ABSTRACT
Previous systematic literature reviews of rotavirus genotype circulation in Europe and the Middle East are limited because they do not include country-specific prevalence data. This study documents country-specific evidence on the prevalence of rotavirus genotypes in Europe and the Middle East to enable more precise epidemiological modeling and contribute to the evidence-base about circulating rotavirus genotypes in the post-vaccination era. This study systematically searched PubMed, Embase and Scopus for all empirical epidemiological studies that presented genotype-specific surveillance data for countries in Europe and the Middle East published between 2006 and 2021. The STROBE checklist was used to assess the quality of included studies. Proportional meta-analysis was conducted using the generic inverse variance method with arcsine transformation and generalized linear-mixed models to summarize genotype prevalence. Our analysis estimated the genotype prevalence by country across three date categories corresponding with rotavirus seasons: 2006–2010, 2011–2015, 2016–2021. A total of 7601 deduplicated papers were identified of which 88 studies were included in the final review. Rotavirus genotypes exhibited significant variability across regions and time periods, with G1P[8], G2P[4], G3P[8], G4P[8], G9P[8], and, to a lesser extent G12P[8], being the most prevalent genotypes through different regions and time-periods. Uncommon genotypes included G3P[9] in Poland, G2P[6] in Iraq, G4P[4] in Qatar, and G9P[4] as reported by the European Rotavirus Network. There was high genotype diversity with routinely identified genotypes being G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8]; there was high variability across time periods and regions. Continued surveillance at the national and regional levels is relevant to support further research and inform public health decision-making.
KEYWORDS: Rotavirus, vaccination, epidemiology, child health, diarrhoea
SUMMARY
This study synthesizes data from rotavirus surveillance studies to characterize genotype-specific prevalence of rotavirus in Europe and the Middle East following the licensure of rotavirus vaccines in 2006. In line with previous pan-European studies, results highlight the lack of a single dominant genotype across this time period. There was high genotype diversity with G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8] being the most commonly identified genotypes through different regions and time-periods.
Background
Rotavirus infections are one of the world’s leading causes of severe diarrhea and dehydration among children under five years of age.1 Transmitted through an oral-fecal route, rotaviruses are highly contagious and estimated to infect nearly all children by the age of five.1
Rotavirus contains a genome composed of 11 segments of double-stranded RNA that encode six structural proteins (VP1–VP4 and VP6–VP7) and six nonstructural proteins (NSP1–NSP6).2,3 Previous studies estimating the genotype-specific prevalence of rotavirus have mainly used glycoprotein (G) and protease-sensitive protein (P) type data.4–6 These genotype classifications are largely based on two outer viral proteins: VP7 and VP4, respectively. G genotypes refer to the glycosylated VP7 protein on the virus’s surface whereas P genotypes refer to the protease-sensitive VP4 spike protein on the virus’s surface.5
Since 2006, two vaccines, Rotarix™ (RV1) and RotaTeq™ (RV5), have been available globally. Rotarix™, a monovalent vaccine developed by GSK Biologicals, Belgium, is derived from a single common genotype of human rotavirus and contains G1P[8]. A full course of Rotarix™ consists of two doses given orally, 4 weeks apart, between 6 and 24 weeks of age.7 RotaTeq™ is a pentavalent vaccine developed by Merck & Co., Inc., Kenilworth, NJ, USA, containing five human bovine reassortants G1, G2, G3, G4 and P[8].8 RotaTeq™ is administered orally and consists of three doses between the ages of 6 and 32 weeks.7
A systematic literature review performed in the early post-licensure period synthesized global longitudinal data from 2006 to 2010, and suggested that, in this period, genotype prevalence data did not show any consistent pattern indicative of selection pressure resulting from vaccine use and that six genotype combinations were mostly responsible for human rotavirus infections globally (G1P[8], G2P[4], G3P[8], G4P[8], G9P[8] and G12P[8]).5 A more recent review showed a transient increase of G2P[4] following recent vaccine introduction.6 However, the latter did not present results by country.
Since the last country-specific systematic review in 2010,5 genotyping studies have been published. In this study, we synthesized available evidence using meta-analysis to provide an updated view of rotavirus genotype circulation in the post-licensure period (2006–2021) by country and region. This systematic literature review and meta-analysis provides the latest rotavirus genotype circulation by country in Europe and the Middle East to enable more precise epidemiological modeling and contribute to the evidence-base about circulating rotavirus genotypes in the post-vaccination era.
Methods
This systematic literature review was undertaken according to the principles of systematic reviews established in the Cochrane Handbook and guidance document published by the Center for Reviews and Dissemination (CRD) of York University, United Kingdom. We used the PRISMA checklist and guidelines for systematic reviews to report results.9
Data sources and searches
We systematically searched PubMed, Embase and Scopus for all empirical epidemiological studies presenting genotype-specific prevalence of rotavirus in countries and regions after rotavirus vaccine licensure in 2006 (search strategies in Appendix I). We also searched Google and Google Scholar to identify gray literature, such as surveillance reports.
The population of interest encompassed all people living in Europe and the Middle East (Table 1). The primary outcome was the prevalence of circulating genotypes after the licensure of rotavirus vaccines. Secondary outcomes included the prevalence of genotypes by region. Studies were eligible for inclusion if they were empirical epidemiological studies, including longitudinal studies, and cross-sectional studies from multiple time points within the same setting. We included all studies meeting these inclusion criteria and that were published between January 2006 and up to 25 August 2021. Records in languages other than English were eligible for inclusion and were translated using Google Translate.10
Table 1.
Inclusion and exclusion criteria.
| PICOTS | Descriptions |
|---|---|
| Inclusion criteria | |
| Population(s) | All populations in Europe and the Middle East |
| Interventions | All rotavirus vaccines |
| Comparators | Not applicable |
| Outcomes |
|
| Timing | All studies with data collection period between 2006 and August 2021 |
| Study design | Empirical epidemiological studies (sample serology testing) |
| Exclusion criteria | |
| Any study with the following characteristic | Studies published before 2006 or that did not contain extractable post-2006 data |
| Less than 30 positive rotavirus samples | |
| Countries not in Europe and the Middle East | |
| Studies lacking at least six consecutive months of surveillance or studies with two cross-sections fewer than not 6 months apart | |
Studies were excluded if their study design deviated from empirical epidemiological research, or they did not provide surveillance data on observed genotype circulation. Additionally, we excluded editorials, published abstracts, conference proceedings, and studies that were inaccessible. For feasibility, and to select the most representative studies, we excluded studies lacking at least six consecutive months of surveillance. To maximize the number of studies included while maintaining an acceptable statistical sample,11 we excluded studies with fewer than 30 rotavirus positive samples. Studies published before 2006 or studies that did not contain extractable post-2006 data were also excluded.
Study selection
An initial screening of titles and abstracts was performed by one reviewer to eliminate studies that did not mention rotavirus genotype prevalence. To minimize bias, two reviewers subsequently evaluated the titles and abstracts of all studies in accordance with the established inclusion and exclusion criteria. Any titles and abstracts that did not clearly meet our exclusion criteria were considered for a full-text review, which was carried out independently by two reviewers. Any discrepancies were resolved through discussion between the two reviewers.
Data extraction
A data extraction form was created to collate relevant data, including circulating genotypes and age distribution data. The form was tested by two researchers on one study to confirm that it effectively captured the relevant data. Subsequently, one reviewer conducted the data extraction, and a second reviewer independently cross-checked the extracted data. Any discrepancies were resolved through discussion.
Quality assessment of included studies
The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) checklist was used to assess the quality of the observational studies included.12 All studies that met the inclusion criteria were reviewed with the STROBE checklist for quality appraisal by one reviewer. Each completed STROBE checklist was further reviewed by a second independent reviewer.
Analyses
We examined rotavirus prevalence in five regions. European regions were categorized as Western Europe, Eastern Europe, Northern Europe, Southern Europe, as defined by the United Nations Statistical Division.13 Countries belonging to the Middle East were defined as per the United Nations World Tourism Organization14 (see Appendix II). To facilitate the comparison of data while maintaining a level of granularity, we summarized the genotype-specific prevalence of rotavirus both at the country and regional levels across three time spans: 2006–2010, 2011–2015, 2016–2021. For studies with two overlapping time periods, we categorized the studies based on rotavirus seasons. For example, Spring 2006–Summer 2010; Autumn 2010–Winter 2015; Spring 2015–25 August 2021.
For each available combination of country (or region), time period, and rotavirus genotype, we conducted meta-analyses to estimate overall prevalence of the genotype. If there was only one prevalence record available, we calculated the overall prevalence based on that single record. For each study, we required information on the total number of samples tested and the number of samples positive for each genotype. The individual yearly estimates of genotype prevalence were incorporated into the meta-analysis. In cases where a study lacked year- or season-specific data and had data overlapping with predefined periods, we assigned the study to the period with the most overlapping years of data collection. To avoid double counting, because investigators may submit national results to regional networks and both may be summarized independently, we assessed multi-country pan-European studies and reports using a narrative synthesis and did not include the data in our meta-analysis. Additionally, studies that reported only G or P types were reported narratively and not included in our meta-analysis.
We conducted proportional meta-analysis using random effects models because we assumed that within each country, region, and time-period the true underlying prevalence will differ between studies conducted in different populations and settings.15 To conduct the meta-analysis, we applied the arcsine transformation to each proportion (p) (arcsin(√p)), used the generic inverse variance method to pool the transformed proportions and back-transformed the pooled estimate to the original scale.16 We estimated the between-study variance using the restricted maximum-likelihood estimator,17 and calculated the 95% confidence intervals for the random effects estimate using the DerSimonian and Laird method.18 To assess the sensitivity of our method, we also conducted a meta-analysis using generalized linear-mixed models,16 estimating the between-study variance using the maximum-likelihood estimator and again calculating the 95% confidence intervals for the random effects estimate using the DerSimonian and Laird method15,17,18 Heterogeneity was evaluated using the I-squared (I2) statistic. We interpreted the I2 statistics following guidelines in the Cochrane Handbook with 0%–40%, 30%–60%, 50%–90% and 75%–100% suggesting unimportant, moderate, substantial and considerable heterogeneity respectively.9 All analyses were conducted using R version 4.2.319 and the meta package version 6.5-0.20
Results
Search results
The search of electronic databases identified 7,601 deduplicated papers (Figure 1). We excluded 7,479 studies during title and abstract screening because they did not include data in the country or region of interest or were not empirical epidemiological studies reporting observed genotype circulation data. In total, 122 records appeared to meet the inclusion criteria during title and abstract screening of which 119 papers were able to be retrieved to proceed with full text screening. Three studies could not be retrieved through a review of databases and online search engines and were subsequently excluded. Other records were excluded following full text screening because: 1) small sample size (<30) (n = 8); 2) not empirical epidemiological studies reporting observed genotype circulation data (n = 8); 3) contained data that could not be extracted post 2006 (n = 7); 4) not a full paper (n = 3); 5) wrong setting (n = 3); 6) wrong study design (n = 2); 7) less than six months consecutive data (n = 1). In total, eighty-seven empirical epidemiological studies met the inclusion criteria and were included in the review in addition to one report identified as gray literature (see Appendix III for a visual representation on data availability). For ease of readability, we present meta-analysis results using the generic inverse variance method with arcsine transformation. Results of this method were largely consistent with meta-analysis results using the generalized linear-mixed models, which are available upon request.
Figure 1.
