Abstract
Our previous studies determined that elevating SOX2 in a wide range of tumor cells leads to a reversible state of tumor growth arrest. Efforts to understand how tumor cell growth is inhibited led to the discovery of a SOX2:MYC axis that is responsible for downregulating c-MYC (MYC) when SOX2 is elevated. Although we had determined that elevating SOX2 downregulates MYC transcription, the mechanism responsible was not determined. Given the challenges of targeting MYC clinically, we set out to identify how elevating SOX2 downregulates MYC transcription. In this study, we focused on the MYC promoter region and an upstream region of the MYC locus that contains a MYC super-enhancer encompassing five MYC enhancers and which is associated with several cancers. Here we report that BRD4 and p300 associate with each of the MYC enhancers in the upstream MYC super-enhancer as well as the MYC promoter region and that elevating SOX2 decreases the recruitment of BRD4 and p300 to these sites. Additionally, we determined that elevating SOX2 leads to increases in the association of SOX2 and H3K27me3 within the MYC super-enhancer and the promoter region of MYC. Importantly, we conclude that the increases in SOX2 within the MYC super-enhancer precipitate a cascade of events that culminates in the repression of MYC transcription. Together, our studies identify a novel molecular mechanism able to regulate MYC transcription in two distinctly different tumor types and provide new mechanistic insights into the molecular interrelationships between two master regulators, SOX2 and MYC, widely involved in multiple cancers.
Keywords: SOX2, c-MYC, p300, BRD4, transcription, super-enhancer, RNA polymerase II, medulloblastoma, colorectal cancer, A-485
The transcription factor c-MYC (MYC) is dysregulated in roughly 70% of human cancers (1). In addition to its direct roles in the transcription of a sizable fraction of the genome, MYC is a master regulator of the cell cycle and a vast array of other essential cellular functions (2). MYC has long been viewed as a high value target in the treatment of cancer, because of its essential roles in cancer formation and progression (3). However, attempts to target MYC have met with considerable difficulties (4). Given the challenges of targeting MYC, efforts to understand how MYC can be downregulated would be highly informative. Recently, we identified an important SOX2:MYC axis that is responsible for the downregulation of MYC when SOX2 is elevated (5). The significance of this SOX2:MYC axis is evident from our efforts to rescue MYC expression. Although there is little or no effect on survival in SOX2-elevated and MYC-downregulated cells, there is massive cancer cell death when MYC levels are rescued in the presence of elevated SOX2 (5). This unexpected finding led to the conclusion that downregulating MYC when SOX2 is elevated serves as an important tumor cell survival strategy.
The critical roles played by SOX2, in both normal and tumor cells, when its levels change have come to light over the past dozen years. During development, high levels of SOX2 limit the proliferation of normal fetal stem cells in multiple tissues (6, 7). Like in normal fetal stem cells, the levels of SOX2 in embryonic stem cells affect their self-renewal. Even a small increase in SOX2 (∼2-fold) in embryonic stem cells rapidly blocks their self-renewal and triggers their differentiation into multiple cell types (8). Importantly, elevated levels of SOX2 have also been identified in subpopulations of tumor cells that possess enhanced tumor-initiating capacity and which are largely quiescent (9, 10, 11). The significance of the relationship between the levels of SOX2 and tumor cell quiescence is notable given the clinical challenge posed by quiescent/slowly proliferating tumor cells (12). These cells serve as a residual tumor cell population that can reinitiate tumor growth, because they are largely resistant to current cancer therapies (12). Not surprisingly, SOX2 has been linked to drug resistance in multiple cancers (9, 13, 14).
Previous work from this laboratory has replicated the growth inhibitory functions of elevated SOX2 in nearly 20 tumor cell lines representing seven types of human cancer (5, 14, 15, 16, 17). This was accomplished by engineering tumor cells for upregulation of SOX2 from a Doxycycline (Dox) inducible promoter. Elevating SOX2 in SOX2-inducible tumor cell lines in vitro invariably leads to growth inhibition (5, 14, 15, 16, 17). When elevated in vivo in three different tumor types, SOX2 induces a reversible state of tumor growth-arrest (17). Importantly, in each case, removal of the inducer (Dox) leads to the rapid return of SOX2 to basal levels and reinitiation of tumor growth at a growth rate virtually identical to that of the control tumors (17). Our efforts to determine how elevated SOX2 inhibits tumor growth led to the finding that elevating SOX2 increases the expression of several 1000 genes while repressing approximately 1500 genes, in particular MYC-target genes (5). Moreover, we observed in multiple tumor cell types that MYC itself is downregulated at both the protein and RNA level when SOX2 is elevated, without a significant decrease in the half-life of MYC protein or its mRNA (5). Significantly, nuclear run-on studies demonstrated that MYC is downregulated at the transcriptional level when SOX2 is elevated (5). However, the molecular mechanism responsible was not determined.
Early studies identified several mechanisms that regulate MYC transcription, including repression at the level of transcriptional elongation (18), positive and negative feedback loops involving MYC itself (19, 20), and mutations in APC signaling (21). Other studies demonstrated that the MYC locus contains multiple TCF4/β-catenin binding sites and that MYC expression is upregulated when serum is added to serum-starved cells, likely due to increases in nuclear levels of β-catenin (22, 23, 24, 25). More recently, a far deeper understanding of MYC expression has begun to emerge with the discovery of multiple enhancers within the 3 Mb MYC locus, including a super-enhancer located far upstream of the MYC promoter (24). This super-enhancer, which is located more than 400 kb upstream of the MYC promoter, consists of a cluster of at least five MYC enhancers (A-E) (24) that are distributed over a region of approximately 50 kb in chromosome 8 (chr8:127214000-127267000). Interestingly, this upstream MYC super-enhancer is within the region of the MYC locus that is associated with multiple cancers, including cancers of the pancreas, thyroid, breast, and colon (26, 27, 28). Attempts made to map the location of the MYC region(s) associated with these tumors employed mouse models in which several regions upstream of the MYC promoter were deleted. These efforts identified a region located between 335 kb and 540 kb upstream of the MYC promoter that substantially increases chemically induced mammary tumorigenesis and increases intestinal polyps when placed on an APCmin background (28). Surprisingly, deletion of the region 2 kb to 540 kb upstream of the MYC promoter did not significantly alter normal development or viability of the mice. Thus, this upstream region is dispensable for healthy cells in vivo, but is required for tumorigenicity (28).
Building on our current understanding of MYC transcriptional regulation, we set out to investigate the molecular mechanisms by which elevated SOX2 downregulates MYC transcription. Toward this goal, we focused on the recruitment of key transcriptional machinery, in particular p300, BRD4, and RNA polymerase II (Pol II), to the MYC super-enhancer and promoter regions within the MYC locus. To help assess whether the findings identified are cell type specific, we performed our studies in two distinctly different human tumor cell types, namely medulloblastoma and colorectal tumor cells. Collectively, the studies described in this report identify a novel molecular mechanism that regulates MYC transcription. Our work also provides new mechanistic insights into the molecular interrelationships between two critical master regulators that underpin a SOX2:MYC axis.
Results
Elevating SOX2 reduces the level of RNA Pol II at MYC promoters and the MYC gene body
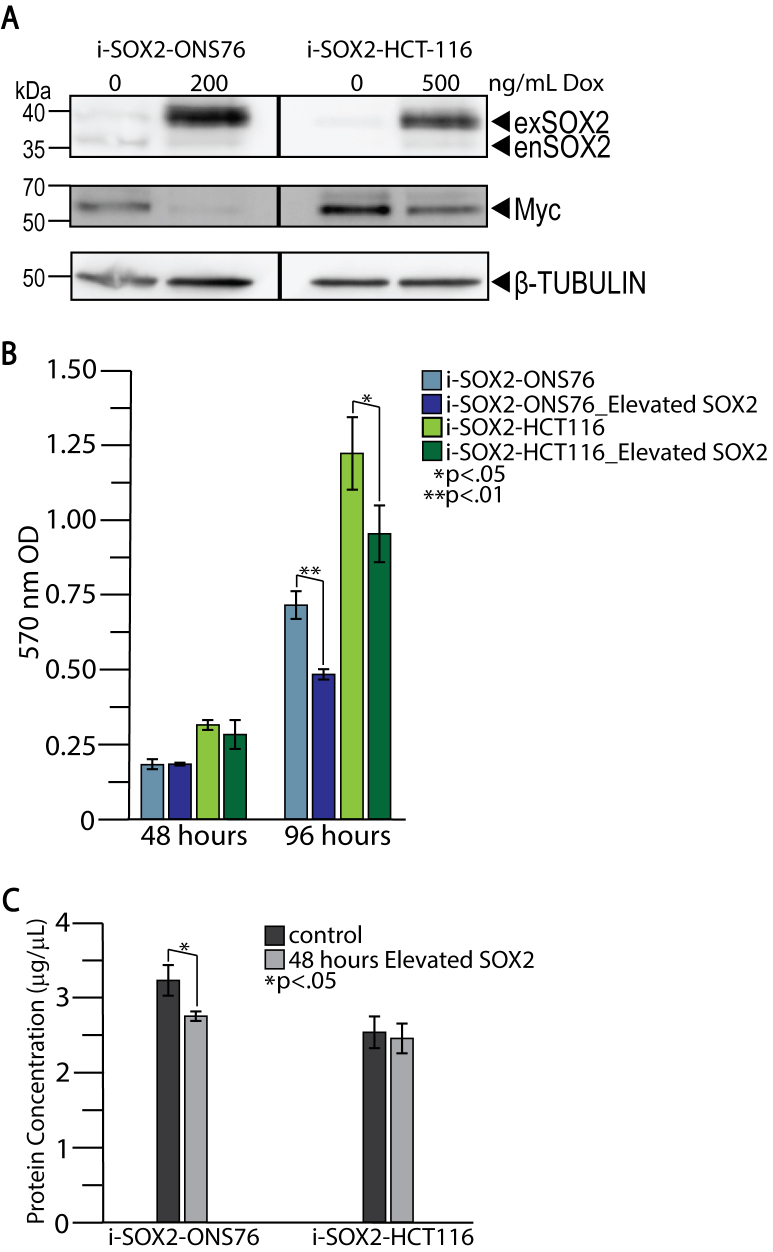
In this study, we confirmed our earlier finding (5) that elevating SOX2 in our SOX2-inducible medulloblastoma cells (i-SOX2-ONS76) and SOX2-inducible colorectal carcinoma cells (i-SOX2-HCT116) decreases MYC expression 48 h after treatment with the SOX2-inducer, Dox (Fig. S1A). At this time point, there was little change in cell proliferation in either cell line (Fig. S1B). In addition, even though elevating SOX2 had begun to downregulate protein translation (5), total protein present in cellular extracts isolated from i-SOX2-ONS76 and i-SOX2-HCT116 cells at the 48-h time point was only reduced ∼16% and 3%, respectively (Fig. S1C). To extend our earlier finding, our current work set out to determine how SOX2 downregulates MYC transcription.
