Abstract
The covalent modification of proteins by the small ubiquitin-like protein SUMO has been implicated in the regulation of numerous biological processes, including nucleocytoplasmic transport, genomic stability, and gene transcription. Sumoylation occurs by a multienzyme process similar to ubiquitination and, in Saccharomyces cerevisiae, is reversed by desumoylating enzymes encoded by the Ulp1 and Smt4/Ulp2 genes. The physiological importance of desumoylation has been revealed by mutations in either gene, which lead to nonoverlapping defects in cell cycle transition and meiosis. Several mammalian Ulp homologues have been identified, but, to date, nothing is known of the phenotypic effects of their loss of function. Here, we describe a random retroviral insertional mutation of one homolog, mouse SENP1/SuPr-2. The mutation causes increased steady-state levels of the sumoylated forms of a number of proteins and results in placental abnormalities incompatible with embryonic development. Our findings provide the first insight into the critical importance of regulating sumoylation in mammals.
Posttranslational modification of proteins by ubiquitin is now well established as a key cellular regulatory mechanism (14, 48). The paradigm is the addition of polyubiquitin chains as a signal for proteasomal degradation, but additional roles in regulating protein function also have recently come to light (16). The function of the ubiquitin-like protein SUMO (small ubiquitin-like modifier; also called SMT3 in Saccharomyces cerevisiae) is less well understood, but sumoylation is emerging as an important control point for numerous biological processes, including nucleocytoplasmic trafficking, transcription, DNA repair and replication, and mitotic and meiotic chromosome behavior (10, 15, 30, 39, 41, 43, 47). A variety of proteins have been shown to be sumoylated, but the specific effects of SUMO modification vary depending on the target protein. For example, sumoylation of IκB indirectly affects its stability by antagonizing ubiquitination (8), whereas sumoylation of RanGAP1 and PML regulates their subcellular localization to the nuclear pore and nuclear bodies, respectively (31, 35, 45). In addition, mammals have four SUMOs: SUMO-1 (also known as GMP-1, PIC1, UBL1, Sentrin, SMT3C, or Smt3h3) and the highly related SUMO-2 (also known as Sentrin-3, SMT3B, or Smt3h2), SUMO-3 (also known as Sentrin-2, SMT3A, or Smt3h1), and the recently discovered SUMO-4 (7). These can modify distinct sets of target proteins and therefore probably serve distinct functions (40).
SUMO is added to lysine residues on target proteins by a three-step process involving E1, E2, and E3 enzymatic activities that show great similarity to the corresponding ubiquitin-specific pathway enzymes. Again like ubiquitination, sumoylation is a dynamic, reversible process with deconjugation influencing overall levels of sumoylated forms of target proteins. The first desumoylating enzyme identified, Ulp1 from yeast, was found to be bifunctional, acting both to deconjugate sumoylated proteins and to carry out proteolytic processing of immature SUMO (26). A second yeast protein, Smt4/Ulp2, and several vertebrate proteins that share homology to Ulp1 and show evidence for desumoylation activity have since been identified (reviewed in references 15, 32, and 43). These include Sentrin protease 1 (SENP1) (13), SUMO-specific protease 1 (SUSP1, also known as SENP6) (19), and Smt3-specific isopeptidase 1 (Smt3IP1, also called SENP3, SUSP3, or SuPr-3) (37). There are also three proteins encoded by splice variants of the same gene, namely: SENP2, also called Axam; Smt3IP2, also called Axam2; and SuPr-1 (6, 17, 36). In yeast, Ulp1 and Smt4/Ulp2 have nonoverlapping biological functions. Mutations in Ulp1 cause cell cycle defects and are lethal (26), whereas Smt4/Ulp2 mutants are viable but do not undergo normal meiosis, among other defects (28, 42). In addition, Ulp1 and Smt4/Ulp2 have distinct subcellular localizations, a characteristic which appears to be a key element in determining their different substrate specificities (27). The vertebrate desumoylating enzymes also have apparent differences in intracellular distribution, and therefore probably also have different substrates and distinct overall biological functions. However, no mutations that would provide insight into these roles have yet been described.
Here, we describe the characterization of a random retroviral insertional mutation in the gene for the mouse ortholog of SENP1, also known as SuPr-2 (6). The mutation causes a net increase in the level of conjugation with SUMO-1, but leaves SUMO-2 and SUMO-3 protein sumoylation levels unaffected. In addition, mutant cells show accumulation of unprocessed SUMO-1. These results confirm a role for SENP1/SuPr-2 in both deconjugation and maturation of SUMO-1. The physiological consequences of loss of SENP1/SuPr-2 function during mouse development are first apparent after midgestation in the placenta. These defects appear incompatible with normal placental function and embryonic viability. Our findings highlight the critical importance of regulating levels of sumoylation in the developing mammalian organism.
MATERIALS AND METHODS
Developmental analysis of SENP1/SuPr-2 mutants.
The origin and genotyping analysis of the proviral mutant mouse strain has been described previously (49). To examine developmental defects, heterozygotes were mated, with the day of plug detection counted as embryonic day 0.5 (e0.5). Fetus and placenta were isolated and fixed in 4% paraformaldehyde at 4°C overnight. The yolk sac was used for PCR genotyping. Fixed samples of defined genotype were then paraffin embedded and serially sectioned at a thickness of 5 μm. Alternate slides were either stained with hematoxylin and eosin (H&E), processed for immunohistological analysis using rabbit polyclonal anti-SUMO-1 antiserum (gift of M. Dasso) and mouse monoclonal anti-RanGAP1 antibody (Zymed), or used for in situ hybridization following the procedure of Moorman et al. (34).
Gene structure, expression, and enhancer analysis.
To extend the 5′ end of expressed sequence tag (EST) W64824, 5′ rapid amplification of cDNA ends (12) was used. Briefly, first-strand cDNA was synthesized from total e12.5 RNA isolated with TRIzol Reagent (Invitrogen), using Superscript II reverse transcriptase (Invitrogen) primed with oligonucleotides TGAGCCAAGGAAACTGTCTGAGG (lying in SENP1/SuPr-2 exon 5) and AAGCAGTGGTATCAACGCAGAGTACGCGGG. Nested PCR was then carried out using first-round primers AAGCAGTGGTATCAACGCAGAGT and CTGGTCAGAAAGCAGAAGCTGC (lying in exon 2) and second-round primers AAGCAGTGGTATCAACGCAGAGTA and AGCGTCCATCTTCACCCCATCAGC (also lying in exon 2). PCR details are available upon request. PCR products were cloned in pGEM-T Easy (Promega) and sequenced.
For reverse transcriptase (RT)-PCR analysis, first-strand cDNA was synthesized from total RNA isolated from e12.5 wild types and mutants, using random hexamers to prime and SuperScript III First-Strand Synthesis System (Invitrogen) according to the manufacturer's instructions. To amplify upstream exon A (UEA)-specific isoforms, primers CTAGCAGCCGGTTCCGGTTCG (located in UEA) and either GTCACCTGAGCCAAGGAAACTG (in exon 5) or CATCAATGTGACTCCGTGGTGTC (in exon 4) were used. To amplify upstream exon B (UEB) isoforms, primer CCTTACCTGGCTCATCGCCTCTTG (in UEB) was used with either CAAGGAAACTGTCTGAGGAAGGG (in exon 5) or the above exon 4-specific primer. To amplify hypoxanthine phosphoribosyltransferase (HPRT), primers GCTGGTGAAAAGGACCTCTC and CACAGGACTAGAACACCTGC were used. PCR conditions are available upon request.
