Abstract
The “rule of six” stipulates that the Paramyxovirus RNA polymerase efficiently replicates only viral genomes counting 6n + 0 nucleotides. Because the nucleocapsid proteins (N) interact with 6 nucleotides, an exact nucleotide-N match at the RNA 3′-OH end (3′-OH congruence) may be required for recognition of an active replication promoter. Alternatively, assuming that the six positions for the interaction of N with the nucleotides are not equivalent, the nucleotide position relative to N may be critical (N phase context). The replication abilities of various minireplicons, designed so that the 3′-OH congruence could be discriminated from the N phase context, were studied. The results strongly suggest that the application of the rule of six depends on the recognition of nucleotides positioned in the proper N phase context.
The active genome of the negative-stranded RNA viruses is found in the form of a nucleocapsid. In the nucleocapsid, the single-stranded RNA is tightly associated with the nucleocapsid proteins (N) in a rod-shaped helical structure. The interaction between the RNA and the N subunits can be so stable that the nucleocapsid structure does not disintegrate in high salt concentrations, even upon density equilibrium centrifugation (35). In such cases, the sugar-phosphate backbone may not be accessible to solvent (29) and the RNA chain may be protected from RNase attack (37). This suggests that the RNA is inside the nucleocapsid structure. However, its exact position is not known, because the RNA-N three-dimensional structure has not been resolved at atomic resolution.
For the Sendai virus (SeV), a member of the Paramyxoviridae family, Respirovirus genus (other genera of the same family include Morbillivirus, Rubulavirus, and Pneumovirus), the nucleocapsid rod, containing an RNA of 15,384 nucleotides (54, 55), is ∼1.0 μm long and has a diameter of ∼20.0 nm (19, 28). A three-dimensional reconstruction at 2.4-nm resolution has been obtained from electron microscopy images (17). The left-handed helix can coexist in three different pitch conformations: 5.3, 6.8, and 37.5 nm. In the most prevalent pitch state, 5.3 nm, there are 13 N subunits per helix turn and each N subunit is proposed to interact with 6 nucleotides.
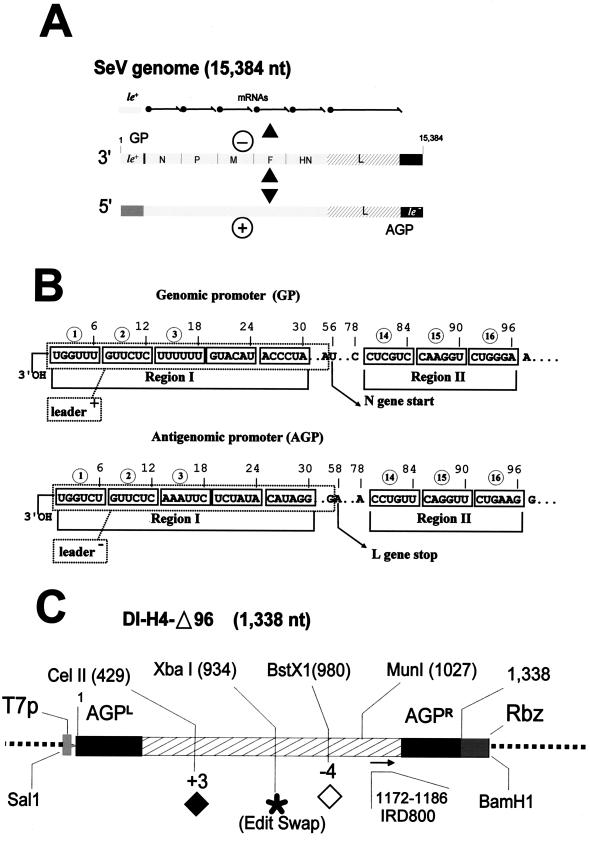
The RNA genome is linearly organized in six successive transcription units separated by a few nucleotides (see Fig. 1A). This coding region is flanked, at the 3′ and 5′ ends, by short extracistronic sequences, of 55 and 57 nucleotides, respectively, for SeV. These sequences (called leader+ and leader− or trailer [see Fig. 1B]) have been predicted to represent promoters for viral RNA synthesis. However, the Paramyxovirus promoters outflank these regions, in contrast with those of the Rhabdoviruses (vesicular stomatitis virus [VSV]) (12, 46, 60). They extend over 96 nucleotides, overlapping the first (N gene) and last (L gene) transcription units (Fig. 1B) (41, 58). At the genome 3′ end, the genomic promoter (GP) operates in transcription and replication, while at the antigenome 3′ end, the antigenomic promoter (AGP) is used for replication only (Fig. 1A). It is noteworthy that the replication promoter, in the present context, refers not only to the 3′-end nucleotides of the template RNA but also to the 5′-end nucleotides of the nascent RNA. This is because the encapsidation efficiency of this nascent RNA is seen as a factor that governs replication (for a review, see reference 35).
FIG. 1.
Structural features of SeV genome, promoters, and minireplicon. (A) Schematic representation of the SeV genome (−) with the plus (le+) and minus (le−) leaders, and the six transcription units (N, P, M, F, HN, and L) portrayed. Above it are shown the products of the transcription initiating at the GP only. Below it is shown the product of replication, the antigenome (+), which in turn serves as a template for genome amplification, starting at the AGP. (B) Detailed representation of GP and AGP primary structure and features. The sequence, portrayed as RNA of the sense template, is written in groups of 6 nucleotides (hexamers, numbered 1 through 16) to account for the fact that the N (small rectangle) contacts 6 nucleotides. Definitions of regions I and II are given in the text. Nucleotides 56 on GP and 58 on AGP, representing, respectively, the start of the N gene and the end of the L gene, are indicated. (C) Schematic representation of the parent minireplicon DNA sequence flanked by the T7 promoter (T7p) and the ribozyme (Rbz) as in plasmid pSP65 (see Materials and Methods). Restriction sites relevant for construction of derivatives, some harboring an insertion (CelII) (⧫), a deletion (BstX1) (◊), or an introduction of the Swap editing site (XbaI) (∗), are shown. The position of the IRD800 oligonucleotide used in the primer extensions is indicated.
In the Paramyxoviridae family, with the possible exception of the members of the Pneumovirus genus (respiratory syncytial virus [RSV]), the replication promoters are composed of two discontinuous elements. The first 20 to 30 nucleotides most proximal to the 3′ end (Region I in Fig. 1B), fairly conserved among members of the same genus, are likely to contain, in addition to the signal for RNA synthesis initiation, the encapsidation signal, active on the 5′ end of the nascent RNA. The second element (Region II in Fig. 1B) spans the internal extremity of the promoter. A motif repeated three times, common for the respiro- and morbilliviruses [(CNNNNN)3, nucleotides 79 to 96] or for the rubulaviruses [(NNNNGC)3, nucleotides 77 to 94], has been shown to be absolutely required in number and location for replication to take place (41, 42, 58). Note that the motifs are repeated, for SeV and simian virus 5 (SV5), with a periodicity of 6 nucleotides. The mechanism underlying the role of this second element is not known, but it is noteworthy that, in the nucleocapsid three-dimensional structure, it is found juxtaposed to the first element, on the second turn of the helix (58). The two elements, then, are on the same face of the helix, composing a surface that may allow the polymerase complex to interact with two helix turns.
The definition of the promoters was achieved thanks to a system of reverse genetics that only recently became available for negative-stranded RNA viruses (1a, 2, 8, 11, 15, 21, 25, 27, 31, 36, 43, 45, 50, 51, 53, 61). By using this system to replicate minigenome templates (minireplicons), the length of the RNA was shown, for the members of the Paramyxoviridae family, to be a major factor that determines the level of genome replication, which is most efficient when the total number of nucleotides is a multiple of 6. This basic requirement, dubbed the “rule of six,” appears to apply with varying stringency. Very strict for SeV, softer for other members of the family such as SV5 (16, 41), it nevertheless seems valid for all the members of the family, with the exception of the Pneumovirus genus (52).
