Abstract
Symmetry breaking, polarity establishment, and spontaneous cell protrusion formation are fundamental but poorly explained cell behaviors. Here, we demonstrate that a biochemical network, where the mutually inhibitory localization of PIP5K and Ras activities plays a central role, governs these processes. First, in resting cells devoid of cytoskeletal activity, PIP5K is uniformly elevated on the plasma membrane, while Ras activity remains minimal. Symmetry is broken by spontaneous local displacements of PIP5K, coupled with simultaneous activations of Ras and downstream signaling events, including PI3K activation. Second, knockout of PIP5K dramatically increases both the incidence and size of Ras-PI3K activation patches, accompanied by branched F-actin assembly. This leads to enhanced cortical wave formation, increased protrusive activity, and a shift in migration mode. Third, high inducible overexpression of PIP5K virtually eliminates Ras-PI3K signaling, cytoskeletal activity, and cell migration, while acute recruitment of cytosolic PIP5K to the membrane induces contraction and blebs in cancer cells. These arrested phenotypes are reversed by reducing myosin II activity, indicating myosin’s involvement in the PIP5K-Ras-centered regulatory network. Remarkably, low inducible overexpression of PIP5K unexpectedly facilitates polarity establishment, highlighting PIP5K as a highly sensitive master regulator of these processes. Simulations of a computational model combining an excitable system, cytoskeletal loops, and dynamic partitioning of PIP5K recreates the experimental observations. Taken together, our results reveal that a bistable, mutually exclusive localization of PIP5K and active Ras on the plasma membrane triggers the initial symmetry breaking. Coupled actomyosin reduction and increased actin polymerization lead to intermittently extended protrusions and, with feedback from the cytoskeleton, self-organizing, complementary gradients of PIP5K versus Ras steepen, raising the threshold of the networks at the rear and lowering it at the front to generate polarity for cell migration.
Introduction
The diverse morphological behaviors displayed by cells all depend on the ability to achieve an asymmetric form spontaneously or in response to an external or internal cue [1–6]. During migration, for example, a cell must generate and maintain a distinct front and back [7–10]. Signaling and cytoskeletal components and activities self-organize precisely into specific front or back regions of the cortex/plasma membrane [2, 6, 11–20]. This complementary organization is conserved across various processes, including macropinocytosis, phagocytosis, cytokinesis, and apical-basal polarity in epithelial cells [21–25]. Our studies focus on the role of PIP5K in regulating this elegant spatiotemporal organization during migration. Still, the insights gained should apply broadly to symmetry-breaking processes.
Symmetry-breaking is a critical initial step, but further events must follow to achieve effective cell migration [6, 21, 26–33]. In human leukocytes and epithelial cells, as well as Dictyostelium amoebae, protrusions underlying cell movement are driven by spontaneously triggered waves of Ras, PI3K, which couple to cytoskeletal events at the front, while PTEN, RhoA, and myosin II assembly retreat from these active areas [22, 26, 34–42]. Additional important aspects of migration are directional sensing and polarity, where cells consistently display the same front-back complementary relationship of all these molecular events [36]. Uncovering the relationship that links the downstream processes to the initial symmetry-breaking events would provide a deep understanding of the basis of dynamic cellular morphology.
The initial symmetry-breaking event in migration, which can occur even in the absence of cytoskeletal activities, is the spontaneous local activation of Ras [35, 43–45]; however, the mechanism that controls Ras activation at specific membrane regions remains obscure. Whatever the mechanism, it must amplify the signal locally and simultaneously restrict it to prevent the activation elsewhere. Recent studies show that PIP2 levels are depleted on the spontaneous protrusions [19]. Globally lowering PI(4,5)P2 activates Ras and increases the size of protrusions in Dictyostelium [46], and causes spreading in human cells [34]. Reasoning that PIP5K could be the central regulator of PIP2 production [47–50], we focused on the roles of this enzyme in fine-tuning Ras activation.
Here, we report remarkably conserved localizations and outsized roles of PIP5Ks in cell migration in amoebae, neutrophils, macrophages, and cancer cells and delineate the function of these enzymes in cellular behavior. A central finding is a bistable, mutually exclusive localization of PIP5Ks and active Ras on the plasma membrane, leading to symmetry-breaking and locally regulated signal transduction network activities. These activities are coupled to and amplified by interactions and feedback from the cytoskeleton. The dramatic effects of deletion or overexpression of the enzymes on protrusions, cell migration, and polarity demonstrate the significance of the spatial-temporal regulation of the PIP5Ks. These findings are incorporated into a new model that seamlessly links symmetry-breaking, cell polarity, and migration.
Results
Lowering PIP2 increases signal transduction, cytoskeletal, and protrusive activities
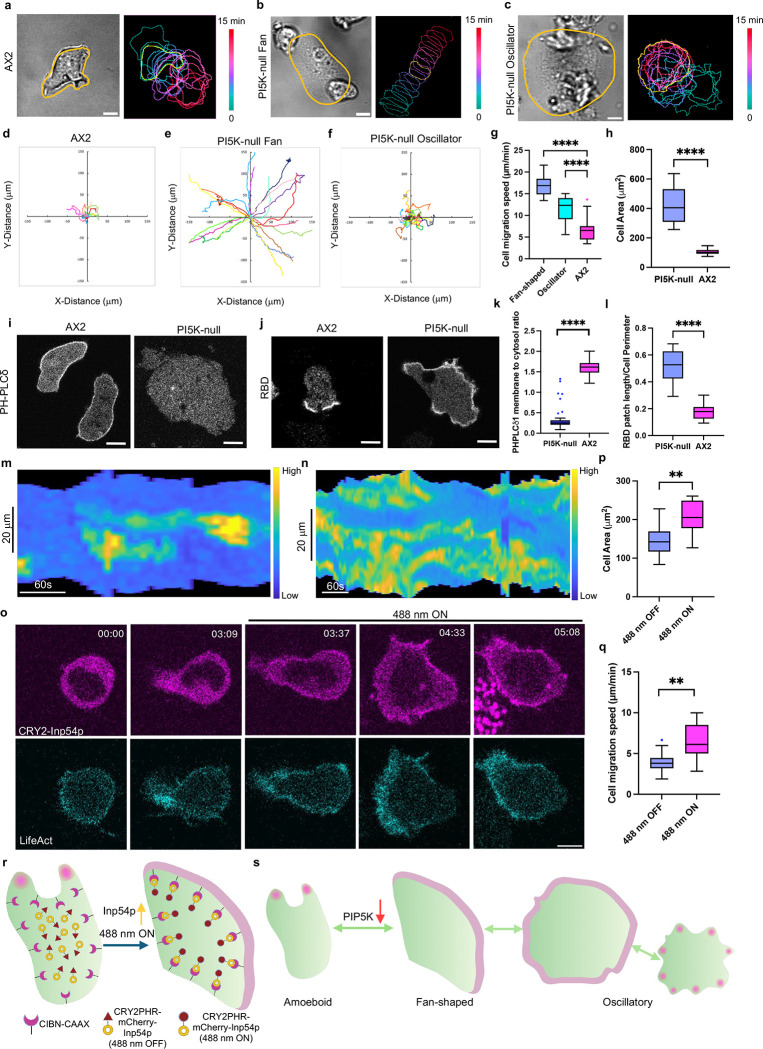
The inconsistency of a report that PIP5K had little effect on random cell migration with our previous reports of strong phenotypes caused by lowering PI(4,5)P2, prompted us to reexamine this issue [51]. More detailed observation of pi5k- (piki-) cells revealed a heterogeneous spectrum of migratory patterns (Figure 1a–h). While a minority of cells retained wild-type amoeboid behavior, most exhibited fan-shaped, keratocyte-like movements or oscillatory spreading/contracting movements, consistent with the previously reported phenotype induced by abrupt reduction of PI(4,5)P2 in wild-type cells [46]. These observations were supported by color-coded temporal overlay profiles (Figure 1a–c). Tracking experiments showed that fan-shaped cells migrate rapidly, with speeds reaching 16.9 ± 2.2 μm min−1 (Figure 1e, 1g), in contrast to the 6.8 ± 2.8 μm min−1 observed in WT cells (Figure 1d, 1g). Since the spreading phases of the oscillatory cells were asymmetric, these cells also displayed increased mobility, moving at a rate of around 11.2 ± 3 μm min−1 (Figure 1f–g). Quantification of cell area of the entire population (Figure 1h) revealed an overall 4-fold increase, primarily due to flattening, although some of the increase was due to a multinuclear phenotype of pi5k- cells (Figure S1c–d). These defects in migration probably explain the inability of pi5k- cells to aggregate and form multicellular structures in a timely fashion (Figure S1e). Most of these studies were carried out on the original pi5k- line generated by homologous recombination [51]. To validate that the phenotypes were attributable to loss of PI5K activity, we used CRISPR-mediated disruption of PIP5K to create an independent loss-of-function cell line. These cells displayed a consistent heterogeneous phenotype, primarily consisting of fan-shaped and oscillatory cell populations, with cells more spread than wild-type cells (Figure S1a–b).
Figure 1. Lowering PIP2 increases signal transduction, cytoskeletal, and protrusive activities.
(a-c) Representative live-cell time-lapse confocal images (left, DIC) and color-coded temporal overlay profiles (right) of Dictyostelium AX2 (WT) cells (a), pi5k- fan-shaped (b), and pi5k- oscillatory cells (c). A linear color map shows that green corresponds to 0 min and red corresponds to 15 min. The yellow outline corresponds to the frame of time-lapse images and cell boundaries. Scale bars represent 5 mm. (d-f) Centroid tracks of cells (nc=22) showing random motility in AX2 (WT) cells (d), pi5k- fan-shaped (e), and pi5k- oscillatory cells (f). Each track lasted at least 15 minutes and was reset to the same origin. (g-h) Box-and-whisker plots of (g) cell migration speed, (h) cell area. nc=22 from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (i-j) Representative live-cell time-lapse confocal images of Dictyostelium AX2 (WT) cells (left), and pi5k- cells (right) expressing PHPLCδ-YFP (biosensor for PI(4,5)P2) (i) or RBD-GFP (biosensor for activated Ras) (j). Scale bars represent 5 mm. (k-l) Box-and-whisker plots of (k) PHPLCd1 membrane-to-cytosol ratio, (l) RBD patch length/Cell Perimeter. nc=22 from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (m-n) Representative membrane kymograph of RBD intensity in AX2 (m) or pi5k- (n) cells, respectively. A linear color map shows that blue has the lowest RBD intensity, whereas yellow has the highest. (o) Time-lapse confocal images of differentiated HL-60 neutrophil expressing CIBN-CAAX, CRY2PHR-mCherry-Inp54p (magenta; upper panel) and LifeAct-miRFP703 (cyan; lower panel), before or after 488 nm laser was switched on globally. Time in min:sec format. Scale bars represent 5 μm. (p-q) Box-and-whisker plots of (p) cell area, (q) cell migration speed correspond to (o). nc=10 from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (r) Cartoon illustrating mechanism of opto-Inp54p global recruitment on differentiated HL-60 neutrophil membrane with the help of CRY2-CIBN optogenetic system. (s) Cartoon illustrating signal transduction activities in amoeboid, fan-shaped and oscillatory cells.
To further understand the basis of these phenotypes, we examined a series of signal transduction and cytoskeletal activities using biosensors. As expected, PH-PLCδ was mainly found on the membrane of WT cells, suggesting a significant level of PI(4,5)P2 [19], but there was no apparent membrane association of the biosensor in most of the pi5k- cells (Figure 1i, 1k, Video S1). Cell boundary-to-cytosol quantification showed an apparent 78% decrease of PH-PLCδ on the membrane in pi5k- (Figure 1k). Curiously, a few outlier cells showed a bright signal comparable to WT cells (Figure 1k); these cells are under investigation. Next, we examined the spatial distribution of Ras activity using a Ras binding domain (RBD) biosensor in WT and pi5k- cells. Compared with WT cells, which have characteristic patches at the cell protrusions, pi5k- cells have much broader RBD patches, nearly a 2.9-fold increase (Figure 1j, 1l, Video S2). These observations were supported by membrane kymograph analyses (Figure 1m–n). Similar results were observed using the PHcrac biosensor to examine PIP3 accumulation between pi5k- cells and WT cells, with a 2.1-fold increase in pi5k- cells (Figure S2a, 2c, Video S2). LimE, a biosensor reflecting newly formed F-actin, also displayed much broader patches in pi5k- cells, with a 3-fold increase in pi5k- cells (Figure S2b, 2d, Video S2). The dynamic behavior of cytoskeletal activities was captured in membrane kymographs (Figure S2h–i), which also revealed that the signaling activities oscillate in oscillatory cells (Figure S1f–g). When cells spread, the signaling activities increase, while they disappear upon cell shrinking. The three biosensors examined are typically associated with the active, “front-state” of the cell, and the elevated levels suggest that pi5k- cells are vastly more activated than wild-type cells.
Next, we examined signal transduction and cytoskeletal activities typically associated with the “back-state” of cells. CynA is a biosensor that reports the level of PI(3,4)P2 [52]. Strikingly, there generally was no detectable signal from CynA biosensor in pi5k- cells. This reduction is indirect since PI(3,4)P2 is not a product of PI5K. (Figure S2f, Video S1). However, in oscillatory cells, CynA did display a transient patch when cells contracted, and this activity disappeared upon cell spreading (Figure S1h), indicating that signaling activities at the cell back can also oscillate. Similar results were observed when we examined cytoskeletal activities at the cell back with Myosin II (Figure S2e, Video S1). These results suggested that cell activities are low at the cell back.
We also observed the same biosensors within ventral waves in the pi5k- cells. As explained above, cellular protrusions consist of spontaneously initiated waves of signal transduction and cytoskeletal activities. The waves are more conveniently observed along the basal surface than in confocal slices. On the ventral surface, biosensors for Ras and PI3K activity and F-actin appear as broad propagating regions, while “back-state” sensors, CynA and myosin II, leave these zones, creating traveling “shadow waves.” As shown in Figure S2n–o, ventral waves are not apparent in single WT cells and are typically visualized in electro-fused giant cells. However, they are readily apparent without electrofusion in the flat, multinucleated pi5k- cells [41, 46, 53–55] (Figure S2j–k, Video S3).
Since the phenotypes of pi5k- cells were similar to those induced by reduction of PI(4,5)P2 in Dictyostelium cells [46], we used recruitment of Inp54p to extend our studies to differentiated HL-60 neutrophils and macrophages. We developed a recruitable Inp54p by fusing it with CRY2PHR-mCherry, which enabled light-induced association with membrane anchor CIBN-CAAX (Figure 1r). Globally recruiting Inp54p to the plasma membrane induced fan-shaped cells and increased F-actin activities at the protrusions reported by the LifeAct biosensor in both differentiated HL-60 neutrophils (Figure 1o, Video S4) and macrophages (Figure S3a, Video S4). Recruiting an empty vector did not cause a cell shape change (Figure S3e, Video S4) [44]. These observations were supported by membrane kymograph analyses (Figure S3j, S3l) and color-coded temporal overlay profiles (Figure S3k, S3m). Across the population, upon Inp54p recruitment, differentiated HL-60 neutrophils and macrophages induced a 1.43-fold and 1.5-fold increase in the cell area and aspect ratio, respectively (Figure 1p, Figure S3b). Cell migration speed also increased from 4.0 ± 1.4 μm min−1 (488 nm OFF) to 6.3 ± 2.2 μm min−1 (488 nm ON) for differentiated HL-60 neutrophils (Figure 1q, S3f–g) and 4.0 ± 1.4 μm min−1 (488 nm OFF) to 6.3 ± 2.2 μm min−1 (488 nm ON) for differentiated HL-60 macrophages (Figure S3c, S3h–i). Although cells became fan-shaped with one broad lamellipodia structure at the cell front, cell polarity quantified by aspect ratio did not change significantly (Figure S2g, S3d).
In summary, lowering PI(4,5)P2 levels by depleting PIP5K or recruiting Inp54p has profound effects on the overall state of cells, generally causing Dictyostelium cells, neutrophils, and macrophages to spread with broader protrusions and increased speed (Figure 1s). Increased “front-state” and reduced “back-state” signal transduction and cytoskeletal activities all suggest that cells are highly activated. These results imply that PI5K, by regulating its product, PI(4,5)P2, is a key negative regulator of cell behavior.