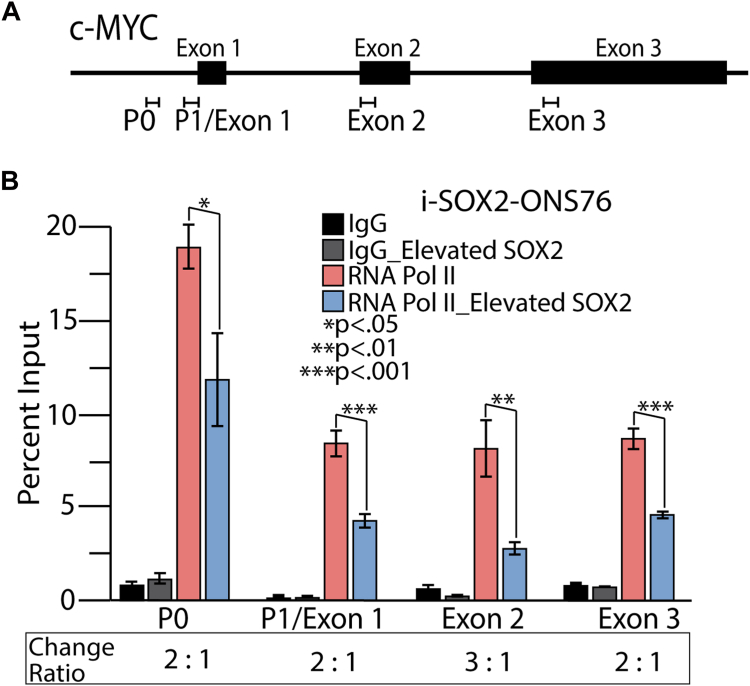
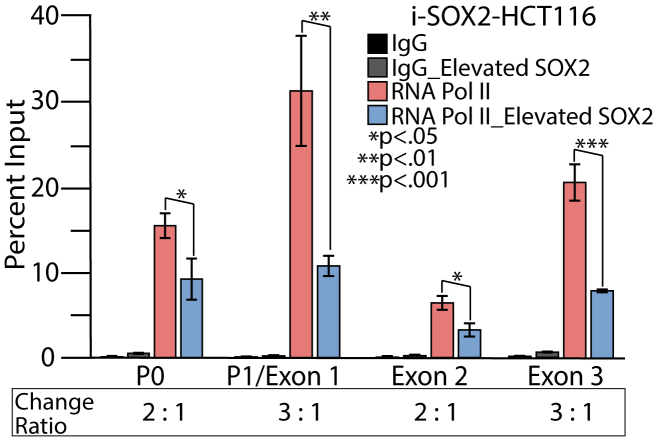
The effect of elevated SOX2 on MYC transcription could occur through multiple mechanistic changes. A common mode of transcriptional regulation near the promoters of MYC is the pausing of RNA Pol II close to the transcription start site without further elongation (18). One way to detect paused RNA Pol II is to perform Chromatin Immunoprecipitation followed by qPCR (ChIP-qPCR) to determine if there is an accumulation of RNA Pol II near the promoter region relative to the remainder of the gene body. As a general comment for this study, RNA Pol II and the other factors and modifications examined were considered to be enriched if the DNA pulled down by the antibody against that factor or histone modification was greater than with the non-specific IgG control antibody. To determine if SOX2 elevation leads to pausing of RNA Pol II at MYC promoters, we conducted RNA Pol II ChIP-qPCR analysis on samples obtained from i-SOX2-ONS76 and i-SOX2-HCT116 cells under control conditions and SOX2-elevated conditions, which were achieved by addition of the inducer, Dox, to the culture medium. For these studies, we interrogated the level of RNA Pol II enrichment throughout the MYC promoter region and the remainder of the gene body (Fig. 1A) using a total of four primer sets: two MYC promoter sets, P0 (also referred to as an upstream promoter) and P1 (designed for this study with some overlap into exon 1), a primer set for the second MYC exon, and a primer set for the third and last MYC exon (Table 1). Our RNA Pol II analysis determined that elevating SOX2 caused a significant reduction in RNA Pol II at both MYC promoter regions, as well as at exons 2 and 3, in both cell lines (Figs. 1B and S2). More specifically, the ratio at each site varied between 2 to 1 and 3 to 1 for the control and the SOX2 elevated cells, in both i-SOX2-ONS76 and i-SOX2-HCT116 cells. Therefore, the downregulation of MYC transcription when SOX2 is elevated is not the result of transcriptional pausing (blockage of elongation).
Figure 1.
RNA Pol II association with the MYC promoter region and the MYC gene body before and after elevation of SOX2.A, the MYC gene within the 3 Mb MYC locus consists of promoter regions designated P0 and P1, as well as three exons, for which primer sets (regions represented by bracketed horizontal lines) were utilized to test for the presence of different transcription machinery by ChIP-qPCR. B, i-SOX2-ONS76 control cells and cells induced for elevated SOX2 with 48 h of 200 ng/ml Doxycycline were processed for ChIP-qPCR with an IgG control antibody and RNA Pol II antibody. Enrichment was determined as compared to the Input and p values by t test for two tails, two sample equal variance, n = 3.
Table 1.
qPCR Primer Sequences
| ChIP primer | Forward sequence 5′ - 3′ | Reverse sequence 5′ - 3′ |
| MYC-P0 | AGGCAACCTCCCTCTCGCCCTA | AGCAGCAGATACCGCCCCTCCT |
| MYC-P1 | ATAATGCGAGGGTCTGGAC | CAGCGAGTTAGATAAAGCCC |
| MYC Exon 2 | CCCTCAACGTTAGCTTCACC | AGCAGCTCGAATTTCTTCCA |
| MYC Exon 3 | CAGATCAGCAACAACCGAAA | GTTTTCCAACTCCGGGATCT |
| MYC-A | AAATCAAGGGCAGGGACCACAG | CAGAATGGCAGAGTGAGGGGACAT |
| MYC-B | CAGGGCTTATTGTTGGGACAGAGTAGT | CGAAAACGCTCACGATTCACAGA |
| MYC-C | AGGGTGGACAAGCACAAGCATTA | ATTGGGACCTTTGGAGGCAAGAAT |
| MYC-D | CTTCTGCTCCCTTCTCTTCTCTCAGT | ACCACCACACTCCATCTTTCCAAC |
| MYC-E | GAGGGCGATAAAAGGGACAAGGA | GATGTTTGCTGGAACGCTGCTC |
| RT-qPCR Primer | ||
| GAPDH | ACAGCGACACCCACTCCTCC | GAGGTCCACCACCCTGTTGC |
| MYC Exon 2 | CCCTCAACGTTAGCTTCACC | AGCAGCTCGAATTTCTTCCA |
MYC, c-MYC; P0, promoter upstream of the MYC gene; P1, promoter near the transcription start site with the primer set amplicon providing some overlap into exon1 of the MYC gene; MYC-A through MYC-E, each of the five enhancers within the MYC super-enhancer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
SOX2 elevation alters the landscape of the MYC promoter region
The reduction of RNA Pol II throughout the MYC gene body when SOX2 is elevated suggests that recruitment and/or stabilization of the RNA Pol II complex could be disrupted by changes in the landscape of the MYC promoter region. The histone modification H3K4me3 (trimethylation at the fourth lysine residue of histone H3) is commonly found in the promoter regions of transcriptionally active genes (29). This histone modification is believed to contribute to RNA Pol II recruitment to gene promoters by helping to establish the pre-initiation complex through H3K4me3 binding by TAF3, which is a subunit of the TFIID complex (30). As we suspected, ChIP-qPCR analysis determined that elevating SOX2 reduced the levels of H3K4me3 at both the P0 and P1 MYC promoters in i-SOX2-ONS76 (Fig. 2A) and at the P1 MYC promoter region in i-SOX2-HCT116 cells (Fig. S3A).
Figure 2.
The levels of histone H3K4me3 and BRD4 association at the P0 and P1 MYC promoters and nuclear protein levels of BRD4 before and after elevation of SOX2. i-SOX2-ONS76 control cells and cells treated with 200 ng/ml Dox for 48 h were processed for ChIP-qPCR with an IgG control antibody and an antibody against (A) H3K4me3 and (B) BRD4. Enrichment was determined as compared to the Input and p values by t test for two tails, two sample equal variance, n = 3. C, Western blot analysis to measure BRD4 protein levels in nuclear extracts prepared from i-SOX2-ONS76 and i-SOX2-HCT116 cells after 48 h treatment at the indicated Dox dosage followed by benzonase treatment of the isolated nuclei. Western blot analysis of BRD4 was repeated and similar results were obtained. Dox, doxycycline.
The reduction of RNA Pol II at MYC promoters when SOX2 is elevated could be due primarily to the reduction of H3K4me3 levels upon SOX2 elevation. However, a change in the presence of critical co-activators within the MYC promoter when SOX2 is elevated could also contribute to reductions in the recruitment of RNA Pol II to MYC promoters. In this regard, it was recently determined that the recruitment of BRD4 to the MYC promoter region decreases when STAT3 is knocked down and MYC transcription is repressed (31). This is noteworthy because the co-activator BRD4 is known to play an important role in the stimulation of P-TEFb, which activates RNA Pol II by CTD phosphorylation at multiple serine residues, including serine 5 and serine 2 (32, 33). When we examined BRD4 association with the MYC promoter region by ChIP-qPCR, we observed a large decrease in BRD4 recruitment when SOX2 was elevated (Figs. 2B and S3B). Elevation of SOX2 led to over 70 to 90% reduction of BRD4 at the P0 and P1 promoter regions in i-SOX2-HCT116 cells (Fig. S3B) and nearly 50% reduction of BRD4 within the P0 promoter region and approximately 75% reduction within the P1 promoter region in i-SOX2-ONS76 cells (Fig. 2B). To extend these findings, we examined whether the nuclear levels of BRD4 were altered after SOX2 was elevated. Western blot analysis of nuclear extracts indicated that there was a small decrease (<20%) in BRD4 nuclear protein levels in i-SOX2-ONS76 cells (Fig. 2C), and a small increase (∼25%) in nuclear BRD4 protein levels in i-SOX2-HCT116 cells after SOX2 was elevated (Fig. S3C). Hence, the substantial reductions in BRD4 association with the MYC promoter when SOX2 is elevated is far greater than the smaller changes in BRD4 nuclear protein levels.