The question of the mechanism underlying the application of this rule now arises. A first interpretation flows from the general picture of the genome replication process. The nucleotides on the nascent strand are encapsidated, concomitantly with replication, in units of 6, with 5′-to-3′-end polarity. At the 3′ end, when the total number of replicated nucleotides is a multiple of 6, the last 6 nucleotides synthesized are perfectly covered by the last N subunit (3′-OH congruence). This 3′-OH congruence may be required for this 3′ end to be recognized as an active promoter. Another possible mechanism integrates the notion that the nucleotides are seen in the context of the N subunit, and that interaction points N1 to N6 are not equivalent. A particular nucleotide, then, part of a signal for the viral polymerase, could be seen as such only in the proper N subunit context, N phase context N1, N2, or N3, etc. The existence of such a phase context is supported by the phase conservation of the repeated motifs identified in the promoters (see above). In SeV, the C is found repeated three times in phase N1 (58), while in SV5, GC in positions N5 and N6 is repeated three times (41, 42). Extending this observation, it is noteworthy that the first nucleotides of the transcription start and stop signals, as well as the P gene editing site, are not found in a random N phase context in a particular genome (33). Finally, experimentally changing the N phase context of nucleotides in the middle of the GP has been shown to affect replication (49).
In the present study, we compare the replication properties of minireplicons in which the 3′-OH congruence has been separated from the 3′-end N phase context. This approach allows the conclusion that the RNA polymerase complex is insensitive to 3′-OH congruence. Moreover, the RNA polymerase is shown to start internally when an irrelevant sequence is added at the 3′ end before the promoter region. Also, initiation at two different sites is observed when two promoters are present in tandem, directly adjacent or separated by intervening sequences. The relevance of these results for the application of the rule of six and for the mechanism of action of the RNA polymerase is discussed.
MATERIALS AND METHODS
Virus and cells.
Cells (A549 and HeLa) were grown in regular minimal essential medium (MEM) supplemented with 5% fetal calf serum (FCS) under a 5% CO2 atmosphere. Recombinant vaccinia virus expressing T7 RNA polymerase (vTF7-3) a gift from Bernard Moss (National Institutes of Health, Bethesda, Md.), has been described by Fuerst and colleagues (20) and was used as specified. vTF7-3 stocks were grown in HeLa cells with titers ranging from 5 × 108 to 109 PFU/ml.
Sequence and plasmids.
The complete SeV RNA primary sequence (15,384 nucleotides) was taken from the work of Shioda et al. (54, 55) and Neubert et al. (44). Except for Fig. 1B (see legend), the sequence is written as DNA according to standard convention, 5′ to 3′, left to right. The plasmids expressing the SeV N protein (pGem-N), P plus C protein (pGem-P/Cwt), P without C protein (pGem-PCstop), and L protein (pGem-L) under the control of the T7 RNA polymerase promoter have been described previously (13, 14, 30).
The structure of the minireplicon SeV DI-H4-(+)ΔDRA/AGP96 (referred to below as DI-H4-Δ96, construct #1) and its cloning into pSP65 have been described by Tapparel et al. (58). As before (4, 58, 59), the 3′ end of the viral genome is called the GP, and the 3′ end of the antigenome is called the AGP. In the case of H4 RNA, which contains the AGP on both the minus- and the plus-strand RNA, AGPL or AGPR respectively denote these two promoters (see Fig. 1C). In all the derivatives used in this study, the 3′ end of the H4 minus-strand RNA containing AGPL was never changed. Modifications, including site-directed base substitutions (G91C or G4T) and extension of AGP with GP (with intervening sequences) or with unrelated sequences at the 3′ end, were made exclusively at the AGPR 3′ end of the DI-H4-Δ96 minireplicon. Creation of constructs #15 and #19, and subsequent modifications to yield constructs #16, #17, and #18 (see Fig. 6), were introduced by fusion PCR methodology (26), using adequate overlapping oligonucleotides at the inner modification sites and external primers containing the MunI site (position 1027 in Fig. 1C) in the minireplicon sequence and the BamHI site at the 3′ end of the ribozyme sequence (3) (Fig. 1C). The intervening sequences between AGP and the ribozyme in constructs #8 and #9 (see Fig. 4) were introduced by taking advantage of an XhoI site (6 nucleotides) introduced first by fusion PCR at the 3′ end of AGP to generate construct #6. These promoter-unrelated sequences originate from the SeV M gene (accession number X53056) between nucleotides 595 and 821. They were PCR amplified with adequate primers flanked by two XhoI sites. From construct #6, a cut at the XhoI site, followed by filling and religation, yielded construct #7. In contrast, a recession at the same site, followed by blunt ligation, yielded construct #21 (see Fig. 7). The +3 addition at the CelII site (see Fig. 1C) in constructs #4, #5, #20, #22, and #23 (Fig. 3 and 7) was made by replacing the small SalI (prior to the T7 promoter [Fig. 1C])/XbaI (position 934) fragment of the pSV DI-H4-Δ96 derivatives with that of derivative pSV-DIH4-01 (3). The −4 deletion at the BstX1 site (see Fig. 1C) in constructs #2, #3, #11, #12, #13, and #14 (Fig. 3 and 5) was created by replacing the large MunI (position 1027)/BamHI (at the end of the ribozyme sequence) fragment of pSV-DI-H4-Δ96 with that of derivative pSV-DIH4-02 (3). The Swap editing site was introduced (constructs #01, #3, #5, #12, and #14) (see Fig. 3 and 5) by cloning the Swap editing cassette (5′-CTAGAACTACTGGTAGGGAATTAAAAAAGGGCTCGTAAACTGCCGTAT-3′) (boldfaced nucleotides, editing site; italicized nucleotides, complementary sequence of the primer used for limited primer extension) (23) flanked with two XbaI sites into the unique XbaI site (position 934) (Fig. 1C) of pSV-DI-H4-Δ96 and its derivatives. The orientation of the editing cassette was such that genome length correction took place upon minus-strand RNA synthesis. The portions of the constructs that had undergone modifications were all extensively sequenced using the LICOR DNA 4000 automated sequencing apparatus.
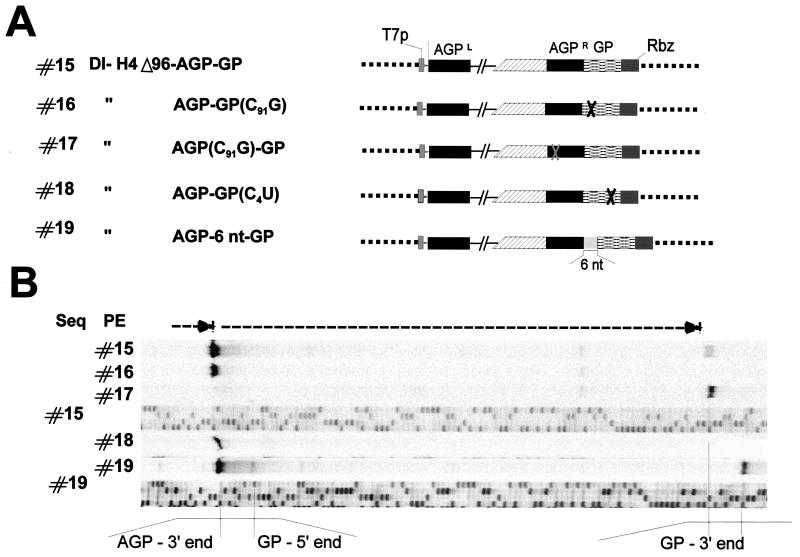
FIG. 6.
Introducing constructs with two promoters. (A) Schematic representation of plasmid constructs expressing minireplicons harboring two promoters in tandem (AGP and GP) at the 3′-OH end of the T7 RNA transcript, as in construct #15. In constructs #16, #17, and #18, the activity of either AGP or GP is obliterated by a base substitution (X) made explicit by the name of the construct [for example, AGP-GP (C91G) indicates that G in position 91 of GP has been substituted for C]. Construct #19 has a 6-nucleotide insertion between the two promoters. All the constructs led to the production of T7 RNA transcripts containing 6n + 0 nucleotides. (B) Digitalized images of primer extensions (PE) performed with RNAs from replication of constructs shown in panel A and of sequences (Seq) obtained with the same primer on the indicated plasmids. Primer extensions were performed with RNA purified from transfection-replication assays involving 4 × 107 cells.
