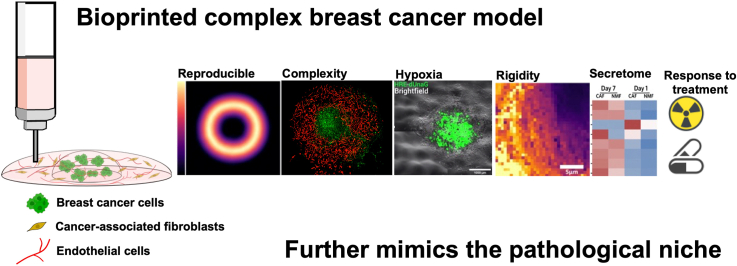
Abstract
Deciphering breast cancer treatment resistance remains hindered by the lack of models that can successfully capture the four-dimensional dynamics of the tumor microenvironment. Here, we show that microextrusion bioprinting can reproducibly generate distinct cancer and stromal compartments integrating cells relevant to human pathology. Our findings unveil the functional maturation of this millimeter-sized model, showcasing the development of a hypoxic cancer core and an increased surface proliferation. Maturation was also driven by the presence of cancer-associated fibroblasts (CAF) that induced elevated microvascular-like structures complexity. Such modulation was concomitant to extracellular matrix remodeling, with high levels of collagen and matricellular proteins deposition by CAF, simultaneously increasing tumor stiffness and recapitulating breast cancer fibrotic development. Importantly, our bioprinted model faithfully reproduced response to treatment, further modulated by CAF. Notably, CAF played a protective role for cancer cells against radiotherapy, facilitating increased paracrine communications. This model holds promise as a platform to decipher interactions within the microenvironment and evaluate stroma-targeted drugs in a context relevant to human pathology.
Keywords: Bioprinting, Cancer microenvironment, Extracellular matrix
Graphical abstract
Highlights
-
•
The bioprinted model further replicates the three-dimensional structure of breast cancer, integrating relevant stromal cells.
-
•
CAFs influence model maturation by remodeling ECM, increasing microvascular structures and enhancing stiffness, mimicking fibrosis.
-
•
The bioprinted model shows that CAFs protect cancer cells from radiotherapy via paracrine signals, offering insights for stroma-targeted therapies.
1. Introduction
Breast cancer continues to impose a critical burden on healthcare systems, maintaining its status as the most diagnosed and lethal cancer among females. In 2020 alone, it accounted for 2.3 million new cases and an estimated 685,000 deaths [1]. Mortality is mainly associated with treatment resistance, favoring local and distant recurrence [2]. Two decades of research increasingly emphasized the influence of the tumor microenvironment (TME) on these mechanisms [3,4]. Notably, cancer-associated fibroblasts (CAF), as major cellular components of the TME, play pivotal roles in radiosensitivity, targeted treatment efficacy, and metastatic risks [[5], [6], [7]]. Similarly, endothelial cells, that compose the TME microvasculature, have shown to modulate breast cancer cell aggressivity through paracrine signaling following therapy [8,9]. Despite advancements in the understanding of the intricacies and interactions within the TME, novel therapies leveraging this intercommunication have often failed to progress toward clinical use in breast cancer [5,[10], [11], [12]].
This failure stems in part from the historical use of 2D culture and in vivo models for cancer drug discovery, gradually reaching its limits. Indeed, and despite successful pre-clinical trials, nearly 90 % of drug candidates fail to obtain approval for clinical use [13]. The inadequacy of complexity in 2D culture and the absence of a 3D structure organization, coupled with the allogenic TME in in vivo models, has hindered their relevance to human physiopathology, thereby escalating false positive rates [14,15]. The TME, composed by a dynamic set of cells and extracellular matrix (ECM), requires models that can capture its intricate complexity to further decipher the breast cancer niche. The rise of 3D culture, as an answer to this problematic, revolutionized cancer research [16]. However, strategies such as spheroids and organoids, while leveraging cells self-organization and ECM secretion, often fall short in consistently reproducing an architecture that faithfully mimics tumor physiology.
Biofabrication has propelled the advancement of 3D tissue cultures by automating the production of spatially organized biological constructs [[17], [18], [19], [20]]. Among multiple biofabrication processes, bioprinting arose as a leading strategy to model breast cancer [[21], [22], [23]]. Microextrusion is the principal technology used in bioprinting [24], notably in the case of cancer, for its ability to combine millimeter-scale tissues and multimaterial printing, hardly achievable with other biofabrication technologies. Advancing toward the 3D bioprinting of stromal elements to recapitulate the TME is now a necessary step toward next generation tumor modeling [15,25]. Such models have been used to investigate the impact of fibroblasts [[26], [27], [28], [29]], endothelial cells [26,28,30], macrophages [31] and adipocytes [[32], [33], [34]] on TME intercommunications.
However, in these studies, authors rarely describe the dynamic evolution of their model properties related to the specificity of CAF. Notably, ECM composition and mechanical properties after the introduction and maturation with stromal cells, have been consistently overlooked, despite the established TME remodeling over time. Understanding this 4th dimension in the presence of CAF appears critical to evaluate tumor progression in the context of treatment response.
In this study, we report a novel bioprinted breast cancer model (BpBCM) that adopts a holistic approach to faithfully replicate the three-dimensional spatial structure of breast cancer, while integrating stromal cells relevant to the TME. We not only establish the reliability of our methodology but also delve into a comprehensive understanding of how CAF influence the fate of the tumor as a dynamic four-dimensional entity. We meticulously characterize the changes occurring throughout the maturation process, shedding light on significant TME remodeling involving the microvasculature, ECM composition and mechanical properties, all specifically driven by CAF. We finally demonstrate that BpBCM can further replicate treatment response and paracrine communication in the TME.
2. Results
2.1. Bioprinting reproducibly generates spatially organized breast cancer models
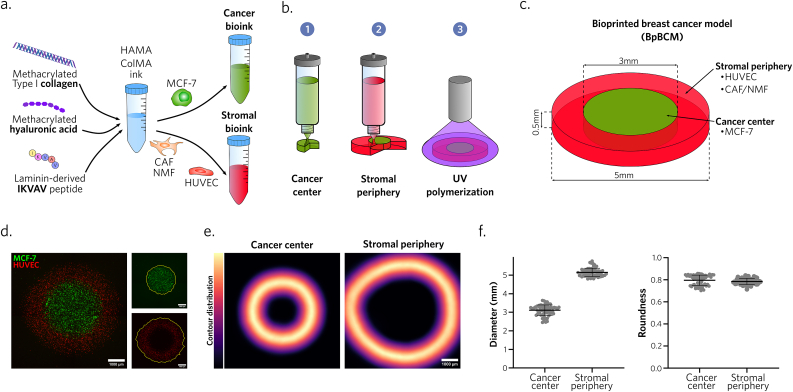
Capturing the complexity of breast cancer ECM is essential to the 3D model development. Notably, type I collagen as well as hyaluronic acid are upregulated in breast cancer, and play crucial roles on tumor progression and invasiveness [35,36]. We mimicked this composition by combining methacrylated collagen (ColMA) and hyaluronic acid (HAMA) based on a previous formulation [37]. We functionalized this hydrogel with the laminin-111 derived peptide IKVAV, already shown to enhance cancer cell migration, growth and metastasis [38,39](Fig. 1a). This functionalization induced a significant increase in metabolic activity of MCF-7 up to 1 mg/mL, while higher concentrations led to a decrease in ink homogeneity unsuitable for bioprinting application (Supplementary Figs. 1b,c,d). Consequently, the concentration was standardized at 1 mg/mL for the remainder of the study. This biomaterial ink displayed expected mechanical properties, with a characteristic shear-thinning behavior, crucial for microextrusion, and a post-polymerization shear storage modulus G’ = 334 ± 82 Pa (Supplementary Figs. 1e and f). We also confirmed that the photo-polymerization protocol used in this study does not induce the apparition γH2Ax labeled DNA damage (Supplementary Fig. 2).
Fig. 1.
Microextrusion bioprinting reproducibly produces millimeter-sized breast cancer models with distinct cancer and stromal areas.
a. Bioink formulation schematic. b. Schematic representation of the 3-step bioprinting process. c. Bioprinted breast cancer model (BpBCM) design. d. BpBCM epifluorescence imaging 4h post-printing with MCF-7 GFP+ (green) in the center and HUVEC mKate+ (red) in the periphery (left). Scale bar: 1000 μm. Cancer center and stromal periphery contour detection (right). e,f. Cancer center and stromal periphery contour distribution (e, scale bar: 1000 μm), and quantifications of contour diameter and roundness (f) (N = 3, n = 62). Shown as mean ± s.d.
Primary breast cancer invasion into the stroma leads to an architecture closely related to its function. To achieve a tumor-like spatial organization, we used microextrusion bioprinting to deposit the bioink encapsulating MCF-7 hormone-responsive breast cancer cells within a 3 mm diameter disk. This first disk was then surrounded with a stromal bioink encapsulating fibroblasts and endothelial cells (Fig. 1b, Supplementary Video 1). This process aimed to obtain a disk shaped BpBCM with a thickness of 500 μm, and a total diameter of 5 mm, containing distinct cancer center and stromal periphery (Fig. 1c). This design aimed to reproduce a relevant tumor size [40], with a cancer-stromal organization relevant to low grade tumors [41].
