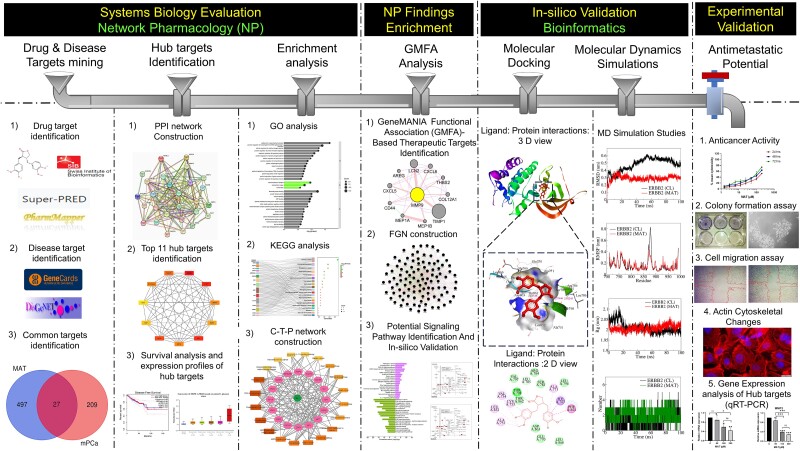
Abstract
Matairesinol (MAT), a plant lignan renowned for its anticancer properties in hormone-sensitive cancers like breast and prostate cancers, presents a promising yet underexplored avenue in the treatment of metastatic prostate cancer (mPC). To elucidate its specific therapeutic targets and mechanisms, our study adopted an integrative approach, amalgamating network pharmacology (NP), bioinformatics, GeneMANIA-based functional association (GMFA), and experimental validation. By mining online databases, we identified 27 common targets of mPC and MAT, constructing a MAT-mPC protein–protein interaction network via STRING and pinpointing 11 hub targets such as EGFR, AKT1, ERBB2, MET, IGF1, CASP3, HSP90AA1, HIF1A, MMP2, HGF, and MMP9 with CytoHuba. Utilizing DAVID, Gene Ontology (GO) analysis highlighted metastasis-related processes such as epithelial–mesenchymal transition, positive regulation of cell migration, and key Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, including cancer, prostate cancer, PI3K-Akt, and MAPK signaling, while the web resources such as UALCAN and GEPIA2 affirmed the clinical significance of the top 11 hub targets in mPC patient survival analysis and gene expression patterns. Our innovative GMFA enrichment method further enriched network pharmacology findings. Molecular docking analyses demonstrated substantial interactions between MAT and 11 hub targets. Simulation studies confirmed the stable interactions of MAT with selected targets. Experimental validation in PC3 cells, employing quantitative real-time reverse-transcription PCR and various cell-based assays, corroborated MAT’s antimetastatic effects on mPC. Thus, this exhaustive NP analysis, complemented by GMFA, molecular docking, molecular dynamics simulations, and experimental validations, underscores MAT’s multifaceted role in targeting mPC through diverse therapeutic avenues. Nevertheless, comprehensive in vitro validation is imperative to solidify these findings.
Keywords: metastatic prostate cancer (mPC), matairesinol, network pharmacology, molecular docking, molecular dynamics, PC3 cells
Graphical Abstract
Graphical Abstract.
Introduction
Prostate adenocarcinoma, the second most prevalent cancer in males globally, presents a significant health burden with increasing incidence projected to reach 2.43 million cases by 2040 [1, 2]. Metastatic prostate cancer (mPC) is associated with bone metastasis and elevated mortality rates. The 5-year survival rate for mPC is merely 32%, highlighting the urgent need for effective treatments [3, 4]. Androgen deprivation therapy (ADT) is standard for castration-dependent mPC but often leads to recurrence and progression to metastatic castration-resistant prostate cancer (mCRPC) [5]. Despite advances, mCRPC remains incurable due to limited therapeutic options and an incomplete understanding of its molecular complexities [6, 7]. To address the inadequacies in available treatments, there is a growing interest in utilizing natural phytochemicals as complementary and alternative medicines alongside existing therapeutic modalities for mPC. Given their substantial efficacy and low toxicity profile, current research endeavors focus on exploring phytochemicals to enhance the overall quality of life for individuals with mPC [8, 9]. Conducting in-depth mechanistic research to explore these phytochemicals’ anticancer and antimetastatic potential is imperative. Such research efforts can significantly contribute to advancing the field of targeted therapy, providing avenues for innovative drug discovery, and playing a crucial role in the specific development of antimetastatic therapeutics.
Natural compounds and their analogs are pivotal in drug discovery, especially in oncology [10]. Natural compounds, notably dietary lignans found in sources like flaxseeds, hold the potential for cancer treatment and metastasis prevention [11, 12]. Various dietary lignans such as lariciresinol, pinoresinol, and medioresinol demonstrate significant regulatory effects on different cancer metastases [13]. Research indicates that dietary lignans, including matairesinol (MAT), can complement cancer treatment alongside traditional chemotherapy approaches. Moreover, it is essential to investigate the mechanisms by which these dietary lignans confer these benefits. A comprehensive understanding of these mechanisms could pave the way for not only the development of adjuvants in cancer management, potentially enhancing the quality of life for individuals undergoing chemotherapy [14], but also the discovery of new antimetastatic therapeutics. MAT, a phytoestrogenic polyphenol found in flaxseeds, sesame seeds, and less quantity in berries like strawberries and blackberries, and some cereals (wheat, rye, oats), shows promise as an anticancer agent [15]. While it exhibits anti-inflammatory, anticancer, and anti-angiogenic properties in various cancers, including prostate cancer, its exact molecular mechanism remains unclear [16]. Further research is needed to elucidate MAT’s mode of action for potential therapeutic applications in prostate cancer.
Network pharmacology (NP) offers a holistic approach for understanding complex diseases by elucidating drug–target interactions and underlying biological mechanisms [17, 18]. Molecular docking and simulation studies provide valuable insights into the binding patterns and stability of protein–drug complexes [19, 20]. In this study, we employed NP, molecular docking, and molecular dynamics (MD) simulation to predict MAT’s mechanism of action against mPC. Additionally, GeneMANIA-based functional association (GMFA) analysis was utilized to enrich NP findings by exploring functional associations of hub targets. In vitro validation confirmed MAT’s anticancer and antimetastatic potential in PC3 cells, highlighting its therapeutic promise.
Methods
Identification of potential targets of MAT against metastatic prostate cancer
GeneCards and DisGeNet databases were utilized to identify human genes associated with mPC, resulting in 236 unique targets. Drug target identification was performed through Swiss Target Prediction, Superpred, and Pharmapper databases, yielding 524 potential MAT-related targets after deduplication. The conversion of protein names to gene symbols was facilitated by the UniProt database. Venn diagram analysis identified 27 unique targets shared between MAT and mPC, forming the basis for further analysis [21–25].
Construction of the PPI network and identification of the top 11 hub targets
The protein–protein interaction (PPI) network for predicted targets of MAT against mPC (PTM-mPC) was constructed using the STRING database [26] with a confidence score ≥0.7 and restricted to “Homo sapiens.” The network data in TSV format were then imported into Cytoscape 3.9.1. The top 11 MAT targets against mPC were identified as hub targets using the CytoHubba plug-in, employing the degree algorithm to assess the biological significance [27]. This approach ensured a robust and high-confidence network analysis to identify key therapeutic targets of MAT.
GO and KEGG enrichment analysis, and elaboration of an integrated network involving compound, targets, and signaling pathways
GO and KEGG pathway enrichment analyses were conducted using the DAVID database [28]. The analysis focused on “H. sapiens” gene annotations, with significance set at P ≤.05 and False Discovery Rate (FDR) cut-off ≤0.05. The top 15 GO terms and 20 KEGG pathways were selected for visualization using the SR plot and Sankey plot, respectively. A “compound–target–pathway (C-T-P) network” involving MAT, KEGG pathways, and associated target genes was constructed using Cytoscape version 3.9.1.
Expression profiles of the top 11 hub targets in both normal and tumor tissues
The gene expression profiles of the top 11 hub targets identified through CytoHubba were assessed using UALCAN, an online database. UALCAN facilitated the evaluation of gene expression levels in prostate adenocarcinoma (PRAD), considering parameters such as normal versus tumor samples, patients’ Gleason scores, and nodal metastasis status [29].
Survival analysis investigating the correlation between gene expression profiles of the top 11 hub genes and patient survival outcomes in prostate adenocarcinoma
Survival probability associations with the expression of the top 11 hub targets were explored using the GEPIA2 web server, leveraging data from the Genotype-Tissue Expression and The Cancer Genome Atlas datasets [30]. Stratified by each hub gene’s expression status, survival analysis focused on PRAD patients and evaluated overall survival (OS) and disease-free survival (DFS). Plots with a median cut-off value of 50% and hazard ratios with a 95% confidence interval were generated. Statistical significance was set at P <.05, and significant correlations between the expression of the top 11 hub genes and the survival probability of PRAD patients were observed.
In-depth analysis of MAT interaction with mPC hub targets by molecular docking and molecular dynamics simulation studies
Molecular docking analyzed MAT’s interaction with its top 11 hub targets from PPI network analysis. MAT’s 3D structure was retrieved from PubChem, while the targets’ protein structures were obtained from the RCSB PDB database (Supplementary File S1-Table S1). HIF1, IGF1, and HGF proteins were excluded due to lack of appropriate PDB structures to maintain precision. AutoDock Vina performed docking, with AutoDock Tools for binding site determination [31]. Ligands underwent energy minimization with the Avogadro Program [32]. Docking exhaustiveness was set to 100. Biovia Discovery Studio software visualized optimized conformations, providing insights into molecular interactions. Docked complexes underwent 100-ns all-atom MD simulation in Gromacs 2020.4 [33, 34]. Partial charges for MAT and cocrystal ligands were generated with the AmberTools23 antechamber module [35]. AMBER99SB-ILDN force field was used for protein topology [36]. Systems were solvated with a TIP3P water model in a dodecahedron unit cell, neutralized with sodium and chloride counter ions. Energy minimization was done until reaching a force constant threshold. Systems were equilibrated at constant particle number N, constant volume V and a constant temperature conditions of 300 K (NVT) and constant pressure using Parrinello–Rahman barostat at 1-atm pressure with a leap-frog integrator. Production phase MD simulations used a 2-fs step size. Trajectories stored at each 10 ps were analyzed for stability with root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and hydrogen bond interactions [37–39].
GeneMANIA-based functional association network analysis of top 11 hub targets to create an expanded potential therapeutic targets database of MAT against mPC
To comprehensively identify genes functionally related to the initial top 11 hub targets of MAT, we employed a novel method within the GeneMANIA framework [40]. This GMFA network analysis approach involved discovering 10 additional genes for each hub gene, prioritizing those with the strongest associations within the gene–gene network [41, 42]. The analysis focused on three key parameters, co-expression, genetic interaction, and physical interaction, significantly enhancing the precision of therapeutic target identification. Co-expression analysis aids in selecting targets involved in shared disease-related processes, while genetic interaction analysis identifies potential targets with intricate network dependencies. Physical interaction analysis helps pinpoint crucial components of disease-associated networks for targeted intervention. By considering these parameters collectively, we achieved a more nuanced understanding of gene function, facilitating the identification of robust therapeutic targets with multifaceted roles in disease processes. Subsequently, we consolidated all newly identified genes with the initially obtained top 11 hub targets, creating a GMFA-based Expanded Database (GMFA-ED) for MAT against mPC. This integration resulted in a total of 112 unique genes in the expanded database, providing a more comprehensive foundation for identifying potential therapeutic targets of MAT against mPC.
GO and KEGG enrichment analysis of GMFA-ED in relation to MAT against mPC
The GMFA-ED dataset underwent extensive GO and KEGG pathway enrichment analyses, yielding the top 20 KEGG pathways and the top 15 terms for each GO category (BP, CC, and MF). A stringent threshold of significance, set at P ≤.05, guided the analysis. These enriched pathways and terms were visually depicted using the SR plot tool. A C-T-P network was constructed to elucidate the intricate relationships within the MAT–targets–pathway network, focusing on KEGG enrichment results from the GMFA-ED dataset and visualizing the interconnected elements with the ShinyGo tool [43].
