Abstract
Plants sense phosphate (Pi) deficiency and initiate signaling that controls adaptive responses necessary for Pi acquisition. Herein, evidence establishes that AtSIZ1 is a plant small ubiquitin-like modifier (SUMO) E3 ligase and is a focal controller of Pi starvation-dependent responses. T-DNA insertional mutated alleles of AtSIZ1 (At5g60410) cause Arabidopsis to exhibit exaggerated prototypical Pi starvation responses, including cessation of primary root growth, extensive lateral root and root hair development, increase in root/shoot mass ratio, and greater anthocyanin accumulation, even though intracellular Pi levels in siz1 plants were similar to wild type. AtSIZ1 has SUMO E3 ligase activity in vitro, and immunoblot analysis revealed that the protein sumoylation profile is impaired in siz1 plants. AtSIZ1-GFP was localized to nuclear foci. Steadystate transcript abundances of Pi starvation-responsive genes AtPT2, AtPS2, and AtPS3 were moderate but clearly greater in siz1 seedlings than in wild type, where Pi is sufficient. Pi starvation induced the expression of these genes to the same extent in siz1 and wild-type seedlings. However, two other Pi starvation-responsive genes, AtIPS1 and AtRNS1, are induced more slowly in siz1 seedlings by Pi limitation. PHR1, a MYB transcriptional activator of AtIPS1 and AtRNS1, is an AtSIZ1 sumoylation target. These results indicate that AtSIZ1 is a SUMO E3 ligase and that sumoylation is a control mechanism that acts both negatively and positively on different Pi deficiency responses.
Keywords: phosphate starvation response, phosphate starvation signaling, sumoylation, phosphorous
Phosphorous is a component of many important biological compounds, and it is an essential macronutrient for all organisms. However, acquisition by plants is problematic because phosphate (Pi) is unevenly distributed and relatively immobile in soils (1). Plants react to Pi limitation by activating numerous adaptive responses that presumably facilitate acquisition of this essential nutrient (2). These responses include biochemical processes that limit metabolic requirements for Pi, secretion of organic acids to the apoplast, and synthesis of enzymes that enable access to phosphorus contained in intracellular organic molecules, insoluble complexes, and organophosphates in the soil. Pi limitation also causes morphological responses that are presumed to be adaptive, including attenuated primary root growth, increased lateral root development, root/shoot mass ratio, lateral root number and length (3), and root/hair production (2). Pi deficiency also induces the expression of genes that facilitate Pi uptake into roots, distribution in planta, and acquisition from organic sources (2).
Several Arabidopsis genetic loci appear to function in Pi signaling and acquisition. Mutations in the PHO3, PSR1, PDR2, and PHR1 genes impair Pi starvation signaling (4–7), whereas pho1, pho2, and pup1 mutations attenuate Pi uptake and distribution within tissues (8–10). The transporters AtPT1 (Pht1;1) and AtPT2 (Pht1;4) facilitate Pi uptake in environments with low (2 μM) and high (500 μM) levels of Pi, respectively (11). Pht2;1 transports Pi into chloroplasts and facilitates in planta Pi distribution (12).
The molecular mechanisms controlling Pi starvation sensing, signaling, and associated target gene activation that result in induced metabolic and phenotypic changes remain largely unresolved. The MYB transcription factor PHR1 is the only known molecular determinant required for Pi starvation-dependent responses (7). However, PHR1 controls a small subset of Pi starvation responses that apparently includes only a few genes whose expression is activated by Pi limitation. Genes controlled by PHR1 genes include AtACP5 (acid phosphatase), AtIPS1/3, At4, and RNS1 (7, 13, 14). PHR1 binds to the P1BS (PHR1-binding sequence) element that exists in the promoters of these genes (7). To a small extent, phr1 plants respond to Pi limitation by accumulating less anthocyanin, fresh weight mass, and intracellular Pi, and they exhibit a lower root/shoot ratio than the wild type (7). Other root architecture modifications typically associated with Pi deficiency, such as increased root hair number and length, are unaffected by the phr1 mutation (7). Together, the available data indicate that Pi starvation sensing and signal control are complex and involve numerous molecular determinants, many of which are not part of the PHR1 regulon and remain to be discovered.
Herein, we establish that mutations in AtSIZ1 enhance sensitivity of Arabidopsis plants to Pi deprivation based on morphological responses, including reduction in primary root elongation and enhanced lateral root development, root/shoot mass ratio, and root hair number and development. Furthermore, transcript abundance of some Pi starvation-responsive genes is greater in siz1 [SAP (scaffold attachment factor, acinus, protein inhibitor of activated signal transducer and activator of transcription) and Miz1 (Msx2-interacting zinc finger), SIZ] plants than in the wild type under Pi sufficient conditions. Interestingly, the induction rates of two genes (AtIPS1 and AtRNS1), whose responses to Pi starvation are controlled by PHR1, are reduced in siz1 plants compared with the wild type. Heat shock-induced sumoylation is less in siz1 plants, and recombinant AtSIZ1 can function as a small ubiquitin-like modifier (SUMO) E3 ligase to mediate sumoylation in vitro. SUMO E3 ligases transfer the SUMO peptide to the substrate in vivo (15) through functions that increase the affinity of the conjugating enzyme (E2) subunit for a specific target, stabilize E2–substrate interaction, orient the substrate acceptor K in the sumoylation motif (ψKXE), and contribute mechanistically to conjugation (15–17). Conjugation of the SUMO superfamily of proteins to a substrate (sumoylation) in both yeast and animals affects both positive and negative regulation by modulating protein–protein and protein–DNA interactions and intracellular targeting and controlling ubiquitination and other covalent modifications of proteins. Recently, functional evidence for a SUMO conjugation pathway in plants has been reported (18–20). Here, we determined that PHR1, which is the only presently identified transcriptional modulator of low Pi-induced responses, is sumoylated by an AtSIZ1-dependent process. Considering these findings and its function as a SUMO E3 ligase with multiple targets, AtSIZ1 appears to be an important regulator of Pi starvation responses in plants.
