Abstract
Deep learning has proven highly effective in various medical imaging scenarios, yet the lack of an efficient distribution platform hinders developers from sharing models with end-users. Here, we describe brainchop, a fully functional web application that allows users to apply deep learning models developed with Python to local neuroimaging data from within their browser. While training artificial intelligence models is computationally expensive, applying existing models to neuroimaging data can be very fast; brainchop harnesses the end user’s graphics card such that brain extraction, tissue segmentation, and regional parcellation require only seconds and avoids privacy issues that impact cloud-based solutions. The integrated visualization allows users to validate the inferences, and includes tools to annotate and edit the resulting segmentations. Our pure JavaScript implementation includes optimized helper functions for conforming volumes and filtering connected components with minimal dependencies. Brainchop provides a simple mechanism for distributing models for additional image processing tasks, including registration and identification of abnormal tissue, including tumors, lesions and hyperintensities. We discuss considerations for other AI model developers to leverage this open-source resource.
Introduction
Neuroimaging has emerged as a powerful tool for studying brain structure, function, and connectivity, offering insights into the underlying neural mechanisms of various cognitive processes and disorders. As we describe next, recent AI models have proved to be more accurate, robust, and rapid than traditional image preprocessing stages, including brain extraction, tissue segmentation, regional parcellation, and anomaly detection. The rate-limiting factor in bringing these AI models to academic and clinical settings is that they are difficult to deploy to end users, often requiring creation of project specific environments by experts in the field. These complexities, coupled with the lack of a platform for deploying AI solutions to common neuroimaging problems, severely limits the impact of these potentially revolutionary tools.
The promise of AI to revolutionize brain imaging
Machine learning models have already proved capable of robustly, rapidly, and objectively solving many labor intensive and error-prone neuroimaging tasks. These benefits are particularly attractive given the burgeoning availability of large open-access datasets that allow teams to aggregate images across diverse populations to make new discoveries. Here we summarize a few notable breakthroughs. Critically, for each advance, we also highlight the barriers that hinder end users from exploiting these tools. Our goal is not to provide an exhaustive list of AI-based medical imaging software, but rather to provide an overview of the breadth of existing, and potential applications, by highlighting a few seminal solutions. We further restrict our review to the applications of machine learning to replace traditional image processing stages including brain extraction, brain segmentation, regional parcellation, coregistration, normalization, and anomaly detection; the prognostic benefits for behavior and disease1–6 are beyond the scope of the present work.
Brain Extraction
Brain extraction, the segmentation of brain tissues from surrounding tissue, fat and bone, plays a vital role in brain imaging. The traditional FSL Brain Extraction Tool (BET) has been widely adopted as a first step towards restricting analyses to cortical regions and additionally aids algorithms responsible for registration of similar modalities (i.e., T1w structural scans) between individuals and co-registration across modalities within a given subject (i.e., T1w to T2-FLAIR)7. Furthermore, accurately extracted brains are better anonymized than medical images de-identified using traditional defacing methods. This is particularly the case in clinical situations where scalp features may be recognizable (dermoid cysts, craniotomy scars, ear shape)8. FSL’s BET has already been ported to web assembly, providing a zero-footprint solution9. Recently, several machine learning-based brain extraction models have been shown to outperform BET. Two noteworthy examples are HD-BET10 and SynthStrip11, the latter of which has proven exemplary when working with disparate image modalities and images of inconsistent quality. However, the deployment of these machine learning tools is hampered by the fact that they require local software installation and perform slowly, or not at all, without access to a local Nvidia graphics card.
Brain Segmentation
FreeSurfer12, FSL FAST13, and SPM14 all provide elegant solutions to the problem of brain segmentation, and each accurately classifies voxels as white matter, gray matter or cerebral spinal fluid. The thousands of citations attributed to each of these programs demonstrates that these measures provide users with access to a powerful biomarker for brain function and disease. Recently, several teams have introduced competitive machine-learning based models, including SynthSeg15. SynthSeg is now included with recent FreeSurfer versions, making model distribution straightforward. However, to gain the full speed benefits, users still need an Nvidia graphics card accompanied by properly installed CUDA drivers.
Brain Morphometry (Parcellation/Volume/Thickness)
Estimates of cortical thickness combined with parcellation into distinct regions, based on any variety of publicly available brain atlases, has proved a powerful method for neuroscientists. Indeed, the seminal FreeSurfer articles12,16–18 have each been cited thousands of times. Recently, a deep learning based implementation of FreeSurfer brain parcellation, FastSurfer19, was developed. This new version produces volume segmentation results that are similar to those produced by FreeSurfer, but in a fraction of the time. Additionally, FastSurfer demonstrated better test-retest reliability than FreeSurfer in estimating cortical thickness in longitudinal studies. However, once again, users wishing to take advantage of this incredible speed-up must have access to a high-performance Nvidia GPU with appropriate CUDA drivers.
Spatial Coregistration and Normalization
Spatial coregistration methods play a crucial role in many neuroimaging pipelines. For example, multi-volume time series such as functional MRI (fMRI) and diffusion-weighted imaging benefit from motion correction, a process in which all volumes are aligned to either the first volume or the mean volume of a series of images. It is also often necessary to coregister images of different modalities, or acquired at different timepoints, from the same individual. For example, aligning a low-resolution fMRI scan to a high-resolution anatomical image is fundamental. Finally, it is common to spatially normalize images, warping each individual’s brain to match the shape and alignment of a common anatomical template, a process that allows for comparisons between individuals as well as application of group-level statistics. EasyReg20 has provided a convolutional neural network that is fast, works across modalities, and does not require pre-processing (such as brain extraction or bias field correction) to operate. An exception to the general rule, this FreeSurfer tool provides reasonable inference speed without requiring a graphics card or other specialized hardware.
Anomaly Detection
Mapping the location and extent of brain injury can aid the diagnosis, prognosis and treatment of the brain2 with seminal work describing the methods for objective analyses highly cited21,22. Within the field of medical imaging, substantial effort has been put into the development of automated algorithms for detecting various anomalies including but not limited to, white matter hyperintensities23, microbleeds24, tumors25, as well as both acute26 and chronic stroke27 lesions. These methods are important, especially within the field of stroke. For example, lesion studies can provide a stronger inference than activation measures by revealing brain regions that are necessary28. However, manually drawing a lesion on a high-resolution scan can often take an hour, with demarcation along the edges being highly subjective29. Seminal work used traditional spatial normalization with outlier detection and clustering to automatically identify chronic30 and acute31 injuries. Both of these methods do depend on the robustness of the spatial normalization, which can be disrupted by the presence of brain injury32, albeit the acute method cleverly uses both the diffusion-unweighted and weighted imaging pair and leverages the fact that acute injury only appears in the latter (so spatial transforms are estimated for an image where the injury is invisible, while the lesion is identified from the image where it is visible). Another limitation of both approaches is that they require Matlab software, which hinders web deployment. More recently, LINDA introduced a random forest machine learning algorithm to identify chronic lesions33 and ADS (Acute-stroke Detection Segmentation) employed a deep learning model to identify acute injury26. However, both of these tools rely on accurate initial normalization using traditional methods34 which do influence the robustness, deployability and performance of these algorithms. The emergence of clinical datasets with gold-standard human drawn lesions27 can allow objective competitions to identify accurate automated lesion identification.