Molecular docking and molecular dynamics simulation studies of MAT with identified PI3K/AKT signaling pathway targets
The methodology mirrors procedures described in molecular docking and MD simulation studies of hub targets with MAT, with emphasis on PI3K/AKT pathway targets identified in the KEGG enrichment analysis of the GMFA-ED dataset. Protein structures corresponding to these targets were retrieved from the RCSB PDB database (Supplementary File S1-Table S2). Comprehensive analyses involving molecular docking and MD simulations were conducted to explore dynamic interactions between MAT and selected protein targets associated with the PI3K/AKT pathway, a potential therapeutic signaling pathway of MAT against mPC.
Experimental validation of MAT’s antimetastatic potential against mPC by in vitro cell culture studies
Drug, reagents, and materials
Cell culture reagents, including Ham’s F12 medium and fetal bovine serum (FBS), were procured from Gibco (USA). MAT (HPLC grade, purity >95%), TRITC-phalloidin, MTT dye, DAPI, DMSO, and an antibiotic solution containing penicillin and streptomycin were sourced from Sigma-Aldrich (St. Louis, MO). RNAiso plus total RNA extraction reagent and the PrimeScript 1st strand cDNA synthesis kit was acquired from Takara Bio USA. All plastic ware necessary for cell culture was obtained from BD Biosciences (CA, USA).
Cell culture and treatment
The PC3 cell line, devoid of androgen receptor (AR) and prostate-specific antigen expression, symbolizes aggressive androgen-independent behavior and metastatic tendencies. Acquired from the National Centre for Cell Science in Pune, India, cells were cultured in Ham’s F12 medium supplemented with 10% FBS, 2 mM l-glutamine, and 100 U/ml penicillin–streptomycin under standard conditions at 37°C in a 5% CO2 incubator, following [44]. MAT, stocked as a 50-mM solution in DMSO at 4°C, was serially diluted with the culture medium for experimentation, with DMSO serving as a negative control at concentrations over 0.5% (v/v).
Assessing the cytotoxic effect of MAT on PC3 prostate cancer cells by MTT assay
PC3 cells were seeded at a density of 1.5 × 103 cells per well in 96-well plates and cultured overnight. MAT at concentrations ranging from 1.56 to 200 μM, prepared as 2× exponential dilutions, was administered for 24, 48, and 72 h. Control groups included untreated cells and cells treated with DMSO, ensuring DMSO concentrations remained below 0.5% (v/v). After the respective treatment periods, 100 μl of 1 mg/ml MTT solution was added per well, followed by a 4-h incubation at 37°C. Purple-colored formazan crystals confirmed cell viability, dissolved in 100 μl DMSO, and quantified for absorbance at 570–630 nm [45]. The results were derived from at least three independent sets (n = 3) of triplicate experiments.
Assessing the inhibitory effect of MAT on clonogenic potential in PC3 prostate cancer cells by anchorage-dependent clonogenic assay
PC3 cells were seeded at a density of 500 cells per well in 6-well plates and cultured overnight. Following incubation, cells were treated with MAT at concentrations of 0, 50, 100, and 200 μM, alongside controls, and incubated for 8 days to allow colony formation. Colonies were then fixed, stained with crystal violet, and quantified by measuring absorbance at 595 nm after extraction with 10% acetic acid [45]. The results were derived from three independent sets (n = 3) of triplicate experiments. This experimental setup enabled a comprehensive assessment of MAT’s impact on the clonogenic potential of PC3 prostate cancer cells.
Investigating the antimetastatic effect of MAT on PC3 prostate cancer cells by cell migration assay
A wound-healing assay was conducted to evaluate cellular migration post-treatment. PC3 cells were seeded at 3 × 105 cells per well in 6-well plates and incubated overnight. Wounds were created, and a medium containing MAT (0, 50, 100, and 200 μM) was added. Wound closure was monitored at 0 and 24 h post-treatment using an inverted microscope, and closure rates were analyzed with ImageJ [45, 46]. The results were derived from three independent sets (n = 3) of triplicate experiments. This method facilitated a thorough assessment of MAT’s impact on cell migration over time.
Evaluating the antimetastatic efficacy of MAT on PC3 prostate cancer cells by fluorescent microscopy–based filopodia and lamellipodia formation
The impact of varying MAT concentrations on actin-based filopodia and lamellipodia formation, essential for cell motility, was evaluated. PC3 cells (1 × 105 cells) were seeded on glass coverslips and treated with MAT (0, 50, 100, and 200 μM) for 24 h. Following treatment, cells underwent fixation, permeabilization, blocking, and staining with TRITC-phalloidin. Prepared slides were analyzed using an Olympus IX73 Inverted Fluorescence Microscope to quantify changes in filopodia and lamellipodia induced by MAT treatment [47]. The results were obtained from three independent sets of experiments, each conducted in triplicate (n = 3). This method provided insight into MAT’s antimetastatic effects on actin cytoskeleton dynamics in PC3 cells.
The qRT–PCR investigations to validate the network pharmacology-predicted therapeutic targets of MAT against mPC in PC3 prostate cancer cells
PC3 cells (3 × 105 cells/well in 6-well plates) were treated with MAT at varying concentrations (0, 50, 100, and 200 μM). Total RNA extraction was carried out using RNAiso plus total RNA extraction reagent (Takara Bio USA), followed by cDNA synthesis with the PrimeScript 1st strand cDNA synthesis kit (Takara Bio USA). Quantitative real-time reverse-transcription PCR (qRT–PCR) was performed on a 7500 fast Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using TB Green Premix Ex Taq II (Tli RNase H Plus; Takara Bio USA) [48], following the manufacturer’s protocol. Data analysis was conducted using the 2−ΔΔCT method with actin normalization, as described by [49]. Primer sequences for the targeted genes are provided in Supplementary File S1-Table S3. The results were obtained from three independent sets of experiments, each conducted in triplicate (n = 3).
Statistical analysis
All experiments were conducted in triplicate and independently repeated three times. The results are expressed as mean ± SD. Statistical analyses were performed using GraphPad Prism 5. Differences between groups were evaluated for statistical significance using one-way analysis of variance followed by multiple comparison, while independent samples were analyzed using the t-test. A P-value of <.05 was considered statistically significant.
Results
Potential targets of MAT against mPC
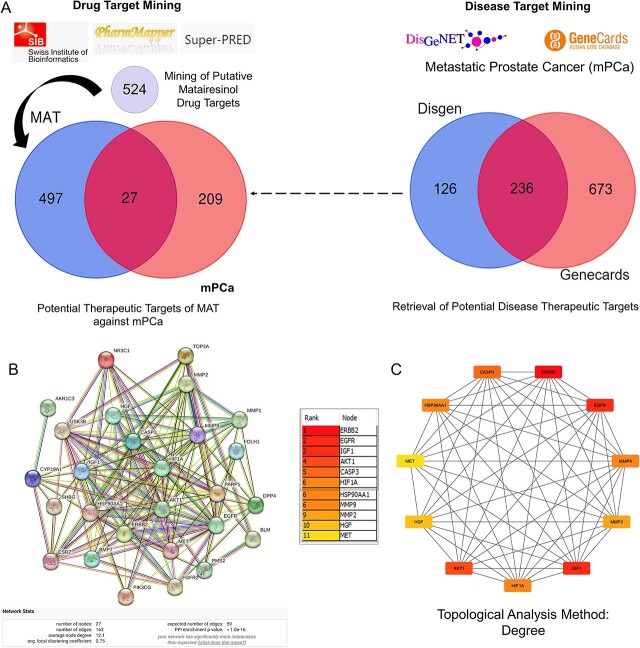
A systematic approach was employed to predict MAT targets against mPC. Initially, MAT targets were retrieved from Swiss Target Prediction, Pharmapper, and Superpred databases, resulting in 524 potential targets. Simultaneously, mPC targets were identified from GeneCards and DisGeNet databases, yielding 236 targets (Supplementary File S2). Intersection analysis revealed 27 potential therapeutic targets of MAT against mPC (PTM-mPC) (Fig. 1A).
Figure 1.
Process of identifying potential targets of MAT against mPC, constructing the PPI network, and determining the top 11 hub targets. (A) Identification of PTM-mPC. (B) Construction and analysis of the interactive PPI network using the STRING database, involving the 27 PTM-mPC. (C) Identification of the top 11 hub targets of MAT against mPC using the CytoHubba plug-in in Cytoscape, applying the degree topological analysis method.
Construction of PPI network and analysis of top 11 hub targets of MAT against mPC
The PPI network comprised 27 PTM-mPC hub targets identified from the intersection of mPC and MAT genes (Fig. 1B). It featured 27 nodes, 163 edges, an average PPI enrichment P-value of <1.0e-16, an average local clustering coefficient of 0.75, and an average node degree of 12.1. Cytoscape analysis further identified the top 11 genes—EGFR, AKT1, ERBB2, MET, IGF1, CASP3, HSP90AA1, HIF1A, MMP2, HGF, and MMP9—via the CytoHubba plug-in (Fig. 1C), ranked by their degree algorithm scores with color-coding indicating node degree.
Enrichment analysis of KEGG pathways and construction of C-T-P network for predicted targets of MAT against mPC
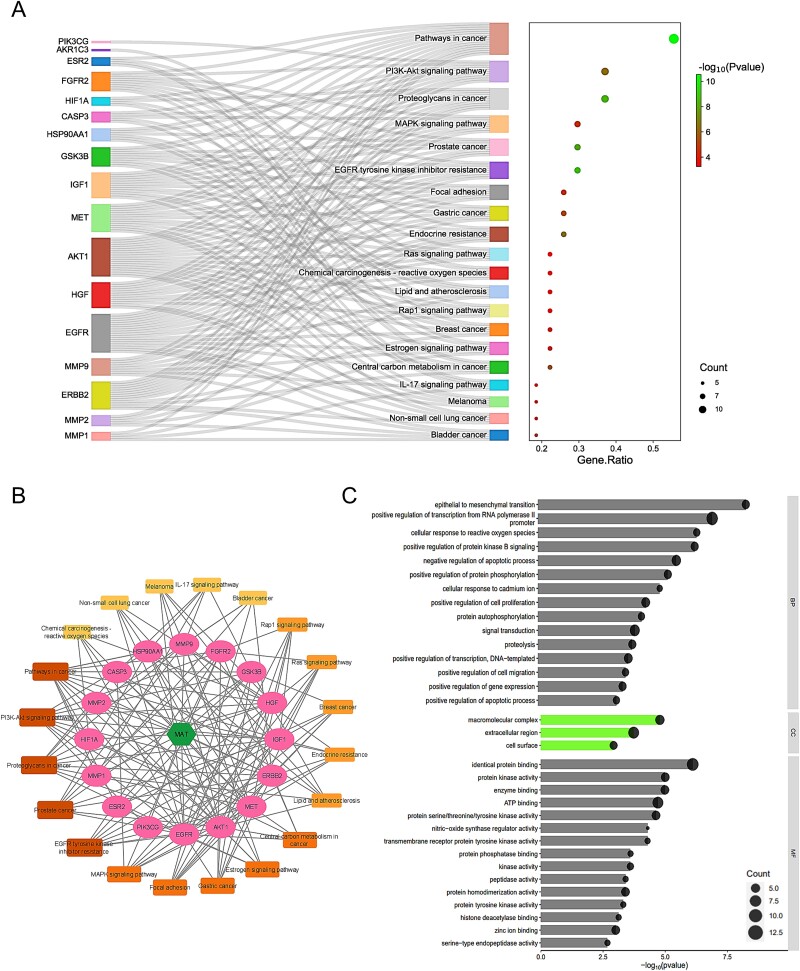
Utilizing DAVID, we analyzed KEGG pathways for the 27 shared targets to understand MAT-induced alterations in mPC progression (Supplementary File S3). We identified 36 pathways (FDR ≤ 0.05, P-value ≤ .05), with the top 20 pathways listed in Fig. 2A. A “compound–target–pathway (C-T-P)” network (Fig. 2B) was constructed to visualize MAT, its targets, and associated pathways. Noteworthy pathways include “Pathways in cancer” (degree: 15), “Proteoglycans in cancer” (degree: 10), “PI3K-Akt signaling pathway” (degree: 10), “MAPK signaling pathway” (degree: 8), “Prostate cancer” (degree: 8), and “EGFR tyrosine kinase inhibitor resistance” (degree: 8). Prostate cancer is strongly linked to eight genes, including GSK3B, HSP90AA1, ERBB2, AKT1, IGF1, MMP9, EGFR, and FGFR2 within the C-T-P network, highlighting the potential association between MAT and cancer-related signaling cascades.
