Abstract
Proliferation of renal tubular epithelial cells (TEC) is essential for restoring tubular integrity and thereby to support renal functional recovery from kidney ischemia/reperfusion (KI/R) injury. Activation of transcriptional factor c-Myc promotes TEC proliferation following KI/R; however, the mechanism regarding c-Myc activation in TEC is incompletely known. Heat shock protein A12A (HSPA12A) is an atypic member of HSP70 family. In this study, we found that KI/R decreased HSPA12A expression in mouse kidneys and TEC, while ablation of HSPA12A in mice impaired TEC proliferation and renal functional recovery following KI/R. Gain-of-functional studies demonstrated that HSPA12A promoted TEC proliferation upon hypoxia/reoxygenation (H/R) through directly interacting with c-Myc and enhancing its nuclear localization to upregulate expression of its target genes related to TEC proliferation. Notably, c-Myc was lactylated in TEC after H/R, and this lactylation was enhanced by HSPA12A overexpression. Importantly, inhibition of c-Myc lactylation attenuated the HSPA12A-induced increases of c-Myc nuclear localization, proliferation-related gene expression, and TEC proliferation. Further experiments revealed that HSPA12A promoted c-Myc lactylation via increasing the glycolysis-derived lactate generation in a Hif1α-dependent manner. The results unraveled a role of HSPA12A in promoting TEC proliferation and facilitating renal recovery following KI/R, and this role of HSPA12A was achieved through increasing lactylation-mediated c-Myc activation. Therefore, targeting HSPA12A in TEC might be a viable strategy to promote renal functional recovery from KI/R injury in patients.
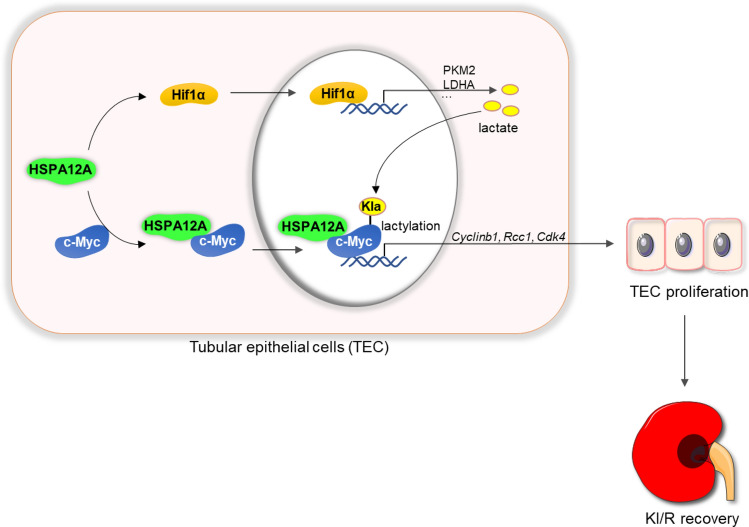
Graphical abstract
HSPA12A facilitated renal functional recovery from kidney ischemia/reperfusion (KI/R) injury. This protective effect of HSPA12A was mediated through directly interacting with c-Myc as well as upregulating the Hif1α-mediated lactate generation, thereby increasing c-Myc lactylation and nuclear localization to drive expression of genes related to cell proliferation, and ultimately promoting TEC proliferation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-024-05427-5.
Keywords: Lactylation, c-Myc, Heat shock protein A12A (HSPA12A), Tubular epithelial cells (TEC), Proliferation, Kidney ischemia/reperfusion (KI/R)
Introduction
Kidney ischemia/reperfusion (KI/R) injury, the leading cause of acute renal injury, occurs when renal blood flow is decreased, such as trauma, stroke, vascular disease, and organ transplantation [1, 2]. KI/R leads to a rapid decline of renal function and higher mortality, and the patients those survived from the acute phase have an increased risk of developing chronic kidney disease. Despite numerous interventions reduce KI/R injury in experimental models, specific treatments are still lack besides supportive care and dialysis in clinic [3, 4]. It is imperative to identify molecular mediators that control recovery from KI/R injury.
The outcome of KI/R injury ranges widely from full recovery to chronic kidney disease [3, 5, 6]. This wide spectrum is closely associated with the degrees of tubular epithelial cells (TEC) injury and the subsequent proliferation of surviving TEC [4, 5]. TEC, which form the tubular epithelium responsible for reabsorption to maintain solute and volume homeostasis, have been considered as sensors, effectors and targets of KI/R injury [5], [7–10]. Due to their high metabolic activity, TEC are highly vulnerable to KI/R [1, 5]. Fortunately, although TEC are typically quiescent under normal conditions in the adult kidney, surviving TEC after KI/R can re-enter the cell cycle and proliferate to restore tubular integrity. Nonetheless, spontaneous proliferation of TEC is often insufficient to restore the cell number and functional integrity of the kidneys upon severe renal injury [4, 5, 11–13]. Therefore, promoting TEC proliferation is a promising therapeutic approach to manage recovery from KI/R injury.
c-Myc is a pivotal regulator for cell proliferation [14]. As a multifunctional transcription factor, c-Myc is thought to influence expression of up to ~ 15% of genes, including the genes associated with cell cycle [15–17]. c-Myc promotes cell proliferation by driving expression of genes for accelerating entry of cells into S-phase of the cell cycle [18]. Besides its well-known ability in promoting proliferation of cancer cells, recent studies demonstrate that c-Myc also plays a role in TEC proliferation after KI/R [19, 20]. However, how c-Myc is activated in TEC after KI/R is incompletely known.
Recent studies have revealed that nuclear localization and activation of some transcription factors, such as Snail1 and YY1, are regulated by their lactylation [21, 22]. Lactylation is a newly identified and p300-mediated post-translational modification that adds lactyl groups to proteins using glycolysis-derived lactate. Intriguingly, metabolism reprograming to aerobic glycolysis is found during cell proliferation [23–25], and lactate derived from glycolysis has been shown to stimulate proliferation of cancer cells by promoting Histone lactylation [26]. c-Myc can be regulated by multiple post-translational modifications such as phosphorylation, acetylation and ubiquitination [27], however, whether c-Myc can be lactylated and the role of c-Myc lactylation in TEC proliferation are completely unknown.
Heat shock protein A12A (HSPA12A) is an atypic member of the HSP70 family [28, 29]. We and others reported that HSPA12A is essential to maintain mood stability and to protect the brain and liver from ischemia and endotoxin insults [30–33]. Particularly, we recently demonstrated that HSPA12A is also a regulator of metabolism especially glycolysis [33, 34]. HSPA12A shows heterogeneity in glycolytic regulation, depending on types of cells and stimuli. As an example, HSPA12A activates glycolysis to protect heart from ischemia/reperfusion injury [35]. By contrast, HSPA12A inhibits glycolysis to suppress metastasis of renal cancer cells, cardiac fibrosis after myocardial infarction, and hepatic inflammation following liver ischemia/reperfusion [36–38]. Interestingly, HSPA12A can regulate glycolysis-mediated lactate production to modulate lactylation of Histone 3 and HMGB1 in cardiomyocytes and hepatocytes, respectively [35, 38]. However, it is unknown whether HSPA12A can modulate glycolysis and c-Myc activation to impact on TEC proliferation after KI/R.
To answer this question, experiments were performed using both mouse and primary TEC models. KI/R decreased HSPA12A expression in kidneys and TEC, while ablation of HSPA12A (Hspa12a−/−) in mice impaired TEC proliferation after KI/R. Further studies revealed that HSPA12A was required for renal functional recovery from KI/R injury, and this action of HSPA12A was mediated through promoting TEC proliferation in a c-Myc lactylation-dependent manner.
Materials and methods
Reagents
MTT [3-(4, 5-Dimethylthiazol-2-yl)-2, and 5-diphenyltetrazolium bromide] was from Sigma-Aldrich (St. Louis, MO). Bovine serum albumin (BSA) was from Roche (Basel, Switzerland). Type II collagenase was from Worthington biochemical Corporation (Lakewood, NJ). RPMI 1640 medium and fetal bovine serum (FBS) were from Biological Industries (Kibbutz Beit Haemek, Israel). High-sig ECL western blotting substrate was from Tanon (Shanghai, China). Protein A-Agarose was from Santa Cruz Biotechnology (Dallas, TX). Cell-Light™ EdU Apollo®567 In Vitro Imaging Kit was from RiboBio (Guangzhou, China). DAPI reagent was from cell signaling Technology (Boston, MA). Kits for measuring lactate content, blood urea nitrogen (BUN) and creatinine (Cr) were from Jiancheng Biotech (Nanjing, China). Nuclear and cytoplasmic protein extraction kits were from Beyotime (Shanghai, China). Insulin-Transferrin-Selenium (ITS) solution was from Gibco (Carlsbad, CA). 2-Deoxy-D-glucose (2-DG), C646, and YC-1 reagents were from MedChemExpress (Monmouth Junction, NJ). Epidermal growth factor (EGF) was from Peprotech (Cranbury, NJ). Oxamate and Propidium Iodide (PI) were from Selleck (Houston, TX). RNase was from Yeasen (Shanghai, China).
Bioinformatic analysis
The heat shock protein RNA-seq data of KI/R kidneys were obtained from Gene Expression Omnibus (GEO) datasets (https://www.ncbi.nlm.nih.gov/gds/, GSE172042) [39].
