Abstract
Loss of S-methyl-5′-thioadenosine phosphorylase (MTAP) expression is a common event in cancer leading to a critical vulnerability of cancer cells towards anti-cancer drugs. Homozygous MTAP deletions result in a complete expression loss that can be detected by immunohistochemistry (IHC). In this study, a tissue microarray containing 17,078 samples from 149 different tumor entities was analyzed by IHC, and complete MTAP loss was validated by fluorescence in situ hybridization. MTAP loss was observed in 83 of 149 tumor categories, including neuroendocrine neoplasms (up to 80%), Hodgkin lymphoma (50.0%), mesothelioma (32.0% to 36.8%), gastro-intestinal adenocarcinoma (4.0% to 40.5%), urothelial neoplasms (10.5% to 36.7%), squamous cell carcinomas (up to 38%), and various types of sarcomas (up to 20%) and non-Hodgkin lymphomas (up to 14%). Homozygous MTAP deletion was found in 90% to 100% of cases with MTAP expression loss in most tumor categories. However, neuroendocrine tumors, Hodgkin lymphomas, and other lymphomas lacked MTAP deletions. MTAP deficiency was significantly linked to unfavorable tumor phenotype in selected tumor entities and the presence of PD-L1 expression on tumor cells, absence of PD-L1 expression on immune cells, and a low density of CD8+ lymphocytes. In summary, MTAP deficiency can occur in various tumor entities and is linked to unfavorable tumor phenotype and noninflamed tumor microenvironment, but is not always related to deletions. MTAP IHC is of considerable diagnostic value for the detection of neoplastic transformation in multiple different applications.
Key Words: MTAP, tissue micro array, immunohistochemistry, FISH, diagnostic marker
The enzyme S-methyl-5′-thioadenosine phosphorylase (MTAP) is essential for the salvage pathway of adenine synthesis.1 The gene is located at 9p21.3, only 30 kb apart from the CDKN2A gene, which is deleted in up to 15% of cancers.2–4 Codeletions of MTAP occur in 80% to 90% of tumors with CDKN2A homozygous deletion.5 Evidence is accumulating that MTAP deficiency results in a critical vulnerability of cancer cells towards drugs targeting several different pathways.6 As adenine synthesis can only be maintained by de novo biosynthesis in MTAP deficient cancer cells, inhibition of enzymes for folate synthesis leads to increased cell death in experimental models and also showed significant anti-cancer efficiency in patients with urinary bladder cancer.6 MTAP deficient cells can also be targeted by drugs inhibiting protein arginine N-methyltransferase 5 (PRMT5) and methionine adenosyltransferase II, alpha (summarized in reference7). PRMT5 is an essential enzyme for the methylation of numerous proteins, thus regulating their activity,8 whereas MAT2A is essential for the synthesis of S-adenosylmethionine, the methyl donor, and substrate of PRMT5.9,10 Upregulation of PRMT5 was found to result in progression and poor prognosis in a variety of malignancies including breast cancer,11 glioblastoma,12 prostate cancer,13 pancreatic cancer,14 bladder cancer,15 leukemia,16 lymphoma,17 ovarian cancer,18 lung cancer,19 and gastric cancer.20 Because PRMT5 is partially inhibited by the intracellular accumulation of the unprocessed MTAP metabolite MTA,21 MTAP deficiency makes cells more susceptible to drugs targeting PRMT5 or methionine adenosyltransferase II, alpha (summarized in reference7). A clinical phase 1 trial published in November 2023 has shown significant size reduction in tumor lesions of patients with MTAP-deleted cancers, including epithelioid malignant mesothelioma, non–small-cell lung cancer, malignant melanoma, and adenocarcinoma with PRMT5 inhibitor MRTX1719.22 Several other clinical trials targeting PRMT5 are ongoing.23 Also of interest, both MTAP deficiency24 and CDKN2A loss25 could be linked to poor response to immune checkpoint inhibitor (CPI) therapy in urothelial carcinoma. 9p21 loss was also associated with poor response to CPI in lung cancer, melanoma, and miscellaneous solid tumors.26
Considering these promising data and the abundance of 9p21 deletions in many cancer types, it is likely that many cancers from different entities may benefit from drugs targeting MTAP deficiency. Immunohistochemical detection of MTAP may represent a suitable surrogate for the detection of homozygous 9p21 deletions because MTAP is rather ubiquitously expressed in normal tissues, and its expression is completely lost in homozygously deleted cells. Because of frequent homozygous 9p21 deletions in pleural mesothelioma, MTAP immunohistochemistry (IHC) is used as a tool for supporting this diagnosis in histologic and cytologic specimens.27 Only few studies have investigated MTAP expression by IHC in other tumor entities and described complete expression loss to occur in subsets of high-grade gliomas,28 pulmonary adenocarcinoma,29 esophageal adenocarcinomas,30 serous borderline tumors and serous carcinomas of the ovary,31 pancreatic intraepithelial neoplasia,32 Ewing sarcoma,33 and osteosarcoma.34 However, most previous studies on nonmesothelial neoplasms employed rather small patient cohorts, and many tumor entities have so far not been analyzed.
To comprehensively determine the prevalence of MTAP protein expression loss in cancer and to assess the potential diagnostic utility of MTAP IHC, we analyzed a preexisting set of tissue microarrays (TMAs) containing more than 17,000 tumor tissue samples from 149 different tumor types and subtypes, as well as 76 non-neoplastic tissue categories for MTAP expression by IHC in this study. All cancers with complete MTAP loss were also validated for MTAP deletions by fluorescence in situ hybridization (FISH).
MATERIALS AND METHODS
Tissue Microarrays
Our normal TMA was composed of 8 samples from 8 different donors from each of 76 different normal tissue types (608 samples on one slide). The cancer TMAs included a total of 17,078 primary tumors from 149 different tumor types and subtypes.
Detailed histopathologic and molecular data were available for cancers of the liver (n = 231), pancreas (n = 598), breast (n = 1208), neuroendocrine tumors (NETs; n = 138), and squamous cell carcinomas (SQCC) of various origin (n = 902). The composition of normal and cancer TMAs is described in the results section. All samples were from the archives of the Institute of Pathology, University Hospital of Hamburg, Germany, the Institute of Pathology, Clinical Center Osnabrueck, Germany, and the Department of Pathology, Academic Hospital Fuerth, Germany. Tissues were fixed in 4% buffered formalin and then embedded in paraffin. The TMA manufacturing process was described earlier in detail.35,36 In brief, one tissue spot (diameter: 0.6 mm) per patient was used. The use of archived remnants of diagnostic tissues for TMA manufacturing, their analysis for research purposes, and the use of patient data were according to local laws (HmbKHG, §12), and the analysis had been approved by the local ethics committee (Ethics Commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration. Data on PD-L1 and CD8+ lymphocyte density were available for subsets of our tumors from a previous study.37
Fluorescence In Situ Hybridization
Five micrometer TMA sections were deparaffinized with xylol, rehydrated through a graded alcohol series, and exposed to heat-induced denaturation for 10 minutes in a water bath at 99 °C in P1 pretreatment solution (Agilent Technologies; #K5799). For proteolytic treatment, slides were added to VP2000 protease buffer (Abbott; #2J.0730) for 200 minutes at 37 °C in a water bath. A commercial 315 kilobases (kb) FISH probe spanning the 194 kb region on chromosome 9p21 where the CDKN2A (27 kb) and the MTAP (135 kb) gene localize was utilized for 9p21 copy number detection (ZytoLight SPEC CDKN2A/CEN 9 Dual Color Probe, Zytovision; #Z-2063). Loss of the 9p21 probe signal in FISH experiments indicates the presence of large deletions, including both CDKN2A and MTAP. Hybridization was performed overnight at 37 °C in a humidified chamber. Posthybridization washes were done according to the manufacturer’s direction at 37 °C (Agilent Technologies; #K5799). Nuclei were counterstained with 125 ng/mL 4’,6-diamino-2-phenylindole in antifade solution (Biozol; #VEC-H-1200). Stained tissues were manually interpreted with an epifluorescence microscope, and copy numbers of 9p21 and centromere 9 were estimated for each tissue spot. The presence of equal numbers of 9p21 and centromere 9 signals in tumor cell nuclei was considered as 9p21 normal. The presence of fewer 9p21 signals than centromere 9 probe signals in at least 60% of tumor cell nuclei or one 9p21 and one centromere 9 signal (monosomy of chromosome 9) in nearly all tumor cell nuclei were considered as heterozygous deletion. The complete absence of 9p21 signals but the presence of centromere 9 signals in the tumor cell nuclei and the presence of unequivocal 9p21 and centromere 9 signals in tumor-adjacent normal cell nuclei were considered homozygous 9p21 deletion.
Tissue spots lacking any detectable 9p21 signals in all (tumor and normal cell nuclei) or lack of any normal cells as an internal control for successful hybridization of the 9p21 probe were excluded from the analysis. These thresholds were based on the results of a previous study in which 100% concordance in PTEN copy number status was achieved using FISH and single nucleotide polymorphism array hybridization in a cohort of 72 prostate cancers.38 Representative FISH findings are shown in Supplemental Figure 1 (Supplemental Digital Content 1, http://links.lww.com/PAS/B920).