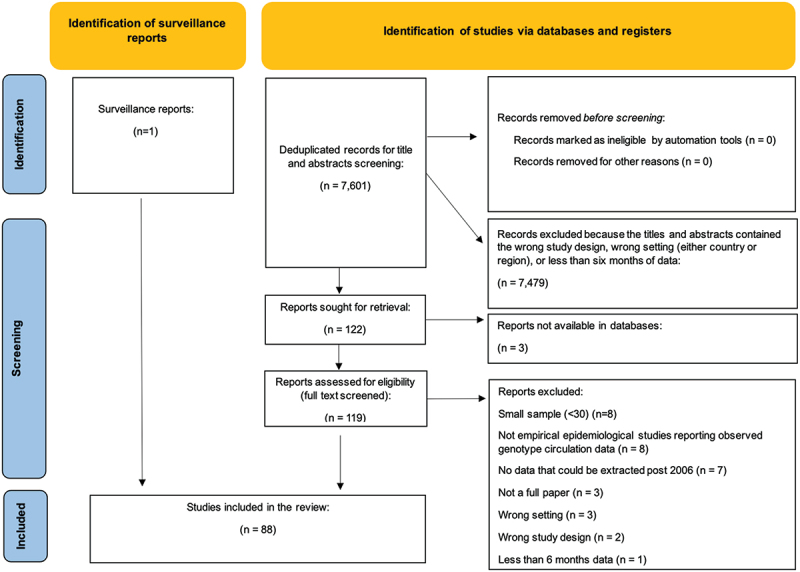
PRISMA flow diagram.
Eastern Europe
For the period 2006–2010, genotype prevalence data were available from 3,114 positive rotavirus samples, taken from 20 records from eight studies, across seven countries (Figure 2a).21–28 Based on within-country meta-analyses, the most common rotavirus genotypes in the region were G4P[8] and G1P[8], representing 30% (95% CI: 17%, 45%) and 27% (95% CI: 17%, 38%) of circulating genotypes, respectively.
Figure 2.
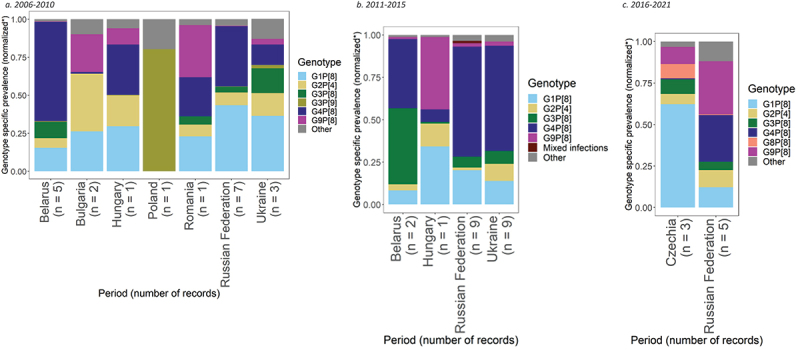
Prevalence of the most common rotavirus genotypes in Eastern Europe.
G4P[8] was the most prevalent genotype in Belarus and Hungary, representing 58% (95% CI: 48%, 68%) and 33% (95% CI: 29%, 38%) of genotypes, respectively. In both countries G1P[8] was the second most common genotype, with a prevalence of 14% (95% CI: 8%, 21%) in Belarus and 30% (95% CI: 26%, 34%) in Hungary. G1P[8] was the most common genotype in the Russian Federation with 42% (95% CI: 20%, 65%), followed by G4P[8] with 38% (95% CI: 15%, 65%). G1P[8] was also the most dominant in Ukraine with 37% (95% CI: 20%, 56%), followed by G3P[8] with 17% (95% CI: 0%, 50%). G2P[4] was the most prevalent genotype in Bulgaria 36% (95% CI: 18%, 57%), followed by G1P[8] with 25% (95% CI: 3%, 59%). In Poland, G3P[9] was the predominant genotype at 80% (95% CI: 67%, 90%). G9P[8] was most common in Romania with 34% (95% CI: 30%, 39%), followed by G4P[8] with 26% (95% CI: 22%, 30%).
For the period 2011–2015, genotype prevalence data were available from 2,887 positive rotavirus samples available from 21 records from eight studies, across four countries (Figure 2b).21,22,25,26,29–32 Based on this data, the most common rotavirus genotype in the region was G4P[8], representing 56% of circulating genotypes (95% CI: 46%, 66%), followed by G1P[8], with 16% prevalence (95% CI: 11%, 23%).
G4P[8] was the most prevalent genotype in the Russian Federation and Ukraine with 63% (95% CI: 50%, 76%) and 59% (95% CI: 47%, 70%) prevalence, respectively. G1P[8] was the second most common genotype in both countries, representing 20% (95% CI: 9%, 33%) of genotypes in the Russian Federation and 13% (95% CI: 7%, 21%) in Ukraine. In Belarus, G3P[8] was the most prevalent genotype with 43% (95% CI: 7%, 85%), followed by G4P[8] with 39% (95% CI: 12%, 71%). In Hungary G9P[8] was the most dominant genotype with 43% (95% CI: 38%, 48%), followed by G1P[8] with 34% (95% CI: 30%, 39%).
Lastly, for the period 2016–2021, genotype prevalence data were available from two countries, representing 3,663 positive rotavirus samples available from 8 records from six studies (Figure 2c).30,33–37 Based on this data, the most common rotavirus genotypes in the region were G1P[8] and G9P[8], showing similar regional prevalences of 25% (95% CI: 10%, 44%) and 21% (95% CI: 9%, 37%), respectively. G1P[8] was the most dominant genotype in Czechia with 57% (95% CI: 38%, 75%), followed by G9P[8] with 10% (95% CI: 2%, 21%). G9P[8] was the most dominant genotype in the Russian Federation with 30% (95% CI: 13%, 51%), followed by G4P[8] with 27% prevalence, respectively (95% CI: 11%, 46%).
The Russian Federation was the only country included in meta-analyses across all three time periods. During these periods the dominant genotype varied. In 2006–2010, it was G1P[8] and G4P[8] with 42% and 38% prevalence, respectively. From 2011–2015, G4P[8] emerged as the dominant genotype with 63% prevalence, followed by G1P[8] with 20%. While from 2016–2021, G9P[8] emerged as the most prevalent genotype with 30% prevalence, followed by G4P[8] with 27%.
Northern Europe
For the period 2006–2010, genotype prevalence data were available from 1,525 positive rotavirus samples, taken from 11 records from six studies, across four countries (Figure 3a).38–43 Based on this data, the most common rotavirus genotypes in the region were G1P[8] and G2P[4], representing 49% (95% CI: 30%, 69%) and 13% (95% CI: 3%, 28%) of genotypes, respectively.
Figure 3.
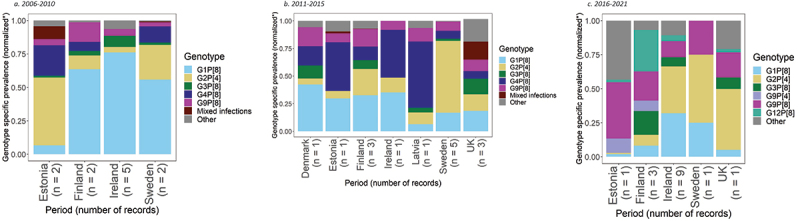
Prevalence of the most common rotavirus genotypes in Northern Europe.
G1P[8] had the highest prevalence in three countries, with 58% prevalence (95% CI: 24%, 89%) in Finland, 68% (95% CI: 51%, 82%) in Ireland and 49% (95% CI: 3%, 97%) in Sweden. G2P[4] was the second most common genotype in Sweden with a prevalence of 23% (95% CI: 0%, 84%). In Estonia, G2P[4] was the most common genotype with a prevalence of 43% (95% CI: 1%, 94%), followed by G4P[8] with 19% (95% CI: 0%, 68%).
For the period 2011–2015, genotype prevalence data were available from 4,232 positive rotavirus samples, taken from 15 records from eight studies, across eight countries (Figure 3b).39,44–50 Based on this data, the most common rotavirus genotypes across all eight countries were G2P[4] and G1P[8], with a pooled prevalence across all analyzed samples of 25% (95% CI: 14%, 38%) and 23% (95% CI: 15%, 31%), respectively.
G1P[8] was the most prevalent genotype in Denmark and Finland, with prevalences of 42% (95% CI: 39%, 46%) and 30% (95% CI: 24%, 35%), respectively. In Denmark, the second most common genotypes were G4P[8] and G9P[8], each with a prevalence of 17% (95% CI: 15%, 20%). In Finland, G2P[4] was the second most common genotype with a prevalence of 22% (95% CI: 17%, 26%). G4P[8] was the most prevalent genotype with a prevalence of 44% (95% CI: 39%, 50%) in Estonia, 43% (95% CI: 27%, 61%) in Ireland, and 60% (95% CI: 56%, 65%) in Latvia. The second most prevalent genotype in both Estonia and Ireland was G1P[8] with prevalences of 30% (95% CI: 25%, 35%) and 35% (95% CI: 20%, 53%), respectively. In the United Kingdom of Great Britain and Northern Ireland, G1P[8] and G2P[4] were the most prevalent genotype with 20% (95% CI: 8%, 35%) and 16% (95% CI: 4%, 34%) prevalence, respectively. In Sweden, G2P[4] was the most prevalent genotype with a prevalence of 56% (95% CI: 34%, 77%).
From 2016 to 2021, genotype prevalence data were available from 1,810 positive rotavirus samples, taken from 15 records from eight studies, across five countries (Figure 3c).39,44–46,48,51–53 Based on this data, the most common rotavirus genotypes in the region were G2P[4] and G1P[8], with a prevalence of 23% (95% CI: 12%, 35%) and 18% (95% CI: 8%, 30%), respectively.
G2P[4] was the most common genotype in Ireland, the United Kingdom of Great Britain and Northern Ireland and Sweden, with prevalences of 29% (95% CI: 15%, 46%), 45% (95% CI: 41%, 48%) and 56% (95% CI: 34%, 77%), respectively. In Ireland and Sweden, the second most common genotype was G1P[8] with a prevalence of 27% (95% CI: 12%, 46%) and 14% (95% CI: 2%, 36%), respectively. In the United Kingdom of Great Britain and Northern Ireland the second most common genotype was G9P[8] with a prevalence of 18% (95% CI: 16%, 21%). In Estonia, G9P[8] was the most common genotype with 41% (95% CI: 32%, 51%). In Finland, G12P[8] was the most prevalent genotype with 28% (95% CI: 7%, 55%), followed by G9P[8] with a prevalence of 20% (95% CI: 15%, 25%).
Three countries, Estonia, Ireland and Finland, were included in meta-analyses across all three time periods. In Estonia, G2P[4] was the dominant genotype from 2006 to 2010 with 43% prevalence, followed by G4P[8] with 19% prevalence. From 2011–2015, G4P[8] emerged as the dominant genotype at 44% prevalence followed by G1P[8] with 30% prevalence. Between 2016–2021, G9P[8] was the most common genotype, with 41%, followed by G4P[4] with 12% prevalence. In Finland, G1P[8] was the most prevalent genotype from 2006 to 2010 at 58% prevalence. Although remaining the most prevalent, the dominance of G1P[8] decreased during 2011–2015 to 30%, while and G2P[4] increased to 22% as the second most prevalent genotype. During 2016–2021, G12P[8] was the most observed genotype at 28%, followed by G9P[8] at 20%. In Ireland, G1P[8] was the most prevalent genotype from 2006 to 2010, with a prevalence of 68%. From 2011 to 2015, G1P[8] dropped to second most prevalent at 35%, behind G4P[8] with a 43% prevalence. From 2016–2021, G2P[4] led with a 29% prevalence.
Southern Europe
For the period 2006–2010, genotype prevalence data were available from 5,628 positive rotavirus samples, taken from 26 records from 14 studies, across five countries (Figure 4a).54–67 Based on this data, the most common rotavirus genotypes in the region were G1P[8] and G9P[8], showing 42% (95% CI: 33%, 52%) and 14% (95% CI: 7%, 23%) prevalence.