The following are the Supplementary data related to this article:
To ensure the reliability and reproducibility of this approach, we conducted spatial segmentation of both GFP-tagged MCF-7 and mKate-tagged human umbilical vein endothelial cells (HUVEC) bioprinted as cancer and stromal bioink, respectively (Fig. 1d). The obtained contour distribution aligned with the intended bioprinting design, with a central diameter of 3.09 ± 0.28 mm and a peripheral diameter of 5.14 ± 0.22 mm (Fig. 1e). Similarly, the circular geometry was respected and reproducible with a roundness score of 0.79 ± 0.05 and 0.78 ± 0.03 for the center and periphery, respectively (Fig. 1f). Such reproducibility was achieved with a medium throughput process, with one bioprinted model per minute, in 24 well plates (Supplementary Figs. 3c and d, Supplementary Video 2). Post-printing viabilities were also confirmed to be >70 % for all cell types (Supplementary Figs. 3a and b).
Altogether, these results indicate that our bioprinting strategy allowed the reproducible production of millimeter-sized BpBCM using an ECM-like bioink encapsulating viable cells relevant to breast cancer TME, easily adaptable to other cell types such as invasive MDA-MB-231 (Supplementary Video 3).
Supplementary data related to this article can be found online at https://doi.org/10.1016/j.bioactmat.2024.08.037
The following are the Supplementary data related to this article:
2.2. BpBCM develop a tumor-like necrotic and hypoxic core
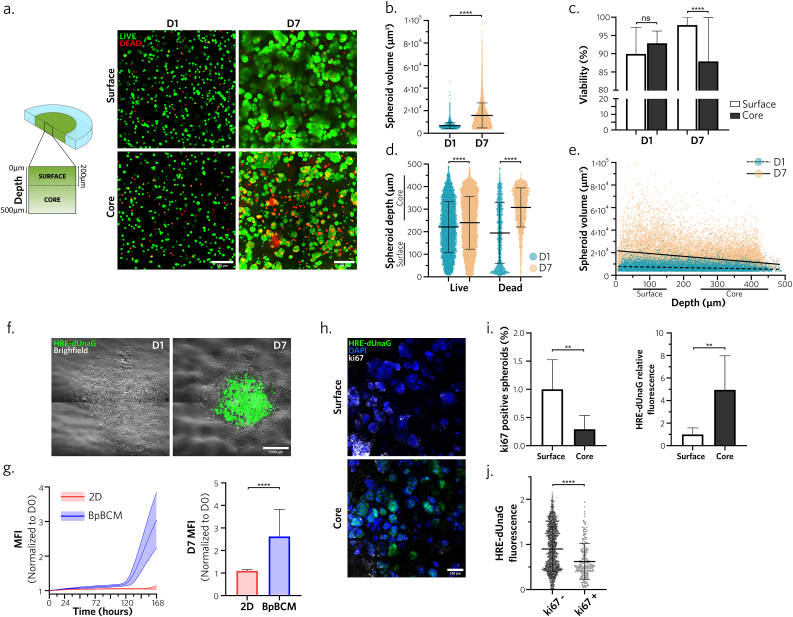
We then assessed the ability of the bioprinted matrix to sustain MCF-7 cells viability and maturation over time. On day 1 post-printing, we observed a homogenous distribution of live cells along the z-axis of the model. Conversely, on day 7, we noted an increased dispersion in terms of spheroid size at different regions of the model (Fig. 2a). As expected, we observed a significant increase in spheroid volume on day 7, compared to day 1 (Fig. 2b). To better discriminate the spatial distribution of both live and dead cells, we segmented the surface of the model, until 200 μm from the contact with media corresponding to the diffusion gradient limit described in solid tumors [42], and the core, deeper in the model (Fig. 2a). Although we could observe a viability superior to 85 %, we detected a significant decrease in viability in the core of the model compared to the surface on day 7, but not on day 1 (Fig. 2c). We also noted a marginal difference in the distribution of live spheroids, which proved insufficient to account for the contrast. On the contrary, the distribution of dead cells was more informative (Fig. 2d), with an accumulation of dead cells at the surface on day 1, attributed to the gel syneresis following photo-polymerization. Interestingly, with the maturation of the model, we observed the accumulation of dead cells at the core of the model (Fig. 2d). A negative correlation between spheroid volume and model depth also appeared on day 7 (Fig. 2e). Together, these results indicated the apparition of a necrotic-like core and a proliferative region at the external surface.
Fig. 2.
BpBCM cancer center develops a necrotic-like hypoxic core with a proliferative surface
a. BpBCM printed with MCF-7 cells in the cancer bioink and an empty ink for the periphery were incubated in Calcein-AM for live cells staining (green) and EthD-1 for dead cells staining (red) on day 1 and day 7. Shown are representative confocal scanning layer in the surface (<200 μm from medium contact) and core (>200 μm from medium contact) areas from 3D confocal acquisitions. Scale bars: 100 μm. b. Live spheroid volume quantifications on day 1 and 7 (N = 3, n = 18). c. Percent of viable cells in the model, in the surface and the core on day 1 and 7 (N = 3, n = 18). d. Live and dead cells depths in the model on day 1 and 7. e. Linear regression between live spheroid volume and depth on day 1(slope = −5.254) and day 7 (slope = −25.10). f. Timelapse imaging of BpBCM printed with MCF-7 transduced with HRE-dUnaG hypoxia reporter (green) merged with brightfield image on day 1 and day 7. Scale bar: 1000 μm. g. Quantification of HRE-dUnaG reporter mean fluorescence intensity over time (left) and at day 7 (right), in 2D MCF-7 HRE-dUnaG cells (red) and printed in the center of BpBCM (blue) (N = 3, n = 15–24). h. Immunofluorescence for ki67 (gray). BpBCM models printed with MCF-7 HRE-dUnaG reporter (green), nuclei stained with DAPI (blue). Scale bar: 100 μm. j. Spheroid positivity to ki67 (left) and HRE-dUnaG reporter fluorescence, and (j) unique spheroid HRE-dUnaG fluorescence in function of ki67 status (N = 3, n = 8). All data represent mean ± s.d., and each datapoint represents a single spheroid (b, d, e, j). P-values were determined with two-tailed Mann-Whitney test (b,g,i,j) or Kruskal-Wallis test followed by Dunn's post-hoc test (c, d). ns, not significant. **p < 0.01; ****p < 0.0001.
We hypothesized that the bioprinted model could support the creation of a hypoxic core, contrasting with the normoxic surface. To assess this question, we transduced MCF-7 cells with the HRE-dUnaG hypoxia reporter [43] and subsequently bioprinted models integrating these cells in the center. Timelapse analysis revealed the emergence of HRE positive cells after 5 days of culture, with incremental fluorescence levels up to day 7 (Fig. 2f and g, Supplementary Video 4). Notably, fluorescence levels were significantly increased in the BpBCM compared to control cells cultured in 2D under 21 % O2. To better understand the spatial distribution of these cells and the impact of hypoxia on cell proliferation, we conducted ki67 immunostaining on day 7 (Fig. 2h). Surface and core displayed opposite phenotype with a significantly higher ki67 index and low HRE-dUnaG fluorescence at the top, compared to high hypoxia reporter signal and low proliferation in the core (Fig. 2i). The opposition between proliferation status and hypoxic phenotype was further confirmed with the significant decrease of HRE-dUnaG signal in ki67 positive spheroids (Fig. 2j).
Supplementary data related to this article can be found online at https://doi.org/10.1016/j.bioactmat.2024.08.037
The following are the Supplementary data related to this article:
2.3. BpBCM stromal maturation is under the control of CAF
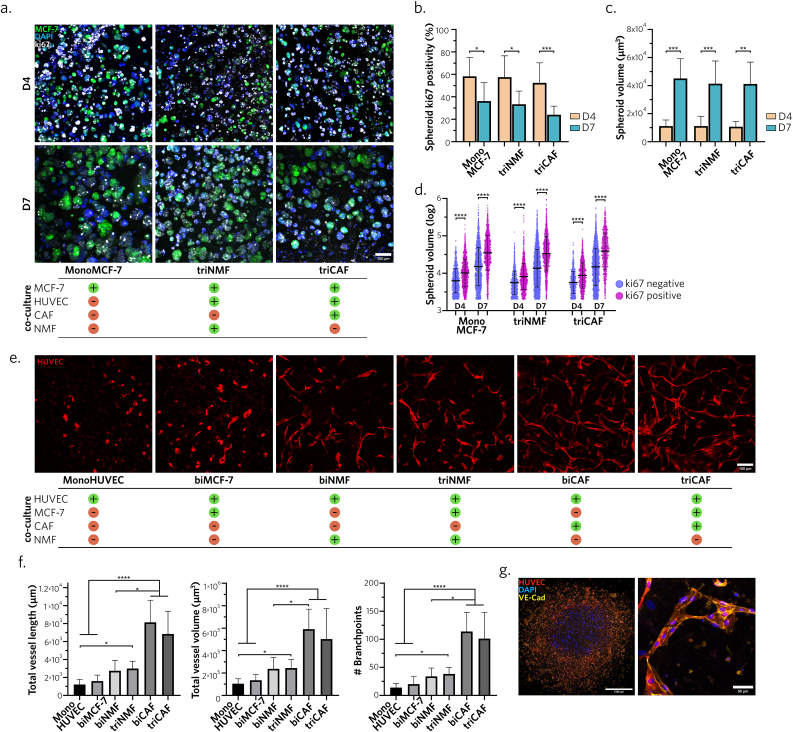
To gain further insights on the impact of stromal cell components on breast cancer cell maturation, we investigated several BpBCM co-culture conditions for MCF-7 growth and proliferation (Fig. 3a). We observed similar ki67 profiles between MCF-7 monoculture and their tri-culture with HUVEC and either CAF (triCAF condition) or normal mammary fibroblasts (NMF, triNMF condition) (Fig. 3b). As expected, with increased cell density over time, we identified a significant decrease in MCF-7 spheroid proliferation from day 4 to day 7 in all conditions (Fig. 3b). Conversely, spheroid volume increased on day 7 in all conditions compared to day 4 (Fig. 3c). Bi-culture conditions, designed to isolate the distinct effect of HUVEC and fibroblasts, yielded similar results (Supplementary Figs. 4a,b,c). As anticipated, ki67 positive spheroids were significantly larger than non-proliferative ones in each condition, on both day 4 and 7 (Fig. 3d). However, no difference appeared between the conditions. Together, these results confirmed that cancer cells proliferation was not hindered in the presence of stroma.
