Highlights
-
•
E. coli OMVs were engineered to display scFv via the SpyTag/SpyCatcher system.
-
•
scFv targeting MUC1 (scFvSM3) was successfully conjugated onto the surface of bioengineered OMVs.
-
•
OMVs presenting scFvSM3 demonstrated specific binding to and internalization into MUC1-expressing cells, highlighting their potential as a cell-specific delivery system.
Keywords: Outer membrane vesicles, Surface display system, Single-chain variable fragment, SpyTag/SpyCatcher system, Cell-specific delivery system
Abstract
Outer membrane vesicles (OMVs), non-replicating spherical liposomes derived from Gram-negative bacteria, are a promising vaccine platform and multifunctional delivery systems. Their ability to be modified via genetic engineering for the incorporation and display of heterologous proteins enhances their functionality. In this study, we demonstrated a bio-ligation approach to display single-chain variable fragments (scFv) on the OMV surface using the SpyTag/SpyCatcher system. SpyTag-fused scFv, expressed by mammalian cells, bound to OMVs with SpyCatcher-fused Lpp'OmpA after a simple incubation. Biophysical analysis indicated that the conjugated OMVs maintained their physicochemical properties. We used an scFv targeting mucin 1 protein (MUC1) for specific cell targeting. Confocal microscopy revealed that conjugated OMVs specifically bound to and were internalized by MUC1-presenting cells, but not by MUC1-deficient cells. In conclusion, this rapid and efficient bio-ligation system facilitates the display of functional scFv on OMV surfaces, offering a promising approach for targeted delivery to MUC1-expressing cancer cells.
1. Introduction
Outer membrane vesicles (OMVs) are spherical vesicles with a bilayer membrane structure that are naturally released by Gram-negative bacteria. OMVs play significant roles in diverse biological processes such as stress responses, communication, host interaction, delivery of toxins, and horizontal gene transfer [1,2]. They have a typical diameter in the range of 20 – 200 nm and are mainly composed of outer membrane proteins (OMPs), lipopolysaccharides (LPS), periplasmic proteins, lipids, nucleic acids, and virulence factors [3]. Naturally derived OMVs from pathogenic bacteria have been used as vaccine candidates against the parental strain. OMVs isolated from Neisseria meningitidis have been a part of formulations to generate vaccines for serogroup B meningococcal disease, so-called ‘4CMenB’ or ‘BEXSERO’ vaccines. This vaccine was authorized by the European Medicines Agency (EMA) in 2013 and the United States Food and Drug Administration (U.S. FDA) in 2015 [4]. This highlights the potential use of OMVs as a safe and effective human vaccine.
Apart from naturally secreted vesicles, genetically modified OMVs produced from non-pathogenic bacteria can be used in various nanobiotechnology applications and can easily be modified to carry desired heterologous proteins. Foreign proteins are incorporated into OMVs by fusing them with either signal peptides or membrane proteins [[5], [6], [7]]. Alternatively, to retain the native structure and function, proteins were produced in suitable expression systems and attached to OMVs using “Plug-and-display technology”, also known as the “SpyTag/SpyCatcher” system. In this system, a protein-protein ligation derived from fibronectin binding protein (FbaB) of Streptococcus pyogenes, consists of a 13-amino acid peptide (SpyTag) and its partner protein (SpyCatcher). The ligation of these two parts occurs between an aspartic acid residue from SpyTag and a lysine residue from SpyCatcher [8]. These two amino acids spontaneously form a specific and irreversible isopeptide bond and this system has been widely used in the surface decoration of various nanoparticles [[9], [10], [11]].
The plug-and-display technology was utilized to develop a surface display system in OMVs by presenting SpyTag or SpyCatcher through genetic fusion with well-known OMV-associated proteins such as cytolysin A (ClyA) and hemoglobin protease (Hbp) [[12], [13], [14], [15]]. Apart from those proteins, Lpp'OmpA can serve as an alternative SpyCatcher-anchored protein. It is a bacterial surface display system widely used in many studies [[16], [17], [18]]. Previously, it was genetically fused to SpyCatcher at the C-terminus, enabling the display of SpyTag-fused proteins of interest on the surface of bacterial cell [19,20]. Truncated OmpA fused with SpyCatcher has also been shown to attach with Staphylococcus aureus (S. aureus) antigens to OMVs [21]. Therefore, Lpp'OmpA can be effectively exploited as an anchor protein for the presentation of desired proteins on the OMV surface.
In this study, OMVs were engineered as cancer-targeting vesicles by displaying a single-chain variable fragment (scFv) against a tumor antigen. The scFv against MUC1 clone SM3 (scFvSM3) was chosen as it is found at elevated levels in various cancers, and its presence is associated with the advancement of the disease [22]. It has shown promising activity in targeting cancer-associated MUC1 and has been used in other therapeutic applications [23,24]. The SpyTag fusion of scFvSM3 was transiently expressed in Chinese hamster ovary (CHO) cells to achieve high expression level while preserving its biological activities [25]. The SpyTag-fused scFvSM3 was subsequently attached to engineered OMVs presenting SpyCatcher produced by E. coli BL21(DE3), resulting in scFvSM3-displaying OMVs. The presence of scFvSM3 on the surfaces of the conjugated OMVs and their physicochemical properties were then studied. The efficacy of the conjugated OMVs was examined, including their cytotoxicity, binding, and internalization by MUC1-presenting cells and MUC1-lacking cells. This bio-ligation system allows us to create novel drug delivery nanocarriers, with the potential to incorporate therapeutic agents, thus advancing the development of bioengineered OMVs as a targeted delivery system.
2. Material and methods
2.1. Plasmid construction
To construct a plasmid for the expression of Lpp'OmpA-SpyCatcher (pLpp'OmpA-SpyCatcher), the genes encoding Lpp'OmpA, GS-linker, SpyCatcher [26], and a C-terminal His-tag with 5′-NdeI and 3′-EcoRI recognition sites were synthesized by Twist Bioscience (USA). This gene fragment was cloned into the pET21a+ vector at those restriction sites. To produce recombinant SpyTag-scFvSM3, pSpyTag-scFvSM3 was constructed by the Gibson assembly technique using NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs, USA). The SpyTag [8], GS-linker, and anti-MUC1 scFv namely SM3 genes with a N-terminal His-tag [27] were constructed in the 5′ EcoRI and 3′ NheI sites on pFUSE-hIgG1-Fc2 vector (Invivogen, USA). Sequences of both recombinant plasmids were validated by Sanger DNA sequencing (BIONICS, Korea).
2.2. Isolation of OMVs
Isolation of OMVs followed a previous study with additional modifications [28]. Briefly, E. coli BL21(DE3) harboring pLpp'OmpA-SpyCatcher was cultured in TB broth supplemented with 100 μg/ml of ampicillin. When the optical density (OD) of 600 nm reached 0.6 – 0.8, the expression of Lpp'OmpA-SpyCatcher was induced by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. The culture was incubated in a shaking incubator for 21 h at 30 °C. After incubation, the bacterial cells and cell debris were removed by centrifugation at 7000 x g for 30 min at 4 °C. The supernatant was then filtered through a Steritop 0.22 µm PES filter unit (Merck, USA) and spun at 150,000 x g for 3 h at 4 °C. The pellet was resuspended in phosphate-buffered saline (PBS) and centrifuged at 15,000 x g at 4 °C for 10 min. After that, OMV-containing supernatant was sterilized by passing through a 0.2 μm Supor® membrane syringe filter (Pall, China), and stored at -20 °C until use.