The presence of BRD4 at genes, such as MYC, is attributed to the action of BRD4 as a reader of histone acetylation (34). Specifically, BRD4 recognizes and binds H3K27ac (acetylation at lysine 27 of histone H3) sites within enhancers and promoters to help recruit other transcriptional machinery (35, 36, 37). This led us to examine whether elevating SOX2 alters the levels of H3K27ac in the MYC promoter region. In the case of i-SOX2-ONS76 cells before SOX2 was elevated (control), we observed H3K27ac at both P0 and P1 within the MYC promoter region. Consistent with the reduction in BRD4 recruitment to the MYC promoter regions, H3K27ac levels decreased at both P0 and P1 when SOX2 was elevated in i-SOX2-ONS76 cells (Fig. 3A). Similar results were observed when SOX2 was elevated in i-SOX2-HCT116 cells. In these cells, H3K27ac was more highly enriched at P0 relative to P1 in the MYC promoter region in the control and elevating SOX2 led to a large decrease in H3K27ac at P0 (Fig. S4A).
Figure 3.
Acetylation of histone H3K27 and binding of p300 at the P0 and P1 promoters of the MYC gene, and p300 nuclear protein levels before and after SOX2 elevation. i-SOX2-ONS76 control cells and cells induced to elevate SOX2 using 200 ng/ml Dox for 48 h were processed for ChIP-qPCR with an IgG control antibody and an (A) H3K27ac antibody, as well as a (B) p300 antibody. Enrichment was determined as compared to the Input and p values by t test for two tails, two sample equal variance, n = 3. C, Western blot analysis of nuclear extracts prepared from i-SOX2-ONS76 control cells and cells cultured for 48 h in the indicated doses of Dox was conducted following the benzonase treatment of the nuclei to ensure inclusion of chromatin-bound protein. Western blot analysis of p300 was repeated and similar results were obtained. Dox, doxycycline.
Next, we examined whether the elevation of SOX2 alters association of an acetyltransferase responsible for the acetylation of H3K27, which is often acetylated at enhancers and promoters by p300 and/or its paralog CBP. Given our use of the sonic hedgehog medulloblastoma cell line i-SOX2-ONS76 in this study, we focused on p300 rather than CBP, because p300 has been found to play a more prominent role than CBP in multiple cancers, including medulloblastoma (38). When we probed by ChIP-qPCR for changes in p300 recruitment to the MYC promoter region after elevating SOX2 in i-SOX2-ONS76 cells, we observed a reduction in p300 recruitment at P0 and noted there was far less recruitment of p300 to the P1 region relative to the P0 region, which reached a reduction of ∼40% (Fig. 3B). A sizable reduction of p300 was also observed at the P0 MYC promoter region when SOX2 was elevated in i-SOX2-HCT116 cells, which reached >50% (Fig. S4B). In regard to the P1 region in control i-SOX2-ONS76 and i-SOX2-HCT116 cells, we observed relatively little association of p300; however, there was a significant decrease in this limited association of p300 within this region when SOX2 was elevated in i-SOX2-HCT116 cells. As part of these studies, we examined whether the decrease in p300 recruitment to the MYC promoter region was due to diminished levels of nuclear p300. Western blot analysis of nuclear extracts determined that there was a small decrease in p300 protein levels within the nuclei of both cell lines (Figs. 3C and S4C). Interestingly, in HCT116 cells, p300 is truncated, but remains functional, and migrates with a molecular weight of approximately 240 kDa, whereas wild-type p300 protein migrates at a molecular weight of approximately 300 kDa (39, 40, 41). This was observed in three other studies (39, 40, 41) and verified in our work using a different p300 antibody than used previously. With a reduction of ∼20% in nuclear p300 protein levels within both cell lines when SOX2 is elevated, we suspect that the lower p300 protein levels only partially explain the much larger decreases (40–50%) in p300 recruitment to the MYC promoter region. Collectively, our studies indicate that elevating SOX2 substantially alters the landscape of the MYC promoter region, including reductions in H3K4me3, H3K27ac, BRD4 and p300, as well as RNA Pol II.
Elevation of SOX2 alters the co-activator landscape of an upstream MYC super-enhancer
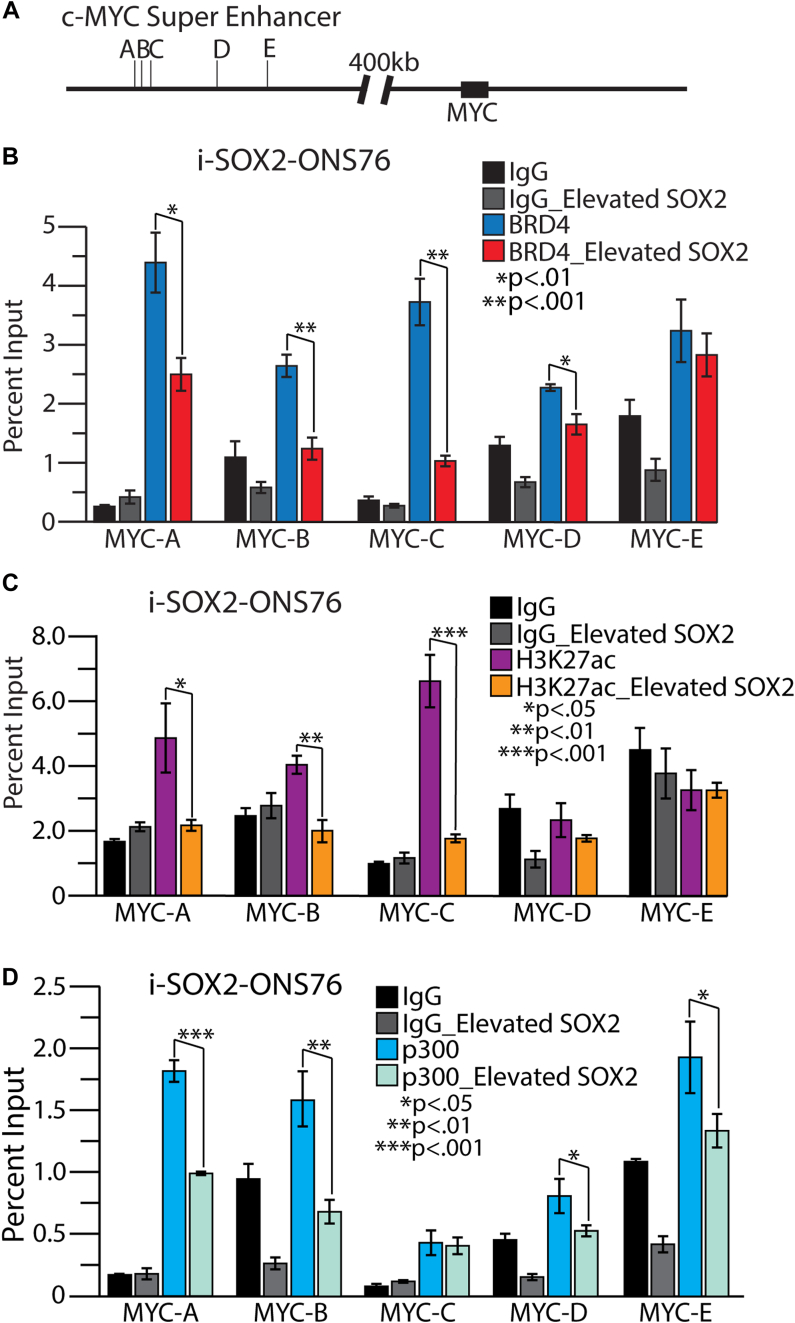
The MYC gene body is located in a 3 Mb protein gene desert on the short arm of chromosome 8. Previous studies identified a region far upstream of the MYC promoter region that is associated with several cancers (26, 27, 28), including a region located between −540 kb and −335 kb upstream of the MYC promoter region that is associated with breast and intestinal cancers (28). Importantly, studies conducted in the colorectal tumor cell line HCT116 previously identified a super-enhancer within this region that is located more than 400 kb upstream of the MYC promoter region (Fig. 4A). This super-enhancer consists of five MYC enhancers, designated as A-E, that are distributed over a region of approximately 50 kb. Given the major impact of super-enhancers on the activity of their associated promoters, we next examined recruitment of two key co-activators, BRD4 and p300. Similar to the role of p300 in other super-enhancers (42, 43, 44), the co-activator p300 is likely to play a major role in the activation of this MYC super-enhancer. One of the key roles of p300 is the acetylation of histone H3 at lysine residue 27 (H3K27ac) within super-enhancers (44, 45). Importantly, H3K27ac helps recruit the epigenetic reader BRD4 to super-enhancers, which, in turn, assists in the assembly of the preinitiation complex and recruitment of RNA Pol II to gene promoters (44, 46).
Figure 4.
BRD4, acetylated histone H3K27, and p300 association with the MYC super-enhancer before and after elevation of SOX2.A, the MYC super-enhancer consists of a cluster of five MYC enhancers, A-E denoted here by vertical lines at the approximate primer set sites, located more than 400 kb upstream of the MYC gene body. i-SOX2-ONS76 control cells and cells cultured for 48 h with 200 ng/ml Doxycycline were processed for ChIP-qPCR with an IgG control antibody and an antibody against (B) BRD4, (C) H3K27ac, and (D) p300. Enrichment was determined as compared to the Input and p values by t test for two tails, two sample equal variance, n = 3.
To begin the examination of the MYC super-enhancer, we employed primer sets previously used to identify each of the MYC enhancers, A-E (Table 1), within the upstream MYC super-enhancer in HCT116 cells (24). Our ChIP-qPCR analysis determined that BRD4 was associated with each of the five MYC enhancers A-E in both control i-SOX2-HCT116 and control i-SOX2-ONS76 cells (Figs. S5A and 4B). Equally important, elevating SOX2 dramatically decreased BRD4 association at all five MYC enhancers in i-SOX2-HCT116 cells, with the largest reductions (>80%) occurring at MYC enhancers A and B and significant decreases (∼50% or less) at enhancers C-E (Fig. S5A). We also observed significant reductions in BRD4 recruitment when SOX2 was elevated in the i-SOX2-ONS76 cells (Fig. 4B). As we noted above, this decrease in BRD4 recruitment to the MYC super-enhancer upon SOX2 elevation is unlikely to be due primarily to lower levels of nuclear BRD4 protein. The decrease in nuclear BRD4 (∼20%) when SOX2 was elevated in the i-SOX2-ONS76 cells (Fig. 2C), was much smaller than the reduction of BRD4 at the MYC enhancers, especially enhancers A-C that reached ∼50% or more. Moreover, elevating SOX2 in the i-SOX2-HCT116 cells led to an increase in the levels of nuclear BRD4 (Fig. S3C).