Fig. 3.
BpBCM stromal compartment favors overtime maturation of cancer cells and microvascular structures
a. Representative ki67 immunostaining (gray), and stained nuclei using DAPI (blue), of BpBCM models printed with MCF-7 GFP+ (green) in co-culture conditions. Scale bar: 100 μm. b,c. Spheroid volume (b) and ki67 spheroid positivity (c) on day 4 and 7 in the different culture conditions (N = 3, n = 10–13). d. Spheroid, either positive or negative for ki67, volume distribution (log scale). e. Representative 3D acquisition maximum intensity projection of the BpBCM stroma after 7 days of culture. BpBCM models were printed with HUVEC mKate+ (red) in the stromal periphery in different co-culture conditions. Scale bar: 100 μm. f. 3D network analysis and quantification of total vessel length, volume and number of branchpoints (N = 3, n = 10–14). g. Immunostaining of triCAF model for VE-Cadherin (yellow), HUVEC mKate+ (red), nuclei (blue), full model imaging (left), and close up on one representative vessel (right). Scale bars: 1000 μm (left), 50 μm (right). All data represent mean ± s.d. Each dot represents a single spheroid (d). P-values were calculated with Kruskal-Wallis test corrected by Dunn's multiple comparison test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Endothelial cells also play a central role in the tumor niche, not only for their vasculature function driving oxygen, nutrients, tumor and immune cells migration [44], but also for their paracrine communication with tumor cells regulating treatment response [9]. In this line, we interrogated the ability of endothelial cells to mature in the BpBCM depending on their stromal partners. We tested bi and tri-culture conditions of HUVECs with or without MCF-7 and CAF or NMF. Interestingly, while HUVEC monoculture or bi-culture with MCF-7 led to poor network maturation, the introduction of fibroblasts in the model increased 3D network complexity (Fig. 3e, Supplementary Fig. 4d). Notably, bi- and triCAF conditions showed a significantly increased total vessel length, volume and number of branch points compared to conditions in the absence of fibroblasts or with NMF, with a nearly 3-fold increase (Fig. 3f). However, the presence of MCF-7 in the cancer center of the BpBCM did not induce significant differences between bi-culture and their corresponding tri-culture conditions. The maturation of microvascular-like networks was also indorsed by the VE-Cadherin expression along cell-cell junctions (Fig. 3g).
Together, these results demonstrate a pathologically relevant maturation of the whole model, for both cancer cells and endothelial cells, with a CAF-mediated modulation of microvascular structures.
2.4. CAF actively remodel ECM composition and modulate its mechanical properties
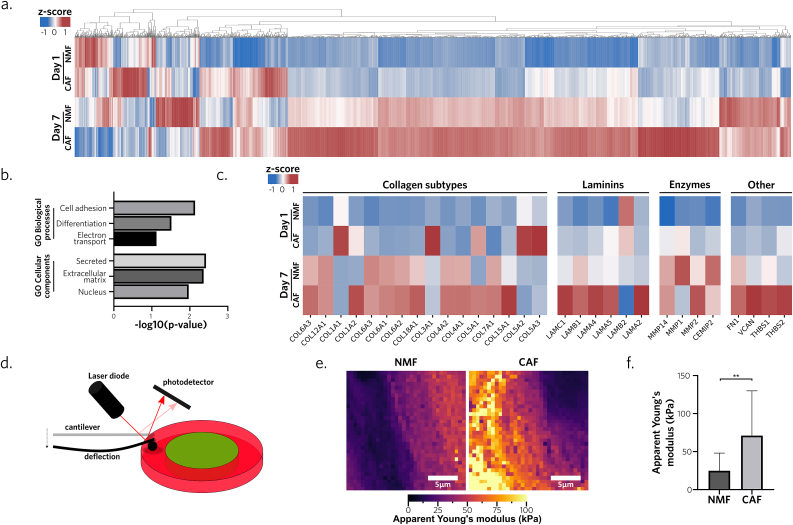
We hypothesized that the interplay with endothelial cells was arising from CAF-mediated secretion and ECM remodeling. The CAF population used in this study was identified to be CD29pos and PDGFRβpos but αSMAneg and FAPneg (Supplementary Fig. 5a), suggesting a subtype S345, showing a secretory rather than contractile phenotype. To assess its protein secretory profile, we conducted a proteome analysis, comparing triCAF and triNMF conditions, evaluated their respective compositions on days 1 and 7, and examined remodeling differences and kinetics. Among the 5226 detected proteins, and the 2408 that were quantified, we observed an increase in protein abundance in triCAF models from day 1 to day 7. It appeared that CAF might overexpress specific proteins compared to the healthy fibroblasts after maturation (Fig. 4a). To better understand this difference, proteins were associated with their corresponding GO biological processes and cellular components [46,47] to conduct an enrichment analysis. Interestingly, the most overexpressed GO terms in triCAF BpBCM compared to triNMF on day 7 were ‘cell adhesion’, ‘secreted’ and ‘extracellular matrix’ (Fig. 4b), suggesting a matrisome remodeling. Among ECM proteins, similar behaviors were observed between NMF and CAF for the secretion of most collagen subtypes (Fig. 4c). Of note, it appeared that CAF degraded the chains COL1A2, COL5A2, COL5A3 during maturation, while secreting higher COL1A2 and COL15A1 compared to NMF. Laminin subunits were shown to be overexpressed by triCAF BpBCM with the exception of LAMB2 (Fig. 4c). Similarly, digestion enzymes were increased at day 7 in both NMF and CAF conditions, with the notable secretion of MMP-2 and MMP-14 (Fig. 4c). Lastly, fibronectin (FN1) was secreted on day 7 by both cell types while other matricellular proteins, namely versican (VCAN), and thrombospondins (THBS-1/2) were specifically overexpressed in CAF condition (Fig. 4c).
Fig. 4.
CAF remodel ECM protein composition and mechanical properties
a. Proteomics heatmap of the relative abundance of 2408 quantified proteins in BpBCM in tri-culture with either CAF or NMF on day 1 and day 7. b. Enrichment analysis of overexpressed GO biological processes and cellular components in triCAF compared to triNMF condition on day 7. Processes are ordered in decreasing p-values. c. Heatmap of extracellular matrix specific proteins: collagen subtypes, laminin chains, restriction enzymes and other fibrillar and matricellular proteins (N = 3). d. Schematic representation of the atomic force microscopy force-spectroscopy protocol. Stromal area was probed using a colloidal probe after 7 days of maturation in triNMF or triCAF conditions. e. Stromal region scan by atomic force microscopy in force spectroscopy representing local apparent Young's modulus. Scale bar: 5 μm. f. Mean region apparent Young's modulus in NMF and CAF BpBCM stroma (N = 2). Shown as mean ± s.d. P values were calculated with Fisher's exact test (b), and Mann-Whitney two-tailed test (f).**P < 0.01.
Based on these changes in matrix composition, notably in fibrillar proteins, we formulated the hypothesis that BpBCM could model the stiffness dynamics observed during tumor progression. We probed the stroma using force spectroscopy atomic force microscopy (Fig. 4d) to measure the substrate stiffness by recording force-distance curves (Supplementary Fig. 5c). Despite similar topographies (Supplementary Fig. 5b), scans of the stromal area revealed higher apparent Young's modulus in the presence of CAF with mean measurements at approximately 71 kPa and 27 kPa, respectively (Fig. 4e, Supplementary Fig. 5d).
These results express CAF ability to drastically remodel the ECM, changing not only the composition but also the mechanical properties of the BpBCM.
