Abstract
Sixteen patient-derived xenografts (PDXs) were analyzed using a mass spectrometry (MS)-based kinase inhibitor pull-down assay (KIPA), leading to the observation that death-associated protein kinase 3 (DAPK3) is significantly and specifically overexpressed in the triple-negative breast cancer (TNBC) models. Validation studies confirmed enrichment of DAPK3 protein, in both TNBC cell lines and tumors, independent of mRNA levels. Genomic knockout of DAPK3 in TNBC cell lines inhibited in vitro migration and invasion, along with down-regulation of an epithelial–mesenchymal transition (EMT) signature, which was confirmed in vivo. The kinase and leucine-zipper domains within DAPK3 were shown by a mutational analysis to be essential for functionality. Notably, DAPK3 was found to inhibit the levels of desmoplakin (DSP), a crucial component of the desmosome complex, thereby explaining the observed migration and invasion effects. Further exploration with immunoprecipitation–mass spectrometry (IP–MS) identified that leucine-zipper protein 1 (LUZP1) is a preferential binding partner of DAPK3. LUZP1 engages in a leucine-zipper domain–mediated interaction that protects DAPK3 from proteasomal degradation. Thus, the DAPK3/LUZP1 heterodimer emerges as a newly discovered regulator of EMT/desmosome components that promote TNBC cell migration.
Keywords: triple-negative breast cancer, DAPK3, migration, invasion, epithelial-to-mesenchymal transition
Significance Statement.
There is an urgent need to identify new therapeutic vulnerabilities and druggable targets for triple-negative breast cancer (TNBC). While contemporary multiomics profiling of clinical tumor samples is a productive but broad-based approach, our kinase inhibitor pull-down assay (KIPA) provides a specific platform for efficiently assessing kinase expression profiles. Herein, KIPA identified DAPK3 as a TNBC-enriched kinase that plays a critical role in regulating cell migration, invasion, and the epithelial-to-mesenchymal transition (EMT). Additionally, LUZP binds to DAPK3 to reduce proteasome degradation. Thus, DAPK3/LUZP1 heterodimer emerges as a new driver of TNBC cell migratory behavior in vitro that should be further investigated as a potential therapeutic target.
Introduction
Triple-negative breast cancer (TNBC) is defined by the absence of estrogen receptor (ER), progesterone receptor, and human epidermal growth factor receptor 2 (HER2) expression. The pathophysiology of TNBC is poorly understood (1). Unfortunately, patients with TNBC, accounting for over 20,000 cases of breast cancer annually in the United States of America alone, experience worse outcomes compared with other breast cancer subtypes, with a 5-year mortality rate of 40% (1). The high mortality rate can be attributed to the metastatic propensity of TNBC and the lack of effective targeted therapies. Remarkably, nearly half of patients with TNBC develop distant metastasis to the brain and visceral organs, with a median survival time of only 13.3 months following a Stage 4 diagnosis (2). The efficacy of systemic chemotherapy is therefore limited, and there is an unmet need to identify new therapeutic vulnerabilities and pathophysiological insights.
Protein kinases play critical roles in cellular processes such as cell cycle progression, apoptosis, differentiation, and signal transduction (3). Dysregulated kinase activity is a likely driver in the majority of human cancers (4). For breast cancer treatment, targeting the HER2 kinase with antibodies or small molecules has been a notable success (5). Additionally, for ER-positive advanced breast cancers, targeting mutations in PIK3CA with alpelisib (6) or PIK3CA/AKT1/PTEN mutations with the AKT inhibitor capivasertib (7) has improved outcomes in advanced disease in combination with second-line endocrine therapy. However, the fact that there are so few examples of successful kinase targeting in breast cancer, and no approved examples in TNBC, underscores our incomplete understanding of the breast cancer kinase dependency landscape.
To address this therapeutic gap, we developed a kinase inhibitor pull-down assay (KIPA) to provide a proteomic screen for kinase expression by incorporating multiple kinase inhibitors to produce broad kinome coverage (8). The KIPA methodology employs a mixture of nine kinase inhibitors that are individually conjugated to Sepharose beads. Native tumor lysates are incubated with the beads, followed by stringent washing steps. The bound kinases are released by trypsin digestion and identified and quantified using mass spectrometry (MS) (8). In this study, the KIPA was performed on 16 breast cancer patient-derived xenografts (PDXs) representing diverse molecular subtypes (9). Analysis of KIPA data identified kinases that are enriched in TNBC over ER-positive and HER2-enriched breast tumors, including death-associated protein kinase 3 (DAPK3).
DAPK3, previously named zipper-interacting protein kinase (ZIPK), is a serine/threonine kinase linked to various cellular functions. DAPK3 is an unusual kinase because it has a C-terminal leucine-zipper domain (10). While DAPK3 has been implicated in diverse cellular processes (11–13), a functional role in TNBC has not been previously reported. This study therefore focuses on the functional significance of DAPK3 in TNBC pathophysiology.
Results
KIPA prioritizes DAPK3 as an understudied kinase enriched in TNBC
We have recently reported an optimized KIPA incorporating nine kinase inhibitors (palbociclib, crizotinib, CZC-8004, axitinib, GSK690693, AZD4547, afatinib, abemaciclib, and FRAX597) (8). The panel of kinase inhibitors was designed for a broad coverage of the kinome. Each inhibitor was individually coupled to ECH-Sepharose beads and deployed as an affinity matrix for kinase enrichment from native tumor lysates. The KIPA was initially performed on 16 WHIM breast cancer PDXs, including seven derived from TNBC, using the data-dependent acquisition (DDA) mode of protein MS (8). To enhance sensitivity and precision, the KIPA was further refined using a parallel reaction monitoring (PRM) mode to target 100 preselected kinases (8). Hierarchical clustering of kinases measured by PRM revealed distinct expression patterns associated with different molecular subtypes (Fig. S1). Notably, established luminal subtype-associated kinases rearranged during transfection proto-oncogene (RET) and insulin-like growth factor 1 receptor (IGF1R) (14, 15) were significantly elevated in luminal PDXs, whereas ERBB2 (HER2) expression was enriched in two HER2-enriched models (WHIM 8 and 35, Fig. S1).
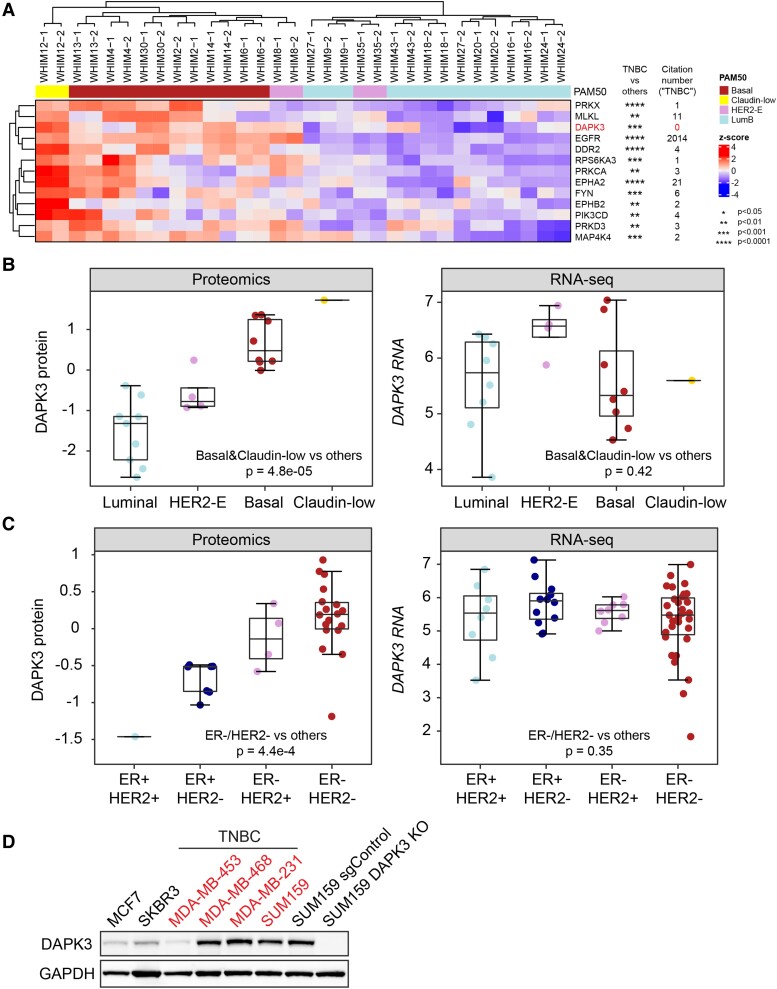
To pinpoint TNBC-specific enriched kinases, Wilcoxon sum-rank tests were performed between TNBC and non-TNBC PDXs, unveiling 13 TNBC-specific kinases (Fig. 1A), including established TNBC-associated kinases, for example, EGFR and discoidin domain receptor tyrosine kinase 2 (DDR2) (17, 18). To prioritize novel TNBC biomarkers and therapeutics targets, the number of publications for each kinase significantly related to TNBC was examined in PubMed. This analysis revealed DAPK3 was one of the most significantly up-regulated kinases with zero relevant citations, justifying further investigation (Fig. 1A).