PIP5Ks localize to back-state regions of the membrane
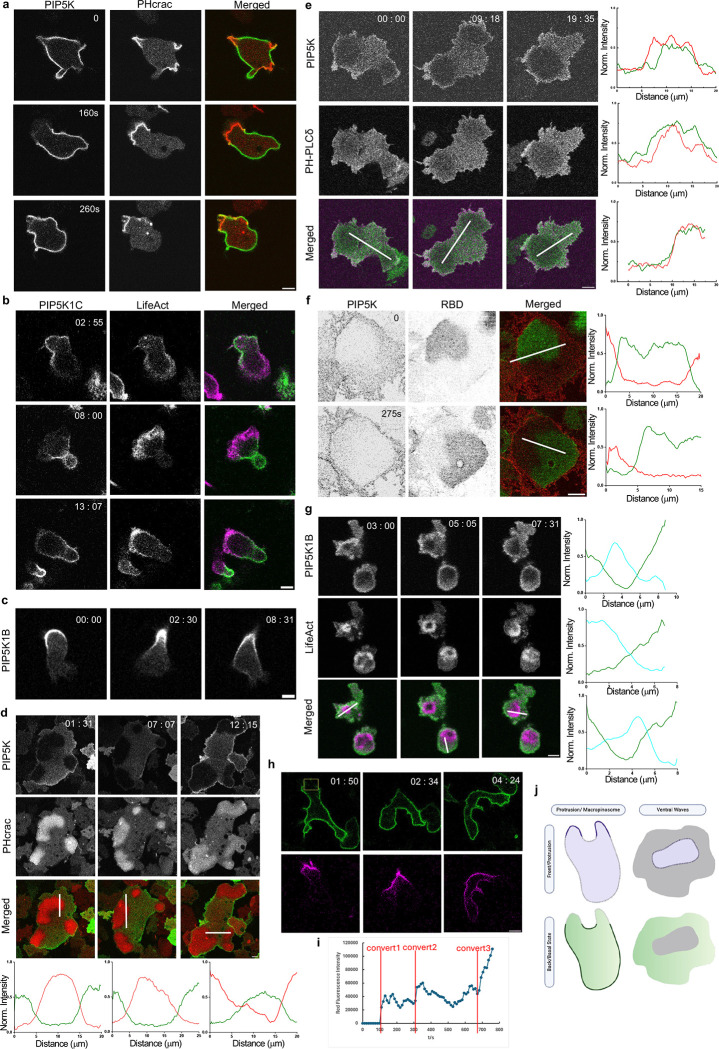
To investigate the localization of PIP5K and whether it could shed light the decreases in PI(4,5)P2 at protrusions [19], we co-expressed PIP5K with a typical front-state biosensor PHcrac in Dictyostelium cells. The distribution of PIP5K was complementary to PHcrac and was depleted from protrusions in migrating single cells (Figure 2a, Video S5). The back-state localization of PIP5Ks was conserved in HL60-differentiated neutrophils. Both PIP5K1B and PIP5K1C isomers displayed a higher signal on the back-state of the membrane in neutrophils (Figure 2b–c, Video S5) during random migration, which was consistent with previously reported PIP5K1B and PIP5K1C back localization during chemotaxis to N-formyl-met-leu-phe (fMLP) [49, 56].
Figure 2. PIP5Ks localize to the back-state regions of the membrane.
(a) Representative live-cell time-lapse images of Dictyostelium cells coexpressing PI5K-GFP and PHcrac–RFP (biosensor for PIP3) during migration showing PIP5K dynamically moves away from protrusions in migrating cells. Time in sec format. Scale bars represent 5 mm. (b-c) Representative live-cell time-lapse images of differentiated HL-60 neutrophils coexpressing PIP5K1C-GFP and LifeAct-iRFP703 (biosensor for F-actin) (b) or expressing PIP5K1B-GFP (c) during migration showing PIP5Ks dynamically moves away from protrusions in migrating cells. Time in min:sec format. Scale bars represent 5 mm. (d-e) Representative live-cell time-lapse images of Dictyostelium cells coexpressing PIP5K-GFP and PHcrac-RFP (biosensor for PIP3) (d) or PIP5K-mCherry and PHPLCδ–GFP (biosensor for PI(4,5)P2) (e) during ventral wave propagation, showing PI5K dynamically localizes to the back-state regions in ventral waves. Line-scan intensity profiles are shown in the bottommost panels. The red line represents PHcrac and PI5K in (d) and (e). The green line represents PIP5K and PHPLCδ. Time in min:sec format. Scale bars represent 5 mm. (f) Representative live-cell images of Dictyostelium cell co-expressing PIP5K-mCherry and RBD-GFP (biosensor for activated Ras), where the substrate-attached ventral surface was imaged, showing consistent complementarity between PIP5K and RBD during 2D wave propagation, even in the absence of actin cytoskeleton. Cells were pre-treated with actin polymerization inhibitor Latrunculin A (final concentration 5μM) for 20min. Line-scan intensity profiles are shown in the bottommost panels. Red line and green line represent PIP5K and RBD, respectively. Time in sec format. Scale bars represent 5 mm. (g) Live-cell images of a differentiated HL-60 macrophage coexpressing PIP5K1B and LifeAct demonstrating dynamic complementary distribution in its ventral waves. Line-scan intensity profiles are shown in the bottommost panels. Green line and Cyan line represent PIP5K1B and LifeAct, respectively. Time in min:sec format. Scale bars represent 5 mm. (h) Representative live-cell time-lapse images of Dictyostelium cells expressing KikGR-PIP5K. Yellow box indicates where the photo conversion happened. Top panel is GFP channel (488nm), while bottom panel is RFP channel (561nm). Time in min:sec format. Scale bars represent 5 mm. (i) Scatter plot of RFP channel fluorescence intensity vs. time. The red line indicated when the conversions happened. (j) Cartoon illustrating the front-back complementarity in migrating cell protrusions and ventral wave propagation.
We then examined ventral wave activities of the same cell lines in Figure 2a and found, consistently, that PI5K was depleted in the front-state regions marked by high PIP3 and high Ras (Figure 2d, S4a, Video S6). The complementary pattern of PIP5K and PIP3 or PIP5K and Ras was maintained dynamically as the waves traveled across the basal surface of the cells. Line scans showed about 80% less PIP5K in the front-state regions compared with the back-state regions in Dictyostelium cells. Next, we co-expressed PIP5K and PI(4,5)P2 biosensor PH-PLCδ and observed ventral wave activities. PIP5K and PH-PLCδ travel together as “shadow” waves; they co-localize at the back-state regions of the membrane (Figure 2e, Video S6). Line scans showed that PIP5K and PH-PLCδ are largely co-localized, suggesting that the redistribution of PIP5K is the basis PI(4,5)P2 is localized at the back-state of the membrane. Reciprocal patterns of PIP5K1B and LifeAct were observed during frustrated phagocytosis in HL-60-differentiated macrophages (Figure 2g, Video S6) and in RAW 264.7 macrophages, as previously reported [54]. These results suggest that the localization of PIP5K to the rear and back-state of the cells is a highly conserved phenomenon.
Previous studies have shown that signal transduction events can be triggered, and the membrane can be spontaneously segregated into front- and back-states in the absence of F-actin [40, 46, 57–59]. To test whether the spatiotemporal separation of PIP5K depends on the existence of actin barrier between front and back states, we treated Dictyostelium cells with Latrunculin A. We observed that on the ventral surface of these cytoskeleton-impaired cells, the asymmetric waves of PI5K can propagate, maintaining consistent complementarity to Ras-rich domains (Figure 2f, Videos S6). Furthermore, the data in Dictyostelium indicates that the localization of PIP5K is independent of the cytoskeletal network.
Typically, when cells are activated with global chemoattractant stimulation, the front-state biosensors, such as PHcrac, transiently translocate to the membrane while back-state proteins, such as PTEN, dissociate [60–64]. We therefore anticipated that, based on its back-state localization, PIP5K would transiently move to the cytosol during stimulation. However, upon adding cAMP to differentiated cells, PIP5K remained on the membrane even as the front-state indicator PHcrac displayed its typical transient translocation to the membrane, demonstrating robust receptor activation. (Figure S4b–c, Video S7). PH-PLCδ biosensor also did not dramatically translocate to the cytosol, although there was a 5% shift upon global stimulation (Figure S4d–e, Video S7).
When PIP5K is depleted in the front region as protrusions form, it remains on the membrane, suggesting that it must redistribute to higher levels elsewhere. To more definitively test this, we designed a KikGR-tagged PIP5K to photo convert a specific region on the plasma membrane. As Figure 2h and Video S8 shows, PIP5K remains on the membrane where the region was converted and gradually diffused along the membrane. The red fluorescence intensity on the membrane increased in three steps corresponding to three conversions in different regions (Figure 2i). Eventually the red fluorescence on the membrane was uniformly distributed. At no time, the red fluorescence was redistributed to the cytosol. This suggests that the amount of PIP5K on the membrane is conserved, and depletion in one region must produce an increase in another region. Since PIP5K controls signaling activities, its redistribution provides a powerful mechanism for symmetry breaking.
To seek regions of PIP5K that control membrane binding and trailing edge accumulation in Dictyostelium cells, we generated a series of truncation constructs to examine their localization (Figure S4f). Despite its robust membrane association, PIP5K does not contain a typical membrane association or transmembrane domain. PIP5K301-718aa was still localized as WT cells, although a small fraction of cells showed a relatively smaller localization at the back-state. (Figure S4g, Video S9). PIP5K316-718aa appeared significantly in the cytosol, and the protein remaining on the membrane was not localized at the back. Upon recruitment to a uniformly localized protein cAR1-iLiD, this fragment also did not relocalize cAR1-iLiD to the back (Figure S4h, Video S8). N-terminal 315aa was not on the membrane and did not relocate cAR1-FKBP-FKBP to the back (Figure S4i, Video S8). These results indicate that a region between 301–316 aa is important for localization to the membrane, but the back localization region was not identified. Back localization may require a coincidental association of two independent sequences within the protein. Future experiments will address these possibilities.
Expressing PIP5Ks suppresses cell protrusions and alters migration
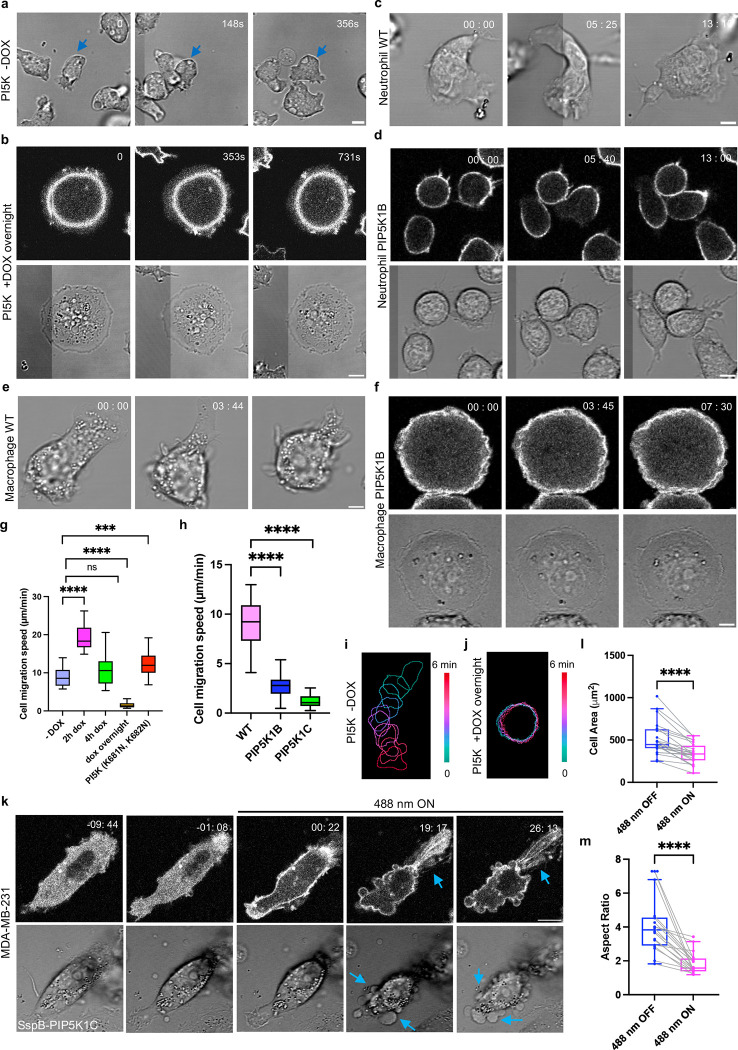
In previous experiments (Figure 2), where PIP5K was expressed from a constitutive vector and the expression level was relatively low, we noticed a slight impairment of migration. Reasoning that the highly expressing cells were selected against and lost before observation, we developed a conditional doxycycline-inducible version of PIP5K. Without doxycycline (DOX) induction, Dictyostelium cells show their normal amoeboid migration behavior (Figure 3a, Video S10). Upon overnight DOX induction, two striking phenotypes appeared. About 60% of the cells became much rounder, lacked protrusions, and failed to migrate (Figure 3b, Video S10). About 40% of the cells displayed many tiny, broken protrusions, resembling short filopodia, along the cell perimeter and migration was also impaired (Figure S5a, Video S10). These observations were supported by color-coded temporal overlay profiles (Figure 3i–l, S5l). Combining data from the “rounded” and “spiky” cells, tracking experiments showed that the migration speed of was reduced to 1.52 ± 0.7 μm min−1 (Figure 3g, S5b), in contrast to the 8.8 ± 2.3 μm min−1 observed in cells without DOX incubation (Figure 3g, S5c). The cells were flatter and occupied about 1.7-fold more surface area (Figure S5d). Z-stack imaging of cells showed that upon PIP5K expression, the z-axis values drop from 12 μm to 4.87 μm (Figure S5g). In addition, FITC-dextran uptake measurements showed that macropinocytosis was significantly reduced, consistent with the nearly absent or tiny protrusions (Figure S5h–j) [37, 65, 66].
Figure 3. Expressing PIP5Ks suppresses cell protrusions and alters migration.
(a-b) Representative live-cell images of Dictyostelium cells expressing doxycycline-inducible PIP5K without DOX induction (a) or with overnight DOX induction (rounded) (b). Time in sec format. Scale bars represent 5 μm. (c-d) Representative live-cell images of differentiated HL-60 neutrophils (WT) (c) and expressing PIP5K1B (rounded) (d). Time in min:sec format. Scale bars represent 5 μm. (e-f) Representative live-cell images of differentiated HL-60 macrophages (WT) (e) and expressing PIP5K1B (f). Time in min:sec format. Scale bars represent 5 μm. (g-h) Box-and-whisker plots of cell migration speed of Dictyostelium cells expressing PIP5K at different DOX incubation time (g), or differentiated HL-60 neutrophils expressing PIP5Ks (h). nc=20 from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Pris 10). (i-j) Color-coded temporal overlay profiles of Dictyostelium cells expressing doxycycline-inducible PIP5K without DOX incubation (i) or with overnight DOX incubation (j). (k) Time-lapse confocal images of MDA-MB-231 cells expressing crimson-SspB-PIP5K1C-P2A-iLiD-CAAX, before or after 488 nm laser was switched on globally. Time in min:sec format. Scale bars represent 10 μm. Blue arrows indicate where retraction fibers or blebs are formed. Cells are pretreated with 10 ng/ml EGF for 10 mins. (l-m) Box-and-whisker plots of (l) cell area, (m) aspect ratio correspond to (m-n). nc=10 from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10).
We could titrate PIP5K activity in several ways. Protein expression levels varied with doxycycline incubation times (Figure S5e). Surprisingly, cells with low PIP5K expression after 2h DOX incubation became very polarized (Figure S5k), with a 1.32-fold increase in aspect ratio (Figure S5f). Tracking experiments showed that these cells migrated much faster than uninduced cells, with speeds reaching 19.2 ± 3 μm min−1 (Figure 3g, S5o). These observations were supported by color-coded temporal overlay profiles (Figure S5m). A similar increased speed was observed in cells expressing a PIP5K mutant (K681N, K682N) predicted to have impaired kinase activity, with speeds reaching 12.5 ± 3.3 μm min−1 (Figure 3g, S5n, S5r, Video S10) [67].
