Abstract
Denaturing gradient gel electrophoresis (DGGE) was used to probe for mutations associated with pyrazinamide (PZA) resistance in the pncA gene of Mycobacterium tuberculosis. DGGE scans for mutations across large regions of DNA and rivals sequencing in its ability to detect DNA alterations. Specific mutations can often be recognized by their characteristic denaturation pattern, which serves as a molecular fingerprint. Five PCR target fragments were designed to scan for DNA alterations across 600 bp of pncA in 181 M. tuberculosis isolates from patients residing in the U.S-Mexico border states of Texas and Tamaulipas, respectively. A region of pncA was observed with a high GC content and a melting temperature approaching 90°C that was initially refractory to denaturation, and a DGGE target fragment was specifically designed to detect mutations in this region. DGGE detected pncA mutations in 82 of 83 PZA-resistant isolates. By contrast, only 1 of 98 PZA-susceptible isolates harbored a detectable DNA alteration. The pncA gene was sequenced from 41 isolates, and 32 DNA alterations in 32 PZA-resistant isolates were identified, including 11 new mutations. DGGE also detected nine isolates whose susceptibility to PZA appeared to be incorrect, and DNA sequencing confirmed these apparent errors in drug susceptibility testing. These results demonstrate the power and usefulness of DGGE in detecting mutations associated with PZA resistance in M. tuberculosis.
Pyrazinamide (PZA) is a front-line drug used in the treatment of tuberculosis (TB). In a typical short-course (6-month) therapy, PZA is administered with rifampin (RIF), isoniazid (INH), and ethambutol (EMB) for the first 2 months of treatment, followed by 4 months of treatment with INH and RIF (1). During the initial acute phase, PZA and RIF are responsible for much of the killing of persisting Mycobacterium tuberculosis bacteria (23, 48). The mode of action of PZA is complex and not fully understood. It requires activation to pyrazinoic acid by a pyrazinamidase/nicotinamidase (PZase) encoded by the pncA gene (48). The activated acid form is excreted and then reabsorbed in a protonated form. It is thought that the acidification of the M. tuberculosis cells by the protonated form represents the primary mechanism by which PZA kills tubercle bacilli (48, 49). PZase inactivation is the primary mechanism for developing resistance to PZA (31, 35, 47, 48). Any mutation in pncA that inactivates the encoded enzyme appears to be sufficient to confer resistance. Consequently, PZA resistance mutations are highly diverse and are found throughout pncA. Cumulative reports indicate that between 72 and 98% of PZA-resistant isolates harbor pncA mutations (33, 46, 49). More recent reports, however, tend to support the upper end of this range (48). Earlier reports may have been subject to inaccurate susceptibility testing using culture methods. Susceptibility testing media must have a pH of 6 for PZA to be active against susceptible cells, and this pH is near the limit for growth of tubercle bacilli. Problems with susceptibility testing are magnified in developing countries with limited resources and a high burden of TB. While newer formulations of media yield more consistent results (32), alternative assays to detect PZA resistance are needed.
Molecular techniques (44) are receiving increased scrutiny as alternatives to traditional culture methods of drug susceptibility testing since they can directly detect drug resistance as DNA mutations (31, 47). Denaturing gradient gel electrophoresis (DGGE) is a simple yet powerful technique that is capable of detecting many mutations over large stretches of DNA (10, 44). With DGGE, mutations within an amplicon are recognized as alterations in melting temperature as the fragment migrates through a gradient of denaturants. GC-rich portions at the 5′ end of the PCR primers serve as the highest melting domains and as clamps that prevent DNA fragments from denaturing completely. Mutational alterations in a fragment result in bands that migrate differently from the wild type. For organisms like M. tuberculosis, having a single gene copy, heteroduplexing of the DNA to a reference DNA (usually wild type) generates additional homoduplex and heteroduplex bands that facilitate mutation detection. DGGE can detect point mutations, insertions, and deletions and is more sensitive than DNA sequencing in identifying mutations in complex DNA samples (11, 40). DGGE has previously been used in the detection of rpoB mutations associated with RIF resistance in M. tuberculosis (20, 33), including in primary clinical samples from infected patients (33).
In this study, DGGE was used for the first time to detect PZA resistance mutations in pncA. Overlapping PCR products were used to scan the pncA open reading frame and flanking noncoding regions in a large set of M. tuberculosis isolates from the U.S.-Mexico border states of Texas and Tamaulipas, respectively. By using DGGE, DNA alterations were detected in 99% of PZA-resistant isolates, while only 1% of PZA-susceptible isolates harbored DNA alterations. Coupled with comparable data for detecting RIF resistance mutations in rpoB (20, 33), these results suggest that DGGE is a sensitive, fast, and inexpensive method for detecting drug resistance mutations in M. tuberculosis.
MATERIALS AND METHODS
Bacterial isolates and susceptibility testing.
For this study, 181 individual patient isolates were identified and cultured on Lowenstein-Jensen medium, and mycobacterial DNA was extracted as previously described (20, 30). In this data set, 118 (65%) M. tuberculosis strains were isolated from patients residing in the U.S. state of Texas, while 63 (35%) strains were isolated from patients residing in the neighboring Mexican state of Tamaulipas (30). Strain genotypes were determined by IS6110 restriction fragment length polymorphism analysis (42) and spoligotyping (5) as previously described (30). Isolates were chosen for this analysis based on genotypic diversity, PZA susceptibility, and susceptibility to other anti-TB drugs (RIF, INH, EMB, and streptomycin). Drug susceptibility testing was performed by the Texas Department of State Health Services Laboratories. Primary susceptibility results were determined by the BACTEC460 radiometric method (32) using 100 μg PZA per ml, although some older cultures were tested using the agar proportion method with Middlebrook 7H10 agar (pH 5.5) and 25 μg PZA per ml. Retesting was performed exclusively by the BACTEC460 method. Classifications of isolates were revised if susceptibility retesting was consistent with DGGE genotyping of pncA and DNA sequencing of pncA and if the clinical laboratory records indicated variable and inconsistent results regarding PZA susceptibility. The susceptibility revisions were done exclusively for DGGE assay development and did not affect patient treatment.
DGGE and heteroduplex analysis.