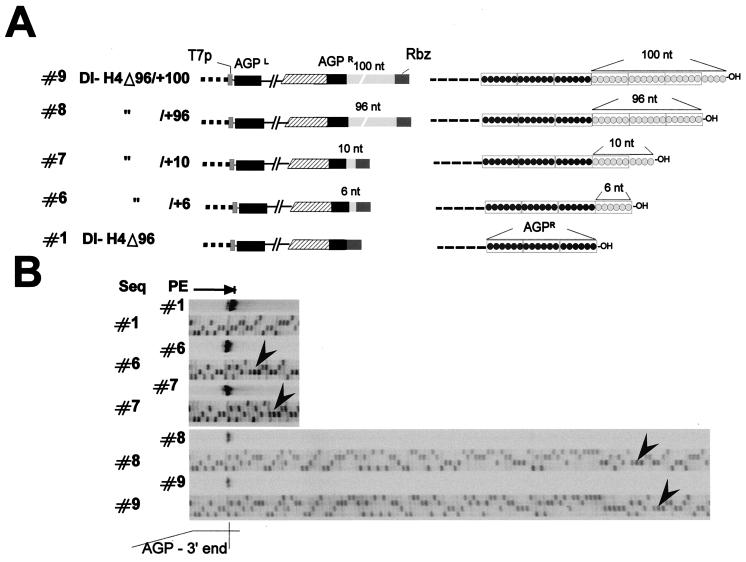
FIG. 4.
Testing the value of the 3′-OH congruence. (A) (Left) Schematic representation of plasmid constructs harboring an extended sequence past the AGPR down to the ribozyme (Rbz) sequence (dark shaded rectangle). T7p, T7 RNA polymerase promoter. (Right) Expected 3′-OH ends of the T7 RNA transcript made by these plasmids. AGPR is portrayed by three hexamers, as in Fig. 3A, to highlight the 3′-OH nature of the RNA. Constructs #6 and #8 (of 6n + 0 nucleotides) exhibit perfect 3′-OH congruence, while constructs #7 and #9 (6n + 4) exhibit dangling nucleotides. (B) Digitalized images of primer extensions (PE) performed with RNAs from replication of constructs #6, #7, #8, and #9, with sequences (Seq) obtained with the same primer on the corresponding plasmids. Arrowheads point to the first nucleotide of the ribozyme sequence. Primer extensions were performed with RNA purified from transfection-replication assays involving 4 × 107 cells except for construct #1 (0.5 × 107 cells).
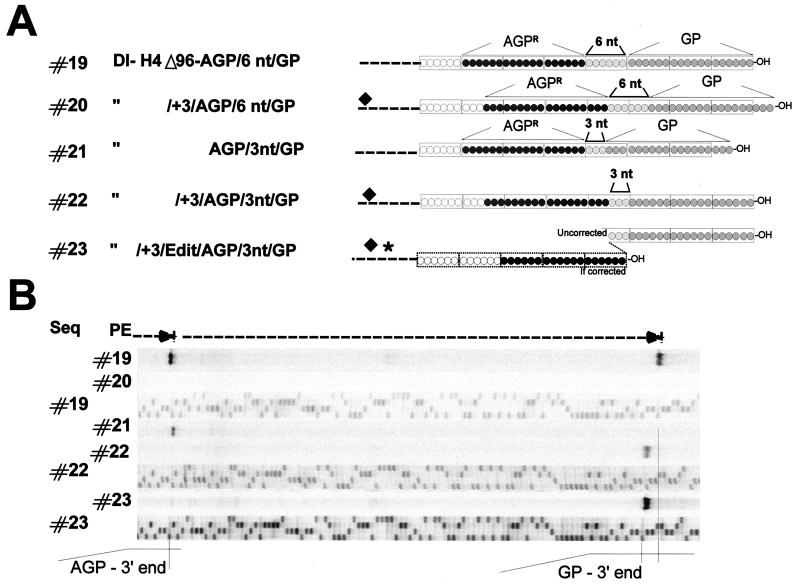
FIG. 7.
Testing the value of a change in the phase context with two promoters in tandem. (A) Schematic representation (as in Fig. 6) of the expected 3′-OH ends of T7 RNA transcripts derived from plasmids carrying two promoters with a 6 (constructs #19 and #20)- or 3 (constructs #21, #22, and #23)-nucleotide insertion between them. Constructs #20, #22, and #23 also contain a 3-nucleotide addition at the CelII site (⧫) (see Fig. 1C). Finally, construct #23 harbors a Swap editing site (✶). For this construct, if initiation takes place at the out-of-phase AGP, the genome length correction generates a new 3′-OH end with a congruent AGP (dotted-line boxes) (see also Fig. 6). (B) Digitalized images of primer extensions (PE) performed with RNAs from replication of constructs shown in panel A and of sequences (Seq) obtained with the same primer on the indicated plasmids. Primer extensions were performed with RNA purified from transfection-replication assays involving 4 × 107 cells.
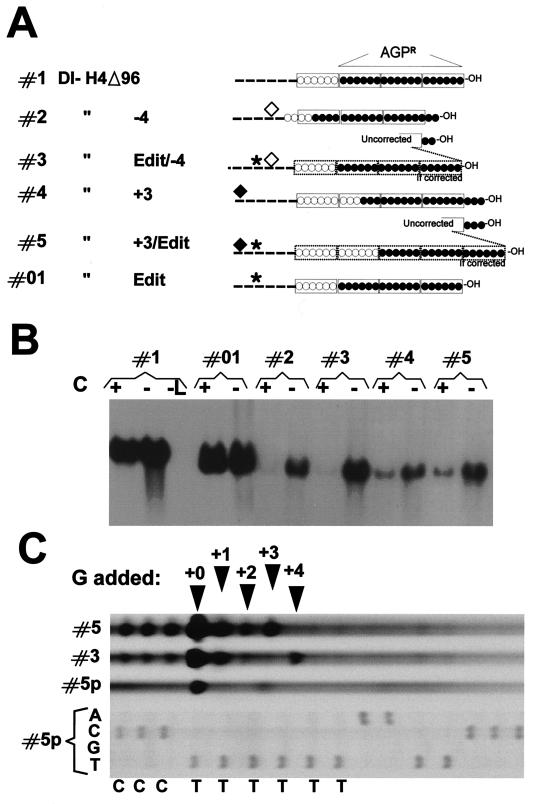
FIG. 3.
Effect of the C proteins on compliance with the rule of six and genome length correction. (A) Schematic partial representations of positive-polarity minigenomes showing the nature of their 3′-OH ends, where AGPR lies, featured with three hexamers, i.e., 3 times 6 nucleotides boxed within rectangles representing the N proteins. Construct #1, of 6n + 0 nucleotides, exhibits perfect congruence of the last 3′-OH nucleotide and the N protein. Construct #2, with a 4-nucleotide deletion (◊) at the BstX1 site (see Fig. 1C), has 6n + 2 nucleotides and is portrayed with 2 nucleotides dangling at the 3′-OH end. Construct #3 is like construct #2 but contains a Swap editing site (✶) inserted at XbaI (see Fig. 1C). During replication, if editing leads to a +4G addition, the genome length is corrected and congruence at the 3′-OH end is recovered (indicated in #3 and #5 as new 3′-OH ends boxed in by dotted lines). Constructs #4 and #5 correspond to constructs #2 and #3 but contain a +3 nucleotide addition (⧫) at the CelII site (see Fig. 1C) rather than the −4 deletion. #01, 6n + 0 construct with the Swap site. (B) Northern blot analysis of the replicated RNAs originating from the constructs presented in panel A. Replication assays were performed with the addition of plasmid pGem-P/Cwt or pGem-PCstop (see Materials and Methods) to generate conditions where the C proteins were present or absent, respectively, during replication (+C or −C). −L, replication assay where the pGem-L plasmid was omitted. (C) Limited primer extension assay (see Materials and Methods and Results) across the editing site of the replicated RNAs from constructs #5 and #3 (lanes #5 and #3) or across the editing site of the plasmid expressing construct #5 (lane #5p). A regular sequence of the same site is presented below to allow positioning of the limited primer extension stops.