In conclusion, AI-augmented image processing has the potential to provide fast, robust and objective solutions for many common and fundamental neuroimaging tasks. Ensuring easy and fast deployment of these approaches is critical for researchers and clinicians wishing to aggregate vast databases of medical images across multiple sites and/or studies.
Mechanisms for Sharing Neuroimaging AI Models
Despite the proven utility of existing AI models, they remain difficult to distribute to other scientists and clinicians. Many models demand specific software and hardware configurations. The level of technical expertise required to install and maintain these configurations is beyond the ability of many researchers and clinicians. We briefly describe four mutually inclusive solutions to distribute machine learning models that fill different niches: native installation, containers, cloud implementations, and edge-based web technologies.
Solution #1: Native Installation
Perhaps the most common method for distributing AI models in the field of neuroimaging is through bare-metal installations. This traditional approach requires users to install the necessary drivers and software environments directly onto their local systems. This typically includes setting up a Python environment to manage and isolate dependencies effectively. Users must then clone the model’s repository from a version control system like GitHub, install required dependencies using package managers like pip, and subsequently download the specific AI models needed for the task (which may be hosted on separate sites due to file size limitations imposed by most online code repositories). This time-consuming process assumes the availability of specific types of hardware (i.e., a x86 architecture CPU and an Nvidia GPU equipped with the appropriate CUDA drivers). Addressing these hardware and emergent software discrepancies places a considerable burden on both the model developers and the end users, with developers being forced to ensure their models are compatible with a range of operating systems/hardware configurations, and end users being forced to buy and maintain specific hardware components. Indeed, maintenance of software ecosystems essential to properly train and use AI models often requires driver/software updates that can break the system, demanding painful and time-consuming complete reinstallations.
Solution #2: Containers
Containers like Docker and Singularity/Apptainer offer valuable solutions for managing and distributing complex software more efficiently. These tools act as encapsulated environments, allowing developers to package their applications along with all of the necessary dependencies and configurations required to use them successfully. By doing so, they mitigate compatibility concerns and streamline the deployment process across different computing environments. Moreover, Docker and Singularity/Apptainer enable precise version control, ensuring that software behaves consistently regardless of the underlying system thereby simplifying software distribution while, at the same time, enhancing reproducibility and reliability of computational tasks. Field-general solutions like Docker and Singularity/Apptainer have thus far dominated the field of neuroimaging-relevant tools. However, infrastructure specific to the deployment of brain-data based inference generation models has recently emerged. Nobrainer (https://github.com/neuronets/nobrainer) pioneered an infrastructure for sharing algorithms from different organizations including SynthMorph35, SynthSeg15 SynthSR36, SynthStrip11, kwyk37, and DeepCSR38. Nobrainer provides a unified container for running all of these models, simplifying the distribution and usage of AI inference. However, as of this writing this container only supports the x86 architecture, is only tuned for Linux, and inference is only accelerated with NVidia graphics cards1. Another more generalized example of this approach is neurodesk39 which allows loading of specific software versions as self-contained packages. For example, one could load either current or previous releases of FreeSurfer to replicate studies that used previously described models from that team (e.g. easyReg, SynthSeg, SynthStrip, SynthSR).
Solution #3: Cloud Computing
For many use cases, cloud computing can provide an elegant method for deploying machine learning inferences. In this case, a centralized computer houses the specific software, versioning and hardware required to compute inferences. The user simply needs to select the images to process, and upload their data. A clear example of this approach is brainlife40 (brainlife.io) which allows users to apply machine learning inferences like SynthSR and SynthStrip to either a user’s personal data or from linked open-access repositories such as OpenNeuro41. Another example is neurodesk which can be deployed as cloud instances39. An important benefit of cloud resources is their ability to scale on demand, whereas other methods are constrained by local hardware. This advantage makes them particularly apt for processing large datasets that would be difficult to analyze in a timely manner using local compute resources. Therefore, cloud computing enhances efficiency by allowing multiple users to concurrently share a pool of resources, eliminating the need for each user to maintain sufficient resources to handle their peak demands independently. However, cloud resources are not ideal for every application. One significant limitation is that images must be shared with another organization. This is not a viable option in many situations for ethical, regulatory, or legal reasons (particularly prior to anonymization measures such as removing the face or scalp from images). Further, cloud computing can incur a penalty for transferring large data between a user’s computer and the remote service. Another serious limitation with current cloud implementations is that the results are not interactive; a strong assumption is that all input data are similar and that the output from the automated models is without error. In practice, both of these assumptions are rarely correct. Interactive visualization and editing tools are required to help confirm and ensure that the inference model processed the images successfully.
Solution #4: Edge-Based Deployment
The final method of distributing neuroimaging machine learning models leverages edge-based web technologies. This method harnesses the power of end users’ local compute resources to process images and generate and save the derivatives. This approach leverages two properties of most inference models: estimating the inferences of existing models is much less computationally taxing than training new models, and application of existing models to existing data is a task that is ideally suited for graphics cards’ advantage for massively parallel operations. Of particular interest, the Open Neural Network Exchange (ONNX) web runtime and TensorFlowJS JavaScript packages allow models to be run using the WebGL and WebGPU libraries which harness the local graphics card regardless of manufacturer, thanks to the browser acting as an operating system removing the need for the user to provide specific hardware and drivers (e.g., Nvidia graphics cards with CUDA libraries). Since the image data is not shared with the cloud, edge computing can address the privacy concerns associated with cloud computing. From the model developer’s perspective, a key advantage of edge computing is that it harnesses the end user’s hardware for computation, allowing it to scale effortlessly with the number of users. This is in contrast to cloud computing, where demands on centralized hardware increase as the number of active users grows, requiring continuous resource allocation and management.
Edge-based deployment is simple for the user, with no software to install. To start using pre-trained deep learning models a user just needs to open a URL in their browser, which in our case takes around 200 ms to load six segmentation models and provide 15 ways to run them. These packages run the models locally within the sandbox of the user’s browser without the need for data exchange with a remote server, as there is no back-end. Since image data remains local, this approach bypasses privacy concerns of cloud solutions and is therefore ideally suited for clinical applications (such as lesion detection) and image de-facing or brain extraction which remove recognizable features to allow subsequent sharing of anonymized images. The brainchop42,43 project from the Nobrainer team showcases this approach, providing models for brain extraction, tissue segmentation and parcelation as shown in Figure 1.