Real-time PCR was carried out with an Optican2 DNA engine (MJ Research) using a SYBR green PCR kit (Applied Biosystems) with primers CTACAAGAAGCCCAGCCTATCGTC and GTCACCTGAGCCAAGGAAACTG, located in exons 3 and 5, respectively. Amplification with the same HPRT-specific primers served as a reference standard. PCR conditions are available upon request. Cycle threshold (CT) values were determined using Optical2 software with the fluorescence threshold set to 0.016.
To assess enhancer activity, fragments from the first intron were cloned into pGL3-Promoter (Promega), transfected into P19 cells using Lipofectamine, and assayed for luciferase activity as described previously (24).
Transgene phenotype rescue.
The complete coding region from W64284, linked to the ROSA26 promoter (22) and polyadenylation sequences from the PGK gene, was introduced into fertilized eggs of the FVB strain by standard methods. Transgene positive founders, identified by Southern blot analysis, were mated with proviral heterozygotes. F1 progeny were intercrossed, and F2 mice were PCR genotyped for the proviral integration as described previously (49) and scored for presence of the transgene by PCR using primers GATTCTGTGTCTTTACTAAAGC and TTGACCAAAGGTTCTTACGTCACC.
Western blotting and antibodies.
To prepare protein extracts, fetus and nonmaternal portions of the placenta were isolated separately, washed in cold phosphate-buffered saline (without Ca and Mg), quickly minced, and then transferred into either sodium dodecyl sulfate (SDS) sample buffer (250 mM Tris HCl [pH 6.8], 4% SDS, 10% glycerol, 2% beta-mercaptoethanol) or NP-40 lysis buffer (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), both supplemented with protease inhibitor cocktail, phenylmethylsulfonyl fluoride, and 10 mM iodoacetamide (prepared fresh at the time of use). Lysates were then passed successively through 19-, 22-, and 25-gauge needles and clarified by centrifugation (13,000 rpm for 30 min at 4°C). Samples in SDS buffer were boiled for 5 min prior to centrifugation. Protein concentrations were determined using a modified Bradford assay (Bio-Rad Laboratories). From 10 to 30 μg of protein extract was then fractionated on 8 to 16% gradient SDS-polyacrylamide gels (Invitrogen), transferred to polyvinylidene difluoride or nitrocellulose membranes using semidry electrophoretic transfer (Hoefer), and processed as described previously (24). The primary antibodies used were rabbit polyclonal anti-SUMO-1 (gift of M. Dasso), mouse monoclonal anti-SUMO-1 (anti-GMP-1; Zymed), rabbit polyclonal anti-SUMO-2 (gift of M. Dasso), rabbit polyclonal anti-SUMO-3 (anti-Sentrin-2; Zymed), mouse monoclonal anti-RanGAP1 (Zymed), and mouse monoclonal anti-green fluorescent protein (anti-GFP) (Clontech). Mouse monoclonal anti-β-actin (Sigma) was used to confirm equivalent protein loading. Detection of secondary antibody-horseradish peroxidase conjugates was done using Super Signal West Pico solution (Pierce). Blots were stripped using Restore Western Blot Stripping buffer (Pierce) according to the manufacturer's specifications.
Mouse embryo fibroblasts.
Mouse embryo fibroblasts (MEFs) were isolated from e12.5 mutant and wild-type fetuses by passing eviscerated torsos successively through 18- and 21-gauge needles. The cell suspension was then placed in a 25-cm2 flask containing Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. After 2 days, cells were transferred to 75-cm2 flasks, cultured to 90% confluency, and then subcultured at 5 × 105 cells per 75-cm2 flask every 3 to 4 days in the absence of antibiotics. MEFs were maintained in reduced oxygen (3%) to allow long-term culture as described previously (38). To prepare protein extracts, MEFs were lysed directly in flasks by adding either SDS sample buffer or NP-40 lysis buffer and proceeding as described above.
Retrovirus production and infection.
An NheI/SmaI fragment containing the GFP-SUMO-1 sequence (20) was inserted at the HindIII site of retroviral vector LNCX (33). High-titer stocks of helper-free retrovirus were generated by transient transfection of Phoenix Ampho packaging cells (21). The LNCX-GFP-SUMO-1 virus-containing supernatants were collected at 6- to 12-h intervals starting 48 h after transfection and were used to infect wild-type and mutant MEFs seeded at 1 × 106 per 10-cm dish in the presence of 8 μg/ml Polybrene. After 24 h, infected cultures were subjected to selection in the presence of 400 μg/ml G418 (Gemini).
RESULTS
Phenotypic analysis of a prenatal lethal insertion mutation reveals placental defects.
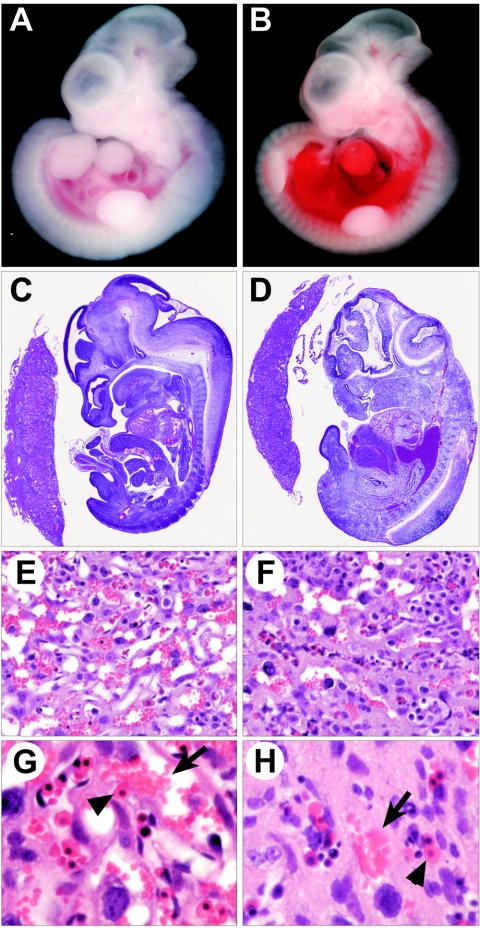
In a screening of mice derived from retrovirus-infected embryonic stem cells, we identified a provirus that, when homozygous, leads to death between e12.5 and e14.5. Gross examination of mutants revealed no conspicuous morphological or patterning defects (Fig. 1A to D), although some collected at e13.5 and e14.5 showed pooling of blood in the trunk region (Fig. 1B and D). Histology also showed no obvious abnormalities at any stage examined until e13.5, when the majority of mutants began showing widespread cell death. The lack of overt defects coupled with the timing of prenatal death, which occurs soon after dependence on placental circulation for gas and nutrient exchange begins, led us to examine the placenta for defects. Up to e12.5, we found no significant change in placental size, as determined by gross analysis (Fig. 1C and D), or in structure, as determined by in situ hybridization with placental layer-specific markers (data not shown). However, at e12.5 and later, we did find defects within the placental labyrinths of mutants. Compared to wild-type labyrinths, which show extensive maternal blood sinuses and intercalated fetal capillaries (Fig. 1E), mutant labyrinths had a significant reduction in both maternal and fetal blood spaces and appeared to contain excess trophoblast (Fig. 1F). Closer examination showed that compared to the wild type, in which fetal and maternal blood supplies are in close proximity (Fig. 1G), mutant fetal vessels were widely separated from maternal blood sinuses (Fig. 1H). These defects no doubt negatively affect the capacity of the mutant placenta to carry out gas and/or nutrient exchange, thus leading to death of the fetus. Importantly, we found these placental abnormalities even in e12.5 and e13.5 mutants that still appeared viable, although they were more pronounced in ones in which the fetus was undergoing widespread cell death.