2.5. CAF modulate BpBCM response to radiotherapy
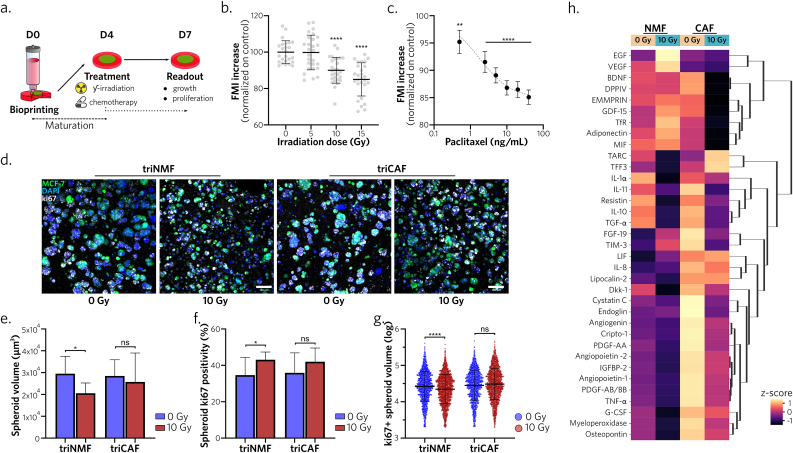
Due to these important changes in model dynamic maturation, we hypothesized that response to treatment could be affected. First, to confirm the model relevance in treatment response evaluation, we exposed triCAF models to increasing doses of radiotherapy and chemotherapy (Fig. 5a). We observed a significant decrease in mean fluorescence intensity, showing a reduction of GFP+ MCF-7 cells after 10Gy and 15Gy irradiations, compared to non-irradiated controls (Fig. 5b, Supplementary Fig. 6a). Response to chemotherapy was also characterized by a dose-effect relationship (Fig. 5c, Supplementary Fig. 6a). Together, these experiments confirm the ability of BpBCM to respond to radio/chemotherapy.
Fig. 5.
BpBCM response to radiotherapy is modulated in the presence of CAF
a. Schematic representation of the treatment protocol. Models matured for 4 days before treatment application and analysis 72h post-treatment. b, c. BpBCM in triCAF co-culture conditions. MCF-7 GFP + Fluorescence Mean Intensity (FMI) after irradiation (b, N = 3, n = 12–16) and paclitaxel (c, N = 3, n = 24–27) 72h post-treatment at specified doses, compared to non treated control. d. Representative immunostaining for ki67 (gray), MCF-7 GFP+ (green), nuclei (blue) of triNMF and triCAF models after 0 or 10 Gy irradiations. Scale bar: 100 μm. e,f. Quantifications of mean spheroid volume (e) and ki67 positivity (f). g. ki67 positive spheroids volume distribution (N = 3, n = 15–16). h. Clustered heatmap of detected cytokines in conditioned medium from day 4 to day 7 (N = 4). All data represent mean ± s.d., each datapoint represents a single spheroid (g). P-values were calculated with Kruskal-Wallis test followed by Dunn's post-hoc test. *P < 0.05; **P < 0.01; ****P < 0.0001; ns: not significant.
To further evaluate CAF impact on treatment response, we applied the same irradiation protocol for triCAF and triNMF conditions and then quantified spheroid volume and proliferative status by ki67 immunostaining (Fig. 5d, Supplementary Fig. 6b). We observed a significant decrease in spheroid volume in the triNMF condition, at 72h post-irradiation, compared to non-irradiated control, while triCAF models showed no significant difference (Fig. 5e). Concurrently, spheroid proliferation % significantly increased in the irradiated triNMF models, but not in the triCAF models (Fig. 5f), and we confirmed that a decreased ki67 positive spheroid volume was observed only for the triNMF condition (Fig. 5g). We attribute the volume reduction for triNMF condition to a treatment induced growth inhibition, while the increased proliferation is likely associated with the inverse relationship between spheroid volume and ki67 expression, as illustrated in Fig. 3b and c.
These findings collectively suggest a differential response to radiotherapy between triNMF and triCAF models, with a potential protective effect associated with the latter.
Lastly, we further evaluated this differential response by assessing the secretion of a 105-cytokine panel through a cytokine array, for triNMF or triCAF models in control or irradiated conditions (Supplementary Figs. 7a and e). We could detect 35 cytokines in all conditions and replicates, and clustered them based on their modulation between the different conditions (Fig. 5h). Overall, the exposure to 10Gy induced a decreased cytokine secretion, attributed to the effect of radiotherapy on metabolic activity. When comparing triNMF or triCAF secretome, in basal conditions without irradiation, we observed significant higher levels for endoglin, IGFBP-2, IL-8, TNFα, lipocalin-2, PDGF-AA and PDGF-AB/BB (Supplementary Fig. 7b). We also demonstrated that CXCL-1 (GROα), a predictor for poor prognosis [48] and linked to radioresistance [49], was overexpressed by CAF compared to NMF and secreted at higher doses in models implementing CAF (Supplementary Figs. 7c and d).
These results indicated the potential of the BpBCM to model treatment response and study paracrine intercommunication in the TME.
3. Discussion
Bioprinting potential for tumor microenvironment modeling is now well established [15,22,23,25,50], notably for breast cancer [[26], [27], [28],30,33,51,52]. In this line, our research indorses the relevance of microextrusion bioprinting in generating reproducible models in a medium-throughput fashion.
In this work, we used a ColMA-HAMA bioink, functionalized with a laminin-derived peptide, recapitulating breast cancer ECM. Other bioprinted breast cancer models preferred the use of decellularized ECM from animal or human breast tissues bioinks [28,52,53], but our findings support that CAF quickly induces major ECM remodeling, overriding the role of initial matrix. Our work describes a novel aspect of the maturation of bioprinted models through matrix recomposition. Notably, laminins remodeling appears relevant to human pathology, with the overexpression of LAMA4/5 and LAMB1/C1 which is consistent with the roles of laminins-411 [54] and −511 [55] in pro-tumor mechanisms. The undetected α1 subunit necessary to laminin-111 assembling, could mimick the downregulation observed in breast cancer tissues [56]. We also observed the crucial implication of CAF in the secretion of versican [57] and thrombospondins [58,59] matricellular proteins, identified during breast cancer progression. Conversely, our results point to a lower remodeling capacity from NMF, even when in co-culture with MCF-7 cells, indicating that the supposed cancer cell-induced phenotype shift, created by placing healthy fibroblasts in contact with cancer cells [60,61], was not sufficient to mimic CAF impact, reinforcing the need for patient derived CAF in 3D models. Our study is based on a subtype S3 CAF cell line, present in juxta-tumoral tissues [45], relevant to our model geometry and poorly described until now. Thus, and with a different methodology, our findings diverge with studies reporting αSMApos CAF cells, nonetheless cultured in 2D, that showed attenuated collagen secretion in medium [62]. Here, we show an increased collagen deposition by CAF, consistent with the fibrotic changes reported during breast cancer progression. Additionally, our results highlights the stiffness increase that follows this ECM deposition, with relative quantifications in the range of patient benign and cancer tissues stiffness measurements, for NMF and CAF, respectively [[63], [64], [65]]. Notably, to our knowledge, we provide the first report of a difference in matrix elastic modulus induced by CAF and NMF, in bioprinted models. This specificity might offer perspectives in the study of CAF-driven fibrosis mechanisms.
Our approach showed the development of a hypoxic and necrotic-like core, exhibited by other models of this size [26,28,66]. While this aspect can be overlooked or lacking in smaller models, it is highly important to tumor angiogenesis, progression [67] and treatment response [68]. In this work, we did not only assess the qualitative apparition of such a gradient, but quantified thoroughly its 3D organization, and relation to the proliferative surface. Our disk-shaped model allowed the apparition of a gradient mimicking tumor pathophysiology [42], while keeping essential readout capacities. In this line, our design accounts for the obstacle posed by stromal tissues, which can impede data collection from sphere-shaped models.
Along the maturation, we were able to keep high proliferation in stromal containing BpBCM, but we did not observe an increase in cancer cell proliferation in the presence of CAF, despite consequent evidence on this subject [[69], [70], [71]]. We hypothesize that the CAF subtype used in our study, different from the αSMApos phenotypes usually used, might not have the capacity to upregulate cancer cell proliferation. These results open new avenues for the study of subtype-specific CAF mechanisms.
Despite the lack of donor pairing between healthy and cancer-associated cells, this CAF line shows pro-angiogenic capacities in accordance with previous studies comparing paired cells [72,73]. ECM composition in fibronectin [74], type XV collagen [75], MMP-2 [74,76] and mechanical properties modulation [77] might partly explain this phenomenon, along with the basal increase in pro-angiogenic cytokines we demonstrated, namely PDGF-AA, -AB/BB [78] and lipocalin-2 [79]. Our findings also indicate that the direct influence of cancer cells might be marginal compared to the indirect effect mediated by CAF.
We lastly found that our model responds to chemotherapy and radiotherapy. CAF-induced radioresistance and paracrine communication in our BpBCM is in accordance with the growing consensus in the field [80,81], suggesting CAF-S3 implication in these mechanisms. We identified the secretion by CAF of cytokines correlated with tumor progression, notably endoglin [82], related to endothelial activation [83], IGFBP2 [84] and IL-8 [85] that can both promote proliferation in cancer and endothelial cells. The observed differential cytokine secretion profiles in our triNMF and triCAF breast cancer models underscores the critical importance of replicating the breast cancer niche in vitro. In future work, our model could incorporate fully patient derived primary cells to assess patient-specific treatment response, as done for glioblastoma [66] and pancreatic cancer [26] notably, as well as other cell types from the TME such as macrophages, lymphocytes and adipocytes.
Overall, our findings contribute to the better understanding of CAF implication in breast cancer physiopathology. This bioprinted model, capturing the holistic complexity of breast cancer tissues, may have a substantial interest for the screening of new drugs targeting the tumor microenvironment to improve standard patient care.
4. Methods
4.1. Cell culture
The human hormone-responsive breast cancer cell line MCF-7 was purchased from ATCC (HTB-22) and grown in Dulbecco's modified Eagle's High Glucose medium (DMEM HG, Gibco) supplemented with 10 % (v/v) Foetal Bovine Serum (Dutscher, S1900-500C). Primary human CAF (HC-6071) and NMF (H-6071) were purchased from Cell Biologics and amplified in a 1:1 mix of supplemented DMEM HG and EGM-2 MV (Lonza). Primary HUVECs were isolated based on previous protocols [86], and grown in EGM-2 MV. Cells were maintained at 37 °C in a humidified incubator with 5 % CO2. Primary cells were used until passage 10. All cells were tested negative for mycoplasma infection following regular PCR testing.
