Abstract
TFIID plays a key role in transcription initiation of RNA polymerase II preinitiation complex assembly. TFIID is comprised of the TATA box binding protein (TBP) and 14 TBP-associated factors (TAFs). A second set of transcriptional regulatory multiprotein complexes containing TAFs has been described (called SAGA, TFTC, STAGA, and PCAF/GCN5). Using matrix-assisted laser desorption ionization mass spectrometry, we identified a novel TFTC subunit, human TAF9Like, encoded by a TAF9 paralogue gene. We show that TAF9Like is a subunit of TFIID, and thus, it will be called TAF9b. TFIID and TFTC complexes in which both TAF9 and TAF9b are present exist. In vitro and in vivo experiments indicate that the interactions between TAF9b and TAF6 or TAF9 and TAF6 histone fold pairs are similar. We observed a differential induction of TAF9 and TAF9b during apoptosis that, together with their different ability to stabilize p53, points to distinct requirements for the two proteins in gene regulation. Small interfering RNA (siRNA) knockdown of TAF9 and TAF9b revealed that both genes are essential for cell viability. Gene expression analysis of cells treated with either TAF9 or TAF9b siRNAs indicates that the two proteins regulate different sets of genes with only a small overlap. Taken together, these data demonstrate that TAF9 and TAF9b share some of their functions, but more importantly, they have distinct roles in the transcriptional regulatory process.
Transcription initiation of protein-encoding genes by RNA polymerase II (Pol II) requires the transcription factor TFIID that is comprised of the TATA binding protein (TBP) and series of TBP-associated factors (TAFs) (1, 6, 57). In human HeLa cells, we showed that different human TFIID complexes containing or lacking TAF10 which exhibit functionally distinct properties exist (6, 7, 25). Cell type-specific TFIID complexes have been found to be composed of core TAFs and cell type-specific TAFs. TAF4b was found to be enriched in differentiated human B lymphocytes, and a unique TAF4b-containing TFIID was isolated from these cells (13). Moreover, during spermatogenesis, TAF7L-containing TFIID complexes have been found (48). Another set of human transcriptional regulatory multiprotein complexes containing TAFs are called TFTC, STAGA, or PCAF/GCN5 (6a, 36, 63). These complexes are functional homologues of the Saccharomyces cerevisiae SAGA complex, and all contain human homologues of the yeast histone acetyltransferase Gcn5 as well as a subset of SPT and ADA proteins, the 400-kDa TRRAP protein, and a number of TAFs (shared TAFs) also found in TFIID (36).
TAF9 was first identified as a TFIID subunit from multiple organisms: human (formerly called hTAFII31 or hTAFII32 [29, 35]), Drosophila melanogaster (formerly dTAFII40 [55]), and yeast (formerly yTaf17p [42]). Later, TAF9 was also identified as a component of different TBP-free TAF complexes containing the GCN5-type histone acetyltransferase, such as the yeast SAGA complex (19), the Drosophila TFTC complex (44), and human TFTC-type complexes (10, 36, 67). Drosophila TAF9 has also been described in the Polycomb group complex and the e(y)2 protein-containing complex (17, 50).
TAF9 was shown to interact directly with the tumor suppressor protein p53, the herpes simplex virus activator VP16, and the basal transcription factor TFIIB, and these interactions were suggested to be important for mediating transcriptional activation (29, 35, 55). TAF9 was shown to bind to the N-terminal region of p53, a region that is also required for interaction with the oncoprotein Mdm2. It has been shown that overexpression of TAF9 inhibits Mdm2-mediated ubiquitination of p53 and increases p53 levels and that TAF9-mediated p53 stabilization results in activation of p53-mediated transcriptional activity and leads to p53-dependent growth arrest in fibroblasts (9, 24).
Crystal structures demonstrate that the amino-terminal portions of Drosophila TAF9 and TAF6 adopt a canonical histone fold (HF) configuration consisting of two short α-helices flanking a long central α-helix. In the crystal structure, the dTAF9/TAF6 HF complex exists as a heterotetramer, resembling the (H3/H4)2 heterotetrameric core of the histone octamer, suggesting that TFIID may contain a histone octamer-like substructure (65). Furthermore, it was suggested that this histone octamer-like structure in TFIID may play a role in DNA wrapping and in the stable positioning of promoter DNA relative to TAFs (46). Our experiments localizing most of the yeast TAFs and TBP in TFIID (33, 34) showed that the nine TAFs, which contain the HF structural motif (15), are located in three distinct lobes of the TFIID structure. The distribution of these TAFs indicates that the previously reported pairwise interactions between HF-containing TAFs occur in the native TFIID complex. The TAF9-TAF6 pair was located in lobes A and B (34). The fact that most of the HF-containing TAF pairs have been found in two distinct lobes suggested that the organization of TFIID is more complex than originally thought and that there may be several histone octamer-like structures present in TFIID (34).
In the past decade, the role of different TAFs has been extensively investigated in a few model organisms. Several independent studies revealed that 3% to 60% of the yeast genes require individual TAFs for transcription depending on the TAF subunit, and in total, approximately 84% of yeast Pol II genes are TAF dependent (20, 53). Studies in several metazoan organisms revealed a wide but rarely universal requirement for TAFs. In Caenorhabditis elegans, individual TAF (i.e., TAF4, TAF5, TAF9, and TAF10) knockdowns by RNA interference affect embryo development and gene expression to different degrees, with TAF4 being required for essentially all early embryonic mRNA transcription, while TAF5, TAF9, and TAF10 show a more restricted requirement for embryonic transcription (58, 59). In Drosophila, TAF1 loss-of-function mutants affect several differentiation processes (62), and dTAF4 and dTAF6 affect Dorsal-mediated transcription (68), whereas dTAF6 seems to be broadly required for somatic and germ cell development (3). Also, in vertebrates, Pol II transcription displays a broad but not complete TAF dependency. In mice, TAF10 is required for early embryo development but not for the survival of trophoblast cells or the retinoic acid-mediated differentiation of F9 cells (38, 40). TAF4b knockout mice are characterized by female infertility (14). Moreover, the different enzymatic activity-containing functional domains of TAF1 modulate expression of a distinct subset of mammalian genes, suggesting that they are important for regulating largely nonoverlapping sets of genes (45).
Certain TAFs have paralogue genes, which often encode TAF-like (L) proteins with cell type- or tissue-specific expression patterns. The first TAF paralogue seems to have appeared in fission yeast where two genes encode TAF5 homologues, and both are present in the same TFIID complex (39). In C. elegans, Drosophila, and the human genome, several TAF paralogue genes encoding TAF-like proteins have been described (4, 16, 22, 57). Interestingly, the Drosophila homologues of TAF4 (Can), TAF5 (Nht), TAF6 (Mia), TAF8 (Sa), and TAF12 (Rye) are expressed exclusively in the testis and seem to regulate the testis-specific gene expression program in primary spermatocytes (21, 22). Moreover, Drosophila TAF10 homologues (TAF10 and TAF10b) are also differentially expressed during Drosophila embryogenesis (16). Also, a retrotranscribed and integrated copy of human TAF1 (TAF1L) and a homologue of TAF7 (TAF7L) are specifically expressed during male germ cell differentiation (48, 60). A human TAF9 paralogue gene was also identified (termed TAF9L), and it was shown that TAF9L can fully restore the function of chicken TAF9 in DT40 TAF9−/− cells, suggesting redundant function for these two proteins. Furthermore, human TAF9L was suggested to play a role in transcriptional repression and/or silencing (11). However, the association of TAF9L with TBP was not demonstrated.
Genome-wide expression analysis in Saccharomyces cerevisiae indicated that depletion of TAF9 causes a decrease in transcription of about 60 to 65% of the genes (41, 53). Thus, Saccharomyces cerevisiae TAF9 (scTAF9) seems to be broadly, but not universally, required for transcription in yeast. However, the fact that either targeted depletion of TAFs or different temperature-sensitive alleles of a single TAF can either result in broad effects on Pol II transcription (30, 49) or affect the transcription of only a subset of genes (20) has generated a yet-unresolved controversy concerning the true role of TAFs in regulation of transcription. Interestingly, depletion of chicken TAF9 DT40 cells resulted in no or only minor changes in total poly(A)+ mRNA transcription, suggesting that chicken TAF9 is not essential for general mRNA transcription in chicken DT40 cells (12).
