Abstract
Inactivity of the human cytomegalovirus (HCMV) major immediate-early regulatory region (MIERR), which is composed of promoter, enhancer, unique region, and modulator, is linked to lack of HCMV replication in latently infected cells and in other nonpermissive cell types, including human embryonal NTera2 carcinoma (NT2) cells. I refined the embryonal NT2 cell model to enable characterization of the unknown mechanistic basis for silencing of HCMV MIERR-dependent transcription and viral replication in nonpermissive cells. These infected NT2 cells contain nonreplicating viral genomes with electrophoretic mobility equivalent to a supercoiled, bacterial artificial chromosome of comparable molecular weight. MIERR-dependent transcription is minimal to negligible. Increasing the availability of positive-acting transcription factors by retinoic acid (RA) treatment after infection is largely insufficient in reactivating the MIERR. In contrast, trichostatin A (TSA), a histone deacetylase inhibitor, reactivates MIERR-dependent transcription. Contrary to prior findings produced from transfected MIERR segments, deletion of the 21-bp repeats and modulator from the MIERR in the viral genome does not relieve MIERR silencing. To demonstrate that MIERR silencing likely results from enhancer inactivity, I examined an HCMV with a heterologous MIERR promoter that is enhancer dependent but exempt from IE2 p86-mediated negative autoregulation. This heterologous promoter, like its neighboring native MIERR promoter, exhibits immediate-early transcriptional kinetics in fibroblasts. In embryonal NT2 cells, the heterologous MIERR promoter is transcriptionally inactive. This silence is relieved by TSA but not by RA. Remarkably, TSA-induced reactivation of MIERR-dependent transcription from quiescent viral genomes is followed by release of infectious virus. I conclude that a mechanism of active repression imposes a block to MIERR-dependent transcription and viral replication in embryonal NT2 cells. Because TSA overcomes the block, viral gene silencing may involve histone deacetylase-based modification of viral chromatin, which might account for the covalently closed circular conformation of quiescent HCMV genomes.
Human cytomegalovirus (HCMV) replicates in wide variety of cell types, including central nervous system (CNS) neurons (reviewed in reference 82). The virus replicates poorly or negligibly in lymphocytes, neutrophils, and immature neurons (61, 65, 73, 80, 82), and it resides latently in monocytes and their precursors (34, 41, 42, 59, 60, 80, 83, 87, 88, 91). The mechanisms that preclude viral replication in nonpermissive cell types are poorly understood. One possible mechanism involves silencing of the viral major immediate-early (MIE) genes, whose products (e.g., IE1 p72 and IE2 p86) are required for initiating viral replication (32, 37, 62, 70, 84). These genes are vigorously transcribed in productive (lytic) infection but are relatively inactive in latently or other nonpermissively infected cell types (41, 42, 45, 59, 66, 80, 88). Hence, MIE gene expression and viral replication may be silenced by the same mechanism(s).
The NTera2/D1 (NT2) cell line, derived from a human teratocarinoma (5), is a useful model in which to examine the coordinate regulation of HCMV replication and MIE gene expression (30, 31, 45, 66). These cells resemble embryonal cells and are able to differentiate predominantly into CNS neurons when induced with retinoic acid (RA) (4, 40, 72). Embryonal NT2 cells do not permit HCMV replication, whereas differentiated cells do (30, 31). These findings are consonant with the differentiation-dependent nature of HCMV replication in cultured primary CNS neurons (73). The lack of HCMV replication in embryonal NT2 cells corresponds to a block in viral MIE gene expression (45, 66). This block is abrogated by RA pretreatment, which induces cellular differentiation prior to infection (45, 66). However, the block persists if RA treatment is delayed to the postinfection (p.i.) period (31). Thus, establishing the RA-induced cellular condition prior to infection is key to promoting viral replication and MIE gene expression. HCMV's fate within embryonal NT2 cells is not fully known, although viral genomes are associated with cell nuclei at 5 h p.i. (66). DNase I mapping of the MIE regulatory region (MIERR) of these viral genomes reveals a hypersensitivity profile that differs considerably from that produced in the RA-pretreated NT2 cells (66). This finding implies that superstructure or chromatin organization of viral genomes differs among the two isogenic cell types.
Previous studies analyzing MIERR segments in in vitro, transfection, and transgenic animal studies indicate that this region controls transcription of its genes through interplay of both positive and negative cis-acting elements (reviewed in references 58 and 61). The MIERR is composed of promoter, enhancer, unique region, and modulator, although the boundaries of these components are inexact (reviewed in references 58 and 61). The enhancer spans base positions -65 to -550, with respect to the +1 start site of MIE RNAs (58). The enhancer's activity varies depending on cell type, cellular differentiation, and signal transduction pathways. Such variability reflects collective differences in the amounts or activities of many types of cellular and viral proteins that act on this region. For instance, cellular NF-κB/rel, CREB/ATF, AP1, SP-1, serum response factor, and ELK-1 can each bind to one or more cognate sites located in the enhancer and consequently stimulate transcription (17, 58, 61). The enhancer also contains three RA response elements (RAREs) (base positions -275, -472, and -544) that augment MIERR segment activity in transfected NT2 cells when occupied by liganded RA receptor (RAR) homodimer or RAR-retinoid X receptor (RXR) heterodimer (8, 9, 29). Viral proteins, such as pp71 and IE1 p72, also act through cis-acting sites to increase enhancer activity (19, 48, 53, 85). While these findings were not confirmed in the context of the HCMV genome, a distal enhancer deletion (-300 to -582) was shown recently to greatly impair MIE gene expression and viral replication at low but not high multiplicity of infection (MOI) (56). This finding suggests that cis-acting sites in the distal enhancer (e.g. serum response factor, ELK-1, NF-κB/rel, CREB/ATF, SP-1, and/or RARE sites) are important for stimulating MIE gene expression under conditions that likely occur in vivo. Recent findings showing that murine CMV MIE gene expression and viral replication are dependent on the MIERR enhancer, whether it be of murine or human CMV origin, also underscore the enhancer's pivotal role in the viral life cycle (7, 33).
The MIERR also has negative cis-acting mechanisms for conditionally silencing its genes. In monocytic THP-1 and embryonal NT2 cells, MIERR activity is repressed by cellular YY1, as judged from transfection studies of MIERR segments and in vitro binding studies (43, 49, 81). YY1 binds to each of three 21-bp repeats located in the distal enhancer (base positions -305, -350, and -475), as well as to one or more sites in the modulator. YY1-mediated repression is alleviated by deletion of the 21-bp repeats en bloc or by cellular differentiation or stimulation (43, 49, 81). Whether these findings hold true in the context of viral infection is unknown. The modulator (-750 to -1140) also inhibits activity of MIERR segments in transfected THP-1 and NT2 cells. This inhibition is partly conferred by 21 recognition sites for a cellular silencing binding protein, which is inactive in the differentiated cellular counterparts (16, 36, 78, 89). However, repression of MIE gene expression in infected THP-1 and NT2 cells is not lessened by removing the modulator from the viral genome (57). The reason for the disparate findings is unclear, although one possible explanation is that there prevails in the viral genome another repressive mechanism(s) that is dysfunctional or missing in transfected MIERR segments. Cellular Gfi-1 is another candidate repressor that binds to two sites in the proximal enhancer and is expressed preferentially in undifferentiated cells, but its role in silencing MIERR activity in THP-1 and NT2 cells is unknown (93). Last, the MIERR is under negative autoregulation by the IE2 p86 protein, which accumulates during lytic infection and binds the cis repression sequence element (+1 to -15) to turn off MIE gene expression (reviewed in references 58 and 61).
This report assesses the suitability of a refined embryonal NT2 cell population as a model in which to characterize the unknown mechanistic basis for silencing of HCMV MIERR-dependent transcription and viral replication in nonpermissive cells. The four objectives were to (i) determine if infected NT2 cells contain quiescent viral genomes having a structure of covalently closed circle (CCC) molecules, which are known to exist in latently infected blood monocytes (13); (ii) determine whether the lack of MIERR-dependent transcription is a result of active repression, absence of positive-acting transcription factors, or both; (iii) resolve whether removal of the virus's 21-bp repeats and modulator can overcome MIERR silencing, as predicted by prior transient transfection studies (36, 43, 49, 67, 78, 81); and (iv) validate this model by demonstrating inducible reactivation of MIERR-dependent transcription and viral replication from preexisting quiescent viral genomes. I define the roles of trichostatin A (TSA), a specific inhibitor of histone deacetylases (HDACs), and RA in the reactivation process.
MATERIALS AND METHODS
Cells, viruses, and infections.
Primary human foreskin fibroblast (HFF) cells were grown in Eagle's minimal essential medium (MEM) supplemented with 10% newborn bovine serum (57). The D1 clone of NTera2 cells (NTera2/D1) at passage 28 was kindly provided by E. Gonczol (31). These cells were maintained in Dulbecco's MEM supplemented with 4 μM glutamine, 4.5 g of glucose per liter, 15% knockout serum replacement (Life Technologies, Rockville, Md.), and 3 to 5% charcoal-treated fetal bovine serum. Differentiation of NT2 cells was induced by addition of 10 μM RA (Sigma, St. Louis, Mo.) to the growth medium for ≥7 days. Recombinant HCMVs rΔ-640/-1108SVgfp, rΔ-300/-1108Egfp, and rΔ-582/-1108Egfp (described by Meier and Pruessner [56]) were proagated and harvested as described previously (56, 57). Virus absorption was carried out for 1.5 h, and cells were subsequently washed thrice with Hanks' balanced salt solution without calcium and magnesium. Titers of HCMVs lacking the MIE distal enhancer cannot be accurately determined by plaque assay. Therefore, titers of these viruses were normalized to known titers of replication-competent viruses on the basis of cytopathic effect (CPE) in HFF cells at 24 h p.i. and amount of viral DNA in HFF cells prior to viral replication (4 to 5 h p.i.).