Next, we extended our studies to differentiated HL-60 neutrophils and macrophages to examine whether overexpressing PIP5Ks inhibits cell migration in leukocytes. We first overexpressed either PIP5K1B or PIP5K1C in differentiated HL-60 neutrophils, as these two isomers, according to previous literature, are important to neutrophil chemotaxis and directed migration [49, 56]. Like Dictyostelium cells, differentiated HL60 WT neutrophils showed a typical flat and polar phenotype (Figure 3c, Video S11). Upon overexpressing either PIP5K1B or PIP5K1C, two phenotypes existed in the basal state populations: round cells with PIP5Ks broadly distributed on the membrane (Figure 3d, S6a, Video S11) or polarized cells with PIP5Ks enriched on the tail (Figure S6b–c, Video S11). These observations were supported by color-coded temporal overlay profiles (Figure S6d–e, S6i–j). Tracking experiments showed that the migration speed of cells overexpressing PIP5Ks is impaired, with speeds reaching 2.92 ± 1.3 μm min−1 for PIP5K1B and 1.23 ± 0.6 μm min−1 for PIP5K1C (Figure 3h, S5q, S6h), in contrast to the 9.0 ± 2.4 μm min−1 observed in WT cells (Figure 3h, S6g). The overlay suggested that even though the polarized cells were active at the leading edge, they did not move well because the tail appeared to adhere more strongly to the surface (Figure S6d–e). We also measured the mean membrane intensity between the polarized and round phenotype cells. Cells with rounder shapes have higher PIP5K expression levels, consistent with the Dictyostelium results (Figure S6n–o), although the round-shaped cells are not as flat as Dictyostelium cells. Cell areas were about 33% and 40% reduced for PIP5K1B and PIP5K1C cells, respectively (Figure S6f). We also overexpressed PIP5K1B and PIP5K1C in differentiated HL-60 macrophages. This led to a very flat and round-shaped phenotype, abolishing protrusions compared with WT macrophages (Figure 3e–f, Figure S6p, Video S12). These observations were supported by color-coded temporal overlay profiles (Figure S6k–m).
Next, we extended our inquiry to epithelial-derived cancer cells by studying MDA-MB-231 cells. Since these cells are very heterogeneous, we developed a system to immediately observe the effects caused by activating PIP5K in an individual cell. We subcloned a previously developed recruitable PIP5K1C [68] into an optogenetic system by fusing it with crimson-SspB, which enabled light-induced association with membrane anchor iLiD-CAAX [69] (Figure S6q). Globally recruiting cytosolic PIP5K1C to the plasma membrane in MDA-MB-231 cells induced cell retraction with retraction fibers at the cell rear, possibly due to the increased contractility (Figure 3k, Video S13). About 5 min after recruitment, cells started to display retraction fibers. Contraction was formed after about 20 minutes (Figure 3k, S7a). Recruiting an empty vector did not cause a cell shape change (Figure S7e, Video S13). Across the population, upon PIP5K1C recruitment, MDA-MB-231 cells induced a 25% and 59% reduction in the cell area and aspect ratio, respectively (Figure 3l–m), while recruiting an empty vector did not induce any cell area or polarity change (Figure S7f–g). These observations were supported by color-coded temporal overlay profiles (Figure S7b–d). In some recruited cells, blebbing was induced in addition to retraction (Figure 3k), indicating these cells potentially had even higher contractility upon PIP5K1C recruitment.
In summary, the phenotypes of overexpressing PIP5Ks are generally conserved across Dictyostelium, leukocytes, and epithelial cells. Cells show different migratory behaviors upon different PIP5K expression levels. We suggest that moderate expression of PIP5Ks tends to polarize cells by quenching extraneous protrusions, while strongly expressing PIP5Ks eliminates protrusive activities and severely impairs cell migration.
Expressing PIP5Ks suppresses signal transduction and cytoskeletal activities
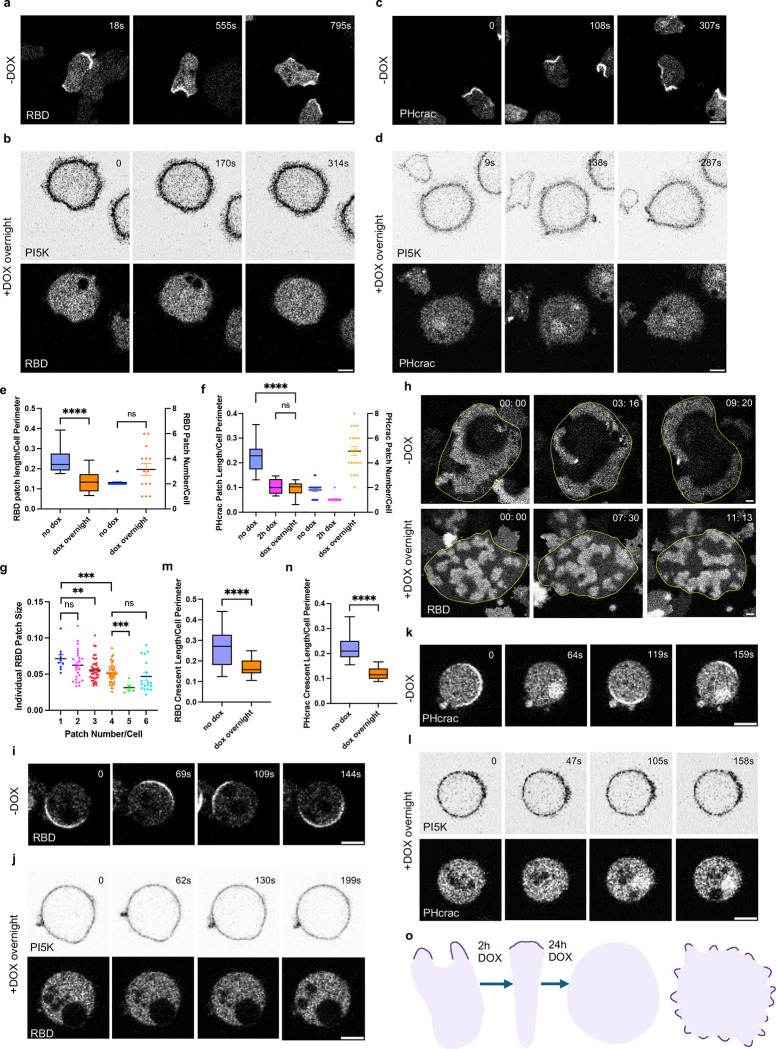
To further understand the basis of the overexpression phenotype, we examined a series of signal transduction and cytoskeletal activities using biosensors. We first examined the spatial distribution of Ras activity using a Ras binding domain (RBD) biosensor in Dictyostelium cells with different DOX incubation times. As expected, cells showed characteristic patches at the cell protrusions without DOX incubation (Figure 4a, Video S14), whereas cells incubated with DOX overnight had much fewer RBD patches. Notably, there were no RBD patches in rounded, non-moving cells (Figure 4b, Video S14). In the “spiky” cells, there were small, narrow RBD patches (Figure S8e, Video S14), although these were increased in number (Figure 4e). These observations were supported by membrane kymograph analyses (Figure S8a–b). There was an inverse correlation between patch size and number (Figure 4g). Combining data from “rounded” and “spiky” cells, there was an overall 44% reduction in total Ras patches (Figure 4b, S8e, 4e). Similar results were observed using the PHcrac biosensor to examine PIP3 activities in cells with different DOX incubation times, with an overall 58% reduction in cells with overnight DOX incubation (Figure 4c–d, 4f, S8f, Video S14). These observations were supported by membrane kymograph analyses (Figure S8c–d). We also observed the ventral wave differences for cells with or without DOX incubation. We found that uninduced cells displayed broad, sustained waves, and cells with overnight DOX incubation showed no waves or smaller, slower-moving waves, indicating low Ras signaling activities (Figure 4h, Video S15).
Figure 4. Expressing PIP5Ks suppresses signal transduction and cytoskeletal activities.
(a-b) Representative live-cell time-lapse confocal images of Dictyostelium AX2 co-expressing RBDGFP (biosensor for activated Ras) and doxycycline-inducible PI5K without DOX induction (a) or with overnight DOX induction (rounded) (b). Time in sec format. Scale bars represent 5 mm. (c-d) Representative live-cell time-lapse confocal images of Dictyostelium AX2 co-expressing PHcrac-YFP (biosensor for PIP3) and doxycycline-inducible PI5K without DOX induction (c) or with overnight DOX induction (rounded) (d). Time in sec format. Scale bars represent 5 mm. (e-f) Box-and-whisker plots of RBD patch size (left axis) and RBD patch number (right axis) (e), or PHcrac patch size (left axis) and PHcrac patch number (right axis) (f). nc=20 from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (g) Box-and-whisker plots individual RBD patch size at different patch numbers. Data is from Figure e. nc=10 for column 1, nc=26 for column 2, nc=42 for column 3, nc=45 for column 4, nc=5 for column 5, and nc=6 for column 6. All results are from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (h) Representative live-cell time-lapse confocal images of Dictyostelium AX2 co-expressing RBD-GFP (biosensor for activated Ras) and doxycycline-inducible PI5K without DOX induction (upper panel) or with overnight DOX induction (lower panel) during ventral wave propagation. Time in min:sec format. Scale bars represent 10 mm. (i-j) Representative live-cell time-lapse confocal images of Dictyostelium AX2 co-expressing RBD-GFP (biosensor for activated Ras) and doxycycline-inducible PI5K without DOX induction (i) or with overnight DOX induction (j), even in the absence of actin cytoskeleton. Cells were pre-treated with actin polymerization inhibitor Latrunculin A (final concentration 5μM) and caffeine (final concentration 4mM) for 20min. Time in sec format. Scale bars represent 5 mm. (k-l) Representative live-cell time-lapse confocal images of Dictyostelium AX2 co-expressing PHcrac-YFP (biosensor for PIP3) and doxycycline-inducible PI5K without DOX induction (k) or with overnight DOX induction (l), even in the absence of actin cytoskeleton. Cells were pre-treated with actin polymerization inhibitor Latrunculin A (final concentration 5μM) and caffeine (final concentration 4mM) for 20min. Time in sec format. Scale bars represent 5 mm. (m-n) Box-and-whisker plots of RBD crescent size (m), or PHcrac crescent size (n). nc=35 from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (o) Cartoon illustrating the localization of front signaling components in cell protrusions and cell morphology at different doxycycline-inducible time points.
We further examined cytoskeletal activities in the PIP5K overexpressing cells. LimE displayed much smaller or no patches in cells following overnight DOX induction, with a 60% reduction in these cells (Figure S8g–i, S9a, Video S16). The dynamic behavior of these activities was captured in t-stack kymographs (Figure S9b–c), indicating that the tiny LimE patches in cells expressing PIP5K are more transient than patches in untreated cells. Since branched actin is activated by a Rac1/Arp2/3 complex/branched actin pathway, we used a Pak1-GBD and ArpC biosensors to examine these activities. The Rac1 activity paralleled localization of LimE (Figure S9d–f, Video S16). Induced cells showed a 70% reduction in Rac1 patch size (Figure S9g). The ArpC patches were also much smaller in PI5K-expressing cells (Figure S9h–i, Video S16).
We have previously reported that cells lacking actin-sequestering actobindins A,B,C (abnABC-) have increased Ras and PI3K activity, suggesting that branched actin is in a positive feedback loop with the these STEN activities [70]. We examined the extent to which the inhibitory effect of PIP5K expression can interrupt this feedback loop by overexpressing PIP5K in abnABC- cells [70]. Without DOX incubation, cells showed broad RBD patches as expected (Figure S10j, Video S17). The RBD patches decreased in length and increased in numbers dramatically upon overnight DOX incubation (Figure S10k, Video S17), indicating competition between the positive feedback from branched actin and inhibition by PIP5K. However, unlike WT, in the abnABC- background there were no completely round cells containing no RBD patches, again suggesting a balance between activation by positive feedback from F-actin and inhibition by PIP5K.
In previous experiments (Figure 3), we showed that cells showed a much more polarized phenotype and migrated faster when incubated only 2h with DOX than without DOX incubation. We examined both Ras and PIP3 signaling activities in these cells and found the Ras and PIP3 signaling activities were confined to tiny patches at the leading edge or undetectable (Figure 4f, S10a–b, Video S14). Similar Ras signaling activities were low or undetectable in fast-moving cells expressing mutant PIP5K (K681N, K682N) (Figure S10c, Video S18). The LimE patches were smaller and confined at the front of these cells (Figure S10d, Video S18). Our lab previously reported that a RasGAP C2GAPB in Dictyostelium cells can also inhibit Ras activities but increase polarity and migration speed [43], which showed a similar phenotype compared to the cells expressing a low level of PI5K activity. To assess whether C2GAPB and PI5K are acting in the same pathway, we co-expressed PIP5K and C2GAPB and found that both cell shape and migration speed mimic expressing PIP5K alone (Figure S11 a–b, Video S19). To investigate whether attenuating Ras activity in a highly activated cell could restore polarity and migration, we overexpressed C2GAPB in pi5k- cells. Indeed, upon expressing C2GAPB, the pi5k- cells changed from fan-shaped or oscillatory cells to more polarized cells, with migration speed increase from 5.1 ± 2.2 μm min−1 (−C2GAPB) to 22 ± 7 μm min−1 (+ C2GAPB) (Figure S11 c–e, Video S19).
PI(4,5)P2 has been considered to promote cell migration by interacting with many actin-binding proteins [71–75]. To assess whether the inhibitory role of PIP5K in our studies is dependent of cytoskeletal dynamics, we treated cells with Latrunculin A to remove all F-actin activities and caffeine to induce the crescent patches of RBD or PHcrac. Without DOX incubation, cells displayed the characteristic crescent RBD or PHcrac patches (Figure 4i, 4K, Video S20). However, with overnight DOX incubation, 60% cells did not have any patches, while the rest, showed a 31% or 45% reduction in the RBD or PHcrac crescent length, respectively (Figure 4j–l, 4m–n). These observations were supported by membrane kymograph analyses (Figure S10e–h). These responses of the STEN patches to PIP5K-mediated inhibition parallel the PI5K-induced conversion of amoeboid cells to “rounded” or “spiky” cells seen in the absence of latrunculin A. These observations suggest that the PIP5K-induced cell shape, migration speed, and cytoskeletal phenotypes reported above can be traced back to the inhibitory effects of PIP5K expression on STEN activities (Figure 4o), although there may also be additional effects on feedback from the cytoskeleton.
Expressing PIP5Ks increases the threshold for STEN activation
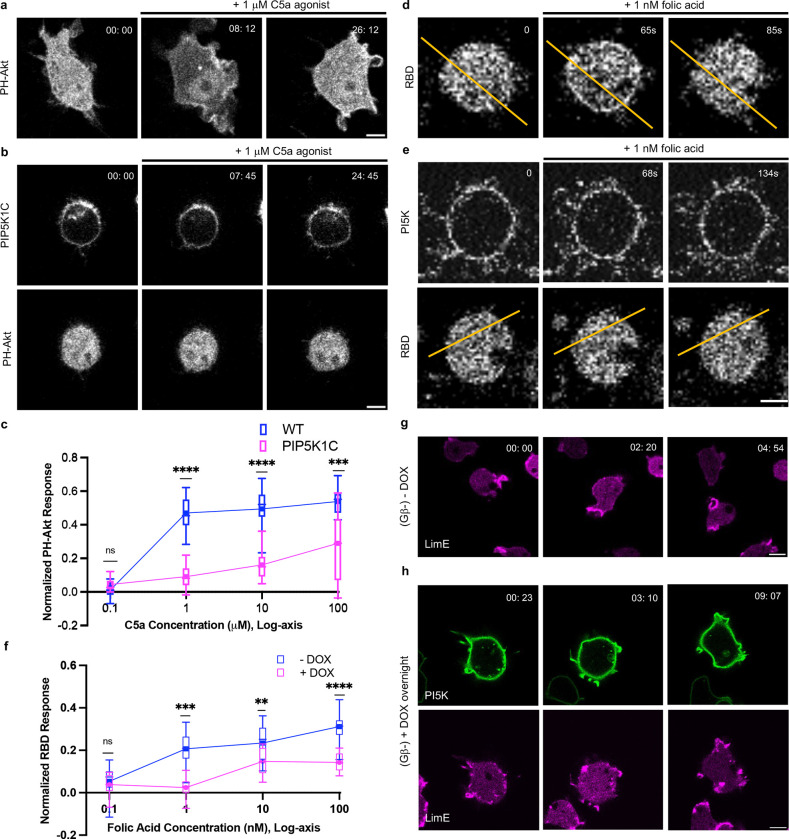
In previous experiments (Figure 4), we showed that expressing PIP5Ks inhibits Ras and PI3K activities and that cells expressing PIP5Ks showed smaller, shorter-lived waves. Reduced spontaneous signaling and curtailed wave activity in an excitable system indicate an increased threshold [20, 31, 34, 40, 41, 46]. To assess the threshold of the network more directly, we monitored the accumulation of PIP3 with biosensor PH-Akt in response to chemoattractant C5a in RAW 264.7 cells with or without PIP5K expression (Figure 5a–c, Video 21). In WT macrophages stimulated with 1 μM C5a, PIP3 transiently increased and cells extended protrusions. In contrast, at this dose, there was no PIP3 increase in cells expressing PIP5K1C; cells stayed rounded and inactivated (Figure 5a–b, Video 21). Compared with WT cells, the dose-response spanning 0.1 μM to 100 μM C5a for PIP5KIC-expressing cells was shifted to higher concentrations (Figure 5c, Video 21). The PH-Akt translocation responses at each C5a concentration are shown in Figure S13. The PH-Akt translocation in cells expressing PIP5K1C was evident only at 100 μM C5a (Figure S12h, Video 21).