Our recent identification of an alternatively spliced TAF6 transcript, which is induced upon different apoptotic stimuli and encodes for TAF6δ that is part of a TFIID-like complex lacking TAF9, showed that the TAF6δ interplay is an important component of a novel signaling pathway that acts as an interface between apoptotic signals and the Pol II transcriptional machinery (5, 18). The notion that TFIID function is important in the regulation of apoptosis was also supported by several previous reports which suggested that various TFIID subunits, including TAF9, may play a role in the regulation of apoptosis (2, 12, 38, 51, 61, 66).
In this study, we have identified TAF9L/DN7 as a bona fide subunit of both TFIID and TFTC, hereafter called TAF9b, and investigated the structural and functional similarities between human TAF9 and TAF9b. We show that TAF9 and TAF9b have specific as well as redundant roles in the regulation of Pol II transcription.
MATERIALS AND METHODS
Plasmid constructions and cell transfections.
The eukaryotic expression plasmids for TAF9, p53, and TAF6δ have been previously described (5); TAF9b cDNA has been provided by the IMAGE Consortium (accession number BC009566) and PCR amplified between nucleotides 23 and 778 with oligonucleotides containing EcoRI at 5′ and NotI at 3′ restriction sites and then cloned into the pXJ41 vector for mammalian expression and pTRIEX (Novagen) for bacterial and insect cell expression. Yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) TAF9b fusions were generated by subcloning TAF9b cDNA from pXJ41 using BamHI and EcoRI in BglII and EcoRI sites of pEYFP-C1 and pECFP-C1 (Clontech), respectively. Subcloning the NheI excised and blunted TAF6 and TAF9 cDNA fragments from pRSET vector (31) into the blunted BamHI in EcoRI sites of pEYFP-C1 and pECFP-C1 vectors, respectively, generated YFP- and CFP-TAF9 as well as YFP- and CFP-TAF6 fusions.
A total of 1.5 × 105 HeLa cells were transfected by using JetPEI (PolyTransfection, France) in 35-mm plates and harvested at the indicated time points after transfection (see below).
Immunization and antibody production.
To generate the anti-TAF9 and TAF9b monoclonal antibodies (MAbs), peptides corresponding to amino acids 132 to 146 of hTAF9 [LQKKASTSAGRITV(C)] and amino acids 132 to 144 of hTAF9b [LIKKGPNQGRLVP(C)] (Fig. 1B, boxed area) were synthesized, coupled to ovalbumin as a carrier, and used for immunization of mice. Immunization and monoclonal antibody production were done essentially as described previously (7).
FIG. 1.
Identification of TAF9b as a component of TFTC by mass spectrometry. (A) TAF9- and TAF9b-specific peptide masses (measured and computed) obtained by MALDI-TOF analysis and their sequences are represented. The accuracy between the measured and computed masses is shown in parts per million (ppm). M-ox, oxidized methionine. (B) Protein sequence comparison between human TAF9 and TAF9b (formerly TAF9L). The amino acid one-letter code is used. Identical amino acids are shown by bars, and double points represent residues with the same physicochemical properties. The peptides used for generating TAF9- or TAF9b (formerly TAF9L)-specific antibodies are boxed. (C) Recombinant TAF9 (rTAF9) (lanes 2 and 4) and TAF9b (rTAF9b) (formerly TAF9L) (lanes 1 and 3) were overexpressed using the baculovirus system in Sf9 cells, cells were lysed, and proteins were separated by SDS-PAGE and tested by Western blot using the antibodies raised against either TAF9 (α-TAF9) (lanes 1 and 2) or TAF9b (α-TAF9b) (lanes 3 and 4), respectively. (D) HeLa cell NE was prepared in the presence of phosphatase inhibitors, and increasing amounts (15, 30, and 60 μg) were separated by SDS-PAGE and tested by Western blot using the antibodies raised against either TAF9 (lanes 1, 2, and 3) or TAF9b (lanes 4, 5, and 6), respectively.
Immunoprecipitation and Western blot analysis.
Routinely, proteins from 200 μg of nuclear extract (NE) were immunoprecipitated (IP) with 50 μl protein G-Sepharose (Pharmacia) and approximately 5 to 8 μg of the different antibodies (as indicated in the figures). Antibody-protein G-Sepharose-bound protein complexes were washed three times with IP buffer (25 mM Tris-HCl, pH 7.9, 10% [vol/vol] glycerol, 0.1% NP-40, 0.5 mM dithiothreitol, 5 mM MgCl2) containing 0.5 M KCl and twice with IP buffer containing 100 mM KCl. After washing, either proteins were eluted by an excess of the corresponding epitope peptide, or protein-G-antibody-bound proteins were directly boiled in sodium dodecyl sulfate (SDS) sample buffer and separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred onto nitrocellulose membrane, and probed with the indicated primary antibodies. Chemiluminescence detection was performed according to the manufacturer's instructions (Amersham). All the other antibodies used were previously described (5, 63). Monoclonal anti-β-actin antibody was purchased from Sigma (A5441).
Apoptosis assays.
Apoptosis induction was obtained and measured as described previously (5) or by measuring sub-G1 DNA content with a Becton Dickinson FACScalibur.
Mass spectrometry.
TFTC subunits were separated on a 10% SDS-PAGE gel. Protein bands were visualized by Coomassie G250 (Bio-Rad) staining, excised, and in-gel digested with trypsin (10). Peptide extracts (0.5 μl) were mixed with an equal volume of saturated α-cyano-4-hydroxycinnamic acid (LaserBio Labs) dissolved in 50% acetonitrile and applied to the target. Mass measurements were carried out on a Bruker Reflex IV matrix-assisted laser desorption-time of flight (MALDI-TOF) spectrometer in the positive-ion reflector mode. The acquisition mass range was 800 to 3,000 Da with the low-mass gate set at 700 Da. Internal calibration was performed using autolytic trypsin peptides (MH+ with m/z = 842.51, 2211.11, and 2807.47). Monoisotopic peptide masses were assigned manually using the Bruker X-TOF software. Database searches were performed using the Profound program (http://prowl.rockefeller.edu/) with the following parameters: NCBI database, proteins of human origin, molecular mass between 20 and 100 kDa, trypsin digestion with one missed cleavage allowed, cysteines modified by carbamidomethylation, methionine oxidation, and mass tolerance of 75 ppm.
FRET measurements.
We used a Leica TCS SP2 (AOBS) confocal microscope operating with a 40-mW argon laser. The laser was tuned to lines of 458 nm and 514 nm. Cells were examined with a 63× 1.40-numerical aperture oil immersion objective and a ×2.5 zoom. Fluorescence resonance energy transfer (FRET) was measured using the acceptor photobleaching method (26). According to this procedure, if two proteins interact and FRET occurs, then photobleaching of the acceptor (YFP) should yield a significant increase in fluorescence of the donor (CFP). We bleached cells in the YFP channel by zooming in and scanning the cell of interest six times using the 514-nm argon laser line at 100% intensity. Before and after the bleaching, CFP and YFP images were collected to assess changes in donor fluorescence that corresponds specifically to the cell with the bleached acceptor. Images were collected at 16% of the 458-nm laser line intensity and 5% of the intensity of the 514-nm laser line. The gain of photon multipliers and the emission intervals were adjusted to eliminate cross talk between the CFP (470 to 500 nm) and YFP (530 to 600 nm) channels. First, we subtracted the background from both CFP and YFP signals measured. To calculate FRET efficiency, we used the EF = (Ipost − Ipre) × 100/Ipost, formula, where EF is the FRET efficiency and the Ipre and Ipost are the mean intensities measured using the Leica software in the CFP channel before and after photobleaching, respectively. As a control, we always performed similar calculations for the nonbleached cells of the image.
Reverse transcriptase PCR (RT-PCR).
Total RNA was isolated with RNAsolv (Biofidal), precipitated, and quantified. One microgram was reverse transcribed with M-MLV reverse transcriptase (Sigma), and the TAF9- and TAF9b-specific PCRs were performed with the following oligonucleotides: at the 5′ end, GCC ACC TGA TAG ATA CTG CTT AAC, common to TAF9 and TAF9b; at the 3′ end for TAF9, GCT AGA TTA CAG ATT ATC ATA GTC; and at the 3′ end for TAF9b, GAG ATC TAA CAG TTT TAA CTG ACG. Other oligonucleotide sequences used in Fig. 8 are available upon request.
FIG. 8.
