Abstract
The heterodimeric sweet taste receptor, TAS1R2/1R3, is a class C G protein-coupled receptor (GPCR) that couples to gustducin (Gt), a G protein (GP) specifically involved in taste processing. This makes TAS1R2/1R3 a possible target for newly developing low caloric ligands that taste sweet to address obesity and diabetes. The activation of TAS1R2/1R3 involves the insertion of the GαP C-terminus of the GP into the GPCR in response to ligand binding. However, it is not known for sure whether the GP inserts into the TAS1R2 or TAS1R3 intracellular region of this GPCR dimer. Moreover, TAS1R2/1R3 can also connect to other GPs, such as Gs, Gi1, Gt3, Go, Gq, and G12. These GPs have different C-termini that may modify GPCR signaling. To understand the possible GP dependence of sweet perception, we use molecular dynamic (MD) simulations to examine the coupling of various GαP C20 termini to TAS1R2/1R3 for various steviol glycoside ligands and an artificial sweetener. Since the C20 could interact with the transmembrane domain (TMD) of either TAS1R2 (TMD2) or TAS1R3 (TMD3), we consider both cases. Without any sweetener, we find that the apo GPCR shows similar Go and Gt selectivities, while all steviol glycoside ligands increase the selectivity of Gt but decrease Go selectivity at TMD2. Interestingly, we find that high sweet rebaudioside M (RebM) and RebD ligands show better interactions of C20 at TMD3 for the Gt protein, but low sweet RebC and hydRebM ligands show better interaction of C20 at TMD2 for the Gt protein. Thus, our MD simulation suggests that TAS1R2/1R3 may couple the GP to either 1R2 or to 1R3 and that it can couple other GPs compared to Gt. This will likely lead to multimodal functions producing multiple patterns of intracellular signaling for sweet taste receptors, depending on the particular sweetener. Directing the GP to one of the other may have beneficial therapeutic outcomes.
Introduction
The three members of the TAS1R taste family form two heteromeric receptors: TAS1R1/1R3 and TAS1R2/1R3. The sweet taste is mediated by TAS1R2/1R3, while the umami taste is mediated by TAS1R1/1R3.1 The sweet taste receptor is considered as a potential novel therapeutic target for treatment of obesity and related metabolic dysfunctions such as diabetes.2 TAS1R2/1R3 belongs to the class C G protein-coupled receptor (GPCR) family, which also includes metabotropic glutamate receptors (mGluRs), γ-aminobutyric acid receptor B (GABABR), calcium-sensing receptors (CaSR), and others. Class C GPCRs are characterized by having a large extracellular Venus flytrap domain (VFD) coupled to a short cysteine-rich domain (CRD) that links the VFD to the seven-helix transmembrane domain (TMD).1 So far, there are 22 approved and marketed drugs that target class C GPCRs on the market.3
We previously carried out molecular dynamics (MD) studies of various stevia ligands to the TAS1R2/1R3 heterodimer to provide predictive atomistic descriptions of the structure, binding, and activation since there is no experimental structure for the human sweet heterodimer.4 This includes rebaudioside M (RebM),5 which has the highest sweetness. We observed that the activation mechanism of the TAS1R2/1R3 heterodimer includes the following four sequential events:
-
1.
A sweet enhancer binds to VFD2 in the dimer to stabilize the closed conformation.
-
2.
The VFD2 in the closed conformation transduces this activation signal to the TM6/TM6 interface between TMD2 and TMD3 via the cysteine-rich domains (CRD) of TAS1R3 (CRD3).
-
3.
Step 2 opens the helix 3–6 intracellular helix on TAS1R3, allowing the GNT3 G protein (GP) to bind.
-
4.
This binding of the GP leads to opening the Gα subunit of the GP to release GDP that leads to GP signaling.
