Abstract
Numerous studies have demonstrated that the hepatitis B virus HBx protein stimulates signal transduction pathways and may bind to certain transcription factors, particularly the cyclic AMP response element binding protein, CREB. HBx has also been shown to promote early cell cycle progression, possibly by functionally replacing the TATA-binding protein-associated factor 250 (TAFII250), a transcriptional coactivator, and/or by stimulating cytoplasmic signal transduction pathways. To understand the basis for early cell cycle progression mediated by HBx, we characterized the molecular mechanism by which HBx promotes deregulation of the G0 and G1 cell cycle checkpoints in growth-arrested cells. We demonstrate that TAFII250 is absolutely required for HBx activation of the cyclin A promoter and for promotion of early cell cycle transit from G0 through G1. Thus, HBx does not functionally replace TAFII250 for transcriptional activity or for cell cycle progression, in contrast to a previous report. Instead, HBx is shown to activate the cyclin A promoter, induce cyclin A–cyclin-dependent kinase 2 complexes, and promote cycling of growth-arrested cells into G1 through a pathway involving activation of Src tyrosine kinases. HBx stimulation of Src kinases and cyclin gene expression was found to force growth-arrested cells to transit through G1 but to stall at the junction with S phase, which may be important for viral replication.
Chronic infection with hepatitis B virus (HBV) is closely associated with the development of hepatocellular carcinoma in humans. Consequently, there has been intense interest in HBV gene products that can alter cellular gene activity. The smallest open reading frame of the mammalian HBVs, including the human virus, encodes a 17-kDa regulatory protein known as HBx (or X protein) (reviewed in references 2 and 92). HBx is essential for productive infection by the mammalian HBVs (12, 95). Studies initially characterized HBx as a transcriptional transactivator of weak to moderate strength. Transcription factors activated by HBx include NF-κB, NF-AT, AP-1, and ATF/CREB (4, 6, 8, 45, 48, 51, 52, 55, 68, 73, 77, 81, 82, 90). In addition to stimulation of RNA polymerase II-directed transcription, HBx also stimulates transcription by RNA polymerases I and III (3, 43, 86, 88). HBx is therefore a modest activator of many types of transcription elements and factors (28, 29, 46, 51, 54, 60, 61, 90). Many of the reported activities of HBx have been shown to result from its ability to activate cytoplasmic signal transduction pathways, particularly the Ras–Raf–mitogen-activated protein kinase (MAPK) pathway (6, 18, 55, 86), the cell stress-induced MEKK1–p38–c-Jun N-terminal kinase (JNK) pathway (8, 73), and the family of Src tyrosine kinases (41). HBx activation of Src may be important for viral replication (40). In vitro, HBx can interact with several components of the transcriptional apparatus, including factors TATA-binding protein (TBP), TFIIB, TFIIH, and the RPB5 subunit of RNA polymerases (14, 60, 61). HBx also possesses nuclear transcription-activating functions (23) that may involve interaction with CREB (51, 59, 77, 90) or possibly a reported coactivator activity (29, 30, 83).
HBx has been shown to stimulate deregulation of early cell cycle checkpoint controls (7, 42, 71). Expression of HBx in cells that have had their growth arrested by serum withdrawal results in their transit through the G1 phase of the cell cycle but without progression into S phase (71). Thus, in the absence of serum, HBx-expressing cells are stalled at the G1-S phase junction, whereas control cells without HBx remain in G0. If serum is provided to growth-arrested cells that express HBx, these cells advance more rapidly through G1 and may enter S phase, in contrast to cells without HBx, particularly if the cell is transformed (7, 42). Although HBx activation of Ras has been shown to be necessary for deregulation of the G0 checkpoint (7), there is little understanding of the mechanism by which HBx promotes early cell cycle progression. In this regard, HBx was also reported to functionally replace TBP-associated factor 250 (TAFII250) in transcriptional activation, induction of cell cycle progression, and inhibition of apoptosis in ts13 cells. Thus, these data implicate both cytoplasmic signal transduction and nuclear transcription functions in the ability of HBx protein to promote cell cycle progression. ts13 cells are from a hamster cell line containing a temperature-sensitive defect in TAFII250 caused by a single amino acid change in the TAFII250 protein sequence (31, 76). The TAFII250 ts13 mutation causes cell cycle arrest and apoptosis when these cells are shifted from the permissive temperature (33°C) to the nonpermissive temperature (39.5°C). TAFII250, the largest TAF identified, interacts with TBP and several other TAFs (reviewed in references 1, 11, and 58), and it is essential for assembly of the TFIID complex (89). It was recently established that TAFII250 targets specific chromatin-bound promoters via multiply acetylated histone H4 proteins. By utilizing its intrinsic histone acetyltransferase (HAT) activity, TAFII250 activates transcription through chromatin remodeling and recruitment of the transcription complex (36). TAFII250 is essential for activation of A- and D-type cyclin genes, a necessary event for progression through the cell cycle (67). Importantly, extensive analyses of ts13 cells have demonstrated that it is the HAT activity of TAFII250, and not other activities, that is impaired at the restrictive temperature (24). Thus, at the restrictive temperature, failure of TAFII250 to induce cyclin A and D genes results in cell cycle arrest and ultimately apoptosis (67), which results from the loss of TAFII250 HAT activity. In addition, the effect of the TAFII250 mutation in ts13 cells on cellular transcription is not global, and only a small number of genes are affected (47, 67, 84). Transcription of the c-fos gene, for example, is not influenced by a shift of ts13 cells to 39.5°C, whereas the activity of the cyclin A promoter is decreased by 8- to 10-fold.
The simian virus 40 large T antigen (SV40 T-Ag) and the human cytomegalovirus (HCMV) major immediate-early proteins (MIEPs) can overcome the transcriptional defect of ts13 cells at the restrictive temperature for TAFII250 (19, 49). Rescue of ts13 cell transcription is partially restored by the IEP86 protein, although near-complete rescue requires expression from the entire major immediate-early transcription unit, implying that at least several proteins are required to overcome the loss of TAFII250 (49, 50). Moreover, studies demonstrate that while SV40 T-Ag and HCMV MIEPs can compensate for some of the functions of TAFII250, it is unlikely that a single viral protein is able to replace the multiple functions of TAFII250. For instance, the HCMV MIEPs cannot rescue the cell cycle defect in ts13 cells grown at the nonpermissive temperature for TAFII250 function, although their expression does prevent apoptosis (50). Genetic evidence also indicates that different MIEP functions prevent apoptosis and stimulate transcription (50). This is consistent with the multiple activities of TAFII250, such as transcriptional activation, cell cycle progression, and effects on apoptosis, involving distinct and separable functions of TAFII250. Thus, the multifunctional nature of TAFII250 could explain why transcriptional activation of cyclin promoters by a rescuing protein does not necessarily result in cell cycle progression or inhibition of apoptosis when ts13 cells are shifted to the nonpermissive temperature. HBx protein was reported to promote cell cycling in ts13 cells incubated at the restrictive temperature for endogenous mutant TAFII250, as well as to overcome the transcriptional defect (27). As HBx has been shown to be an inducer of apoptosis in many cells (9, 16, 39, 66, 70, 74, 78, 79), it is unclear how expression of HBx in a TAFII250 mutant cell line can block the antiapoptotic effect that results from the loss of TAFII250 activity.
Studies have characterized the temperature-responsive component of the cyclin A promoter, demonstrating that this element contains an ATF/CREB transcription factor binding site which is involved in its activation (85). ATF and CREB factors and the ATF–CREB-responsive element (CRE) site in cyclin A and D promoters can be induced via a Src kinase signaling pathway (44). HBx can activate Src kinases (40, 41), implicating an established mechanism of action in HBx induction of cyclin gene activity. Moreover, HBx is widely reported to bind to and activate ATF/CREB (reviewed in reference 2). Thus, there are potentially multiple mechanisms by which HBx promotes early cell cycling. In this report, we characterized the mechanism by which HBx promotes cycling of growth-arrested cells. We examined whether HBx induction of cell cycling occurs through its nuclear functions by replacing TAFII250 or through its cytoplasmic functions by activating Src kinases. We show that the induction of the cyclin A promoter by HBx in ts13 cells, which is essential for cell transit through G1, is dependent on activation of the Src family of tyrosine kinases. HBx could not functionally replace the loss of TAFII250 activity for activation of the cyclin A promoter or for early cell cycle progression. Furthermore, HBx induction of the cyclin A promoter through cytoplasmic activation of Src kinases is shown to be involved in the release of cells from quiescence and their transit through G1 to the junction with S phase. HBx is shown to be located predominately in the cytoplasm of ts13 cells regardless of temperature, consistent with an absolute requirement for induction of cytoplasmic signaling cascades but inconsistent with an essential requirement for nuclear transcription functions. In fact, an HBx protein that was engineered to concentrate in the nucleus by fusion to a nuclear localization signal (NLS) failed to substitute for TAFII250 function or to promote cell cycle progression. Finally, HBx is shown to activate the endogenous cyclin A promoter and cyclin A–cyclin-dependent kinase 2 (cdk2) complexes in wild-type (TAFII250 containing) cells through cytoplasmic activation of Src signaling pathways.