Figure 1:
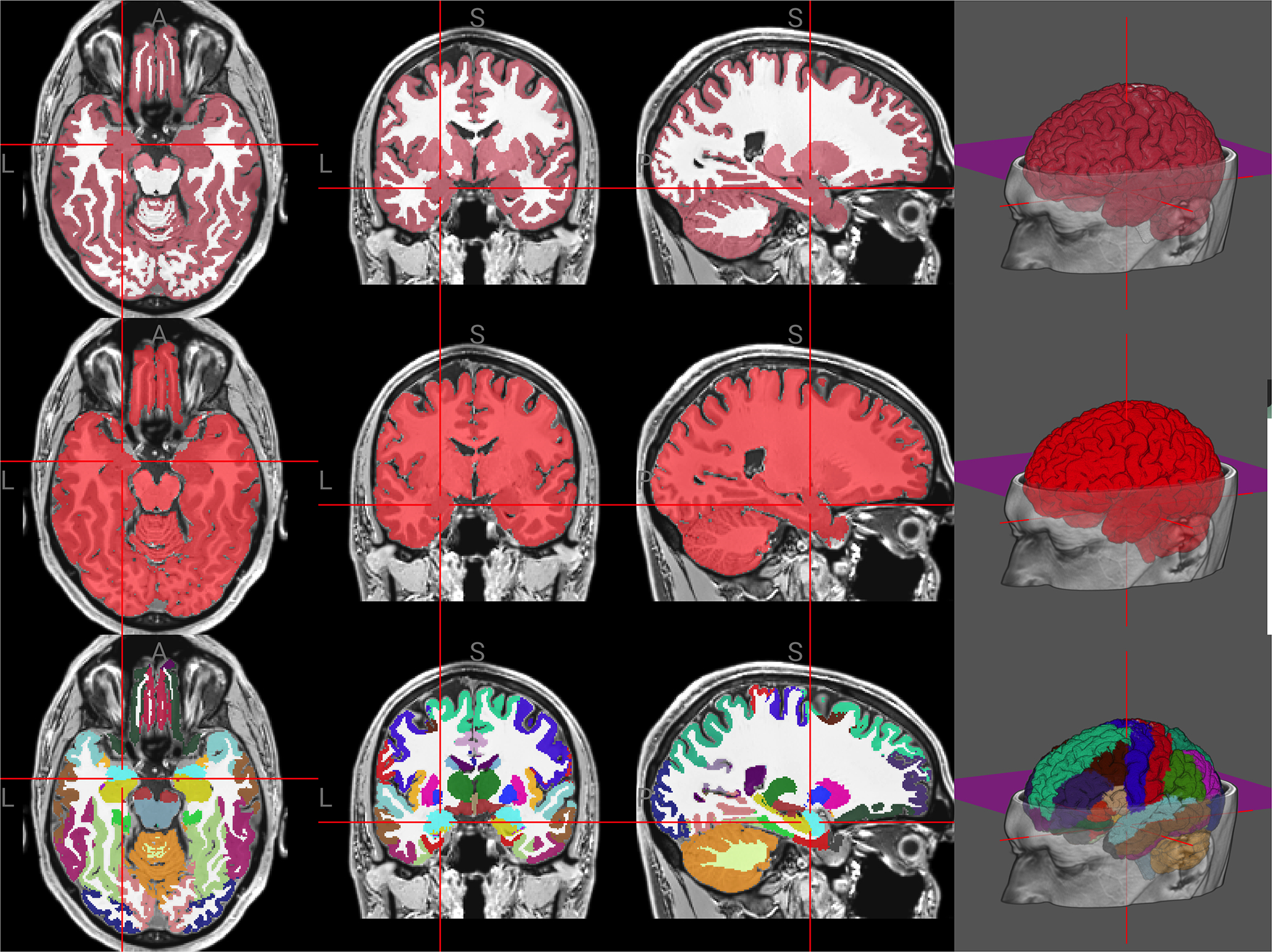
Examples for each of the three model families currently supported by brainchop. The segmentation (top) identifies gray (red) and white (white) matter. The brain extraction (middle) creates a mask of the brain voxels (red). The parcellation classifies 104 cortical regions. The colors and regions come from the FreeSurfer Color Look Up Table.
Rather than assume that all inferences are successful, brainchop allows users to interactively view the input and segmented images, ensuring model accuracy. In this way, brainchop is an ideal framework for customization and user-driven model tuning that could eventually allow end-users to further refine/tailor AI models to their specific needs or the needs of the research community. One way this could be accomplished is by including the ability to adjust model parameters, like threshold levels, segmentation boundaries, and coregistration points directly within the visualization/feedback module. For instance, in cases where automated image segmentation fails due to atypical anatomy or poor image quality, users could manually refine the segmentation boundaries. This interactive approach could be used to iteratively provide AI model developers with error feedback, that could then be used to justify change in the AI model, perhaps resulting in an improved ability to process both normal data and handle occasional edge-cases. We envision that this on-the-fly feedback could be part of a virtuous cycle that would allow AI systems to continuously ‘learn’ and improve their performance over time. It is worth noting that visualization is useful for machine learning regardless of the form of distribution (e.g., native, container, cloud or edge), and a web-based visualization module could be shared across all these methods. Indeed, this could allow crowdsourced training for huge datasets, leveraging the diversity of aggregated datasets and editors.
Mission Statement
Our primary objective is to provide a method to deploy neuroimaging AI models that is simple, efficient, ensures data safety, and provides users with sufficient feedback to catch and fix errors. This method should allow developers to quickly and easily deploy their AI models and allow users to confidently apply these solutions to their images. We believe the best way to achieve this objective is through edge-based computing which protects privacy by using local hardware, has zero footprint (no specialized software to install) and is universal (works on any operating system and graphics card). To achieve this aim, we comprehensively refined and optimized brainchop, bringing it from a prototype to a robust tool that can be directly used by end users with existing models and harnessed by other teams with their own models. These refinements include removing dependencies, improving performance, and adding features such as the ability to manually edit AI-generated native space segmentations. The Methods section describes our implementation and the Results section describes the performance improvements relative to the initial prototype.