Materials and Methods
Plant Materials and Growth Conditions. The Arabidopsis T-DNA population in the Col-0 gl1 sos3-1 background and identification of mutations that suppress sos3-1 Na+ hypersensitivity were described in ref. 21. Unless indicated otherwise, 3-day-old seedlings were transferred to basal medium containing 1× Murashige and Skoog (MS) micronutrients, 1/5× or 1/20× macronutrients without KH2PO4, 3% sucrose, 2.5 mM Mes, B5 vitamins, 1.2% agar, and without or with various amounts of KH2PO4 or NaH2PO4. The Pi content of the basal medium without a Pi supplement was 10.6 ± 0.75 μM.
Three-week-old plants were removed from Metro-mix (Scotts, Marysville, OH), roots were carefully washed with water, and plants were transferred to modified Hoagland's solution (without aeration) containing 1 mM KH2PO4 (22). After 4 days of adaptation to solution culture, plants were transferred to a modified Hoagland's solution with 0.01 or 1 mM KH2PO4 (22).
Molecular Genetic Analysis of siz1 T-DNA Insertion Alleles. The genomic sequence flanking the T-DNA left border in sos3-1 siz1-1 plants (the Col-0 gl1 background) was determined by thermal asymmetric interlaced PCR analysis as described in ref. 21. siz1-2 and siz1-3 alleles in the Col-0 background were identified in the Salk Institute Genome Analysis Laboratory database, and T3 seeds were provided by the Salk Institute laboratory through the Arabidopsis Biological Resource Center at Ohio State University.
Analysis of Sumoylation Profiles. Total protein from seedlings incubated at 24°C or at 40°C for 30 min were extracted as described in ref. 20 and separated by SDS/PAGE. The gel blot was probed with the AtSUMO1 antibody (kindly provided by G. Coupland, Max Planck Institute, Cologne, Germany) and detected by using ECL plus (Amersham Pharmacia).
Purification of Proteins for the in Vitro SUMO Conjugation Assay. To construct pGST-SIZ1, a cDNA fragment of AtSIZ1 ORF was inserted in-frame into the pGEX-2TK vector (Amersham Pharmacia). To construct PHR1-expressing plasmids, a cDNA fragment encoding wild-type PHR1 (pT7-PHR1) or a mutated PHR1 [AA782A to AGA and AA1115A to AGA by site-directed mutagenesis pT7-PHR1(2KR); ref. 23] was inserted into pET21a vector (Novagen). The wild-type protein is designated as T7-PHR1, and the variant is designated as T7-PHR1(2KR), harboring a mutation (K261R and K372R) in each of the two predicted sumoylation motifs in PHR1. Recombinant protein synthesis from pGST-SIZ1, pT7-PHR1, pT7-PHR1(2KR), or other expression vectors (kindly provided by Y. Kikuchi, Tokyo University, Tokyo) and the in vitro sumoylation assay were performed as described in ref. 23.
Supporting Information. For additional methods, see Supporting Methods, which is published as supporting information on the PNAS web site.
Results
T-DNA Insertional Mutations in AtSIZ1 Suppress the Salt-Sensitive Phenotype of sos3-1 and Cause Hyperresponsiveness to Pi Limitation. T2 lines from a T-DNA insertion population (pSKI015) in the Arabidopsis Col-0 gl1 sos3-1 background were screened to identify second-site mutations that enhance NaCl tolerance (21). The siz1-1 mutation suppressed NaCl sensitivity of sos3-1 seedlings, but to a lesser extent than did hkt1 alleles (data not shown and ref. 21). Decreasing the macronutrient concentration in the MS salt formulation to 1/20× resulted in a substantial reduction in primary root growth of sos3-1 siz1-1 seedlings (data not shown). Normal root growth was restored by inclusion in the medium of 1.25 mM PO43–, either as KH2PO4 or NaH2PO4, but not by inclusion of any other macronutrient at the concentration in the full-strength MS salt formulation, e.g., 20 mM KCl, 19 mM KNO3, 3 mM CaCl2, or 1.6 mM MgSO4 (data not shown).
