Abstract
The biological aspects of infancy within late Upper Palaeolithic populations and the role of southern refugia at the end of the Last Glacial Maximum are not yet fully understood. This study presents a multidisciplinary, high temporal resolution investigation of an Upper Palaeolithic infant from Grotta delle Mura (Apulia, southern Italy) combining palaeogenomics, dental palaeohistology, spatially-resolved geochemical analyses, direct radiocarbon dating, and traditional anthropological studies. The skeletal remains of the infant – Le Mura 1 – were directly dated to 17,320-16,910 cal BP. The results portray a biological history of the infant’s development, early life, health and death (estimated at ~72 weeks). They identify, several phenotypic traits and a potential congenital disease in the infant, the mother’s low mobility during gestation, and a high level of endogamy. Furthermore, the genomic data indicates an early spread of the Villabruna-like components along the Italian peninsula, confirming a population turnover around the time of the Last Glacial Maximum, and highlighting a general reduction in genetic variability from northern to southern Italy. Overall, Le Mura 1 contributes to our better understanding of the early stages of life and the genetic puzzle in the Italian peninsula at the end of the Last Glacial Maximum.
Subject terms: Archaeology, Biological anthropology, Archaeology, Genetics
Here, the authors present paleogenomics, dental histology, geochemistry, radiocarbon dating, and bioarchaeological analysis of an Upper Palaeolithic infant from Grotta delle Mura (southern Italy). These data depict the health and development of the individual and point to regional population turnover at the time.
Introduction
The end of the Last Glacial Maximum (LGM) is a crucial period of great transformation, characterised by the beginning of a general climate improvement1 with new human expansions throughout southern Europe and Italy2,3 and apparent changes in material culture, technology, population structures and settlement patterns4–6. Despite recent works2,3,7–9, the role of southern refugia is still not fully understood, and even less is known for the biological aspects related to early life history of late Upper Palaeolithic individuals. The study of both human life history (e.g., ontogenetic patterns, biological profile, health) and population dynamics (e.g., mobility patterns, social behaviour and organisation, evolutionary trajectories) is of great importance for the exploration and understanding of the impact the different glacial refugia, such as southern Italy, had on the cultural and genetic contribution of post-LGM populations, as well as of the demographic trajectories that occurred in the rapidly changing environment of late Upper Palaeolithic Europe. In this respect, infant human fossil remains have the potential to provide a wide range of data on human biological and cultural patterns, early life history, and the relationship between different human groups. Nevertheless, high-resolution multidisciplinary investigations into the biological profile, life history and ancient DNA (aDNA) of Upper Palaeolithic infants have so far been limited4,10–13.
The infant of Grotta delle Mura (Apulia, southern Italy; Fig. 1) – known as Le Mura 1 – was discovered in the homonymous cave in 1998, carefully covered by rock slabs, with no grave goods14 (Supplementary Note 1). Based on stratigraphic association (Fig. 1b), the inhumated remains were attributed to the final Epigravettian (Supplementary Note 1).
Fig. 1. Contextual information for the skeletal remains of Le Mura 1.
a Position of Grotta delle Mura within the Italian peninsula (from NASA Visible Earth project ‒ credits to Jacques Descloitres, MODIS Rapid Response Team, NASA/GSFC); b Stratigraphy and complete cultural sequence at the site; the infant’s location within the stratigraphy is highlighted (see Supplementary Note 1); c Photograph of the infant burial during excavation (Photo by Mauro Calattini).
This study presents an in-depth analysis of the skeletal remains by combining traditional anthropological studies, direct AMS radiocarbon dating, dental palaeohistology, spatially resolved elemental and isotopic analyses by LA-(MC-)ICPMS, and palaeogenomics for an enhanced and high temporal resolution investigation into the chrono-technological attribution, physiological development, biological profile, early life history, genetic relatedness, and ancestry of this Upper Palaeolithic infant with poor contextual information, contributing – at the same time – to our better understanding of the genetic puzzle of the Italian peninsula and its role as glacial refugia at the end of the Last Glacial Maximum.
Results
The skeletal remains
The skeletal remains were in a good state of preservation and mostly intact. From the length of the femur’s diaphysis, the infant’s height was estimated at around 82 cm15, whereas the stage of dental and skeletal development suggests an age at death between approximately 7.5 months and 1.5 years of age16 (Supplementary Note 2). Notably, a likely fracture of the left clavicula was recorded and interpreted as a possible consequence of complications during birth (Supplementary Note 2). Two vertebral arches (a cervical and a lumbar) were selected and sampled for direct AMS radiocarbon dating of the infant. Two separate radiocarbon dates were obtained (Supplementary Data 1) and combined using R_ Combine in OxCal 4.417 to determine the 14C age of the remains, which were then calibrated to provide a calibrated calendar age range of 17,320–16,910 cal BP (95.4% probability) (Supplementary Data 1).
Palaeohistology
The deciduous upper left first incisor (ULdi1; Fig. 2a) and the permanent upper right first molar (URM1; Fig. 2b) were selected based on known odontogenesis patterns for modern humans16,18,19 for palaeohistological and spatially resolved geochemistry analyses to cover the broadest period of the infant’s development, from the foetal stage to its premature death.
Fig. 2. Photographic record and virtual reconstruction of the sectioned teeth.

a Photographic record (left) and virtual reconstruction (right) of the ULdi1. In the centre, virtual representation of the sectioning plane. b Photographic record (left) and virtual reconstruction (right) of the URM1. In the centre, virtual representation of the sectioning plane.
The ULdi1 presented no occlusal wear (stage 020), one caries, and two hypoplastic linear defects21 on the buccal aspect of the crown. The root is at stage R ¾ of formation22 (Supplementary Note 3). On the histological thin section (Fig. 3a), the Neonatal Line (NNL; marking the moment of birth23) and four accentuated lines (ALs; indicators of physiological stress events24) are clearly identifiable (Fig. 3b). The crown growth parameters (Supplementary Data 2) register a mean daily secretion rate (DSR; the daily amount of enamel matrix secreted by the ameloblasts25) within inner enamel and along the enamel dentine junction (EDJ) of 2.86 ± 0.24 µmd−1 (n = 124) on the buccal side and 2.74 ± 0.23 µmd−1 (n = 160) on the lingual side, in line with reported ranges from archaeological and contemporary humans26,27. The enamel extension rate (EER; the rate at which ameloblasts differentiate along the EDJ, determining the speed of longitudinal crown formation28), which in the buccal aspect of cuspal enamel (at a 78.84 µmd−1 rate during the first ~20-days of formation) is sensibly higher than the lingual (Supplementary Data 2), also falls within reported ranges26,28–30. The construction of an individual-specific odontochronology (see methods), based on the position of the NNL, indicates that crown initiation (Ci) began in utero around 30 weeks before birth and the crown reached completion (Crc) around 12 weeks after birth, for a total crown formation time (CFT) of 42 weeks, portraying an early and fast formation in comparison to published ranges and modern European averages16,26,29. The accentuated lines were recorded at −21, −11, −5, and +3 weeks from birth, with the latter two (−5 and +3 weeks from birth) corresponding to the formation of hypoplastic defects on the enamel surface.
Fig. 3. Histological thin section of the ULdi1.
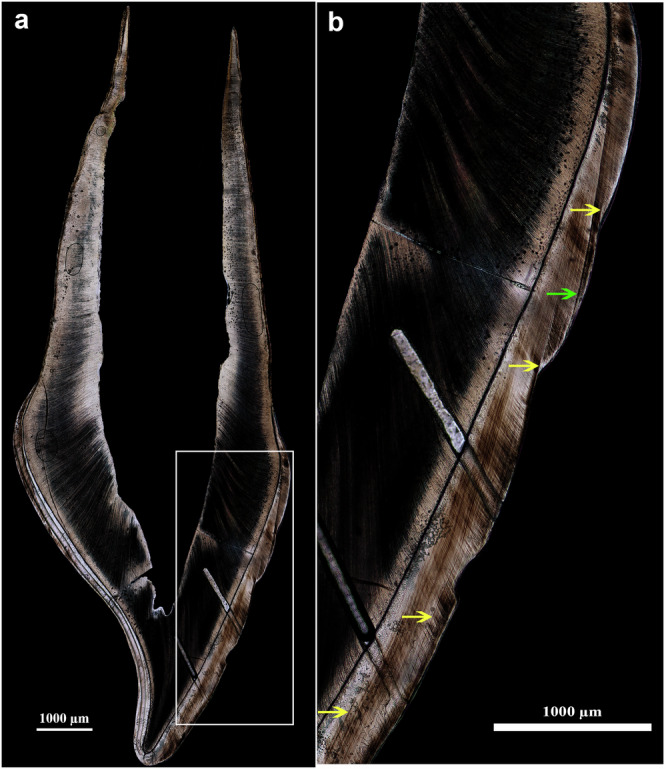
a Histological thin section of ULdi1 (buccal aspect on right, lingual aspect on left; laser ablation tracks still visible along EDJ on lingual enamel and following prism orientation on buccal enamel and dentine); b Close-up view of the buccal lateral aspect (yellow arrows indicate ALs marking systemic stress events; the green arrow points to the NNL, marking the moment of birth in the tooth’s enamel).
The URM1, with a large Carabelli’s tubercle (grade 531), is approximately at Cr ½ stage of formation32. No enamel hypoplastic defect is visible on its surface (Supplementary Note 3). On the thin section, a clear NNL is not identifiable, hence, in line with average timing of odontogenesis reported in literature16,19,25, birth was set at crown initiation. The crown growth parameters of the paracone (Supplementary Data 1) register a mean DSR within inner enamel and along the EDJ of 3.11 ± 0.26 µmd−1 (n = 149) – with fewer peak variations and generally higher values compared to the ULdi1 – which is within previously reported values33,34. The initial EER in cuspal enamel is of 35.18 µmd−1 (within the first ~20-days of formation), with values slowing down and flattening out along the tooth’s growth. Five ALs were identified at 17, 20, 40, 45, and 52 weeks after birth. Since the crown of the URM1 was not complete, the total CFT could not be estimated. However, its stage of formation (Cr ½32) at as early as ~70 weeks confirms, as for the previous tooth, the tendency to a slightly precocious development compared to modern European averages16,18. The cessation of enamel secretion caused by the infant’s death was used for an informed estimate of the age at death on a histological/chronological basis at ~72 weeks (i.e. ~16.5 months) from birth, once a minimum of 14 additional days were considered for the loss of the last formed immature enamel35; a timing that also fits well with the incisor’s root formation stage (R ¾)16,22.
Spatially resolved geochemistry
Spatially resolved geochemical analyses of both ULdi1 and URM1 were performed by LA-(MC-)ICPMS along the EDJ at ~100 µm from it (LA tracks as in Fig. 3 and Supplementary Fig. 1). Descriptive statistics of the elemental content are reported in Supplementary Data 3, whereas detailed elemental content is reported in Supplementary Data 4. Elemental profiles display a strong diagenetic overprint due to elevated U and Mn concentrations, with the two teeth seemingly showing different post-depositional pathways, likely due to their different stages of mineralization (i.e., ULdi1 is fully mineralized, while URM1 is only partially mineralized).
Along the entire EDJ path of the URM1, U concentrations are strongly elevated (~22 µg/g on average), indicative of an overprint of the biogenic signal. In contrast, [U] of ULdi1 is lower (~0.5 µg/g on average, but still elevated relative to enamel free of diagenetic imprint; Supplementary Fig. 2). Indeed, the Sr/Ca ratio displays a remarkable correlation with both U and Mn profiles along the EDJ (fitted generalized additive model (GAM) with Sr/Ca dependent [U] and [Mn] independent variables R2 = 0.65, approximate significance of smooth terms: Mn p < 0.018, U p < 0.001), suggesting that the original in-vivo signal is again masked by diagenetic alterations (Supplementary Fig. 3). In both teeth, Ba seems to be preserved even worse than Sr, showing a stronger correlation with U (ULdi1 fitted GAM R2 = 0.80, approximate significance of smooth terms: Mn p = 0.36, U p < 0.001). The low-U portions ([U] <0.1 ppm) of ULdi1 EDJ yield 87Sr/86Sr-ratios between 0.7082 and 0.7084. Obtaining reliable 87Sr/86Sr ratios for URM1 is hampered by the high Rb content (85Rb/86Sr up to 0.9, with an average of 0.1); however, the lowest Rb/Sr portions of the profile (ca. 500 µm in length) show a mean 87Sr/86Sr ratio of 0.7082, consistent with ULdi1 (Supplementary Fig. 4, Supplementary Data 5).
Palaeogenomics
Palaeogenomic analysis was carried out on the left petrous bone. Through deep shotgun sequencing, we reconstructed 75.26% of the genome with a mean coverage of ~2.21X (Table 1). Deamination and fragmentation patterns were typical of ancient remains (up to ~17% damage at both 5′ and 3′ ends of the reads and an average fragment length of 47.72 bp)36 and the overall modern human DNA contamination was very low (mtDNA contamination: 3; X-chromosome: 0.52% (SE = 1.299559 × 10−3); ContamLD: −0.076 (SE 0.013)) (Table 1, Supplementary Fig. 5, Supplementary Data 6 and 7). By comparing the number of aligned reads on the sex chromosomes and autosomes, the infant was determined as male (Supplementary Data 8). The mitochondrial genome belongs to the U2’3’4’7’8’9 haplogroup and the Y-chromosome is assigned to the I2a haplogroup (sub-clade I2a1a1) (Supplementary Discussion, Supplementary Data 9), some of the most common mtDNA and Y lineages among the Italian hunter-gatherers associated to the Epigravettian culture and belonging to the Villabruna genetic cluster2,37–40.
Table 1.
Mapping results
| Sequencing | # of Raw Reads prior C&M | Merged Reads (%) | Mapped Reads after DeDup | Human DNA (%) | Depth of Coverage | Genome coverage 1X (%) | CtoT (%) | GtoA (%) | Average fragment length (bp) | SNPs sites covered in 1240 K panel |
|---|---|---|---|---|---|---|---|---|---|---|
| Light-shotgun | 10834630 | 68.68 | 523365 | 22.46 | 0.0078 | 0.77 | 17.37 | 17.19 | 47.78 | – |
| Deep-shotgun | 8.59E + 09 | 68.39 | 1.44E + 08 | 22.00 | 2.21 | 75.26 | 16.87 | 16.81 | 47.72 | 1080593 |
Results of raw sequencing data, pre-processing and mapping on human reference genome (Hg19) results are reported for both shallow shotgun and deep sequencing.
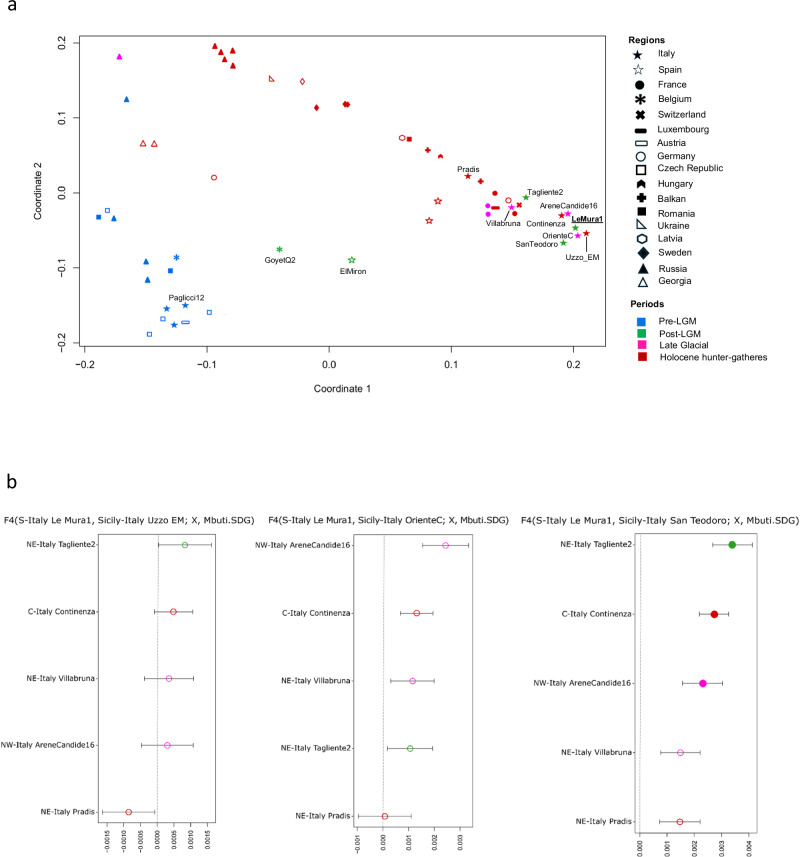
Investigating the genetic relationships between Upper Palaeolithic and Mesolithic hunter-gatherers from Eurasia (Supplementary Discussion, Supplementary Data 10, 11, and 12), Le Mura 1 fell within the Western hunter-gatherer (WHG) population (Principal Component Analysis, Supplementary Fig. 6a), clustering closer to the Late Upper Palaeolithic and Mesolithic specimens from Sicily, the Upper Palaeolithic specimen from Arene Candide cave (AC16) and the Mesolithic individuals from Grotta Continenza (Multi-Dimensional Scaling, Fig. 4a and Supplementary Fig. 6b). Conversely, it exhibits lesser affinity with coeval and later specimens from north-eastern Italy, as confirmed by outgroup f3-statistics (f3(Le Mura 1, X; Mbuti), where X represents alternative individuals listed in Supplementary Data 10 (Supplementary Fig. 7), and f4-statistics analysis (Fig. 4b, Supplementary Discussion, Supplementary Data 13 and 14). In particular, f4-statistics in the form f4(Le Mura 1, Group3; Group1/Group2, Mbuti), where Group 3 are Late Upper Palaeolithic and Mesolithic specimens from Sicily, and Group1/Group2 are alternative Epigravettian/Mesolithic specimens respectively from northern and central Italy (as defined in ref. 3), show that Le Mura 1 is mostly symmetrically related to the later Sicilian hunter-gatherers (Fig. 4b; Supplementary Discussion and Supplementary Data 13). Furtherly, qpWave and qpAdm were utilised to model potential ancestral sources for the Le Mura individual (Supplementary Data 15, 16, and Supplementary Discussion). These analyses revealed that our genome from southern Italy can be modelled as deriving the entirety of its ancestry from Villabruna-like ancestry.
Fig. 4. MDS plot and genetic affinity among post-LGM Italian hunter-gatherers.
a Multi-Dimensional Scaling (MDS) plot of European hunter-gatherers based on 1 − f3(Le Mura 1, X; Mbuti.SDG). The dimensions are calculated using the reported specimen and previously published hunter-gatherer groups or individuals listed in Supplementary Data 6. Only labels of the populations mentioned in the main text are reported (all labels are shown in Supplementary Fig. 6a); b f4-statistics comparing the affinity of Le Mura 1 with Late UP and Mesolithic individuals from Italy. Filled and empty circles represent the significant (Z-score > |3|) and non-significant Z-score respectively, associated with the statistics. Error bars show 1 × s.e.m. (black) or 3 × s.e.m. (grey) of the f4 values estimated from 5 cM-block jackknife analysis. The values of the f4-statistics suggest no recent interactions between tested individuals and Le Mura 1 (See Supplementary Data 9). The higher genetic affinity was observed between Le Mura 1 and the later Sicilian hunter-gatherers. In both figures, samples dated to the Pre-LGM are coloured in blue, Post-LGM in green, Late Glacial in magenta and Holocene hunter-gatherers in red. Source data are provided as a Source Data file.
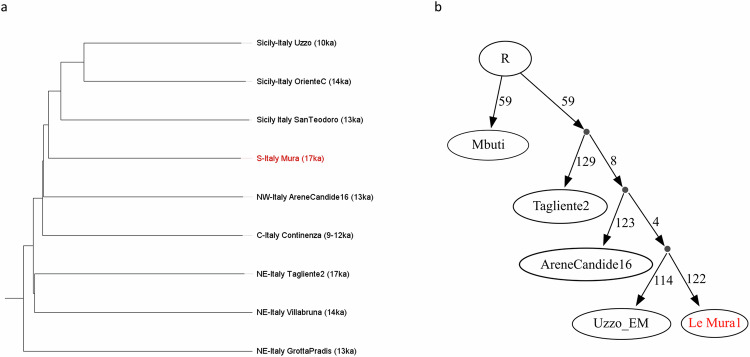
Phylogenetic reconstruction using pairwise 1- outgroup f3 genetic distance matrix indicates that, despite its ancient date, the Le Mura 1 genome exhibits a derived placement that branches off after the split of northern (Pradis 1, Tagliente 2, Villabruna) and central Italian (AC16, Continenza group) hunter-gatherers, but before the lineage separating Sicilian individuals (Oriente C, San Teodoro 2, early Mesolithics from Uzzo cave) (Fig. 5a; Supplementary Discussion). A simplified version of the same phylogenetic relationships was specifically tested via qpGraph (Fig. 5b and Supplementary Discussion). The analysis provided statistical support for a model in which the Le Mura 1 genome closely related to a derived population in the southernmost region of Italy that settled Sicily possibly by crossing a temporary land-bridge before the end of the LGM7,41,42 (Supplementary Data 17).
Fig. 5. Population structure of the post-LGM Italian hunter-gatherers.
a Neighbour-joining tree of Italian Epigravettian and Mesolithic individuals based on pairwise 1-outgroup f3 genetic distance matrix; b Phylogenetic relationship graph of the main post-LGM Italian individuals modelled using qpGraph. Source data are provided as a Source Data file.
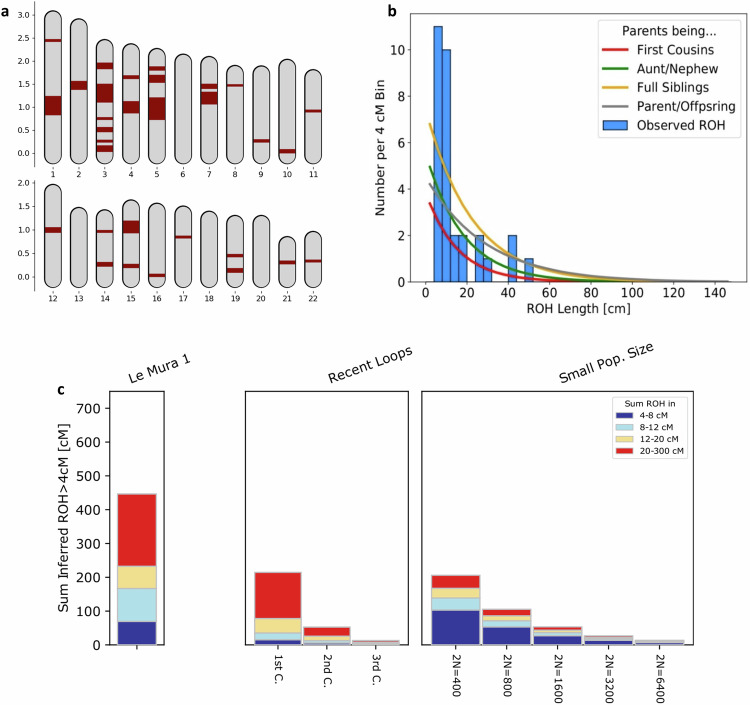
Pairwise mismatch rates (PMR) between the Italian Mesolithic and Epigravettian individuals confirm a general decline of the genetic variability along a North to South cline, with a more pronounced reduction between north-eastern regions and the rest of the peninsula, and later through the spread into Sicily. The pattern indeed suggests that Le Mura 1 belongs to a population that exhibited a higher degree of genetic variability than the Sicilian group (Supplementary Data 18; Supplementary Fig. 8). To better investigate this issue, we performed genome-wide analysis of long and shorter runs of homozygosity (ROH)43. Le Mura 1 carries less than 100 centiMorgans (cM) of short ROH segments (sROH 4–8 cM) (Supplementary Data 19), similarly to the Epigravettian-related individuals from northern Italy3. Moreover, the amount of long ROH (sROH>20 = 213,69 cM) segmentsobserved suggests close kinship between the infant’s parents (e.g., first cousins) (Fig. 6).
Fig. 6. ROH in Mura’s autosomes.
a ROH positions across the 22 autosomes are marked in red, with map length in Morgan. b Distribution of observed ROH in Le Mura 1 with the expected densities of ROH for certain degrees of parental relationship calculated in Ringbauer et al. c Le Mura 1’s ROH represented by vertical bars, where the length of each bar corresponds to the four classes of ROH (distinguished by different colours). On the right, legend of expected ROH for offspring of close kin or for small populations, calculated in Ringbauer et al. Source Data for are found in Supplementary Data 19.
Using f4-statistics, we assessed the genetic proximity of Italian Epigravettian-related groups to El Mirόn or GoyetQ238, each representing two distinct genetic clusters associated with the Magdalenian culture. Aware of the potential bias introduced by genotyping strategies, Le Mura 1 seems more closely related to El Mirόn than specimens from north-eastern Italy. However, among all the Italian Epigravettians from west-central and southern Italy, El Mirόn has a stronger genetic connection with the Epigravettian and early Mesolithic individuals from Sicily, rather than with Le Mura 1 and Grotta Continenza (Supplementary Data 20). Moreover, an extra genetic affinity was found between Pop1 individuals and GoyetQ2 when Pop2 is Tagliente2 (f4(Pop1, Pop2; GoyetQ2, Mbuti)) (Supplementary Data 20) (Pop1 and Pop2 are samples listed in Supplementary Data 20). This is true except for Le Mura 1 (Le Mura 1, RIP; GoyetQ2, Mbuti f4 = 0.00172 Z = 2.216) which shows no additional genetic relationship with individuals related to GoyetQ2.
In addition, possible traces of local genetic continuity between Gravettian individuals and early post-LGM Epigravettian populations were assessed using f4-statistics, though no significant genetic signal of Gravettian-related ancestry was found even when comparing pre- and post-LGM genomes from southern Italy (Supplementary Data 21).
The estimation of Neanderthal DNA proportion using f4-ratio statistics, using high-quality Neanderthal genome from Altai and Vindija 33.1944 (Supplementary Discussion), estimated 2.25% (Standard Error (SE): 0.4%) Neanderthal DNA in the infant (Supplementary Figs. 9 and 10), like other post-LGM and present-day Europeans (2.2–2.7%)45. However, applying methods described in ref. 46, with a total of 194,076 SNPs (out of 221,482 informative variants), a late admixture with Neanderthals along the lineage of Le Mura 1 was detected dating back 814 +/− 122.77 generations, which was estimated to 40,623 +/− 3560 years BP assuming 29 years per generation47, or more recently if a 2.68/3.39-year shorter generation interval is assumed, as recently suggested for West Eurasian populations48 (Supplementary Discussion, Supplementary Data 22 and Supplementary Fig. 11).
Additionally, functional and phenotypically interesting SNP sites were targeted among those annotated in SNPedia49, with a unique variant allele reported in dbSNP (build 153)50 (Supplementary Discussion). Based on data from the 41 SNPs in the HIrisPlex-S panel51, the infant most likely had blue eyes, dark skin and curly dark brown/almost black hair (Supplementary Data 23). Additionally, we screened polymorphisms at two SNPs loci linked to lactase persistence (rs4988235 and rs182549) on the MCM6 gene, identifying in Le Mura 1 the ancestral alleles (Supplementary Data 24) that suggest the inability to digest milk in adulthood, in line with previous observations in hunter-gatherer populations52. Finally, a list of candidate SNPs associated with several genetic disorders were analysed and two heterozygous mutations in two different genes (TNNT2 and MYBPC3) encoding sarcomeric proteins, linked to familial hypertrophic cardiomyopathy (FHC or HCM) were observed with the highest genotype likelihood value (loglikeratios to the most likely, scaled as log10, = 0) (Supplementary Data 24)53,54.
Discussion
The direct AMS dating and its combination with traditional anthropological, histological, geochemical, and genomic analyses of the infant of Grotta delle Mura opens a broader perspective on the human population in southern Italy at the end of the LGM.
The palaeohistological analysis offers a picture of the development and odontogenic bio-rhythms during the foetal stage and early infancy in the Upper Palaeolithic, which is of primary importance in understanding potential changes that have occurred in the timing and modality of dental development during gestation and its stage at birth. While both DSRs and EERs fall within the reported ranges for the same tooth classes from archaeological and contemporary humans26–30,33,34 suggesting comparable rates of tissue secretion across both the crown’s thickness and length, the biochronological reconstruction of the development highlights an earlier and abbreviated period of toot formation (Ci at ~30 weeks before birth, Crc at ~12 weeks after birth, and CFT of ~42 weeks) when compared to the published range of values and to modern European averages16,18,26,29 (Supplementary Data 2). This supports previous observations of generally faster dental and skeletal development in pre-industrial infants and an Upper Palaeolithic foetus also from Apulia12,26,55, aligning with the faster spectrum of modern variability. Such findings advise caution regarding the use of modern standards for archaeological and paleoanthropological specimens.
The identification of nine ALs indicates that the infant experienced at least as many different physiological stress events (two of which developed hypoplastic defects). Their cause cannot be ascertained due to the non-specificity of the stress markers; however, their number draws a picture of severe physical stresses throughout his development and life. The ALs in prenatal enamel (21, 11 and 5 weeks before birth) – unusual during the sheltered intrauterine period, though not unprecedented12,13,56 – may reflect the mother’s health during pregnancy. Nevertheless, the detection of two variants in the TNNT2 and MYBPC3 genes, suggesting a potential congenital heart defect (FHC or HCM) – cause of arrhythmias, syncopes and sudden cardiac arrest and death in young adults and children57,58 – may be able to account for the peculiarly high number of stresses, and potentially affecting his death, which was refined on a histological/chronological basis at ~16.5 months from birth.
Due to the strong post-depositional elemental alteration that affected relevant portions of the EDJ tracks, we cannot retrieve robust dietary in-vivo information from the Sr/Ca profiles, and even less so from Ba/Ca data. Yet, Sr isotopes from the unaltered enamel portions where [U] is less than 0.1 ppm, can inform about the mobility of the mother. Little is directly known about sub-seasonal human mobility for the Late Upper Palaeolithic, including Italian sites. The individual from Grotte di Pradis (northern Italy)10 shows a remarkably variable intra-tooth Sr isotopic composition, possibly linked to seasonal mobility patterns of the human group. We need to stress that Grotte di Pradis is characterized by quite heterogeneous geology, thus hampering a direct comparison with southern Italian contexts, where the geological bedrock is more homogenous and mainly composed of carbonate rocks. Three Epigravettian individuals from Grotta Paglicci (also in Apulia)4 display highly-radiogenic Sr isotopic compositions in their deciduous teeth, suggesting they were born non-locally. On the other hand, the Sr isotopic signature of Le Mura 1 is fully compatible with the local bioavailable Sr (see4 and Supplementary Fig. 4), suggesting that the mother of the individual was likely local/low-mobile during the last months of her pregnancy.
Genome-wide analysis improved the information on the infant’s biological profile (Supplementary Data 8, 23 and 24), determining the individual as male and with a phenotypic combination (blue eyes, dark skin and curly dark brown, almost black hair) that was likely very common among WHGs, since it has been inferred for several late Upper Palaeolithic and Mesolithic individuals in south-western and central Europe3,39,59,60. Moreover, the infant also appears to be the offspring of close kin parents (Fig. 6). A recent study43 attributes similar ROH lengths prevalently to first cousin unions and recorded two other cases in the European Upper Palaeolithic (i.e., El Mirón and Villabruna, in the Iberic and Italian peninsula, respectively). In ethnographic literature, the union amongst first cousins – especially cross-cousins – with the effect to reinforce clan/group ties, has recurrently been documented61,62. However, this type of union has not commonly been observed across most ancient populations and is not associated with particular regions or chrono-cultural groups, though few cases of high consanguinity were especially observed among members of farming societies43,63. In this instance, the small group size, as suggested by the reduced genetic diversity across the Italian peninsula, might have contributed to the likelihood of it occurring.
The genomic data from Le Mura 1 are also important to investigate post LGM human population dynamics in southern Italy. Le Mura 1 falls within the variability of the WHGs and shares the highest genetic affinity with Mesolithic and Late Epigravettian individuals from central Italy and Sicily, rather than with the coeval specimen from northern Italy (Tagliente2) (Fig. 4a; Supplementary Figs. 6a and 7), confirming the presence of a geographic rather than chrono-cultural pattern of genetic variability in Italy. The presence of Villabruna ancestry in southern Italy at least 17,000 years BP, together with a highly derived genome of Le Mura 1 among the Italian Epigravettian-related groups (Fig. 5), suggests the spread of Villabruna-like components along the Italian peninsula during or immediately after the LGM2,3. Our data, therefore, corroborate the hypothesis of an early expansion of Villabruna ancestry in southern Europe well before the Bølling-Allerød warming event3, and the lack of Gravettian-related ancestry in Le Mura 1 directly shows that a population turnover effectively occurred in southern Italy, where the abrupt techno-typological transition from the late Gravettian to the early Epigravettian technocomplex64 was likely influenced by a demographic process linked with the spread of Villabruna-like ancestry. However, genome-wide data from the Italian peninsula dated to the LGM (~24,000–19,000 cal BP), essential to explore in detail the genetic transformation observed between pre- and post-LGM groups and to investigate the role of southern European refugia during the LGM, are not currently available.
Consistent with a progressive north-to-south reduction of the genetic variability previously recorded in the Italian Epigravettian and Mesolithic hunter-gatherers3, Le Mura 1 exhibits a lower PMR value than those found in Epigravettian and Mesolithic individuals from northern and central Italy (Supplementary Data 18), marking a population decline along the Italian peninsula favoured by its geographical features. Progressing southwards, the ancestral population did not admix with populations carrying distinct ancestries, as suggested by qpAdm and qpWave results, which are consistent with a one-way model where Villabruna is the unique ancestry component in Le Mura 1. This scenario is further supported by mitochondrial genome data, with individuals from both Grotta delle Mura and Riparo Tagliente carrying mitochondrial DNA sequences of the generally rare U2’3’4’7’8’9 clade (Supplementary Discussion and Supplementary Data 25), but which is the most represented lineage among hunter-gatherers from Sicily2,7,65–67. In contrast, later Epigravettian and Mesolithic individuals from northern and central Italy - such as Villabruna, Arene Candide 16, Pradis 1, and Continenza 8 - dating from 14,000 BP3,38,66,68, harbour more differentiated mitochondrial DNA sequences of the U5b lineage (Supplementary Fig. 12). The particular distribution and the TMRCA of the U2’3’4’7’8’9 sequences among Italian hunter-gatherers of 20,610 years BP (95% HPD: 17,817–23,743) (Supplementary Fig. 12; Supplementary Data 26, 27 and 28) thus appear compatible with its spread across the peninsula via a singular population movement. Overall, we can hypothesise that the ancestry associated with the Epigravettian culture reached Italy much earlier than 17,000 years BP through a rapid expansion of a single source population, most likely starting from north-east (as suggested by Pradis 1 and Tagliente2) to then reach the most southern regions and Sicily. As shown here, Le Mura 1 is basal to Sicilian hunter-gatherers (Fig. 5a), with which it also shares a phylogenetically related mitogenome sequence (Supplementary Data 25, Supplementary Fig. 12). Le Mura 1 could therefore represent an ancestral population, characterised by a limited group size and increased inbreeding – as evidenced by the close genetic relationship between his parents (Fig. 6) – that was widespread in the southern regions of the Italian peninsula during the latter phase of the LGM and contributed to the peopling of Sicily. However, more genomic data from Epigravettian individuals of continental southern Italy are necessary to better investigate this issue.
Previous mitochondrial and genome-wide analyses on late Upper Palaeolithic hunter-gatherers from the Mediterranean area have shown an early connection between different LGM refugia in southern Europe3,8,38,40,66,67. We showed that El Mirόn has a higher genetic affinity with individuals from central-western and southern Italy, including Le Mura 1, than with Epigravettian individuals from north-eastern Italy (i.e., Tagliente2, Villabruna and Pradis 1) (Supplementary Discussion, Supplementary Data 18). This affinity is even higher in the Epigravettian and early Mesolithic individuals from Sicily, confirming a deep connection between glacial refugia during the LGM that mainly involved the Tyrrhenian area of southern Italy8,66,67. Additionally, no extra genetic affinity was observed between the Magdalenian GoyetQ2 from Belgium (central Europe) and the oldest Italian Epigravettian specimens (Le Mura 1 and Tagliente2), whereas a higher amount of shared genetic drift was detected between GoyetQ2 and late Epigravettian and early Mesolithic individuals from Italy (Supplementary Data 18). Genetic exchange between those different hunter-gatherer groups (i.e., Magdalenian-related groups from central Europe and the most recent Epigravettian-related groups from Italy) could suggest increased mobility between Italy and central Europe across the Alps after 17,000 years BP, thanks to an initial climatic amelioration after the LGM. Even if more in-depth studies are needed, these contacts seem to be also testified by some parallelisms in techno-economic tendencies69 and by the sharing of genetically closely related domestic dogs70.
On a different level, although no additional proportion of Neanderthal DNA was detected in our sample compared to other post-LGM and present-day Europeans38 (Supplementary Figs. 9 and 10), the tentative estimation of a Neanderthal late admixture along the lineage of Le Mura 1 at around 40,000 years BP (Supplementary Data 22) could have important implications on the genetic relationship between early modern humans and late Neanderthals in Europe. However, more genome-wide data of post-LGM individuals from southern Europe are needed to confirm the significance of this relatively late admixture.
Overall, the implementation of an advanced multidisciplinary high-resolution approach within the Italian panorama, directly portrays the history of development, early life, health and death of a late Upper Palaeolithic infant, zooming in on the dental growth patterns during foetal development and early infancy, and contributing to the observation of variations in the timing of odontogenic development between pre-industrial and modern societies12,26,55,71. In concert, it hints at social dynamics within the group, highlighting the mother’s low mobility during pregnancy and parental consanguinity. Furthermore, the great genomic preservation contributed to our better understanding of the genetic puzzle in the Italian peninsula following the Last Glacial Maximum, confirming a population turnover around the time of the cultural transition from the Gravettian to the Epigravettian technocomplex.
The infant from Grotta delle Mura represents an exceptional testimony to the early phases of life and peopling of southern Italy, with several implications for the interpretation of social and population dynamics at the end of the LGM. These findings reinforce the importance of this period in the peopling history of the Italian peninsula and the need for further biological data from early Epigravettian-associated individuals.
Methods
The analytical set-up included the sampling of two vertebral arches (cervical and lumbar) for AMS radiocarbon dating, the upper left first deciduous incisor (ULdi1) and the upper right first permanent molar (URM1) for histomorphometric and histochemical analyses, and the sampling of the left petrous bone for aDNA analysis. Destructive sampling was conducted under the permissions granted to the involved institutions by the Soprintendenza Archeologia, belle arti e paesaggio per la città metropolitana di Bari within the Ministry of Cultural Heritage and Activities and Tourism (MiBACT) (protocol number 4997).
AMS radiocarbon analysis
Both vertebral samples were pretreated at MPI-EVA Leipzig. Sample R-EVA 3275, which involved a small amount of bone material (Supplementary Data 1), followed the protocol described in ref. 72. Whereas R-EVA 3276 followed the standard procedure described in ref. 73. Both samples were demineralized in 0.5 M HCl at room temperature until no CO2 effervescence could be observed. 0.1 M NaOH was then added for 30 min to remove humics. The NaOH step was followed by further rinsing with 0.5 M HCl for 15 min. The samples were then gelatinized, following74, in a pH3 solution at 75 °C for 20 h. The resulting gelatine was first filtered in an Eeze-Filter™ (Elkay Laboratory Products (UK) Ltd.) to remove small (<8 µm) particles and then through a 30 kDa ultrafilter (Sartorius “Vivaspin 15”)75. Prior to use, the filter was cleaned to remove carbon-containing humectants73. The samples were lyophilized for 48 hours, and those with a satisfactory collagen yield (>1%) and a C:N ratio between 2.9 and 3.5 were selected for dating73,76. The collagen was combusted and converted to graphite using the AGE system77 before being measured at the AMS at the Klaus-Tschira-Labor für Physikalische Altersbestimmung (Curt-Engelhorn- Zentrums für Archaometrie), Mannheim, Germany78. Radiocarbon dates were calibrated in OxCal v4.417 using the IntCal20 calibration curve79.
Palaeohistology
Virtual acquisition of dental specimens
Prior to processing, the dental specimens were micro-CT scanned at the IRCCS Istituto Ortopedico Rizzoli (Bologna) by using a Skyscan 1072 (Bruker Corp.). The data acquisitions were performed at a voltage of 50 kV, 197 µA, and a voxel resolution of 16.45 µm for the ULdi1 and of 12.50 µm for the URM1. The X-ray beam was filtered with by 1 mm aluminium. The exposure time was set at 5936 ms, and the image averaged on 2 frames, with a 180° rotation and a rotation step of 0.9°. The images were reconstructed with a Beam Hardening Correction of 25% and a Ring Artifact Correction of 1 using NRecon software (Bruker Corp.).
Following processing, though before extraction from epoxy, the specimens were micro-CT scanned at the University of Ferrara by using a microfocus X-ray tube (Hamamatsu L9421-02), a rotation stage and a flat panel detector (Hamamatsu C14400DK-51). The data acquisitions were performed at a voltage of 80 kVp, 80 μA and a focal spot size of nearly 5 μm. The X-ray beam was filtered by 1 mm aluminium. A set of 720 images over an angular scan of 360° was acquired of each specimen with an integration time of 100 ms. The planar images were dark and flat corrected and reconstructed using a home-made software in a 3D volume with an isotropic voxel of 30 μm.
Specimen processing
The dental elements were processed and analysed at the Laboratory of Osteoarchaeology and Palaeoanthropology of the University of Bologna (Bones Lab), in Ravenna. The sectioning protocol adapted the method described in ref. 11, consisting of a composite embedding-cutting-mounting procedure, and involved a thorough cleaning of all tool surfaces with absolute ethanol in-between each step of the cutting-mounting process. Each specimen was first coated with Crystalbond 509TM (Aremco Products, Inc.) and then embedded in EpoThinTM 2 epoxy resin (Buehler). The favourable cutting plane for both specimens was identified on a 3-dimensional (3D) virtual reconstruction, which was generated from the micro-CT images. The ULdi1 was sectioned along its central labio-lingual plane and the URM1 along the bucco-lingual plane of its paracone by using an IsoMetTM low speed saw mounted with a 0.3 mm thick IsoMetTM Diamond Wafering Blade (Buehler). One of the resulting blocklets from each tooth was then selected and gently grinded with P2500 abrasive paper and polished with 1 μm MetaDiTM Polycrystalline Diamond Suspension (Buehler) to eliminate any saw marks, rinsed with demineralised water, and left to dry. The dry blocklets were then attached to a microscope glass slide with EpoThinTM 2 epoxy resin (Buehler) and left overnight at room temperature for full polymerisation. At which point a second sectioning was performed to obtain single longitudinal sections of ~300 μm of thickness. Both sections were then grinded to a thickness of ~150 μm and polished with 1 μm MetaDiTM Polycrystalline Diamond Suspension (Buehler), rinsed with demineralised water and left to dry at room temperature. After LA-(MC-)ICPMS, the thin sections were furtherly grinded and polished to a more optimal thickness of ~100 μm for histological analysis.
Micrographic documentation of the thin sections at the different stages of processing was acquired at 100× magnification under transmitted polarised light with an Axioscope 7 microscope mounting a 208 colour camera (Zeiss). The micrographs were automatically taken and assembled in a single image using the mosaic setting of ZenCore 3 software (Zeiss).
Finally, the blocklets containing the remaining portions of both teeth – after being micro-CT-scanned (see above) – were shorty treated with acetone to remove the Crystalbond 509TM coating (Aremco Products Inc.), allowing the extraction of the teeth portions from the blocklets of epoxy resin and their restoration through the use of reverse engineering and rapid prototyping by adapting the method described in ref. 74 (below).
Histomorphometric analysis
The micrographs of the thin sections were opened with Adobe Photoshop (Adobe Inc.), on which a chronological framework based on the dental enamel microstructures was constructed adapting methods described in literature30,80. A first segment was traced following the local prism orientation from the dentine horn to an isochronous line (Retzius line) in the enamel, which was then followed back down to the EDJ (enamel dentine junction). From this intersection, a new segment was traced following the local prism orientation to another isochronous line, which was also followed back down to the EDJ. This zig-zag motion was repeated to the crown cervix. All prism segments were traced within 100 μm for the ULdi1 and within 200 μm of length for the URM1. Following, the image with the chronological framework was opened on ImageJ 1.54b software81, where all measurements were performed (e.g., prism segments, EDJ lengths, lengths of Laser Ablation tracks) The time (in days) expressed by each prism length was estimated dividing its value by the local mean of inner enamel DSRs, which in turn was calculated by measuring visible consecutive cross-striations and dividing each length by the corresponding number of cross striations. The total crown formation time is equal to the sum of the selected prism segments. The local EER was calculated by dividing the EDJ length between each two selected prism segments by the number of days represented by the corresponding prism segment.
Spatially resolved geochemical analyses
Spatially resolved elemental and strontium (Sr) isotopic analysis, by Laser Ablation (Multi-Collector) Inductively Coupled Plasma Mass Spectrometry (LA-MC-ICPMS) was carried out on the same thin sections used for paleohistological investigation at the Frankfurt Isotope and Element Research Center (FIERCE) at Goethe University Frankfurt, Germany, closely following analytical protocols described by Müller et al.82 and Müller and Anczkiewicz83, respectively. The tracks were performed in continuous profiling mode, always following the enamel-dentine junction from apex to cervix (within 100 μm distance) and along selected prisms. The micrographs of the thin sections displaying the tracks left by the ablation were superimposed onto the micrographs used for the histomorphometric analysis and onto the chronological framework itself. This allowed the temporal attribution of multiple points along the ablation tracks. The distance along the ablation track of each temporally-known point was recorded through ImageJ 1.54b81 and the translation of the spatial information into enamel (secretion) chronologies and the smoothing of the elemental and isotopic profiles were carried out using R software environment for statistical computing (v 4.2.3)84 and a local polynomial regression fitting85.
Restoration of dental specimens
The here-described protocol for physical restoration, adapted from ref. 86, involved a composite procedure consisting in: a) the µCT scanning of the original specimens; b) histological sectioning; c) the µCT scanning of the specimens after sectioning; d) the building of the digital 3D surfaces of the specimens before and after sectioning; e) the reconstruction of the digital models of the extracted portions by subtracting the 3D surfaces of the preserved portions (after sectioning) from the surfaces of the intact specimens (before sectioning) by using reverse engineering techniques; f) the prototyping of the extracted portions to be integrated; g) the application of the prototypes through the use of compatible and reversible adhesives (Supplementary Fig. 13).
The following is a detailed description of the work plan applied:
The teeth were measured using microcomputed tomography scanners.
The ULdi1 was sectioned along its central labio-lingual plane and the URM1 along the bucco-lingual plane of its paracone by using an IsoMetTM low speed saw mounted with a 0.3 mm thick IsoMetTM Diamond Wafering Blade (Buehler). Single longitudinal thin sections of ~300 μm of thickness were obtained.
An additional computed tomographic scan was performed on the teeth following the sampling.
µCT image data (pre- and post-sectioning) were segmented semi-automatically using Avizo Lite 9.2.0 software (Thermo Fisher Scientific) to render the pre- and post-sectioning 3D digital models, which were then imported in Geomagic Design X (3D Systems) for cleaning processes and for the correction of incidental defects (e.g., filling of small holes) to create fully closed surfaces.
The digital surfaces (of the specimens before and after sectioning) were overlaid using the overlay algorithms found in Geomagic Design X software. To extract the missing portion of the tooth, best-fit planes were generated on the surfaces of the sectioning cut present on the preserved fragment model (post-sectioning). The same planes were then used to divide the digital model of the whole tooth into three portions: the sectioned portion and the two preserved portions. The result is a distinct three-dimensional model of the missing portion. In the case of URM1, some channels were created through the model to allow expansion of the glue when the printed prototype would be applied to the original portions of the tooth; this was not possible for ULdi1 due to constraints linked to the model’s size.
Exact replicas of the sectioned portions were reproduced with rapid prototyping technology (LCD Stereolithography (SLA)) using a Photon MONO X 3D printer (Anycubic). The prototype was produced using Anycubic resin with a layer thickness of 0.05 mm, UV Matrix 405 nm LED lighting sources and slicing Chitubox V1.9.1 software.
Finally, the printed replicas were applied between the preserved portions of the original specimens by using compatible and reversible glues (specifically UHU extra gel Polyvinylester) (Supplementary Figs. 14, 15).
Paleogenomics
Laboratory work and bioinformatics analysis
Genetic analysis was carried out at the Laboratory of Anthropology (University of Florence), in facilities exclusively dedicated to aDNA analysis and following strict guidelines to minimise the risk of modern human DNA contamination87,88. Moreover, negative controls were processed alongside each experimental step and they did not exhibit human DNA (Supplementary Data 6). After removal of the environmental contaminants from the outer layer of the bone by brushing and UV light, approximately 50 milligrams of bone powder was recovered from the inner part of the petrous bone and used for DNA extraction83. A double-strand and double indexed library89,90 with partial uracil-DNA glycosylase91 was generated to reduce the impact of deaminations and improve the subsequent analyses. Library underwent shallow shotgun sequencing and mitochondrial DNA capture92 to assess the overall endogenous DNA content and preservation. The same library was subsequently sequenced on an entire S2 flow cell of the Illumina NovaSeq 6000 with 2 × 50 + 8 + 8 cycles.
Bioinformatic analysis of shotgun data: reads processing, mapping, variant calling and sex identification
After demultiplexing, raw reads were pre-processed and filtered for sequencing quality and fragments length using EAGER pipeline93. Merged reads longer than 30 bp were then mapped to the hg19 human reference genome using BWA v.0.6.2 (parameters: -n 0.01, -l 16500)94; reads with mapping quality below 30 were discarded. PCR duplicates were removed using DeDup34. The analysis of the first shallow whole genome shotgun sequencing showed a total human DNA content of 22.46%, with a deamination rate of 17% on both ends of the reads and an average fragment length of <50 bp (Table 1 and Supplementary Data 6), confirming the presence of authentic ancient DNA molecules.
Data from deep sequencing were used for subsequent analysis after trimming of the 4 bp at both termini of the sequences. Pseudo-haploid genotype was called by randomly choosing an allele at each site using pileupCaller tool95 with base and mapping quality set to 30. A total of 1,080,593 SNPs overlapping with the 1240 K SNPs Array were recovered. Then our data was merged with previously published modern and ancient genotype data collected on 1240 K dataset (AADR v44.3)96,97 available at https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data, plus the addition of useful ancient individuals2,3,40,67,98 and with ancient individuals from the Edmond Data Repository of the Max Planck Society available at https://edmond.mpdl.mpg.de/dataset.xhtml?persistentId=doi:10.17617/3.Y1KJMF99.
Genetic sex was determined comparing the coverages on the sex chromosomes and autosomes. We also calculated the ratio of the reads aligning to the Y chromosome to the reads mapping to either X and Y chromosomes as described in ref. 100. Additionally, the ratio of X chromosome-mapped reads to the autosomal coverage was estimated (Supplementary Data 8)101. In all cases, Le Mura 1 is attributed to male sex.
Authentication of the results
The authenticity of the data was further assessed in silico with several methods. Firstly, post-mortem degradation (i.e., short sequence length and high percentage of nucleotide misincorporation at the end of reads) was evaluated with mapDamage2.0102. Exploiting the male genetic sex attribution of the specimen, nuclear contamination was estimated on deep shotgun data by measuring levels of heterozygosity of the X chromosome103, using the ANGSD pipeline104. Moreover, the degree of mitochondrial DNA contamination was used as an indirect estimate of the nuclear contamination level together with the mtDNA/nDNA ratio105 in shotgun sequencing (see Supplementary Data 6 and 25).
Finally, we applied contamLD method106 based on breakdown of linkage disequilibrium when there are human contaminations in the ancient sample. Only reads with mapping quality ≥30 were considered for the analysis. Linkage pattern was determined using the panel of 1240 K sites downloaded from https://reichdata.hms.harvard.edu/pub/datasets/release/contamLD/. For Le Mura 1 individual, the Finnish in Finland (FIN) panel - representing the present-day European population with the most shared genetic drift, determined by the outgroup-f3 statistics analysis - yielded a contamination estimate of −0.076 (SE 0.013). Negative value indicates a very low contamination level, in agreement with X chromosome contamination estimate obtained (Supplementary Data 7).
Uniparental markers analysis
Raw reads from the mtDNA capture library was pre-processed and mapped against the revised Cambridge Reference Sequence, rCRS (GenBank Accession Number NC_012920) using BWA94 integrated in EAGER (CircularMapper)93, with the same parameters as above. Only reads with mapping quality >30 were used for estimating deamination rates and assembling the endogenous consensus sequence using schmutzi107 setting: --lengthDeam 2, --uselength. For contamination estimation, we first applied ContDeam which computes contamination level using deamination patterns: 7.5% of contamination was obtained (Supplementary Data 25). This estimate is refined in subsequent iterations of the schmutzi program to promote endogenous consensus calling. Additionally, for the consensus sequence reconstruction, quality cutoff was set at 20 and bases with individual likelihood lower than this value were considered as unassigned positions (Ns). In the last iterations, present-day human contamination was evaluated using a database of 256 Eurasian mitochondrial genomes distributed with schmutzi tool107. Complete mitochondrial genome was reconstructed with a depth of coverage of ~120X and with a final modern human contamination level estimated to 3% (Supplementary Data 25). Mitochondrial haplogroup was determined with Haplogrep2108,109 on the command line, using fasta file as input. Le Mura 1 was assigned to the U2’3’4’7’8’9 lineage.
A Maximum Parsimony (MP) tree was constructed to infer the phylogenetic position of the mitochondrial consensus sequence of Le Mura 1. MUSCLE software110 was used to perform a multiple alignment between the 54 present-day mtDNAs111 and 76 Upper Palaeolithic and Mesolithic Eurasian specimens (Supplementary Data 26). A Neanderthal mtDNA genome (Feldhofer 2, FM865408112,) was included as phylogenetic out-group. From the aligned sequences, we removed poly-C regions and AC-indels (positions 16180–16193, 303–315, 515–524 and 573–576) and mutational hotspot (nucleotide position 16519). Maximum Parsimony tree was built using MEGAX113, with Subtree Pruning and Regrafting (SPR) algorithm and 99% partial deletion, as proposed in ref. 114. The phylogeny was tested with the bootstrap method and 1000 replications.
The age node of the U2’3’4’7’8’9 mitochondrial lineage was estimated using a Bayesian statistic approach. The dataset used consist of 54 modern mtDNAs and 67 ancient mitogenomes (including Feldhofer 2 as out-group) dated between 45,000 to 6500 BP (Supplementary Data 27), aligned by MUSCLE software110. After removal of all positions with gaps, a total of 16544 sites were retained and analysed with jModelTest v2.1.7115,116. Hasegawa–Kishino–Yano with invariant sites and gamma distributed rates (HKY + G + I) was identified as the best-fitting model of evolution according to the AIC and BIC criteria and was selected in the Bayesian analysis with BEAST v1.10.4117. Estimated mutation rate (2.57 × 10−8 mutations/site/year, 95% HPD: 2.21 × 10−8–2.94 × 10−8) was set and a Bayesian Skyline coalescent with 20 as group number with a strict molecular clock model were used as priors114. The tip dates of the ancient samples were indicated according to their radiocarbon calibrated BP ages and considering their confidence intervals as uniform priors. Tip dates of zero were selected for present-day mtDNA sequences. MCMC was performed with 50,000,000 iterations and trees were sampled every 1000 generations. Effective sample sizes (ESS) values and the adequate convergence of the MCMC chains were checked using Tracer v1.7.1118. A Maximum Clade Credibility (MCC) tree was generated with TreeAnnotator v.2.6.2119 after discarding the first 10% of iterations as burn-in, visualized and edited by FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Y chromosome haplogroup was inferred from deep shotgun data using Yleaf v2.2 software120. A total of 22,965 Y-SNP markers covered by at least 2 reads (-r) with mapping quality above 30 (-q) were used for haplogroup classification, assigning the most derived lineage. To avoid miscalling, the automated haplogroup predictions were subsequently manually verified, checking the marker positions and their state (ancestral or derived) in the phylogenetic tree in the ISOGG database v.15.73.
Population genetic analysis
The genetic relationship between Le Mura 1 and 115 Eurasian Upper Palaeolithic and Mesolithic hunter-gatherers previously published2,3,37,39–41,59,60,98,121–134 (with at least 30,000 SNPs covered at least once following38) (Supplementary Data 10) was computed by calculating an outgroup f3-statistics (Supplementary Data 10 and 11) using the qp3Pop program from ADMIXTOOLS (v7.0.1)135. The pairwise dissimilarity (1-f3) matrix was applied in the Multi-Dimensional-Scaling (MDS) analysis, computed using the cmdscale() function of R.
The Principal Component Analysis (PCA) was carried out on 58 present-day western Eurasian populations of the 1240 K HO dataset (v44.3) using the smartpca software (EIGENSOFT v7.2.1) with lsqproject and shrinkmode set to YES, to minimize the effect of missing data in ancient individuals. Le Mura 1 and other ancient comparison individuals (Supplementary Data 12) were projected on the calculated PCs (Supplementary Fig. 6a).
The genetic relationships between Le Mura 1 and the other Italian Epigravettian hunter-gatherers were explored by reconstructing phylogeny using pairwise 1- outgroup f3 genetic distance matrix. Individuals of this cultural transect were grouped into three different genetic clusters according to their geographical origin, as suggested in ref. 3. The basal “Group 1” that includes the samples from North-Eastern Italy: Tagliente 2, date to 17 ka; Villabruna, 14 ka and Pradis, 13 ka; “Group 2” consisting of the AC16 (13 ka) from Arene Candide cave located in northeastern Italy the Mesolithic hunter-gatherers from Grotta Continenza (12–9 ka) in central Italy; and the most derived “Group 3” comprising Sicilian hunter-gatherers from Grotta d’Oriente, OC dated to 14 ka, 3 individuals from San Teodoro cave (15–13 ka) year-old, and two early Mesolithic specimens from Uzzo cave, dated to 10 ka. Several f4 statistics (Supplementary Data 13, 14, 20 and 21) were calculated using the qpDstat program from ADMIXTOOLS (v7.0.1)136 with the parameter f4mode: YES on 1240 K panel to increase the number of SNPs used in the analyses. The corresponding standard errors were calculated with the default block jacknife.
Admixture graph was reconstructed using the qpGraph 7365 tool in ADMIXTOOLS 7.0.1 package with default parameters and also using “useallsnps” set to NO (Fig. 5b and Supplementary Data 17).
The software hapROH version 0.4a143 was used to detect ROH segments from the Le Mura 1’s pseudo-haploid genotypes. We used default parameters that are optimized to detect runs of homozygosity longer than 4 centimorgan (cM) on pseudo-haploid data with at least 400 K SNPs on 1240 K panel using 5008 haplotypes of present-day human (from 1000 Genomes) as reference. We report values (Supplementary Data 19) for sROH 4–8 cM and sROH >20 cM that are representative of background relatedness and close kin unions respectively43.
Neanderthal ancestry estimation
We computed D and f4-ratio statistics to test whether Le Mura 1 admixed with archaic humans and to estimate the proportion of Neanderthal ancestry, respectively. We applied ADMIXTOOLS (v7.0.1)136 to genotypes related to the 1240 K SNP panel (v44.3)124,137,138. Other ancient and present-day modern individuals with known ancestry were chosen as terms for comparison due to their close dating with respect to Le Mura 1 (i.e., Tagliente 2 and Villabruna) or their significantly low or high proportion of archaic ancestry (14 Mbuti individuals and Oase 1 for Neanderthal ancestry, 29 Papuan and 2 Australian individuals for Denisovan ancestry) (Supplementary Figs. 10 and 11). Following previous approaches136, the D statistics was calculated in the form D(X, Yoruba; Archaic, Ancestral) where “Archaic” was alternately the high-coverage Neanderthal Vindija 33.19 or Denisova 3 genome139,140, X represents Le Mura 1, and more generally each of the samples/populations tested, chimp was used as ancestral reference, and “Yoruba” refers to 24 Yoruba individuals from the AADR resource96,97 (https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data, version 44.3). The f4-ratio was computed in the form f4(A,O;X,C)/f4(A,O;B,C) where X represents each tested sample/population, A and B correspond to Altai46 and Vindija Neanderthals, respectively, C refers to Yoruba, and chimp was used as outgroup (“O”).
Dating of Neanderthal admixture
We used the method by Moorjani et al.46 for dating of Neanderthal admixture in Le Mura 1 specimen, as well as in all the Italian hunter-gatherers genotyped so far and Ust’-Ishim as a direct comparison (Supplementary Data 22). The method allows to calculate covariance of informative sites for Neanderthal ancestry (the so called “ascertainment0”, supplied with the method46) in bins of 0.001 centiMorgan (cM), and then fits an exponential distribution to the slope of covariance to estimate the date since admixture. Reads with a minimum mapping and base quality of 30 were considered. We used genetic distances from the shared African American map93. The exponential distribution was fit with the rexpfit.r script from ADMIXTOOLS136 with the affine term option to the range 0.02–1 cM for Italian individuals, while the extended range of 0.02–10 cM was chosen for Ust’-Ishim, yielding more plausible estimates for a sample closer in time to the Neanderthal admixture, as noted before46. Finally, we computed the corrected date of gene flow both in generations and years (assuming 29 years per generation47) by applying the genetic map correction and standard error estimation, as previously reported46,141.
Neanderthal and Denisova introgressed segments along the genome
We used admixfrog (v0.6.2)142 to infer Neanderthal introgressed regions along the genome of the Le Mura 1 individual (Supplementary Fig. 4). As potential sources of ancestry we set the diploid high-coverage Vindija 33.19, Altai Neanderthal, Denisova 3 genomes44,140, and the genomes of the African 1000 Genomes individuals (108 Yoruba from phase 3)143. We also used ancestral alleles derived from Ensembl Compara 143,144 release 59, based on a 6-way primate alignment. The analysis was focused on the “Archaic admixture” panel124, containing ~1.7 million SNPs informative of Neanderthal and Denisovan ancestry. The reference file was generated specifying the African American recombination map145 for re-scaling physical distances (in base pairs) into genetic distances (in cM). The input file was produced by admixfrog-bam, setting deamination cutoff to 4, minimum mapping quality to 30, minimum length size to 30. Admixfrog was then run with parameters “--run-penalty 0.4 --bin-size 10,000”. When overlapping introgressed regions were called for the heterozygous and homozygous state, the longer region was chosen.
Phenotypic SNPs analysis
Variant calling of ~1200 SNPs related to phenotypic traits including pigmentation, metabolism, and blood type, was performed using ANGSD (v0.933)135. Targeted sites were chosen among those annotated in SNPedia49, with a unique variant allele reported in dbSNP (build 153)50, including 41 SNPs from the HIrisPlex-S panel49 (Supplememtary Data 23). Allele frequencies were estimated from genotype likelihoods according to the GATK model135, assuming both major and minor alleles to be known from dbSNP50. Genotype calling was based on calculating the genotype posterior probability using an estimate of the population frequency as prior. We set minimum mapping and base quality to 30, and posterior probability cutoff to 99%. Annotations for each SNP identified in the Le Mura 1 sample was retrieved from SNPedia49 using the R package SNPediaR (v1.16.0)146. Moreover, we used the HIrisPlex-S51,147–149 online webtool to predict eye, hair, and skin colour from called genotypes.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was funded by PRIN grant number 20177PJ9XF of the Italian Ministry of Research “1000 Ancient Italian Genomes: Evidence from ancient biomolecules for unravelling past human population Dynamics (AGED)” to D.C., and by the European Union grant – Next Generation EU – PNRR M4C2 – Investimento 1.3. PE5-Change. A.N. received funding from the European Research Council (ERC) under the European Union’s Horizon Europe Research and Innovation Programme (Grant Agreement no. 101077348 — MOTHERS; https://erc-mothers.eu/). S.T. received funding from the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (grant agreement No. 803147 RESOLUTION, https://site.unibo.it/resolution-erc/en). F.L. is supported by the European Union’s Horizon Europe Research and Innovation programme under the Marie Skłodowska-Curie IF action (Grant Agreement no. 101104566 — AROUSE). FIERCE is financially supported by the Wilhelm and Else Heraeus Foundation and by the Deutsche Forschungsgemeinschaft (DFG: INST 161/921-1 FUGG, INST 161/923-1 FUGG and INST 161/1073-1 FUGG), which is gratefully acknowledged. This is FIERCE contribution No. 169. We would like to extend our gratitude to the Sovrintendenza di Bari for granting the necessary authorizations to conduct the analyses presented in this study. Additionally, we express our appreciation to the University of Siena for financing the extensive years of excavation at the site, which provided the foundational data for our research.
Author contributions
D.C., S.B., S.R., O.A.H. and A.Mod. conceived the study; M.C. excavated the remains; S.R. and L.M. focused on the study and restoration of the osteological material; A.Mod., S.V.,V.Z. and M.L. performed or supervised laboratory work; C.C. and M.A.D. performed bioinformatics and population genetics analysis, with input from C.P., H.Y., M.L., A.Mod., and D.C.; S.T. performed the radiocarbon analysis; O.A.H. performed the palaeohistological analysis, with input from A.N. and L.B.; W.M. performed the geochemical analysis; W.M., F.L., A.N., and L.B. elaborated and interpreted the geochemical analyses; A.V. and M.Rom. restored the dental specimens sectioned for palaeohistological analysis; G.D.D. and F.Ba. conducted µCT scanning of the dental specimens; G.O. performed the morphological analysis of the deciduous incisor and permanent molar crowns; F.Bo., M.Ros., A.Mor., and A.R., provided valuable contextual background; O.A.H. and A.Mod. primarily wrote the manuscript, with significant inputs from S.B., M.L., D.C., C.P., A.N., L.B., W.M., F.L., S.T. and all other co-authors; O.A.H., A.Mod. C.C., M.A.D., F.L., S.R., V.Z., S.V., A.V., M.Rom., H.Y., F.Bo., L.M., M.Ros., G.D.D., F.Ba., G.O., E.B., E.D., A.Mor., A.R., S.T., W.M., M.C., A.N., C.P., M.L., L.B., S.B., D.C. reviewed and revised the manuscript; The senior authors of this study are S.R., S.B., and D.C.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All software packages used for analysis are publicly available and cited in the Methods section or in the Supplementary Information. Source Data for Figs. 4, 5, and 6 can be found in the Supplementary Data file. Source Data for Supplementary Figs. 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 and 12 can be found in the Supplementary Data file. Additional source data are provided as a Source Data file. The specimen is deposited at the University of Siena (Siena, Italy) with free access following a research application by contacting Stefano Ricci at stefano.ricci@unisi.it. The genomic data (BAM format) generated in this study have been deposited in the European Nucleotide Archive database under accession code PRJEB66279. The genotype data of the previously published samples used for analyses were downloaded from the Edmond Data Repository of the Max Planck Society (https://edmond.mpdl.mpg.de/dataset.xhtml?persistentId=doi:10.17617/3.Y1KJMF) and from the Allen Ancient DNA Resource of the David Reich Lab (https://reichdata.hms.harvard.edu/pub/datasets/amh_repo/curated_releases/index_v44.3.html). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Owen Alexander Higgins, Alessandra Modi, Costanza Cannariato.
Contributor Information
Owen Alexander Higgins, Email: owenalexanderhiggins@gmail.com.
Alessandra Modi, Email: alessandra.modi@unifi.it.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51150-x.
References
- 1.Lambeck, K., Rouby, H., Purcell, A., Sun, Y. & Sambridge, M. Sea level and global ice volumes from the Last Glacial Maximum to the Holocene. Proc. Natl Acad. Sci. USA111, 15296–15303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bortolini, E. et al. Early Alpine occupation backdates westward human migration in Late Glacial Europe. Curr. Biol.31, 2484–2493.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Posth, C. et al. Palaeogenomics of Upper Palaeolithic to Neolithic European hunter-gatherers. Nature615, 117–126 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lugli, F. et al. Strontium and stable isotope evidence of human mobility strategies across the Last Glacial Maximum in southern Italy. Nat. Ecol. Evol.3, 905–911 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Wren, C. D. & Burke, A. Habitat suitability and the genetic structure of human populations during the Last Glacial Maximum (LGM) in Western Europe. PLoS One10.1371/journal.pone.0217996 (2019). [DOI] [PMC free article] [PubMed]
- 6.Ruiz-Redondo, A. et al. Mid and Late Upper Palaeolithic in the Adriatic Basin: Chronology, transitions and human adaptations to a changing landscape. Quat. Sci. Rev.276, 107319 (2022). [Google Scholar]
- 7.Modi, A. et al. Paleogenetic and morphometric analysis of a Mesolithic individual from Grotta d’Oriente: An oldest genetic legacy for the first modern humans in Sicily. Quat. Sci. Rev.248, 106603 (2020). [Google Scholar]
- 8.Villalba-Mouco, V. et al. Survival of Late Pleistocene Hunter-Gatherer Ancestry in the Iberian Peninsula. Curr. Biol.29, 1169–1177.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Villalba-Mouco, V. et al. A 23,000-year-old southern Iberian individual links human groups that lived in Western Europe before and after the Last Glacial Maximum. Nat. Ecol. Evol.7, 597–609 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lugli, F. et al. Tracing the mobility of a Late Epigravettian (~ 13 ka) male infant from Grotte di Pradis (Northeastern Italian Prealps) at high-temporal resolution. Sci. Rep.10.1038/s41598-022-12193-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nava, A. et al. Early life of Neanderthals. Proc. Natl Acad. Sci. USA117, 28719–28726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nava, A. et al. Virtual histological assessment of the prenatal life history and age at death of the Upper Paleolithic fetus from Ostuni (Italy). Sci. Rep.7, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teschler-Nicola, M. et al. Ancient DNA reveals monozygotic newborn twins from the Upper Palaeolithic. Commun. Biol.3, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calattini, M. Scoperta di una sepoltura paleolitica a Grotta delle Mura (BA). Rassegna di archeologia: preistorica e protostorica 37–45 10.1400/248076 (2002).
- 15.Stloukal, M. & Hanakova, H. Die Longe der langskonochen altslavischer Bevolkerungen unter besonderer Bercksichtigung von Wachstumsfragen. Homo29, 53–69 (1978). [Google Scholar]
- 16.AlQahtani, S. J., Hector, M. P. & Liversidge, H. M. Brief communication: The London atlas of human tooth development and eruption. Am. J. Phys. Anthropol.142, 481–490 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Bronk Ramsey, C. Bayesian analysis of radiocarbon dates. Radiocarbon51, 337–360 (2009). [Google Scholar]
- 18.Reid, D. J. & Dean, M. C. Variation in modern human enamel formation times. J. Hum. Evol.50, 329–346 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Hillson, S. Dental Anthropology (Cambridge University Press, 1996).
- 20.Molnar, S. Human tooth wear, tooth function and cultural variability. Am. J. Phys. Anthropol.34, 175–189 (1971). [DOI] [PubMed] [Google Scholar]
- 21.Hutchinson, D. L. & Spencer Larsen, C. Determination of Stress Episode Duration from Linear Enamel Hypoplasias: A Case Study from St. Catherines Island, Georgia. Hum. Biol.60, 93–110 (1988). [PubMed] [Google Scholar]
- 22.Moorrees, C. F. A., Fanning, E. A. & Hunt, E. E. Formation and resorption of three deciduous teeth in children. Am. J. Phys. Anthropol.21, 205–213 (1963). [DOI] [PubMed] [Google Scholar]
- 23.Sabel, N. et al. Neonatal lines in the enamel of primary teeth—A morphological and scanning electron microscopic investigation. Arch. Oral. Biol.53, 954–963 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Nava, A., Frayer, D. W. & Bondioli, L. Longitudinal analysis of the microscopic dental enamel defects of children in the Imperial Roman community of Portus Romae (necropolis of Isola Sacra, 2nd to 4th century CE, Italy). J. Archaeol. Sci. Rep.23, 406–415 (2019). [Google Scholar]
- 25.Reid, D. J., Beynon, A. D. & Ramirez Rozzi, F. V. Histological reconstruction of dental development in four individuals from a medieval site in Picardie, France. J. Hum. Evol.35, 463–477 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Nava, A. et al. New regression formula to estimate the prenatal crown formation time of human deciduous central incisors derived from a Roman Imperial sample (Velia, Salerno, Italy, I-II cent. CE). PLoS One12, e0180104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahoney, P. Incremental enamel development in modern human deciduous anterior teeth. Am. J. Phys. Anthropol.147, 637–651 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Dean, C. Extension rates and growth in tooth height of modern human and fossil hominin canines and molars. Front. Oral Biol. 13, 68–73 (2009). [DOI] [PubMed]
- 29.Mahoney, P. Dental fast track: Prenatal enamel growth, incisor eruption, and weaning in human infants. Am. J. Phys. Anthropol.156, 407–421 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Guatelli-Steinberg, D., Floyd, B. A., Dean, M. C. & Reid, D. J. Enamel extension rate patterns in modern human teeth: Two approaches designed to establish an integrated comparative context for fossil primates. J. Hum. Evol.63, 475–486 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Ortiz, A., Skinner, M. M., Bailey, S. E. & Hublin, J. J. Carabelli’s trait revisited: An examination of mesiolingual features at the enamel-dentine junction and enamel surface of Pan and Homo sapiens upper molars. J. Hum. Evol.63, 586–596 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Moorrees, C. F. A., Fanning, E. A. & Hunt, E. E. Age variation of formation stages for ten permanent teeth. J. Dent. Res.42, 1490–1502 (1963). [DOI] [PubMed] [Google Scholar]
- 33.Mahoney, P. Intraspecific variation in M1 enamel development in modern humans: implications for human evolution. J. Hum. Evol.55, 131–147 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Aris, C. Enamel growth rate variation of inner, mid, and outer enamel regions between select permanent tooth types across five temporally distinct British samples. Arch Oral Biol137, (105394, 2022). [DOI] [PubMed]
- 35.Antoine, D., Hillson, S. & Dean, M. C. The developmental clock of dental enamel: A test for the periodicity of prism cross-striations in modern humans and an evaluation of the most likely sources of error in histological studies of this kind. J. Anat.214, 45–55 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawyer, S., Krause, J., Guschanski, K., Savolainen, V. & Pääbo, S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS One7, (e34131, 2012). [DOI] [PMC free article] [PubMed]
- 37.Jones, E. R. et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun.6, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu, Q. et al. The genetic history of Ice Age Europe. Nature534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scorrano, G. et al. Bioarchaeological and palaeogenomic portrait of two Pompeians that died during the eruption of Vesuvius in 79 AD. Sci. Rep.12, 1–12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Catalano, G. et al. A mitogenome sequence of an Equus hydruntinus specimen from Late Quaternary site of San Teodoro Cave (Sicily, Italy). Quat. Sci. Rev.236, 106280 (2020). [Google Scholar]
- 42.Antonioli, F. et al. Timing of the emergence of the Europe-Sicily bridge (40-17 cal ka BP) and its implications for the spread of modern humans. Geol. Soc. Spec. Publ.411, 111–144 (2016). [Google Scholar]
- 43.Ringbauer, H., Novembre, J. & Steinrücken, M. Parental relatedness through time revealed by runs of homozygosity in ancient DNA. Nat. Commun.12, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prüfer, K. et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petr, M., Pääbo, S., Kelso, J. & Vernot, B. Limits of long-term selection against Neandertal introgression. Proc. Natl Acad. Sci. USA116, 1639–1644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moorjani, P., Amorim, C. E. G., Arndt, P. F. & Przeworski, M. Variation in the molecular clock of primates. Proc. Natl Acad. Sci. USA113, 10607–10612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fenner, J. N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol.128, 415–423 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Coll Macià, M., Skov, L., Peter, B. M. & Schierup, M. H. Different historical generation intervals in human populations inferred from Neanderthal fragment lengths and mutation signatures. Nat. Commun.12, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cariaso, M. & Lennon, G. SNPedia: A wiki supporting personal genome annotation, interpretationand analysis. Nucleic Acids Res.40, (D1308-12, 2012). [DOI] [PMC free article] [PubMed]
- 50.Sherry, S. T. et al. DbSNP: The NCBI database of genetic variation. Nucleic Acids Res.29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaitanya, L. et al. The HIrisPlex-S system for eye, hair and skin colour prediction from DNA: Introduction and forensic developmental validation. Forensic. Sci. Int. Genet.35, 123–135 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Allentoft, M. E. et al. Population Genomics of Bronze Age Eurasia. Nature522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Christiaans, I. et al. Founder mutations in hypertrophic cardiomyopathy patients in the Netherlands. Neth. Heart J.18, 254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richard, P. et al. Hypertrophic Cardiomyopathy. Circulation107, 2227–2232 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Tompkins, R. L. Human population variability in relative dental development. Am. J. Phys. Anthropol.99, 79–102 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Hodgkins, J. et al. An infant burial from Arma Veirana in northwestern Italy provides insights into funerary practices and female personhood in early Mesolithic Europe. Sci Rep8, (23735, 2021). [DOI] [PMC free article] [PubMed]
- 57.Brenner, B., Seebohm, B., Tripathi, S., Montag, J. & Kraft, T. Familial hypertrophic cardiomyopathy: Functional variance among individual cardiomyocytes as a trigger of fhc-phenotype development. Front. Physiol.5, (392, 2014). [DOI] [PMC free article] [PubMed]
- 58.Bagnall, R. D. et al. A Prospective Study of Sudden Cardiac Death among Children and Young Adults. N. Engl. J. Med.374, 2441–2452 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Olalde, I. et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature507, 225–228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Günther, T. et al. Population genomics of Mesolithic Scandinavia: Investigating early postglacial migration routes and high-latitude adaptation. PLoS Biol.16, e2003703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lévi-Strauss, C. Les Structures Élémentaires de La Parenté (Mouton de Gruyter, 1967).
- 62.Bittles, A. H. Consanguinity, genetic drift, and genetic diseases in populations with reduced numbers of founders. in Vogel and Motulsky’s Human Genetics: Problems and Approaches (Fourth Edition) 507–528 (Springer-Verlag, 2010). 10.1007/978-3-540-37654-5_19.
- 63.Ceballos, F. C. et al. Human inbreeding has decreased in time through the Holocene. Curr. Biol.31, 3925–3934.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Palma di Cesnola, A. Il Paleolitico Superiore in Italia: Introduzione (Allo Studio, 1993).
- 65.Catalano, G. et al. Late Upper Palaeolithic hunter-gatherers in the Central Mediterranean: New archaeological and genetic data from the Late Epigravettian burial Oriente C (Favignana, Sicily). Quat. Int.537, 24–32 (2020). [Google Scholar]
- 66.Modi, A. et al. More data on ancient human mitogenome variability in Italy: new mitochondrial genome sequences from three Upper Palaeolithic burials. Ann. Hum. Biol.48, 213–222 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Yu, H. et al. Genomic and dietary discontinuities during the Mesolithic and Neolithic in Sicily. iScience25, 104244 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu, Q. et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol.23, 553–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Naudinot, N. et al. Between Atlantic and Mediterranean: Changes in technology during the Late Glacial in Western Europe and the climate hypothesis. Quat. Int.428, 33–49 (2017). [Google Scholar]
- 70.Boschin, F. et al. The first evidence for Late Pleistocene dogs in Italy. Sci. Rep.10, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Šešelj, M. Brief communication: An analysis of dental development in Pleistocene Homo using skeletal growth and chronological age. Am. J. Phys. Anthropol.163, 531–541 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Fewlass, H. et al. Pretreatment and gaseous radiocarbon dating of 40-100 mg archaeological bone. Sci. Rep.9, 5342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Talamo, S., Fewlass, H., Maria, R. & Jaouen, K. ‘Here we go again’: the inspection of collagen extraction protocols for 14 C dating and palaeodietary analysis. Sci. Technol. Archaeol. Res.7, 62–77 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Longin, R. New method of collagen extraction for radiocarbon dating. Nature230, 241–242 (1971). [DOI] [PubMed] [Google Scholar]
- 75.Brown, T. A., Nelson, D. E., Vogel, J. S. & Southon, J. R. Improved Collagen Extraction by Modified Longin Method. Radiocarbon30, 171–177 (1988). [Google Scholar]
- 76.Van Klinken, G. J. Bone collagen quality indicators for palaeodietary and radiocarbon measurements. J. Archaeol. Sci.26, 687–695 (1999). [Google Scholar]
- 77.Wacker, L., Němec, M. & Bourquin, J. A revolutionary graphitisation system: Fully automated, compact and simple. Nucl. Instrum. Methods Phys. Res B268, 931–934 (2010). [Google Scholar]
- 78.Kromer, B., Lindauer, S., Synal, H. A. & Wacker, L. MAMS - A new AMS facility at the Curt-Engelhorn-Centre for Achaeometry, Mannheim, Germany. Nucl. Instrum. Methods Phys. Res. B294, 11–13 (2013). [Google Scholar]
- 79.Reimer, P. J. et al. The IntCal20 Northern Hemisphere Radiocarbon Age Calibration Curve (0-55 cal kBP). Radiocarbon62, 725–757 (2020). [Google Scholar]
- 80.Birch, W. & Dean, M. C. A method of calculating human deciduous crown formation times and of estimating the chronological ages of stressful events occurring during deciduous enamel formation. J. Forensic Leg. Med.22, 127–144 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed]
- 82.Müller, W. et al. Enamel mineralization and compositional time-resolution in human teeth evaluated via histologically-defined LA-ICPMS profiles. Geochim. Cosmochim. Acta255, 105–126 (2019). [Google Scholar]
- 83.Müller, W. & Anczkiewicz, R. Accuracy of laser-ablation (LA)-MC-ICPMS Sr isotope analysis of (bio)apatite-a problem reassessed. J. Anal. Spectrom.31, 259–269 (2016). [Google Scholar]
- 84.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2023).
- 85.Cleveland, W. S., Grosse, E. & Shyu, W. M. Local Regression Models. in Statistical Models in S (eds. Chambers, J. M. & Hastie, T. J.) (Routledge, 1992). 10.1201/9780203738535-8.
- 86.Vazzana, A. et al. High-accuracy methodology for the integrative restoration of archaeological teeth by using reverse engineering techniques and rapid prototyping. J Archaeol Sci Rep44, (103511, 2022).
- 87.Willerslev, E. & Cooper, A. Ancient DNA. Proc. R. Soc. B Biol. Sci.272, 16 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gilbert, M. T. P., Bandelt, H. J., Hofreiter, M. & Barnes, I. Assessing ancient DNA studies. Trends Ecol. Evol.20, 541–544 (2005). [DOI] [PubMed] [Google Scholar]
- 89.Kircher, M., Sawyer, S. & Meyer, M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res.40, e3 (2012). [DOI] [PMC free article] [PubMed]
- 90.Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc.5, pdb.prot5448 (2010). [DOI] [PubMed]
- 91.Rohland, N., Harney, E., Mallick, S., Nordenfelt, S. & Reich, D. Partial uracil – DNA – glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. B Biol. Sci.370, 20130624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maricic, T., Whitten, M. & Pääbo, S. Multiplexed DNA Sequence Capture of Mitochondrial Genomes Using PCR Products. PLoS One5, e14004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peltzer, A. et al. EAGER: efficient ancient genome reconstruction. Genome Biol.17, 60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schiffels, S. GitHub - stschiff/sequenceTools. https://github.com/stschiff/sequenceTools (2020).
- 96.Mallick, S. et al. The Allen Ancient DNA Resource (AADR) a curated compendium of ancient human genomes. Sci. Data11, 1–10 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mallick, S. & Reich, D. The Allen Ancient DNA Resource (AADR): A curated compendium of ancient human genomes. David Reich Lab Dataverse, V810.7910/DVN/FFIDCW (2023). [DOI] [PMC free article] [PubMed]
- 98.Prüfer, K. et al. A genome sequence from a modern human skull over 45,000 years old from Zlatý kůň in Czechia. Nat. Ecol. Evol.5, 820–825 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu, H. Paleogenomics of Upper Paleolithic to Neolithic European hunter-gatherers. EDMOND, V310.17617/3.Y1KJMF (2022).
- 100.Skoglund, P., Storå, J., Götherström, A. & Jakobsson, M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci.40, 4477–4482 (2013). [Google Scholar]
- 101.Mittnik, A., Wang, C.-C., Svoboda, J. & Krause, J. A Molecular Approach to the Sexing of the Triple Burial at the Upper Paleolithic Site of Dolní Věstonice. PLoS One11, e0163019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jónsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. F. & Orlando, L. MapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rasmussen, M. et al. An aboriginal Australian genome reveals separate human dispersals into Asia. Science334, 94–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Korneliussen, T. S., Albrechtsen, A. & Nielsen, R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinforma.15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Furtwängler, A. et al. Ratio of mitochondrial to nuclear DNA affects contamination estimates in ancient DNA analysis. Sci. Rep.8, 14075 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nakatsuka, N. et al. ContamLD: Estimation of ancient nuclear DNA contamination using breakdown of linkage disequilibrium. Genome Biol.21, 199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Renaud, G., Slon, V., Duggan, A. T. & Kelso, J. Schmutzi: Estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol.16, 224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weissensteiner, H. et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res.44, W58–W63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van Oven, M. PhyloTree Build 17: Growing the human mitochondrial DNA tree. Forensic Sci Int Genet Suppl Ser5, e392–e394 (2015).
- 110.Edgar, R. C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res.32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ingman, M., Kaessmann, H., Pääbo, S. & Gyllensten, U. Mitochondrial genome variation and the origin of modem humans. Nature408, 708–713 (2000). [DOI] [PubMed] [Google Scholar]
- 112.Briggs, A. W. et al. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science325, 318–321 (2009). [DOI] [PubMed] [Google Scholar]
- 113.Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms | Molecular Biology and Evolution | Oxford Academic. Mol. Biol. Evol.35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Posth, C. et al. Pleistocene mitochondrial genomes suggest a single major dispersal of non-africans and a late glacial population turnover in Europe. Curr. Biol.26, 827–833 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Guindon, S. & Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol.52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- 116.Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods9, 10.1038/nmeth.2109 (2012). [DOI] [PMC free article] [PubMed]
- 117.Suchard, M. A. et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol.4, vey016 (2018). [DOI] [PMC free article] [PubMed]
- 118.Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol.67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bouckaert, R. et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol15, e1006650 (2019). [DOI] [PMC free article] [PubMed]
- 120.Ralf, A., Montiel González, D., Zhong, K. & Kayser, M. Yleaf: Software for Human Y-Chromosomal Haplogroup Inference from Next-Generation Sequencing Data. Mol. Biol. Evol.35, 1291–1294 (2018). [DOI] [PubMed] [Google Scholar]
- 121.Gamba, C. et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun.5, 5257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mathieson, I. et al. The genomic history of southeastern Europe. Nature555, 197–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Raghavan, M. et al. Upper palaeolithic Siberian genome reveals dual ancestry of native Americans. Nature505, 87–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fu, Q. et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fu, Q. et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature514, 445–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sikora, M. et al. Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers. Science (1979)358, 659–662 (2017). [DOI] [PubMed] [Google Scholar]
- 127.Skoglund, P. et al. Genomic diversity and admixture differs for stone-age Scandinavian foragers and farmers. Science (1979)344, 747–750 (2014). [DOI] [PubMed] [Google Scholar]
- 128.Antonio, M. L. et al. Ancient Rome: A genetic crossroads of Europe and the Mediterranean. Science366, 708–714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Seguin-Orlando, A. et al. Genomic structure in Europeans dating back at least 36,200 years. Science346, 1113–1118 (2014). [DOI] [PubMed] [Google Scholar]
- 130.González-Fortes, G. et al. Paleogenomic Evidence for Multi-generational Mixing between Neolithic Farmers and Mesolithic Hunter-Gatherers in the Lower Danube Basin. Curr. Biol.27, 1801–1810.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.De Barros Damgaard, P. et al. 137 ancient human genomes from across the Eurasian steppes. Nature557, 369–374 (2018). [DOI] [PubMed] [Google Scholar]
- 132.Mathieson, I. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mittnik, A. et al. The genetic prehistory of the Baltic Sea region. Nat. Commun.9, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jones, E. R. et al. The Neolithic Transition in the Baltic Was Not Driven by Admixture with Early European Farmers. Curr. Biol.27, 576–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McKenna, A. et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res.20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Patterson, N. et al. Ancient admixture in human history. Genetics192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haak, W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature522, 207–211 (2015). [DOI] [PMC free article] [PubMed]
- 138.Lazaridis, I. et al. Genomic insights into the origin of farming in the ancient Near East. Nature536, 419–424 (2016). [DOI] [PMC free article] [PubMed]
- 139.Mafessoni, F. et al. A high-coverage neandertal genome from chagyrskaya cave. Proc. Natl Acad. Sci. USA117, 15132–15136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sankararaman, S., Patterson, N., Li, H., Pääbo, S. & Reich, D. The Date of Interbreeding between Neandertals and Modern Humans. PLoS Genet.8, e1002947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Peter, B. M. 100,000 years of gene flow between Neandertals and Denisovans in the Altai mountains. bioRxiv, 10.1101/2020.03.13.990523 (2020).
- 143.Auton, A. et al. A global reference for human genetic variation. The 1000 Genomes Project Consortium. Nature526, 68-74 (2015). [DOI] [PMC free article] [PubMed]
- 144.Herrero, J. et al. Ensembl comparative genomics resources. Database2016, bav096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hinch, A. G. et al. The landscape of recombination in African Americans. Nature476, 170–175 (2011). [DOI] [PMC free article] [PubMed]
- 146.Montaner, D. SNPediaR: Query data from SNPedia. R package version 1.30.0, https://github.com/genometra/SNPediaR (2024).
- 147.Walsh, S. et al. DNA-based eye colour prediction across Europe with the IrisPlex system. Forensic Sci. Int. Genet.6, 330–340 (2012). [DOI] [PubMed] [Google Scholar]
- 148.Walsh, S. et al. Developmental validation of the HIrisPlex system: DNA-based eye and hair colour prediction for forensic and anthropological usage. Forensic Sci. Int. Genet.9, 150–161 (2014). [DOI] [PubMed] [Google Scholar]
- 149.Walsh, S. et al. Global skin colour prediction from DNA. Hum. Genet.136, 847–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All software packages used for analysis are publicly available and cited in the Methods section or in the Supplementary Information. Source Data for Figs. 4, 5, and 6 can be found in the Supplementary Data file. Source Data for Supplementary Figs. 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 and 12 can be found in the Supplementary Data file. Additional source data are provided as a Source Data file. The specimen is deposited at the University of Siena (Siena, Italy) with free access following a research application by contacting Stefano Ricci at stefano.ricci@unisi.it. The genomic data (BAM format) generated in this study have been deposited in the European Nucleotide Archive database under accession code PRJEB66279. The genotype data of the previously published samples used for analyses were downloaded from the Edmond Data Repository of the Max Planck Society (https://edmond.mpdl.mpg.de/dataset.xhtml?persistentId=doi:10.17617/3.Y1KJMF) and from the Allen Ancient DNA Resource of the David Reich Lab (https://reichdata.hms.harvard.edu/pub/datasets/amh_repo/curated_releases/index_v44.3.html). Source data are provided with this paper.