Abstract
The hypoxia inducible factor (HIF) plays an important role in the progression of a number of pathophysiological processes including tumorigenesis. In addition to several well characterized oxygen-dependent modes of regulation, the function of the HIF transcription factor can also be influenced through the action of other regulatory pathways. Misregulation of these factors resulting in inappropriate HIF expression or activity can contribute to the progression of human cancers through the induction of genes promoting angiogenesis, glycolysis, cell survival, and metastasis, among other processes. The candidate tumor suppressor protein inhibitor of growth family member 4 (ING4) has recently been implicated as a repressor of angiogenesis and tumor growth through association with NF-κB. Here we demonstrate that suppression of ING4 further induces HIF transcriptional activity as well. ING4 directly associates with the HIF prolyl hydroxylase, an Fe(II)-dependent oxygenase previously shown to mediate HIF stability as a function of oxygen availability. However, rather than affecting HIF's stability, ING4 mediates HIF's activity. These data support a model in which, in addition to regulating HIF stability, HIF prolyl hydroxylases can modulate HIF function through the recruitment of ING4, a likely component of a chromatin-remodeling complex.
Keywords: prolyl hydroxylase, inhibitor of growth
To maintain adequate levels of aerobic respiration, mammalian cells must be able to sense the status of O2 availability and respond to changes in O2 levels when they fall below critical levels. A key component of the hypoxic response pathway is the hypoxia inducible factor (HIF). Decreased O2 within a cell results in HIF induction and the subsequent transcription of a number of target genes that promote adaptation to this environmental stress. The list of HIF targets includes genes affecting metabolism, O2 delivery, angiogenesis, and cellular survival (1). Consequently, HIF plays an important role in physiological and pathophysiological states in which O2 supply is limiting (1, 2).
HIF is an obligate heterodimer composed of a HIF-β subunit that is essentially insensitive to O2 availability and a HIF-α subunit whose accumulation and activity is acutely responsive to O2 levels (reviewed in refs. 1–3). Briefly, under normoxic conditions HIF-α is targeted for rapid proteosomal degradation after association with a protein ubiquitin ligase complex containing the product of the von Hippel–Lindau tumor suppressor gene (pVHL). pVHL fails to bind the α-subunit under hypoxic conditions, allowing HIF to accumulate. Differential recognition of HIF-α by VHL is mediated by hydroxylation of conserved proline residues within the HIF-α O2-dependent degradation domain (4). Under normoxic conditions, HIF prolyl hydroxylases (HPHs; also called PHDs or EGLNs) efficiently modify these residues (5–7), a prerequisite for VHL binding to HIF-α (8, 9). The HPH active site contains Fe(II) coordinated by a His-Xaa-Asp... His triad. These enzymes in turn bind 2-oxoglutarate, HIF-α, and O2 to effect hydroxylation of both the HIF-α subunit and 2-oxoglutarate. Hydroxylated 2-oxoglutarate undergoes decarboxylation to produce succinate and CO2. Because these enzymes require O2 as a substrate for the hydroxylation reaction, they have been implicated as direct O2 sensors in the hypoxic response pathway (10). Independent of protein stability, coactivator recruitment by the C-terminal transactivation domain of HIF is also regulated by posttranslational hydroxylation (11) by another O2-dependent hydroxylase, factor inhibiting HIF 1 (FIH-1) (12–14).
The hypoxic response pathway has been recognized as an important contributor to a number of cancers, because increased levels of HIF are often associated with increased tumor aggressiveness, therapeutic resistance, and mortality (15). HIF can be induced as a result of intratumoral hypoxia stemming from rapid growth and/or inefficient O2 delivery from tortuous vasculature. Alternatively, HIF function can be altered in an O2-independent manner because of genetic alterations that activate signaling pathways or inactivate tumor suppressors (15). Understanding of the mechanisms by which oncogenes or tumor suppressor genes affect HIF function sheds light not only on how their misregulation promotes cancer but also reveals physiologically relevant mechanisms by which cells fine-tune HIF activity under the borderline hypoxic conditions encountered by most “normoxic” tissues (16–18).
Recently, inhibitor of growth family member 4 (ING4) has emerged as a strong candidate tumor suppressor protein, repressing tumor growth and angiogenesis (19) and the loss of contact inhibition (20). ING4 is a member of a family of proteins characterized by a highly conserved C-terminal plant homeodomain (PHD)-like zinc-finger domain and implicated in a variety of processes including oncogenesis, apoptosis, DNA repair, and cell cycle control (21). The best-studied ING family members reside in the nucleus where they positively or negatively regulate gene expression through interactions between their N terminus and chromatin-remodeling complexes containing histone acetyltransferases, histone deacetyltransferases, or factor acetyltransferases (21).
Although ING4's precise mode of action has yet to be elucidated, ING4 has been shown to interact with the RelA subunit of NF-κB to suppress the expression of angiogenesis-related genes including IL-6, IL-8, and Cox-2 (19). Here we show that ING4 also serves to suppress expression of HIF target genes under hypoxic conditions. Furthermore, HPH-2 directly interacts with ING4, providing a possible mechanism for ING4 recruitment to HIF. Interestingly, ING4 association with HPH-2 does not seem to affect hydroxylase activity or HIF stability. Instead, our data are consistent with a model in which ING4, recruited by HPH-2 to HIF under hypoxic conditions, acts as an adapter protein to recruit transcriptional repressors to mediate HIF activity.
Materials and Methods
Recombinant Protein Expression and Purification. Protein coding sequences for HPH-2 (GenBank accession no. AF229245) and ING4 (GenBank accession no. NM_016162) were amplified by PCR and confirmed by sequencing. Recombinant FIH-1 was prepared as described (13). The sequence encoding HPH-2 residues 181–426 (HPH-2C) was subcloned into the pHIS parallel vector (22) and purified by following the same procedure used for FIH-1 (13).