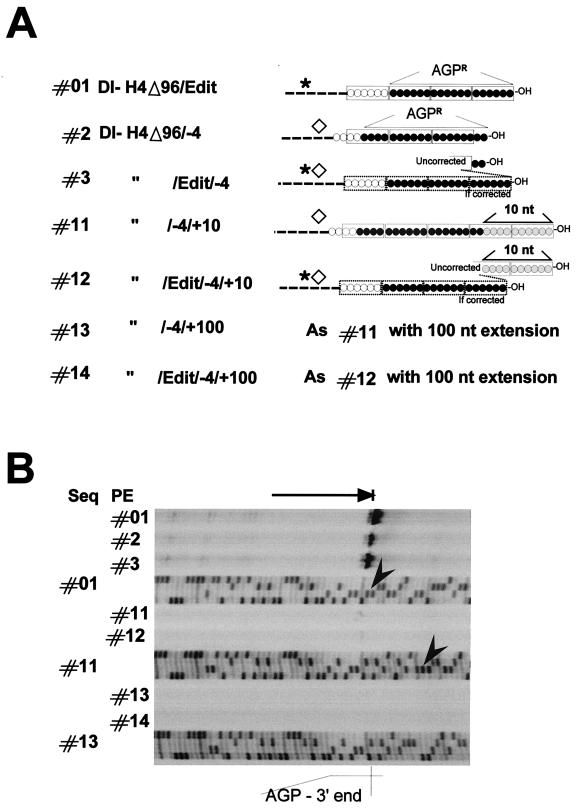
FIG. 5.
Testing the value of a change in the phase context with unrelated sequence at the 3′-OH end. (A) Schematic representation of the expected 3′-OH ends of T7 RNA transcripts derived from the indicated plasmids. Constructs #2, #3, and #11 through #14 contain a 4-nucleotide deletion at the BstX1 site (◊) (see Fig. 1). This deletion is associated with the presence of the Swap editing site (✶) (see Fig. 1) in constructs #3, #12, and #14. Sequence extensions of 10 or 100 nucleotides past AGPR are also present in constructs #11 and #12 or #13 and #14, respectively. For all these constructs, AGPR is portrayed by three hexamers (as in Fig. 3A) to highlight the 3′-OH congruence (as in #01, #11, #12, #13, and #14) or the change in promoter phase context (as in #2 and #3). Note that the presence of the editing site in constructs #3, #12, and #14 can correct the RNA length by a 4-nucleotide addition, if replication starts at the out-of-phase AGPR. This is indicated by the corrected new 3′-OH end, featured in #3 and #12 (dotted-line boxes). (B) Digitalized images of primer extensions (PE) performed with RNAs from replication of constructs shown in panel A and of sequences (Seq) obtained with the same primer on the corresponding plasmids. When appropriate, arrowheads point to the first nucleotide of the ribozyme sequence. Primer extensions were performed with RNA purified from transfection-replication assays involving 4 × 107 cells.
Replication system and recovery of encapsidated RNA.
The replication system has been described previously (4) and was used here with few modifications (Fig. 2). In brief, A549 cells (about 107, in 9-cm-diameter petri dishes) were infected with the recombinant vaccinia virus vTF7-3 (multiplicity of infection 3) expressing T7 RNA polymerase. One hour later, the cells were transfected with 5 μg of pGem-N, 5 μg of pGem-PCstop (or −PCwt), 1.0 μg of pGem-L, and 5 μg of the plasmid carrying the minireplicon sequence, by incubation of the cells (for 3 h at 33°C) with 3 ml of MEM containing the mixed plasmids and 15 μl of Transfectase (BRL). Seven milliliters of MEM plus 5% FCS was then added. Forty hours later, cells were disrupted in 300 μl of lysis buffer (0.6% NP-40, 50 mM Tris-HCl [pH 8.0], 10 mM NaCl). The cytoplasmic extract was clarified (at 3,000 × g for 15 min at 4°C), and the encapsidated RNA was purified by banding to equilibrium in CsCl density gradients. The collected nucleocapsids were pelleted and resuspended in TE-SDS (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.1% sodium dodecyl sulfate). After chloroform-phenol extraction, the RNA was precipitated with ethanol in the presence of 20 μg of tRNA carrier and kept at −20°C.
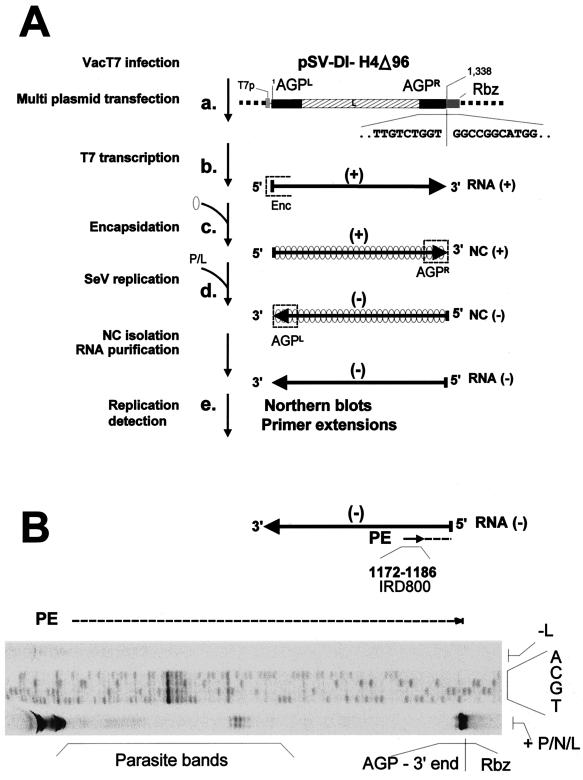
FIG. 2.
Schematic representation of the minireplicon replication system and primer extension analysis. (A) (a) A549 cells are infected with vTF7-3 and transfected with plasmids expressing the SeV minireplicon RNA and the N, P, and L proteins as described in Materials and Methods. The sequence of the minireplicon 3′ end is given along with that of the adjacent ribozyme (Rbz) on the plasmid to set the expected size of the primer extension product. (b) The T7 RNA polymerase transcript representing the positive sense minireplicon RNA is produced with the encapsidation nucleation site at the 5′ end (Enc). (c) Encapsidation of the T7 transcript by the N proteins produces a positive-strand minigenome containing AGPR, the promoter first recognized by the SeV RNAp (P/L), at its 3′ end. (d) SeV RNAp copies the plus-strand minigenome to yield the minus-strand minigenome, which can in turn serve as a template for minigenome amplification (data not shown). The replicated minireplicon RNAs are then purified as nucleocapsids. (e) The encapsidated minus-strand RNAs (specific for active replication) are finally detected with a plus-strand ribo- or oligoprobe in Northern blots or primer extensions, respectively. (B) Primer extension (PE) of the replicated RNA. The IRD800 primer used for the extension and the sequencing is indicated. A digitalized image of primer extension is presented. Lane +P/N/L, complete replication reaction. The strong signal at the AGP 3′ end, corresponds to the extension of a product with the expected size, as indicated by the plasmid sequence shown (see the expected sequence in panel A). The parasite bands correspond to reverse transcription early stops caused by the arrest at hybrid structures resulting from 3′–5′ end complementarity of the RNA. These bands can be minimized by RNA fragmentation (see Materials and Methods). Moreover, they are not observed when 3′–5′ end self-complementation is not possible, as in constructs with AGP at the 5′ end and GP at the 3′ end (see for example constructs #19, #22, and #23 [Fig. 7]). Lane −L, negative control, primer extension run from a replication assay where the pGem-L plasmid was omitted. Primer extensions were performed with RNA purified from transfection-replication assays involving 0.5 × 107 cells.
Analysis of encapsidated RNA.