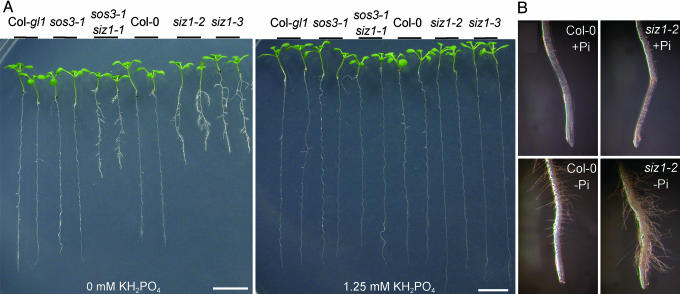
sos3-1 siz1-1 seedlings exhibited substantially more pronounced prototypical Pi starvation root architecture responses than sos3-1 or wild-type seedlings (Fig. 1A). On Pi-limited medium, sos3-1 siz1-1 seedlings exhibited an inhibition of primary root growth (Fig. 1 A; see also Fig. 6A, which is published as supporting information on the PNAS web site), an increase in lateral root development and length (Figs. 1A and 6 B–D), an increase in root hair number and length (Fig. 1B), and higher root/shoot fresh weight ratio (Table 1, which is published as supporting information on the PNAS web site) than wild type. sos3-1 siz1-1 plants grown on Pi-deficient medium for 25 days also accumulated more anthocyanin in leaves than did the wild type, and many of the siz1 leaves were necrotic (data not shown). sos3-1 siz1-1 plants grown in the greenhouse or growth chamber always exhibited reduced shoot and root biomass relative to the wild type that was not evident when these plants were grown in vitro. Despite this phenomenon, the relative increase in root/shoot biomass was greater for sos3-1 siz1-1 plants than the wild type after transfer to Pi-deficient medium in hydroponic solution (Table 2, which is published as supporting information on the PNAS web site). sos3-1 siz1-1 plants grown hydroponically began anthocyanin accumulation sooner than the wild type after transfer to Pi-deficient medium (data not shown).
Fig. 1.
Arabidopsis plants harboring siz1 mutations are hyperresponsive to Pi limitation in root architecture development. (A) Photographs are of representative wild-type [Col-0 or Col-0 gl1 (Col-gl1)], sos3-1, sos3-1 siz1-1, siz1-2, and siz1-3 seedlings 11 days after transfer onto medium containing 1/20× MS macronutrients without or with KH2PO4 supplement. sos3-1 and sos3-1 siz1-1 are in the Col-0 gl1 background, and siz1-2 and siz1-3 are in the Col-0 background. (B) Photographs are of wild-type and siz1-2 seedling roots 7 days after transfer onto medium containing 1/20× MS macronutrients without (–Pi) or with 1.25 mM KH2PO4 (+Pi).
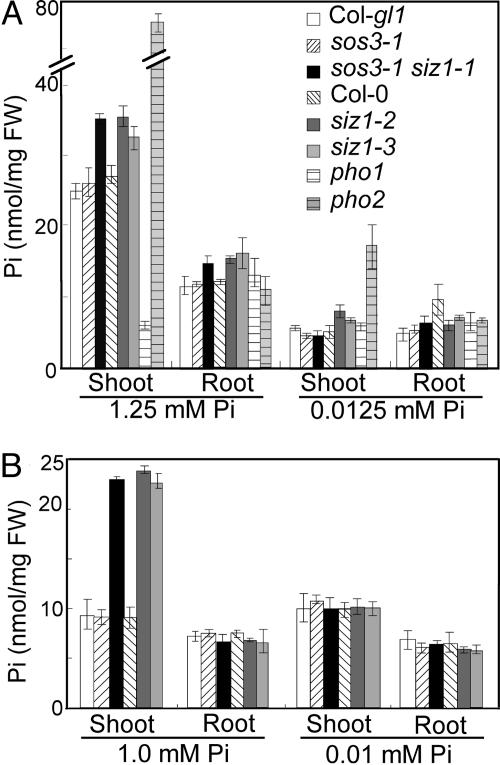
sos3-1 siz1-1 seedlings and plants grown in Pi-sufficient medium (Fig. 2A) and hydroponic solution (Fig. 2B), respectively, accumulated more intracellular Pi (Piint) in shoots than did the wild type or sos3-1, but Piint in roots of these plants was similar. Symptoms of Pi toxicity were not detected in shoots of sos3-1 siz1-1 seedlings, as is the case in shoots of Pi hyperaccumulating pho2 (9) that was included as a control in this experiment (Fig. 2). Piint levels in sos3-1 siz1-1, sos3-1, and wild-type seedlings and plants were similar when grown in Pi-deficient medium or solution (Fig. 2) even though sos3-1 siz1-1 seedlings exhibited responses to Pi limitation earlier and to a greater extent than the wild type (Figs. 1 and 6).
Fig. 2.
Piint in the shoot and the root of wild-type and siz1 seedlings and plants is comparable. Shown are seedlings grown for 11 days on medium containing 1/5× MS macronutrients with 0.0125 or 1.25 mM KH2PO4 (A) or plants grown for 2 weeks in hydroponic solution with 0.01 or 1 mM KH2PO4 (B) were harvested, mean values ± SE, n = 6. pho1 and pho2 seedlings were used as controls in A (8, 9).
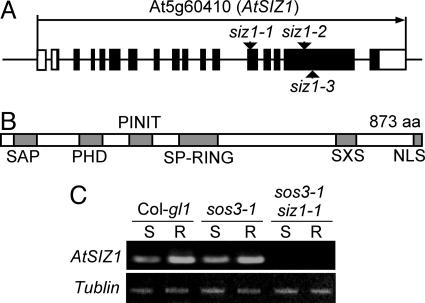
AtSIZ1 Encodes a Putative SUMO E3 Ligase. Thermal asymmetric interlaced PCR analysis of sos3-1 siz1-1 plants identified a T-DNA insertion in the 13th exon of At5g60410 (GenBank accession no. NM_125434) at +986 bp from the translation initiation codon as determined by analysis of the full-length cDNA sequence (R22197; Fig. 3A). At5g60410 is annotated as AtSIZ1, and this mutant allele is designated as siz1-1. Using the GenBank database, the AtSIZ1 product is predicted to be a peptide of 833 aa, which is an apparent ortholog of Saccharomyces cervisiae Siz1 and Siz2 and human protein inhibitor of activated signal transducer and activator of transcription (PIAS) proteins (Fig. 3B; see also Fig. 7, which is published as supporting information on the PNAS web site) (19). AtSIZ1 contains five predicted domains conserved in Siz/PIAS SUMO E3 ligases. These domains include a SAP domain that is implicated in chromatin organization, a Siz/PIAS-RING domain that is necessary for SUMO E3 ligase activity, a “PINIT” motif for nuclear retention (24), a “SXS” motif that promotes binding to SUMO (25), and a putative nuclear localization sequence (Fig. 3B). AtSIZ1 also has a plant homeodomain-finger (PHD) domain that is not present in Siz/PIAS proteins. The PHD domain is associated with chromatin remodeling complexes (26) or functions as an ubiquitin E3 ligase (27).