We previously used metadynamics (metaD) to examine diverse sweeteners: sucralose, Ace-K, saccharin, and rebaudioside A (Reb-A),6 to understand the observed synergic effects for all binary combinations, which includes cases with structural similarity (saccharin and Ace-K) and dissimilarity (sucralose and Ace-K, Reb-A and sucralose, and Ace-K).7 Our metaD simulations led to results consistent with the experiment.7 Thus, the metaD results for partial synergic (sucralose and Ace-K) or full synergic (Reb-A and Ace-K and Reb-A and sucralose) cases show more negative binding free-energies than single cases, while the metaD results for the suppression case (saccharin and Ace-K) show less negative free energy than single cases.7
Recently, our docking results found preferred binding sites that differ depending on the ligand modifications of steviol glycosides. Thus, rubusoside (Rubu) binds best at VFD3, while RebB binds best at TMD3, and the others prefer VFD2. This provides an explanation of the mixed data from the radioligand binding experiments (submitted). We also observed GPCR allostery experimentally using the label-free frequency locking optical whispering evanescent resonator (FLOWER) method. The C20 carboxy terminus of the Gα protein can bind to the intracellular region of either the TMD of TAS1R2 (TMD2) or the TMD of TAS1R3 (TMD3), which can alter the GPCR affinity to the high-affinity state for steviol glycosides (submitted).
GPCRs are a diverse family of cell surface receptors that can activate different intracellular signaling pathways through the coupling with a specific GP. The selectivity of GPCR-GP coupling is a complex and dynamic process, depending on numerous factors including the type of GPCR and the specific GP involved.
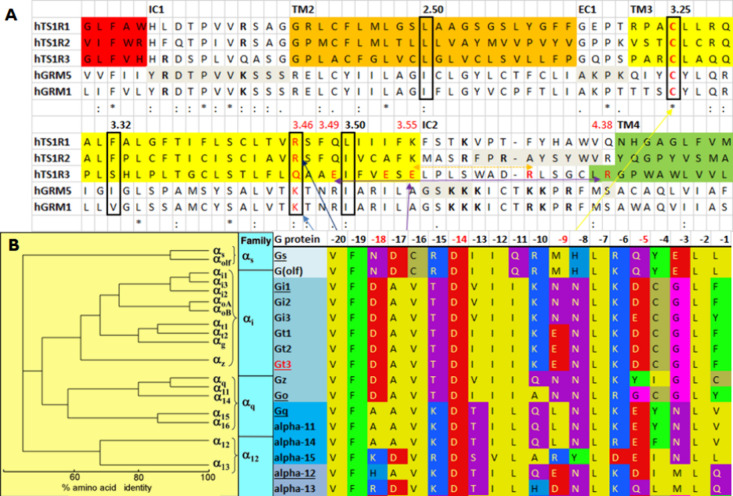
The human genome encodes 16 α-subunits for GP, which are grouped into four subfamilies based on their sequence homology and functional similarity, Gs, Gi/o, Gq/11, and G12/13, as shown in Figure 1.8 Different GPCRs can selectively couple to one or more of these GP families. For example,
-
1.
Gs proteins stimulate adenylate cyclase, leading to an increase in cAMP.
-
2.
Gi/o proteins inhibit adenylate cyclase, leading to a decrease in cAMP.
-
3.
Gq/11 proteins activate phospholipase C, leading to the production of inositol trisphosphate (IP3) and diacylglycerol (DAG).
Figure 1.
(A) Sequence alignment of the taste TAS1R family with metabotropic glutamate receptors (mGluRs). (B) Sequence alignments of all G proteins. Six C20 peptides (Gs, Gi1, Gt3, Go, Gq, and G12) were selected for the study.
This specificity of GPCR-GP coupling plays a crucial role in the functional diversity of these receptors. However, some GPCRs can activate multiple types of GPs depending on the cellular context and conditions.
Sweet taste receptors in the tongue couple to gustducin, a GP specifically involved in taste processing, but they also couple to other GPs.9 Relative to TMD2, TMD3 does not effectively couple to transducin α. Based on the experiments, TMD3 did not significantly activate any of the other Gα subunits tested.9 The TMDs of hT1R1 and hT1R2 catalyzed GDP/GTPγS exchange on Gαi1/gus51, Gαo, and Gαi1.9
In this paper, we report the results of MD simulations to determine GP C20 coupling properties of the sweet taste receptor for multiple C20 peptides at TMD2 or TMD3 with six steviol glycosides (high sweet: RebM, RebD, and isoRebM; middle sweet: Rubu; and low sweet: RebC and hydRebM) and the Ace-K artificial sweetener. To understand C20 GP coupling selectivity, we selected the six C20 peptides in Figure 1 (Gs, Gi1, Gt3, Go, Gq, and G12). To study the ligand-dependent GP coupling of the TAS1R2/1R3 heterodimer, we calculated the binding energy between C20 and either TAS1R2 or TAS1R3 through MD simulations. This study increases our understanding of the mechanisms and factors that determine GP coupling selectivity to the heterodimeric sweet taste receptor, which sets the stage for developing novel therapeutic applications for the treatment of obesity and related metabolic dysfunctions such as diabetes and novel sweet enhancers that can enable lower sugar usage levels while retaining the sweet taste.