Plasmids.
Plasmids pUS3α, p1.6, pIE1, pactin, and p4EM have been reported previously (56). The 250-kbp bacterial artificial chromosome (BAC) vector containing a human genome fragment was a generous gift from Brian Schutte. pGFP consists of the BsrG1-BsrF1 fragment (blunted with Klenow enzyme) of pΔMSVgfp (56) cloned into the SmaI site of pGEM-4Z (Promega, Madison, Wis.). p71 was derived by subcloning the XbaI (blunted with Klenow enzyme)-KpnI (blunted with T4 polymerase) fragment of pCMV71 into the BamHI (blunted with Klenow enzyme) and BglII (blunted with T4 polymerase) sites, respectively, of pSG5 (Stratagene, La Jolla, Calif.).
Gardella gel analysis.
Gardella gel analysis was performed by a modified method (26). Cells were washed with phosphate-buffered saline and gently resuspended in sample buffer composed of 15% Ficoll, 1× Tris-borate-EDTA, and xylene cyanol. Isolated virions or bacteria (containing BAC) were mixed with uninfected cells in sample buffer. Samples were loaded into wells of a horizontal 0.8% agarose running gel in 1× Tris-borate-EDTA buffer. The wells were separated from the stacking gel by a distance of 2 to 3 mm. The stacking gel was made of 0.7% agarose, 1 mg of self-digested pronase per ml, and 2% sodium dodecyl sulfate. Electrophoresis was performed at a constant voltage at room temperature for 2 h (0.8 V/cm) and then at 4°C (5 V/cm) overnight. The running gel was subsequently subjected to Southern blot analysis and autoradiography as described previously (57). The labeled HCMV-specific probe was derived as a 1.6-kbp fragment of p1.6 (T probe) (56), which corresponds to HCMV open reading frames (ORFs) IRL3 and IRL4 (nucleotide positions 185496 to 187110). The BAC was multiprimed 32P labeled for use as a probe. Because of low-level nonspecific hybridization of the HCMV-specific probe to components of uninfected cells that are electrophoretically immobile, another method was used to verify that the heightened intensity of hybridization signal from immobile infected cell components was due to the presence of HCMV DNA. The Gardella gel-immobilized DNAs of infected and uninfected NT2 cells were excised and isolated using β-Agarase I (New England Biolabs, Beverly, Mass.) according to the manufacturer's directions. The isolated DNA samples were digested with HindIII, fractionated by standard agarose gel electrophoresis, and subjected to Southern blot hybridization using the HCMV-specific probe. The findings clearly revealed that Gardella gel-immobilized DNA derived from HCMV-infected cells contained HCMV DNA, whereas that of uninfected cells did not (data not shown).
RNA analysis.
Whole-cell RNA of uninfected or infected cells was isolated according to the method of Chomczynski and Sacchi (21). For inhibition of protein synthesis, 100 μM anisomycin (Sigma) was added to the growth medium 30 min before, during, and after viral absorption. When indicated, RA (10 μM) or TSA (100 ng/ml) was added to or omitted from growth medium at 2 or 24 h after initiation of infection. RNase protection assays (RPAs) were performed as described previously (56). IE1-, US3-, and actin-specific riboprobes were made from pIE1, pUS3α, and pactin, as reported previously (56). The green fluorescent protein (GFP)- and pp71-specific riboprobes were made with T7 polymerase from EcoRI-linearized pGFP and XbaI-linearized p71 templates, respectively. Protected RNA products were analyzed on 6% polyacrylamide-urea gels.
Flow cytometric analysis and microscopy.
Samples containing live cells were analyzed on a FACScan flow cytometer using CellQuest software (Becton Dickinson, Franklin Lakes, N.J.). Cells were resuspended in phosphate-buffered saline containing propidium iodide (50 μg/ml). Ten thousand live cell events (as determined by forward and side scatter) were acquired for analysis. Flow sorting of live NT2 cells was performed with a Coulter EPICS 753 cytometer (Beckman Coulter, Inc.). One thousand live cells either emitting or lacking GFP fluorescence were collected in growth medium. These cells were cocultivated with subconfluent adherent HFF cells in a 12-well dish (102 NT2 cells per 105 HFF cells in each well) containing Dulbecco's MEM supplemented with 10% fetal bovine serum. CPE was monitored by light microscopy. At 21 days p.i., the wells were examined by inverted Zeiss 510 laser scanning confocal microscope.
RESULTS
Evidence of HCMV CCC-like genomes in embryonal NT2 cells.
Previous reports indicate that HCMV replication and MIE gene expression are undetectable in most infected NT2 cells (30, 31, 57, 66, 79). The NT2 cell minority in which these viral activities manifest is thought to arise from “breakthrough” cellular differentiation, which develops at a variable rate depending on cell density (30), cell aggregation (20), and quality of fetal bovine serum (J. Meier, unpublished observation). Such cellular inhomogeneity likely resulted in initial findings of low levels of lytic viral DNA replication in NT2 cells (data not shown), which were grown under conditions reported previously (30, 31, 57, 66, 79). To prevent breakthrough cellular differentiation, NT2 cells were grown in an embryonic stem cell culture condition as specified in Materials and Methods. Under this growth condition, the confounding variable of lytic viral DNA replication was no longer evident when assayed by Southern blotting (data not shown). Abundance of viral MIE RNA as determined by RPA was also markedly decreased by this change (data not shown). Whether the modified growth condition actually enhanced the utility of these cells for analyzing mechanisms that preclude viral replication was the subject of further investigation.
The first objective was to determine if the refined embryonal NT2 cell population would still allow HCMV entry. To obviate the possible confounder of virus entering a dead-end pathway and not reaching the nucleus, I opted to ascertain the structural configuration(s) of viral DNA within the cells. This strategy is predicated on prior knowledge indicating that linear genomes of the virion circularize after entering the cell nucleus, which occurs in either lytic or latent viral infection (1, 13, 54, 55). Such a strategy also permits the identification of lytic replicative intermediates, such as linear concatemers or branched forms, should they be synthesized.
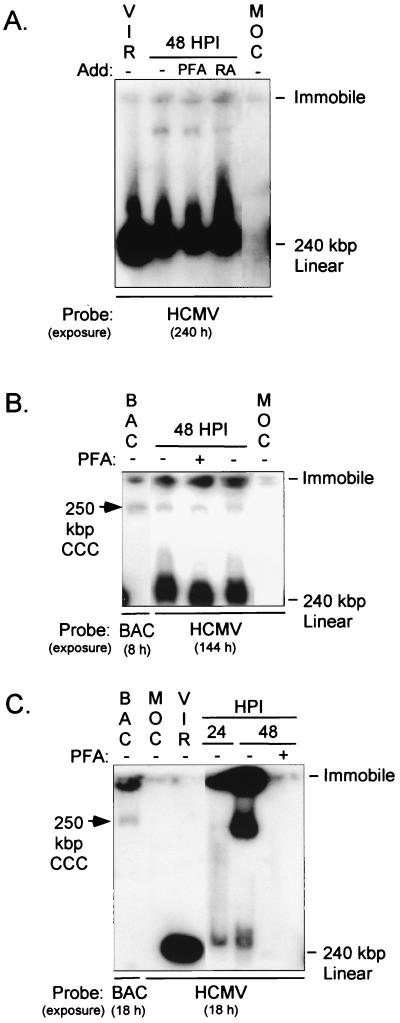
Gardella gel electrophoresis was used to help resolve the various types of viral genome structures that may exist in embryonal NT2 cells. In the starting analysis, a recombinant HCMV, rΔ-640/-1108SVgfp (56), was used for the purpose of also monitoring the proportion of cells initiating the lytic viral life cycle. This virus's MIERR modulator (-640 to -1108) is replaced with a simian virus 40 (SV40) early transcription unit and GFP ORF. A deletion of the modulator was shown previously to have no effect on MIE gene expression and viral replication in NT2 cells (57). The SV40 early promoter exhibits early/late transcriptional kinetics (57), indicating its dependency on HCMV IE events for activity. Thus, GFP production indirectly reflects activation of the lytic life cycle. Monitoring by fluorescent microscopy of GFP-positive NT2 cells at 48 h p.i. (MOI of 10 to 50) revealed that less than 0.01% of cells initiated the lytic viral life cycle (data not shown). The same infected cells were subjected to Gardella gel electrophoresis and Southern blot analysis using an HCMV-specific probe. Isolated virions were mixed with uninfected cells to mark the position of the 240-kbp linear genome. As shown in Fig. 1A, three types of viral genomic structures are evident based on electrophoretic mobility. The most abundant structure migrates at rate comparable to that of a 240-kbp linear genome. A lesser amount of viral genomes are immobile, and their presence was confirmed by an independent method described in Materials and Methods. Immobility is a known feature of large relaxed open circular (OC) or linear or branched concatemeric molecules. The third viral genomic structure is low in abundance, migrates slower than unit-length linear genomes, and is not contained in virions. These features are reminiscent of the CCC viral genomes in latently infected monocytes (13). The viral DNA polymerase inhibitor phosphonoformic acid (PFA) does not affect the abundance of the three viral genomic structures. The addition of RA after viral absorption to induce cellular differentiation also does not appreciably alter abundance of these viral genomes. Thus, formation of the three viral genomic structures is not dependent on the viral DNA polymerase and is not likely a manifestation of breakthrough spontaneous differentiation.