Immunohistochemistry
A TMA containing bladder carcinomas with 9p21 wild type (n = 20), heterozygous deletions (n = 20), and homozygous deletions (n = 20) was used to titrate the anti-MTAP antibodies to obtain maximal staining in non-neoplastic cells while background staining was still lacking in homozygously deleted cancers (data not shown). The optimal protocol was as follows. Freshly prepared 2.5 μm TMA sections were applied on 1 day in 1 experiment in a Dako Omnis automated stainer (Agilent Technologies) using the EnVision FLEX, High pH Kit (Agilent Technologies, #GV800). Slides were deparaffinized with Clearify agent (Agilent Technologies, #GC810) and exposed to heat-induced antigen retrieval for 30 minutes at 97 °C in target retrieval solution, high pH reagent (part of Agilent kit #GV800). Primary antibody specific for MTAP (recombinant rabbit monoclonal, MSVA-741R, MS Validated Antibodies GmbH, #5293-741R) was applied at ambient temperature for 30 minutes at a dilution of 1:50. Endogenous peroxidase activity was blocked with peroxidase blocking-reagent (part of Agilent kit #GV800) for 3 minutes. Bound antibody was visualized using the EnVision FLEX, High pH kit reagents DAB+ Chromogen and Substrate Buffer (parts of Agilent kit #GV800) and EnVision FLEX + Rabbit LINKER (Agilent Technologies; #GV809) according to the manufacturer’s directions. The sections were counterstained with hemalaun. For tumor tissues, the average staining intensity of unequivocally neoplastic cells was estimated as 0, 1+, 2+, and 3+. For the classification of a tumor as completely negative (0, complete loss of MTAP), the presence of unequivocal MTAP staining in tumor adjacent normal tissue was required. Tumors with a complete absence of MTAP staining in cancerous cells and a lack of stromal cells with unequivocal MTAP staining were considered “noninformative.” For the purpose of antibody validation, the normal tissue TMA was also analyzed by the mouse monoclonal MTAP antibody 2G4 (Abnova; # H00004507-M01) with a manual protocol. These slides were deparaffinized with xylol, rehydrated through a graded alcohol series, and exposed to heat-induced antigen retrieval for 5 minutes in an autoclave at 121 °C in pH 7.8 tris-EDTA-citrate puffer. Endogenous peroxidase activity was blocked with Dako Peroxidase Blocking Solution (Agilent Technologies; #S2023) for 10 minutes. The MTAP antibody 2G4 was applied at 37 °C for 60 minutes at a dilution of 1:40. The sections were counterstained with hemalaun.
Statistics
Statistical calculations were performed with JMP17 software (SAS). Contingency tables and the χ2 test were performed to search for associations between MTAP immunostaining and tumor phenotype and PD-L1 immunostaining. Analysis of variance analysis was used to search for associations between MTAP immunostaining and the density of CD8+ lymphocytes (tumor microenvironment).
RESULTS
Technical Issues
A total of 13,067 (76.5%) of 17,078 tumor samples were interpretable in our tumor TMA analysis. Noninterpretable samples demonstrated a lack of unequivocal tumor cells, loss of the entire tissue spot, or a complete absence of MTAP staining in both tumor cells and adjacent stroma cells. Sufficient numbers of samples (≥4) of each normal tissue type were evaluable.
MTAP in Normal Tissue
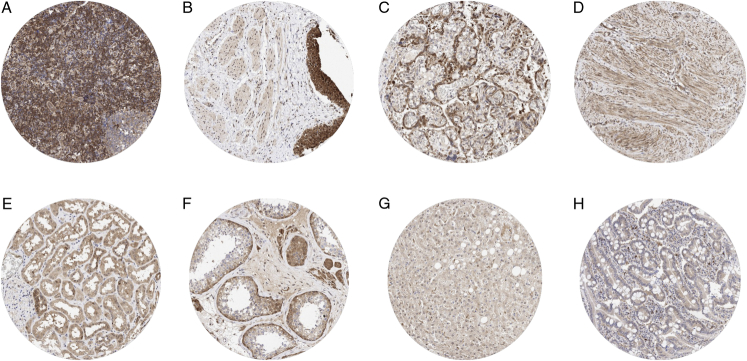
MTAP immunostaining revealed a nuclear and/or cytoplasmic positivity at variable intensity of most cell types. Particularly strong staining occurred in the ovarian stroma, endothelial cells, inflammatory cells, cytotrophoblast and chorion cells of the placenta, urothelium, epithelial cells of the epididymis and the seminal vesicles, breast glands, salivary glands, spermatogonia, adrenal gland, thyroid, and the adenohypophysis. MTAP staining was particularly low or even absent in hepatocytes, heart muscle, fat tissue, and the brain. Representative images are shown in Figure 1. All these findings were obtained by using the recombinant rabbit monoclonal antibody MSVA-741R and the mouse monoclonal 2G4 antibody (Supplemental Fig. 2, Supplemental Digital Content 2, http://links.lww.com/PAS/B921) and, therefore, considered to be specific. By using the antibody 2G4, a strong additional staining of goblet cells of the entire intestinal mucosa was observed. This staining was considered an antibody-specific cross-reactivity of antibody 2G4.
FIGURE 1.
MTAP immunostaining of normal tissues. The panels show MTAP immunostaining of variable intensity for almost every cell type in each tissue. MTAP staining was particularly intense in interfollicular lymphocytes of a lymph node (A), urothelial cells in the urinary bladder (B), and in cytotrophoblast cells of the placenta (C). A moderate to strong MTAP staining was also seen in myometrium cells of the uterus (D), tubular cells of the kidney (E), and in spermatogonia and Leydig cells of the testis (F). MTAP staining was only weak in maturing spermatids of the testis (F), hepatocytes of the liver (G), and in colorectal epithelial cells (H).
MTAP in Cancer
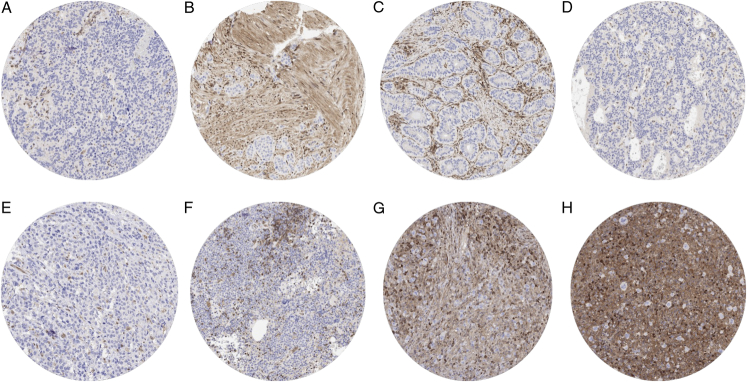
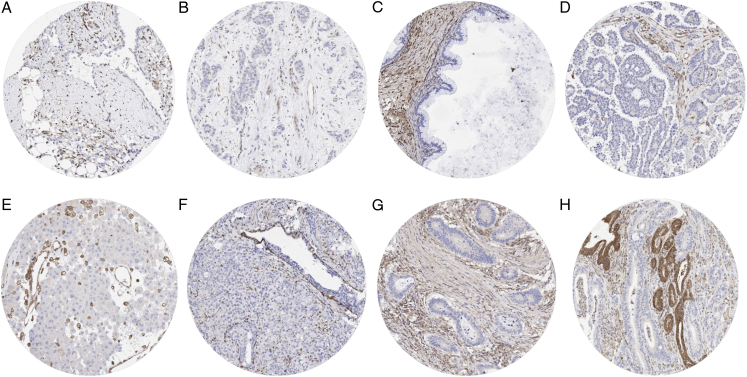
MTAP immunostaining in tumors was of various intensities and included a variable mixture of cytoplasmic and nuclear staining. MTAP immunostaining was considered weak (1+) in 6,339 (48.5%), moderate in 4,802 (36.7%), and strong in 993 (7.6%) of tumors. A complete loss (0) of MTAP staining was seen in 933 (7.1%) of tumors. At least one case with a complete MTAP expression loss was observed in 83 (55.7%) of 149 tumor categories (Table 1). Representative images of MTAP deficient tumors lacking 9p21 deletions and harboring 9p21 deletions are shown in Figures 2 and 3. MTAP staining was most commonly lost in Hodgkin lymphoma (50%), neuroendocrine neoplasms of various sites (up to 80%), mesothelioma (32.0% to 36.8%), biliopancreatic adenocarcinomas (17.9% to 40.5%), urothelial neoplasms (10.5% to 36.7%), malignant melanoma (15.7% to 28.4%), mucinous carcinoma of the ovary (29.2%), SQCC of various organs of origin (up to 38%), various types of sarcomas (up to 20%), various types of non-Hodgkin lymphoma (up to 14%), adenocarcinoma of the lung (12.0%), and in gastric adenocarcinoma (4.0% to 11.9%).
TABLE 1.