Primer sets used to scan for pncA mutations by DGGE are listed in Table 1. DGGE primers were designed using the SG Primer program (43). Primers contained 18 to 20 bp of unique M. tuberculosis sequence to amplify the region of interest. One primer in each pair also included a 40- to 50-bp GC clamp that functions as a highest melting domain. The GC clamps have a melting temperature of ∼95°C and prevent the strands of a PCR fragment from separating completely. The melting curve of pncA was generated using the Poland algorithm (http://www.biophys.uni-duesseldorf.de/local/POLAND/poland.html) as modified by Steger (37) to simulate the transition curve of double-stranded DNA in 19 mM NaCl (12).
TABLE 1.
Sequences of primers used in this study
| PCR fragmenta | Forward primerb | Reverse primerb | bp (codons)c |
|---|---|---|---|
| A | 5′-gcgcgGCGTCGGTAGGCAAACTG-3′ | 5′-cgcccgccgcgccccgcgcccggcccgccg- cccccgcccgTGTGCCGGAGAAGTGGTC-3′ | −50-182 (−27 bp-53) |
| B | 5′-cgcccgccgccgcccgccgcgccccgcgcccgtcccgc- cgcccccgcccgGTCGTGGCAACCAAGGAC-3′ | 5′-gcgcgAGAACACCGCCTCGATTG-3′ | 131-284 (52-86) |
| C | 5′-CATTGCGTCAGCGGTACT-3′ | 5′-cgcccgccgccgcccgccgcgccccgcgccc- gtcccgccgcccccgcccgaaataataaaTG- GCAATACCGACCACAT-3′ | 212-403 (80-126) |
| D | 5′-cgcccgccgccgcccgccgcgccccgcgcccgtcccgccgcccc- cgcccgaaataataaaCACGCCACTGCTGAATTG-3′ | 5′-ACAGTTCATCCCGGTTCG-3′ | 340-588 (121-187) |
| E | 5′-cgtcccgcGCGGGCGTTGATCATCGTCG-3′ | 5′-cgcccgccgcgccccgcgcccggcccgccg- cccccgcccgCGGTGTGCCGGAGAAGTGGT-3′ | 3-186 (9-53) |
| SEQ | 5′-CGGCGTCATGGACCCTATATC-3′ | 5′-CCGCCAACAGTTCATCCC-3′ | −81-594 |
Primers sets amplify the PCR fragments for DGGE; SEQ, primers used to amplify pncA for sequencing.
Clamp sequences are in lowercase, and pncA-specific sequences are in uppercase.
The 5′ nucleotides of forward and reverse primers relative to the pncA open reading frame; −50, base pairs upstream of pncA initiation codon. The codons scanned by each PCR product represent the range of whole codons between the 3′ ends of primers and include a gap of 2 to 5 bp.
PCR amplification of M. tuberculosis genomic DNA and heteroduplexing to H37Rv were performed as previously described (20). Samples were loaded onto denaturing gradient gels, 40 to 90% urea:formamide/8% acrylamide (19:1) with TAE (40 mM Tris, 20 mM acetic Acid, 1 mM EDTA, pH 8) (primer sets A to D), or 50 to 90% urea:formamide/8% acrylamide with TAE (primer set E). Electrophoresis of samples occurred at 60°C for 16 to 20 h at 100 V in an OptiAnalyzer 8000 electrophoresis tank (CBS Scientific, Solana Beach, CA) containing TAE buffer (21, 43, 45). After electrophoresis, gels were stained with ethidium bromide in TAE for 15 to 20 min, and DNA was visualized on a UV transilluminator. DNA samples that did not harbor mutations were visualized as a single band. DNA samples that harbored mutations generated up to four bands (two homoduplex and two heteroduplex) that denatured at distinctive concentrations of denaturant.
DNA sequencing of pncA.
Initial sequencing of pncA alleles (see Fig. 2) was performed as described previously (36). Subsequent sequencing was performed as described elsewhere (20) using the SEQ primers described in Table 1. DNA sequencing was performed by Lark Technologies (Houston, TX), an FDA-approved sequencing facility. DNA changes relative to pncA from H37Rv (Rv2043c) were noted. Previous reports of PZA resistance mutations in pncA (3, 4, 6, 8, 9, 14-19, 22, 24-29, 31, 34-36, 38, 39, 46, 47) were used to identify unique alterations for this report.
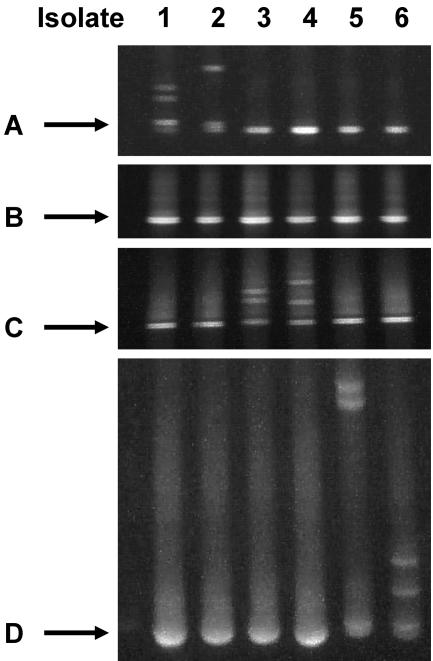
FIG. 2.
Detection of pncA mutations by PCR fragments A through D. Isolates were scanned for mutations associated with PZA resistance in pncA using DGGE PCR products A, B, C, and D. Mutations were detected only within a single PCR product, and this allowed for the localization of mutations within the pncA open reading frame. Isolate (mutation): 1 (G17S, GGC→AGC); 2 (I5-frameshift, ATC→ATG TC…); 3 (Y103stop, TAC→TAG); 4 (V93L, GTG→CTG); 5 (T160TPE, ACA GCG→ACA CCT GAA GCG); 6 (V180F, GTC→TTC).
RESULTS
DGGE detection of pncA mutations.
Four primer sets, A through D, were initially designed to scan for mutations in pncA (Fig. 1). An additional primer set, E, was later added to detect a particular subset of pncA mutations (see below). Collectively, these primer sets amplified the entire pncA open reading frame and approximately 75 bp of noncoding flanking sequence. These primer pairs were tested using six independent M. tuberculosis PZA-resistant isolates from which pncA mutations had been identified by DNA sequencing. All mutations were correctly detected within the appropriate PCR fragment (Fig. 2). The primer sets were then used to screen for nucleotide polymorphisms in PZA-resistant and -susceptible isolates. These independent strains were isolated from the U.S.-Mexico border states of Texas and Tamaulipas, respectively (30). All isolates were scanned for DNA polymorphisms in regions A through D. Positive isolates had detectable alterations in at least one PCR fragment. Since the scanned regions overlapped, some mutations were detected in multiple fragments (Tables 2 and 3).