FIG. 1.
HCMV genomes in uninduced and RA-induced NT2 cells differ in structure. (A) Analysis of HCMV rΔ-640/-1108SVgfp genomes in uninduced NT2 cells at 48 h p.i. rΔ-640/-1108SVgfp was isolated by centrifugation through a sorbital cushion, and uninduced NT2 cells were grown in stem cell conditions (see Materials and Methods). Mock (MOC)- or rΔ-640/-1108SVgfp-infected cells (MOI of 50) were washed thrice after viral absorption (1.5 h). RA (10 μM), PFA (400 μg/ml), or nothing (−) was added (Add) to the growth medium. Less than 0.01% of infected NT2 cells without additive emitted green fluorescence at 48 h p.i. (HPI). Cells (106/sample) were harvested, washed, and subjected to Gardella gel electrophoresis and Southern blot analysis using a 32P-labeled HCMV-specific probe. The autoradiogram was exposed for 240 h. Isolated WT virions (VIR) were mixed with uninfected cells (106) to determine mobility of the 240-kbp linear genome among cellular chromosomes. Positions of immobile and 240-kbp linear viral genomes are shown. (B) Comparison of mobility of a 250-kbp BAC with WT genomes in uninduced NT2 cells. Infection was performed as described for panel A except that the source of WT was filtered (0.4-μm-pore-size-filter) crude viral stock (MOI of 10). After viral adsorption, PFA (400 μg/ml) was added to or omitted from the growth medium. At 48 h p.i. (HPI), WT- and mock (MOC)-infected cells (106/sample) were subjected to Gardella gel electrophoresis and Southern blot analysis using a 32P-labeled BAC- or HCMV-specific probe. Isolated virions (VIR) and bacteria containing a 250-kbp BAC were each mixed with uninfected cells (106) to mark positions of 240-kbp linear and 250-kbp CCC molecules, respectively. Autoradiography exposure was 8 and 144 h for BAC- and HCMV-specific probes, respectively. (C) Analysis of WT genomes in RA-induced NT2 cells at 24 and 48 h p.i. (HPI). NT2 cells were pretreated with RA (10 μM) for 7 days prior to infection; RA was omitted during infection. WT- and mock (MOC)-infected RA-induced NT2 cells (106/sample) were subjected to Gardella gel electrophoresis and Southern blot analysis as described for panel B. After viral absorption, PFA (400 μg/ml) was added to or omitted from the growth medium. Isolated virions (VIR) or bacteria containing a 250-kbp BAC were prepared for analysis as described for panel B. Autoradiography exposure was 18 h for BAC- and HCMV-specific probes.
The viral genomes with possible CCC-like structure were further characterized by comparing their electrophoretic mobility to that of a 250-kbp BAC with known CCC conformation. In this experiment, wild-type HCMV (WT) was used instead of rΔ-640/-1108SVgfp to ensure that deletion of the MIERR modulator did not influence our earlier findings. WT- and mock-infected NT2 cells were harvested at 48 h p.i. and subjected to Gardella gel electrophoresis and Southern blot analysis, using either an HCMV- or BAC-specific probe. The findings again reveal the three viral genomic structures of different mobilities (Fig. 1B). Remarkably, one of the genomic structures migrates at a rate equivalent to a 250-kbp BAC. In contrast, the rate of mobility of a 180- or 280-kbp BAC differs considerably from that of the viral CCC-like molecule (data not shown). Once again, the abundance of the viral CCC-like molecule is unaltered by PFA. Thus, these findings indicate some of the 240-kbp viral genomes in NT2 cells form a CCC-like structure that migrates with mobility equivalent to a 250-kbp BAC having CCC conformation.
To examine whether pretreatment of the embryonal NT2 cells with RA for 7 days would allow viral DNA replication, RA-induced NT2 cells were infected with WT virus and subjected to Gardella gel electrophoresis and Southern blot analysis at 24 and 48 h p.i. Parallel analyses of virions and BAC-containing bacteria mixed with uninfected cells marked the positions of 240-kbp linear and 250-kbp CCC DNA molecules, respectively. The findings shown in Fig. 1C provide clear evidence of de novo synthesis of viral DNA by 48 h p.i. Based on electrophoretic mobility, two types of replicative intermediates can be ascertained. The immobile genomic structures are most abundant. They are likely the large concatemers that arise from rolling circle replication and subsequently undergo cleavage into unit-length linear genomes. The other form of replicative intermediate has mobility suggestive of a CCC-like molecule, which may be a product of theta replication of preexisting OC templates. Unlike the viral CCC-like molecule in untreated NT2 cells, the formation of the CCC-like replicative intermediate is abrogated by PFA.
In summary, these findings reveal that a refined embryonal NT2 cell population prohibits viral genome replication. Treatment of the cells with RA prior to infection can overcome this restriction, whereas treatment with RA after infection cannot. In untreated embryonal NT2 cells, some of the nonreplicating viral genomes possess CCC-like structure, implying that they have successfully entered the cell nucleus.
TSA-induced reactivation of the MIERR in embryonal NT2 cells.
These findings (Fig. 1A), as well as those of others (31), revealed RA's ineffectiveness in rescuing viral replication in previously infected embryonal NT2 cells. This outcome was linked to RA's failure to activate viral MIE gene expression (31) despite RA's success in stimulating activity of transfected MIERR segments contained in reporter plasmids (8, 9, 29). On this basis, I postulated that the virus's MIERR in these cells is quickly silenced to render it refractory to the actions of relevant positive-acting transcription factors. HDAC-based repression was considered as a possible silencing mechanism, given its prominent role in gene silencing in other organisms, including other herpesviruses (39). I therefore examined whether inhibition of HDACs by TSA after viral infection could reactivate transcription from a quiescent MIERR.
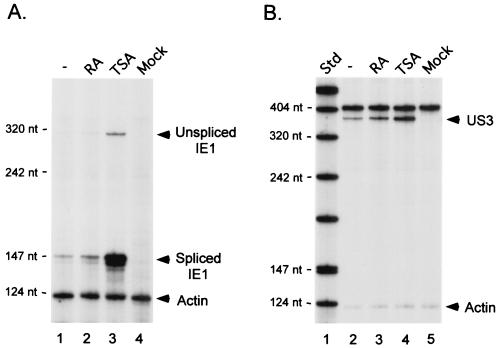
RA or TSA was added to or omitted from growth medium of embryonal NT2 cells following absorption for 1.5 h with WT (MOI of 5). RNA was isolated at 24 h p.i. and subjected to RPA of viral IE1, which originates from the MIERR. Concomitant analysis of cellular actin RNA controlled for sample-to-sample variation. Production of viral US3 RNA made by another IE kinetic class gene located approximately 21 kbp from the MIERR was also assessed. As shown in Fig. 2A, the IE1 RNA of the MIERR is in extremely low amount in untreated cells since it was barely detectable by RPA even when an extended interval of autoradiographic exposure was used. Addition of RA had a minimal (1.8-fold) effect on IE1 RNA abundance compared to untreated cells. In contrast, TSA induced a 19.6-fold increase in amount of both spliced and unspliced IE1 RNAs. The abundance of RNA from the viral IE US3 gene was increased only 3.2-fold by TSA compared to untreated cells (Fig. 2B). RA had a negligible effect on US3 RNA abundance.
FIG. 2.
Effect of TSA or RA on viral MIERR- and US3 promoter-dependent transcription in preinfected embryonal NT2 cells. (A) Analysis of IE1 RNA in WT- and mock-infected NT2 cells at 24 h p.i. RA (10 μM) or TSA (100 ng/ml) was added to or omitted from growth medium at 1.5 h p.i. as described in Materials and Methods. Isolated RNA (25 μg/sample) was subjected to RPA using both viral IE1- and cellular actin-specific riboprobes. Arrows indicate positions of protected unspliced and spliced IE1 and actin RNAs. (B) Analysis of US3 RNA at 24 h p.i. Isolated RNAs used for panel A were also subjected to RPA, with US3- and actin-specific riboprobes. Std, standard; nt, nucleotides.
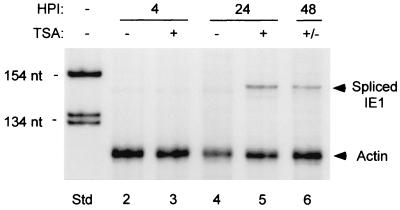
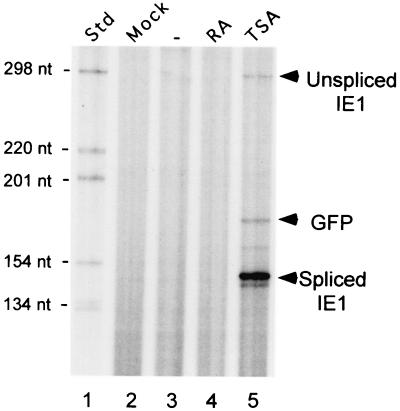
Timing and durability of TSA-induced reactivation of MIERR-dependent transcription were also examined. TSA was added to or omitted from growth medium of embryonal NT2 cells for up to 24 h p.i. following absorption of 1.5 h with WT (MOI of 5). RNA was isolated at 4, 24, and 48 h p.i. and subjected to RPA of viral IE1 RNA. There was not a rapid response to TSA since IE1 RNA was not evident at 4 h p.i. (Fig. 3), nor was there detectable TSA-inducible IE1 RNA at 8 h p.i. (data not shown). A marked increase in IE1 RNA production was reproducibly evident by 24 h of TSA exposure, although the magnitude of this increase varied between experiments. The continual presence of TSA was not required for sustaining IE1 RNA levels for at least another 24 h (Fig. 3).