Figure 4.
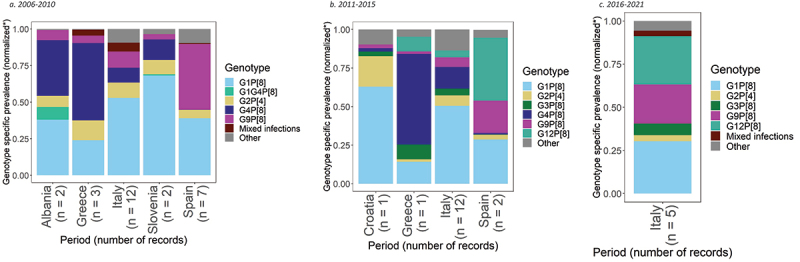
Prevalence of the most common rotavirus genotypes in Southern Europe.
In Albania, G1P[8] and G4P[8] were the most prevalent genotypes with 37% (95% CI: 31%, 44%) and 37% (95% CI: 25%, 51%) prevalence, respectively. G1P[8] was also the most prevalent genotype in Italy and Slovenia with 47% (95% CI: 39%, 56%) and 66% (95% CI: 46%, 84%) prevalence. In Greece, G4P[8] was the most prevalent genotype with 49% prevalence (95% CI: 10%, 88%), followed by G1P[8] with 22% (95% CI: 4%, 50%). While in Spain, G9P[8] was the most prevalent genotype with 44% (95% CI: 15%, 75%) prevalence, followed by G1P[8] with 38% (95% CI: 14%, 65%).
For the period 2011–2015, genotype prevalence data were available from 4,929 positive rotavirus samples, taken from 16 records from nine studies, across four countries (Figure 4b).56,60,64,68-73 Based on this data, the most common rotavirus genotypes in the region were G1P[8] and G4P[8], with prevalences of 42% (95% CI: 30%, 55%) and 12% (95% CI: 5%, 21%), respectively.
In Croatia and Italy, G1P[8] was the most common genotype with a prevalence of 63% (95% CI: 59%, 66%) and 46% (95% CI: 31%, 61%), respectively. The second most common genotype was G2P[4] in Croatia with 20% prevalence (95% CI: 17%, 23%) and G4P[8] in Italy with 13% (95% CI: 6%, 21%). In Greece, G4P[8] was the most prevalent genotype with 59% (95% CI: 50%, 67%), followed by G1P[8] with 14% (95% CI: 9%, 22%). In Spain, G12P[8] was the most common genotype with a prevalence of 36% (95% CI: 2%, 83%), followed by G1P[8] with 26% (95% CI: 17%, 35%) prevalence.
Italy was the only country in Southern Europe with data for the period 2016–2021. Genotype prevalence data were available from 2,429 positive rotavirus samples, taken from five records from three studies64,74,75 (4C). During this period, G1P[8] was the most prevalent genotype with 29% (95% CI: 19%, 40%), followed by G12P[8] with 27% (95% CI: 15%, 41%) prevalence.
Italy was the only country in Southern Europe to be included in the meta-analyses for all three time periods. Through the different time-periods, G1P[8] became less dominant, with the relative proportion of this genotype declining from 47% during 2006–2010, to 46% in 2011–2015, and 29% in 2016–2021. The second most prevalent genotype varied from G9P[8] in 2006–2010, to G4P[8] from 2011–2015, and G12P[8] with a prevalence of 27%, from 2016–2021.
Western Europe
For the period 2006–2010, genotype prevalence data were available from 3,561 positive rotavirus samples from 12 records from seven studies, across four countries (Figure 5a).76–82 Across all rotavirus samples retrieved for this region, the most common rotavirus genotypes were G1P[8] and G2P[4], with a prevalence of 39% (95% CI: 27%, 53%) and 16% (95% CI: 7%, 28%), respectively. G2P[4] was the most prevalent genotype observed in Austria with 43% (95% CI: 24%, 64%) prevalence and Belgium with 43% (95% CI: 34%, 54%) prevalence. G1P[8] was the second most prevalent genotype in both countries with 26% (95% CI: 11%, 46%) prevalence in Austria, and 27% (95% CI: 13%, 45%) in Belgium. G1P[8] was most dominant in France with 59% prevalence (95% CI: 54%, 64%), followed by G9P[8] with 21% (95% CI: 15%, 28%). In Germany, G3P[8] was the most common genotype with a prevalence of 24% (95% CI: 0%, 100%), followed by G1P[8] with 19% (95% CI: 0%, 64%).
Figure 5.
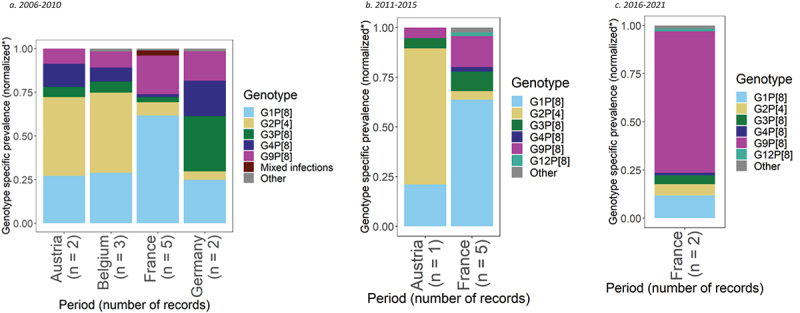
Prevalence of the most common rotavirus genotypes in Western Europe.
For 2011–2015, genotype prevalence data were available from 4,989 positive rotavirus samples from six records from three studies, across two countries, Austria and France (Figure 5b).78,82,83 Based on within-country meta-analyses or individual prevalence records, the most common rotavirus genotypes were G1P[8] and G9P[8], showing a prevalence of 57% (95% CI: 44%, 68%) and 14% (95% CI: 6%, 23%), respectively. In France, G1P[8] had a prevalence of 61% (95% CI: 53%, 69%), followed by G9P[8] with 15% (95% CI: 7%, 26%). G2P[4] was the most prevalent genotype in Austria with a prevalence of 68% (95% CI: 43%, 87%), followed by G1P[8] with 21% (95% CI: 6%, 46%).
For the period 2016–2021, genotype prevalence data were available from 1,202 positive rotavirus samples in two records from one study in France (Figure 5c).83 Based on this data, the dominant rotavirus genotype was G9P[8] with 72% (95% CI: 59%, 84%) prevalence. G1P[8] was the second most common genotype at only 12% (95% CI: 3%, 24%) prevalence.
One country, France, was included in meta-analyses across all three time periods. G1P[8] was the dominant genotype from 2006 to 2010, representing 59% (95% CI: 54%, 64%) of genotypes and 61% (95% CI: 53%, 69%) from 2011 to 2015. However, from 2016 to 2021, G9P[8] emerged as the dominant genotype with a prevalence of 72% (95% CI: 59%, 84%), with G1P[8] genotype prevalence decreasing to 12%.
Middle East
For the period 2006–2010, genotype prevalence data were available from 957 positive rotavirus samples, taken from seven records from seven studies, across seven countries (Figure 6a).84–90 Across these samples, the most common rotavirus genotypes reported were G1P[8] and G2P[4], with prevalences of 35% (95% CI: 14%, 59%) and 14% (95% CI: 2%, 36%), respectively.
Figure 6.
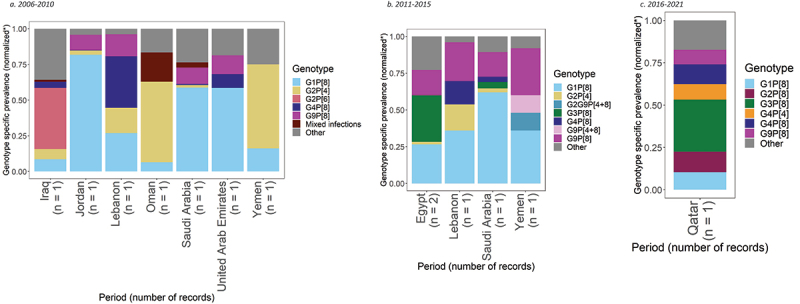
Prevalence of the most common rotavirus genotypes in the Middle East.
G1P[8] was the most prevalent genotype in three countries: Jordan, Saudi Arabia and United Arab Emirates, representing 82% (95% CI: 76%, 87%), 59% (95% CI: 49%, 68%), and 59% (95% CI: 52%, 65%) of genotypes, respectively. G2P[4] was the most prevalent genotype in Oman and Yemen, with prevalences of 56% (95% CI: 47%, 66%) and 59% (95% CI: 46%, 71%). In Iraq, G2P[6] was the most prevalent genotype with 43% (95% CI: 31%, 55%) prevalence. In Lebanon, G4P[8] was the most prevalent genotype with a prevalence of 36% (95% CI: 28%, 45%), followed by G1P[8] with a prevalence of 27% (95% CI: 20%, 35%).
For the period 2011–2015, genotype prevalence data were available from 687 positive rotavirus samples, taken from five records from five studies, across four countries (Figure 6b).90–94 For these samples, the most common rotavirus genotypes were G1P[8] and G9P[8], showing prevalences of 36% (95% CI: 22%, 51%) and 20% (95% CI: 14%, 28%).
G1P[8] and G9P[8] were the most prevalent genotypes in Lebanon, Saudi Arabia and Yemen. G1P[8] and G9P[8] prevalence was 36% (95% CI: 31%, 41%) and 26% (95% CI: 22%, 31%) in Lebanon, 62% (95% CI: 52%, 71%) and 17% (95% CI: 10%, 25%) in Saudi Arabia, and 36% (95% CI: 18%, 57%) and 32% (95% CI: 15%, 54%) in Yemen, respectively. In Egypt, G3P[8] was the most prevalent genotype with a prevalence of 26% (95% CI: 8%, 50%), followed by G1P[8] at 22% (95% CI: 15%, 30%).
Qatar was the only country which included data for the period 2016–2021. Genotype prevalence data were available from 231 positive rotavirus samples, taken from one record in one study.95 In Qatar, G3P[8] was the most prevalent genotype with a prevalence of 31% (95% CI: 25%, 37%). G2P[8] and G4P[8] were the second most prevalent genotypes, each representing 12% (95% CI: 8%, 17%).