Figure 5. Expressing PIP5Ks increases the threshold for STEN activation.
(a-b) Representative live-cell time-lapse confocal images of responses of PH-Akt-mCherry to global simulation C5a agonist at 1 μM in RAW 264.7 WT cells (a) or cells overexpressing PIP5K1C (b). Time in min:sec format. Scale bars represent 5 mm. (c) Normalized PH-Akt responses (a drop of cytosolic intensity) to different doses of C5a agonist for WT (blue) and PIP5K1C cells (magenta). From the lowest to highest concentration, nc=15 for each column from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001, ***P ≤ 0.001, nsP = 0.0408, (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (d-e) Representative live-cell time-lapse confocal images of responses of RBD-GFP to global simulation folic acid at 1 nM in Dictyostelium AX2 expressing doxycycline-inducible PI5K without DOX induction (d) or with overnight DOX induction (e). Time in sec format. Scale bars represent 5 mm. (f) Normalized RBD responses (a drop of cytosolic intensity) to different doses of folic acid for WT (blue) and PIP5K1C cells (magenta). From the lowest to highest concentration, nc=15 for each column from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, nsP = 0.2854, (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (g-h) Representative live-cell time-lapse confocal images of Dictyostelium Gb- cells co-expressing RBD-GFP and doxycycline-inducible PI5K without DOX induction (g) or with overnight DOX induction (h). Time in min:sec format. Scale bars represent 5 mm.
Similar results were observed in Dictyostelium cells stimulated with folic acid (FA). At 1 nM FA, untreated cells showed about 25% RBD translocation response (Figure 5d). Following induction of PIP5K by overnight incubation, there was no RBD translocation response at 1 nM (Figure 5e). The line scan results support these observations (Figure S13a–b). Compared to untreated cells, the dose-response curve from 0.1 nM to 100 nM FA for PI5K-expressing cells was shifted to higher concentrations (Figure 5f). Figure S12c–d shows typical images of the 5% and 20% PH-Akt or RBD responses. Dividing the cells into < 5% or > 20% response groups showed clearly that there many fewer PIP5K-expressing cells in the > 20% response set even with the highest concentrations of C5aR and FA (Figure S13e–f).
Since both the C5a receptor (C5aR) in macrophages and the folic acid receptor (fAR1) in Dictyostelium are G-protein-coupled receptors (GPCRs) [76, 77], we wondered whether the inhibitory effects of PIP5Ks are still evident when GPCR is uncoupled from the signal transduction network. In Dictyostelium cells lacking Gβ, conditional expression of PIP5K, spontaneous LimE patches still decreased in size and increased in numbers as previously observed in WT cells (Figure 5g–h, Video S22, and see Figure S8g–i). Considering that LimE is a downstream indicator of network activity, these data indicate the inhibitory role of PI5K can act downstream of GPCR signaling.
Since expressing PI5K caused severe impairment of random migration and shifted the dose-response curve to higher concentrations, we expected that directional migration would also be inhibited. Indeed, untreated Dictyostelium cells mounted a chemotactic response towards folic acid, while the insensitive, rounded PIP5K-expressing cells largely failed to migrate towards higher concentrations of (Figure S13g, S14a, Video 23). There was a weak response in the “spiky” or less affected cells. Overall, cells expressing PIP5K have less directional migration speed and polarity (Figure S14a–c).
In summary, expressing PIP5Ks inhibits signal transduction activities in response to chemoattractant stimulation, but this inhibitory role does not require G-protein, indicating that it acts at a point downstream. We suggest that the PIP5K inhibitory effect raises the threshold of the network and reduces its response to internal spontaneous activation as well as receptor inputs. We might reasonably expect responses to other external cues, such as electric fields or shear forces, to also be attenuated.
Inhibiting myosin II activity counteracts PIP5K-induced phenotypes
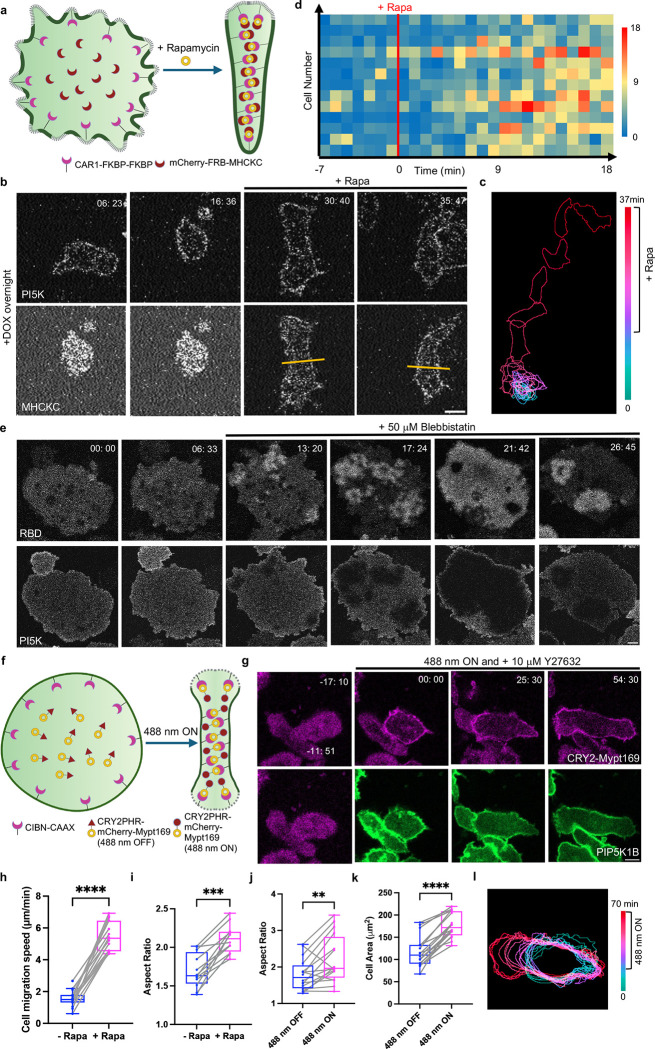
In previous experiments (Figures 3 and 4), we showed that expressing PIP5Ks can eliminate signal transduction and cytoskeletal activities and cell migration. We hypothesized that the inhibitory effects of PIP5Ks on cell morphology and migration might be related to increased Myosin II activities [78] and asked the extent to which lowering Myosin II would counteract PIP5K expression and restore migration. To test this, we co-expressed PIP5K and a recruitable Myosin heavy chain kinase C (MHCKC) previously shown to abruptly decrease Myosin II bipolar thick filament assembly [70] (Figure 6a). In PIP5K-expressing “rounded” cells, global recruitment of MHCKC to the plasma membrane caused cells to polarize and migrate faster (Figure 6b, Video S24). These observations were illustrated by color-coded temporal overlay profiles and line scans (Figure 6c, S15a–b). Across the population, upon MHCKC recruitment, migration speed increased from 1.6 ± 0.6 μm min−1 to 5.4 ± 0.9 μm min−1 (Figure 6h, S15c–d). The aspect ratio also increased by 1.23-fold (Figure 6i). A heat map of individual cell migration speed at each time illustrates the increase in migration speed after MHCKC recruitment, which took about 10 mins to establish (Figure 6d).
Figure 6. Inhibiting myosin II activity counteracts PIP5K-induced phenotypes.
(a) Cartoon illustrating mechanism of MHCKC global recruitment on Dictyostelium membrane with the help of FKBP-FRB CID system in cells expressing doxycycline-inducible PI5K with overnight DOX incubation. (b) Representative live-cell time-lapse confocal images of Dictyostelium AX2 co-expressing CAR1-FKBP-FKBP, mCherry-FRB-MHCKC, and doxycycline-inducible PI5K with overnight DOX induction before and after 5 μM rapamycin treatment. Time in min:sec format. Scale bars represent 5 mm. (c) Color-coded temporal overlay profile corresponds to (b). (d) Heatmap quantification of cell migration speed at individual time points (time frame 1min/frame) of Dictyostelium AX2 co-expressing CAR1-FKBP-FKBP, mCherry-FRB-MHCKC, and doxycycline-inducible PI5K with overnight DOX induction before and after 5 μM rapamycin treatment. nc=13 from at least 3 independent experiments. (e) Representative live-cell time-lapse images of Dictyostelium cells coexpressing RBD-GFP and doxycycline-inducible PI5K with overnight DOX induction during ventral wave propagation, before and after 50 μM blebbistatin treatment. Time in min:sec format. Scale bars represent 5 mm. (f) Cartoon illustrating mechanism of opto-Mypt169 global recruitment on differentiated HL-60 neutrophil membrane with the help of CRY2-CIBN optogenetic system in cells expressing PIP5K1B. (g) Representative live-cell time-lapse confocal images of differentiated HL-60 neutrophils expressing CIBN-CAAX, CRY2PHR-mCherry-Mypt169 (magenta) and PIP5K1B-GFP (green), before or after 488 nm laser was switched on globally. Time in min:sec format. Scale bars represent 5 μm. Cells are pretreated with 10 μM Y27632 for 10 mins. (h-i) Box-and-whisker plots of cell migration speed (h) or aspect ratio (i) correspond to Figure 6b. nc=15 (e) or nc=12 (f) from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001, ***P = 0.0005 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (j-k) Box-and-whisker plots of aspect ratio (j) or cell area (k) correspond to Figure 6g. nc=16 (g-h) from at least 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001, **P = 0.0049 (Mann-Whitney test. Compare ranks). The median is at the center, and whiskers and outliers are graphed according to Tukey’s convention (GraphPad Prism 10). (l) Color-coded temporal overlay profile corresponds to Figure 6g.
Corresponding results were observed in propagating waves of Ras activity on the ventral surface of fused giant cells. As shown in Figure 6e and Video S25, in this example, expressing PIP5K completely suppressed waves of Ras activity. Strikingly, upon adding Myosin II inhibitor, Blebbistatin, the RBD waves started to burst and gradually increased until the entire surface displayed activity. Activity then decreased and reappeared. These results suggest that the mutual antagonism between Ras and PIP5K activities can be modulated by Myosin II. These results are consistent with an independent study from our laboratory demonstrating negative feedback from Myosin II to Ras [70].
We then extended these studies to differentiated HL-60 neutrophils and RAW 264.7 macrophage-like cells and assessed the effects of Myosin II reduction caused by recruitment of protein phosphatase 1 regulatory subunit 12A, Mypt1 in cells expressing PIP5Ks. We took advantage of a recruitable Mypt169, which enabled light-induced association with the membrane (Figure 6f) [79]. Globally recruiting Mypt169 to the plasma membrane in WT cells as the control induced cell spreading and polarity in differentiated HL-60 macrophages (Figure S15e–f, Video S26). Differentiated HL-60 neutrophils co-expressing PIP5K1B and the recruitable Mypt169 in initially displayed the same rounded phenotype as those expressing PIP5K1B alone (Figure 6g, Video S26). Globally recruiting Mypt169 to the plasma membrane, combined with Rock inhibitor Y27632, induced cell spreading and a polarized cell shape (Figure 6g, Video S26). These observations were supported by color-coded temporal overlay profiles (Figure 6l). Cell migration did not show a significant change (Figure 6l). Across the population, upon Mypt169 recruitment, differentiated HL-60 neutrophils induced a 1.53-fold and 1.23-fold increase in the cell area and polarity, respectively (Figure 6j–k). Similarly, globally recruiting Mypt169 to the plasma membrane in RAW 264.7 cells co-expressing PIP5K1C induced cell spreading and protrusion formation (Figure S15g, Video S26). These observations were supported by color-coded temporal overlay profiles (Figure S15h).
Since there is a shift from front towards “back” activities in PIP5K overexpressing cells, we reasoned that there might be a selective inhibition of branched actin relative to linear, cortical actin. ABD120 is a protein that labels total F-actin in Dictyostelium cells [80], while LimE selectively labels branched actin. We expressed both constructs in a PIP5K-overexpressing cell line and used the ABD120-LimE patch ratio to indicate the relative level of inhibition. Before inducing PIP5K, ABD120 is primarily labeled uniformly on the plasma membrane, with some enrichment at the protrusions, whereas LimE is localized almost exclusively to protrusions (Figure S16a, Video S27). Upon inducing PIP5K, cells changed to a rounder shape, and LimE patches disappeared (Figure S16b, Video S27). However, ABD120 was still uniformly distributed on the plasma membrane, resulting in a higher ABD120-LimE ratio (Figure S16e), suggesting a selective inhibition of branched actin. Consistent results were observed when staining cells with phalloidin. In uninduced cells, phalloidin mainly stained at the protrusions, while in cells expressing PIP5K, phalloidin staining colocalized with PIP5K around the perimeter (Figure S16c–d). Note that earlier (Figure 4), we found that PIP5K-expressing cells that showed the “spiky” phenotype changed to the “rounded” phenotype with CK666, further indicating that round-shaped cells have less branched actin (Figure S16f, Video S28).
The stochastic, reaction-diffusion model recreates experimental observations
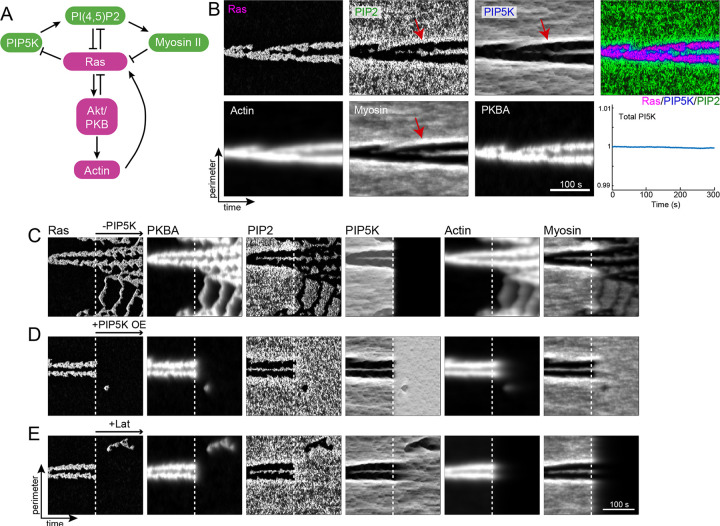
We next sought a simple mathematical model that could account for the extensive observations of the effects loss- and gain-of-function of PIP5Ks on cell migration and polarity and explain the observed symmetry breaking in the absence of cytoskeletal activity (Methods). The previously reported core of the model included interactions between Ras, PIP2, and PKBA, which gave rise to excitable behavior [36, 46]. Based on our new findings, we adjusted the inhibitory interaction from Ras to PIP2 to act through PIP5K. This, together with the negative regulation of Ras by PI(4,5)P2, closed a double-negative feedback loop, functionally equivalent to a positive feedback loop as in the original core model (Fig. 7A). However, based on the new findings, we made a critical change by implementing this loop with a dynamic partitioning scheme, where PIP5K remained on the membrane and was pushed away from regions of high Ras ([18]; Methods). We included two further feedback loops representing the interactions of the cytoskeleton with Ras activity, which were supported by an independently published study as well as the current findings. The first involved the positive contribution of actin to Ras activity, mediated through PKBA [40]. The second was the contribution of myosin, regulated by PI(4,5)P2, also in a double-negative feedback motif [70]. Simulations of this system on a one-dimensional periodic domain, representative of the cell boundary, gave rise to long-lived periods of localized, elevated Ras activity (Fig. 7B). Kymographs of the other model components showed that most signaling events either colocalized with Ras (e.g., Actin and PKBA) or formed a corresponding “shadow wave” where the signaling molecule was absent (e.g., PI(4,5)P2, PIP5K, and myosin). Notably, because of the dynamic partitioning, PIP5K was highest at the boundary between the wave and shadow wave (red arrow in Fig. 7B). In this case, the enzyme could not stay in the region of high Ras, but could not leave the membrane and it was elevated elsewhere. As shown the total PIP5K across the perimeter remained constant over time. As PIP5K regulated PI(4,5)P2 and subsequently myosin, we observed these bands of high activity in both markers.