FIG. 3.
Timing and durability of TSA-induced reactivation of MIERR-dependent transcription in embryonal NT2 cells. (A) Analysis of IE1 RNA in WT-infected NT2 cells at 4, 24, and 48 h p.i. (HPI). TSA (100 ng/ml) was added to or omitted from growth medium at 1.5 h p.i. and removed from the medium at 24 h p.i. RNA (25 μg/sample) was isolated at the indicated times p.i. and subjected to RPA using both viral IE1- and cellular actin-specific riboprobes. Arrows indicate positions of protected spliced IE1 and actin RNAs. nt, nucleotides; Std, standard
Thus, TSA reactivates MIERR-dependent transcription in embryonal NT2 cells. The kinetics of this response are consistent with those of TSA-mediated reactivation of other viral or cellular genes that are known to be subject to histone-mediated repression (15, 18). The reactivation of the MIERR continues after withdrawal of TSA treatment. In contrast, RA has a minimal to negligible effect on MIERR-dependent transcription in these cells.
MIERR-dependent transcription is not increased by removal of the 21-bp repeats and modulator.
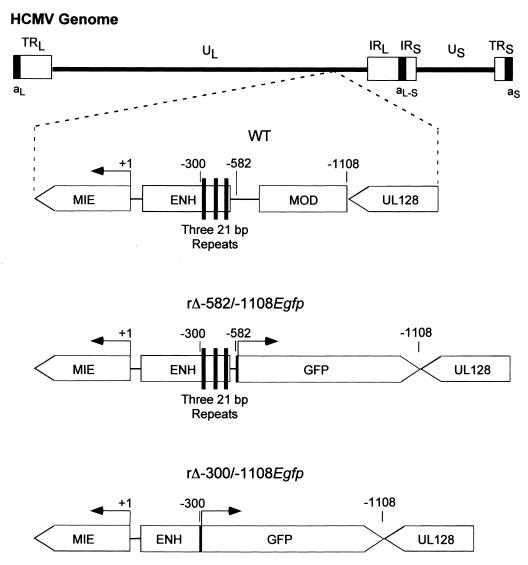
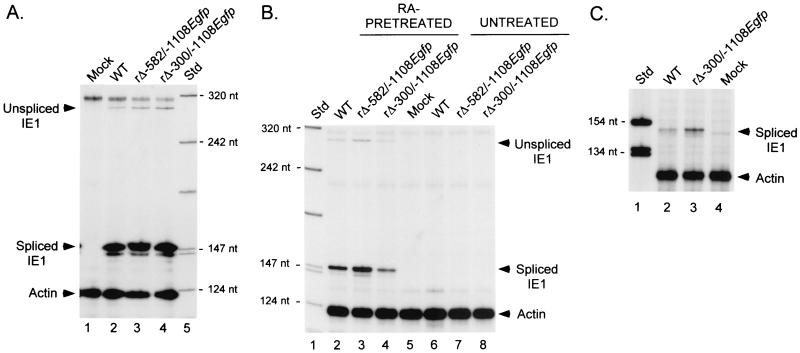
Previous studies of transfected MIERR segments found the distal enhancer containing three 21-bp repeats and the modulator to repress transcriptional activity in NT2 cells but not in their RA-induced descendants. Given the reiteration of putative cis-acting repressors within the MIERR, such functional redundancy might explain our prior finding of failure to alleviate repression by selective deletion of the virus's modulator (56). To test this hypothesis, I determined whether repression could be lessened by combined deletion of the virus's distal enhancer and modulator, as was demonstrated with a similar deletion of transfected MIERR segments (43, 49, 81). A recombinant HCMV, termed rΔ-300/-1108Egfp, was shown recently to lack both distal enhancer and modulator (-300 to -1108) (56). This virus has an adenovirus E1b TATA box and GFP cassette at the site of the deletion. rΔ-582/-1108Egfp has the same insertion at the site of the modulator deletion but retains its distal enhancer (56) (Fig. 4). Both viruses were shown recently to be comparable to WT in function of their MIERRs in HFF cells at an MOI of ≥1 (56). However, rΔ-300/-1108Egfp differed from the other viruses in exhibiting deficient MIERR-dependent transcription at low MOI (56). To avoid this conditional deficiency, embryonal NT2 cells were infected at an MOI of 3.
FIG. 4.
Schematic diagram of HCMVs lacking the MIERR distal enhancer and modulator or modulator alone. The MIERR of WT virus is composed of promoter (+1 to -64), enhancer (ENH; -65 to -550), unique region (-551 to -749), and modulator (MOD; -750/-1140). HCMVs rΔ-300/-1108Egfp and rΔ-582/-1108Egfp have deletions in the MIERR from base positions -300 to -1108 and -582 to -1108, respectively (56). Each of them also has adenovirus E1b TATA box, gfp ORF, and SV40 early intron and polyadenylation signal inserted at the site of deletion. Three copies of the 21-bp repeats (black bars) are located in the MIERR distal enhancer (-300 to -550), which was deleted from rΔ-300/-1108Egfp. Positions of MIE and putative UL128 genes (open boxes) are shown. Depicted above is the HCMV genome and its unique long (UL) and short (US), internal repeat long (IRL) and short (IRS), terminal repeat long (TRL) and short (TRS), and a-sequence (as) components.
RA-pretreated and untreated embryonal NT2 cells were infected in parallel with equivalent titers of WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp. At 24 h p.i., viral IE1 RNA production was assessed by RPA and compared to the actin RNA control. As shown in Fig. 5A, the IE1 RNAs of WT, rΔ-582/-1108Egfp, and rΔ-300/-1108Egfp are produced in equivalent amounts in HFF cells at an MOI of 1, confirming the similarity in input viral titers (56). All three viruses also make substantial amounts of IE1 RNA in RA-pretreated NT2 cells despite omission of RA after the infections (Fig. 5B). rΔ-300/-1108Egfp makes slightly less IE1 RNA (1.5 to 2.5-fold) in these cells compared to WT and rΔ-582-1108Egfp. In untreated embryonal NT2 cells, none of the viruses produces IE1 RNA in amounts that are detectable by the RPA (Fig. 5B). Addition of TSA after viral absorption overcomes this repression regardless of whether the distal enhancer and modulator are present (Fig. 5C). Moreover, the latter finding reveals that rΔ-300/-1108Egfp's inability in producing detectable IE1 RNA in untreated embryonal NT2 cells is not because of lower efficiency in viral entry when compared to WT infection.
FIG. 5.
MIERR-dependent transcription is not increased by deletion of the 21-bp repeats and modulator. (A) Analysis of viral IE1 RNAs in WT-, rΔ-582/-1108Egfp-, rΔ-300/-1108Egfp-, or mock-infected HFF cells at 8 h p.i. Infections (MOI of 3) were performed in parallel with equivalent input viral titers as described in Materials and Methods. Isolated RNA was subjected to RPA, using IE1- and actin-specific riboprobes. Arrows point to positions of protected unspliced and spliced IE1 and actin RNAs. Std, standard; nt, nucleotides. (B) Analysis of viral IE1 RNAs in WT-, rΔ-582/-1108Egfp-, rΔ-300/-1108Egfp-, or mock-infected RA-pretreated or untreated NT2 cells at 24 h p.i. Viral stocks used for panel A were used to infect NT2 cells (MOI of 3). Isolated RNA (25 μg/sample) was subjected to RPA, using both IE1- and actin-specific riboprobes. (C) TSA-induced reactivation of IE1 RNA production in NT2 cells preinfected with WT or rΔ-300/-1108Egfp. Infections (MOI of 3) were performed in parallel with viral inocula used for experiments shown in panels A and B. TSA (100 ng/ml) was added 1.5 h p.i. RNA (25 μg/sample) was isolated at 24 h p.i. and subjected to RPA, using IE1- and actin-specific riboprobes.
These findings indicate that deletion of the virus's distal enhancer and modulator does not appreciably increase MIERR-dependent transcription in embryonal NT2 cells. The ability of TSA to reactivate a MIERR lacking these components suggests that another mechanism(s) of repression may be operating in embryonal NT2 cells.
Enhancer inactivity accounts for lack of MIERR-dependent transcription.
To determine whether lack of MIERR-dependent transcription was a result of enhancer inactivity, I examined the function of a heterologous promoter that is also enhancer dependent but is not subject to negative autoregulation by IE2 p86. HCMV rΔ-300/-1108Egfp was used for this purpose for two reasons. First, this virus has only a heterologous TATA box as its promoter that is fused to a downstream GFP gene (Fig. 4). While this simple promoter replaces the distal one-third of the enhancer, a major portion of the enhancer remains located between both heterologous and native promoters, which are positioned back-to-back. Second, the heterologous promoter exhibits IE transcriptional kinetics, inferring that its function is dependent on enhancer activity.