ING4 was expressed in Escherichia coli as a Gβ1 fusion protein. Cells were lysed in buffer containing 20 mM Tris·HCl (pH 8.0), 50 mM NaCl, 5 mM 2-mercaptoethanol, 100 μM ZnCl2, and 1:200 Protease Inhibitor Mixture (Sigma) and purified over Source Q resin (Amersham Pharmacia). The Gβ1 tag was removed by TEV protease digestion followed by ion exchange chromatography using Mono S resin (Amersham Pharmacia). ING4 was stored in buffer containing 50 mM NaPO4 (pH 5.9), 300 mM NaCl, 1.0 mM 2-mercaptoethanol, and 10 μM ZnCl2 at 4°C.
Yeast Two-Hybrid Assay. The yeast strain L40 was transformed with vectors encoding HPH-2C fused to the LexA DNA-binding domain (DBD) (HPH-2C/pVJL10) and ING4 fragments fused to the GAL4 activation domain (ING4/pGAD-NotII). Positive protein–protein interactions were denoted by LacZ reporter expression.
Western Blot Analysis. Cells were resuspended in SDS sample buffer, and proteins were resolved by SDS/PAGE before Western blot analysis. Polyclonal antiserum was obtained from rabbits immunized with HPH-2C, ING4, or the first 140 amino acids of the HIF-β subunit. The HPH-2 antiserum failed to detect endogenous HPH-1 or HPH-3. The ING4 antiserum displayed minimal cross-reactivity with the other ING family members, although weak detection of ING1 and ING5 was seen after overexpression. Mouse monoclonal antibodies to p-ATF-2, annexin I (Santa Cruz Biotechnology), and HIF-1α (BD Transduction Laboratories, Lexington, KY) were purchased. Immune complexes were detected by enhanced chemiluminescence using peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch).
Cell Culture. HeLa cell lines were maintained in Dulbecco's modified Eagle's medium (HyQ DME, HyClone) containing high glucose and supplemented with 10% FBS (Gemini Biological Products, Calabasas, CA) in the presence of 5.0% CO2 at 37°C. Cells were maintained under hypoxic conditions at 37°C within a humidified hypoxic chamber (Coy Laboratory Products, Ann Arbor, MI) filled with 1% O2/5.0% CO2 and balanced with N2. Luciferase assays were performed according to the method of ref. 23.
Coimmunoprecipitation Assay. HeLa cells were transfected with ING4 in the p3XFLAG-CMV10 vector (Sigma) by using Lipofectamine plus reagent (Invitrogen). After 24 h, cells were lysed with 20 mM Tris·HCl (pH 7.5), 100 mM NaCl, 5 mM 2-mercaptoethanol, 1% Nonidet P-40, and Protease Inhibitor Mixture (Sigma). Lysates were precleared with Protein A-agarose beads (Roche Diagnostics) and incubated in the presence of preimmune serum or HPH-2C immune serum overnight at 4°C. Antibody–protein complexes were precipitated with Protein A-agarose beads, washed extensively with lysis buffer, and eluted with SDS loading buffer. Coimmunoprecipitated 3XFLAG-tagged ING4 was detected by Western blot analysis using anti-FLAG M2 antibody (Sigma).
GST Pull-Downs. 35S-labeled proteins were prepared in the TNT Coupled Reticulocyte Lysate System (Promega). Lysates containing 35S-labeled proteins were incubated with GST fusion proteins immobilized on glutathione Sepharose 4B resin (Amersham Pharmacia) for 1 h at 4°C. After several washes with buffer containing 20 mM Tris·HCl (pH 7.5), 200 mM NaCl, 1 mM DTT, and 0.5% Nonidet P-40, bound proteins were eluted by boiling in SDS sample buffer and resolved by SDS/PAGE.
Subcellular Fractionation. HeLa cells were washed with ice-cold PBS and harvested by scraping. Cell pellets were resuspended in 10 mM Hepes-KOH (pH 7.5), incubated on ice for 10 min, and centrifuged at 2,400 × g at 4°C for 5 min. Pellets were washed with 25 mM Hepes-KOH (pH 7.5), 3 mM MgCl2, 1 mM DTT, 0.1% Nonidet P-40, and 1:200 Protease Inhibitor Mixture and resuspended in the same buffer. Cells were lysed by passage through a 25-gauge needle and centrifuged for 30 s. The cytosolic supernatant was removed, and the nuclear pellets were washed three times before resuspension in 20 mM Hepes-KOH (pH 7.5), 400 mM KCl, 1 mM DTT, and 10% glycerol and incubated at 4°C for 30 min. The samples were centrifuged at 200,000 × g for 45 min at 4°C to separate the extracted nuclear proteins from insoluble material.
[14C]-2-Oxoglutarate Decarboxylation Assay. HPH activity was measured by a modified [14C]-2-oxoglutarate decarboxylation assay derived from ref. 24. Briefly, a 1-ml reaction containing 3.0 μM peptide substrate (HIF-1α residues 556–574) was incubated with recombinant HPH-2C enzyme in the presence of 50 mM Tris·HCl (pH 8.0), 2 mg/ml BSA, 0.2 mg/ml catalase, 5 mM KCl, 1.5 mM MgCl2, 1.0 mM DTT, 64 μM [14C]-2-oxoglutarate [14.6 nCi/nmol specific activity (1 Ci = 37 GBq)], 50 μM ascorbate, and 20 μM FeSO4 in a sealed 15-ml tube. After a 1-h incubation at room temperature with gentle shaking, the pH of the solution was lowered to ≈2 with diluted HClO4. Released [14C]CO2 was captured by 3M Whatman paper saturated with 10 M NaOH and measured by scintillation counting.