Fig. 3.
AtSIZ1 locus is At5g60410 as determined by thermal asymmetric interlaced PCR and T-DNA insertion diagnostic PCR. (A) Gene organization of AtSIZ1 and schematic representation of T-DNA insertion alleles are illustrated (arrowheads). Filled boxes are exons, and open rectangles are 5′ or 3′ untranslated regions. (B) Diagram illustrates the domain organization of AtSIZ1 (see Results). (C) AtSIZ1 is expressed in the root (R) and the shoot (S) of the wild type but not in these organs of siz1 seedlings.
F1 and F2 analyses of progeny derived from a backcross with sos3-1 (sos3-1 × sos3-1 siz1-1) indicated that siz1-1 is a monogenic recessive mutation [F2 analysis, n = 407 for, χ2 = 1.25, P = 0.26 for a 3:1 (normal:short primary root on Pi-deficient medium) segregation ratio]. Seedlings harboring additional mutations in AtSIZ1, siz1-2 (SA LK_065397), and siz1-3 (SALK_034008) (Fig. 3A) exhibited similar root architecture responses to Pi deficiency as did sos3-1 siz1-1 seedlings, and these responses were suppressed by the addition of either KH2PO4 or NaH2PO4 (Figs. 1 and 6). Furthermore, siz1-2 and siz1-3 seedlings and plants accumulated comparable Piint and developed a higher root/shoot mass ratio in response to Pi deprivation, as did sos3-1 siz1-1 (Fig. 2 and Tables 1 and 2). These data establish that the responses to Pi limitation exhibited by sos3-1 siz1-1 seedlings are due to the siz1-1 mutation. Primary root growth analysis on Pi-deficient medium of F1 progeny from the cross between siz1-2 and siz1-3 indicated that the mutations are allelic (data not shown). AtSIZ1 mRNA was more abundant in roots than in shoots of wild-type and sos3-1 seedlings (Fig. 3C) but was not detected in sos3-1 siz1-1 seedlings (Fig. 3C) or siz1-2 or siz1-3 seedlings (data not shown).
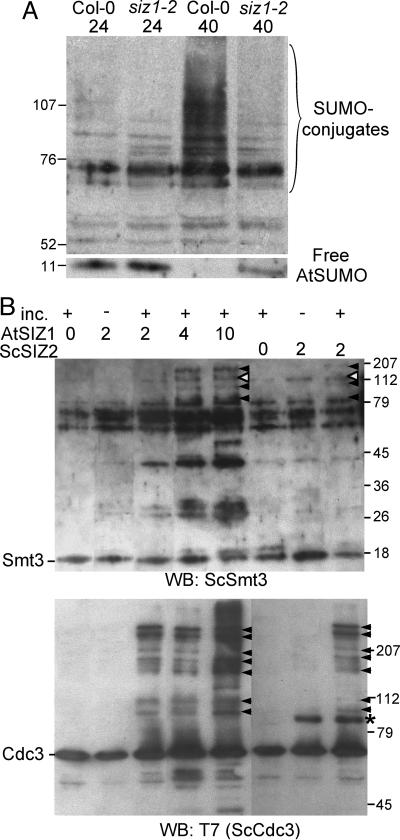
In Vitro and in Vivo Data Indicate That AtSIZ1 Functions as a SUMO E3 Ligase. An immunoblot of total protein by using anti-AtSUMO1 (20) indicated no difference in accumulation of SUMO or SUMO conjugation products in siz1-2 and wild-type seedlings that are grown at 24°C (Fig. 4A). In contrast, esd4 seedlings accumulate numerous sumoylated products, whereas the amounts of free SUMO peptides are depleted (data not shown and ref. 20). Heat shock increased SUMO conjugation in wild-type plants (19), but the extent of heat shock-induced sumoylation was substantially less in siz1-2 plants (Fig. 4A). Together these results provide evidence that AtSIZ1 is involved in the sumoylation of Arabidopsis proteins.
Fig. 4.
In vitro and in vivo assays indicate that AtSIZ1 is a SUMO E3 ligase. (A) In planta sumoylation profiles of wild-type and siz1-2 seedlings that were grown for 10 days on 1× MS medium at 24°C (24) and then exposed to a 30-min heat shock of 40°C (40). (B) Analysis of in vitro sumoylation profiles indicates that AtSIZ1 facilitates production of ScSmt3-ScCdc3 conjugates. The indicated amount of GST-AtSIZ1 (0, 2, 4, or 10 μg) or ScSIZ2 (0 or 2 μg) was added to the reaction mixture and incubated with 10 mM ATP at 37°C for 0 (inc. –) or 90 min (inc. +) as described in ref. 23. Immunoblot analysis was performed with anti-ScSmt3 (Upper) or anti-T7 (detecting T7-ScCdc3; Lower). Filled arrowheads indicate bands corresponding to ScSmt3-ScCdc3 conjugates. Open arrowheads identify the AtSIZ1-ScSmt3 and ScSiz2-ScSmt3 conjugates. An asterisk identifies the T7-ScSIZ2-His protein. Numerals identify the migration position and molecular mass of the markers (kilodaltons).