MTAP Immunostaining in Human Tumors
| MTAP immunostaining | |||||||
|---|---|---|---|---|---|---|---|
| Tumor category | Tumor entity | on TMA (n) | Analyzable (n) | 0 (%) | 1+ (%) | 2+ (%) | 3+ (%) |
| Tumors of the skin | Basal cell carcinoma of the skin | 89 | 68 | 2.9 | 45.6 | 38.2 | 13.2 |
| Benign nevus | 29 | 23 | 0.0 | 4.3 | 26.1 | 69.6 | |
| Squamous cell carcinoma of the skin | 145 | 103 | 9.7 | 68.9 | 19.4 | 1.9 | |
| Malignant melanoma | 65 | 51 | 15.7 | 19.6 | 49.0 | 15.7 | |
| Malignant melanoma lymph node metastasis | 86 | 67 | 28.4 | 32.8 | 23.9 | 14.9 | |
| Merkel cell carcinoma | 2 | 1 | 0.0 | 100.0 | 0.0 | 0.0 | |
| Tumors of the head and neck | Squamous cell carcinoma of the larynx | 109 | 81 | 24.7 | 56.8 | 18.5 | 0.0 |
| Squamous cell carcinoma of the pharynx | 60 | 59 | 11.9 | 28.8 | 49.2 | 10.2 | |
| Oral squamous cell carcinoma (floor of the mouth) | 130 | 119 | 11.8 | 44.5 | 37.8 | 5.9 | |
| Pleomorphic adenoma of the parotid gland | 50 | 38 | 0.0 | 60.5 | 36.8 | 2.6 | |
| Warthin tumor of the parotid gland | 104 | 63 | 0.0 | 60.3 | 39.7 | 0.0 | |
| Adenocarcinoma, NOS (papillary cystadenocarcinoma) | 14 | 6 | 0.0 | 100.0 | 0.0 | 0.0 | |
| Salivary duct carcinoma | 15 | 3 | 66.7 | 33.3 | 0.0 | 0.0 | |
| Acinic cell carcinoma of the salivary gland | 181 | 53 | 1.9 | 62.3 | 35.8 | 0.0 | |
| Adenocarcinoma NOS of the salivary gland | 109 | 29 | 10.3 | 65.5 | 20.7 | 3.4 | |
| Adenoid cystic carcinoma of the salivary gland | 180 | 33 | 6.1 | 27.3 | 63.6 | 3.0 | |
| Basal cell adenocarcinoma of the salivary gland | 25 | 12 | 0.0 | 33.3 | 41.7 | 25.0 | |
| Basal cell adenoma of the salivary gland | 101 | 26 | 0.0 | 26.9 | 65.4 | 7.7 | |
| Epithelial-myoepithelial carcinoma of the salivary gland | 53 | 18 | 0.0 | 44.4 | 55.6 | 0.0 | |
| Mucoepidermoid carcinoma of the salivary gland | 343 | 149 | 4.0 | 65.8 | 29.5 | 0.7 | |
| Myoepithelial carcinoma of the salivary gland | 21 | 10 | 0.0 | 70.0 | 30.0 | 0.0 | |
| Myoepithelioma of the salivary gland | 11 | 8 | 0.0 | 37.5 | 62.5 | 0.0 | |
| Oncocytic carcinoma of the salivary gland | 12 | 2 | 0.0 | 100.0 | 0.0 | 0.0 | |
| Polymorphous adenocarcinoma, low grade, of the salivary gland | 41 | 2 | 0.0 | 0.0 | 50.0 | 50.0 | |
| Pleomorphic adenoma of the salivary gland | 53 | 22 | 4.5 | 45.5 | 40.9 | 9.1 | |
| Tumors of the lung, pleura and thymus | Adenocarcinoma of the lung | 196 | 167 | 12.0 | 64.7 | 21.6 | 1.8 |
| Squamous cell carcinoma of the lung | 80 | 59 | 16.9 | 62.7 | 20.3 | 0.0 | |
| Mesothelioma, epithelioid | 40 | 25 | 32.0 | 24.0 | 40.0 | 4.0 | |
| Mesothelioma, biphasic | 29 | 19 | 36.8 | 42.1 | 15.8 | 5.3 | |
| Thymoma | 29 | 21 | 0.0 | 52.4 | 47.6 | 0.0 | |
| Lung, NET | 29 | 28 | 53.6 | 46.4 | 0.0 | 0.0 | |
| Tumors of the female genital tract | Squamous cell carcinoma of the vagina | 78 | 54 | 18.5 | 38.9 | 37.0 | 5.6 |
| Squamous cell carcinoma of the vulva | 157 | 119 | 21.0 | 48.7 | 27.7 | 2.5 | |
| Squamous cell carcinoma of the cervix | 136 | 119 | 1.7 | 57.1 | 35.3 | 5.9 | |
| Adenocarcinoma of the cervix | 23 | 22 | 4.5 | 31.8 | 59.1 | 4.5 | |
| Endometrioid endometrial carcinoma | 338 | 296 | 0.7 | 61.5 | 35.5 | 2.4 | |
| Endometrial serous carcinoma | 86 | 67 | 1.5 | 38.8 | 41.8 | 17.9 | |
| Carcinosarcoma of the uterus | 57 | 52 | 0.0 | 59.6 | 34.6 | 5.8 | |
| Endometrial carcinoma, high grade, G3 | 13 | 12 | 0.0 | 16.7 | 75.0 | 8.3 | |
| Endometrial clear cell carcinoma | 9 | 7 | 0.0 | 85.7 | 14.3 | 0.0 | |
| Endometrioid carcinoma of the ovary | 130 | 96 | 1.0 | 39.6 | 44.8 | 14.6 | |
| Serous carcinoma of the ovary | 580 | 443 | 1.1 | 38.1 | 47.9 | 12.9 | |
| Mucinous carcinoma of the ovary | 101 | 65 | 29.2 | 44.6 | 18.5 | 7.7 | |
| Clear cell carcinoma of the ovary | 51 | 32 | 6.3 | 62.5 | 31.3 | 0.0 | |
| Carcinosarcoma of the ovary | 47 | 40 | 7.5 | 40.0 | 42.5 | 10.0 | |
| Granulosa cell tumor of the ovary | 44 | 42 | 2.4 | 38.1 | 45.2 | 14.3 | |
| Leydig cell tumor of the ovary | 4 | 4 | 0.0 | 0.0 | 100.0 | 0.0 | |
| Sertoli cell tumor of the ovary | 1 | 1 | 0.0 | 0.0 | 100.0 | 0.0 | |
| Sertoli Leydig cell tumor of the ovary | 3 | 3 | 0.0 | 33.3 | 66.7 | 0.0 | |
| Steroid cell tumor of the ovary | 3 | 3 | 0.0 | 0.0 | 100.0 | 0.0 | |
| Brenner tumor | 41 | 39 | 5.1 | 12.8 | 66.7 | 15.4 | |
| Tumors of the breast | Invasive breast carcinoma of no special type | 499 | 454 | 1.5 | 48.0 | 40.5 | 9.9 |
| Lobular carcinoma of the breast | 192 | 166 | 0.0 | 34.3 | 54.8 | 10.8 | |
| Medullary carcinoma of the breast | 23 | 19 | 5.3 | 21.1 | 42.1 | 31.6 | |
| Tubular carcinoma of the breast | 20 | 11 | 0.0 | 27.3 | 54.5 | 18.2 | |
| Mucinous carcinoma of the breast | 29 | 26 | 0.0 | 26.9 | 65.4 | 7.7 | |
| Phyllodes tumor of the breast | 50 | 47 | 0.0 | 27.7 | 57.4 | 14.9 | |
| Tumors of the digestive system | Adenomatous polyp, low-grade dysplasia | 50 | 42 | 0.0 | 33.3 | 64.3 | 2.4 |
| Adenomatous polyp, high-grade dysplasia | 50 | 48 | 0.0 | 27.1 | 60.4 | 12.5 | |
| Adenocarcinoma of the colon | 2483 | 1892 | 0.8 | 37.3 | 41.8 | 20.1 | |
| Gastric adenocarcinoma, diffuse type | 215 | 151 | 4.0 | 64.9 | 31.1 | 0.0 | |
| Gastric adenocarcinoma, intestinal type | 215 | 168 | 11.9 | 44.0 | 38.7 | 5.4 | |
| Gastric adenocarcinoma, mixed type | 62 | 57 | 5.3 | 57.9 | 29.8 | 7.0 | |
| Adenocarcinoma of the esophagus | 83 | 45 | 13.3 | 46.7 | 28.9 | 11.1 | |
| Squamous cell carcinoma of the esophagus | 76 | 37 | 27.0 | 32.4 | 37.8 | 2.7 | |
| Squamous cell carcinoma of the anal canal | 91 | 78 | 5.1 | 43.6 | 46.2 | 5.1 | |
| Cholangiocarcinoma | 58 | 43 | 16.3 | 51.2 | 32.6 | 0.0 | |
| Gallbladder adenocarcinoma | 51 | 42 | 40.5 | 42.9 | 16.7 | 0.0 | |
| Gallbladder Klatskin tumor | 42 | 34 | 11.8 | 61.8 | 17.6 | 8.8 | |
| HCC | 312 | 294 | 9.2 | 60.9 | 24.8 | 5.1 | |
| Ductal adenocarcinoma of the pancreas | 659 | 506 | 32.4 | 42.1 | 22.3 | 3.2 | |
| Pancreatic/ampullary adenocarcinoma | 98 | 78 | 17.9 | 51.3 | 29.5 | 1.3 | |
| Acinar cell carcinoma of the pancreas | 18 | 16 | 18.8 | 50.0 | 18.8 | 12.5 | |
| GIST | 62 | 55 | 3.6 | 56.4 | 38.2 | 1.8 | |
| Appendix, NET | 25 | 14 | 35.7 | 42.9 | 21.4 | 0.0 | |
| Colorectal, NET | 12 | 10 | 70.0 | 10.0 | 20.0 | 0.0 | |
| Ileum, NET | 53 | 47 | 78.7 | 19.1 | 2.1 | 0.0 | |
| Pancreas, NET | 101 | 89 | 11.2 | 71.9 | 12.4 | 4.5 | |
| Colorectal, NEC | 14 | 13 | 7.7 | 38.5 | 46.2 | 7.7 | |
| Ileum, NEC | 8 | 5 | 80.0 | 0.0 | 20.0 | 0.0 | |
| Gallbladder, NEC | 4 | 4 | 0.0 | 25.0 | 75.0 | 0.0 | |
| Pancreas, NEC | 14 | 12 | 8.3 | 58.3 | 33.3 | 0.0 | |