All transductions were based on lentiviral infection. HUVECs and MCF-7 were transduced for mKate and GFP, respectively, under the control of the EF1a promoter (Flash Therapeutics). MCF-7 were also transduced with a dUnaG fluorochrome under the control of HRE promoter, as previously described [43] (Vector Builder).
4.2. CAF cytometry phenotyping
The cancer-associated fibroblasts were assessed for the expression of CD29 (Biolegend, 303015, 5 μL/106 cells), α-smooth muscle actin (αSMA, R&D Systems, IC1420T, 5μL/106 cells), and Fibroblast activation protein (FAP, R&D Systems, FAB3715G, 0.25μL/106 cells) by flow cytometry. Cells cultured in flasks were detached by incubation in 0.05 % trypsin-EDTA, and 2.106 cells were resuspended in PBS before fixation in 4 % paraformaldehyde (PFA) (v/v) for 15 min. After rinsing in PBS, the cells were incubated for 30 min in PBS supplemented with 3 % (w/v) bovine serum albumin (BSA). Cells were then rinsed in PBS and resuspended at 1.106 cells/mL before being analyzed by flow cytometry on the AccuriTM C6 Flow cytometer (BD Biosciences). Data were analyzed on floreada.io (https://floreada.io). Doublets and cellular debris were filtered out by a first gate based on FSC-H and SSC-H, before plotting fluorescence intensity histograms.
4.3. Bioink formulation
Methacrylated type 1 bovine collagen (ColMA) and methacrylated hyaluronic acid (HAMA) were obtained as previously described [37]. A 6 mg/mL ColMA solution, diluted in 0.02 M acetic acid, and a 30 mg/mL of HAMA, diluted in DMEM HG, were combined to obtain final concentrations of 2 mg/mL and 10 mg/mL, respectively. pH and osmolarity were neutralized using NaOH 0.1 M and 10X PBS (Gibco). When indicated, the laminin-derived peptide Cys-(Beta-Ala)-IleLys-Val-Ala-Val-(Beta-Ala)-Cys (IKVAV, Biomatik) was combined with the previous formulation, diluted to a final concentration of 1 mg/mL from a stock solution at 25 mg/mL in 0.02 M acetic acid. The photonitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP, TCI Chemicals) was added to a final concentration of 1 mg/mL. The ink was prepared extemporaneously before printing and kept at 4 °C protected from light.
Cancer bioink was obtained by adding MCF-7 at a concentration of 5.106 cells/mL to the biomaterial ink, and stromal bioink by the addition of fibroblasts and/or HUVECs at a concentration of 5.106 cells/mL each. HUVECs concentration was increased to 20.106 cells/mL for microvascular maturation experiments.
4.4. Bioprinting
This work was performed on the commercial 3D Discovery bioprinting platform (RegenHU). A design comprising a 1.2 mm circle surrounded by a 3.3 mm diameter circle was elaborated on the integrated computer assisted design software (BioCAD, RegenHU).
The printing procedure started with a pneumatic microextrusion printhead loaded with cancer bioink printing the first circle, followed by a 5 s UV photo-polymerization. Then, the second printhead loaded with stromal bioink followed the surrounding circle, with a final photo-polymerization of 30s, equivalent to a fluence of 2.37 kJ/m2. Microextrusion printheads were mounted with a 0.41 mm diameter conical nozzle. Velocity was set at 25 mm/s and pneumatic pressure at 2–6 kPa. Models were printed onto 12 mm coverslips in 24-well plates. For convenience, a third inkjet printhead was used occasionally to dispense media in wells.
4.5. Model maintenance and treatment
Models were maintained in EGM2-MV at 37 °C in humid conditions with 5 % CO2. Models were treated on day 4 post-printing. Radiotherapy was conducted by ƴ-irradiation (IONISOS) in a single dose at a rate of 3.07 Gy/min, with total doses of 5 Gy (1min37s), 10 Gy (3min15s), and 15 Gy (4min52s). Paclitaxel diluted in DMSO at 2.5–50 ng/mL was added into the medium and kept from day 4 to day 7.
4.6. Stainings
Live/Dead stainings (Invitrogen) based on calcein-AM (1 μL/mL) and EthD-1 (4 μL/mL) were incubated with models for 25 min and rinsed with PBS directly before imaging. For immunohistology, models were fixed in paraformaldehyde 4 %, permeabilized in 0.05 % (v/v) Triton X100 and blocked with 2.5 % (w/v) Bovine Serum Albumin (BSA, VWR Life Science). Models were incubated overnight in rabbit anti-ki67 antibody (1 μg/mL, ab15580, Abcam) in 1 % BSA and 0.1 % Triton X100, and rinsed before another overnight incubation with DAPI (1 μg/mL, Fisher Scientific) and secondary donkey anti-rabbit antibody coupled with Alexa Fluor 647 (2.5 μg/mL, ab150075, Abcam). The same protocol was used with a primary rabbit antibody directed toward VE-Cadherin (1:400, D87F2, Ozyme) for endothelial cells maturation study. Models were rinsed and kept at 4 °C in the dark in a Fructose-Glycerol solution, used to homogenize refraction indexes and improve imaging resolution as previously described [87]. 2D MCF-7 cells irradiated with UV were stained using a rabbit antibody directed to human γH2Ax, coupled with Alexa Fluor 647 (1:500, ab195189, Abcam) for 2h following the same fixation, permeabilization and blockage protocols.
4.7. Imaging
Models were imaged over time on the Lumascope LS720 timelapse microscope (Etaluma) in humid conditions at 37 °C with 5 % CO2. Confocal microscopy (SPE7, Leica Microsystems) was used to acquire single 500 μm thick stacks in the center of the model for Live/Dead experiment. For ki67 and microvascular acquisitions we performed 200 μm thick stacks in 3 distinct fields per model, in the central and stromal area, respectively. Tilescan images of the model were acquired using a Stellaris confocal microscope (Leica Microsystems).
4.8. Mechanical testing
Ink viscometry was tested with a Kinexus pro + rheometer (Malvern Instruments). Rotational shear-viscosity measurements were performed in flow mode with shear rate ranging from 0.01 to 500 s-1. Measurements were performed in triplicate and at 25 °C.
Ink rheology was tested with the same instrument on fully photo-polymerized hydrogels stabilized at 37 °C before and during measurements. Frequency sweeps at 0.1 % constant strain with an angular frequency of 0.1–100 rad/s were performed to determine storage and loss moduli (G’ and G”, respectively).
Atomic Force Microscopy scans were done on day 7 fixed models in stromal areas. AFM experiments were performed on a Bruker Resolve AFM mounted on a Zeiss Axio Observer inverted microscope. The hydrogels matrix was deposited in a 50 mm glass bottom dish (Willco Wells HBST-5040). For apparent Young Modulus measurements, custom-made colloidal probes were used. To prepare them, NPO10-C tipless AFM probes (Bruker) were put in contact with a bit of Norland NOA 63 UV curable glue, deposited only at the end of the cantilever. Next, the cantilever was approached to a silica microsphere (diameter = 7.75 μm) to stick a single sphere at the end of the cantilever. The probe was then put under UV illumination overnight to cure the glue. AFM measurements with these probes were performed in PeakForce Tapping mode with Quantitative Nanomechanical analysis (PFQNM). The probe spring constant was calibrated by performing a force-distance curve on a clean glass surface combined with thermal noise method. The sample was visualized with the optical microscope to differentiate between the two zones of the model. The probe was then positioned on the region of interest and the sample was probed at several locations. PeakForce setpoint was typically set to few nN with typical PeakForce amplitude of 500–1000 nm and with a peak force frequency of 0.25 kHz. The sample was scanned with 256 x 256 resolution and a scan rate of 0.1 Hz. All PeakForce curves of an image were saved and later analyzed using the software Nanoscope Analysis (Bruker). For the fit of the apparent Young Modulus, a Hertz spherical indenter model was used. The baseline was corrected, the tip radius was modified to match the dimension of the sphere and the fitting was done on the extended curve, only for indentation below 2 μm. The apparent Young Modulus values fitted were then extracted and plotted after exclusion of aberrant values and filtering of extreme values in the highest and lowest 2.5 % of the datasets. Over 5 different area were measured from 3 to 5 independent samples, with over 4000 force curves obtained for each different sample.
4.9. Protein extraction
Extraction protocol was adapted from Hill et al., 2015 [88]. Briefly, we pooled 120 models rinsed with PBS 1X and mixed in High Saline solution buffer (50 mM Tris-HCL, 0.25 % CHAPS Sigma-Aldrich, 3 M NaCl, protease inhibitor at 10 μL/mL Sigma-Aldrich, pH 7.4), repeated 3 times to solubilize the cellular fraction proteins. The extracellular fraction that we used in further analysis was obtained after another solubilization in Urea Buffer (8 M urea, 100 mM Amonium Bicarbonate Sigma-Aldrich, 25 mM Tris(2-carboxyethyl)phosphine hydrochloride Sigma-Aldrich, pH 8.0) followed by an overnight digestion in 100 mM CNBr (Sigma-Aldrich) solubilized in 86 % trifluoroacetic acid. After rinsing in H2O and lyophilization, a last solubilization in urea buffer recovered extracellular matrix proteins, to complete the extracellular fraction. Samples were kept at −80 °C until quantifications.