Fig. 1.
DAPK3 is significantly enriched in the TNBC subtype at the protein level, but not at the mRNA level. A) Hierarchical clustering heatmap shows the abundance of TNBC-enriched kinases measured by the KIPA–PRM assays, across 16 breast cancer WHIM PDXs (two replicates for each PDX). The kinase levels are normalized across all samples by z-scores. The P-values are determined by Wilcoxon rank-sum tests comparing protein levels for TNBC PDXs to non-TNBC PDXs. The citation number of each kinase in the field of TNBC was obtained using R package RISmed (16). The citation number of DAPK3 also includes its older gene name (ZIPK). B) The protein (Left) and mRNA (Right) levels of DAPK3 in different molecular subtypes of breast cancer WHIM PDXs, measured by the iTRAQ proteomics and RNA-Seq, respectively. The boxplots show IQR with median marked in center. The whiskers indicate 1.5 × IQR. P-values are determined by Wilcoxon rank-sum test between TNBC (basal and Claudin-low subtype) and non-TNBC (luminal and HER2-enriched subtype). C) The protein (Left) and mRNA (Right) levels of DAPK3 in different subtypes of breast cancer cell lines, measured by TMT-based proteomics and RNA-Seq, respectively. P-values are determined by Wilcoxon rank-sum test between ER-negative/HER2-negative cell lines and other subtypes. D) Immunoblotting shows the protein levels of DAPK3 in TNBC and non-TNBC cell lines. SUM159 DAPK3 KO #3 cells (in below experiments) are used as the negative control. GAPDH serves as a loading control. The representative image is based on three independent experiments.
To validate these findings, DAPK3 protein levels were examined using isobaric tags for relative and absolute quantitation (iTRAQ) proteomics data on the same set of PDXs analyzing existing datasets (19). DAPK3 protein levels were significantly enriched in basal and claudin-low PDXs (Wilcoxon test P = 4.8e − 5), consistent with the KIPA results (Fig. 1B). Interestingly, DAPK3 mRNA levels remained unchanged across subtypes (Wilcoxon test P = 0.42, Fig. 1B). This finding was corroborated by tandem mass tag (TMT)-based proteomics and RNA-Seq data of breast cancer cell lines from the DepMap dataset (20), where DAPK3 levels were significantly elevated at the protein level in ER-negative/HER2-negative cell lines (Wilcoxon test P = 4.4e − 4) but not at the mRNA level (Wilcoxon test P = 0.35, Fig. 1C). These observations suggest that DAPK3 is dominantly regulated at the posttranscriptional level. Immunoblotting further confirmed high DAPK3 protein expression in the majority of TNBC cell lines as compared to non-TNBC lines (Figs. 1D and S2A). These findings collectively position DAPK3 as an underexplored kinase in the context of TNBC, warranting further investigation into its functional significance and therapeutic potential.
Genomic knockout of DAPK3 inhibits migration and invasion of TNBC cells in vitro
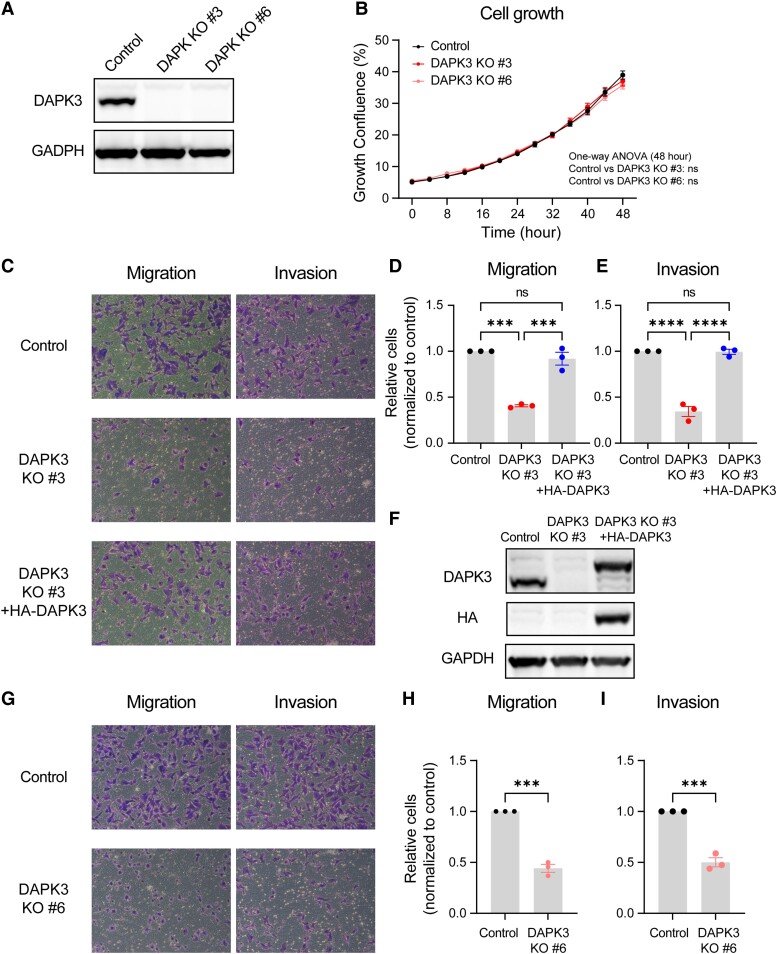
To investigate the functional role of DAPK3 in TNBC, the CRISPR/Cas9 method was used to knock out DAPK3 in SUM159 cells. Two single clones, designated as DAPK3 KO #3 and #6, were identified with the highest knockout efficiency (Fig. 2A). The growth rates of SUM159 DAPK3 KO cells showed no significant difference compared with control cells, indicating that DAPK3 did not obviously affect in vitro proliferation (Fig. 2B). Additionally, SUM159 DAPK3 KO #3 cells showed approximately similar cleaved PARP and LC3B levels as control cells, suggesting that DAPK3 did not majorly affect in vitro apoptosis (21) or autophagy (22) (Fig. S2B).