FIG. 1.
Phenotype of the retroviral insertional mutant. (A) Wild-type e12.5 fetus and (B) mutant littermate showing accumulated blood in the trunk region. (C) H&E-stained, mid-sagittal section of an e12.5 wild-type fetus and placenta and (D) that of a mutant littermate, showing no significant change in placental size. (E) Close-up of a wild-type placental labyrinth and (F) that of a mutant littermate showing a less porous structure. (G) Higher magnification of a wild-type labyrinth and (H) that of a mutant littermate showing wide separation of fetal (arrowhead) and maternal (arrow) blood.
Proviral insertion is in the SENP1/SuPr-2 gene.
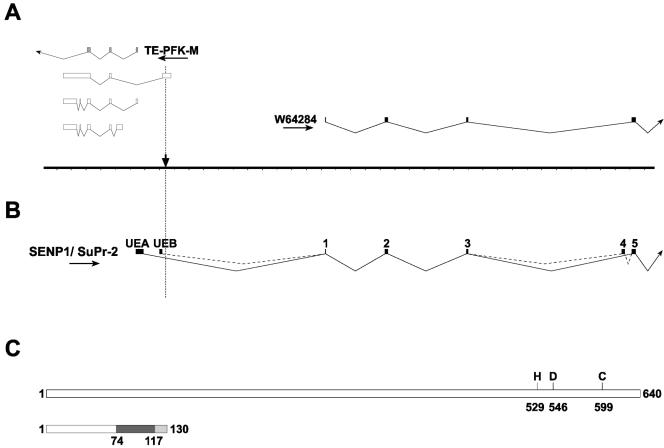
We recently reported the isolation and initial characterization of the genomic region surrounding the provirus (49). This locus is highly complex, with multiple transcripts initiating in both orientations. These include a novel isoform of muscle phosphofructokinase (PFK-M) expressed only in testes and embryos (TE-PFK-M) and three short overlapping polyadenylated transcripts with which it shares exons. In addition, the 5′ end of EST W64284, which is transcribed in the opposite orientation, was found on the other side of the proviral integration (Fig. 2A). BLAST searches with the complete sequence for W64284, which we determined, showed significant similarity to the human gene for the SUMO protease SENP1 (13). Recently, the sequences of three putative mouse SUMO proteases have been described, with SuPr-2 identified as the likely mouse ortholog of SENP1 (6). Importantly, the published protein sequence of SuPr-2 is identical to our deduced protein sequence of W64284.
FIG. 2.
Structure of the SENP1/SuPr-2 genomic region and transcriptional isoforms. (A) Transcripts in the area surrounding the proviral insertion (downward-pointing arrow) include the EST W64824 as well as the oppositely transcribed TE-PFK-M and three small overlapping mRNAs. Hatch marks on the line graph are 1 kb apart. (B) Full-length SENP1/SuPr-2 transcripts initiate at upstream exon A (UEA) or B (UEB) and show alternative splicing of exon 4. The ATG initiation codon is in exon 1 (original 5′ end of W64824). (C) Full-length SENP1/SuPr-2 protein is 640 amino acids. Key amino acid residues involved in the active site are marked. Translation of the alternatively spliced transcripts results in the addition of 43 amino acids encoded by exon 4 (from position 74 to 117, shown in dark gray). The shift in the reading frame in exon 5 (shown in light gray) results in a stop codon after amino acid 130.
Using 5′ rapid amplification of cDNA ends (12) on a midgestation cDNA library, we identified an upstream noncoding exon (UEA) lying on the opposite side of the proviral integration site (Fig. 2B). RT-PCR analysis of fetal cDNA and examination of several ESTs subsequently deposited in the database confirmed this structure and also identified a second alternatively used upstream noncoding exon (UEB). Thus, it is likely that the retroviral insertion is within the first intron of the mouse SENP1/SuPr-2 gene.
Proviral insertion disrupts SENP1/SuPr-2 expression.
As a first step in determining whether SENP1/SuPr-2 is affected in mutants, we examined mRNA expression during normal development. Using RT-PCR analysis with primers corresponding to sequences in UEA and in a downstream exon, we detected SENP1/SuPr-2 mRNA expression starting at about e9.5 (data not shown). This study also revealed a second, larger transcript present at only very low levels, which further analysis showed to be due to alternative inclusion of an intervening exon (Fig. 2B, exon 4). Transcripts initiating in UEB also show alternative splicing of this exon. Examination of the EST database revealed a small number of entries corresponding to the larger transcript, but the majority lacks this exon. Due to its inclusion, these larger transcripts have a frame shift in the following exon (exon 5), resulting in a stop codon, and thus encode truncated translational isoforms lacking the more carboxyl-terminal protease active site (Fig. 2C).
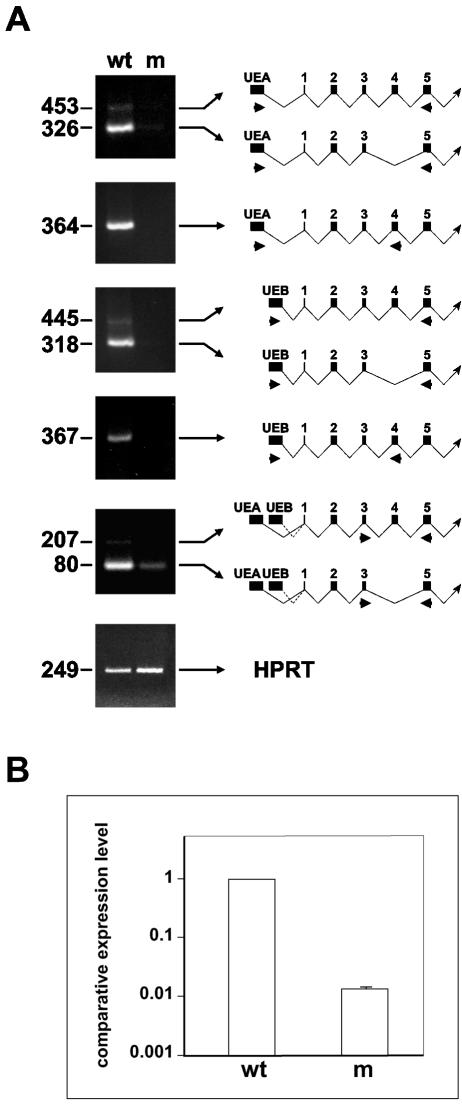
To examine mRNA expression in mutants, RT-PCR was carried out using the above primer sets to detect transcripts initiating in UEA or UEB, supplemented with additional primer sets specific for the minor transcripts. Analysis of e12.5 mutants and wild-type littermates revealed a significant reduction of all transcripts in mutants (Fig. 3A). To obtain a more quantitative estimate of the total level of SENP1/SuPr-2 expression in the mutant compared to the wild type, we carried out real-time RT-PCR using a primer pair that amplifies all of the transcripts. This analysis showed an over 50-fold decrease in transcription in the mutant (Fig. 3B).