Human kidney samples
Human kidney samples, which used for characterizing HSPA12A expression in renal TEC, were collected from patients who had undergone nephrectomy in the First Affiliated Hospital of Nanjing Medical University (Nanjing, China). The collected kidney tissues were 3 cm away from tumor lesion. The Ethical Board of First Affiliated Hospital of Nanjing Medical University approved these studies (#2019-SR-489). Patients gave informed consent at the time of recruitment. All the human studies were conducted according to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
Animals
Hspa12a knockout (Hspa12a−/−) mice were generated using the loxP and Cre recombinant system as described in our previous studies [40–42]. To remove the Hspa12a gene, the chimeric mice were crossed with EIIa-Cre transgenic mice. The mice were bred at the Model Animal Research Center of Nanjing University and were maintained in the Animal Laboratory Resource Facility of the same institution. All experiments conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication, 8th Edition, 2011). The animal care and experimental protocols were approved by Nanjing University’s Committee on Animal Care (GX55). All experiments conformed to international guidelines on the ethical use of animals.
Mice (C57BL/6 background) were randomly assigned to all analyses. Investigators were blinded to the histological analysis, but not blinded to animal handling, sampling, and raw data collection.
Kidney ischemia–reperfusion (KI/R) surgery
KI/R was induced in male mice (8–10 weeks of age) using previous methods [43, 44]. Briefly, mice were anesthetized by inhalation of 1.5–2% isoflurane and maintained core body temperature at 37 ± 0.2 °C throughout the surgery. After anesthesia, mice were subjected to right renal removing and left renal pedicles clamping to induce ischemia. After 40 min of ischemia, the clamps were removed to allow blood reperfusion. Warmed normal saline was given intraperitoneally two hours after surgery. In sham-operated mice, the same procedure was performed except for left renal pedicles clamping. Twenty-four hours after reperfusion, blood was sampled and kidneys were collected for the indicated measurements.
Isolation, culture, and treatments of primary renal tubular epithelial cells (TEC)
Isolation and culture. Primary TEC were isolated from 6-8-week-old male C57BL/6 mice, as described previously [44]. Briefly, the kidneys were peeled off, minced, digested with Type II collagenase (1 mg/ml) for 25 min at 37 °C, and washed with RPMI 1640 medium. Then, the kidney digests were washed through two sieves (mesh diameters of 150 and 70 μm). Finally, tubules were suspended in RPMI 1640 supplemented with 10% FBS, ITS (10 μg/ml of insulin, 5.5 μg/ml of transferrin, and 5 ng/ml of selenium), EGF (10 ng/ml), penicillin (100 units/ml), and streptomycin (100 μg/ml). Following growth for 3–4 days to reach 60%–80% confluency in culture dishes, the TEC were collected and grown in the growth medium mentioned above. All cultures were free of mycoplasma contamination.
Hypoxia/reoxygenation (H/R). To mimic KI/R process in vitro, H/R was performed in primary TEC cultures. To induce hypoxia, TEC were changed to serum- and glucose-free medium and incubated in 94% N2, 5% CO2, and 1% O2 in a humidified incubator at 37 °C. After exposed to hypoxia for 12 h, TEC were reoxygenated by incubation with normal growth medium in normoxic condition for another 12 h.
Gene overexpression or knockdown. To overexpress HSPA12A, TEC were infected with HSPA12A-expressing recombinant adenovirus (Hspa12aO/E) or normal control vectors as controls (NC). This adenoviral vector, which containing a three flag-tagged coding region of mouse Hspa12a (NM_175199), was generated by GeneChem (Shanghai, China) as described in our previous studies [40, 45]. To knockdown c-Myc expression, TEC were transfected with c-Myc-targeting siRNA (c-MycK/D) or the corresponding scramble negative controls (NC) using siRNA-mate (Genepharma, China). The siRNA sequences were shown in Supplemental Table 1.
Inhibitor treatments. The following inhibitors were employed during H/R procedure for the indicated purposes: C646 (10 μM) was used to inhibit c-Myc lactylation; Oxamate (Oxa, 5 mM) and 2-DG (5 mM) were used to inhibit lactate generation; YC-1 (10 μM) was used to inhibit Hif1α.
Biochemical measurements
In mouse experiments, serum and kidneys were collected for measuring renal function (blood urea nitrogen, BUN; creatinine, Cr) and lactate levels respectively, using the assay kits according to the manufacture’s instruction. In TEC experiments, culture medium was collected for measuring lactate levels using the assay kits.
Histological analysis and immunofluorescence staining
Histological analysis. Renal tubular injury was evaluated by hematoxylin–eosin (H&E) on paraffin-embedded sections following KI/R according to previous methods [24, 46]. Briefly, the tubular injury was scored on H&E-stained sections based on the percentage of tubules with tubular dilation and intertubular hemorrhage as followings: 0, no damage; 1, 10–25%; 2, 26–50%; 3, 51–75%; and 4, > 75% [47].
Immunofluorescence staining. This was performed on 4% PFA-fixed cells or frozen kidney sections as described previously using appropriate antibodies (Supplemental Table 2). Briefly, after incubation with the indicated primary antibodies (1: 100) overnight at 4 °C, Cy3- or Alexa Fluor™ 488-conjugated secondary antibody was applied to the samples to visualize the staining. DAPI reagent was used to counterstain the nuclei. The staining was observed using a fluorescence microscope (IX73, Olympus, Tokyo, Japan). Four to six random fields were observed on each sample. Quantification was performed using Cellsens Dimention software (Olympus, Tokyo, Japan).
MTT and EdU incorporation assay
MTT assay was performed in TEC at the indicated time points after H/R according to previous methods [48].
For EdU incorporation assay, TEC were incubated with EdU for 2 h after H/R. EdU incorporation was analyzed using the assay kit, and TEC proliferation was expressed as the percentage of EdU+ cells over total cells.
Flow cytometric analysis
TEC were collected, washed with PBS for twice, re-suspended with 300 µl of PBS, and fixed with 700 µl of pre-cooled 75% ethanol overnight at 4 °C. Following centrifugation and washed with PBS, the cells were digested with RNase (5 mg/ml) and exposed to PI staining (5 mg/ml) for 30 min on ice in dark. The DNA contents at each stage of the cell cycle were measured by FACSCalibur (Becton Dickinson). All data were analyzed with the FlowJo Software (Becton Dickinson).
Immunoblotting and immunoprecipitation-immunoblotting
Immunoblotting. Protein extracts were prepared from mouse kidneys or TEC. Immunoblotting was performed according to our previous methods [34, 49]. To control for lane loading, the membranes were probed with anti–GAPDH or anti–α-Tubulin antibody for total proteins and with anti–H3 or anti–LaminA/C antibody for nuclear proteins.
Immunoprecipitation-immunoblotting. For analysing c-Myc lactylation, the anti–c-Myc immunoprecipitation was performed from H/R TEC and followed by immunoblotting with anti–L-lactyl-lysine (Klac) antibody. For analysing the interaction between HSPA12A and c-Myc or Hif1α, the anti–Flag-tagged HSPA12A immunoprecipitation was performed from H/R TEC and followed by immunoblotting with anti–c-Myc or anti–Hif1α antibody according to our previous methods [34, 45, 49].
Glycolysis-derived ATP measurement
ATP rate was examined using a Seahorse XFe24 Extracellular Flux Analyzer (Agilent). Experiments were performed following manufacturer’s protocols. ATP rate was assessed using Seahorse XF Real-Time ATP Rate Assay Kit. In brief, TEC were seeded at 2 × 104 cells per well. After reaching 70–90% confluency, TEC were equilibrated for 1 h in XF assay medium supplemented with glucose (10 mM), sodium pyruvate (1 mM), and glutamine (2 mM) in a non-CO2 incubator. ATP rate was monitored at baseline and throughout sequential injections of oligomycin (1.5 µM), followed by rotenone or antimycin A (0.5 µM) each. ATP rate was shown in pmol/min, and the data were analyzed by Seahorse XFe24 Wave software.
Quantitative real-time PCR (RT-PCR)
RT-PCR was performed using our previous methods [45]. Briefly, total RNA was prepared and an amount of 2 µg was used for cDNA synthesis using the oligo (dT) primer. Expressions of the indicated genes were estimated by RT-PCR using the SYBR Green Master (Roche, Indianapolis, IN). Actin served as internal controls. 2-ΔΔCT methodology was used to analyze the gene expression. The related primer sequences were listed in Supplemental Table 3.
Statistical analysis
Data represent as mean ± standard deviation (SD). All data sets were first tested for normality and homogeneity before choosing the statistical test. For data that consistent with normality and homogeneity of variance, groups were compared using Student’s two-tailed paired t-test, one-way ANOVA or two-way ANOVA followed by Tukey’s test as a post-hoc test. For data that not consistent with normality and homogeneity of variance, Mann–Whitney U test and Brown-Forsythe and Welch test were used for analyses. P < 0.05 was considered statistically significant.