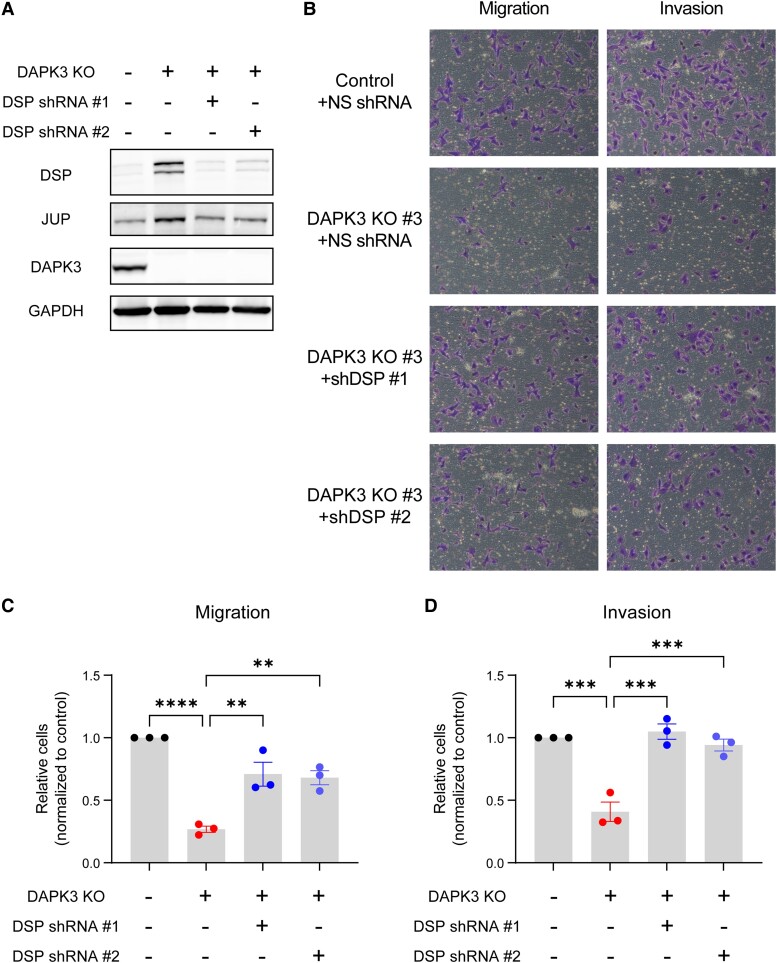
Fig. 2.
DAPK3 knockout decreases migration and invasion of SUM159 cells. A) Immunoblotting shows the protein levels of DAPK3 in SUM159 control, DAPK3 KO #3, and DAPK3 KO #6 cells. GAPDH serves as a loading control. The representative image is based on three independent experiments. B) The confluence of SUM159 control, DAPK3 KO #3, and DAPK3 KO #6 cells at indicated time points acquired by the IncuCyte imaging system. The data are the representative of three independent experiments. NS, not significant (one-way ANOVA with Dunnett's multiple comparisons test, comparing the confluency of DAPK3 KO #3 or #6 with control group at the last time point measured). C) Representative transwell migration and invasion assay images (10 × magnification) from SUM159 control, DAPK3 KO #3 cells, and DAPK3 KO #3 cells with HA-tagged DAPK3 overexpression. SUM159 DAPK3 KO #3 cells have decreased migration and invasion potential compared with control cells. DAPK3 overexpression rescues the migration and invasion phenotypes in DAPK3 KO #3 cells. D), E) ImageJ-based quantification results of C. Data represent the mean ± SEM from three independent experiments. NS, not significant, ****P < 0.0001, ***P < 0.001, one-way ANOVA with Tukey's multiple comparisons test. F) Immunoblotting shows the protein levels of DAPK3 and HA-tagged DAPK3 in SUM159 control, DAPK3 KO #3 cells, and DAPK3 KO #3 cells with HA-tagged DAPK3 overexpression. GAPDH serves as a loading control. The representative image is based on three independent experiments. G) Representative transwell migration and invasion assay images (10 × magnification) from SUM159 control and DAPK3 KO #6 cells. SUM159 DAPK3 KO #6 cells have decreased migration and invasion potential compared with control cells. H), I) ImageJ-based quantification results of G. Data represent the mean ± SEM from three independent experiments. NS, not significant, ***P < 0.001, Student's t test.
To determine the impact of DAPK3 on migration and invasion, transwell assays were conducted on both DAPK3 control and knockout cells. Strikingly, DAPK3 KO #3 and #6 cells exhibited a significant reduction in both migratory and invasive abilities when compared with control cells (Fig. 2C–E and G–I). To exclude off-target effects of CRISPR/Cas9-mediated knockout, HA-tagged DAPK3 cDNA was reintroduced into DAPK3 KO #3 cells to “rescue” the migration/invasion phenotype. The expression levels of exogenous DAPK3 protein were verified using both DAPK3 and HA antibodies (Fig. 2F). Ectopic expression of DAPK3 successfully rescued the migration and invasion phenotypes caused by DAPK3 knockout, providing evidence for direct involvement of DAPK3 in these cellular processes (Fig. 2C–E).
Complementary wound healing assays further supported these findings, indicating that DAPK3 KO #3 cells exhibited reduced cell motility compared with control cells (Fig. S2C and D). Additionally, CRSIPR-mediated knockout of DAPK3 in another TNBC cell line, MDA-MB-231, also demonstrated significant inhibition of cell migration, supporting a role for DAPK3 in this cellular process (Fig. S2E–G).
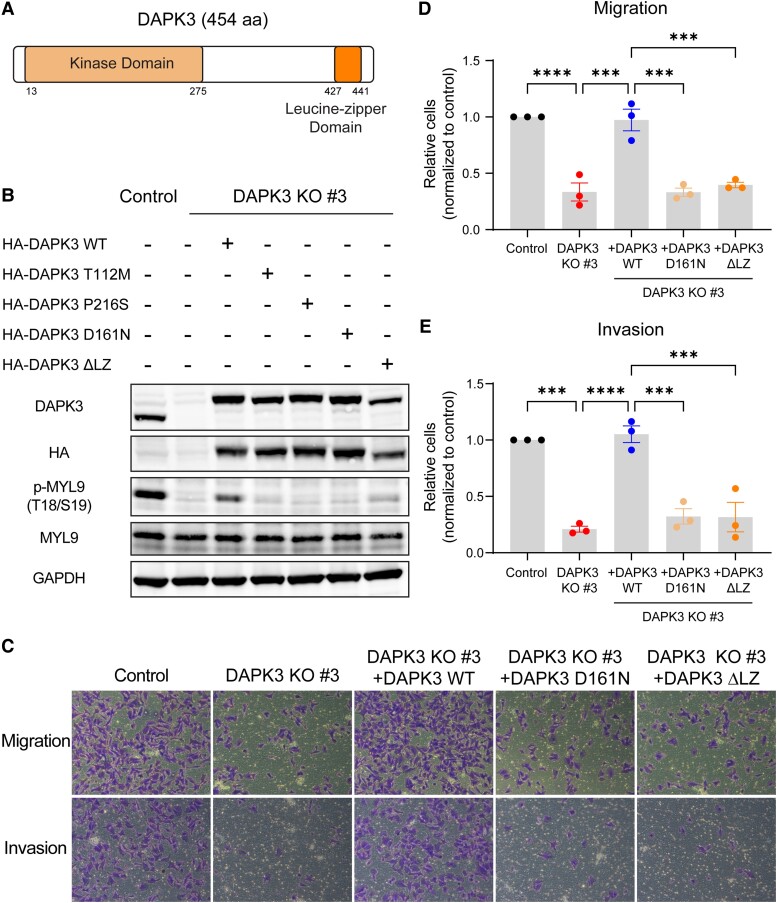
Both the kinase and leucine-zipper domains of DAPK3 are required for functionality
DAPK3 is a kinase of 454 amino acids containing an N-terminal kinase domain that is similar in other DAPK family members and a C-terminal leucine-zipper domain of unknown function (Fig. 3A) (23). Previous investigations have determined that DAPK3 directly phosphorylates myosin light chain 9 (MYL9) and regulates smooth muscle contraction through MYL9 phosphorylation (24, 25). Consistent with these findings, DAPK3 knockout in SUM159 cells significantly diminished the phosphorylation of MYL9 at threonine 18 (T18) and serine 19 (S19), which was rescued by ectopic expression of HA-tagged wild-type (WT) DAPK3 (Fig. 3B). This effect was similarly observed in MDA-MB-231 cells upon DAPK3 knockout (Fig. S2G).