MATERIALS AND METHODS
Cell culture.
ts13 cells were maintained at 33°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing penicillin (100 U/ml) and streptomycin (100 μg/ml), supplemented with 10% fetal bovine serum. Chang cells were maintained in DMEM at 37°C as described above.
Plasmids and viruses.
The HBx-expressing plasmids, pAd-CMVX, pAd-HBxFlag, pAd-HBxo, pAd-HBxNLSFlag, and pAd-HBxSLNFlag, the TAFII250-expressing plasmid, the Csk-expressing plasmid (pCaCsk), and the cyclin A promoter-driven luciferase reporter plasmid (pCyA-Luc) have been described previously (23, 41, 84). High-concentration stocks of plasmids were purified using the Concert High Purity Plasmid Maxiprep System (Life Technologies) according to the manufacturer's instructions. Replication-defective recombinant adenovirus (Ad) vectors have been described previously (23). Ad-HBx and Ad-HBxo express wild-type HBx and an HBx mutant mRNA that lacks all potential translation initiation codons and does not synthesize HBx protein, respectively. HBx was inserted in place of Ad region E1. Ad vectors were propagated and titers were determined on 293 cells, which complement the loss of the E1 transcription unit.
Transfection, luciferase assays, and survival curves.
All transient transfections were performed by both standard calcium phosphate transfection methods and lipid methods using Lipofectamine-Plus (Life Technologies) according to the manufacturer's instructions. Both methods gave similar results, and only those from the Lipofectamine-Plus transfections are reported here. Luciferase reporter assays were performed using the Promega luciferase assay system according to the manufacturer's instructions. To analyze HBx-induced ts13 survival at the nonpermissive temperature, cells were cotransfected with 0.5 μg of a green fluorescent protein (GFP)-expressing plasmid (pGFP) and amounts of pAd-CMV control vector or pAd-CMVX (23) ranging from 1 to 10 μg/5 × 106 cells. After 24 h of recovery to ensure protein expression, cells were either maintained at 33°C (permissive for TAFII250) or shifted to 39.5°C (restrictive). At indicated times after the temperature shift, relative cell survival was calculated from the mean of surviving GFP-expressing cells, collected by viewing 10 fields at 40× power using a standard UV light microscope outfitted with GFP filters.
Flow cytometry.
These studies were carried out as previously described (7), with minor changes as described below. Briefly, Chang cells were made quiescent by cultivation in DMEM without serum for 30 h, infected with replication-defective Ad vectors expressing HBx (Ad-CMVX) or a mutant devoid of all potential HBx AUG codons, known as HBxo (Ad-CMVXo), at 25 PFU per cell, and maintained in DMEM in the absence of serum for up to 24 h. Cells were lysed in a solution containing 0.1% Nonidet P-40, 50 μg of propidium iodide per ml, 100 μg of DNase-free RNase A per ml, 5 mM NaCl, and 10 mM trisodium citrate. Flow cytometry was performed without modification as described previously (34). Results shown are typical of three independent trials.
Kinase assays.
For the cyclin A-cdk2 complex assay, serum-starved Chang cells were transfected with plasmid DNAs for 24 h and lysed, and extracts were prepared as described previously (7). Cyclin A was specifically immunoprecipitated with a commercial monoclonal antibody to cyclin A (Upstate Biotechnology, Lake Placid, N.Y.), and equal fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% gel) and immunoblotted with the same antibody, followed by visualization with the enhanced chemiluminescence system (ECL; Amersham). The remaining immunoprecipitate was added to in vitro kinase reactions containing 5 μCi (1 Ci = 37 GBq) of [γ-32P]ATP and 50 mg of histone H1 per ml in 50-μl reaction volumes as described previously (7, 33). Proteins were resolved by SDS-PAGE (12% gel) and quantitated and detected by phosphorimage analysis. Data represent typical results from three independent experiments. The Src kinase assay was carried out as described previously (41), with minor changes. Briefly, cells were lysed in buffer containing 1% Nonidet P-40, 150 mM NaCl, 20 mM Tris-HCl (pH 8), 2.5 mM EDTA, 1 mM Na3VO4, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, and 10 μg of leupeptin per ml. c-Src was immunoprecipitated from equal amounts of extract using a commercial monoclonal antibody (Upstate Biotechnology), and equal amounts were resolved by SDS-PAGE (12% gel) and immunoblotted with the same antibody. Equal fractions of the remaining immunoprecipitate were resuspended in kinase buffer (20 mM HEPES [pH 7.4], 10 mM MnCl2) with 0.2 μg of acid-denatured enolase (Sigma), 20 μCi of [γ-32P]ATP, and 10 μM ATP, as previously described (41). Samples were resolved by SDS-PAGE (12% gel) and subjected to phosphorimage analysis. Data shown are typical of three independent experiments.
Indirect immunofluorescence.
Immunofluorescent antibody staining of ts13 cells transfected with HBx (pAd-HBxFlag) was performed as previously described (23). Briefly, cells were grown on collagen-coated coverslips, transfected with either pAd-HBxFlag, pAd (DNA vector control), pAd-HBxNLSFlag, or pAd HBx SLNFlag, and allowed to recover for 24 h. For fixation and permeabilization, the medium was removed and cells were washed twice with phosphate-buffered saline (PBS) and then treated in 95% ethanol plus 5% acetic acid overnight at −20°C or in 70% acetone plus 30% methanol for 10 min at −20°C. Fixed and permeabilized cells were washed with PBS and then blocked in PBS plus 1% nonfat dry milk for 30 min at 37.5°C. Cells were incubated with anti-Flag M1 antibody (Kodak) for 1 h at 37°C, washed four times with PBS, and then incubated with secondary donkey anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibody. Cells were visualized and photographed using a Zeiss Axiophot fluorescence photomicroscope. Both fixation methods gave identical results.
RESULTS
HBx requires TAFII250 activity for cyclin A promoter transcription.
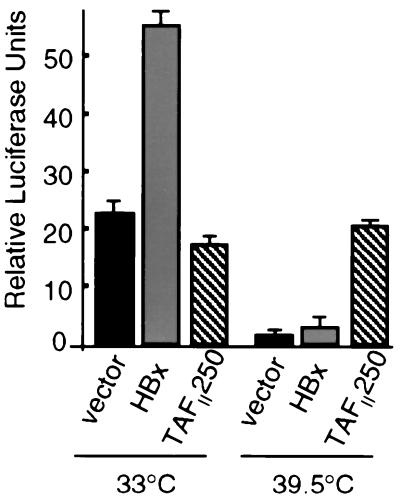
Studies first examined the ability of HBx to stimulate transcription of a cyclin A promoter-luciferase reporter construct in ts13 cells at the permissive temperature for TAFII250 transcriptional activity (33°C). Cells were transiently transfected with a plasmid expressing wild-type HBx or control plasmid DNA and were maintained at the permissive temperature. Cotransfection with pGFP demonstrated equal transfection efficiencies in all samples (∼70%; data not shown). HBx was found to reproducibly induce the ectopic cyclin A promoter two- to threefold at the permissive temperature (Fig. 1). These results are consistent in magnitude with the widely reported weak to moderate transactivation activity of HBx in a variety of cells and with different promoters (typically two- to sixfold). To evaluate whether HBx can rescue the ts13 transcription defect resulting from inactivation of TAFII250 at the restrictive temperature, cells were transfected for 5 h at 33°C with expression plasmids for HBx and the cyclin A promoter-controlled luciferase reporter and were maintained at the nonrestrictive temperature (33°C) or shifted to 39.5°C. At the restrictive temperature for TAFII250 activity, HBx only marginally stimulated transcription of the transfected cyclin A promoter just slightly above that of vector alone (Fig. 1). Importantly, HBx did not even stimulate the cyclin A promoter to the basal levels of transcription that were observed at the permissive temperature. In fact, the experiment could not be continued past 30 h of transfection due to cell death, indicating the failure of HBx to rescue TAFII250 function. Transfection of cells with the HBx expression plasmid, ranging from 0.25 to 10 μg per 107 cells (a 40-fold concentration range), also failed to recover cyclin A promoter activity at the restrictive temperature (data not shown). Identical results were obtained when cells were transfected at 33°C and then shifted to 39.5°C 1 day later or when they were transfected at 39.5°C and maintained at the nonpermissive temperature. Consequently, there were no experimental conditions in which HBx replaced TAFII250 function for cyclin A promoter activity. In contrast, cotransfection of ts13 cells at the restrictive temperature (39.5°C) with the cyclin A promoter-reporter construct and a wild-type TAFII250 expression vector recovered almost full cyclin A promoter transcriptional activity compared to control cells expressing functional endogenous TAFII250 at the permissive temperature. Since about 70% of the cells were transfected (data not shown), these data demonstrate that ectopic expression of TAFII250 can rescue the transcription defect of ts13 cells within 24 h at 39.5°C. Ectopic expression of wild-type TAFII250 in ts13 cells at the permissive temperature actually reduced cyclin A promoter activity slightly, possibly as a result of squelching due to supraphysiological expression levels. HBx did not lose function at 39.5°C, since at 39.5 and 33°C it stimulated transcription equally well of a reporter controlled by a minimal promoter and four AP-1 transcription factor binding sites in Chang cells (Table 1). In addition, HBx is stably synthesized at 39.5°C (shown later in Fig. 2). Collectively, these data demonstrate that HBx requires TAFII250 activity for cyclin A promoter activation, suggesting that nuclear HBx functions are either insufficient or unnecessary to promote early cell cycling, which was investigated next.