Methods
Design Considerations
Here we describe refinements since brainchop version 2, which we have previously described42,43. The Results section provides objective measures for the cumulative benefit of these optimizations. The major performance difference between version 2 and 3 is implementing 16-bit instead of 32-bit textures for the models which significantly improved the performance by reducing memory usage and data transfer times. In contrast, the version 4 optimizations focused on new features, reducing idle time, reducing time spent on concurrent tasks and the image processing stages that occur before and after the model inferences. Therefore, it is worth noting that the changes from version 2 to 3 impact the actual inference time of the model, while the changes from version 3 to 4 largely focus on processing operations that occur before and after the inference. Therefore, the version 4 optimizations will have smaller proportional benefits as the inference model complexity increases (though it is worth noting that the introduction of web workers can reduce time spent idle or waiting for concurrent tasks).
The new user interface is shown in Figure 2. The user can drag and drop a voxel-based medical image of the head, in any of the supported formats (with NiiVue already providing support for the NIfTI, NRRD, MRtrix MIF, AFNI HEAD/BRIK, MGH/MGZ, ITK MHD, ECAT7, and DICOM formats) and select any of the pre-specified image processing models from the drop down menu. The results are shown as an interactive overlay on top of the source image, with sliders allowing the user to independently adjust the visibility of the source and classified image. A button allows the user to save the model as a NIfTI-format image. Alternatively, a button allows the user to save the entire scene (background image, overlay, drawings, contrast settings, crosshair location and annotations) as a single file (using NiiVue’s document format with the `.nvd` extension) that can be viewed with any NiiVue instance. As we note, this provides a virtuous cycle between model developers and users, providing a mechanism for sharing edge cases where models do not act as expected. A checkbox allows users to select whether models are run on an independent thread (web worker) or on the main thread. Simple drawing tools allow the user to modify segmentation models, with the ability to edit model results. To provide a minimal user interface, these tools currently only provide binary operations, but the underlying NiiVue visualization system can support more complicated drawings (e.g., using different pen colors to edit specific classes for the segmentation and parcelation models). We also provide a diagnostics model, which provides text-based details on the user’s system and the execution of the most recent model. These details can help troubleshoot unexpected behavior. The user interface is built using pure HTML, rather than using a widget framework (e.g., Angular, React, or Vue). This minimal, framework agnostic approach aids users who wish to embed these models in their preferred framework, as the web page directly interacts with the modular NiiVue and brainchop functions.
Figure 2:
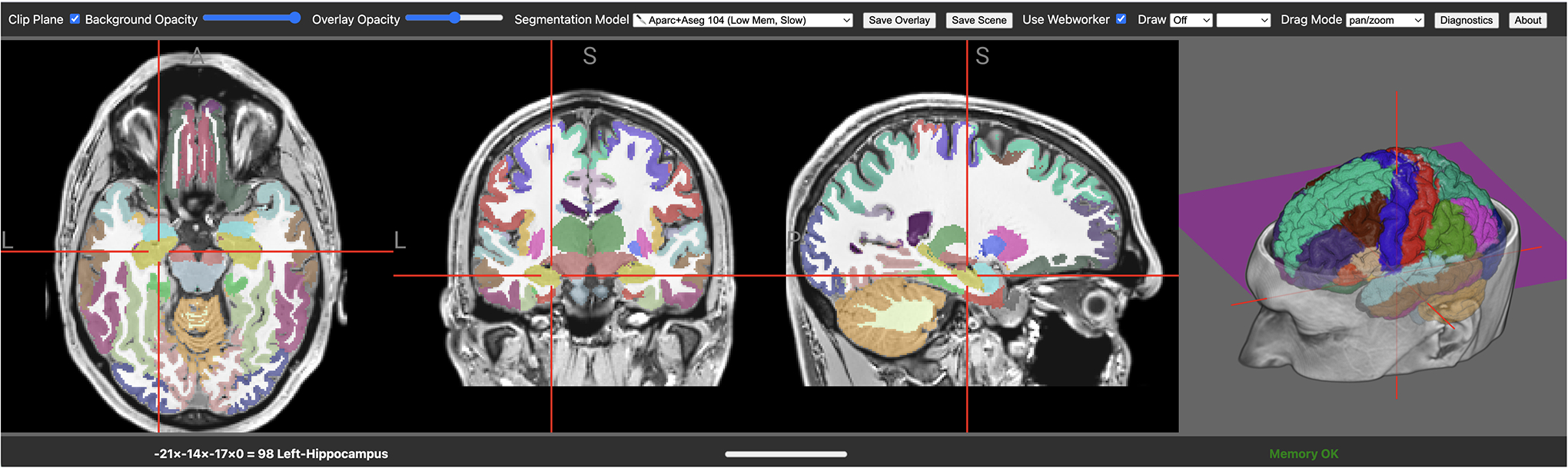
The brainchop web page (https://neuroneural.net/brainchop/) allows users to select between tissue segmentation, brain extraction and parcelation models from a drop down menu. Users can drag and drop their own images. The integrated visualization allows users to interact with the images. Note that the name of the region selected by the crosshairs is shown in the status bar in the bottom left. Users can save the resulting classification results as NIfTI images or edit them as required prior to saving them which allows for improved quality control.
Segmentation accuracy and out-of-sample robustness are the driving objective measures for selecting between competing neuroimaging machine learning models. However, considerations regarding the speed and resource demands of the inference models have clear implications. Users at well-resourced institutions working with huge datasets would clearly prefer fast but demanding solutions. On the other hand, catering to these users excludes many potential users. Brainchop supports both groups by having both fast but demanding, as well as slow but lean variations for some of the more complex models. The current brainchop models are all based on MeshNet44 models that are renowned for their modest computational requirements. These models were converted to TensorFlow.JS (TFJS)45. Beyond the basic model inference, we also provide TensorFlowJS filters for attenuating noisy voxels to improve segmentation accuracy. Both models and filters currently use the WebGL2 TFJS backend that leverages the graphics card of the user’s computer (and can be extended to the WebGPU backend if this matures to support 3D convolutions). Indeed, the recent releases of brainchop conduct more of the computations on the graphics card, improving the speed.
Input Image Harmonization
Raw neuroimaging data is often acquired with a range of resolutions and voxel sizes. However, machine learning models are typically trained on images of a specific resolution. Similar to many other neuroimaging AI tools, brainchop requires that the input images are 256×256×256 voxels with a 1mm isotropic resolution. The original brainchop used the Python code from FastSurfer19 to reslice input images of any resolution to these dimensions. However, this choice added a large number of dependencies including matplotlib, cycler, six, fonttools, kiwisolver, pillow, python-dateutil, pytz, scipy, nibabel, and numpy all of which needed to be emulated via pyodide. As we demonstrate in the Results section, downloading these packages to conform an initial image is slow (particularly penalizing users with limited internet bandwidth), retaining these packages for subsequent images holds on to local memory, and the emulation for reslicing is slow. To address this, we ported these routines to pure JavaScript.