| Tumors of the urinary system | Noninvasive papillary urothelial carcinoma, pTa G2 low-grade | 87 | 76 | 10.5 | 27.6 | 25.0 | 36.8 |
| Noninvasive papillary urothelial carcinoma, pTa G2 high-grade | 80 | 66 | 28.8 | 16.7 | 45.5 | 9.1 | |
| Noninvasive papillary urothelial carcinoma, pTa G3 | 126 | 101 | 18.8 | 26.7 | 42.6 | 11.9 | |
| Urothelial carcinoma, pT2-4 G3 | 735 | 573 | 29.0 | 30.4 | 28.8 | 11.9 | |
| Squamous cell carcinoma of the bladder | 22 | 21 | 38.1 | 52.4 | 4.8 | 4.8 | |
| Small cell NEC of the bladder | 5 | 4 | 0.0 | 0.0 | 50.0 | 50.0 | |
| Sarcomatoid urothelial carcinoma | 25 | 12 | 8.3 | 33.3 | 50.0 | 8.3 | |
| Urothelial carcinoma of the kidney pelvis | 62 | 49 | 36.7 | 26.5 | 28.6 | 8.2 | |
| Clear cell renal cell carcinoma | 1287 | 1109 | 0.6 | 62.2 | 36.4 | 0.7 | |
| Papillary renal cell carcinoma | 368 | 318 | 1.3 | 45.3 | 47.5 | 6.0 | |
| Clear cell (tubulo) papillary renal cell carcinoma | 26 | 21 | 0.0 | 52.4 | 42.9 | 4.8 | |
| Chromophobe renal cell carcinoma | 170 | 146 | 0.0 | 73.3 | 26.0 | 0.7 | |
| Oncocytoma of the kidney | 257 | 222 | 0.0 | 49.5 | 49.5 | 0.9 | |
| Tumors of the male genital organs | Adenocarcinoma of the prostate, Gleason 3+3 | 83 | 67 | 0.0 | 47.8 | 43.3 | 9.0 |
| Adenocarcinoma of the prostate, Gleason 4+4 | 80 | 65 | 0.0 | 32.3 | 60.0 | 7.7 | |
| Adenocarcinoma of the prostate, Gleason 5+5 | 85 | 70 | 0.0 | 38.6 | 57.1 | 4.3 | |
| Adenocarcinoma of the prostate (recurrence) | 258 | 159 | 1.9 | 42.8 | 49.1 | 6.3 | |
| Small cell NEC of the prostate | 2 | 1 | 0.0 | 100.0 | 0.0 | 0.0 | |
| Seminoma | 682 | 454 | 0.4 | 72.5 | 24.2 | 2.9 | |
| Embryonal carcinoma of the testis | 54 | 44 | 0.0 | 40.9 | 50.0 | 9.1 | |
| Leydig cell tumor of the testis | 31 | 31 | 0.0 | 22.6 | 77.4 | 0.0 | |
| Sertoli cell tumor of the testis | 2 | 2 | 0.0 | 50.0 | 50.0 | 0.0 | |
| Sex cord stromal tumor of the testis | 1 | 1 | 0.0 | 100.0 | 0.0 | 0.0 | |
| Spermatocytic tumor of the testis | 1 | 1 | 0.0 | 0.0 | 100.0 | 0.0 | |
| Yolk sac tumor | 53 | 40 | 0.0 | 62.5 | 30.0 | 7.5 | |
| Teratoma | 53 | 40 | 0.0 | 45.0 | 55.0 | 0.0 | |
| Squamous cell carcinoma of the penis | 92 | 86 | 2.3 | 64.0 | 25.6 | 8.1 | |
| Tumors of endocrine organs | Adenoma of the thyroid gland | 113 | 99 | 0.0 | 58.6 | 39.4 | 2.0 |
| Papillary thyroid carcinoma | 391 | 335 | 0.0 | 41.2 | 57.3 | 1.5 | |
| Follicular thyroid carcinoma | 154 | 104 | 0.0 | 55.8 | 42.3 | 1.9 | |
| Medullary thyroid carcinoma | 111 | 74 | 5.4 | 83.8 | 10.8 | 0.0 | |
| Parathyroid gland adenoma | 43 | 37 | 0.0 | 35.1 | 56.8 | 8.1 | |
| Anaplastic thyroid carcinoma | 45 | 39 | 5.1 | 43.6 | 48.7 | 2.6 | |
| Adrenal cortical adenoma | 48 | 46 | 0.0 | 41.3 | 58.7 | 0.0 | |
| Adrenal cortical carcinoma | 27 | 27 | 0.0 | 59.3 | 33.3 | 7.4 | |
| Pheochromocytoma | 51 | 44 | 2.3 | 86.4 | 11.4 | 0.0 | |
| Paraganglioma | 41 | 36 | 0.0 | 72.2 | 27.8 | 0.0 | |
| Tumors of hematopoietic and lymphoid tissues | Hodgkin lymphoma | 103 | 50 | 50.0 | 46.0 | 4.0 | 0.0 |
| B-SLL, B-cell type (B-CLL) | 50 | 48 | 0.0 | 39.6 | 41.7 | 18.8 | |
| DLBCL | 113 | 108 | 4.6 | 56.5 | 33.3 | 5.6 | |
| Follicular lymphoma | 88 | 86 | 7.0 | 60.5 | 31.4 | 1.2 | |
| T-cell non-Hodgkin lymphoma | 25 | 22 | 13.6 | 50.0 | 36.4 | 0.0 | |
| Mantle cell lymphoma | 18 | 18 | 0.0 | 33.3 | 61.1 | 5.6 | |
| Marginal zone lymphoma | 16 | 14 | 14.3 | 35.7 | 28.6 | 21.4 | |
| DLBCL in the testis | 16 | 14 | 7.1 | 85.7 | 7.1 | 0.0 | |
| Burkitt lymphoma | 5 | 4 | 0.0 | 50.0 | 50.0 | 0.0 | |
| Tumors of soft tissue and bone | Granular cell tumor | 23 | 15 | 0.0 | 0.0 | 93.3 | 6.7 |
| Leiomyoma | 50 | 41 | 0.0 | 90.2 | 9.8 | 0.0 | |
| Leiomyosarcoma | 94 | 70 | 5.7 | 57.1 | 37.1 | 0.0 | |
| Atypical lipomatous tumor/well-differentiated liposarcoma | 18 | 11 | 0.0 | 54.5 | 45.5 | 0.0 | |
| Dedifferentiated liposarcoma | 54 | 42 | 0.0 | 73.8 | 26.2 | 0.0 | |
| Myxoid liposarcoma | 18 | 14 | 0.0 | 64.3 | 35.7 | 0.0 | |
| Pleomorphic liposarcoma | 3 | 1 | 0.0 | 100.0 | 0.0 | 0.0 | |
| Pleomorphic rhabdomyosarcoma | 3 | 1 | 0 | 100 | 0 | 0 | |
| MPNST | 15 | 13 | 30.8 | 53.8 | 15.4 | 0.0 | |
| Myofibrosarcoma | 26 | 24 | 0.0 | 41.7 | 54.2 | 4.2 | |
| Angiosarcoma | 42 | 26 | 0.0 | 50.0 | 46.2 | 3.8 | |
| Angiomyolipoma | 91 | 60 | 0.0 | 85.0 | 15.0 | 0.0 | |
| Dermatofibrosarcoma protuberans | 21 | 16 | 6.3 | 50.0 | 43.8 | 0.0 | |
| Ganglioneuroma | 14 | 12 | 0.0 | 91.7 | 8.3 | 0.0 | |
| Kaposi sarcoma | 8 | 4 | 0.0 | 100.0 | 0.0 | 0.0 | |
| Neurofibroma | 117 | 104 | 0.0 | 90.4 | 9.6 | 0.0 | |
| Sarcoma, NOS | 74 | 63 | 14.3 | 46.0 | 39.7 | 0.0 | |
| Ewing sarcoma | 23 | 10 | 0.0 | 50.0 | 30.0 | 20.0 | |
| Alveolar rhabdomyosarcoma | 2 | 1 | 0.0 | 100.0 | 0.0 | 0.0 | |
| Pleomorphic rhabdomyosarcoma | 2 | 2 | 50.0 | 50.0 | 0.0 | 0.0 | |
| Embryonal rhabdomyosarcoma | 3 | 2 | 0.0 | 0.0 | 50.0 | 50.0 | |
| Schwannoma | 122 | 113 | 0.9 | 59.3 | 38.1 | 1.8 | |
| Synovial sarcoma | 12 | 10 | 0.0 | 10.0 | 30.0 | 60.0 | |
| Osteosarcoma | 19 | 12 | 0.0 | 66.7 | 25.0 | 8.3 | |
| Chondrosarcoma | 15 | 10 | 20.0 | 40.0 | 40.0 | 0.0 | |
| Rhabdoid tumor | 5 | 5 | 0.0 | 40.0 | 60.0 | 0.0 | |
| Solitary fibrous tumor | 17 | 17 | 0.0 | 41.2 | 52.9 | 5.9 | |
Immunostaining score “0” indicates complete loss of MTAP expression.
CLL indicates chronic lymphocytic lymphoma; DLBCL, diffuse large B-cell lymphoma; GIST, gastrointestinal stromal tumor; MPNST, malignant peripheral nerve sheath tumor; NEC, neuroendocrine carcinoma; NOS, not otherwise specified; SLL, small lymphocytic lymphoma.
FIGURE 2.
MTAP loss in tumors lacking 9p21 deletion. MTAP staining was completely absent in tumor cells of a NET of the lung (A), a NET of the ileum (B), a colorectal NET (C), a pancreatic NET (D), a T-cell non-Hodgkin lymphoma (E), a marginal cell lymphoma (F) and in 2 cases of Hodgkin lymphoma (G and H). Distinct MTAP positivity of intermingled stroma cells serves as a positive internal control.
FIGURE 3.
MTAP loss in tumors with homozygous 9p21 deletion. MTAP staining was completely lacking in tumor cells of an (epithelioid) malignant mesothelioma (A), a urothelial carcinoma of the renal pelvis (B), a mucinous ovarian carcinoma (C), a serous high-grade ovarian carcinoma (D), a malignant melanoma (E), a recurrent adenocarcinoma of the prostate (F), a colorectal adenocarcinoma (G), and a ductal adenocarcinoma of the pancreas (H). Distinct MTAP positivity of intermingled stroma cells and of non-neoplastic epithelial cells serves as a positive internal control.
