Abstract
Previously, we have demonstrated that bridge proteins comprised of avian leukosis virus (ALV) receptors fused to epidermal growth factor (EGF) can be used to selectively target retroviral vectors with ALV envelope proteins to cells expressing EGF receptors. To determine whether another type of ligand incorporated into an ALV receptor-containing bridge protein can also function to target retroviral infection, the TVA-VEGF110 bridge protein was generated. TVA-VEGF110 consists of the extracellular domain of the TVA receptor for ALV subgroup A (ALV-A), fused via a proline-rich linker peptide to a 110-amino-acid form of vascular endothelial growth factor (VEGF). This bridge protein bound specifically to its cell surface receptor, VEGFR-2, and efficiently mediated the entry of an ALV-A vector into cells. These studies indicate that ALV receptor-ligand bridge proteins may be generally useful tools for retroviral targeting approaches.
The ability to target viral infection only to specific cell types remains one of the formidable challenges to the use of retroviral vectors for gene therapy. We are developing avian leukosis virus (ALV) receptor-ligand bridge proteins as tools to deliver retroviral vectors to specific cell types. The feasibility of this approach was demonstrated using bridge proteins containing the mature form of human epidermal growth factor (EGF) fused to the extracellular domains of either the TVA receptor or the TVBS3 receptor for subgroups B and D of ALV. These bridge proteins mediated the highly selective infection of cells that express EGF receptors (3, 23). Recent work by another group has demonstrated adenovirus targeting by using a similar type of bridge protein consisting of the extracellular domain of the coxsackievirus and adenovirus receptor fused to EGF (8).
In the present study, we have tested whether vascular endothelial growth factor (VEGF) can also function as a retroviral targeting ligand when it is introduced into the context of a TVA-containing bridge protein. VEGF is a member of the cysteine-knot growth factor superfamily and is produced as an antiparallel disulfide-linked homodimer with symmetrical receptor-binding sites located at opposite ends of the molecule (27). Alternative splicing of a common primary mRNA transcript generates differently sized ligand isoforms: VEGF121, VEGF145, VEGF165, VEGF189, and VEGF206 (27). The murine VEGF110 form that was used in this study consists of the N-terminal 110 amino acids of VEGF165, with the C-terminal heparin-binding domain (7) removed to reduce nonspecific binding of the bridge protein to cell surfaces.
Three different types of VEGF receptors have been identified: VEGFR-1, VEGFR-2, and VEGFR-3 (27). VEGF receptors are selectively expressed on the surfaces of endothelial cells (27). In addition to these three receptors, the NRP-1 protein that is a receptor for collapsins and semaphorins is also a receptor for VEGF165 (27). Compared to VEGF165, VEGF110 has the same binding affinity for VEGFR-2, a lower affinity for VEGFR-1, and does not bind to NRP-1 (15, 25).
VEGF is important to test as a potential ligand for retroviral targeting because it binds to receptors that are expressed on tumor vasculature. Solid tumors require the presence of a network of blood vessels to obtain oxygen and nutrients for their growth (10). To induce formation of new blood vessels, a process termed angiogenesis, tumors express a variety of growth factors, one of which is VEGF (5, 9, 12, 13, 14, 18, 22, 26). VEGF is known to specifically induce growth and migration of endothelial cells as well as to cause permeability of blood vessels, and inhibitors of VEGF signaling retard tumor growth in mice (11, 16, 19–21).
