Summary
A novel algorithm, AlphaMissense, has been shown to have an improved ability to predict the pathogenicity of rare missense genetic variants. However, it is not known whether AlphaMissense improves the ability of gene-based testing to identify disease-influencing genes. Using whole-exome sequencing data from the UK Biobank, we compared gene-based association analysis strategies including sets of deleterious variants: predicted loss-of-function (pLoF) variants only, pLoF plus AlphaMissense pathogenic variants, pLoF with missense variants predicted to be deleterious by any of five commonly utilized annotation methods (Missense (1/5)) or only variants predicted to be deleterious by all five methods (Missense (5/5)). We measured performance to identify 519 previously identified positive control genes, which can lead to Mendelian diseases, or are the targets of successfully developed medicines. These strategies identified 0.85 million pLoF variants and 5 million deleterious missense variants, including 22,131 likely pathogenic missense variants identified exclusively by AlphaMissense. The gene-based association tests found 608 significant gene associations (at p < 1.25 × 10−7) across 24 common traits and diseases. Compared with pLoFs plus Missense (5/5), tests using pLoFs and AlphaMissense variants found slightly more significant gene-disease and gene-trait associations, albeit with a marginally lower proportion of positive control genes. Nevertheless, their overall performance was similar. Merging AlphaMissense with Missense (5/5), whether through their intersection or union, did not yield any further enhancement in performance. In summary, employing AlphaMissense to select deleterious variants for gene-based testing did not improve the ability to identify genes that are known to influence disease.
Keywords: AlphaMissense, gene burden test, UK Biobank, ExWAS
AlphaMissense, a new algorithm, improved missense variant pathogenicity prediction, yet its effectiveness in enhancing gene-based testing to identify disease-influencing genes remains unclear. By comparing it with commonly used variant annotation methods across 24 common traits and diseases, we demonstrate that using AlphaMissense to select deleterious variants offers limited improvement in identifying disease-influencing genes.
Main text
Rare genetic variants are important contributors to human diseases. They contribute to most Mendelian disorders, and their effect sizes upon common diseases are larger than those attributed to common variants.1,2,3,4 Importantly, associated rare genetic variants are often coding and can therefore be directly attributed to a gene. Loss-of-function rare variants can offer insights into the direction of genetic effect on disease outcome. However, studying rare causal variants is challenging since most of the genetic variation in the genome is both rare and benign. Thus, gene-based analysis is usually employed to improve statistical power by aggregating multiple rare variants across a gene into one test to improve statistical power to detect disease associations.5
Previous gene-based multi-variant tests like exome-wide association studies (ExWAS) have successfully identified disease-influencing genes, like WNT1 [MIM: 164820] for osteoporosis [MIM: 166710],6 and drug-targeting genes, such as PCSK9 [MIM: 607786] for low-density lipoprotein (LDL)-cholesterol levels.7 Nevertheless, the power of ExWAS relies heavily on the prior identification of variants with a likely functional impact5 to reduce the number of irrelevant genetic variants included in the tests. While predicted loss-of-function (pLoFs) rare variants are most likely to contribute to gene-based tests, deleterious missense variants can also increase statistical power as they tend to be more common. However, to use deleterious missense variants, one must understand which of the missense variants is most likely to influence protein function—a process referred to as variant annotation. Moreover, all deleterious missense variant annotation strategies must strike a balance between false positive and false negative identification of such variants.8,9
Recent advances in missense variant effect prediction have made progress toward resolving this problem. AlphaMissense, a recently described method based on an unsupervised language model, combines protein structural context with evolutionary conservation and has claimed to achieve over 90% precision when predicting the known clinical impact of missense variants.9 Additionally, their variant pathogenicity annotations improved the prediction of gene essentiality for cell survival and fitness.
However, it is not known whether the improvements observed in AlphaMissense’s ability to predict the deleteriousness of missense variants results in improved association testing between genes and diseases. If this improvement were striking, it could help to identify new causes of disease and consequently drug targets for needed drug development. Using the UK Biobank whole-exome sequencing (WES) data,10,11 we tested the ability of AlphaMissense variant annotation to improve the ability to identify positive control genes (known to influence disease)12,13 through collapsing gene-based tests on 12 continuous traits and 12 diseases. We compared its performance to other leading algorithms. The results empirically test the ability of AlphaMissense to improve the identification of known disease-influencing genes. The information for the tested diseases and traits can be found in Tables S1 and S2, and the list of positive control genes is in Table S3.