FISH Validation
FISH validation was performed for all cancers with complete MTAP loss by IHC. It showed that the concordance rate between MTAP IHC and FISH varied greatly between tumor entities (Fig. 4). The concordance rate was 100% in mesothelioma, urothelial carcinoma and SQCC of the urinary bladder, adenocarcinoma and SQCC of the esophagus, malignant peripheral nerve sheath tumor, sarcoma not otherwise specified, as well as in several other entities with low numbers of tumors analyzed. It was 90% to 99% in many other tumor types, including gallbladder adenocarcinoma, mucinous carcinoma of the ovary, malignant melanoma (including nodal metastases), SQCC of the oral cavity, and hepatocellular carcinoma (HCC). Most notably, there were various other tumor entities with common MTAP loss by IHC but an almost complete lack of concomitant 9p21 deletions. These included Hodgkin lymphoma, several other lymphoma types, as well as NETs and carcinomas of various sites of origin. A graphical representation of the ranking order of cancers with MTAP deficiency by IHC and their 9p21 status obtained by FISH is given in Figure 4. If neuroendocrine and hematological neoplasms were excluded, a homozygous 9p21 deletion occurred in 96.7% of the tumors with a complete MTAP expression loss.
FIGURE 4.
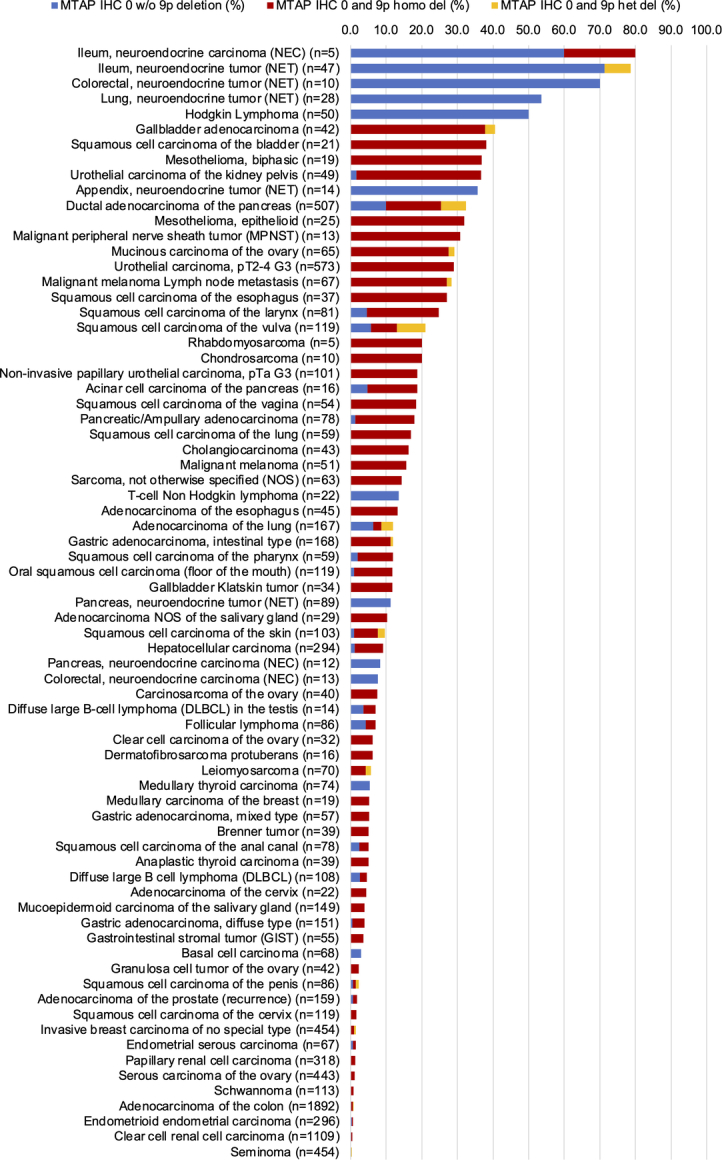
Concordance rate of MTAP IHC and 9p21 copy number analysis.
MTAP Deficiency and Tumor Phenotype
The relationship between MTAP deficiency and clinically important histopathologic and molecular tumor features in carcinomas from different sites is shown in Table 2. MTAP deficiency was linked to the presence of nodal metastasis (P = 0.0349) and high grade (P = 0.0156) in HCC, advanced pT stage in NETs (P = 0.0166), as well as high grade (P = 0.0034), advanced pT stage (P = 0.0299) and absence of human papillomavirus (HPV) infection (P < 0.0001) in SQCC of different organs of origin. Markedly higher rates of MTAP deficiency in SQCC lacking HPV infection were also observed in most subgroups of tumors from individual organs, although statistical significance was only reached for SQCC of the cervix (P = 0.0028) and the anal canal (P = 0.045; Table 3). The frequency of MTAP deficiency further increased with pT and grade in invasive breast cancer of no special type and in pancreatic adenocarcinoma, although not all these differences reached statistical significance. Across all tumor entities, tumors with a complete MTAP loss were more often PD-L1 positive (P = 0.0125), had fewer PD-L1 positive macrophages (P = 0.0008), and fewer cytotoxic (CD8+) lymphocytes in their microenvironment (P= 0.0042; Table 4).
TABLE 2.
MTAP Immunostaining and Tumor Phenotype
| Entity | Pathologic parameters | n | MTAP status (%)* | P | ||
|---|---|---|---|---|---|---|
| Loss | Retained | |||||
| HCC | Tumor stage | pT1 | 77 | 6.5 | 93.5 | 0.0911 |
| pT2 | 83 | 10.8 | 89.2 | — | ||
| pT3-pT4 | 59 | 18.6 | 81.4 | — | ||
| Nodal stage | pN0 | 74 | 10.8 | 89.2 | 0.0349 | |
| pN+ | 42 | 26.2 | 73.8 | — | ||
| Grade | G1 | 36 | 0.0 | 100.0 | 0.0156 | |
| G2 | 128 | 10.9 | 89.1 | — | ||
| G3 | 52 | 13.5 | 86.5 | — | ||
| NETs† | Tumor stage | pT1 | 28 | 35.7 | 64.3 | 0.0166 |
| pT2 | 24 | 37.5 | 62.5 | — | ||
| pT3 | 36 | 47.2 | 52.8 | — | ||
| pT4 | 32 | 71.9 | 28.1 | — | ||
| Nodal stage | pN0 | 45 | 37.8 | 62.2 | 0.0946 | |
| pN+ | 59 | 54.2 | 45.8 | — | ||
| Pancreatic adenocarcinoma | Tumor stage | pT1 | 7 | 14.3 | 85.7 | 0.6582 |
| pT2 | 59 | 28.8 | 71.2 | — | ||
| pT3 | 316 | 29.7 | 70.2 | — | ||
| pT4 | 24 | 37.5 | 62.5 | — | ||
| Nodal stage | pN0 | 81 | 33.3 | 66.7 | 0.4091 | |
| pN+ | 325 | 28.6 | 71.4 | — | ||
| Grade | G1 | 16 | 18.8 | 81.3 | 0.3964 | |
| G2 | 282 | 31.2 | 68.8 | — | ||
| G3 | 88 | 26.1 | 73.9 | — | ||
| Invasive breast cancer of no special type | Tumor stage | pT1 | 162 | 0.0 | 100.0 | 0.1019 |
| pT2 | 194 | 1.5 | 98.5 | — | ||
| pT3-pT4 | 35 | 2.9 | 97.1 | — | ||
| Nodal stage | pN0 | 198 | 0.5 | 99.5 | 0.8765 | |
| pN+ | 98 | 0.6 | 99.4 | — | ||
| Grade | G1 | 13 | 0.0 | 100.0 | 0.0268 | |
| G2 | 221 | 0.0 | 100.0 | — | ||
| G3 | 161 | 2.5 | 97.5 | — | ||
| SQCCs‡ | Tumor stage | pT1 | 235 | 10.6 | 89.4 | 0.0299 |
| pT2 | 250 | 10.0 | 90.0 | — | ||
| pT3 | 121 | 19.0 | 81.0 | — | ||
| pT4 | 125 | 17.6 | 82.4 | — | ||
| Nodal stage | pN0 | 276 | 12.0 | 88.0 | 0.0615 | |
| pN+ | 285 | 17.5 | 82.5 | — | ||
| Grade | G1 | 30 | 0.0 | 100.0 | 0.0034 | |
| G2 | 341 | 15.0 | 85.0 | — | ||
| G3 | 234 | 9.8 | 90.2 | — | ||
| HPV | Negative | 275 | 18.2 | 81.8 | <0.0001 | |
| Positive | 233 | 5.6 | 94.4 | — | ||
MTAP loss: no MTAP immunostaining, MTAP retained: MTAP 1+ to 3+ immunostaining.
Neuroendocrine tumors of the appendix, colorectum, ileum, pancreas, and lung.
Squamous cell carcinomas of the oral cavity (n = 119), pharynx (n = 59), larynx (n = 82), esophagus (n = 36), lung (n = 59), cervix (n = 1139), vagina (n = 54), vulva (n = 100), penis (n = 77), skin (n = 68), and the anal canal (n = 78).
HPV indicates human papillomavirus.
TABLE 3.