RNA Interference. Cells were plated onto 24-well plates (3 × 104 cells per well) 16 h before transfection, and 200 nM small interfering RNA (siRNA) duplexes (Dharmacon, Lafayette, CO) were transfected by using Oligofectamine reagent (Invitrogen). Media were replaced 16–20 h after transfection. Cells were maintained either under normoxic conditions (20% O2) for 72 h after transfection or under normoxic conditions for 57 h followed by 12–15 h of hypoxia (1% O2). Total RNA preparation and Northern blot analysis were performed according to the methods of ref. 23. For transient transfections, HeLa cells were transfected with the 3XHRE-tk-Luc reporter construct 51 h after siRNA transfection by using Lipofectamine Plus (Invitrogen). siRNA duplexes were composed of the following oligonucleotides: ING4#1, UGAGGGACCUAGACCAAAGTT and CUUUGGUCUAGGUCCCUCATT; ING4#2, GAACGGAAGAAGAAAUAGATT and UCUAUUUCUUCUUCCGUUCTT; and GFP, GGCUACGUCCAGGAGCGCACC and UGCGCUCCUGGACGUAGCCUU.
Chromatin Immunoprecipitation Assay. Briefly, a HeLa cell line stably transfected with the 3XHRE-tk-Luc reporter construct was maintained under hypoxic conditions (1% O2) for 15 h. Crosslinking, lysate preparation, immunoprecipitations, and DNA purification were performed according to the experimental procedures described in ref. 25. A 227-bp region of the 3XHRE promoter was amplified by using the following primers: 5′-AGTGCAGGTGCCAGAACATT-3′ and 5′-CGGTAGGTCGAGAGGTCAGA-3′. PCR products were separated by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide. Relative band intensities were quantitated with quantity one (Bio-Rad) software.
Results
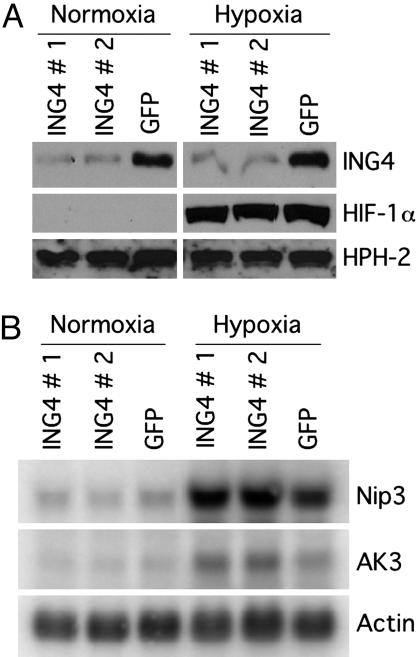
ING4 Represses Hypoxic Induction of HIF Target Genes. siRNAs were used to efficiently suppress expression of ING4 in HeLa cells as determined by Western blot analysis (Fig. 1A). The remaining protein signal could be due to low levels of unsuppressed ING4 or potential cross-reactivity with endogenous ING5. After siRNA treatment, cells were incubated under normoxic conditions or subjected to hypoxia to promote induction of HIF-responsive genes. When cells were treated with a control siRNA targeting GFP, two well characterized HIF target genes, Nip3 (23) and adenylate kinase 3 (AK3) (26), were both induced in response to hypoxia as assessed by Northern blot analysis (Fig. 1B). When ING4 expression was suppressed by either of two ING4-targeted siRNAs, the levels of both of these endogenous HIF target genes under hypoxic conditions typically increased an additional 2- to 3-fold relative to the GFP siRNA control-treated cells (Fig. 1B). These increases in HIF-responsive gene expression were not due to an increase in nuclear HIF-1α levels (Fig. 1A), suggesting that HIF activity, rather than HIF stability, was being suppressed by ING4. As expected, no effect of ING4 suppression was observed for these HIF target genes in conditions (normoxia) under which HIF-1α does not accumulate (Fig. 1A). Consistent with the ING4-dependent phenotype observed for NF-κB-responsive genes (19), there is an inverse relationship between ING4 levels and expression of HIF target genes in cells in which HIF has been induced by hypoxia.
Fig. 1.
siRNA suppression of ING4 enhances HIF target gene expression under hypoxic conditions. (A) Western blot analysis indicating relative protein levels of HPH-2, ING4, and nuclear HIF-1α under normoxic (20% O2) or hypoxic (1% O2) conditions for 12 h after treatment of HeLa cells with siRNA duplexes specific for ING4 (ING4#1 or ING4#2) or GFP control. HIF-2α was undetectable in Western blots of HeLa cell extracts. (B) Northern blot analysis of the endogenous HIF target genes Nip3 and AK3 after siRNA-mediated suppression of ING4. Actin mRNA levels are shown to confirm equivalent RNA loading. All results are representative of multiple experiments.