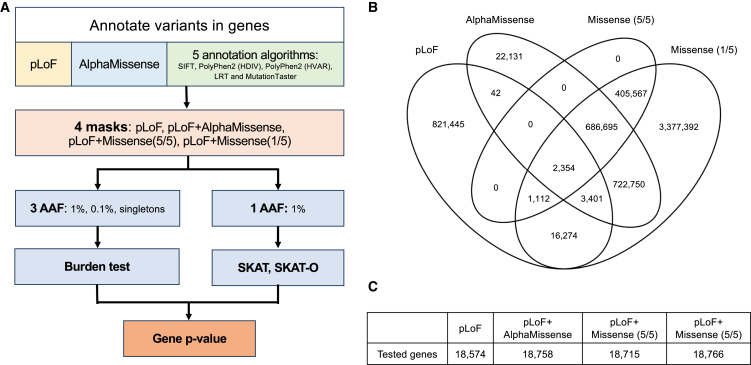
Starting with 19,606 genes, for every exon, we annotated deleterious variants into four categories: pLoF, AlphaMissense, Missense (5/5), and Missense (1/5). Then we assembled four sets of predicted deleterious variants (i.e., masks): (1) pLoF, (2) pLoF with AlphaMissense, (3) pLoF with Missense (5/5), and (4) pLoF with Missense (1/5). Each mask provided a list of variants for genes in gene-based association analysis. Last, we retained the smallest p values from the five different combinations of alternative allele frequency and statistical test method for the association between each gene and each tested trait or disease under different masks (Figure 1A).
Figure 1.
Overview of ExWAS analysis and comparison of variants annotations by different methods
(A) Strategy to obtain single p value for each gene.
(B) Overlap of annotated likely deleterious variants by different ExWAS mask settings. (C) Number of tested genes with different mask settings.
Of 26 million variants from UK Biobank WES data, we identified 0.85 million pLoF variants and 5 million predicted deleterious missense variants by AlphaMissense or any of the five commonly used annotation methods (i.e., SIFT,14 PolyPhen2 [HDIV],15 PolyPhen2 [HVAR],16 MutationTaster,17 and LRT18). Specifically, AlphaMissense classified 1.4 million variants as “likely pathogenic,” including 22,131 identified exclusively by AlphaMissense. Missense (1/5) captured over 98% of AlphaMissense predicted “likely pathogenic” variants while Missense (5/5) covered 48% of AlphaMissense predicted “likely pathogenic” variants (Figure 1B). Moreover, our results showed that among the masks evaluated, Missense (1/5) labeled the highest number of deleterious variants per gene on average (267 variants per gene), followed by AlphaMissense (74 variants per gene), Missense (5/5) (56 variants per gene), and pLoF (43 variants per gene) (Table S4). Despite the considerable variance in the number of annotated variants across different annotation categories, 99% of genes were tested in all masks (Figure 1C).