MTAP Immunostaining and HPV Status in SQCCs of Various Origins
| Tumor entity | HPV status | n | MTAP status (%)* | P | |
|---|---|---|---|---|---|
| Loss | Retained | ||||
| Oral squamous cell carcinoma | Negative | 66 | 15.2 | 84.8 | 0.0578 |
| Positive | 12 | 0.0 | 100.0 | — | |
| Squamous cell carcinoma of the pharynx | Negative | 22 | 13.6 | 86.4 | 0.8368 |
| Positive | 34 | 11.8 | 88.2 | — | |
| Squamous cell carcinoma of the larynx | Negative | 48 | 31.3 | 68.8 | 0.7177 |
| Positive | 8 | 25.0 | 75.0 | — | |
| Squamous cell carcinoma of the cervix | Negative | 9 | 22.2 | 77.8 | 0.0028 |
| Positive | 67 | 0.0 | 100.0 | — | |
| Squamous cell carcinoma of the vagina | Negative | 15 | 26.7 | 73.3 | 0.4638 |
| Positive | 13 | 15.4 | 84.6 | — | |
| Squamous cell carcinoma of the vulva | Negative | 45 | 24.4 | 75.6 | 0.4485 |
| Positive | 24 | 16.7 | 83.3 | — | |
| Squamous cell carcinoma of the penis | Negative | 29 | 3.4 | 96.6 | 0.7537 |
| Positive | 45 | 2.2 | 97.8 | — | |
| Squamous cell carcinoma of the skin | Negative | 36 | 8.3 | 91.7 | 0.6787 |
| Positive | 1 | 0.0 | 100.0 | — | |
| Squamous cell carcinoma of the anal canal | Negative | 5 | 20.0 | 80.0 | 0.0450 |
| Positive | 29 | 0.0 | 100.0 | — | |
MTAP loss: no MTAP immunostaining, MTAP retained: MTAP 1+ to 3+ immunostaining.
HPV indicates human papillomavirus.
TABLE 4.
MTAP Immunostaining Versus PD-L1 Immunostaining and CD8+ T-cell Density
| PD-L1 positive (% of tumors) | CD8+ density (cells/mm2) | |||||
|---|---|---|---|---|---|---|
| MTAP status* | n | Tumor cells | n | Immune cells | n | Mean±SE |
| Loss | 559 | 19.9 | 61.9 | 321 | 179.2±28.2 | |
| Retained | 7262 | 15.7 | 71.8 | 3915 | 263.3±8.1 | |
| P | — | 0.0125 | 0.0008 | — | 0.0042 | |
MTAP loss: no MTAP immunostaining, MTAP retained: MTAP 1+ to 3+ immunostaining.
DISCUSSION
The highly standardized analysis of 13,067 tumors from 149 different tumor entities provided an overview of the prevalence of “MTAP deficiency” across different tumor entities and identified Hodgkin lymphoma, neuroendocrine neoplasms of various sites of origin, mesothelioma, biliopancreatic adenocarcinoma, urothelial carcinoma, malignant melanoma, mucinous carcinoma of the ovary, and SQCC from various organs as the most commonly MTAP negative tumors. For most of these tumor entities, 9p21 deletions have previously been described, but the reported prevalence often varied greatly due to the use of different methods, and the rate of homozygous deletions was not described in most of these studies. For example, the prevalence of 9p21 deletions ranged at least from 25% to 88% in malignant mesothelioma,39–41 from 18% to 70% in pancreatic adenocarcinoma,42–44 from 14% to 96% in urothelial carcinoma45,46, from 16% to 84% (6% to 41% homozygous) in malignant melanoma,47–51 from 25% to 100% (14% to 18% homozygous) in ovarian cancer,52–54 from 28% to 68% in HCC,55,56 and from 6% to 18% in prostatic adenocarcinoma.57,58
In a recent study on more than 2,600 urothelial neoplasms, we found a very high sensitivity and specificity of our MTAP IHC assay for the detection of homozygous 9p21 deletions (Gorbokon et al, manuscript submitted). In this study, 98.4% of 252 urothelial tumors with a complete MTAP expression loss had a homozygous 9p21 deletion, and only 3.9% of 258 tumors with homozygous 9p21 deletions showed MTAP immunostaining. Other authors had also applied MTAP IHC to tumor cohorts with predefined 9p21 deletion status and found sensitivities between 59% and 100% and specificities between 96% and 100% for detection of homozygous deletions in mesothelioma, astrocytoma, and meningioma (Supplemental Table 1, Supplemental Digital Content 3, http://links.lww.com/PAS/B922). Our FISH validation of all MTAP IHC negative cases revealed that such a near-complete FISH/IHC concordance does not apply to all tumor entities. Especially in neuroendocrine neoplasms, Hodgkin lymphomas, and in several other lymphoma entities, including T-cell lymphoma, loss of MTAP expression was never or only rarely due to a 9p21 deletion. Considering that within these tumor entities, a fraction of tumors also showed high-level MTAP expression and that normal precursor cells such as pancreatic islet cells or lymphocytes exhibited a distinct MTAP positivity, MTAP negativity in these tumors cannot be caused by particularly low MTAP levels in their cells of origin. In concordance with our data, Woollard et al59 identified MTAP expression loss in a significant fraction of T-cell lymphomas that lacked 9p21 deletions. As a potential cause for MTAP downregulation, these authors observed MTAP promotor hypermethylation and subsequent MTAP downregulation in one T-cell lymphoma cell line. Hellerbrand et al60 also found promotor hypermethylation to cause MTAP downregulation in HCC cell lines lacking genomic 9p21 deletions.
Although MTAP deficiency regularly occurs in the absence of 9p21 deletions in some tumor entities, we conclude from our data that homozygous MTAP deletion may represent the sole mechanism for MTAP deficiency in non-hematological and non-neuroendocrine cancers. Methodological issues may have caused many or even all of the discrepant cases in tumor entities with high rates of 9p21 deletions. As IHC and FISH analysis had not been executed on consecutive TMA sections, it appears likely that small tumor cell groups with unequivocal MTAP deficiency could not be confirmed by FISH in a fraction of cases because the respective cell population were not present on the respective FISH-stained sections. That small MTAP deficient tumor cell groups can easily be detected by IHC (Supplemental Fig. 3, Supplemental Digital Content 4, http://links.lww.com/PAS/B923) further emphasized the high utility of MTAP IHC for the detection of MTAP deficiency.
Several studies had earlier suggested a treatment-agnostic impact of MTAP deficiency on the clinical course of cancer. For example, a relationship between MTAP deficiency/9p21 deletions and unfavorable tumor features has earlier been described in cohorts of 99 non–small-cell lung cancer,61 75 oral SQCC,62 35 head and neck SQCC,63 48 adenosquamous carcinoma of the pancreas,64 140 HCC,65 22 thymic carcinomas,66 146 gastrointestinal stromal tumors,67 113 Ewing sarcoma family of tumors,33 and 62 bladder cancers.68 In our own previous study on 636 patients undergoing cystectomy for muscle-invasive urothelial bladder cancer, both MTAP expression loss and 9p21 deletions were—independent of pT and pN stage—associated with poor outcome (Gorbokon et al, manuscript submitted). Significant associations between MTAP deficiency and unfavorable tumor features in HCC, NETs, and SQCC in this study are also in line with an increased aggressiveness of MTAP-deficient cancers.
A direct impact of MTAP deficiency on the antitumoral immune response has been suggested as a possible explanation for a prognostic role of 9p21 deletions and/or MTAP expression loss.69 The significant correlation of MTAP deficiency with PD-L1 expression in tumor cells and a low density of CD8+ lymphocytes across all analyzed tumor entities is in line with this assumption, and also consistent with earlier reports describing a noninflamed microenvironment and poor response to CPI in MTAP deficient/9p21 deleted cancers.26,70 The strong inverse correlation of MTAP deficiency with HPV infection in SQCC may suggest that MTAP deficiency is specifically not required in cancer cells having HPV-induced cancer-driving pathways activated. An inverse correlation between MTAP/9p21 loss and HPV infection has earlier been found in SQCC of the urinary bladder71 and in oropharyngeal carcinomas.72
Our data also suggest a considerable diagnostic utility of MTAP IHC in surgical pathology. The complete MTAP expression loss in 32.0% of our epithelioid and in 36.8% of the biphasic mesotheliomas supports the established role of MTAP IHC for the distinction of malignant mesothelioma from benign mesothelial conditions. Earlier studies had reported an MTAP loss in 12% to 77% of epithelioid mesotheliomas.73,74 It is assumed that homozygous 9p21 deletions do not occur in benign mesothelial proliferations.75 Our data support several further potential diagnostic applications for MTAP IHC in the distinction of neoplastic from benign changes. In urothelial and skin samples, MTAP loss could support the diagnosis of urothelial dysplasia or melanoma. Both diagnoses are often difficult to establish based on morphology alone.76,77 More generally, a distinct MTAP loss in suspicious cells could support the diagnosis of a malignant process in small biopsies or cytologic specimens from various sites including the pancreas, bile ducts, soft tissues, the lung, or lymphatic tissues.
Given the potential clinical importance of MTAP expression analysis and the large scale of our study, emphasis was placed on a thorough validation of our assay. To assure the specificity of our immunostaining, our antibody was validated by a comparison of its staining pattern with a different independent antibody, as suggested by the International Working Group for Antibody Validation.78 Our exceedingly large panel of 76 different normal tissues assures that virtually all proteins occurring in cells of adult humans were subjected to the cross-reactivity screening. That all nuclear and cytoplasmic staining patterns observed by MSVA-741R were also seen by 2G4 is strong evidence for the specificity of MSVA-741R. The additional staining of gastrointestinal mucus-producing goblet cells by 2G4, which was not observed by MSVA-741R, represents an antibody-specific cross-reactivity of 2G4 (Supplemental Fig. 2, Supplemental Digital Content 2, http://links.lww.com/PAS/B921). It cannot be excluded that this cross-reactivity may obscure MTAP expression loss in some mucus-producing tumors.