Methods
The full structure of the TAS1R2/1R3 heterodimer was predicted using GEnSeMBLE10 for the TMD helixes and homology modeling for the VFD and CRD. The detailed procedures and results are described in our previous paper.4
Generation of the C20 Bound Structure of the TAS1R2/1R3 Heterodimer at TMD2 or TMD3
Various initial positions of GNAT3 C20 were considered based on the experimental structures of GPCR-GP complexes, class C mGlu2R-Gi (PDB ID: 7MTS(11) and 7E9G(12)), mGlu4R-Gi (PDB ID: 7E9H(12)), GABAB2R-Gi (PDB ID: 7EB2),13 and class A OPRM-Gi (PDB ID: 6DDF).14 We also included our predicted structure of the GNAT3-TAS1R3/1R3 homodimer complex.15 After alignment of the GPCR backbone, all residues of the Gα protein (Gt3) except the C-terminal 20 amino acids were deleted followed by minimization of the fixed GPCR after the side chain refinement. The other five C20 peptides (Gs, Gi1, Go, Gq, and G12) were mutated from predicted Gt3.
MD Simulations
The C20 bound structure of the TAS1R2/1R3 heterodimer at either TMD2 or TMD3 was inserted into a POPC lipid bilayer using periodic boundary conditions while including full solvation with water at the physiological salt concentration (0.9% concentration of NaCl, 154 mM). The full calculation included ∼192,000 atoms per periodic cell.
Initially, the disulfide bridges in TAS1R2 were constrained: C59–C102, C359–C363, and C405–C410 in the VFD, C495–C514, C499–C517, C520–C535, and C538–C551 in the CRD, C233–C513 between the VFD and CRD, and C6303.25–C720 between the TMD and EC2. The disulfide bridges in TAS1R3 were also constrained: C62–C103, C351–C370, C373–C375, and C410–C415 in the VFD, C499–C518, C503–C521, C524–C538, and C541–C554 in the CRD, C236–C517 between the VFD and CRD, and C6333.25 and C722 between the TMD and EC2.
The POPC lipid available in VMD was used to insert the protein into a lipid–water box, where lipids within 1 Å of the protein and water within 2 Å of the protein were removed.
For electrostatic calculations, we used the particle mesh Ewald (PME) method.16 The charge of the system was balanced through replacing waters with Na+ and Cl– ions.
After inserting the TAS1R2/1R3 heterodimer, including loops, into the box containing the periodic POPC membrane, water, and ions, we initially fixed the protein and minimized the lipid, water, and ion atoms for 1000 steps. We then equilibrated the NPT dynamics for 500 ps while continuing to keep the protein fixed. This allows the lipid and water to readjust to the protein. Then, we minimized the full system without constraints for 1000 steps and then performed NPT dynamics heating from 20 to 310 K at 1 atm pressure (1.01325 bar, atmospheric pressure at sea level) without constraints. The cutoff distance for noncovalent interactions was set to 12 Å. This predicted structure was then equilibrated at 310 K temperature for 20 ns based on the root-mean-square deviation (RMSD) convergence of protein backbones using the NAMD 2.9 (nanoscale molecular dynamics) program.17 We used the CHARMM36 force field parameters for the protein, the TIP3 model for water,18 and the CHARMM27 force field parameters for the lipids.19
Results and Discussion
The sequence alignment of the taste TAS1R family with mGluR1 and mGluR5 in Figure 1A shows that the TMD2 is more similar than the TMD3 when compared to the cytoplasmic end of TM3 in class C mGluR1 and mGluR5, especially for K/R3.46. K/R3.46 participates in the formation of an ionic lock with E6.30 in class C GPCRs.4 The conserved R/K of 3.46 in class C GPCR is also conserved at TAS1R1 and TAS1R2 but not at TAS1R3. Instead of the positively charged residue at 3.46, TAS1R3 has Q. In addition, E3.49, E3.53, and E3.55 are at the cytoplasmic end of TMD3 of TAS1R3, which are not negatively charged residues in TAS1R1 or 1R2 and mGluRs. The whole TMD sequence identities to TMD2 of TAS1R2/1R3 are 22.13 and 23.03% for mGluR1 and mGluR5, respectively, while the whole TMD sequence identities of TMD3 of TAS1R2/1R3 are 20.69 and 17.29% for mGluR1 and mGluR5, respectively.