Threads and Web Workers
The original brainchop ran all computations on the web page’s main thread, which impacted performance and interactivity. The current version of brainchop allows the user to specify whether the models run on the main thread (using timers and callback functions to return results) or independently on a web worker thread (using asynchronous calls when necessary and messaging to return results to the main thread). This feature required numerous changes as we found that contemporary web workers have constrained heap size relative to the main thread. To address this, we preallocated arrays of known sizes rather than dynamically growing arrays. Fortuitously, this led to speed benefits. We maintain code for both the main thread and web worker for two reasons. First, a web worker requires access to an OffScreen canvas that has only recently been introduced in the WebKit-based browsers such as Safari and is not yet supported by TensorFlowJS (so at the time of this writing our models must run on the main thread for these browsers). Second, there is no intuitive way to predict whether a given task will perform better on the main thread or using a web worker. Pragmatically testing the Chrome and Firefox browsers we have observed that some models are faster with web workers while others are faster on the main thread in a complex manner that interacts with the choice of browsers. We speculate that this reflects differences in resources provided to these different threads. Regardless, these differences might change with future web browser updates, so providing both methods allows the developers and users to choose the fastest solution for their situation.
Connected Components
The results of many AI image segmentation models benefit from refinement for connected components. For example, it is often necessary to ensure that voxels in a given area are contiguous with each other. For instance, proper consideration of connected components prevents erroneous identification of two areas (connected by a narrow bridge) as a single contiguous region. We developed a fast, pure javascript solution based on the algorithm of Thurfjell and colleagues46. Specifically, we ported the C code from SPM’s bwlabel function47. Crucially, since parcellations can include many classes (for example, our FreeSurfer parcellation model generates 104 distinct regions), we modified the algorithm to identify the largest connected components of all classes in a single pass.
Image Visualization
The prior releases of brainchop used the WebGL1-based Papaya for visualizations. Unfortunately, the development of Papaya has been suspended and WebGL1 does not support 3D textures that can aid interactive volume rendering. Therefore, brainchop also depended on a WebGL2-based ThreeJS volume rendering module. To address this, we upgraded brainchop to use the WebGL2-based NiiVue which supports volume loading (allowing brainchop to import images in the NIfTI, NRRD, MRtrix MIF, AFNI HEAD/BRIK, MGH/MGZ, ITK MHD, ECAT7, and DICOM formats), planar visualization and volume rendering using a single context (reducing resource usage) without requiring Papaya or ThreeJS. While our reference implementation uses NiiVue for our visualization, our modular code can be embedded in other web-capable viewers including BioImage Suite Web48, the OHIF viewer49, and VTK.js (https://kitware.github.io/vtk-js/i). Since NiiVue has already been adopted by the AFNI, brainlife, FreeSurfer, FSL and OpenNeuro teams and is supported by an active grant (NIH RF1MH121885), sustained development is ensured. Additionally, NiiVue provides several important capabilities to enhance brainchop. First, NiiVue provides drawing tools that allow users to edit models, for example removing mis-classified tissue before saving the result. In the future, we envision dynamic models, where a user can correct a single slice and the model uses this feedback for other slices. Second, NiiVue provides an ability to not only save NIfTI-format images, but also annotate images and save the entire scene. This allows one user to interactively adjust the contrast, crosshair position and write comments that they can send as an email attachment to another user. A nice feature of web pages is that they are required to live in a sandbox, without access to a computer’s file system and restricted memory. Therefore, web pages provide relatively safe attachments. This can help create a virtuous cycle between model users and model developers, allowing end users to document edge cases.
Refactoring
The original brainchop code was monolithic, with the machine learning code interleaved with Papaya specific visualization calls as well as diagnostics. The desire to support NiiVue as well as the move to support image processing using either web workers (which must communicate with the main thread via serialized objects) and the main thread (which can pass data directly via callbacks) encouraged us to modularize the code. Separating the visualization from the image processing can allow future developers to replace NiiVue with another visualization tool, or even remove the visualization entirely (for example, running the image processing from the command line using node.js). Likewise, brainchop functions for acquiring machine specific diagnostic data (which can help resolve machine specific issues) are now provided in a separate file.
License
Our implementation uses the permissive and open BSD 2-Clause (NiiVue) and MIT (brainchop) licenses. We see these licenses as being universal donors, allowing inclusion in all other projects and not restricting contributions from researchers at different institutions50.
Installation
Anyone with access to a web browser can use brainchop (https://brainchop.org/). This provides drag-and-drop support for any voxel-based format that NiiVue supports (e.g. NIfTI, NRRD, MRtrix MIF, FreeSurfer MGH, ITK MHD, DICOM). Developers can easily clone the main repository to create forks that support their own models (https://github.com/neuroneural/brainchop) and if they wish they can make pull requests to contribute to the core functions. The source code is also available at github (https://github.com/neuroneural/brainchop) with a command to host a local hot-reloadable web page (ǹpm run dev`) that can run on the Linux, Windows, and MacOS operating system. The hot-reloadable page automatically refreshes when any of the source files are modified, allowing developers to interactively modify the underlying code.
Results
The impetus for our optimization of brainchop was to improve compatibility (support for constrained hardware), enhance interactivity (with web workers running in the background), reduce dependencies (easing deployment) and adding features (e.g., the ability to edit model predictions). The benefits of these modifications are impossible to objectively quantify. However, a consequence of these changes is that the resulting models are faster and require less resources. Here we quantify the improvements in these metrics.
We evaluated the performance of brainchop release 4.0 (which incorporates all the features described in the previous section) with brainchop release 2.1 (which incorporates feedback from the review of our earlier publications42), as well as release 3.2.1. For brevity, we refer to these releases as versions 2, 3 and 4 respectively, though we note that each version has multiple releases that each introduce incremental improvements. All testing used a T1-weighted 3D gradient echo with inversion recovery scan (TI = 750ms, TR = 7.25ms, TE = 3.1ms) acquired using a UIH scanner (https://github.com/neurolabusc/dcm_qa) with the raw image having an interpolated resolution of 460×512 in the sagittal plane (230×256mm field of view) with 160 1mm thick slices with an in-plane acceleration factor of 2.5. This image was chosen as no images from this manufacturer were included in any of the training datasets. All tests were conducted on a 16GB Apple MacBook Pro with an Apple M2 Pro CPU and integrated GPU running MacOS with the Chrome browser version 124 as well as an AMD Ryzen 7950X3D CPU with 128GB of RAM and aNVIDIA RTX 4070 Ti 12GB graphics card running Linux with the Firefox v 128 browser. All tests were run 3 times with the median time reported.