TAF9 and TAF9b have only minimal overlapping functions. (A) RNA from TAF9, TAF9b, and Luc siRNA-treated cells was used to probe an oligonucleotide microarray representing 25,000 genes. The graph represents the results of three independent microarray experiments. Numbers are expressed as percentages of the 5,974 genes (100%) classified as “present” (see Results). The color code of the graph is represented at the bottom of the panel. (B) Quantitative RT-PCR carried out using RNA samples prepared from TAF9, TAF9b, and control Luc siRNA-treated cells (indicated below each column). For the amplifications, oligonucleotides corresponding to four genes which were found regulated only by TAF9 (upper panel) or TAF9b (lower panel) in the microarray experiments were used. The y axis represents arbitrary units compared to the same standard in each panel. Each qRT-PCR was repeated three times, and the standard deviations are shown. The NCBI accession number of each cDNA is indicated above the corresponding panel. Complete microarray data are available at http://www.ncbi.nlm.nih.gov/projects/geo/.
RNA interference.
Double-stranded siRNAs were produced with a Silencer siRNA kit (Ambion) according to the manufacturer's instructions. Target sequences were obtained with the Dharmacon siDesign engine using the less-stringent parameters. A total of 1.5 × 105 HeLa cells were transfected by using Lipofectamine 2000 (Invitrogen) in 35-mm plates. Sequences of the specific siRNAs are available upon request.
Microarray analysis.
A 25,000-gene human oligonucleotide microarray covering most of the known human transcripts has been used for transcriptome analysis. This oligonucleotide collection is a public resource (Réseau National des Génopoles, France, and MRC, United Kingdom). Information about these oligonucleotides is available at http://medcal.ipmc.cnrs.fr:8080/mediante. Oligonucleotides (synthesized by Proligo) were diluted to 50 μM in 50% dimethyl sulfoxide and 100 mM phosphate buffer and spotted using a μGridII arrayer (BioRobotics) onto amino silane-coated slides (UltraGAPS TM; Corning).
Total RNA from HeLa cells treated with siRNA targeting TAF9, TAF9b, or luciferase (Luc) was extracted (Trizol, Invitrogen). Total RNA (220 ng) was amplified by linear PCR, and the amplification products were labeled by Cy3 and Cy5 and purified using Nucleospin Extract II columns (Macherey Nagel). Labeled cDNAs were hybridized in a Discovery station using ChipHybe 80 hybridization buffer at 42°C for 8 h without any final stringency washes (Ventana Medical System hybridization automate, reagents, and microarray hybridization procedure). All the protocols used are available at http://www-microarrays.u-strasbg.fr/Chips/index.html. After slides scanning (ScanArray4000; Perkin-Elmer), the obtained TIF images were quantified using Imagene 5 (BioDiscovery), and the raw data were normalized using the Lowess method. For each different RNA sample, a flip-flop experiment was performed using the RNA obtained from luciferase RNA interference-treated HeLa cells as a reference. The normalized ratio of RNA sample to reference RNA (log2) was calculated, and this ratio was directly used for the comparison of different samples.
RESULTS
Identification of human TAF9b (formerly TAF9L) as a bona fide subunit of TFIID and TFTC.
To further characterize TFTC subunits, the complex (63) was analyzed by MALDI mass spectrometry (10). As expected, a ∼35-kDa protein species was identified as TAF9 (previously called hTAFII31 or hTAFII32 [29, 35]) with a 41% coverage. However, MALDI peptide mass fingerprinting analysis showed that out of 16 peptides, 4 were TAF9 specific (Fig. 1A) and 9 were common with a human factor that has previously been named TAF9Like (TAF9L; GenBank accession number NP_057059) or neuronal cell death-related protein (DN7). TAF9 and TAF9L proteins are 82% identical (Fig. 1B) and encoded by two different human paralogue genes. Interestingly, accurate analysis of the TAF9 MALDI peptide mass fingerprinting also revealed the presence of three TAF9L-specific peptides (Fig. 1A). These results indicated that TAF9L comigrates with TAF9 and that TAF9L is a component of TFTC.
To confirm the presence of TAF9L in TFTC, we raised anti-peptide mouse MAbs against TAF9L (see Materials and Methods and Fig. 1B). In parallel, we also raised specific MAbs against hTAF9. When these antibodies were tested on recombinant TAF9 and TAF9L proteins, they showed high specificity and no cross-reaction (Fig. 1C). A further test on a HeLa NE prepared in the presence of phosphatase inhibitors showed that anti-TAF9 and anti-TAF9L antibodies recognized protein species migrating around 32 kDa, with a slightly different molecular weight (Fig. 1D). Moreover, the anti-TAF9L antibody recognized a single band, while the anti-TAF9 antibody recognized two bands (Fig. 1D). When phosphatase inhibitors were omitted during the NE preparation, the slower-migrating TAF9 band disappeared (data not shown), indicating a possible in vivo phosphorylation of TAF9.
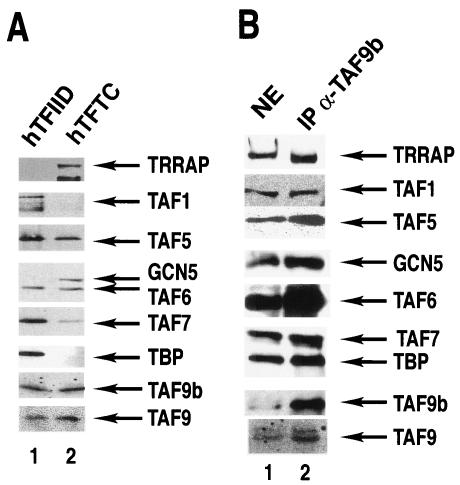
We then verified the presence of TAF9L in highly purified human TFTC and tested whether it is present in TFIID complexes (63) by Western blot analysis using the anti-TAF9L-specific MAb. In good agreement with the MALDI spectrometry data, TAF9L was present in TFTC, and in addition, we demonstrate that it is also present in highly purified TFIID preparations (Fig. 2A). Thus, according to the novel TAF nomenclature (57), the TAF9L protein will hereafter be called TAF9b. Interestingly, both TAF9b and TAF9 were detected in either TFIID or TFTC preparations.
FIG. 2.
TAF9b is a bona fide TAF present in both TFIID and TFTC. (A) TFIID and TFTC complexes were purified (63) and separated by SDS-PAGE, and the presence of different subunits was analyzed by Western blot using indicated antibodies. (B) HeLa cell NE was prepared in the presence of phosphatase inhibitors, and TAF9b-containing complexes were immunoprecipitated (IP) with a mouse MAb raised against TAF9b (IP α-TAF9b). Antibody-bound proteins were eluted in SDS-loading buffer, separated by SDS-PAGE, and analyzed by Western blot with indicated monoclonal antibodies.
To prove unequivocally that TAF9b is a subunit of both TFTC and TFIID, the anti-TAF9b MAb has been used to immunoprecipitate proteins which associate with TAF9b in different multiprotein complexes (Fig. 2B). The analysis of the proteins which coimmunoprecipitated with TAF9b revealed that TFTC-specific subunits (i.e., GCN5 and TRRAP) and TFIID-specific subunits (TBP and TAF1) as well as common subunits (TAF5, TAF6, TAF9, and TAF10) associate with TAF9b. These results together show that TAF9b is a stable subunit of both TFTC and TFIID. The fact that TAF9b copurifies with TBP in an anti-TBP immunoprecipitation (Fig. 2A) and TBP coprecipitates with TAF9b (Fig. 2B) indicates that TAF9b is a bona fide TAF. Furthermore, the immunoprecipitation using an anti-TAF9b antibody also copurified TAF9 (Fig. 2B), even in the presence of ethidium bromide (data not shown), strongly suggesting that in HeLa cells, there are TAF-containing complexes (TFIID and/or TFTC) in which TAF9 and TAF9b are simultaneously present.
TAF9 and TAF9b are both coexpressed in multiple cell lines.