GST-AtSIZ1 could substitute for T7-ScSiz2-His to stimulate sumoylation in an in vitro assay (23) that included S. cerevisiae E1 (GST-ScAos1 + GST-ScUba2), E2 (ScUbc9), SUMO substrate (T7-ScCdc3-His), and SUMO (His-ScSmt3) and used anti-ScSmt3 to detect sumoylated products (Fig. 4B Upper). GST-AtSIZ1 (112 kDa) or ScSiz2 (82 kDa) was detected (Fig. 4B) on the anti-ScSmt3 immunoblot because each protein is sumoylated. Immunoblot analysis by using anti-T7 antibody (detecting T7-ScCdc3-His) indicated that AtSIZ1 mediates ScSmt3 cojugation to ScCdc3, as does ScSiz2 (Fig. 4B Lower). ScCdc3 contains seven sumoylation sites, and these different products are evident on the blot (Fig. 4B Lower and ref. 23). Together, the immunoblot analysis of the in vivo sumoylation products and the in vitro assay results establish that AtSIZ1 functions as a SUMO E3 ligase in Arabidopsis.
AtSIZ1-GFP was detected specifically in the same compartment as the red fluorescent protein control (ref. 28 and Fig. 8A, which is published as supporting information on the PNAS web site), indicating that AtSIZ1 is localized to the nucleus. Higher magnification revealed that AtSIZ1 localizes predominantly to punctuate structures in the nucleus (Fig. 8B). Expression of AtSIZ1-GFP suppressed both the hyperresponses of siz1-2 seedlings to Pi starvation and the reduced shoot and root biomass phenotypes of siz1-2 plants (data not shown). These results indicate that AtSIZ1-GFP is functional and that AtSIZ1 compartmentalizes to nuclear speckles, as do PIAS proteins (29).
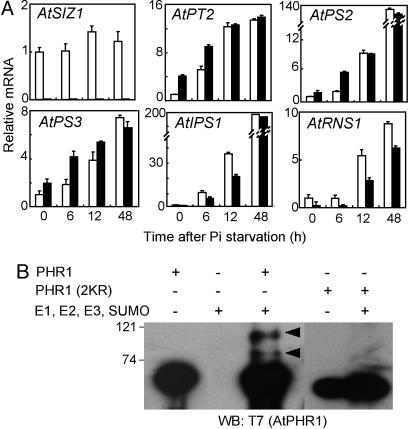
siz1 Mutations Cause an Alteration in Pi Starvation-Dependent Signaling. AtPT2 (Pht1;4), AtPS2 (acid phosphatase; ref. 30), and AtPS3 (glycerol-3-phosphate permease, Raghothama, unpublished data), but not AtIPS1 or AtRNS1, transcript abundances were greater in siz1 than in wild-type seedlings when both were grown in Pi-sufficient medium (Fig. 5A). However, Pi starvation induced the expression of AtPT2, AtPS2, and AtPS3 to a comparable level in both the wild-type and siz1 seedlings after 48 h in Pi-deficient medium (Fig. 5A). Pi starvation induction of AtIPS1 and AtRNS1 transcript abundances occurred more slowly in siz1 than in wild-type seedlings (Fig. 5A) but eventually increased to levels similar to the wild type after an extended period of Pi limitation (i.e., 48 h or 72 h; data not shown). Together, these results indicate that the function of AtSIZ1 in Pi starvation signaling is different for these two specific subsets of target genes. AtPT2, AtPS2, and AtPS3 are negatively regulated by AtSIZ1 in Pi-sufficient conditions, and AtIPS1 and AtRNS1 are positively regulated by AtSIZ1 during the initial stages of induction by Pi limitation (for primer sequences for these genes, see Table 3, which is published as supporting information on the PNAS web site).
Fig. 5.
Pi-responsive transcript abundance is altered in siz1 plants, and AtSIZ1 facilitates sumoylation of PHR1. (A) Pi starvation-responsive gene mRNA levels in Col-0 (wild type, open bars) and siz1-2 (filled bars) seedlings were determined by quantitative PCR. Seedlings were grown in liquid medium containing 1/5× MS macronutrients and 1× MS micronutrients with 1.25 mM KH2PO4 for 7 days and then transferred to medium without KH2PO4. Samples were collected immediately after transfer to medium without Pi (0 h) or after various time points in this medium (6, 12, or 48 h). (B) AtSIZ1 mediates in vitro sumoylation of PHR1. T7-PHR1 or T7-PHR1(2KR) were used as substrates for sumoylation in the reaction mixture containing ScAos1 (E1), ScUba2 (E1), ScUbc9 (E2), ScSmt3 (SUMO, 23), and AtSIZ1 (E3) proteins. PHR1 proteins were detected with anti-T7 antibody. Arrowheads indicate the position of sumoylated PHR1 proteins.