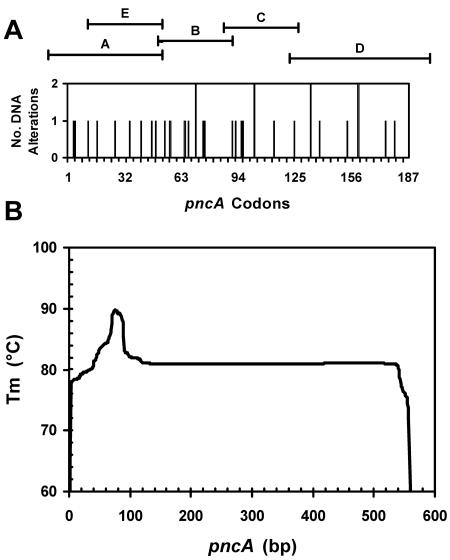
FIG. 1.
PCR products for DGGE and properties of pncA. A. Distribution of altered codons in pncA that were sequenced, including synonymous alterations. Codons that were identified more than once include identical mutations from independent strains as well as different alterations at the same codon. See Table 2 for details of mutations. The regions of pncA that are scanned by DGGE PCR products A through E are shown. B. Melting profile of pncA. The melting transition of the pncA open reading frame is simulated by the modified Poland application. The nucleotide length of pncA (561 bp) is aligned to the codon length of pncA (187 codons) in panel A above.
TABLE 2.
Sequenced pncA mutations
| Specimen no.a | Codonb | Base changec | Amino acid changed | DGGE fragment(s)e | Fig:lanef |
|---|---|---|---|---|---|
| 2778 | 4 | TTG→TCG | Leu→Ser | A | 3:1 |
| 4067 | 5 | ATC→A^TG TC... | Ile→Met + fs | A | 2:2, 3:2 |
| 5242 | 12 | GAC→GCC | Asp→Ala | A, E | 3:3 |
| 2381 | 17 | GGC→AGC | Gly→Ser | A, E | 2:1, 3:4 |
| 3579 | 27 | CTG→CCG | Leu→Pro | (A), E | 3:5 |
| 2609 | 35 | CTG→CCG | Leu→Pro | (A), E | 3:6 |
| 2886 | 41 | TAC→TAG | Tyr→Stop | (A), E | 3:7 |
| 3772 | 47 | ACC→GCC | Thr→Ala | (A), E | 3:8 |
| 1908 | 49 | GAC→GCC | Asp→Ala | (A), E | 3:9 |
| 75 | GGT→GGC | Gly→Gly | B | 4:5 | |
| 1417 | 54 | CCG→CTG | Pro→Leu | E, B | 3:10, 4:6 |
| 5745 | 57 | CAC→GAC | His→Asp | B | 4:3 |
| 4508i | 65 | TCC→TCT | Ser→Ser | B | 4:2 |
| 4862 | 67 | TCG→CCG | Ser→Pro | B | 4:7 |
| 2753 | 71 | CAT→TAT | His→Tyr | B | 4:1 |
| 3377 | 71 | CAT→GAT | His→Asp | B | 4:4 |
| 5482 | 76 | ACT→CCT | Thr→Pro | B | 4:8 |
| 2091 | 91 | GAG→TAG | Glu→stop | C | 4:15 |
| 4086 | 93 | GTG→CTG | Val→Leu | C | 2:4, 4:12 |
| 4074 | 96 | AAG→ACG | Lys→Thr | C | 4:13 |
| 820 | 97 | GGT→AGT | Gly→Ser | C | 4:14 |
| 2319 | 103 | TAC→TAG | Tyr→stop | C | 2:3, 4:11 |
| 3455 | 103 | TAC→cAC | Tyr→His | C | 4:10 |
| 5171 | 114 | 114̂IS6110 | fs | C | 4:16 |
| 570 | 125 | GTC→TTC | Val→Phe | C, Dg | 4:17 |
| 1902 | 134 | GCC→GTC | Ala→Val | D | 4:23 |
| 5330 | 134 | GCC→GTC | Ala→Val | D | 4:26 |
| 1419 | 139 | GTG→GCG | Val→Ala | D | 4:21 |
| 1969 | 154 | AGG→GGG | Arg→Gly | D | 4:24 |
| 4694 | 160-161 | ACA GCG→AĈA CCT GAA GCG | Thr Ala→ThrPro Glu Ala | D | 2:5, 4:20 |
| 5387 | 160 | ACA→CCA | Thr→Pro | D | 4:22 |
| 4259 | 175 | ATG→GTG | Met→Val | D | 4:25 |
| 4547 | 180 | GTC→TTC | Val→Phe | D | 2:6, 4:19 |
| 4720 | None | None | None | None |
All isolates are resistant to pyrazinamide except 4508; isolate 5745 is M. bovis.
Codons of pncA (Rv2043c) open reading frame.
Base change of altered codon with altered nucleotide(s) underlined; ^, insertion; lowercase letter, minor nucleotide mixed with wild type as determined by DNA sequence.
Amino acid alteration of mutation is indicated; fs, frameshift; stop, nonsense mutation; ^, insertion.
DNA alteration was identified within DGGE fragments A to E as indicated; (A), mutation is within fragment A but was not detected.
Fig:lane, figure and lane in which the denaturation profile of the mutation is shown.
DGGE profile is not shown.
TABLE 3.
Summary of pncA mutation detection by DGGE and DNA sequencing
| PZA phenotype | Assayb | Mutation detection by DGGE PCR fragment(s)a
|
||||||
|---|---|---|---|---|---|---|---|---|
| A + E | B | C | D | Multiple | Deletion | None | ||
| Resistant (83 [98.8%])d | DGGE + sequencing | 8 | 5c | 7 | 8 | 3 | 1 | |
| DGGE only | 20 | 13 | 1 | 10 | 6 | 1 | ||
| Total | 28 | 18 | 8 | 18 | 9 | 1 | 1 | |
| Susceptible (98 [1.0%])d | DGGE + sequencing | 1 | 8 | |||||
| DGGE only | 89 | |||||||
| Total | 1 | 97 | ||||||
Mutations were observed by DGGE using PCR fragments A to E as indicated; A + E, mutations detected within fragments A or E or both as described in the text and in Table 2; multiple, mutations detected in more than one fragment; deletion, pncA was not amplified from isolate.
DGGE + sequencing, mutation confirmed by DGGE and DNA sequencing of pncA; DGGE only, mutation detected by DGGE only.