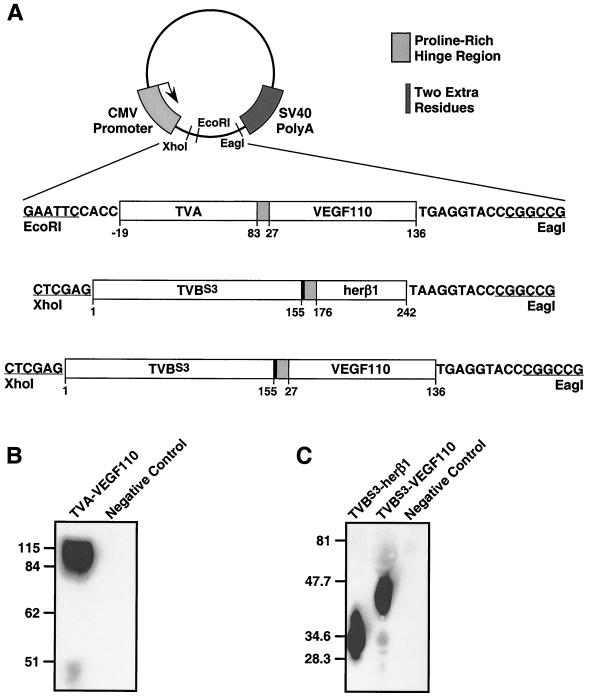
The TVA-VEGF110 protein consists of the extracellular domain of TVA fused via a proline-rich hinge region to murine VEGF110 (Fig. 1A). Additional bridge proteins were also generated, consisting of the extracellular domain of TVBS3 fused via the same hinge region to either VEGF110 or the EGF-like region of human heregulin-β1 (herβ1), respectively (Fig. 1A). Production of each bridge protein in the extracellular supernatant of transiently transfected human 293 cells was confirmed after sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with a subgroup A- or a subgroup B-specific surface (SU)-immunoglobulin fusion protein (SU-rIg) to detect TVA- and TVB-containing bridge proteins, respectively, as described previously (3, 23). Under nonreducing conditions, TVA-VEGF110 migrated as an 84- to 115-kDa protein species (Fig. 1B), consistent with it being a disulfide-linked dimer like VEGF (see Fig. 1A legend for a description of the expected molecular mass of this protein). Under reducing conditions, the TVBS3-containing bridge proteins migrated at positions that were consistent with their expected monomeric molecular masses (Fig. 1C).
FIG. 1.
Construction and expression of retroviral receptor-ligand bridge proteins. (A) Recombinant genes encoding each bridge protein were generated by PCR-based methods and introduced into the pCI-plasmid expression vector (Promega) as shown. The numbering schemes for the amino acid residues of TVA, TVBS3, and heregulinβ1 were taken from references 2 and 6 and GenBank accession number B43273, respectively. The VEGF110 residues are described under GenBank accession number A44881. The positions of a proline-rich hinge region (PPPELLGGP) and of a 2-amino-acid insertion (His-Gly) that resulted during the construction of the TVBS3-containing bridge proteins are indicated. The TVA-VEGF110 monomer was expected to have a molecular mass ranging from 33 to 52 kDa because the primary amino acid sequence predicts a 22.4-kDa protein but the extracellular domain of TVA is subjected to extensive posttranslational modifications which add an additional 21 to 30 kDa to its apparent molecular mass (1, 2). Monomeric forms of TVBS3-VEGF110 and TVBS3-herβ1 were expected to have molecular masses of 37 and 33 kDa, respectively, based on their primary amino acid sequences (28 and 24 kDa, respectively) and the presence of three putative N-linked glycosylation sites in each protein. (B and C) Production of bridge proteins. Forty-five-microliter aliquots of extracellular supernatant taken from transfected human 293 cells that expressed the bridge proteins, or from nontransfected cells (negative controls), were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis under either nonreducing (B) or reducing (C) conditions. The proteins were then transferred to a nitrocellulose membrane and were probed with subgroup A-specific (panel B) or subgroup B-specific (panel C) SU-rIg fusion proteins and then with a horseradish peroxidase-conjugated secondary antibody, as described previously (3, 23). The bridge proteins were then detected by enhanced chemiluminescence.
Flow cytometry was performed to analyze the binding of TVA-VEGF110 to porcine aortic endothelial (PAE) cells that express few or no VEGF receptors (17, 25, 28) or to transduced PAE cells expressing mouse VEGFR-2 (PAE-VEGFR-2 cells). PAE-VEGFR-2 cells were generated by transduction of PAE cells with a VSV-G pseudotyped murine leukemia virus (MLV) vector encoding VEGFR-2 [MLV(VSV-G)-VEGFR-2]. The pseudotyped virus was produced from human 293T cells plated at 60% confluence on 100-mm tissue culture plates. These cells were transiently transfected with 5 μg of pMD.G plasmid encoding the VSV-G protein, 15 μg of pMMD.gagpol plasmid encoding MLV Gag and Gag-Pol structural proteins (24), and 15 μg of pSFG.Flk1 plasmid encoding VEGFR-2 (unpublished data). A 30%-confluent well of a six-well plate of PAE cells was then incubated with 1 ml of MLV(VSV-G)-VEGFR-2 in the presence of 8 μg of Polybrene per ml. The resultant VEGFR-2-expressing cells were then isolated by flow cytometric sorting after incubation with supernatants containing TVA-VEGF110 and SUA-rIgG, and then with a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (data not shown).