In the exome-wide gene-based analysis, we first checked the genomic inflation factors of the p values for each mask and test method combination. In general, no strong genomic inflation was observed (value range: 0.96–1.39) except for standing height (value range: 1.11–1.94) (Table S5). This is not surprising as height is a well-known highly polygenic trait.19
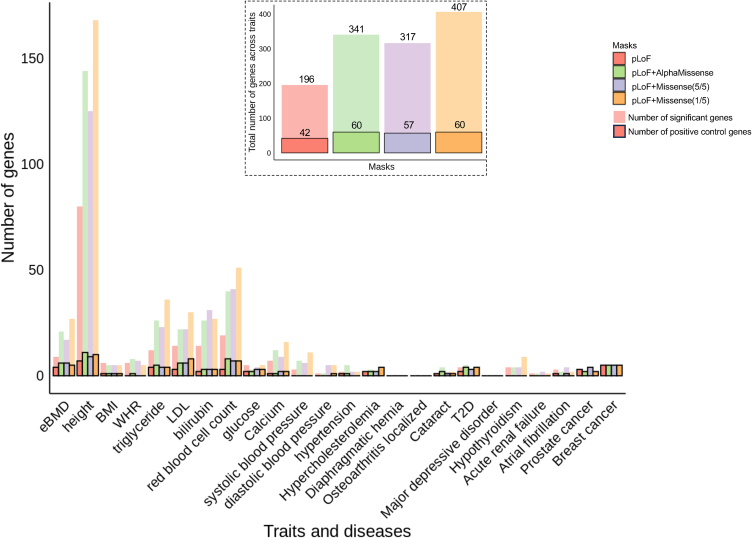
In total, our gene-based association tests found 608 significant gene associations (p < 1.25 × 10−7) across 24 common traits and diseases. We found that adding predicted deleterious missense variants to masks led to the identification of at least 60% more significant gene-trait associations and about 30% more positive control genes as compared with pLoF-only mask (Figures 2 and S1A; Table S6). Despite different numbers of associations identified, 114 significant associations and 30 positive control genes were captured using any of the masks, which accounts for between 27% and 57% and 50% and 71% of the findings, respectively, of each mask (Figures S1B and S1C). Comparing across masks, pLoF with AlphaMissense and pLoF with Missense (5/5) resulted in more significant associations and positive control genes than the pLoF-only mask. Meanwhile, these two masks also identified a slightly higher proportion of positive control genes (18% for pLoF with Missense (5/5), and 17.6% for pLoF with AlphaMissense) than pLoF with Missense (1/5) mask (14.7%). Between these two preferred masks, pLoF with AlphaMissense identified largely similar or slightly higher numbers of significant gene-trait and gene-disease associations compared with pLoF with Missense (5/5) (Figure 2). The proportion of positive control genes identified using pLoF with AlphaMissense was similar to pLoF with Missense (5/5) when using variants <1% or 0.1%, and were in fact slightly better when using singletons only (Figure S2; Table S7). Additionally, the pLoF with AlphaMissense and pLoF with Missense (5/5) masks shared 245 (71% and 76%) significant association findings and 46 (77% and 81%) of identified positive control genes (Figures S1B and S1C). Furthermore, to evaluate the impact of including additional predicted deleterious variants on the effect sizes for a specific mask, we compared the median absolute estimated effects per gene of the 114 significant associations identified across all four masks. As shown in Figure S3, these effects were closer to the null when more variants were included across different alternative allele frequency categories, with the greatest change occurring in pLoF with Missense (1/5) (Table S8). The median effect sizes of pLoF with AlphaMissense and pLoF with Missense (5/5) are similar, although both are smaller than those observed in pLoF only.
Figure 2.
Significant gene associations identified in exome-wide gene burden analysis across 12 traits and 12 diseases
The bars without outlines indicate the numbers of significant genes (p < 1.25 × 10−7) identified in each trait and disease by different masks. The bars with outlines indicate the number of significant genes that are also positive control genes for each trait and disease identified by different masks. The inset figure shows the total number of significant genes and positive controls identified by each mask across 21 tested traits and diseases that have at least one known positive control gene. eBMD, estimated bone mineral density; BMI, body mass index; WHR, waist-hip circumference ratio; LDL, serum low-density lipoproteins; T2D, type 2 diabetes.
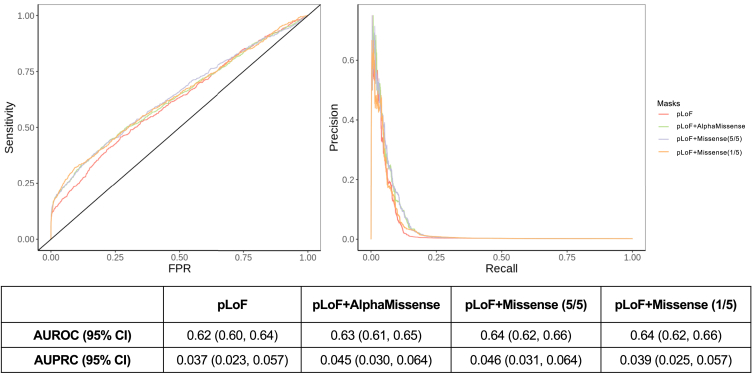
Next, to evaluate whether different masks enhanced the distinction between positive control genes and non-positive control genes by offering more divergent p values, we evaluated the performance of using different masks in classifying these genes by calculating the receiver operating characteristic curve (ROC) and precision-recall curve (PRC). Upon comparison, we observed that all four masks have statistically indistinguishable area under the receiver-operator curves (AUROC) (Figure 3, left panel). However, pLoF with Missense (5/5) and pLoF with AlphaMissense have a higher estimated area under the precision-recall curves (AUPRC) than the other two masks despite the fact that all the 95% confidence intervals of AUPRCs overlapped (Figure 3, right panel). Similar AUROC and AUPRC patterns can be observed across tested traits, but we did observe that specific masks could perform better for certain traits and diseases (Figure S4). Additionally, we tested whether using different aggregating methods for counting alleles, in burden tests, across genetic sites within genes changed the mask performance. Using the maximum number of alternative alleles across sites (the default approach) and using the sum of the number of alternative alleles in gene-based association analyses performed similarly (Figure S5).