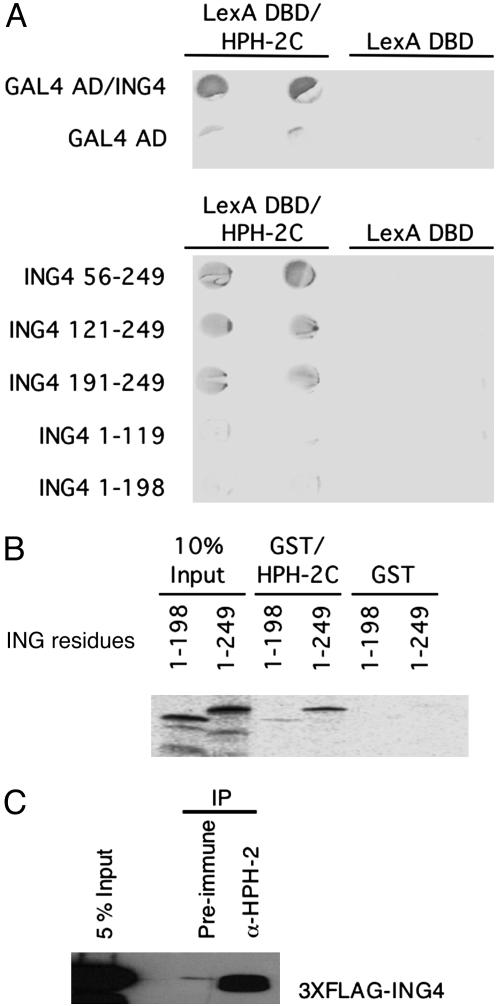
HPH-2 Interacts with ING4. Although we hypothesized that ING4 might interact directly with HIF, we were unable to detect such an interaction in a variety of assays (data not shown). However, we did detect an interaction between ING4 and a factor known to regulate HIF-α stability, HPH-2. An interaction between these two proteins was initially observed in a yeast two-hybrid screen in which the LexA DNA-binding domain (DBD) was fused to the catalytic hydroxylase domain of HPH-2 (HPH-2C). By itself, the LexA DBD/HPH-2C fusion protein was not capable of activating reporter genes driven by the LexA promoter (Fig. 2A). When the yeast cells were cotransformed with a construct expressing ING4 fused to the GAL4 activation domain, activation of the LexA-responsive LacZ reporter gene was observed, indicative of a positive interaction between HPH-2C and ING4 (Fig. 2A).
Fig. 2.
ING4 interacts with HPH-2 through its C-terminal PHD (residues 191–249). (A Upper) Yeasts were transformed with vectors encoding HPH-2C fused to the LexA DNA-binding domain (DBD) and ING4 fused to the GAL4 activation domain. Positive protein–protein interactions are denoted by LacZ reporter expression. Transformations were performed in duplicate. (Lower) Yeast two-hybrid analysis of HPH-2C interactions with N- and C-terminal truncations of ING4 are shown. (B) 35S-labeled full-length ING4 (residues 1–249) or ING4 lacking the PHD (residues 1–198) were incubated with immobilized GST or the GST/HPH-2C fusion protein. Bound 35S-labeled ING4 was visualized after SDS/PAGE. (C) Lysates from HeLa cells transfected with N-terminal 3XFLAG-tagged ING4 were immunoprecipitated with anti-HPH-2 serum or preimmune serum as a control. ING4 that coimmunoprecipitated with HPH-2 was detected by Western blot analysis using an anti-FLAG M2 antibody. IP, immunoprecipitate.
To identify the region of ING4 that associates with HPH-2C, a series of N- and C-terminal deletion constructs were assayed. As shown in Fig. 2A, residues 191–249 of ING4, encompassing the PHD, are sufficient to interact with HPH-2C. Almost no interaction was observed with the first 198 residues of ING4. Consistent with these results, HPH-2C was able to interact strongly with full-length ING4 but only weakly with ING4 lacking the PHD in in vitro GST pull-down assays (Fig. 2B). An interaction between HPH-2 and ING4 could also be detected in a coimmunoprecipitation assay (Fig. 2C).
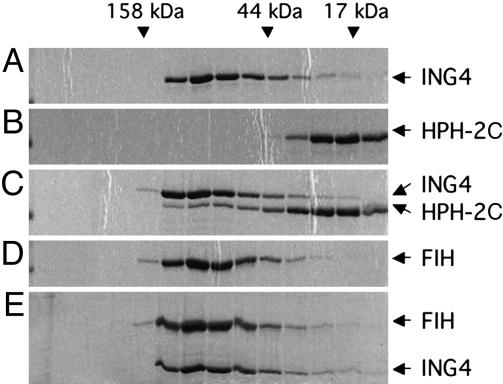
To further demonstrate a direct association between ING4 and HPH-2 in vitro, both proteins were expressed in bacteria and purified to near homogeneity. As shown in Fig. 3, HPH-2C migrated as predicted for a 27-kDa monomer on a size exclusion column (Fig. 3B), whereas the behavior of ING4 (predicted molecular mass of 29 kDa) was consistent with that of a multimeric species (Fig. 3A). When the two proteins were incubated together, they were observed to migrate together in larger complexes (Fig. 3C). In contrast, incubation of ING4 with another regulatory HIF hydroxylase, FIH-1, did not result in the appearance of a larger protein complex (Fig. 3E). Together, these independent lines of evidence demonstrate a direct association between ING4 and HPH-2.
Fig. 3.
Purified ING4 and HPH-2C directly associate in vitro. One hundred micrograms of purified recombinant ING4, HPH-2C, and/or FIH-1 proteins was incubated alone or together before resolution on a Superdex 200 10/30 column (Amersham Pharmacia Biotech). Eluted fractions were resolved by SDS/PAGE and visualized by Coomassie blue staining. The elution profile of protein standards is given at the top.
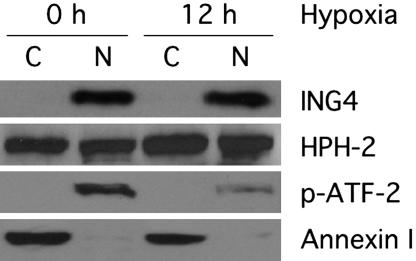
Both HPH-2 and ING4 Reside in the Nucleus. The results above indicate that HPH-2 directly interacts with ING4. In cells, however, ING family members are predicted to reside within the nucleus (27). However, localization studies performed with overexpressed GFP–HPH protein fusions indicate that HPH-2 resides primarily in the cytoplasm (28, 29). To determine whether endogenous ING4 and HPH-2 both reside within the same subcellular compartment, we examined their distribution in HeLa cell extracts (Fig. 4). After incubation, cells were lysed, and soluble cytosolic proteins were separated from insoluble cellular material, including intact nuclei, which was then subjected to high-salt extraction of nuclear proteins. As predicted, ING4 was found exclusively in the fraction containing extracted nuclear proteins. Contrary to the GFP–HPH-2 fusion results, endogenous HPH-2 was equally distributed in both fractions. These data provide evidence that a substantial portion of HPH-2 does reside in the nucleus, where it might promote O2-dependent degradation of nuclear HIF as well as associate with nuclear factors such as ING4.