Results
Ischemia/reperfusion downregulates HSPA12A expression in kidneys and TEC
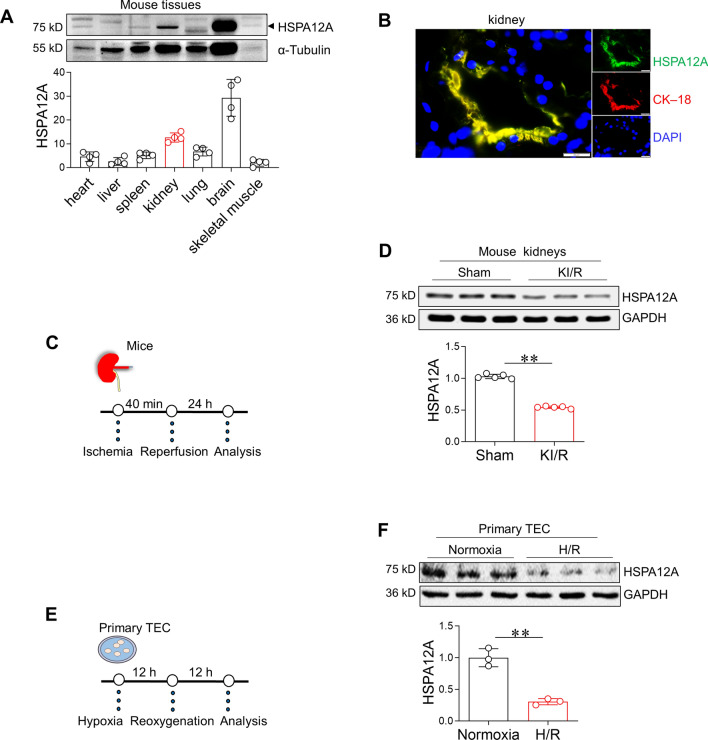
The protein levels of HSPA12A were compared among the kidney and other tissues in mice. As shown in Fig. 1A, HSPA12A expression was the second high in kidney among seven mouse tissues, including kidney, liver, spleen, lung, skeletal muscle, heart, and brain. Notably, HSPA12A colocalized with the epithelial cell marker cytokeratin–18 (CK–18) in kidney (Fig. 1B), indicating that TEC express HSPA12A.
Fig. 1.
KI/R downregulates HSPA12A in kidneys and TEC. A HSPA12A expression in different tissues. The indicated tissues were collected from adult C57BL/6 mice. Immunoblotting for HSPA12A was performed. Blots for α-Tubulin served as loading controls. n = 4. B Characterization of HSPA12A in TEC of kidneys. Frozen sections of human kidney were prepared for immunostaining against HSPA12A (green). CK–18 was used to indicate TEC (red). DPAI was counter stained nuclei (blue). Scale bar = 20 μm. C Experimental setting of mice. D Effects of KI/R on HSPA12A expression in kidneys. Following KI/R or sham surgery, kidneys were collected to examine HSPA12A expression by immunoblotting. Blots for GAPDH served as loading controls. Data are mean ± SD, **P < 0.01 by Student’s two-tailed unpaired t-test. n = 5. E Experimental setting of primary TEC. F Effects of H/R on HSPA12A expression in TEC. Primary TEC were subjected to H/R followed by examination of HSPA12A expression by immunoblotting. Data are mean ± SD, **P < 0.01 by Student’s two-tailed unpaired t-test. n = 3
High expression level of HSPA12A in the kidney prompted us to investigate whether HSPA12A plays a role in KI/R. To this end, we analyzed publicly available data of mouse KI/R (Gene Expression Omnibus: GSE172042), and found that among all 29 examined Hsps in kidneys, the level of Hspa12a mRNA was the highest under normal condition but was the most decreased following KI/R (Fig. S1A–C). To conform these results, we induced KI/R (40 min/24 h) in mice and analysed expression of several HSPs in kidneys. KI/R only decreased expression of HSPA12A but not decreased expression of HSP32, HSP47, HSP60, and HSP70 in kidneys compared to sham controls (Fig. 1C–D, S2). To determine whether KI/R-induced downregulation of HSPA12A occurs in TEC, primary TEC were challenged with H/R (12 h/12 h) to mimic the KI/R process in vitro (Fig. 1E). The primary TEC were verified by CK–18 immunofluorescence staining (Fig. S3). Similar with results obtained from kidneys, HSPA12A expression was lower in H/R-challenged TEC than in normoxic controls, and this lowered HSPA12A levels persisted to 12 h after H/R (Fig. 1F, S4).
HSPA12A ablation impairs TEC proliferation and renal functional recovery following KI/R
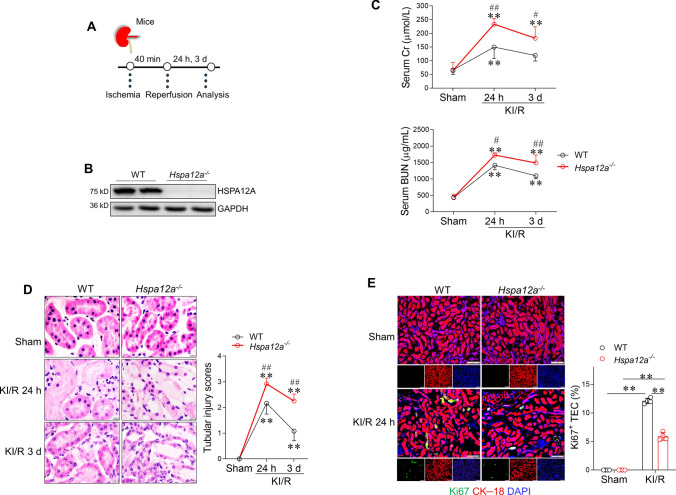
To investigate the biological role of HSPA12A downregulation in KI/R injury, Hspa12a knockout (Hspa12a−/−) mice and their wild-type (WT) littermates were employed in the experiments (Fig. 2A). Kidneys of Hspa12a−/− mice showed no HSPA12A expression (Fig. 2B). KI/R increased blood BUN and Cr levels in both genotypes compared with their sham controls; however, Hspa12a−/− mice displayed higher BUN and Cr levels than WT mice after reperfusion for 24 h and 3 d (Fig. 2C), suggesting that HSPA12A ablation impaired the renal functional recovery from KI/R injury. In supporting this, KI/R-induced disruption of tubular structural integrity was severer in Hspa12a−/− mice than in WT mice (Fig. 2D).
Fig. 2.
HSPA12A ablation impairs TEC proliferation and renal functional recovery following KI/R. A Experimental setting of mice. B Ablation of HSPA12A in mouse kidneys. HSPA12A expression in Hspa12a−/− and WT mouse kidneys were examined using immunoblotting. Note the absence of HSPA12A expression in Hspa12a−/− kidneys. C Renal function. Serums were collected from mice for measurement of BUN and Cr levels after KI/R. Data are mean ± SD, **P < 0.01 vs. their sham controls, #P < 0.05 and ##P < 0.01 vs. the time-matched WT groups, analyzed by two-way ANOVA followed by Tukey’s test. n = 5. D Histological alterations. Kidneys were prepared for paraffin-embedded sectioning. Histological alterations were examined using H&E staining. Tubular injury was scored. Data are mean ± SD, **P < 0.01 vs. their sham controls, ##P < 0.01 vs. the time-matched WT groups, analyzed by two-way ANOVA followed by Tukey’s test. n = 6. Scale bar = 20 μm. E TEC proliferation in kidneys. Cell proliferation was examined by Ki67 immunostaining (green). CK–18 was used to indicate TEC (red). DPAI was counter stained nuclei (blue). TEC proliferation was expressed as the percentage of Ki67+/CK–18+ cells. Scale bar = 50 μm. Data are mean ± SD, **P < 0.01 by two-way ANOVA followed by Tukey’s test. n = 4
Proliferation of surviving TEC is both a hallmark of and a pivotal contributor to renal recovery from KI/R injury [5, 50, 51]. To examine whether TEC proliferation is involved in delayed renal functional recovery in Hspa12a−/− mice, kidneys were co-immunostained for the proliferative marker Ki67 and the TEC marker CK–18. KI/R increased the number of Ki67+ TEC in kidneys, but this increase was diminished in Hspa12a−/− mice compared with WT mice (Fig. 2E). These findings indicate that HSPA12A was required for TEC proliferation and renal functional recovery after KI/R.