4.10. LC MS/MS proteomics
Three independent extracellular fractions from triNMF and triCAF conditions extracted on day 1 and day 7 were analyzed by mass spectrometry to compare the protein composition. Proteins were loaded on a 10 % acrylamide SDS-PAGE gel and visualized by Colloidal Blue staining. Migration was stopped when samples had just entered the resolving gel and the unresolved region of the gel was cut into only one segment. The steps of sample preparation and protein digestion by trypsin were performed as previously described [89]. NanoLC-MS/MS analyses were performed using an Ultimate 3000 RSLC Nano-UPHLC system (Thermo Scientific, USA) coupled to a nanospray Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer (Thermo Fisher Scientific, California, USA). Each peptide extracts were loaded on a 300 μm ID x 5 mm PepMap C18 precolumn (Thermo Scientific, USA) at a flow rate of 10 μL/min. After a 3 min desalting step, peptides were separated on a 50 cm EasySpray column (75 μm ID, 2 μm C18 beads, 100 Å pore size, ES903, Thermo Fisher Scientific) with a 4–40 % linear gradient of solvent B (0.1 % formic acid in 80 % ACN) in 57 min. The separation flow rate was set at 300 nL/min. The mass spectrometer operated in positive ion mode at a 1.9 kV needle voltage. Data were acquired using Xcalibur 4.4 software in a data-dependent mode. MS scans (m/z 375–1500) were recorded at a resolution of R = 120000 (@ m/z 200), a standard AGC target and an injection time in automatic mode, followed by a top speed duty cycle of up to 3 s for MS/MS acquisition. Precursor ions (2–7 charge states) were isolated in the quadrupole with a mass window of 1.6 Th and fragmented with HCD@28 % normalized collision energy. MS/MS data were acquired in the Orbitrap cell with a resolution of R = 30000 (@m/z 200), a standard AGC target and a maximum injection time in automatic mode. Selected precursors were excluded for 60 s.
4.11. Secretome quantifications
For each condition, samples were collected 72 h post-treatment and mediums from 4 models of the same condition were pooled to obtain the desired final volume. Proteome profiler Human XL Cytokine Array Kit (R&D Systems) was used to quantify cytokine secretion following the supplier's protocol. Briefly, membranes were incubated in a blocking buffer before an overnight incubation at 4 °C in conditioned mediums. Membranes were then washed and revealed. Signal was acquired on the ImageQuant LAS 4000 Mini (GE Healthcare) with incrementation from 1 to 10 min. The experiment was replicated 4 times with different membrane batches and normalization for further quantifications was based on background signal.
CXCL-1 quantification in conditioned media was performed using the ProQuantumTM Human GROα immunoassay Kit (Invitrogen, A42896), following the supplier's instructions. Briefly, medium conditioned from day 4 to day 7 of the model culture were mixed with antibody-conjugates and fluorescence levels were compared to a standard for quantifications.
4.12. Metabolic activity evaluation
Printed models were incubated in 500 μL of EGM2-MV with 10 % resazurin (v/v) for 3 h on day 0 and day 7. The fluorescence of the conditioned media was then measured (excitation: 530 nm; emission: 590 nm; Victor X3, PerkinElmer), before rinsing the models in PBS and renewing their medium.
4.13. Gene expression
The total RNA extraction was performed using the RNeasy® Plus Micro Kit (Qiagen) according to the manufacturer's protocol. The final concentration and purity of the RNA (OD260/280) were determined using a NanoPhotometer® P 330 (Implen GmbH, Munich, Germany). RNA was transcribed into cDNA using the Maxima Reverse Transcriptase Kit (Thermo ScientificTM, Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer's protocol and using 300 ng of total RNA. RT-qPCR experiments were carried out using a CFX ConnectTM real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) and analyzed with CFX ManagerTM software, version 3.0 (Bio-Rad Laboratories).
The expression of target genes was quantified using cycle threshold (Ct) values, and relative mRNA expression levels were calculated as follows: 2^(Ct reference gene - Ct target gene). The reference genes used were human ribosomal protein lateral stalk subunit P0 (Rplp0) and glyceraldehyde 3-phosphate dehydrogenase (Gapdh).
The primers used were the following: Gapdh: GGCATTGCTCTCAATGACAA/TGTGAGGGAGATGCTCAGTG; Rplp0: CACTGGCTGAAAAGGTCAAGG/GTGTGAGGGGCTTAGTCGAA; CXCL-1: AGGCAGGGGAATGTATGTGC/AAGCCCCTTTGTTCTAAGCCA.
4.14. Bioinformatics analysis
Image analysis was computed using custom Python 3.8 [90] algorithms available in supplementary materials. Briefly, image 3D segmentation was achieved with pyclesperanto library [91] for label counting and quantification. Microvascular network analysis was computed using the Qiber3D package [92]. FIJI [93] and Imaris 8.0 (Oxford Instruments) were used for visual representation. For image quantifications, 3 distinct acquisitions from different areas in the model were meaned to obtain one value per model when possible.
Protein identification and Label-Free Quantification (LFQ) were done in Proteome Discoverer 3.0. CHIMERYS node using prediction model inferys_2.1 fragmentation was used for protein identification in batch mode by searching against a Uniprot Homo sapiens database (82408 entries, release September 2023) and an UniProt Bos taurus database (37500 entries, release November 2023). Two missed enzyme cleavages were allowed for the trypsin. Peptide lengths of 7–30 amino acids, a maximum of 3 modifications, charges between 2 and 4 and 20 ppm for fragment mass tolerance were set. Oxidation (M) and carbamidomethyl (C) were respectively searched as dynamic and static modifications by the CHIMERYS software. Peptide validation was performed using Percolator algorithm [94] and only “high confidence” peptides were retained corresponding to a 1 % false discovery rate at peptide level. Minora feature detector node (LFQ) was used along with the feature mapper and precursor ions quantifier. The normalization parameters were selected as follows: (1) Unique peptides, (2) Precursor abundance based on intensity, (3) Normalization mode: total peptide amount from Homo sapiens, (4) Protein abundance calculation: summed abundances, (5) Protein ratio calculation: pairwise ratio based and (6) Missing values are replaced with random values sampled from the lower 5 % of detected values. Quantitative data were considered for master proteins, quantified by a minimum of 2 unique peptides, a fold changes ≥2 and a statistical p-value adjusted using Benjamini-Hochberg correction for the FDR lower than 0.05. Data were further analyzed and displayed using a custom Python algorithm adapted from Schnesser et al. [95]. Specifically, the enrichment analysis was done by matching identified proteins Entry names with GO Database's annotations [46,47] (Uniprot Homo sapiens database, November 2023). Fisher's exact test was conducted to compute the p-values for overexpression of biological processes and cellular components.
Cytokine array spot intensities quantification was done using a previously developed FIJI macro [96] and raw data were normalized and processed in Python. Other data types were also pre-processed and formatted in Python before statistical analysis.
4.15. Statistical analysis
Mean ± standard deviation is displayed for all data. Statistical comparisons were performed in GraphPad Prism 8.0. Kurskal-Wallis non-parametric one-way ANOVA test was used for multiple hypothesis testing, followed by a post-hoc Dunn's test. Mann-Whitney non-parametric t-test was used for single hypothesis testing. Differences are considered significant for p < 0.05.
Data availability
Data supporting the results are available within the article and its Supplementary Information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [97] partner repository with the dataset identifier PXD050584. The raw and analyzed datasets generated during the study are available from the corresponding authors on reasonable request.
Ethics approval and consent to participate.
Not applicable in this work.
CRediT authorship contribution statement
Theo Desigaux: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Leo Comperat: Visualization, Validation, Methodology, Investigation, Conceptualization. Nathalie Dusserre: Writing – review & editing, Validation, Resources, Methodology, Investigation. Marie-Laure Stachowicz: Validation, Resources, Methodology, Investigation. Malou Lea: Resources, Methodology, Investigation. Jean-William Dupuy: Software, Methodology, Investigation, Formal analysis, Data curation. Anthony Vial: Methodology, Investigation, Formal analysis, Data curation. Michael Molinari: Validation, Methodology. Jean-Christophe Fricain: Validation, Supervision, Resources, Project administration, Methodology, Funding acquisition. François Paris: Writing – review & editing, Validation, Supervision, Resources, Project administration, Methodology, Funding acquisition, Data curation, Conceptualization. Hugo Oliveira: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
All authors declare that there is no conflict of interest.