2.3. Purification of SpyTag-scFvSM3
SpyTag-scFvSM3 was transiently expressed in ExpiCHO-S™ cells (Gibco™, USA) using a PEI-mediated transfection method. ExpiCHO-S™ cells were maintained in HyClone HyCell TransFx-C transfection media (Cytiva, USA) on a shaking incubator at 125 rpm under a humidified atmosphere at 37 °C and 8 % CO2. The day before transfection, ExpiCHO-S™ cells were seeded at a density of 0.5 × 106 cells/ml in 30 ml of media and cultured for 24 h. After that, 30 µg of pSpyTag-scFvSM3, prepared using plasmid maxi kits (QIAGEN, Germany), and 90 µg of 40 kDa PEI MAX (Polyscience, USA) were mixed in media and incubated at room temperature for 10 min. The DNA/PEI complexes were slowly added to the culture. After five days, the conditioned medium was spun at 5000 x g for 30 min at 4 °C, filtered through a 0.2 µm Supor® membrane syringe filter (Pall, China) to remove intact cells and cell debris. Recombinant SpyTag-scFvSM3 was purified from the resulting supernatant by using a Histrap™ FF 1 ml column (Cytiva, USA) equipped in ÄKTA start (Cytiva, USA). The column was previously equilibrated with binding buffer (50 mM sodium phosphate buffer, 300 mM NaCl, 20 mM imidazole, pH 8.0) and loaded with the filtered supernatant at a flow rate of 0.5 ml/min. The unbound proteins were washed with binding buffer and recombinant SpyTag-scFvSM3 was then eluted with elution buffer (50 mM sodium phosphate buffer, 300 mM NaCl, 250 mM imidazole, pH 8.0). The protein-containing fractions were pooled, concentrated, and diafiltrated into storage buffer (50 mM sodium phosphate buffer, 150 mM NaCl, pH 8.0) using an Amicon® 10 kDa MWCO ultra-4 centrifugal filter unit (Merck, USA). The protein sample was kept at -20 °C until use.
2.4. Ligation of SpyTag:SpyCatcher in OMV samples
Prior to SpyTag:SpyCatcher ligation, the protein concentration of OMVs-Lpp'OmpA-SpyCatcher and SpyTag-scFvSM3 was determined by a bicinchoninic acid (BCA) assay using a Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, USA) following the manufacturer's protocol. SpyTag-scFvSM3 was incubated with OMVs-Lpp'OmpA-SpyCatcher at a weight ratio of 2:1 at 4 °C for 21 h. Aggregates were removed by filtration using a 0.2 µm Supor® membrane syringe filter (Pall, China), and unreacted SpyTag-scFvSM3 was removed by an Amicon® 100 kDa MWCO ultra-4 centrifugal filter unit (Merck, USA). The conjugated OMV samples were stored at −20 °C until further experiments.
2.5. SDS-PAGE and Western blot analysis
Samples were mixed with 5X SDS-PAGE protein loading buffer (Enzmart, Thailand), boiled at 95 °C for 10 min, then resolved on a 10 % SDS-polyacrylamide gel. The resulting gels were either stained by Coomassie blue or subjected to Western blotting. To identify proteins by Western blot analysis, proteins in the gel were transferred onto a nitrocellulose membrane. The membranes were washed with tris-buffered saline with 0.05 % Tween-20 (TBST) and blocked with blocking buffer (5 % skim milk in TBST) for 2 h at room temperature with agitation. Afterwards, His-tagged proteins were probed with 1:10,000 dilution of 6x-His Tag monoclonal antibody (MA1-21315; Invitrogen, USA) in blocking buffer for 1 h at room temperature with gentle rocking. The blot was washed with TBST three times for 15 min intervals and subsequently incubated with a 1:50,000 dilution of horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody (BioLegend, USA) for 30 min at room temperature with agitation. After an extensive washing step, horseradish peroxidase reaction was developed using Immobilon Forte Western HRP substrate (Merck, USA). Subsequently, the resulting membrane was visualized using the ImageQuant™ LAS 4000 (GE HealthCare, USA).
2.6. Proteinase K protection assay
About 7 µg of conjugated OMVs were digested by proteinase K (0.1 mg/ml) for 15 min at 37 °C in the presence or absence of 1 % Triton X-100. The proteolytic reaction was quenched by adding phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 10 µM and incubating on ice for 15 min. After that, the digested SpyTag:SpyCatcher product was detected by Western blotting.
2.7. Dynamic light scattering (DLS)
OMVs samples (60 µg/ml) were prepared in PBS and analyzed using a Zetasizer Nano ZS instrument (Malvern Panalytical, UK). The DLS measurement followed this protocol: 173° backscatter, temperature = 25 °C, run duration = 60 s, number of runs = 5, number of measurements = 3. Size measurements and data analysis were performed using Zetasizer Software (version 8.01).
2.8. Nanoparticle tracking analysis (NTA)
OMVs samples were diluted in PBS to a final protein concentration of 2.4 ng/ml and analyzed by NanoSight NS300 (Malvern Panalytical Ltd., UK) using an sCMOS camera with a Blue488 laser. Five replicates of 60 s video captures were recorded at 25 °C using a camera level of 15, a slide shutter of 1206, a slider gain of 245, and a detection threshold of 7. Size distribution profiles and particle concentration were calculated using NTA 3.4 Build 3.4.003.
2.9. Transmission electron microscopy (TEM)
100 µl of OMVs samples (0.1 mg/ml) were dropped onto a copper grid for 15 min. The grids were washed with water, stained with 2 % uranyl acetate for 30 s, dried in a humidity-controlled incubator and then imaged using a Hitachi HT7700 transmission electron microscope (Hitachi High-Technologies, USA) at 120 kV. The size of the vesicles from one hundred particles of each OMVs sample was measured by ImageJ (https://imagej.nih.gov/ij/index.html). In the gold-labeled TEM experiment, SpyTag-scFvSM3 was labeled by 10 nm gold nanoparticles using a gold conjugation kit (Abcam, UK) according to the manufacturer's protocol. Then, 2 µg of OMVs-Lpp'OmpA-SpyCatcher were mixed with 200 ng of gold-labeled SpyTag-scFvSM3 at 4 °C for 21 h. The labeled particles were visualized using protocol as described above.
2.10. Cell culture
MUC1-expressing human breast cancer cells, MCF-7 and MDA-MB-231, were maintained in Dulbecco's Modified Eagle Medium high glucose (DMEM; Gibco, USA) supplemented with 10 % of heat-inactivated Fetal Bovine Serum (FBS; Gibco, USA) in a humidified atmosphere of 5 % CO2 and 37 °C. MUC1-lacking cell, the human liver HepG2 cell was cultured in DMEM plus 10 % of heat-inactivated FBS and 1 % of MEM Non-Essential Amino Acids (MEM NEAA; Gibco, USA). The cells were passaged when confluency reached 70 - 80 % by enzymatic dissociation using TrypLE™ Express Enzyme (Thermo Fisher Scientific, USA) and cultured at least three times post-thawing before conducting experiments.
2.11. Immunofluorescence assay
A total of 25,000 cells were seeded into an 8-well chamber slide (SPL, Korea) and grown overnight under standard condition. After that, the cells were fixed with 4 % paraformaldehyde in PBS and washed twice with ice-cold PBS. Nonspecific binding was blocked by adding 3 % bovine serum albumin (BSA) in PBS for 1 h at 25 °C. Then, either 5 μg of purified SpyTag-scFvSM3 or 10 μg of OMVs samples were added to the well and incubated for 2 h at 4 °C. Unbound samples were washed with ice-cold PBS, and His-tagged proteins were probed with a 1:500 dilution of 6x-His Tag monoclonal antibody (MA1-21315; Invitrogen, USA) overnight at 4 °C. Then, a washing procedure was performed and 1:1000 of goat anti-Mouse IgG (H + L) cross-adsorbed secondary antibody, Alexa Fluor™ 647 (A-21235; Invitrogen, USA) was added to the wells for 30 mins at 25 °C. After washing, nuclei were stained by DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) for 15 min at 25 °C. The slide was washed and mounted with 10 % glycerol in PBS and observed under a Zeiss Microscopy LSM 900 (Carl Zeiss, USA).