FIG. 3.
Reduced expression of SENP1/SuPr-2 transcriptional isoforms in mutants. (A) RT-PCR analysis of e12.5 wild-type (wt) or mutant (m) littermates. The size of the RT-PCR products in base pairs is at the left. The schematics to the right of each panel indicate the corresponding cDNA structure and location of primers used for the reactions. The upper five panels show PCR of SENP1/SuPr-2, and the lower panel shows HPRT results as the control. (B) Real-time RT-PCR analysis of SENP1/SuPr-2. Relative expression levels were normalized to HPRT.
Proviral insertion affects a SENP1 enhancer.
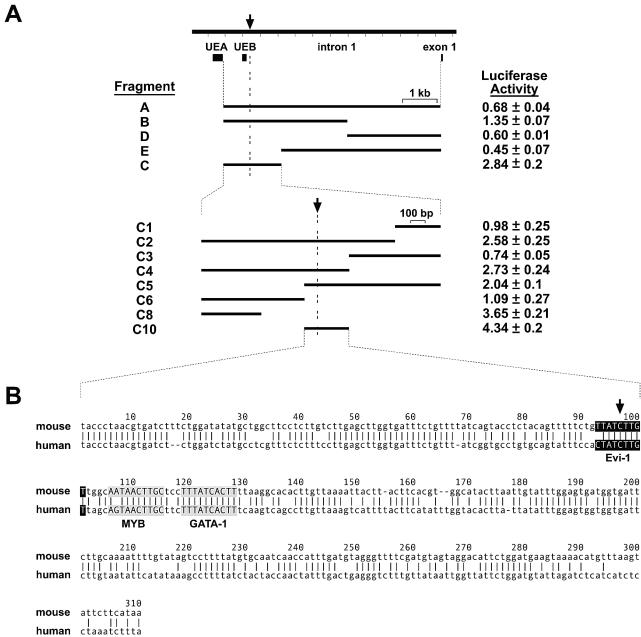
To gain insight into the mechanism by which proviral insertion affects SENP1/SuPr-2 expression, we asked whether a regulatory sequence is disrupted. This was a good possibility in that the integration is not within coding DNA, and other proviruses causing insertional mutations have been shown to disrupt intronic regulatory elements (5; M.R.K., unpublished results). Using a luciferase reporter-based assay, we analyzed for enhancer activity a series of fragments encompassing the entire first intron (Fig. 4A). These fragments were introduced into the pGL3 promoter vector and transfected into P19 embryonal carcinoma cells. Constructs containing a 1.5-kb fragment that surrounds the proviral preintegration site showed an almost threefold increase in reporter levels compared to the empty vector. Deletion analysis of this fragment identified two separate regions with enhancer activity in this assay, with the 310-bp region immediately surrounding the preintegration site showing a greater-than-fourfold increase in luciferase activity. This region has several binding sites for transcription factors, including one directly at the site of integration (Fig. 4B). Thus, the significant reduction in SENP1/SuPr-2 expression can be explained by the physical disruption of an intron enhancer element by the integrated provirus.
FIG. 4.
Identification of SENP1/SuPr-2 transcriptional regulatory domains. (A) The entire first intron and fragments A to E were analyzed for enhancer activity. Luciferase activity levels are relative to the activity of the empty pGL3-promoter vector (set to 1). Fragment C showed the highest activity and was further divided (C1 to C10). Fragment C10 (containing the proviral preintegration site; arrow) showed maximal activity. (B) The sequence of fragment C10 is shown aligned with the orthologous human region. The alignment was done using the AVID algorithm with output generated by the VISTA set (http://www.gsd.lbl.gov/vista/index.html). Conserved binding sites for the transcription factors Evi-1 (ectopic viral integration site 1-encoded factor; shown as white letters on a black background), c-Myb (MYB; black letters on gray background), and GATA-1 (black letters on gray background), which were determined using the Transfac Matrix and Factor databases created by Biobase (http://www.gene-regulation.com/pub/databases.html), are shown.
A SENP1/SuPr-2 transgene rescues the proviral mutation.
Given the complexity of the SENP1/SuPr-2 genomic region, the integrated provirus could affect the expression of multiple genes. To determine if the phenotype is due specifically to effects on SENP1/SuPr-2 expression, we carried out a transgene phenotype rescue experiment. Using a SENP1/SuPr-2 cDNA driven by regulatory elements from the ubiquitously expressed Rosa26 gene (22, 50), several transgenic strains that varied from approximately 10 to over 100 copies of the transgene were generated. Representative strains with low (10), moderate (10 to 50), and high (50 to 100) transgene copy number were bred to mice heterozygous for the provirus. The resulting F1 animals carrying both the transgene and provirus were then intercrossed to generate F2 animals. As shown in Table 1, F2 animals homozygous for the provirus and carrying the SENP1/SuPr-2 transgene did not undergo prenatal lethality and were found at the expected Mendelian ratio. Indeed, these animals have proven to be fully viable and fertile. Transgene rescue occurred regardless of transgene copy number, indicating that even at low copy number, the transgene is expressed above the required threshold. These results provide conclusive evidence that the mutant phenotype is due solely to transcriptional down regulation of SENP1/SuPr-2 and not to effects of proviral integration on the other transcripts originating in this region.
TABLE 1.
A SENP1/SuPr-2 transgene rescues homozygous mutantsa
| Genotype | Transgene copy no.
|
|||
|---|---|---|---|---|
| Low | Intermediate | High | Total (%) | |
| Proviral heterozygote | 24 | 10 | 22 | 56 (53) |
| Wild type | 12 | 5 | 9 | 26 (24) |
| Proviral homozygote | 11 | 5 | 8 | 24 (23) |
The numbers of offspring obtained from intercrosses between proviral heterozygotes in which one parent was transgene positive (with indicated SENP1/SuPr-2 transgene copy number) are shown.
Mutation of SENP1/SuPr-2 reduces SUMO-1 deconjugation.