To extend the finding of reduced recruitment of BRD4 to the MYC super-enhancer when SOX2 was elevated, we examined the presence of H3K27ac within MYC enhancers A-E. As others have shown previously in HCT116 cells (24, 45), our studies in control i-SOX2-HCT116 cells show the enrichment of H3K27ac at all five enhancers within the MYC super-enhancer (Fig. S5B). Similarly, we observed enriched H3K27ac present at the A through C enhancer cluster in control i-SOX2-ONS76 cells (Fig. 4C). Interestingly, when SOX2 was elevated in the i-SOX2-HCT116 cells, H3K27ac levels were significantly reduced at enhancers B, C and D, with no significant changes observed at enhancers A or E (Fig. S5B). Similarly, H3K27ac was significantly reduced at the A through C enhancers, but exhibited no change at D or E, in the i-SOX2-ONS76 cells following SOX2 elevation (Fig. 4C). Together, these results indicate that elevating SOX2 reduces H3K27ac levels within the MYC super-enhancer in both cell lines, which provides a likely explanation for at least some of the decreases in recruitment of BRD4 within the MYC super-enhancer when SOX2 is elevated (Figs. S5A and 4B).
Given the reduction of H3K27ac within the super-enhancer, we examined the status of p300 enrichment at the MYC super-enhancer in i-SOX2-ONS76 and i-SOX2-HCT116 cells and whether elevating SOX2 in these cells alters p300 association with the MYC super-enhancer. Although it is widely accepted that p300 is present at super-enhancers of transcriptionally active genes (44), its recruitment to the MYC super-enhancer, in particular the five MYC enhancers A-E, has not been examined. Our ChIP-qPCR analysis detected significant enrichment of p300 at each of the five enhancers within the MYC super-enhancer in control i-SOX2-HCT116 cells (Fig. S5C) and control i-SOX2-ONS76 cells (Fig. 4D). Importantly, elevating SOX2 in i-SOX2-HCT116 cells significantly decreased p300 enrichment at all five MYC enhancers (Fig. S5C). In the case of enhancers A and B, the reduction reached nearly 50% and an even larger reduction was observed at enhancer E, >65%. Similarly, we observed a significant reduction in p300 recruitment in i-SOX2-ONS76 cells when SOX2 was elevated. In these cells, there was a significant decrease in p300 recruitment to MYC enhancers A, B, D, and E, with no change at C (Fig. 4D). In i-SOX2-ONS76 cells the largest reductions were at MYC enhancers A and B, reaching 45 and 55%, respectively. Like the reduction of BRD4 within the super-enhancer, the reduction of p300 within the super-enhancer appears to be greater than the overall reduction of the nuclear levels of p300 protein. Altogether, our studies indicate that elevating SOX2 in both i-SOX2-HCT116 and i-SOX2-ONS76 cells substantially alters the co-activator landscape of the MYC super-enhancer.
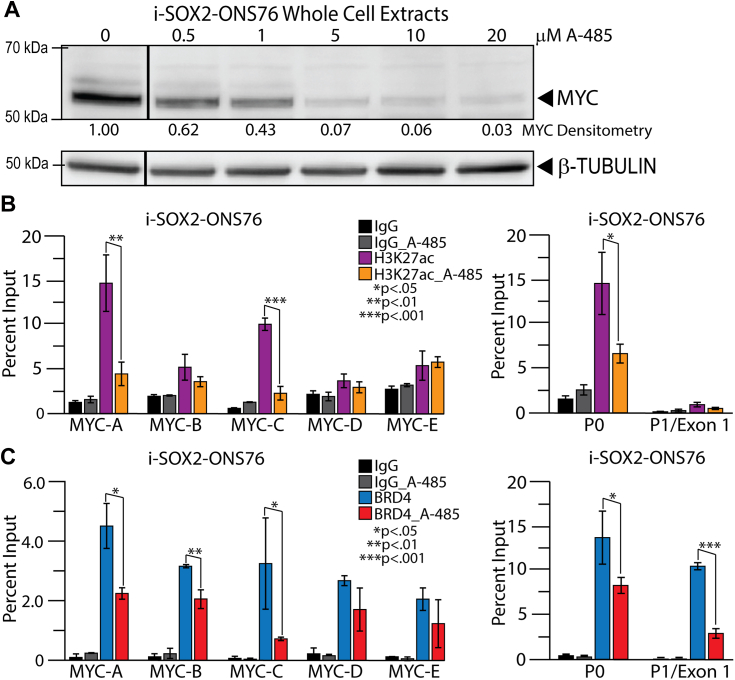
Inhibiting the acetyltransferase activity of p300 induces changes in the MYC super-enhancer and its promoter region that parallel the effects observed when SOX2 is elevated
The decreases in p300 recruitment to the MYC super-enhancer when SOX2 is elevated could trigger the overall changes in the MYC super-enhancer described above and, ultimately, the changes at the MYC promoter. To test this possibility, we examined how inhibiting p300 affects MYC expression, the MYC super-enhancer, and the MYC promoter region. For this purpose, we used A-485, which inhibits the acetyl transferase activity of both p300 and CBP (47). Although previous findings reported that A-485 leads to the downregulation of the expression of many genes (47), we only examined its effects on MYC expression. As expected, Western blot analysis revealed that A-485 induced a dose-dependent decrease in MYC protein expression in both i-SOX2-ONS76 and i-SOX2-HCT116 cells (Figs. 5A and S6A). Next, we investigated the effects of A-485 on H3K27ac in i-SOX2-ONS76 and i-SOX2-HCT116 cells before and after elevating SOX2. In the case of i-SOX2-HCT116 cells, we observed a dramatic reduction in the levels of H3K27ac at all five MYC enhancers within the super-enhancer and in the MYC promoter region (Fig. S6B). Although the effects of A-485 on H3K27ac were not as large in i-SOX2-ONS76 cells, we observed large reductions in H3K27ac at MYC enhancers A and C and in the MYC promoter region (Fig. 5B). To extend these studies, we examined the effects of A-485 on the association of BRD4 with the MYC super-enhancer and the MYC promoter region. In both i-SOX2-ONS76 and i-SOX2-HCT116 cells, treatment with A-485 reduced BRD4 levels at MYC enhancers A-C and at both P0 and P1 within the MYC promoter region (Figs. 5C and S6C). Together, these studies indicate that treatment with A-485 leads to decreases in H3K27ac and BRD4 recruitment at the MYC super-enhancer and MYC promoter regions, similar to what we observed when SOX2 was elevated. Importantly, the findings with A-485 provide strong support for the premise that p300, and possibly CBP, is a critical player in SOX2-mediated repression of MYC transcription. They also suggest that the reduction in p300/CBP recruitment to the MYC super-enhancers triggers a cascade of events that leads to alterations in the MYC super-enhancer and, ultimately, to the downregulation of MYC transcription.
Figure 5.
MYC protein expression, histone modification, and BRD4 association with the MYC super-enhancer before and after inhibition of p300 in i-SOX2-ONS76 cells.A, control cells and cells inhibited for 19 h with A-485 at increasing concentrations were processed for Western blot analysis. An unrelated lane, between the first and second lane in the figure, was spliced out to simplify the figure as it contained a cell sample of Dox treatment without A-485 that served as additional confirmation of MYC downregulation upon SOX2 elevation, which we provided with another experiment in Fig. S1A. Control cells and cells inhibited with 10 μM A-485 were processed for ChIP-qPCR with an IgG control antibody and antibodies to determine the status of (B) H3K27ac and (C) BRD4 binding. Enrichment was determined as compared to the Input and p values by t test for two tails, two sample equal variance, n = 3.
Elevating SOX2 increases the association of both SOX2 and the histone modification H3K27me3 within the MYC locus
To extend our study of how SOX2 downregulates MYC transcription, we examined whether elevating SOX2 altered the association of SOX2 and the levels of H3K27me3 (trimethylation at lysine 27 of histone H3) within the MYC locus. In the case of SOX2, in both i-SOX2-ONS76 and i-SOX2-HCT116 control cells, we observed by ChIP-qPCR relatively little association of endogenous SOX2 within the MYC super-enhancer or promoter region. However, when SOX2 was elevated, we observed significant SOX2 association at each of the MYC enhancers within the MYC super-enhancer in both i-SOX2-ONS76 and i-SOX2-HCT116 cells (Figs. 6A and S7A). We also observed increases in SOX2 within the MYC promoter region in both cell lines (Figs. 6A and S7A). These findings raised the possibility that when bound to the MYC enhancer and/or promoter regions, SOX2 contributes directly to repression of the MYC gene by recruitment of transcriptional repressors.
Figure 6.
Association of SOX2 and histone H3K27me3 at the MYC locus before and after SOX2 elevation. i-SOX2-ONS76 control cells and cells induced to elevate SOX2 using 200 ng/ml Dox for 48 h were processed for ChIP-qPCR with an IgG control antibody and antibodies against (A) SOX2 and (B) H3K27me3. Enrichment was determined as compared to the Input and p values by t test for two tails, two sample equal variance, n = 3. C, RNA isolation and RT-qPCR analysis of MYC levels of i-SOX2-ONS76 control cells and cells in the absence and presence of either or both 200 ng/ml Dox and 10 μM EPZ-6438 treatment for 48 h. D, i-SOX2-ONS76 cells induced to elevate SOX2 using 200 ng/ml Dox alone or in combination with 10 μM EPZ-6438 for 48 h were processed for ChIP-qPCR with an IgG control antibody and an antibody against H3K27me3. Enrichment was determined as compared to the Input and p values by t test for two tails, two sample equal variance, n = 3. Dox, doxycycline.