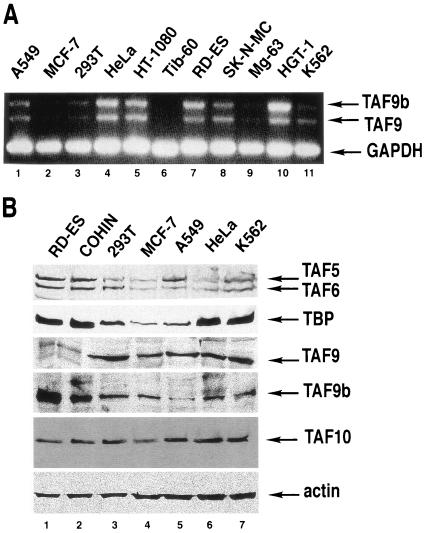
Next, we investigated whether TAF9 and TAF9b are similarly expressed at either the RNA or protein level in different human cell lines. To determine the relative ratio of TAF9 and TAF9b mRNA, RT-PCRs were carried out by using a common 5′ and two different specific 3′ oligonucleotides which amplify either a 550-bp fragment on TAF9b cDNA or a 445-bp band on TAF9 cDNA. RT-PCR analysis revealed that both TAF9b and TAF9 were expressed at the RNA level in the different cell lines tested; however, expression levels and the ratio between the two mRNAs are variable from cell line to cell line (Fig. 3A). These findings were in good agreement with data from the Symatlas databank concerning TAF9 and TAF9b mRNA expression (http://symatlas.gnf.org/SymAtlas).
FIG. 3.
Both TAF9 and TAF9b are expressed at the mRNA and protein levels in several human cell lines. (A) RNA was prepared from the indicated human cell lines, and RT-PCR analysis was carried out with common 5′ primers specific to TAF9 and TAF9b mRNA and with different 3′ primers specific for each transcript. In addition, primers amplifying the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcript were used to normalize the amount of RNA used. Tib60 is a mouse cell line and thus should be considered a negative specificity control in this experiment. PCR products were analyzed on a 1.5% agarose gel. (B) Whole-cell extracts were prepared from the indicated cell lines, and then protein samples were analyzed by Western blot with the indicated antibodies. Protein levels were normalized using an anti-β-actin monoclonal antibody.
We then performed Western blot analysis on whole-cell extracts from certain cell lines tested by RT-PCR (Fig. 3B). In all of the tested protein extracts, both TAF9b and TAF9 were detected, and their ratio appeared to be relatively constant, with two exceptions. We found that in the two Ewing's sarcoma cell lines (RD-ES and COHIN), TAF9 is expressed at much lower levels than TAF9b (Fig. 3B, lanes 1 and 2). No clear correlation has been found between mRNA and protein levels detected, indicating that their expression is regulated at multiple steps (Fig. 3A and B). Surprisingly, the protein expression levels of different TAFs and TBP seem also to be highly variable between the different cell lines tested (Fig. 3B).
In vitro and in living cells, TAF9b interacts with TAF6 similarly to TAF9.
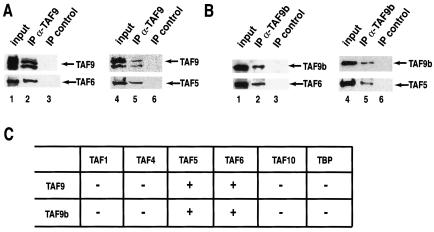
TAF9 has been reported to interact in vitro with TAF6 (23) and TAF5 (54). To verify whether TAF9b interacts with these and other TAF subunits in a way similar to TAF9, TAF9b was coexpressed with a series of TAFs, including TAF5 and TAF6, in insect Sf9 cells using the baculovirus system (Fig. 4A). In parallel, TAF9 was also coexpressed with the same TAFs as a control. Using either TAF9- or TAF9b-specific antibodies to coimmunoprecipitate the interacting proteins, we show that TAF9b interacts with its putative histone fold dimerization partner, TAF6, and with the WD-40 repeat containing TAF5 similarly as TAF9 (Fig. 4A and B). Moreover, TFIID subunits which do not interact in vitro with TAF9 do not interact with TAF9b either (Fig. 4C and data not shown).
FIG. 4.
TAF9b interacts with the same partners, as does TAF9, in vitro. (A) TAF9 or (B) TAF9b was coexpressed with the indicated TAFs (TAF5 and TAF6) in Sf9 cells using the baculovirus expression system. From these cells, protein extracts were prepared, and either TAF9 (A) or TAF9b (B) was immunoprecipitated (IP) with the corresponding antibodies. From the same extracts, a control IP was performed with an unrelated antibody. Resin-bound proteins were then washed, boiled, separated by SDS-PAGE, and analyzed by Western blot using the indicated antibodies. (C) Summary of the interaction study carried out using the baculovirus expression system between either TAF9 or TAF9b and the indicated TFIID subunits.
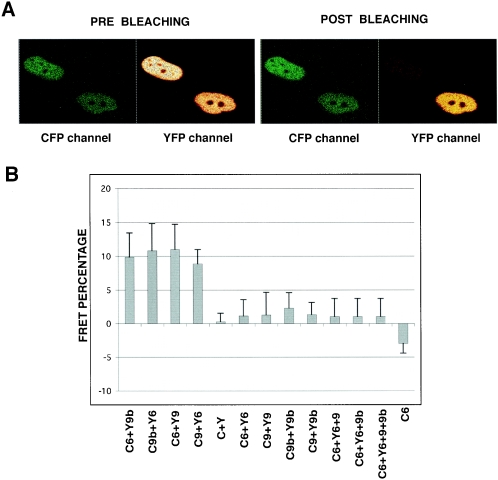
Next, we aimed to verify whether these in vitro TAF9b/TAF6 and TAF9/TAF6 interactions also exist in living cells. To this end, we used FRET and the acceptor photobleaching method. HeLa cells were transfected with constructs expressing the YFP-TAF6/CFP-TAF9, CFP-TAF6/YFP-TAF9, YFP-TAF6/CFP-TAF9b, and CFP-TAF6/YFP-TAF9b pairs, and mean FRET efficiencies were measured in 60 cells from three different experiments for each combination. Both the TAF6/TAF9 and TAF6/TAF9b interactions resulted in similar FRET efficiencies, at around 11% (Fig. 5B). These FRET efficiencies were the same when the two fluorophores were exchanged on the different pairs (Fig. 5B, compare C6 plus Y9b to C9b plus Y6 and C6 plus Y9 to C9 plusY6), indicating that the nature of the fluorescent protein tags did not influence the interactions. When measuring FRET with the control CFP/YFP pair, where the two fluorescent proteins are not supposed to interact, we obtained only 0.4% efficiency (Fig. 5B). In a further control experiment, when cells were transfected with the CFP-TAF6 construct alone, we obtained negative numbers that are the effect of photobleaching of the CFP fluorescence due to imaging (C6 in Fig. 5), pointing to the fact that the FRET efficiencies of the TAF6-TAF9 and TAF6-TAF9b pairs may have been underestimated. In contrast to the TAF6-TAF9 and TAF6-TAF9b pairs, the combinations among YFP-TAF6/CFP-TAF6,YFP-TAF6/CFP-TAF6/TAF9/TAF9b, CFP-TAF9b/YFP-TAF9b, CFP-TAF9/YFP-TAF9, and CFP-TAF9/YFP-TAF9b did not result in significant FRET efficiency (around 1 to 2%), suggesting that, at least with this method, no (TAF6-TAF9)2, (TAF6-TAF9b)2, or TAF62-TAF9-TAF9b heterotetramerstructures similar to that of histones H3-H4 can be observed. Note, however, that the absence of a FRET signal between two proteins does not necessarily mean that the proteins do not interact, because the orientation of the CFP or YFP tags on the TAFs may not be optimal for FRET detection in the case of the heterotetramer. In the latter experiments, when more than two proteins have been overexpressed, the third and fourth partners were expressed as nonfluorescent proteins (Fig. 5). These data together indicate that TAF6-TAF9 and TAF6-TAF9b pairs form in vivo.
FIG. 5.