Does not include synonymous G75G polymorphism; H57D polymorphism of M. bovis is detected within this fragment.
Total number of resistant isolates scanned is shown in parentheses; percentage of isolates with detectable DNA alterations is shown in brackets.
Once analysis was completed, the DGGE assay was able to detect pncA alterations in 82 of 83 PZA-resistant isolates. On the other hand, only 1 of 98 PZA-susceptible isolates harbored a DGGE-detectable DNA alteration. A portion of these alterations are shown in Fig. 3 and 4. Only denaturing gradient gels in which alterations were detected are shown. Each DNA alteration generated a distinct denaturation pattern of bands (corresponding to the homoduplex wild type, homoduplex mutant, and two heteroduplex fragments) that was characteristic of the DNA alteration. Isolates with identical denaturation patterns in the same PCR product (e.g., Fig. 4, lanes 23 and 26) were confirmed by DNA sequencing to harbor the same mutation.
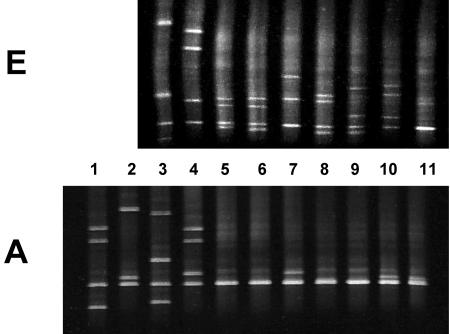
FIG. 3.
Detection of pncA mutations by fragments A and E. Isolates in lanes 1 to 11 were scanned for mutations with PCR fragments A and E as indicated. The presence of multiple bands on the denaturing gel indicates that DNA alterations were detected within the fragment. The wild type (H37Rv), which did not contain any mutations by definition, generated a single band (lane 11). Mutations in lanes 5 to 10 did not generate multiple bands within fragment A but did generate multiple bands within fragment E. Isolates in lanes 1 and 2 are not within the region scanned by fragment E. See Table 2 for details of mutations and altered codons. Mutations were detected only within one PCR fragment except where indicated. Lane (altered amino acid): 1 (L4S); 2 (I52̂bp); 3 (D12A); 4 (G17S); 5 (L27P); 6 (L35P); 7 (Y41stop); 8 (T47A); 9 (D49A); 10 (P54L; also detected within fragment B, Fig. 4, lane 6); 11 (wild type, H37Rv).
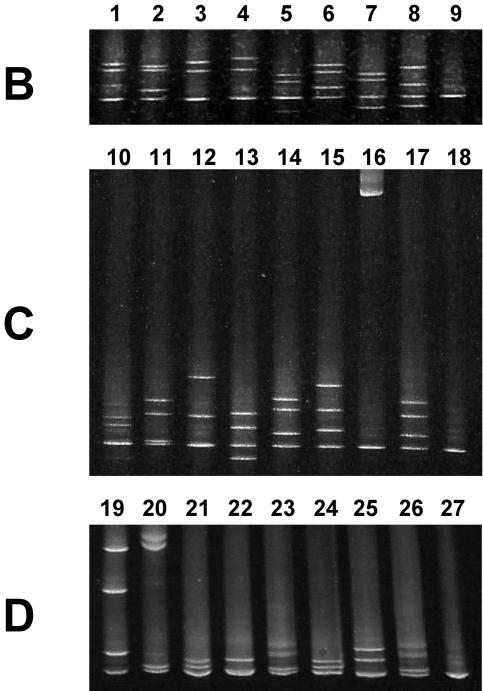
FIG. 4.
Detection of pncA mutations by fragments B to D. Isolates in lanes 1 to 27 were scanned for mutations with PCR products B, C, and D as indicated. The presence of multiple bands on the denaturing gel indicates that DNA alterations were detected within the fragment. See Table 2 for details of mutations and altered codons. Mutations were detected only within one PCR fragment except where indicated. Lane(s) (altered amino acid): 1 (H71Y); 2 (S65S); 3 (H57D); 4 (H71E); 5 (G75G); 6 (P54L; also detected within fragment E); 7 (S67P); 8 (T76P); 9, 18, and 27 (wild type, H37Rv); 10 (Y103H); 11 (Y103stop); 12 (V93L); 13 (K96T); 14 (G97S); 15 (E91stop); 16 (IS6110 insertion at codon 114); 17 (V125F; also detected within fragment D, not shown); 19 (V180F); 20 (T160TPE); 21 (V139A); 22 (T160P); 23 (A134V); 24 (R154G); 25 (M175V); 26 (A134V).
Sequencing of pncA mutations.
The pncA gene was sequenced from 41 isolates. Isolates were chosen from three categories: PZA-susceptible isolates with DGGE-detectable mutations, PZA-resistant isolates without DGGE-detectable mutations, and PZA-resistant isolates with DGGE-detectable mutations. Mutations selected for sequencing were distributed throughout pncA (Fig. 1A) and do not represent a random distribution of mutations within the gene (Table 3). DNA alterations were identified in one PZA-susceptible isolate and in 31 PZA-resistant isolates (Tables 2 and 3). The PZA-susceptible isolate harbored a synonymous polymorphism (TCC→TCT) at codon 65, which encodes a Ser residue. Another synonymous polymorphism at codon 75 (GGT→GGC), both encoding Gly residues, was observed in a PZA-resistant isolate that also harbored a D49A mutation. Interestingly, the latter alteration was not detected within fragment A even though it was within the region scanned by the PCR product. Other mutations in this same region were also not detected within the fragment (see below). One isolate contained an IS6110 insertion after codon 114. The insertion was apparent from agarose gels of the PCR-amplified products from this isolate, and it also resulted in an aberrant migration pattern on the denaturing gels (Fig. 4, lane 16). Two isolates harbored small insertions. One contained a 2-bp insertion after the first nucleotide of codon five and resulted in a frameshift mutation. The other was a 6-bp insertion after the second nucleotide of codon 160 that would result in the insertion of two amino acids (Pro and Glu) within the protein. Interestingly, another isolate contained a T160P mutation. The remaining isolates harbored single nucleotide changes. Three mutations resulted in nonsense mutations at codons 41, 91, and 103. Two strains with differing genetic backgrounds contained the same mutation (GCC→GTC), which encoded an A134V alteration, and the denaturation patterns from these isolates were identical (Fig. 4, lanes 23 and 26). One DNA sample yielded both wild-type and mutant sequence at codon 103; the mutant sequence encodes a Tyr-to-His alteration. As reported previously, DGGE can detect mixtures of DNA that might arise from mixed strains or emerging drug resistance (20, 33). Although these mixed cultures are usually confirmed by DNA sequencing, we have recently reported procedures to confirm the presence of DNA mixtures by DGGE (20). The remaining isolates harbored single nucleotide changes that are predicted to result in single amino acid changes within the open reading frame. Of the 33 DNA alterations within pncA reported here, 11 have not been reported previously (3, 4, 6, 8, 9, 14-19, 22, 24-29, 31, 34-36, 38, 39, 46, 47), although other alterations in the same codon have been previously reported for five of these novel alterations.