Overexpression of HSPA12A promoted TEC proliferation upon H/R
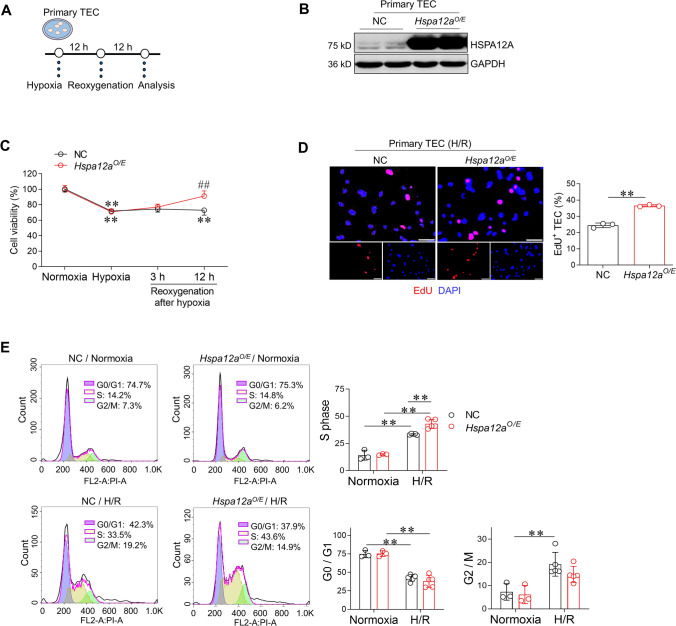
To determine the direct effects of HSPA12A on TEC proliferation, HSPA12A was overexpressed in primary TEC (Hspa12aO/E) and then these cells were challenged with H/R (Fig. 3A–B). The hypoxia-induced decrease in cell viability did not differ between Hspa12aO/E TEC and their negative controls (NC) TEC; however, after reoxygenation, the viability of Hspa12aO/E TEC gradually recovered while that of NC TEC remained at low levels up to 12 h after reoxygenation (Fig. 3C), suggesting that HSPA12A promoted proliferation of TEC that survived from hypoxia. Indeed, EdU incorporation assay demonstrated more proliferation in Hspa12aO/E groups than NC groups after H/R (Fig. 3D). Moreover, cell cycle analysis by flow cytometry showed that H/R increased TEC entry into S-phase, but this increase was greater in Hspa12aO/E TEC than in NC TEC (Fig. 3E). Collectively, these results demonstrate that HSPA12A promotes proliferation of TEC that survived from hypoxia.
Fig. 3.
Overexpression of HSPA12A promotes TEC proliferation after H/R. A Experimental setting of primary TEC. B Overexpression of HSPA12A in TEC. Overexpression of HSPA12A (Hspa12aO/E) was examined using immunoblotting in primary TEC after infection with HSPA12A-adenovirus. The TEC infected with the control vector served as negative controls (NC). C MTT assay. MTT assay was performed at the indicated time points. Data are mean ± SD, **P < 0.01 vs. their Normoxia groups, ##P < 0.01 vs. the time-matched NC controls, analyzed by two-way ANOVA followed by Tukey’s test. n = 4. D EdU incorporation. TEC proliferation was indicated by EdU incorporation following H/R. DPAI was counter stained nuclei. Data are mean ± SD, **P < 0.01 by Student’s two-tailed unpaired t- test. n = 3. Scale bar = 50 μm. E Cell cycle distribution. Flow cytometry was used to indicate cell cycle distribution of TEC. Data are mean ± SD, **P < 0.01 by two-way ANOVA followed by Tukey’s test. n = 3 for normoxia groups and n = 5 for H/R groups
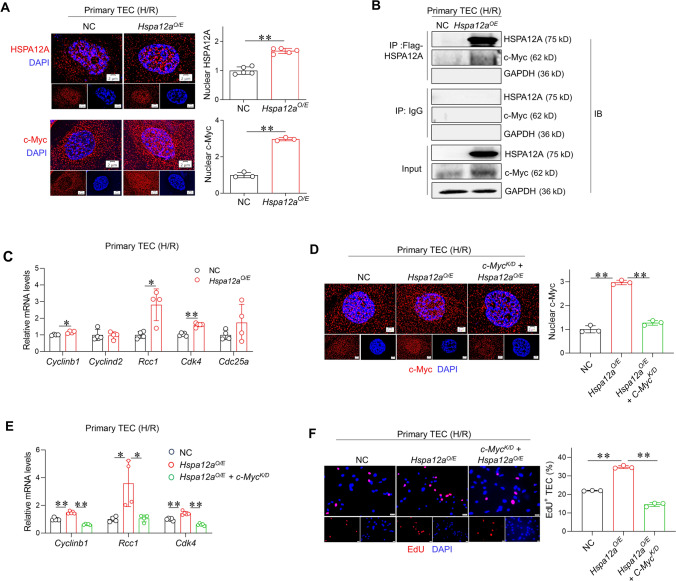
HSPA12A interacts with c-Myc and increases its nuclear localization in TEC upon H/R
c-Myc is important for cell proliferation by controlling expression of its target genes that related to cell cycle, such as Cyclinb1, Cyclind2, Rcc1, Cdk4 and Cdc25a [16, 52–58]. Given that c-Myc is a nuclear transcription factor, we analyzed whether its nuclear localization was affected by HSPA12A. Interestingly, H/R increased nuclear localization of both c-Myc and HSPA12A (Fig. 4A). Also, c-Myc showed higher nuclear abundance in Hspa12aO/E TEC than in NC TEC following H/R (Fig. S5). Notably, an immunoprecipitation-immunoblotting assay showed that c-Myc protein was recovered in HSPA12A immunocomplexes prepared from H/R-treated TEC (Fig. 4B). Though no difference of c-Myc mRNA levels were found between Hspa12aO/E TEC than in NC TEC following H/R, higher c-Myc protein levels were detected in H/R- Hspa12aO/E TEC than in H/R-NC TEC (Fig. S6A-B). Also, less colocalization of c-Myc with ubiquitin was found in Hspa12aO/E TEC than in NC TEC following H/R (Fig. S6C). The findings indicate that HSPA12A binds to c-Myc and increases its nuclear localization in TEC after H/R.
Fig. 4.
HSPA12A increases nuclear c-Myc localization to mediate the promotion of TEC proliferation after H/R. A Nuclear localization of HSPA12A and c-Myc. HSPA12A and c-Myc nuclear localization in TEC was visualized by immunostaining after H/R. DPAI was counter stained nuclei. Data are mean ± SD, **P < 0.01 by Student’s two-tailed unpaired t-test. Scale bar = 2 μm. n = 5 for HSPA12A groups and n = 3 for c-Myc groups. B Interaction of c-Myc with HSPA12A. Anti-Flag-HSPA12A immunoprecipitates, which prepared from H/R-treated TEC, were immunoblotted with c-Myc. Input or IgG-immunoprecipitates served as positive or negative controls. Note that c-Myc was recovered in Flag-HSPA12A immunoprecipitates. C mRNA levels. mRNA levels of the indicated genes were analyzed in H/R-treated TEC by RT-PCR. Data are mean ± SD, *P < 0.05, **P < 0.01 by Student’s two-tailed unpaired t- test or Mann–Whitney U test. n = 5 for Cdk4 groups and n = 4 for all the other groups. D c-Myc localization in nuclei. Following c-Myc knockdown, c-Myc abundance in nuclei of TEC were examined by immunostaining after H/R. Data are mean ± SD, **P < 0.01 by one-way ANOVA followed by Tukey’s test. Scale bar = 2 μm. n = 3/group. E mRNA levels. mRNA levels of the indicated genes were analyzed after c-Myc knockdown by RT-PCR. *P < 0.05 and **P < 0.01 by one-way ANOVA followed by Tukey’s test or Brown-Forsythe and Welch test. n = 4 for Rcc1 groups and n = 5 for all the other groups. F TEC proliferation. EdU incorporation was performed following c-Myc knockdown in H/R-treated TEC. Data are mean ± SD, **P < 0. 01 by one-way ANOVA followed by Brown-Forsythe and Welch test. n = 3. Scale bar = 50 μm
c-Myc mediates the HSPA12A-induced promotion of TEC proliferation
When HSPA12A increased c-Myc nuclear localization in H/R TEC, it also upregulated mRNA levels of c-Myc target genes related to cell proliferation, including Cyclinb1, Rcc1 and Cdk4 (Fig. 4C). This suggests a possible involvement of c-Myc in HSPA12A-promoted proliferation of TEC. To test this possibility, c-Myc was knockdown by transfection with siRNA that targeted to c-Myc (Fig. 4D, S7). Notably, following c-Myc knockdown, the increases of Cyclinb1, Rcc1 and Cdk4 mRNA levels and EdU+ TEC in Hspa12aO/E TEC were diminished after H/R (Fig. 4E–F). The data indicate that HSPA12A promoted TEC proliferation in a c-Myc dependent manner.
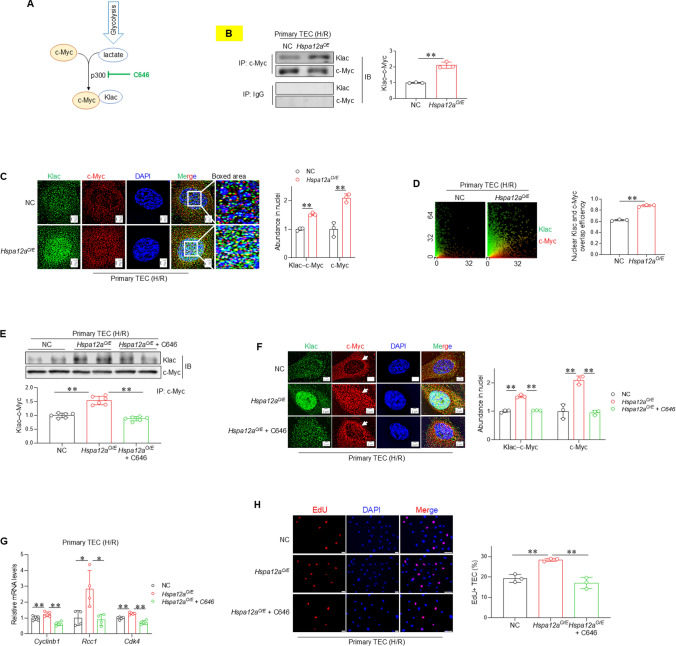
HSPA12A promotes c-Myc lactylation in TEC following H/R
Next, we investigated how HSPA12A promotes c-Myc nuclear localization of TEC. Recent evidence demonstrates that nuclear localization of some transcription factors, such as Snail1 and YY1, are controlled by their lactylation [21, 59]. Lactylation is a newly identified, p300-mediated post-translational modification that adds lactyl groups to proteins using glycolysis-derived lactate (Fig. 5A). Immunoprecipitation of c-Myc followed by immunoblotting for L-lactyl-lysine (Klac) showed that the level of c-Myc lactylation was higher in Hspa12aO/E TEC than in NC TEC after H/R (Fig. 5B). Meanwhile, more lactylated c-Myc (Klac–c-Myc) and total c-Myc were localized in nuclei of Hspa12aO/E TEC than NC TEC (Fig. 5C). Also, higher overlap efficiency of Klac and c-Myc was found in nuclei of Hspa12aO/E TEC than NC TEC after H/R (Fig. 5D). Thus, HSPA12A increased levels of both lactylated c-Myc and total c-Myc in nuclei of TEC after H/R.