FIG. 1.
HBx stimulation of cyclin A promoter requires TAFII250 function. ts13 cells grown at 33°C were transiently transfected with control vector, HBx expression vector, or wild-type TAFII250 expression vector plus a cyclin A promoter-luciferase reporter construct. Cells were maintained at the permissive temperature (33°C) or shifted 5 h later to the nonpermissive temperature (39.5°C) for TAFII250 activity as described previously (24). At 24 h posttransfection, transcriptional activity was measured by relative luciferase light units and normalized for transfection efficiency and protein concentration. Data represent the means of at least three independent experiments, with calculated standard errors shown.
TABLE 1.
HBx requirement for TAFII250 in AP-1-directed transcription
| Temperature (°C) | Construct | AP-1 luciferase activity (103) |
|---|---|---|
| 33 | ΔBSa | 6.2 ± 0.4 |
| HBx | 13.3 ± 0.5 | |
| 39.5 | ΔBS | 7.0 ± 0.3 |
| HBx | 12.3 ± 0.4 |
ΔBS, vector alone.
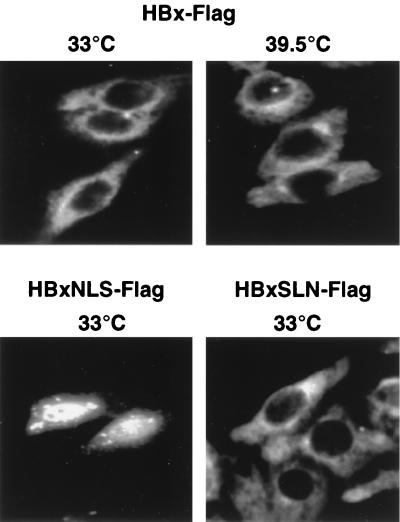
FIG. 2.
HBx remains predominantly cytoplasmic in ts13 cells regardless of temperature. ts13 cells were grown on coverslips at 33°C, transiently transfected with a plasmid expressing HBxFlag, which is an HBx protein containing a C-terminal Flag epitope that behaves identically to wild-type HBx, or HBxFlag containing an NLS (HBxNLS) or a control plasmid expressing a mutant NLS (HBxSLN) (23). Cells were maintained at 33°C or shifted to 39.5°C for 24 h, fixed and permeabilized on coverslips, and reacted with M2 anti-Flag antibodies followed by FITC-conjugated secondary antibody to visualize HBxFlag. Immunofluorescence photomicrographs (magnification, ×400) are shown for cells representative of each field. Control cells with vector alone did not demonstrate antibody staining (23, 74; data not shown).
HBx protein engineered to concentrate in the nucleus requires TAFII250 function.
Most studies have found HBx protein to be largely, but not exclusively, in the cytoplasm of a variety of cell types, whether expressed by transient transfection or in the context of viral infection (e.g., see references 20, 21, and 23). The cellular location of HBx in ts13 cells was investigated first using an HBx construct containing a C-terminal foreign Flag epitope. HBxFlag was shown previously to behave identically to unmodified wild-type HBx (23). ts13 cells were transfected with the HBxFlag expression plasmid or control vector DNA, and then indirect immunofluorescence analysis was carried out on cells fixed to coverslips and stained with anti-Flag antibodies followed by FITC-conjugated secondary antibodies. Although there was a low level of nuclear staining, the majority of HBx was found in the cytoplasm in all cells observed (Fig. 2). Furthermore, there was no change in wild-type HBx intracellular distribution when ts13 cells were shifted to the nonpermissive temperature for TAFII250 activity 24 h prior to fixation (Fig. 2). An HBx variant engineered to contain an N-terminal NLS from SV40 T-Ag (HBxNLS) (23) was concentrated in the nucleus but retained some cytoplasmic distribution. An HBx control which contains a defective NLS sequence (HBxSLN) (23) remained cytoplasmic. HBxNLS and HBxSLN have been characterized extensively in a variety of cell types (e.g., see references 23 and 77). Shifting of the cells to 39.5°C did not alter the intracellular distribution of HBxNLS or HBxSLN proteins from that observed at 33°C (data not shown). It can be concluded, therefore, that HBx proteins are synthesized and retain their typical intracellular distribution at both 33 and 39.5°C.
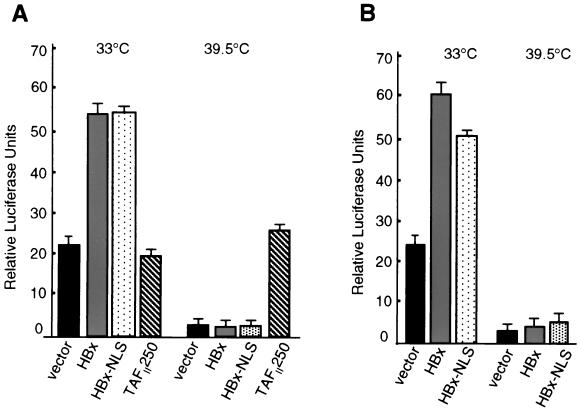
Studies were then conducted to examine whether HBx failed to functionally replace TAFII250 at 39.5°C because too little HBx accumulates in the nucleus in ts13 cells. Cells at the permissive or restrictive temperature were transfected with wild-type HBx, HBxNLS, or wild-type TAFII250 expression plasmids, and the effect on cyclin A promoter activity was determined (Fig. 3A). At the restrictive temperature, ectopic expression of wild-type TAFII250 recovered normal levels of cyclin A promoter transcriptional activity compared to the nonrestrictive temperature. There was no recovery of cyclin A promoter activity at the restrictive temperature by HBx or HBxNLS proteins. Thus, nuclear HBx protein does not functionally replace TAFII250 transcriptional activity at the restrictive temperature. Studies performed at the nonrestrictive temperature for TAFII250 activity demonstrate similar activation by HBxNLS and HBx. Since some HBxNLS remains cytoplasmic whereas the nuclear level of HBx is increased strongly, these results indicate that even with increased levels of HBx in the nucleus, like wild-type HBx, TAFII250 function is still required for activation of the cyclin A promoter. Because titration analysis of HBx showed that even 10-fold-lower levels could activate transcription, we suspect that the small amount of cytoplasmic HBxNLS in ts13 cells is sufficient for activation of the cyclin A promoter. These results also suggest that nuclear and cytoplasmic HBx functions are involved in early cell cycle progression, but the nuclear function does not include a previously reported TAFII250 activity which could not be reproduced here.
FIG. 3.
Nuclear HBx does not complement for loss of TAFII250 in cyclin A or CRE-dependent promoter activity. (A) ts13 cells were transiently transfected with a cyclin A promoter-luciferase reporter construct and a vector expressing wild-type HBx, an HBx engineered to concentrate largely but not completely in the nucleus by inclusion of the SV40 T-Ag NLS (HBxNLS) (23), or wild-type TAFII250. (B) Cells were transfected as described above but with a 4× multimerized CRE-basal promoter-luciferase reporter construct. Cells were maintained at 33°C or shifted to the nonpermissive temperature for TAFII250 (39.5°C) for 24 h, and transcriptional activity was determined as described in the legend for Fig. 1. Results represent the means of at least three independent experiments, with calculated standard errors shown.
Studies were therefore performed to determine whether the nuclear HBx function involves activation of the transcription factor CREB, since HBx has been shown to bind and activate CREB in vitro and since CREB activation is required for stimulation of the cyclin A promoter. Cotransfection of ts13 cells at the permissive temperature with a CRE-luciferase reporter and wild-type HBx (Fig. 3B) demonstrated two- to threefold activation of CREB-dependent transcription by wild-type HBx. Similar analysis using HBxNLS, which is largely but not entirely distributed in the nucleus, demonstrated slightly lower activation of the CREB-dependent reporter. This is consistent with a previous report which suggested that HBx activation of CREB might involve both cytoplasmic and nuclear HBx functions (90). At the restrictive temperature for TAFII250, neither wild-type HBx nor HBxNLS proteins stimulated CREB-dependent transcription (Fig. 3B). This is consistent with an essential requirement for TAFII250 function in transcriptional activation by HBx. Studies presented later demonstrate that HBx activation of Src kinase signaling is involved in CREB activation as well.