Brainchop 4 allows the user to select between 15 models, providing three families of operation: tissue segmentation, brain extraction, and parcellation. The performance of these models on the test image is shown in Figure 2. Specific models vary in terms of the number of segmentation classes (e.g., number of regions for a parcellation) and hardware demands (e.g., the FreeSurfer 104 region parcellation provides both a slow, low memory model as well as a faster, higher memory model). For evaluation, we tested one exemplar from each family. We chose the `Tissue GWM (light)` segmentation model that identifies white and gray matter throughout the brain. The representative brain extraction model was Èxtract the Brain (FAST)`. Finally, the FreeSurfer 104 region model was the representative parcellation model, using the `Low Memory` variation for the MacOS computer (which used integrated graphics) and the `High Memory` variation for the Linux computer (which had a discrete graphics card).
While all subsequent measures focus on the time to perform tasks, it is worth noting that our revisions also dramatically reduce memory demands. In particular, the older version of brainchop downloads and runs Python code in emulation to conform data, with this code cached in memory to accelerate future runs. After running this stage, a brainchop web page reports consuming around 270MB of memory, and after several runs of models this can exceed 500MB of memory usage. In contrast, the new pure JavaScript conform function is very compact, and, by forcing web workers to terminate when the process is completed, we can ensure thorough garbage collection, with memory usage reported around 8MB when not actively calculating a model.
Memory differences are also observed for the 104 region parcellation. This model failed with brainchop 2 using the MacOS computer. This model succeeded with brainchop 3, which we believe reflects the improvements in memory usage. However, brainchop 4 dramatically reduces heap memory usage relative to version 3 (as previously noted, adding web workers required optimization of heap usage). Specifically, version 4 has 173 times less peak heap usage than version 3 (7.7 vs 1338.2MB).
Figure 3 illustrates the performance improvement of brainchop versions 3 and 4 relative to brainchop 2. This figure illustrates performance on the conform function as well as the time to compute the segmentation, extraction and parcellation models.
Figure 3:
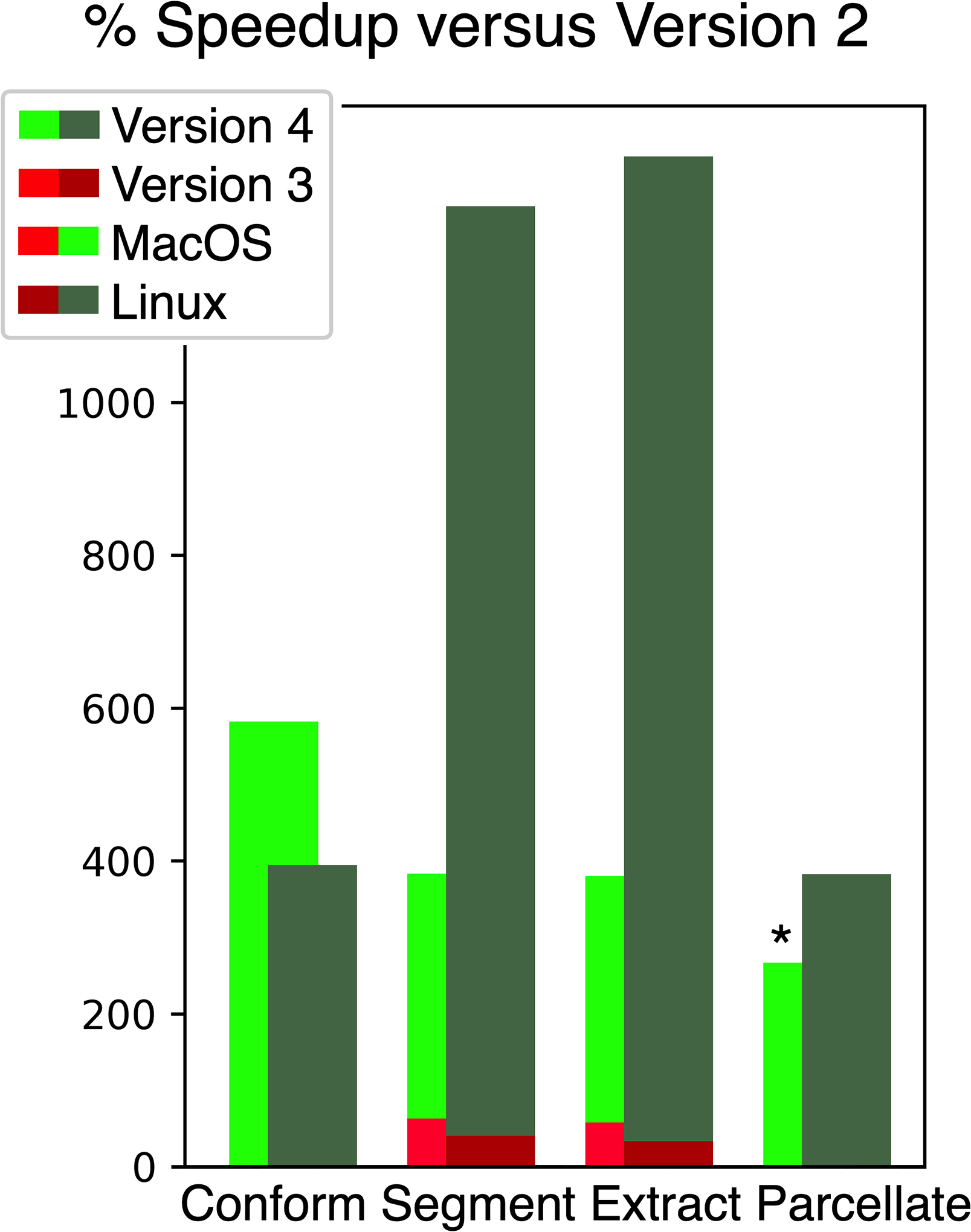
Acceleration of brainchop 4 (green) and brainchop 3 (red) relative to brainchop 2. This reflects the cumulative effect of all optimizations on the total time to apply different image processing steps. The MacOS computer (bright bars) used the Chrome browser and an integrated graphics card (Apple M2 Pro). The Linux computer (dark bars) used the Firefox browser with a discrete graphics card (AMD 7950X3D with Nvidia 4070 Ti). The acceleration is shown as percent, so a 100% speedup reflects half the time to complete an operation. The asterisk notes that parcellation crashed with version 2 on the MacOS computer, and therefore the bright green bar illustrates the speedup of version 4 versus 3.
The first stage with all processing was to conform the data to be 256×256×256 voxels with a 1mm resolution using an unsigned 8-bit data type. This timing is excluded from all subsequent tests, which used the conformed image as input. Because the time to download the large Python libraries depends on internet bandwidth, and the fact that this step is only required for the first run, we only report the time to compute the conform stage. This stage uses identical code for brainchop versions prior to version 4, so only one set of comparisons is provided. On the MacOS computer, version 4’s native code was 6.8 times (676ms vs 4611ms; or 582%) faster than the emulated Python, while conforming was 4.9 times (739ms vs 3655ms) faster on Linux.