The replicated RNA was characterized by Northern blotting and/or primer extension. Northern blotting was performed as described previously using an RNA probe of the same polarity (5′ ex-probe) (38) as the T7 defective interfering (DI) RNA transcript (plus strand) to detect the minus-strand RNA resulting from replication (4, 59). For primer extension, sodium carbonate-treated RNA (see “RNA fragmentation” below) was resuspended in 12 μl of TE containing 1 pmol of IRD800 oligonucleotide (positions 1172 to 1186) (see Fig. 2B). After being boiled for 1 min, the mixture was cooled on ice for 3 min before addition of 4 μl of 0.8 M NaCl. After annealing for 5 min at 42°C, 34 μl of primer extension mix (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, 5 mM dithiothreitol [DTT], 0.5 mM deoxynucleoside triphosphates [dNTPs], 12.5 U of RNasin [Amersham-Promega], 4 U of Moloney murine leukemia virus [M-MLV] reverse transcriptase [RTase] [Promega]) was added, and the reaction mixture was further incubated for 1 h at 42°C. The reaction was stopped by addition of 20 μg of tRNA and ethanol precipitation. For analysis, the product of the reaction was resuspended in 2 μl of H2O and 1 μl of stop dye (90% formamide, 2 mM EDTA, xylene cyanol, and bromophenol blue) and was then boiled for 1 min. A 1.5-μl sample was analyzed on a LICOR DNA 4000 automated sequencing apparatus according to the manufacturer's instructions, along with a sequence obtained from plasmids expressing the minireplicon RNA and amplified with the same primer. The replicated RNA obtained from amounts of transfected cells ranging from 5 × 106 to 4 × 107 was treated in the same way. The use of different cell equivalent amounts of RNA is required to compensate for differences in replication efficiencies exhibited by modified templates (see Results and figure legends). In that respect, the primer extension signal was found not to be totally proportional to the input RNA, due to built-in properties of the automated sequencer, as well as to different degrees of potential intramolecular hybridization hindering the primer extension reactions, despite attempts to fragment the RNA (see below). For these reasons, the results can be interpreted only semiquantitatively.
RNA fragmentation.
To minimize intramolecular hybridization due to the inverse complementary sequences of AGPL and AGPR, purified nucleocapsid RNAs were fragmented as follows (7). After resuspension in TE and preheating at 50°C, an identical volume of preheated 100 mM Na2Co3 was added and incubation at 50°C was continued for 30 min. The reaction was stopped by lowering the pH by addition of 100 mM (final concentration) Tris-HCl (pH 7.5), followed by ethanol precipitation. The period of incubation was experimentally set so as to find the peak of primer extension efficiency (before extensive RNA degradation took place [data not shown]).
Limited primer extension.
Following replication, the nucleocapsid RNAs were amplified by reverse transcription-PCR (RT-PCR) with primers (20 oligomers) flanking the Swap editing cassette (see above). One-tenth of a classical reverse transcription reaction mixture (M-MLV RTase [Promega]), using one of the primers, was amplified using 20 pmol of each primer in 30 cycles of a standard 100-μl PCR using 2.5 U of Taq polymerase (Gibco) in an MWG-Biotech Primus apparatus. The purified PCR product (1.5% agarose gel) was recovered by elution from the gel, followed by ethanol precipitation in the presence of 20 μg of tRNA. For limited primer extension, the DNA fragment was resuspended in 12 μl of TE containing 1 pmol of an IRD800 oligonucleotide (5′-CTAGATACGGCAGTTT-3′) complementary to the 3′ side of the Swap editing cassette (italicized nucleotides in sequence under “Sequence and plasmids” above) so that the extension through the run of six A's of the editing site (boldfaced nucleotides in sequence under “Sequence and plasmids” above) was blocked by the presence of ddTTP (for details see reference 23). After boiling for 1 min, the mixture was cooled on ice for 3 min before addition of 4 μl of 0.8 M NaCl. After annealing for 5 min at 42°C, 34 μl of the limited primer extension mix (as described under “Analysis of the encapsidated RNA” above except for the presence of 0.5 mM ddTTP in place of the dTTP) was added and the reaction mixture was further incubated for 10 min at 42°C. Then the four dNTPs (0.5 mM each) were added, and incubation was prolonged for 10 more min at 42°C to chase the stalled complexes. The reaction was finally stopped by addition of 20 μg of tRNA followed by ethanol precipitation. For analysis, the products were resuspended in 2 μl of H2O plus 1 μl of stop dye (see above) and boiled for 1 min before being loaded on a 12.5% agarose sequencing gel. Similar extensions using plasmids encoding the RNA templates were performed as controls.
RESULTS
The replication system.
The replication system used is schematically represented in Fig. 2A (see Materials and Methods for details). A few points are noteworthy. All the minireplicons were derived from the copy-back parent DI-H4Δ96, which contains the same AGP promoter at the 3′ ends of the minus-strand (AGPL) and the plus-strand (AGPR) RNAs (Fig. 2Aa). The T7 RNA transcript is of positive polarity (Fig. 2Ab), so that the first promoter recognized by SeV RNA polymerase (SeV RNAp) is AGPR (Fig. 2Ac). Only AGPR, then, or, more generally, the context of the plus-strand RNA 3′ end was modified in order to investigate its ability to prime replication (Fig. 2Ad). In the end, the replicated minus-strand RNA was detected (Fig. 2Ae) by Northern blotting for quantitative recording of replication or by primer extension for precise mapping of the replicated RNA 5′ end.
The rule of six and the C proteins.
Paramyxovirus P genes are remarkable for the complexity of their genetic information. Apart from producing the P protein, an essential component of the P/L viral RNA polymerase complex, this gene contains additional overlapping open reading frames (ORF). In SeV, these ORF encode, among others, a set of four C proteins, with staggered N termini but a common C terminus (reviewed in reference 34). Expression of this set of C proteins during replication of SeV minireplicons imposes greater selectivity on the SeV RNAp function (57): in general, the tolerance of the SeV RNAp for conditions that do not precisely fulfil those of the wild-type template is higher in the absence than in the presence of the C proteins. Figure 3 illustrates this phenomenon. As previously assessed, the presence or absence of C had no effect on the efficiency of replication of a copy-back minireplicon which complied with the rule of six (compare Fig. 3B, lanes #1 and #01, +C and −C conditions). In contrast, replication of constructs #2 and #4, of 6n + 2 (result of −4 deletion) and 6n + 3 nucleotides, respectively, was highly affected. Almost undetected under +C conditions (∼50- to 100-fold lower than that of the parent template, #1), their replication was favored in the absence of C (Fig. 3B, lanes #2 and #4, +C and −C). An engineered editing site (Swap), responsible for a 1- to 4-guanine insertion at the canonical SeV A6G3 editing sequence (23), was then introduced into constructs #2 and #4 to generate derivatives #3 and #5 (Fig. 3A; see also Fig. 1C and Materials and Methods). Upon replication, at least partial genome length correction can occur at the editing site (24, 57) to generate RNAs with 6n + 0 nucleotides. Depending on the frequency of +4 or +3 nucleotide addition, the replication products of constructs #3 and #5 were expected to replicate better than their corresponding parents, since the nucleotide length was potentially corrected. Indeed they did so (Fig. 3B; compare lanes #2 and #3 and lanes #4 and #5, especially under −C conditions) (57). To better demonstrate genome length correction, the region spanning the editing site of the RNAs resulting from #3 and #5 replication was amplified by RT-PCR (see the legend to Fig. 3C; also Materials and Methods), and a limited primer extension, across the editing site sequence (template strand, 3′-GGGAAAAAA-5′), was performed in the presence of ddTTP. G insertion was then monitored by the presence of a product extending past the G3 limit (for details, see Materials and Methods; also reference 32). Figure 3C shows that during construct #5 replication, in which a +3 insertion was required to reach 6n + 0, insertions of 1, 2, and 3 nucleotides took place at about equal frequency. In construct #3 (of 6n + 2 nucleotides), a +4 insertion was observed, along with a +1 insertion. Note that, as expected, no extension past the last G was observed when the plasmid expressing construct #5 was used as the template (lane #5p). These results show that RNAs with corrected lengths had been amplified during replication, although not exclusively.
The conclusions of this experiment were twofold. First, the experiment demonstrated that although the SeV RNAp was less selective in the absence than in the presence of C, the rule of six still applied, since, when templates with nucleotide lengths that were not multiples of 6 replicated, they did so less efficiently than those whose lengths were multiples of 6. The absence of C could thus be seen as a condition under which the spectrum of observation was widened, so that replication events that would not be detected in the presence of C could now be observed. Such conditions were subsequently used with that understanding. Second, this experiment showed that the Swap editing site was effective for genome length correction, a property that was exploited later.
The 3′-OH congruence of the template promoter is not critical for replication.