2.12. Internalization assay
Internalization of OMVs samples into MUC1 positive and negative cells was examined by a confocal microscope. In brief, OMV samples were labeled with Vybrant™ DiO cell-labeling solution (Invitrogen, USA) by incubating at 37 °C for 30 mins. Excess dyes were removed by washing with PBS four times using an Amicon® 100 kDa MWCO ultra-4 centrifugal filter unit (Merck, USA). After that, 25,000 of the cells were grown overnight in an 8-well chamber slide (SPL, Korea). 50 μg/ml of DiO-labeled OMVs samples were added to the wells and incubated at 37 °C for 4 h. After incubation, the cells were washed with ice-cold PBS and fixed with 4 % paraformaldehyde in PBS. After washing, the nuclei were stained with DAPI for 15 min at 25 °C. The slide was washed, covered with 10 % glycerol in PBS and images were captured by a Zeiss Microscopy LSM 900 (Carl Zeiss, USA).
3. Results
3.1. SpyCatcher incorporated into OMVs via Lpp'OmpA fusion
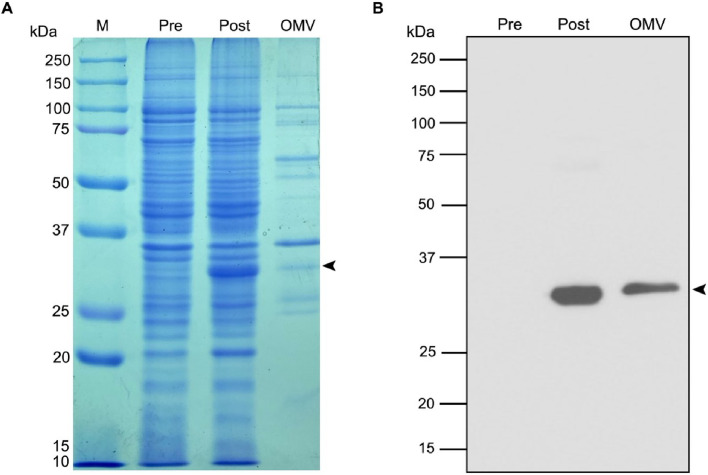
A simple strategy to decorate the surface of E. coli OMVs with scFvSM3, utilizing the SpyTag/SpyCatcher ligation system, is presented. The SpyCatcher displayed on OMVs and the SpyTag fused with scFvSM3 (SpyTag-scFvSM3) were connected via an irreversible isopeptide bond resulting in scFvSM3-displaying OMVs (Fig. 1). The SpyCatcher, the first component of the ligation system, was designed to be genetically fused to Lpp'OmpA. The gene encoding the SpyCatcher sequence was fused at the C-terminus of Lpp'OmpA to ensure the surface display of SpyCatcher. With the first nine residues of Braun's lipoprotein signal peptides (Lpp’) in front of OmpA genes, the expression of the chimeric protein consisting of Lpp'OmpA and SpyCatcher was expected to translocate to the E. coli outer membrane. This chimeric protein should then be incorporated into OMVs secreted by E. coli. The expression and incorporation of the chimeric protein were examined by SDS-PAGE and Western blot analysis with an anti-His-tag antibody. After induction with IPTG, the presence of Lpp'OmpA-SpyCatcher was confirmed as a protein band at its theoretical size of 29.9 kDa, as shown in the post-induced cell lysate stained with Coomassie blue (Fig. 2A). This was also confirmed by immunoblotting technique (Fig. 2B). Furthermore, OMV samples isolated by differential centrifugation were examined by Western blotting. Lpp'OmpA-SpyCatcher was shown to be localized in isolated recombinant OMVs since the band corresponding to Lpp'OmpA-SpyCatcher proteins was also detected (Fig. 2B). Consequently, these observations prove that Lpp'OmpA-SpyCatcher was successfully expressed and transported to the recombinant OMVs.
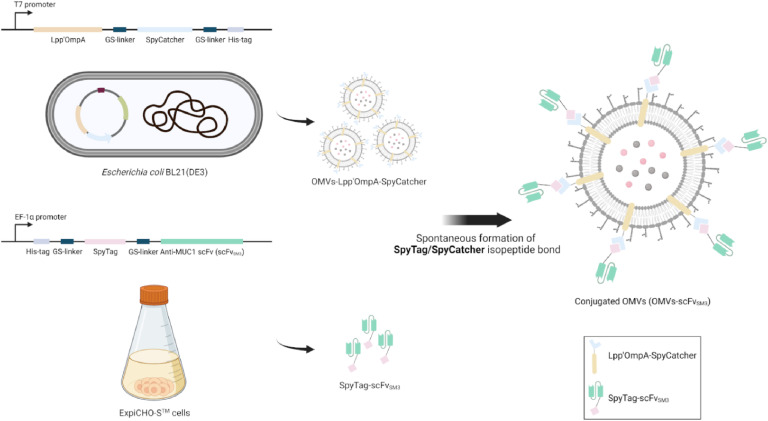
Fig. 1.
Schematic illustration of the display platform for scFv targeting MUC1 (scFvSM3) on the surface of E. coli-derived OMVs utilizing the SpyTag/SpyCatcher system. This scheme was created using BioRender.
Fig. 2.
Expression and localization of Lpp'OmpA-SpyCatcher on OMVs. (A) Coomassie blue-stained SDS-PAGE gel and (B) anti-His-tag Western blotting of cell lysate collected before (Pre), after (Post) IPTG induction, and purified OMVs-Lpp'OmpA-SpyCatcher (OMV). Lane M represents protein standards (Precision Plus Protein™ All Blue Prestained Protein Standards; Bio-rad, USA). An arrow indicates the band of Lpp'OmpA-SpyCatcher corresponding to its theoretical size of 29.9 kDa.
3.2. scFvSM3 protein attached to OMVs via SpyTag/SpyCatcher
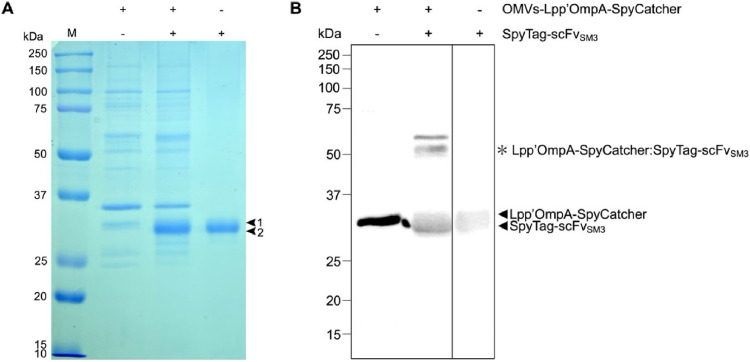
SpyTag-scFvSM3 fusion gene was inserted into pFUSE vector containing an IL2 signal sequence. The incorporation of this signal sequence facilitated the secretion of recombinant proteins from CHO cells. SpyTag-scFvSM3 fusion protein was then purified and identified. Fig. 3A shows the purified fraction in lane 3 as a major protein band corresponding to its predicted molecular weight of 29.4 kDa. This is consistent with the Western blotting result where only the corresponding protein band was observed in lane 3 (Fig. 3B). This suggests that SpyTag-scFvSM3 was successfully expressed in the CHO expression system. The purified SpyTag-scFvSM3 was then coupled to OMVsLpp'OmpA-SpyCatcher by co-incubation, as seen in lane 2. Since OMVs contain many other proteins, the conjugated product (Lpp'OmpA-SpyCatcher:SpyTag-scFvSM3) was not clearly shown in Coomassie blue-stained SDS-PAGE gel in lane 2 (Fig. 3A). However, a protein band shift at around 59.3 kDa in lane 2 was observed in the immunoblotting (Fig. 3B). This band is expected to be the conjugated product (Lpp'OmpA-SpyCatcher:SpyTag-scFvSM3) because its size agrees with the combined molecular weight of Lpp'OmpA-SpyCatcher (29.9 kDa) and SpyTag-scFvSM3 (29.4 kDa). Additionally, the conjugation between Lpp'OmpA-SpyCatcher and SpyTag-scFvSM3 was shown to be specific to their partners, as the band shift was not observed in a mixture of OMVs-Lpp'OmpA-SpyCatcher and an irrelevant protein (Supplemental Fig. S1).