Fig. 3.
Both the kinase and leucine-zipper domains of DAPK3 are required for its functionality. A) Schematic of the domain structure of WT DAPK3 protein (NP_001339). B) Immunoblotting shows the protein levels of DAPK3, HA-tagged DAPK3, total MYL9, and phosphorylated-MYL9 (T18/S19) in SUM159 control cells, DAPK3 KO #3 cells, DAPK3 KO #3 cells with HA-tagged DAPK3 WT overexpression, DAPK3 KO #3 cells with HA-tagged DAPK3 mutants (T112M, P216S, D161N, and ΔLZ) overexpression. GAPDH serves as a loading control. The representative image is based on three independent experiments. C) Representative transwell migration and invasion assay images (10 × magnification) from SUM159 control cells, DAPK3 KO #3 cells, DAPK3 KO #3 cells with HA-tagged DAPK3 WT overexpression, DAPK3 KO #3 cells with HA-tagged DAPK3 mutants (D161N and ΔLZ) overexpression. While DAPK3 WT overexpression rescues the migration and invasion phenotypes caused by DAPK3 KO, neither DAPK3 D161N nor DAPK3 ΔLZ mutants rescue the phenotypes. D), E) ImageJ-based quantification results of C. Data represent the mean ± SEM from three independent experiments. ****P < 0.0001, ***P < 0.001, one-way ANOVA with Tukey's multiple comparisons test.
Three nonsynonymous somatic point mutations within the DAPK3 kinase domain (T112M, P216S, and D161N) have been reported to reduce or ablate the kinase activity of DAPK3 (26). Stable overexpression of each of these kinase-dead mutants individually in DAPK3 KO #3 cells revealed an inability to restore MYL9 phosphorylation when compared with WT DAPK3 (Fig. 3B). Furthermore, in a reversion experiment, DAPK3 D161N, as an exemplar kinase-dead mutation, failed to rescue the migration and invasion phenotypes observed with WT DAPK3 (Fig. 3C–E).
A unique structural feature of DAPK3, in comparison with other DAPK family members, is a C-terminal leucine-zipper domain (Fig. 3A), which has been reported to be necessary for the activation of the DAPK3 kinase (11). To investigate the functional consequences of the leucine-zipper domain of DAPK3 in TNBC, a DAPK3 deletion mutant (ΔLZ) lacking the entire leucine-zipper domain (deleted for amino acids 427–441) was generated. Overexpression of the DAPK3 ΔLZ mutant in DAPK3 KO #3 cells did not restore MYL9 phosphorylation (Fig. 3B). Additionally, transwell assays demonstrated that the DAPK3 ΔLZ mutant failed to rescue the migration and invasion phenotypes observed in DAPK3 knockout cells (Fig. 3C–E). These findings underscore the indispensability of both the kinase and leucine-zipper domains for the catalytic activity of DAPK3 on the downstream substrate MYL9, as well as for DAPK3-mediated TNBC migration and invasion in vitro.
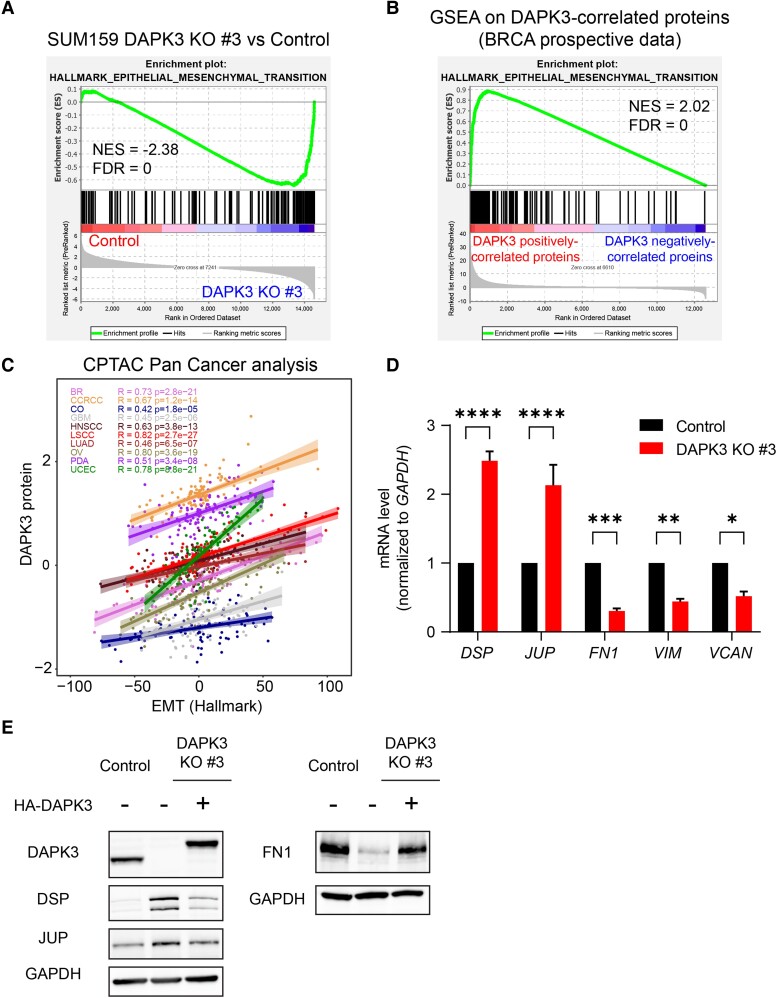
DAPK3 up-regulates epithelial–mesenchymal transition markers and reduces the level of desmosome components
To elucidate the mechanism whereby DAPK3 regulates migration and invasion in TNBC cells, RNA-sequencing analysis was conducted using SUM159 DAPK3 KO #3 and control cells (Fig. S3A). The epithelial–mesenchymal transition (EMT) pathway was the most down-regulated pathway in DAPK3 KO #3 compared with control cells after gene set enrichment analysis (GSEA) using Hallmark gene sets (false discovery rate [FDR] = 0, Figs. 4A and S3B). The correlation between DAPK3 and the EMT pathway signature was further supported by the enrichment of DAPK3-positively correlated genes in the EMT pathway in the prospective human breast cancer proteogenomic CPTAC dataset (27) (FDR = 0, Fig. 4B). Furthermore, a positive correlation between DAPK3 protein levels and EMT hallmark pathway score (29) was observed across multiple cancer types (Fig. 4C), highlighting the close and broad association between DAPK3 and the EMT pathway.
Fig. 4.