Figure 2.
GO, KEGG enrichment analysis, and compound–targets–pathways network of MAT targets against mPC. (A) Sankey diagram for KEGG enrichment analysis of top 20 signaling pathways of MAT against mPC. (B) Compound–targets–pathways network illustrating the interactions between MAT and its targets in mPC. (C) GO enrichment analysis showing top BP, CC, and MF functional attributes of MAT’s targets against mPC.
Enrichment analysis of GO for predicted targets of MAT against mPC
GO enrichment analysis using DAVID (Supplementary File S3) uncovered 150 BP, 20 CC, and 35 MF terms meeting the P-value ≤.05 threshold. Application of an FDR filter (FDR ≤ 0.05) yielded 34 BP, 3 CC, and 16 MF terms. The top 15 BP and MF terms, along with 3 CC terms, are shown in Fig. 2C. BP enrichment included pathways like transcription regulation, signal transduction, apoptosis, cell proliferation, and epithelial–mesenchymal transition. CC terms indicated gene presence in cellular regions like the membrane and extracellular space. MF analysis revealed key functions such as protein binding, ATP binding, and kinase activity. This analysis enhances understanding of MAT’s therapeutic impact against mPC.
Comparative analysis of gene expression patterns with a focus on the top 11 hub targets of MAT against mPC in normal and tumor tissues
The gene expression analysis of the top 11 hub targets in PRAD patients was conducted using publicly available datasets like UALCAN. MMP-9, AKT1, and CASP3 showed significantly increased expression across primary tumors, high Gleason scores (6–10), and metastatic nodal expression compared to normal tissue. Additionally, AKT1, ERBB2, HSP90AA1, and MMP9 displayed increasing expression trends with elevated Gleason scores, albeit AKT1 and ERBB2 showed a decrease in Gleason score 10 (Fig. 3). These findings illuminate distinct expression patterns of hub targets across various PRAD progression stages.
Figure 3.
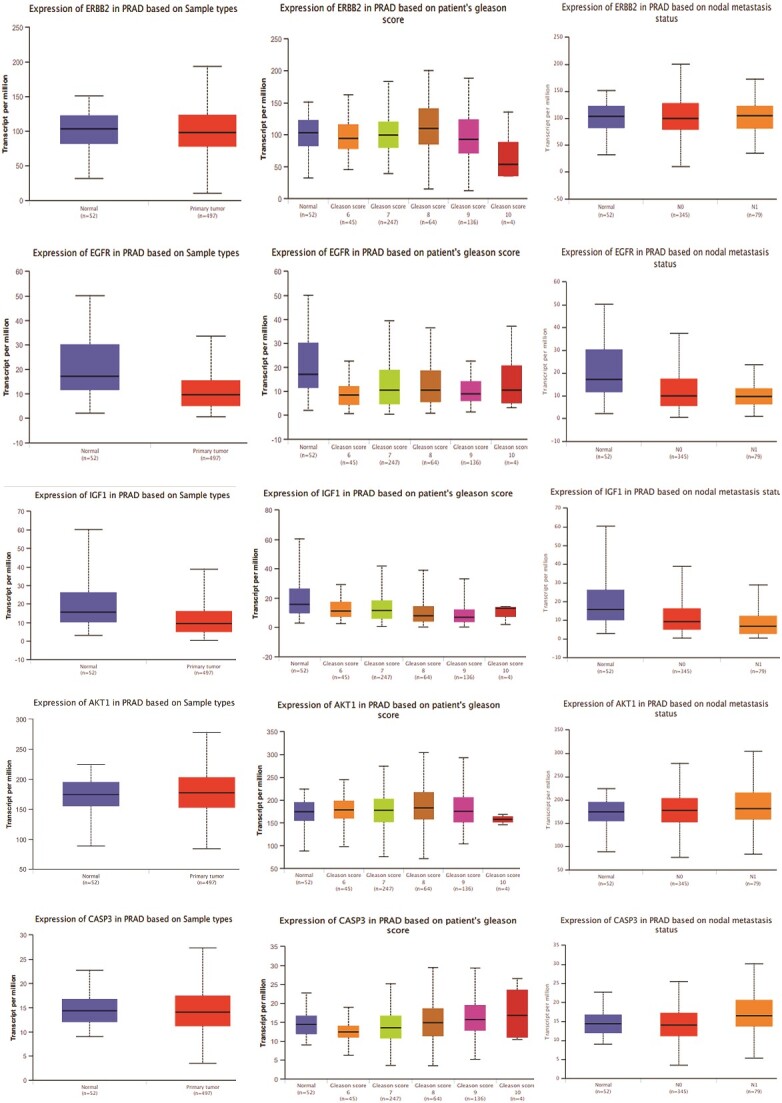
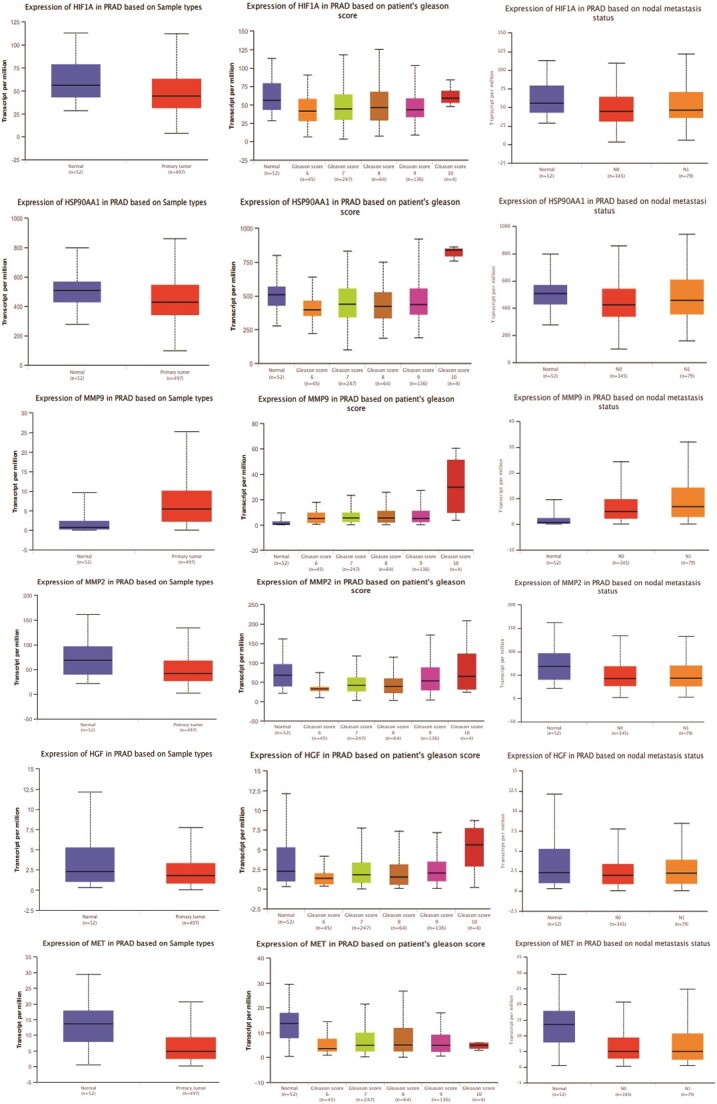
Gene expression patterns of top 11 hub targets across normal and tumor tissues of PRAD based on sample types, patient’s Gleason score, and nodal metastasis status obtained from the UALCAN platform.
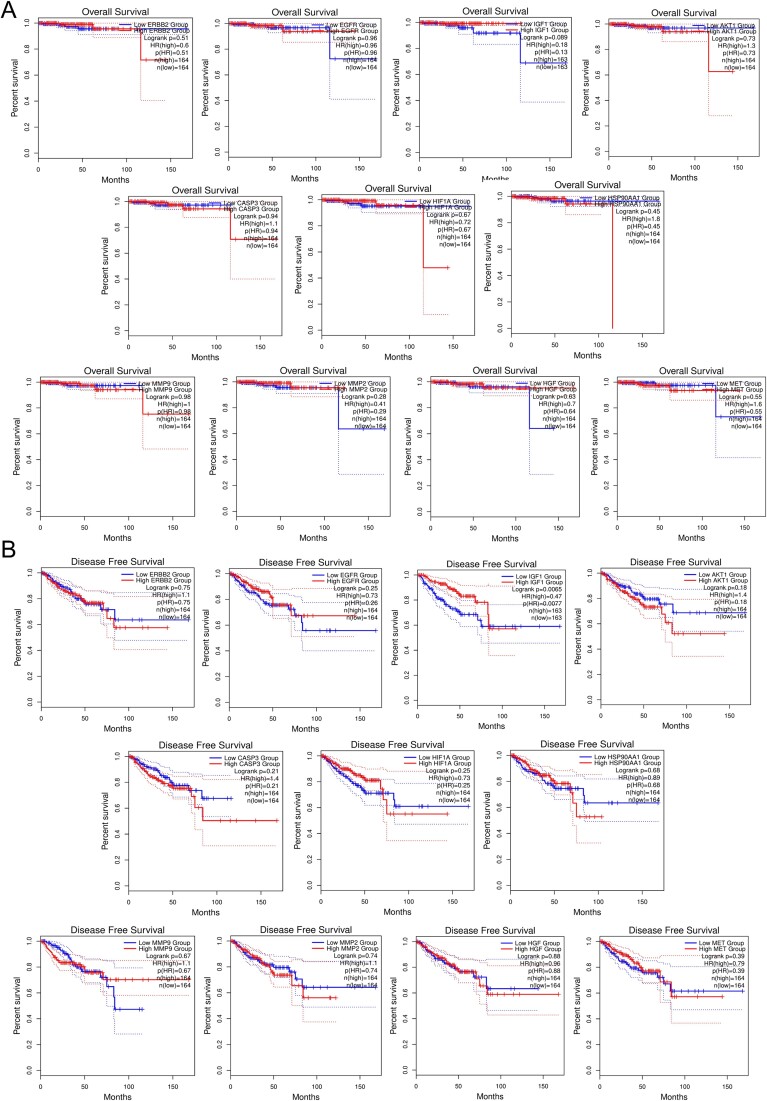
Survival analysis of top 11 hub genes in PRAD: implications for patient prognosis and therapeutic strategies
Survival plot analysis assesses gene expression patterns and patient survival outcomes, crucial for prognosis and treatment strategies. Using GEPIA2, we analyzed OS and relapse-free survival (RFS) probabilities of the top 11 hub targets (Fig. 4). Plots with a median cut-off value of 50% and hazard ratios with a 95% confidence interval were generated. Notably, HSP90AA1, MMP2, AKT1, CASP3, and ERBB2 exhibited reduced survival rates with increasing time in RFS analysis. Conversely, elevated expression of HIF1A, HSP90AA1, AKT1, MMP9, and ERBB2 significantly reduced overall survival as prostate cancer progressed. These findings suggest hub gene expression levels significantly impact survival outcomes in prostate adenocarcinoma patients.
Figure 4.
Survival analysis predicting the relationship between gene expression patterns of top 11 hub targets and patient survival outcomes in PRAD. (A) OS analysis. (B) DFS analysis.