To study the potential role of transcriptional repressors in the downregulation of MYC transcription, we examined whether the SOX2-induced reduction of H3K27ac within the MYC locus is accompanied by increases in H3K27me3, a histone modification associated with gene repression (48). We examined H3K27me3 because of the substantial rise in SOX2 association within the MYC super-enhancer and promoter region and our previous finding that SOX2 interacts with the Polycomb Repressor Complex 2 (PRC2) subunit, RBBP4, in medulloblastoma cells (49). Interestingly, ChIP-qPCR detected very low levels of H3K27me3 within the MYC super-enhancer at MYC enhancers A-E in the control cells; whereas there was a substantial increase in H3K27me3 at each of the MYC enhancers within the super-enhancer when SOX2 was elevated in both i-SOX2-ONS76 and i-SOX2-HCT116 cells (Fig. 6B and S7B). In addition, there was little or no H3K27me3 within the promoter region in the control cells, but there were increases within the MYC promoter region in both SOX2-inducible cell lines when SOX2 was elevated (Fig. 6B and S7B).
The increases in H3K27me3 raised the interesting possibility that this histone modification helps repress MYC transcription when SOX2 is elevated. This possibility was examined in the i-SOX2-ONS76 cells by interfering with the activity of EZH2, which is the catalytic subunit of the PRC2 complex that can generate H3K27me3 (48). For this purpose, we initially tested whether the EZH2 inhibitor, EPZ-6438, could reverse the downregulation of MYC expression when SOX2 is elevated. Significantly, EPZ-6438 led to only a small reversal in the downregulation of MYC mRNA when SOX2 was elevated in i-SOX2-ONS76 cells (Fig. 6C). The failure to more fully reverse the downregulation of MYC expression could have resulted from the inability of EPZ-6438 to block the H3K27me3 within the MYC locus. To address this possibility, we examined whether H3K27me3 within the MYC super-enhancer was blocked when EPZ-6438 was added to the SOX2 elevated i-SOX2-ONS76 cells. Importantly, under these conditions, H3K27me3 levels were strongly decreased at each of the MYC enhancers A-E, and there was a decrease of H3K27me3 within the MYC promoter region (Fig. 6D). Collectively, our studies suggest that increases in the levels of H3K27me3 within the MYC super-enhancer and promoter region when SOX2 is elevated does not appear to be sufficient on their own to repress MYC expression.
Discussion
This report provides new insights into the mechanistic relationship between two master regulators, SOX2 and MYC, that play a myriad of roles in cellular physiology before and after birth, as well as in cancer. SOX2 is expressed in over 20 different cancers, and it has been implicated in the cancer stem cell populations of many of these cancers (12), while MYC is dysregulated in approximately 70% of all human cancers (1). In this report, we describe a novel molecular mechanism by which MYC transcription can be regulated. We show that elevating SOX2 substantially alters the landscape of both an upstream MYC super-enhancer and the MYC promoter region, in the course of downregulating MYC transcription. We focused specifically on SOX2 and MYC because elevating SOX2 in vivo leads to a reversible state of tumor growth arrest (17) and because the failure to downregulate MYC when SOX2 is elevated in these cells leads to a drastic reduction in tumor cell survival (5).
Our initial studies determined that elevating SOX2 reduces enrichment of RNA Pol II at both the MYC promoter region and the MYC gene body. Elevating SOX2 also led to decreases in H3K4me3 at the MYC promoter region. This histone modification is typically associated with active promoters. In addition, we observed a significant decrease in recruitment of BRD4 and p300 to the promoter region of the MYC gene. The decrease in BRD4 within the promoter region of MYC is significant because it likely contributes to the reduction in RNA Pol II at the MYC promoter (44). Interestingly, our findings revealed no evidence that elevating SOX2 leads to pausing of RNA Pol II at the MYC promoter. In both cell lines used in our work, elevating SOX2 leads to a reduction of RNA Pol II at exons 1, 2 and 3. Thus, the mechanism used by elevated SOX2 to downregulate MYC transcription appears to differ substantially from the mechanism that downregulates MYC transcription due to a buildup of RNA Pol II within exon 1 relative to exons 2 and 3 when HL60 are induced to differentiate (18).
To extend these studies, we examined the recruitment of BRD4 and p300 to five MYC enhancers located within an upstream MYC super-enhancer. Like our results with the promoter region, we determined that elevating SOX2 disrupts the landscape of the MYC super-enhancer. Specifically, we determined that the association of BRD4 at the MYC super-enhancer was substantially reduced at four of the five MYC enhancers in i-SOX2-ONS76 cells and at each of the MYC enhancers in i-SOX2-HCT116 cells. Significantly, our findings suggest that the reduction in BRD4 recruitment to the MYC super-enhancer is likely due, at least in part, to the reduction in H3K27ac at three of the MYC enhancers within the MYC super-enhancer in both i-SOX2-HCT116 and i-SOX2-ONS76 cells. Importantly, we also determined that elevating SOX2 significantly reduced the recruitment of p300 to each of the five MYC enhancers in i-SOX2-HCT116 cells and four of the MYC enhancers in i-SOX2-ONS76 cells, which likely explains the decrease in H3K27ac within the MYC super-enhancer. Interestingly, our studies suggest that the decreases in p300 recruitment to the MYC super-enhancer and promoter region result in part due to decreases in the expression of p300 when SOX2 is elevated. However, the reduction in nuclear p300 protein expression of ∼20% is less than half the average 40 to 50% reduction in p300 recruitment to the five MYC enhancers in i-SOX2-ONS76 and i-SOX2-HCT116 cells. This appears to be the case, even when decreases in p300 are adjusted for the small decreases in total protein in i-SOX2-ONS76 cells (∼16%) and i-SOX2-HCT116 cells (∼3%). We believe this also applies to BRD4. Thus, additional mechanisms are likely to contribute to the decreases in p300 and BRD4 recruitment to the MYC super-enhancer and promoter region.
The significance of our findings is highlighted by the changes observed at the MYC super-enhancer when SOX2 is elevated. Although previous studies had identified this super-enhancer, relatively little was known about how it might be regulated. The MYC super-enhancer was presumed to be populated by p300 and BRD4, like other active super-enhancers. Our studies are the first to show that both co-activators are associated with each of the five MYC enhancers within this super-enhancer and, very importantly, that elevating SOX2 reduces the recruitment of both co-activators to the MYC super-enhancer. Furthermore, the MYC super-enhancer examined in our study is located within a region of the MYC locus, between −540 kb and −335 kb relative to the MYC promoter, that has been implicated in at least two cancers, breast and intestinal cancer (28), and quite possibly several other cancers (26, 27). This is potentially significant because this region of the MYC gene locus is dispensable for normal cells during development and after birth yet enhances cellular transformation in models of breast and intestinal cancers (28).
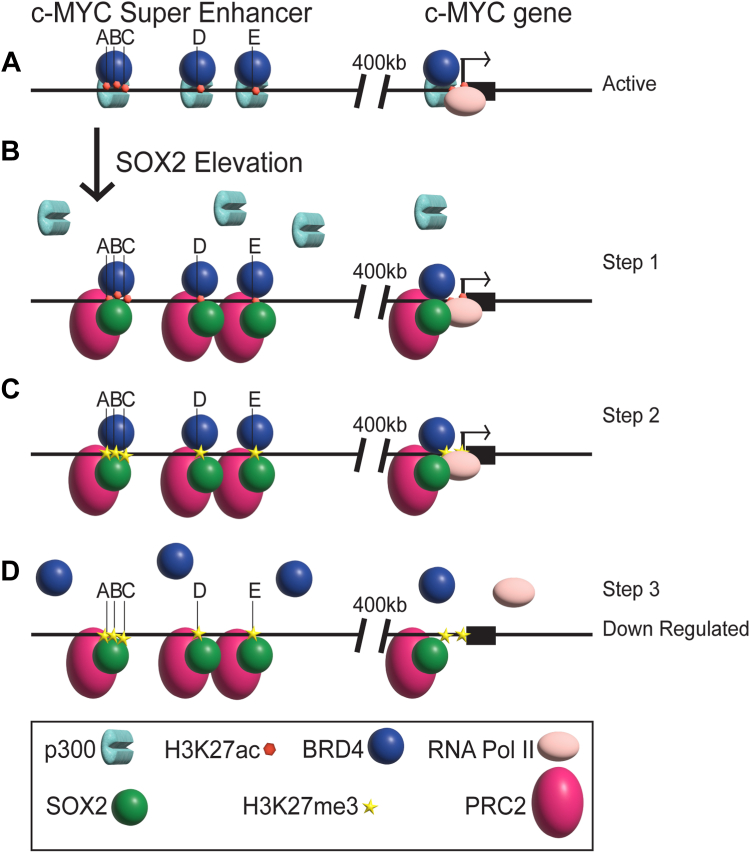
Collectively, our studies suggest a likely sequence of events that leads to changes in the MYC super-enhancer and the MYC promoter when SOX2 is elevated (Fig. 7). Specifically, our studies are consistent with a model in which elevating SOX2 initially leads to significant accumulation of SOX2 at each of the MYC enhancers located within the MYC super-enhancer and at the MYC promoter region. We speculate that SOX2 accumulation within the MYC locus contributes to the reduction in the recruitment of p300 within the MYC locus, possibly due to SOX2 recruitment of repressor complexes, such as PRC2 to the MYC locus. We further posit that the reduction in recruitment of p300, and possibly CBP, to the MYC super-enhancer is responsible for the reduction of H3K27ac within the super-enhancer, which, in turn, leads to a reduction in the recruitment of BRD4 to the MYC super-enhancer. The reduction of p300 and BRD4 to the MYC super-enhancer results in functional disruption of the super-enhancer, which then disrupts critical steps required for the formation of the preinitiation complex and recruitment of RNA Pol II to the MYC promoter. In support of this model, we have demonstrated that treatment with the p300/CBP inhibitor, A-485, which blocks the acyltransferase activity of these co-activators, leads to a reduction in H3K27ac at all five of the MYC enhancers of the super-enhancer in i-SOX2-HCT116 cells and two of the five MYC enhancers in i-SOX2-ONS76 cells. This inhibitor also induced a reduction in BRD4 at three of the MYC enhancers in both i-SOX2-HCT116 and i-SOX2-ONS76 cells, as well as a reduction in BRD4 in the MYC promoter region in these cells. Thus, we propose that the decreases in p300/CBP recruitment after SOX2 accumulates at both the MYC super-enhancer and the MYC promoter region is a key step in the downregulation of MYC transcription.
Figure 7.