Figure 3.
Performance curves (ROC and PRC) for all four masks to identify positive controls genes across all 21 tested traits and diseases with positive control genes. The 95% confidence interval (CI) was determined using 1,000 bootstrap replicates.
Considering that performance was better when pLoF variants were combined with either Missense (5/5) or AlphaMissense annotated deleterious variants, we further investigated whether merging AlphaMissense and Missense (5/5) annotations before combining with the pLoF variants could improve their ability to classify positive control genes. We tested two designs: using pLoF variants and variants predicted to be deleterious by (1) both AlphaMissense and Missense (5/5) or by (2) either AlphaMissense or Missense (5/5). Utilizing deleterious variants predicted by either method identified slightly more significant associations (372 pairs), although the proportion of positive control genes remained similar (17.7%) (Table S9; Figure S6). In contrast, using the overlapping predictions led to fewer significant associations (287 pairs) but a marginally higher proportion of positive control genes (18.5%). The AUROC and AUPRC of these two new mask definitions are similar to other masks (Figure S7). Overall, little improvement was observed by merging Missense (5/5) with AlphaMissense.
Last, we compared the performance of AlphaMissense vs. Missense (5/5) without any pLoF variants. As shown in Figure S8, AlphaMissense predicted deleterious variants identified slightly more significant genes and positive control genes than Missense (5/5) (42 vs. 37, respectively). However, the proportion of positive control genes identified is similar (24% for Missense (5/5) vs. 22% for AlphaMissense). Interestingly, 108 significant gene-disease and gene-trait associations (70% for Missense (5/5) and 55% for AlphaMissense) and 28 (76% and 67%) of identified positive control genes overlapped, suggesting that most findings were captured by both Missense (5/5) and AlphaMissense (Table S10).
Gene-based tests offer an elegant way to study the effect of rare coding variants on human traits by improving statistical power. However, the best way to combine genetic variants into gene sets is still not fully determined, simply because there are usually many irrelevant genetic variants in each gene set that may dilute any signal from the set of causal variants. Hence, such analyses usually rely on algorithms to predict which variants are likely to be loss-of-function or missense variants with deleterious effects. As gene-based analyses are restricted to a likely deleterious subset of variants to increase this signal to noise ratio, the success of these analyses rests partially on the performance of the predictions. The emergence of a language model-based variant effect prediction methods, AlphaMissense, has been suggested to be able to improve gene-based association. However, our results showed that AlphaMissense did not importantly outperform the current state-of-the-art masks in gene-based association analyses using whole-exome data.
There are multiple reasons why the inclusion of “likely pathogenic” missense variants, as annotated by AlphaMissense, does not lead to significant improvements. First, the masks used in our analysis always included pLoF variants, which already contribute significantly to the associations observed between genes and traits. Furthermore, the addition of AlphaMissense’s predicted pathogenic missense variants expands the analyzed gene pool by only 184 genes (when added to pLoFs) or 33 genes (when added to pLoF and Missense (5/5)) beyond those tested using pLoF-only masks. This modest increase in the number of genes tested offers limited scope for enhancing the performance of gene-based association tests. Last, as noted earlier in this report, other missense annotation methods largely capture the same “likely pathogenic” variants identified by AlphaMissense. Given that all gene-based tests then summarize information across all analyzed variants in a gene (in various ways), the small number of different prediction variants may not render a large difference in the associated genes.