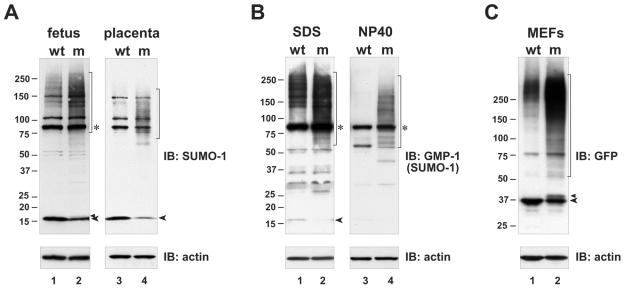
The dramatically reduced SENP1/SuPr-2 mRNA levels found in the mutant should result in a significant reduction in SENP1/SuPr-2 protein levels and in desumoylation activity. Although suitable antiserum against mouse SENP1/SuPr-2 is not yet available to test protein levels, we were able to examine the overall level of SUMO conjugation by Western blotting using anti-SUMO specific antisera. Protein extracts from individual fetuses and their associated placentas, for which the genotype had been determined by analysis of yolk sac material, were prepared first in buffer containing high SDS to eliminate nonspecific postlysis desumoylation by any of the other related SUMO proteases. Previous studies have shown that desumoylation activity continues following lysis in less stringent buffer conditions (8). As shown in Fig. 5A and B, analysis with two different anti-SUMO-1 antisera revealed a significant increase in high-molecular-weight SUMO-1 conjugates in the mutant fetus and placenta compared to the wild type and a corresponding decrease in free mature SUMO-1. These findings indicate a substantial reduction in desumoylation in the mutant, leading to higher steady-state levels of the sumoylated forms of target proteins and decreased replenishment of the pool of free SUMO-1. The latter result suggests that, normally, the majority of the free SUMO-1 pool comes from desumoylation rather than from de novo expression. Similar conclusions were reached based on results obtained from expressing a dominant negative form of SENP1 in established cell lines (4).
FIG. 5.
Increased conjugation and decreased maturation of SUMO-1 in SENP1/SuPr-2 mutants. In panels A and B, immunoblots (IB) of lysates from e12.5 wild-type (wt) and mutant (m) fetus and placenta were analyzed with either polyclonal anti-SUMO-1 antiserum (A) or monoclonal anti-SUMO-1 antiserum (anti-GMP-1; Zymed) (B). Only fetal samples are shown in panel B, with samples in lanes 1 and 2 lysed in SDS buffer and samples in lanes 3 and 4 lysed in NP-40 buffer. High-molecular-weight SUMO conjugates are marked by brackets. Asterisks mark a prominent 90-kDa band presumed to be SUMO-RanGAP1. Unconjugated fully processed SUMO-1 is marked by arrowheads. In panel A, the upper arrowhead marks what is presumed to be unprocessed SUMO-1. In panel B, a very long exposure is shown for the left panel to allow free SUMO-1 to be visualized. (C) Immunoblot of whole cell lysates from wild-type (wt) and mutant (m) MEFs infected with GFP-SUMO-1 virus analyzed with anti-GFP antiserum. High-molecular-weight GFP-SUMO-1 conjugates are marked by the bracket. The lower arrowhead marks unconjugated fully processed GFP-SUMO-1. The upper arrowhead marks unprocessed GFP-SUMO-1. Lower panels show reblotting with anti-β-actin (actin) to confirm equivalent sample loading.
Extracts also were prepared in standard NP-40 lysis buffer. As expected, the analysis of wild-type samples revealed a loss of high-molecular-weight conjugates (Fig. 5B, compare lane 1 to lane 3), confirming that lysis in this less stringent denaturing buffer fails to eliminate desumoylation activity and also indicating that a far greater number of SUMO conjugates are potential substrates than are actually desumoylated under physiological conditions. Surprisingly, SENP1/SuPr-2 mutant samples lysed in NP-40 buffer retain a significant level of high-molecular-weight SUMO conjugates (Fig. 5B, lane 4), although other SUMO proteases should be present and still active in these lysates, as in wild-type samples. No reduction in the level of these conjugates was seen, even after prolonged incubation at 37°C (data not shown). The inability of these proteins to be desumoylated by other SUMO proteases suggests they may be targeted uniquely by SENP1/SuPr-2. These results were somewhat surprising, as it has been proposed that SUMO proteases are generally active towards conjugated proteins, and specificity is a by-product of localization to the same subcellular compartments (27). Our results indicate that this nonspecific activity may be more limited.
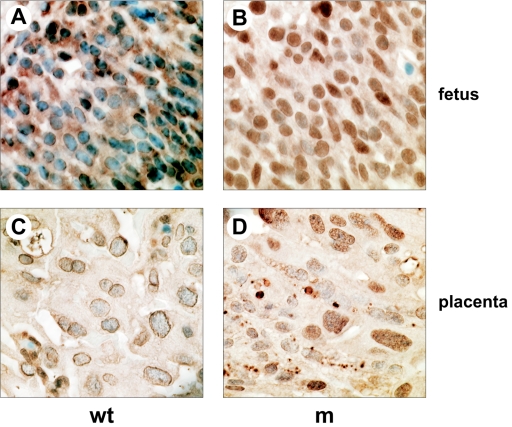
To provide additional evidence for reduced desumoylation, we carried out immunohistological staining of paraffin sections of e12.5 wild-type and mutant fetus and placenta using anti-SUMO-1 antiserum (Fig. 6). With wild-type fetus and placenta sections, we found strong staining around the nucleus as well as diffuse cytoplasmic staining (Fig. 6A and C). With mutant sections, the staining pattern showed a dramatic shift to strong intranuclear staining (Fig. 6B and D). The intensity of the anti-SUMO-1 staining was such that it occluded the nuclear counterstaining seen for the wild type. These results are consistent with either an increase in the level of sumoylation of nuclear proteins or accumulation of sumoylated proteins within the nucleus due to retargeting to, or retention within, the nuclear compartment.
FIG. 6.
Increased nuclear SUMO-1 staining in the SENP1/SuPr-2 mutant fetus and placenta. Immunohistochemistry on paraffin sections using anti-SUMO-1 polyclonal antiserum. (A) A representative field of an e12.5 wild-type (wt) fetal brain is shown. (B) A comparable region of the brain of a mutant (m) littermate is shown. The lower two panels show representative fields of wild-type (C) and mutant (D) placental labyrinth.
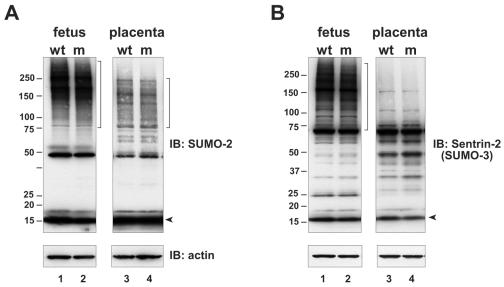
We also examined the level of SUMO-2 and SUMO-3 conjugation in the SENP1/SuPr-2 mutant, using two different antisera that are presumed to cross-react with these two highly related SUMO types (and perhaps also with the newly discovered SUMO-4). Although the specificities of these antisera were somewhat different, neither revealed any significant changes in the levels of sumoylated proteins or in free SUMO (Fig. 7A and B). Thus, SENP1/SuPr-2 does not appear to play a major role in SUMO-2 or SUMO-3 deconjugation or processing, at least during development.
FIG. 7.