Fig. 4.
Both ING4 and HPH-2 are found among salt-extracted nuclear proteins. HeLa cells were incubated under normoxic (20% O2) or hypoxic (1% O2) conditions for 12 h. Soluble cytoplasmic (C) and salt-extracted nuclear (N) proteins were separated and analyzed by Western blot analysis with antibodies raised against ING4 or HPH-2. Antibodies to p-ATF-2 or annexin I were used to assess the integrity of the nuclear and cytoplasmic samples, respectively.
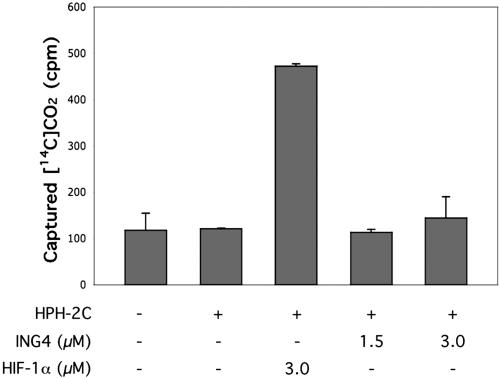
ING4 Is Not an HPH-2 Substrate. In addition to ING4, two-hybrid interactions can be observed between HPH-2C and the known HPH substrates, HIF-1α and HIF-2α (data not shown). Therefore, we speculated that the positive interaction could reflect an enzyme–substrate relationship between HPH-2 and ING4. Known targets for HPHs contain a conserved LXXLAP motif (30). Although this sequence is not present in any of the ING family members, mutations can be tolerated adjacent to the proline residue in peptide substrates (31). To test whether ING4 might be a hydroxylation substrate for HPH-2, we employed a hydroxylase assay using [14C]-2-oxoglutarate as a cosubstrate (24). [14C]-2-oxoglutarate is efficiently hydroxylated by HPH-2C only in the presence of a substrate such as the HIF-1α peptide. Hydroxylated [14C]-2-oxoglutarate then undergoes decarboxylation to release [14C]CO2, which can be captured and quantified. The levels of emitted [14C]CO2 therefore reflect the relative utilization of a putative polypeptide substrate in the hydroxylation reaction (Fig. 5). Unlike the results obtained with a HIF-1α substrate, incubation of HPH-2C with ING4 did not result in increased levels of [14C]CO2 production, indicating that ING4 is not a HPH-2 substrate.
Fig. 5.
ING4 is not an HPH-2 substrate in vitro. Substrate utilization by HPH-2C was assessed by [14C]CO2 generation in the presence of either a control HIF-1α peptide substrate or a recombinant ING4 protein. Assays were performed in triplicate.
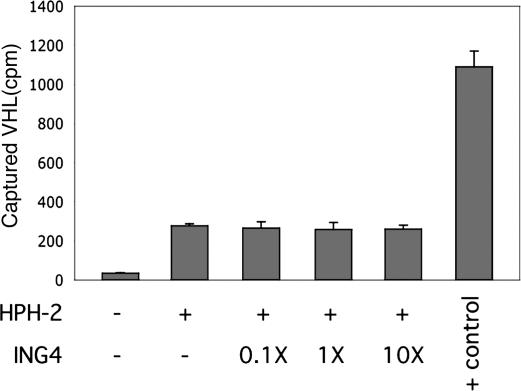
ING4 Does Not Affect the HPH-2 Activity or HIF Stability. Because ING4 did not appear to be a HPH-2 substrate nor to affect HPH-2 or HIF accumulation (Fig. 1A), we reasoned that ING4 association with HPH-2 might directly affect hydroxylase activity. To test this hypothesis, hydroxylation of a HIF peptide substrate by HPH-2C was followed in the presence of increasing amounts of purified ING4. The ability of HPH-2C to hydroxylate HIF-α in this assay was assessed by subsequent interaction of the peptide substrate with 35S-labeled VHL in a pull-down assay (6). As shown in Fig. 6, recombinant ING4 neither stimulated nor inhibited hydroxylation of the peptide substrate by HPH-2C. The lack of an effect of ING4 on hydroxylase activity in vitro is consistent with the lack of change in nuclear HIF-1α protein levels when ING4 levels are suppressed by siRNA (Fig. 1A).
Fig. 6.
ING4 does not affect HPH-2C activity in vitro. Hydroxylase activity of recombinant HPH-2C was measured in a 35S-labeled VHL pull-down assay in the presence of a HIF-1α peptide substrate and increasing amounts of recombinant ING4 (the amount of ING4 protein relative to HPH-2C is indicated). Reaction conditions were chosen to provide approximately half-maximal substrate hydroxylation to observe either stimulation or inhibition of hydroxylase activity. 35S-labeled VHL binding to a fully hydroxylated peptide (+ control) is shown as a reference. Assays were performed in triplicate.