In summary, our data provide a comprehensive overview of the prevalence of MTAP deficiency in cancer and identify cancer entities that might most benefit from cancer drugs targeting MTAP deficiency. It will be interesting to study the potential intratumoral heterogeneity of MTAP loss in affected tumor types, as this could challenge the success of such therapies, and whether tumor entities with MTAP expression loss in the absence of deletion will equally well respond to these drugs. Our findings also highlight the diagnostic utility of MTAP IHC as a marker for neoplastic transformation in various different applications, including the distinction of mesothelioma, urothelial dysplasia, or melanoma from benign conditions.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Melanie Steurer, Laura Behm, Inge Brandt, Sünje Seekamp, Sascha Eghtessadi, Silvia Schnöger, Heike Jordan, Vivian Modi, and Lucy Joan Veloz Ramirez for excellent technical assistance.
Footnotes
The use of archived remnants of diagnostic tissues for manufacturing of TMAs and their analysis for research purposes as well as patient data analysis has been approved by local laws (HmbKHG, §12) and by the local ethics committee (Ethics commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration.
N.G., N.W., R.S., M.K., G.S., and L.B.: conception, design, data collection, data analysis, and manuscript writing. N.W., M.L., S.D.R., S.K., V.R., F.V., F.L., C.F., A.M.L., A.M., R. Schlichter, T.K., A.H., E.B., S.S., A.H.M., P.L., D.D., S.M., F.J., T.S.C., T.H., F.G.U., and C.B.: pathology data analysis, data interpretation, and collection of samples. N.G., R. Simon, M.K., and C.H.M.: data analysis. N.G., R.S., G.S., and M.K.: study supervision.
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest and Source of Funding: The recombinant rabbit monoclonal antibody, clone MSVA-741R was provided from MS Validated Antibodies GmbH, Hamburg, Germany (owned by a family member of G.S.). For the remaining authors none were declared.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.ajsp.com.
Contributor Information
Natalia Gorbokon, Email: n.gorbokon@uke.de.
Niklas Wößner, Email: niklasw1711@gmail.com.
Maximilian Lennartz, Email: m.lennartz@uke.de.
Sebastian Dwertmann Rico, Email: s.dwertmann-rico@uke.de.
Simon Kind, Email: s.kind@uke.de.
Viktor Reiswich, Email: v.reiswich@uke.de.
Florian Viehweger, Email: f.viehweger@uke.de.
Florian Lutz, Email: f.lutz@uke.de.
Christoph Fraune, Email: c.fraune@uke.de.
Andreas M. Luebke, Email: luebke@uke.de.
Claudia Hube-Magg, Email: c.hube@uke.de.
Anne Menz, Email: a.menz@uke.de.
Ria Schlichter, Email: r.schlichter@uke.de.
Till Krech, Email: t.krech@uke.de.
Andrea Hinsch, Email: a.hinsch@uke.de.
Eike Burandt, Email: e.burandt@uke.de.
Guido Sauter, Email: g.sauter@uke.de.
Ronald Simon, Email: r.simon@uke.de.
Stefan Steurer, Email: s.steurer@uke.de.
Andreas H. Marx, Email: Andreas.Marx@klinikum-fuerth.de.
Patrick Lebok, Email: p.lebok@uke.de.
David Dum, Email: d.dum@uke.de.
Sarah Minner, Email: s.minner@uke.de.
Frank Jacobsen, Email: f.jacobsen@uke.de.
Till S. Clauditz, Email: t.clauditz@uke.de.
Thilo Hackert, Email: t.hackert@uke.de.
Faik G. Uzunoǧlu, Email: f.uzunoglu@uke.de.
Lukas Bubendorf, Email: Lukas.Bubendorf@usb.ch.
Christian Bernreuther, Email: c.bernreuther@uke.de.
Martina Kluth, Email: m.kluth@uke.de.
REFERENCES
- 1.Della Ragione F, Carteni-Farina M, Gragnaniello V, et al. Purification and characterization of 5’-deoxy-5’-methylthioadenosine phosphorylase from human placenta. J Biol Chem. 1986;261:12324–12329. [PubMed] [Google Scholar]
- 2.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aaltonen Lauri A, Abascal F, Abeshouse A, et al. Consortium ITP-CAoWG. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison PW, Amode MR, Austine-Orimoloye O, et al. Ensembl 2024. Nucleic Acids Res. 2023;52:D891–D899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Chen ZH, Savarese TM. Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-alpha1, interferon-beta1, and other 9p21 markers in human malignant cell lines. Cancer Genet Cytogenet. 1996;86:22–28. [DOI] [PubMed] [Google Scholar]
- 6.Alhalabi O, Chen J, Zhang Y, et al. MTAP deficiency creates an exploitable target for antifolate therapy in 9p21-loss cancers. Nat Commun. 2022;13:1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bray C, Balcells C, McNeish IA, et al. The potential and challenges of targeting MTAP-negative cancers beyond synthetic lethality. Front Oncol. 2023;13:1264785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65:8–24. [DOI] [PubMed] [Google Scholar]
- 9.Lu SC, Mato JMS. Adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray B, Antonyuk SV, Marina A, et al. Crystallography captures catalytic steps in human methionine adenosyltransferase enzymes. Proc Natl Acad Sci U S A. 2016;113:2104–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang K, Zielinska AE, Shaaban AM, et al. PRMT5 Is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 2017;21:3498–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan F, Alinari L, Lustberg ME, et al. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014;74:1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng X, Shao G, Zhang HT, et al. Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene. 2017;36:1223–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin Y, Hu Q, Xu J, et al. PRMT5 enhances tumorigenicity and glycolysis in pancreatic cancer via the FBW7/cMyc axis. Cell Commun Signal. 2019;17:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Shao G, Shao J, et al. PRMT5-activated c-Myc promote bladder cancer proliferation and invasion through up-regulating NF-kappaB pathway. Tissue Cell. 2022;76:101788. [DOI] [PubMed] [Google Scholar]
- 16.Radzisheuskaya A, Shliaha PV, Grinev V, et al. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat Struct Mol Biol. 2019;26:999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh CM, Bezzi M, Low DH, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96–100. [DOI] [PubMed] [Google Scholar]
- 18.Bao X, Zhao S, Liu T, et al. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J Histochem Cytochem. 2013;61:206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Z, Gao S, Zhang F, et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem J. 2012;446:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M, Yao B, Gui T, et al. PRMT5-dependent transcriptional repression of c-Myc target genes promotes gastric cancer progression. Theranostics. 2020;10:4437–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kryukov GV, Wilson FH, Ruth JR, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engstrom LD, Aranda R, Waters L, et al. MRTX1719 Is an MTA-cooperative PRMT5 inhibitor that exhibits synthetic lethality in preclinical models and patients with MTAP-deleted cancer. Cancer Discov. 2023;13:2412–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feustel K, Falchook GS. Protein arginine methyltransferase 5 (PRMT5) inhibitors in oncology clinical trials: a review. J Immunother Precis Oncol. 2022;5:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mo Q, Li R, Adeegbe DO, et al. Integrative multi-omics analysis of muscle-invasive bladder cancer identifies prognostic biomarkers for frontline chemotherapy and immunotherapy. Commun Biol. 2020;3:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adib E, Nassar AH, Akl EW, et al. CDKN2A alterations and response to immunotherapy in solid tumors. Clin Cancer Res. 2021;27:4025–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han G, Yang G, Hao D, et al. 9p21 loss confers a cold tumor immune microenvironment and primary resistance to immune checkpoint therapy. Nat Commun. 2021;12:5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beasley MB, Galateau-Salle F, Dacic S. Pleural mesothelioma classification update. Virchows Arch. 2021;478:59–72. [DOI] [PubMed] [Google Scholar]
- 28.Menezes WP, Silva VAO, Gomes INF, et al. Loss of 5’-methylthioadenosine phosphorylase (MTAP) is frequent in high-grade gliomas; nevertheless, it is not associated with higher tumor aggressiveness. Cells. 2020;9:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jing W, Zhu H, Liu W, et al. MTAP-deficiency could predict better treatment response in advanced lung adenocarcinoma patients initially treated with pemetrexed-platinum chemotherapy and bevacizumab. Sci Rep. 2020;10:843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell EL, Leoni LM, Canto MI, et al. Concordant loss of MTAP and p16/CDKN2A expression in gastroesophageal carcinogenesis: evidence of homozygous deletion in esophageal noninvasive precursor lesions and therapeutic implications. Am J Surg Pathol. 2005;29:1497–1504. [DOI] [PubMed] [Google Scholar]
- 31.Nilforoushan N, Moatamed NA. Evaluation of MTAP immunohistochemistry loss of expression in ovarian serous borderline tumors as a potential marker for prognosis and progression. Ann Diagn Pathol. 2020;48:151582. [DOI] [PubMed] [Google Scholar]
- 32.Hustinx SR, Leoni LM, Yeo CJ, et al. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: evidence of homozygous deletion in a noninvasive precursor lesion. Mod Pathol. 2005;18:959–963. [DOI] [PubMed] [Google Scholar]
- 33.Abrahao-Machado LF, Antunes B, Filippi RZ, et al. Loss of MTAP expression is a negative prognostic marker in Ewing sarcoma family of tumors. Biomark Med. 2018;12:35–44. [DOI] [PubMed] [Google Scholar]