Fig. 3.
Evaluation of SpyTag/SpyCatcher conjugation from OMVs-Lpp'OmpA-SpyCatcher and SpyTag-scFvSM3. (A) SDS-PAGE analysis and (B) Immunoblot analysis targeting His-tag proteins of OMVs to verify the SpyTag/SpyCatcher conjugation. The components identified are Lpp'OmpA-SpyCatcher (arrow 1) and SpyTag-scFvSM3 (arrow 2). The conjugated product (Lpp'OmpA-SpyCatcher:SpyTag-scFvSM3) is indicated by an asterisk (*) at the calculated size of 59.3 kDa.
3.3. Physicochemical properties of scFvSM3-displaying OMVs revealed similar characteristics to unconjugated OMVs
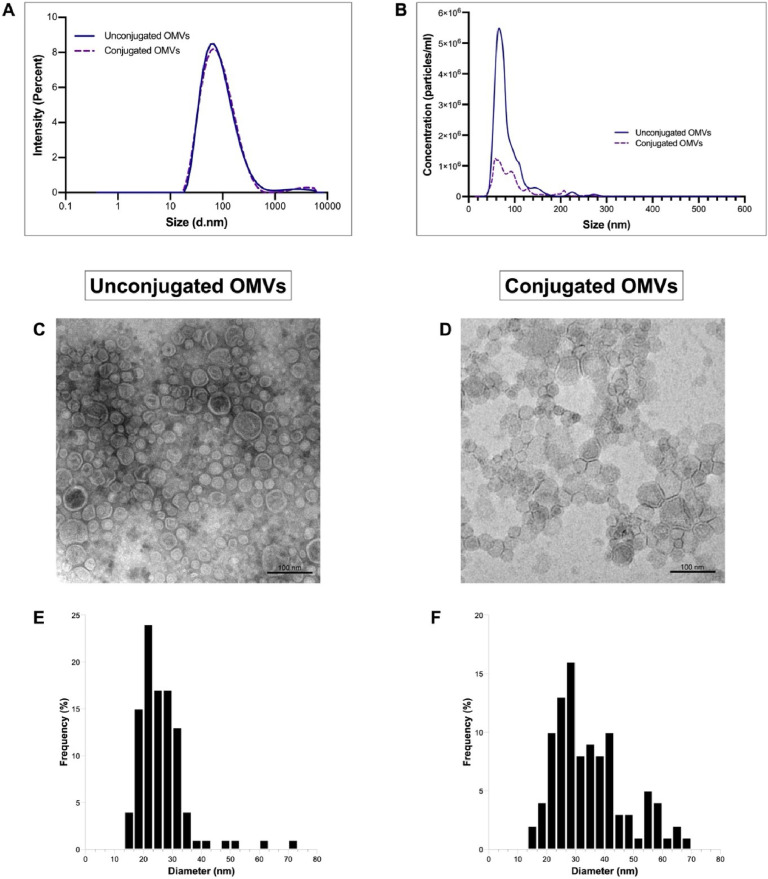
The physicochemical properties of isolated and conjugated OMVs were investigated using DLS, NTA and TEM. Fig. 4A shows a similar size distribution profile between unconjugated (without scFvSM3) and conjugated OMVs (with scFvSM3) as observed by DLS. The Z-average and polydispersity index (PDI) of both were also very similar, indicating moderate polydispersity. Additionally, an increase in the diameter of conjugated OMVs was observed by nanoparticle tracking analysis (Fig. 4B), likely due to the decoration of scFvSM3 on the surface of OMVs. Table 1 summarizes the results of different physicochemical characterization techniques. TEM images show that both OMV samples displayed a spherical morphology with a unique bilayer structure (Fig. 4C and D). The size of these intact vesicles ranged from 16 to 71 nm, with average diameters of 26.38 nm for unconjugated OMVs and 35.07 nm for conjugated OMVs (Fig. 4E and F).
Fig. 4.
Characterization of OMVs using DLS, NTA, and TEM. (A) Dynamic Light Scattering (DLS) and (B) Nanoparticle Tracking Analysis (NTA) showing size distribution of unconjugated OMVs (blue solid line) and conjugated OMVs (purple dashed line) (C) TEM images of unconjugated OMVs and (D) conjugated OMVs with scale bars representing 100 nm. (E) Size distribution of unconjugated OMVs and (F) conjugated OMVs as measured by TEM.
Table 1.
Physicochemical characterization of the OMV samples determined by DLS and NTA.
| Sample | DLS |
NTA | |
|---|---|---|---|
| Z-average (d.nm) | Polydispersity index (PDI) | Mean hydrodynamic size (nm) | |
| Unconjugated OMVs | 69.44 ± 0.80 | 0.278 | 84.0 ± 1.7 |
| Conjugated OMVs | 68.70 ± 0.65 | 0.287 | 94.7 ± 4.2 |
3.4. scFvSM3 protein displayed on the OMV surface
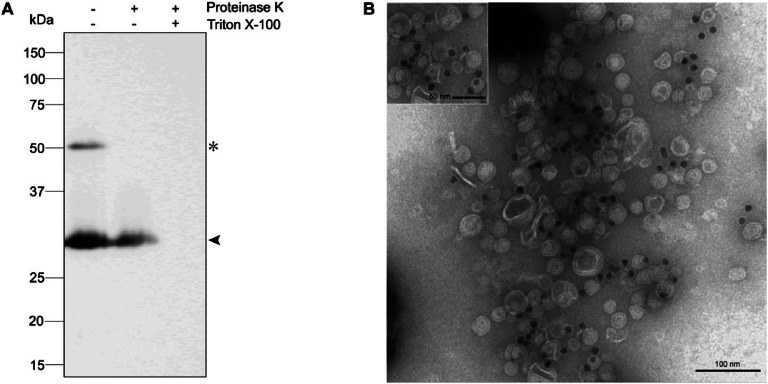
To confirm whether scFvSM3 was attached to the surface of OMVs after SpyTag/SpyCatcher conjugation, proteinase K protection assay and gold-labeled TEM analysis were performed. The proteinase K protection assay determined whether the scFvSM3 protein resided in the lumen or on the surface of the OMVs. Surface proteins are susceptible to proteinase K digestion while luminal proteins are not due to the inability of proteinase K to translocate across the vesicle lipid bilayer. Proteolytic digestion of luminal proteins is only feasible when the lipid bilayer is solubilized by detergents. As illustrated in Fig. 5A, after proteinase K treatment without Triton X-100 detergent, conjugated product was degraded while only Lpp'OmpA-SpyCatcher remained. Moreover, Lpp'OmpA-SpyCatcher was not detected after treatment with both proteinase K and Triton X-100. This suggests that scFvSM3 is located solely on the surface of OMVs. Consistent with the proteinase K protection assay, gold-labeled electron microscopy analysis indicated that scFvSM3 was displayed on the surface of OMVs. This analysis also determined the location of scFvSM3 proteins after SpyTag:SpyCatcher coupling. Gold-labeled SpyTag-scFvSM3 was incubated with OMVs-Lpp'OmpA-SpyCatcher and TEM images revealed black dots representing SpyTag-scFvSM3 situated on the OMV surface (Fig. 5B). Fig. 5B clearly shows a higher density of black dots surrounding OMV particles when compared to the background, confirming that scFvSM3 was specifically attached to the OMV surface.
Fig. 5.
