Stuhlmann F., Jäschke A. Characterization of an RNA active site: interactions between a Diels–Alderase ribozyme and its substrates and products. J. Am. Chem. Soc. 2002;124:3238–3244. doi: 10.1021/ja0167405. [DOI] [PubMed] [Google Scholar]
- 6.Chun S., Jeong S., Kim J., Chong B., Park Y., Park H., Yu J. Cholesterol esterase activity by in vitro selection of RNA against a phosphate transition-state analogue. J. Am. Chem. Soc. 1999;121:10844–10845. [Google Scholar]
- 7.Famulok M., Szostak J.W. In vitro selection of specific ligand-binding nucleic acids. Angew. Chem. Int. Ed. Engl. 1992;31:979–988. [Google Scholar]
- 8.Osborne S.E., Ellington A.D. Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 9.Hermann T., Patel D.J. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 10.Osborne S.E., Matsumura I., Ellington A.D. Aptamers as therapeutic and diagnostic reagents: problems and prospects. Curr. Opin. Chem. Biol. 1997;1:5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 11.Jayasene S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 12.Jhaveri S., Rajendran M., Ellington A.D. In vitro selection of signaling aptamers. Nat. Biotechnol. 2000;18:1293–1297. doi: 10.1038/82414. [DOI] [PubMed] [Google Scholar]
- 13.Clark S., Remcho V.T. Electrochromatographic retention studies on a flavin-binding RNA aptamer sorbent. Anal. Chem. 2003;75:5692–5696. doi: 10.1021/ac030156s. [DOI] [PubMed] [Google Scholar]
- 14.Nolte A., Klussmann S., Bald R., Erdmann V.A., Fürste J.B. Mirror-design of l-oligonucleotide ligands binding to l-arginine. Nat. Biotechnol. 1996;14:1116–1119. doi: 10.1038/nbt0996-1116. [DOI] [PubMed] [Google Scholar]
- 15.Lebruska L.L., Maher L.J. Selection and characterization of an RNA decoy for transcription factor NF-κB. Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 16.Pagratis N.C., Bell C., Chang Y., Jennings S., Fitzwater T., Jellinek D., Dang C. Potent 2′-amino-, and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 1997;15:68–73. doi: 10.1038/nbt0197-68. [DOI] [PubMed] [Google Scholar]
- 17.Lato S.M., Ozerova N.D.S., He K., Sergueeva Z., Shaw B.R., Burke D.H. Boron-containing aptamers to ATP. Nucleic Acids Res. 2002;30:1401–1407. doi: 10.1093/nar/30.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jhaveri S., Olwin B., Ellington A.D. In vitro selection of phosphorothiolated aptamers. Bioorg. Med. Chem. Lett. 1998;8:2285–2290. doi: 10.1016/s0960-894x(98)00414-4. [DOI] [PubMed] [Google Scholar]
- 19.Naka T., Nishizono N., Minakawa N., Matsuda A. Investigation of the stereoselective coupling of thymine with meso-thiolane-3,4-diol-1-oxide derivative via the Pummerer reaction. Tetrahedron Lett. 1999;40:6297–6300. [Google Scholar]
- 20.Naka T., Minakawa N., Abe H., Kaga D., Matsuda A. The stereoselective synthesis of 4′-β-thioribonucleosides via the Pummerer reaction. J. Am. Chem. Soc. 2000;122:7233–7243. [Google Scholar]
- 21.Minakawa N., Kato Y., Uetake K., Kaga D., Matsuda A. An improved large scale synthesis of 1,4-anhydro-4-thio-d-ribitol. Tetrahedron. 2003;59:1699–1702. [Google Scholar]
- 22.Bellon L., Barascut J.-L., Maury G., Divita G., Goody R., Imbach J.-L. 4′-Thio-oligo-β-d-ribonucleotides: synthesis of β-4′-thio-oligouridylates nuclease resistance base pairing properties and interaction with HIV-1 reverse transcriptase. Nucleic Acids Res. 1993;21:1587–1593. doi: 10.1093/nar/21.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshika S., Minakawa N., Matsuda A. Synthesis and physical and physiological properties of 4′-thioRNA: application to post-modification of RNA aptamer toward NF-κB. Nucleic Acids Res. 2004;32:3815–3825. doi: 10.1093/nar/gkh705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandrova L.A., Semizarov D.G., Krayevsky A.A., Walker R.T. 4′-Thio-5-ethyl-2′-deoxyuridine 5′-triphosphate (TEDUTP): synthesis and substrate properties in DNA-synthesizing systems. Antiviral Chem. Chemother. 1996;7:237–242. [Google Scholar]
- 25.Minakawa N., Kaga D., Kato Y., Endo K., Tanaka M., Sasaki T., Matsuda A. Synthesis and structural elucidation of 1-(3-C-ethynyl-4-thio-β-d-ribofuranosyl)cytosine (4′-thioECyd) J. Chem. Soc. Perkin Trans. I. 2002:2182–2189. [Google Scholar]
- 26.Kubik M.F., Stephens A.W., Schneider D., Marlar R.A., Tasset D. High-affinity RNA ligands to human α-thrombin. Nucleic Acids Res. 1994;22:2619–2626. doi: 10.1093/nar/22.13.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zucker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 28.Hancox E.L., Walker R.T. Some reactions of 4′-thionucleosides and their sulfones. Nucleosides Nucleotides. 1996;15:135–148. [Google Scholar]
- 29.Ludwig J., Eckstein F. Rapid and efficient synthesis of 5′-O-(1-thiotriphosphates), 5′-triphosphate and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- 30.Kuzmine I., Gottlieb P.A., Martin C.T. Structure in nascent RNA leads to termination of slippage transcription by T7 RNA polymerase. Nucleic Acids Res. 2001;29:2601–2606. doi: 10.1093/nar/29.12.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Y.W., Steitz T.A. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 32.Liu C., Martin C.T. Promoter clearance by T7 RNA polymerase. J. Biol. Chem. 2002;277:2725–2731. doi: 10.1074/jbc.M108856200. [DOI] [PubMed] [Google Scholar]
- 33.Kujau M.J., Siebert A., Wölfl S. Design of sequences that improve the efficiency of the enzymatic synthesis of 2′-aminopyrimidine RNA for in vitro selection. J. Biochem. Biophys. Methods. 1997;35:141–151. doi: 10.1016/s0165-022x(97)00039-0. [DOI] [PubMed] [Google Scholar]
- 34.Shimayama T., Nishikawa F., Nishikawa S., Taira K. Nuclease-resistant chimeric ribozymes containing deoxyribonucleotides and phosphorothioate linkages. Nucleic Acids Res. 1993;21:2605–2611. doi: 10.1093/nar/21.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 36.Kudoh T., Fukuoka M., Ichikawa S., Maruyama T., Ogawa Y., Hashii M., Higashida H., Kunerth S., Weber K., Guse A.H., et al. Synthesis of stable and cell-type selective analogues of cyclic ADP-ribose, a Ca2+-mobilizing second messenger. Structure-activity relationship of the N1-ribose moiety. J. Am. Chem. Soc. 2005 doi: 10.1021/ja050732x. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.