- 34.Miyazaki S, Nishioka J, Shiraishi T, et al. Methylthioadenosine phosphorylase deficiency in Japanese osteosarcoma patients. Int J Oncol. 2007;31:1069–1076. [PubMed] [Google Scholar]
- 35.Bubendorf L, Nocito A, Moch H, et al. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol. 2001;195:72–79. [DOI] [PubMed] [Google Scholar]
- 36.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. [DOI] [PubMed] [Google Scholar]
- 37.Moller K, Knoll M, Bady E, et al. PD-L1 expression and CD8 positive lymphocytes in human neoplasms: a tissue microarray study on 11,838 tumor samples. Cancer Biomark. 2023;36:177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krohn A, Diedler T, Burkhardt L, et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol. 2012;181:401–412. [DOI] [PubMed] [Google Scholar]
- 39.Neragi-Miandoab S, Sugarbaker DJ. Chromosomal deletion in patients with malignant pleural mesothelioma. Interact Cardiovasc Thorac Surg. 2009;9:42–44. [DOI] [PubMed] [Google Scholar]
- 40.Chiosea S, Krasinskas A, Cagle PT, et al. Diagnostic importance of 9p21 homozygous deletion in malignant mesotheliomas. Mod Pathol. 2008;21:742–747. [DOI] [PubMed] [Google Scholar]
- 41.Takeda M, Kasai T, Enomoto Y, et al. 9p21 deletion in the diagnosis of malignant mesothelioma, using fluorescence in situ hybridization analysis. Pathol Int. 2010;60:395–399. [DOI] [PubMed] [Google Scholar]
- 42.Gutierrez ML, Munoz-Bellvis L, Abad Mdel M, et al. Association between genetic subgroups of pancreatic ductal adenocarcinoma defined by high density 500 K SNP-arrays and tumor histopathology. PLoS One. 2011;6:e22315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franko J, Krasinskas AM, Nikiforova MN, et al. Loss of heterozygosity predicts poor survival after resection of pancreatic adenocarcinoma. J Gastrointest Surg. 2008;12:1664–1672; discussion 72-73. [DOI] [PubMed] [Google Scholar]
- 44.Ribeiro A, Peng J, Casas C, et al. Endoscopic ultrasound-guided fine needle aspiration with fluorescence in situ hybridization analysis in 104 patients with pancreatic mass. J Gastroenterol Hepatol. 2014;29:1654–1658. [DOI] [PubMed] [Google Scholar]
- 45.Baud E, Catilina P, Boiteux JP, et al. Human bladder cancers and normal bladder mucosa present the same hot spot of heterozygous chromosome-9 deletion. Int J Cancer. 1998;77:821–824. [DOI] [PubMed] [Google Scholar]
- 46.Orlow I, LaRue H, Osman I, et al. Deletions of the INK4A gene in superficial bladder tumors. Association with recurrence. Am J Pathol. 1999;155:105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. [DOI] [PubMed] [Google Scholar]
- 48.Gerami P, Scolyer RA, Xu X, et al. Risk assessment for atypical spitzoid melanocytic neoplasms using FISH to identify chromosomal copy number aberrations. Am J Surg Pathol. 2013;37:676–684. [DOI] [PubMed] [Google Scholar]
- 49.Rakosy Z, Vizkeleti L, Ecsedi S, et al. Characterization of 9p21 copy number alterations in human melanoma by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2008;182:116–121. [DOI] [PubMed] [Google Scholar]
- 50.Gammon B, Beilfuss B, Guitart J, et al. Enhanced detection of spitzoid melanomas using fluorescence in situ hybridization with 9p21 as an adjunctive probe. Am J Surg Pathol. 2012;36:81–88. [DOI] [PubMed] [Google Scholar]
- 51.Franchi A, Alos L, Gale N, et al. Expression of p16 in sinonasal malignant melanoma. Virchows Arch. 2006;449:667–672. [DOI] [PubMed] [Google Scholar]
- 52.Saretzki G, Hoffmann U, Rohlke P, et al. Identification of allelic losses in benign, borderline, and invasive epithelial ovarian tumors and correlation with clinical outcome. Cancer. 1997;80:1241–1249. [DOI] [PubMed] [Google Scholar]
- 53.Aravidis C, Panani AD, Kosmaidou Z, et al. Detection of numerical abnormalities of chromosome 9 and p16/CDKN2A gene alterations in ovarian cancer with fish analysis. Anticancer Res. 2012;32:5309–5313. [PubMed] [Google Scholar]
- 54.Schultz DC, Vanderveer L, Buetow KH, et al. Characterization of chromosome 9 in human ovarian neoplasia identifies frequent genetic imbalance on 9q and rare alterations involving 9p, including CDKN2. Cancer Res. 1995;55:2150–2157. [PubMed] [Google Scholar]
- 55.Piao Z, Park C, Lee JS, et al. Homozygous deletions of the CDKN2 gene and loss of heterozygosity of 9p in primary hepatocellular carcinoma. Cancer Lett. 1998;122:201–207. [DOI] [PubMed] [Google Scholar]
- 56.Nishida N, Nishimura T, Ito T, et al. Chromosomal instability and human hepatocarcinogenesis. Histol Histopathol. 2003;18:897–909. [DOI] [PubMed] [Google Scholar]
- 57.Barros EAF, Pontes-Junior J, Reis ST, et al. Correlation between chromosome 9p21 locus deletion and prognosis in clinically localized prostate cancer. Int J Biol Markers. 2017;32:e248–e254. [DOI] [PubMed] [Google Scholar]
- 58.Brys M, Migdalska-Sek M, Pastuszak-Lewandoska D, et al. Diagnostic value of DNA alteration: loss of heterozygosity or allelic imbalance-promising for molecular staging of prostate cancers. Med Oncol. 2013;30:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woollard WJ, Kalaivani NP, Jones CL, et al. Independent loss of methylthioadenosine phosphorylase (MTAP) in primary cutaneous T-cell lymphoma. J Invest Dermatol. 2016;136:1238–1246. [DOI] [PubMed] [Google Scholar]
- 60.Hellerbrand C, Muhlbauer M, Wallner S, et al. Promoter-hypermethylation is causing functional relevant downregulation of methylthioadenosine phosphorylase (MTAP) expression in hepatocellular carcinoma. Carcinogenesis. 2006;27:64–72. [DOI] [PubMed] [Google Scholar]
- 61.Su CY, Chang YC, Chan YC, et al. MTAP is an independent prognosis marker and the concordant loss of MTAP and p16 expression predicts short survival in non-small cell lung cancer patients. Eur J Surg Oncol. 2014;40:1143–1150. [DOI] [PubMed] [Google Scholar]
- 62.Chen C, Zhang Y, Loomis MM, et al. Genome-wide loss of heterozygosity and DNA copy number aberration in HPV-negative oral squamous cell carcinoma and their associations with disease-specific survival. PLoS One. 2015;10:e0135074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graveland AP, Golusinski PJ, Buijze M, et al. Loss of heterozygosity at 9p and p53 immunopositivity in surgical margins predict local relapse in head and neck squamous cell carcinoma. Int J Cancer. 2011;128:1852–1859. [DOI] [PubMed] [Google Scholar]
- 64.Jiang Y, Wu Y, Zhang L, et al. Loss of chromosome 9p21 is associated with a poor prognosis in adenosquamous carcinoma of the pancreas. Precis Clin Med. 2023;6:pbad030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirovski G, Stevens AP, Czech B, et al. Down-regulation of methylthioadenosine phosphorylase (MTAP) induces progression of hepatocellular carcinoma via accumulation of 5’-deoxy-5’-methylthioadenosine (MTA). Am J Pathol. 2011;178:1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aesif SW, Aubry MC, Yi ES, et al. Loss of p16(INK4A) expression and homozygous CDKN2A deletion are associated with worse outcome and younger age in thymic carcinomas. J Thorac Oncol. 2017;12:860–871. [DOI] [PubMed] [Google Scholar]
- 67.Huang HY, Li SH, Yu SC, et al. Homozygous deletion of MTAP gene as a poor prognosticator in gastrointestinal stromal tumors. Clin Cancer Res. 2009;15:6963–6972. [DOI] [PubMed] [Google Scholar]
- 68.Gallucci M, Vico E, Merola R, et al. Adverse genetic prognostic profiles define a poor outcome for cystectomy in bladder cancer. Exp Mol Pathol. 2007;83:385–391. [DOI] [PubMed] [Google Scholar]
- 69.Chang WH, Hsu SW, Zhang J, et al. MTAP deficiency contributes to immune landscape remodelling and tumour evasion. Immunology. 2023;168:331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gjuka D, Adib E, Garrison K, et al. Enzyme-mediated depletion of methylthioadenosine restores T cell function in MTAP-deficient tumors and reverses immunotherapy resistance. Cancer Cell. 2023;41:1774–87 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghelani GH, Zerdan MB, Jacob J, et al. HPV-positive clinically advanced squamous cell carcinoma of the urinary bladder (aBSCC): a comprehensive genomic profiling (CGP) study. Urol Oncol. 2023;41:486 e15–e23. [DOI] [PubMed] [Google Scholar]
- 72.Sepiashvili L, Waggott D, Hui A, et al. Integrated omic analysis of oropharyngeal carcinomas reveals human papillomavirus (HPV)-dependent regulation of the activator protein 1 (AP-1) pathway. Mol Cell Proteomics. 2014;13:3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naso JR, Tessier-Cloutier B, Senz J, et al. Significance of p53 immunostaining in mesothelial proliferations and correlation with TP53 mutation status. Mod Pathol. 2022;35:77–81. [DOI] [PubMed] [Google Scholar]
- 74.Lynggard LA, Panou V, Szejniuk W, et al. Diagnostic capacity of BAP1 and MTAP in cytology from effusions and biopsy in mesothelioma. J Am Soc Cytopathol. 2022;11:385–393. [DOI] [PubMed] [Google Scholar]
- 75.Takeda M, Kasai T, Enomoto Y, et al. Genomic gains and losses in malignant mesothelioma demonstrated by FISH analysis of paraffin-embedded tissues. J Clin Pathol. 2012;65:77–82. [DOI] [PubMed] [Google Scholar]
- 76.Filosa A, Filosa G. Melanoma diagnosis: the importance of histopathological report. Dermatopathology (Basel). 2018;5:41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lopez-Beltran A, Montironi R, Vidal A, et al. Urothelial dysplasia of the bladder: diagnostic features and clinical significance. Anal Quant Cytopathol Histpathol. 2013;35:121–129. [PubMed] [Google Scholar]
- 78.Uhlen M, Bandrowski A, Carr S, et al. A proposal for validation of antibodies. Nat Methods. 2016;13:823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.