The location of SpyTag-scFvSM3 on the OMVs after SpyTag/SpyCatcher conjugation was identified using two methods: (A) Proteinase K protection assay: Western blotting results using an anti-His-tag antibody of conjugated OMVs after digestion by proteinase K in a presence or an absence of Triton X-100 were observed. An arrow indicates the protein band of Lpp'OmpA-SpyCatcher, and an asterisk (*) indicates the protein band of the conjugated product. (B) Gold-labeled TEM: 200 ng of 10 nm gold-labeled SpyTag-scFvSM3 (black dots) were added to 2 µg of OMVs-Lpp'OmpA-SpyCatcher and subsequently observed by TEM. Scale bars are presented in each image.
3.5. scFvSM3-displaying OMVs could specifically bind and facilitate internalization to MUC1-presenting cells
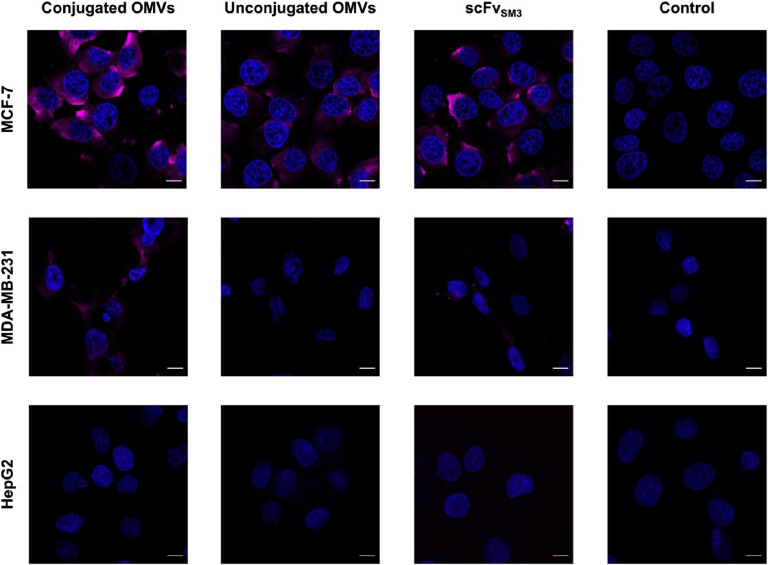
To verify the binding of scFvSM3-displaying OMVs to MUC1-expressing cells, MCF-7 cells were employed as an in vitro model because they abundantly express MUC1 on the cell surface and have been previously used in MUC1-related studies [29,30]. Additionally, MDA-MB-231 cells, also MUC1-positive, were utilized [31,32]. HepG2 cells, which lack MUC1 expression, served as the negative control. The binding of OMVs and scFvSM3 to the cells was assessed by immunofluorescence assay followed by confocal microscopy. The LSCM results showed a fluorescence signal in MCF-7 and MDA-MB-231 cells bound by OMVs-scFvSM3 and purified SpyTag-scFvSM3 (Fig. 6), indicating that scFvSM3 maintained its binding activity to MUC1 after conjugation to OMVs. In contrast, no fluorescence signal was observed in HepG2 cells, suggesting that scFvSM3 on the conjugated OMVs specifically binds to MUC1-presenting cells. However, unconjugated OMVs non-specifically bound to MCF-7 cells, as indicated by a faint fluorescence signal. This signal could originate from the interaction of anti-His tag antibody with Lpp'OmpA-SpyCatcher on the unconjugated OMVs. Additionally, the weak signal of bound OMVs-scFvSM3 and scFvSM3 in MDA-MB-231 cells is likely due to the low expression of membrane-associated MUC1 (32).
Fig. 6.
Binding analysis of scFvSM3-displaying OMVs to MCF-7, MDA-MB-231, and HepG2 cells as evaluated by confocal microscopy. The cells were incubated with either unconjugated OMVs, conjugated OMVs, or soluble SpyTag-scFvSM3. Samples were detected using a mouse anti-His-tag antibody followed by an anti-mouse IgG antibody conjugated with Alexa Fluor™ 647 (pink). Nuclei were stained with DAPI (blue). Scale bar represents 10 µm.
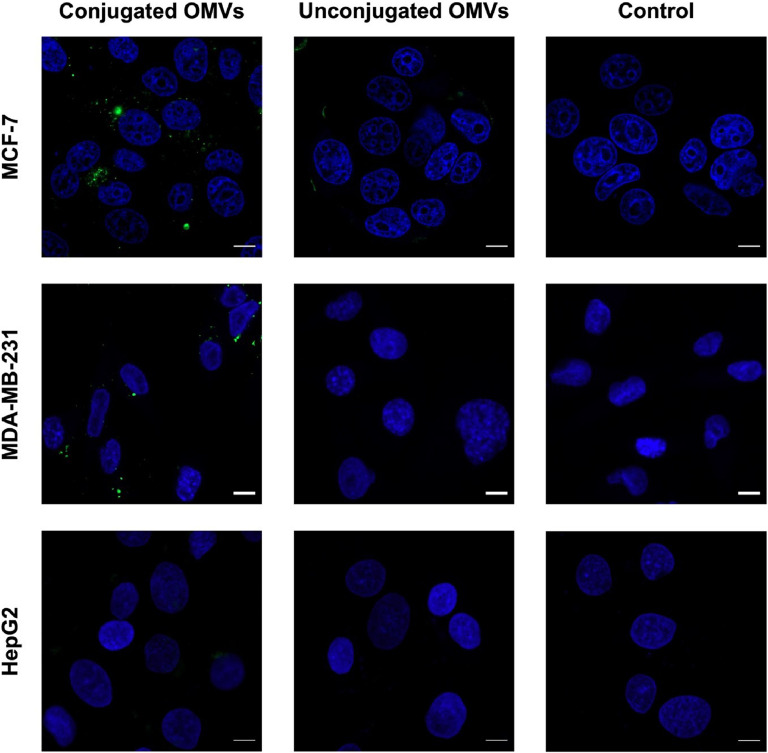
Prior to the internalization analysis, a cytotoxicity assay was performed to determine the non-toxic concentration of conjugated OMVs for MUC1+ (MCF-7 and MDA-MB-231) and MUC1- (HepG2) cells. The MTT assay indicated that OMVs-scFvSM3 (1.56 – 50 µg/ml) caused a slight decrease in cell viability as the OMV concentration increased (Supplemental Fig. S2). Since 50 µg/ml of OMVs-scFvSM3 maintained more than 80 % cell viability (MCF-7: 80.57±1.38 %; MDA-MB-231: 90.20±20.55 %; HepG2: 85.85±15.09 %), this non-toxic concentration was chosen for the uptake assay. The activity of MUC1-mediated internalization of OMVs-scFvSM3 into MUC1+ and MUC1- cells was then examined. The assay involved a 4-hour incubation of cells with DiO-labeled OMVs, which emit a green fluorescence signal. Confocal imaging showed that the green fluorescence signal was observed throughout MCF-7 and MDA-MB-231 cells treated with OMVs-scFvSM3 (Fig. 7), while no signal was detected in HepG2 cells treated with the same. The result implies that the internalization of OMVs-scFvSM3 is specific to MUC1-presenting cells. Furthermore, a minimal signal was detected in MCF-7 cells treated with unconjugated OMVs. Notably, the fluorescence signal from unconjugated OMVs was localized on the surface of MCF-7 cells, indicating no uptake of OMVs without scFvSM3. These results demonstrate that scFvSM3 facilitates the endocytosis of conjugated OMVs into MUC1+ cells.
Fig. 7.
LSCM images showing cellular internalization of conjugated OMVs by MCF-7, MDA-MB-231, and HepG2 cells. The cells were treated with DiO-labeled OMVs samples (green) for 4 h at 37 °C followed by nuclei staining with DAPI (blue). Scale bar indicates 10 µm.