Studies excluded from meta-analysis
Twelve studies were excluded from the meta-analysis because they: 1) reported only G or P types separately;96–102 2) did not report the number of samples;103 3) were pan-European studies;104–106 and 4) included data from gray literature.107
Among seven studies that reported only G types, G1 was the most identified type in three European settings from 2006–2010, including Denmark, Norway and Spain with reported prevalences of 39%, 55%, and 50%, respectively.96,97,99 G12 was most commonly identified in one study in Spain, representing 30% of genotypes reported from 2010 to 201898 and G4 was most commonly identified in one study in Greece with a prevalence of 60% from 2008 to 2010.100 Two studies reporting G and P types separately also showed P[8] as the predominant P type, with prevalences of 85% and 75% from 2006 to 2010, respectively.97,100 In the Middle East, G1 was the most commonly identified genotype in two studies in Egypt, with prevalence varying from 44% from 2008 to 2010 to 55%.101,102 One study, which was excluded from the meta-analysis for not reporting the number of samples reported that G1P[8], G2P[4], G4P[8] and G9P[8] represented 84–96% of genotypes from 2005 to 2013.103
Three pan-European peer-reviewed studies in addition to the 2019 EuroRotaNet annual report were included for narrative synthesis. The EuroRotaNet surveillance network was established in 2007 and monitors rotavirus genotype diversity and year-to-year to identify genotype fluctuations across Europe. Two of the manuscripts summarized early results from member countries of EuroRotaNet;104,105 the third manuscript examined data from Czechia, Germany, Italy, Poland, Spain and the United Kingdom.106 In the two studies investigating data from EuroRotaNet member countries, G1P[8] was the most prevalent aggregated genotype in every year from 2006 to 2009. Iturriza-Gómara 2009 showed G1P[8] accounted for 43% of genotypes from 2005 to 2006, 42% from 2006 to 2007, and 62% from 2007 to 2008.104 Iturriza-Gómara 2010 showed G1P[8] accounted for 43% of genotypes from 2006 to 2007, 53% from 2007 to 2008, and 46% from 2008 to 2009.105 The third study, using data from European countries from 2005–2007, showed that genotype distribution varied between countries.106 G9P[8] was the most common type in Poland and Spain, G1P[8] was predominant in Czechia and Italy, and G4P[8] and G1P[8] were both prevalent in Germany. The 2019 EuroRotaNet annual report summarized information from all previous seasons and showed that G1P[8] was the most common identified genotype until 2015/2016.107 In 2018/2019, G1P[8] was identified in 9% of samples, and G3P[8], G9P[8] and G2P[4] were detected in 19%–25% of samples. Seven genotypes circulated with a prevalence > 1%, including G1P[8], G4P[8], G2P[4], G9P[8], G3P[8], G12P[8], and G9P[4]. The report also shows that dominance of a single genotype has become rarer in recent years, while the relative proportion of less common genotypes has increased.107
Discussion
We performed a meta-analysis of rotavirus genotypes circulating in Europe and the Middle East between 2006 and 2021, providing an update to previously published systematic reviews and providing data for countries not included in EuroRotaNet. Our results highlight the lack of a single dominant genotype across time period, with G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8] frequently identified as the most prevalent genotypes. Because our results summarized individual country results, they can be used to characterize genotype circulation by country and are complementary to EuroRotaNet data.
Results from our meta-analysis are consistent with surveillance reports and pan-European studies,104–107 which characterize emerging genotype diversity across time in the post-licensure period and the decrease in prevalence of G1P[8]. According to EuroRotaNet annual report, G1P[8] genotype was consistently the most prevalent genotype between 2006–2007 and 2014–2015, ranging from a maximum of 62% in 2007–2008 to 31% in 2014–2015.107 From 2015–2016, G9P[8] was found in 34% of single genotype infections characterized. In subsequent years, dominant genotypes also included G3P[8] and G2P[4].107
Our study included two EuroRotaNet member countries that had data across all three time periods: France and Finland. Consistent with EuroRotaNet data, our analysis shows G1P[8] was the most prevalent genotype in France from 2006–2015, whereas from 2016 to 2021, G9P[8] emerged as the most prevalent genotype. Similarly, in Finland a decrease of G1P[8] was observed from 2011 to 2021, as the prevalence of G2P[4], G9P[8], and G12P[8] increased. Lastly, our results for the Middle East are consistent with a recent review in the Middle East and North Africa, which showed that G1P[8], G9P[8], and G2P[4] were the most common genotypes from 1980 to 2019.108 Of note, our analysis shows G3P[8] as the most prevalent genotype from 2016 to 2021 in Qatar.
While we have not captured vaccination data in our review, we find that the prevalent genotypes remain consistent through 2006–2021, with routinely identified genotypes being G1P[8], G2P[4], G3P[8], G4P[8], G9P[8] and, to a smaller extent, G12P[8], with their relative dominance and prevalence varying from year to year. Consistent with EuroRotaNet data, our analysis demonstrates high genotype diversity, with lack of dominance by a particular genotype over all time-periods. Our meta-analysis reveals two uncommon genotypes, G3P[9] and G2P[6], in Poland and Iraq, respectively. However, these findings were derived from small sample sizes and should be interpreted cautiously (68 samples in Poland and 98 samples in Iraq). Of note, G9P[4] has also been identified by EuroRotaNet;109 since the inception of the network (from 2006–2007 season to 2020–2021), its overall prevalence has been 1%.
This is important for rotavirus vaccination policy as it suggests that vaccination programs have had limited or no impact on the the prevalence of uncommon strains from the pre-vaccination period. Additionally, to date, limited to no evidence of vaccine induced selective pressure has been found, with a recent review noting a, potentially transient, increased prevalence of G2P[4] in post-introduction scenarios.5 The findings echo those of an earlier systematic review, which did not find a consistent pattern indicative of vaccine related selective prevention, while indicating the need to monitor the increased detection rate of G2P[4] genotype in some countries following RV1 vaccination.6 Of note, rotavirus vaccines have long been observed to possess cross-protective effects, with RV1 being suggested to be marginally less effective against fully heterotypic G2P[4].6,110–112
G1P[8], G2P[4], G3P[8], G4P[8], G9P[8], and G12P[8] have frequently been identified worldwide.5 Of note, strains uncommon in Europe and the Middle East have been identified elsewhere. Data from 2001 to 2017 in Latin America show G9P[4] was identified in Colombia with a prevalence of ~15%, while G1P[4] was identified in Nicaragua with a prevalence of 13%;113 in Africa, G2P[6] (4.2%) and G3P[6] (3.7%) were identified Ethiopia, Zambia and Zimbabwe through a study across Eastern and Southern Africa from 2010 to 2015;114 while in Asia G12P[6] was identified in various surveillance studies from 2001 to 2021, including Bangladesh (no prevalence specified), Nepal (41%–45%) and India (30%), respectively.115,116
Our analysis is limited by the uneven representation of countries across different timeframes and variation in the number of samples per region and time period. Some countries, such as the Russian Federation, Estonia, Finland, Ireland, and France are consistently represented, allowing for more comprehensive temporal analyses. In contrast, other countries appear only in certain periods, making trend analyses challenging.
Additionally, this study included any country regardless of vaccination introduction or uptake. Our analysis did not differentiate between countries where vaccines were available through national immunization plans or only through the private market and did not include the type of vaccine available in each country. In 2016, it was reported that 17 countries in Europe had introduced the universal rotavirus vaccines and 11 countries in the Eastern Mediterranean Region, with the vaccine type varying across countries.117 In many studies, where vaccination was only available through the private market, vaccination uptake rates were below 10%.25,42,72,74,76,78,83,118,119 By contrast, in countries with vaccination available through national immunization plans, multiple studies reported coverage levels above 90%.45–47,79 We did not analyze how varying coverage rates impacted rotavirus genotype circulation. However, the results from the current study can be compared with genotype prevalence in the pre-licensure data.5,120 As mentioned, results are consistent with those reported by EuroRotaNet, and suggest that, while the relative prevalence of genotypes varies in any given year (either during the pre and post licensure area), no new genotypes emerged in the post-licensure era.
Lastly, this study did not include sub-group analyses of genotype prevalence among specific age groups due to considerable variation of reporting in the included studies. Previous studies have shown genotype diversity was higher in older age groups121 and it is possible that we may be reporting results that apply to specific age-groups in some countries. Of note, EuroRotanet results are aggregated by country, regardless of age-group.
Finally, we did not include changes in the “DS-1-like” or “Wa-like” genotype constellation of the strains as that information is not widely reported. However, a recent study in Belgium has noted an increase in G3P[8] (traditionally associated with a “Wa-like” constellation) with a DS-1-like constellation post-vaccine introduction.122 Such changes warrant further consideration in future studies.
Going forward, rotavirus genotype surveillance studies would be complemented by reporting comprehensive information on genotype constellations, and genetic diversity of rotavirus in vaccinated and non-vaccinated individuals over time.
Conclusion
Results show high variability among dominant genotypes in Europe and the Middle East in the post-licensure period, with G1P[8], G2P[4], G3P[8], G4P[8], G9P[8] and G12P[8] being the most commonly identified genotypes. Consistent with EuroRotaNet data, there was high temporal and regional variability, with dominant genotypes varying by period and country, even among neighboring countries.
Supplementary Material
Acknowledgments
We thank Hannah Wood and Mick Arbor at the University of York for their technical support in developing the search strategy. The authors also thank Stasha Mamotra, Elizabeth Salazar, Tejaswini Vachaspathi, Adriana Mancilla and Anastasia Döldös for Triangulate Health Ltd who supported screening and data extraction. The authors also thank Chrissy Bishop for her support in providing quality appraisal and review, as well as Deeksha Parashar and Arnold Hagens for their support in developing maps for this manuscript.
Biographies
Tim Jesudason is a global health consultant specializing in evidence synthesis, health economics, and communications. He has a strong background in research focused on the control of infectious diseases and childhood illnesses. Tim holds bachelor’s degrees in International Relations from the University of Melbourne and in Philosophy from Victoria University of Wellington, as well as a Master of Public Health from the London School of Hygiene & Tropical Medicine.
Dr. Oluwaseun Sharomi is a Principal Scientist and Health Economist at Merck, where he has led modeling activities for RotaTeq and is currently leading and supporting modeling activities for Epstein-Barr Virus and Pneumococcal Disease. Prior to joining Merck, Dr. Sharomi served as an Associate Director of Modeling at CHEORS and as an Assistant Professor in the Department of Mathematics at Khalifa University in the United Arab Emirates. Dr. Sharomi’s work focuses on using mathematical modeling approaches and analysis to understand the transmission dynamics and control of emerging and re-emerging diseases of public health interest. He has designed, analyzed, and simulated novel mathematical models for the spread of various diseases, including respiratory diseases (H1N1, Mycobacterium tuberculosis), sexually transmitted infections (Chlamydia, HIV, HPV, syphilis), and their coinfections (e.g., HIV-TB coinfection). Dr. Sharomi has used these models to provide realistic assessments of various intervention strategies, such as vaccines (e.g., for H1N1 and HPV) and drug treatments (e.g., for HIV). His statistical skills include proficiency in parameter estimation, uncertainty and sensitivity analysis of model parameters, optimal control analysis, and fitting models to data. Dr. Sharomi’s expertise also encompasses the design and analysis of mathematical models of infectious diseases, simulation of electrical activity in myocardial tissue, parallel computing, problem-solving environments, and numerical algorithms and software for exascale computer architectures.
Kelly Fleetwood is a statistician with 20 years’ experience across industry and academia. She has worked in statistical consultancy providing input to a broad range of projects in the life sciences, from gene expression studies to evaluations of the cost-effectiveness of new drugs. She currently works for the University of Edinburgh where her research uses large health datasets to better understand the relationship between physical and mental health conditions. Her areas of expertise include meta-analysis and the analysis of observational data, including electronic health records. Kelly holds a bachelor’s degree in Mathematics from the University of Queensland, Australia and a master’s degree in Statistics from the University of Sheffield, UK.
Alex Lapting Cheuk is a health data scientist at the London Borough of Tower Hamlets (Health Determinants Research Collaboration), where she investigates the health impacts of social policies by linking Local Authority administrative data with NHS health records. Alex’s research interests include social epidemiology, behavioral science in public health policy, and the economic evaluation of health and social interventions.
Maria Bermudez is a senior analyst with a Master’s degree in Health Policy, Planning, and Financing, as well as a medical degree. She has a background in health technology assessment, health policy analysis, and clinical medicine. Maria has worked with a wide range of organizations in both the public and private sectors and currently works with Triangulate Health Ltd on projects involving health policy analysis, systematic reviews, and stakeholder engagement. Her areas of interest include public health policies and population health in low- and middle-income countries.
Hannah Schirrmacher is a health economist with a background in development economics. She has several years of consultancy experience at Triangulate Health Ltd and the Office of Health Economics, where she has worked on research projects related to vaccines, advanced therapy medicinal products, and broader value in health technology assessments. Her research experience includes literature reviews, qualitative research methods, and economic modeling, and she has published in leading journals such as Value in Health, Vaccines, and PharmacoEconomics. Hannah has presented her research at several conferences, including the Health Economics Study Group and the International Health Economics Association. Her areas of interest include global health, infectious diseases, and public health policies.