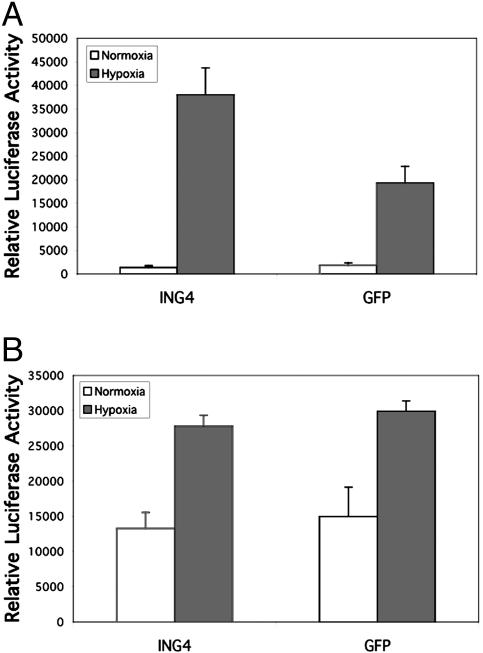
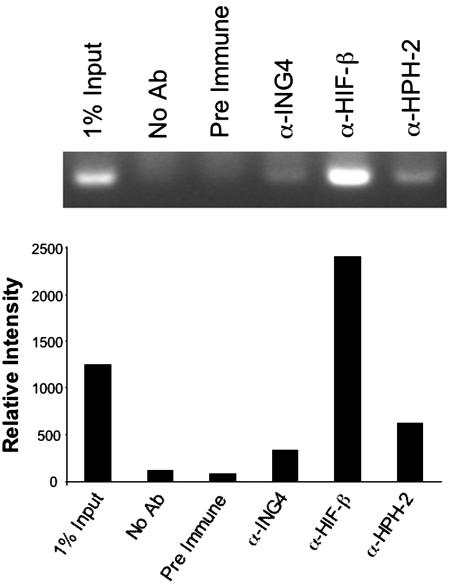
ING4 Suppresses HIF Activity in a Chromatin-Dependent Manner. Because other ING family members interact with chromatin-remodeling factors (21), we speculated that ING4 might likewise suppress HIF activity by recruiting factors that restrict accessibility of transcriptional machinery to the DNA. To begin to test this model, we compared the effect of siRNA-mediated ING4 suppression on the hypoxic induction of a HIF-responsive luciferase reporter driven by multiple HIF responsive elements, 3XHRE-tk-Luc (32), that was either stably (Fig. 7A) or transiently (Fig. 7B) transfected. After siRNA treatment, stably transfected cells were incubated under hypoxic conditions to promote the induction of the luciferase reporter gene. When cells were treated with a control GFP siRNA, relative luciferase activity was induced almost 10-fold in response to hypoxia (Fig. 7A). When ING4 expression was suppressed, luciferase reporter activity under hypoxic conditions typically increased an additional 2-fold relative to the GFP-targeted siRNA controls. This result is entirely consistent with the data presented in Fig. 1B for endogenous HIF target genes. Consistent with our hypothesis, ING4, HPH-2, and a HIF subunit were each found to associate with the 3XHRE element under hypoxic conditions in a chromatin immunoprecipitation assay (Fig. 8).
Fig. 7.
ING4 affects HIF activity in a chromatin-dependent manner. (A) Suppression of ING4 increases expression of a stably transfected HIF-driven luciferase reporter gene. A HeLa cell line stably expressing the HIF-responsive 3XHRE-tk-Luc reporter gene was transfected with siRNA duplexes specific for either ING4 (duplex ING4#1) or GFP followed by incubation under normoxic (20% O2) or hypoxic (1% O2) conditions for 15 h. Similar results were obtained with other independently isolated stably transfected HeLa cell lines (data not shown). (B) siRNA suppression of ING4 does not affect hypoxic induction of a transiently transfected HIF reporter gene. Wild-type HeLa cells were transfected with siRNA duplexes specific for either ING4 or a GFP control followed by the 3XHRE-tk-Luc HIF reporter construct and incubation under normoxic or hypoxic conditions. All assays were performed in triplicate, and the results are representative of multiple experiments.
Fig. 8.
ING4, HPH-2, and HIF associate with the HRE promoter. HeLa cells stably transfected with the 3XHRE-tk-Luc reporter construct were incubated under hypoxic (1% O2) conditions for 15 h followed by crosslinking, sonication, and immunoprecipitation using antibodies specific for ING4, HIF-β, or HPH-2. (Upper) Primers flanking the 3XHRE promoter element were used to amplify associated chromatin DNA by PCR. (Lower) Relative band intensities are indicated. No Ab, no antibody control; Pre Immune, preimmune serum control.
In contrast to the results observed with the stably transfected reporter, hypoxic induction of the transiently transfected reporter was unaffected by the ING4-targeted siRNA (Fig. 7B). These results are consistent with a model in which ING4 suppresses HIF activity through the recruitment of chromatin-remodeling factors, because such factors would not be expected to affect transcription from transiently transfected (nonchromosomal) DNA.
Discussion
The HIF-dependent hypoxic response pathway is present in virtually every cell in the body and participates in physiological processes as well as disease states such as ischemia and cancer (1, 2, 15). The discovery of the hydroxylases that mediate HIF-α stability and coactivator recruitment have provided a mechanism by which HIF function may be directly regulated by O2. Although the availability of O2 to serve as a hydroxylase substrate plays a major role in determining the status of HIF induction in a given cell (10), both HIF expression and hydroxylase function are likely subject to additional layers of regulation involving signaling pathways, post-translational modification, feedback loops, protein–protein interactions, and protein localization (1). The extensive and dynamic regulation of HIF reflects not only its critical biological role but also the need to fine-tune HIF induction under physiologically relevant O2 concentrations encountered under “normoxic” conditions in which the HIF transcription factors are often partially induced. Although the relationship between O2 availability and hydroxylase activity may serve as a general determinant of HIF induction, multiple layers of regulation are likely required to subtly adapt HIF function in a variety of dynamic microenvironments.
The physiological importance and potential therapeutic utility of the hypoxic response pathway have driven the search for additional regulatory components. To that end, we investigated the candidate tumor suppressor ING4. An inverse relationship between ING4 expression and tumor growth and vascular volume in human glioblastomas and mouse xenograft models has been observed. Whereas two-hybrid and coimmunoprecipitation experiments have revealed an interaction between ING4 and NF-κB (19), the precise mechanism by which ING4 suppresses NF-κB function has not been determined. Although the HIF transcription factors are also frequently induced in many cancers, where they, too, promote tumorigenesis, in the aforementioned cell culture studies no change in expression levels of proangiogenic HIF target genes (i.e., VEGF) was observed after manipulation of ING4 (19). However, it must be noted that these studies were not performed under hypoxic conditions where one would expect to see HIF induction. Suspecting that ING4 might also suppress HIF-mediated effects on tumor growth and angiogenesis, we examined the effects of ING4 on the levels of HIF target gene induction under both normoxic and hypoxic conditions.