Three of eighty-three isolates were monoresistant to PZA, and since this is a property of Mycobacterium bovis, which is naturally resistant to PZA, these isolates were investigated in more detail. The pncA gene of M. bovis (Mb2069c) encodes an Asp residue at codon 57, and it appears that this residue prevents the M. bovis PZase from activating PZA. M. tuberculosis pncA normally encodes a His residue at codon 57, and the encoded PZase can activate PZA and render the cells susceptible to the drug. An H57D mutation has also been identified in PZA-resistant M. tuberculosis isolates (31, 47, 48). Codon 57 is amplified by primer set B, and all three monoresistant isolates harbored pncA alterations within this PCR product. One isolate displayed a distinct denaturation pattern (Fig. 4, lane 2), and DNA sequencing revealed that its pncA encoded a P54L alteration (Table 2). The other two isolates displayed the same denaturation patterns (Fig. 4, lane 3), and DNA sequencing of pncA from one isolate revealed the H57D alteration. Strain genotyping indicated that both of these isolates had an IS6110 RFLP pattern (1.001) and spoligotype (676773777777600) characteristic of M. bovis. An investigation of the clinical laboratory records confirmed that these two isolates were M. bovis. Confirming earlier reports, PZA monoresistance alone is not a unique characteristic of M. bovis isolates (14). The DGGE assay is capable of detecting pncA polymorphisms that distinguish M. tuberculosis from M. bovis.
Resolving problem isolates.
Problems were encountered in the initial analysis of 18 isolates whose PZA susceptibility did not match their DGGE pattern. These problems were resolved by retesting these isolates for PZA susceptibility, by DNA sequencing their pncA alleles, and by further DGGE analysis. Initially, 11 isolates were identified as PZA resistant but did not harbor DGGE-detectable mutations. Susceptibility was revised from resistant to susceptible for six of these isolates, and DNA sequencing confirmed that they lacked alterations in pncA. A single isolate was confirmed as resistant to PZA but lacked any alteration within pncA. Finally, four isolates were confirmed as resistant to PZA, and mutations within pncA were identified by sequencing. The mutations resided in a region of pncA amplified by primer set A and spanned codons 27 to 47 (Fig. 1, lanes 5 to 8 and 10). Only the nonsense mutation at codon 41 resulted in a partial denaturation pattern. An additional isolate harbored a defect at codon 49 that was also not detected (lane 9). This isolate also contained a synonymous alteration at codon 75 that was detected within fragment B. Inspection of the pncA gene revealed a domain of the gene containing a high GC content. While most of pncA has a melting temperature around 81 to 82°C, this region had a melting temperature peak close to 90°C (Fig. 1B). The undetected mutations were associated with this high melting domain. We hypothesized that these mutations were not detected because the high melting temperature of this region prevented the heteroduplexes from melting properly during electrophoresis. To address this problem, an additional primer set was designed. The fragment amplified by primer set E is slightly shorter than fragment A and contains a different combination of clamps to fine tune denaturation (Table 1). The electrophoretic conditions were also modified to detect denaturation products (see Materials and Methods). All of the mutations not detected in fragment A were detected in fragment E (Fig. 1, lanes 5 to 10). Once this was achieved, every sequenced pncA mutation was detected within one of the DGGE amplicons (Fig. 3 and 4). Only one PZA-resistant isolate, described above, lacked a mutation in pncA as assayed by DGGE and DNA sequencing.
Seven PZA-susceptible isolates were identified with DGGE mutations during the initial screening. One isolate remained PZA susceptible upon retesting and harbored the synonymous S65S polymorphism within pncA, as described above. PZA susceptibility results were revised from susceptible to resistant for three isolates, and DNA sequencing indicated that they harbored alterations in pncA indicative of drug resistance. These alterations included amino acid changes L4S, H71Y, and H71D (Table 2). The L4S mutation has been reported previously, and PZA-resistant isolates harboring this amino acid alteration lack PZase activity (26). The two amino acid changes at codon 71 have not been reported previously, but another alteration at this codon, H71R, has been reported in PZA-resistant isolates lacking PZase activity (26-28). Intuitively, isolates with these amino acid alterations have been defined as PZA resistant. Three other isolates remained PZA susceptible upon retesting. DNA sequencing indicated that these isolates lacked alterations in pncA, and repeat DGGE assays confirmed the absence of polymorphisms.
DISCUSSION
In this study, DGGE was used to screen M. tuberculosis isolates from the U.S.-Mexico border region for nucleotide polymorphisms in pncA that impart resistance to the anti-TB drug PZA. The five DGGE PCR products were used to scan for polymorphisms in approximately 600 bp of the M. tuberculosis genome that encompass the pncA gene. Using DGGE, we detected DNA alterations in 98.8% (82 of 83) of PZA-resistant isolates and in only 1% (one of 98) of PZA-susceptible isolates. These results by DGGE rival the theoretical maximum attained by DNA sequencing (31, 44, 47) and suggest that DGGE can be a viable, more economic alternative to sequencing in detecting pncA mutations. DGGE can be performed on clinical samples, such as sputum (33), and can be completed rapidly with high throughput methodology (21). DGGE also detected apparent reporting errors in PZA susceptibility testing by culture methods. Based on the initial results reported here, we can compare the diagnostic criteria of the DGGE genotypic assay with those of the culture phenotypic assay of PZA susceptibility. Initial culture testing had a sensitivity of 96.4% (80 of 83) in predicting PZA resistance and a specificity of 93.9% (92 of 98) in predicting PZA susceptibility. The sensitivity of the DGGE assay was 98.8% (82 of 83), and the specificity was 95% (94 of 98). Since the values for the DGGE PZA assay exceed those of the culture assay, this suggests that the DGGE genotypic assay may be a suitable alternative to the phenotypic culture assay for predicting PZA susceptibility. Blinded studies will be necessary, however, to confirm these predictions.