Christian Hauck holds a Bachelor of Arts in Political Science and Global Studies and a Master of Arts in International Public Policy, with specializations in Human Security and Global Governance. Christian worked as a research analyst with Triangulate Health, where he primarily focused on qualitative analysis, report writing, and data analysis. During his time at Triangulate, Christian was involved in developing health policy recommendations related to smoking cessation and assisted the team in conducting a cost-effectiveness analysis on incontinence in Europe. His research interests include healthcare policy, non-communicable diseases, health technology assessments, and global issues surrounding tuberculosis. Christian has completed courses and obtained certifications through the academic consortium R for Health Technology Assessment, exploring the use of R for cost-effectiveness analysis.
Jelle Matthijnssens serves as a professor at the KU Leuven in Belgium, and has performed research on rotavirus for two decades. He developed a comprehensive classification system encompassing all 11 rotavirus gene segments. This classification system has been instrumental to study of rotavirus genetic diversity, reassortment, zoonosis, evolution, and epidemiology. He is a member of EuroRotaNet, runs the national reference center (NRC) for rotavirus in Belgium together with prof. Marc Van Ranst, and has been member and chair of the Reoviridae family study group of the ICTV for many years. In recent years his research has shifted towards viral metagenomics for which he optimized and developed the NetoVIR protocol to purify viral particles from biological samples to be analyzed using deep sequencing, in combination with dedicated bio-informatics pipelines. This technique is being used to investigating a wide range of scientific question ranging from: how does the gut virome development of infants? How does the gut virome associate with human disease? What is the role of the virome in honeybee health and disease? What is the role of insect specific viruses in mosquito vector competence? or can viral metagenomics be used on environmental samples such as indoor air?
Daniel Hungerford is an infectious disease epidemiologist with expertise in gastrointestinal infections and vaccines, in relation to health inequalities. Previously, he worked for the Health Protection Agency and then Public Health England, specialising in infectious disease surveillance and outbreak investigation; and the LJMU, Centre for Public Health, developing the Trauma and Injury Intelligence Group surveillance system. He is a Senior lecturer in the NIHR Health Protection Research Unit in Gastrointestinal Infections at the University of Liverpool and has recently completed the NIHR Future Focused Leaders – Emerging Leaders Programme. He currently leads the European Rotavirus Surveillance Network (EuroRotaNet) and is Theme lead for Measuring the Value of Vaccines at the Centre for Global Vaccine Research. His main research interests involve the use of “real world” big data for vaccine evaluations and effectiveness studies, focusing on respiratory and diarrhoeal disease.
David Tordrup is a health economist with a background in life sciences. He has led multiple projects on pharmaceutical policies, financing, health technology assessment (HTA), and reimbursement, working with global clients, including biopharma companies and multilateral organizations. He serves as an external expert for the European Commission and the Journal of Pharmaceutical Policy and Practice and is currently a doctoral researcher at the WHO Collaborating Centre for Pharmaceutical Policy and Regulation. With over a decade of experience, he has managed and implemented numerous pharmaceutical and health economics research projects, publishing extensively in reputable international journals.
Cristina Carias is a Technological Physics Engineer with a PhD in Strategy, Entrepreneurship, and Technological Change, and advanced training in Econometrics. She has experience in both the public and private sectors, and has worked on health economics, outcomes research, emergency preparedness, and public health modeling.
Funding Statement
This work was supported by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA.
Disclosure statement
SO and CC are employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA and shareholders of Merck & Co., Inc., Rahway, NJ, USA, a manufacturer of RotaTeq©, a rotavirus vaccine. Triangulate Health Ltd (that employed MB, ALC, CH, TJ, HS and funded KF) received funding from Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA to conduct the analysis. DH has received consulting fees from Merck & Co., Inc., Rahway, NJ, USA, and institutional grants from Seqirus UK Ltd, Merck & Co., Inc., Rahway, NJ, USA, and GlaxoSmithKline Biologicals. JM has received consulting fees from GlaxoSmithKline and Merck & Co., Inc., Rahway, NJ, USA.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website at https://doi.org/10.1080/21645515.2024.2389606
References
- 1.Rotavirus vaccines WHO position paper: January 2013 - recommendations. Vaccine. 2013;31(52):6170–15. doi: 10.1016/j.vaccine.2013.05.037. [DOI] [PubMed] [Google Scholar]
- 2.Esona MD, Steele D, Kerin T, Armah G, Peenze I, Geyer A, Page N, Nyangao J, Agbaya V, Trabelsi A, et al. Determination of the G and P types of previously nontypeable rotavirus strains from the African rotavirus network, 1996–2004: identification of unusual G types. J Infect Dis. 2010 Sep 1;202(SUPPL. 1):49–54. doi: 10.1086/653552. [DOI] [PubMed] [Google Scholar]
- 3.Fields BN, Knipe DM, Howley PM.. Fields virology. Vol. 2. 5th ed. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. p. 1917–1974. [Google Scholar]
- 4.Bányai K, László B, Duque J, Steele AD, Nelson EAS, Gentsch JR, Parashar UD. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30:A122–A130. doi: 10.1016/j.vaccine.2011.09.111. [DOI] [PubMed] [Google Scholar]
- 5.Dóró R, László B, Martella V, Leshem E, Gentsch J, Parashar U, Bányai K. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: is there evidence of strain selection from vaccine pressure? Infect, Genet And Evol. 2014 Dec 1;28:446–461. doi: 10.1016/j.meegid.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amin AB, Cates JE, Liu Z, Wu J, Ali I, Rodriguez A, Panjwani J, Tate JE, Lopman BA, Parashar UD, et al. Rotavirus genotypes in the postvaccine era: a systematic review and meta-analysis of global, regional, and temporal trends by rotavirus vaccine introduction. J Infect Dis [Internet]. [accessed 2023 Sep 21;229(5):1460–1469. https://academic.oup.com/jid/advance-article/doi/10.1093/infdis/jiad403/7280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aballéa S, Millier A, Quilici S, Carroll S, Petrou S, Toumi M. A critical literature review of health economic evaluations of rotavirus vaccination. Hum Vaccin Immunother. 2013. June; 9(6):1272–1288. doi: 10.4161/hv.24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotirum S, Vutipongsatorn N, Kongpakwattana K, Hutubessy R, Chaiyakunapruk N. Global economic evaluations of rotavirus vaccines: a systematic review. Vaccine. 2017;35(26):3364–3386. doi: 10.1016/j.vaccine.2017.04.051. [DOI] [PubMed] [Google Scholar]
- 9.Higgins J, Thomas J, Chandler J, Cumpston M, Li T, Page M, Cochrane handbook for systematic reviews of interventions. 2022. www.training.cochrane.org/handbook.
- 10.Google Translate . Google [Internet]. 2023. https://translate.google.ca/.
- 11.World Health Organization. Rotavirus . Internet]. 2018. Sep accessed 2024 Jan 9]. https://www.who.int/publications/m/item/vaccine-preventable-diseases-surveillance-standards-rotavirus.
- 12.STROBE. STROBE STATEMENT . Internet]. 2007. accessed 2021 Jul 13]. https://www.strobe-statement.org/index.php?id=available-checklists.
- 13.United Nations Statistics Division. United Nations . United Nations Statistics Division: methodology. 2023. cited 2023 Oct 25]. https://unstats.un.org/unsd/methodology/m49/.
- 14.Healy N, Carvao S. World tourism organization. In: Jafari J, Xiao H, editors. Encyclopedia of tourism. Cham: Springer; 2016. p. 1027–1028. [Google Scholar]
- 15.Barker TH, Migliavaca CB, Stein C, Colpani V, Falavigna M, Aromataris E, Munn Z. Conducting proportional meta-analysis in different types of systematic reviews: a guide for synthesisers of evidence. BMC Med Res Methodol. 2021;21(1):189. doi: 10.1186/s12874-021-01381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarzer G, Chemaitelly H, Lj A-R, Rücker G. Seriously misleading results using inverse of freeman-tukey double arcsine transformation in meta-analysis of single proportions. Res Synth Methods. 2019;10(3):476–483. doi: 10.1002/jrsm.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viechtbauer W. Bias and efficiency of meta-analytic variance estimators in the random-effects Model. J Educ Behavioral Stat. 2005 Sep 23;30(3):261–93. doi: 10.3102/10769986030003261. [DOI] [Google Scholar]
- 18.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986. Sep;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 19.R Core Team . R: a language and environment for statistical computing. R version 4.2.3; Vienna, Austria: Foundation for Statistical Computing; 2023. [Google Scholar]
- 20.Balduzzi S, Rücker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evidence Based Ment Health. 2019;22(4):153–160. doi: 10.1136/ebmental-2019-300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhirakovskaia E, Tikunov A, Tymentsev A, Sokolov S, Sedelnikova D, Tikunova N. Changing pattern of prevalence and genetic diversity of rotavirus, norovirus, astrovirus, and bocavirus associated with childhood diarrhea in Asian Russia, 2009–2012. Infect Genet Evol. 2019 Jan 1;67:167–182. doi: 10.1016/j.meegid.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Zhirakovskaya EV, Aksanova RK, Gorbunova MG, Tikunov AY, Kurilshchikov AM, Sokolov SN, Netesov SV, Tikunova NV. Genetic diversity of group a rotavirus isolates found in Western Siberia in 2007–2011. Mol Genet Microbiol Virol. 2012;27(4):174–183. doi: 10.3103/S0891416812040076. [DOI] [PubMed] [Google Scholar]
- 23.Mladenova Z, Korsun N, Geonova T, Iturriza-Gómara M. Molecular epidemiology of rotaviruses in Bulgaria: annual shift of the predominant genotype. Eur J Clin Microbiol Infect Dis. 2010. May. 29(5):555–562. doi: 10.1007/s10096-010-0895-1. [DOI] [PubMed] [Google Scholar]