Consistent with our hypothesis, suppression of ING4 in HeLa cells led to a significant and reproducible increase in the induction of endogenous HIF target genes under hypoxic conditions. A similar result was obtained by using a stably transfected reporter gene driven by a minimal HIF-responsive promoter, supporting the contention that the effects of ING4 were mediated through HIF rather than other transcriptional regulators. Although transcriptional targets of NF-κB have been reported to promote HIF induction by increasing HIF-1α stability (33), we do not believe that ING4 indirectly regulates HIF through NF-κB by such a mechanism. In our studies, no change in HIF-1α stability was observed after ING4 suppression, and the effects of ING4 suppression were most pronounced under hypoxic conditions, unlike the effects of NF-κB on HIF, which have been observed only under normoxia (33).
Although ING4 seems to be directly recruited to NF-κB, we were unable to detect a direct association between ING4 and HIF. Instead, ING4 associated directly with a known HIF regulator, HPH-2, in multiple assays. ING4 was not a substrate for HPH-2, nor did it affect HPH activity like the recently reported HPH-interacting factor OS-9 (34). Instead, this study provides evidence that a substantial fraction of HPH-2 resides in the nucleus where it can both mediate HIF degradation upon reoxygenation as well as associate with ING4 to affect HIF activity.
ING family members have been shown to promote chromatin remodeling through the recruitment of factors that promote or suppress transcriptional activation (21). When O2 levels are high, HPH-2 hydroxylates proline residues within the O2-dependent degradation domain and is subsequently displaced by the product of VHL (pVHL), leading to rapid HIF-α degradation and perhaps transcriptional repression through pVHL-mediated recruitment of histone deacetylases (12). When O2 levels are low, HIF-α recognition by HPH-2 may instead promote recruitment of chromatin-remodeling factors to the promoter regions of HIF target genes via ING4. In the case of HIF-responsive genes in HeLa cells, these putative ING4-associated factors would seem to modestly suppress transcription. This could be due to local changes in the histone acetylation state or perhaps to changes in the acetylation state of nonhistone proteins including HIF itself. In support of a chromatin-remodeling model, suppression of ING4 did not affect the expression of a transiently transfected HIF reporter that is not subject to regulation as a function of chromatin structure.
We have thus far focused our attention on ING4 association with HPH-2. Given the high conservation among the C-terminal regions of the three HPH proteins, it is possible that ING4 could also interact with the other members of the HPH family. Likewise, because the PHD is highly conserved among members of the ING family, HPHs may also interact with other ING proteins that could in turn recruit different sets of proteins to HIF and HIF-dependent promoters. Therefore, the precise consequence(s) of ING association with HPHs requires additional study and will likely be complicated by differences in function, protein recruitment, and expression patterns among the ING and HPH family members.
Together, these data demonstrate that the candidate tumor suppressor protein ING4 represses the function of other factors mediating tumor growth and angiogenesis in addition to NF-κB. Furthermore, we describe a direct interaction between ING4 and a HIF regulatory factor, suggesting a mechanism for ING4 recruitment to HIF as well as an additional role for the HIF prolyl hydroxylases in mediating HIF function under hypoxic conditions. Lastly, we provide evidence that the repressive effects of ING4 depend on the context of transcribed target genes, implying that ING4's mode of action may depend on its ability to recruit chromatin-remodeling factors. Although candidate chromatin-remodeling factors have been proposed to interact with ING4 (35), additional studies will be required to identify the physiological complexes in which ING4 resides.
Acknowledgments
We thank B. Horazdovsky, C. Dann, P. Erbel, and J. Rutter for helpful advice. This work was supported by a Burroughs Wellcome Fund Career Award, the University of Texas Southwestern President's Research Council, the Robert A. Welch Foundation, and an American Cancer Society Young Investigator Award (to R.K.B.) and was conducted in a facility constructed with support from Research Facilities Improvement Program Grant C06 RR-15437. R.K.B. is the Michael L. Rosenberg Scholar in Medical Research.
Abbreviations: HIF, hypoxia inducible factor; VHL, von Hippel–Lindau tumor suppressor gene; HPH, HIF prolyl hydroxylase; FIH-1, factor inhibiting HIF-1; ING, inhibitor of growth family member; PHD, plant homeodomain; siRNA, short interfering RNA.