To discriminate between the importance of the 3′-OH congruence and the phase context for the application of the rule of six, it was necessary to separate the two properties. This was achieved in a first step by inserting sequence extensions at the minireplicon extremity, further than the AGPR sequence. Depending on whether the extra sequence was 6n + 0 nucleotides long, the 3′-OH congruence of the nucleocapsid was or was not conserved, under conditions where the N phase context of AGPR remained unaltered (see Fig. 4). To monitor replication of constructs in which the 3′-OH no longer corresponded with AGPR, the precise 5′ end of the replicated RNA had to be identified. Primer extensions were therefore performed using a primer of positive polarity directing the reverse extension to the end of the replicated RNA. The sequencing of the plasmid carrying the minireplicon was done in parallel with the same primer, to precisely localize the size of the extended product. Figure 2B illustrates the essential features of the primer extension and shows the analysis of the products obtained in a replication assay of SV-DI-H4Δ96. Under conditions of positive replication (+P/N/L), a main product was seen corresponding to the extension ending at the right end of AGPR. This product was not seen when L was omitted. The phenomenon of parasite bands, whose presence depended on a positive replication assay, refers to extension products blocked by panhandle RNA structures due to inverted complementary sequences at both RNA extremities (AGPL and AGPR; see Materials and Methods). Constructs with extra 3′-OH sequences were therefore analyzed by this method, which proved very reproducible but had the drawback of being only semiquantitative (see Materials and Methods). Figure 4A shows, to the left, a schematic representation of the constructs, with the light shaded boxes representing the unrelated extra sequences (see Materials and Methods for details). To the right, the 3′-OH N phase context is portrayed (see the legend). Constructs #6 and #8, in contrast to constructs #7 and #9, each contained an extra sequence that was 6n + 0 nucleotides long and therefore harbored a correct 3′-OH congruence at the end of the template nucleocapsid. Replication of these constructs started at AGPR regardless (Fig. 4B). Although there appeared to be a penalty in replication for the 3′-OH extensions, a penalty proportional to the length of the extensions (compare the replication obtained with a control RNA [construct #1], present in an eightfold-lower RNA amount, with that of the other four constructs, and compare constructs #6 and #7 with constructs #8 and #9), whether the 3′-OH congruence was kept or not did not seem to matter (compare replication of constructs #6 and #8 with that of their corresponding partners #7 and #9). From this experiment we would conclude that 3′-OH congruence did not represent a pertinent signal for the SeV RNAp. It is noteworthy that in the four constructs with extensions, AGPR is placed internally to the end of the template, and that promoter recognition was not affected by this configuration. With this unexpected flexibility of the replication system (this is, for example, not allowed for VSV minireplicon replication [47]), it became possible to analyze constructs in which the promoter phase context was altered while 3′-OH congruence was maintained.
In Fig. 5A, the phase context of such constructs is schematized, either as such or after a putative genome size correction carried out by an inserted editing site, in case the SeV RNAp starts at an out-of-phase AGPR (constructs #3 and #12). The analysis of constructs #01, #2, and #3 was a repetition of the experiment for which results are shown in Fig. 3. It served to illustrate the same results obtained, this time, by the primer extension method. The two constructs #11 and #13, having, respectively, 10 and 100 nucleotides of extra sequences, also contained an internal −4 deletion, which changed the promoter phase context. The templates, however, harbored a precise congruence at their 3′-OH end, due to the number of nucleotides in the extra sequences (6n + 4). Both constructs replicated at the limit of detection or below, potentially indicating that an out-of-phase AGPR was not functional in these constructs. It is, however, important to remember that if AGPR had been used, the replicated RNA would have been 6n + 2 nucleotides long and could, therefore, have been hampered in replication. For this reason, the Swap editing site, which could correct the genome length in case AGPR was used, was introduced (constructs #12 and #14). Despite this potential genome size correction, replication was still not detected (Fig. 5B, lanes #12 and #14), arguing that replication had not taken place at the out-of-phase AGPR.
Introducing double promoters.
Adding unrelated extra sequences allowed comparison of the in- and out-of-phase configurations of the template 3′-OH end. However, this approach itself introduced a penalty for replication (see comments on Fig. 4B, corresponding Northern blots [data not shown]). Unrelated extra sequences were then replaced by a bona fide promoter sequence added in tandem (Fig. 6). GP, containing the 96 nucleotides required, was added to the right of AGPR (Fig. 6A, #15). Interestingly, construct #15 led to the replication of two populations of RNA (Fig. 6B, lane #15), a minor one starting at GP and a major one starting at AGPR. The two promoters appeared to act independently, since a nucleotide substitution (C91G) known to abolish promoter activity (58) on one of the promoters did not kill the activity of the other (construct #16 or #17). Even when the killing substitution (C4U, found in this study to impair promoter activity [data not shown]) was very close to the GP 3′-OH end (construct #18), the internal AGPR activity was still present. Finally, the insertion of 6 nucleotides between the two promoters (construct #19) led to the efficient utilization of both promoters. Note that all these constructs contained 6n + 0 nucleotides and that the promoters were found in the normal N phase context.
The availability of two active promoters in tandem, separated by nucleotides, allowed repetition of the N-phase-context analysis without the penalty of detrimental irrelevant sequences preceding the internal promoter (Fig. 7). Construct #19 in Fig. 7 was a repetition of construct #19 in Fig. 6, meant to show a functional double promoter. Construct #20, derived from construct #19, had a 3-nucleotide addition at CelII (see Fig. 1 and the legend to Fig. 7) and as such harbored two out-of-phase promoters, combined with the absence of 3′-OH congruence. Not surprisingly, this construct did not lead to detectable replication. Construct #21, on the other hand, had only 3 nucleotides added between AGPR and GP, with the consequence that only GP was out of phase, with no 3′-OH congruence. In this case, only GP was nonfunctional. AGPR, in phase, led to replication, showing again that AGPR activity did not depend on 3′-OH congruence (as well as on GP being active (Fig. 6B, lanes #16 and #18). Combining the 3-nucleotide insertion at CelII, which put AGPR out of phase, with the insertion of 3 nucleotides between AGPR and GP, which allowed GP to stay in phase, created a template where the out-of-phase promoter (AGPR) was preceded by a functional promoter (construct #22). Construct #22 showed no evidence of AGPR activity, and the only replicated RNA started at GP. The introduction of the Swap editing sequence, to potentially correct the phase context of AGPR, still did not lead to detectable replication (construct #23), arguing that no initiation had taken place at the out-of-phase AGPR. Note that this lack of replication at AGPR was concomitant with efficient initiation at GP.
DISCUSSION
In conclusion, in cases where the N phase context of the promoter was conserved, the 3′-OH congruence of the template was not critical for replication. On the other hand, when the N phase context was changed so as to obliterate replication, the 3′-OH congruence could not restore replication. Therefore, it appears that the SeV RNAp is likely to be sensitive to the nucleotide N phase context. The observance of the rule of six, then, must depend on the recognition of certain nucleotides properly positioned, in the promoter region, with regard to the N interaction points (pockets).
The first 12 nucleotides of the promoter, conserved in GP and AGP, are obvious candidates for recognition in the N phase context, although it is not yet clear whether every nucleotide has a significant role. A nucleotide addition at position 13, shifting the phase of nucleotides 1 to 12 by 1, was shown to abolish expression of a reporter gene, possibly by preventing replication of the minireplicon (22). Other phase shifts were accompanied by a nucleotide deletion, making the interpretation of the results difficult. For VSV and RSV (48; reference 62 and references therein), detailed studies have come to the conclusion that not all the nucleotides in the promoter sequence are relevant for replication. Whether these results apply to SeV (and the Paramyxoviridae family) is open to question, since RSV and VSV do not conform to the rule of six (or to the rule of any number [52]), a fact that may indicate that the nucleotides are not seen in the context of N (see below). This assertion, however, is counterbalanced by the finding that the chemical probe reactivities of the bases in the VSV nucleocapsid are not uniform. Some are protected and others are hyperreactive, implying a discriminate association of certain nucleotides with N (29).