Molecular docking and molecular dynamics simulation studies of the top 11 hub targets of MAT against mPC
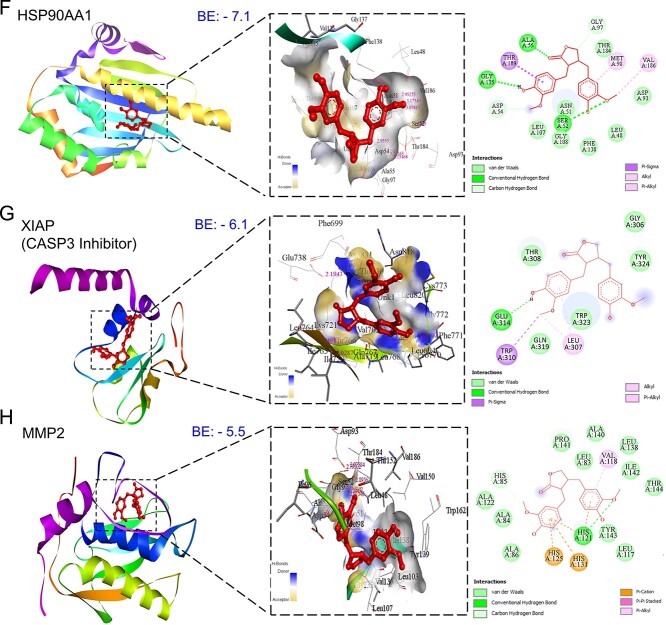
Conducting molecular docking, we explored MAT’s interactions with the top 11 mPC targets to understand their binding affinities and therapeutic implications. Lower binding energy values were observed for MAT with ERBB2 (−8.6), AKT1 (−8.5), MMP-9 (−8.4), MET (−7.8), and EGFR (−7.6), indicating strong affinities. Notably, MAT showed high affinity toward tyrosine kinase membrane receptors ERBB2, EGFR, and MET, suggesting potential inhibition of tyrosine kinase activity. Additionally, MAT interacted with HSP90AA1 and MMP-2 with binding energies of −7.1 and −5.5 kcal/mol, respectively (Fig. 5). Docking analyses against IGF1R and MET, receptors for IGF1 and HGF growth factors, were conducted due to the absence of suitable PDB IDs for cocrystal binding at the inhibition site. Additionally, we performed docking of MAT against XIAP, an inhibitor of CASP3, to investigate MAT’s possible modulation of CASP3 activation, with binding energy (−6.1) considering the lack of suitable PDB IDs supporting the CASP3 activation site [50]. Detailed results, including binding energies and specific amino acids involved, are provided in Supplementary File S1-Table S4.
Figure 5.
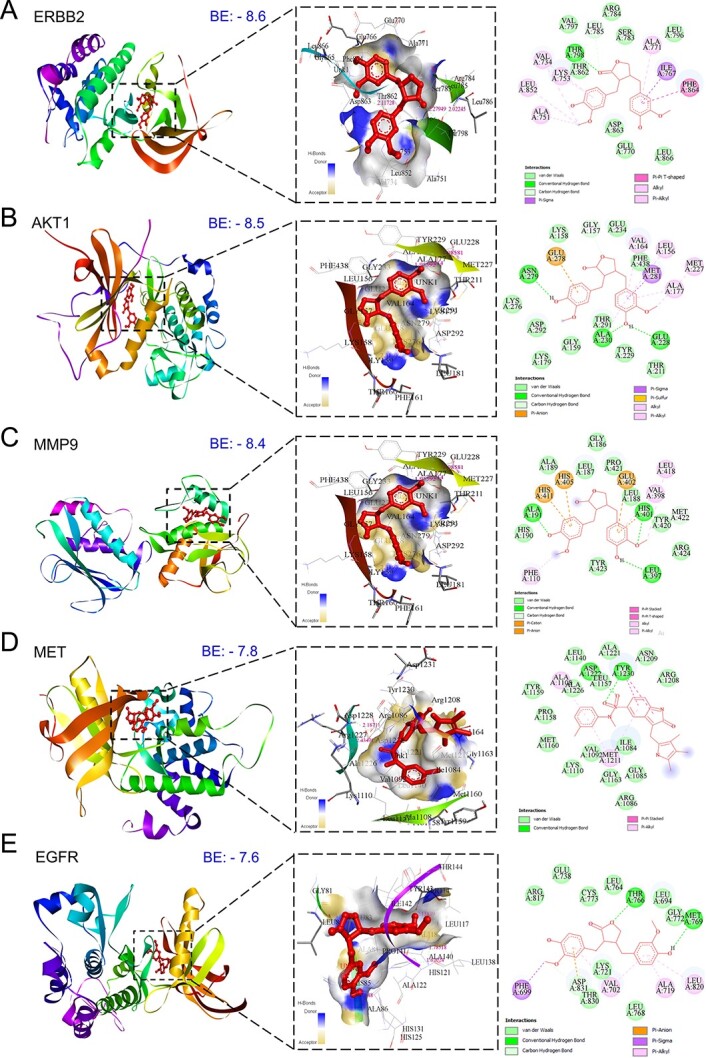
Three-dimensional and two-dimensional docking patterns and interactions of MAT with the hub targets. (A) ERBB2, (B) AKT1, (C) MMP9, (D) MET, (E) EGFR (F) HSP90AA1, (G) XIAP, and (H) MMP2.
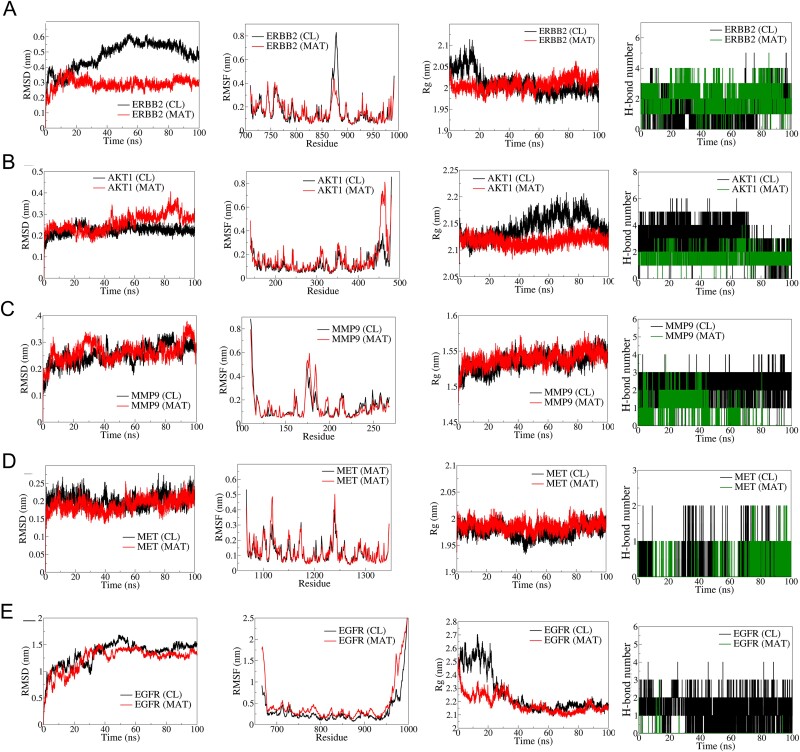
The top five hub targets, ERBB2, AKT1, MMP9, MET, and EGFR, were selected for MD simulation based on their binding affinity and biological relevance in mPC (Fig. 6). RMSD analysis showed better stability in the ERBB2-MAT complex (average RMSD: 0.3 nm) compared to the ERBB2-CL complex (average RMSD: 0.5 nm). RMSF analysis indicated lower fluctuations in MAT-bound ERBB2. Rg remained stable ~2 nm for the ERBB2-MAT complex. AKT1-MAT exhibited stable RMSD until 50 ns but deviated thereafter. RMSF showed larger fluctuations in the N-terminal domain of AKT1-MAT. Rg was stable ~2.2 nm for AKT1-MAT. MMP9 complexes showed similar RMSD and RMSF, with stable Rg ~1.525 nm. MET-MAT had slightly lower RMSD compared to MET-CL, with similar RMSF and stable Rg. EGFR complexes had higher RMSD and RMSF, with stable Rg ~2.2 nm. CL formed three consistent hydrogen bonds with EGFR, while MAT formed occasional hydrogen bonds.
Figure 6.
Results of molecular dynamics (MD) simulation analysis illustrating root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and number of H bonds for five distinct MAT–proteins and cocrystal ligand–protein complexes: (A) ERBB2, (B) AKT1, (C) MMP9, (D) MET, and (E) EGFR.
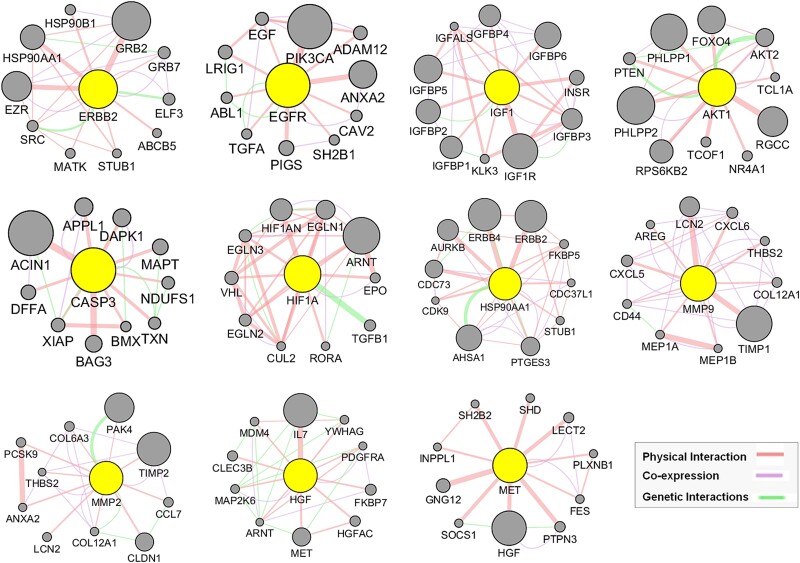
GeneMANIA-based functional association network analysis of top 11 hub targets of MAT against mPC
Utilizing our previously established GeneMANIA analysis method [41], we expanded the top 11 hub targets’ gene list by identifying 10 additional genes per hub target (Fig. 7), resulting in the GMFA expanded database (GMFA-ED) (Supplementary File S4). This dataset comprises 112 genes after removing duplicates, facilitating comprehensive gene–gene interaction analysis. The GMFA approach integrates co-expression, genetic interaction, and physical interaction parameters to capture diverse genes relevant to disease processes. The GMFA-ED was instrumental in subsequent GO and KEGG enrichment analyses, providing a robust foundation for exploring functional implications associated with the identified genes.
Figure 7.
GeneMANIA functional association (GMFA) network analysis illustrating functionally related genes associated with the top 11 hub targets of MAT and the creation of the expanded potential target database against mPC (GMFA-ED).