4. Discussion
The development of bioengineered OMVs has gained wide attention in various nanotechnology applications due to their favorable characteristics, including intrinsic adjuvant-like properties, high stability, and ease of modification [33]. Sorting or presenting foreign proteins, such as antigen, antibody fragment, and other functional molecules into OMVs is crucial for modifying OMVs for desired applications [15,34,35]. In this study, we expressed exogenous anti-MUC1 scFv in suitable expression system to retain their function and structure to fabricate it onto modified OMVs via a bio-ligation system for utilization as a targeted delivery system.
Two common approaches to targeting foreign proteins into OMVs involve incorporating them into either the lumen or the surface of the vesicles [36,37]. Applications such as targeted drug delivery and vaccines require foreign proteins to be exposed on the OMV surfaces. Anchor or carrier proteins inserted into OMVs play a crucial role in presenting these foreign proteins. Cytolysin A (ClyA) and hemoglobin protease (Hbp) have been reported to serve as carrier proteins for heterologous protein presentation [12,15,38]. A heterologous protein gene can be directly fused with the clyA gene in a plasmid. Once gene expression is induced, the proteins are presented on the OMV surface [37]. Using this strategy, heterologous proteins such as GFP [37], a viral influenza antigen M2e4xHet [39], and a HER2 antibody [40], have been expressed as a fusion protein with ClyA. Additionally, ClyA and Hbp have been modified to fuse with either SpyCatcher or SpyTag [[12], [13], [14], [15]], allowing them to capture antigens and nanobodies for display on OMVs. In this study, we selected the Lpp’ signal peptide combined with truncated OmpA as the anchor protein, as this system has been shown to effectively display foreign proteins on the OMV membrane [21,41,42]. Lpp'OmpA together with SpyCatcher binds to the SpyTag fusion partner and subsequently attaches the proteins. We designed SpyCatcher coupling with Lpp'OmpA because SpyCatcher, being larger than SpyTag, would not be obstructed by the OMV membrane environment. This design allows SpyCatcher to be more accessible and to bind more easily to heterologous proteins fused with SpyTag. This approach was also demonstrated by displaying S. aureus antigens on OMVs, which induced potent immune responses and protected against S. aureus lethal challenge in a mouse model [21]. Although ClyA and Hbp have been linked to SpyCatcher previously, we hypothesize that Lpp'OmpA offer advantages as its smaller size (∼16 kDa) and simple structure allows for higher density expression of SpyCatcher. Furthermore, previous studies have noted potential toxicity associated with the use of ClyA in SpyCatcher fusions [43], making Lpp'OmpA an attractive alternative carrier. In addition, the SpyTag/SpyCatcher-mediated surface display system provides the advantage of enabling the attachment of multiple proteins onto the OMVs simultaneously. This display system has been shown to fuse either two heterologous proteins [14] or several antigens on OMVs [21]. This feature facilitates the development of a multivalent vaccine platform and allows the presentation of multiple different targeting molecules per OMV.
Another main obstacle in protein presentation on OMVs is the inability to load proteins with post-translational modifications using bacterial protein presentation systems, in which target proteins are genetically fused to bacterial anchor proteins. For example, presenting SARS-CoV-2 RBD in OMVs via fusion of cytolysin A (ClyA) showed low expression levels and incorrect RBD conformation [44]. In addition, recombinant OMVs presenting anti-digoxin scFv fused with Lpp'OmpA failed to bind digoxin [45]. These studies suggest that the success of loading target proteins into OMVs via co-expression with bacterial membrane proteins depends on the correct structure and function of target proteins. Proteins with complex post-translational modifications cannot be properly expressed and incorporated into OMVs using bacteria alone. Therefore, in our approach, the scFv was initially expressed in mammalian cells and then attached to OMVs expressed in bacteria. This method has been demonstrated previously in displaying mammalian cell culture-derived SARS-CoV-2 RBD on OMVs, where RBD-OMVs triggered neutralizing antibody and protection against SARS-CoV-2 in golden Syrian hamsters [15]. We displayed scFvSM3 on SpyCatcher-presenting OMVs after confirming SpyTag/SpyCatcher conjugation by Western blot analysis. The binding of OMVs containing scFvSM3 to MUC1 on the surface of MUC1-presenting cells was confirmed by immunofluorescence assay, indicating that the proper conformation of scFvSM3 was produced by CHO cells. As a result, antigens and antibody fragments that require post translational modification for functional activity can be expressed in other host cells prior to OMV attachment. The shape and integrity of OMVs did not change even after conjugation, making this a suitable approach for OMV decoration. The size and morphology of recombinant OMVs in this study were consistent with other studies [3,41,42].
Numerous studies have demonstrated the potential of bioengineered OMVs as an attractive cell-specific delivery system by generating modified OMVs presenting targeting molecules such as affibodies and nanobodies [34,40]. In the present study, scFvSM3 was selected as the protein of interest due to its capability to internalize conjugated molecules upon binding with the MUC1 antibody [46,47]. While affibodies and nanobodies can be directly fused to ClyA and presented on the surface of OMVs, scFvs offer greater target cell diversity owing to the variety of available scFvs [48]. Using confocal microscopy, OMVs-scFvSM3 showed evident internalization to target cells, while OMVs without scFvSM3 lacked this ability. MUC1-mediated endocytosis of OMVs-scFvSM3 was confirmed by the binding between scFvSM3 and MUC1, resulting in successful uptake into MUC1-presenting cells (MCF-7 and MDA-MB-231). Moreover, no binding or uptake of OMVs-scFvSM3 was observed in MUC1-deficient cells (HepG2), suggesting a specific delivery of OMVs-scFvSM3 to MUC1-expressing cells. Although non-specific binding of unconjugated OMVs was observed in MCF-7 cells, it did not facilitate the uptake of the OMVs into these cells, as unconjugated OMVs were observed on the cell surface. This implies the importance of scFvSM3 for internalization into the cells. Collectively, scFvSM3 conjugated on the OMVs facilitated binding and subsequent internalization into MUC1-presenting cells, suggesting a promising application as a cell-specific delivery system.
The modification of OMVs with scFv could enable them to be loaded with therapeutic agents such as siRNA/miRNA and anti-tumor drugs, as these therapeutic molecules have been successfully incorporated into OMVs [40,49,50]. The additional beneficial characteristics of OMVs in cancer treatment, including their rigid structure and nanoscale particle size that reduce drug leakage and the enhanced permeability and retention (EPR) effect that promotes penetration into tumor tissue, further support the use of OMVs as a favorable anti-cancer drug delivery cargo [33].
In this study, we demonstrated a simple and rapid approach to produce bioengineered OMVs that requires only cultivation and ultracentrifugation, followed by simple incubation for conjugation. This protocol facilitates large-scale production and development of bioengineered OMVs for the decoration of proteins obtained from other expression systems while their structure and function. The promoter used for the expression of Lpp'OmpA in this study is not tightly controlled compared to promoters used in other studies. The leaky expression of Lpp'OmpA from the promoter induced toxicity in the host cells and resulted in a suboptimal number of Lpp'OmpA displayed on the OMVs [26,51]. Therefore, it would be beneficial to replace the plasmid carrying the Lpp'OmpA-SpyCatcher hybrid with another plasmid carrying a more tightly controlled promoter to enhance the display of Lpp'OmpA and consequently increase the number of SpyCatcher and its partner after conjugation. Despite these considerations, this study provides a valuable contribution toward the development of bioengineered OMVs as a promising targeted drug delivery system.
5. Conclusions
OMVs are vesicles derived from Gram-negative bacteria and are valuable forvarious nanotechnology applications due to their favorable properties. To enhance their potential, parental bacteria can be bioengineered to produce and transport desired proteins within the vesicles. In this study, we incorporated a bio-ligation system, SpyTag/SpyCatcher, in combination with the Lpp'OmpA bacterial display system to display scFv on the surface of OMVs. Anti-MUC1 scFv was selected to modify ordinary OMVs for targeted cargo delivery to MUC1-aberrantly expressed cancer cells. This study demonstrates that the convenient and efficient use of bioengineered OMVs represents a promising approach for cell-specific delivery systems.