Fig. 5.
c-Myc lactylation mediates the HSPA12A-increased c-Myc nuclear localization in TEC following H/R. A Scheme illustrates the process of c-Myc lactylation. p300 inhibitor C646 was used to inhibit c-Myc lactylation. B c-Myc lactylation. The anti–c-Myc immunoprecipitates were prepared from H/R-treated TEC followed by immunoblotted for L-lactyl-lysine (Klac). The IgG immunoprecipitates served as negative controls. Data are mean ± SD, **P < 0.01 by Student’s two-tailed unpaired t- test. n = 3. C Nuclear localization of lactylated and total c-Myc. Immunostaining for lactylation (Klac, green) and c-Myc (red) was performed in TEC after H/R. DAPI was used to stain nuclei (blue). Data are mean ± SD, **P < 0.01 by Student’s two-tailed unpaired t- test. Scale bar = 2 μm. n = 3. D Overlap efficiency of Klac and c-Myc in nuclei. This overlap efficiency was quantified from the images of (B). Data are mean ± SD, **P < 0.01 by Student’s two-tailed unpaired t- test. n = 3. TEC were challenged with H/R in the presence or absence of C646. The following experiments were performed subsequently. E c-Myc lactylation. Anti–c-Myc immunoprecipitates prepared from H/R-treated TEC were immunoblotted for lactylation antibody (Klac). Data are mean ± SD, **P < 0.01 by one-way ANOVA followed by Brown-Forsythe and Welch test. n = 6. F Nuclear localization of lactylated and total c-Myc. Immunostaining for lactylation (Klac, green) and c-Myc (red) was performed in H/R-treated TEC. DAPI was used to stain nuclei (blue). Data are mean ± SD, ** < 0.01 by one-way ANOVA followed by Tukey’s test. Scale bar = 2 μm. n = 3. Arrows showed reduced nuclear but increased perinuclear localization of c-Myc following inhibition of c-Myc lactylation. G mRNA levels. The mRNA expression of indicated genes were analyzed by RT-PCR. *P < 0.05 and **P < 0. 01 by one-way ANOVA followed by Tukey’s test. n = 4 for Rcc1 groups and n = 5 for all the other groups. H TEC proliferation. Cell proliferation was indicated by EdU incorporation following H/R. Data are mean ± SD, **P < 0.01 by one-way ANOVA followed by Tukey’s test. n = 3. Scale bar = 50 μm
c-Myc lactylation mediates the HSPA12A-induced promotion of c-Myc nuclear localization and TEC proliferation
To determine the role of c-Myc lactylation in HSPA12A-induced increase of c-Myc nuclear localization, lactylation was inhibited by the p300 inhibitor C646 (Fig. 5A). As expected, C646 treatment abolished the increase of c-Myc lactylation in Hspa12aO/E TEC (Fig. 5E–F). Following inhibition of c-Myc lactylation by C646, the increase of nuclear c-Myc in Hspa12aO/E TEC were also abolished (Fig. 5F, S8). Notably, c-Myc accumulated perinuclearly of TEC following inhibition of c-Myc lactylation by C646 (Fig. 5F). Also, inhibition of c-Myc lactylation abolished the HSPA12A-promoted TEC proliferation, as reflected by measurements of mRNA levels of proliferation-related genes (Cyclinb1, Rcc1 and Cdk4) and EdU incorporation (Fig. 5G–H). Thus, c-Myc lactylation was required for its nuclear localization to mediate the HSPA12A-promoted TEC proliferation after H/R.
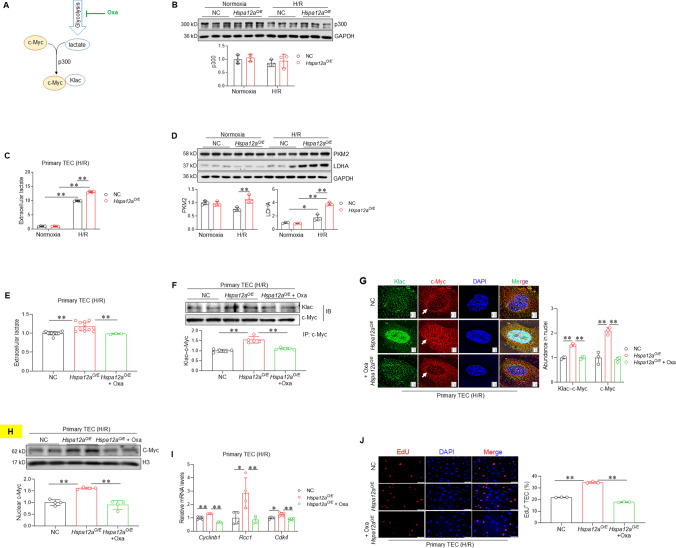
HSPA12A increases c-Myc lactylation by upregulating lactate generation
Lactylation is a p300-mediated modification that uses glycolysis-derived lactate (Fig. 6A). Although HSPA12A increased c-Myc lactylation, it did not change the p300 expression level in TEC under either normoxic or H/R conditions (Fig. 6B). This suggests that HSPA12A might increase c-Myc lactylation by modulating the generation of lactate, an end product of glycolysis. Indeed, the H/R-induced lactate generation was higher in Hspa12aO/E TEC than that in NC TEC (Fig. 6C). The glycolysis-derived ATP was also greater in Hspa12aO/E TEC than in NC TEC after H/R (Fig. S9A). In line with this, expression levels of glycolysis-related genes (PKM2, LDHA, GLUT1, GLUT4 and MCT4) were higher in Hspa12aO/E TEC than in NC TEC after H/R (Fig. 6D, S9B). In agreement, knockout of HSPA12A in mice (Hspa12a−/−) decreased the levels of serum lactate and protein expression of PKM2, LDHA, GLUT1, GLUT4 and MCT4 following KI/R (Fig. S10A–B).
Fig. 6.
HSPA12A promotes c-Myc lactylation and nuclear localization via upregulating lactate generation. A Brief Scheme of lactylation. This scheme illustrates that c-Myc lactylation is mediated by p300 using the glycolysis-derived lactate. LDHA inhibitor Oxa was used to block lactate generation. B p300 expression. The expression of p300 in primary TEC was examined by immunoblotting. Data are mean ± SD, analyzed by two-way ANOVA followed by Tukey’s test. n = 3. C Extracellular lactate. Lactate contents in culture medium of TEC were measured. Data are mean ± SD, **P < 0.01 by two-way ANOVA followed by Tukey’s test. n = 4. D Expression of glycolysis genes. The expression of PKM2 and LDHA were examined by immunoblotting in primary TEC. Data are mean ± SD, *P < 0.05 and **P < 0. 01 by two-way ANOVA followed by Tukey’s test. n = 3. TEC were challenged with H/R in the presence or absence of Oxa. The following experiments were performed subsequently. E Extracellular lactate following Oxa treatment. Lactate contents in culture medium of H/R-treated TEC were measured following Oxa treatment. Data are mean ± SD, **P < 0.01 by one-way ANOVA followed by Brown-Forsythe and Welch test. n = 3 for Oxa group and n = 12 for all the other groups. F c-Myc lactylation. Anti–c-Myc immunoprecipitates, which prepared from H/R-treated TEC in the presence or absence of Oxa, were immunoblotted for lactylation antibody (Klac). Data are mean ± SD, **P < 0. 01 by one-way ANOVA followed by Tukey’s test. n = 5. G Nuclear localization of lactylated and total c-Myc. Immunostaining for lactylation (Klac, green) and c-Myc (red) was performed. DAPI was used to stain nuclei (blue). Data are mean ± SD, **P < 0. 01 by one-way ANOVA followed by Tukey’s test. Scale bar = 2 μm. n = 3. H c-Myc in nuclear fractions. Nuclear fractions were prepared for examining c-Myc protein abundance by immunoblotting. Blots for H3 served as loading controls. Data are mean ± SD, **P < 0.01 by one-way ANOVA followed by Tukey’s test. n = 4. I mRNA expression. mRNA levels of the indicated genes were examined by RT-PCR. *P < 0.05 and **P < 0.01by one-way ANOVA followed by Tukey’s test or Brown-Forsythe and Welch test. n = 4 for Rcc1 groups and n = 5 for all the other groups. J TEC proliferation. Cell proliferation was examined by EdU incorporation. Data are mean ± SD, **P < 0. 01 by one-way ANOVA followed by Brown-Forsythe and Welch test. n = 3. Scale bar = 50 μm
To clarify the role of lactate generation in HSPA12A-induced increase of c-Myc lactylation, H/R-challenged TEC were treated with the LDHA inhibitor Oxamate (Oxa) to block lactate generation (Fig. 6A). When Oxa abolished the increase of lactate generation in Hspa12aO/E TEC (Fig. 6E), the increase of c-Myc lactylation in these cells was also abolished (Fig. 6F–G). Remarkably, following inhibition of c-Myc lactylation by Oxa, the increase of nuclear c-Myc was also abolished in H/R-treated Hspa12aO/E TEC (Fig. 6G–H). Moreover, inhibition of c-Myc lactylation by Oxa attenuated the increased proliferation of Hspa12aO/E TEC, as reflected by measurements of mRNA levels of Cyclinb1, Rcc1 and Cdk4 (Fig. 6I) and EdU incorporation (Fig. 6J). Similar results were obtained following treatment with another glycolysis inhibitor, 2-DG, which also removed the increases of lactate generation and proliferation of Hspa12aO/E TEC after H/R (Fig. S11A-C).