HBx requires TAFII250 activity and cytoplasmic functions to promote cell viability and cycling.
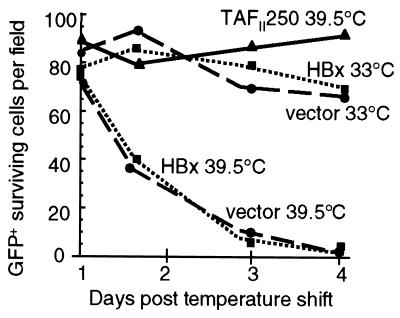
We sought to determine whether HBx promotion of ts13 cell viability and cycling involves nuclear HBx functions that require or are independent of TAFII250 activity. Cells were cotransfected with a GFP expression vector and pAd-CMVX or control plasmid DNA and then maintained at the permissive temperature (33°C) or shifted to a restrictive temperature (39.5°C) for endogenous TAFII250 activity. Under these conditions, all GFP-expressing cells were cotransfected with either TAFII250, HBx, or vector DNA at ∼70% efficiency, regardless of temperature (data not shown). At different times following the temperature shift, cells were observed by light microscopy, and the number of viable GFP-expressing cells was quantified as described in Material and Methods. Approximately 90% of the transfected ts13 cells containing wild-type TAFII250 and GFP survived at the nonpermissive temperature, indicating inhibition of cell death (Fig. 4). Cells expressing HBx were identical to control cells transfected with vector alone at the nonpermissive temperature, demonstrating widespread cell death. Thus, HBx did not delay the onset or rate of cell death and therefore did not promote cell cycling in the absence of TAFII250 activity. Identical results were obtained with HBxNLS, which concentrates in the nucleus (data not shown). We attempted to isolate surviving colonies of ts13 cells that were transformed by HBx at 39.5°C. Cells were transformed with vector alone or plasmids expressing HBx, HBxNLS, or wild-type TAFII250, at concentrations ranging from 1 to 10 μg of plasmid per 5 × 106 cells. The numbers of surviving transformants after 2 weeks of selection at 39.5°C are shown in Table 2. HBx, HBxNLS, and vector DNA all gave rise to fewer than two colonies per 107 cells, a transformation frequency of 2 × 10−7. In comparison, cells transformed with TAFII250 averaged 7 × 106 transformants per 107 cells, a transformation efficiency almost equal to the efficiency of transfection, indicating that the majority of cells transfected with TAFII250 could be rescued. In addition, attempts to produce stably selected ts13 cells expressing HBx under a regulated promoter failed due to cell death, likely resulting from leaky synthesis of HBx protein (data not shown). Collectively, these results demonstrate that nuclear HBx functions are not sufficient to promote cell viability and cycling in the absence of TAFII250 activity.
FIG. 4.
Effect of HBx on ts13 cell viability and growth in the absence of TAFII250 activity. ts13 cells at 33°C were transiently transfected with vector alone or with an HBx or wild-type TAFII250 expression vector. Duplicate plates of cells containing either vector alone or HBx were maintained at the permissive (33°C) temperature for endogenous TAFII250 activity. Cell viability and growth were determined at days 1 to 4 after the shift to the nonpermissive temperature based on cotransfected cells expressing a GFP marker, as described in Materials and Methods. Analysis of cell viability commenced at 24 h following the shift of samples to the nonpermissive temperature. Data represent a typical experiment that did not differ from three other studies by more than 10% and are displayed as the mean numbers of surviving cells per field obtained by the average of 10 fields per time point. An equal number of cells per field was plated at the start of the experiment.
TABLE 2.
Effect of HBx and TAFII250 on ts13 cell viability
| Construct | No. of colonies | Transformation efficiency |
|---|---|---|
| Vector alone | <2 | 2 × 10−7 |
| HBx | <2 | 2 × 10−7 |
| HBxNLS | <2 | 2 × 10−7 |
| pTAFII250 | 7 × 106 | 7 × 106 |
HBx stimulates the cyclin A promoter, promotes formation of cyclin A-cdk2 complexes, and deregulates early cell cycle checkpoints in a Src kinase-dependent manner.
It was shown previously that activation of transcription by HBx in part involves its cytoplasmic stimulation of Src kinase signaling and the Ras-Raf-MAPK/JNK pathway (6, 8, 40, 41, 73, 86). Moreover, HBx was shown previously to stimulate cyclin E and A synthesis by acting on cytoplasmic signal transduction pathways (7). The failure of HBx to stimulate cell cycling and the cyclin A promoter in the absence of TAFII250 activity at 39.5°C, even when relocated to the nucleus (Fig. 1 and 3), implicated HBx activation of signaling pathways in this action. We therefore characterized HBx activation of the endogenous cyclin A promoter and cyclin A-cdk2 complexes. Previous studies have shown that HBx can deregulate the G0 cell cycle control checkpoint in a variety of cell types with arrested growth and can facilitate transit into or through G1 to the early S phase transition (7, 42, 71). We first show that HBx promotes transit of G0-G1-arrested cells through G1 to the G1-S junction in the absence of serum. It was not possible to conduct these studies in ts13 cells, which apoptose when growth is arrested by serum depletion (67), particularly with expression of HBx (data not shown). Consequently, Chang cells, a human liver cell line which can have its growth arrested by serum withdrawal without inducing apoptosis, were used.
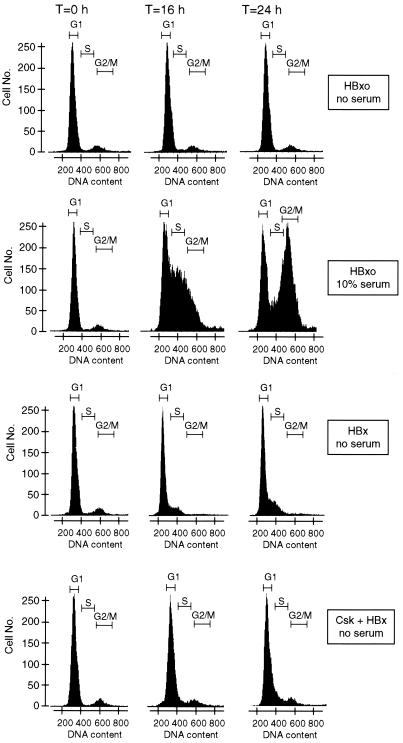
Quiescent cells (cells in G0 and at the G0-G1 junction) were accumulated by serum withdrawal. Cells were then transduced by a replication-defective Ad vector lacking the Ad E1 region, which in its place expresses the HBx gene, or by a control HBxo gene which expresses an mRNA that cannot synthesize HBx protein (7, 23, 40, 73). Studies have shown that these vectors remain genetically silent for the time course of these experiments (73). Cells were lysed at various times after transduction with Ad vectors, and flow cytometry was performed on propidium iodide-treated nuclei (Fig. 5). In the absence of serum stimulation, cells expressing the HBxo gene did not exit from the G0-G1 fraction upon the termination of these studies (24 h posttransfection). Control cells expressing HBxo in the presence of serum proliferated more rapidly and entered S phase during this same time course. In comparison, HBx-expressing cells in the absence of serum showed a gradual increase in accumulation at the G1 junction with early S phase which was striking by 24 h of expression. Although cells expressing HBx in the absence of serum did not enter S phase and replicate, there was an evident increase in cell number at the rightward G1 transition with S phase. This likely represents the initiation of DNA decompaction in the nucleus which precedes entry into S phase and causes an increased fluorescent signal (62). The disappearance of the small fraction of cells in the G2-M phase in the HBx-expressing sample at time zero probably represents cells that cycled through to G1 phase and then stopped (71).
FIG. 5.
Effect of HBx on the cell cycle. Change cells were accumulated largely in the G0-G1 phase by 30 h of maintenance in serum-free medium and then transduced with replication-defective Ad vectors with the wild-type HBx or mutant HBxo gene substituted for region E1. HBxo is a control that cannot synthesize HBx protein. Control cells containing HBxo were supplemented with 10% serum at the time of Ad transduction. Flow cytometry was performed using propidium iodide staining of nuclei. Histograms of nuclei are shown, obtained from cells at time zero (immediately following Ad transduction) and at 16 and 24 h. The DNA content of the nuclei was determined within 2 h of cell lysis by flow cytometry using the MODFIT program. Data are presented from a single experiment which did not vary by more than 10% in three trials.