Brainchop 4 is dramatically faster than brainchop 2. On the MacOS computer, tissue segmentation was 4.2 times faster (2144ms vs 9001ms), brain extraction was 4.2 times faster (1962ms vs 9251ms) and the 104 model parcellation only ran using the optimized code (23196ms). For the Linux computer, tissue segmentation was 13.2 times faster (898ms vs 11820ms), brain extraction was 13.9 times faster (911ms vs 12646ms) and the parcellation was 7.0 times faster (1627ms vs 12428ms)
Brainchop 4 is also reliably faster than brainchop 3. On the MacOS computer, tissue segmentation was 2.6 times faster (2144ms vs 5519ms), brain extraction was 2.7 times faster (1962ms vs 5788ms) and the 104 model parcellation was 37 times faster (23196ms vs 85048). For the Linux computer, tissue segmentation was 9.3 times faster (898ms vs 8387ms), brain extraction was 10.3 times faster (911ms vs 9419ms) and the parcellation was 4.8 times faster (1627ms vs 8444ms).
Discussion
The brainchop web page provides fast and robust brain extraction, tissue classification and parcellation with a simple drag and drop interface. The models work across hardware and software, merely requiring any modern web browser. All computations are conducted locally, protecting the privacy of the user’s data. By leveraging the user’s graphics card, most models run in a few seconds. The graphical interface lets the user inspect the results. The user can also edit errors, save the resulting images and provide diagnostics back to the developers. Taken together, this showcases an end-to-end ecosystem for deploying image processing AI models for voxel-based neuroimaging data.
Furthermore, brainchop provides a framework for other developers to extend. Developers can fork the project to distribute their own models, or contribute new models to enhance the core brainchop distribution. In particular, we look forward to models that can provide image registration and anomaly detection, as well as those that are not modality dependent.
We recognize that most neuroimaging AI model development teams use Python-based model training frameworks like PyTorch and TensorFlow, and utilize the mature and well supported set of Python libraries such as numpy and nibabel. One of our primary goals was to provide optimized, high performance JavaScript helper functions to allow these developers to easily bring their models to web pages. In the future, we hope to expand these functions to meet the needs of the community. For example, we have already introduced a function to conform data, but some may also want methods to reverse this process (to transform a classification image back to the native space of the source image).
Limitations
Our objective is to create an ecosystem that will help other model developers disseminate their work to end users. Our initial models are purposefully restricted to tissue segmentation, brain extraction and brain parcellation, but this need not be the case as brainchop continues to evolve. As noted previously, other groups have described models for spatial registration, and mapping abnormalities (acute stroke, chronic stroke, tumor, white matter hyperintensities) that could be converted too, but these are not available as edge applications. Furthermore, all of our current models require T1-weighted MR scans as input, and will fail with other modalities. Of course models could be trained to use other modalities, especially given the demonstrated multi-modality robustness of models like SynthStrip11. Another limitation is that many of the technologies we are using are emergent, and there are clear early-stage difficulties. First, while our current code runs across operating systems (Linux, Windows, MacOS) and graphics card vendors (Apple, AMD, Intel, NVidia) during our development we identified interactions, such that a specific graphics card would work on one operating system and not another. A nice aspect of having live demos is that it was easy for the vendor to duplicate our problem, which is a necessary first step towards resolution of core issues that create compatibility issues for TFJS users. Second, as we note above, some web worker implementations and access to the OffScreen canvas remain immature which impacts performance. Despite using web standards, we note that at the time of this writing many tablets and phones do not currently support our TFJS models. Also, it is worth noting that efforts to improve the privacy of web browsers can interfere with frameworks like ONNX and TFJS that use the graphics card for computation. For example, Firefox features an advanced function named `privacy.resistFingerprinting` which falsely reports artificially constrained graphics card capabilities so the variabilities in hardware cannot be used to identify a user. With this security feature set, frameworks will report that the hardware is insufficient to run typical machine learning models.
Future Directions
The current brainchop distribution has a deliberate minimalism, providing a basic recipe for delivering AI models to users. However, we have a clear vision for upcoming forks that can address specific niches. Specifically, we are actively working with teams that are using ONNX51, an interoperable format for many training frameworks, to support a diverse variety of models and TinyGrad (an emerging lightweight deep learning library) rather than only supporting TFJS. These solutions all rely on the same core helper functions, but leverage the specific strengths of each of these platforms. NiiVue also supports `boostlets` that can allow a user to interactively select a region of an image and apply a filter, with initial functions already supporting the Segment Anything Model52. As noted, we envision a future in which dynamic models will learn in real time as users make corrections, propagating this knowledge downstream. To ensure privacy, web pages are intentionally constrained. However, the modular design of NiiVue and brainchop make it easy to embed in desktop applications. Indeed, the NiiVue project already includes electron applications and Apple applications (built using Swift) that embed the visualization into a desktop application. This can aid niches such as processing BIDS datasets (where features like the inheritance principle are incompatible with a web page’s restricted permissions). While our current models offer solutions for brain extraction, tissue segmentation, and region parcellation, we envision teams leveraging these core functions for additional applications. For instance, future models could be utilized for anomaly detection, lesion mapping, spatial registration, and quantifying structural and functional brain connectivity.
Acknowledgements
MM, SSG were supported by the NIH grant RF1-MH121885. SMP was in part supported by NIH 2R01-EB006841 and NSF 2112455. CD, CR and RNN are supported by NIH RF1-MH133701 and P50-DC01466. TH is supported by core funding from the Wellcome Trust (203139/Z/16/Z and 203139/A/16/Z). Special thanks to Alex Fedorov, Mike Doan and Thu Le for helping with model training.