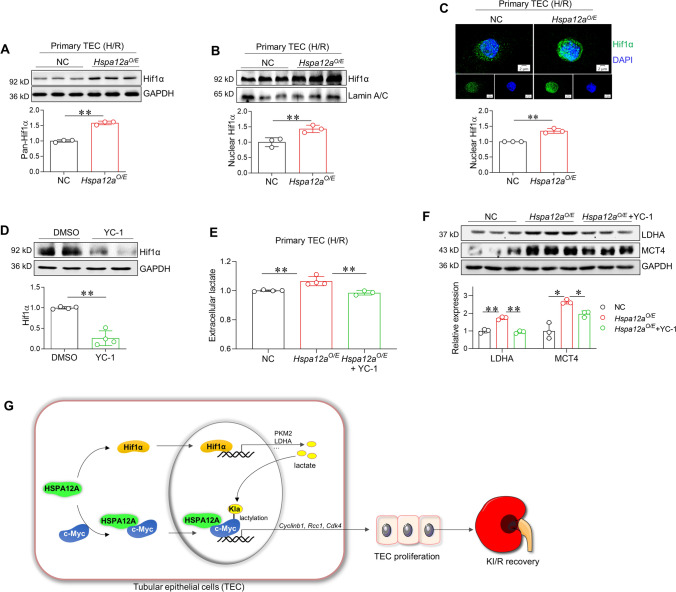
HSPA12A upregulates lactate generation of TEC in a Hif1α-dependent manner
Finally, we sought to determine how HSPA12A upregulates lactate generation. Hif1α is an important transcription factor for activation of glycolysis through inducing expression of glycolytic genes [60]. Though no direct interaction between HSPA12A and Hif1α was detected in TEC (Fig.S12A-B), higher levels of total Hif1α and nuclear Hif1α proteins were found in Hspa12aO/E TEC than in NC TEC after H/R (Fig. 7 A-B). Immunostaining confirmed more Hif1α localized in nuclei in Hspa12aO/E TEC than in NC TEC after H/R (Fig. 7C). By contrast, knockout of HSPA12A in mice decreased nuclear Hif1α protein levels in kidneys compared to WT controls following KI/R (Fig. S12C-D). When inhibition of Hif1α using YC-1 (Fig. 7D), the increases of lactate generation (Fig. 7E), glycolysis-related gene expression (Fig. 7F), and proliferation (Fig. S13) of Hspa12aO/E TEC were also diminished.
Fig. 7.
HSPA12A upregulates lactate generation in TEC via activation of Hif1α. A Hif1α expression. The expression of Hif1α was examined by immunoblotting in H/R-treated TEC. Data are mean ± SD, **P < 0. 01 by Student’s two-tailed unpaired t- test. n = 3. B–C. Nuclear Hif1α abundance. Nuclear content of Hif1α in H/R-treated TEC was examined by immunoblotting (B) and immunostaining (C, Scale bar = 2 μm). Blots for LaminA/C served as loading controls (B). DAPI was used to stain nuclei (C). Data are mean ± SD, *P < 0.05 and **P < 0.01 by Student’s two-tailed unpaired t- test. n = 3. D Inhibition of Hif1α by YC-1. The efficiency of YC-1 on Hif1α inhibition was examined by immunoblotting. Data are mean ± SD, **P < 0.01 by Student’s two-tailed unpaired t- test. n = 4. E Extracellular lactate. Lactate contents in culture medium of TEC were measured following H/R in the presence or absence of YC-1. Data are mean ± SD, *P < 0.05 by one-way ANOVA followed by Brown-Forsythe and Welch test. n = 4. F Glycolysis genes expression. Expression of LDHA and MCT4 were examined in H/R-treated TEC in the presence or absence of YC-1. Data are mean ± SD, *P < 0.05 and **P < 0.01 by one-way ANOVA followed by Tukey’s test or Brown-Forsythe and Welch test. n = 3. G Mechanistic scheme. By binding with c-Myc to help its nuclear localization as well as activating the Hif1α-dependent lactate generation, HSPA12A promotes c-Myc lactylation, thereby sequestrates c-Myc in nuclei to drive the proliferation-related genes expression, and ultimately facilitates TEC proliferation after H/R. Also, inhibition of c-Myc lactylation leads to c-Myc localization perinuclearly. Thus, HSPA12A promotes TEC proliferation to facilitate renal functional recovery from KI/R injury through lactylation-mediated c-Myc nuclear localization
Discussion
The significant finding of this study is that HSPA12A was essential for functional recovery from KI/R injury. HSPA12A achieved this action by promoting TEC proliferation through increasing c-Myc lactylation (Fig. 7G).
KI/R is the most common cause of acute kidney injury, which has high morbidity and is an instigator and multiplier for dysfunctions in lung, heart, liver, and neurologic system [3, 5, 61]. KI/R injury is also unavoidable in patients who undergo kidney transplantation or experience major cardiac events such as infarction or surgery [2]. However, effective treatments for managing the recovery from KI/R injury are stilling lacking [3, 4]. In this study, KI/R downregulated HSPA12A in kidneys and TEC, suggesting that HSPA12A might play a role in KI/R injury and recovery. Indeed, ablation of HSPA12A impaired renal functional recovery from KI/R. Our study provides clear evidence that HSPA12A is essential for recovery of renal function from KI/R injury.
TEC are important for renal structural integrity and physiological function. Due to their high energy demand, TEC are sensitive to KI/R insults. Fortunately, although TEC have a limited regenerative capacity under normal condition in adult kidney, the TEC survived from KI/R retain an ability to proliferate for repopulation and repairment of damaged tubules. Thus, TEC proliferation is a hallmark for KI/R, and promoting TEC proliferation is considered a promising therapeutic approach to limit KI/R injury [4, 18, 51]. In this study, we found that HSPA12A ablation impaired TEC proliferation following KI/R in mice, while HSPA12A overexpression promoted TEC proliferation and facilitated TEC entry S-phase of the cell cycle after reoxygenation. Thus, targeting HSPA12A in TEC is an alternative therapeutic approach to facilitate recovery from KI/R injury via promoting TEC proliferation.
Proliferation of TEC involves multiple genes, and these genes are regulated by various transcription factors, including c-Myc [18]. c-Myc functions as a critical regulator of cell proliferation by driving expression of genes related to cell cycle, protein biosynthesis, and other processes [14–17]. There is evidence that c-Myc promotes cell entry S-phase of the cell cycle to facilitate cell proliferation [19, 20]. However, it is largely unknown regarding the regulation of c-Myc activation to exert its transcriptional regulatory roles in TEC after KI/R. In this study, we made the following findings: (1) HSPA12A directly interacted with c-Myc in TEC; (2) HSPA12A and c-Myc simultaneously showed more nuclear localization in TEC following H/R; and (3) knockdown of c-Myc abrogated the HSPA12A-promoted proliferation of TEC and expression of proliferation-related genes. These results indicate that HSPA12A interacts with c-Myc to increase its nuclear localization, thereby drives proliferation-related genes expression, and ultimately facilitates TEC proliferation after H/R.
Lactylation is a recently characterized, p300-mediated post-translational modification that adds lactyl groups to lysine residues of proteins using glycolysis-derived lactate [62]. Evidence shows that lactylation modulates the nuclear localization and transcriptional effects of some transcription factors, such as Snail1 and YY1 [21, 22]. Though transcription factor c-Myc can be regulated by variant types of post-translational modifications such as phosphorylation, acetylation, and ubiquitination [27], whether c-Myc can be lactylated and the role of c-Myc lactylation in TEC proliferation are absolutely unknown. In this study, we found the following results: (1) HSPA12A increased c-Myc lactylation in TEC after H/R; (2) inhibition of c-Myc lactylation removed the HSPA12A-induced increases of c-Myc nuclear localization, proliferation-related genes expression, and TEC proliferation after H/R. The results indicate that HSPA12A increased c-Myc nuclear localization and TEC proliferation through promoting c-Myc lactylation.