Funding
This research was financially supported by the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund) from Chulalongkorn University [grant number: GCUGR1125633029M] and the Thailand Science research and Innovation Fund Chulalongkorn University [HEA663300002]. S.L. was supported by Chulalongkorn University for Research Assistant Scholarship [grant number: GCUGE17].
CRediT authorship contribution statement
Sedthawut Laotee: Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Data curation. Wanatchaporn Arunmanee: Writing – review & editing, Supervision, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
MUC1-expressing cell lines, including the human breast cancer MCF-7 and MDA-MB-231, were kindly provided by Dr. Nonthaneth Nalinratana and Assoc. Prof. Dr.Supannikar Tawinwung, respectively. The human liver cancer cell line HepG2 (MUC1-negative cells) was a gift from Dr. Patcharawalai Whongsiri. We would like to thank Dr. Chavee Laomeephol and Miss Suchada Srifa for their guidance in the confocal imaging study. We also acknowledge the Research Instrument Center of the Faculty of Pharmaceutical Sciences, Chulalongkorn University for providing research facilities.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.btre.2024.e00854.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Kulkarni H.M., Jagannadham M.V. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology (Reading) 2014;160(Pt 10):2109–2121. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- 2.Jan A.T. Outer membrane vesicles (OMVs) of Gram-negative bacteria: a perspective update. Front. Microbiol. 2017;8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwechheimer C., Kuehn M.J. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 2015;13(10):605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rappuoli R., Pizza M., Masignani V., Vadivelu K. Meningococcal B vaccine (4CMenB): the journey from research to real world experience. Expert. Rev. Vaccines. 2018;17(12):1111–1121. doi: 10.1080/14760584.2018.1547637. [DOI] [PubMed] [Google Scholar]
- 5.Huang W., Wang S., Yao Y., Xia Y., Yang X., Li K., et al. Employing Escherichia coli-derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against Acinetobacter baumannii infection. Sci. Rep. 2016;6:37242. doi: 10.1038/srep37242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartolini E., Ianni E., Frigimelica E., Petracca R., Galli G., Berlanda Scorza F., et al. Recombinant outer membrane vesicles carrying Chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J. ExtraCell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grandi A., Fantappiè L., Irene C., Valensin S., Tomasi M., Stupia S., et al. Vaccination With a FAT1-derived B cell epitope combined with tumor-specific B and T cell epitopes elicits additive protection in cancer mouse models. Front. Oncol. 2018;8:481. doi: 10.3389/fonc.2018.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakeri B., Fierer J.O., Celik E., Chittock E.C., Schwarz-Linek U., Moy V.T., et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U S. A. 2012;109(12):E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brune K.D., Leneghan D.B., Brian I.J., Ishizuka A.S., Bachmann M.F., Draper S.J., et al. Plug-and-display: decoration of virus-like particles via isopeptide bonds for modular immunization. Sci. Rep. 2016;6:19234. doi: 10.1038/srep19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S., Han B.K., Kim Y.H., Ahn J.Y. SpyCatcher-SpyTagged ApxIA toxoid and the immune-modulating yeast ghost shells. J. Biomed. Nanotechnol. 2020;16(11):1644–1657. doi: 10.1166/jbn.2020.2992. [DOI] [PubMed] [Google Scholar]
- 11.Li D., Zhang H., Yang L., Chen J., Zhang Y., Yu X., et al. Surface display of classical swine fever virus E2 glycoprotein on gram-positive enhancer matrix (GEM) particles via the SpyTag/SpyCatcher system. Protein Expr. Purif. 2020;167 doi: 10.1016/j.pep.2019.105526. [DOI] [PubMed] [Google Scholar]
- 12.Cheng K., Zhao R., Li Y., Qi Y., Wang Y., Zhang Y., et al. Bioengineered bacteria-derived outer membrane vesicles as a versatile antigen display platform for tumor vaccination via plug-and-display technology. Nat. Commun. 2021;12(1):2041. doi: 10.1038/s41467-021-22308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Berg van Saparoea H.B., Houben D., de Jonge M.I., Jong W.S.P., Luirink J. Display of recombinant proteins on bacterial outer membrane vesicles by using protein ligation. Appl. Environ. Microbiol. 2018;84(8) doi: 10.1128/AEM.02567-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Berg van Saparoea H.B., Houben D., Kuijl C., Luirink J., Jong W.S.P. Combining protein ligation systems to expand the functionality of semi-synthetic outer membrane vesicle nanoparticles. Front. Microbiol. 2020;11:890. doi: 10.3389/fmicb.2020.00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang L., Driedonks T.A.P., Jong W.S.P., Dhakal S., Bart van den Berg van Saparoea H., Sitaras I., et al. A bacterial extracellular vesicle-based intranasal vaccine against SARS-CoV-2 protects against disease and elicits neutralizing antibodies to wild-type and Delta variants. J. ExtraCell Vesicles. 2022;11(3):e12192. doi: 10.1002/jev2.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres-Bañaga R., Mares-Alejandre R.E., Terán-Ramírez C., Estrada-González A.L., Muñoz-Muñoz P.L.A., Meléndez-López S.G., et al. Functional display of an amoebic chitinase in Escherichia coli expressing the catalytic domain of EhCHT1 on the bacterial cell surface. Appl. Biochem. Biotechnol. 2020;192(4):1255–1269. doi: 10.1007/s12010-020-03389-5. [DOI] [PubMed] [Google Scholar]
- 17.Francisco J.A., Campbell R., Iverson B.L., Georgiou G. Production and fluorescence-activated cell sorting of Escherichia coli expressing a functional antibody fragment on the external surface. Proc. Natl. Acad. Sci. U. S. A. 1993;90(22):10444–10448. doi: 10.1073/pnas.90.22.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wendel S., Fischer E.C., Martínez V., Seppälä S., Nørholm M.H. A nanobody:GFP bacterial platform that enables functional enzyme display and easy quantification of display capacity. Microb. Cell Fact. 2016;15:71. doi: 10.1186/s12934-016-0474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francisco J.A., Earhart C.F., Georgiou G. Transport and anchoring of beta-lactamase to the external surface of Escherichia coli. Proc. Natl. Acad. Sci. U S. A. 1992;89(7):2713–2717. doi: 10.1073/pnas.89.7.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stathopoulos C., Georgiou G., Earhart C.F. Characterization of Escherichia coli expressing an Lpp'OmpA(46-159)-PhoA fusion protein localized in the outer membrane. Appl. Microbiol. Biotechnol. 1996;45(1–2):112–119. doi: 10.1007/s002530050657. [DOI] [PubMed] [Google Scholar]
- 21.Sun J., Lin X., He Y., Zhang B., Zhou N., Huang J.D. A bacterial outer membrane vesicle-based click vaccine elicits potent immune response against Staphylococcus aureus in mice. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1088501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horm T.M., Schroeder J.A. MUC1 and metastatic cancer: expression, function and therapeutic targeting. Cell Adh. Migr. 2013;7(2):187–198. doi: 10.4161/cam.23131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burchell J., Gendler S., Taylor-Papadimitriou J., Girling A., Lewis A., Millis R., et al. Development and characterization of breast cancer reactive monoclonal antibodies directed to the core protein of the human milk mucin. Cancer Res. 1987;47(20):5476–5482. [PubMed] [Google Scholar]