The C residues in the repeated motif C1NNNNN (or the NNNNC5G6 residues of the corresponding Rubulavirus SV5 motif [42]), on the other hand, are likely to constitute such signals. Indeed, the appearance of these motifs three times in the same context speaks for the importance of the phase in their recognition by the viral RNAp (vRNAp). The start-stop transcription sequence motifs, as well as the editing sites (D. Kolakofsky, personal communication), have been proposed to represent other examples of such signals, on the basis of their nonrandom positioning with regard to the phase context (33). Recently, a recombinant measles virus (a Paramyxovirus of the Morbillivirus genus) series with modifications resulting in shifting of these regions into all possible phases exhibited an abnormal pattern of readthrough at the M/F and F/H gene boundaries (1). Similarly, all mutants showed slightly altered P transcript editing behavior, suggesting that the nucleotides participating in these signals might be recognized differently depending on the N phase context. The difference between the editing patterns of constructs #5 and #3 (no +2 or +3 addition in #3 [Fig. 3C]) may also add to the argument, since the Swap editing sequence is not in a similar N phase context in the two constructs (N6 and N3 for constructs #5 and #3, respectively).
In the absence of precise structural information on the N-RNA complex, one is reduced to mere speculations as to how the signal recognition operates. The nucleotide signal could be properly accessible to the vRNAp only when in the proper N pocket. Alternatively, the nucleotide could induce a conformational change in the N protein only when in a defined pocket. This second possibility is particularly attractive for the (C1NNNNN)3 motif, since the C's in phase context N1 appear to signal independently of the primary sequence environment (N2 to N6). If it is not repeated, the signal must obviously be formed by more than one nucleotide, since the probability is increased that any nucleotide is going to be presented many times in each N phase context. If the signal is made by a local conformational change in the nucleocapsid, this change should propagate through more than one N subunit. The picture could be more complicated, considering that the signal recognition mechanism might have to be flexible, if, for instance, it is confirmed that the N phase context is relevant for transcription. Indeed, the transcriptional start-stop signals remain ignored during replication. The vRNAp could be made insensitive to the signals, subsequent to a structural modification (phosphorylation or conformational change induced by encapsidation of the nascent RNA chain or by interaction with a viral cofactor, etc.). Alternatively, the signal could be masked to the vRNAp by a modification of the N-RNA complex itself, although a biochemical or conformational difference between the transcription and replication templates has not been considered so far. At early times of infection, the nucleocapsids could conform better to transcription. Later, following interaction with a viral partner that accumulates with time, or following a slow chemical modification, they could conform better to replication. The C proteins, shown to modulate the selectivity of the vRNAp for the replication signals, may in fact act by modifying the template. Indeed, the different C protein-to-template ratios required to inhibit replication at AGP or at GP—inhibition seen here as the price of selectivity—argue for a stoichiometric rather than an enzymatic role of the C's (57). It is noteworthy in this context that the polR mutants of VSV, producing more leader-N junction readthrough transcripts, harbor a mutation in N (6). This is compatible with the idea that the modification of the template could be responsible for the lower stringency with which the signal is observed. Finally, nucleocapsid subpopulations with different sensitivities to RNase or harboring a cleaved N have been reported (5, 10, 39, 40).
The ability to add a sequence extension at the 3′-OH end of the T7 RNA template without losing the ability to replicate is surprising. Similar extensions are not tolerated for VSV and RSV, in contrast to 5′-end extensions (guanosine residues generally added to improve T7 RNA polymerase activity) (9, 47, 52; D. Garcin, unpublished data). Whether the discrepancy between SeV and VSV or RSV reflects a significant difference in the template structure or in the template recognition mechanism is open to question. The vRNAp could recognize the nucleocapsid 3′-OH end as such, regardless of the RNA sequence, or regardless of 3′-OH congruence. After binding, the vRNAp would gain access to the nucleotides and scan the template in search of the promoter sequence to start RNA synthesis. Evidence for forward, and even backward, scanning by negative-stranded RNA virus polymerases has been provided (18, 56). Alternatively, the vRNAp could bind internally to the promoter region, a mechanism that implies a vRNAp capable of nucleotide sequence discrimination before binding. We propose that, when the rule of six applies, the phase context provides the vRNAp with all the information needed to bind the template at the promoter and to initiate replication, without the need for 3′-OH-end recognition. The 3′-OH-end recognition would then be the prerogative of the templates (VSV and RSV) lacking the phase context conditions and thus requiring the information given by the 3′-OH end. The question of access to internal replication promoters, although it is academic considering that the replication promoters are naturally present at the nucleocapsid 3′-OH end, may still be of interest for understanding of the general manner in which the vRNAp operates. The achievement of the present work, namely, the use of double promoter constructs, may lead to the design of more-appropriate experiments to contribute to the understanding of transcription initiation or of the mechanism of defective interfering RNA generation.
The conditions used to perform these experiments, i.e., the absence of the C proteins, may raise concerns about the validity of the results, since the C proteins are known to influence SeV RNAp activity by increasing selectivity (57). However, even if the absence of C tones down the stringency of the rule of six, the rule still applies (see Fig. 3B). In the absence of C, observations not possible in the presence of C, where constructs of 6n + x (x ≠ 0) nucleotides do not replicate to detectable levels, can now be made. Extending sequences past the promoter (Fig. 4) and adding two promoters in tandem (Fig. 7) represent other artificial conditions. These clearly impose more penalties on the replicating system, so that a distinction can now be perceived between 3′-OH congruence and a change in the N phase context. This distinction was, for instance, not possible in Fig. 3, where the constructs still replicated efficiently. Thus, the application of both sets of conditions appears to widen the window of observation so that more recognition steps can be identified.
ACKNOWLEDGMENTS
We are indebted to Caroline Tapparel, who, before leaving at the end of her Ph.D. studies, introduced the double promoter constructs, and to Dominique Garcin and Daniel Kolakofsky for critical discussions and critical reading of the manuscript. D.V. is very grateful to Geneviève Mottet-Osman for constant support and technical expertise and to Nathalie Fouillot-Coriou for the demonstration of scientific rigor.
L.R. is the recipient of a grant from the Swiss National Foundation for Scientific Research.
REFERENCES
- 1.Billeter M A, et al. Reverse genetics with measles virus to define viral protein and genome function. In: Mahy B W J, Kolakofsky D, editors. NSV: 11th International Conference on Negative Strand Viruses. 2000. p. 131. [Google Scholar]
- 1a.Boyer J-C, Haenni A-L. Infectious transcripts and cDNA clones of RNA viruses. Virology. 1994;198:415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- 2.Bridgen A, Elliott R M. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci USA. 1996;93:15400–15404. doi: 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calain P, Roux L. Functional characterisation of the genomic and antigenomic promoter of the Sendai virus. Virology. 1995;211:163–173. doi: 10.1006/viro.1995.1464. [DOI] [PubMed] [Google Scholar]
- 5.Chanda P K, Banerjee A K. Two distinct populations of vesicular stomatitis virus ribonucleoprotein cores with differential sensitivities to micrococcal nuclease. Biochem Biophys Res Commun. 1979;91:1337–1345. doi: 10.1016/0006-291x(79)91213-0. [DOI] [PubMed] [Google Scholar]
- 6.Chuang J L, Jackson R L, Perrault J. Isolation and characterization of vesicular stomatitis virus PolR revertants: polymerase readthrough of the leader-N gene junction is linked to an ATP-dependent function. Virology. 1997;229:57–67. doi: 10.1006/viro.1996.8418. [DOI] [PubMed] [Google Scholar]
- 7.Coffin J M, Billeter M A. A physical map of the Rous sarcoma virus genome. J Mol Biol. 1976;100:293–318. doi: 10.1016/s0022-2836(76)80065-4. [DOI] [PubMed] [Google Scholar]