Figure 7. Stochastic, reaction-diffusion model recreates experimental observations.
(a) Schematic demonstrating the interactions modeled. (b) Kymographs of each of the six model elements for a wild-type 300 second simulation. The red arrows in the three “rear” signals (PIP2, PI5K and myosin) show the accumulation of these elements at the boundary between front and back. The bottom right panel shows the sum over the complete perimeter of PIP5K as a function of time. (c-e) Kymographs of simulations involving perturbations. In all cases, the simulations started with WT parameters, and the perturbation was made after 150 s (marked by the white, dotted lines). Specifically, PIP5K levels were lowered (c) or increased (d); actin was eliminated (e). As in b, all kymographs show 300 s of simulated time.
We then used the model to recreate several perturbation experiments. Following the elimination of PIP5K, we observed Ras and actin activity increase greatly, transitioning from localized streaks to high levels that extended almost throughout the cell perimeter (Fig. 7C). In contrast, modeling PIP5K overexpression by increasing the PIP5K levels eliminated the Ras streaks completely (Fig. 7D). Simulations of latrunculin treatment showed that although the duration and persistence of the streak diminished, the model could still generate persistent streaks (Fig. 7E). Overall, the simulations strongly agreed with the experiments.
Discussion
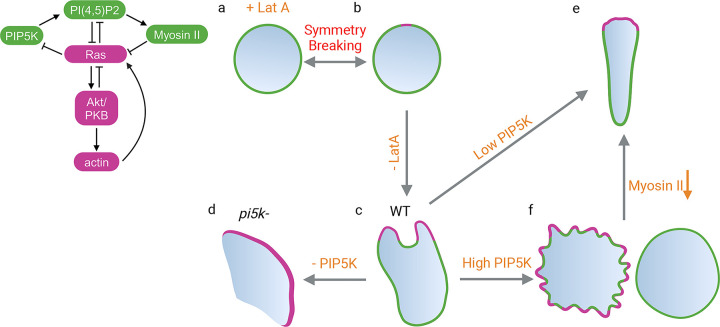
Our studies reveal that spatiotemporal regulation of PIP5K on the membrane is a primary determinant of polarity, protrusion formation, and resulting mode of migration (Figure 8). The phenotypes we observed are consistent with the kymographs generated in the computational simulations. In the absence of cytoskeletal activities (Figure 8a), cells are largely in a “back” state where PIP5K is elevated and Ras activity is low. Local spontaneous depletions of PIP5K and coordinated activations of Ras build up, breaking symmetry (Figure 8b). Coupled myosin reduction and increased actin polymerization lead to intermittently extended protrusions of relatively defined size and cell polarity (Figure 8c and S17). When PIP5K is absent in pi5k- cells, Ras is highly activated and signal transduction events spread across most of the cortex. Myosin is depleted, actin is polymerized, protrusions are wide, and cells switch their migratory behaviors from amoeboid to fan-shaped and oscillatory phenotypes (Figure 8d and S17). When PIP5K is slightly overexpressed in WT, signal transduction activities and protrusions are curtailed and cells, can polarize and migrate more rapidly (Figure 8e and S17). When PIP5K is increased further, Ras, PI3K, and F-actin activities are confined to small patches, or extinguished, and the entire cortex is shifted toward the back state. Protrusions become smaller and spikier, or disappear entirely and cells stop migration. (Figure 8f and S17). In summary, different phenotypes can be explained as lying along a spectrum from low-to-high PIP5K expression levels, which are inversely related to high-to-low signal transduction network activities (Figure S17).
Figure 8. Schematic illustration showing the effects of PIP5Ks on symmetry-breaking and cell morphology.
Schematic demonstrating the interactions modeled. Each cell morphology's front components and front localizations are highlighted in magenta, while back components and back localizations are highlighted in green. At rest state, back components, such as PIP5K will be uniformly distributed on plasma membrane in Lat A treated cell (a). Symmetry breaking will happen at one spot on the membrane and PIP5K moves away from the spot (b). Upon removing Lat A, the cytoskeletal activities will be restored and cell polarizes, becoming WT (c). Deleting PIP5K in WT cells will induce fan-shaped cells (d). Expressing low level PIP5K will promote cell polarity (e). Expressing high-level PIP5K will either induce fiopodias or completely shut down cells (f). This phenotype can be reversed to (e) through lowering myosin II activities.
As indicated in Figure 8, our results suggested that PIP5K is a negative regulator of signal transduction and cytoskeletal activities, which was not previously recognized. We propose that the chemotaxis defects previously reported in pi5k- cells can be traced to removal of the normal PIP5K-mediated constraint on Ras, leading to elevated, unregulated signal transduction and cytoskeletal activities which interfere with the cell’s ability to selectively direct protrusions towards chemoattractants. We found that although the overall speed of the pi5k- cells on average does not decrease, the actual behaviors of the cells as fans and oscillators are very defective, which was overlooked in the previous report [51]. In neutrophils and macrophages, we found that lowering PIP2 by recruiting Inp54p induced more protrusions, fan-shaped movement, and increased actin polymerization, consistent with the phenotype of the pi5k- cells and previous results in Dictyostelium [46]. Furthermore, previous reports that a dominant negative PIP5K1C interferes with the uropod in neutrophils and disrupts chemotaxis may also have been related to the loss of PIP5K function, leading to excessive Ras and associated protrusive activity [56]. The effects of PIP5K overexpression or recruitment in Dictyostelium, neutrophils, macrophages, and cancer-derived MDA-MB-231 cells provided further evidence that the enzyme is a negative regulator: 1) Cell protrusions and migration speed are severely suppressed; 2) Ras, PI3K, and F-actin activities are strongly inhibited. 3) The dose-response curves for Ras and PI3K activation by chemoattractant are shifted to higher concentrations. The effects of overexpression of PIP5K could be due to elevating PIP2 levels or preventing it from going down, as it and other back localized components normally do as protrusions form.
PIP5K plays a crucial role in symmetry breaking and polarity establishment. As migrating Dictyostelium cells and human neutrophils migrate, they occasionally shift between more circular and polarized profiles whereupon PIP5K shifts between a uniform localization and a back-to-front gradient. Since the enzyme is tightly associated with the membrane (confirmed in centrifugation assay), it must diffuse along the membrane to the back [18]. Protrusions are formed at the nascent front as PIP5K moves away from a local region. As it does not shuttle to the cytosol, PIP5K leaving the front accumulates at the rear, suppressing Ras, PI3K, and branched actin polymerization activities at the back and promoting a shift toward actomyosin. Thus, the spatiotemporal relocalization of PIP5K provides a way to activate the cell front and suppress the rear simultaneously, an elegant mechanism for symmetry breaking and self-organizing polarity. The regulation of signal transduction by PIP5K can occur without cytoskeletal activities in the presence of latrunculin, but there are significant interactions with actomyosin. In untreated cells, feedback from actomyosin is an essential part of the suppressive mechanism at the back since inhibiting myosin II can overcome the PIP5K-mediated suppression and reactivate the signal transduction activities.
Various models have been put forth suggesting that cell polarity arises from the competition of local positive and global negative feedback loops. Whereas positive feedback helps to amplify initial cellular asymmetries, negative feedback prevents other sites from developing, ensuring that the cell has a singular region of high activity, thus enabling persistent directional motion. Most of these models suggest that the positive feedback loop incorporates actin-mediated activation of various signaling molecules [81, 82]. The origin of the negative feedback is less clear, though mechanisms by which membrane or cortical tension inhibit protrusions have been suggested [17, 29, 83]. Computational models of these complementary motifs generate simulations of cell motility that closely resemble the shape and tracks of real cells [84, 85]. The dynamic partitioning of PIP5K reported here suggests an alternative actin-independent mechanism by which the same molecule can provide both localized positive feedback and global negative feedback. At the regions of high Ras activity, the removal of PIP5K lowers the levels of the Ras-inhibiting PIP2. This double-negative feedback loop provides localized positive feedback. Additionally, the dispersal of this membrane-bound PIP5K to regions away from high Ras activity, increases PIP2 levels and lowers the probability that Ras excitability is triggered elsewhere – thus completing a global negative feedback.
There are numerous reported interactions of PIP2 with various actin-binding proteins, but as some are reported to be activators and others are inhibitors, the overall effects of altering PIP2 in cells are challenging to understand. Some of the studies suggest that PIP2 sequesters critical actin-binding proteins. This mechanism would align with our proposal that PIP5K is a negative regulator: Van Rheenen et al. reported that EGF-induces PIP2 reduction in metastatic tumor cells and promotes actin polymerization [86]; Sengelaub et al. reported that in metastatic breast cancer cells, lowering PI(4,5)P2 levels releases cofilin to enhance actin turnover and cell migration [87]. Alternative studies suggested that PIP2 is an activator: It can directly interact with WASP to promote Arp2/3 complex nucleation [74] and it can bind to proteins like talin, vinculin, and focal adhesion kinases (FAK), to promote focal adhesion formation [88–93]. Our study indicates that inhibition to Ras by PIP5K, transmitted downstream, shifts the balance from branched F-actin to actomyosin and that the inhibitory role of PIP5Ks occurs in cells that do not form stable focal adhesions. Thus, the overall direction of the behavior in living cells favors PIP5K as a negative regulator regardless of potential direct interactions of PI(4,5)P2 with individual actin-binding proteins.
The counterintuitive effects of PIP5K inhibition on cell migration can be explained by the Ras-PIP5K mutual inhibitory feedback loop [46], with additional feedback loops from the cytoskeletal network, shown in Figure 7 and 8, but further studies are needed to sort out the detailed mechanism. Even without the cytoskeletal network, there is a strong reciprocal connection between Ras and PIP5K. It has been reported that Ras can directly interact with PIP5K [94]; if that is the case, the interaction must push Ras towards the inactive form to be consistent with our observations. It is possible that PIP5K or PIP2 interacts with Ras GAPs or competes with Ras GEFs to inhibit Ras activities. We are currently testing these possibilities. In the presence of the cytoskeletal network, PIP2 may regulate the interaction between the membrane and the actomyosin cortex [38]. While raising PIP2 would increase the interaction, lowering PIP2 may loosen the linkage, and could initiate a shift from actomyosin to branched actin. Such a mechanism would be consistent with the model of front and back-promoting feedback loops we recently reported [46, 70].
Our results suggest that phenotypes resulting from loss-of-function or gain-of-function alterations of PIP5K in tissue will likely depend on the initial state of the affected cells and may be difficult to predict. Without this study, PIP5K might be considered an oncogene since it is the substrate for producing second messengers, such as PIP3, DAG, and IP3, which are classic tumor promoters. The depletion of PIP5K and its product PIP2 from the front of the cells is puzzling since these messengers are localized at the front. However, even when PIP2 is lowered by as much as 90%, PIP3 is more elevated, and when PIP5K is overexpressed, Ras and PI3K activity are severely inhibited. Based on this, one might expect PIP5K to act like a tumor suppressor and consider activating or increasing PIP5K as a therapeutic mechanism to suppress activated oncogenic Ras. Our results show that a slight increase in PIP5K and a decrease in PIP3 activity can enhance polarity, while a large rise in PIP5K and elimination in PIP3 completely shut down the movement. Therefore, if PIP5K were to be considered a deterrent to Ras-mediated growth, caution would be needed to monitor the effects on cell migration.
Methods
Preparation of reagents and inhibitors
Rapamycin (Millipore Sigma, 553210) was dissolved in DMSO to prepare the 5 mM stock solution. Then, 1 μl aliquots were diluted 1:100 in the development buffer (DB) to a 10x concentration (50 μM). CK666 (Millipore Sigma, 182515) was dissolved in DMSO to prepare the 100 mM stock solution. Then, 0.5 μl aliquots were diluted 1:100 in the warm development buffer (DB) to a 10x concentration (1 mM). 237 μM latrunculin A in ethanol (Cayman, 10010630) was diluted in the development buffer (DB) to prepare the 50 μM (10x) stock solution. Folic acid (Sigma-Aldrich, 329823065) was dissolved in sterile water, with the addition of 2 M NaOH, to prepare the 1.25 mM stock solution. Caffeine (Millipore-Sigma,1085003) was dissolved to 1 M in sterile water and then diluted to a 10x concentration (40 mM) in the development buffer (DB). Fibronectin (Sigma-Aldrich, F4759-2MG) was dissolved in 2ml sterile water, followed by dilution in 8ml PBS to prepare the 200 μg ml−1 stock solution. N-Formyl-Met-Leu-Phe (fMLP; Sigma-Aldrich, 47729) was dissolved in dimethylsulfoxide (DMSO) (Sigma-Aldrich, D2650) to prepare the 50 mM stock solution. Then, 1 μl aliquots were diluted in RPMI to a 10x concentration (2 μM). Phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, P8139) was dissolved in DMSO to prepare the 1 mM stock solution. Then, 1 μl aliquots were diluted in RPMI to a 10x concentration (320 nM). FKP-(D-Cha)-Cha-r (ChaCha peptide, Anaspec; 65121) was dissolved in PBS to prepare the 2.5 mM stock solution. Then, 1 μl aliquots were diluted in DMEM to a 10x concentration (320 nM). A stock solution of EGF (Sigma-Aldrich, E9644) was prepared by dissolving it in 10 mM acetic acid to a final concentration of 1 mg ml−1. . The anti-BSA mouse monoclonal antibody was acquired from Sigma-Aldrich (SAB4200688, clone BSA-33). Blebbistatin (Peprotech, 8567182) was dissolved in DMSO to prepare the 50mM stock solution. Then, 1 μl aliquots were diluted 1:100 in the development buffer (DB) to a 10x concentration (500 μM). 5 mM Y-27632 (Sigma-Aldrich, 688001) was dissolved in DMSO to prepare the 5 mM stock solution. Then, 1 μl aliquots were diluted in RPMI to a 10x concentration (100 μM). Hygromycin B (Thermo Fisher Scientific, 10687010) or G418 sulfate (Thermo Fisher Scientific, 10131035) was purchased as 50 mg ml−1 stock solution. Then, 10 μl Hygromycin B or G418 sulfate was added in a 10ml cell culture. Blasticidin S (Sigma-Aldrich, 15205) or puromycin (Sigma-Aldrich, P8833) was dissolved in sterile water to prepare the stock solutions of 10 mg ml−1 or 2.5 mg ml−1, respectively. Then, 10 μl Blasticidin S or 4 μl puromycin was added in a 10ml cell culture. Doxycycline hyclate (Sigma, D9891-1G) was dissolved in sterile water to prepare the stock solution of 5 mg ml−1. Then, 100 μl Doxycycline hyclate was added in a 10ml cell culture before the experiment. TRITC-dextran (Sigma-Aldrich, T1162) was dissolved in sterile water to prepare the 50 mg ml−1 stock solution. Then, the stock solution was further diluted to 2 mg ml−1 in the HL5 culture medium before the experiment. All stock solutions were aliquoted and stored at −20 °C.
Cells and plasmids
WT Dictyostelium discoideum cells of the AX2 strain (dictyBase, strain ID DBS0235521) were obtained from the Kay laboratory (MRC Laboratory of Molecular Biology) and cultured in HL5 medium (laboratory stock) at 22 °C [95] for a maximum of 2 months after thawing from frozen stock. Pi5k- (piki-) cells were purchased from Dictybase (strain ID DBS0350270). pikI− cells were grown on a Klebsiella aerogenes lawn on an SM plate and transferred to HL5 medium supplemented with heat-killed Klebsiella aerogenes before the experiment. Gβ- cells were generated in the Devreotes Lab previously [96]. abnABC- cells were generated previously in the Devreotes Lab [97]. Growth-stage cells were used for all the experiments. GFP-arpC cells were purchased from Dictybase (strain ID DBS2036065).
The human HL-60 cell line (ATCC CCL-240; RRID:CVCL_0002) was obtained from the Weiner laboratory (UCSF) and cultured in RPMI 1640 medium (Gibco, 22400–089) supplemented with 15% heat-inactivated fetal bovine serum (Thermo Fisher, 16140071)4,75. To obtain migration-competent neutrophils, WT or stable lines were differentiated in the presence of 1.3% DMSO with a total of 1.5 million cells over 5–7 days [98]. Differentiated cells are an effective model to study human neutrophils [99]. To differentiate HL-60 cells into macrophages, cells were incubated with 32 nM PMA for 48–72h [100]. Cells were grown in humidified conditions at 5% CO2 and 37 °C.