Acknowledgements
Authors thank the financial support of INSERM (France) and "Fondation ARC pour la recherche sur le cancer". We thank Dr. Erapaneedi for supplying HRE-dUnaG reporter plasmid sequence. Authors lastly thank the ART bioprint team for the useful discussion that supported this work.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2024.08.037.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sung H., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Luque-Bolivar A., Pérez-Mora E., Villegas V.E., Rondón-Lagos M. Resistance and overcoming resistance in breast cancer. Breast Cancer. 2020;12:211–229. doi: 10.2147/BCTT.S270799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Place A.E., Jin Huh S., Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res. 2011;13:227. doi: 10.1186/bcr2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández-Nogueira P., et al. Cancer-associated fibroblasts in breast cancer treatment response and metastasis. Cancers. 2021;13:3146. doi: 10.3390/cancers13133146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De P., Aske J., Dey N. Cancer-associated fibroblast functions as a road-block in cancer therapy. Cancers. 2021;13:5246. doi: 10.3390/cancers13205246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z., Tang Y., Tan Y., Wei Q., Yu W. Cancer-associated fibroblasts in radiotherapy: challenges and new opportunities. Cell Commun. Signal. 2019;17:47. doi: 10.1186/s12964-019-0362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang H.J., et al. Endothelial cells under therapy-induced senescence secrete CXCL11, which increases aggressiveness of breast cancer cells. Cancer Lett. 2020;490:100–110. doi: 10.1016/j.canlet.2020.06.019. [DOI] [PubMed] [Google Scholar]
- 9.Ketteler J., et al. Caveolin-1 regulates the ASMase/ceramide-mediated radiation response of endothelial cells in the context of tumor–stroma interactions. Cell Death Dis. 2020;11:228. doi: 10.1038/s41419-020-2418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang P., et al. Monotherapy and combination therapy using anti-angiogenic nanoagents to fight cancer. Adv. Mater. 2021;33 doi: 10.1002/adma.202005155. [DOI] [PubMed] [Google Scholar]
- 11.De Vlieghere E., Verset L., Demetter P., Bracke M., De Wever O. Cancer-associated fibroblasts as target and tool in cancer therapeutics and diagnostics. Virchows Arch. 2015;467:367–382. doi: 10.1007/s00428-015-1818-4. [DOI] [PubMed] [Google Scholar]
- 12.Kümler I., Christiansen O.G., Nielsen D.L. A systematic review of bevacizumab efficacy in breast cancer. Cancer Treat Rev. 2014;40:960–973. doi: 10.1016/j.ctrv.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Sun D., Gao W., Hu H., Zhou S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B. 2022;12:3049–3062. doi: 10.1016/j.apsb.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamura Y., et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015;33:1837–1843. doi: 10.3892/or.2015.3767. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y.S., et al. Bioprinting the cancer microenvironment. ACS Biomater. Sci. Eng. 2016;2:1710–1721. doi: 10.1021/acsbiomaterials.6b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong E.L.S., Harrington D.A., Farach-Carson M.C., Yu H. Heralding a new paradigm in 3D tumor modeling. Biomaterials. 2016;108:197–213. doi: 10.1016/j.biomaterials.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groll J., et al. Biofabrication: reappraising the definition of an evolving field. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/1/013001. [DOI] [PubMed] [Google Scholar]
- 18.Moroni L., et al. Biofabrication strategies for 3D in vitro models and regenerative medicine. Nat. Rev. Mater. 2018;3:21–37. doi: 10.1038/s41578-018-0006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang G., Chen Y.-C., Lu H., Jin D. Advances in spheroids and organoids on a chip. Adv. Funct. Mater. 2023;33 [Google Scholar]
- 20.Law A.M.K., et al. Advancements in 3D cell culture systems for personalizing anti-cancer therapies. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.782766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sztankovics D., et al. 3D bioprinting and the revolution in experimental cancer model systems—a review of developing new models and experiences with in vitro 3D bioprinted breast cancer tissue-mimetic structures. Pathol. Oncol. Res. 2023;29 doi: 10.3389/pore.2023.1610996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belgodere J.A., et al. Engineering breast cancer microenvironments and 3D bioprinting. Front. Bioeng. Biotechnol. 2018;6:66. doi: 10.3389/fbioe.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shukla P., Yeleswarapu S., Heinrich M., Prakash J., Pati F. Mimicking tumor microenvironment by 3D bioprinting: 3D cancer modeling. Biofabrication. 2022 doi: 10.1088/1758-5090/ac6d11. [DOI] [PubMed] [Google Scholar]
- 24.Pedroza-González S.C., Rodriguez-Salvador M., Pérez-Benítez B.E., Alvarez M.M., Santiago G.T. Bioinks for 3D bioprinting: a scientometric analysis of two decades of progress. Int J Bioprint. 2021;7:333. doi: 10.18063/ijb.v7i2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahcecioglu G., Basara G., Ellis B.W., Ren X., Zorlutuna P. Breast cancer models: engineering the tumor microenvironment. Acta Biomater. 2020;106:1–21. doi: 10.1016/j.actbio.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langer E.M., et al. Modeling tumor phenotypes in vitro with three-dimensional bioprinting. Cell Rep. 2019;26:608–623.e6. doi: 10.1016/j.celrep.2018.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang T., et al. Engineering bioprintable alginate/gelatin composite hydrogels with tunable mechanical and cell adhesive properties to modulate tumor spheroid growth kinetics. Biofabrication. 2019;12 doi: 10.1088/1758-5090/ab3a5c. [DOI] [PubMed] [Google Scholar]
- 28.González-Callejo P., et al. 3D bioprinted breast tumor-stroma models for pre-clinical drug testing. Mater Today Bio. 2023;23 doi: 10.1016/j.mtbio.2023.100826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang G., et al. Unidirectional intercellular communication on a microfluidic chip. Biosens. Bioelectron. 2021;175 doi: 10.1016/j.bios.2020.112833. [DOI] [PubMed] [Google Scholar]
- 30.Swaminathan S., Cranston A.N., Clyne A.M. A three-dimensional in vitro coculture model to quantify breast epithelial cell adhesion to endothelial cells. Tissue Eng. C Methods. 2019;25:609–618. doi: 10.1089/ten.tec.2019.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grolman J.M., Zhang D., Smith A.M., Moore J.S., Kilian K.A. Rapid 3D extrusion of synthetic tumor microenvironments. Adv. Mater. 2015;27:5512–5517. doi: 10.1002/adma.201501729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinson B.T., et al. Laser direct-write based fabrication of a spatially-defined, biomimetic construct as a potential model for breast cancer cell invasion into adipose tissue. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa6bad. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., et al. 3D bioprinting of breast cancer models for drug resistance study. ACS Biomater. Sci. Eng. 2018;4:4401–4411. doi: 10.1021/acsbiomaterials.8b01277. [DOI] [PubMed] [Google Scholar]
- 34.Chaji S., Al-Saleh J., Gomillion C.T. Bioprinted three-dimensional cell-laden hydrogels to evaluate adipocyte-breast cancer cell interactions. Gels. 2020;6 doi: 10.3390/gels6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kauppila S., Stenbäck F., Risteli J., Jukkola A., Risteli L. Aberrant type I and type III collagen gene expression in human breast cancer in vivo. J. Pathol. 1998;186:262–268. doi: 10.1002/(SICI)1096-9896(1998110)186:3<262::AID-PATH191>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Wu M., et al. A novel role of low molecular weight hyaluronan in breast cancer metastasis. Faseb. J. 2015;29:1290–1298. doi: 10.1096/fj.14-259978. [DOI] [PubMed] [Google Scholar]
- 37.Oliveira H., et al. Extracellular matrix (ECM)-derived bioinks designed to foster vasculogenesis and neurite outgrowth: characterization and bioprinting. Bioprinting. 2021;22 [Google Scholar]
- 38.Tashiro K., et al. A synthetic peptide containing the IKVAV sequence from the A chain of laminin mediates cell attachment, migration, and neurite outgrowth. J. Biol. Chem. 1989;264:16174–16182. [PubMed] [Google Scholar]
- 39.Kikkawa Y., et al. Laminin-111-derived peptides and cancer. Cell Adhes. Migrat. 2013;7:150–159. doi: 10.4161/cam.22827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welch H.G., Prorok P.C., O'Malley A.J., Kramer B.S. Breast-cancer tumor size, overdiagnosis, and mammography screening effectiveness. N. Engl. J. Med. 2016;375:1438–1447. doi: 10.1056/NEJMoa1600249. [DOI] [PubMed] [Google Scholar]
- 41.Seo E.S., et al. Decoding spatial organization maps and context-specific landscapes of breast cancer and its microenvironment via high-resolution spatial transcriptomic analysis. 2023. 2023.10.25.563904 Preprint at. [DOI]
- 42.Helmlinger G., Yuan F., Dellian M., Jain R.K. Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat. Med. 1997;3:177–182. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 43.Erapaneedi R., Belousov V.V., Schäfers M., Kiefer F. A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO J. 2016;35:102–113. doi: 10.15252/embj.201592775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobierajska K., Ciszewski W.M., Sacewicz-Hofman I., Niewiarowska J. In: Tumor Microenvironment: Non-hematopoietic Cells. Birbrair A., editor. Springer International Publishing; Cham: 2020. Endothelial cells in the tumor microenvironment; pp. 71–86. [DOI] [Google Scholar]
- 45.Costa A., et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479.e10. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 46.Ashburner M., et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The Gene Ontology Consortium et al. The gene ontology knowledgebase in 2023. Genetics. 2023;224 doi: 10.1093/genetics/iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou A., et al. Elevated CXCL1 expression in breast cancer stroma predicts poor prognosis and is inversely associated with expression of TGF-β signaling proteins. BMC Cancer. 2014;14:781. doi: 10.1186/1471-2407-14-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., et al. Cancer-associated fibroblasts promote irradiated cancer cell recovery through autophagy. EBioMedicine. 2017;17:45–56. doi: 10.1016/j.ebiom.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datta P., Ayan B., Ozbolat I.T. Bioprinting for vascular and vascularized tissue biofabrication. Acta Biomater. 2017;51:1–20. doi: 10.1016/j.actbio.2017.01.035. [DOI] [PubMed] [Google Scholar]