Although pncA was not sequenced from all 181 isolates, the concordance between DNA sequencing and DGGE was 100% for the 41 isolates from which pncA was sequenced. Of the 32 isolates with DGGE-detected DNA alterations, all 32 yielded pncA alterations upon sequencing. Furthermore, all of the codon changes in these isolates were localized to the correct region of pncA probed by the PCR fragments. In addition, all nine isolates without DGGE-detectable alterations were shown to lack pncA alterations by DNA sequencing. Similar concordance was observed between DGGE and DNA sequencing for RIF resistance mutations associated with rpoB (20). Altogether, we identified 32 distinct alterations in pncA, 11 of which have not been reported previously (3, 4, 6, 8, 9, 14-19, 22, 24-29, 31, 34-36, 38, 39, 46, 47). DNA alterations were distributed throughout pncA (Table 3), and representatives detected within the four major DGGE PCR products (A to D) were sequenced to establish a distribution of DNA alterations throughout the gene (Fig. 1). This strategy proved useful when testing whether the four primer sets were sufficient for detecting all pncA mutations in these isolates.
Initial analysis identified 18 isolates whose DGGE profiles did not match their PZA susceptibilities, including 11 PZA-resistant isolates without DGGE patterns and 7 PZA-susceptible isolates with DGGE patterns. The PZA susceptibility profiles of half of these isolates were reassigned upon retesting, and DNA sequencing confirmed that these changes were justified. Six isolates without DNA alterations in pncA were classified as PZA susceptible upon retesting, and three isolates with pncA polymorphisms were revised to PZA resistant. The altered residues of the latter isolates were characteristic of PZA resistance. One isolate remained PZA resistant, but defects in pncA could not be detected by DGGE or by sequencing. A small number of PZA-resistant isolates do not harbor mutations in pncA, and these isolates also usually retain PZase activity (31, 47, 48). PZase activity was not tested, and so this correlation remains to be confirmed for this isolate. Similarly, one PZA-susceptible isolate harbored a S67S synonymous polymorphism.
Problems with the initial DGGE assays were apparent for seven isolates. This included three PZA-susceptible isolates that were incorrectly assigned mutations based on the initial DGGE screen but lacked pncA alterations by sequencing or by repeat DGGE assays. In addition, four PZA-resistant isolates harbored pncA mutations that were not detected within the initial four DGGE PCR products. All four DNA alterations were located in a region scanned by fragment A; a fifth mutation was also identified in this region that was not detected. These mutations were clustered in a region of pncA that had an unusually high melting temperature due to a high GC content. The melting temperature of this region peaked around 90°C, which was about 10°C higher than that of the rest of the gene (Fig. 1B). Indeed, the GC clamps attached to the ends of the amplicons have a peak melting temperature of 95°C, which is only slightly higher than this naturally occurring M. tuberculosis domain. The GC clamps are attached to prevent complete separation of the DNA strands during electrophoresis. These results suggested that this region was not melting properly in the denaturing gradient gels, and therefore the mutations were not detected. Similar problems are expected for any molecular technique that probes for DNA alterations in pncA, probably including sequencing. For instance, Mohamed et al. (25) reported problems detecting a 1-bp deletion within codon 24 using denaturing high-performance liquid chromatography. Branch migration inhibition (18) and single-stranded conformation polymorphism (6, 34) have also been used to detect pncA mutations. These three techniques work on the same basic principles as DGGE in that they detect DNA mismatches that represent mutations. In theory, any molecular technique that relies on denaturation, hybridization, or DNA conformation (44) may face problems in detecting pncA alterations in this region. Once problems associated with this high-melting region were understood, an additional DGGE primer set was used to successfully detect mutations in this region. Mutations from five PZA-resistant isolates were localized to this region from a total of 83 PZA-resistant isolates scanned. Based on these results, it is estimated that 6% of PZA-resistant isolates harbor mutations in this region.
Mutations that inactivate the PZase encoded by pncA are distributed throughout the gene, and no major hot spots have been observed. Alterations, such as T47A and H57D, are commonly observed among groups of PZA-resistant isolates, but these mutations do not represent a significant proportion of the total resistant isolates. The pncA gene can be divided into eight regions that are scanned by the five DGGE PCR products, including four regions in which these fragments overlap. Based on the denaturation patterns that are characteristic of each mutation and on the location of a mutation to one of the aforementioned subdomains, it is possible to predict mutations by DGGE (e.g., Fig. 4, lanes 23 and 26). This property could be used in several ways. First, it is possible to detect the same mutation in different isolates based on their identical denaturation pattern. Second, it can be used to identify synonymous DNA alterations that are not associated with drug resistance. These are infrequent in pncA, although a few have been observed (3, 4, 6, 8, 9, 14-19, 22, 24-29, 31, 34-36, 38, 39, 46, 47). By recognizing these polymorphisms by their denaturation fingerprint, it should be possible to improve the specificity of a DGGE clinical assay to predict PZA susceptibility. Third, it could be used to track outbreaks and transmission of PZA-resistant strains. Fourth, polymorphisms can be used to type strains by multilocus sequence typing (41) or comparable techniques. Multilocus sequence typing can be performed using DGGE because of its high throughput and low cost (13). Finally, as shown here, DGGE fingerprints can also be used to help distinguish M. bovis isolates from other members of the M. tuberculosis complex (14).
The results obtained here are similar to those obtained when DGGE was used to probe rpoB for mutations associated with RIF resistance (20, 33). Using two DGGE PCR products, we were able to detect mutations in 99% of RIF-resistant isolates, which were also collected from the U.S.-Mexico border region (20). By contrast, only 2% of RIF-susceptible isolates produced detectable polymorphisms. RIF resistance mutations in rpoB are highly localized to an 81-bp stretch of rpoB between codons 507 and 533, but mutations outside this region exist as well. The dispersed nature of mutations in both genes makes it expensive to identify mutations by DNA sequencing (2, 7, 44). From the analyses of rpoB and pncA polymorphisms from our data sets, we have estimated that mutation detection by DNA sequencing is 15- to 50-fold more expensive than detection by DGGE. The lower range includes material costs only, while the upper range includes equipment, data analysis, and labor costs. Expense, sensitivity, and mutation detection present problems for other molecular assays since multiple probes (such as oligonucleotides) must be used. Although multiple primer sets are used for DGGE, they are capable of scanning large (50 to 500 bp) stretches of DNA and are capable of detecting all alterations in the amplicon.