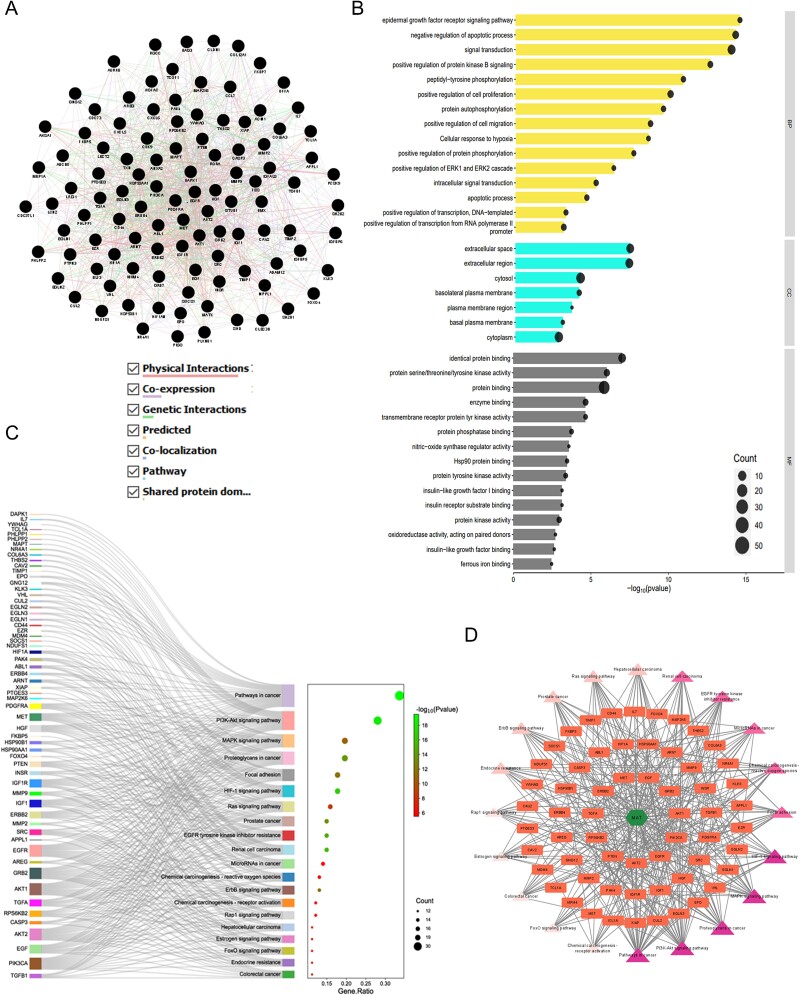
GO and KEGG enrichment analyses, C-T-P network construction, and comparative analysis of GMFA-ED enrichment in relation to MAT against mPC
The GMFA-ED dataset revealed an expanded gene network of 112 genes, forming a functional gene network (FGN) (Fig. 8A). Gene ontology and pathway analyses conducted using the DAVID database (Supplementary File S5) unveiled significant enrichments in BP, including pathways crucial for mPC progression, such as protein kinase B signaling, epidermal growth factor receptor signaling, ERK1 and ERK2 cascade regulation, and cell migration. Moreover, enriched CC associated with plasma membrane proteins, such as basolateral plasma membrane and basal plasma membrane, underscored their relevance in cancer metastasis. The MF enriched after GMFA-ED analysis indicated close associations with growth factor receptor-associated signaling, including protein kinase activity, transmembrane receptor protein tyrosine kinase activity, protein phosphatase binding, protein tyrosine kinase activity, insulin receptor substrate binding, insulin-like growth factor I binding, and insulin-like growth factor binding (Fig. 8B).
Figure 8.
GO, KEGG enrichment analysis, and compound–targets–pathways network of MAT targets identified in GMFA-ED data set. (A) FGN of all 112 genes, identified as new potential targets of MAT against mPC. (B) The bar-dot plot of the top 15 GO-BP, GO-CC, and GO-MF terms with enriched targets of MAT. (C) Sankey diagram for KEGG enrichment analysis of top 20 signaling pathways of MAT against mPC. (D) Compound–targets–pathways network illustrating the interactions between MAT and its GMFA-based predicted targets in mPC.
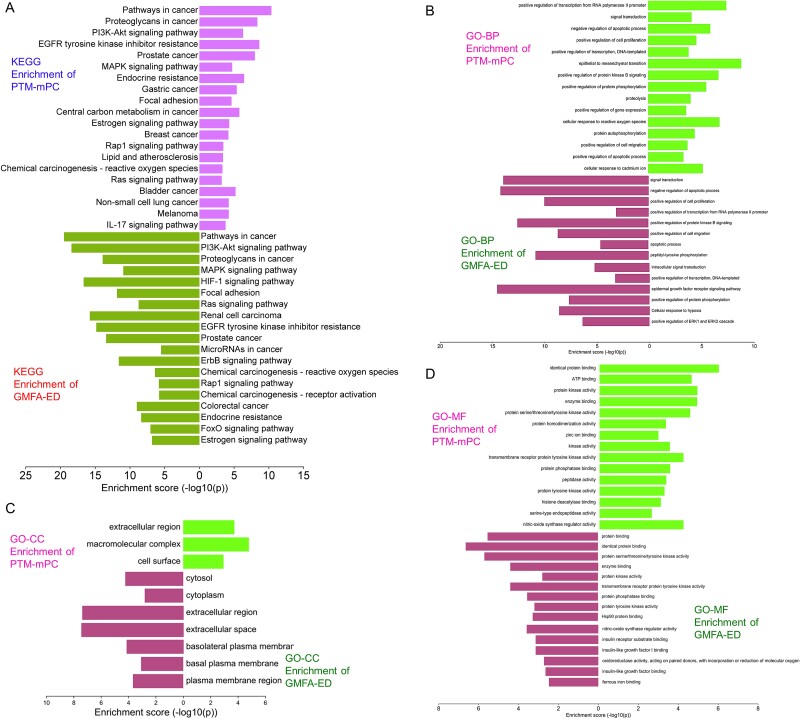
KEGG pathway analysis post-GMFA revealed enrichment in mPC-associated signaling, highlighting pathways such as HIF1A, ErbB, FOXO, and microarray in cancer. PI3K/Akt signaling exhibited extensive enrichment, emphasizing crucial players compared to pre-GMFA-ED analysis. The MAT–Targets–Pathways network, constructed using the top 20 signaling pathways from GMFA-ED, unveiled significant interactions (Fig. 8C). This network, with 82 nodes (MAT, 61 targets, and 20 pathways) and 392 edges, demonstrated MAT’s pronounced engagement through various targets within pivotal signaling pathways (Fig. 8D), including Pathways in cancer (degree = 36), PI3K/Akt pathway (degree = 28), Proteoglycans in cancer (degree = 21), MAPK signaling pathway (degree = 20), HIF-1 signaling pathway (degree = 19), and Focal adhesion (degree = 18). These findings underscore the collective functioning of these pathways in mediating MAT’s antimetastatic therapeutic effects against mPC. A visual comparison of GO and KEGG enrichment analyses before and after GMFA implementation (PTM-mPC versus GMFA-ED) is illustrated in Fig. 9. The GO and KEGG enrichment analyses of GMFA-ED unveiled a comprehensive landscape of enriched BP, CC, and MF associated with MAT’s potential targets against mPC. These enrichments were more pertinent compared to the initial enrichment of hub targets (PTM-mPC), providing insights into the intricate molecular mechanisms governing MAT’s potential antimetastatic effects in mPC.
Figure 9.
Comparison of GO and KEGG enrichment of PTM-mPC versus GMFA-ED targets of MAT against mPC. (A) KEGG enrichment, (B) BP terms of GO enrichment, (C) CC terms of GO enrichment, (D) MF terms of GO enrichment.
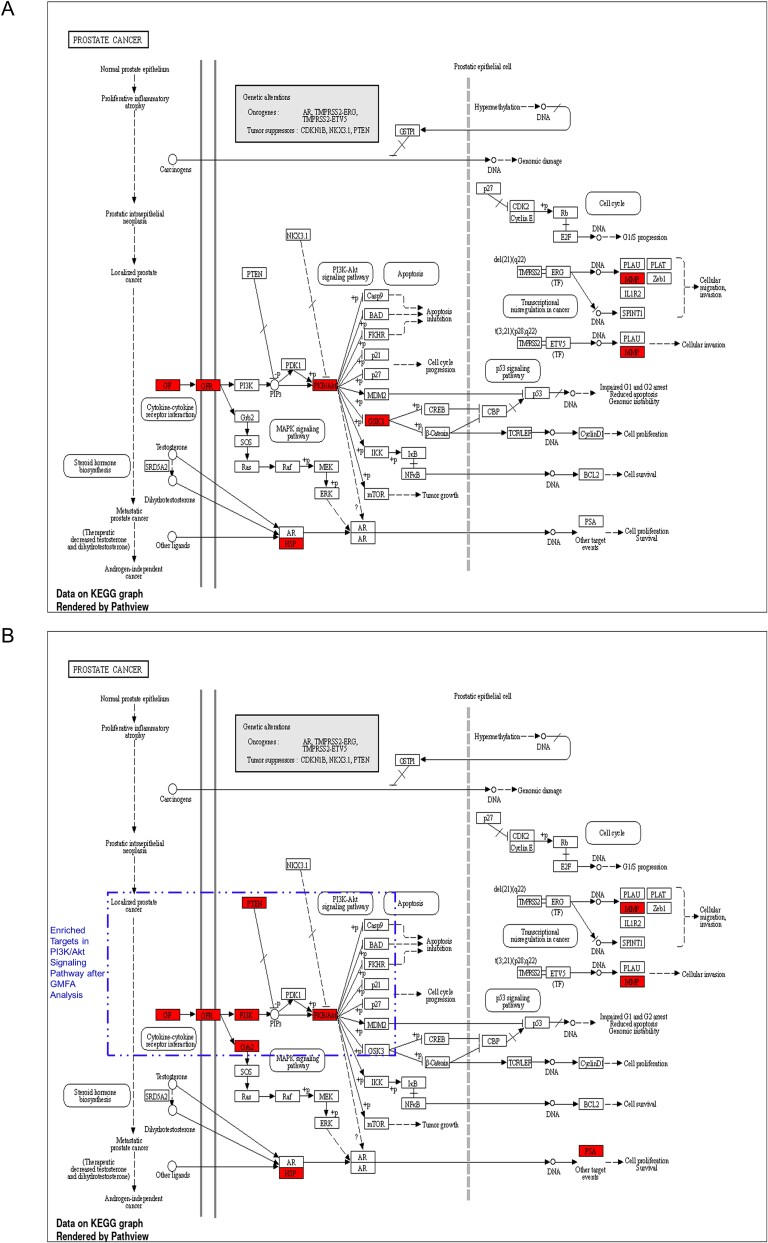
In the context of prostate cancer, the KEGG pathway analysis following the enrichment of GMFA-ED identified a substantial enrichment of genes within the PI3K-Akt signaling pathway. Notably, this enrichment encompassed six genes: GF, GFR, PI3K, Grb2, PTEN, and Akt. It is important to highlight that this enrichment was not observed in the initial analysis of PTM-mPC, except for GF, GFR, and Akt, which were previously identified as targets of MAT. This emphasizes the enhanced and more specific insights gained through the GMFA-ED analysis in unraveling the involvement of the PI3K-Akt signaling pathway in the potential antimetastatic effects of MAT in prostate cancer (Fig. 10).
Figure 10.
Comparison of prostate cancer KEGG pathway enrichment between PTM-mPC and GMFA-ED. (A) KEGG pathway enrichment analysis of PTM-mPC targets for prostate cancer. (B) KEGG pathway enrichment analysis of GMFA-ED targets for prostate cancer with significant enrichment in the PI3K/Akt signaling pathway following the GMFA analysis, indicating the identification of additional relevant therapeutic targets of MAT against mPC.
Molecular docking and molecular dynamics simulation studies of predicted PI3K/AKT signaling pathway targets of MAT
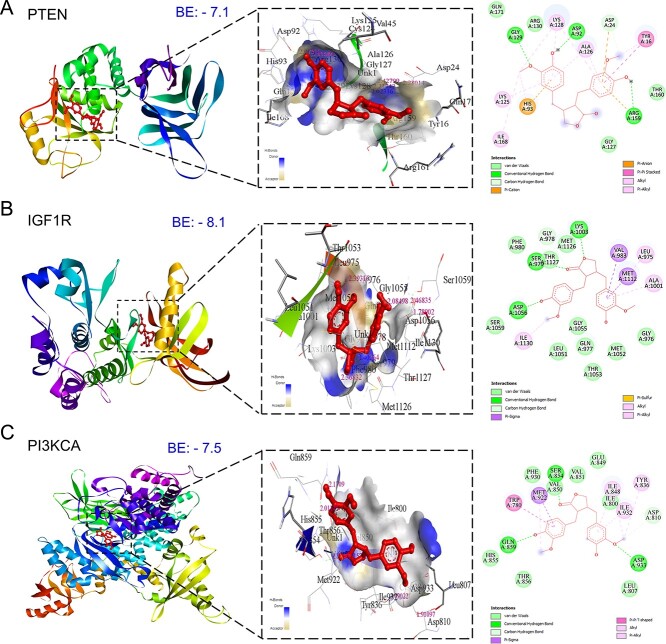
Key components of the PI3K/Akt signaling pathway, pivotal in mPC, were selected for comprehensive molecular docking and simulation study. Activation of PI3K signaling, triggered by growth factors like IGF1, HGF, and EGF, leads to downstream activation of the AKT-driven cascade [51]. MAT exhibited a notable affinity for PI3K/Akt pathway components (Fig. 11), demonstrating lower binding energies with IGF1R (−8.1 kcal/mol), PI3KCA (−7.5 kcal/mol), PTEN (−7.1 kcal/mol), and AKT1 (Supplementary File S1-Table S5). It is noteworthy that other targets within the PI3K/AKT signaling pathway were previously investigated for molecular docking and MD simulation as part of the analysis of the top 10 hub targets identified from PTM-mPC.