RAW 264.7 macrophage-like cells were obtained from the N. Gautam laboratory (Washington University School of Medicine in St. Louis). RAW 264.7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 4,500 mg l−1 glucose, l-glutamine, sodium pyruvate and sodium bicarbonate (Sigma-Aldrich, D6429), supplemented with 10% heat-inactivated fetal bovine serum (ThermoFisher Scientific, 16140071) and 1% penicillin–streptomycin (ThermoFisher Scientific, 15140122). Cells were grown in humidified conditions at 5% CO2 and 37 °C.
All DNA oligonucleotides were purchased from Sigma-Aldrich. Dictyostelium PIP5K (below named PI5K) was PCR amplified from genome DNA and cloned in pCV5-GFP expression plasmid to generate PI5K-GFP (pCV5). Two-point mutations K681N and K582N were introduced to PI5K-GFP (pCV5) to generate PI5K (K581N, K682N)-GFP (pCV5). PI5K-GFP was then PCR amplified and cloned into doxycycline-inducible pDM335 plasmid (dictyBase, ID no. 523) using BglII/SpeI restriction digestion to generate mRFPmars-PI5K (pDM335). mRFPmars-PI5K was than PCR amplified and cloned into doxycycline-inducible pDM359 plasmid (dictyBase, ID no. 518) using BglII/SpeI restriction digestion to generate mRFPmars-PI5K (pDM359). mRFPmars was then replaced with EGFP to generate EGFP-PI5K (pDM359). EGFP was then replaced with KikGR to generate KikGR-PI5K (pDM359). DNA sequences encoding the 1-315aa, or 316-718aa, or 369-718aa or 1-718aa amino acids of PI5K, were PCR-amplified from PI5K-GFP (pCV5) and cloned into mCherry-FRB-MCS (pCV5) expression plasmid previously created in the Devreotes Lab using NheI/XhoI restriction digestion to generate mCherry–FRB-PI5K (1-315aa) (pCV5), mCherry–FRB- PI5K (316-718aa) (pCV5), mCherry–FRB-PI5K (369-718aa) (pCV5) and mCherry–FRB-PI5K (1-718aa) (pCV5). DNA sequences encoding the 316-718aa amino acids of PI5K, were PCR amplified and cloned into mRFPmars-SspBR73Q-MCS (pCV5) using NheI/NotI restriction digestion to generate mRFPmara-SspBR73Q-PI5K (316-718aa). DNA sequences encoding the 301-718aa amino acids of PI5K, were PCR amplified and cloned into pCV5-GFP expression plasmid to generate PI5K (301-718aa)-GFP (pCV5). PH-PLCδ-YFP (pCV5), CynA-KikGR (KF2), RBD-EGFP (pDM358), PHcrac-RFP (pDRH), LimEΔcoil-mCherry (pDM181), PHcrac-YFP (pDM358), LimEΔcoil-RFP (pDRH), cAR1-FKBP-FKBP (pDM358 or pCV5), LimE-GFP-FKBP-FKBP (pDM358), mCherry-FRB-MHCKC (pCV5), and PKBR1N150-iLiD (pDM358) were previously created in the Devreotes Lab. Myosin II-GFP (pDM181) was obtained from the Robinson laboratory (School of Medicine, JHU). PAK1(GBD)-YFP (pDEXH) was a gift from the C. Huang lab (JHU). RBD-RFP (pDM358) GFP-ABD120 (pDXA) was purchased from Dictybase (ID no. 472).
For transit transfection in RAW 264.7 macrophage-like cells and MDA-MB-231 cells, PHAkt-mCherry and CIBN-CAAX (Addgene #79574) were from Devreotes Lab. EGFP-PIP5K1B (Addgene #202722), GFP-PIP5K1gama90 (Addgene #22299) , CRY2-mCherry-MYPT169 (Addgene #178526), TagBFP-FKBP-PIP5K1C (Addgene #220090) (recruitable PIP5K1C), and mCherry-NES-SSPB-MCS-iLiD-CAAX (Addgene #173869) were purchased from Addgene. mCherry was replaced with Crismon fluorescent protein, and a point mutation R73Q was introduced to SapB to generate Crismon-NES-SapBR73Q-MCS-iLiD-CAAX. Recruitable PIP5K1C was PCR amplified and cloned into the XhoI/EcoRI sites of the Crismon-NES-SSPBR73Q-MCS-iLiD-CAAX expression plasmid to generate Crismon-NES-SspBR73Q-PIP5K1C-iLiD-CAAX.
For stable lines in HL-60 cells, stable lines co-expressing LifeAct–miRFP703 (pLJM1) (Addgene #201750) and CIBN-CAAX (pLJM1) (Addgene #201749) or expressing RFP-PHAkt (pFUW2) were previously created in the Devreotes Lab [44]. EGFP-PIP5K1B and EGFP-PIP5K1C were PCR amplified and cloned into the BspEI/SalI sites of the PiggyBac transposon plasmid to generate EGFP-PIP5K1B (pPB) and EGFP-PIP5K1C (pPB). Two-point mutations K359N, K260K or K407N, K408N were introduced to EGFP-PIP5K1B (pPB) or EGFP-PIP5K1C (pPB), to generate EGFP-PIP5K1B (K359N, K360N) and EGFP-PIP5K1C (K407N, K408N), respectively. Recruitable PIP5K1C were PCR amplified and cloned into the BspEI/NotI sites of the PiggyBac transposon plasmid to generate CRY2PHR-mCherry-PIP5K1C (pPB). MYPT169 was PCR amplified and cloned into the BsiWI/NotI sites of the mCherry-CRY2PHR (pHR) expression plasmid , which was previously modified from construct mCherry-CRY2PHR (pHR) (Addgene #201750) by introducing two restriction enzyme sites BstZ17I and BsiWI, to generate mCherryCRY2PHR (pHR). Constructs were verified by diagnostic restriction digestion and sequenced at the JHMI Synthesis and Sequencing Facility.
Transfection
Dictyostelium AX2 cells were transfected using a standard electroporation protocol. Briefly, for each transfection, 5 × 106 cells were pelleted, washed twice with ice-cold H-50 buffer (20 mM HEPES, 50 mM KCl, 10 mM NaCl, 1 mM MgSO4, 5 mM NaHCO3, 1 mM NaH2PO4, pH adjusted to 7.0) and subsequently resuspended in 100 μl ice-cold H-50 buffer. 2 μg of total DNA was mixed with the cell suspension, which was then transferred to an ice-cold 0.1-cm-gap cuvette (Bio-Rad, 1652089) for two rounds of electroporation at 850 V and 25 μF with an interval of 5 s (Bio-Rad Gene Pulser Xcell Electroporation Systems). After a 10 min incubation on ice, the electroporated cells were transferred to a 10-cm Petri dish containing HL-5 medium. The cells were selected by the addition of hygromycin B (50 μg ml−1) and/or G418 (20–30 μg ml−1) after 24h as per the antibiotic resistance of the vectors.
RAW 264.7 cells and MDA-MB-231 cells were transitly transfected by nucleofection in an Amaxa Nucleofector II device using Amaxa Cell line kit V (Lonza, VACA-1003) following a pre-existing protocol [101]. For each transfection, 3 × 106 cells were pelleted, resuspended in 100 μl supplemented Nucleofector Solution V. A total of 2–3 μg of DNA mixture was added and immediately transferred to a Lonza cuvette for electroporation using the program setting D-032 and X-013 for RAW 264.7 cells and MDA-MB-231 cells, respectively. Pre-warmed pH-adjusted culture medium (500 μl) was added to the electroporated cells in the cuvette. The cell suspension was then transferred to a 1.5 ml vial and incubated at 37 °C and 5% CO2 for 10 min and 30min for RAW 264.7 cella and MDA-MB-231 cells, respectively. Next, 50–100 μl solution containing cells was transferred to a coverslip chamber and allowed to adhere for 1 h. Finally, approximately 400 μl of pre-warmed pH-adjusted culture medium was added to each chamber and the cells were further incubated for 4–6 h before imaging for RAW 264.7 cells. MDA-MB-231 cells were imaged the next day.
Stable expression lines in HL-60 cells were generated by a combination of 3rdgeneration lentiviral- and PiggyBac™ transposon-integration based approaches [44, 102]. For transposon integration in HL-60 cell line, 5 μg transposon plasmid was co-electroporated with an equal amount of transposase expression plasmid into two million cells using Neon™ transfection kit (Invitrogen; MPK10025B). Cells and DNA mix were resuspended in buffer ‘R’ before electroporation in 100 μl pipettes at 1350V for 35 ms in Neon™ electroporation system (Invitrogen; MPK5000). Cells were resuspended in mixed culture medium in a 6-well plate and allowed to recover for 24 hours. Post-recovery, transfected cells were selected in presence of 10 mg/mL blasticidine S for 5–6 days. Once blasticidine S was removed, resistant cells were transferred to 48-well cell culture plate (Sarstedt; 83.3923), and further grown over 3–4 weeks into stable cell lines. Stable cells were maintained throughout in Blasticidine S.
For virus transfection, 2nd generation virus was prepared in HEK293T cells grown to about 80% confluency in 10 cm cell culture dish (Greiner Bio-One; 664160). For each reaction, a mixture of 2 μg pMD2.G (Addgene #12259), 8 μg psPAX2 (Addgene #12260), and 10 μg mCherry-CRY2PHR-MYPT19 (pHR) construct were transfected using Lipofectamine 3000 as per manufacturer’s instructions (Invitrogen; L3000–008). After 96 hours, virus containing culture medium was harvested at 3000 rpm for 20 mins at 4°C. In a 6-well plate (Greiner Bio-One; 657160), entire viral medium was added to 4 × 106 HL-60 cells (seeded at a density of 0.25 × 106 cells/mL) in presence of 10 μg /mL polybrene (Sigma; TR1003). After 24 hours, viral medium was removed, and cells were introduced to a mix of fresh and conditioned (mixed) culture medium. For selecting expressors, infected cells were sorted after 5 days, and subsequently, grown to confluency. For high-speeding sorting, we used 561 nm excitation laser to sort for mCherry expression using SH800S cell sorter (Sony). Briefly, cells were pelleted, resuspended in sorting buffer (1x PBS, Ca2+/Mg2+ free; 0.9% heat-inactivated FBS; 2% penicillin-streptomycin) at a density of 15 × 106 cells/ml, and sorted using 100 mm microfluidic sorting chip. High expressors (top 1%-5%) were collected in fresh culture medium (containing 2% penicillin-streptomycin) and grown to confluency.
Confocal microscopy and Live-cell imaging
All Dictyostelium experiments were performed on a 22 °C stage. All RAW 264.7, MDA-MB-231 and neutrophil experiments were performed inside a 37 °C chamber with 5% CO2 supply. All time-lapse live-cell imaging experiments were performed using one of the following microscopes: (1) Zeiss LSM 780-FCS Single-point, laser scanning confocal microscope (Zeiss Axio Observer with 780-Quasar; 34-channel spectral, high-sensitivity gallium arsenide phosphide detectors), (2) Zeiss LSM800 GaAsP Single-point, laser scanning confocal microscope with wide-field camera, (3) Nikon Eclipse Ti-E dSTROM total internal reflection fluorescence (TIRF) microscope (Photometrics Evolve EMCCD camera). The Zeiss 780 confocal microscope was controlled using ZEN Black software; Zeiss 800 confocal microscope was controlled using ZEN Blue software; the Nikon TIRF was controlled using Nikon NIS-Element software. The 40X/1.30 or 63X/1.4 Plan-Neofluar oil objective (with proper digital zoom) was used in all microscopes. The 488 nm (Ar laser) excitation was used for GFP and YFP, 561 nm (solid-state) excitation was used for RFP and mCherry, and 633nm (diode laser) excitation was used for miRFP703 in Zeiss 780 and 800 confocal microscopes. The 488nm (Ar laser) excitation was used for GFP and 561 nm (0.5W fiber laser) excitation was used for mCherry and RFP in Nikon TIRF.
To prepare imaging, vegetative Dictyostelium or differentiated macrophages were adhered on an eight-well coverslip chamber for 40 min. Differentiated neutrophils, pre-treated with heat-killed Klebsiella aerogenes, were adhered to fibronectin-coated chambers for 40 min. Next, fresh DB (5 mM Na2HPO4 and 5 mM KH2PO4 supplemented with 2 mM MgSO4 and 0.2 mM CaCl2) or RPMI 1640 medium was added to attached cells and used for imaging. To induce PI5K expression in Dictyostelium, doxycycline (50 μg ml−1) was added 8 h before imaging. For macropinocytosis assay, cells were incubated with 2 mg/ml TRITC-dextran for 10 min, washed twice with DB and imaged subsequently. All live- or fix- cell imaging was acquired with 0.3–0.8% (Dictyostelium) or 1–8% (HL-60) laser intensity. In single cells, confocal imaging was performed at a middle plane of the cells. To visualize cortical/ventral waves in Dictyostelium and RAW 264.7 cells, confocal microscopes were focused on the very bottom of the cell to capture the substrate-attached ventral surface of the cell. For the inhibitor experiments, neutrophils were treated with 10 μM Y-27632 for at least 10 min before imaging. In Dictyostelium, 50 μM blebbistatin or 100 μM CK666 was added during imaging, while 5 μM latrunculin A and 4 mM caffeine were added at least 20 min before imaging.
Chemically induced dimerization
The plasmids and experimental details of the chemically induced dimerization system have mostly been previously described [40, 46]. Here, to generate Figure S4i, S5a–c, and 6a–d data, the two following combinations were used to express the systems: (1) cAR1-FKBP-FKBP (pDM358) as the membrane anchor, mCherry–FRB-PI5K (1-315aa) (pCV5) as the recruitee; (2) LimE-GFP-FKBP-FKBP (pDM358) as the membrane anchor, mCherry–FRB- PI5K (1-718aa) (pCV5), or mCherry–FRB- PI5K (316-718aa) (pCV5), or mCherry–FRB- PI5K (369-718aa) (pCV5), as the recruitee; (3) cAR1-FKBP-FKBP (pCV5) as the membrane anchor, mCherry–FRB-MHCKC (pCV5) as the recruitee. Dictyostelium cells in the growth phase were transferred to an eight-well coverslip chamber and incubated for 10–15 min to allow them to adhere well. The HL-5 medium was then replaced with 450 μl DB buffer. Image acquisition was started 15–20 min after the medium change. After imaging a certain number of frames, rapamycin was added gently to the chamber (to a final concentration of 5 μM) during image acquisition to facilitate the recruitment of Inp54p to the plasma membrane.
Optogenetics
Optogenetics experiments were performed using slightly modified protocols [103] for Dictyostelium, HL-60 cells or MDA-MB-231 cells. Throughout image acquisition, a solid-state laser (561 nm excitation and 579–632 nm emission) was used for visualizing proteins or recruitable effectors fused to an mCherry, mRFPmars, or crismon tag, whereas a diode laser (633 nm excitation and 659–709 nm emission) was used to capture miRFP703 expression. Images were acquired for 5–10 min, before which a 450/488 nm excitation laser was switched on globally to activate recruitment and 10–20 min after. Image acquisition and photoactivation were performed at 7s intervals. Using the T-PMT associated with the red channel, we acquired DIC images. For recruiting MYPT169 or PIP5K1C in RAW264.7, MDA-MB-231, and HL-60 cells, 10 μM C5aR agonist, 10 ng/ml EGF, and 200 nM fMLP were added at least 15mins before imaging, respectively.
Frustrated phagocytosis and osmotic shock
We slightly modified a pre-existing protocol to visualize ventral waves in PMA-differentiated macrophages [54]. Briefly, Nunc Lab-Tek eight-well coverslip chambers were pre-washed with 30% nitric acid, coated with 1 mg ml−1 BSA for 3 h, washed with PBS and finally incubated with 5 μg ml−1 anti-BSA (1:200 dilution) for 2 h. The chambers were finally washed twice with PBS to remove excess antibodies. Before imaging, transfected PMA-differentiated macrophage cells were starved in suspension in 1×Ringer’s buffer (150 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 20 mM HEPES and 2 g l−1 glucose, pH 7.4) for 30 min. Next, these cells were introduced to the opsonized chambers and allowed to spread on the antibody-coated surface for 5–10 min, and then hypotonic shock was applied using 0.5×Ringer’s solution before imaging.
For development on non-nutrient agar, cells were washed with developmental buffer (DB) and plated on solidified DB agar at a density of 5.2 × 105 cells/cm2. For most experiments, development in suspension was carried out by pulsing cells resuspended in DB at 2 × 107 cells/ml with 50 to 100 nM cAMP every 6 min starting from 1 hour. For the experiments presented in Fig. 6 (A to D) and fig. S10 (A to C), crac− or GFP-GtaC/crac− cells were pulsed with different concentrations of cAMP or at different intervals starting from 2 hours. For development in the presence of sp-cAMPS, 1 μM was included in DB or DB agar.