Studies next determined whether HBx activation of Src kinases is important for deregulation of early cell cycle checkpoints. Chang cells were transfected with a plasmid expressing the kinase Csk, which phosphorylates and downregulates Src kinases. Csk was chosen for these studies because it very specifically blocks the family of Src kinases. At 8 h following transfection, cells were transduced with an Ad vector expressing HBx or HBxo. Approximately 70% of the cells were found to be transfected, whereas 95% were transduced by the Ad vector (data not shown). Previous studies demonstrated that Csk does not inhibit the expression of HBx controlled by the CMV promoter (40, 41). Expression of Csk in HBx-expressing cells reduced the fraction of cells that accumulated at the G1-S phase junction by about half at 24 h in the absence of serum. Various chemical inhibitors of Src kinases were found to provide equivalent results and will be published elsewhere. Cells expressing both Csk and HBxo were identical to the HBxo controls in the absence of serum (data not shown). Since Src kinases are stimulated by HBx acting in the cytoplasm (41), which is required for HBx promotion of cell cycling as shown here, we did not evaluate the effect of HBxNLS. These results demonstrate that HBx deregulates early cell cycle checkpoints in a Src kinase-dependent manner, an event that includes synthesis of cyclin A and activation of cyclin A-cdk2 complexes.
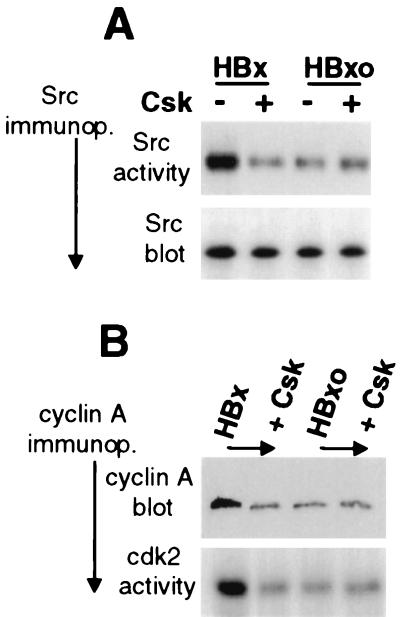
Studies therefore examined the role of cytoplasmic HBx activities in deregulation of early cell cycle control. Since many cytoplasmic functions of HBx can be accounted for by its activation of Src signaling, the role of Src activation in the stimulation of the endogenous cyclin A promoter was investigated. Serum-starved Chang cells were transfected with HBx or HBxo expression vectors, with or without cotransfection of a plasmid expressing the kinase Csk. Cell extracts were prepared 30 h later, c-Src was specifically immunoprecipitated, and HBx activation was shown by the ability of immunoprecipitated Src to phosphorylate the substrate enolase in vitro (Fig. 6A). HBx induced a fourfold activation of Src compared to cells transfected with the HBxo vector which was prevented by coexpression with Csk (Fig. 6A). Equal amounts of Src were assayed, as shown by the immunoblot analysis of the immunoprecipitates. Studies then determined whether HBx stimulates endogenous cyclin A gene expression through a Src kinase pathway in growth-arrested Chang cells. Cells were assayed for cyclin A protein levels and cyclin A-cdk2 activity 24 h after transfection, corresponding to the accumulation of cells in G1-early S phase (Fig. 5). Compared to the HBxo control, HBx induced about a fivefold increase in cyclin A protein levels in these cells, which was blocked by coexpression of Csk (Fig. 6B). The activity of cyclin A-cdk2 complexes was determined by immunoprecipitation of cyclin A followed by an in vitro assay of cdk2 phosphorylation of the substrate histone H1 (7) (Fig. 6B). Compared to the HBxo control, HBx induced a fivefold stimulation of cyclin A-cdk2 complexes in serum-starved cells by 24 h after transfection, which was prevented by coexpression with Csk. These data therefore demonstrate that HBx stimulation of Src kinases is an important cytoplasmic component of deregulation of early cell cycle control.
FIG. 6.
HBx stimulates endogenous cyclin A promoter and cyclin A-cdk2 complexes through a Src kinase pathway. Quiescent serum-starved Chang cells were transiently transfected at ∼70% efficiency (data not shown) with vectors expressing HBx or HBxo, with and without cotransfection of a plasmid expressing Csk, a negative regulator of Src kinases. At 24 h posttransfection, cell lysates were prepared. (A) pp60 c-Src was immunoprecipitated from equal amounts of cell lysates using a specific monoclonal antibody, and equal fractions were resolved by SDS-PAGE (12% gel) and immunoblotted with the same antibody (Src blot) or assayed for in vitro phosphorylation activity with [γ-32P]ATP and the substrate enolase (Src activity). (B) Cyclin A was immunoprecipitated using a specific monoclonal antibody from equal amounts of lysate, and equal fractions of the immunoprecipitate were resolved by SDS-PAGE (15% gel) and immunoblotted for cyclin A protein (cyclin A blot) or assayed for associated cdk2 activity by in vitro phosphorylation of histone H1, a substrate of cdk2, using [γ-32P]ATP. Phosphorylated enolase and histone H1 were resolved by SDS-PAGE (12% gel), autoradiographed, and quantitated by digital densitometry. Results shown are typical of three independent experiments which did not vary by more than 20%.
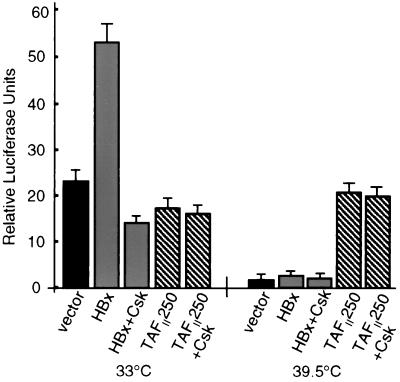
Studies next showed that HBx activation of Src signaling is also vital for stimulation of the ectopic cyclin A promoter-reporter in ts13 cells. Cells were cotransfected with vectors expressing HBx or TAFII250, plus the cyclin A promoter-reporter, with or without cotransfection of a plasmid expressing Csk. Cells maintained at the permissive temperature for the ts13 mutation in TAFII250 showed a consistent increase in cyclin A promoter activity of two- to threefold with HBx expression, which was blocked by overexpression of Csk (Fig. 7). At the restrictive temperature for TAFII250, HBx stimulated the cyclin A promoter only very slightly, and this stimulation was also blocked by Csk expression. As a control, cells maintained at the restrictive temperature and expressing wild-type TAFII250 were shown to be unaffected by overexpression of Csk. Furthermore, Csk did not downregulate transcription of a CMV promoter-controlled reporter construct (data not shown), excluding inhibition of HBx activity by a general decrease in transcription (Fig. 7). Thus, HBx stimulates the cyclin A promoter and deregulates early cell cycle control through a pathway that requires TAFII250 function and activation of Src kinases.
FIG. 7.
HBx stimulation of cyclin A promoter through a Src kinase pathway. ts13 cells grown at the permissive temperature (33°C) or the restrictive temperature (39.5°C) were transiently transfected with the cyclin A promoter-luciferase reporter construct and control vector, HBx, or wild-type TAFII250, with or without a cotransfected Csk expression vector. Cells were then maintained at 33°C or shifted to 39.5°C 5 h after transcription. In some studies, cells were shifted to 39.5°C 24 h after transfection at 33°C, with identical results (data not shown). At 24 h posttransfection, transcriptional activity was measured by relative luciferase light units and normalized for transfection efficiency and protein concentration. Data represent the means of at least three independent experiments, with calculated standard errors shown.
DISCUSSION
The HBV HBx protein possesses a wide variety of activities, such as activation of cytoplasmic signal transduction pathways, induction or sensitization of cells to apoptosis, loss of early cell cycle control checkpoints, possibly direct interaction with several components of the transcription apparatus, and a weak to moderate activation of transcription directed by RNA polymerases I, II, and III. Given the variety of activities attributed to HBx and the fact that its ability to activate transcription is at best moderate, there has been considerable difficulty in firmly establishing the mechanism(s) through which HBx activates transcription. The in vitro demonstration of interactions between HBx and components of the transcriptional machinery such as RBP5 or TFIIH or the transcription factor ATF/CREB has led to the suggestion that HBx functions directly in the nucleus (14, 51, 60, 61, 90). Transcriptional activation by HBx, according to this model, is a consequence of direct interaction with any of several transcription factors in the nucleus. While most evidence points to a largely cytoplasmic location for HBx, some of the protein is typically in the nucleus, providing support for this model (20, 21, 23, 63, 72). Moreover, HBx is thought to activate the transcription factor CREB, at least in part by direct interaction in the nucleus (90). Another large body of evidence also supports a cytoplasmic function for HBx in transcriptional activation. Activation of NF-κB, NF-AT, and AP-1 by HBx as well as stimulation of transcription directed by RNA polymerases, I, II, and III involve HBx activation of cytoplasmic signaling (6–8, 15, 17, 18, 23, 32, 38, 40, 41, 43, 48, 55, 73, 74, 86–88). Furthermore, most studies on HBx cellular location have demonstrated that most of the protein is located in the cytoplasm, regardless of whether it is synthesized during transient transfection or in the context of woodchuck hepatitis virus or human HBV infection of liver cells in vivo (20, 21, 23, 35, 63, 72). This is in accord with the observation that cytoplasmic HBx activation of Src signaling, in particular, strongly stimulates viral replication in cultured cells (40).