AtIPS1 and AtRNS1 are members of the PHR1 regulon (7). The MYB transcription factor PHR1, which binds to a P1BS element in the promoter of AtIPS1 and is necessary for transcription of AtIPS1 and AtRNS1 (7), contains two predicted sumoylation sites. AtSIZ1 facilitated the sumoylation of PHR1 (Fig. 5B). Mutations to residues in the predicted sumoylation sites, K261R and K372R, in PHR1 prevented sumoylation of PHR1 (Fig. 5B). These results further indicate that sumoylation of PHR1 positively controls the expression of AtIPS1 and AtRNS1.
Discussion
Genetic, physiological, molecular, and biochemical data presented here establish an important function for the plant SUMO E3 AtSIZ1 in the signaling system that coordinates plant responses to Pi starvation. More than any other identified locus or molecular genetic determinant yet described, AtSIZ1 influences a greater number of processes that are considered to be adaptation responses to low Pi availability, including root architecture changes, gene expression modulation, and anthocyanin accumulation (4–12). Our results establish that the MYB transcription factor PHR1, the only defined molecular controller of Pi deficiency responses (7), is a sumoylation target of AtSIZ1. However, AtSIZ1 affects several Pi starvation responses that are not controlled by PHR1, indicating that sumoylation is a more encompassing process in the control of adaptation to Pi deficiency.
AtSIZ1 can replace ScSiz2 to sumoylate the substrate ScCdc3 in an in vitro assay, providing evidence of its SUMO E3 ligase activity (Fig. 4B and ref. 23). Reduction of heat shock-induced SUMO conjugation in seedlings with a defective AtSIZ1 gene demonstrates that AtSIZ1 facilitates sumoylation in planta (Fig. 4A). AtSIZ1 localizes to nuclear speckles (Fig. 8), as do mammalian Siz/PIAS proteins, which are in structures that resemble the promyelocytic leukemia nuclear bodies (29). Together these results indicate that AtSIZ1 is a plant member of the Siz/PIAS category of SUMO E3.
AtSIZ1 is annotated as the only member of a single-gene family in Arabidopsis (19); however, three independent dysfunctional siz1 T-DNA insertion alleles are not lethal. Apparently, an alternative SUMO E3 performs the necessary functions of AtSIZ1, or AtSIZ1 is not essential for survival, even though sumoylation through AtSIZ1 controls critical processes in plants, as established by the results presented herein. Mutations in both ScSiz genes that are present in yeast also are not lethal (16). Although no additional gene encoding a Siz/PIAS E3 is evident in the Arabidopsis genome based on database analysis, it should be noted that ESD4 was not predicted to be a SUMO protease by bioinformatics (19, 20), further suggesting that there are unidentified functional Arabidopsis loci that can substitute for the function of AtSIZ1. Alternatively, two other categories of SUMO E3 are known to exist in animals: RanBP2 (Ran binding protein 2) and Pc2 (polycomb protein 2). RanBP2 functions as an E3 that attaches SUMO to RanGAP, a process that is involved in nucleocytoplasmic trafficking (15). At3g18610 and At2g34150 encoding AtRanGAP1 and AtRanGAP2, respectively, are likely to be sumoylated by a yet to be identified RanBP2-like E3 ligase (31). The Pc2-type SUMO E3 is a member of the polycomb group (PcG) of proteins that form large multimeric corepressor complexes (PcG bodies) that facilitate gene silencing (15, 32). Mammalian Pc2 contains a CHROMO (chromatin organization modifier) domain, and sumoylation is a control process involved in the function of PcG complexes (32). Arabidopsis LHP1/TFL2 has a CHROMO domain and functions in the PcG complex to repress floral homeotic genes and modulate plant development (33). LHP1 is also localized to discrete rounded subnuclear foci throughout the nucleoplasm similar to Pc2 in Drosophila (33).
AtSIZ1 Is a Repressor of the Low Pi-Induced Responses. siz1 seedlings and plants exhibit exaggerated symptoms that are associated with Pi deficiency, including reduced primary root growth and increased lateral root and root hair number and length, root/shoot mass ratio, and anthocyanin accumulation (Figs. 1 and 6 and refs. 34 and 35). Pi limitation reduces primary root growth by attenuating cell division and promoting lateral root density and length, root architecture changes that are considered to facilitate Pi acquisition (3, 34). These results indicate that AtSIZ1 is a negative regulator of Pi starvation-dependent signaling that controls root architecture and anthocyanin accumulation. Interestingly, these hyperresponses to low extracellular Pi occur even though Pint is somewhat greater (sufficient medium) or equivalent (deficient medium) in the shoots of siz1 seedlings and plants compared with the wild type (Fig. 2). However, shoot Piint accumulation (sufficient medium) in siz1 seedlings is not equivalent to pho2 plants that hyperaccumulate Pi and exhibit symptoms of Pi toxicity (9).
Transcript abundances of the Pi starvation responsive genes AtPT2, AtPS2, and AtPS3 are greater in siz1 compared with the wild-type seedlings grown under Pi sufficient conditions (Fig. 5A). However, Pi deprivation still increases the abundances of transcripts of these genes in siz1 seedlings to the same extent as observed in the wild type. This capacity for siz1 seedlings and plants to still respond to Pi limitation by increasing the expression of these genes, by changing root architecture, and by accumulating anthocyanin is indicative that the function of AtSIZ1 may be downstream of the Pi starvation sensing mechanism (Fig. 5). Other genetic loci, including PHR1, PDR2, PHO1-3, Pht1;1, Pht1;4, and PSR1 (4–12), are not individually required to regulate all of the Pi starvation responses that are affected by AtSIZ1, indicating that AtSIZ1 may be a critical regulator that is upstream of these other determinants.