No change in SUMO-2 or SUMO-3 conjugation levels in SENP1/SuPr-2 mutants. Immunoblots (IB) of lysates from e12.5 wild-type (wt) and mutant (m) fetus and placenta were analyzed with either (A) polyclonal anti-SUMO-2 antiserum or (B) polyclonal anti-SUMO-3 antiserum (anti-Sentrin-2; Zymed). The two antisera are presumed to cross-react with both SUMO-2 and SUMO-3. Arrowheads mark unconjugated, fully processed SUMO-2 or SUMO-3. Lower panels show reblotting with anti-β-actin (actin) to confirm equivalent sample loading.
SENP1/SuPr-2 mutants show decreased SUMO-1 processing.
In some experiments, we could detect in mutants an anti-SUMO-1 reactive band with a molecular weight slightly higher than that of free SUMO (Fig. 5A, lane 2). This was presumed to be the unprocessed form of SUMO-1, which has a 4-amino-acid carboxyl-terminal extension. It was not clear whether this form is detectable only in the mutant because of the overall decrease in levels of free SUMO or is unique to the mutant due to decreased processing. To explore further whether there is indeed a normal role for SENP1/SuPr-2 in the maturation of the SUMO-1 precursor, we derived primary MEFs from SENP1/SuPr-2 mutant embryos and wild-type littermates at day 12.5 of gestation and examined the processing of a GFP-tagged SUMO-1 (20) stably introduced by retroviral infection. This GFP-SUMO-1 construct encodes an elongated carboxyl-terminal tail of 12 amino acids, allowing the unprocessed and mature forms to be more readily distinguished. Western blots of lysates prepared from infected wild-type MEF cells showed a 37-kDa band representing the fully processed free GFP-SUMO-1 as well as higher-molecular-weight anti-GFP reactive species representing proteins conjugated with GFP-SUMO-1 (Fig. 5C, lane 1). Mutant MEFs showed a significant increase in high-molecular-weight GFP-SUMO-1 conjugates and a corresponding reduction in free mature GFP-SUMO-1 (Fig. 5C, lane 2), similar to our findings with endogenous SUMO-1 in the developing fetus and placenta (Fig. 5A). Strikingly, there was also a significant accumulation of an anti-GFP reactive species with slightly higher molecular weight than mature GFP-SUMO-1, consistent with it representing the unprocessed form (Fig. 5C, lane 2). Together, this data confirms the role of SENP1/SuPr-2 in the desumoylation of conjugated proteins and also provides firm evidence that SENP-1/SuPr-2 activity contributes to the maturation of SUMO-1.
RanGAP1 sumoylation in SENP1/SuPr-2 mutants.
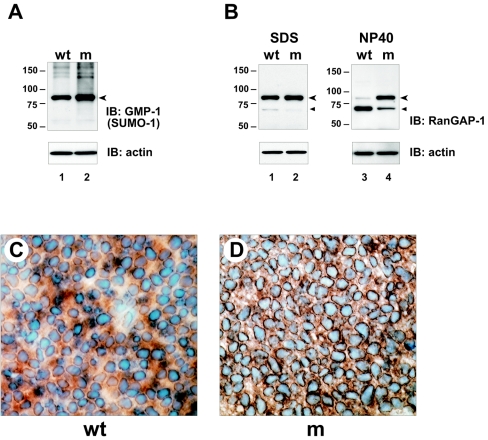
Studies in cell lines have shown that the major target for sumoylation in higher eukaryotes is the RanGTPase-activating protein RanGAP1, which is approximately 70 kDa when unconjugated and 90 kDa when sumoylated (29). By Western blot analysis of fetal and placental lysates, we found that one of the strongest, if not the strongest, anti-SUMO-1 reactive band was 90 kDa, consistent with it being SUMO-RanGAP1 (Fig. 5A and B; Fig. 8A). Analysis with anti-RanGAP1 antisera also detected a strong band at 90 kDa, which was presumed to be SUMO-conjugated RanGAP1, and a weak 70-kDa band, which was presumed to represent the unsumoylated form (Fig. 8B, lane 1). Thus, as in cell lines, RanGAP1 is the major target for sumoylation also in the developing fetus and placenta and is found predominantly in the conjugated form.
FIG. 8.
RanGAP1 sumoylation in SENP1/SuPr-2 mutants. (A) Immunoblots (IB) of lysates from e12.5 wild-type (wt) and mutant (m) fetus were analyzed with monoclonal anti-SUMO-1 antiserum (anti-GMP-1; Zymed). The arrowhead marks the prominent 90-kDa band presumed to be SUMO-RanGAP1. (B) Immunoblots of lysates from e12.5 wild-type (wt) and mutant (m) fetus were analyzed with monoclonal anti-RanGAP1 antiserum. RanGAP1 is approximately 70 kDa; SUMO-RanGAP1 is approximately 90 kDa. Samples in lanes 1 and 2 were lysed in SDS buffer, and samples in lanes 3 and 4 were lysed in NP-40 buffer. (A and B) Lower panels show reblotting with anti-β-actin (actin) to confirm equivalent sample loading. (C) Immunohistochemistry on paraffin sections using anti-RanGAP1 antiserum. A representative field of an e12.5 wild-type (wt) fetal brain is shown. (B) A comparable region of the brain of a mutant (m) littermate is shown.
An examination of SENP1/SuPr-2 mutants revealed a small increase, approximately 20% based on densitometric analysis, in steady-state levels of the sumoylated form of RanGAP1 in the fetus as detected either with anti-SUMO-1 (Fig. 8A) or anti-RanGAP1 antisera (Fig. 8B). Analysis with anti-RanGAP1 also revealed a compensatory reduction in unconjugated RanGAP1. However, similar changes in the placenta were not evident (data not shown). The increase in SUMO-RanGAP1 levels in the mutant fetus indicates that it is normally a target for SENP1/SuPr-2, but the rather modest change points to only a very low normal desumoylation rate. We also examined RanGAP1 sumoylation in samples lysed in NP-40 buffer. Wild-type samples showed an almost complete loss of the conjugated form, as expected due to continued desumoylation activity in this buffer (Fig. 8B, lane 3). However, mutant samples lysed in NP-40 buffer showed almost no loss of the sumoylated form, retaining the same high ratio of SUMO-RanGAP1 to unconjugated RanGAP1 as seen with SDS lysis buffer (Fig. 8B, lane 4). This limited desumoylation in NP-40 buffer by other SUMO proteases provides additional evidence that SUMO-RanGAP1 may be targeted predominantly by SENP1/SuPr-2, but under physiological conditions this desumoylation is tightly regulated.
We also carried out RanGAP1 immunostaining of sections of wild-type and mutant fetuses. Because anti-RanGAP1 antiserum detects both the unconjugated and sumoylated forms, this approach did not allow a direct determination of any increase in sumoylation. However, because SUMO-RanGAP1 localizes to the nuclear pore complex (29, 31), an increase in perinuclear staining provides indirect evidence for increased RanGAP1 sumoylation in mutants. With wild-type samples, we found strong perinuclear and cytoplasmic staining for RanGAP1, consistent with a significant fraction of the protein normally being sumoylated and associated with the nuclear pore complex (Fig. 8C). In most tissues of the mutant fetus, there was an apparent increase in staining around nuclei, consistent with an accumulation of the sumoylated form of RanGAP1 (Fig. 8D). However, no changes in the placenta were seen (data not shown). Together, these findings provide evidence for a role for SENP1/SuPr-2 desumoylating activity in the normal low-level turnover of SUMO-RanGAP1, but only in the fetal compartment during development.