Figure 11.
Three-dimensional and two-dimensional docking patterns and interactions of MAT with the targets of the PI3K/Akt signaling pathway. (A) PTEN, (B) IGF1R, and (C) PI3KCA.
The top three complexes from the docking study underwent MD simulations. The RMSD analysis of PI3KCA backbone atoms showed a slightly higher average RMSD for the PI3KCA-MAT complex compared to the PI3KCA-CL complex, ~0.45 nm (Fig. 12). RMSF analysis revealed significant fluctuations in side chain atoms of residues within the loop region (250–500) in both complexes. MAT formed 5 consistent hydrogen bonds throughout, while CL formed ~3 hydrogen bonds, occasionally reaching a maximum of 5. For IGF1R complexes, RMSD deviated significantly in the IGF1R-MAT complex, whereas it remained stable but slightly higher in the IGF1R-CL complex. RMSF was similar for both complexes except for terminal residues and residues ~950–1000. Rg initially deviated for both complexes, stabilizing ~2.5 nm. MAT formed 4 consistent hydrogen bonds, while CL formed a maximum of 5 in the first 10 ns and ~2 occasionally thereafter. In PTEN complexes, RMSD stabilized after ~50 ns, averaging ~0.3 nm, with fluctuations in side chain atoms similar to apo and PTEN-MAT complex. Rg stabilized ~2.25 nm after initial deviations, and MAT formed ~2 occasional hydrogen bonds throughout.
Figure 12.
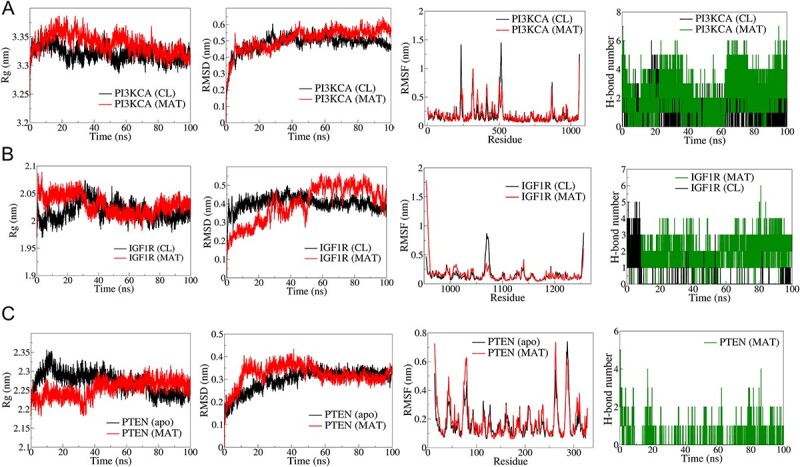
Results of molecular dynamics (MD) simulation analysis illustrating radius of gyration (Rg), root mean square deviation (RMSD), root mean square fluctuation (RMSF), and number of H bonds for three distinct MAT–proteins and cocrystal ligand–protein complexes: (A) PI3KCA, (B) IGF1R, (C) PTEN.
Experimental validation of network pharmacology and bioinformatics-driven exploration of the antimetastatic potential of MAT in mPC
MAT exhibited anticancer activity on PC3 cells
The antiproliferative effect of MAT on PC3 cells was assessed through MTT assays at 24, 48, and 72 h. The outcomes, illustrated in logarithmic concentration curves in terms of mean percent cytotoxicity and denoted as IC50 values (Fig. 13A), revealed a dose-dependent reduction in cell viability following MAT treatment over the specified time intervals (24, 48, and 72 h), substantiating the anticancer potential of MAT against metastatic PC3 prostate cancer cells.
Figure 13.
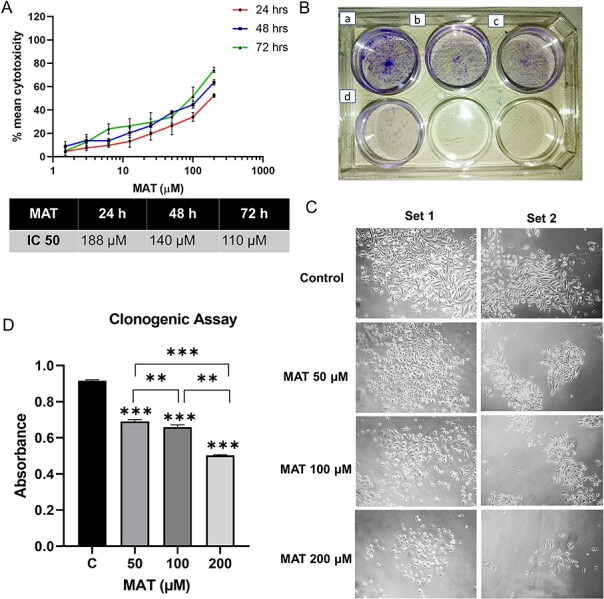
Impact of MAT on the proliferation and clonogenic abilities of PC3 prostate cancer cells. (A) MAT’s effects on PC3 prostate cancer cell viability were assessed through mean percent cytotoxicity in MTT assay. (B) Anchorage-dependent clonogenic assay revealing decreased colony numbers following MAT treatment. Panels depict (a) control, (b) MAT 50 μM, (c) MAT 100 μM, and (d) MAT 200 μM. (C) Inverted phase microscope images capturing morphological changes, including reduced colony size and fewer cells per colony. (D) Quantification of colony numbers through crystal violet staining and absorbance measurement at 595 nm. Values are presented as mean ± SD from three independent experiments (n = 3) with *P ≤ .05, **P ≤ .01, and ***P ≤ .001, denoting statistical significance.
MAT inhibited the clonogenic ability of PC3 cells
Using the anchorage-dependent clonogenic assay, we evaluated MAT’s effect on PC3 cells’ clonogenic potential, indicative of their uncontrolled proliferation. MAT’s impact is visually depicted (Fig. 13B), showing reduced colony and cell numbers with increasing MAT concentration (Fig. 13C). Quantitative analysis revealed a dose-dependent decrease in clonogenicity, with significant reductions at 50 μM (1.196-fold), 100 μM (1.44-fold), and 200 μM (2.097-fold) MAT doses (Fig. 13D).
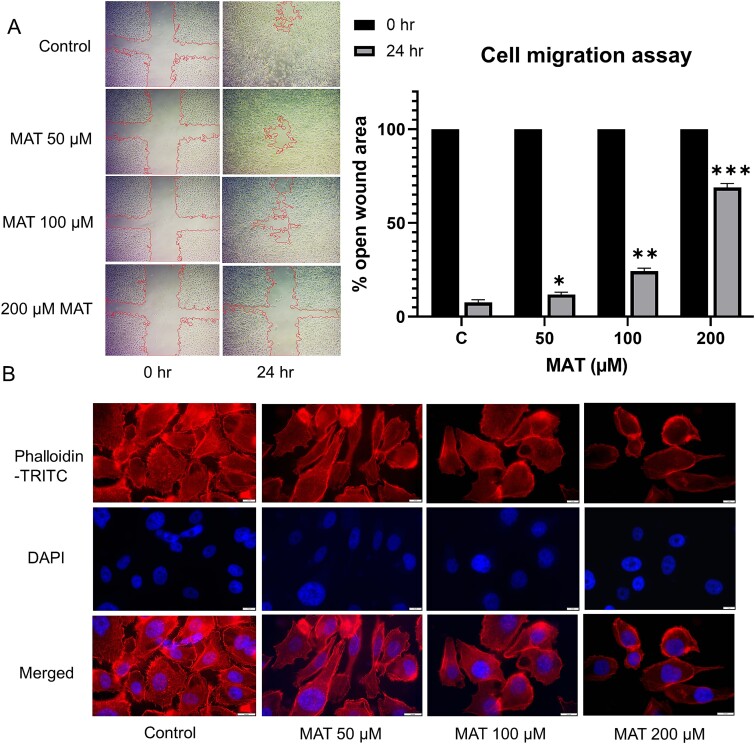
MAT inhibited the migration of PC3 prostate cancer cells
The wound-healing assay assessed PC3 cell migration, revealing a dose-dependent inhibition with MAT treatment (Fig. 14A). Untreated cells exhibited the highest migration rate, with a percent open wound area decreasing to 9% after 24 h. In contrast, MAT treatments at 50, 100, and 200 μM inhibited migration, resulting in percent open wound areas of 10.99%, 24.48%, and 71.33%, respectively, as quantified by ImageJ software analysis.
Figure 14.
Antimetastatic effect of MAT on PC3 prostate cancer cells: (A) MAT’s dose-dependent influence on cell migration was evaluated by quantifying the percent open wound area in the migration assay, visualized using an inverted microscope with a 4× objective at 24-h intervals. (B) TRITC-phalloidin fluorescence staining illustrates the dose-dependent reduction of lamellipodia and filopodia formation post-MAT treatment. Data represent mean ± SD of three independent experiments (n = 3), with *P ≤ .05, **P ≤ .01, and ***P ≤ .001, indicating statistical significance.
MAT reduced the actin-based lamellipodia and filopodia formation in PC3 prostate cancer cells
Actin-rich structures, namely, filopodia and lamellipodia, located on cell surfaces, play a pivotal role in metastatic progression by facilitating ECM remodeling. The dynamics of the actin cytoskeleton orchestrate cell motility and invasion, contributing substantially to the metastatic cascade in cancer. Utilizing fluorescence staining with phalloidin dye, our study demonstrated a pronounced reduction in both filopodia and lamellipodia formation in a dose-dependent manner following 24-h MAT treatment (Fig. 14B).
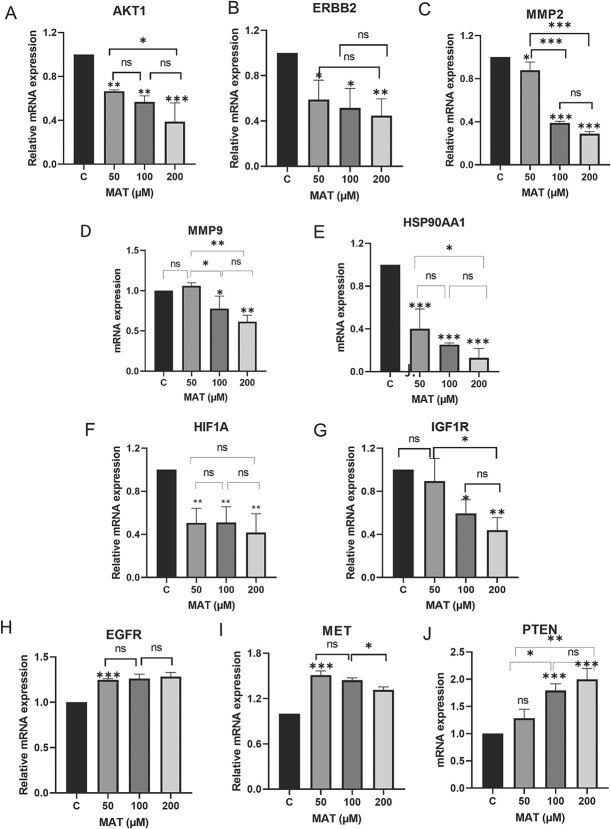
MAT downregulated the hub targets at the mRNA level analyzed by qPCR
MAT treatment significantly altered mRNA expression levels of key genes in PC3 cells (Fig. 15). Notably, MAT reduced mRNA levels of AKT1, ERBB2, MMP2, MMP9, HSP90AA1, HIF1A, and IGF1R, while increasing PTEN expression, a regulator of PI3K/AKT signaling. EGFR and MET mRNA levels remained largely unaffected. These findings support MAT’s targeting of tyrosine kinase receptors, particularly ERBB2 and IGF1R, and its modulation of the PI3K/AKT pathway, indicating its potential therapeutic efficacy against MMP-associated metastatic prostate cancer.