As above mentioned, lactylation is a p300-mediated post-translational modification using glycolysis-derived lactate [62]. However, when HSPA12A promoted c-Myc lactylation in TEC, HSPA12A did not change p300 expression levels. This suggests that HSPA12A may promote c-Myc lactylation through increasing glycolysis-derived lactate generation. Indeed, previous studies demonstrated that metabolic reprogramming, especially enhanced aerobic glycolysis, has been observed in rapidly proliferating cells [23]. Activation of glycolysis promotes proliferation of TEC [63]. In this study, we found the following results: (1) HSPA12A promoted lactate generation by activating Hif1α-mediated aerobic glycolysis; (2) inhibition of lactate generation abolished the HSPA12A-induced increases of c-Myc lactylation, c-Myc nuclear localization, proliferation-related genes expression, and proliferation of TEC after H/R. Our results indicate that HSPA12A promoted TEC proliferation through increasing c-Myc lactylation by coordinating glycolysis and c-Myc activation.
In conclusion, our study revealed that HSPA12A facilitated renal functional recovery after KI/R through promoting TEC proliferation in a c-Myc lactylation dependent manner. The findings suggest that targeted HSPA12A in TEC is a viable target to promote renal functional recovery from KI/R injury.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
Z.D., L.L. and C.L. developed the study concept and experimental design; Y.L., X.M., X.Z., X.C., Q.K., H.C., Q.M. and L.G. performed all animal study procedures and most of the in vitro experiments; Z.D., L.L. and Y.L. interpreted the data and wrote the manuscript. Z.D involved in study supervision.
Funding
This work was supported by the National Natural Science Foundation of China (82170295, 82170851, 82000296, 82002023, 81970692 and 81870234), by Jiangsu Province’s Outstanding Medical Academic Leader program (15), and by a project funded by Collaborative Innovation Center for Cardiovascular Disease Translational Medicine, and Jiangsu Provincial Key Discipline of Medicine.
Data availability
All data in this study will be made available to other researchers upon reasonable request from the corresponding author.
Declarations
Conflict of interest
The authors declared that no conflict of interest exists.
Ethics approval
The Ethical Board of First Affiliated Hospital of Nanjing Medical University approved these studies (#2019-SR-489). All the human studies were conducted according to the principles set out in the WMA Declaration of Helsinki. The animal care and experimental protocols were approved by Nanjing University’s Committee on Animal Care (GX55). All experiments conformed to international guidelines on the ethical use of animals.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
All authors consent to the publication of the article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yunfan Li and Xinxu Min have contributed equally to this work.
References
- 1.Thadhani R, Pascual M, Bonventre JV (1996) Acute renal failure. N Engl J Med 334(22):1448–1460 10.1056/NEJM199605303342207 [DOI] [PubMed] [Google Scholar]
- 2.Hao J, Wei Q, Mei S, Li L, Su Y, Mei C et al (2017) Induction of microRNA-17-5p by p53 protects against renal ischemia-reperfusion injury by targeting death receptor 6. Kidney Int 91(1):106–118 10.1016/j.kint.2016.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raup-Konsavage WM, Wang Y, Wang WW, Feliers D, Ruan H, Reeves WB (2018) Neutrophil peptidyl arginine deiminase-4 has a pivotal role in ischemia/reperfusion-induced acute kidney injury. Kidney Int 93(2):365–374 10.1016/j.kint.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monteiro M, Ramm S, Chandrasekaran V, Boswell S, Weber E, Lidberg K et al (2018) A high-throughput screen identifies DYRK1A inhibitor ID-8 that stimulates human kidney tubular epithelial cell proliferation. J Am Soc Nephrol 29(12):2820–2833 10.1681/ASN.2018040392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang-Panesso M, Kadyrov F, Lalli M, Wu H, Ikeda S, Kefaloyianni E et al (2019) FOXM1 drives proximal tubule proliferation during repair from acute ischemic kidney injury. J Clin Investig 129(12):5501–5517 10.1172/JCI125519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu M, Shi M, Gillings N, Flores B, Takahashi M, Kuro-O M et al (2017) Recombinant α-Klotho may be prophylactic and therapeutic for acute to chronic kidney disease progression and uremic cardiomyopathy. Kidney Int 91(5):1104–1114 10.1016/j.kint.2016.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffield J, Park K, Hsiao L, Kelley V, Scadden D, Ichimura T et al (2005) Restoration of tubular epithelial cells during repair of the postischemic kidney occurs independently of bone marrow-derived stem cells. J Clin Investig 115(7):1743–1755 10.1172/JCI22593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humphreys B, Valerius M, Kobayashi A, Mugford J, Soeung S, Duffield J et al (2008) Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell 2(3):284–291 10.1016/j.stem.2008.01.014 [DOI] [PubMed] [Google Scholar]
- 9.Humphreys BD, Czerniak S, DiRocco DP, Hasnain W, Cheema R, Bonventre JV (2011) Repair of injured proximal tubule does not involve specialized progenitors. Proc Natl Acad Sci USA 108(22):9226–9231 10.1073/pnas.1100629108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin F, Moran A, Igarashi P (2005) Intrarenal cells, not bone marrow-derived cells, are the major source for regeneration in postischemic kidney. J Clin Invest 115(7):1756–1764 10.1172/JCI23015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonventre JV (2003) Dedifferentiation and proliferation of surviving epithelial cells in acute renal failure. J Am Soc Nephrol 14(Suppl 1):S55-61 10.1097/01.ASN.0000067652.51441.21 [DOI] [PubMed] [Google Scholar]
- 12.Basile D, Bonventre J, Mehta R, Nangaku M, Unwin R, Rosner M et al (2016) Progression after AKI: understanding maladaptive repair processes to predict and identify therapeutic treatments. J Am Soc Nephrol 27(3):687–697 10.1681/ASN.2015030309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkatachalam M, Weinberg J, Kriz W, Bidani A (2015) Failed tubule recovery, AKI-CKD transition, and kidney disease progression. J Am Soc Nephrol 26(8):1765–1776 10.1681/ASN.2015010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adhikary S, Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6(8):635–645 10.1038/nrm1703 [DOI] [PubMed] [Google Scholar]
- 15.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F (2006) The c-Myc target gene network. Semin Cancer Biol 16(4):253–264 10.1016/j.semcancer.2006.07.014 [DOI] [PubMed] [Google Scholar]
- 16.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J et al (2003) Genomic targets of the human c-Myc protein. Genes Dev 17(9):1115–1129 10.1101/gad.1067003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menssen A, Hermeking H (2002) Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA 99(9):6274–6279 10.1073/pnas.082005599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers N, Zhang Z, Wang J, Thomson A, Isenberg J (2016) CD47 regulates renal tubular epithelial cell self-renewal and proliferation following renal ischemia reperfusion. Kidney Int 90(2):334–347 10.1016/j.kint.2016.03.034 [DOI] [PubMed] [Google Scholar]
- 19.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB (2004) Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer 4(7):562–568 10.1038/nrc1393 [DOI] [PubMed] [Google Scholar]
- 20.Levens D (2002) Disentangling the MYC web. Proc Natl Acad Sci U S A 99(9):5757–5759 10.1073/pnas.102173199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan M, Yang K, Wang X, Chen L, Gill PS, Ha T et al (2023) Lactate promotes endothelial-to-mesenchymal transition via Snail1 lactylation after myocardial infarction. Sci Adv 9(5):eadc9465 10.1126/sciadv.adc9465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Fan W, Li N, Ma Y, Yao M, Wang G et al (2023) YY1 lactylation in microglia promotes angiogenesis through transcription activation-mediated upregulation of FGF2. Genome Biol 24(1):87 10.1186/s13059-023-02931-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lunt S, Vander HM (2011) Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 27:441–464 10.1146/annurev-cellbio-092910-154237 [DOI] [PubMed] [Google Scholar]
- 24.Liu N, Wang H, Han G, Cheng J, Hu W, Zhang J (2018) Enhanced proliferation and differentiation of HO-1 gene-modified bone marrow-derived mesenchymal stem cells in the acute injured kidney. Int J Mol Med 42(2):946–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan R, Geng H, Singha P, Saikumar P, Bottinger E, Weinberg J et al (2016) Mitochondrial pathology and glycolytic shift during proximal tubule atrophy after ischemic AKI. J Am Soc Nephrol 27(11):3356–3367 10.1681/ASN.2015020177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Zhang H, Gao P (2022) Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell 13(12):877–919 10.1007/s13238-021-00846-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatma H, Maurya S, Siddique H (2022) Epigenetic modifications of c-MYC: Role in cancer cell reprogramming, progression and chemoresistance. Semin Cancer Biol 83:166–176 10.1016/j.semcancer.2020.11.008 [DOI] [PubMed] [Google Scholar]