- 8.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins P L, Mink M A, Stec D S. Rescue of synthetic analogs of respiratory syncytial genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663–9667. doi: 10.1073/pnas.88.21.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compans R W, Holmes K V, Dales S, Choppin P W. An electron microscopic study of moderate and virulent virus-cell interactions of the parainfluenza virus SV5. Virology. 1966;30:411–426. doi: 10.1016/0042-6822(66)90119-x. [DOI] [PubMed] [Google Scholar]
- 11.Conzelmann K-K. Genetic manipulation of non-segmented negative-strand RNA viruses. J Gen Virol. 1996;77:381–389. doi: 10.1099/0022-1317-77-3-381. [DOI] [PubMed] [Google Scholar]
- 12.Conzelmann K-K, Schnell M J. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol. 1994;68:713–719. doi: 10.1128/jvi.68.2.713-719.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curran J, Boeck R, Kolakofsky D. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 1991;10:3079–3085. doi: 10.1002/j.1460-2075.1991.tb07860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curran J, Marq J-B, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 15.Durbin A P, Hall S L, Siew J W, Whitehead S S, Collins P L, Murphy B R. Recovery of infectious human parainfluenza virus type 3 from cDNA. Virology. 1997;235:323–332. doi: 10.1006/viro.1997.8697. [DOI] [PubMed] [Google Scholar]
- 16.Durbin A P, Siew J W, Murphy B R, Collins P L. Minimum protein requirements for transcription and RNA replication of a minigenome of human parainfluenza virus type 3 and evaluation of the rule of six. Virology. 1997;234:74–83. doi: 10.1006/viro.1997.8633. [DOI] [PubMed] [Google Scholar]
- 17.Egelman E H, Wu S-S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fearns R, Collins P L. Model for polymerase access to the overlapped L gene of respiratory syncytial virus. J Virol. 1999;73:388–397. doi: 10.1128/jvi.73.1.388-397.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finch J T, Gibbs A J. Observations on the structure of the nucleocapsids of some paramyxoviruses. J Gen Virol. 1970;6:141–150. doi: 10.1099/0022-1317-6-1-141. [DOI] [PubMed] [Google Scholar]
- 20.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harty R N, Palese P. Mutations within noncoding terminal sequences of model RNAs of Sendai virus: influence on reporter gene expression. J Virol. 1995;69:5128–5131. doi: 10.1128/jvi.69.8.5128-5131.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hausmann S, Garcin D, Morel A S, Kolakofsky D. Two nucleotides immediately upstream of the essential A6G3 slippery sequence modulate the pattern of G insertions during Sendai virus mRNA editing. J Virol. 1999;73:343–351. doi: 10.1128/jvi.73.1.343-351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hausmann S, Jacques J P, Kolakofsky D. Paramyxovirus RNA editing and the requirement for hexamer genome length. RNA. 1996;2:1033–1045. [PMC free article] [PubMed] [Google Scholar]
- 25.He B, Paterson R G, Ward C D, Lamb R A. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 26.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman M A, Banerjee A K. An infectious clone of human parainfluenza virus type 3. J Virol. 1997;71:4272–4277. doi: 10.1128/jvi.71.6.4272-4277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosaka Y, Shimizu K. Lengths of the nucleocapsids of Newcastle disease and mumps viruses. J Mol Biol. 1968;35:369–373. doi: 10.1016/s0022-2836(68)80031-2. [DOI] [PubMed] [Google Scholar]
- 29.Iseni F, Baudin F, Blondel D, Ruigrok R W. Structure of the RNA inside the vesicular stomatitis virus nucleocapsid. RNA. 2000;6:270–281. doi: 10.1017/s135583820099109x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kast W M, Roux L, Curran J, Blom H J J, Voordouw A C, Meloen R H, Kolakofsky D, Melief C J M. Protection against lethal Sendai virus infection by in vivo priming of virus-specific cytotoxic T lymphocytes with a free synthetic peptide. Proc Natl Acad Sci USA. 1991;88:2283–2287. doi: 10.1073/pnas.88.6.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells. 1996;1:569–579. doi: 10.1046/j.1365-2443.1996.d01-261.x. [DOI] [PubMed] [Google Scholar]
- 32.Kolakofsky D, Curran J, Pelet T, Jacques J P. Paramyxovirus P gene mRNA editing. In: Benne R, editor. RNA editing. London, England: Ellis Horwood; 1993. pp. 105–123. [Google Scholar]
- 33.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolakofsky D, Vidal S, Curran J. Paramyxovirus RNA synthesis and P gene expression. In: Kingsbury D W, editor. The Paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 215–233. [Google Scholar]
- 35.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1177–1204. [Google Scholar]
- 36.Lawson N D, Stillman E A, Whitt M A, Rose J K. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynch S, Kolakofsky D. The ends of the RNA within Sendai virus defective interfering nucleocapsids are not free. J Virol. 1978;28:584–589. doi: 10.1128/jvi.28.2.584-589.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mottet G, Roux L. Budding efficiency of Sendai virus nucleocapsids: influence of size and ends of the RNA. Virus Res. 1989;14:175–188. doi: 10.1016/0168-1702(89)90037-3. [DOI] [PubMed] [Google Scholar]
- 39.Mountcastle W E, Compans R W, Caliguiri L A, Choppin P W. Nucleocapsid protein subunits of simian virus 5, Newcastle disease virus, and Sendai virus. J Virol. 1970;6:677–684. doi: 10.1128/jvi.6.5.677-684.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mountcastle W E, Compans R W, Lackland H, Choppin P W. Proteolytic cleavage of subunits of the nucleocapsid of the paramyxovirus simian virus 5. J Virol. 1974;14:1253–1261. doi: 10.1128/jvi.14.5.1253-1261.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy S K, Parks G D. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the Paramyxovirus simian virus 5. Virology. 1997;232:145–157. doi: 10.1006/viro.1997.8530. [DOI] [PubMed] [Google Scholar]
- 42.Murphy S K, Parks G D. RNA replication for the paramyxovirus simian virus 5 requires an internal repeated (CGNNNN) sequence motif. J Virol. 1999;73:805–809. doi: 10.1128/jvi.73.1.805-809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagai Y, Kato A. Paramyxovirus reverse genetics is coming of age. Microbiol Immunol. 1999;43:613–624. doi: 10.1111/j.1348-0421.1999.tb02448.x. [DOI] [PubMed] [Google Scholar]
- 44.Neubert W J, Eckerskorn C, Homann H E. Sendai virus NP gene codes for a 524-amino-acid NP protein. Virus Genes. 1991;5:25–32. doi: 10.1007/BF00571728. [DOI] [PubMed] [Google Scholar]
- 45.Palese P, Zheng H, Engelhardt O G, Pleschka S, Garcia-Sastre A. Negative-strand RNA viruses: genetic engineering and applications. Proc Natl Acad Sci USA. 1996;93:11354–11358. doi: 10.1073/pnas.93.21.11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pattnaik A K, Ball L A, LeGrone A, Wertz G W. The termini of VSV DI particles are sufficient to signal encapsidation, replication and budding to generate infectious particles. Virology. 1995;206:760–764. doi: 10.1016/S0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 48.Peeples M E, Collins P L. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J Virol. 2000;74:146–155. doi: 10.1128/jvi.74.1.146-155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 50.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Christiansen G, Billeter M A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts A, Rose J K. Recovery of negative-strand RNA viruses from plasmid DNAs: a positive approach revitalizes a negative field. Virology. 1998;247:1–6. doi: 10.1006/viro.1998.9250. [DOI] [PubMed] [Google Scholar]
- 52.Samal S K, Collins P L. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schnell M J, Mebatsion T, Conzelmann K-K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shioda T, Hidaka Y, Kanda T, Shibuta H, Nomoto A, Iwasaki K. Sequence of 3,687 nucleotides from the 3′ end of Sendai virus genome RNA and the predicted amino acid sequences of viral NP, P and C proteins. Nucleic Acids Res. 1983;11:7317–7330. doi: 10.1093/nar/11.21.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shioda T, Iwasaki K, Shibuta H. Determination of the complete nucleotide sequence of the Sendai virus genome RNA and the predicted amino acid sequences of the F, HN, and L proteins. Nucleic Acids Res. 1986;14:1545–1563. doi: 10.1093/nar/14.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stillman E A, Whitt M A. The length and sequence composition of vesicular stomatitis virus intergenic regions affect mRNA levels and the site of transcript initiation. J Virol. 1998;72:5565–5572. doi: 10.1128/jvi.72.7.5565-5572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tapparel C, Roux L. The efficiency of Sendai virus genome replication: the importance of the RNA primary sequence independent of terminal complementarity. Virology. 1996;225:163–171. doi: 10.1006/viro.1996.0584. [DOI] [PubMed] [Google Scholar]
- 60.Wertz G W, Whelan S P J, LeGrone A, Ball L A. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc Natl Acad Sci USA. 1994;91:8587–8591. doi: 10.1073/pnas.91.18.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whelan S P J, Ball L A, Barr J N, Wertz G W. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whelan S P J, Wertz G W. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J Virol. 1999;73:297–306. doi: 10.1128/jvi.73.1.297-306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]