The various Cryo-EM structures of GPCR-GP complexes show that class C GPCRs adopt a distinct mode of GP coupling compared to class A, B, and F GPCRs.13 The C-terminal α5 helices for class A, B, and F GPCRs couple through nearly the same intracellular cavity, closer to intracellular loop 3 (ICL3) for class A and B GPCRs or closer to ICL1 for class F GPCRs from the cytoplasmic view, which all reach the same depth into the TMDs of the receptor.13 However, the C-terminal α5 of Gi coupled to class C GPCRs is inserted less deeply. Consequently, the C-terminal end of the α5 of Gαi in the class C GPCR-GP complex does not penetrate into the central cavity between TM3 and TM6 but rather inserts into a cavity located at the periphery, interacting with ICL2. Thus, based on the sequence similarity of the intracellular side of TMD3, which interacts with the C-terminal α5 helices of GP, we expect that when the GP binds to TMD2, it might interact in a noncanonical way.
There are four main families of GPs: Gs, Gi/o, Gq/11, and G12/13, as shown in Figure 1B. We selected six C20 peptides (Gs, Gi1, Gt3, Go, Gq, and G12), with three from the Gi subfamily, to understand the selectivity of C20 to the heterodimeric sweet taste receptor, TAS1R2/1R3. We generated the initial position of GNAT3 (Gt3) C20 using the experimental structures of GPCR-GP complexes, class C mGlu2R-Gi (PDB ID: 7MTS(11) and 7E9G(12)), mGlu4R-Gi (PDB ID: 7E9H(12)) and GABAB2R-Gi (PDB ID: 7EB2)13 in the noncanonical way, and class A OPRM-Gi (PDB ID: 6DDF)14 in the canonical way. We also included our predicted structure of the GNAT3-TAS1R3/1R3 homodimer complex in the canonical way (Figure S1).15
After 20 ns of MD simulation with backbone constraints, the TAS1R2/1R3-C20 complex with GT3 C20 bound at TMD3 (TMD3-C20) shows better interaction energies of Gt3 C20 with TAS1R than the TAS1R2/1R3-C20 complex within C20 bound at TMD2 (TMD2-C20), as shown in Figure S3A. The average MD interaction energy for TMD3-Gt3 C20 is −255.40 kcal/mol and for TMD2-Gt3 C20 is −200.66 kcal/mol. Overall, the RMSD values of the whole protein are similar for both (3.75 Å for TMD2-Gt3 C20 vs 3.77 Å for TMD3-Gt3 C20). However, the RMSD of Gt3 C20 for TMD3-C20 (5.27 Å) is much higher than that of TMD2-C20 (3.20 Å), as shown in Figure S3B,C.
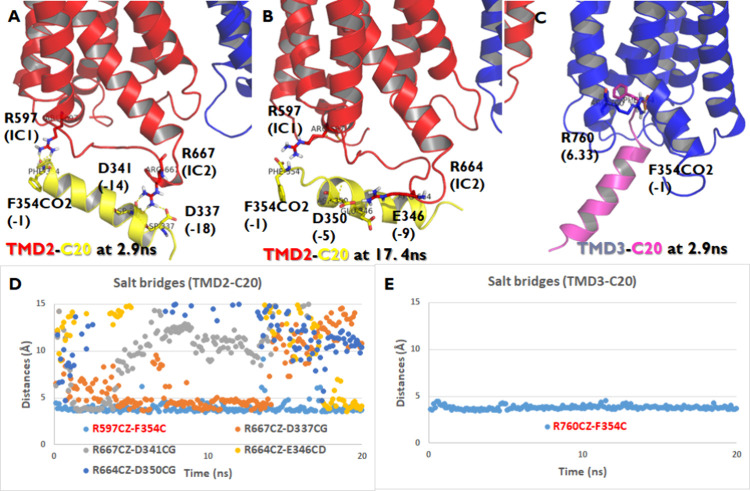
The noncanonical interaction between TMD2 and Gt3 C20 upon GP coupling was maintained through various salt bridges (Figure 2A,B,D): one stable salt bridge between R597 (IC1) and the C-terminal carboxyl, one medium stable salt bridge between R667 (IC2) and D337 (−18), and three weak salt bridges between R667 (IC2) and D341 (−14), between R664 (IC2) and E346 (−9), and between R664 (IC2) and D350 (−5) from 20 ns MD with backbone constraints. A conserved R/K in IC1 of TAS1R2 but not TAS1R3 made a stable salt bridge between R597 (IC1) and the C-terminal carboxyl end during MD. However, the IC2 of 1R2 and 1R3 has fewer positively charged residues than mGluRs (Figure 1A). Figure 2A and Figure 2B show the alternative salt bridges at 2.9 and 17.4 ns, respectively, which have low interaction energies during the MD trajectories. The canonical GP coupling of TMD3-Gt C20 has one stable salt bridge between R760 (6.33) and the C-terminal carboxyl end at 2.9 ns, which leads to the lowest interaction energy during MD (Figure 2C,E).