Because coregulation of transcriptional activators is a major function of mammalian PIAS family proteins (17, 29), repression of low Pi-induced root architecture and anthocyanin accumulation, and AtPT2, AtPS2, and AtPS3 expression responses that are controlled by AtSIZ1 likely involves negative regulation of transcription factor(s) through AtSIZ1-mediated sumoylation. Two regions of the AtPT2 promoter bind nuclear protein factors from Pi-sufficient but not Pi-deficient plants (36), indicating that these factors may play a role in the repression of AtPT2 expression. However, these regulatory factors have yet to be identified, so direct evidence that sumoylation is necessary for the function of a repressor complex that controls low Pi signaling cannot be assessed.
AtSIZ1 Acts also as a Positive Regulator of Some Pi Starvation Responses. Pi starvation-dependent transcript accumulation of AtIPS1 and AtRNS1 occurs at a reduced rate in siz1 seedlings compared with wild type (Fig. 5A). Transcriptional activation of these genes has been attributed to the interaction of the MYB transcription factor PHR1 with the P1BS element in the promoter of these genes (7). Recently, additional evidence based on experimentation by using HvPht1;1 has supported the possibility that the P1BS element has a transcriptional activation function (37). Our determination that AtSIZ1 participates in the sumoylation of PHR1 (Fig. 5B) indicates that sumoylation may activate PHR1, which provides a mechanism for reduced Pi starvation induction of AtIPS1 and AtRNS1 expression in siz1 seedlings. Sumoylation functions to activate transcription factors in animals, including heat shock factors (38). After prolonged Pi deprivation, AtIPS1 and AtRNS1 transcript abundances are similar in siz1 and wild type (Fig. 5A for AtIPS1 and data not shown for AtRNS1), indicating that AtSIZ1 has a transient function in this process. However, this mechanism does not explain how AtSIZ1 affects PHR1 modulation of anthocyanin or Pi accumulation or root/shoot mass ratio. Other genetic loci are implicated in Pi starvation signaling, such as PDR2, PSR1, and PHO3 (4–6). The pdr2 mutation causes inhibition of primary root cell division and subsequent loss of meristematic activity (6). PDR2 may function in modulating meristematic activity and controlling root architecture when Pi is limiting. However, the sequences of these genes have not been identified, and, therefore, it is not possible to establish by more direct evidence whether sumoylation is involved in their functions.
Taken together, our results confirm that multiple signaling pathways are involved in the control of Pi starvation responses (Fig. 5). AtIPS1 and AtRNS1 are positively controlled by AtSIZ1 through sumoylation of the transcription factor PHR1. However, PHR1 and its sumoylation apparently do not have a major effect on low Pi-induced phenotypic responses or on the expression of AtPT2, AtPS2, and AtPS3, which are involved in adaptation and are negatively controlled by AtSIZ1. Alternatively, PHR1 function in Arabidopsis may be redundant. In any case, it is clear that a major function AtSIZ1 in Pi-starvation responses involves the control of a repressor system that attenuates gene expression and developmental responses that are only derepressed (activated) by Pi starvation. The role of AtSIZ1 as an important regulator of plant adaptation to Pi deficiency constitutes a previously uncharacterized function of sumoylation. It is clear that further research is necessary to define all of the mechanisms by which AtSIZ1 controls Pi deficiency responses.
Supplementary Material
Acknowledgments
We thank Ms. Wanda Hunter and Ms. Tomoko Miura for assistance and the Arabidopsis Biological Research Center and the Salk Institute for T-DNA-tagged lines and a cDNA clone. This work was supported by National Science Foundation Plant Genome Award DBI-98-13360, grants from the Biogreen 21 program, the Environmental Biotechnology National Core Research Center (R15-2003-012-01002-0), the Plant Diversity Research Center, the Ministry of Sports and Technology (PF0330401-00), and the BK21 program, and JSPS-Junior Scientist Research Fellowship 3117. This work is Purdue University Agricultural Research Program Paper 2005-17626.
Author contributions: K.M., R.A.B., D.-J.Y., and P.M.H. designed research; K.M., A.R., A.S., S.Y., A.S.K., D.B., Y.D.K., and J.-B.J. performed research; R.A.B., D.-J.Y., and P.M.H. contributed new reagents/analytic tools; K.M., A.R., K.G.R., R.A.B., D.-J.Y., and P.M.H. analyzed data; and K.M., R.A.B., D.-J.Y., and P.M.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MS, Murashige and Skoog; Pi, phosphate; PIAS, protein inhibitor of activated signal transducer and activator of transcription; Piint, intracellular Pi; SUMO, small ubiquitin-like modifier.