TAF9b interacts with TAF6, similarly to TAF9, in vivo. Acceptor photobleaching of cells transfected with the indicated combinations of vectors expressing YFP (Y) and CFP (C) fusion proteins with TAF6 (6), TAF9 (9), and TAF9b (9b). (A) Representative CFP-TAF6 (donor) channel and YFP-TAF9b (acceptor) channel images before and after bleaching are shown. Note the dramatic drop in YFP fluorescence and the increase in CFP fluorescence in the cell after bleaching by the 514-nm laser line (the cell in the upper left corner of the panels). The cell in the lower right corner of the panels was not bleached and serves as a control. (B) Mean values of FRET efficiencies following quantification of the increase of the CFP fluorescence intensity after bleaching in 60 cells from three different experiments. To calculate FRET efficiency in living cells, the following formula was used: EF = (Ipost − Ipre) × 100/Ipost. The mean values for the FRET efficiency of TAF6-TAF9 (C6+Y9 or Y6+C9) and TAF6-TAF9b (C6+Y9b or Y6+C9b) combinations (first four bars) are similar to each other and significantly different from the empty CFP-YFP vector combination (C+Y) or CFP-TAF6 alone (C6), suggesting that TAF6-TAF9 and TAF6-TAF9b interact in living cells. In contrast, no significant interactions were detected between YFP-TAF6 and CFP-TAF6 (Y6+C6), CFP-TAF9 and YFP-TAF9 (C9+Y9), CFP-TAF9b and YFP-TAF9b (C9b+Y9b), and CFP-TAF9 and YFP-TAF9b as well as among YFP-TAF6; CFP-TAF6 and TAF9 (C6+Y6+9); YFP-TAF6, CFP-TAF6, and TAF9b (C6+Y6+9b); and YFP-TAF6, CFP-TAF6, TAF9, and TAF9b (C6+Y6+9+9b). In the three latter transfections, TAF9 and TAF9b vectors expressed these two proteins without their fluorescent protein tags.
TAF9 and TAF9b are differentially regulated during apoptosis induction.
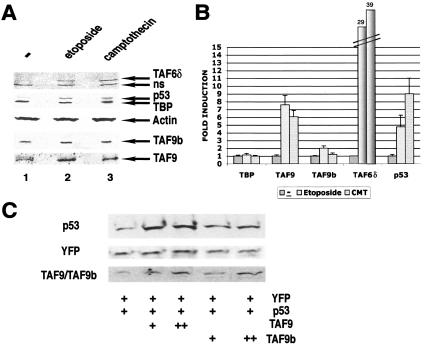
Since elevated levels of TAF9b RNA have been measured in apoptotic PC12 rat cells (2), we tested whether TAF9 or TAF9b protein levels are also induced by apoptotic stimuli in human HeLa cells. To induce apoptosis, VP16-etoposide or camptothecin was used, and induction was confirmed by testing p53 and TAF6δ levels by Western blot analysis (5) (Fig. 6A). In parallel, we have monitored whether we can detect an increase in TAF9 or TAF9b protein levels in these apoptotic cells (Fig. 6A). After both apoptotic stimuli, TAF9 protein expression levels increased about 6- to 8-fold, while TAF9b levels increased only about 1.5-fold (Fig. 6A and B), suggesting that their function is differentially required during apoptosis.
FIG. 6.
TAF9 and TAF9b are differently induced by apoptotic stimuli and involved in p53 stabilization. (A) HeLa cells were treated with etoposide VP16 (Et-VP16; 68 μM) or camptothecin (CMT; 15 μM). Nuclear extracts were prepared after no treatment (−) or the indicated treatment for 6 h and analyzed by SDS-PAGE followed by immunoblotting with the indicated antibodies. The samples were normalized by loading the same amount of actin. ns, nonspecific. (B) Three independent experiments as shown in panel A were densitometrically scanned, and the induction (n-fold) of the expression of different indicated proteins (compared to actin) is represented in the graph. The induction of TAF6δ is indicated on the top of the corresponding bars. Error bars are shown. (C) MCF7 cells were transfected with an expression vector for p53 alone or cotransfected with increasing amounts of TAF9 or TAF9b expression vectors. Transfection efficiency was monitored by cotransfecting pEYFP. Total cell extracts were prepared after 48 h and analyzed by SDS-PAGE followed by immunoblotting for the presence of p53 and YFP. TAF9 and TAF9b were detected with a polyclonal antibody that recognizes both proteins.
TAF9b overexpression has a weaker effect on p53 stabilization than TAF9.
To further investigate functional differences between TAF9 and TAF9b, we used a previously described assay system where it was shown that overexpressed TAF9 is able to protect p53 from degradation due to the fact that it competes with MDM2 for p53 binding (9, 24). As shown in Fig. 6C, TAF9b overexpression has a weaker effect on p53 stabilization than TAF9 in MCF7 and HeLa cells (data not shown). These results further suggest that despite the 82% identity between these two proteins, they have different functions.
TAF9 and TAF9b are essential for cell viability.
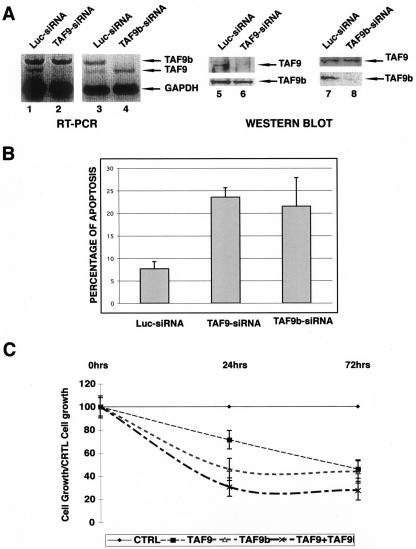
Previously, it was shown that TAF9b (formerly TAF9L) is essential for cell growth in human cells (11) and that TAF9 depletion leads to apoptotic cell death in chicken cells (12). To further characterize the functional differences between human TAF9 and TAF9b during normal cell growth and in the same system, we specifically knocked down either TAF9 or TAF9b expression by using siRNAs. When TAF9 or TAF9b siRNAs were transfected in HeLa cells, we obtained a specific reduction of the targeted mRNAs and the consequent protein levels without affecting the expression of the other homologue (Fig. 7A and B). Moreover, the almost complete elimination of one or the other factor resulted in a significant induction (about threefold) of apoptosis over control siRNA transfection (Fig. 7B). When the number of cells was scored in samples which had been treated with either control, TAF9, TAF9b, or TAF9 plus TAF9b siRNAs, we observed that 24 h after transfection, the TAF9b siRNA reduced the growing cell population by 54% (Fig. 7C). This reduction was almost as efficient as the TAF9 and TAF9b siRNAs together (60%), while the TAF9 siRNA had only a mild effect (25%) on cell survival (Fig. 7C). However, at the 72-h time point, both TAF9 and TAF9b siRNAs induced apoptosis in a similar way compared to the control (58%). Interestingly, at this time point, the TAF9 and TAF9b siRNAs together had a stronger effect (70%) on cell death than the individual siRNAs alone (Fig. 7C). These results suggest that both genes are essential for cell viability, and the results obtained with the double TAF9 and TAF9b siRNA treatment suggest that their roles are only partially redundant.
FIG. 7.
Specific down-regulation of TAF9 and TAF9b mRNAs by siRNA indicates that both factors are necessary for cell survival. (A) HeLa cells were transfected with control siRNA (Luc-siRNA; lanes 1, 3, 5, and 7) and siRNA specific for either TAF9 (TAF9-siRNA; lane 2 and 6) or TAF9b (TAF9b-siRNA; lanes 4 and 8). Forty-eight hours after transfection, cells were harvested, and either RNA or protein extracts were prepared. The RT-PCRs were carried out and analyzed as described in the legend of Fig. 3A. Primers amplifying the GAPDH transcript were used to normalize the amount of RNA used. Twenty micrograms of total protein extracts from each sample was analyzed by Western blots with the indicated antibodies. (B) Percentage of apoptosis induced by transfection of different siRNAs (as indicated) was determined by flow cytometry 48 h after transfection measuring sub-G1 DNA content. The error bars give the standard deviations of three independent transfections. (C) The effect on cell growth induced by transfection of control (CTRL), TAF9, TAF9b, and TAF9 plus TAF9b siRNAs was determined by measuring sub-G1 DNA content by flow cytometry at 0, 24, and 72 h after transfection. The graph represents the ratios obtained by dividing the number of control siRNA (Luc-siRNA)-treated cells at the different time points with that of the different indicated siRNA-treated samples. Error bars give the standard deviations of two independent experiments.
TAF9 and TAF9b have only minimal overlapping functions.