AlphaMissense may provide useful and clarifying information in scenarios where understanding single variant effects is crucial. For example, AlphaMissense could be particularly helpful in pinpointing actionable genetic sites within known disease-influencing genes. This may be particularly useful for individuals with Mendelian diseases without major structural disruptions in the genetic region.20,21 Additionally, since AlphaMissense integrates protein structure context into its predictions of variant effects, it should be more effective when identifying deleterious variants for diseases where protein malfunction arises from changes in protein conformation. AlphaMissense could also be advantageous in predicting pharmacogenetic effects that involve protein-drug interactions.22
We recognize that while pLoF and missense variant annotations should not be affected by genetic ancestry, we only performed our analyses in European genetic ancestry individuals from the UK Biobank, and we only examined 24 traits. Hence, these results will need replication in other populations once sample sizes allow this. Second, the UK Biobank cohort is a relatively healthy cohort. The number of individuals with diseases is low, which can limit the statistical power to identify disease-related genes, which may make it more difficult to compare the performance of different masks in ExWAS. Third, using different masks resulted in differing performance characteristics when identifying positive control genes across various traits. Further analyses are needed to test whether a specific type of genetic effect on disease can be better represented by one of these annotation algorithms. Last, there are other annotation masks that we have not tested, and that may perform differently. Nevertheless, we compared our results to the best currently available annotations,10 and we have established that any future work should make comparisons to AlphaMissense.
In summary, we found that most of the “likely pathogenic” missense variants identified by AlphaMissense were also generally predicted to be deleterious by at least one of five commonly used variant annotation methods. Using masks combining AlphaMissense with pLoF did not outperform the state-of-the-art missense annotation tools for gene-based studies.
Data and code availability
Individual-level genotype, exome sequencing, and phenotype data are available to approved researchers via the UK Biobank. The main ExWAS summary statistics generated during this study are available at GWAS Catalog (GCST90446417-GCST90446464).
UK Biobank exome data were analyzed using Regenie 3.2.1. All other data analysis was performed using R (v.4.1.2). Additional codes are available at Github (https://github.com/richardslab/ExWAS_using_AlphaMissense).
Acknowledgments
We appreciate the individuals who participated in UK Biobank. This research has been conducted using UK Biobank data under application ID 27449.
The Richards research group is supported by the Canadian Institutes of Health Research (CIHR: 365825, 409511, 100558, 169303), the McGill Interdisciplinary Initiative in Infection and Immunity (MI4), the Lady Davis Institute of the Jewish General Hospital, the Jewish General Hospital Foundation, the Canadian Foundation for Innovation, the NIH Foundation, Cancer Research UK, Genome Québec, the Public Health Agency of Canada, McGill University, and the Fonds de Recherche du Québec - Santé (FRQS). J.B.R. is supported by an FRQS Mérite Clinical Research Scholarship. Support from Calcul Québec and Compute Canada is acknowledged. TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. Y.C. is supported by an FRQS doctoral training fellowship and the Lady Davis Institute/TD Bank Studentship Award. G.B.-L. is supported by scholarships from the FRQS, the CIHR, and Québec’s Ministry of Health and Social Services. T.S. is supported by Fund for the Promotion of Joint International Research (Fostering Joint International Research; 23KK0301) by the Japan Society for the Promotion of Science.
Declaration of interests
J.B.R. is the CEO of 5 Prime Sciences (www.5primesciences.com), which provides research services for biotech, pharma, and venture capital companies for projects unrelated to this research. He has served as an advisor to GlaxoSmithKline and Deerfield Capital. J.B.R.’s institution has received investigator-initiated grant funding from Eli Lilly, GlaxoSmithKline, and Biogen for projects unrelated to this research. Y.C. is an employee of 5 Prime Sciences. T.S. has received an endowment unrelated to this research from Eli Lilly.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100344.
Web resources
UK Biobank data: https://www.ukbiobank.ac.uk.
GWAS Catalog resources: https://www.ebi.ac.uk/gwas/.
VEP software: https://github.com/Ensembl/ensembl-vep.
Regenie software: https://github.com/rgcgithub/regenie.