DISCUSSION
Here, we describe a prenatal lethal insertional mutation in the mouse SENP1/SuPr-2 gene caused by exogenous proviral integration into the first intron, which is presumed to disrupt an enhancer element. While there is not a complete block in transcription, perhaps because another nearby enhancer element is undisrupted, SENP1/SuPr-2 mRNA expression levels are sufficiently reduced to cause SENP1/SuPr-2 activity to fall below the threshold required for normal function. This mutant provides the first example in mammals of the genetic disruption of a component of the SUMO pathway and reveals the critical importance of proper regulation of this protein conjugation system for normal mouse development.
The mutation results in the accumulation of the sumoylated forms of a number of proteins both in the fetus and placenta. Thus, a wide variety of cellular processes potentially could be affected, although the first phenotypic defects are seen in the placenta just after midgestation. At this stage, the fetus begins to exceed the size for simple diffusion and starts to depend on placental circulation for gas and nutrient exchange. In the developing placenta, fetal blood vessels grow up into the labyrinth layer and intercalate into sinuses that fill with maternal blood. Specialized trophoblast cells lining these spaces mediate the necessary interaction between the fetal and maternal blood supplies (1, 9). In the mutant, the placenta appears to develop normally up to approximately e11.5, but by e12.5, the normally porous structure of the labyrinth is absent. There is an increase of apparently undifferentiated trophoblast, which may contribute to there being fewer blood spaces and an overall decrease in fetal blood flow into the placenta. The conspicuous increase in blood in mutant fetuses may be a secondary consequence of the reduced circulation into the placenta, with blood normally allocated to the placental compartment instead being retained within the fetus. Reduced blood flow into the labyrinth, together with an observed increase in the physical separation of the fetal and maternal blood supplies, no doubt severely restricts placental function and compromises fetal survival. An important question is whether these defects are intrinsic to the placenta or arise due to defective instructive signaling from fetal derived cells within the labyrinth. Relevant to this, our preliminary attempts to rescue mutants with either a SENP1/SuPr-2 transgene under the control of the labyrinth-specific promoter of the CYP19 gene (18), or by aggregation of mutant with tetraploid embryos (25) have failed. Thus, it is quite possible that the primary defect is in the fetal compartment.
Our major focus now is to identify SENP1/SuPr-2 targets and ascertain those candidate substrates whose altered sumoylation levels might underlie the mutant phenotype. Our analysis of RanGAP1 has revealed an increase of SUMO-RanGAP1 levels in the fetus and a compensatory loss of unconjugated RanGAP1 as well as increased perinuclear accumulation, providing evidence that sumoylated RanGAP1 is normally an in vivo target. This result was unexpected given that in tissue culture cells, transfected human SENP1 was reported to lack any activity on SUMO-RanGAP1 (13). One explanation for this discrepancy may be the very tightly regulated and extremely low desumoylation rate for SUMO-RanGAP1 revealed in our study. The small amount of SUMO-RanGAP1 available for deconjugation would be handled by endogenous SENP1 activity, and no amount of overexpression of SENP1 would alter the overall levels of the modified form.
A key issue is whether increased steady-state sumoylation of RanGAP1 contributes to the mutant phenotype. The lack of any detectable changes in SUMO-RanGAP1 levels in the placenta might argue against such a role. However, if placental failure in SENP1/SuPr-2 mutants results from fetal defects, the increase in SUMO-RanGAP1 might be contributing in whole or part by causing some adverse effect on nucleocytoplasmic transport, perhaps specifically in the import of sumoylated proteins, as has been postulated previously (39). Thus, the significant increase in nuclear SUMO-1 staining observed with the mutant may be due to altered levels of SUMO-RanGAP1 at the nuclear pore as well as to the lack of SENP1/SuPr-2 desumoylating activity in the nucleus itself.
The nuclear accumulation of SUMO in the mutant is consistent with most known sumoylated proteins being either components of nuclear bodies, transcription factors or cofactors, or proteins involved in maintaining genome integrity (reviewed in reference 43). SUMO modification has been shown to negatively regulate the activity of most known sumoylated transcription factors (10, 47). Thus, overall higher levels of sumoylation might mimic loss of function mutations in a wide variety of transcription factors. In this regard, it is interesting that a targeted null mutation of ARNT, a subunit of the heteromeric transcription factor hypoxia-inducible factor (HIF) recently shown to be sumoylated and to have an altered subnuclear localization and reduced transcriptional activity when modified (46), results in abnormal placental labyrinth development (2, 23). The HIF-1α subunit of HIF also has been shown recently to be sumoylated, which appears to stabilize HIF-1α and thereby increase HIF transcriptional activity (3, 44). Interestingly, null mutation of pVHL, which normally destabilizes HIF-1α by targeting it for proteasomal degradation, results in increased levels of HIF-1α and leads to placental failure (11). Thus, abnormal sumoylation levels of components of the HIF pathway might also lead to placental defects.
Further evaluation of ARNT and HIF-1α as potential SENP1/SuPr-2 substrates and the identification of other candidates, which may include known and previously unknown SUMO target proteins, should be facilitated by exploiting the unique attributes of this mutant mouse strain. For most SUMO targets, the sumoylated forms exist at low levels and/or are sumoylated only transiently and are thus hard to detect. The significant increase in steady-state levels of sumoylated proteins in the mutant, and the use of relaxed stringency lysis conditions that allow continued SUMO protease activity and thus enrich for conjugated proteins specifically targeted by SENP1/SuPr-2, should now permit proteomic approaches to determine their identities and allow an assessment of their potential roles in the mutant phenotype. Regardless of the underlying mechanism, it is clear that perturbations in sumoylation due to deficiency of SENP1/SuPr-2 function illustrate the critical importance of maintaining SUMO homeostasis.
Acknowledgments
We thank Allison Galica for technical support, Lionel Feigenbaum and the NCI Transgenic Core for generation of transgenic mice, Philippe Soriano for ROSA26 constructs, Yongsok Kim for the original GFP-SUMO-1 construct, Mary Dasso for helpful discussions and anti-SUMO-1 and anti-SUMO-2 antisera, and Allan M. Weissman and Stan Lipkowitz for critical comments on the manuscript. Analysis of real-time PCR results utilized software developed by the Gene Expression Laboratory, SAIC, NCI—Frederick.
Animal care was provided in accordance with the procedures outlined in the “Guide for the care and use of laboratory animals” (NIH publication no. 86-23, 1985).
This study has been funded in part by federal funds from the National Cancer Institute under contract no. NO1-CO-12400.