To assay for specific binding of TVA-VEGF110 to VEGF receptor-expressing cells, 3.5 × 105 PAE-VEGFR-2 cells and the same number of control PAE cells were incubated for 1 h at 4°C with different amounts of a TVA-VEGF110-containing supernatant that was supplemented with a control 293 cell-conditioned medium to a total volume of 500 μl. The cells were then washed with ice-cold phosphate-buffered saline (PBS) (containing 2% fetal bovine serum) and then incubated with SUA-rIgG and an FITC-conjugated secondary antibody and subjected to flow cytometric analysis as described before (23). Because the bound TVA-VEGF110 protein was detected with a soluble SU reagent, these studies also established whether the bridge protein can bind simultaneously to cell surface VEGF receptors and to ALV subgroup A (ALV-A) SU. Indeed, TVA-VEGF110 bound in a dose-dependent manner to PAE-VEGFR-2 cells (Fig. 2A) but reproducibly bound only weakly to PAE cells (Fig. 2B), perhaps indicating that these cells do in fact express a small number of VEGF receptor(s). These binding studies supported the idea that TVA-VEGF110 can serve as a bridge between cell surface VEGFR-2 and ALV-A SU.
FIG. 2.
TVA-VEGF110 binds specifically to cells that express VEGFR-2. PAE-VEGFR-2 cells (A) and PAE cells (B) were incubated with increasing amounts of a TVA-VEGF110-containing extracellular supernatant. (C) Prior to incubation with TVA-VEGF110, PAE-VEGFR-2 cells were incubated with extracellular supernatant that contained either TVBS3-VEGF110, TVBS3-herβ1, or no TVB-ligand bridge protein. Following these treatments the TVA-VEGF110 protein that was bound to the cells was detected by flow cytometric analysis using a subgroup A-specific SU-rIg fusion protein and an FITC-conjugated secondary antibody as described previously (23). These experiments were performed three times with similar results, and results of a representative example are shown.
To formally show that TVA-VEGF110 binds to VEGFR-2, competition binding experiments were performed in the presence of heterologous bridge proteins that either contained the same (TVBS3-VEGF110) or different (TVBS3-herβ1) ligand moieties (Fig. 1). The competition binding experiments were performed by incubating 3.5 × 105 PAE-VEGFR-2 cells at 4°C for 1 h with 490 μl of extracellular supernatants that contained equivalent amounts (as judged by quantitative chemiluminescence using a Bio-Rad FluorS instrument) of either TVBS3-herβ1 or TVBS3-VEGF110. A 10-μl aliquot of a TVA-VEGF110-containing supernatant was then added and the cells were incubated at 4°C for an additional hour. The cells were then washed in PBS and analyzed by flow cytometry using SUA-rIgG and the FITC-conjugated antibody as before. TVA-VEGF110 binding was blocked by preincubation with TVBS3-VEGF110 but not with TVBS3-herβ1 (Fig. 2C). These data confirm that TVA-VEGF110 binds specifically to VEGFR-2 expressed at the surface of PAE-VEGFR-2 cells.
To determine whether TVA-VEGF110 can mediate ALV-A entry when bound to VEGFR-2, approximately 105 PAE-VEGFR-2 cells were incubated for 1 h at 4°C with increasing amounts of a TVA-VEGF110-containing supernatant that was made up to a total volume of 500 μl with control supernatant taken from nontransfected human 293 cells. The cells were then washed with ice-cold medium and incubated with 500 μl of ice-cold medium containing 5 μl of a 100-fold concentrated stock of an ALV-A vector RCASBP(A)-EGFP encoding the enhanced green fluorescent protein, which was prepared as described elsewhere (24).