Cell differentiation
For Dictyostelium cell development, 1 × 108 cells in the exponential growth phase were collected from suspension culture and pelleted as previously described [104, 105]. After washing with DB twice, the cells were resuspended in 4 ml DB and shaken at 105 r.p.m. for 1 h. The cells were then pulsed with 100 nM cAMP (5s pulse every 6 min) using a time-controlled peristaltic pump for 5–6 h with continual shaking. This allowed the cells to develop and polarize. After development, Around 10–15 μL of differentiated cells (in DB media) was collected were transferred from the shaker to an eight-well coverslip chamber, resuspended thoroughly in 450 μl DB and incubated for 20–30 min before imaging. For development on non-nutrient agar, cells were washed with developmental buffer (DB) and plated on solidified DB agar at a density of 5.2 × 105 cells/cm2.
Global receptor activation assay
Cells were first differentiated as described in cell differentiation protocol. Cells were subsequently incubated with 5 μM Latrunculin A for around 20 min before imaging. Using a confocal laser scanning microscope, 10–20 frames were first acquired to record the basal activity of the proteins, then cAMP was added to the chamber (to a final concentration of 1 μM) to activate all the cAR1 receptors, and the image acquisition was continued. An imaging frequency of 3 s/frame was maintained throughout the experiment.
For vegetative Dictyostelium cells, cells were seeded on the eight-well coverslip chamber and incubated for 20–30 min. Then, the HL5 medium was replaced with DB and incubated cells with an additional 20 minutes before the imaging. Cells were subsequently incubated with 5 μM Latrunculin A for around 20 min before imaging. Using a confocal laser scanning microscope, 10–20 frames were first acquired to record the basal activity of the proteins, then folic acid was added to the chamber (to a final concentration of 0.1, 1, 10, or 100 nM) to activate all the fAR1 receptors, and the image acquisition was continued. An imaging frequency of 3 s/frame was maintained throughout the experiment.
For RAW 264.7 cells, transfected cells were previously incubated on the eight-well coverslip chamber for 4–6h. The culture medium was replaced with HBSS buffer supplemented with 1 g/L glucose and incubate with another 30 min before the imaging. Using a confocal laser scanning microscope, 10–20 frames were first acquired to record the basal activity of the proteins, then C5aR agonist was added to the chamber (to a final concentration of 0.1, 1, 10, or 100 μM) to activate all the C5a receptors, and the image acquisition was continued. An imaging frequency of 15 s/frame was maintained throughout the experiment.
2D Chemotaxis Assay
We slightly modified a pre-existing protocol to perform chemotaxis assay for vegetative Dictyostelium cells [106]. Briefly, cells were seeded on the 1-well Nunc Lab-Tek chamber as a drop at the center and incubated for 20–30 min. Then 2 mL DB buffer was added to the chamber and cells were dispersed by pipetting multiple times. A Femtotip microinjection needle (Eppendorf) was loaded with 10 μM of filtered folic acid solution and then connected to a FemtoJet microinjector (Eppendorf). The microinjector was operated in continuous injection mode with a compensation pressure of 15 hPa. To initiate the chemotaxis, the micropipette was brought to the (x,y,z) coordinate of cells using a programmed micromanipulator. The imaging was continued with an acquisition frequency of 30 s/frame.
CRISPR/Cas9 to generate pi5k- cell line
We slightly modified a pre-existing protocol for using CRISPR/Cas9 system to target specific genes and generate KO mutants in Dictyostelium cells [107]. gRNA guide sequences targeting pi5k gene were designed using Cas-Designer. Customized gRNA oligos were purchased from Sigma-Aldrich. This selected gRNA sequence AAACAACTCATTGGATTCAGATGC (forward) was then cloned into pTM1285 plasmid obtained from the Robinson laboratory (School of Medicine, JHU). AX2 cells were transformed with pTM1285 plasmid containing pi5k gRNA using electroporation, using the transfection protocol described above. After 8–16 h of transformation, we replaced the media with HL-5 media containing G418 (20–30 μg ml−1) and cultured the transformed cells for 3 more days. After that, floating dead cells were washed off with HL-5 medium. Remaining live cells were plated on SM plates with cultured Klebsiella aerogenes and incubated at room temperature for 3–4 d until visible, isolated plaques formed. These individual plaques were separately transferred to 96-well plates containing HL-5 medium in the absence of G418. After approximately 2–3 d when colonies grew, genomic DNA was extracted for PCR verification and then the rest cells were transferred to 6-well and then 10-cm Petri dishes and grown to confluency.
Multinucleation assay
Cells that have been in suspension at 200 rpm for 2–3 d were seeded at ~70% confluency on glass coverslips in HL-5 medium and allowed to adhere for 20 min. Next, HL-5 medium was replaced with ice-cold fixative solution (2% PFA + 0.01% Triton X-100 in 1.5× HL-5 medium) for 10 min. Cells were then washed quickly with 1× PBS, and the nuclei were stained with 10 μg/ml Hoechst for 10 min. Afterwards, cells were washed three times, 10 min each time, with 1× PBT (1× PBS + 0.01% Triton X-100). Before visualization, cells were mounted on coverslips using Invitrogen SlowFade™ Gold antifade reagent. Cells were imaged using Zeiss LSM 780-FCS Single-point, laser scanning confocal microscope with 405 nm laser.
Immunofluorescence
Cells were seeded on glass coverslips in HL-5 medium and allowed to adhere for 20 min. Next, HL-5 medium was replaced with fixative solution (2% buffered paraformaldehyde with 0.25% Glutaraldehyde and 0.1% Triton X-100 in HL-5 medium) for 10min at room temperature (RT), then quenched in 1mg/mL Sodium Borohydride for 10min, then washed three times with 1x PBT (1x PBD supplemented with 0.5% BSA and 0.05% Triton X-100), each time for 3min. Cells were then blocked in blocking buffer (3% BSA in 1× PBT) for 1 h at room temperature. Blocking buffer was removed and cells were washed with 1x PBT quicky then incubated with Invitrogen Alexa Fluor ™ 647 phalloidin (1: 200 dilution) for 1h at RT. Cells were then washed three times with 1x PBT, each time for 3min. Before visualization, cells were mounted on coverslips using Invitrogen SlowFade™ Gold antifade reagent. Cells were imaged using Zeiss LSM 780-FCS Single-point, laser scanning confocal microscope with 405 nm laser.
Image Analysis
All images were analysed with Fiji/ImageJ 1.52i (NIH), Python v.3.10, and MATLAB 2019b (MathWorks) software. We utilized GraphPad Prism 10 (GraphPad), OriginPro v.9.0 (Originlab Corporation) and Microsoft Excel (Microsoft) for plotting our results.
Kymographs.
For the membrane kymographs, the cells were segmented against the background following standard image-processing steps with custom code written in MATLAB 2019b. The kymographs were created from the segmented cells as previously described, where consecutive lines over time were aligned by minimizing the sum of the Euclidean distances between the coordinates in two consecutive frames using a custom-written MATLAB function [19]. A linear colourmap was used for the normalized intensities in the kymographs. In coloured kymographs, the lowest intensity is indicated by blue and the highest by yellow. Line kymographs that accompanied ventral waves were generated in Fiji/ImageJ by drawing a thick line with a line width of 8–12 pixels and processing the entire stack in the ‘KymographBuilder’ plugin.
t-stacks.
The ImageJ (NIH) software was used for image processing and analysis. For t-stacks, TIRF time-lapse videos were opened and converted to greyscale in ImageJ. The ImageJ 3D Viewer plugin was then used to stack the frames from the video. The resampling factor is set to 1 to avoid blurring of activities between frames.
Line-scan intensity profile.
Line scans were performed in Fiji/ImageJ (NIH) by drawing a straight-line segment (using line tool) inside the cells with a line width of 8–12 pixels to obtain an average intensity value. The intensity values along that particular line were obtained in the green and red channels using the ‘Plot Profile’ option. The values were saved and normalized in OriginPro 9.0 (OriginLab). The intensity profiles were graphed and finally smoothened using the adjacent-averaging method, using proper boundary conditions. The plots were then normalized by dividing by the maximum value. For a particular line scan, the green and red profiles were smoothed and processed using the exact same parameters to maintain consistency.
Membrane to cytosol ratio quantification.
The cytosol of cells was first outlined with freehand selection tool in Fiji/ImageJ (NIH). The mean intensity of the cytosol was measured using analyze>>measure function. Then make band function was applied to the outline and proper band size was applied according to the cells. Then mean membrane intensity was measured using the same method. Using mean membrane to cytosol intensity to divide mean cytosol intensity, we can get the membrane to cytosol ratio for each biosensor. Each data was then plotted as box-and-whisker graphs in GraphPad Prism 10.
Cell migration analysis
Analysis was performed by segmenting Dictyostelium or neutrophil cells in Fiji/ImageJ 1.52i software. For this, image stack was thresholded using the ‘Threshold’ option. ‘Calculate threshold for each image’ box was unchecked, and range was not reset. Next, cell masks were created by size-based thresholding using the ‘Analyzed particles’ option. To optimize binarized masks, ‘Fill holes’, ‘Dilate’, and ‘Erode’ were done several times. For creating temporal color-coded cell outlines, ‘Outline’ was applied on binarized masks, followed by ‘Temporal-Color Code’ option. Next, ‘Centroid’ and ‘Shape descriptors’ boxes were checked in ‘Set Measurements’ option under ‘Analyze’ tab. This provided us with values for centroid coordinates and aspect ratio. Mean and SEM from replicates of aspect ratio values were determined and plotted in GraphPad Prism 10. The starting point for centroid values was set to zero for each track, and these new coordinates were plotted in Microsoft Excel to generate migration tracks. Velocity was calculated by computing displacement between two consecutive frames. Displacement was then divided by time interval to obtain speed for each cell. These speed values were then time-averaged over all frames to produce data points for cell speed which were plotted as box-and-whisker graphs in GraphPad Prism 10.
Reaction-diffusion model
Core excitable network.
The simulations are based on a model, whose core consists of three interacting species, Ras, PI(4,5)P2, and PKB, that together form an excitable network [46]. The concentrations of each of these molecules is described by stochastic, reaction-diffusion partial differential equations (RD-PDEs):
In each of these equations, the final term represents the diffusion of the species, where is the respective diffusion coefficient and is the spatial Laplacian (in one dimension). The second-to-last terms represent the molecular noise. Our model assumes a Langevin approximation in which the size of the noise is based on the reaction terms [108]. For example, in the case of PKB, the noise is given by
where is a zero mean, unit variance, Gaussian, white noise process. In the simulations, the size of this noise was adjusted with the empirical parameter .
PIP5K incorporation through dynamic partitioning.
To this model we include the contribution of PIP5K, and base it on a dynamic partition principle [18]. In this case, PI5PK exists on the membrane in two states with corresponding low and high diffusivities ( and ). The conversion from low to high diffusivity is mediated by Ras. Together, they obey the following two RD-PDEs:
Note that we do not include noise in these equations. That the total concentration is conserved can be seen by noting that the sum of the reaction terms is equal to zero. Thus, the only changes in concentration appear through the diffusion terms. Integrating over the complete domain and using the fact that boundary conditions are periodic guarantee that the total concentration of PI5PK is constant. The main effect of PI5PK is to increase the production of PIP2:
Cytoskeleton feedback loops.
Finally, we incorporated two other terms. The first represents feedback from branched actin onto Ras. Since the actin cytoskeleton is not directly modeled, we model the origin of this feedback from PKB, which is upstream to the cytoskeleton [40] and incorporated one actin term:
The second feedback loop accounts for the inhibitory regulation by actomyosin on Ras that originates from PIP2:
Together they modify the equations for :
Modeling perturbations.
The perturbations in Fig. 7c–d were done by modifying the decay and production rates of the relevant species during the simulation. Specifically:
Eliminating PIP5K: , for both types of PIP5K
Increasing PIP5K: , for both types of PIP5K.
Eliminating myosin: and .
Eliminating actin: and .
Implementing the simulations.
Simulations were run on MATLAB 2023b (MathWorks, Natick, MA) on custom-code based on the Itô solution in the Stochastic Differential Equation toolbox (http://sdetoolbox.sourceforge.net). The simulations aim to recreate the membrane fluorescence observed in single-cell confocal images. The dimension is therefore smaller, assuming a cell radius of 5 μm and a spacing of 0.25 μm, resulting in points along the perimeter, and periodic boundary conditions. Initial conditions were set by solving the steady-state value of the core model. The PIP5K values are normalized. All plots shown are after a 200 second interval so that the effect of the initial values is minimized. For the kymographs, contrast for each species was done through MATLAB’s imadjust command.
| Parameter | Value | Units | Parameter | Value | Units |
|---|---|---|---|---|---|
|
| |||||
| 0.122 | 2.85 | ||||
| 1.14 | 440 | ||||
| 0.0144 | |||||
| 2134 | 1.75 | ||||
| 0.0744 | 0.864 | ||||
| 0.025 | 0.05 | ||||
| 160 | |||||
| 0.05 | 0.05 | ||||
| 0.05 | 0.05 | ||||
| 0.03 | 0.03 | ||||
| 0.09 | |||||
| 0.005 | 0.0025 | ||||
| 0.0025 | 0.5 | ||||
| 0.0833 | |||||
Statistics and reproducibility
Statistical analyses were executed using unpaired or paired two-tailed nonparametric tests on GraphPad Prism 10. Results are expressed as mean ± s.d. from at least three independent experiments. NS, P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ***P ≤ 0.0001. Tukey’s convention was used to plot box-and-whisker plots. Statistical test details are indicated in the figure legends. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications4,35,79. Each representative image or image series is from n > 3 independent experiments. Any variations observed between treatment groups are not attributed to sampling bias. No data were excluded from the analyses.
Supplementary Material
Acknowledgments
We thank all members of the Devreotes, Iglesias, and Robinson laboratories (Schools of Medicine and Engineering, JHU) for helpful discussions and for providing resources. We thank Jonathan Kuhn and Yiyan Lin (Devreotes Lab) for providing constructs. We acknowledge Orion Weiner (UCSF) for providing HL-60 cell line. We thank Sean Collins (UC Davis) for providing transposon plasmids. We thank Stephen Gould (School of Medicine, JHU) for help with instrumentation. We appreciate services of DictyBase and Addgene for providing plasmids. This work was supported by NIH grant R35 GM118177 (to P.N.D.), DARPA HR0011-16-C-0139 (to P.A.I. and P.N.D.), AFOSR MURI FA95501610052 (to P.N.D.), as well as NIH grant S10OD016374 (to S. Kuo of the JHU Microscope Facility).
Footnotes
Competing Interests
The authors declare no competing interests.
Statistics and reproducibility
Statistical analyses were executed using unpaired or paired two-tailed nonparametric tests on GraphPad Prism 10. Results are expressed as mean ± s.d. from at least three independent experiments. NS, P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ***P ≤ 0.0001. Tukey’s convention was used to plot box-and-whisker plots. Statistical test details are indicated in the figure legends. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications4,35,79. Each representative image or image series is from n > 3 independent experiments. Any variations observed between treatment groups are not attributed to sampling bias. No data were excluded from the analyses.
Data availability
All data are provided in the main or supplementary text. Requests for additional information on this work are to be made to the corresponding author.