Footnotes
References
- 1.Nenning K-H & Langs G Machine learning in neuroimaging: from research to clinical practice. Radiologie (Heidelb) 62, 1–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore MJ, Demeyere N, Rorden C & Mattingley JB Lesion mapping in neuropsychological research: A practical and conceptual guide. Cortex 170, 38–52 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monsour R, Dutta M, Mohamed A-Z, Borkowski A & Viswanadhan NA Neuroimaging in the Era of Artificial Intelligence: Current Applications. Fed. Pract 39, S14–S20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sui J, Jiang R, Bustillo J & Calhoun V Neuroimaging-based Individualized Prediction of Cognition and Behavior for Mental Disorders and Health: Methods and Promises. Biol. Psychiatry 88, 818–828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tejavibulya L et al. Predicting the future of neuroimaging predictive models in mental health. Mol. Psychiatry 27, 3129–3137 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plis SM et al. Deep learning for neuroimaging: a validation study. Front. Neurosci 8, 229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith SM Fast robust automated brain extraction. Hum. Brain Mapp 17, 143–155 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cali RJ et al. The Influence of Brain MRI Defacing Algorithms on Brain-Age Predictions via 3D Convolutional Neural Networks. Conf. Proc. IEEE Eng. Med. Biol. Soc 2023, 1–6 (2023). [DOI] [PubMed] [Google Scholar]
- 9.Harmouche A et al. WebMRI: Brain extraction and linear registration in the web browser. Bildgebung 15, 31–36 (2023). [Google Scholar]
- 10.Isensee F et al. Automated brain extraction of multisequence MRI using artificial neural networks. Hum. Brain Mapp 40, 4952–4964 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoopes A, Mora JS, Dalca AV, Fischl B & Hoffmann M SynthStrip: skull-stripping for any brain image. Neuroimage 260, 119474 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale AM, Fischl B & Sereno MI Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9, 179–194 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Brady M & Smith S Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Ashburner J & Friston KJ Unified segmentation. Neuroimage 26, 839–851 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Billot B et al. SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining. Med. Image Anal 86, 102789 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desikan RS et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Fischl B FreeSurfer. Neuroimage 62, 774–781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischl B et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Henschel L et al. FastSurfer - A fast and accurate deep learning based neuroimaging pipeline. Neuroimage 219, 117012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iglesias JE A ready-to-use machine learning tool for symmetric multi-modality registration of brain MRI. Sci. Rep 13, 6657 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rorden C, Karnath H-O & Bonilha L Improving lesion-symptom mapping. J. Cogn. Neurosci 19, 1081–1088 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Bates E et al. Voxel-based lesion-symptom mapping. Nat. Neurosci 6, 448–450 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Sundaresan V, Zamboni G, Rothwell PM, Jenkinson M & Griffanti L Triplanar ensemble U-Net model for white matter hyperintensities segmentation on MR images. Med. Image Anal 73, 102184 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan P et al. Cerebral Microbleed Automatic Detection System Based on the ‘Deep Learning’. Front. Med 9, 807443 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boaro A et al. Deep neural networks allow expert-level brain meningioma segmentation and present potential for improvement of clinical practice. Sci. Rep 12, 15462 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C-F et al. Deep learning-based detection and segmentation of diffusion abnormalities in acute ischemic stroke. Commun. Med 1, 61 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liew S-L et al. A large, curated, open-source stroke neuroimaging dataset to improve lesion segmentation algorithms. Sci Data 9, 320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rorden C & Karnath H-O Using human brain lesions to infer function: a relic from a past era in the fMRI age? Nat. Rev. Neurosci 5, 813–819 (2004). [DOI] [PubMed] [Google Scholar]
- 29.de Haan B, Clas P, Juenger H, Wilke M & Karnath H-O Fast semi-automated lesion demarcation in stroke. Neuroimage Clin 9, 69–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seghier ML, Ramlackhansingh A, Crinion J, Leff AP & Price CJ Lesion identification using unified segmentation-normalisation models and fuzzy clustering. Neuroimage 41, 1253–1266 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mah Y-H, Jager R, Kennard C, Husain M & Nachev P A new method for automated high-dimensional lesion segmentation evaluated in vascular injury and applied to the human occipital lobe. Cortex 56, 51–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brett M, Leff AP, Rorden C & Ashburner J Spatial normalization of brain images with focal lesions using cost function masking. Neuroimage 14, 486–500 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Pustina D et al. Automated segmentation of chronic stroke lesions using LINDA: Lesion identification with neighborhood data analysis. Hum. Brain Mapp 37, 1405–1421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avants BB et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann M et al. SynthMorph: Learning Contrast-Invariant Registration Without Acquired Images. IEEE Trans. Med. Imaging 41, 543–558 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iglesias JE et al. SynthSR: A public AI tool to turn heterogeneous clinical brain scans into high-resolution T1-weighted images for 3D morphometry. Sci Adv 9, eadd3607 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClure P et al. Knowing What You Know in Brain Segmentation Using Bayesian Deep Neural Networks. Front. Neuroinform 13, 479876 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruz RS et al. DeepCSR: A 3D deep learning approach for cortical surface reconstruction. in 2021 IEEE Winter Conference on Applications of Computer Vision (WACV) (IEEE, 2021). doi: 10.1109/wacv48630.2021.00085. [DOI] [Google Scholar]
- 39.Renton AI et al. Neurodesk: an accessible, flexible and portable data analysis environment for reproducible neuroimaging. Nat. Methods (2024) doi: 10.1038/s41592-023-02145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayashi S et al. brainlife.io: a decentralized and open-source cloud platform to support neuroscience research. Nat. Methods (2024) doi: 10.1038/s41592-024-02237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markiewicz CJ et al. The OpenNeuro resource for sharing of neuroscience data. Elife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masoud M, Hu F, & Plis S (2023). Brainchop: In-browser MRI volumetric segmentation and rendering. Journal of Open Source Software, 8(83), 5098. 10.21105/joss.05098 [DOI] [Google Scholar]
- 43.Masoud M, Reddy P, Hu F, & Plis S (2023). Brainchop: Next Generation Web-Based Neuroimaging Application. arXiv preprint arXiv:2310.16162 [Google Scholar]
- 44.Fedorov A et al. End-to-end learning of brain tissue segmentation from imperfect labeling 10.1109/IJCNN.2017.7966333. [DOI] [Google Scholar]
- 45.Developers T TensorFlow (Zenodo, 2024). doi: 10.5281/ZENODO.4724125. [DOI] [Google Scholar]
- 46.Thurfjell L, Bengtsson E & Nordin B A new three-dimensional connected components labeling algorithm with simultaneous object feature extraction capability. CVGIP Graph. Models Image Process 54, 357–364 (1992). [Google Scholar]
- 47.Andersson JLR, Skare S & Ashburner J How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 20, 870–888 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Papademetris X et al. BioImage Suite: An integrated medical image analysis suite: An update. Insight J 2006, 209 (2006). [PMC free article] [PubMed] [Google Scholar]
- 49.Ziegler E et al. Open Health Imaging Foundation Viewer: An Extensible Open-Source Framework for Building Web-Based Imaging Applications to Support Cancer Research. JCO Clin Cancer Inform 4, 336–345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rorden C et al. niimath and fslmaths: replication as a method to enhance popular neuroimaging tools. Aperture Neuro 4, (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dao T, Ye X, Rorden C, Eckstein K, Haehn D, Varade S, & Bollmann S Developing a secure, browser-based, and interactive image segmentation system for medical images (2024).
- 52.Gaibor E et al. Boostlet.js: Image processing plugins for the web via JavaScript injection. arXiv [cs.CV] (2024). [Google Scholar]