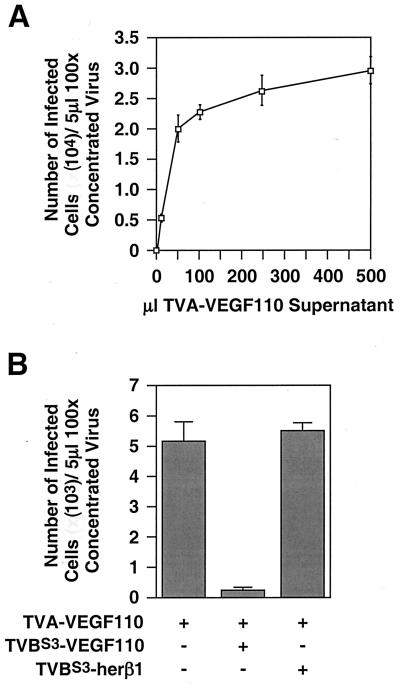
Approximately 72 h after viral challenge, the cells were washed with PBS and removed from plates with Ca2+- and Mg2+-free PBS containing 1 mM EDTA and 7 μM propidium iodide. The infected cells were then identified by flow cytometry and dead cells that had taken up propidium iodide were excluded from the analysis by electronic gating. These studies showed that TVA-VEGF110 rendered PAE-VEGFR-2 cells susceptible to ALV-A infection in a dose-dependent manner (Fig. 3A).
FIG. 3.
TVA-VEGF110 mediates ALV-A infection when bound to VEGFR-2. (A) PAE-VEGFR-2 cells were incubated with increasing amounts of extracellular supernatant containing TVA-VEGF110 and then challenged with 5 μl of a 100-fold concentrated stock of RCASBP(A)-EGFP. The total number of infected cells obtained was then calculated by flow cytometry as described previously (24). The average data obtained from an experiment that was performed in triplicate are shown with standard deviations indicated with error bars. (B) PAE-VEGFR-2 cells were incubated with equivalent amounts of TVBS3-VEGF110 or TVBS3-herβ1 or with no TVB-ligand bridge protein, prior to the addition of 10 μl of TVA-VEGF110. The cells were then challenged with RCASBP(A)-EGFP as in panel A and analyzed by flow cytometry. The results of an experiment performed in triplicate are shown with standard deviations of the data indicated with error bars.
To determine the efficiency and specificity of TVA-VEGF110-dependent infection, parental PAE cells and PAE-TVAsyn cells which express a transmembrane form of TVA were also challenged with the ALV-A vector. PAE-TVAsyn cells were generated by transducing PAE cells with an MLV vector encoding a synthetic transmembrane form of the TVA receptor (2). The RCASBP(A)-EGFP titer that was obtained with these cells was approximately 7.5 × 107 infectious units/ml of 100-fold-concentrated virus (defined as 100%, Table 1). Strikingly, TVA-VEGF110-mediated infection of PAE-VEGFR-2 cells was only 11.3-fold less than the level seen with the control TVA-expressing cells (Table 1). Furthermore, in the absence of TVA-VEGF110, only low levels of ALV infection were observed (Table 1), consistent with the previously published “background” levels of ALV infection seen in various mammalian cell types (4). The addition of TVA-VEGF110 to the control PAE cells did lead to a slight enhancement of viral entry (Table 1), a result which again indicates that these cells may express a low number of VEGF receptors (Fig. 2B).
TABLE 1.
TVA-VEGF110-dependent infection of PAE-VEGFR-2 cells
| Cell type | Efficiency of ALV-A infection (%)a
|
|
|---|---|---|
| Without TVA-VEGF110 | With TVA-VEGF110 | |
| PAE-TVAsyn | 100 | Not done |
| PAE | 0.01 | 0.09 |
| PAE-VEGFR-2 | 0.02 | 8.72 |
RCASBP(A)-EGFP titer relative to that in PAE-TVAsyn cells (7.5 × 107 infectious units/ml, defined as 100%).
To confirm that the TVA-VEGF110–VEGFR-2 interaction is necessary for the enhanced viral entry seen with PAE-VEGFR-2 cells, we attempted to block this infection by incubating these cells at 4°C for 30 min with equivalent amounts of TVBS3-VEGF110 or TVBS3-herβ1 prior to adding the TVA-containing bridge protein as before (Fig. 2C). The cells were then challenged with RCASBP(A)-EGFP and analyzed by flow cytometry as described above. TVA-VEGF110-dependent infection of PAE-VEGFR-2 cells was inhibited by TVBS3-VEGF110 but not by TVBS3-herβ1, thereby confirming that the VEGF110-VEGFR-2 interaction is essential for bridge protein-enhanced viral entry (Fig. 3B).