Reference
- 1.Sasaki A.T., et al. , G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol, 2007. 178(2): p. 185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiner O.D., et al. , Spatial control of actin polymerization during neutrophil chemotaxis. Nat Cell Biol, 1999. 1(2): p. 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai H. and Devreotes P.N., Moving in the right direction: how eukaryotic cells migrate along chemical gradients. Semin Cell Dev Biol, 2011. 22(8): p. 834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., et al. , Identifying network motifs that buGer front-to-back signaling in polarized neutrophils. Cell Rep, 2013. 3(5): p. 1607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swaney K.F., Huang C.H., and Devreotes P.N., Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys, 2010. 39: p. 265–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parent C.A., Making all the right moves: chemotaxis in neutrophils and Dictyostelium. Curr Opin Cell Biol, 2004. 16(1): p. 4–13. [DOI] [PubMed] [Google Scholar]
- 7.SenGupta S., Parent C.A., and Bear J.E., The principles of directed cell migration. Nat Rev Mol Cell Biol, 2021. 22(8): p. 529–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saha S., Nagy T.L., and Weiner O.D., Joining forces: crosstalk between biochemical signalling and physical forces orchestrates cellular polarity and dynamics. Philos Trans R Soc Lond B Biol Sci, 2018. 373(1747). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shellard A. and Mayor R., All Roads Lead to Directional Cell Migration. Trends Cell Biol, 2020. 30(11): p. 852–868. [DOI] [PubMed] [Google Scholar]
- 10.Ridley A.J., et al. , Cell migration: integrating signals from front to back. Science, 2003. 302(5651): p. 1704–9. [DOI] [PubMed] [Google Scholar]
- 11.Hadjitheodorou A., et al. , Directional reorientation of migrating neutrophils is limited by suppression of receptor input signaling at the cell rear through myosin II activity. Nat Commun, 2021. 12(1): p. 6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghabache E., et al. , Coupling traction force patterns and actomyosin wave dynamics reveals mechanics of cell motion. Mol Syst Biol, 2021. 17(12): p. e10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., et al. , Excitable networks controlling cell migration during development and disease. Semin Cell Dev Biol, 2020. 100: p. 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladoux B., Mege R.M., and Trepat X., Front-Rear Polarization by Mechanical Cues: From Single Cells to Tissues. Trends Cell Biol, 2016. 26(6): p. 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagorda A. and Parent C.A., Eukaryotic chemotaxis at a glance. J Cell Sci, 2008. 121(Pt 16): p. 2621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rickert P., et al. , Leukocytes navigate by compass: roles of PI3Kgamma and its lipid products. Trends Cell Biol, 2000. 10(11): p. 466–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houk A.R., et al. , Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell, 2012. 148(1–2): p. 175–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee T., et al. , A dynamic partitioning mechanism polarizes membrane protein distribution. Nat Commun, 2023. 14(1): p. 7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee T., et al. , Spatiotemporal dynamics of membrane surface charge regulates cell polarity and migration. Nature Cell Biology, 2022. 24(10): p. 1499–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C.H., et al. , An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat Cell Biol, 2013. 15(11): p. 1307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghose D., Elston T., and Lew D., Orientation of Cell Polarity by Chemical Gradients. Annu Rev Biophys, 2022. 51: p. 431–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pal D.S., et al. , The excitable signal transduction networks: movers and shapers of eukaryotic cell migration. The International Journal of Developmental Biology, 2019. 63(8–9-10): p. 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schink K.O., Tan K.W., and Stenmark H., Phosphoinositides in Control of Membrane Dynamics. Annu Rev Cell Dev Biol, 2016. 32: p. 143–171. [DOI] [PubMed] [Google Scholar]
- 24.Shewan A., Eastburn D.J., and Mostov K., Phosphoinositides in cell architecture. Cold Spring Harb Perspect Biol, 2011. 3(8): p. a004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrasco-Rando M., et al. , An acytokinetic cell division creates PIP2-enriched membrane asymmetries leading to slit diaphragm assembly in Drosophila nephrocytes. Development, 2023. 150(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu M. and Liu J., Mechanobiology in cortical waves and oscillations. Curr Opin Cell Biol, 2021. 68: p. 45–54. [DOI] [PubMed] [Google Scholar]
- 27.Cramer L.P., Forming the cell rear first: breaking cell symmetry to trigger directed cell migration. Nat Cell Biol, 2010. 12(7): p. 628–32. [DOI] [PubMed] [Google Scholar]
- 28.Teruel M.N. and Meyer T., Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction. Cell, 2000. 103(2): p. 181–4. [DOI] [PubMed] [Google Scholar]
- 29.De Belly H., et al. , Cell protrusions and contractions generate long-range membrane tension propagation. Cell, 2023. 186(14): p. 3049–3061 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carvalho K., et al. , Actin polymerization or myosin contraction: two ways to build up cortical tension for symmetry breaking. Philos Trans R Soc Lond B Biol Sci, 2013. 368(1629): p. 20130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong Y., et al. , Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc Natl Acad Sci U S A, 2010. 107(40): p. 17079–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X. and Harrison A., A general principle for spontaneous genetic symmetry breaking and pattern formation within cell populations. J Theor Biol, 2021. 526: p. 110809. [DOI] [PubMed] [Google Scholar]
- 33.van Haastert P.J.M., Symmetry Breaking during Cell Movement in the Context of Excitability, Kinetic Fine-Tuning and Memory of Pseudopod Formation. Cells, 2020. 9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhan H., et al. , An Excitable Ras/PI3K/ERK Signaling Network Controls Migration and Oncogenic Transformation in Epithelial Cells. Developmental Cell, 2020. 54(5): p. 608–623.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X., et al. , Mutually inhibitory Ras-PI(3,4)P(2) feedback loops mediate cell migration. Proc Natl Acad Sci U S A, 2018. 115(39): p. E9125–E9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devreotes P.N., et al. , Excitable Signal Transduction Networks in Directed Cell Migration. Annual Review of Cell and Developmental Biology, 2017. 33(1): p. 103–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buckley C.M., et al. , Coordinated Ras and Rac Activity Shapes Macropinocytic Cups and Enables Phagocytosis of Geometrically Diverse Bacteria. Curr Biol, 2020. 30(15): p. 2912–2926 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bisaria A., et al. , Membrane-proximal F-actin restricts local membrane protrusions and directs cell migration. Science, 2020. 368(6496): p. 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai T.Y., et al. , EGicient Front-Rear Coupling in Neutrophil Chemotaxis by Dynamic Myosin II Localization. Dev Cell, 2019. 49(2): p. 189–205 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao Y., et al. , Wave patterns organize cellular protrusions and control cortical dynamics. Mol Syst Biol, 2019. 15(3): p. e8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerhardt M., et al. , Actin and PIP3 waves in giant cells reveal the inherent length scale of an excited state. J Cell Sci, 2014. 127(Pt 20): p. 4507–17. [DOI] [PubMed] [Google Scholar]
- 42.Matsuoka S. and Ueda M., Mutual inhibition between PTEN and PIP3 generates bistability for polarity in motile cells. Nat Commun, 2018. 9(1): p. 4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y., et al. , Ras suppression potentiates rear actomyosin contractility-driven cell polarization and migration. Nat Cell Biol, 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pal D.S., et al. , Actuation of single downstream nodes in growth factor network steers immune cell migration. Dev Cell, 2023. 58(13): p. 1170–1188 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolch W., Berta D., and Rosta E., Dynamic regulation of RAS and RAS signaling. Biochem J, 2023. 480(1): p. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao Y., et al. , Altering the threshold of an excitable signal transduction network changes cell migratory modes. Nature Cell Biology, 2017. 19(4): p. 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Bout I. and Divecha N., PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci, 2009. 122(Pt 21): p. 3837–50. [DOI] [PubMed] [Google Scholar]
- 48.Ling K., et al. , Type I gamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin traGicking via a direct interaction with mu 1B adaptin. J Cell Biol, 2007. 176(3): p. 343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lacalle R.A., et al. , Type I phosphatidylinositol 4-phosphate 5-kinase controls neutrophil polarity and directional movement. J Cell Biol, 2007. 179(7): p. 1539–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Sayegh T.Y., et al. , Phosphatidylinositol-4,5 bisphosphate produced by PIP5KIgamma regulates gelsolin, actin assembly, and adhesion strength of N-cadherin junctions. Mol Biol Cell, 2007. 18(8): p. 3026–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fets L., Nichols J.M., and Kay R.R., A PIP5 kinase essential for eGicient chemotactic signaling. Curr Biol, 2014. 24(4): p. 415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swaney K.F., et al. , Novel protein Callipygian defines the back of migrating cells. Proc Natl Acad Sci U S A, 2015. 112(29): p. E3845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flemming S., et al. , How cortical waves drive fission of motile cells. Proc Natl Acad Sci U S A, 2020. 117(12): p. 6330–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masters T.A., Sheetz M.P., and Gauthier N.C., F-actin waves, actin cortex disassembly and focal exocytosis driven by actin-phosphoinositide positive feedback. Cytoskeleton (Hoboken), 2016. 73(4): p. 180–96. [DOI] [PubMed] [Google Scholar]
- 55.Zhan H., et al. , Self-organizing glycolytic waves fuel cell migration and cancer progression. bioRxiv, 2024. [Google Scholar]
- 56.Lokuta M.A., et al. , Type Igamma PIP kinase is a novel uropod component that regulates rear retraction during neutrophil chemotaxis. Mol Biol Cell, 2007. 18(12): p. 5069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shin D.Y., et al. , Sphingomyelin metabolism underlies Ras excitability for eGicient cell migration and chemotaxis. Cell Struct Funct, 2023. 48(2): p. 145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fukushima S., Matsuoka S., and Ueda M., Excitable dynamics of Ras triggers spontaneous symmetry breaking of PIP3 signaling in motile cells. J Cell Sci, 2019. 132(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arai Y., et al. , Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc Natl Acad Sci U S A, 2010. 107(27): p. 12399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sobczyk G.J., Wang J., and Weijer C.J., SILAC-based proteomic quantification of chemoattractant-induced cytoskeleton dynamics on a second to minute timescale. Nat Commun, 2014. 5: p. 3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sasaki A.T., et al. , Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol, 2004. 167(3): p. 505–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iijima M. and Devreotes P., Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell, 2002. 109(5): p. 599–610. [DOI] [PubMed] [Google Scholar]
- 63.Parent C.A., et al. , G protein signaling events are activated at the leading edge of chemotactic cells. Cell, 1998. 95(1): p. 81–91. [DOI] [PubMed] [Google Scholar]
- 64.Janetopoulos C., et al. , Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc Natl Acad Sci U S A, 2004. 101(24): p. 8951–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X., et al. , Reverse fountain flow of phosphatidylinositol-3,4-bisphosphate polarizes migrating cells. EMBO J, 2021. 40(4): p. e105094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veltman D.M., et al. , A plasma membrane template for macropinocytic cups. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kunz J., et al. , The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol Cell, 2000. 5(1): p. 1–11. [DOI] [PubMed] [Google Scholar]
- 68.Weckerly C.C., et al. , Nir1-LNS2 is a novel phosphatidic acid biosensor that reveals mechanisms of lipid production. bioRxiv, 2024.
- 69.Inaba H., Miao Q., and Nakata T., Optogenetic control of small GTPases reveals RhoA mediates intracellular calcium signaling. J Biol Chem, 2021. 296: p. 100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuhn J., et al. , Complementary Cytoskeletal Feedback Loops Control Signal Transduction Excitability and Cell Polarity. bioRxiv, 2024. [Google Scholar]
- 71.Zhao H., Hakala M., and Lappalainen P., ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophys J, 2010. 98(10): p. 2327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bae Y.H., et al. , Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc Natl Acad Sci U S A, 2010. 107(50): p. 21547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Papayannopoulos V., et al. , A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell, 2005. 17(2): p. 181–91. [DOI] [PubMed] [Google Scholar]
- 74.Rozelle A.L., et al. , Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr Biol, 2000. 10(6): p. 311–20. [DOI] [PubMed] [Google Scholar]
- 75.Janmey P.A. and Stossel T.P., Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature, 1987. 325(6102): p. 362–4. [DOI] [PubMed] [Google Scholar]
- 76.Pan M., et al. , A G-protein-coupled chemoattractant receptor recognizes lipopolysaccharide for bacterial phagocytosis. PLoS Biol, 2018. 16(5): p. e2005754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li X.X., Clark R.J., and Woodrue T.M., C5aR2 Activation Broadly Modulates the Signaling and Function of Primary Human Macrophages. J Immunol, 2020. 205(4): p. 1102–1112. [DOI] [PubMed] [Google Scholar]
- 78.Ren Y., et al. , Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin I. Curr Biol, 2009. 19(17): p. 1421–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamamoto K., et al. , Optogenetic relaxation of actomyosin contractility uncovers mechanistic roles of cortical tension during cytokinesis. Nat Commun, 2021. 12(1): p. 7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shibata K., et al. , Actin binding domain of filamin distinguishes posterior from anterior actin filaments in migrating Dictyostelium cells. Biophys Physicobiol, 2016. 13: p. 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Inoue T. and Meyer T., Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PLoS One, 2008. 3(8): p. e3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srinivasan S., et al. , Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J Cell Biol, 2003. 160(3): p. 375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kabacoe C., et al. , Dynacortin facilitates polarization of chemotaxing cells. BMC Biol, 2007. 5: p. 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi C., et al. , Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells. PLoS Comput Biol, 2013. 9(7): p. e1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neilson M.P., et al. , Chemotaxis: a feedback-based computational model robustly predicts multiple aspects of real cell behaviour. PLoS Biol, 2011. 9(5): p. e1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Rheenen J., et al. , EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol, 2007. 179(6): p. 1247–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sengelaub C.A., et al. , PTPRN2 and PLCbeta1 promote metastatic breast cancer cell migration through PI(4,5)P2-dependent actin remodeling. EMBO J, 2016. 35(1): p. 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chinthalapudi K., Rangarajan E.S., and Izard T., The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation. Proc Natl Acad Sci U S A, 2018. 115(41): p. 10339–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thapa N., et al. , PIPKIgamma and talin couple phosphoinositide and adhesion signaling to control the epithelial to mesenchymal transition. Oncogene, 2017. 36(7): p. 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goni G.M., et al. , Phosphatidylinositol 4,5-bisphosphate triggers activation of focal adhesion kinase by inducing clustering and conformational changes. Proc Natl Acad Sci U S A, 2014. 111(31): p. E3177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cai X., et al. , Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol Cell Biol, 2008. 28(1): p. 201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gilmore A.P. and Burridge K., Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4–5-bisphosphate. Nature, 1996. 381(6582): p. 531–5. [DOI] [PubMed] [Google Scholar]
- 93.Fukami K., et al. , alpha-Actinin and vinculin are PIP2-binding proteins involved in signaling by tyrosine kinase. J Biol Chem, 1994. 269(2): p. 1518–22. [PubMed] [Google Scholar]
- 94.Adhikari H. and Counter C.M., Interrogating the protein interactomes of RAS isoforms identifies PIP5K1A as a KRAS-specific vulnerability. Nat Commun, 2018. 9(1): p. 3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Watts D.J. and Ashworth J.M., Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem J, 1970. 119(2): p. 171–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lilly P., et al. , A G-protein beta-subunit is essential for Dictyostelium development. Genes Dev, 1993. 7(6): p. 986–95. [DOI] [PubMed] [Google Scholar]
- 97.Lampert T.J., et al. , Shear force-based genetic screen reveals negative regulators of cell adhesion and protrusive activity. Proc Natl Acad Sci U S A, 2017. 114(37): p. E7727–E7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Millius A. and Weiner O.D., Manipulation of neutrophil-like HL-60 cells for the study of directed cell migration. Methods Mol Biol, 2010. 591: p. 147–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rincon E., Rocha-Gregg B.L., and Collins S.R., A map of gene expression in neutrophil-like cell lines. BMC Genomics, 2018. 19(1): p. 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rovera G., Santoli D., and Damsky C., Human promyelocytic leukemia cells in culture diGerentiate into macrophage-like cells when treated with a phorbol diester. Proc Natl Acad Sci U S A, 1979. 76(6): p. 2779–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meshik X., O'Neill P.R., and Gautam N., Optogenetic Control of Cell Migration. Methods Mol Biol, 2018. 1749: p. 313–324. [DOI] [PubMed] [Google Scholar]
- 102.Bell G.R.R., et al. , Optogenetic control of receptors reveals distinct roles for actin- and Cdc42-dependent negative signals in chemotactic signal processing. Nat Commun, 2021. 12(1): p. 6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pal D.S., et al. , Optogenetic modulation of guanine nucleotide exchange factors of Ras superfamily proteins directly controls cell shape and movement. Frontiers in Cell and Developmental Biology, 2023. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kamimura Y., Tang M., and Devreotes P., Assays for chemotaxis and chemoattractant-stimulated TorC2 activation and PKB substrate phosphorylation in Dictyostelium. Methods Mol Biol, 2009. 571: p. 255–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cai H., et al. , Nucleocytoplasmic shuttling of a GATA transcription factor functions as a development timer. Science, 2014. 343(6177): p. 1249531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aufderheide K.J. and Janetopoulos C., Migration of Dictyostelium discoideum to the Chemoattractant Folic Acid. Methods Mol Biol, 2016. 1407: p. 25–39. [DOI] [PubMed] [Google Scholar]
- 107.Sekine R., Kawata T., and Muramoto T., CRISPR/Cas9 mediated targeting of multiple genes in Dictyostelium. Sci Rep, 2018. 8(1): p. 8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gillespie D.T., The chemical Langevin equation. Journal of Chemical Physics, 2000. 113(1): p. 297–306. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided in the main or supplementary text. Requests for additional information on this work are to be made to the corresponding author.