DAPK3 significantly correlates with the EMT pathway signature. A) Enrichment plot of Hallmark EMT pathway from the GSEA for DAPK3 KO #3 compared with SUM159 control cells. NES, normalized enrichment score; FDR, false discovery rate. B) Enrichment plot of Hallmark EMT pathway from the GSEA in the prospective CPTAC breast cancer dataset (27). GSEA input was a ranked list of signed -log10P-values from Pearson’s correlation between DAPK3 protein levels and all the gene levels measured by the RNA-Seq. C) Scatter plot shows the correlation between DAPK3 protein levels and Hallmark EMT score (from IPAS calculation (28)) in the CPTAC Pan-Cancer dataset (29). D) mRNA expression levels of DSP, JUP, FN1, VIM, and VCAN in SUM159 control and DAPK3 KO #3 cells. Values were normalized to GAPDH mRNA, and relative expression level was calculated as fold change to SUM159 control cells. Data represent the mean ± SEM from three independent experiments. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, two-way ANOVA with Sidak's multiple comparisons test. E) Immunoblotting shows the protein levels of DAPK3, DSP, JUP, and FN1 in SUM159 control cells, DAPK3 KO #3 cells, and DAPK3 KO #3 cells with HA-tagged DAPK3 WT overexpression. GAPDH serves as a loading control. The representative image is based on three independent experiments.
Differential expression analysis identified 1,925 up-regulated genes and 2,171 down-regulated genes in DAPK3 KO #3 cells (P < 0.05, Fig. S3A). Among the differentially expressed genes, several mesenchymal markers (ZEB1, ZEB2, CDH2, FN1, VCAN, and VIM) were decreased in DAPK3 KO #3 cells, while two epithelial markers (JUP and DSP) were elevated (Fig. S3C). qRT-PCR validation confirmed the up-regulation of DSP and JUP mRNA levels and the down-regulation of FN1, VIM, and VCAN mRNA levels in DAPK3 KO #3 cells (Fig. 4D). At the protein level, DAPK3 knockout led to the up-regulation of the epithelial markers desmoplakin (DSP) and JUP, which could be reversed by HA-tagged WT DAPK3 overexpression (Fig. 4E). Conversely, protein levels of an assayed mesenchymal marker, FN1, were down-regulated in DAPK3 KO cells and could also be reversed by HA-tagged WT DAPK3 overexpression (Fig. 4E).
DAPK3 up-regulates EMT markers in TNBC cells grown as a xenograft
To determine whether the gene expression changes associated with DAPK3 knockout observed in vitro persisted in vivo, SUM159 DAPK3 KO #3 or control cells were injected into the mammary fat pad of NOD/SCID gamma mice (NSG mice) (Fig. S4A). Up-regulated DSP protein levels were selectively observed in engrafted tumor cells in the DAPK3 knockout condition, confirming that the regulatory effects of DAPK3 knockout persisted in vivo (Fig. S4B). To further compare the in vitro versus in vivo effects of DAPK3 loss, cell extracts were subjected to proteomic profiling using label-free MS. GSEA confirmed down-regulation of the EMT pathway signature in the DAPK3 KO #3 knockout condition persisted primary xenografts (Fig. S4C and D). However, several metabolic pathways, including oxidative phosphorylation, adipogenesis, and fatty acid metabolism, were observed to be significantly up-regulated by DAPK3 knockout only in vitro (Fig. S4C and D).
DAPK3 modulates migration and invasion in TNBC cells through modulation of desmoplakin
DSP and plakoglobin (JUP) serve as integral components of the desmosome complex, a cell–cell adhesion structure crucial for maintaining epithelial homeostasis. Increasing evidence underscores the significant involvement of desmosomes in cancer progression and metastasis (30). Loss of desmosome components, including JUP and DSP, has been observed in various cancers (31–34). For example, reduced JUP expression is correlated with poorer clinical outcomes in nonsmall cell lung cancer (35) and decreased DSP expression is associated with breast cancer metastasis (33). A mouse model of pancreatic neuroendocrine tumors demonstrated that genetic deletion of DSP increased tumor microinvasion independent of adherens junction status, highlighting the potential tumor-suppressing functions of DSP (36). These functions may be related to the involvement of DSP in cell adhesive desmosome complexes and regulation by the Wnt/β-catenin signaling pathway (34).
To investigate the roles of DSP in DAPK3-modulated migration and invasion phenotypes in TNBC cells, DSP was knocked down using two different shRNAs in SUM159 DAPK3 KO #3 and control cells (Figs. 5A and S5A). Interestingly, DSP knockdown resulted in decreased JUP protein levels (Fig. 5A). Transwell assays revealed that DSP knockdown rescued the impaired migratory and invasive abilities caused by DAPK3 knockout in SUM159 cells (Fig. 5B–D). DSP knockdown in SUM159 control cells had no significant effects on migration and invasion (Fig. S5B–D). These findings suggest that DAPK3 modulates migration and invasion of TNBC cells through regulation of DSP, indicating an intricate interplay between DAPK3, desmosome complex components, and the invasive phenotype in TNBC.
Fig. 5.
DSP knockdown rescues the migration and invasion deficits caused by DAPK3 knockout in SUM159 cells. A) Immunoblotting shows the protein levels of DAPK3, DSP, and JUP in SUM159 control cells, DAPK3 KO #3 cells, and DAPK3 KO #3 cells with DSP knockdown mediated by two different shRNAs. GAPDH serves as a loading control. The representative image is based on three independent experiments. B) Representative transwell migration and invasion assay images (10× magnification) from SUM159 control cells, DAPK3 KO #3 cells, and DAPK3 KO #3 cells with DSP knockdown. DSP knockdown by two different shRNAs increases migration and invasion potential compared with nonsilencing shRNA in DAPK3 KO #3 cells. C), D) ImageJ-based quantification results of B. Data represent the mean ± SEM from three independent experiments. ****P < 0.0001, ***P < 0.001, **P < 0.01, one-way ANOVA with Tukey's multiple comparisons test.
The identification of the DAPK3 interactome by IP–MS
Herein, it has been established that the functionality of DAPK3 requires both the kinase and leucine-zipper domains. Given the role of the leucine-zipper domain in facilitating protein–protein interactions (PPIs), which was first studied in transcription factors (37), the identification of proteins interacting with DAPK3 was the next focus of study. To identify the DAPK3 interactome, DAPK3 antibodies were employed for immunoprecipitation in SUM159 control and DAPK3 KO #3 cells. The bound proteins were separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), stained with Coomassie blue, and each gel was divided into six slices. Peptides were generated through in-gel trypsin digestion and analyzed using liquid chromatography tandem–mass spectrometry (LC–MS/MS) (Fig. 6A). A nonspecific protein list was downloaded from the CRAPome database (38), and those nonspecific proteins were disregarded from the analysis. The potential DAPK3 interactome was defined as proteins with intensity-based absolute quantification (iBAQ) levels at least 2-fold higher in control cells, or proteins confidently identified only in control cells but not DAPK3 KO #3 cells. Subsequently, a total of 58 candidate proteins potentially interacting with DAPK3 in SUM159 cells were identified (Table S1). As expected, DAPK3 itself emerged as the most abundant protein detected in this analysis, affirming the specificity of the antibody used in the pulldown.