The content of the manuscript does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
REFERENCES
- 1.Adamson, S. L., Y. Lu, K. J. Whiteley, D. Holmyard, M. Hemberger, C. Pfarrer, and J. C. Cross. 2002. Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev. Biol. 250:358-373. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, D. M., M. Gertsenstein, A. Nagy, M. C. Simon, and E. Maltepe. 2000. Placental cell fates are regulated in vivo by HIF-mediated hypoxia responses. Genes Dev. 14:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, S.-H., J.-W. Jeong, J. A. Park, S.-H. Kim, M.-K. Bae, S.-J. Choi, and K.-W. Kim. 2004. Sumoylation increases HIF-1α stability and its transcriptional activity. Biochem. Biophys. Res. Commun. 324:394-400. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, D., and P. O'Hare. 2004. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J. Biol. Chem. 279:692-703. [DOI] [PubMed] [Google Scholar]
- 5.Barker, D. D., H. Wu, S. Hartung, M. Breindl, and R. Jaenisch. 1991. Retrovirus-induced insertional mutagenesis: mechanism of collagen mutation in Mov13 mice. Mol. Cell. Biol. 11:5154-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. [DOI] [PubMed] [Google Scholar]
- 7.Bohren, K. M., V. Nadkarni, J. H. Song, K. H. Gabbay, and D. Owerbach. 2004. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 279:27233-27238. [DOI] [PubMed] [Google Scholar]
- 8.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 9.Georgiades, P., A. C. Ferguson-Smith, and G. J. Burton. 2002. Comparative developmental anatomy of the murine and human definitive placentae. Placenta 23:3-19. [DOI] [PubMed] [Google Scholar]
- 10.Gill, G. 2003. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 13:108-113. [DOI] [PubMed] [Google Scholar]
- 11.Gnarra, J. R., J. M. Ward, F. D. Porter, J. R. Wagner, D. E. Devor, A. Grinberg, M. R. Emmert-Buck, H. Westphal, R. D. Klausner, and W. M. Linehan. 1997. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc. Natl. Acad. Sci. USA 94:9102-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong, B., and R. Ge. 2000. Using the SMART cDNA system to map the transcription initiation site. BioTechniques 28:846-848, 850-852. [DOI] [PubMed] [Google Scholar]
- 13.Gong, L., S. Millas, G. G. Maul, and E. T. Yeh. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355-3359. [DOI] [PubMed] [Google Scholar]
- 14.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, E. S. 2002. Ubiquitin branches out. Nat. Cell Biol. 4:E295-E298. [DOI] [PubMed] [Google Scholar]
- 17.Kadoya, T., H. Yamamoto, T. Suzuki, A. Yukita, A. Fukui, T. Michiue, T. Asahara, K. Tanaka, M. Asashima, and A. Kikuchi. 2002. Desumoylation activity of Axam, a novel Axin-binding protein, is involved in downregulation of beta-catenin. Mol. Cell. Biol. 22:3803-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamat, A., and C. R. Mendelson. 2001. Identification of the regulatory regions of the human aromatase P450 (CYP19) gene involved in placenta-specific expression. J. Steroid Biochem. Mol. Biol. 79:173-180. [DOI] [PubMed] [Google Scholar]
- 19.Kim, K. I., S. H. Baek, Y. J. Jeon, S. Nishimori, T. Suzuki, S. Uchida, N. Shimbara, H. Saitoh, K. Tanaka, and C. H. Chung. 2000. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem. 275:14102-14106. [DOI] [PubMed] [Google Scholar]
- 20.Kim, Y. H., C. Y. Choi, and Y. Kim. 1999. Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc. Natl. Acad. Sci. USA 96:12350-12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 22.Kisseberth, W. C., N. T. Brettingen, J. K. Lohse, and E. P. Sandgren. 1999. Ubiquitous expression of marker transgenes in mice and rats. Dev. Biol. 214:128-138. [DOI] [PubMed] [Google Scholar]
- 23.Kozak, K. R., B. Abbott, and O. Hankinson. 1997. ARNT-deficient mice and placental differentiation. Dev. Biol. 191:297-305. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, A., V. Novoselov, A. J. Celeste, N. M. Wolfman, P. ten Dijke, and M. R. Kuehn. 2001. Nodal signaling uses activin and transforming growth factor-beta receptor-regulated Smads. J. Biol. Chem. 276:656-661. [DOI] [PubMed] [Google Scholar]
- 25.Kupriyanov, S., and H. Baribault. 1998. Genetic control of extraembryonic cell lineages studied with tetraploid↔diploid chimeric concepti. Biochem. Cell Biol. 76:1017-1027. [PubMed] [Google Scholar]
- 26.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 27.Li, S. J., and M. Hochstrasser. 2003. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160:1069-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, S. J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20:2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 30.Matunis, M. J. 2002. On the road to repair: PCNA encounters SUMO and ubiquitin modifications. Mol. Cell 10:441-442. [DOI] [PubMed] [Google Scholar]
- 31.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melchior, F., M. Schergaut, and A. Pichler. 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28:612-618. [DOI] [PubMed] [Google Scholar]
- 33.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7:980-982, 984-986, 989-990. [PMC free article] [PubMed] [Google Scholar]
- 34.Moorman, A. F., A. C. Houweling, P. A. de Boer, and V. M. Christoffels. 2001. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J. Histochem. Cytochem. 49:1-8. [DOI] [PubMed] [Google Scholar]
- 35.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishida, T., F. Kaneko, M. Kitagawa, and H. Yasuda. 2001. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat Axam, which is an Axin-binding protein promoting β-catenin degradation. J. Biol. Chem. 276:39060-39066. [DOI] [PubMed] [Google Scholar]
- 37.Nishida, T., H. Tanaka, and H. Yasuda. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423-6427. [DOI] [PubMed] [Google Scholar]
- 38.Parrinello, S., E. Samper, A. Krtolica, J. Goldstein, S. Melov, and J. Campisi. 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichler, A., and F. Melchior. 2002. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3:381-387. [DOI] [PubMed] [Google Scholar]
- 40.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz, D. C., and M. Hochstrasser. 2003. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28:321-328. [DOI] [PubMed] [Google Scholar]
- 42.Schwienhorst, I., E. S. Johnson, and R. J. Dohmen. 2000. SUMO conjugation and deconjugation. Mol. Gen. Genet. 263:771-786. [DOI] [PubMed] [Google Scholar]
- 43.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 44.Shao, R., F. P. Zhang, F. Tian, P. Anders Friberg, X. Wang, H. Sjoland, and H. Billig. 2004. Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1α in adult mouse brain and heart in vivo. FEBS Lett. 569:293-300. [DOI] [PubMed] [Google Scholar]
- 45.Sternsdorf, T., K. Jensen, and H. Will. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139:1621-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tojo, M., K. Matsuzaki, T. Minami, Y. Honda, H. Yasuda, T. Chiba, H. Saya, Y. Fujii-Kuriyama, and M. Nakao. 2002. The aryl hydrocarbon receptor nuclear transporter is modulated by the SUMO-1 conjugation system. J. Biol. Chem. 277:46576-46585. [DOI] [PubMed] [Google Scholar]
- 47.Verger, A., J. Perdomo, and M. Crossley. 2003. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 4:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 49.Yamada, S., H. Nakajima, and M. R. Kuehn. 2004. Novel testis- and embryo-specific isoforms of the phosphofructokinase-1 muscle type gene. Biochem. Biophys. Res. Commun. 316:580-587. [DOI] [PubMed] [Google Scholar]
- 50.Zambrowicz, B. P., A. Imamoto, S. Fiering, L. A. Herzenberg, W. G. Kerr, and P. Soriano. 1997. Disruption of overlapping transcripts in the ROSA βgeo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94:3789-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]