- 28.Han Z, Truong Q, Park S, Breslow J (2003) Two Hsp70 family members expressed in atherosclerotic lesions. Proc Natl Acad Sci USA 100(3):1256–1261 10.1073/pnas.252764399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroes R, Panksepp J, Burgdorf J, Otto N, Moskal J (2006) Modeling depression: social dominance-submission gene expression patterns in rat neocortex. Neuroscience 137(1):37–49 10.1016/j.neuroscience.2005.08.076 [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Lu T, Gui Y, Zhang X, Cao X, Li Y et al (2023) HSPA12A controls cerebral lactate homeostasis to maintain hippocampal neurogenesis and mood stabilization. Transl Psychiatry 13(1):280 10.1038/s41398-023-02573-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao Y, Kong Q, Li R, Zhang X, Gui Y, Li Y et al (2018) Heat shock protein A12A encodes a novel prosurvival pathway during ischaemic stroke. Biochim Biophys Acta 1864:1862–1872 10.1016/j.bbadis.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Du S, Kong Q, Zhang X, Jiang S, Cao X et al (2020) HSPA12A attenuates lipopolysaccharide-induced liver injury through inhibiting caspase-11-mediated hepatocyte pyroptosis via PGC-1α-dependent acyloxyacyl hydrolase expression. Cell Death Differ 27(9):2651–2667 10.1038/s41418-020-0536-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du S, Zhang X, Jia Y, Peng P, Kong Q, Jiang S et al (2023) Hepatocyte HSPA12A inhibits macrophage chemotaxis and activation to attenuate liver ischemia/reperfusion injury via suppressing glycolysis-mediated HMGB1 lactylation and secretion of hepatocytes. Theranostics 13(11):3856–3871 10.7150/thno.82607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min X, Zhang X, Li Y, Cao X, Cheng H, Li Y et al (2020) HSPA12A unstabilizes CD147 to inhibit lactate export and migration in human renal cell carcinoma. Theranostics 10(19):8573–8590 10.7150/thno.44321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wansu Y, Qiuyue K, Surong J, Yunfan L, Zhaohe W, Qian M et al (2024) HSPA12A maintains aerobic glycolytic homeostasis and Histone3 lactylation in cardiomyocytes to attenuate myocardial ischemia/reperfusion injury. JCI Insight. 10.1172/jci.insight.169125 10.1172/jci.insight.169125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xinxu M, Xiaojin Z, Yunfan L, Xiaofei C, Hao C, Yuehua L et al (2020) HSPA12A unstabilizes CD147 to inhibit lactate export and migration in human renal cell carcinoma. Theranostics 10(19):8573 10.7150/thno.44321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian M, Xiaojin Z, Jinna Y, Qiuyue K, Hao C, Wansu Y et al (2024) HSPA12A acts as a scaffolding protein to inhibit cardiac fibroblast activation and cardiac fibrosis. J Adv Res 10:8573–8590 [DOI] [PubMed] [Google Scholar]
- 38.Shuya D, Xiaojin Z, Yunxiao J, Peipei P, Qiuyue K, Surong J et al (2023) Hepatocyte HSPA12A inhibits macrophage chemotaxis and activation to attenuate liver ischemia/reperfusion injury via suppressing glycolysis-mediated HMGB1 lactylation and secretion of hepatocytes. Theranostics 13(11):3856–3871 10.7150/thno.82607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aomatsu A, Kaneko S, Yanai K, Ishii H, Ito K, Hirai K et al (2022) MicroRNA expression profiling in acute kidney injury. Transl Res 244:1–31 10.1016/j.trsl.2021.11.010 [DOI] [PubMed] [Google Scholar]
- 40.Kong Q, Li N, Cheng H, Zhang X, Cao X, Qi T et al (2019) HSPA12A Is a novel player in nonalcoholic steatohepatitis via promoting nuclear PKM2-mediated M1 macrophage polarization. Diabetes 68(2):361–376 10.2337/db18-0035 [DOI] [PubMed] [Google Scholar]
- 41.Mao Y, Kong Q, Li R, Zhang X, Gui Y, Li Y et al (2018) Heat shock protein A12A encodes a novel prosurvival pathway during ischaemic stroke. Biochim Biophys Acta Mol Basis Dis 1864(5 Pt A):1862–1872 10.1016/j.bbadis.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Chen X, Qi T, Kong Q, Cheng H, Cao X et al (2019) HSPA12A is required for adipocyte differentiation and diet-induced obesity through a positive feedback regulation with PPARγ. Cell Death Differ 26(11):2253–2267 10.1038/s41418-019-0300-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kishi S, Campanholle G, Gohil VM, Perocchi F, Brooks CR, Morizane R et al (2015) Meclizine preconditioning protects the kidney against ischemia-reperfusion injury. EBioMedicine 2(9):1090–1101 10.1016/j.ebiom.2015.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers NM, Zhang ZJ, Wang JJ, Thomson AW, Isenberg JS (2016) CD47 regulates renal tubular epithelial cell self-renewal and proliferation following renal ischemia reperfusion. Kidney Int 90(2):334–347 10.1016/j.kint.2016.03.034 [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Chen X, Qi T, Kong Q, Cheng H, Cao X et al (2019) HSPA12A is required for adipocyte differentiation and diet-induced obesity through a positive feedback regulation with PPARgamma. Cell Death Differ 26(11):2253–2267 10.1038/s41418-019-0300-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D, Wang Y, Zou X, Shi Y, Liu Q, Huyan T et al (2020) FOXO1 inhibition prevents renal ischemia-reperfusion injury via cAMP-response element binding protein/PPAR-γ coactivator-1α-mediated mitochondrial biogenesis. Br J Pharmacol 177(2):432–448 10.1111/bph.14878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Liu X, Gu Y, Gao Y, Jankowski V, Was N et al (2023) DNA binding protein YB-1 is a part of the neutrophil extracellular trap mediation of kidney damage and cross-organ effects. Kidney Int 104(1):124–138 10.1016/j.kint.2023.02.032 [DOI] [PubMed] [Google Scholar]
- 48.Li R, Ma H, Zhang X, Li C, Xiong J, Lu T et al (2015) Impaired autophagosome clearance contributes to local anesthetic bupivacaine-induced myotoxicity in mouse myoblasts. Anesthesiology 122(3):595–605 10.1097/ALN.0000000000000568 [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Du S, Kong Q, Zhang X, Jiang S, Cao X et al (2020) HSPA12A attenuates lipopolysaccharide-induced liver injury through inhibiting caspase-11-mediated hepatocyte pyroptosis via PGC-1alpha-dependent acyloxyacyl hydrolase expression. Cell Death Differ 27(9):2651–2667 10.1038/s41418-020-0536-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menke J, Iwata Y, Rabacal W, Basu R, Yeung Y, Humphreys B et al (2009) CSF-1 signals directly to renal tubular epithelial cells to mediate repair in mice. J Clin Investig 119(8):2330–2342 10.1172/JCI39087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allison S (2020) Contribution of dedifferentiated proximal tubule cells to repair in acute kidney injury. Nat Rev Nephrol 16(2):65 10.1038/s41581-019-0235-3 [DOI] [PubMed] [Google Scholar]
- 52.Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M et al (2001) Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev 15(16):2042–2047 10.1101/gad.907901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ et al (2000) Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A 97(5):2229–2234 10.1073/pnas.050586197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren X, Jiang K, Zhang F (2020) The Multifaceted Roles of RCC1 in Tumorigenesis. Front Mol Biosci 7:225 10.3389/fmolb.2020.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang B, Gong Q, Chen F (2023) CDC25A inhibition suppresses cell proliferation and induces G(1)/S-phase cell cycle arrest in nasopharyngeal carcinoma. Mol Med Rep. 10.3892/mmr.2023.12996 10.3892/mmr.2023.12996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zörnig M, Evan GI (1996) Cell cycle: on target with Myc. Curr Biol 6(12):1553–1556 10.1016/S0960-9822(02)70769-0 [DOI] [PubMed] [Google Scholar]
- 57.Llombart V, Mansour M (2022) Therapeutic targeting of “undruggable” MYC. EBioMedicine 75:103756 10.1016/j.ebiom.2021.103756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi Y, Hutchinson H, Hall D, Zalewski A (1993) Downregulation of c-myc expression by antisense oligonucleotides inhibits proliferation of human smooth muscle cells. Circulation 88(3):1190–1195 10.1161/01.CIR.88.3.1190 [DOI] [PubMed] [Google Scholar]
- 59.Yang K, Fan M, Wang X, Xu J, Wang Y, Tu F et al (2022) Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ 29(1):133–146 10.1038/s41418-021-00841-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Semenza G (2023) Hypoxia-inducible factors: roles in cardiovascular disease progression, prevention, and treatment. Cardiovasc Res 119(2):371–380 10.1093/cvr/cvac089 [DOI] [PubMed] [Google Scholar]
- 61.Kormann R, Kavvadas P, Placier S, Vandermeersch S, Dorison A, Dussaule J et al (2020) Periostin promotes cell proliferation and macrophage polarization to drive repair after AKI. J Am Soc Nephrol 31(1):85–100 10.1681/ASN.2019020113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y et al (2019) Metabolic regulation of gene expression by histone lactylation. Nature 574(7779):575–580 10.1038/s41586-019-1678-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao H, Luo J, Zhang Y, Mao X, Wen P, Ding H et al (2020) Tuberous sclerosis 1 (Tsc1) mediated mTORC1 activation promotes glycolysis in tubular epithelial cells in kidney fibrosis. Kidney Int 98(3):686–698 10.1016/j.kint.2020.03.035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this study will be made available to other researchers upon reasonable request from the corresponding author.