Figure 2.
Interaction surface of the TAS1R2/TAS1R3-C20 complex when C20 binds at the TMD of TAS1R2 (TMD2-C20) or TAS1R3 (TMD3-C20). (A, B) The salt bridges are in TMD2-C20. (C) Salt bridges in TMD3-C20. (D) Salt bridge trajectory of TMD2-C20: one stable salt bridge between R597 (IC1) and the C-terminal carboxyl end, one medium stable salt bridge between R667 (IC2) and D337 (−18), and three weak salt bridges between R667 (IC2) and D341 (−14), between R664 (IC2) and E346 (−9), and between R664 (IC2) and D350 (−5) from 20 ns MD with backbone constraints. (E) The salt bridge trajectory of TMD3-C20 was with one stable salt bridge between R760 (6.33) and the C-terminal carboxyl end.
To analyze the interactions between the TAS1R heterodimer and various C20 peptides, we computed the average salt bridge distances for TAS1R with six C20 peptides (Gs, Gi1, Gt3, Go, Gq, and G12) using 20 ns MD simulations (Table 1). The TAS1R2/TAS1R3-C20 complex was studied under conditions where C20 binds either to TAS1R2’s transmembrane domain (TMD2-C20) or TAS1R3's transmembrane domain (TMD3-C20). Among these peptides, the Gt C20 peptide forms the most salt bridges (five out of six peptides), whereas the G12 C20 peptide shows the weakest interaction, with only one salt bridge. The other peptides (Go, Gi, Gs, and Gq C20) each form two salt bridges. Specifically, the Go C20 peptide exhibits stable interactions between the side chains of R667CZ (IC2) and D341CG (−14) and between R597CZ (IC1) and the C-terminal end of F354C (−1). Thus, the pattern of salt bridges observed in the TAS1R2/TAS1R3-C20 complex varies depending on the specific GP subtype, highlighting distinct interaction profiles.
Table 1. Average Salt Bridge (SB) Distance (AVE) between TAS1R and Six C20 Peptides (Gs, Gi1, Gt3, Go, Gq, and G12) from 20 ns MD for the TAS1R2/TAS1R3-C20 Complex When C20 Binds at the TMD of TAS1R2 (TMD2-C20) or TAS1R3 (TMD3-C20).