Ras and Src signal transduction pathways, both of which are activated by HBx, are critical effectors for progression of cells to the G1-early S phase transition of the cell cycle. Transit of quiescent cells through G1 involves stimulated synthesis of cyclin D followed by cyclin E, both of which associate with and activate cdk-4, -6, and -2 (reviewed in reference 65). Transit of cells to the S phase junction involves synthesis of cyclin A, which associates with and activates cdk2. Ras and Src signaling are involved in multiple events for progression to the G1-S transition (65), which include activation of Fos and Jun (AP-1) and ATF/CREB family members (5, 10, 37, 44, 53, 56, 64, 91; reviewed in reference 80). Studies have demonstrated that a key target of these signaling events for G1 progression is activation of cyclin D and A promoters by ATF/CREB transcription factors (69, 75, 85) in conjunction with specific activation of TAFII250 (85). The critical function of TAFII250 for activation of cyclin D and A promoters was shown to be its intrinsic HAT activity, as the ts13 cell mutation renders TAFII250 HAT defective at the restrictive temperature, prevents D and A cyclin gene transcription, and blocks progression to the G1-early S transition (24).
In this study, we explored the functions of HBx in promoting early cell cycling, including its purported functional replacement of TAFII250 (27). Since TAFII250 couples transcriptional activation to cell proliferation, functional substitution by HBx would be unprecedented, given that it is only 17 kDa in size and possesses no known enzymatic activities. It would also be expected to reflect a novel and underscribed proto-oncogenic activity of HBx, because continuous expression of HBx is presumably required during virus replication (21). We showed that HBx does not functionally replace known activities of TAFII250. HBx did not stimulate the cyclin A promoter in the absence of TAFII250 activity (Fig. 1), regardless of whether the nuclear levels of HBx were elevated (Fig. 2 and 3), and it did not rescue cells from death induced by the absence of TAFII250 activity (Table 1; Fig. 4). However, it was found that HBx required TAFII250 function to stimulate the cyclin A promoter (Fig. 1, 6, and 7). HBx also required TAFII250 function to induce quiescent cells to transit from G1 to the S phase (Fig. 5 and 6), since switching cells to the restrictive temperature for TAFII250 blocked cell cycling. Since the only defect at the restrictive temperature is in TAFII250 function and since TAFII250 function is essential for cell cycle progression (24, 49, 50, 67, 75, 84, 85), these data demonstrate that HBx does not override the requirement for TAFII250 in stimulating cell cycling. These data are consistent with other previous reports demonstrating induction of early cell cycling by HBx (and presumably activation of the cyclin D promoter as well) (6, 7, 42, 71), possibly to the G1-early S phase junction (71). In addition, HBx activation of Src signaling, which is coupled to activation of Ras (41), was shown to be important for progression of cells through G1 and activation of the cyclin A promoter (Fig. 6 and 7).
The results reported here are consistent with critical cytoplasmic functions of HBx in the stimulation of early cell cycling. HBx stimulation of Src signaling was found to be essential and is consistent with the established critical importance of Src and Ras in promoting progression of cells through G1. The inability of HBx to functionally replace TAFII250 during 24 to 30 h of expression is also consistent with the established longevity of the TAFII250-TFIID complex. Most studies demonstrate that this complex is very stable. In ts13 cells at the restrictive temperature for TAFII250, almost 24 h was required for replacement of the mutant form of TAFII250 by ectopically expressed wild-type protein (19, 49). Thus, the reported rescue by HBx of ts13 TAFII250 within 5 h of the inactivating temperature shift, if correct, would be unprecedented (27). However, HBx has been shown to bind in vitro to the CREB transcription factor (4, 51, 77, 90) and to stimulate CREB-dependent transcription in vivo (59, 90). Furthermore, an ATF/CREB element can function in transcriptional activation of cyclin D and A genes. Taken together, these observations raised the possibility that perhaps HBx might circumvent TAFII250 function by activating CREB, leading to cell cycling and prevention of cell death. Nevertheless, neither wild-type HBx nor a variant engineered to concentrate in the nucleus blocked cell death or displayed any TAFII250-independent activity, including transcriptional activation of the cyclin A promoter and CREB-dependent transcription at the restrictive temperature for TAFII250 (Fig. 3). Although nuclear HBx was not sufficient to activate cell cycling, studies need to determine its involvement in promotion of early cell cycling, possibly by direct interaction between HBx and CREB.
HBx stimulation of the cyclin A promoter and G1 cell cycle progression may represent a significant activity in the HBV life cycle. HBx expression during infection by HBV may stimulate quiescent hepatocytes, not to divide but to transit the G1-to-S-phase transition. A similar function is well established for regulatory proteins of many oncogenic viruses. It is thought that release of cells from quiescence may aid in viral replication by expanding the pool of deoxynucleoside triphosphates (dNTPs), which is significantly restricted during G0 and is elevated during the G1 transition (22). Moreover, dNTP pools are much lower in the cytoplasm, where HBV replicates, than in the nucleus, which could present a considerable impediment to HBV replication in quiescent cells. Transit through G1 would therefore increase dNTP metabolism and elevate the available pool for HBV replication. In contrast, if HBx were to function as a TAFII250-like protein, promoting cells into cycling, this would be deleterious, as studies have shown greatly impaired HBV replication in cells during S phase (57). Importantly, for classic retroviruses, depletion of dNTP pools, particularly dCTP, has been shown to be responsible for arrest of viral reverse transcription in G0 cells, which is relieved by transit into the G1-S transition or by supplementation of G0-arrested cells with high levels of dNTPs (13, 25, 26, 93, 94). HBx has been shown to be required for HBV replication (12, 95), which also utilizes reverse transcription of the viral RNA genome. In striking similarity to classic retroviruses, HBx promotes HBV reverse transcription and second-strand DNA synthesis in cultured cells through a pathway that involves HBx activation of Src kinases (40). Thus, our data support a role for HBx stimulation of early cell cycle transit, in part through Src kinase and TAFII250 stimulation of cyclin promoters and activation of cdks, so as to promote viral replication.
ACKNOWLEDGMENTS
This work was supported by NIH grants CA56533 (R.J.S.), CA74476 (M.B.), and GM51314 (N.T.) and by American Cancer Society grant RPG-98-201-CCG (E.H.W.).