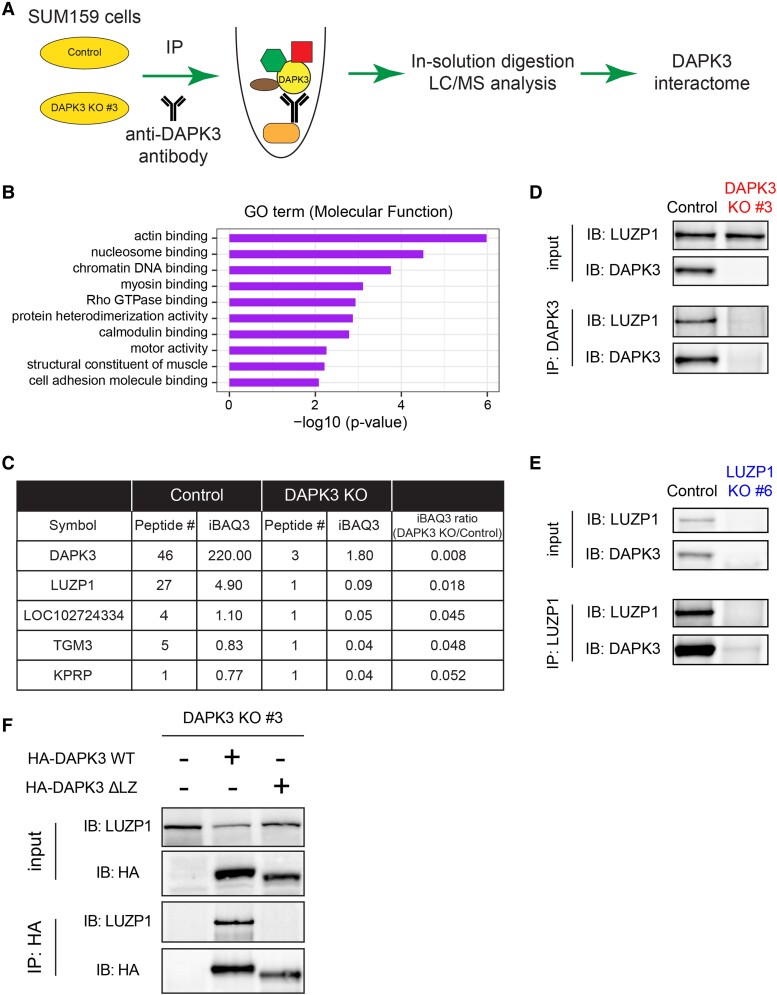
Fig. 6.
IP–MS identifies that LUZP1 is one of the top interacting proteins of DAPK3. A) Schematic representation of IP–MS workflow using a DAPK3 antibody in SUM159 control and DAPK3 KO #3 cells. B) Top 10 MF terms of GO overrepresentation analysis with a total of 73 DAPK3-interacting proteins identified from IP–MS analysis. P-values are determined by the overrepresentation analysis. C) Top 5 interacting proteins of DAPK3 identified from IP–MS analysis. The proteins are ranked based on the ratio of iBAQ levels in DAPK3 KO #3 cells compared with control cells. The full list of proteins in the DAPK3 immunoprecipitants is shown in Table S1. D) Immunoblotting shows the protein levels of LUZP1 and DAPK3 in whole-cell lysates and co-IP samples of anti-DAPK3 antibody obtained from SUM159 control and DAPK3 KO #3 cells. The representative image is based on two independent experiments. E) Immunoblotting shows the protein levels of LUZP1 and DAPK3 in whole-cell lysates and co-IP samples of antiLUZP1 antibody obtained from SUM159 control and LUZP1 KO #6 cells. The representative image is based on two independent experiments. F) Immunoblotting shows the protein levels of LUZP1 and HA-tagged DAPK3 in whole-cell lysates and co-IP samples of anti-HA antibody obtained from SUM159 DAPK3 KO #3 cell, DAPK3 KO #3 cells with HA-tagged DAPK3 WT overexpression, and DAPK3 KO #3 cells with HA-tagged DAPK3 ΔLZ overexpression. The representative image is based on two independent experiments.
The 58 proteins comprising the DAPK3 interactome underwent gene ontology (GO) overrepresentation analysis to elucidate their functional associations (Fig. 6B). The DAPK3-interacting proteins were found to be enriched in actin-binding pathway (P = 1.08e − 6, Fig. 6B), supporting a role for DAPK3 in actin cytoskeletal pathway physiology, which is pivotal for cell movement and motility.
Leucine-zipper protein 1 interacts with DAPK3 and is positively correlated with DAPK3 at the protein level across cell lines
The members of the DAPK3 interactome were ranked based on the ratio of the iBAQ levels in DAPK3 KO #3 cells compared with control cells (Fig. 6C). Among these proteins, leucine-zipper protein 1 (LUZP1) stood out as one of the top interaction partners for DAPK3 and it is the only protein with a leucine-zipper domain, suggesting that the interaction between DAPK3 and LUZP1 is likely to be leucine-zipper-mediated. LUZP1 was therefore knocked out in SUM159 cells using the CRISPR/Cas9 method, and two clones (LUZP1 KO #6 and #8) were identified with the most complete knockout. Endogenous DAPK3 protein was immunoprecipitated using a DAPK3 antibody in SUM159 control cells and in DAPK3 KO #3 cells as the negative control. Immunoblotting analysis revealed the presence of LUZP1 protein in the immunoprecipitation fraction of SUM159 control cells but not DAPK3 KO #3 cells, indicating a specific interaction between LUZP1 and DAPK3 (Fig. 6D). Similarly, endogenous LUZP1 protein was immunoprecipitated using a LUZP1 antibody in SUM159 control and LUZP1 KO #6 cells, and DAPK3 protein was detected in immunoprecipitation fraction, further confirming the interaction in a reciprocal manner (Fig. 6E). In SUM159 DAPK3 KO #3 cells with re-expressed HA-tagged WT DAPK3, exogenous DAPK3 protein was immunoprecipitated using a HA antibody, and LUZP1 protein was detected in immunoprecipitation fraction (Fig. 6F). However, the interaction between DAPK3 and LUZP1 was abolished in the presence of the DAPK3 ΔLZ mutant (Fig. 6F), indicating that the leucine-zipper domain of DAPK3 is required for the interaction with LUZP1.
The correlation between DAPK3 and LUZP1 expression levels was also examined. First, correlation analyses were conducted between the levels of DAPK3 protein and other proteins in the WHIM PDX proteomics dataset (Fig. S6A). This unbiased analysis revealed that LUZP1 exhibited one of the strongest positive correlations with DAPK3 among ∼10,000 proteins examined (Pearson R = 0.90, P = 1.27e − 8, Fig. S6A and B). However, the correlation between DAPK3 and LUZP1 at the mRNA level was not significant (Pearson R = 0.06, P = 0.79, Fig. S6B). Similarly, strong positive correlations between DAPK3 and LUZP1 at the protein level were also observed in breast cancer cell lines (DepMap dataset) (20), Pearson R = 0.81, P = 4.6e − 8, Fig. S6C) and in human basal-like breast cancer samples (the CPTAC Prospective Breast Cancer dataset (27), Pearson R = 0.88, P = 2.2e − 10, Fig. S6D). However, the correlations between DAPK3 and LUZP1 at the mRNA level were either not significant or less so in each dataset (Fig. S6C and D), suggesting a dominant posttranscriptional regulation mechanism controlling the levels of these two proteins. Moreover, LUZP1 was found to be significantly enriched in the TNBC subtype at both the protein and the mRNA levels in multiple datasets, including the WHIM PDX dataset (Fig. S7A), the DepMap cell line dataset (Fig. S7B), and in human primary breast cancer samples (METABRIC (39) and TCGA (40) datasets, Fig. S7C and D). These findings highlight the potential relevance of LUZP1 in TNBC and further support the association with DAPK3 in breast cancer.