Taken together, the studies presented in this report clearly demonstrate that targeted ALV-A vector entry can be achieved through the TVA-VEGF110–VEGFR-2 interaction. TVA-VEGF110 bound specifically to cells that express VEGFR-2 and mediated efficient infection of these cells by an ALV-A vector. This system for viral targeting represents an attractive model for the development of retroviral vectors that can be targeted to tumor vasculature. Furthermore, these findings, coupled with the demonstration of retroviral targeting via bridge proteins containing the EGF ligand (3, 23) or a single-chain antibody raised against a tumor-specific form of the EGF receptor (24), indicate that ALV receptor-containing bridge proteins may be generally useful reagents for cell-type-specific retroviral targeting.
Acknowledgments
We thank members of the Young laboratory for helpful discussions and John Naughton for help with preparing the figures. We also thank John Daly for assistance with flow cytometry and Mark Federspiel and Matt van Brocklin for providing the RCASBP(A)-EGFP virus.
This work was supported by a grant from the U.S. Department of the Army (DAMD-17-98-1-8488) and by NIH grant CA70810 from the National Cancer Institute.
REFERENCES
- 1.Bates P, Young J A T, Varmus H E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 2.Belanger C, Zingler K, Young J A T. Importance of cysteines in the LDLR-related domain of the subgroup A avian leukosis and sarcoma virus receptor for viral entry. J Virol. 1995;69:1019–1024. doi: 10.1128/jvi.69.2.1019-1024.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerger A L, Snitkovsky S, Young J A. Retroviral vectors preloaded with viral receptor-ligand bridge protein are targeted to specific cell types. Proc Natl Acad Sci USA. 1999;96:9867–9872. doi: 10.1073/pnas.96.17.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bova-Hill C, Olsen J C, Swanstrom R. Genetic analysis of the Rous sarcoma virus subgroup D env gene: mammal tropism correlates with temperature sensitivity of gp85. J Virol. 1991;65:2073–2080. doi: 10.1128/jvi.65.4.2073-2080.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brekken R A, Huang X, King S W, Thorpe P E. Vascular endothelial growth factor as a marker of tumor endothelium. Cancer Res. 1998;58:1952–1959. [PubMed] [Google Scholar]
- 6.Brojatsch J, Naughton J, Rolls M M, Zingler K, Young J A. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 7.Cohen T, Gitay-Goren H, Sharon R, Shibuya M, Halaban R, Levi B Z, Neufeld G. VEGF121, a vascular endothelial growth factor (VEGF) isoform lacking heparin binding ability, requires cell-surface heparan sulfates for efficient binding to the VEGF receptors of human melanoma cells. J Biol Chem. 1995;270:11322–11326. doi: 10.1074/jbc.270.19.11322. [DOI] [PubMed] [Google Scholar]
- 8.Dmitriev I, Kashentseva E, Rogers B E, Krasnykh V, Curiel D T. Ectodomain of coxsackievirus and adenovirus receptor genetically fused to epidermal growth factor mediates adenovirus targeting to epidermal growth factor receptor-positive cells. J Virol. 2000;74:6875–6884. doi: 10.1128/jvi.74.15.6875-6884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dvorak H F, Sioussat T M, Brown L F, Berse B, Nagy J A, Sotrel A, Manseau E J, Van de Water L, Senger D R. Distribution of vascular permeability factor (vascular endothelial growth factor) in tumors: concentration in tumor blood vessels. J Exp Med. 1991;174:1275–1278. doi: 10.1084/jem.174.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folkman J. Tumor angiogenesis. Adv Cancer Res. 1985;43:175–203. doi: 10.1016/s0065-230x(08)60946-x. [DOI] [PubMed] [Google Scholar]