REFERENCES
- 1.Albright S R, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 2.Andrisani O, Barnabas S. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis. Int J Oncol. 1999;15:1–7. doi: 10.3892/ijo.15.2.373. [DOI] [PubMed] [Google Scholar]
- 3.Aufiero B, Schneider R J. The hepatitis B virus X-gene product trans-activates both RNA polymerase II and III promoters. EMBO J. 1990;9:497–504. doi: 10.1002/j.1460-2075.1990.tb08136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnabas S, Hai T, Andrisani O M. The hepatitis B virus X protein enhances the DNA binding potential and transcription efficacy of bZip transcription factors. J Biol Chem. 1997;272:20684–20690. doi: 10.1074/jbc.272.33.20684. [DOI] [PubMed] [Google Scholar]
- 5.Barone M V, Courtneidge S A. Myc but not Fos rescue of PDGF signalling block caused by kinase-inactive Src. Nature. 1995;378:509–512. doi: 10.1038/378509a0. [DOI] [PubMed] [Google Scholar]
- 6.Benn J, Schneider R J. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signalling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benn J, Schneider R J. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci USA. 1995;92:11215–11219. doi: 10.1073/pnas.92.24.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benn J, Su F, Doria M, Schneider R J. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70:4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergametti F, Prigent S, Luber B, Benoit A, Tiollais P, Sarasin A, Transy C. The proapoptotic effect of hepatitis B virus HBx protein correlates with its transactivation activity in stably transfected cell lines. Oncogene. 1999;18:2860–2871. doi: 10.1038/sj.onc.1202643. [DOI] [PubMed] [Google Scholar]
- 10.Broome M A, Hunter T. Requirement for c-Src catalytic activity and the SH3 domain in platelet-derived growth factor BB and epidermal growth factor mitogenic signaling. J Biol Chem. 1996;271:16798–16806. doi: 10.1074/jbc.271.28.16798. [DOI] [PubMed] [Google Scholar]
- 11.Burley S, Roeder R. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 12.Chen H-S, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen I S, Temin H M. Establishment of infection by spleen necrosis virus: inhibition in stationary cells and the role of secondary infection. J Virol. 1982;41:183–191. doi: 10.1128/jvi.41.1.183-191.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong J-H, Yi M-K, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chirillo P, Falco M, Puri P L, Artini M, Balsano C, Levrero M, Natoli G. Hepatitis B virus pX activates NF-κB-dependent transcription through a Raf-independent pathway. J Virol. 1996;70:641–646. doi: 10.1128/jvi.70.1.641-646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chirillo P, Pagano S, Natoli G, Puri P L, Burgio V L, Balsano C, Levrero M. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc Natl Acad Sci USA. 1997;94:8162–8167. doi: 10.1073/pnas.94.15.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong Y-S, Yao Y-L, Yang W-M, Kuzhandaivelu N, Seto E. The hepatitis B virus X-associated protein, XAP3, is a protein kinase C-binding protein. J Biol Chem. 1997;272:16482–16489. doi: 10.1074/jbc.272.26.16482. [DOI] [PubMed] [Google Scholar]
- 18.Cross J C, Wen P, Rutter W J. Transactivation by hepatitis B virus X protein is promiscuous and dependent on mitogen activated cellular serine/threonine kinases. Proc Natl Acad Sci USA. 1993;90:8078–8082. doi: 10.1073/pnas.90.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damania B, Alwine J. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 20.Dandri M, Petersen J, Stockert R J, Harris T M, Rogler C E. Metabolic labeling of woodchuck hepatitis B virus X protein in naturally infected hepatocytes reveals a bimodal half-life and association with the nuclear framework. J Virol. 1998;72:9359–9364. doi: 10.1128/jvi.72.11.9359-9364.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dandri M, Schirmacher P, Rogler C E. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J Virol. 1996;70:5246–5254. doi: 10.1128/jvi.70.8.5246-5254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denhardt D T, Edwards D R, Parfett C L. Gene expression during the mammalian cell cycle. Biochim Biophys Acta. 1986;865:83–125. doi: 10.1016/0304-419x(86)90024-7. [DOI] [PubMed] [Google Scholar]
- 23.Doria M, Klein N, Lucito R, Schneider R J. Hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunphy E L, Johnson T, Auerbach S S, Wang E H. Requirement for TAFII250 acetyltransferase activity in cell cycle progression. Mol Cell Biol. 2000;20:1134–1139. doi: 10.1128/mcb.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goulaouic H, Subra F, Mouscadet J F, Carteau S, Auclair C. Exogenous nucleosides promote the completion of MoMLV DNA synthesis in G0-arrested Balb c/3T3 fibroblasts. Virology. 1994;200:87–97. doi: 10.1006/viro.1994.1166. [DOI] [PubMed] [Google Scholar]
- 26.Harel J, Rassart E, Jolicoeur P. Cell cycle dependence of synthesis of unintegrated viral DNA in mouse cells newly infected with murine leukemia virus. Virology. 1981;110:202–207. doi: 10.1016/0042-6822(81)90022-2. [DOI] [PubMed] [Google Scholar]
- 27.Haviv I, Matza Y, Shaul Y. pX, the HBV-encoded coactivator, suppresses the phenotypes of TBP and TAFII250 mutants. Genes Dev. 1998;12:1217–1226. doi: 10.1101/gad.12.8.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haviv I, Shamay M, Doitsch G, Shaul Y. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol Cell Biol. 1998;18:1562–1569. doi: 10.1128/mcb.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haviv I, Vaizel D, Shaul Y. pX, the HBV-encoded coactivator, interacts with components of the transcription machinery and stimulates transcription in a TAF-independent manner. EMBO J. 1996;15:3413–3420. [PMC free article] [PubMed] [Google Scholar]
- 30.Haviv I, Vaizel D, Shaul Y. The X protein of hepatitis B virus coactivates potent activation domains. Mol Cell Biol. 1995;15:1079–1085. doi: 10.1128/mcb.15.2.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashida T, Sekiguchi T, Noguchi E, Sunamoto H, Ohba T, Nishimoto T. The CCG1/TAFII250 gene is mutated in thermosensitive G1 mutants of the BHK21 cell line derived from golden hamster. Gene. 1994;141:267–270. doi: 10.1016/0378-1119(94)90583-5. [DOI] [PubMed] [Google Scholar]
- 32.Henkler F, Lopes A R, Jones M, Koshy R. Erk-independent partial activation of AP-1 sites by the hepatitis B virus HBx protein. J Gen Virol. 1998;79:2737–2742. doi: 10.1099/0022-1317-79-11-2737. [DOI] [PubMed] [Google Scholar]
- 33.Hermann C P, Krais S, Montenarch M. Association of casein kinase II with immunopurified p53. Oncogene. 1991;6:877–886. [PubMed] [Google Scholar]
- 34.Ignatius M J, Chandler C R, Shooter E M. Nerve growth factor-treated neurite bearing PC12 cells continue to synthesize DNA. J Neurosci. 1985;5:343–351. doi: 10.1523/JNEUROSCI.05-02-00343.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacob J R, Ascenzi M A, Roneker C A, Toshkov I A, Cote P J, Gerin J L, Tennant B C. Hepatic expression of the woodchuck hepatitis virus X-antigen during acute and chronic infection and detection of a woodchuck hepatitis virus X-antigen antibody response. Hepatology. 1997;26:1607–1615. doi: 10.1002/hep.510260632. [DOI] [PubMed] [Google Scholar]
- 36.Jacobson R H, Ladurner A G, King D S, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 37.Johnson D, Frame M C, Wyke J A. Expression of the v-Src oncoprotein in fibroblasts disrupts normal regulation of the CDK inhibitor p27 and inhibits quiescence. Oncogene. 1998;16:2017–2028. doi: 10.1038/sj.onc.1201727. [DOI] [PubMed] [Google Scholar]
- 38.Kekule A S, Lauer U, Weiss L, Luber B, Hofschneider P H. Hepatitis B virus transactivator HBx uses a tumor promoter signalling pathway. Nature. 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 39.Kim H, Lee H, Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem. 1998;273:381–385. doi: 10.1074/jbc.273.1.381. [DOI] [PubMed] [Google Scholar]
- 40.Klein N, Bouchard M, Wang L-H, Kobarg C, Schneider R J. Src kinases involved in hepatitis B virus replication. EMBO J. 1999;18:5019–5027. doi: 10.1093/emboj/18.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein N P, Schneider R J. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol Cell Biol. 1997;17:6427–6436. doi: 10.1128/mcb.17.11.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koike K, Moriya K, Yotsuyanagi H, Iino S, Kurokawa K. Induction of cell cycle progression by hepatitis B virus HBx gene expression in quiescent mouse fibroblasts. J Clin Investig. 1994;94:44–49. doi: 10.1172/JCI117343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwee L, Lucito R, Aufiero B, Schneider R J. Alternate translation initiation on hepatitis B virus X mRNA produces multiple polypeptides that differentially transactivate class II and III promoters. J Virol. 1992;66:4382–4389. doi: 10.1128/jvi.66.7.4382-4389.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee R J, Albanese C, Stenger R J, Watanabe G, Inghirami G, Haines III G K, Webster M, Muller W J, Brugge J S, Davis R J, Pestell R G. pp60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. J Biol Chem. 1999;274:7341–7350. doi: 10.1074/jbc.274.11.7341. [DOI] [PubMed] [Google Scholar]
- 45.Levrero M, Balsano C, Natoli G, Avantaggiati M L, Elfassi E. Hepatitis B virus X protein transactivates the long terminal repeats of human immunodeficiency virus types 1 and 2. J Virol. 1990;64:3082–3086. doi: 10.1128/jvi.64.6.3082-3086.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Y, Nomura T, Cheong J, Dorjsuren D, Iida K, Murakami S. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and RNA polymerase II subunit 5. J Biol Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Gibson C, Hirschhorn R, Rittling S, Baserga R, Mercer W. Expression of thymidine kinase and dihydrofolate reductase genes in mammalian ts mutants of the cell cycle. J Biol Chem. 1985;260:3269–3274. [PubMed] [Google Scholar]