To further investigate gene regulation by TAF9 and TAF9b, we performed gene expression profiling in siRNA knockdown cells. HeLa cells were treated, in triplicate, with control Luc siRNA, TAF9 siRNA, or TAF9b siRNA. The treatment was carried out for 30 h to be able to analyze the expression of genes, which are primarily affected by the knockdowns, and to obtain good quality RNA that is not affected by the induced cell death (Fig. 7). Total RNA was extracted, retrotranscribed, labeled, and used to probe an oligonucleotide-based microarray representing 25,000 genes (see Materials and Methods). In order to reduce false positives due to oscillations of weak signals, we qualified only those genes whose average signal intensities of Cy3 and Cy5 as log2 were over 10 in the luciferase samples as “present.” After this passage, we were able to qualify 5,974 genes as “present.” We then compared TAF9 siRNA- and TAF9b siRNA-treated samples to Luc siRNA-treated samples in order to obtain up- and down-regulated genes and to eliminate genes that were not specifically affected by the siRNA treatment. Out of 5,974 “present” genes, a total of 1,081 were affected (18%). Out of this total, 222 genes (3.7%) were affected by the TAF9 knockdown; 689 genes (11.6%) were affected by the TAF9b knockdown, and 160 genes (2.7%) were common between the TAF9 and TAF9b knockdowns. Out of all 387 TAF9 knockdown-affected genes, 177 (2.9%) were up-regulated and 210 (3.6%) were down-regulated; out of the 854 TAF9b knockdown-affected genes, 150 (2.5%) were up-regulated, whereas 704 (11.8%) were down-regulated. Affected genes do not fall into any specific category or pathway, suggesting that TAF9 and TAF9b are not restricted to any specific gene expression program and that they rather function in a more general manner. The two most interesting groups of the genes are those whose expression is only either TAF9 or TAF9b dependent. We chose some of these genes to perform quantitative RT-PCR (qRT-PCR) (Fig. 8B) to validate the microarray results. The qRT-PCR data confirmed the microarray data (Fig. 8B), further suggesting that only TAF9- or TAF9b-regulated genes exist and that the two proteins regulate only partially overlapping transcription programs. In agreement with several previous studies (11, 20, 27, 28, 32, 53), our results also demonstrate that TAF depletion (either TAF9 or TAF9b) causes both up- and down-regulation of gene expression.
DISCUSSION
TAFs have been identified in biochemically stable complexes with TBP, whereas TAF-like (TAFL) proteins are those which are present in multiprotein complexes without TBP or which were not yet shown to be associated in stable complexes with TBP (57). In our study, we demonstrate that the previously described TAF9L factor (11) is a bona fide member of the TAF family since it is associates with TBP in TFIID complexes. Thus, according to the existing nomenclature, it is called TAF9b. In contrast to most of the other known TAF homologues, which usually replace their counterparts during defined spatiotemporal developmental events (14, 21, 48, 60), TAF9b is ubiquitously expressed, together with TAF9, in all cell types tested. We show here that in the same cell type, TAF-containing complexes in which TAF9 and TAF9b are simultaneously present exist. In the light of this finding and despite the high degree of identity between the two proteins, it is surprising that their functions are only partially overlapping (see below).
How are TAF9 and TAF9b positioned in the TFIID/TFTC structure?
Immunoprecipitations using anti-TAF9b antibodies demonstrated that at least some of the TAF9b-containing complexes also contain TAF9. These data suggest that in certain TAF-containing complexes, both proteins can be present and that their presence is not mutually exclusive in TFIID and TFTC complexes. We have shown previously that in Saccharomyces cerevisiae TFIID, of which the genome contains only one TAF9 gene, TAF9 can be present in two copies, one in lobe A and one in lobe B (34). Moreover, in the SAGA complex, which is the yeast counterpart of the human TFTC-type complexes, TAF9 seems to be present in two copies as well, one in domain II and another one in domain IV (56, 64). These findings open the interesting possibility that in mammals, where two TAF9 paralogue genes exist in the different genomes, TAF9 would be present in lobe A (or domain II) and TAF9b would be present in lobe B (or domain IV) of TFIID (of TFTC), or vice versa. However, it is also conceivable that in mammalian cells, different TFIID and TFTC complexes may exist that contain (i) only TAF9, (ii) only TAF9b, (iii) TAF9 in lobe A (or domain II) and TAF9b in lobe B (or domain IV in TFTC), and (iv) TAF9b in lobe A (or domain II) and TAF9 in lobe B (domain IV in TFTC). Despite our efforts to identify the precise location of TAF9 and TAF9b in human TFIID using electron microscopy, we could not distinguish among the above-mentioned four possibilities. Concerning the overall structure of the different TAF-containing complexes, our FRET experiments seem to exclude the formation of a heterotetramer between TAF6-TAF9 and/or TAF6-TAF9b in living cells, suggesting that in TFIID and TFTC complexes, TAF6-TAF9 and TAF6-TAF9b exist as heterodimers in the distinct lobes. Nevertheless, in the future, it will be important to understand how the incorporation of the TAF9/TAF6 and TAF9b/TAF6 heterodimers in the different domains of the distinct TAF complexes is regulated in spite of the high degree of identity between TAF9 and TAF9b (82%). As TAF9 was found to be phosphorylated (reference 37 and this study), it is possible that the incorporation of TAF9 (and TAF9b) in the different domains of the complexes is regulated by distinct posttranslational modifications. If TAF9 and TAF9b are present in different complexes or domains of the same complex, the above-described functional differences between TAF9 and TAF9b suggest that the complexes or domains containing one or the other protein will have distinct functions.
As in the in vitro and in vivo interaction tests, where TAF9 and TAF9b interacted with their partners (TAF6 and TAF5) in a comparable way, it is conceivable that TAF9/TAF6 and TAF9b/TAF6 heterodimers will have very similar interactions within the different lobes or domains of the TAF-containing complexes. However, they may have differential interactions outside of the respective complexes with general transcription factors, transcription activators, and/or promoter sequences. The Drosophila HF pair TAF9/TAF6 has been shown to bind to downstream promoter elements (8). More recently, the DNA-binding domain of human TAF9 has been assigned to an evolutionarily conserved region (between amino acids 70 and 147 of TAF9), and it was shown that the HF domain-mediated interaction between TAF9 and TAF6 enhanced the DNA binding activity of this heterodimer (52). Thus, in the future, it will be important to test whether TAF9b-containing complexes would have a different DNA binding specificity than that of TAF9-containing complexes.
TAF-containing complexes with only TAF9 or only TAF9b.
The fact that the ratio between endogenous TAF9 and TAF9b is far from being 1:1 in Ewing's sarcoma cell lines suggests that at least in these cells, different TAF-containing complexes exist, and those which contain both TAF9 and TAF9b are less abundant than those which contain only TAF9b. In addition, TAFs and TBP do not show a uniform expression pattern in different cells but are expressed at very different levels in distinct cell types or tissues (references 40 and 47 and this study). Thus, our results are in good agreement with recent data suggesting that TFIID and TFTC are not unique complexes but rather a family of complexes that can be modulated depending the transcriptional requirements of the different cell types that compose an organism (5, 6, 43).
Eighty-two percent identical TAF homologues with only partially overlapping functions.
Interestingly, two TAF9 paralogue genes were found only in mammalian genomes and not in fully sequenced invertebrate or other vertebrate genomes, i.e., Drosophila, zebra fish, Xenopus laevis, and chicken (our unpublished observations). At present, however, it is difficult to determine when the TAF9 gene duplication occurred and whether it is a mammal-specific event or it appeared at an earlier vertebrate evolutionary stage. The general rule for the retention of a duplicated gene copy is either divergence in expression pattern and/or differences in functions. As TAF9 and TAF9b are coexpressed in the different cell lines tested, it would rather suggest that the duplication of the TAF9 gene in mammals served a new function. TAF9 and TAF9b have been the subject of different evolutionary pressures, and this led to a series of mutations in TAF9b which resulted in partially novel functions. While the TAF9 functions seem to be conserved from yeast to humans, TAF9b might have evolved to serve functions that may tolerate more diversity, suggesting that TAF9b is not necessarily involved in exactly the same regulatory pathways as TAF9.
Nevertheless, the relatively high similarity between human TAF9 and TAF9b would suggest rather redundant functions. However, when the different results obtained with these two proteins are compared, and in agreement with a new function that was retained following gene duplication in mammals, the following important functional differences were found: (i) distinct apoptotic stimuli do not induce the expression of the two proteins to the same levels, although they induce similar levels of apoptosis, suggesting that they are differentially required during induction of apoptotic pathways (Fig. 6A); (ii) exogenous expression of the two proteins does not protect p53 in the same way from Mdm2-mediated degradation (Fig. 6B); (iii) knocking down either TAF9 or TAF9b led to cell death, but the double knockdown has a stronger effect than the individual knockdowns alone (Fig. 7); and (iv) the analysis of genes affected by the two different knockdowns revealed that only a rather limited subset of genes is regulated by both TAF9 and TAF9b (Fig. 8). All these differences together indicate that the two proteins do not fulfill identical roles in the cell but also serve different functions.