- 11.Goldman C K, Kendall R L, Cabrera G, Soroceanu L, Heike Y, Gillespie G Y, Siegal G P, Mao X, Bett A J, Huckle W R, Thomas K A, Curiel D T. Paracrine expression of a native soluble vascular endothelial growth factor receptor inhibits tumor growth, metastasis, and mortality rate. Proc Natl Acad Sci USA. 1998;95:8795–8800. doi: 10.1073/pnas.95.15.8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holash J, Maisonpierre P C, Compton D, Boland P, Alexander C R, Zagzag D, Yancopoulos G D, Wiegand S J. Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF. Science. 1999;284:1994–1998. doi: 10.1126/science.284.5422.1994. [DOI] [PubMed] [Google Scholar]
- 13.Ke L, Qu H, Nagy J A, Eckelhoefer I A, Masse E M, Dvorak A M, Dvorak H F. Vascular targeting of solid and ascites tumours with antibodies to vascular endothelial growth factor. Eur J Cancer. 1996;32A:2467–2473. doi: 10.1016/s0959-8049(96)00391-7. [DOI] [PubMed] [Google Scholar]
- 14.Keck P J, Hauser S D, Krivi G, Sanzo K, Warren T, Feder J, Connolly D T. Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science. 1989;246:1309–1312. doi: 10.1126/science.2479987. [DOI] [PubMed] [Google Scholar]
- 15.Keyt B A, Berleau L T, Nguyen H V, Chen H, Heinsohn H, Vandlen R, Ferrara N. The carboxyl-terminal domain (111-165) of vascular endothelial growth factor is critical for its mitogenic potency. J Biol Chem. 1996;271:7788–7795. doi: 10.1074/jbc.271.13.7788. [DOI] [PubMed] [Google Scholar]
- 16.Kim K J, Li B, Winer J, Armanini M, Gillett N, Phillips H S, Ferrara N. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 17.Kroll J, Waltenberger J. The vascular endothelial growth factor receptor KDR activates multiple signal transduction pathways in porcine aortic endothelial cells. J Biol Chem. 1997;272:32521–32527. doi: 10.1074/jbc.272.51.32521. [DOI] [PubMed] [Google Scholar]
- 18.Leung D W, Cachianes G, Kuang W J, Goeddel D V, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 19.Millauer B, Shawver L K, Plate K H, Risau W, Ullrich A. Glioblastoma growth inhibited in vivo by a dominant-negative Flk-1 mutant. Nature. 1994;367:576–579. doi: 10.1038/367576a0. [DOI] [PubMed] [Google Scholar]
- 20.Olson T A, Mohanraj D, Roy S, Ramakrishnan S. Targeting the tumor vasculature: inhibition of tumor growth by a vascular endothelial growth factor-toxin conjugate. Int J Cancer. 1997;73:865–870. doi: 10.1002/(sici)1097-0215(19971210)73:6<865::aid-ijc17>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Ramakrishnan S, Olson T A, Bautch V L, Mohanraj D. Vascular endothelial growth factor-toxin conjugate specifically inhibits KDR/flk-1-positive endothelial cell proliferation in vitro and angiogenesis in vivo. Cancer Res. 1996;56:1324–1330. [PubMed] [Google Scholar]
- 22.Shweiki D, Itin A, Soffer D, Keshet E. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature. 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- 23.Snitkovsky S, Young J A T. Cell-specific viral targeting mediated by a soluble retroviral receptor-ligand fusion protein. Proc Natl Acad Sci USA. 1998;95:7063–7068. doi: 10.1073/pnas.95.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snitkovsky S, Niederman T M J, Carter B S, Mulligan R C, Young J A T. A TVA–single-chain antibody fusion protein mediates specific targeting of a subgroup A avian leukosis virus vector to cells expressing a tumor-specific form of the epidermal growth factor receptor. J Virol. 2000;74:9540–9545. doi: 10.1128/jvi.74.20.9540-9545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soker S, Takashima S, Miao H Q, Neufeld G, Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 26.Tanigawa N, Amaya H, Matsumura M, Shimomatsuya T. Correlation between expression of vascular endothelial growth factor and tumor vascularity, and patient outcome in human gastric carcinoma. J Clin Oncol. 1997;15:826–832. doi: 10.1200/JCO.1997.15.2.826. [DOI] [PubMed] [Google Scholar]
- 27.Veikkola T, Karkkainen M, Claesson-Welsh L, Alitalo K. Regulation of angiogenesis via vascular endothelial growth factor receptors. Cancer Res. 2000;60:203–212. [PubMed] [Google Scholar]
- 28.Waltenberger J, Claesson-Welsh L, Siegbahn A, Shibuya M, Heldin C H. Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor. J Biol Chem. 1994;269:26988–26995. [PubMed] [Google Scholar]