- 24.László B, Czellár E, Deák J, Juhász Á, Kovács J, Kónya J, Mészáros J, Mészner Z, Mihály I, Molnár P, et al. Post vaccination rotavirus surveillance in Hungary, in 2007. Orv Hetil. 2009 Aug 1;150(31):1443–1450. doi: 10.1556/oh.2009.28690. [DOI] [PubMed] [Google Scholar]
- 25.Chernyshova LI, Radionova NM, Demchyshyna IV, Kotlik LS, Sadkova OB, Samoilovich EO, Semeiko GV, Daniels DS, Cohen AL, Aliabadi N. Observations on the epidemiology of rotavirus infection among hospitalized children younger than 5 years in 2 Ukrainian hospitals, 2007–2015. Vaccine. 2018 Dec 14;36(51):7798–7804. doi: 10.1016/j.vaccine.2017.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semeiko GV, Yermalovich MA, Poliakova N, Mijatovic-Rustempasic S, Kerin TK, Wasley A, Videbaek D, Gentsch JR, Bowen MD, Samoilovich EO. Rotavirus genotypes in Belarus, 2008–2012. Infect, Genet Evol. 2014 Dec 1;28:480–485. doi: 10.1016/j.meegid.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piekarska A, Kacerka A, Majda-Stanisławska E, Jóźwiak B, Sidorkiewicz M. Predominance of genotype P[9]G3 in rotavirus gastroenteritis in Polish children. Archiv Med Sci. 2015 June 1;11(3):577–583. doi: 10.5114/aoms.2015.50229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anca A, Furtunescu FL, Pleșca D, Streinu-Cercel A, Rugină S, Holl K. Hospital-based surveillance to estimate the burden of rotavirus gastroenteritis in children below five years of age in Romania. Germs [Internet]. 2014;4(2):30–40. www.germs.ro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobzin YV, Kharit SM, Goveia MG, O’Brian MA, Podkolzin AT, Blokhin BM, Bekhtereva MK, Rudakova AV, Tikunova NV. Burden of childhood rotavirus disease in the outpatient setting of the Russian federation. Pediatr Infect Disease J. 2017 Dec 15;36(5):472–476. doi: 10.1097/INF.0000000000001472. [DOI] [PubMed] [Google Scholar]
- 30.Denisyuk NB. The results of rotavirus group a antigen types monitoring in the Orenburg region during 2013-2016. Jurnal Infektologii. 2017;9(3):92–97. doi: 10.22625/2072-6732-2017-9-3-92-97. [DOI] [Google Scholar]
- 31.Kiseleva V, Faizuloev E, Meskina E, Marova A, Oksanich A, Samartseva T, Bakhtoyarov G, Bochkareva N, Filatov N, Linok A, et al. Molecular-genetic characterization of human Rotavirus a strains circulating in Moscow, Russia (2009–2014). Virol Sin. 2018 Aug 1;33(4):304–313. doi: 10.1007/s12250-018-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dóró R, Mihalov-Kovács E, Marton S, László B, Deák J, Jakab F, Juhász Á, Kisfali P, Martella V, Melegh B, et al. Large-scale whole genome sequencing identifies country-wide spread of an emerging G9P[8] rotavirus strain in Hungary, 2012. Infect, Genet Evol. 2014 Dec 1;28:495–512. doi: 10.1016/j.meegid.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Morozova OV, Sashina TA, Epifanova NV, Zverev VV, Kashnikov AU, Novikova NA. Phylogenetic comparison of the VP7, VP4, VP6, and NSP4 genes of rotaviruses isolated from children in Nizhny Novgorod, Russia, 2015–2016, with cogent genes of the rotarix and RotaTeq vaccine strains. Virus Genes. 2018 Apr 1;54(2):225–235. doi: 10.1007/s11262-017-1529-9. [DOI] [PubMed] [Google Scholar]
- 34.Ivashechkin AA, Yuzhakov AG, Grebennikova TV, Yuzhakova KA, Kulikova NY, Kisteneva LB, Smetanina SV, Bazarova MV, Almazova MG. Genetic diversity of group a rotaviruses in Moscow in 2018-2019. Arch Virol. 2020 Mar 1;165(3):691–702. doi: 10.1007/s00705-020-04534-5. [DOI] [PubMed] [Google Scholar]
- 35.Yuzhakov A, Yuzhakova K, Kulikova N, Kisteneva L, Cherepushkin S, Smetanina S, Bazarova M, Syroeshkin A, Grebennikova T. Prevalence and genetic diversity of group a rotavirus genotypes in Moscow (2019–2020). Pathogens. 2021 June 1;10(6):674. doi: 10.3390/pathogens10060674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sashina TA, Morozova OV, Epifanova NV, Kashnikov AU, Leonov AV, Novikova NA. Molecular monitoring of the rotavirus (reoviridae: sedoreovirinae: rotavirus: rotavirus A) strains circulating in Nizhny Novgorod (2012-2020): detection of the strains with the new genetic features. Vopr Virusol. 2021;66(2):140–151. doi: 10.36233/0507-4088-46. [DOI] [PubMed] [Google Scholar]
- 37.Moutelíková R, Sauer P, Heroldová MD, Holá V, Prodelalová J. Emergence of rare bovine–human reassortant DS-1-Like rotavirus a strains with G8P[8] genotype in human patients in the Czech Republic. Viruses. 2019 Nov 1;11(11):1015. doi: 10.3390/v11111015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinder M, Tran AN, Bennet R, Brytting M, Cassel T, Eriksson M, Frithiof D, Gothefors L, Storsaeter J, Trollfors B, et al. Burden of severe rotavirus disease leading to hospitalization assessed in a prospective cohort study in Sweden. Scand J Infect Dis. 2014;46(4):294–302. doi: 10.3109/00365548.2013.876511. [DOI] [PubMed] [Google Scholar]
- 39.Beck-Friis T, Andersson M, Gustavsson L, Lindh M, Westin J, Andersson LM. Burden of rotavirus infection in hospitalized elderly individuals prior to the introduction of rotavirus vaccination in Sweden. J Clin Virol. 2019 Oct 1;119:1–5. doi: 10.1016/j.jcv.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Cashman O, Collins PJ, Lennon G, Cryan B, Martella V, Fanning S, Staines A, O’SHEA H. Molecular characterization of group a rotaviruses detected in children with gastroenteritis in Ireland in 2006–2009. Epidemiol Infect. 2012. Feb. 140(2):247–259. doi: 10.1017/S0950268811000306. [DOI] [PubMed] [Google Scholar]
- 41.Collins PJ, Mulherin E, O’Shea H, Cashman O, Lennon G, Pidgeon E, Coughlan S, Hall W, Fanning S. Changing patterns of rotavirus strains circulating in Ireland: re-emergence of G2P[4] and identification of novel genotypes in Ireland. J Med Virol. 2015 May 1;87(5):764–773. doi: 10.1002/jmv.24095. [DOI] [PubMed] [Google Scholar]
- 42.Räsänen S, Lappalainen S, Halkosalo A, Salminen M, Vesikari T. Rotavirus gastroenteritis in Finnish children in 2006- 2008, at the introduction of rotavirus vaccination. Scand J Infect Dis. 2010. Jan. 43(1):58–63. doi: 10.3109/00365548.2010.508462. [DOI] [PubMed] [Google Scholar]
- 43.Soeorg H, Tamm E, Huik K, Pauskar M, Mägi D, Pruudel K, Vainomäe L, Moosar L, Kirss K, Torm S, et al. Group a rotavirus genotypes circulating prior to implementation of a national immunization program in Estonia. Hum Vaccin Immunother. 2012. Apr. 8(4):465–469. doi: 10.4161/hv.19135. [DOI] [PubMed] [Google Scholar]
- 44.Yandle Z, Coughlan S, Drew RJ, O’Flaherty N, O’Gorman J, De Gascun C. Circulating rotavirus genotypes in the Irish paediatric population prior to the introduction of the vaccination programme. Ir J Med Sci. 2017 Nov 1;186(4):1003–1007. doi: 10.1007/s11845-017-1604-1. [DOI] [PubMed] [Google Scholar]
- 45.Markkula J, Hemming-Harlo M, Savolainen-Kopra C, Al-Hello H, Vesikari T. Continuing rotavirus circulation in children and adults despite high coverage rotavirus vaccination in Finland. J Infect. 2020 Jan 1;80(1):76–83. doi: 10.1016/j.jinf.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Hungerford D, Allen DJ, Nawaz S, Collins S, Ladhani S, Vivancos R, Iturriza-Gómara M. Impact of rotavirus vaccination on rotavirus genotype distribution and diversity in England, September 2006 to August 2016. Eurosurveillance [Internet]. 2019;24(6). www.eurosurveillance.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukhopadhya I, Murdoch H, Berry S, Hunt A, Iturriza-Gomara M, Smith-Palmer A, Cameron JC, Hold GL. Changing molecular epidemiology of rotavirus infection after introduction of monovalent rotavirus vaccination in Scotland. Vaccine. 2017 Jan 3;35(1):156–163. doi: 10.1016/j.vaccine.2016.11.028. [DOI] [PubMed] [Google Scholar]
- 48.Kõivumägi K, Soeorg H, Toompere K, Kallas E, Jõegeda EL, Lass E, Huik K, Lutsar I. Rotavirus strain surveillance in Estonia after introduction of rotavirus universal mass vaccination. Pediatr Infect Disease J. 2021;40(5):489–494. doi: 10.1097/INF.0000000000003039. [DOI] [PubMed] [Google Scholar]
- 49.Laizane G, Ķivite A, Grope I, Smane L, Miklaševics E, Ozoliņa L, Gardovska, D.. Clinical characterisation of rota virus infection associated with most commonly circulating genotypes in children hospitalised in children’s university hospital: a cross-sectional study in Latvia. Proc Latv Acad Scie, Sec B: Nat, Exact, Appl Sci; 2019. Aug 1;73(4):312–316. https://sciendo.com/issue/PROLAS/73/4. [Google Scholar]
- 50.Midgley S, Böttiger B, Jensen TG, Friis-Møller A, Person LK, Nielsen L, Barzinci S, Fischer TK. Human group a rotavirus infections in children in Denmark; detection of reassortant G9 strains and zoonotic P[14] strains. Infect Genet Evol. 2014;27:114–120. doi: 10.1016/j.meegid.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 51.Yandle Z, Coughlan S, Dean J, Tuite G, Conroy A, De Gascun CF. Group a rotavirus detection and genotype distribution before and after introduction of a national immunisation programme in Ireland: 2015–2019. Pathogens. 2020 June 1;9(6):1–15. doi: 10.3390/pathogens9060449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yandle Z, Coughlan S, Dean J, Hare D, De Gascun CF. Indirect impact of rotavirus vaccination on viral causes of acute gastroenteritis in the elderly. J Clin Virol. 2021 Apr 1;137:104780–104785. doi: 10.1016/j.jcv.2021.104780. [DOI] [PubMed] [Google Scholar]
- 53.Barsoum Z. Paediatric rotavirus gastroenteritis: a prospective study of regional prevalent genotypes, genotype correlation with disease severity and viral co-infection in county Mayo, Ireland, in the year following rotavirus vaccine introduction in Ireland. J Virol Methods. 294 2021 Aug 1;294:114179. doi: 10.1016/j.jviromet.2021.114179. [DOI] [PubMed] [Google Scholar]
- 54.Kota M, Bino S, Delogu R, Simaku A, Neza B, Ruggeri FM, Fiore L. Epidemiology of rotavirus diarrhoea in Albania. Arch Virol. 2014 Sep 1;159(9):2491–2495. doi: 10.1007/s00705-014-2093-4. [DOI] [PubMed] [Google Scholar]
- 55.Cilla G, Montes M, Gomariz M, Piñeiro L, Pérez-Trallero E. Rotavirus genotypes in children in the Basque country (northern Spain) over a 13-year period (July 1996–June 2009) (July 1996-June 2009). Eur J Clin Microbiol Infect Dis. 2010. Aug;29(8):955–960. doi: 10.1007/s10096-010-0951-x. [DOI] [PubMed] [Google Scholar]