Knockdown of TAF9 and TAF9b revealed that they are both indispensable for human HeLa cell survival and that each plays an essential role (Fig. 7). Gene expression profiling revealed that the two proteins control the expression of different sets of genes and that, surprisingly, there is only a small subset of genes which are regulated by both TAF9 and TAF9b. Whether the different functions of TAF9 and TAF9b reflect their distinct (i) structural interactions within TFIID and/or TFTC complexes, (ii) interactions with transcription factors outside of the complexes, and/or (iii) DNA binding properties has to be further investigated. Unfortunately, our antibodies against TAF9 and TAF9b do not work in chromatin IP and thus do not allow us to verify whether the identified TAF9- or TAF9b-dependent genes are indeed bound by only one or both factors. The finding that there are genes regulated only by TAF9 or TAF9b suggests that in HeLa cells, TAF9 cannot structurally and/or functionally replace TAF9b and vice versa. Surprisingly, human TAF9b (formerly hTAF9L) could restore the viability of the chicken DT40 TAF9−/− cells (11), suggesting that in chicken cells, only one TAF9 gene exists and that at this stage of evolution, TAF9 and TAF9b functions were not yet separated. Our results also demonstrate that TAF depletion (either TAF9 or TAF9b) causes both up- and down-regulation of gene expression, further suggesting that TAFs can act as both positive and negative regulatory factors. This finding together with other TAF knockdown studies carried out mainly in yeast (11, 20, 27, 28, 32, 53) raise an interesting hypothesis that TAFs, when binding to promoters in the different TAF-containing complexes (TFIID and TFTC type), would be involved at certain promoters as repressors and at others as positive cofactors of transcription.
Acknowledgments
We are grateful to M. Oulad-Abdelghani for generating specific TAF9 and TAF9b antibodies, to E. Martinez and R. G. Roeder for the polyclonal antibody that recognizes both TAF9 and TAF9b, to I. Kolb-Cheynel for help with the recombinant baculoviruses, to P. Schultz and C. Ruhlman for the EM study, to A. Jànoshàzi for training to do the FRET experiments, and to T. Hilton, M. Demény, S. Brancorsini, and N. Clarke for critical reading of the manuscript. We also thank the IGBMC cell culture facility for providing cells.
M.F. is supported by a European Community Marie Curie individual fellowship (MEIF-CT-2003-501146), and E.S. is supported by an EMBO long-term fellowship. This work was supported by funds from INSERM, CNRS, Hôpital Universitaire de Strasbourg, Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, the Font Nationale de La Science ACI, INTAS (01-0211), and European Community RTN (HPRN-CT-2000-00087, HPRN-CT-2000-00088, and HPRN-CT-2004-504228), STREP (LSHG-CT-2004-502950), and AICR (03-084) grants.
REFERENCES
- 1.Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1-13. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, T., T. Koike, T. Nakano, K. Shibahara, H. Nishimura, H. Kikuchi, and T. Honjo. 1997. Rat TAFII31 gene is induced upon programmed cell death in differentiated PC12 cells deprived of NGF. Biochem. Biophys. Res. Commun. 234:230-234. [DOI] [PubMed] [Google Scholar]
- 3.Aoyagi, N., and D. A. Wassarman. 2001. Developmental and transcriptional consequences of mutations in Drosophila TAFII60. Mol. Cell. Biol. 21:6808-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoyagi, N., and D. A. Wassarman. 2000. Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: diversity in TFIIA and TFIID components contributes to gene-specific transcriptional regulation. J. Cell Biol. 150:F45-F50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, B., E. Scheer, and L. Tora. 2001. Identification of hTAF(II)80 delta links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8:591-600. [DOI] [PubMed] [Google Scholar]
- 6.Bell, B., and L. Tora. 1999. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res. 246:11-19. [DOI] [PubMed] [Google Scholar]
- 6a.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274:18285-18289. [DOI] [PubMed] [Google Scholar]
- 7.Brou, C., S. Chaudhary, I. Davidson, Y. Lutz, J. Wu, J. M. Egly, L. Tora, and P. Chambon. 1993. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 12:489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke, T. W., and J. T. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buschmann, T., Y. Lin, N. Aithmitti, S. Y. Fuchs, H. Lu, L. Resnick-Silverman, J. J. Manfredi, Z. Ronai, and X. Wu. 2001. Stabilization and activation of p53 by the coactivator protein TAFII31. J. Biol. Chem. 276:13852-13857. [DOI] [PubMed] [Google Scholar]
- 10.Cavusoglu, N., M. Brand, L. Tora, and A. Van Dorsselaer. 2003. Novel subunits of the TATA binding protein free TAFII-containing transcription complex identified by matrix-assisted laser desorption/ionization-time of flight mass spectrometry following one-dimensional gel electrophoresis. Proteomics 3:217-223. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Z., and J. L. Manley. 2003. In vivo functional analysis of the histone 3-like TAF9 and a TAF9-related factor, TAF9L. J. Biol. Chem. 278:35172-35183. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Z., and J. L. Manley. 2000. Robust mRNA transcription in chicken DT40 cells depleted of TAFII31 suggests both functional degeneracy and evolutionary divergence. Mol. Cell. Biol. 20:5064-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dikstein, R., S. Zhou, and R. Tjian. 1996. Human TAFII105 is a cell type-specific TFIID subunit related to hTAFII130. Cell 87:137-146. [DOI] [PubMed] [Google Scholar]
- 14.Freiman, R. N., S. R. Albright, S. Zheng, W. C. Sha, R. E. Hammer, and R. Tjian. 2001. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293:2084-2087. [DOI] [PubMed] [Google Scholar]
- 15.Gangloff, Y., C. Romier, S. Thuault, S. Werten, and I. Davidson. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26:250-257. [DOI] [PubMed] [Google Scholar]
- 16.Georgieva, S., D. B. Kirschner, T. Jagla, E. Nabirochkina, S. Hanke, H. Schenkel, C. de Lorenzo, P. Sinha, K. Jagla, B. Mechler, and L. Tora. 2000. Two novel Drosophila TAFIIs have homology with human TAFII30 and are differentially regulated during development. Mol. Cell. Biol. 20:1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgieva, S., E. Nabirochkina, F. J. Dilworth, H. Eickhoff, P. Becker, L. Tora, P. Georgiev, and A. Soldatov. 2001. The novel transcription factor e(y)2 interacts with TAFII40 and potentiates transcription activation on chromatin templates. Mol. Cell. Biol. 21:5223-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill, G. 2001. Death signals changes in TFIID. Mol. Cell 8:482-484. [DOI] [PubMed] [Google Scholar]
- 19.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates, and J. L. Workman. 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 20.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 21.Hiller, M., X. Chen, M. J. Pringle, M. Suchorolski, Y. Sancak, S. Viswanathan, B. Bolival, T. Y. Lin, S. Marino, and M. T. Fuller. 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131:5297-5308. [DOI] [PubMed] [Google Scholar]
- 22.Hiller, M. A., T. Y. Lin, C. Wood, and M. T. Fuller. 2001. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann, A., C. M. Chiang, T. Oelgeschlager, X. Xie, S. K. Burley, Y. Nakatani, and R. G. Roeder. 1996. A histone octamer-like structure within TFIID. Nature 380:356-359. [DOI] [PubMed] [Google Scholar]
- 24.Jabbur, J. R., A. D. Tabor, X. Cheng, H. Wang, M. Uesugi, G. Lozano, and W. Zhang. 2002. Mdm-2 binding and TAF(II)31 recruitment is regulated by hydrogen bond disruption between the p53 residues Thr18 and Asp21. Oncogene 21:7100-7113. [DOI] [PubMed] [Google Scholar]
- 25.Jacq, X., C. Brou, Y. Lutz, I. Davidson, P. Chambon, and L. Tora. 1994. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell 79:107-117. [DOI] [PubMed] [Google Scholar]
- 26.Karpova, T. S., C. T. Baumann, L. He, X. Wu, A. Grammer, P. Lipsky, G. L. Hager, and J. G. McNally. 2003. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 209:56-70. [DOI] [PubMed] [Google Scholar]