- 48.Lucito R, Schneider R J. Hepatitis B virus X protein activates transcription factor NF-κB without a requirement for protein kinase C. J Virol. 1992;66:983–991. doi: 10.1128/jvi.66.2.983-991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukac D, Harel N, Tanese N, Alwine J. TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol. 1997;71:7227–7239. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukac D M, Alwine J C. Effects of human cytomegalovirus major immediate-early proteins in controlling the cell cycle and inhibiting apoptosis: studies with ts13 cells. J Virol. 1999;73:2825–2831. doi: 10.1128/jvi.73.4.2825-2831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maguire H F, Hoeffler J P, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 52.Mahe Y, Mukaida N, Kuno K, Akiyama M, Ikeda N, Matshushima K, Murakami S. Hepatitis B virus X protein transactivates human interleukin-8 gene through acting on nuclear factor κB and CCAAT/enhancer-binding protein-like cis elements. J Biol Chem. 1991;266:13759–13763. [PubMed] [Google Scholar]
- 53.Mou S, Linnekin D. Lyn is activated during late G1 of stem-cell-factor-induced cell cycle progression in haemopoietic cells. Biochem J. 1999;342:163–170. [PMC free article] [PubMed] [Google Scholar]
- 54.Murakami S, Cheong J-H, Ohno S, Matsushima K, Kaneko S. Transactivation of human hepatitis B virus X protein, HBx, operates through a mechanism distinct from protein kinase C and okadaic acid activation pathways. Virology. 1994;199:243–246. doi: 10.1006/viro.1994.1119. [DOI] [PubMed] [Google Scholar]
- 55.Natoli G, Avantaggiati M L, Chirillo P, Puri P L, Ianni A, Balsano C, Levrero M. Ras- and raf-dependent activation of c-jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene. 1994;9:2837–2843. [PubMed] [Google Scholar]
- 56.Nebigil C G, Launay J M, Hickel P, Tournois C, Maroteaux L. 5-Hydroxytryptamine 2B receptor regulates cell-cycle progression: cross-talk with tyrosine kinase pathways. Proc Natl Acad Sci USA. 2000;97:2591–2596. doi: 10.1073/pnas.050282397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ozer A, Khaoustov V I, Mearns M, Lewis D E, Genta R M, Darlington G J, Yoffee B. Effect of hepatocyte proliferation and cellular DNA synthesis on hepatitis B virus replication. Gastroenterology. 1996;110:1519–1528. doi: 10.1053/gast.1996.v110.pm8613059. [DOI] [PubMed] [Google Scholar]
- 58.Pennisi E. Matching the transcription machinery to the right DNA. Science. 2000;288:1372–1373. doi: 10.1126/science.288.5470.1372. [DOI] [PubMed] [Google Scholar]
- 59.Perini G, Oetjen E, Green M R. The hepatitis B pX protein promotes dimerization and DNA binding of cellular basic region/leucine zipper proteins by targeting the conserved basic region. J Biol Chem. 1999;274:13970–13977. doi: 10.1074/jbc.274.20.13970. [DOI] [PubMed] [Google Scholar]
- 60.Qadri I, Conaway J W, Conaway R C, Schaack J, Siddiqui A. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc Natl Acad Sci USA. 1996;93:10578–10583. doi: 10.1073/pnas.93.20.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qadri I, Maguire H F, Siddiqui A. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rabinovitch P S. DNA content histogram and cell-cycle analysis. Methods Cell Biol. 1994;41:263–296. doi: 10.1016/s0091-679x(08)61723-9. [DOI] [PubMed] [Google Scholar]
- 63.Rahmani Z, Huh K W, Lasher R, Siddiqui A. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J Virol. 2000;74:2840–2846. doi: 10.1128/jvi.74.6.2840-2846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roche S, Koegl M, Barone M V, Roussel M F, Courtneidge S A. DNA synthesis induced by some but not all growth factors requires Src family protein tyrosine kinases. Mol Cell Biol. 1995;15:1102–1109. doi: 10.1128/mcb.15.2.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roussel M F. Key effectors of signal transduction and G1 progression. Adv Cancer Res. 1997;74:1–24. doi: 10.1016/s0065-230x(08)60763-0. [DOI] [PubMed] [Google Scholar]
- 66.Schuster R, Gerlich W H, Schaefer S. Induction of apoptosis by the transactivating domains of the hepatitis B virus X gene leads to suppression of oncogenic transformation of primary rat embryo fibroblasts. Oncogene. 2000;19:1173–1180. doi: 10.1038/sj.onc.1203417. [DOI] [PubMed] [Google Scholar]
- 67.Sekiguchi T, Makashima T, Hayashida T, Kuraoka A, Hashimoto S, Tsuchida N, Shibata Y, Hunter T, Nishimoto T. Apoptosis is induced in BHK cells by the tsBN462/13 mutation in CCG1/TAFII250 subunit of the TFIID basal transcription factor. Exp Cell Res. 1995;218:490–498. doi: 10.1006/excr.1995.1183. [DOI] [PubMed] [Google Scholar]
- 68.Seto E, Mitchell P J, Yen T S B. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcription factors. Nature. 1990;344:72–74. doi: 10.1038/344072a0. [DOI] [PubMed] [Google Scholar]
- 69.Shimizu M, Nomura Y, Suzuki H, Ichikawa E, Takeuchi A, Suzuki M, Nakamura T, Nakajima T, Oda K. Activation of the rat cyclin A promoter by ATF2 and Jun family members and its suppression by ATF4. Exp Cell Res. 1998;239:93–103. doi: 10.1006/excr.1997.3884. [DOI] [PubMed] [Google Scholar]
- 70.Shintani Y, Yotsuyanagi H, Moriya K, Fujie H, Tsutsumi T, Kanegae Y, Kimura S, Saito I, Koike K. Induction of apoptosis after switch-on of the hepatitis B virus X gene mediated by the Cre/loxP recombination system. J Gen Virol. 1999;80:3257–3265. doi: 10.1099/0022-1317-80-12-3257. [DOI] [PubMed] [Google Scholar]
- 71.Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Brechot C. Hepatitis B virus X mutants present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999;18:4848–4859. doi: 10.1038/sj.onc.1202867. [DOI] [PubMed] [Google Scholar]
- 72.Sirma H, Weil R, Rosmorduc O, Urban S, Israel A, Kremsdorf D, Brechot C. Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene. 1998;16:2051–2063. doi: 10.1038/sj.onc.1201737. [DOI] [PubMed] [Google Scholar]
- 73.Su F, Schneider R J. Hepatitis B virus HBx protein activates transcription factor NF-κB by acting on multiple cytoplasmic inhibitors of rel-related proteins. J Virol. 1996;70:4558–4566. doi: 10.1128/jvi.70.7.4558-4566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Su F, Schneider R J. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by TNFα. Proc Natl Acad Sci USA. 1997;94:8744–8749. doi: 10.1073/pnas.94.16.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki-Yagawa Y, Guermah M, Roeder R G. The ts13 mutation in the TAFII250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Talavera A, Basilico C. Temperature sensitive mutants of BHK cells affected in cell cycle progression. J Cell Physiol. 1977;92:425–436. doi: 10.1002/jcp.1040920310. [DOI] [PubMed] [Google Scholar]
- 77.Tarn C, Bilodeau M L, Hullinger R L, Andrisani O M. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J Biol Chem. 1999;274:2327–2336. doi: 10.1074/jbc.274.4.2327. [DOI] [PubMed] [Google Scholar]
- 78.Terradillos O, Billet O, Renard C-A, Levy R, Molina T, Briand P, Buendia M A. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 79.Terradillos O, Pollicino T, Lecoeur H, Tripodi M, Gougeon M L, Tiollais P, Buendia M A. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene. 1998;17:2115–2123. doi: 10.1038/sj.onc.1202432. [DOI] [PubMed] [Google Scholar]
- 80.Thomas S M, Brugge J S. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 81.Twu J-S, Lai M-Y, Chen D-S, Robinson W S. Activation of protooncogene c-jun by the X protein of hepatitis B virus. Virology. 1993;192:346–350. doi: 10.1006/viro.1993.1041. [DOI] [PubMed] [Google Scholar]
- 82.Twu J-S, Robinson W S. Hepatitis B virus X gene can transactivate heterologous viral sequences. Proc Natl Acad Sci USA. 1989;86:2046–2050. doi: 10.1073/pnas.86.6.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Unger T, Shaul Y. The X protein of the hepatitis B virus acts as a transcription factor when targeted to its responsive element. EMBO J. 1990;9:1889–1895. doi: 10.1002/j.1460-2075.1990.tb08315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang E, Tjian R. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 85.Wang E, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H-D, Trivedi A, Johnson D L. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol Cell Biol. 1997;17:6838–6846. doi: 10.1128/mcb.17.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang H-D, Yuh C-H, Dang C V, Johnson D L. The hepatitis B virus X protein increases the cellular level of TATA-binding protein which mediates transactivation of RNA polymerase III genes. Mol Cell Biol. 1995;15:6720–6728. doi: 10.1128/mcb.15.12.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H D, Trivedi A, Johnson D L. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol Cell Biol. 1998;18:7086–7094. doi: 10.1128/mcb.18.12.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weinzierl R, Dynlacht B, Tjian R. Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature. 1993;362:511–517. doi: 10.1038/362511a0. [DOI] [PubMed] [Google Scholar]
- 90.Williams J S, Andrisani O M. The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc Natl Acad Sci USA. 1995;92:3819–3823. doi: 10.1073/pnas.92.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xie W, Fletcher B S, Andersen R D, Herschman H R. v-src induction of the TIS10/PGS2 prostaglandin synthase gene is mediated by an ATF/CRE transcription response element. Mol Cell Biol. 1994;14:6531–6539. doi: 10.1128/mcb.14.10.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yen T S B. Hepadnaviral X protein: review of recent progress. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 93.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 94.Zack J A, Haislip A M, Krogstad P, Chen I S. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]