- 51.Berger Fridman I., et al. High-throughput microfluidic 3D biomimetic model enabling quantitative description of the human breast tumor microenvironment. Acta Biomater. 2021;132:473–488. doi: 10.1016/j.actbio.2021.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blanco-Fernandez B., et al. Bioprinting decellularized breast tissue for the development of three-dimensional breast cancer models. ACS Appl. Mater. Interfaces. 2022;14:29467–29482. doi: 10.1021/acsami.2c00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mollica P.A., et al. 3D bioprinted mammary organoids and tumoroids in human mammary derived ECM hydrogels. Acta Biomater. 2019;95:201–213. doi: 10.1016/j.actbio.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross J.B., Huh D., Noble L.B., Tavazoie S.F. Identification of molecular determinants of primary and metastatic tumour re-initiation in breast cancer. Nat. Cell Biol. 2015;17:651–664. doi: 10.1038/ncb3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pouliot N., Kusuma N. Laminin-511: a multi-functional adhesion protein regulating cell migration, tumor invasion and metastasis. Cell Adhes. Migrat. 2013;7:142–149. doi: 10.4161/cam.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gudjonsson T., et al. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J. Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kischel P., et al. Versican overexpression in human breast cancer lesions: known and new isoforms for stromal tumor targeting. Int. J. Cancer. 2010;126:640–650. doi: 10.1002/ijc.24812. [DOI] [PubMed] [Google Scholar]
- 58.Yee K.O., et al. The effect of thrombospondin-1 on breast cancer metastasis. Breast Cancer Res. Treat. 2009;114:85–96. doi: 10.1007/s10549-008-9992-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin-Manso G., et al. sFRP-1 binds via its netrin-related motif to the N-module of thrombospondin-1 and blocks thrombospondin-1 stimulation of MDA-MB-231 breast carcinoma cell adhesion and migration. Arch. Biochem. Biophys. 2011;509:147–156. doi: 10.1016/j.abb.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rønnov-Jessen L., Petersen O.W., Koteliansky V.E., Bissell M.J. The origin of the myofibroblasts in breast cancer. Recapitulation of tumor environment in culture unravels diversity and implicates converted fibroblasts and recruited smooth muscle cells. J. Clin. Invest. 1995;95:859–873. doi: 10.1172/JCI117736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mishra P.J., et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68:4331–4339. doi: 10.1158/0008-5472.CAN-08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu Z., et al. Cancer-associated fibroblasts from invasive breast cancer have an attenuated capacity to secrete collagens. Int. J. Oncol. 2014;45:1479–1488. doi: 10.3892/ijo.2014.2562. [DOI] [PubMed] [Google Scholar]
- 63.Wu H., et al. A preliminary comparative study of Young's modulus versus shear modulus in the diagnosis of breast cancer. Ultrasound Q. 2019;35:88. doi: 10.1097/RUQ.0000000000000434. [DOI] [PubMed] [Google Scholar]
- 64.Krouskop T.A., Wheeler T.M., Kallel F., Garra B.S., Hall T. Elastic moduli of breast and prostate tissues under compression. Ultrason. Imag. 1998;20:260–274. doi: 10.1177/016173469802000403. [DOI] [PubMed] [Google Scholar]
- 65.Samani A., Zubovits J., Plewes D. Elastic moduli of normal and pathological human breast tissues: an inversion-technique-based investigation of 169 samples. Phys. Med. Biol. 2007;52:1565. doi: 10.1088/0031-9155/52/6/002. [DOI] [PubMed] [Google Scholar]
- 66.Yi H.-G., et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019;3:509–519. doi: 10.1038/s41551-019-0363-x. [DOI] [PubMed] [Google Scholar]
- 67.Semenza G.L. The hypoxic tumor microenvironment: a driving force for breast cancer progression. Biochim. Biophys. Acta Mol. Cell Res. 2016;1863:382–391. doi: 10.1016/j.bbamcr.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ward C., et al. New strategies for targeting the hypoxic tumour microenvironment in breast cancer. Cancer Treat Rev. 2013;39:171–179. doi: 10.1016/j.ctrv.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 69.Giorello M.B., Borzone F.R., Labovsky V., Piccioni F.V., Chasseing N.A. Cancer-associated fibroblasts in the breast tumor microenvironment. J. Mammary Gland Biol. Neoplasia. 2021 doi: 10.1007/s10911-020-09475-y. [DOI] [PubMed] [Google Scholar]
- 70.Buchsbaum R.J., Oh S.Y. Breast cancer-associated fibroblasts: where we are and where we need to go. Cancers. 2016;8 doi: 10.3390/cancers8020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu D., et al. Cancer-associated fibroblasts in breast cancer: challenges and opportunities. Cancer Commun. 2022;42:401–434. doi: 10.1002/cac2.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koch M.K., et al. Stromal fibroblasts regulate microvascular-like network architecture in a bioengineered breast tumour angiogenesis model. Acta Biomater. 2020;114:256–269. doi: 10.1016/j.actbio.2020.07.036. [DOI] [PubMed] [Google Scholar]
- 73.Orimo A., et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 74.Mongiat M., Andreuzzi E., Tarticchio G., Paulitti A. Extracellular matrix, a hard player in angiogenesis. Int. J. Mol. Sci. 2016;17:1822. doi: 10.3390/ijms17111822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eklund L., et al. Lack of type XV collagen causes a skeletal myopathy and cardiovascular defects in mice. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1194–1199. doi: 10.1073/pnas.031444798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ren F., et al. Overexpression of mmp family members functions as prognostic biomarker for breast cancer patients: a systematic review and meta-analysis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0135544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sewell-Loftin M.K., et al. Cancer-associated fibroblasts support vascular growth through mechanical force. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-13006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pandey P., et al. New insights about the PDGF/PDGFR signaling pathway as a promising target to develop cancer therapeutic strategies. Biomed. Pharmacother. 2023;161 doi: 10.1016/j.biopha.2023.114491. [DOI] [PubMed] [Google Scholar]
- 79.Yang J., McNeish B., Butterfield C., Moses M.A. Lipocalin 2 is a novel regulator of angiogenesis in human breast cancer. Faseb. J. 2013;27:45–50. doi: 10.1096/fj.12-211730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steer A., Cordes N., Jendrossek V., Klein D. Impact of cancer-associated fibroblast on the radiation-response of solid xenograft tumors. Front. Mol. Biosci. 2019;6 doi: 10.3389/fmolb.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krisnawan V.E., Stanley J.A., Schwarz J.K., DeNardo D.G. Tumor microenvironment as a regulator of radiation therapy: new insights into stromal-mediated radioresistance. Cancers. 2020;12:2916. doi: 10.3390/cancers12102916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang J., Zhang L., Lin Q., Ren W., Xu G. Prognostic value of endoglin-assessed microvessel density in cancer patients: a systematic review and meta-analysis. Oncotarget. 2017;9:7660–7671. doi: 10.18632/oncotarget.23546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rakocevic J., et al. Endothelial cell markers from clinician's perspective. Exp. Mol. Pathol. 2017;102:303–313. doi: 10.1016/j.yexmp.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 84.Wei L.F., et al. IGFBP2 in cancer: pathological role and clinical significance. Oncol. Rep. 2021;45:427–438. doi: 10.3892/or.2020.7892. (Review) [DOI] [PubMed] [Google Scholar]
- 85.Waugh D.J.J., Wilson C. The interleukin-8 pathway in cancer. Clin. Cancer Res. 2008;14:6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 86.Bordenave L., et al. Endothelial cell compatibility testing of three different Pellethanes. J. Biomed. Mater. Res. 1993;27:1367–1381. doi: 10.1002/jbm.820271104. [DOI] [PubMed] [Google Scholar]
- 87.Dekkers J.F., et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019;14:1756–1771. doi: 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- 88.Hill R.C., Calle E.A., Dzieciatkowska M., Niklason L.E., Hansen K.C. Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering*[S] Mol. Cell. Proteomics. 2015;14:961–973. doi: 10.1074/mcp.M114.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Campion O., et al. LRP-1 matricellular receptor involvement in triple negative breast cancer tumor angiogenesis. Biomedicines. 2021;9:1430. doi: 10.3390/biomedicines9101430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van Rossum G., Drake F.L. CreateSpace; Scotts Valley, CA: 2009. Python 3 Reference Manual. [Google Scholar]
- 91.Haase R. Image processing filters for grids of cells analogous to filters processing grids of pixels. Front. Comput. Sci. 2021;3 [Google Scholar]
- 92.Jaeschke A., Eckert H., Bray L.J. Qiber3D—an open-source software package for the quantitative analysis of networks from 3D image stacks. GigaScience. 2022;11 doi: 10.1093/gigascience/giab091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schindelin J., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 95.Schessner J.P., Voytik E., Bludau I. A practical guide to interpreting and generating bottom-up proteomics data visualizations. Proteomics. 2022;22 doi: 10.1002/pmic.202100103. [DOI] [PubMed] [Google Scholar]
- 96.Klemm A.H. Semi-automated analysis of dot blots using ImageJ/Fiji. F1000Res. 2020;9:1385. [Google Scholar]
- 97.Deutsch E.W., et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2023;51:D1539–D1548. doi: 10.1093/nar/gkac1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the results are available within the article and its Supplementary Information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [97] partner repository with the dataset identifier PXD050584. The raw and analyzed datasets generated during the study are available from the corresponding authors on reasonable request.
Ethics approval and consent to participate.
Not applicable in this work.