- 27.Kirchner, J., S. L. Sanders, E. Klebanow, and P. A. Weil. 2001. Molecular genetic dissection of TAF25, an essential yeast gene encoding a subunit shared by TFIID and SAGA multiprotein transcription factors. Mol. Cell. Biol. 21:6668-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirschner, D. B., E. vom Baur, C. Thibault, S. L. Sanders, Y. G. Gangloff, I. Davidson, P. A. Weil, and L. Tora. 2002. Distinct mutations in yeast TAFII25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns. Mol. Cell. Biol. 22:3178-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klemm, R. D., J. A. Goodrich, S. Zhou, and R. Tjian. 1995. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc. Natl. Acad. Sci. USA 92:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komarnitsky, P. B., B. Michel, and S. Buratowski. 1999. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 13:2484-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroll, D. J., H. Abdel-Malek Abdel-Hafiz, T. Marcell, S. Simpson, C. Y. Chen, A. Gutierrez-Hartmann, J. W. Lustbader, and J. P. Hoeffler. 1993. A multifunctional prokaryotic protein expression system: overproduction, affinity purification, and selective detection. DNA Cell Biol. 12:441-453. [DOI] [PubMed] [Google Scholar]
- 32.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 33.Leurent, C., S. Sanders, M. Demény, K. A. Garbett, C. Ruhlmann, A. P. Weil, L. Tora, and P. Schultz. 2004. Mapping key functional sites within yeast TFIID. EMBO J. 23:719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leurent, C., S. Sanders, C. Ruhlmann, V. Mallouh, P. A. Weil, D. B. Kirschner, L. Tora, and P. Schultz. 2002. Mapping histone fold TAFs within yeast TFIID. EMBO J. 21:3424-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, H., and A. J. Levine. 1995. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc. Natl. Acad. Sci. USA 92:5154-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez, E. 2002. Multi-protein complexes in eukaryotic gene transcription. Plant Mol. Biol. 50:925-947. [DOI] [PubMed] [Google Scholar]
- 37.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metzger, D., E. Scheer, A. Soldatov, and L. Tora. 1999. Mammalian TAF(II)30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J. 18:4823-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsuzawa, H., H. Seino, F. Yamao, and A. Ishihama. 2001. Two WD repeat-containing TATA-binding protein-associated factors in fission yeast that suppress defects in the anaphase-promoting complex. J. Biol. Chem. 276:17117-17124. [DOI] [PubMed] [Google Scholar]
- 40.Mohan, W. S., II, E. Scheer, O. Wendling, D. Metzger, and L. Tora. 2003. TAF10 (TAFII30) is necessary for TFIID stability and early embryogenesis in mice. Mol. Cell. Biol. 23:4307-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moqtaderi, Z., M. Keaveney, and K. Struhl. 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2:675-682. [DOI] [PubMed] [Google Scholar]
- 42.Moqtaderi, Z., J. D. Yale, K. Struhl, and S. Buratowski. 1996. Yeast homologues of higher eukaryotic TFIID subunits. Proc. Natl. Acad. Sci. USA 93:14654-14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller, F., and L. Tora. 2004. The multicoloured world of promoter recognition complexes. EMBO J 23:2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muratoglu, S., S. Georgieva, G. Pápai, E. Scheer, I. Enünlü, O. Komonyi, I. Cserpán, L. Lebedeva, E. Nabirochkina, A. Udvardy, L. Tora, and I. Boros. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23:306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Brien, T., and R. Tjian. 2000. Different functional domains of TAFII250 modulate expression of distinct subsets of mammalian genes. Proc. Natl. Acad. Sci. USA 97:2456-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oelgeschlager, T., C. M. Chiang, and R. G. Roeder. 1996. Topology and reorganization of a human TFIID-promoter complex. Nature 382:735-738. [DOI] [PubMed] [Google Scholar]
- 47.Perletti, L., J. C. Dantonel, and I. Davidson. 1999. The TATA-binding protein and its associated factors are differentially expressed in adult mouse tissues. J. Biol. Chem. 274:15301-15304. [DOI] [PubMed] [Google Scholar]
- 48.Pointud, J. C., G. Mengus, S. Brancorsini, L. Monaco, M. Parvinen, P. Sassone-Corsi, and I. Davidson. 2003. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J. Cell Sci. 116:1847-1858. [DOI] [PubMed] [Google Scholar]
- 49.Sanders, S. L., E. R. Klebanow, and P. A. Weil. 1999. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274:18847-18850. [DOI] [PubMed] [Google Scholar]
- 50.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 51.Sekiguchi, T., T. Nakashima, T. Hayashida, A. Kuraoka, S. Hashimoto, N. Tsuchida, Y. Shibata, T. Hunter, and T. Nishimoto. 1995. Apoptosis is induced in BHK cells by the tsBN462/13 mutation in the CCG1/TAFII250 subunit of the TFIID basal transcription factor. Exp. Cell Res. 218:490-498. [DOI] [PubMed] [Google Scholar]
- 52.Shao, H., M. Revach, S. Moshonov, Y. Tzuman, K. Gazit, S. Albeck, T. Unger, and R. Dikstein. 2005. Core promoter binding by histone-like TAF complexes. Mol. Cell. Biol. 25:206-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen, W. C., S. R. Bhaumik, H. C. Causton, I. Simon, X. Zhu, E. G. Jennings, T. H. Wang, R. A. Young, and M. R. Green. 2003. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 22:3395-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tao, Y., M. Guermah, E. Martinez, T. Oegelschlager, S. Hasegawa, R. Takada, T. Yamamoto, M. Horikoshi, and R. G. Roeder. 1997. Specific interactions and potential functions of human TAFII100. J. Biol. Chem. 272:6714-6721. [DOI] [PubMed] [Google Scholar]
- 55.Thut, C. J., J. L. Chen, R. Klemm, and R. Tjian. 1995. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science 267:100-104. [DOI] [PubMed] [Google Scholar]
- 56.Timmers, H. T., and L. Tora. 2005. SAGA unveiled. Trends Biochem. Sci. 30:7-10. [DOI] [PubMed] [Google Scholar]
- 57.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 58.Walker, A. K., and T. K. Blackwell. 2003. A broad but restricted requirement for TAF-5 (human TAFII100) for embryonic transcription in Caenorhabditis elegans. J. Biol. Chem. 278:6181-6186. [DOI] [PubMed] [Google Scholar]
- 59.Walker, A. K., J. H. Rothman, Y. Shi, and T. K. Blackwell. 2001. Distinct requirements for C. elegans TAF(II)s in early embryonic transcription. EMBO J. 20:5269-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, P. J., and D. C. Page. 2002. Functional substitution for TAF(II)250 by a retroposed homolog that is expressed in human spermatogenesis. Hum. Mol. Genet. 11:2341-2346. [DOI] [PubMed] [Google Scholar]
- 61.Wang, S., A. J. Dibenedetto, and R. N. Pittman. 1997. Genes induced in programmed cell death of neuronal PC12 cells and developing sympathetic neurons in vivo. Dev. Biol. 188:322-336. [DOI] [PubMed] [Google Scholar]
- 62.Wassarman, D. A., N. Aoyagi, L. A. Pile, and E. M. Schlag. 2000. TAF250 is required for multiple developmental events in Drosophila. Proc. Natl. Acad. Sci. USA 97:1154-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 64.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15:199-208. [DOI] [PubMed] [Google Scholar]
- 65.Xie, X., T. Kokubo, S. L. Cohen, U. A. Mirza, A. Hoffmann, B. T. Chait, R. G. Roeder, Y. Nakatani, and S. K. Burley. 1996. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380:316-322. [DOI] [PubMed] [Google Scholar]
- 66.Yamit-Hezi, A., and R. Dikstein. 1998. TAFII105 mediates activation of anti-apoptotic genes by NF-kappaB. EMBO J. 17:5161-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yanagisawa, J., H. Kitagawa, M. Yanagida, O. Wada, S. Ogawa, M. Nakagomi, H. Oishi, Y. Yamamoto, H. Nagasawa, S. B. McMahon, M. D. Cole, L. Tora, N. Takahashi, and S. Kato. 2002. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol. Cell 9:553-562. [DOI] [PubMed] [Google Scholar]
- 68.Zhou, J., J. Zwicker, P. Szymanski, M. Levine, and R. Tjian. 1998. TAFII mutations disrupt dorsal activation in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 95:13483-13488. [DOI] [PMC free article] [PubMed] [Google Scholar]