| system | salt bridges between TAS1R and C20 | AVE(0–20 ns) |
|---|---|---|
| TMD3-GtC20 | R760CZ(6.33)-F354C(−1) | 3.81 |
| TMD2-GoC20 | R667CZ(IC2)-D341CG(−14) | 4.30 |
| R597CZ(IC1)-F354C(−1) | 4.02 | |
| TMD2-GtC20 | R597CZ(IC1)-F354C(−1) | 3.91 |
| R664CZ(IC2)-E346CD(−9) | 15.52 | |
| R667CZ(IC2)-D337CG(−18) | 7.21 | |
| R667CZ(IC2)-D341CG(−14) | 11.66 | |
| R664CZ(IC2)-D350CG(−5) | 14.18 | |
| TMD2-GiC20 | R667CZ(IC2)-D337CG(−18) | 6.02 |
| R597CZ(IC1)-F354C(−1) | 7.03 | |
| TMD2-GsC20 | R597CZ(IC1)-L354C(−1) | 12.10 |
| R597CZ(IC1)-E352CD(−3) | 4.56 | |
| TMD2-GqC20 | R667CZ(IC2)-D341CG(−14) | 19.65 |
| R597CZ(IC1)-V354C(−1) | 7.98 | |
| TMD2-G12C20 | R597CZ(IC1)-Q354C(−1) | 7.97 |
Various steviol glycosides [RebM, RebD, isoRebM, rubusoside (Rubu), RebC, and hydRebM] and Ace-K artificial sweetener were tested for C20 selectivity using MD simulations. Without any sweeteners, the apo sweet taste receptor shows similar Go and Gt selectivity. All Gi family members (Go, Gi, and Gt) have much better interaction energy than the other GPs (Gs, Gq, and G12) for apo. Our simulations are consistent with the experimental receptor-Gα subunit selectivity. The reported experiments showed that the TMD of TAS1R2 activated Gαo, Gαi1, and transducing α, three members of the Gαi/o family of Gα subunits.9 Therefore, the TAS1Rs are likely to couple in vivo to multiple members of the Gαi/o family, several of which are expressed in taste cells.9 However, when we add all steviol glycosides at the orthosteric binding site, VFD2, the sweet taste receptors decrease the Go selectivity at TMD2 while increasing the Gt selectivity (Table 2). Except for isoRebM, RebC, hydRebM, and Ace-K, Gt at TMD3 shows better interactions in most cases (Table 2). Interestingly, for the Gt peptide, high sweet RebM and RebD showed better interaction of C20 at TMD3, but low sweet RebC and hydRebM showed better interaction of C20 at TMD2. Most steviol glycosides display low selectivity of Gs C20. However, the Ace-K artificial sweetener prefers to bind to Gs at TMD2. Supporting this, when four orthosteric non-nutritive sweeteners, Ace-K, saccharin, sucralose, and glycyrrhizin, were tested for signaling via calcium and cAMP pathways,20 Ace-K was found to bind to Gs not Gt, showing that Ace-K and sucralose could be considered unbiased ligands because they stimulated calcium and cAMP equivalently (presumed to be via Gq and Gs, respectively). Thus, our MD simulations find that sweet taste receptors couple to other GPs as well as Gt. This will lead to multimodal functions producing multiple patterns of intracellular signals for sweet taste receptors depending on sweeteners.
Table 2. Calculated C20 Binding Energies (kcal/mol) to TMD2 and TMD3 of the Sweet Taste Receptor 1R2/1R3 for Multiple C20 Peptidesa.
| TMD3 |
TMD2 |
|||||||
|---|---|---|---|---|---|---|---|---|
| sweeteners | GtC20 | GoC20 | GtC20 | GiC20 | GsC20 | GqC20 | G12C20 | G protein coupling selectivity at TMD2 |
| -(apo) | –255.40 | –193.99 | –200.66 | –183.19 | –124.97 | –110.20 | –72.65 | Gt≥Go>Gi ≫ Gs > Gq ≫ G12 |
| RebM | –226.15 | –104.94 | –191.90 | –171.31 | –88.43 | –112.65 | –73.24 | Gt> Gi ≫ Gq > Go > Gs > G12 |
| RebD | –270.03 | –69.35 | –148.96 | –145.46 | –131.37 | –111.91 | –166.92 | G12> Gt > Gi > Gs > Gq ≫ Go |
| isoRebM | –191.55 | –76.56 | –112.04 | –214.95 | –107.36 | –111.91 | –229.45 | G12> Gi > Gt > Gq > Gs ≫ Go |
| Rubu | –365.06 | –109.48 | –112.30 | –122.62 | –101.70 | –188.38 | –229.45 | G12≫ Gq ≫ Gi > Gt > Go > Gs |
| RebC | –249.91 | –139.71 | –272.42 | –167.50 | –68.48 | –111.05 | –93.73 | Gt≫ Gi > Go > Gq > G12 > Gs |
| hydRebM | –208.27 | –132.62 | –239.34 | –207.22 | –101.94 | –179.87 | –98.94 | Gt> Gi ≫ Gq > Go > Gs > G12 |
| Ace-K | –179.30 | –126.15 | –140.13 | –108.40 | –183.75 | –162.59 | –119.45 | Gs> Gq > Gt > Go > G12 > Gi |
We selected various steviol glycosides [rebaudioside M (RebM), RebD, isoRebM, rubusoside (Rubu), RebC, and hydRebM] and Ace-K artificial sweetener for this study of C20 selectivity. The strongest interactions are displayed in bold face. The sweeter ligands prefer TMD3, while the less sweet ligands prefer TMD2.