Finally, DGGE assays revealed several instances of apparent errors in drug susceptibility testing. These included both false-positive and false-negative errors. These observations point out the inherent problems with drug susceptibility testing. This problem is more apparent with PZA since the pH of the culture assay must be around 6, which is near the limits for growth of mycobacteria. In our analysis of PZA and RIF resistance by DGGE, the number of apparent culture testing errors was comparable to the number of DGGE errors for both false-positive and false-negative scoring of susceptibility. For both RIF and PZA, it will be interesting to perform blinded studies comparing our DGGE assays to culture methods to predict drug susceptibility.
Acknowledgments
This work was supported by grants from the Robert J. Kleberg, Jr., and Helen C. Kleberg Foundation and the Advanced Research Program/Advanced Technology Program from the Texas Higher Education Coordinating Board.
These experiments were initiated in the laboratory of Rebecca A. Cox, and we thank her for her support, encouragement, and enthusiasm. We are also grateful to Denise Dunbar for coordinating the pyrazinamide MIC testing and to Brent Calder for help in obtaining the Poland melting curve data.
REFERENCES
- 1.American Thoracic Society, CDC., and the Infectious Diseases Society of America. 2003. Treatment of tuberculosis. Morb. Mortal. Wkly. Rep. 52:1-77. [Google Scholar]
- 2.Bounpheng, M., S. McGrath, D. Macias, N. van Orsouw, Y. Suh, D. Rines, and J. Vijg. 2003. Rapid, inexpensive scanning for all possible BRCA1 and BRCA2 gene sequence variants in a single assay: implications for genetic testing. J. Med. Genet. 40:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, T. J., O. Tansel, and G. L. French. 2000. Simultaneous identification and typing of multidrug-resistant Mycobacterium tuberculosis isolates by analysis of pncA and rpoB. J. Med. Microbiol. 49:651-656. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, S. J., L. Thibert, T. Sanchez, L. Heifets, and Y. Zhang. 2000. pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob. Agents Chemother. 44:528-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins. J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. N. Quitugua, N. Rastogi, D. van Soolingen, and V. Wright. 2001. Spacer oligonucleotide typing of Mycobacterium tuberculosis: recommendations for standardized nomenclature. Int. J. Tuberc. Lung Dis. 5:216-219. [PubMed] [Google Scholar]
- 6.Davies, A. P., O. J. Billington, T. D. McHugh, D. A. Mitchison, and S. H. Gillespie. 2000. Comparison of phenotypic and genotypic methods for pyrazinamide susceptibility testing with Mycobacterium tuberculosis. J. Clin. Microbiol. 38:3686-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhana, R. K., N. J. van Orsouw, I. Sigalas, C. Eng., and J. Vijg. 1998. Critical factors in the performance and cost of two-dimensional gene scanning: RB1 as a model. BioTechniques 25:664-675. [DOI] [PubMed] [Google Scholar]
- 8.Endoh, T., A. Yagihashi, N. Uehara, D. Kobayashi, N. Tsuji, M. Nakamura, S. Hayashi, N. Fujii, and N. Watanabe. 2002. Pyrazinamide resistance associated with pncA gene mutation in Mycobacterium tuberculosis in Japan. Epidemiol. Infect. 128:337-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escalante, P., S. Ramaswamy, H. Sanabria, H. Soini, X. Pan, O. Valiente-Castillo, and J. M. Musser. 1998. Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tuber. Lung Dis. 89:111-118. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, S. G., and L. S. Lerman. 1979. Analysis of point mutations by denaturing gradient gel electrophoresis. Proc. Natl. Acad. Sci. USA 80:1579-1583. [Google Scholar]
- 11.Garcia-Delgado, M., C. J. Gonzalez-Navarro, M. C. Napal, C. Baldonado, J. L. Vizmanos, and A. Gullon. 1998. Higher sensitivity of denaturing gradient gel electrophoresis than sequencing in the detection of mutations in DNA from tumor samples. BioTechniques 24:72-76. [DOI] [PubMed] [Google Scholar]
- 12.Gotoh, O. 1983. Prediction of melting profiles and local helix stability for sequenced DNA. Adv. Biophys. 16:1-52. [DOI] [PubMed] [Google Scholar]
- 13.Gurtler, V., H. D. Barrie, and B. C. Mayall. 2002. Denaturing gradient gel electrophoresis multilocus sequence typing of Staphylococcus aureus isolates. Electrophoresis 23:3310-3320. [DOI] [PubMed] [Google Scholar]
- 14.Hannan, M. M., E. P. Desmond, G. P. Morlock, G. H. Mazurek, and J. T. Crawford. 2001. Pyrazinamide-monoresistant Mycobacterium tuberculosis in the United States. J. Clin. Microbiol. 39:647-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou, L., D. Osei-Hyiaman, Z. Zhang, B. Wang, A. Yang, and K. Kano. 2000. Molecular characterization of pncA gene mutations in Mycobacterium tuberculosis clinical isolates from China. Epidemiol. Infect. 124:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, T-S., S. S-J. Lee, H-Z. Tu, W-K. Huang, Y-S. Chen, C-K. Huang, S-R. Wann, H-H. Lin, and Y-C. Liu. 2003. Correlation between pyrazinamide activity and pncA mutations in Mycobacterium tuberculosis isolates in Taiwan. Antimicrob. Agents Chemother. 47:3672-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemaitre, N., W. Sougakoff, C. Truffot-Pernot, and V. Jarlier. 1999. Characterization of new mutations in pyrazinamide-resistant strains of Mycobacterium tuberculosis and identification of conserved regions important for the catalytic activity of the pyrazinamidase PncA. Antimicrob. Agents Chemother. 43:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, Y. P., M. A. Behr, P. M. Small, and N. Kurn. 2000. Genotypic determination of Mycobacterium tuberculosis antibiotic resistance using a novel mutation detection method, the branch migration inhibition M. tuberculosis antibiotic resistance test. J. Clin. Microbiol. 38:3656-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marttila, H. J., M. Marjamaki, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1999. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from northwestern Russia. Antimicrob. Agents Chemother. 43:1764-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCammon, M. T., J. S. Gillette, D. P. Thomas, S. V. Ramaswamy, E. A. Graviss, B. N. Kreiswirth, J. Vijg, and T. N. Quitugua. 2005. Detection of rpoB mutations associated with rifampin resistance in Mycobacterium tuberculosis using denaturing gradient gel electrophoresis. Antimicrob. Agents Chemother. 49:2200-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGrath, S. B., M. Bounpheng, L. Torres, M. Calavetta, C. B. Scott, Y. Suh, D. Rines, N. van Orsouw, and J. Vijg. 2001. High-speed, multicolor fluorescent two-dimensional gene scanning. Genomics 78:83-90. [DOI] [PubMed] [Google Scholar]