Model of co-activator and histone modification status at the MYC super-enhancer and MYC promoter region when SOX2 is elevated.A, The actively transcribed state of the MYC gene in tumor cells; presence of p300, H3K27ac, and BRD4 within the MYC super-enhancer, and p300, BRD4, H3K27ac along with RNA Pol II at the MYC promoter region. B, step1: SOX2 elevation leads to a decrease in p300 association and newly associated SOX2, along with other complex proteins (such as PRC2). C, step 2: lower levels of H3K27ac due to the decrease in p300, while H2K27me3 increased in its stead, likely due to the new presence of SOX2’s recruited partner complexes, such as PRC2. D, step 3: decreased recruitment of BRD4 to the MYC super-enhancer and promoter region, and less RNA Pol II at the MYC promoter region, leading to repression of MYC transcription.
Significantly, our studies also demonstrate that elevating SOX2 leads to substantial increases in H3K27me3 within the MYC super-enhancer and promoter regions. Although we did not directly demonstrate that the increases in H3K27me3 are due to recruitment of PRC2 to the MYC locus, we determined that the levels of H3K27me3 within the MYC super-enhancer and promoter region are dramatically reduced when the SOX2-elevated cells are treated with the EZH2 inhibitor EPZ-6438. In this connection, we previously demonstrated by proteomic analysis of SOX2 protein complexes that SOX2 associates with the PRC2 subunit RBBP4 in medulloblastoma cells (49). Thus, SOX2 may be directly responsible for recruitment of PRC2 to the MYC locus when SOX2 is elevated. Importantly, our work tested whether H3K27me3 contributes to the downregulation of MYC transcription when SOX2 is elevated. The failure of the EZH2 inhibitor to fully reverse the downregulation of MYC expression in i-SOX2-ONS76 cells suggests that H3K27me3 accumulation within the MYC super-enhancer and promoter is not sufficient on its own to downregulate MYC transcription when SOX2 is elevated. Thus, it is possible that elevating SOX2 leads to the recruitment of more than one transcriptional repressor complex to the MYC locus that plays an important role in the downregulation of MYC transcription. Alternatively, elevating SOX2 may interfere with the recruitment of other required transcriptional machinery, such as Med1, which is typically observed within active super-enhancers (44, 50).
Our findings with the medulloblastoma and colorectal tumor cells used in this study lead us to suggest that high SOX2 levels are likely to downregulate MYC transcription by the same overall mechanism in other SOX2-positive human cancers. However, subtle differences may be responsible for the downregulation of MYC transcription by high SOX2 levels in some of these cancers. In various tumor cell types, MYC transcription is believed to be dependent on other super-enhancers located within the MYC 3 Mb locus that are used in a tissue specific manner (51). For tumor types where different super-enhancers are utilized to regulate MYC transcription, we speculate that these alternative super-enhancers are also likely to experience an increase in SOX2 recruitment and a decrease in the recruitment of both p300 and BRD4 when SOX2 is elevated. In the future, this could be tested in various tumor cell types. In this regard, we have shown previously that elevating SOX2 reduces MYC expression in five different tumor cell lines, representing three different tumor types, including prostate tumor cells (5).
Finally, our studies suggest that one way to target MYC expression in tumor cells is to block the action of p300/CBP, especially in tumors that depend on MYC super-enhancers to drive MYC transcription. Recently, a different p300/CBP inhibitor, CCS1477, has entered clinical trials for several cancers, including metastatic breast cancer, non-small cell lung cancer, and metastatic castration-resistant prostate cancer (52). In the case of prostate cancer, CCS1477 has been shown to substantially slow the growth of patient-derived xenografts and to decrease the expression of MYC. Moreover, biopsies of patients with metastatic castration-resistant prostate cancer who were treated with CCS1477 exhibited a decrease in both MYC and Ki-67 expression (52). Given the difficulty of directly targeting MYC in cancer, our studies suggest that p300/CBP inhibitors could be promising drugs for the treatment of other tumors, including medulloblastoma and colorectal cancer, if these inhibitors prove to be effective and well tolerated.
In summary, the work described in this study demonstrates that elevating SOX2 radically alters the landscape of a MYC super-enhancer, as well as the landscape of the MYC promoter region. Significantly, we demonstrate that elevating SOX2 leads to the accumulation of SOX2 within the MYC locus and reduces the recruitment of both p300 and BRD4 to the individual MYC enhancers within a far upstream MYC super-enhancer as well as the MYC promoter region. The impact of our findings is highlighted by the location of this super-enhancer within the region of the MYC locus that is associated with at least two different cancers (28). Altogether, our work provides new molecular insights into the SOX2:MYC axis that plays a pivotal role in the repression of MYC transcription when SOX2 levels rise in tumor cells (5).
Experimental procedures
Cell culture conditions
Cell lines engineered for inducible expression of SOX2, i-SOX2-ONS76 and i-SOX2-HCT116 (5, 17), were maintained at 37 °C and 5% CO2 while cultured in Dulbecco’s Modified Eagle Medium (12100046, Life Technologies) supplemented with 10% HyClone Characterized Fetal Bovine Serum (SH30910.03, Cytiva, Fisher Scientific) and 1% Pen-Strep (P3032-10MU & S9137-25G, SIGMA). Validation of cell line authenticity was performed previously (5, 17). Initially, cells were seeded in 100 mm dishes at 0.8 × 106 each for 24 h. The following day media was changed and for experiments involving SOX2 elevation, Dox (631311, Clonetech) was premixed in the media at concentrations of 200 ng/ml for i-SOX2-ONS76 cells or 500 ng/ml for i-SOX2-HCT116 cells to result in ∼40 to 50% growth inhibition (17). The control and experimental cells were cultured for an additional 48 h before processing. Experiments not involving SOX2 elevation were cultured for an initial 24 h, media was changed, and another 24 h passed before the DMSO (D8050-01, US Biologicals) control or small molecule A-485 (S8740, Selleck Chem) was diluted in 1 ml media and added drop-wise to plates that were rocked briefly to result in concentrations indicated in the figures. The cells were cultured for an additional 19 h before processing. DMSO was used to dissolve A-485, thus the controls were treated with the same concentration of DMSO and the same amount of time as experimental cell cultures. Combination experiments involving SOX2 induction using Dox in combination with the EZH2 inhibitor, EZP-6438 (S7128, Selleck Chem), were conducted in the same manner as the experiments with Dox alone (above), except 10 μM EZP-6438 was added at the same time to half of the Dox-treated plates and the other Dox-treated plates had DMSO only added as a control.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed utilizing the MAGnify Chromatin Immunoprecipitation System (492024, Thermo Fisher Scientific). Following experimental cell culture conditions described above in 100 mm plates, cells were fixed in 5 ml of media containing a 1% final concentration of formaldehyde (F8775, SIGMA) for 10 min at room temperature to cross-link proteins to DNA. The reaction was neutralized with 0.5 ml of 1.25 M Glycine from the kit (applied dropwise and rocked briefly to mix and then set still) for 10 min at room temperature, and the cells were washed with 1X PBS twice. Approximately 6 × 106 cells for each condition were pelleted and resuspended in lysis buffer with protease inhibitors from the kit and the chromatin was sheared to a length of ∼200 to 500 bp using a Bioruptor Pico sonication device (Diagenode) with sonication cycles consisting of 30 s “on” and 30 s “off” for 9 min. Sonicated DNA was diluted with the dilution buffer provided in the kit and 10 percent of the sheared chromatin was removed and set aside prior to immunoprecipitation to serve as Input DNA for data normalization. We added a two-hour 4 °C immunoprecipitation pre-clearing step to our procedure, which involved chromatin incubation with a kit-provided Rabbit IgG antibody at a 1:100 dilution associated to A/G agarose magnetic beads. Then we resumed the protocol provided with the MAGnify Chromatin Immunoprecipitation System. The pre-cleared chromatin was transferred to its appropriate control and experimental immunoprecipitation tubes that had been mixed for association of Dynabeads A/G agarose magnetic beads with either Rabbit IgG control antibody (1:200) or the experimental antibodies: RNA Pol II (Millipore, 05–623, 1:50), H3K4me3 (Millipore, 07–473, 1:100), BRD4 (Cell Signaling Technology [CST], 13440, 1:50), H3K27ac (CST, 8173, 1:100), p300 (CST, 54062, 1:50), H3K27me3 (CST 9733S, 1:50), SOX2 (Active Motif 39823, 1:50). Overnight immunoprecipitation was conducted at 4°C with end-over-end tube rotation. The next day, three 5-min washes of the chromatin-bound magnetic antibody-conjugated beads with IP Buffer 1, and two washes with IP Buffer 2 from the kit were conducted at 4 °C with end-over-end tube rotation (each time placing the tubes in a magnetic rack (20–400, Millipore) and removing the liquid to discard the washes without disturbing the beads). Cross-linking was reversed (for IP samples and Input samples) using Reverse Crosslinking Buffer and Proteinase K from the kit and heating the tubes at 55 °C for 15 min in a heat block. For the IP samples, the tubes were placed in the magnetic rack and each liquid sample was transferred to a new tube for additional heating, along with the Input sample tubes, at 65 °C for 15 min. After the reverse crosslinked samples cooled on ice for 5 min, DNA was bound to beads and washed by adding DNA Purification Magnetic Beads supplied in the kit with DNA Purification Buffer to each of the sample tubes (including Inputs) and mixing the tube contents followed by incubation at room temperature for 5 min. With DNA bound to the DNA Purification Magnetic Beads, the tubes were placed in the magnetic rack and allowed to sit for 1 min before, avoiding the beads, the liquid was removed and discarded. The tubes were taken off the magnetic rack and DNA Wash Buffer from the kit was added, mixed gently with the beads, and incubated at room temperature for 5 min, followed by placing the tubes on the magnetic rack again, and removing and discarding the liquid while avoiding the beads. This was repeated once. Next, the tubes containing only beads were removed from the magnetic rack and Elution Buffer from the kit was added and mixed gently with the beads by pipetting up and down. The tubes were incubated at 55 °C for 20 min in a heat block. The tubes were inverted carefully a couple of times to collect liquid condensation from the sides and cap of the tube, and the samples were spun down briefly. The DNA elutes were transferred to new tubes after the beads were pulled down while in the magnetic rack for at least 1 min. These eluted Input DNA and ChIP DNA templates were stored at −20 °C to allow for further analysis.