- 24.You F., Jiang L., Zhang B., Lu Q., Zhou Q., Liao X., et al. Phase 1 clinical trial demonstrated that MUC1 positive metastatic seminal vesicle cancer can be effectively eradicated by modified anti-MUC1 chimeric antigen receptor transduced T cells. Sci. China Life Sci. 2016;59(4):386–397. doi: 10.1007/s11427-016-5024-7. [DOI] [PubMed] [Google Scholar]
- 25.Vendel M.C., Favis M., Snyder W.B., Huang F., Capili A.D., Dong J., et al. Secretion from bacterial versus mammalian cells yields a recombinant scFv with variable folding properties. Arch. Biochem. Biophys. 2012;526(2):188–193. doi: 10.1016/j.abb.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 26.Gallus S., Peschke T., Paulsen M., Burgahn T., Niemeyer C.M., Rabe K.S. Surface display of complex enzymes by in situ SpyCatcher-SpyTag interaction. Chembiochem. 2020;21(15):2126–2131. doi: 10.1002/cbic.202000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojas-Ocáriz V., Compañón I., Aydillo C., Castro-Loṕez J., Jiménez-Barbero J., Hurtado-Guerrero R., et al. Design of α-S-Neoglycopeptides derived from MUC1 with a flexible and solvent-exposed sugar moiety. J. Org. Chem. 2016;81(14):5929–5941. doi: 10.1021/acs.joc.6b00833. [DOI] [PubMed] [Google Scholar]
- 28.Alves N.J., Turner K.B., Daniele M.A., Oh E., Medintz I.L., Walper S.A. Bacterial nanobioreactors–directing enzyme packaging into bacterial outer membrane vesicles. ACS. Appl. Mater. Interfaces. 2015;7(44):24963–24972. doi: 10.1021/acsami.5b08811. [DOI] [PubMed] [Google Scholar]
- 29.Xi X., Wang J., Qin Y., Huang W., You Y., Zhan J. Glycosylated modification of MUC1 maybe a new target to promote drug sensitivity and efficacy for breast cancer chemotherapy. Cell Death. Dis. 2022;13(8):708. doi: 10.1038/s41419-022-05110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaretsky J.Z., Barnea I., Aylon Y., Gorivodsky M., Wreschner D.H., Keydar I. MUC1 gene overexpressed in breast cancer: structure and transcriptional activity of the MUC1 promoter and role of estrogen receptor alpha (ERalpha) in regulation of the MUC1 gene expression. Mol. Cancer. 2006;5:57. doi: 10.1186/1476-4598-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh M.D., Luckie S.M., Cummings M.C., Antalis T.M., McGuckin M.A. Heterogeneity of MUC1 expression by human breast carcinoma cell lines in vivo and in vitro. Breast. Cancer Res. Treat. 1999;58(3):255–266. doi: 10.1023/a:1006345301364. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell S., Abel P., Madaan S., Jeffs J., Chaudhary K., Stamp G., et al. Androgen-dependent regulation of human MUC1 mucin expression. Neoplasia. 2002;4(1):9–18. doi: 10.1038/sj.neo.7900194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J., Su Y., Wang Z. Engineering bacterial membrane nanovesicles for improved therapies in infectious diseases and cancer. Adv. Drug Deliv. Rev. 2022;186 doi: 10.1016/j.addr.2022.114340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Q., Ma X., Cheng K., Liu G., Li Y., Yue Y., et al. Engineered bacterial outer membrane vesicles as controllable two-way adaptors to activate macrophage phagocytosis for improved tumor immunotherapy. Adv. Mater. 2022;34(40) doi: 10.1002/adma.202206200. [DOI] [PubMed] [Google Scholar]
- 35.Gujrati V., Prakash J., Malekzadeh-Najafabadi J., Stiel A., Klemm U., Mettenleiter G., et al. Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat. Commun. 2019;10(1):1114. doi: 10.1038/s41467-019-09034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fantappiè L., de Santis M., Chiarot E., Carboni F., Bensi G., Jousson O., et al. Antibody-mediated immunity induced by engineered Escherichia coli OMVs carrying heterologous antigens in their lumen. J. ExtraCell Vesicles. 2014;3 doi: 10.3402/jev.v3.24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen D.J., Osterrieder N., Metzger S.M., Buckles E., Doody A.M., DeLisa M.P., et al. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc. Natl. Acad. Sci. U S. A. 2010;107(7):3099–3104. doi: 10.1073/pnas.0805532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gnopo Y.M.D., Watkins H.C., Stevenson T.C., DeLisa M.P., Putnam D. Designer outer membrane vesicles as immunomodulatory systems - reprogramming bacteria for vaccine delivery. Adv. Drug Deliv. Rev. 2017;114:132–142. doi: 10.1016/j.addr.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Rappazzo C.G., Watkins H.C., Guarino C.M., Chau A., Lopez J.L., DeLisa M.P., et al. Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza a challenge in BALB/c mice. Vaccine. 2016;34(10):1252–1258. doi: 10.1016/j.vaccine.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 40.Gujrati V., Kim S., Kim S.H., Min J.J., Choy H.E., Kim S.C., et al. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 2014;8(2):1525–1537. doi: 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
- 41.Alves N.J., Turner K.B., DiVito K.A., Daniele M.A., Walper S.A. Affinity purification of bacterial outer membrane vesicles (OMVs) utilizing a His-tag mutant. Res. Microbiol. 2017;168(2):139–146. doi: 10.1016/j.resmic.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Thapa H.B., Müller A.M., Camilli A., Schild S. An intranasal vaccine based on outer membrane vesicles against SARS-CoV-2. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.752739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murase K., Cytolysin A. ClyA): a bacterial virulence factor with potential applications in nanopore technology, vaccine development, and tumor therapy. Toxins. (Basel) 2022;14(2) doi: 10.3390/toxins14020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wo J., Lv Z.Y., Sun J.N., Tang H., Qi N., Ye B.C. Engineering probiotic-derived outer membrane vesicles as functional vaccine carriers to enhance immunity against SARS-CoV-2. iScience. 2023;26(1) doi: 10.1016/j.isci.2022.105772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J.Y., Doody A.M., Chen D.J., Cremona G.H., Shuler M.L., Putnam D., et al. Engineered bacterial outer membrane vesicles with enhanced functionality. J. Mol. Biol. 2008;380(1):51–66. doi: 10.1016/j.jmb.2008.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu H., Gan L., Han Y., Da Y., Xiong J., Hong S., et al. Site-specific labeling of an anti-MUC1 antibody: probing the effects of conjugation and linker chemistry on the internalization process. RSC. Adv. 2019;9(4):1909–1917. doi: 10.1039/c8ra09902b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamann P.R., Hinman L.M., Beyer C.F., Lindh D., Upeslacis J., Shochat D., et al. A calicheamicin conjugate with a fully humanized anti-MUC1 antibody shows potent antitumor effects in breast and ovarian tumor xenografts. Bioconjug. Chem. 2005;16(2):354–360. doi: 10.1021/bc049794n. [DOI] [PubMed] [Google Scholar]
- 48.Asaadi Y., Jouneghani F.F., Janani S., Rahbarizadeh F. A comprehensive comparison between camelid nanobodies and single chain variable fragments. Biomark. Res. 2021;9(1):87. doi: 10.1186/s40364-021-00332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui C., Guo T., Zhang S., Yang M., Cheng J., Wang J., et al. Bacteria-derived outer membrane vesicles engineered with over-expressed pre-miRNA as delivery nanocarriers for cancer therapy. Nanomedicine. 2022;45 doi: 10.1016/j.nano.2022.102585. [DOI] [PubMed] [Google Scholar]
- 50.Kuerban K., Gao X., Zhang H., Liu J., Dong M., Wu L., et al. Doxorubicin-loaded bacterial outer-membrane vesicles exert enhanced anti-tumor efficacy in non-small-cell lung cancer. Acta Pharm. Sin. B. 2020;10(8):1534–1548. doi: 10.1016/j.apsb.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weyant K.B., Oloyede A., Pal S., Liao J., Jesus M.R., Jaroentomeechai T., et al. A modular vaccine platform enabled by decoration of bacterial outer membrane vesicles with biotinylated antigens. Nat. Commun. 2023;14(1):464. doi: 10.1038/s41467-023-36101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.