References
- 1.Holford, I. C. R. (1997) Aust. J. Soil Res. 35, 227–239. [Google Scholar]
- 2.Raghothama, K. G. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 665–693. [DOI] [PubMed] [Google Scholar]
- 3.López-Bucio, J., Cruz-Ramírez, A. & Herrera-Estrella, L. (2003) Curr. Opin. Plant Biol. 6, 280–287. [DOI] [PubMed] [Google Scholar]
- 4.Zakhleniuk, O. V., Raines, C. A. & Lloyd, J. C. (2001) Planta 212, 529–534. [DOI] [PubMed] [Google Scholar]
- 5.Chen, D. L., Delatorre, C. A., Bakker, A. & Abel, S. (2000) Planta 211, 13–22. [DOI] [PubMed] [Google Scholar]
- 6.Ticconi, C. A., Delatorre, C. A., Lahner, B., Salt, D. E. & Abel, S. (2004) Plant J. 37, 801–814. [DOI] [PubMed] [Google Scholar]
- 7.Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A. & Paz-Ares, J. (2001) Genes Dev. 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirier, Y., Thoma, S., Somerville, C. & Schiefelbein, J. (1991) Plant Physiol. 97, 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delhaize, E. & Randall, P. J. (1995) Plant Physiol. 107, 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trull, M. C. & Deikman, J. (1998) Planta 206, 544–550. [DOI] [PubMed] [Google Scholar]
- 11.Shin, H., Shin, H.-S., Dewbre, G. R. & Harrison, M. J. (2004) Plant J. 39, 629–642. [DOI] [PubMed] [Google Scholar]
- 12.Versaw, W. K. & Harrison, M. J. (2002) Plant Cell 14, 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martín, A. C., del Pozo, J. C., Iglesias, J., Rubio, V., Solano, R., de la Peña, A., Leyva, A. & Paz-Ares, J. (2000) Plant J. 24, 559–567. [DOI] [PubMed] [Google Scholar]
- 14.Bariola, P. A., Howard, C. J., Taylor, C. B., Verburg, M. T., Jaglan, V. D. & Green, P. J. (1994) Plant J. 6, 673–685. [DOI] [PubMed] [Google Scholar]
- 15.Melchior, F., Schergaut, M. & Pichler, A. (2003) Trends Biochem. Sci. 28, 612–618. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, E. S. (2004) Annu. Rev. Biochem. 73, 355–382. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt, D. & Müller, S. (2003) Cell. Mol. Life Sci. 60, 2561–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lois, L. M., Lima, C. D. & Chua, N. H. (2003) Plant Cell 15, 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurepa, J., Walker, J. M., Smalle, J., Gosink, M. M., Davis, S. J., Durham, T. L., Sung, D. Y. & Vierstra, R. D. (2003) J. Biol. Chem. 278, 6862–6872. [DOI] [PubMed] [Google Scholar]
- 20.Murtas, G., Reeves, P. H., Fu, Y.-F., Bancroft, I., Dean, C. & Coupland, G. (2003) Plant Cell 15, 2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rus, A., Sharkhuu, A., Reddy, M., Lee, B.-h., Matsumoto, T. K., Koiwa, H., Zhu, J.-K., Bressan, R. A. & Hasegawa, P. M. (2001) Proc. Natl. Acad. Sci. USA 98, 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, C., Muchhal, U. S., Uthappa, M., Kononowicz, A. K. & Raghothama, K. G. (1998) Plant Physiol. 116, 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi, Y., Toh-E, A. & Kikuchi, Y. (2003) J. Biochem. 133, 415–422. [DOI] [PubMed] [Google Scholar]
- 24.Duval, D., Duval, G., Kedinger, C., Poch, O. & Boeuf, H. (2003) FEBS Lett. 554, 111–118. [DOI] [PubMed] [Google Scholar]
- 25.Minty, A., Dumont, X., Kaghad, M. & Caput, D. (2000) J. Biol. Chem. 275, 36316–36323. [DOI] [PubMed] [Google Scholar]
- 26.Bochar, D. A., Savard, J., Wang, W., Lafleur, D. W., Moore, P., Cote, J. & Shiekhattar, R. (2000) Proc. Natl. Acad. Sci. USA 97, 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coscoy, L. & Ganem, D. (2003) Trends Cell Biol. 13, 7–12. [DOI] [PubMed] [Google Scholar]
- 28.Lee, Y. J., Kim, D. H., Kim, Y. W. & Hwang, I. (2001) Plant Cell 13, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seeler, J. S. & Dejean, A. (2003) Nat. Rev. Mol. Cell Biol. 4, 690–699. [DOI] [PubMed] [Google Scholar]
- 30.Baldwin, J. C., Karthikeyan, A. S. & Raghothama, K. G. (2001) Plant Physiol. 125, 728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose, A. & Meier, I. (2001) Proc. Natl. Acad. Sci. USA 98, 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kagey, M. H., Melhuish, T. A. & Wotton, D. (2003) Cell 113, 127–137. [DOI] [PubMed] [Google Scholar]
- 33.Gaudin, V., Libault, M., Pouteau, S., Juul, T., Zhao, G., Lefebvre, D. & Grandjean, O. (2001) Development (Cambridge, U.K.) 128, 4847–4858. [DOI] [PubMed] [Google Scholar]
- 34.Abel, S., Ticconi, C. A. & Delatorre, C. A. (2002) Physiol. Plant. 115, 1–8. [DOI] [PubMed] [Google Scholar]
- 35.Trull, M. C., Guiltinan, M. J., Lynch, J. P. & Deikman, J. (1997) Plant Cell Environ. 20, 85–92. [Google Scholar]
- 36.Mukatira, U. T., Liu, C., Varadarajan, D. K. & Raghothama, K. G. (2001) Plant Physiol. 127, 1854–1862. [PMC free article] [PubMed] [Google Scholar]
- 37.Schünmann, P. H. D., Richardson, A. E., Vickers, C. E. & Delhaize, E. (2004) Plant Physiol. 136, 4205–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hilgarth, R. S., Hong, Y., Park-Sarge, O.-K. & Sarge, K. D. (2003) Biochem. Biophys. Res. Comm. 303, 196–200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.