Quantitative polymerase chain reaction (qPCR) for ChIP DNA
For ChIP analysis, enrichment of immunoprecipitated DNA was determined using RT2 SYBR Green qPCR Mastermix (330502, Qiagen) in 25 μl qPCR reactions prepared in triplicate in 96-well white hard-shell PCR Plates (AB1400WL, Life Technologies). Plates were covered with Microseal ‘B’ Film (MSB1001, BioRad), and qPCR was carried out with the Applied Biosystems QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific). The primer sets for each site examined are provided in Table 1. The ChIP and Input DNA were used at a constant volume of 5 μl in each 25 μl qPCR reaction with 1 μl primer mix (from a pre-mixed stock of 10 μM each of forward and reverse primers listed in Table 1), 6.5 μl Nuclease-free water, and 12.5 μl RT2 SYBR Green Fluor qPCR Mastermix. Enrichment was determined using the Percent Input Method, while taking into account our dilution factor of 10, for an Adjusted Input to 100% = Ct Input - (Log10/2) = Ct Input – 3.322; Delta Ct = Ct (Adjusted Input) - Ct (Test Sample); and Percent Input = 10∗2∧(Delta Ct), as described by Thermo Fisher Scientific with their provided example utilizing a dilution factor of 100.
Extract preparation and western blotting
Nuclear and Cytoplasmic protein extracts from untreated and Dox-treated i-SOX2-ONS76 and i-SOX2-HCT116 cells were prepared using the Pierce NE-PER nuclear and cytoplasmic extraction kit (78833, Pierce, Thermo Fisher Scientific), as described previously (8), supplemented with HALT protease inhibitor cocktail (87786, Thermo Fisher Scientific), 1 mM PMSF, 1 mM Na2VO4, and 1 mM NaF. Following the isolation and lysis of nuclei during this fractionation, the chromatin-bound proteins were isolated as well utilizing a Benzonase treatment step (53). The proteins from this step and the nuclear protein isolation step were combined to result in total nuclear protein extracts. For whole cell protein isolation, RIPA buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.4, 1% IGEPAL, 0.25% Sodium Deoxycholate, and 1 mM EDTA) was supplemented with the protease inhibitors listed above and then utilized for protein isolation, conducted as described in the Santa Cruz western blotting RIPA buffer monolayer cell processing protocol. Protein concentrations were obtained following BCA assays using the Micro BCA Protein Assay Kit and the provided protocol (23235, Pierce, Thermo Fisher). To detect larger proteins, such as p300 and BRD4, 20 μg of nuclear protein samples were loaded in a 6% Tris-Glycine gel (XP00062BOX, Life Tech), and for whole cell protein 40 μg were loaded in 4 to 20% Tris-Glycine gels (XP04202BOX, Life Tech). Proteins within each gel were transferred to methanol-activated PVDF membranes (IPVH20200, SIGMA) and blocked with 5% milk before washing and incubating overnight at 4 °C with the appropriate primary antibody in 3% BSA. The presence of alkaline phosphatase conjugated secondary antibody was detected using the enhanced chemiflourescence (ECF) kit (RPN5785, Amersham Biosciences) and scanned on a ChemiDoc MP Imaging System (BioRad). To detect the loading control protein on the same membrane as the initial experimental proteins, we followed the protocol for the Restore Western Blot Stripping Buffer (46430, Pierce, Thermo Fisher Scientific) between each antibody. The primary antibodies used were BRD4 (CST, 13440S, 1:1000); p300 (CST, 54062S, 1:1000); Lamin B1 (CST, 12586S, 1:1000); c-MYC (CST, 18583S, 1:1000); SOX2 (CST, 3579S, 1:1000); and b-TUBULIN (CST, 2146S, 1:1000); and the secondary anti-rabbit antibody was from CST (7054S, 1:5000). Each protein band pixel density at low intensity (with background signals removed) was normalized to a loading control that was found repeatedly to be present at a similar level across samples, which was measured through Adobe Photoshop software. All antibodies used in this study were selected based on their use in previously published work and shown to identify protein bands of the correct molecular weight.
RNA isolation, cDNA synthesis, and RT-qPCR
RNA isolation was achieved by utilizing the RNeasy Mini Kit (74104, Qiagen). Following experimental cell culture conditions described above in 100 mm plates, after 48 h of Dox induction, media was removed from both control and Dox-induced cells. These cells were lysed by directly adding 350 μl Buffer RLT from the RNeasy Mini Kit to each plate. The cells were scraped and transferred to a 1.5 ml Eppendorf tube, followed by the addition of 1 volume of 200 proof ethanol to the lysate. The tubes were mixed well, and up to 700 μl of the sample was transferred to a RNeasy Mini spin column placed in a 2 ml collection tube. The spin columns were centrifuged for 30 s at 12,000g and the flow-through was discarded. Next, 700 μl Buffer RW1 from the RNeasy Mini Kit was added to the RNeasy spin column which was centrifuged for 30 s at 12,000g and the flow-through was discarded. Added 500 μl Buffer RPE to the RNeasy spin column and centrifuged for 30 s at 12,000g, followed by discarding the flow-through and repeating once more with another 500 μl Buffer RPE, but the second time centrifugation was conducted for 2 min at 12,000g. Then the RNeasy spin column was placed in a new collection tube and centrifuged at full speed for 1 min to dry the membrane. Finally, the RNeasy spin column was placed in a new 1.5 ml Eppendorf tube and 30 μl of 55 °C pre-warmed RNase-free water was added directly to the membrane followed by centrifugation for 1 min at 12,000g to elute the RNA. The RNA concentration was measured using a NanoDrop One Spectrophotometer (Thermo Fisher Scientific).
To synthesize cDNA, 1 μg of RNA was utilized for each sample with the Accuscript High Fidelity first Strand cDNA Synthesis Kit (200820, Agilent Technologies). A total volume of 20 μl was used for each reaction in 0.5 ml tubes, and the volume of RNase-free water was adjusted as needed to compensate for any variation in volume of the 1 μg of RNA. The first kit components added to each tube of RNase-free water and 1 μg of RNA included 2 μl of 10X Accuscript RT buffer, 0.5 μl oligo(dT) primer and 1.5 μl random primers, as well as 0.8 μl dNTP mix from the kit. The tubes were incubated at 65 °C for 5 min. After incubation, the tubes were cooled to room temperature for 5 min. The remaining kit components for the reaction were added to each tube, in order, as follows: 2 μl of 100 mM DTT, 1 μl of Accuscript RT, and 0.5 μl RNase block. Using a thermocycler, the reactions were incubated at 25 °C for 10 min to allow primer extension, followed by 42 °C for 60 min of synthesis, and then cDNA synthesis was terminated by incubation at 70 °C for 15 min. Stored unused cDNA at −20 °C.
For the RT-qPCR reactions using the above cDNA template, a total reaction volume of 25 μl per well was used, and all sample reactions were conducted in triplicate with the same plates and machinery utilized in our ChIP-qPCR reactions described above. Each 25 μl reaction consisted of 1 μl of sample cDNA, 1 μl primer mix (from a pre-mixed stock of 10 μM each of forward and reverse RT-qPCR primers listed in Table 1), 10.5 μl Nuclease-free water, and 12.5 μl RT2 SYBR Green Fluor qPCR Mastermix (330514, Qiagen). Normalization was achieved using primers against GAPDH and the comparative CT method (2∧-ΔΔCT) was used to represent relative gene expression information.
MTT analysis
Cells were seeded in 12-well plates at 4000 cells per ml DME + 10% FBS + 1% P/S in triplicate for analysis at two time points. The next day experimental i-SOX2-ONS76 cells were induced with 200 ng/ml Dox in fresh media, experimental i-SOX2-HCT116 cells with 500 ng/ml Dox, and control cells for both cell lines simply provided fresh media. After 48 h, half of the experiment was analyzed at the 48-h time point and the other half of the cells were cultured for another 48 h after media was changed for all the remaining wells with the experimental wells receiving media containing Dox at the appropriate concentration. For analysis, 5 mg/ml tetrazolium dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was prepared in 1X PBS. The MTT reagent was filtered through a 0.22 μM sterile filter. Additionally, a stop solution solvent was prepared using 20% w/v SDS dissolved in dimethylformamide and water (50:50), and the pH was adjusted to 4.7 using 2.5% of 80% acetic acid and 2.5% of 1N-HCl. The media was removed from each well and 1 ml of fresh DME with 10% FBS and 1% P/S pre-mixed with MTT reagent mix (800 μl DME + 200 μl MTT reagent) was added to each well and to an additional blank well. The reaction was incubated for 2 h at 37 °C. After incubation, 800 μl of stop solution solvent was added to each well and incubated overnight at 37 °C. The next day, the absorbance of each well was measured at a fixed wavelength of 570 nM visible light on Beckman DU640B spectrophotometer.
Data availability
All data are included in the manuscript.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Adam Karpf is thanked for reading this manuscript and for providing helpful feedback. Samikshan Dutta, Virginia Commonwealth University, is thanked for suggesting ChIP protocol improvements. We also thank Sandipan Brahma for the suggestion to examine the status of H3K27me3. Finally, Heather Rizzino is thanked for editorial assistance.
Author contributions
B. D. O. G., D. V. G., G. S. Y., D. W. C., and A. R. writing–review and editing; B. D. O. G. and A. R. writing–original draft; B. D. O. G. visualization; B. D. O. G. and D. V. G. validation; B. D. O. G. and D. V. G. investigation; B. D. O. G. and D. V. G. formal analysis; B. D. O. G., G. S. Y., D. W. C., and A. R. conceptualization; A. R. supervision; A. R. project administration; A. R. funding acquisition.
Funding and additional information
This work was funded by grants from the National Cancer Institute (CA267022), the Nebraska Department of Health and Human Services (2023-51 and 2024-41), and the Edna Ittner Pediatric Research Support Fund.
Reviewed by members of the JBC Editorial Board. Edited by Brian D. Strahl
Supporting information
Supplemental Fig S1.
Supplemental Fig S2.
Supplemental Fig S3.
Supplemental Fig S4.

Supplemental Fig S5.
Supplemental Fig S6.
Supplemental Fig S7.