- 56.Cilla G, Montes M, Gomariz M, Alkorta M, Iturzaeta A, Perez-Yarza EG, PEREZ-TRALLERO E. Rotavirus genotypes in children in the Basque country (north of Spain): rapid and intense emergence of the G12[P8] genotype. Epidemiol Infect. 2013;141(4):868–874. doi: 10.1017/S0950268812001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Téllez Castillo CJ, Montava Vilaplana R, Fernández Jiménez M, Ribes Fernández JM, Buesa Gómez J. Predominio del genotipo G9 de rotavirus en Valencia y Castellón entre 2005 y 2007. An Pediatr (Engl Ed). 2009. Jan. 72(1):49–54. doi: 10.1016/j.anpedi.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 58.Gutierrez-Gimeno MV, Martin- Moreno JM, Díez-Domingo J, Asensi-Botet F, Hernandez-Marco R, Correcher-Medina P, Sánchez-Fauquier, A.. Nosocomial rotavirus gastroenteritis in Spain: a multicenter prospective study. Pediatr Infect Dis J. 2010;29(1):23–27. doi: 10.1097/INF.0b013e3181b3603a. [DOI] [PubMed] [Google Scholar]
- 59.AnnaRita P, Grassi T, Donia D, De Donno A, Idolo A, Alfio C, Alessandri C, Alberto S, Divizia M. Detection and molecular characterization of human rotaviruses isolated in Italy and Albania. J Med Virol. 2010;82(3):510–518. doi: 10.1002/jmv.21700. [DOI] [PubMed] [Google Scholar]
- 60.Medici MC, Tummolo F, Martella V, Arcangeletti MC, De Conto F, Chezzi C, Magrì A, Fehér E, Marton S, Calderaro A, et al. Whole genome sequencing reveals genetic heterogeneity of G3P[8] rotaviruses circulating in Italy. Infect, Genet And Evol. 2016 June 1;40:253–261. doi: 10.1016/j.meegid.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 61.De Donno A, Grassi T, Bagordo F, Idolo A, Cavallaro A, Gabutti G, Collaborative Group for the surveillance of Rotavirus Infection antonella. dedonno@ unile. it . Emergence of unusual human rotavirus strains in Salento, Italy, during 2006–2007. BMC Infect Dis. 2009 Apr 15;9(1). doi: 10.1186/1471-2334-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finamore E, Vitiello M, Kampanaraki A, Rao M, Galdiero M, Galdiero E, Bevilacqua P, Gallo MA, Galdiero M. G2 as an emerging rotavirus strain in pediatric gastroenteritis in southern Italy. Infection. 2011. Apr. 39(2):113–119. doi: 10.1007/s15010-011-0102-z. [DOI] [PubMed] [Google Scholar]
- 63.Ruggeri FM, Delogu R, Petouchoff T, Tcheremenskaia O, De Petris S, Fiore L, RotaNet‐Italy Study Group . Molecular characterization of rotavirus strains from children with diarrhea in Italy 2007-2009. J Med Virol. 2011 Sep 1;83(9):1657–1668. doi: 10.1002/jmv.22163. [DOI] [PubMed] [Google Scholar]
- 64.de Waure C, Sarnari L, Chiavarini M, Ianiro G, Monini M, Alunno A, Camilloni B. 10-year rotavirus infection surveillance: epidemiological trends in the pediatric population of Perugia province. Int J Environ Res Public Health. 2020 Feb 1;17(3):1008. doi: 10.3390/ijerph17031008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steyer A, Bajželj M, Žnuderl K, Berce I, Drinovec B, Harlander T, Orešič, N, Ravnik, M, Štorman, A, Trkov, M, et al. Molecular Epidemiology of rotavirus during rotavirus vaccine introduction in Slovenia. Slovenian Med J [Internet]. 2009;78:381–386. http://www.eurorota.net/. [Google Scholar]
- 66.Trimis G, Koutsoumbari I, Kottaridi C, Palaiologou N, Assimakopoulou E, Spathis A, Lebessi E, Konstantopoulos A, Kafetzis D, Karakitsos P, et al. Hospital-based surveillance of rotavirus gastroenteritis in the era of limited vaccine uptake through the private sector. Vaccine. 2011 Oct 6;29(43):7292–7295. doi: 10.1016/j.vaccine.2011.07.092. [DOI] [PubMed] [Google Scholar]
- 67.Koukou D, Grivea I, Roma E, Tsioni H, Trimis G, Galanakis E, Farmaki E, Iosifidis E, Michos A, Siamopoulou‐Mavridou A, et al. Frequency, clinical characteristics, and genotype distribution of rotavirus gastroenteritis in Greece (2007–2008). J Med Virol. 2011. Jan. 83(1):165–169. doi: 10.1002/jmv.21945. [DOI] [PubMed] [Google Scholar]
- 68.Vrdoljak M, Gužvinec M, Trkulja V, Butić I, Ivić I, Krželj V, Tonkić M, Hegeduš Jungvirth M, Payerl Pal M, Tešović G, et al. Distribution of rotavirus genotypes in three Croatian regions among children ≤5 years of age (2012–2014). Int J Infect Dis. 2019 Dec 1;89:3–9. doi: 10.1016/j.ijid.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 69.Pérez-Ortín R, Santiso-Bellón C, Vila-Vicent S, Carmona-Vicente N, Rodríguez-Díaz J, Buesa J. Rotavirus symptomatic infection among unvaccinated and vaccinated children in Valencia, Spain. BMC Infect Dis. 2019 Nov 27;19(1). doi: 10.1186/s12879-019-4550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ianiro G, Delogu R, Fiore L, Monini M, Ruggeri FM, Pagani E, Moroder L, Binda S, Pellegrinelli L, Mignacca A, et al. Group a rotavirus genotypes in hospital-acquired gastroenteritis in Italy, 2012–14. J Hosp Infect. 2017 Jul 1;96(3):262–267. doi: 10.1016/j.jhin.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 71.Ianiro G, Delogu R, Bonomo P, Fiore L, Ruggeri FM, and “RotaNet‐Italy Study Group ”. Molecular analysis of group a rotaviruses detected in adults and adolescents with severe acute gastroenteritis in Italy in 2012. J Med Virol. 2014;86(6):1073–1082. doi: 10.1002/jmv.23871. [DOI] [PubMed] [Google Scholar]
- 72.Camilloni B, Alunno A, Nunzi E, Sarnari L, Ianiro G, Monini M. Hospital-acquired rotavirus acute gastroenteritis in 10 consecutive seasons in Umbria (Italy). J Med Virol. 2020 Dec 1;92(12):3202–3208. doi: 10.1002/jmv.25878. [DOI] [PubMed] [Google Scholar]
- 73.Koukou D, Chatzichristou P, Trimis G, Siahanidou T, Skiathitou AV, Koutouzis EI, Syrogiannopoulos GA, Lourida A, Michos AG, Syriopoulou VP, et al. Rotavirus gastroenteritis in a neonatal unit of a Greek tertiary hospital: clinical characteristics and genotypes. PLOS ONE. 2015 Jul 27;10(7):e0133891. doi: 10.1371/journal.pone.0133891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rovida F, Vecchio Nepita E, Giardina F, Piralla A, Campanini G, Baldanti F. Rotavirus molecular epidemiology in hospitalized patients, Northern Italy, 2015-2018. New Microbiologica. 2020;43:1–5. [PubMed] [Google Scholar]
- 75.Ianiro G, Micolano R, Di Bartolo I, Scavia G, Monini M, Pagani E, RotaNet-Italy Study Group . Group a rotavirus surveillance before vaccine introduction in Italy, September 2014 to August 2017. Eurosurveillance. 2019 Apr 11;24(15). doi: 10.2807/1560-7917.ES.2019.24.15.1800418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Rougemont A, Kaplon J, Pillet S, Mory O, Gagneur A, Minoui-Tran A, Meritet J-F, Mollat C, Lorrot M, Foulongne V, et al. Molecular and clinical characterization of rotavirus from diarrheal infants admitted to pediatric emergency units in France. Pediatr Infect Dis J. 2011;30(2):118–124. doi: 10.1097/INF.0b013e3181ef034e. [DOI] [PubMed] [Google Scholar]
- 77.Aupiais C, de Rougemont A, Menager C, Vallet C, Brasme JF, Kaplon J, Pothier P, Gendrel D. Severity of acute gastroenteritis in infants infected by G1 or G9 rotaviruses. J Clin Virol. 2009. Nov. 46(3):282–285. doi: 10.1016/j.jcv.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 78.De Rougemont A, Kaplon J, Fremy C, Legrand-Guillien MC, Minoui-Tran A, Payan C, Vabret A, Mendes-Martins L, Chouchane M, Maudinas R, et al. Clinical severity and molecular characteristics of circulating and emerging rotaviruses in young children attending hospital emergency departments in France. Clin Microbiol Infect. 2016 Aug 1;22(8):737.9–15. doi: 10.1016/j.cmi.2016.05.025. [DOI] [PubMed] [Google Scholar]
- 79.Matthijnssens J, Zeller M, Heylen E, De Coster S, Vercauteren J, Braeckman T, Van Herck K, Meyer N, PirÇon J-Y, Soriano-Gabarro M, et al. Higher proportion of G2P[4] rotaviruses in vaccinated hospitalized cases compared with unvaccinated hospitalized cases, despite high vaccine effectiveness against heterotypic G2P[4] rotaviruses. Clin Microbiol Infect. 2014 Oct 1;20(10):O702–O710. doi: 10.1111/1469-0691.12612. [DOI] [PubMed] [Google Scholar]
- 80.Zeller M, Rahman M, Heylen E, De Coster S, De Vos S, Arijs I, Novo L, Verstappen N, Van Ranst M, Matthijnssens J, et al. Rotavirus incidence and genotype distribution before and after national rotavirus vaccine introduction in Belgium. Vaccine. 2010 Nov 3;28(47):7507–7513. doi: 10.1016/j.vaccine.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 81.Pietsch C, Schuster V, Liebert UG. A hospital based study on inter- and intragenotypic diversity of human rotavirus a VP4 and VP7 gene segments, Germany. J Clin Virol. 2011. Feb. 50(2):136–141. doi: 10.1016/j.jcv.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 82.Paulke-Korinek M, Kollaritsch H, Aberle SW, Zwazl I, Schmidle-Loss B, Vécsei A, Kundi M. Sustained low hospitalization rates after four years of rotavirus mass vaccination in Austria. Vaccine. 2013 May 31;31(24):2686–2691. doi: 10.1016/j.vaccine.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 83.Kaplon J, Grangier N, Pillet S, Minoui-Tran A, Vabret A, Wilhelm N, Prieur N, Lazrek M, Alain S, Mekki Y, et al. Predominance of G9P[8] rotavirus strains throughout France, 2014–2017. Clin Microbiol Infect. 2018 June 1;24(6):660.1–4. doi: 10.1016/j.cmi.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 84.Al Awaidy SA, Bawikar S, Al Busaidy S, Baqiani S, Al Abedani I, Varghese R, Abdoan H, Al Abdoon H, Bhatnagar S, Al Hasini K, et al. Considerations for introduction of a rotavirus vaccine in Oman: rotavirus disease and economic burden. J Infect Dis. 2009. Nov. 200(SUPPL. 1):248–253. doi: 10.1086/605339. [DOI] [PubMed] [Google Scholar]
- 85.Salem K, Bdour S, Zeller M, Van Ranst M, Matthijnssens J. Genotypes of rotavirus strains circulating in Amman, Jordan, in 2006/07 and their significance for the potential effectiveness of future rotavirus vaccination. Arch Virol. 2011. Sep. 156(9):1543–1550. doi: 10.1007/s00705-011-1028-6. [DOI] [PubMed] [Google Scholar]
- 86.Khalil M, Azhar E, Kao M, Al-Kaiedi N, Alhani H, Al Olayan I, Pawinski R, Gopala K, Kandeil W, Anis S, et al. Gastroenteritis attributable to rotavirus in hospitalized Saudi Arabian children in the period 2007–2008. Clin Epidemiol. 2015 Feb 11;7:129–137. doi: 10.2147/CLEP.S69502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Howidi M, Balhaj G, Yaseen H, Gopala K, Van Doorn LJ, DeAntonio R. Burden and genotyping of rotavirus disease in the United Arab Emirates: a multicenter hospital-based surveillance. Hum Vaccin Immunother. 2014 Aug 1;10(8):2284–2289. doi: 10.4161/hv.29386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahmed S, Klena J, Albana A, Alhamdani F, Oskoff J, Soliman M, Heylen E, Teleb N, Husain T, Matthijnssens J, et al. Characterization of human rotaviruses circulating in Iraq in 2008: atypical G8 and high prevalence of P[6] strains. Infect, Genet Evol. 2013. June. 16:212–217. doi: 10.1016/j.meegid.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 89.Dbaibo G, Rajab M, Inati A, Mikhael R, Choueiry E, Al-Tannir M, Salam O, Ramakrishnan G, DeAntonio R. Hospital-based surveillance study of rotavirus gastroenteritis in children under 5 years of age in Lebanon. Trials Vaccinol. 2013;2(1):25–30. doi: 10.1016/j.trivac.2013.08.002. [DOI] [Google Scholar]