Conclusions
We observed that binding C20 GP to TMD2 interacts in the noncanonical way like class C GPCRs, while binding to TMD3 interacts in the canonical way like class A GPCRs. To study C20 selectivity, we carried out MD simulations with six various steviol glycosides [RebM, RebD, isoRebM, rubusoside (Rubu), RebC, and hydRebM] and Ace-K artificial sweetener with C20 binding at TMD2 or TMD3. With no sweetener, the apo protein shows similar Go and Gt selectivity, while all Reb sweeteners increase the selectivity of Gt, gustducin, which is specifically involved in taste processing, but decrease Go selectivity at TMD2 (Table 2). Interestingly, for Gt protein, high sweet RebM and RebD both showed better interactions of C20 at TMD3, whereas low sweet RebC and hydRebM showed better interactions of C20 at TMD2.
Thus, our MD simulation suggests that sweet taste receptors can couple GPs other than Gt and that coupling to both TMD2 and TMD3 may play important roles in perception. This will lead to multimodal functions producing multiple patterns of intracellular signals for sweet taste receptors depending on the sweetener. Understanding these trends and differences may lead to new sweeteners that taste sweet but have a low caloric content and novel treatments for obesity and diabetes.
Acknowledgments
This research was supported by grants from the Cargill Corp. and from NIH (R01HL155532).
Glossary
Abbreviations Used:
- Ace-K
acesulfame-K
- CRD
cysteine-rich domains
- GEnSeMBLE
G protein-coupled receptor ensemble of structures in a membrane bilayer environment
- GPCRs
G protein-coupled receptors
- HB
hydrogen bonds
- MD
molecular dynamics
- POPC
2-oleoyl-1-palmitoyl-sn-glycero-3-phosphocholine
- RebM
rebaudioside M
- RMSD
root-mean-square deviation
- SCREAM
side chain rotamer excitation analysis method
- TMD
transmembrane domains
- VFD
Venus flytrap domains
- VFD2
VFD of TAS1R2
- VFD3
VFD of TAS1R3
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpcb.4c04610.
Figure S1. Various initial positions of the C-terminal α-helices of the C20 G protein peptide at the cytoplasmic TMD of taste receptor TAS1R2 in closed and 1R3 in open between helix 3 and 6 before running MD simulation, Figure S2. The best initial positions of the C-terminal α-helices of the C20 G protein peptide at the cytoplasmic TMD of taste receptor TAS1R2 in closed and TAS1R3 in open between helix 3 and 6, and Figure S3. Interaction energy between C20 and TAS1R2 or TAS1R3 and the RMSD of the TAS1R2/1R3-C20 complex when C20 binds at TMD2-C20 or TMD3-C20 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ohta M.; Sasa S.; Inoue A.; Tamai T.; Fujita I.; Morita K.; Matsuura F. Characterization of Novel Steviol Glycosides from Leaves of Stevia rebaudiana Morita. J. Appl. Glycosci. 2010, 57, 199–209. 10.5458/jag.57.199. [DOI] [Google Scholar]
- Sigoillot M.; Brockhoff A.; Meyerhof W.; Briand L. Sweet-taste-suppressing compounds: current knowledge and perspectives of application. Appl. Microbiol. Biotechnol. 2012, 96, 619–630. 10.1007/s00253-012-4387-3. [DOI] [PubMed] [Google Scholar]
- Odoemelam C. S.; Percival B.; Wallisa H.; Changb M.-W.; Ahmadc Z.; Scholeya D.; Burtona E.; Williams I. H.; Kamerlin C. L.; Wilson P. B. G-Protein coupled receptors: structure and function in drug discovery. RSC Adv. 2020, 10, 36337–36348. 10.1039/D0RA08003A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Chen Y.; Abro R.; Goddard W. III; Guthrie B. Activation mechanism of the G protein-coupled sweet receptor heterodimer with sweeteners and allosteric agonists. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (10), 2568–2573. 10.1073/pnas.1700001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölwer-Rieck U. The Leaves of Stevia rebaudiana (Bertoni), Their Constituents and the Analyses Thereof: A Review. J. Agric. Food Chem. 2012, 60 (4), 886–895. 10.1021/jf2044907. [DOI] [PubMed] [Google Scholar]