Figure 15.
Effects of MAT on mRNA expression levels of hub targets identified as potential therapeutic targets of MAT against mPC. Data represent mean ± SD of three independent experiments (n = 3) conducted in triplicates, with *P ≤ .05, **P ≤ .01, and ***P ≤ .001, indicating statistical significance.
Discussion and conclusion
In the landscape of mCRPC treatment, conventional options like enzalutamide or docetaxel-based chemotherapy offer initial benefits but often falter in improving patient survival rates over time [52]. Consequently, exploring complementary therapies is crucial. Dietary supplements, in combination with chemotherapy, have shown promise in enhancing therapeutic effects while minimizing toxicity [16]. MAT, a dietary lignan, holds potential due to its known anticancer properties. However, understanding its mechanisms in mPC is essential for clinical use. To address this research gap, we employed a multidisciplinary approach, integrating NP, bioinformatics, GMFA network analysis, and experimental validation. NP, GMFA, and bioinformatics analyses unveiled potential therapeutic targets and pathways associated with MAT’s antimetastatic effects. Experimental validation strengthened these findings, enhancing the reliability of our results.
In NP studies, the PPI network analysis revealed EGFR, AKT1, ERBB2, MET, IGF1, CASP3, HSP90AA1, HIF1A, MMP2, HGF, and MMP9 as top 11 hub targets of MAT against mPC. EGFR and ERBB2 are crucial in castration-independent PC progression, with aberrant activity promoting mCRPC growth and CTC survival in the bone microenvironment [53, 54]. Active mutations in AKT1 and PIK3CA genes elevate PI3K-Akt signaling in mCRPC [55]. Growth factors like HGF, IGF1, and VEGF-A in the tumor microenvironment drive bone metastasis in advanced PC, with the HGF/MET axis activating metastasis-promoting downstream signalings such as Ras/Raf/MAPK, PI3K/Akt, and Wnt/β-catenin [56, 57]. MMP-2 and MMP-9 serve as novel markers in metastatic PC, strongly associated with tumor invasiveness [58, 59]. CASP3 loss is linked to advanced PC, making it a prognostic marker [60]. Elevated HSP90AA1 levels correlate with PC metastatic potential, with Hsp90-rich extracellular vesicles (EVs) promoting epithelial-to-mesenchymal transition (EMT) [61]. Increased HIF1A induces castration resistance and ADT unresponsiveness in PC, making HIF1A inhibition a crucial therapeutic strategy [62]. Our analysis identifies these molecular targets of mPC as therapeutic targets of MAT, elucidating their roles in MAT’s antimetastatic potential in mPC.
On the other hand, the KEGG enrichment analysis revealed critical pathways associated with mPC, including prostate cancer, PI3K-Akt signaling pathway, EGFR tyrosine kinase inhibitor resistance, MAPK signaling pathway, IL-17 signaling pathway, and TNF signaling pathway. Given the hyperactivation and dysregulation of the PI3K/AKT pathway during the transition to hormone-sensitive mCRPC, components of the PI3K-Akt signaling cascade emerge as key therapeutic targets [51]. Notably, PTEN, which competes with PI3K, plays a central role in regulating AKT [63]. The Ras-MAPK signaling pathway exhibits promising therapeutic potential by influencing cell invasion and migration processes in mCRPC [16]. Resistance to EGFR tyrosine kinase inhibitors such as erlotinib presents a common challenge in CRPC treatment, often triggered by overexpression of hepatocyte growth factor (HGF) and activation of the HGF-induced MET/PI3K/AKT pathway [64]. GO enrichment analysis unveiled metastasis-specific biological processes, including EMT and positive regulation of cell migration. EMT, a pivotal step in metastatic progression, is closely linked to resistance to ADT and docetaxel treatment [65]. The cell surface is the major cellular component found in CC enrichment, which is the prime site for metastasis-linked events such as EMT transition and cytoskeleton changes. Cell surface receptors interact with the extracellular matrix (ECM) components and ECM-bound factors to mediate cell adhesion and cell signaling and thus efficiently activate EMT-associated metastasis [66]. Molecular functions primarily associated with these processes include transmembrane receptor protein tyrosine kinase activity, protein phosphatase activity, and peptidase activity, indicating MAT’s interaction with membrane receptors and downstream signaling pathways of tyrosine kinase receptors.
Our approach enriched NP findings through GMFA network analysis of the top 11 hub targets, incorporating co-expression, physical interaction, and genetic interaction. Co-expression analysis unveiled correlated expression patterns, indicating functional associations. Physical interactions suggested involvement in shared protein complexes, aiding target recognition. Genetic interactions provided insights into genes linked to similar pathways, offering therapeutic opportunities. This integrated approach comprehensively explored MAT’s therapeutic targets in cancer. GMFA-ED exhibited enriched metastatic cancer-related BP and MF, as well as CC. Enriched KEGG pathways like PI3K/AKT signaling, HIF1A signaling, ErbB signaling, and microRNA in cancer highlight their roles in mPC progression [67]. Various microRNAs, acting as metastasis biomarkers, either promote or inhibit PC metastasis [68]. Targeting ErbB receptors and the PI3K/AKT axis in androgen-independent prostate cancer cells shows promise as a therapy for drug-resistant CRPC [69]. Our molecular docking and simulation studies confirmed MAT’s potential to target PI3K/AKT signaling in mPC, suggesting its efficacy in combating mPC progression.
To comprehensively validate our findings regarding the molecular targets and mechanisms of MAT against mPC, we conducted a series of in vitro experiments to explore the antimetastatic potential of MAT against PC3 cells. Initially, we performed the MTT assay to assess the anticancer activity of MAT and determine its IC50 concentration in PC3 cells. Subsequently, we investigated the anti-migratory and anti-clonogenic effects of MAT using migration and clonogenic assays, respectively. Additionally, we examined the inhibitory impact of MAT on filopodia and lamellipodia formation in the cytoskeleton through fluorescence microscopy. Furthermore, qPCR analysis was employed to evaluate MAT’s ability to regulate mRNA levels of the top target genes identified during network pharmacology analysis. Our in vitro experimental validation corroborated the anticancer activity and antimetastatic potential of MAT by suppressing clonogenicity, migratory abilities, and cytoskeletal modifications implicated in metastatic progression. Thus, our study conclusively demonstrates the antimetastatic efficacy of MAT against mPC, not only by predicting key therapeutic molecular targets and mechanisms through network pharmacology analysis but also by confirming these targets through bioinformatics molecular docking and MD simulation studies. While our in vitro validation highlights MAT’s antimetastatic potential in PC3 cells, further investigations at the molecular and cellular levels are essential to gain deeper insights into the underlying mechanisms of MAT in mPC. This holistic approach aims to establish MAT as a promising antimetastatic therapeutic candidate for mPC.
Key Points
MAT appears to influence key hub genes (AKT1, MMP2, MMP9, HIF1A, MET) in mPC, suggesting its potential multimodal approach against metastasis.
KEGG analysis highlights PI3K-Akt and MAPK pathways as potential therapeutic targets for MAT in mPC treatment.
GO enrichment identifies MAT’s role in inhibiting metastasis-related processes like epithelial-to-mesenchymal transition and cell migration regulation.
GMFA-based further enrichment of network pharmacology findings reveals specific and extended therapeutic targets of MAT against mPC, enhancing its efficacy.
In vitro experimental validation confirms MAT’s anticancer and antimetastatic potential by suppressing clonogenicity, migratory abilities, and cytoskeletal modifications crucial in mPC progression.
Supplementary Material
Acknowledgements
This work was supported by the Department of Science and Technology (DST), Government of India, in the form of a DST-INSPIRE fellowship to R.R. The infrastructural support was provided by the Interactive Research School for Health Affairs (IRSHA), BVDU, Pune.
Rama Rajadnya, first author and Ph.D. student, Biotechnology, IRSHA, BVDU, Pune. Aniket Mali, corresponding author, Assoc. Prof and Head of the Department. Akanksha Mahajan, Amrita Ulhe, and Nidhi Sharma, Ph.D. students, IRSHA, BVDU, Pune. Mahabaleshwar Hegde, Prof. Innovative Nutrition-IRSHA. Rajesh Patil, Principal and Prof. (Pharmaceutical Chemistry), Sinhgad College of Pharmacy, Pune.
Contributor Information
Rama Rajadnya, Cancer Biology, Interactive Research School for Health Affairs (IRSHA), Bharati Vidyapeeth (Deemed to be University), Pune-Satara Road, Dhankawadi, Pune, Maharashtra 411043, India.
Nidhi Sharma, Cancer Biology, Interactive Research School for Health Affairs (IRSHA), Bharati Vidyapeeth (Deemed to be University), Pune-Satara Road, Dhankawadi, Pune, Maharashtra 411043, India.
Akanksha Mahajan, Cancer Biology, Interactive Research School for Health Affairs (IRSHA), Bharati Vidyapeeth (Deemed to be University), Pune-Satara Road, Dhankawadi, Pune, Maharashtra 411043, India.
Amrita Ulhe, Cancer Biology, Interactive Research School for Health Affairs (IRSHA), Bharati Vidyapeeth (Deemed to be University), Pune-Satara Road, Dhankawadi, Pune, Maharashtra 411043, India.
Rajesh Patil, Department of Pharmaceutical Chemistry, Sinhgad Technical Education Society, Sinhgad College of Pharmacy, Vadgaon (BK), Off Sinhgad Road, Pune, Maharashtra 411041, India.
Mahabaleshwar Hegde, Innovative Nutrition, Interactive Research School for Health Affairs (IRSHA), Bharati Vidyapeeth (Deemed to be University), Pune-Satara Road, Dhankawadi, Pune, Maharashtra 411043, India.
Aniket Mali, Cancer Biology, Interactive Research School for Health Affairs (IRSHA), Bharati Vidyapeeth (Deemed to be University), Pune-Satara Road, Dhankawadi, Pune, Maharashtra 411043, India.
Conflict of interest: None declared.
Funding
None declared.
Data availability
For access to any research-related data, kindly reach out to the corresponding author.
Author contributions
Rama Rajadnya (Methodology, Investigation, Formal analysis, Data curation, Visualization, Writing—original draft), Nidhi Sharma (Methodology, Investigation), Akanksha Mahajan (Methodology, Visualization), Rajesh Patil (Methodology, Investigation, Formal Analysis, Visualization, Validation, Writing—review of the final draft), Mahabaleshwar Hegde (Supervision, Reviewing of the draft), and Aniket Mali (Conceptualization, Supervision, Methodology, Investigation, Visualization, Formal Analysis, Data curation, Validation, Writing—review of the final draft).