LUZP1 protects DAPK3 from proteasome-mediated degradation
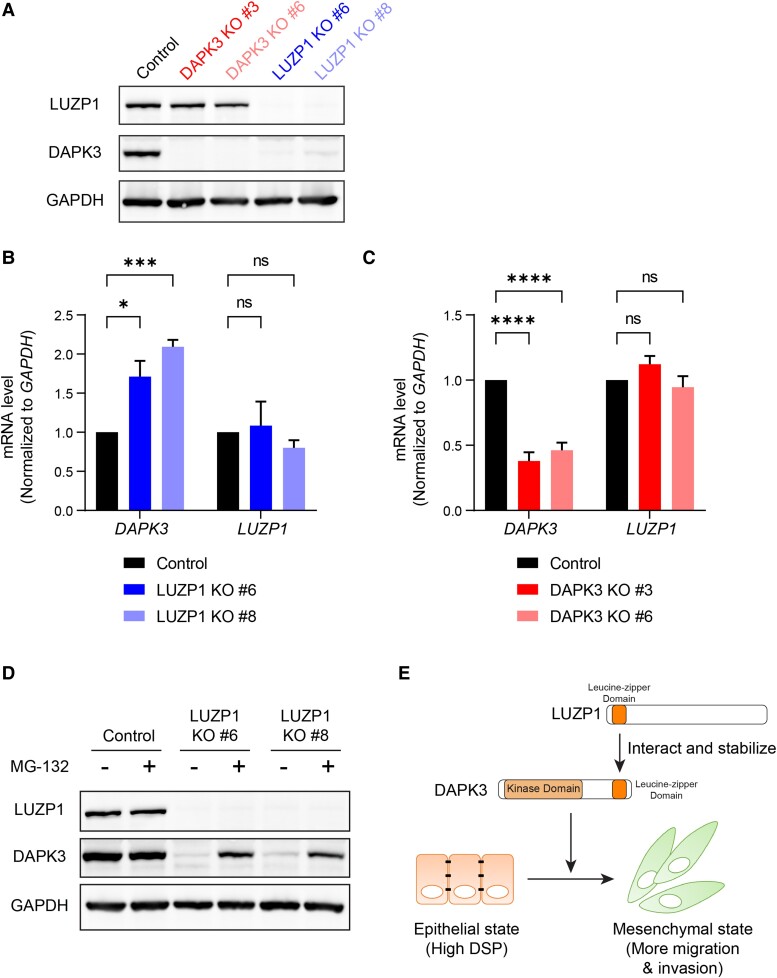
To investigate the regulatory impact of LUZP1 on DAPK3, LUZP1 knockout was performed in SUM159 cells. The successful knockout of LUZP1 was validated in clones #6 and #8, as evidenced by the absence of LUZP1 protein (Fig. 7A). LUZP1 knockout in both clones resulted in a marked reduction in DAPK3 protein levels (Fig. 7A). However, the knockout of LUZP1 did not result in a decrease in DAPK3 mRNA levels (Fig. 7B), indicating that the regulatory mechanism of LUZP1 on DAPK3 occurs at the posttranscriptional level. Notably, LUZP1 knockout significantly increased the mRNA levels of DAPK3 (Fig. 7B), suggesting a possible compensatory effect. Conversely, the knockout of DAPK3 did not exhibit any significant alteration in the protein and mRNA levels of LUZP1 (Fig. 7A and C), indicating that the regulation between LUZP1 and DAPK3 is unidirectional. To test whether loss of LUZP1 may make DAPK3 more susceptible to proteasome-mediated degradation, a proteasome inhibitor (MG-132) was used to treat LUZP1 KO cells. Strikingly, MG-132 treatment of LUZP1 knockout clones (#6 and #8) of SUM159 cells restored DAPK3 protein levels (Fig. 7D). This observation suggests that in the absence of LUZP1, DAPK3 protein level is regulated by the 26S proteasome.
Fig. 7.
LUZP1 knockout markedly decreased DAPK3 protein levels, but did not change DAPK3 mRNA levels. A) Immunoblotting shows the protein levels of LUZP1 and DAPK3 in SUM159 control, DAPK3 KO #3, DAPK3 KO #6, LUZP1 KO #6, and LUZP1 KO #8 cells. GAPDH serves as a loading control. The representative image is based on three independent experiments. B), C) mRNA expression levels of DAPK3 (B) and LUZP1 (C) in SUM159 control, DAPK3 KO #3, DAPK3 KO #6, LUZP1 KO #6, and LUZP1 KO #8 cells. Values were normalized to GAPDH mRNA, and relative expression level was calculated as fold change to SUM159 control cells. Data represent the mean ± SEM from three independent experiments. NS, not significant, ****P < 0.0001, ***P < 0.001, *P < 0.05, two-way ANOVA with Dunnett's multiple comparisons test. D) Immunoblotting shows the protein levels of LUZP1 and DAPK3 in SUM159 control, LUZP1 KO #6, and LUZP1 KO #8 cells with or without MG-132 treatment (10 μM for 16 h). GAPDH serves as a loading control. The representative image is based on three independent experiments. E) Schematic presentation of the functional role of DAPK3 in TNBC cells, which involves interacting with LUZP1, to drive a mesenchymal cell state.
Discussion
This study integrated multiple proteogenomic discovery approaches to identify DAPK3 as a kinase frequently overexpressed in TNBC. DAPK3 has been implicated in both tumor-suppression (11, 26, 41–44) and oncogenesis (45) in various cancers and has recently been linked to tumor-intrinsic immunity through the STING-IFN-β pathway (46). However, a role in TNBC remained unexplored. Our study reveals that DAPK3, through both kinase and leucine-zipper domains, modulates migration and invasion in vitro and drives an EMT pathway in TNBC cells that persists in vivo (Fig. 7E).
Desmosomes, noncanonical cell–cell adhesion complexes associated with intermediate filaments, have implications in cancer progression and metastasis (30–36). In this study, the knockout of DAPK3 in TNBC cells led to a significant increase in DSP at both the protein and mRNA levels. Furthermore, knockdown of DSP enhanced migration and invasion, revealing a regulatory axis in TNBC involving DAPK3 and desmosomal components.
PPIs often provide insights into functional relationships between proteins involved in specific cellular processes, including those driving tumor progression. Previous studies have explored protein–protein interaction networks in various cancer types, identifying potential therapeutic targets (47–51). In our study, IP–MS was used to investigate the interactome of DAPK3, which led to the identification of LUZP1 as one of the strongest interacting partners of DAPK3. Consistent with our findings, another group recently reported that transiently expressed FLAG-tagged LUZP1 co-immunoprecipitated DAPK3 in HEK293T cells (52); however, no follow-up functional studies were reported. LUZP1 contains three leucine-zipper motifs at its N-terminus (53). LUZP1 has been reported to be involved in embryonic brain development (53–55), transcriptional regulation (56) and actin cytoskeletal regulation (57). However, LUZP1 has not previously studied in TNBC. Reciprocal IP–immunoblotting validated the DAPK3/LUZP1 interaction. Correlation analyses across multiple datasets highlight LUZP1 as a TNBC-associated protein.
Unlike the proteomics data, transcriptomics profiling in multiple breast cancer datasets did not prioritize DAPK3 as a TNBC-enriched candidate gene/protein. This observation likely explains why DAPK3 has remained understudied. The DAPK3 example emphasizes the importance of integrated multiomics analyses to comprehensively evaluate the proteogenomic landscape of breast cancer (27, 58–60). In multiple breast cancer datasets, a high positive correlation between DAPK3 and LUZP1 was observed at the protein level, but not at the mRNA level. Furthermore, while LUZP1 knockout eliminated the expression of DAPK3 protein, DAPK3 mRNA levels were not reduced. These findings collectively suggest that the relationship between DAPK3 and LUZP1 occurs at the posttranscriptional level. In contrast to DAPK3, the enrichment of LUZP1 in the TNBC subtype is evident at both the protein and mRNA levels. One possible assumption is that the expression of LUZP1 in TNBC is elevated due to transcriptional regulation. The high LUZP1 level subsequently stabilizes DAPK3 protein expression, enabling DAPK3 to modulate EMT.