References
- 1.Dang C.V. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jha R.K., Kouzine F., Levens D. MYC function and regulation in physiological perspective. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1268275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duffy M.J., O’Grady S., Tang M., Crown J. MYC as a target for cancer treatment. Cancer Treat. Rev. 2021;94 doi: 10.1016/j.ctrv.2021.102154. [DOI] [PubMed] [Google Scholar]
- 4.Madden S.K., De Araujo A.D., Gerhardt M., Fairlie D.P., Mason J.M. Taking the Myc out of cancer: toward therapeutic strategies to directly inhibit c-Myc. Mol. Cancer. 2021;20:3. doi: 10.1186/s12943-020-01291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metz E.P., Wilder P.J., Popay T.M., Wang J., Liu Q., Kalluchi A., et al. Elevating SOX2 downregulates MYC through a SOX2:MYC signaling Axis and induces a slowly cycling proliferative state in human tumor cells. Cancers. 2022;14:1946. doi: 10.3390/cancers14081946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagey D.W., Klum S., Kurtsdotter I., Zaouter C., Topcic D., Andersson O., et al. SOX2 regulates common and specific stem cell features in the CNS and endoderm derived organs. Plos Genet. 2018;14 doi: 10.1371/journal.pgen.1007224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagey D.W., Muhr J. Sox2 acts in a dose-dependent fashion to regulate proliferation of cortical progenitors. Cell Rep. 2014;9:1908–1920. doi: 10.1016/j.celrep.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Kopp J.L., Ormsbee B.D., Desler M., Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem. Cells. 2008;26:903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 9.Vanner R.J., Remke M., Gallo M., Selvadurai H.J., Coutinho F., Lee L., et al. Quiescent Sox2+ cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell. 2014;26:33–47. doi: 10.1016/j.ccr.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malladi S., Macalinao D.G., Jin X., He L., Basnet H., Zou Y., et al. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell. 2016;165:45–60. doi: 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeda K., Mizushima T., Yokoyama Y., Hirose H., Wu X., Qian Y., et al. Sox2 is associated with cancer stem-like properties in colorectal cancer. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-36251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuebben E.L., Rizzino A. The dark side of SOX2: cancer - a comprehensive overview. Oncotarget. 2017;8:44917–44943. doi: 10.18632/oncotarget.16570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie X.P., Laks D.R., Sun D., Ganbold M., Wang Z., Pedraza A.M., et al. Quiescent human glioblastoma cancer stem cells drive tumor initiation, expansion, and recurrence following chemotherapy. Dev. Cell. 2022;57:32–46.e8. doi: 10.1016/j.devcel.2021.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuebben E.L., Wilder P.J., Cox J.L., Grunkemeyer J.A., Caffrey T., Hollingsworth M.A., et al. SOX2 functions as a molecular rheostat to control the growth, tumorigenicity and drug responses of pancreatic ductal adenocarcinoma cells. Oncotarget. 2016;7:34890–34906. doi: 10.18632/oncotarget.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox J.L., Wilder P.J., Desler M., Rizzino A. Elevating SOX2 levels deleteriously affects the growth of medulloblastoma and glioblastoma cells. PloS One. 2012;7 doi: 10.1371/journal.pone.0044087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metz E.P., Wilder P.J., Dong J., Datta K., Rizzino A. Elevating SOX2 in prostate tumor cells upregulates expression of neuroendocrine genes, but does not reduce the inhibitory effects of enzalutamide. J. Cell Physiol. 2020;235:3731–3740. doi: 10.1002/jcp.29267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metz E.P., Wuebben E.L., Wilder P.J., Cox J.L., Datta K., Coulter D., et al. Tumor quiescence: elevating SOX2 in diverse tumor cell types downregulates a broad spectrum of the cell cycle machinery and inhibits tumor growth. BMC Cancer. 2020;20:941. doi: 10.1186/s12885-020-07370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley D.L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 19.Kitaura H., Galli I., Taira T., Iguchi-Ariga S.M.M., Ariga H. Activation of c- myc promoter by c- myc protein in serum starved cells. FEBS Lett. 1991;290:147–152. doi: 10.1016/0014-5793(91)81246-5. [DOI] [PubMed] [Google Scholar]
- 20.Penn L.J., Brooks M.W., Laufer E.M., Land H. Negative autoregulation of c-myc transcription. EMBO J. 1990;9:1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He T.-C., Sparks A.B., Rago C., Hermeking H., Zawel L., Da Costa L.T., et al. Identification of c- MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 22.Yochum G.S., Cleland R., Goodman R.H. A genome-wide screen for β-catenin binding sites identifies a downstream enhancer element that controls c -Myc gene expression. Mol. Cell Biol. 2008;28:7368–7379. doi: 10.1128/MCB.00744-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yochum G.S., Sherrick C.M., MacPartlin M., Goodman R.H. A β-catenin/TCF-coordinated chromatin loop at MYC integrates 5′ and 3′ Wnt responsive enhancers. Proc. Natl. Acad. Sci. U. S. A. 2010;107:145–150. doi: 10.1073/pnas.0912294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yochum G.S. Multiple Wnt/ß-catenin responsive enhancers align with the MYC promoter through long-range chromatin loops. PloS One. 2011;6 doi: 10.1371/journal.pone.0018966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konsavage W.M., Yochum G.S. The myc 3′ Wnt-responsive element suppresses colonic tumorigenesis. Mol. Cell Biol. 2014;34:1659–1669. doi: 10.1128/MCB.00969-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M., Wang Z., Obazee O., Jia J., Childs E.J., Hoskins J., et al. Three new pancreatic cancer susceptibility signals identified on chromosomes 1q32.1, 5p15.33 and 8q24.21. Oncotarget. 2016;7:66328–66343. doi: 10.18632/oncotarget.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahasrabudhe R., Estrada A., Lott P., Martin L., Polanco Echeverry G., Velez A., et al. The 8q24 rs6983267G variant is associated with increased thyroid cancer risk. Endocr. Relat. Cancer. 2015;22:841–849. doi: 10.1530/ERC-15-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dave K., Sur I., Yan J., Zhang J., Kaasinen E., Zhong F., et al. Mice deficient of Myc super-enhancer region reveal differential control mechanism between normal and pathological growth. eLife. 2017;6 doi: 10.7554/eLife.23382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C.T., et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 30.Lauberth S.M., Nakayama T., Wu X., Ferris A.L., Tang Z., Hughes S.H., et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohrer K.A., Song H., Akbar A., Chen Y., Pramanik S., Wilder P.J., et al. STAT3 inhibition attenuates MYC expression by modulating Co-activator recruitment and suppresses medulloblastoma tumor growth by augmenting cisplatin efficacy in vivo. Cancers. 2023;15:2239. doi: 10.3390/cancers15082239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itzen F., Greifenberg A.K., Bösken C.A., Geyer M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42:7577–7590. doi: 10.1093/nar/gku449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujinaga K., Huang F., Peterlin B.M. P-TEFb:The master regulator of transcription elongation. Mol. Cell. 2023;83:393–403. doi: 10.1016/j.molcel.2022.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawson M.A., Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 35.Mujtaba S., Zeng L., Zhou M.-M. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 36.Khoueiry P., Ward Gahlawat A., Petretich M., Michon A.M., Simola D., Lam E., et al. BRD4 bimodal binding at promoters and drug-induced displacement at Pol II pause sites associates with I-BET sensitivity. Epigenetics Chromatin. 2019;12:39. doi: 10.1186/s13072-019-0286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behera V., Stonestrom A.J., Hamagami N., Hsiung C.C., Keller C.A., Giardine B., et al. Interrogating histone acetylation and BRD4 as mitotic bookmarks of transcription. Cell Rep. 2019;27:400–415.e5. doi: 10.1016/j.celrep.2019.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durbin A.D., Wang T., Wimalasena V.K., Zimmerman M.W., Li D., Dharia N.V., et al. EP300 selectively controls the enhancer landscape of MYCN -amplified neuroblastoma. Cancer Discov. 2022;12:730–751. doi: 10.1158/2159-8290.CD-21-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ionov Y., Matsui S.-I., Cowell J.K. A role for p300/CREB binding protein genes in promoting cancer progression in colon cancer cell lines with microsatellite instability. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1273–1278. doi: 10.1073/pnas.0307276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazarova D.L., Wong T., Chiaro C., Drago E., Bordonaro M. p300 influences butyrate-mediated WNT hyperactivation in colorectal cancer cells. J. Cancer. 2013;4:491–501. doi: 10.7150/jca.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krubasik D., Iyer N.G., English W.R., Ahmed A.A., Vias M., Roskelley C., et al. Absence of p300 induces cellular phenotypic changes characteristic of epithelial to mesenchyme transition. Br. J. Cancer. 2006;94:1326–1332. doi: 10.1038/sj.bjc.6603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raisner R., Kharbanda S., Jin L., Jeng E., Chan E., Merchant M., et al. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 2018;24:1722–1729. doi: 10.1016/j.celrep.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 43.Narita T., Ito S., Higashijima Y., Chu W.K., Neumann K., Walter J., et al. Enhancers are activated by p300/CBP activity-dependent PIC assembly, RNAPII recruitment, and pause release. Mol. Cell. 2021;81:2166–2182.e6. doi: 10.1016/j.molcel.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rennoll S. Regulation of MYC gene expression by aberrant Wnt/β-catenin signaling in colorectal cancer. World J. Biol. Chem. 2015;6:290. doi: 10.4331/wjbc.v6.i4.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crump N.T., Ballabio E., Godfrey L., Thorne R., Repapi E., Kerry J., et al. BET inhibition disrupts transcription but retains enhancer-promoter contact. Nat. Commun. 2021;12:223. doi: 10.1038/s41467-020-20400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lasko L.M., Jakob C.G., Edalji R.P., Qiu W., Montgomery D., Digiammarino E.L., et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550:128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y., Yang Q. The roles of EZH2 in cancer and its inhibitors. Med. Oncol. 2023;40:167. doi: 10.1007/s12032-023-02025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cox J.L., Wilder P.J., Gilmore J.M., Wuebben E.L., Washburn M.P., Rizzino A. The SOX2-interactome in brain cancer cells identifies the requirement of MSI2 and USP9X for the growth of brain tumor cells. Plos One. 2013;8 doi: 10.1371/journal.pone.0062857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361 doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuijers J., Manteiga J.C., Weintraub A.S., Day D.S., Zamudio A.V., Hnisz D., et al. Transcriptional dysregulation of MYC reveals common enhancer-docking mechanism. Cell Rep. 2018;23:349–360. doi: 10.1016/j.celrep.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welti J., Sharp A., Brooks N., Yuan W., McNair C., Chand S.N., et al. Targeting the p300/CBP Axis in lethal prostate cancer. Cancer Discov. 2021;11:1118–1137. doi: 10.1158/2159-8290.CD-20-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gillotin S. Isolation of chromatin-bound proteins from subcellular fractions for biochemical analysis. Bio Protoc. 2018;8 doi: 10.21769/BioProtoc.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript.