Summary
Anaplastic lymphoma kinase (ALK) inhibitor crizotinib has dramatic effect in non-small cell lung cancer patients with ALK rearrangement. However, most patients eventually develop resistance. To discover therapeutic targets to overcome crizotinib resistance (CR), we generated patient-derived xenograft CR mice and subjected them to phosphorylation profiling, together with CR mice treated with ASP3026 or alectinib. We identified 100 proteins with different phosphorylation status in CR mice. Among them, AXL phosphorylation was increased in CR mice, which could not be reversed by ASP3026 or alectinib. Importantly, the combined treatment of crizotinib and AXL inhibitor in CR mice significantly inhibited tumor growth, compared to crizotinib alone. We also found that SHC1 phosphorylation was increased in CR mice and SHC1 knockdown sensitized ALK-driven cells to crizotinib. Our study provides a new view of signaling pathways leading to CR, suggesting AXL and SHC1 as potential targets for combination therapy to overcome CR.
Subject areas: Drugs, Molecular biology, Cancer
Graphical abstract

Highlights
-
•
The first large-scale phosphoproteomic profiling on crizotinib-resistant PDX mice
-
•
100 proteins of phosphorylation status were changed in crizotinib-resistant mice
-
•
Phosphorylation of AXL and SHC1 was highly increased in crizotinib-resistant mice
-
•
The combining of AXL inhibitor or siSHC1 with crizotinib enhanced crizotinib effect
Drugs; Molecular biology; Cancer
Introduction
Lung cancer is a leading cause of cancer-related death worldwide, and non-small cell lung cancer (NSCLC) accounts for approximately 85% of all new lung cancer cases. The identification of ALK rearrangement in NSCLC patients has sparked the development of ALK inhibitors for the treatment of ALK-rearranged NSCLC.1 Crizotinib, the first-generation of ALK inhibitor, has been approved by the US Food and Drug Administration (FDA) to treat ALK-driven NSCLC patients.2,3 However, most patients inevitably acquired drug resistance. Around 30% of crizotinib-resistant NSCLC patients develop secondary resistance-related mutations in the tyrosine kinase domain of ALK.4,5 Therefore, the second-generation ALK inhibitors, ceritinib, alectinib, and brigatinib, have been developed to overcome crizotinib resistance.6 Alectinib showed substantial efficacy against crizotinib-resistant ALK secondary mutations including the gatekeeper L1196M in vitro and in vivo; however, it was less effective against the G1202R mutant.7,8,9 A third-generation ALK inhibitor, lorlatinib, is developed and effective against all known clinically acquired ALK mutations, including the highly resistant G1202R mutant.10,11 Besides the aforementioned inhibitors, there are other ALK inhibitors that are under development. One example is ASP3026, a novel ALK inhibitor with antitumor activity against various solid tumors including NSCLC. ASP3026 has inhibitory effects on multiple kinases such as ALK, ROS1, and SRC which differ from the kinase inhibition profile of crizotinib.12,13 Recently, phase 1 studies both in the US and in Japan showed that NSCLC patients bearing ALK rearrangement might benefit from ASP3026, with a tolerable safety profile.14
There is limited understanding of how acquired resistance develops. The possible mechanisms of resistance to crizotinib include (1) ALK signaling-dependent activation, such as ALK secondary mutations, and copy-number amplification, the most common point mutations being L1196M and G1269A15,16,17,18, and (2) activation of alternative survival signaling pathways or bypass pathways, including epidermal growth factor receptor (EGFR),19,20,21 HER2,22 KRAS,23 SRC,24,25 and c-Met26 activations. Other mechanisms such as epithelial-mesenchymal transition (EMT) have also been implicated in crizotinib resistance.27,28 Recently, ALK signaling-independent AXL overexpression was shown to be involved in the acquired resistance to first- and second-generation ALK inhibitors in an NSCLC cell line harboring EML4-ALK.29 However, the exact mechanism of crizotinib resistance is still unknown in a large proportion of patients.
Previous studies have shown that the responses of patient-derived xenograft (PDX) mouse models to tyrosine kinase inhibitors (TKIs) closely approximate the clinical outcomes observed in the donors who were treated with similar TKIs.24 In this study, we generated a PDX mouse model of crizotinib resistance, as well as a crizotinib-resistant NSCLC cell line, and used phosphoproteomics to profile the up-regulated phosphorylated proteins in crizotinib-resistant PDX mice to search for the new molecules involved in the development of crizotinib resistance.
Results
Establishment of crizotinib-resistant PDX mouse model
To investigate the mechanisms underlying crizotinib resistance, we established the crizotinib-resistant PDX mouse model with human ALK-rearranged lung cancer tissues. A total of 630 NSCLC tissue samples collected from the Second Xiangya Hospital were screened by western blotting and multiple reverse-transcription PCR (RT-PCR) for ALK overexpression, and 20 of them were positive for ALK rearrangement (Figure 1A; Table S2). cDNAs synthesized from 20 positive samples for ALK rearrangement were subjected to sequencing analysis, and 3 EML4-ALK variants were identified (Figure 1B; Table S2). Among them, EML4-ALK variant 3 was reported to be the most frequent ALK variant associated with drug resistance.30 Therefore, tumor tissues from patient T5 (carrying EML4-ALK variant 3) were implanted subcutaneously into 3 severe combined immunodeficiency (SCID) Beige mice (P1). Only one mouse had developed tumors. Tumor tissues from this mouse were then cut into 1–2 mm fragments and implanted into new hosts for next passages (P2–P5, Figure S1). mRNAs extracted from tumor tissues of patient T5 and PDX mice were subjected to RT-PCR. The transcript rearrangement of EML4 exon 6 to ALK exon 20 (variant 3) was identified in both the patient’s tumor and PDX tumor tissues (Figures 1B and 1C). Furthermore, the histological characteristics of the patient’s tumor were highly similar to those of the PDX tumors (Figure 1D).
Figure 1.
ALK protein expression and the type of EML4-ALK variant in tumor tissues from NSCLC patients and PDX mice
(A) Western blot analysis of protein lysates from tumor tissues of representative NSCLC patients using antibody against ALK and GAPDH was used as a loading control. Positions of EML4-ALK variants are indicated.
(B) Sequencing analysis of EML4-ALK variants in tumor tissues from representative NSCLC patients. ALK fragments were amplified using primer pairs listed in Table S1. The sequence characteristics of different EML4-ALK variants are indicated by arrows, which mark the starting point of exon 20 of ALK (V1: EML4-ALK variant 1, V2: EML4-ALK variant 2, V3: EML4-ALK variant 3).
(C) RT-PCR analysis of EML4-ALK (variant 3) expression in tumor tissues from the PDX mice (P1–P5) and donor patient (T5, P0). The PCR primers used to amplify the EML4 and ALK cDNA are listed in Table S1. Amplification of GAPDH was used as a qualitative control of the cDNA samples.
(D) Immunohistochemical staining of tumor tissues from the patient (T5, P0) and PDX mice (P1–P5) with ALK antibody (ALK (D5F3) XP Rabbit mAb). The ALK protein was stained in red.
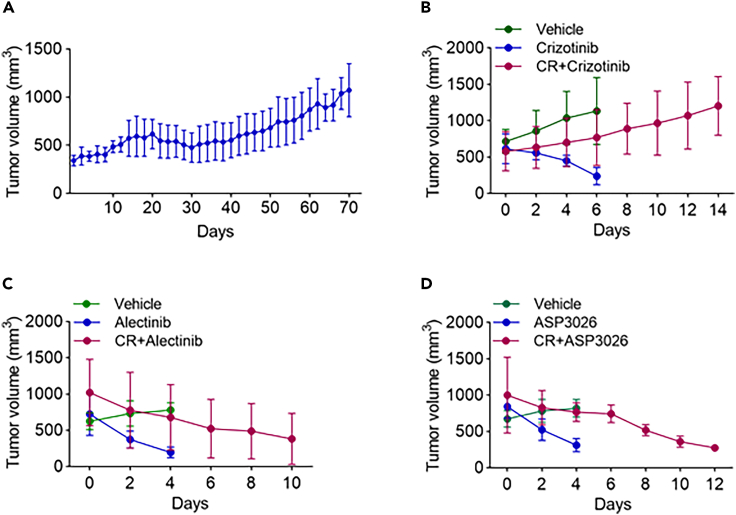
P4–P5 PDX mice were treated with crizotinib following the protocol in the STAR Methods. The last circles of these treatments shown in Figure 2A displayed a slow growth rate and indicated acquired resistance had been achieved. To further evaluate the effects of different ALK inhibitors on crizotinib-resistant tumors, these crizotinib-resistant PDX mice were treated with crizotinib (100 mg/kg/day), alectinib (60 mg/kg/day), or ASP3026 (100 mg/kg/day) for almost 2 weeks. Xenograft crizotinib-resistant tumors grew from 580.9 ± 268.9 mm3 to 1,204.3 ± 404.2 mm3 in the crizotinib group (Figure 2B) but shrank from 1,020.1 ± 460 mm3 to 382.2 ± 352 mm3 in the alectinib group (Figure 2C) and from 1,000.7 ± 523.3 mm3 to 274.3 ± 6.7 mm3 in the ASP3026 group (Figure 2D).
Figure 2.
The effects of different ALK inhibitors on tumor growth in crizotinib-resistant PDX mice
(A) The tumor growth curve of the last round of crizotinib treatment (100 mg/kg/day) in 14 crizotinib-resistant PDX mice. Tumor sizes were measured every other day.
(B) The tumor growth curves of crizotinib-resistant PDX mice further treated with crizotinib (100 mg/kg/day) and crizotinib-sensitive PDX mice treated with crizotinib or vehicle (N = 4). Tumor volumes were measured every other day.
(C) The tumor growth curves of crizotinib-resistant PDX mice when treated with alectinib (60 mg/kg/day) and crizotinib-sensitive PDX mice when treated with alectinib and vehicle (N = 4).
(D) The tumor growth curves of crizotinib-resistant PDX mice when treated with ASP3026 (100 mg/kg/day) and crizotinib-sensitive PDX mice when treated with ASP3026 and vehicle (N = 4).
Proteomic study on crizotinib-resistance tumors
Immunoblotting of tumors from PDX mice harboring EML4-ALK variant 3 under different conditions with a phosphotyrosine-specific antibody (P-Tyr-100) showed heterogeneous reactivity especially in the molecular weight range characteristic of receptor tyrosine kinases (Figure 3A). Compared to the control and crizotinib-sensitive mice, tyrosine phosphorylation was significantly increased in tumor tissues from crizotinib-resistant mice, and the increase was reversed by alectinib or ASP3026, suggesting their effectiveness of overcoming crizotinib resistance.
Figure 3.
PhosphoScan profiling of tumor tissues from crizotinib-resistant PDX mouse model
(A) Immunoblotting with P-Tyr-100 of crizotinib-resistant PDX tumor tissues treated with the indicated inhibitors and crizotinib-sensitive PDX tumor tissues treated with crizotinib or vehicle (#1 and #2 represent tumor tissues from two different mice in the same group; CS, crizotinib-sensitive; CR, crizotinib-resistant; Con, control; Criz, crizotinib; Ale, alectinib; ASP, ASP3026).
(B) PhosphoScan analysis of the proteins whose phosphorylation levels were increased by more than 2.5-folds (Table S3) in crizotinib-resistant PDX tumors compared to crizotinib-sensitive PDX tumors by protein type (N = 2).
(C) Confirmation of MS/MS results by immunoblotting with indicated antibodies. β-actin was used as the loading control (#1and #2 represent tumor tissues from two different mice in the same group; CS, crizotinib-sensitive; CR, crizotinib-resistant; Con, control; Criz, crizotinib; Ale, alectinib; ASP, ASP3026).
To identify signaling pathways involved in crizotinib resistance, we carried out the PhosphoScan profiling of tyrosine phosphorylation in crizotinib-resistant PDX tumors, crizotinib-sensitive tumors, and control tumors,31 as well as tumors from crizotinib-resistant PDX mice treated with alectinib or ASP3026 by liquid chromatography-tandem mass spectrometry (LC-MS/MS). PhosphoScan identified 113 tyrosine phosphorylation sites in 100 proteins. Spectral counts for all phosphotyrosine sites identified in PhosphoScan are listed in Table S3. Consistent with the immunoblotting results (Figure 3A), we observed elevated tyrosine phosphorylation of many receptor tyrosine kinases (RTKs) in crizotinib-resistant PDX mice, including ALK, AXL, discoidin domain receptor 1 (DDR1), ephrin receptor kinase A (EphA), EGFR, and Her2.
We next analyzed the distribution of protein tyrosine phosphorylation in PDX tumors based upon protein function classification. As shown in Figure 3B, the main protein classes involved in crizotinib resistance were adhesion or extracellular matrix proteins, adaptor, and scaffold proteins.
The partial verification of LC-MS/MS results by immunoblotting is shown in Figure 3C. In the control PDX mice tumors, ALK phosphorylation and expression level were decreased upon crizotinib treatment, while, in crizotinib-resistant PDX mice tumors, although crizotinib treatment showed no effect on ALK phosphorylation and ALK expression level, alectinib or ASP3026 treatment resulted in the decreased ALK phosphorylation and ALK expression. It seemed that ASP3026 had stronger effect than alectinib in terms of ALK phosphorylation inhibition. Consistent with the PhosphoScan results, we also found that the phosphorylation of SHC1 and SRC was also increased in crizotinib-resistant tumors and then decreased upon alectinib or ASP3026 treatment, with the stronger effect of ASP3026. On the other hand, the expression of AXL was significantly increased in crizotinib-resistant tumors, and this increase could not be reversed by alectinib or ASP3026 treatment. Considering that AXL mediated TKI resistance,29 this result indicates that an AXL-dependent pathway could compromise the effectiveness of new generation of ALK inhibitors.
New ALK inhibitors affect different signaling pathways leading to crizotinib resistance
PhosphoScan identified extensive tyrosine phosphorylation sites that changed downstream of ALK in crizotinib-resistant PDX tumors (Table S4). As shown in Figure 4A and Table S4, phosphorylation levels of 100 proteins changed by more than 2.5-folds in crizotinib-resistant xenograft tumors. Among them, the phosphorylation of 49 proteins was reversed by ASP3026 and 23 proteins by alectinib (Figure 4A). Phosphorylation of a set of 20 proteins was decreased by both ASP3026 and alectinib (Figure 4B). This core set of proteins included SRC and Her2, which are known to be involved in the development of crizotinib resistance. Previously unrecognized proteins involved in crizotinib resistance such as CUB domain-containing protein 1 (CDCP1), Ephrin type-A receptor 2 and 3 (EphA2, 3), pyruvate kinase 2 (PKM2), and protein kinase C delta (PKCD) were also identified. On the other hand, ASP3026 and alectinib had different effects on the phosphorylation of 32 proteins (Figure 4A; Table S4), suggesting that they could overcome crizotinib resistance through different signaling pathways. As expected, ALK phosphorylation was increased in crizotinib-resistant tumors (Figure 4C; Table S4). Interestingly, we found the ASP3026 had stronger effect than alectinib to inhibit SHC1 phosphorylation of SHC1, a known downstream substrate of ALK, in crizotinib-resistant PDX tumors (Figure 4C; Table S4). Besides, the phosphorylation levels of other RTKs, such as AXL, DDR1, Her2, EphA2, and EphA3, were also increased in crizotinib-resistant PDX tumors. While alectinib and ASP3026 had no effect on AXL and DDR1 phosphorylation in crizotinib-resistant PDX tumors (Figure 4C; Table S4), the phosphorylation of Her2, EphA2, and EphA3 was decreased by alectinib and ASP3026 treatments (Figures 4B and 4C; Table S4), suggesting both alectinib and ASP3026 could overcome crizotinib resistance through Her2-, EphA2-, and EphA3-mediated pathways.
Figure 4.
The cellular signaling networks involved in crizotinib resistance (N = 2)
(A) Venn diagram of 100 proteins whose phosphorylation levels were increased by more than 2.5-folds in crizotinib-resistant PDX tumors compared to crizotinib-sensitive PDX tumors. The numbers of proteins whose phosphorylation were reversed by alectinib or ASP3026 are shown.
(B) 20 proteins whose phosphorylation in crizotinib-resistant tumors were reversed by both ASP3026 and alectinib (CS, crizotinib-sensitive; CR, crizotinib-resistant; Ale, alectinib; ASP, ASP3026).
(C) Top 30 proteins whose increased phosphorylation in crizotinib-resistant tumors were affected by ASP3026 or alectinib (CS, crizotinib-sensitive; CR, crizotinib-resistant; Ale, alectinib; ASP, ASP3026).
AXL inhibition reversed crizotinib resistance in NCI-H3122-CR23 cells
As shown in Figures 3C and 4C, both AXL expression level and its phosphorylation were increased in crizotinib-resistant tumors, suggesting that AXL upregulation could be involved in crizotinib resistance. To further evaluate the effects of AXL on crizotinib resistance, we established a crizotinib-resistant NCI-H3122-CR23 cell line with an ALK kinase domain mutation F1174C (Figures 5A–5C) by combining both high exposure and stepwise methods as described in the STAR Methods. As shown in Figure 5A, IC50 values of crizotinib were 0.37 μmol/L for NCI-H3122 cells and 1.82 μmol/L for NCI-H3122-CR23 cells. We next examined the effect of the AXL inhibitor bemcentinib on the NCI-H3122-CR23 cells. As shown in Figure 5E, when NCI-H3122-CR23 cells were treated with crizotinib and bemcentinib together, it enhanced the inhibitory effect of crizotinib on the proliferation of NCI-H3122-CR23 cells in a concentration-dependent manner. Consistently, the protein expression level of AXL in NCI-H3122-CR23 cells was decreased after bemcentinib treatment in a concentration-dependent manner (Figure 5D).
Figure 5.
The effects of bemcentinib and siSHC1 on NCI-3122 cells and crizotinib-resistant PDX tumors
(A) Cell viability assays of NCI-H3122 and NCI-H3122-CR23 cells treated with crizotinib (Criz) for 72 h. The data are presented as mean ± SD from three independent experiments.
(B) Immunoblotting with P-Tyr-100 of NCI-H3122 and NCI-H3122-CR23 cells treated with 1 μmol/L crizotinib for 4 h. Tubulin was used as the loading control.
(C) Sequencing analysis of EML4-ALK showing a kinase domain heterozygous mutation F1174C in NCI-H3122-CR23. TTC in exon 23 of the ALK kinase region is replaced by TGC.
(D) Immunoblotting of AXL in NCI-H3122-CR23 cells treated with different concentrations of bemcentinib (Bemcen) and NCI-H3122 with Veh (vehicle) for 72 h. Cell lysates were probed with the indicated antibodies. Tubulin was used as the loading control.
(E) The viability of NCI-H3122-CR23 measured by MTS assay after treatment of bemcentinib (Bemcen) in combination with crizotinib (Criz) for 72 h. Results are representative of three independent experiments. The data are presented as mean ± SD. ∗∗p ≤ 0.01. ∗∗∗p ≤ 0.001.
(F) The inhibition effect by the combined treatment of crizotinib and bemcentinib in the crizotinib-resistant PDX mouse model. Tumor volume fold changes and TIR (tumor inhibition ratio) were indicated with drugs. TIR was calculated after the 25-day treatment. ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p ≤ 0.001.
(G) Immunoblotting of AXL and ALK in crizotinib-resistant PDX tumors treated with drugs indicated under the same conditions as in Figure 5F. Tissue lysates were probed with the indicated antibodies. Tubulin was used as the loading control. Veh, Vehicle; Criz, crizotinib; Criz and Bemcen, crizotinib and bemcentinib.
(H) Immunoblotting of NCI-H3122 cells treated with 1 μmol/L crizotinib for 4 h. Cell lysates were probed with the indicated antibodies. Veh, vehicle; Criz, Crizotinib.
(I) SHC1 mRNA levels in NCI-H3122 cells transfected with SHC1 siRNAs (siSHC1) for 72 h. Cells mRNA were amplificated with the primers presented in Table S1. Cont siRNA, negative control virus vector; #1 and #2 were two different interfering sequences of SHC1 mRNA.
(J) Immunoblotting of NCI-H3122 cells transfected with SHC1 siRNAs (siSHC1) #2 for 72 h; cell lysates were probed with the indicated antibodies.
(K) Cell viability assay of NCI-H3122 cells. NCI-H3122 cells were transfected with SHC1 siRNAs (siSHC1) #2 for 72 h and then treated with the indicated concentration of crizotinib for 72 h. Control cells were transfected with scrambled siRNA. Results are representative of three independent experiments. The data are presented as mean ± SD. Cont siRNA, negative control virus vector; Criz, crizotinib. ∗∗p < 0.01. ∗∗∗∗p = 0.000.
AXL inhibition enhances crizotinib sensitivity in PDX mice
We then tested the effect of AXL inhibitor bemcentinib on the crizotinib-resistant PDX mouse model. As shown in Figure 5F, after 25 days of treatment, the tumor volume increased 7.12 ± 1.76-folds in control, 4.05 ± 1.05-folds in crizotinib, 5.37 ± 1.54-folds in bemcentinib, and 2.12 ± 0.63-fold in combined treatment. The synergistic index of the combined treatment of crizotinib and bemcentinib was 1.22 (see the STAR Methods for the calculation), indicating that there was a synergistic effect when crizotinib 25 mg/kg/day and bemcentinib 50 mg/kg/day were used in combination (Figure 5F). Immunoblotting analysis showed that AXL expression level increased when using crizotinib alone, while, when bemcentinib and crizotinib were combined, AXL expression level decreased (Figure 5G).
Knockdown of SHC1 sensitizes NCI-H3122 cells to crizotinib
Previous studies have shown that ALK provided a direct docking site for SH2 domain-containing transforming protein (SHC1) and that recruitment and phosphorylation of SHC1 activated the Ras/extracellular signal-regulated kinase (ERK) pathway and promoted tumorigenesis.32,33 Consistent with these observations, we found hyperphosphorylation of SHC1 in the crizotinib-resistant PDX tumors bearing ALK phosphorylation (Figure 4C).
To investigate whether SHC1 could be involved in crizotinib resistance, we measured crizotinib sensitivity in EML4-ALK-driven cell line NCI-H3122 transfected with SHC1 small interfering RNA (siRNA) directed against SHC1. When NCI-H3122 cells were treated with crizotinib, the phosphorylation of ALK and SHC1 phosphorylation was decreased (Figure 5H), which was consistent with the results from PDX tumors. However, unlike crizotinib treatment in PDX tumors (Figure 3C), crizotinib treatment in NCI-H3122 cells did not affect the expression level of ALK (Figure 5H), which might be explained by the difference between PDX tumor tissues and cancer cell lines, or the duration of crizotinib treatment (144 h for PDX tumors vs. 4 h for NCI-H3122 cells). We then examined the effect of siSHC1 on the crizotinib-inhibited proliferation of NCI-H3122 cells. As shown in Figures 5I–5K, although SHC1 knockdown did not affect cell viability (not shown and Figure 5K, column 1 and 5), it significantly enhanced the inhibitory effect of crizotinib on the proliferation of NCI-H3122 cells (Figure 5K).This suggests that destabilization of SHC1 could sensitize ALK-positive cancer cells to crizotinib treatment.
Discussion
In NSCLC patients with oncogenic ALK rearrangements, crizotinib provides substantial therapeutic benefit. However, most patients develop resistance. Robust, but not durable, response to crizotinib suggests the new therapy to inhibit ALK activity should be at multiple levels. In the present study, we provide a large-scale profiling of cellular signaling networks involved in crizotinib resistance in a PDX mouse model established using tumor samples from patient T5 with EML4-ALK. The histological characteristics and genotypes of PDX tumors were similar to those of the donor patient’s primary tumor samples, validating the use of PDX tumors to investigate the signaling pathways of crizotinib resistance in NSCLC.
We sought to understand the mechanisms of acquired resistance to crizotinib by inducing resistance in this PDX mouse model harboring EML4-ALK gene rearrangement through continual crizotinib treatment. ALK mutations were not detected (data not shown) in the crizotinib-resistant xenograft tumor, suggesting that ALK downstream targets might contributed to acquired crizotinib resistance. A recent study showed that SRC family kinases were involved in sustaining mitogen-activated protein kinase signaling in lung cancer cells treated with crizotinib and that SRC family kinase inhibitor in combination with crizotinib treatment was effective in inhibiting the growth of crizotinib-resistant lung cancer.25 In addition, BRAF mutations were shown to attenuate the antitumor effects of crizotinib.34 Together, these results suggest that, under continuous crizotinib treatment, cellular signaling pathways independently contribute to tumor cell proliferation and survival through a mechanism downstream of ALK, which overrides the effect of crizotinib treatment.
We identified 100 proteins with tyrosine phosphorylation increased more than 2.5-folds once crizotinib resistance was developed. We mapped an extensive signaling network operating downstream of ALK, with the majority of the substrates functioning in various related cellular activities. We identified many inputs from ALK to adhesion and extracellular matrix proteins, receptors, adaptors, and scaffolds acting at the interphase between the cytoskeleton and plasma membrane, suggesting a possible role of cell-cell contact and motility in the development of crizotinib resistance. Other prominent targets involved in crizotinib resistance included protein kinases. Among them, EGFR, Her2, and Src were previously shown to be involved in crizotinib resistance, validating our approach of protein phosphorylation profiling to understand the mechanism of crizotinib resistance.
Among these 100 proteins which could be involved in crizotinib resistance, when the new generation of ALK inhibitors were used, 49 proteins had their phosphorylation inhibited by ASP3026 treatment and 23 by alectinib treatment, suggesting new ALK inhibitor ASP3026 could have broader effect than alectinib in overcoming crizotinib resistance. Interestingly, the phosphorylation of 20 core proteins was inhibited by both new ALK inhibitors, including EphA2, EphA3, Her2, PKCD, PKM2, SRC, and Yes, suggesting that these 20 proteins, especially these 7 kinases, could be important in the development of crizotinib resistance.
Although AXL itself is not thought to be a potent tumor driver, its overexpression is known to be involved in drug resistance to cancer therapy for various types of cancer. Recently, many preclinical and clinical studies using AXL inhibitors are being conducted, especially in NSCLC.35,36 Since the effect of AXL inhibitor alone is not yet satisfactory, the recent efforts have focused on the combination therapies of AXL inhibitors and with EGFR TKIs or immunotherapy to overcome drug resistance.35,36 In the present study, we found that AXL expression level was increased in crizotinib-resistant PDX mice and that the combination of AXL inhibitor with crizotinib reversed crizotinib resistance in NCI-H3122-CR23 cells and in the crizotinib-resistant PDX mouse model. Importantly, the upregulation of AXL could not be overcome by these second or new generation of ALK inhibitors, alectinib and ASP3026, suggesting a potential AXL-mediated crizotinib resistance could be a challenge to develop more effective ALK inhibitors in the future.
We also found that selective targeting of the scaffold protein SHC1 could act together with crizotinib to further inhibit oncogenic ALK signaling. SHC1 knockdown enhanced the inhibitory effect of crizotinib on the proliferation of NCI-H3122 cells, suggesting that targeting both kinase activity and its downstream signaling may confer additional clinical benefit. To our knowledge, there have been no previous reports demonstrating that SHC1 knockdown sensitizes ALK-positive cancer cells to an ALK inhibitor.
In terms of the development of new ALK inhibitors, we noticed that, among the 100 proteins with phosphorylation status changed on the progression in crizotinib-resistant PDX tumors, only 52 had their phosphorylations reversed by ASP3026 or alectinib. For almost half of the proteins, 48 of them, including AXL, it was shown that their phosphorylations were not affected by the treatment of both drugs. Importantly, some proteins like AXL and DDR1 have been shown to be involved in drug resistance for RTK inhibitors,35,36 but their increased phosphorylation in crizotinib-resistant PDX tumors was not affected by the second or new generation of ALK inhibitors, like alectinib and ASP3026. A further study will be performed to demonstrate the role of these proteins in the development of crizotinib resistance, which could provide new targets or new combination therapy to overcome crizotinib resistance. Our study highlights the importance of developing more effective and safe treatment by using new ALK inhibitors, either as a single agent or in combination, to overcome crizotinib resistance.
In summary, we provide a large-scale profiling of cellular signaling networks involved in crizotinib resistance in a PDX mouse model, and our results suggest AXL and SHC1 as the potential targets for combination therapy for NSCLC patients with ALK rearrangement to overcome crizotinib resistance. However, our PDX mouse model only selected one type of ALK arrangement from a patient, which only represents a subset of patients with development of crizotinib resistance. The overall cellular signaling network involved in crizotinib resistance would be more complicated. Nevertheless, our approach provides a new view of crizotinib resistance, revealing many previously unidentified proteins that merit further consideration as targets for combination therapy.
Limitations of the study
In this article, our PDX mouse model only selected one type of ALK arrangement from a patient, which only represents a subset of patients with development of crizotinib resistance. For PhosphoScan analysis, only 2 tumor tissue samples were mixed and analyzed from each treatment condition. Also, due to the lack of site-specific phospho AXL antibody, the phosphorylation status of AXL was not confirmed by western blotting.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ling Tan (dr.tanling@csu.edu.cn).
Materials availability
This study did not generate new unique materials.
Data and code availability
-
•
The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
The DNA sequences data have been deposited at China National Center for Bioinformation, which are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
Acknowledgments
This study was supported by a grant from the National Natural Science Foundation of China (81272584).We thank Qiuguo Wang for technical assistance in manuscript preparation.
Author contributions
The work reported in the paper has been performed by the authors, unless clearly specified in the text. Y.H., Y.L., and L.T.: conception, study design, and protocol. Y.Z., W.L., M.C., Y.H., G.Q., and Z.X.: data collection and assessment. Y.H.: KEGG analysis of proteins from PhosphoScan profiling. Y.H., Y.L., and L.T.: writing. Y.L. and X.Z.: project oversight and supervision. Y.H., Y.L., L.T., and X.Z.: critical revisions for important intellectual content. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-p-ALK | Cell Signaling Technology | Cat#3983 |
| Rabbit monoclonal anti-ALK | Cell Signaling Technology | Cat#3633 |
| Rabbit monoclonal anti-Axl | Cell Signaling Technology | Cat#8661 |
| Rabbit monoclonal anti-SHC | Abcam | Cat#ab33770 |
| Rabbit polyclonal anti-p-SHC1 | Cell Signaling Technology | Cat#2434 |
| Rabbit monoclonal anti-Scr | Cell Signaling Technology | Cat#2109 |
| Rabbit polyclonal anti-p-Scr | Cell Signaling Technology | Cat#2101 |
| Mouse monoclonal anti- Phospho-Tyrosine (p-Tyr-100) | Cell Signaling Technology | Cat#9411 |
| Rabbit polyclonal anti-β-Actin | Cell Signaling Technology | Cat#4967 |
| Rabbit monoclonal anti-GAPDH | Cell Signaling Technology | Cat#2118 |
| Rabbit polyclonal anti-β-Tubulin | Cell Signaling Technology | Cat#2146 |
| Chemicals, peptides, and recombinant proteins | ||
| Crizotinib | Selleck Chemicals | Cat#S1068 |
| ASP3026 | Selleck Chemicals | Cat#S8054 |
| Bemcentinib | Selleck Chemicals | Cat#S2841 |
| Alectinb | Selleck Chemicals | Cat#S5232 |
| Critical commercial assays | ||
| CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) | Promega | Cat#G3580 |
| RevertAid First Strand cDNA Synthesis Kit | ThermoFisher | Cat#K1622 |
| Platinum™ Multiplex PCR Master Mix | ThermoFisher | Cat#4464268 |
| RNeasy Mini Kit | Qiagen | Cat#74104 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat#69504 |
| Experimental models: Cell lines | ||
| NCI-H3122 | CoBier Bioscience (Nanjing, Jiangsu, China) | RRID:CVCL_5160 |
| Experimental models: Organisms/strains | ||
| SCID Beige Mouse: CB17.Cg-PrkdcscidLystbg-J/Crl | Vital River Laboratory Animal Technology, Beijing, China | Cat#405 |
| Oligonucleotides | ||
| Primers for ALK MEL4 GAPDH SHC1, See Table S1 | This paper | N/A |
| siRNA targeting sequence: SHC1 #2: TGACCATCAGTACTATAAT | This paper | N/A |
| Deposited data | ||
| Mass spectrometry proteomics data | ProteomeXchange Consortium:iProX | PXD054055 |
| EML4-ALK DNA sequencing data | China National Center for Bioinformation https://www.cncb.ac.cn/ | GB0004519 |
| NCI-H3122CR23 DNA sequencing data | China National Center for Bioinformation https://www.cncb.ac.cn/ | GB0004523 |
Experimental model and study participant details
Patient selection
This study was approved by the Institutional Review Board of the Second Xiangya Hospital (Ethical Approval Number 357), and written informed consent was obtained from all patients prior to participation in the present study. The sample of PDX mouse model was obtained from a 44-year-old male patient T5 (with EML4–ALK arrangement, Mongoloid) who was diagnosed with stage IV lung adenocarcinoma and underwent treatment at our hospital.
Cell lines
The NCI-H3122 cell line (RRID:CVCL_5160, harboring EML4-ALK variant 1 E13;A20) was purchased from CoBier Bioscience (Nanjing, Jiangsu, China), and has been authenticated using STR profiling within the last three years. NCI-H3122 cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS, Gibco) at 37°C in 5% CO2. All experiments were performed with mycoplasma-free NCI-H3122 cells.
Mice
3- to 4-week-old female SCID Beige mice were purchased from Beijing Vital River Laboratory. They were maintained in the specific pathogen-free facility at Experimental Animal Center of The Second Xiangya Hospital. The animal use protocol has been reviewed and approved by the Institutional Animal Care and Use Committee of The Second Xiangya Hospital (Ethical Approval Number 2013101).
Method details
Reagents
Crizotinib, alectinib, ASP3026 and bemcentinib were purchased from Selleck Chemicals (Houston, TX, USA). All antibodies used in this study were purchased from Cell Signaling Technology (Danvers, MA, USA), except SHC1, which is from Abcam (Cambridge, England, UK).
Establishment of crizotinib-resistant PDX mouse model
The flow chart to establish crizotinib-resistant PDX mouse model is shown in Figure S1. The animal use protocol listed below has been reviewed and approved by the Institutional Animal Care and Use Committee of The Second Xiangya Hospital (Ethical Approval Number 2013101). Primary tumor tissues were collected from a 44-year-old male patient (T5). Tissues were cut into 1-2 mm pieces and implanted subcutaneously into both flanks of three 3- to 4-week-old female SCID Beige mice (P1, Vital River Laboratory Animal Technology, Beijing, China). Remaining tissues were subjected to immunohistochemistry staining, western blot and Sanger sequencing. Tumors from bearing mice were measured twice weekly with a caliper, and tumor volumes were calculated using the formula: volume = (length × width2)/2. When tumors reached about 1 cm in length, they were then removed from the mice, cut into pieces and reimplanted into new hosts as described above for next passages (P2, P3, P4 and P5), but P4 and P5 implanted subcutaneously into only a flank. P4-P5 PDX mice after their tumors reached 800mm3 were treated with crizotinib initially 50mg/kg/day and finally 100mg/kg/day orally once per day. Tumor sizes were measured every other day. When the tumor sizes of the crizotinib-treated mice shrank from 800 mm3 to 100 mm3, the drug administration was suspended until the tumor sizes reached 800mm3 again. After 5-6 circles of these treatments, the tumors in the crizotinib-treated mice exhibited a slow growth rate despite crizotinib (100mg/kg/day) treatment for >50 days, indicating acquired resistance had been achieved. In summary, crizotinib 50mg/kg/day was used for 21-30 days, and 100mg/kg/day was used for 239-289 days.
Histopathlogical examination
Tumor samples from both the patient and xenograft mice were fixed in 10% buffered formalin within 15 min after resection/collection. Tissues were then subjected to routine hematoxylin & eosin staining and immunohistochemistry staining with anti-ALK antibody (ALK (D5F3®) XP® Rabbit mAb #3633, Cell Signaling Technology). Histopathological diagnoses were confirmed by a qualified pathologist.
PhosphoScan analysis of tumor tissues
The peptides were prepared from 0.2 grams of frozen mixed tumor tissues (2 samples for each treatment) by homogenization, trypsin digestion, and Sep-pak C18 column purification as described in the protocol from Cell Signaling Technology. Peptides containing phosphotyrosine were isolated by immunoprecipitation with a monoclonal antibody (P-Tyr-100 mAb) against phosphotyrosine, concentrated on reverse-phase micro tips and analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS). Phosphotyrosine peptide profiling by PhosphoScan was done by Cell Signaling Technology with a global false discovery rate of less than 5% using peptide prophet. 2.5-fold was considered the threshold for significant change.
Western blot
Frozen tumor tissues were minced in liquid nitrogen and resuspended in cell lysis buffer (#9803, Cell Signaling Technology). Tissues or cells suspension was then sonicated and cleared by centrifugation. Immunoblotting analysis was carried out according to the manufacturer's standard protocol (www.cellsignal.com).
Reverse transcription-PCR and DNA sequencing
Total RNA was extracted from tumor tissues or cells using RNeasy Mini Kit (Qiagen, Hilden, German). For reverse transcription-PCR (RT-PCR), first-strand cDNA was synthesized from 2μg of total RNA using RevertAid First Strand cDNA Synthesis Kit (#K1622, ThermoFisher, Waltham, MA, USA). Multiple PCR was done using Platinum™ Multiplex PCR Master Mix (# 4464268, ThermoFisher). Primer pairs used to amplify ALK rearrangement transcripts are listed in Table S1.
For DNA sequencing, DNA was extracted from cells or 15 mg of tumor tissues using DNeasy Blood & Tissue Kit (#69504, Qiagen) and ALK fragments were amplified using primer pairs listed in Table S1. DNA sequencing was done by Beijing Genomics Institute (Beijing, China).
Construction of crizotinib-resistant cell line and cell viability assay
NCI-H3122 cells were seeded at 70% confluence in 25-cm flasks in RPMI-1640 (#22400-089, Gibco) supplemented with 10% FBS (#10099-141, Gibco). Crizotinib was added at a starting concentration of 30 nmol/L, and cells were maintained in fresh drug-containing medium changed every 72 hours. Cells were passaged once they reached 70% confluence. After every two passages at a given concentration of drug, the concentration of crizotinib was increased in half-log intervals until a final concentration of 1μmol/L was achieved. The resulting pool of crizotinib-resistant cells (designated NCI-H3122-CR23) was maintained in RPMI-1640 supplemented with 10% FBS and 1μmol/L crizotinib up to 2 months before NCI-H3122-CR23 cells were treated with crizotinib or bemcentinib plus crizotinib. Cell proliferation inhibition was assessed by MTS assay (#G3580, Promega, Madison, WI, USA). All experimental points were set up in 6 to 12 replicate wells and all experiments were repeated at least 3 times. The proliferation inhibition rate was calculated as 1-(OD of treatment well / OD of control well) %.All experiments were performed with mycoplasma-free cells.
The combined treatment of crizotinib and bemcentinib in crizotinib-resistant PDX mouse model
Take size 1-2 mm pieces of frozen tumor tissues in liquid nitrogen from P5 of crizotinib-resistant mice, and implanted subcutaneously into one flanks of twenty 3- to 4-week-old female SCID Beige mice (Vital River Laboratory Animal Technology, Beijing, China). Tumors from bearing mice were measured twice weekly with a caliper. When tumors reached about 4-8mm in length (after implanted for 34 days), mice were randomized to the following four cohorts (N =5, for each group): vehicle control, crizotinib (25mg/kg/day), bemcentinib (50mg/kg/day) or both crizotinib (25mg/kg/day) and bemcentinib (50mg/kg/day). During 25 days of the drug treatment, tumor volumes (V) were calculated once a week by caliper measurements of the width (W) and length (L) of each tumor (V=W2 ×L/2), statistical analysis was performed based on volume fold change compared to day 0 of initial treatment. The tumor inhibition ratio calculated as 1- (volume fold changes of each group/ volume fold changes of vehicle group) % after the 25-day treatment period. The Bliss independent model method was used to calculate the synergistic index CI of the combination of two drugs, with the formula CI=EAB/ (EA+EB-EA∗EB). EA or EB was the inhibition ratio of drug A or drug B alone and EAB was the inhibition ratio of drug A and B combination. CI>1 represents a synergistic effect, CI=1 represents an additive effect, and CI<1 represents an antagonistic effect. Protein extraction and western blot analyses were performed using tumor tissues from mice after the 25-day treatment period.
siRNA transfection and cell viability assay
NCI-H3122 cells were maintained in RPMI-1640 supplemented with 10% FBS and transfected with SHC1 siRNA (target sequence TGACCATCAGTACTATAAT) following the manufacture’s protocol (Genechem, Shanghai, China). Control cells were transfected with a scrambled control siRNA (control sequence TTCTCCGAACGTGTCACGT) from Genechem. At 72 hr after the transfection, crizotinib was added at the concentrations of 0, 33, 66 and 100 nmol/L, respectively. Viability of cells was measured by MTS assay after crizotinib added for 72 hr and the proliferation inhibition rate was calculated as described above.
Quantification and statistical analysis
Data were expressed as the mean ± standard deviation (SD) of three independent experiments and evaluated with One-Way ANOVA, Independent Sample T test and Univariate Analysis of Variable. P value of < 0.05 was considered to be significant.
Published: August 30, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110846.
Contributor Information
Yu Li, Email: yulihunan@126.com.
Ling Tan, Email: dr.tanling@csu.edu.cn.
Supplemental information
References
- 1.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S.i., Watanabe H., Kurashina K., Hatanaka H., et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 2.Shaw A.T., Kim D.W., Nakagawa K., Seto T., Crinó L., Ahn M.J., De Pas T., Besse B., Solomon B.J., Blackhall F., et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl. J. Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 3.Solomon B.J., Mok T., Kim D.W., Wu Y.L., Nakagawa K., Mekhail T., Felip E., Cappuzzo F., Paolini J., Usari T., et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 4.Shaw A.T., Engelman J.A. ALK in lung cancer: past, present, and future. J. Clin. Oncol. 2013;31:1105–1111. doi: 10.1200/JCO.2012.44.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi Y.L., Soda M., Yamashita Y., Ueno T., Takashima J., Nakajima T., Yatabe Y., Takeuchi K., Hamada T., Haruta H., et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 6.Wu J., Savooji J., Liu D. Second- and third-generation ALK inhibitors for non-small cell lung cancer. J. Hematol. Oncol. 2016;9:19. doi: 10.1186/s13045-016-0251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakamoto H., Tsukaguchi T., Hiroshima S., Kodama T., Kobayashi T., Fukami T.A., Oikawa N., Tsukuda T., Ishii N., Aoki Y. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell. 2011;19:679–690. doi: 10.1016/j.ccr.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Kodama T., Tsukaguchi T., Yoshida M., Kondoh O., Sakamoto H. Selective ALK inhibitor alectinib with potent antitumor activity in models of crizotinib resistance. Cancer Lett. 2014;351:215–221. doi: 10.1016/j.canlet.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Shaw A.T., Gandhi L., Gadgeel S., Riely G.J., Cetnar J., West H., Camidge D.R., Socinski M.A., Chiappori A., Mekhail T., et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol. 2016;17:234–242. doi: 10.1016/S1470-2045(15)00488-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou H.Y., Friboulet L., Kodack D.P., Engstrom L.D., Li Q., West M., Tang R.W., Wang H., Tsaparikos K., Wang J., et al. PF-06463922, an ALK/ROS1 Inhibitor, Overcomes Resistance to First and Second Generation ALK Inhibitors in Preclinical Models. Cancer Cell. 2015;28:70–81. doi: 10.1016/j.ccell.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw A.T., Felip E., Bauer T.M., Besse B., Navarro A., Postel-Vinay S., Gainor J.F., Johnson M., Dietrich J., James L.P., et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol. 2017;18:1590–1599. doi: 10.1016/S1470-2045(17)30680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori M., Ueno Y., Konagai S., Fushiki H., Shimada I., Kondoh Y., Saito R., Mori K., Shindou N., Soga T., et al. The selective anaplastic lymphoma receptor tyrosine kinase inhibitor ASP3026 induces tumor regression and prolongs survival in non-small cell lung cancer model mice. Mol. Cancer Ther. 2014;13:329–340. doi: 10.1158/1535-7163.MCT-13-0395. [DOI] [PubMed] [Google Scholar]
- 13.Li T., LoRusso P., Maitland M.L., Ou S.H.I., Bahceci E., Ball H.A., Park J.W., Yuen G., Tolcher A. First-in-human, open-label dose-escalation and dose-expansion study of the safety, pharmacokinetics, and antitumor effects of an oral ALK inhibitor ASP3026 in patients with advanced solid tumors. J. Hematol. Oncol. 2016;9:23. doi: 10.1186/s13045-016-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono A., Murakami H., Seto T., Shimizu T., Watanabe S., Takeshita S., Takeda K., Toyoshima J., Nagase I., Bahceci E., et al. Safety and Antitumor Activity of Repeated ASP3026 Administration in Japanese Patients with Solid Tumors: A Phase I Study. Drugs R. 2021;21:65–78. doi: 10.1007/s40268-020-00331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuccotto F., Ardini E., Casale E., Angiolini M. Through the "gatekeeper door": exploiting the active kinase conformation. J. Med. Chem. 2010;53:2681–2694. doi: 10.1021/jm901443h. [DOI] [PubMed] [Google Scholar]
- 16.Azam M., Seeliger M.A., Gray N.S., Kuriyan J., Daley G.Q. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat. Struct. Mol. Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doebele R.C., Pilling A.B., Aisner D.L., Kutateladze T.G., Le A.T., Weickhardt A.J., Kondo K.L., Linderman D.J., Heasley L.E., Franklin W.A., et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin. Cancer Res. 2012;18:1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michels S.Y.F., Scheel A.H., Wündisch T., Heuckmann J.M., Menon R., Puesken M., Kobe C., Pasternack H., Heydt C., Scheffler M., et al. ALKG1269A mutation as a potential mechanism of acquired resistance to crizotinib in an ALK-rearranged inflammatory myofibroblastic tumor. npj Precis. Oncol. 2017;1:4. doi: 10.1038/s41698-017-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki T., Koivunen J., Ogino A., Yanagita M., Nikiforow S., Zheng W., Lathan C., Marcoux J.P., Du J., Okuda K., et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051–6060. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katayama R., Shaw A.T., Khan T.M., Mino-Kenudson M., Solomon B.J., Halmos B., Jessop N.A., Wain J.C., Yeo A.T., Benes C., et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyawaki M., Yasuda H., Tani T., Hamamoto J., Arai D., Ishioka K., Ohgino K., Nukaga S., Hirano T., Kawada I., et al. Overcoming EGFR Bypass Signal-Induced Acquired Resistance to ALK Tyrosine Kinase Inhibitors in ALK-Translocated Lung Cancer. Mol. Cancer Res. 2017;15:106–114. doi: 10.1158/1541-7786.MCR-16-0211. [DOI] [PubMed] [Google Scholar]
- 22.Wilson F.H., Johannessen C.M., Piccioni F., Tamayo P., Kim J.W., Van Allen E.M., Corsello S.M., Capelletti M., Calles A., Butaney M., et al. A functional landscape of resistance to ALK inhibition in lung cancer. Cancer Cell. 2015;27:397–408. doi: 10.1016/j.ccell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hrustanovic G., Olivas V., Pazarentzos E., Tulpule A., Asthana S., Blakely C.M., Okimoto R.A., Lin L., Neel D.S., Sabnis A., et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat. Med. 2015;21:1038–1047. doi: 10.1038/nm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crystal A.S., Shaw A.T., Sequist L.V., Friboulet L., Niederst M.J., Lockerman E.L., Frias R.L., Gainor J.F., Amzallag A., Greninger P., et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science (New York, N.Y.) 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida R., Sasaki T., Minami Y., Hibino Y., Okumura S., Sado M., Miyokawa N., Hayashi S., Kitada M., Ohsaki Y. Activation of Src signaling mediates acquired resistance to ALK inhibition in lung cancer. Int. J. Oncol. 2017;51:1533–1540. doi: 10.3892/ijo.2017.4140. [DOI] [PubMed] [Google Scholar]
- 26.Dagogo-Jack I., Yoda S., Lennerz J.K., Langenbucher A., Lin J.J., Rooney M.M., Prutisto-Chang K., Oh A., Adams N.A., Yeap B.Y., et al. MET Alterations Are a Recurring and Actionable Resistance Mechanism in ALK-Positive Lung Cancer. Clin. Cancer Res. 2020;26:2535–2545. doi: 10.1158/1078-0432.CCR-19-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H.R., Kim W.S., Choi Y.J., Choi C.M., Rho J.K., Lee J.C. Epithelial-mesenchymal transition leads to crizotinib resistance in H2228 lung cancer cells with EML4-ALK translocation. Mol. Oncol. 2013;7:1093–1102. doi: 10.1016/j.molonc.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gainor J.F., Dardaei L., Yoda S., Friboulet L., Leshchiner I., Katayama R., Dagogo-Jack I., Gadgeel S., Schultz K., Singh M., et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov. 2016;6:1118–1133. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamichi S., Seike M., Miyanaga A., Chiba M., Zou F., Takahashi A., Ishikawa A., Kunugi S., Noro R., Kubota K., Gemma A. Overcoming drug-tolerant cancer cell subpopulations showing AXL activation and epithelial-mesenchymal transition is critical in conquering ALK-positive lung cancer. Oncotarget. 2018;9:27242–27255. doi: 10.18632/oncotarget.25531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J.J., Zhu V.W., Yoda S., Yeap B.Y., Schrock A.B., Dagogo-Jack I., Jessop N.A., Jiang G.Y., Le L.P., Gowen K., et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J. Clin. Oncol. 2018;36:1199–1206. doi: 10.1200/JCO.2017.76.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rush J., Moritz A., Lee K.A., Guo A., Goss V.L., Spek E.J., Zhang H., Zha X.M., Polakiewicz R.D., Comb M.J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 32.Degoutin J., Vigny M., Gouzi J.Y. ALK activation induces Shc and FRS2 recruitment: Signaling and phenotypic outcomes in PC12 cells differentiation. FEBS Lett. 2007;581:727–734. doi: 10.1016/j.febslet.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., Wang P., Wu F., Li M., Sharon D., Ingham R.J., Hitt M., McMullen T.P., Lai R. Aberrant expression of the transcriptional factor Twist1 promotes invasiveness in ALK-positive anaplastic large cell lymphoma. Cell. Signal. 2012;24:852–858. doi: 10.1016/j.cellsig.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 34.Xie L. Crizotinib resistance: BRAF mutation implications for therapeutic strategies. Cancer Treat. Res. Commun. 2021;28 doi: 10.1016/j.ctarc.2021.100391. [DOI] [PubMed] [Google Scholar]
- 35.Sang Y.B., Kim J.H., Kim C.G., Hong M.H., Kim H.R., Cho B.C., Lim S.M. The Development of AXL Inhibitors in Lung Cancer: Recent Progress and Challenges. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.811247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scaltriti M., Elkabets M., Baselga J. Molecular Pathways: AXL, a Membrane Receptor Mediator of Resistance to Therapy. Clin. Cancer Res. 2016;22:1313–1317. doi: 10.1158/1078-0432.CCR-15-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
The DNA sequences data have been deposited at China National Center for Bioinformation, which are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request