- Ripken D.; van der Wielen N.; Wortelboer H. M.; Meijerink J.; Witkamp R. F.; Hendriks H. F. J. Steviol glycoside rebaudioside A induces glucagon-like peptide-1 and peptide YY release in a porcine ex vivo intestinal model. J. Agric. Food Chem. 2014, 62 (33), 8365–70. 10.1021/jf501105w. [DOI] [PubMed] [Google Scholar]
- Jang J.; Kim S.-K.; Guthrie B.; Goddard W. A. III Synergic Effects in the Activation of the Sweet Receptor GPCR Heterodimer for Various Sweeteners Predicted Using Molecular Metadynamics Simulations. J. Agric. Food Chem. 2021, 69 (41), 12250–12261. 10.1021/acs.jafc.1c03779. [DOI] [PubMed] [Google Scholar]
- Hann V.; Chazot P. L. G-proteins. Curr. Anaesth. Crit. Care. 2004, 15, 79–81. 10.1016/j.cacc.2004.02.004. [DOI] [Google Scholar]
- Sainz E.; Cavenagh M. M.; LopezJimenez N. D.; Gutierrez J. C.; Battey J. F.; Northup J. K.; Sullivan S. L. The G-protein coupling properties of the human sweet and amino acid taste receptors. Dev Neurobiol. 2007, 67 (7), 948–59. 10.1002/dneu.20403. [DOI] [PubMed] [Google Scholar]
- a Bray J. K.; Abrol R.; Goddard W. A. III; Trzaskowski B.; Scott C. E. SuperBiHelix method for predicting the pleiotropic ensemble of G-protein-coupled receptor conformations. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E72–8. 10.1073/pnas.1321233111. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Abrol R.; Bray J. K.; Goddard W. A. III Bihelix: Towards de novo structure prediction of an ensemble of G-protein coupled receptor conformations. Proteins 2011, 80, 505–518. 10.1002/prot.23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seven A. B.; Barros-Álvarez X.; de Lapeyrière M.; Papasergi-Scott M. M.; Robertson M. J.; Zhang C.; Nwokonko R. M.; Gao Y.; Meyerowitz J. G.; Rocher J.-P.; et al. G-protein activation by a metabotropic glutamate receptor. Nature 2021, 595, 450–454. 10.1038/s41586-021-03680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Han S.; Cai X.; Tan Q.; Zhou K.; Wang D.; Wang X.; Du J.; Yi C.; Chu X.; et al. Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature 2021, 594, 583–588. 10.1038/s41586-021-03495-2. [DOI] [PubMed] [Google Scholar]
- Shen C.; Mao C.; Xu C.; Jin N.; Zhang H.; Shen D.-D.; Shen Q.; Wang X.; Hou T.; Chen Z.; et al. Structural basis of GABAB receptor–Gi protein coupling. Nature 2021, 594, 594–598. 10.1038/s41586-021-03507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehl A.; Hu H.; Maeda S.; Zhang Y.; Qu Q.; Paggi J. M.; Latorraca N. R.; Hilger D.; Dawson R.; Matile H.; et al. Structure of the μ-opioid receptor–Gi protein complex. Nature 2018, 558, 547–552. 10.1038/s41586-018-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafi A.; Kim S.-K.; Chou K. C.; Guthrie B.; Goddard W. A. III Structure of Fully Activated Tas1R3/1R3 Homodimer Bound to G Protein and Natural Sugars: Structural Insights into G Protein Activation by a Class C Sweet Taste Homodimer with Natural Sugars. J. Am. Chem. Soc. 2021, 143 (40), 16824–16838. 10.1021/jacs.1c08839. [DOI] [PubMed] [Google Scholar]
- Darden T.; York D.; Pedersen L. Smooth Particle Mesh Ewald: An Efficient Method for Particle-Particle Particle-Mesh Ewald PME simulations. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar]
- Phillips J. C.; Braun R.; Wang W. G. J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kalé L.; Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell J. A. D.; Bashford D.; Bellott M.; Dunbrack J. R. L.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Feller S. E.; Yin D.; Pastor R. W.; MacKerell J A. D. Molecular Dynamics Simulation of Unsaturated Lipids at Low Hydration: Parametrization and Comparison with Diffraction Studies. Biophys. J. 1997, 73, 2269–2279. 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. J.; Grant J. N.; Moon J. I.; So S. S.; Finch A. M. Critically evaluating sweet taste receptor expression and signaling through a molecular pharmacology lens. FEBS Journal 2021, 288, 2660–2672. 10.1111/febs.15768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.