References
- 1.Wenger, R. H. (2002) FASEB J. 16, 1151–1162. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell, P. H. & Ratcliffe, P. J. (2002) Semin. Cell Dev. Biol. 13, 29–37. [DOI] [PubMed] [Google Scholar]
- 3.Bruick, R. K. (2003) Genes Dev. 17, 2614–2623. [DOI] [PubMed] [Google Scholar]
- 4.Huang, L. E., Gu, J., Schau, M. & Bunn, H. F. (1998) Proc. Natl. Acad. Sci. USA 95, 7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epstein, A. C. R., Gleadle, J. M., McNeil, L. A., Hewitson, K. S., O'Rourke, J., Mole, D. R., Mukherji, M., Metzen, E., Wilson, M. I., Dhanda, A., et al. (2001) Cell 107, 43–54. [DOI] [PubMed] [Google Scholar]
- 6.Bruick, R. K. & McKnight, S. L. (2001) Science 294, 1337–1340. [DOI] [PubMed] [Google Scholar]
- 7.Ivan, M., Haberberger, T., Gervasi, D. C., Michelson, K. S., Günzler, V., Kondo, K., Yang, H., Sorokina, I., Conaway, R. C., Conaway, J. W. & Kaelin, W. G. (2002) Proc. Natl. Acad. Sci. USA 99, 13459–13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., M., A. J., Lane, W. S. & Kaelin, W. G. (2001) Science 292, 464–468. [DOI] [PubMed] [Google Scholar]
- 9.Jaakkola, P., Mole, D. R., Tian, Y.-M., Wilson, M. I., Gielbert, J., Gaskell, S. J., von Kriegsheim, A., Hebestreit, H. F., Mukherji, M., Schofield, C. J., et al. (2001) Science 292, 468–472. [DOI] [PubMed] [Google Scholar]
- 10.Hirsila, M., Koivunen, P., Gunzler, V., Kivirikko, K. I. & Myllyharju, J. (2003) J. Biol. Chem. 278, 30772–30780. [DOI] [PubMed] [Google Scholar]
- 11.Lando, D., Peet, D. J., Whelan, D. A., Gorman, J. J. & Whitelaw, M. L. (2002) Science 295, 858–861. [DOI] [PubMed] [Google Scholar]
- 12.Mahon, P. C., Hirota, K. & Semenza, G. L. (2001) Genes Dev. 15, 2675–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lando, D., Peet, D. J., Gorman, J. J., Whelan, D. A., Whitelaw, M. L. & Bruick, R. K. (2002) Genes Dev. 16, 1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hewitson, K. S., McNeill, L. A., Riordan, M. V., Tian, Y. M., Bullock, A. N., Welford, R. W., Elkins, J. M., Oldham, N. J., Bhattacharya, S., Gleadle, J. M., et al. (2002) J. Biol. Chem. 277, 26351–26355. [DOI] [PubMed] [Google Scholar]
- 15.Semenza, G. L. (2003) Nat. Rev. Cancer 3, 721–732. [DOI] [PubMed] [Google Scholar]
- 16.Hochachka, P. W. (1999) Proc. Natl. Acad. Sci. USA 96, 12233–12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Csete, M., Walikonis, J., Slawny, N., Wei, Y., Korsnes, S., Doyle, J. C. & Wold, B. (2001) J. Cell. Physiol. 189, 189–196. [DOI] [PubMed] [Google Scholar]
- 18.Stroka, D. M., Burkhardt, T., Desbaillets, I., Wenger, R. H., Neil, D. A., Bauer, C., Gassmann, M. & Candinas, D. (2001) FASEB J. 15, 2445–2453. [DOI] [PubMed] [Google Scholar]
- 19.Garkavtsev, I., Kozin, S. V., Chernova, O., Xu, L., Winkler, F., Brown, E., Barnett, G. H. & Jain, R. K. (2004) Nature 428, 328–332. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S., Chin, K., Gray, J. W. & Bishop, J. M. (2004) Proc. Natl. Acad. Sci. USA 101, 16251–16256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campos, E. I., Chin, M. Y., Kuo, W. H. & Li, G. (2004) Cell. Mol. Life Sci. 61, 2597–2613. [DOI] [PubMed] [Google Scholar]
- 22.Sheffield, P., Garrard, S. & Derewenda, Z. (1999) Protein Expression Purif. 15, 34–39. [DOI] [PubMed] [Google Scholar]
- 23.Bruick, R. K. (2000) Proc. Natl. Acad. Sci. USA 97, 9082–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kivirikko, K. I. & Myllyla, R. (1982) Methods Enzymol. 82, 245–304. [DOI] [PubMed] [Google Scholar]
- 25.Wang, F., Zhang, R., Beischlag, T. V., Muchardt, C., Yaniv, M. & Hankinson, O. (2004) J. Biol. Chem. 279, 46733–46741. [DOI] [PubMed] [Google Scholar]
- 26.O'Rourke, J. F., Pugh, C. W., Bartlett, S. M. & Ratcliffe, P. J. (1996) Eur. J. Biochem. 241, 403–410. [DOI] [PubMed] [Google Scholar]
- 27.Feng, X., Hara, Y. & Riabowol, K. (2002) Trends Cell Biol. 12, 532–538. [DOI] [PubMed] [Google Scholar]
- 28.Huang, J., Zhao, Q., Mooney, S. M. & Lee, F. S. (2002) J. Biol. Chem. 277, 39792–39800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzen, E., Berchner-Pfannschmidt, U., Stengel, P., Marxsen, J. H., Stolze, I., Klinger, M., Huang, W. Q., Wotzlaw, C., Hellwig-Burgel, T., Jelkmann, W., et al. (2003) J. Cell Sci. 116, 1319–1326. [DOI] [PubMed] [Google Scholar]
- 30.Masson, N., Willam, C., Maxwell, P. H., Pugh, C. W. & Ratcliffe, P. J. (2001) EMBO J. 20, 5197–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, J., Zhao, Q., Mooney, S. M. & Lee, F. S. (2002) J. Biol. Chem. 277, 39792–39800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian, H., McKnight, S. L. & Russell, D. W. (1997) Genes Dev. 11, 72–82. [DOI] [PubMed] [Google Scholar]
- 33.Jung, Y.-J., Isaacs, J. S., Lee, S., Trepel, J. & Neckers, L. (2003) FASEB J. 17, 2115–2117. [DOI] [PubMed] [Google Scholar]
- 34.Baek, J. H., Mahon, P. C., Oh, J., Kelly, B., Krishnamachary, B., Pearson, M., Chan, D. A., Giaccia, A. J. & Semenza, G. L. (2005) Mol. Cell 17, 503–512. [DOI] [PubMed] [Google Scholar]
- 35.Shiseki, M., Nagashima, M., Pedeux, R. M., Kitahama-Shiseki, M., Miura, K., Okamura, S., Onogi, H., Higashimoto, Y., Appella, E., Yokota, J. & Harris, C. C. (2003) Cancer Res. 63, 2373–2378. [PubMed] [Google Scholar]