- 22.Mestdagh, M., P. A. Fonteyne, L. Realini, R. Rossau, G. Jannes, W. Mijs, K. A. L. De Smet, F. Portaels, and E. Van den Eeckhout. 1999. Relationship between pyrazinamide resistance, loss of pyrazinamidase activity, and mutations in the pncA locus in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:2317-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchison, D. A. 2004. The search for new sterilizing anti-tuberculosis drugs. Front. Biosci. 9:1059-1072. [DOI] [PubMed] [Google Scholar]
- 24.Miyagi, C., N. Yamane, B. Yogesh, H. Ano, and T. Takashima. 2004. Genetic and phenotypic characterization of pyrazinamide-resistant Mycobacterium tuberculosis complex isolates in Japan. Diagn. Microbiol. Infect. Dis. 48:111-116. [DOI] [PubMed] [Google Scholar]
- 25.Mohamed, A. M., D. R. Bastola, G. P. Morlock, R. C. Cooksey, and S. H. Hinrichs. 2004. Temperature-mediated heteroduplex analysis for detection of pncA mutations associated with pyrazinamide resistance and differentiation between Mycobacterium tuberculosis and Mycobacterium bovis by denaturing high-performance liquid chromatography. J. Clin. Microbiol. 42:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morlock, G. P., J. T. Crawford, W. R. Butler, S. E. Brim, D. Sikes, G. H. Mazurek, C. L. Woodley, and R. C. Cooksey. 2000. Phenotypic characterization of pncA mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, S. K., J. Y. Lee, C. L. Chang, M. K. Lee, H. C. Son, C. M. Kim, H. J. Jang, H. K. Park, and S. H. Jeong. 2001. pncA mutations in clinical Mycobacterium tuberculosis isolates from Korea. BMC Infect. Dis. 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Portugal, I., L. Barreiro, J. Moniz-Pereira, and L. Brum. 2004. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates in Portugal. Antimicrob. Agents Chemother. 48:2736-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Post, F. A., P. A. Willcox, B. Mathema, L. M. Steyn, K. Shean, S. V. Ramaswamy, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and G. Kaplan. 2004. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J. Infect. Dis. 190:99-106. [DOI] [PubMed] [Google Scholar]
- 30.Quitugua, T. N., B. J. Seaworth, S. E. Weis, J. P. Taylor, J. S. Gillette, I. I. Rosas, K. C. Jost, Jr., D. M. Magee, and R. A. Cox. 2002. Transmission of drug-resistant tuberculosis in Texas and Mexico. J. Clin. Microbiol. 40:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 32.Salfinger, M., L. B. Reller, B. Demchuk, and Z. T. Johnson. 1989. Rapid radiometric method for pyrazinamide susceptibility testing of Mycobacterium tuberculosis. Res. Microbiol. 140:301-309. [DOI] [PubMed] [Google Scholar]
- 33.Scarpellini, P., S. Braglia, P. Carrera, M. Cedri, P. Cichero, A. Colombo, R. Crucianelli, A. Gori, M. Ferrari, and A. Lazzarin. 1999. Detection of rifampin resistance in Mycobacterium tuberculosis by double gradient-denaturing gradient gel electrophoresis. Antimicrob. Agents Chemother. 43:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662-667. [DOI] [PubMed] [Google Scholar]
- 35.Scorpio, A., P. Lindholm-Levy, L. Heifets, R. Gilman, S. Siddiqi, M. Cynamon, and Y. Zhang. 1997. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41:540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sreevatsan, S., X. Pan, Y. Zhang, B. Kreiswirth, and J. M. Musser. 1997. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob. Agents Chemother. 41:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steger, G. 1994. Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res. 22:2760-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, Y., A. Suziki, A. Tamaru, C. Katsukawa, and H. Oda. 2002. Rapid detection of pyrazinamide-resistant Mycobacterium tuberculosis by a PCR-based in vitro system. J. Clin. Microbiol. 40:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tracevska, T., I. Jansone, V. Baumanis, A. Nodieva, O. Marga, and G. Skenders. 2004. Spectrum of pncA mutations in multidrug-resistant Mycobacterium tuberculosis isolates obtained in Latvia. Antimicrob. Agents Chemother. 48:3209-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trulzsch, B., K. Krohn, P. Wonerow, and R. Paschke. 1999. DGGE is more sensitive for the detection of somatic point mutations than direct sequencing. BioTechniques. 27:266-268. [DOI] [PubMed] [Google Scholar]
- 41.Urwin, R., and M. C. J. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 42.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Orsouw, N. J., R. K. Dhanda, R. D. Rines, W. M. Smith, I. Sigalas, C. Eng, and J. Vijg. 1998. Rapid design of denaturing gradient-based two-dimensional electrophoretic gene mutational scanning tests. Nucleic Acids Res. 26:2398-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Victor, T. C., P. D. van Helden, and R. Warren. 2002. Prediction of drug resistance in M. tuberculosis: molecular mechanisms, tools and applications. IUBMB Life 53:231-237. [DOI] [PubMed] [Google Scholar]
- 45.Vijg, J., and N. J. van Orsouw. 1999. Two-dimensional gene scanning: exploring human genetic variability. Electrophoresis 20:1239-1249. [DOI] [PubMed] [Google Scholar]
- 46.Wade, M. M., D. Volokhov, M. Peredelchuk, V. Chizhikov, and Y. Zhang. 2004. Accurate mapping of mutations of pyrazinamide-resistant Mycobacterium tuberculosis strains with a scanning-frame oligonucleotide microarray. Diagn. Microbiol. Infect. Dis. 49:89-97. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y., and A. Telenti. 2000. Genetics of drug resistance in Mycobacterium tuberculosis, p. 235-254. In F. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 48.Zhang, Y., and D. Mitchison. 2003. The curious characteristics of pyrazinamide: a review. Int. J. Tuberc. Lung Dis. 7:6-21. [PubMed] [Google Scholar]
- 49.Zhang, Y., M. M. Wade, A. Scorpio, H. Zhang, and Z. Sun. 2003. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J. Antimicrob. Chemother. 52:790-795. [DOI] [PubMed] [Google Scholar]