Supplemental information
References
- 1.1000 Genomes Project Consortium. Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B., et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson G. Rare and common variants: twenty arguments. Nat. Rev. Genet. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner D.J., Nadig A., Jagadeesh K.A., Dey K.K., Neale B.M., Robinson E.B., Karczewski K.J., O'Connor L.J. Polygenic architecture of rare coding variation across 394,783 exomes. Nature. 2023;614:492–499. doi: 10.1038/s41586-022-05684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S., Abecasis G.R., Boehnke M., Lin X. Rare-Variant Association Analysis: Study Designs and Statistical Tests. Am. J. Hum. Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou S., Sosina O.A., Bovijn J., Laurent L., Sharma V., Akbari P., Forgetta V., Jiang L., Kosmicki J.A., Banerjee N., et al. Converging evidence from exome sequencing and common variants implicates target genes for osteoporosis. Nat. Genet. 2023;55:1277–1287. doi: 10.1038/s41588-023-01444-5. [DOI] [PubMed] [Google Scholar]
- 7.Cohen J.C., Boerwinkle E., Mosley T.H., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 8.Miosge L.A., Field M.A., Sontani Y., Cho V., Johnson S., Palkova A., Balakishnan B., Liang R., Zhang Y., Lyon S., et al. Comparison of predicted and actual consequences of missense mutations. Proc. Natl. Acad. Sci. USA. 2015;112:E5189–E5198. doi: 10.1073/pnas.1511585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng J., Novati G., Pan J., Bycroft C., Žemgulytė A., Applebaum T., Pritzel A., Wong L.H., Zielinski M., Sargeant T., et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science. 2023;381:eadg7492. doi: 10.1126/science.adg7492. [DOI] [PubMed] [Google Scholar]
- 10.Backman J.D., Li A.H., Marcketta A., Sun D., Mbatchou J., Kessler M.D., Benner C., Liu D., Locke A.E., Balasubramanian S., et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature. 2021;599:628–634. doi: 10.1038/s41586-021-04103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Hout C.V., Tachmazidou I., Backman J.D., Hoffman J.D., Liu D., Pandey A.K., Gonzaga-Jauregui C., Khalid S., Ye B., Banerjee N., et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature. 2020;586:749–756. doi: 10.1038/s41586-020-2853-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forgetta V., Jiang L., Vulpescu N.A., Hogan M.S., Chen S., Morris J.A., Grinek S., Benner C., Jang D.K., Hoang Q., et al. An effector index to predict target genes at GWAS loci. Hum. Genet. 2022;141:1431–1447. doi: 10.1007/s00439-022-02434-z. [DOI] [PubMed] [Google Scholar]
- 13.Mountjoy E., Schmidt E.M., Carmona M., Schwartzentruber J., Peat G., Miranda A., Fumis L., Hayhurst J., Buniello A., Karim M.A., et al. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat. Genet. 2021;53:1527–1533. doi: 10.1038/s41588-021-00945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 15.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;Chapter 7 doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 18.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J., Weedon M.N., Purcell S., Lettre G., Estrada K., Willer C.J., Smith A.V., Ingelsson E., O'Connell J.R., Mangino M., et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 2011;19:807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staklinski S.J., Scheben A., Siepel A., Kilberg M.S. Utility of AlphaMissense predictions in Asparagine Synthetase deficiency variant classification. bioRxiv. 2023 doi: 10.1101/2023.10.30.564808. Preprint at. [DOI] [Google Scholar]
- 21.Utsuno Y., Hamada K., Hamanaka K., Miyoshi K., Tsuchimoto K., Sunada S., Itai T., Sakamoto M., Tsuchida N., Uchiyama Y., et al. Novel missense variants cause intermediate phenotypes in the phenotypic spectrum of SLC5A6-related disorders. J. Hum. Genet. 2023;69:69–77. doi: 10.1038/s10038-023-01206-5. [DOI] [PubMed] [Google Scholar]
- 22.Park Y., Lauschke V. Towards more accurate pharmacogenomic variant effect predictions. Pharmacogenomics. 2023;24:841–844. doi: 10.2217/pgs-2023-0187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual-level genotype, exome sequencing, and phenotype data are available to approved researchers via the UK Biobank. The main ExWAS summary statistics generated during this study are available at GWAS Catalog (GCST90446417-GCST90446464).
UK Biobank exome data were analyzed using Regenie 3.2.1. All other data analysis was performed using R (v.4.1.2). Additional codes are available at Github (https://github.com/richardslab/ExWAS_using_AlphaMissense).