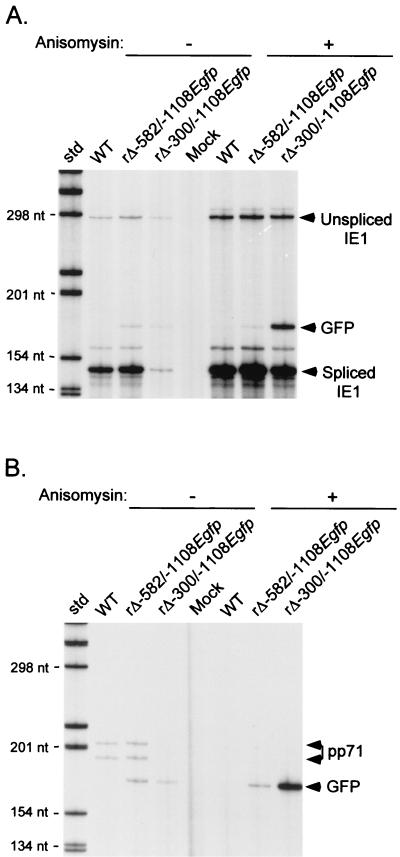
To demonstrate the IE kinetics of rΔ-300/-1108Egfp's heterologous promoter, HFF cells were infected with rΔ-300/-1108Egfp, rΔ-582/-1108Egfp, or WT at an MOI of 0.1 in the presence or absence of anisomycin, a protein synthesis inhibitor. IE1 and GFP RNAs produced from native and heterologous promoters, respectively, were analyzed by RPA at 8 h p.i. RNA of the viral early/late pp71 (UL82) gene was also assessed to determine the completeness of protein synthesis inhibition. As shown in Fig. 6, the IE1 and GFP RNAs of rΔ-300/-1108Egfp are made in abundance despite inhibition of protein synthesis, indicating that both promoters function with IE kinetics. The amount of protected GFP RNA present at 8 h p.i. is similar to that of unspliced IE1 RNA but less than that of spliced IE1 RNA. A perceived difference in levels of protected GFP and IE1 RNAs could reflect dissimilarity in probes, transcriptional activities, or posttranscriptional events. Notably, the GFP RNA of rΔ-582/-1108Egfp is barely detectable in the presence of anisomycin. This finding differs slightly from previous findings of other promoter types implanted in this region, which do not appreciably yield IE transcription (52, 57). Since the infection was performed at low MOI, the activities of rΔ-300/-1108Egfp's native and heterologous MIERR promoters are lessened by the distal enhancer deletion. However, anisomycin restores activities to both promoters, suggesting a compensatory mechanism that acts through elements shared by these neighboring promoters. In contrast, anisomycin effectively inhibits activation of the early/late kinetic class viral pp71 gene (Fig. 6B).
FIG. 6.
The heterologous MIERR promoter of rΔ-300/-1108Egfp exhibits IE transcriptional kinectics. (A) Analysis of viral IE1 and GFP RNAs in WT-, rΔ-582/-1108Egfp-, rΔ-300/-1108Egfp- or mock-infected HFF cells at 8 h p.i. in the presence or absence of anisomycin (100 μM). Infections were performed in parallel at an MOI of 0.05. Isolated RNA (20 μg/sample) was subjected to RPA, using both IE1- and GFP-specific riboprobes. Arrows indicate positions of protected unspliced and spliced IE1 and GFP RNAs. (B) Analysis of pp71 (UL82) and GFP RNAs of these viruses at 8 h p.i. RNAs used for Fig. 5A were also subjected to RPA, with pp71- and GFP-specific riboprobes. Arrows indicate positions of protected pp71 and GFP RNAs. std, standard; nt, nucleotides.
I determined whether the function of rΔ-300/-1108Egfp's heterologous promoter mirrored that of the native MIERR promoter in untreated and TSA- and RA-treated NT2 cells. RA or TSA was added to or omitted from growth medium following viral absorption for 1.5 h (MOI of 3). GFP and IE1 RNAs were analyzed by RPA at 24 h p.i. Prior to this analysis, samples were adjusted for equivalence in cellular actin RNA by RPA. Figure 7 reveals that both GFP and IE1 RNAs are nondetectable in untreated and RA-treated cells, but their amounts are greatly increased by TSA. GFP and IE1 RNA production could not be analyzed in the presence of anisomycin because of rapid drug-induced apoptosis (data not shown).
FIG. 7.
TSA reactivates both heterologous and native MIERR promoters of rΔ-300/-1108Egfp in embryonal NT2 cells. (A) Analysis of IE1 and GFP RNAs in WT- and mock-infected embryonal NT2 cells at 24 h p.i. RA (10 μM) or TSA (100 ng/ml) was added to or omitted from growth medium at 1.5 h p.i. Isolated RNA was subjected to RPA, using both IE1- and GFP-specific riboprobes. Samples were normalized for the amount of cellular actin RNA as determined by RPA (data not shown). Arrows indicate positions of protected unspliced and spliced IE1 and GFP RNAs. std, standard; nt, nucleotides.
These findings show that rΔ-300/-1108Egfp has two IE kinetic class MIERR promoters that share an enhancer containing binding sites for NF-κB/rel, CREB/ATF, and RAR-RXR. Because the heterologous promoter is not subject to IE2 p86-mediated negative autoregulation, its lack of activity in untreated embryonal NT2 cells is likely a reflection of enhancer inactivity. Like the neighboring native promoter, the heterologous promoter's silence is relieved considerably by TSA but not by RA.
Delayed TSA treatment still reactivates the MIERR and produces infectious virus.
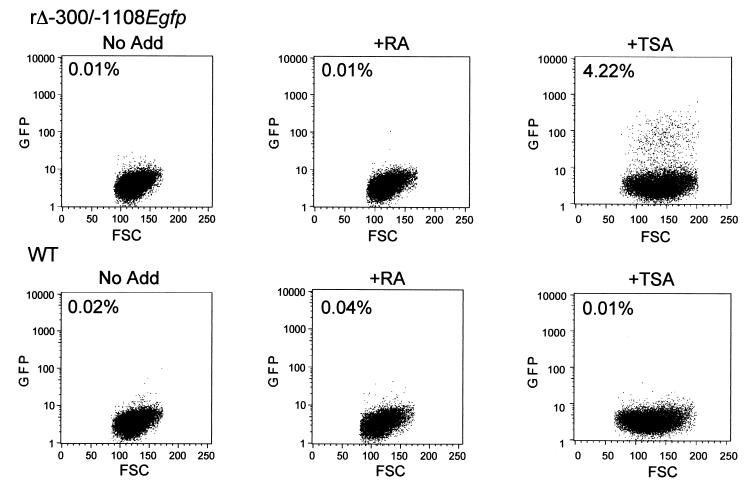
I examined whether delaying the addition of TSA to embryonal NT2 cells at 24 h p.i. would still allow induction of MIERR activity. rΔ-300/-1108Egfp was exploited for this purpose since the GFP it expresses is a marker of MIERR-dependent transcription and can be easily assessed by flow cytometry. WT-infected NT2 cells were analyzed in parallel to control for autofluorescence. The TSA treatment period of 48 h allowed time for accumulation of GFP. An RA treatment group was included for comparison and to corroborate prior RNA findings. As shown in Fig. 8, less than 1 of 104 of the untreated rΔ-300/-1108Egfp-infected NT2 cells exhibit fluorescence at 72 h p.i. This rate is not significantly altered by RA treatment. In contrast, TSA treatment results in a greater than 400-fold increase in number of cells emitting GFP fluorescence, which comprises 4.22% of the TSA-treated cell population. This finding was not inflated by difference in cell viability, which differed minimally among the groups based on propidium iodide exclusion. Although TSA halts cell growth (data not shown), this alone cannot explain the marked increase in proportion of cells expressing GFP at 48 h posttreatment. Comparable results were obtained with rΔ-582/-1108Egfp, indicating that the findings are not unique to a virus with a distal enhancer deletion (data not shown).
FIG. 8.
Percentages of infected embryonal NT2 cells with reactivated MIERR as determined by flow cytometry. rΔ-300/-1108Egfp - and WT-infected embryonal NT2 cells (MOI of 3) were analyzed by flow cytometry at 72 h p.i. RA (10 μM) or TSA (100 ng/ml) was added to or omitted (No Add) from growth medium at 24 h p.i. Ten thousand live cell events in each experimental group were acquired for analysis. For rΔ-300/-1108Egfp, the proportion of TSA-treated and untreated infected cells excluding propidium iodide varied by 21%. FSC, forward scatter.
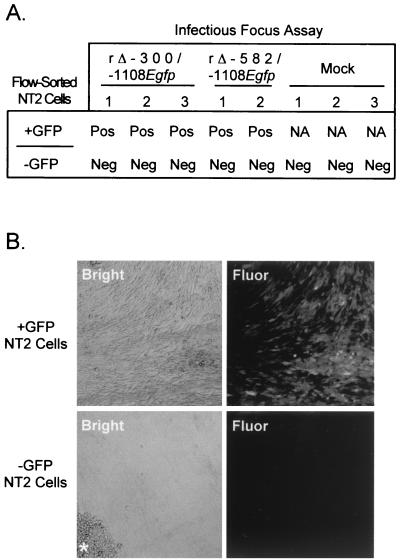
To determine whether the TSA-induced reactivation of viral IE gene expression was followed by production of infectious virus, embryonal NT2 cells were infected with rΔ-300/-1108Egfp or rΔ-582/-1108Egfp (MOI of 3) for 24 h and then treated with TSA for 48 h. Live infected cells were flow sorted on the basis of GFP fluorescence. These infected GFP-positive or -negative NT2 cells were placed in contact with HFF cells at a ratio of 1:103. The HFF monolayer was monitored sequentially for 30 days for evidence of CPE and GFP fluorescence. Analyses reveal that all of the replicate samples containing GFP-expressing NT2 cells yield infectious foci in the HFF monolayer (Fig. 9A). The discrete foci of HFF cells exhibiting CPE also fluoresce because of GFP production, thus providing evidence of transfer of virus among cells (Fig. 9B). Only a small portion of the initial GFP-positive NT2 cell population produces infectious foci (2 to 5 infectious foci per 100 NT2 cells), suggesting an inefficiency in either release of infectious virus or completion of the lytic viral life cycle. rΔ-300/-1108Egfp may be less efficient than rΔ-582/-1108Egfp in producing infectious virus because it yields fewer infectious foci (data not shown). In contrast, the infected NT2 cells not expressing GFP cannot produce CPE or transfer GFP fluorescence (Fig. 9). Unsorted, infected NT2 cells that did not receive TSA treatment also failed to produce infectious foci (data not shown). Clonal expansion of infected NT2 cells lacking GFP fluorescence was a frequent occurrence, yielding tightly packed mounds of nonfluorescing NT2 cells scattered throughout the HFF monolayer (Fig. 9B). GFP-positive NT2 cells very infrequently formed such colonies, and the few colonies that arose lacked GFP-fluorescing cells. This finding is consistent with the known function of viral proteins (e.g., IE2 and UL69 proteins) of the lytic program to block cell cycle progression (14, 23, 35, 38, 51, 64, 92). Last, the capacity for TSA-induced reactivation to produce infectious virus is not limited to these recombinant viruses because WT HCMV also possesses this ability (data not shown).
FIG. 9.
Infectious focus assay of TSA-induced preinfected NT2 cells. (A) Scoring of infectious focus assays for rΔ-300/-1108Egfp- and rΔ-582/-1108Egfp-infected NT2 cells (MOI of 3). Infected embryonal NT2 cells were induced with TSA (100 ng/ml) at 24 h p.i. and flow sorted into GFP-positive (+GFP) and -negative (−GFP) groups at 72 h p.i. Mock, +GFP, or −GFP NT2 cells (102) were cocultivated with subconfluent HFF cells (105). The cultures were monitored for evidence of infectious foci for 30 days by light and fluorescent microscopes. Infectious focus assays were performed in triplicate except for rΔ-582/-1108Egfp, which was tested in duplicate. Infectious focus assays were scored as positive (Pos) or negative (Neg). NA, not applicable. (B) Confocal microscopy of infectious focus. Embryonal NT2 cells were infected with rΔ-300/-1108Egfp, treated with TSA (100 ng/ml) at 24 h p.i., and flow sorted into GFP-positive (+GFP) and -negative (−GFP) groups at 72 h p.i. Segregated infected NT2 cells were cocultivated with HFF cells as described above. Inverted confocal microscopy was performed a 21 days of culture. The asterisk marks the location of a colony of NT2 cells within the HFF monolayer.
Thus, TSA reactivates preexisting quiescent viral genomes to yield infectious virus. The reemergence of transmissible virus coincides with the reactivation of the MIERR.
DISCUSSION
The findings presented here increase our understanding of regulatory mechanisms that control HCMV MIERR-dependent transcription and, consequently, viral replication. Silencing of viral promoters, such as the MIERR, is a key determinant of viral latency (58, 80) yet is a major impediment to gene therapy (10). NT2 cells that are restricted to embryonal cell growth conditions provide a tractable model in which to define mechanisms of MIERR silencing. The confounding variable of superimposed breakthrough lytic viral life cycle events is greatly reduced in this refined cell population. After infection, HCMV genomes enter nuclei of these embryonal NT2 cells but fail to carry out MIERR-dependent transcription (Fig. 1 to 3 and 5 to 8). This finding reinforces those reported previously, using standard culture conditions (45, 66). In the refined embryonal NT2 cell population, there were minimal to negligible amounts of MIERR-dependent transcription (Fig. 2, 3, 5, 6, and 7) and no detectable viral DNA polymerase-dependent genome replication (Fig. 1). These cells also contain a nonreplicating viral genome structure that comigrates electrophoretically with a 250-kbp BAC with superhelical twists (Fig. 1), implying that some HCMV genomes have CCC conformation. Recently, HCMV genomes with electrophoretic mobility equivalent to that of 230-kbp bacterial megaplasmid were also detected in latently infected monocytes of healthy donors (13). The results do not determine whether the viral CCC genomes in embryonal NT2 and latently infected monocytes cells are equivalent in structure but do support this concept. The HCMV CCC genomes presumably have superhelical twists, based both on their electrophoretic mobility and on prior reports showing that relaxed or nicked circles do not enter the gel (11, 22, 54). Supercoiling of HCMV genomes may be a consequence of nucleosome formation, since CCC genomes of other DNA viruses, including Epstein-Barr virus (24, 39, 77), exhibit supercoiling for this reason (28, 63).
On the basis of the findings, I conclude that a mechanism of active repression rapidly imposes a block to MIERR-dependent transcription in embryonal NT2 cells. Three considerations support this conclusion. (i) Increasing the availability of positive-acting transcription factors is largely insufficient in reactivating MIERR-dependent transcription. For instance, the MIERR is poorly or negligibly activated by functioning liganded RARs even though three RAREs are present in its enhancer (Fig. 2, 6, and 7). Also, NF-κB quickly translocates to the NT2 cell nucleus in response to RA (76) yet fails to activate the MIERR, which contains four NF-κB/rel binding sites in its enhancer (58). These findings differ from those of transfected reporter plasmids in which RA can readily activate MIERR segments in the NT2 cells (8, 9, 29). Nevertheless, pretreatment of NT2 cells with RA allows MIERR-dependent transcription from the viral genome (Fig. 5), consistent with previous reports (30, 45, 66). Although positive-acting transcription factors by themselves are ineffective in initiating MIERR reactivation, they likely still contribute to the process. This notion may explain why the IE US3 promoter, which has a much less extensive enhancer system (58), appears to support less transcription when reactivated by TSA (compare Fig. 2A and B). (ii) A heterologous MIERR promoter that is exempt from IE2 p86-mediated negative autoregulation, yet has enhancer binding sites for RAR and NF-κB/rel, is also not susceptible to reactivation by RA (Fig. 6 and 7). Thus, the lack of MIERR-dependent transcription in embryonal NT2 cells likely results from inactivation of the enhancer. (iii) A specific inhibitor of HDAC-based repression can effectively reactivate MIERR-dependent transcription from either a native or heterologous promoter (Fig. 2, 3, 6, and 7). Although these findings do not distinguish whether relief of MIERR silencing is a direct or indirect result of removing acetyl groups from histones, nonetheless they add to a growing body of circumstantial evidence suggesting that chromatin can form on viral genomes in these cells.
How might the MIERR be repressed in the embryonal cells? Conventional wisdom would support the notion of involvement of specific transcription factors that bind to cognate sites in the MIERR to confer repression, perhaps through modification of chromatin structure. A prior report of differences among DNase I hypersensitivity profiles of the MIERR of viral genomes in embryonal and RA-pretreated NT2 cells is consistent with this view (66). Previous studies of transfected MIERR segments in embryonal NT2 cells reveal that transcriptional repression is alleviated by removal of the modulator or distal enhancer, which contains the three 21-bp repeats (36, 43, 49, 67, 78, 81). The binding in vitro of cellular transcription factors, such as YY1 and/or silencing binding protein, to sites clustered in these regions correlated with the MIERR silencing (16, 36, 49, 78, 89). The findings presented here raise uncertainty about the significance of these observations since removal of both modulator and 21-bp repeats or modulator alone from the virus does not relieve MIERR silencing in embryonal NT2 cells (Fig. 5). The latter finding matches that for a different modulator-negative HCMV studied previously in embryonal NT2 cells grown under standard conditions (57). While the modulator and 21-bp repeats are not required for MIERR silencing, I do not discount the possibility of these elements functioning as accessory transcriptional repressors. A redundancy in silencing mechanisms is conceivable given the diversity and repetition of mechanisms involved in MIERR activation during lytic infection (56, 58). Whether other cellular transcription factors (e.g., Gfi-1) act on the MIERR to render it inactive remains to be determined. A prevailing paradigm of gene silencing by specific transcriptional repressors invokes the intermediary role of HDACs. HDACs are generally part of multiprotein complexes that are recruited to specific genes by transcription factors (44, 69, 90). They rid histone tails of acetyl groups to change chromatin conformation in a manner that results in gene silencing. Some HDAC-containing complexes possess ATP-dependent chromatin remodeling activity that can also control gene expression (90). Not all HDAC-containing complexes function through transcription factors that recognize specific DNA sequences; some are recruited by methyl-CpG binding proteins to repress transcription of methylated genes or promoters (12, 68). This type of transcriptional repression is also relieved by TSA (12, 68), although not every kind of methylation-mediated repression is abrogated by HDAC inhibitors (15, 50). The MIERR is enriched in CpG dinucleotides, making it potentially vulnerable to methylation and the attendant effects. Notably, DNA methylation has been found to be important in Epstein-Barr virus promoter silencing during viral latency (71, 86), and it efficiently targets newly integrated retroviral DNA in embryonal cells (10). Further studies are needed to determine if CpG methylation plays a role in MIERR silencing in embryonal NT2 cells. Finally, I cannot exclude the possibility of viral genomes failing to enter a preferred nuclear compartment that fosters transcription. It has been shown that in HCMV-infected fibroblasts, viral replication initiates from subnuclear ND10 (or PML oncogenic) domains, where input viral genomes and IE proteins initially localize (2). Perhaps the observed paucity of such domains in embryonal NT2 cells (47) is a significant factor in the silencing of viral genomes in these cells.
TSA-induced reactivation of MIERR-dependent transcription is accompanied by production of infectious virus (Fig. 9). This observation adds to previous findings suggesting that the MIE gene products play a pivotal role in initiating lytic viral infection (32, 37, 62, 70, 84). However, TSA-induced MIERR reactivation appears to be inefficient (Fig. 8), and the likelihood of this reactivation occurring decreases as the interval of time from infection to TSA treatment increases (data not shown). While the reason for this inefficiency is unknown, it is possible that not all cells contain competent viral genomes, reflecting a problem with viral genome entry or retention. The low abundance of CCC-like HCMV genomes in the overall cell population at 48 h p.i. (Fig. 1) would support this concept. Alternatively, a subset of viral genomes may be modified to a TSA-resistant form in a time-dependent fashion. Notably, the silencing of integrated retroviral genomes in embryonal cells involves various mechanisms that differ in their timing of maximal activity (3, 50). For example, the methylation density on proviral genomes increases over time (27, 46, 50), and genes with high methylation density may not be susceptible to activation by TSA (15, 50). Conceivably, HCMV gene silencing may involve mechanisms that have also hindered gene therapy using modified herpes simplex virus genomes, in which the activities of implanted heterologous promoters, such as the HCMV MIE promoter-enhancer, are gradually extinguished by an unknown mechanism(s) (74, 75). Whether any of these considerations apply to HCMV genome silencing in embryonal NT2 cells remains to be determined.
This study may also provide insight into a functional component of the MIERR that may shield upstream viral genes from regulatory mechanisms conferring IE transcriptional kinetics, which are based ostensibly in the enhancer. The distal enhancer (-300 to -582) may hold this blocking function since its removal imparts IE transcriptional kinetics to an upstream gene (Fig. 6). This regulatory function differs from the upstream element that represses UL127 promoter activity, as removal of the upstream element (-582 to -693) does not give way to IE kinetic class transcription (6, 52). However, I detected a slight amount of IE transcription from an E1b TATA box that is located at -582 (Fig. 6), raising the possibility that this level of transcription may be amplified by placement of this particular heterologous promoter closer to enhancer elements. Therefore, additional studies are required to determine whether the distal enhancer region does indeed contain an enhancer blocker for maintaining the unidirectional characteristic of IE transcription from the MIERR.
In closing, my findings indicate that the embryonal NT2 cell is a useful model in which to elucidate mechanisms that govern repression or derepression of HCMV MIERR activity and viral replication. Such information will also improve our understanding of how transgenes are silenced and will guide future studies in determining the mechanisms by which HCMV latency is controlled in monocytes and their precursors.
ACKNOWLEDGMENTS
I am grateful to Mark Stinski for critical reading of the manuscript. I thank Mark Stinski, members of the Stinski laboratory, and Jim McCoy for helpful discussions of this work. I thank Jonathan Pruessner and Michael Keller for excellent assistance with the Gardella gel and cultures, respectively.
This work was supported by the National Institutes of Health (grant R29-AI-40130), American Cancer Society (Institutional Research Grant IN-122R), and March of Dimes (grant FY99-549).
REFERENCES
- 1.Adler S T, McVoy M A. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J Virol. 1994;68:1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J-H, Jang W J, Hayward G S. The human cytomegalovirus IE2 and UL112–113 proteins accumulate in viral DNA replication compartments that initiate from periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J Virol. 1999;73:10458–10471. doi: 10.1128/jvi.73.12.10458-10471.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akugan E, Ziegler M, Grez M. Determinants of retrovirus gene expression in embryonal carcinoma cells. J Virol. 1991;65:382–388. doi: 10.1128/jvi.65.1.382-388.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews P W. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984;103:285–293. doi: 10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- 5.Andrews P W, Damjanov I, Simon D, Banting G S, Carlin C, Dracopoli N C, Fogh J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Investig. 1984;50:147–162. [PubMed] [Google Scholar]
- 6.Angulo A, Kerry D, Haung H, Borst E-M, Razinsky A, Wu J, Hobom U, Messerle M, Ghazal P. Identification of a boundry domain adjacent to the potent human cytomegalovirus enhancer that represses transcription of the divergent UL127 promoter. J Virol. 2000;74:2826–2839. doi: 10.1128/jvi.74.6.2826-2839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angulo A, Suto C, Boehm M F, Heyman R A, Ghazal P. Retinoid activation of retinoic acid receptors but not of retinoid X receptors promotes cellular differentiation and replication of human cytomegalovirus in embryonal cells. J Virol. 1995;69:3831–3837. doi: 10.1128/jvi.69.6.3831-3837.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angulo A, Suto C, Heymen R A, Ghazal P. Characterization of the sequences of the human cytomegalovirus enhancer that mediate differential regulation by natural and synthetic retinoids. Mol Endocrinol. 1996;10:781–793. doi: 10.1210/mend.10.7.8813719. [DOI] [PubMed] [Google Scholar]
- 10.Bestor T H. Gene silencing as a threat to the success of gene therapy. J Clin Investig. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beverly S M. Characterization of the ‘unusual’ mobility of large circular DNAs in pulse field-gradient electrophoresis. Nucleic Acids Res. 1988;16:925–939. doi: 10.1093/nar/16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird A, Wolffe A P. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 13.Bolovan-Fritts C A, Mocarski E S, Wiedeman J A. Peripheral blood CD14+ cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood. 1999;93:394–398. [PubMed] [Google Scholar]
- 14.Bresnahan W A, Boldogh A I, Thompson E A, Albrecht T. human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 15.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Synergy of demethylation and histone deacetylase inhibition in the reexpression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 16.Chan Y, Tseng W, Hayward G. Two distinct upstream regulatory domains containing multicopy cellular transcription factor binding sites provide basal repression and inducible enhancer characteristics to the immediate-early IES (US3) promoter from human cytomegalovirus. J Virol. 1996;70:5312–5328. doi: 10.1128/jvi.70.8.5312-5328.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan Y-J, Chiou C-J, Huang Q, Hayward G S. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediate both basal enhancement and phorbol ester responsiveness or primate cytomegalovirus major immediate-early promoters in monocyte and T-lymphocyte cell types. J Virol. 1996;70:8590–8605. doi: 10.1128/jvi.70.12.8590-8605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W Y, Townes T M. Molecular mechanism for silencing virally transduced gene involves histone deacetylation and chromatin condensation. Proc Natl Acad Sci USA. 2000;97:377–382. doi: 10.1073/pnas.97.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung W M W, Fu W Y, Hui W S, Ip N Y. Production of human CNS neurons from embryonal carcinoma cells using a cell aggregation method. BioTechniques. 1999;26:946–954. doi: 10.2144/99265rr04. [DOI] [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Decker L L, Klaman L D, Thorley-Lawson D A. Detection of the latent form of Epstein-Barr virus DNA in the blood of healthy individuals. J Virol. 1996;70:3286–3289. doi: 10.1128/jvi.70.5.3286-3289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dittmer D, Mocarscki E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dyson P J, Farrell P J. Chromatin structure of Epstein-Barr virus. J Gen Virol. 1985;66:1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- 25.Fowler K B, Stagno S, Pass R F, Britt W J, Boll T J, Alford C A. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N Engl J Med. 1992;326:663–167. doi: 10.1056/NEJM199203053261003. [DOI] [PubMed] [Google Scholar]
- 26.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaustch J W, Wilson M C. Delayed de novo methylation in teratocarcinoma suggests additional tissue-specific mechanisms for controlling gene expression. Nature. 1986;301:32–37. doi: 10.1038/301032a0. [DOI] [PubMed] [Google Scholar]
- 28.Germond J E, Hirt B, Oudet P, Gross-Bellard M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci USA. 1975;72:1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghazal P, DeMattei C, Giulietti E, Kliewer S A, Umesono K, Evans R M. Retinoic acid receptors initiate induction of the cytomegalovirus enhancer in embryonal cells. Proc Natl Acad Sci USA. 1992;89:7630–7634. doi: 10.1073/pnas.89.16.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonczol E, Andrews P W, Plotkin S A. Cytomegalovirus infection of human teratocarcinoma cells. J Gen Virol. 1985;66:509–515. doi: 10.1099/0022-1317-66-3-509. [DOI] [PubMed] [Google Scholar]
- 31.Gonczol E, Andrews P W, Plotkin S A. Cytomegalovirus replicates in differentiated but not in undifferentiated human embryonal carcinoma cells. Science. 1984;224:159–161. doi: 10.1126/science.6322309. [DOI] [PubMed] [Google Scholar]
- 32.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grzimek N K A, Podlech J, Steffens H P, Holtappels R, Schmalz S, Reddehase M J. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J Virol. 1999;73:5043–5055. doi: 10.1128/jvi.73.6.5043-5055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn G, Jores R, Mocarski E S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayashi L M, Blankenship C, Shenk T. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc Natl Acad Sci USA. 2000;97:2692–2696. doi: 10.1073/pnas.050587597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang T H, Oka T, Asai T, Okada T, Merrills B W, Gerston R H, Witson R H, Itakura K. Repression by a differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 1996;24:1695–1701. doi: 10.1093/nar/24.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jault F M, Jault J M, Ruchiti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenkins P J, Binne U K, Farrell P J. Histone acetylation and reactivation of Epstein-Barr virus from latency. J Virol. 2000;74:710–720. doi: 10.1128/jvi.74.2.710-720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones-Villeneuve E M V, Rudnicki M A, Harris J F, McBurney M W. Retinoic acid-induced neural differentiation of embryonal carcinoma cells. Mol Cell Biol. 1983;3:2271–2279. doi: 10.1128/mcb.3.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo K J X, Xu J, Mocarski E S. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kothari S, Baillie J, Sissons J G, Sinclair J H. The 21bp repeat element of the human cytomegalovirus major immediate early enhancer is a negative regulator of gene expression in undifferentiated cells. Nucleic Acids Res. 1991;19:1767–1771. doi: 10.1093/nar/19.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kui M-H, Allis C D. Roles of histone acetyltransferases and deactylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 45.LaFemina R, Hayward G S. Constitutive and retinoic acid-inducible expression of cytomegalovirus immediate-early genes in human teratocarcinoma cells. J Virol. 1986;58:434–440. doi: 10.1128/jvi.58.2.434-440.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laker C, Meyer J, Schopen A, Friel J, Herberlein C, Ostertag W, Stocking C. Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation. J Virol. 1998;72:339–348. doi: 10.1128/jvi.72.1.339-348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Leo C, Zhu J, Wu X, O'Neil J, Park E-J, Chen J D. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu R, Baillie J, Sissons J G, Sinclair J H. The transcription factor YY1 binds to negative regulatory elements in the human cytomegalovirus major immediate early enhancer/promoter and mediates repression in non-permissive cells. Nucleic Acids Res. 1994;22:2453–2459. doi: 10.1093/nar/22.13.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorincz M C, Schubeler D, Goeke S C, Walters M, Groudine M, Martin D A. Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol Cell Biol. 1999;20:842–850. doi: 10.1128/mcb.20.3.842-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu M, Shenk T. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J Virol. 1999;73:676–683. doi: 10.1128/jvi.73.1.676-683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lundquist C A, Meier J L, Stinski M F. A strong transcriptional negative regulatory region between the human cytomegalovirus UL127 gene and the major immediate-early enhancer. J Virol. 1999;73:9032–9052. doi: 10.1128/jvi.73.11.9039-9052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1505. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McVoy M A, Adler S P. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J Virol. 1994;68:1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McVoy M A, Nixon D E, Adler S P. Circularization and cleavage of guinea pig cytomegalovirus genomes. J Virol. 1997;71:4209–4217. doi: 10.1128/jvi.71.6.4209-4217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meier J L, Pruessner J. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J Virol. 2000;74:1602–1613. doi: 10.1128/jvi.74.4.1602-1613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meier J L, Stinski M F. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J Virol. 1997;71:1246–1255. doi: 10.1128/jvi.71.2.1246-1255.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meier J L, Stinski M F. Regulation of cytomegalovirus immediate early genes. Intervirology. 1996;39:331–342. doi: 10.1159/000150504. [DOI] [PubMed] [Google Scholar]
- 59.Mendelson M, Monard S, Sissons P, Sinclair J. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol. 1996;77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 60.Minton E J, Tysoe C, Sinclair J H, Sissons J G. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mocarski E. Cytomegalovirus and its replication. In: Fields B N, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 62.Mocarski E S, Kemble G, Lyle J, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1 149aa is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muller U, Zentgraf H, Eicken I, Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978;201:406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- 64.Murphy E A, Streblow D N, Nelson J A, Stinski M S. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J Virol. 2000;74:7108–7118. doi: 10.1128/jvi.74.15.7108-7118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nelson J, Fish K, Ibanez C, Depto A, Ghazal P, Moses A, Jupp R. Dependence of cytomegalovirus replication on monocyte differentiation. In: Michelson S, Plotkin S A, editors. Multidisciplinary approach to understanding cytomegalovirus disease. New York, N.Y: Elsevier Science; 1993. pp. 77–86. [Google Scholar]
- 66.Nelson J A, Groudine M. Transcriptional regulation of the human cytomegalovirus major immediate-early gene is associated with induction of DNase I-hypersensitive sites. Mol Cell Biol. 1986;6:452–461. doi: 10.1128/mcb.6.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson J A, Reynolds-Kohler C, Smith B. Negative and positive regulation by a short segment in the 5′-flanking region of the human cytomegalovirus major immediate-early gene. Mol Cell Biol. 1987;7:4125–4129. doi: 10.1128/mcb.7.11.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ng H-H, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 69.Ng H N, Bird A. Histone deacetylases: silencers for hire. Trends Biochem Sci. 2000;25:121–125. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- 70.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paulson E J, Speck S H. Differential methylation of Epstein-Barr virus latency promoters facilitates viral persistence in healthy seropositive individuals. J Virol. 1999;73:9959–9968. doi: 10.1128/jvi.73.12.9959-9968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pleasure S J, Lee V M-Y. NTera 2 cells: a human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J Neurosci. 1993;35:585–602. doi: 10.1002/jnr.490350603. [DOI] [PubMed] [Google Scholar]
- 73.Poland S D, Bambrick L L, Dekaban G A, Rice G P A. The extent of human cytomegalovirus replication in primary neurons is dependent on host cell differentiation. J Infect Dis. 1994;170:1267–1271. doi: 10.1093/infdis/170.5.1267. [DOI] [PubMed] [Google Scholar]
- 74.Preston C M, Nicholl M J. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J Virol. 1997;71:7807–7813. doi: 10.1128/jvi.71.10.7807-7813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Segars J H, Nagata T, Bours V, Medin J A, Franzoso G, Blanco J C G, Drew P D, Becker K G, An J, Tang T, Stephany D A, Neel B, Siebenlist U, Ozato K. Retinoic acid induction of major histocompatibility complex class I genes in NTera-2 embryonal carcinoma cells involves induction of NF-κB (p50–p65) and retinoic acid receptor β-retinoid X receptor β heterodimers. Mol Cell Biol. 1993;13:6157–6169. doi: 10.1128/mcb.13.10.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shaw J E, Levinger L F, Carter C. Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J Gen Virol. 1979;29:657–665. doi: 10.1128/jvi.29.2.657-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shelbourn S L, Kothari S K, Sissons J G P, Sinclair J H. Repression of human cytomegalovirus gene expression associated with a novel immediate early regulatory region binding factor. Nucleic Acids Res. 1989;17:9165–9171. doi: 10.1093/nar/17.22.9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shelbourn S L, Sissons J G P, Sinclair J H. Expression of oncogenic ras in human teratocarcinoma cells induces partial differentiation and for human cytomegalovirus infection. J Gen Virol. 1989;70:367–374. doi: 10.1099/0022-1317-70-2-367. [DOI] [PubMed] [Google Scholar]
- 80.Sinclair J, Sissons P. Cytomegalovirus: latent and persistent infection of monocytes and macrophages. Intervirology. 1996;39:293–301. doi: 10.1159/000150501. [DOI] [PubMed] [Google Scholar]
- 81.Sinclair J H, Baillie J, Bryant L A, Taylor-Wiedeman J A, Sissons J G. Repression of human cytomegalovirus major immediate early gene expression in a monocytic cell line. J Gen Virol. 1992;73:433–435. doi: 10.1099/0022-1317-73-2-433. [DOI] [PubMed] [Google Scholar]
- 82.Sinzger C, Jahn G. Human cytomegalovirus cell tropisim and pathogenesis. Intervirology. 1996;39:302–319. doi: 10.1159/000150502. [DOI] [PubMed] [Google Scholar]
- 83.Soderberg-Naucler N C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus from by allogenic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 84.Stenberg R M. The human cytomegalovirus major immediate-early gene. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]
- 85.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involve common and unique protein domains. J Virol. 1990;1990:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tao Q, Robertson K D, Manns A, Hildesheim A, Ambinder R F. The Epstein-Barr virus major latent promoter Qp is constitutively active, hypomethylated, and methylation sensitive. J Virol. 1998;72:7075–7083. doi: 10.1128/jvi.72.9.7075-7083.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taylor-Wiedeman J, Sissons J G, Borysiewicz L K, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 88.Taylor-Wiedeman J A, Sissons J G P, Sinclair J H. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thrower A R, Bullock G C, Bissell J E, Stinski M F. Regulation of a human cytomegalovirus immediate-early gene (US3) by a silencer/enhancer combination. J Virol. 1996;70:91–100. doi: 10.1128/jvi.70.1.91-100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vignali M, Hassan A H, Neely K E, Workman J L. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.von Laer D, Meyer-Koenig U, Serr A, Finke J, Kanz L, Fauser A A, Neumann-Haefelin D, Brugger W, Hufert F T. Detection of cytomegalovirus DNA in CD34+ cells from blood and bone marrow. Blood. 1995;86:4086–4090. [PubMed] [Google Scholar]
- 92.Wiebusch L, Hagemeier C. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J Virol. 1999;73:9274–9283. doi: 10.1128/jvi.73.11.9274-9283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zweidler-McKay P A, Grimes H L, Flubacher M M, Tsichlis P N. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]