- 90.Al-Kamarany MA, Al-Areqi L, Mujally A, Alkarshy F, Nasser A, Jumaan AO. Diarrheal diseases hospitalization in Yemen before and after rotavirus vaccination. Scientifica (Cairo). 2016;2016:1–6. doi: 10.1155/2016/8485417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aly M, Al Khairy A, Al Johani S, Balkhy H. Unusual rotavirus genotypes among children with acute diarrhea in Saudi Arabia. BMC Infect Dis. 2015 Apr 17;15(1). doi: 10.1186/s12879-015-0923-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ali Z, Harastani H, Hammadi M, Reslan L, Ghanem S, Hajar F, Sabra A, Haidar A, Inati A, Rajab M, et al. Rotavirus genotypes and vaccine effectiveness from a sentinel, hospital-based, surveillance study for three consecutive rotavirus seasons in Lebanon. PLOS ONE. 2016 Aug 1;11(8):1–18. doi: 10.1371/journal.pone.0161345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shoeib ARS, Hull JJ, Jiang B. Rotavirus G and P types in children with acute diarrhea in Cairo, Egypt, 2011-2012. J Egypt Public Health Assoc. 2015;90(3):121–124. doi: 10.1097/01.EPX.0000470849.84604.13. [DOI] [PubMed] [Google Scholar]
- 94.Saudy N, Elshabrawy WO, Megahed A, Foad MF, Mohamed AF. Genotyping and clinicoepidemiological characterization of rotavirus acute gastroenteritis in Egyptian children. Pol J Microbiol. 2016;65(4):433–442. doi: 10.5604/17331331.1227669. [DOI] [PubMed] [Google Scholar]
- 95.Mathew S, Al Ansari K, Al Thani AA, Zaraket H, Yassine HM. Epidemiological, molecular, and clinical features of rotavirus infections among pediatrics in Qatar. Eur J Clin Microbiol Infect Dis [Internet]. 2021;40(6):1177–1190. 10.1007/s10096-020-04108-y. [DOI] [PubMed] [Google Scholar]
- 96.Fischer TK, Rungoe C, Jensen CS, Breindahl M, Jørgensen TR, Nielsen JP, Jensen L, Malon M, Brændholt V, Fisker N, et al. The burden of rotavirus disease in Denmark 2009–2010. Pediatr Infect Dis J. 2011;30(7):e126–e129. doi: 10.1097/INF.0b013e3182145277. [DOI] [PubMed] [Google Scholar]
- 97.Vainio K, Nordbø SA, Njølstad G, Størvold G, Døllner H, Midgaard C, Bosse FJ, Rognlien AGW, Rojahn A, Wathne K-O, et al. Detection and characterization of group a rotaviruses in children hospitalized with acute gastroenteritis in Norway, 2006–2008. J Med Virol. 2009;81(10):1839–1844. doi: 10.1002/jmv.21576. [DOI] [PubMed] [Google Scholar]
- 98.Arana A, Jere KC, Chaguza C, Montes M, Alkorta M, Iturriza-Gomara M, Cilla G. Molecular epidemiology of G12 rotavirus strains during eight consecutive epidemic seasons in the Basque country (north of Spain), 2010–2018. Infect, Genet And Evol. 2019 Jul 1;71:67–75. doi: 10.1016/j.meegid.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 99.Sánchez-Fauquier A, Montero V, Colomina J, González-Galán V, Aznar J, Aisa ML, Gutierrez C, Sainz de Baranda C, Wilhelmi I. Global study of viral diarrhea in hospitalized children in Spain: results of structural surveillance of viral gastroenteritis net work (vigess-net) 2006–2008. J Clin Virol. 2011. Dec. 52(4):353–358. doi: 10.1016/j.jcv.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 100.Konstantopoulos A, Tragiannidis A, Fouzas S, Kavaliotis I, Tsiatsou O, Michailidou E, Spanaki A, Mantagos S, Kafetzis D, Papaevangelou V, et al. Burden of rotavirus gastroenteritis in children <5 years of age in Greece: hospital-based prospective surveillance (2008–2010). BMJ Open. 2013;3(12):1–7. doi: 10.1136/bmjopen-2013-003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hashem S, Shoman SA, Mohamed AF. Isolation and molecular genotyping of group a rotavirus strains circulating among Egyptian infants and children. EJMM. 2012;21(3):11–20. doi: 10.12816/0004888. [DOI] [Google Scholar]
- 102.El-Senousy WM, Abu Senna ASM, Mohsen NA, Hasan SF, Sidkey NM. Clinical and environmental surveillance of rotavirus common genotypes showed high prevalence of common P genotypes in Egypt. Food Environ Virol. 2020 June 1;12(2):99–117. doi: 10.1007/s12560-020-09426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Steyer A, Sagadin M, Kolenc M, Poljšak-Prijatelj M. Molecular characterization of rotavirus strains from pre- and post-vaccination periods in a country with low vaccination coverage: the case of Slovenia. Infect, Genet And Evol. 2014 Dec 1;28:413–425. doi: 10.1016/j.meegid.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 104.Iturriza-Gómara M, Dallman T, Bányai K, Böttiger B, Buesa J, Diedrich S, Fiore L, Johansen K, Korsun N, Kroneman A, et al. Rotavirus surveillance in Europe, 2005–2008: Web-enabled reporting and real-time analysis of genotyping and epidemiological data. J Infect Dis. 2009. Nov. 200(SUPPL. 1):215–221. doi: 10.1086/605049. [DOI] [PubMed] [Google Scholar]
- 105.Iturriza-Gómara M, Dallman T, Bányai K, Böttiger B, Buesa J, Diedrich S, Fiore L, Johansen K, Koopmans M, Korsun N, et al. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiol Infect. 2010. June. 139(6):895–909. doi: 10.1017/S0950268810001810. [DOI] [PubMed] [Google Scholar]
- 106.Diez-Domingo J, Baldo JM, Patrzalek M, Pazdiora P, Forster J, Cantarutti L, Pirçon J-Y, Soriano-Gabarró M, Meyer N. Primary care-based surveillance to estimate the burden of rotavirus gastroenteritis among children aged less than 5 years in six European countries. Eur J Pediatr. 2011. Feb. 170(2):213–222. doi: 10.1007/s00431-010-1289-1. [DOI] [PubMed] [Google Scholar]
- 107.Iturriza-Gómara M, Hungerford D. EuroRotaNet ANNUAL REPORT 2019 [Internet]. 2020.
- 108.Elbashir I, Aldoos NF, Mathew S, Al Thani AA, Emara MM, Yassine HM. Molecular epidemiology, genetic diversity, and vaccine availability of viral acute gastroenteritis in the middle East and North Africa (MENA) region. J Infect Public Health. 2022 Nov 1;15(11):1193–1211. doi: 10.1016/j.jiph.2022.09.001. [DOI] [PubMed] [Google Scholar]
- 109.Hungerford D. EUROROTANET ANNUAL REPORT 2021 [Internet]. Liverpool; 2023. http://www.eurorotanet.com. [Google Scholar]
- 110.Cates JE, Amin AB, Tate JE, Lopman B, Parashar U. Do rotavirus strains affect vaccine effectiveness? A systematic review and meta-analysis. Pediatr Infect Disease J. 2021 Dec 21;40(12):1135–1143. doi: 10.1097/INF.0000000000003286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Amin AB, Tate JE, Waller LA, Lash TL, Lopman BA. Monovalent rotavirus vaccine efficacy against different rotavirus genotypes: a pooled analysis of phase II and III trial data. Clin Infect Dis. 2023 Feb 8;76(3):e1150–e1156. doi: 10.1093/cid/ciac699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Leshem E, Parashar U. Use of surveillance data to assess the impact of vaccination on circulating rotavirus strains. J Pediatr Infect Dis Soc. 2015 Dec 1;4(4):e90–e92. doi: 10.1093/jpids/piu114. [DOI] [PubMed] [Google Scholar]
- 113.Degiuseppe JI, Stupka JA. Genotype distribution of group a rotavirus in children before and after massive vaccination in Latin America and the Caribbean: systematic review. Vaccine. 2020. Jan. 38(4):733–740. doi: 10.1016/j.vaccine.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 114.Seheri LM, Magagula NB, Peenze I, Rakau K, Ndadza A, Mwenda JM, Weldegebriel G, Steele AD, Mphahlele MJ. Rotavirus strain diversity in Eastern and Southern African countries before and after vaccine introduction. Vaccine. 2018. Nov. 36(47):7222–7230. doi: 10.1016/j.vaccine.2017.11.068. [DOI] [PubMed] [Google Scholar]
- 115.Kawai K, O’Brien MA, Goveia MG, Mast TC, El Khoury AC. Burden of rotavirus gastroenteritis and distribution of rotavirus strains in Asia: a systematic review. Vaccine. 2012. Feb. 30(7):1244–1254. doi: 10.1016/j.vaccine.2011.12.092. [DOI] [PubMed] [Google Scholar]
- 116.Jesudason T, Oluwaseun S, Bermudez M, Schirrmacher H, Hauck C, Hungerford D, Systematic literature review on the prevalence of rotavirus genotypes in Asia between 2015-2021, editor. European Society for Paediatric Infectious Diseases 42nd Annual ESPID Meeting Abstract Book [Internet]. European society for paediatric infectious diseases. Copenhagen: European Society for Paediatric Infectious Diseases; 2023. cited 2024 Jul 5]. p. 486–486. https://info.kenes.com/Flip/ESPID24_ESPID24/?_ga=2.208332624.338596466.1720181106-8444816.1720181088. [Google Scholar]
- 117.Abou-Nader AJ, Sauer MA, Steele AD, Tate JE, Atherly D, Parashar UD, Santosham M, Nelson EAS. Global rotavirus vaccine introductions and coverage: 2006 – 2016. Hum Vaccin Immunother. 2018 Sep 2;14(9):2281–2296. doi: 10.1080/21645515.2018.1470725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.GündeşliOğlu ÖÖ, Kocabaş E, Haytoğlu Z, TiMurtaş Dayar G, Kiliç Çil M, Durmaz R. Rotavirus prevalence and genotype distribution in children with acute gastroenteritis in Adana Province. Bull Of Microbiol. 2018 Apr 1;52(2):156–165. doi: 10.5578/mb.66648. [DOI] [PubMed] [Google Scholar]
- 119.Moussa A, Ben Hadj Fredj M, Fodha I, Benhamida-Rebaï M, Kacem S, Argoubi A, Bennour H, Boujaafar N, Trabelsi A. Distribution of rotavirus VP7 and VP4 genotypes circulating in Tunisia from 2009 to 2014: emergence of the genotype G12. J Med Microbiol. 2016 Sep 1;65(9):1028–1037. doi: 10.1099/jmm.0.000305. [DOI] [PubMed] [Google Scholar]
- 120.Hungerford D. EUROROTANET ANNUAL REPORT 2022 [Internet]. 2022. Feb. https://www.eurorotanet.com/wp-content/uploads/2024/03/EuroRotaNet_annual_report-2022_20240228_Final-v1.0.pdf.
- 121.Hungerford D, Vivancos R. EuroRotaNet network members. In-season and out-of-season variation of rotavirus genotype distribution and age of infection across 12 European countries before the introduction of routine vaccination, 2007/08 to 2012/13. Eurosurveillance [Internet]. 2016 Jan 14 cited 2024 Jan 31;21(2):1–12. 10.2807/1560-7917.ES.2016.21.2.30106. [DOI] [PubMed] [Google Scholar]
- 122.Karatas M, Bloemen M, Cuypers L, Wollants E, Van Ranst M, Matthijnssens J. 14-year experience of national reference center for rotavirus a in Belgium: unusual dominance of equine-like G3P[8] genotype after the pandemic. Res Sq [Internet]. 2024 Apr 26 cited 2024 Jul 5]. https://www.researchsquare.com/article/rs-4320299/v1. 1–23.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