References
- 1. Aliebrahimi S, Kouhsari M, Marotte D. et al. Irradiation of localized prostate cancer in the elderly: a systematic literature review. Clin Transl Radiat Oncol 2022;35:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sung H, Ferlay J, Siegel RL. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3. Wong SK, Mohamad NV, Giaze TR. et al. Prostate cancer and bone metastases: the underlying mechanisms. Int J Mol Sci 2019;20:2587. 10.3390/ijms20102587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. La Manna F, Karkampouna S, Zoni E. et al. Metastases in prostate cancer. Cold Spring Harb Perspect Med 2019;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller DR, Ingersoll MA, Teply BA. et al. Combination treatment options for castration-resistant prostate cancer. Prostate Cancer 2021;9:59–80. [PMC free article] [PubMed] [Google Scholar]
- 6. Dong L, Zieren RC, Xue W. et al. Metastatic prostate cancer remains incurable, why? Asian J Urol 2019;6:26–41. 10.1016/j.ajur.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sekhoacha M, Riet K, Motloung P. et al. Prostate cancer review: genetics, diagnosis, treatment options, and alternative approaches. Molecules 2022;27:5730. 10.3390/molecules27175730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hao Q, Wu Y, Vadgama JV. et al. Phytochemicals in inhibition of prostate cancer: evidence from molecular mechanisms studies. Biomolecules 2022;12:1306. 10.3390/biom12091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bai B, Chen Q, Jing R. et al. Molecular basis of prostate cancer and natural products as potential chemotherapeutic and chemopreventive agents. Front Pharmacol 2021;12:1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dzobo K, Centre I, Icgeb B. et al. The role of natural products as sources of therapeutic agents for innovative drug discovery. Compr Pharmacol 2022;408–22. 10.1016/B978-0-12-820472-6.00041-4. [DOI] [Google Scholar]
- 11. De Silva SF, Alcorn J. Flaxseed lignans as important dietary polyphenols for cancer prevention and treatment: chemistry, pharmacokinetics, and molecular targets. Pharmaceuticals 2019;12:21–38. 10.3390/ph12020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ezzat SM, Shouman SA, Elkhoely A. et al. Anticancer potentiality of lignan rich fraction of six flaxseed cultivars. Sci Rep 2018;8:544. 10.1038/s41598-017-18944-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jang WY, Kim MY, Cho JY. Antioxidant, anti-inflammatory, anti-menopausal, and anti-cancer effects of lignans and their metabolites. Int J Mol Sci 2022;23:15482. 10.3390/ijms232415482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee B, Kim KH, Jung HJ. et al. Matairesinol inhibits angiogenesis via suppression of mitochondrial reactive oxygen species. Biochem Biophys Res Commun 2012;421:76–80. 10.1016/j.bbrc.2012.03.114. [DOI] [PubMed] [Google Scholar]
- 15. Rodríguez-García C, Sánchez-Quesada C, Toledo E. et al. Naturally lignan-rich foods: a dietary tool for health promotion? Molecules 2019;24:917. 10.3390/molecules24050917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Z, Zhang Z, Ye X. et al. Based on network pharmacology and molecular docking to predict the mechanism of Huangqi in the treatment of castration-resistant prostate cancer. PloS One 2022;17:e0263291. 10.1371/journal.pone.0263291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shah A, Patel V, Jain M. et al. Network pharmacology and systems biology in drug discovery. CADD Informatics Drug Discov 2023;231–52. 10.1007/978-981-99-1316-9_10. [DOI] [Google Scholar]
- 18. Sakle NS, More SA, Mokale SN. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: an updated prototype in drug discovery. Sci Rep 2020;10:17217. 10.1038/s41598-020-74251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Basu A, Sarkar A, Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci Rep 2020;10:17699. 10.1038/s41598-020-74715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aliebrahimi S, Montasser Kouhsari S, Ostad SN. et al. Identification of phytochemicals targeting c-met kinase domain using consensus docking and molecular dynamics simulation studies. Cell Biochem Biophys 2018;76:135–45. 10.1007/s12013-017-0821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stelzer G, Rosen N, Plaschkes I. et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 2016;54:1–30. [DOI] [PubMed] [Google Scholar]
- 22. Piñero J, Bravo À, Queralt-Rosinach N. et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017;45:D833–9. 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X, Shen Y, Wang S. et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res 2017;45:W356–60. 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47:W357–64. 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gallo K, Goede A, Preissner R. et al. SuperPred 3.0: drug classification and target prediction - a machine learning approach. Nucleic Acids Res 2022;50:W726–31. 10.1093/nar/gkac297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szklarczyk D, Gable AL, Nastou KC. et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021;49:D605–12. 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chin C, Chen S, Wu H. et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sherman BT, Hao M, Qiu J. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022;50:W216–21. 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chandrashekar DS, Karthikeyan SK, Korla PK. et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia 2022;25:18–27. 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang Z, Kang B, Li C. et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556–60. 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trott O, Olson AJ. Software news and update AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455–61. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hanwell MD, Curtis DE, Lonie DC. et al. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Chem 2012;4:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berendsen HJC, Spoel D, Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 1995;91:43–56. 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 34. Abraham MJ, Murtola T, Schulz R. et al. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015;1-2:19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 35. Case DA, Aktulga HM, Belfon K. et al. AmberTools. J Chem Inf Model 2023;63:6183–91. 10.1021/acs.jcim.3c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lindorff-Larsen K, Piana S, Palmo K. et al. Improved side-chain torsion potentials for the amber ff99SB protein force field. Proteins Struct Funct Bioinform 2010;78:1950–8. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 1981;52:7182–90. 10.1063/1.328693. [DOI] [Google Scholar]
- 38. Mark P, Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem A 2001;105:9954–60. 10.1021/jp003020w. [DOI] [Google Scholar]
- 39. Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys 2007;126:014101. 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 40. Franz M, Rodriguez H, Lopes C. et al. GeneMANIA update 2018. Nucleic Acids Res 2018;46:W60–4. 10.1093/nar/gky311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mahajan A, Sharma N, Ulhe A. et al. From dietary lignans to cancer therapy: integrative systems analysis of enterolactone’s molecular targets and signaling pathways in combatting cancer stem cells in triple-negative breast cancer. Food Biosci 2024;58:103732. 10.1016/j.fbio.2024.103732. [DOI] [Google Scholar]
- 42. Ulhe A, Sharma N, Mahajan A. et al. Decoding the therapeutic landscape of alpha-linolenic acid: a network pharmacology and bioinformatics investigation against cancer-related epigenetic modifiers. J Biomol Struct Dyn 2023;1–26. 10.1080/07391102.2023.2293267. [DOI] [PubMed] [Google Scholar]
- 43. Ge SX, Jung D, Jung D. et al. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020;36:2628–9. 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mahajan M, Suryavanshi S, Bhowmick S. et al. Matairesinol, an active constituent of HC9 polyherbal formulation, exhibits HDAC8 inhibitory and anticancer activity. Biophys Chem 2021;273:106588. 10.1016/j.bpc.2021.106588. [DOI] [PubMed] [Google Scholar]
- 45. Mali AV, Joshi AA, Hegde MV. et al. Enterolactone suppresses proliferation, migration and metastasis of MDA-MB-231 breast cancer cells through inhibition of uPA induced plasmin activation and MMPs-mediated ECM remodeling. Asian Pacific J Cancer Prev 2017;18:905–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alves-Silva JM, Pedreiro S, Cavaleiro C. et al. Effect of Thymbra capitata (L.) Cav. on inflammation, senescence and cell migration. Nutrients 2023;15:1930. 10.3390/nu15081930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dubey S, Jaiswal B, Gupta A. TIP60 acts as a regulator of genes involved in filopodia formation and cell migration during wound healing. J Biol Chem 2022;298:102015. 10.1016/j.jbc.2022.102015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xian ZY, Liu JM, Chen QK. et al. Inhibition of LDHA suppresses tumor progression in prostate cancer. Tumor Biol 2015;36:8093–100. 10.1007/s13277-015-3540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen C, Wu J, Hua Q. et al. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 2019;15:1–12. 10.1186/s13007-019-0455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mavra A, Petrou CC, Vlasiou MC. Ligand and structure-based virtual screening in combination, to evaluate small organic molecules as inhibitors for the XIAP anti-apoptotic protein: the Xanthohumol hypothesis. Molecules 2022;27:4825. 10.3390/molecules27154825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shorning BY, Dass MS, Smalley MJ. et al. The PI3K-AKT-mTOR pathway and prostate cancer: at the crossroads of AR, MAPK, and WNT signaling. Int J Mol Sci 2020;21:4507. 10.3390/ijms21124507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He L, Fang H, Chen C. et al. Metastatic castration-resistant prostate cancer: academic insights and perspectives through bibliometric analysis. Medicine (Baltimore) 2020;99:e19760. 10.1097/MD.0000000000019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Day KC, Hiles GL, Kozminsky M. et al. HER2 and EGFR overexpression support metastatic progression of prostate cancer to bone. Cancer Res 2017;77:74–85. 10.1158/0008-5472.CAN-16-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou T, Chen T, Lai B. et al. FBXW2 inhibits prostate cancer proliferation and metastasis via promoting EGFR ubiquitylation and degradation. Cell Mol Life Sci 2022;79:268. 10.1007/s00018-022-04320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Herberts C, Murtha AJ, Fu S. et al. Activating AKT1 and PIK3CA mutations in metastatic castration-resistant prostate cancer. Eur Urol 2020;78:834–44. 10.1016/j.eururo.2020.04.058. [DOI] [PubMed] [Google Scholar]
- 56. Lee C, Whang YM, Campbell P. et al. Dual targeting c-met and VEGFR2 in osteoblasts suppresses growth and osteolysis of prostate cancer bone metastasis. Cancer Lett 2018;414:205–13. 10.1016/j.canlet.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 57. Whang YM, Jung SP, Kim MK. et al. Targeting the hepatocyte growth factor and c-met signaling axis in bone metastases. Int J Mol Sci 2019;20:384. 10.3390/ijms20020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oguić R, Mozetič V, Cini Tešar E. et al. Matrix metalloproteinases 2 and 9 immunoexpression in prostate carcinoma at the positive margin of radical prostatectomy specimens. Patholog Res Int 2014;2014:1–8. 10.1155/2014/262195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fan X, Zhou J, Bi X. et al. L-theanine suppresses the metastasis of prostate cancer by downregulating MMP9 and snail. J Nutr Biochem 2021;89:108556. 10.1016/j.jnutbio.2020.108556. [DOI] [PubMed] [Google Scholar]
- 60. Acar V, Couto Fernandez FL, Buscariolo FF. et al. Immunohistochemical evaluation of PARP and Caspase-3 as prognostic markers in prostate carcinomas. Clin Med Res 2021;19:183–91. 10.3121/cmr.2021.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sager RA, Khan F, Toneatto L. et al. Targeting extracellular Hsp90: a unique frontier against cancer. Front Mol Biosci 2022;9:982593. 10.3389/fmolb.2022.982593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Terzic J, Abu el Maaty MA, Lutzing R. et al. Hypoxia-inducible factor 1A inhibition overcomes castration resistance of prostate tumors. EMBO Mol Med 2023;15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Torrealba N, Vera R, Fraile B. et al. TGF-β/PI3K/AKT/mTOR/NF-kB pathway. Clinicopathological features in prostate cancer. Aging Male 2021;23:801–11. 10.1080/13685538.2019.1597840. [DOI] [PubMed] [Google Scholar]
- 64. Nagano T, Tachihara M, Nishimura Y. Mechanism of resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and a potential treatment strategy. Cells 2018;7:212. 10.3390/cells7110212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chaves LP, Melo CM, Saggioro FP. et al. Epithelial–mesenchymal transition signaling and prostate cancer stem cells: emerging biomarkers and opportunities for precision therapeutics. Genes (Basel) 2021;12:1900. 10.3390/genes12121900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Winkler J, Werb Z, Metcalf KJ. Tumour progression and metastasis. Nat Commun 2020;11:5120. 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Abu el Maaty MA, Terzic J, Keime C. et al. Hypoxia-mediated stabilization of HIF1A in prostatic intraepithelial neoplasia promotes cell plasticity and malignant progression. Sci Adv 2022;8:eabo2295. 10.1126/sciadv.abo2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Oh-hohenhorst SJ, Lange T. Role of metastasis-related microRNAs in prostate cancer. Cancers (Basel) 2021;13:4492. 10.3390/cancers13174492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Adediran S, Wang L, Khan MA. et al. Co-targeting ErbB receptors and the PI3K/AKT axis in androgen-independent taxane-sensitive and taxane-resistant human prostate cancer cells. Cancers (Basel) 2022;14:4626. 10.3390/cancers14194626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For access to any research-related data, kindly reach out to the corresponding author.