While migration and invasion abilities of TNBC cells in vitro were significantly impaired by DAPK3 knockout, there was no discernible difference in lung metastasis when DAPK3 KO and control cells were grown as xenografts in the mammary fat pad of NSG mice despite persisted inhibition of EMT-related gene expression. However, xenografting cancer cell lines into the mammary fat pad bypasses the earliest steps in the metastatic process, for example, invasion though the wall of the mammary duct. More sophisticated in vivo models of invasion and metastasis are therefore warranted. The observation from proteomic analysis that the DAPK3-modulated components of metabolic processes were observed in vitro but not in vivo highlights the dramatic differences in the growth milieu in these experiments. Prior research has suggested that the metabolic setting may play a permissive role in the metastatic process (61). The inconsistency between in vitro and in vivo phenotypes may also be attributed to the complex in vivo tumor microenvironment, which involves interactions with stromal cells and extracellular matrix components (62, 63). In sum, our proteogenomic studies of TNBC identified the LUZP1–DAPK3 axis as a regulator of EMT in TNBC models. Further investigation of DAPK3 and LUZP1 in TNBC pathophysiology is therefore warranted.
Materials and methods
Cell lines and culture methods
All human cancer cell lines were obtained from the American Type Culture Collection (ATCC) and were confirmed negative for mycoplasma. SUM159 cells were maintained in Ham's F-12 medium with 5% fetal bovine serum (FBS), 10 mM HEPES, 5 μg/mL insulin, and 1 μg/mL hydrocortisone. MDA-MB-231, MCF7, MDA-MB-468, MDA-MB-453, and HEK293T cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS. SK-BR-3 cells were maintained in McCoy's 5a medium supplemented with 10% FBS. Hs578T cells were maintained in DMEM supplemented with 10 μg/mL insulin and 10% FBS. MG-132 (purchased from Sigma) was added to cells at 10 μM for 16 h to inhibit the 26S proteasome.
Kinobeads preparation and kinase enrichment by the KIPA
Kinase inhibitors were conjugated with local laboratory generated ECH-Sepharose 4B using the carbodiimide coupling method, as described (8). Protein lysates were extracted from frozen PDX tumors and protein concentrations were determined by the Bradford assay (Bio-Rad), as described (8). The KIPA was performed as described (8). For each KIPA pulldown, 50 μg of native protein lysate was used.
Proteomic data acquisition and processing
KIPA samples were analyzed by MS as previously described (8). Both the DDA and the PRM modes were used in parallel, as described (8).
Generation of DAPK3 and LUZP1 knockouts in TNBC cells and single clone isolations
The CRISPR/Cas9 system was used by electroporation with sgRNA-Cas9 ribonucleoprotein complexes (RNPs) to generate DAPK3 or LUZP1 knockout (KO) clones in SUM159 and MDA-MB-231 cells using the Invitrogen Neon Transfection System (Thermo Fisher Scientific). The optimized electroporation condition was 1,400 V, 30 ms, and two pulses. To electroporate Cas9-sgRNA RNPs, 50 pmol sgRNA was incubated with 20 pmol Cas9 protein (Integrated DNA Technologies, IDT) for 10–20 min at room temperature and then electroporated. Single clones were isolated from SUM159 DAPK3 or LUZP1 KO pool cells by serial dilution in 96-well plates. The knockout level in different clones was confirmed by immunoblotting.
The target sequence of the negative control sgRNA is 5′-TTGATGGCAACAATATCCAC-3′, the target sequence of DAPK3 sgRNA (IDT, Hs.Cas9.DAPK3.1.AA) is 5′-CGTGAACTACGACTTCGACG-3′, and the target sequence of LUZP1 sgRNA (IDT, Hs.Cas9.LUZP1.1.AA) is 5′-AATGACCTACGGATTGAGGA-3′. IDT designed the DAPK3 and LUZP1 sgRNA to introduce frameshift mutations at the target site, which will reduce the protein level but not necessarily the mRNA level.
Molecular cloning to generate DAPK3 WT and mutant constructs
The open reading frame of the human DAPK3 cDNA (NM_001348.3) was synthesized and cloned into pGenDONR donor vector (GenScript). DAPK3 kinase-dead mutants were generated by converting the nucleotide at position 335 from C to T (T112M); nucleotide at position 481 from G to A (D161N); and nucleotide at position 646 from C to T (P216S) using the QuikChange II site-directed mutagenesis kit (Agilent). Leucine-zipper domain-deleted mutant (ΔLZ) was generated by removing 15 amino acids (427–441) using the above mutagenesis kit. Detailed sequence information about mutagenesis primers used is listed below:
| C335T (T112M)-Forward | 5′-CTCGTCCTCCATCAGCGACTCCTTCTCCGC-3′ |
| C335T (T112M)-Reverse | 5′-GCGGAGAAGGAGTCGCTGATGGAGGACGAG-3′ |
| C646T (P216S)-Forward | 5′-CGCCCAGGAACGAGGATGCACCGCT-3′ |
| C646T (P216S)-Reverse | 5′-AGCGGTGCATCCTCGTTCCTGGGCG-3′ |
| G481A (D161N)-Forward | 5′-GCGCGATGCCGAAGTTGATGAGCTTGATTCG-3′ |
| G481A (D161N)-Reverse | 5′-CGAATCAAGCTCATCAACTTCGGCATCGCGC-3′ |
| ΔLZ-Forward | 5′-AGGCGCTGGCCAAGCAAGAGCAGGAGAAG-3′ |
| ΔLZ-Reverse | 5′-CTTCTCCTGCTCTTGCTTGGCCAGCGCCT-3′ |
A modified pHAGE-PGK-HA-hygromycin vector (plasmid HIV-1 Alex Gustavo George Enhanced, a lentiviral vector based on an HIV-1-based backbone) was used for cloning by Gateway recombination cloning technology (Invitrogen, #11791-020). The pGenDONR-DAPK3 WT or pGenDONR-DAPK3 mutants were cloned into the pHAGE-PGK-HA-hygromycin destination vector according to the manufacturer's instructions. All constructs used or generated in this study were validated by the Sanger sequencing.
Lentiviral production and stable cell line generation
Lentiviruses were produced by co-transfection of pHAGE-PGK-HA-hygromycin-DAPK3 lentiviral vectors with the packaging plasmids pMD2.G (Addgene #12259) and psPAX2 (Addgene #12260) into HEK293T cells using Lipofectamine 2000 (Invitrogen, #11668-027). Culture media containing viruses were harvested after 48 h, filtered, and added to SUM159 DAPK3 KO #3 cells in the presence of 8 μg/ml polybrene (Santa Cruz Biotechnology, sc-134220). Stably infected cells were selected by 200 µg/mL hygromycin (Santa Cruz Biotechnology, sc-29067) for 2 days. Expression of DAPK3 WT and mutant proteins was validated using immunoblotting.
shRNA knockdown
Nonsilencing pGIPZ control shRNA was purchased from the BCM Advanced Cell Engineering and 3D Models core. pGIPZ shRNAs targeting DSP were purchased from Horizon Discovery. The target sequence of nonsilencing control shRNA is 5′-ATCTCGCTTGGGCGAGAGTAAG-3′. The target sequence of shRNA targeting DSP #1 is 5′-TTGCTTGTGAATCATTCTG-3′. The target sequence of shRNA targeting DSP #2 is 5′-TAATCCTTAATTGAATTGG-3′.
Lentivirus was produced by co-transfection of shRNA constructs with the packaging plasmids pMD2.G (Addgene #12259) and psPAX2 (Addgene #12260) into HEK293T cells using Lipofectamine 2000 (Invitrogen, #11668-027). Culture media containing viruses were harvested after 48 h, filtered, and added to cells in the presence of polybrene (Santa Cruz Biotechnology, sc-134220). Stably infected cells were selected by 2 µg/mL puromycin (Sigma, P8833) for 2 days. Expression of DSP protein was validated using immunoblotting.
Migration assay
Transwell chambers with 8 μm pores were purchased from Corning (Cat #353097). SUM159 or MDA-MB-231 cells were harvested and resuspended in serum-free media with 0.2% bovine serum albumin (BSA) at concentrations of 250,000 cells/mL; then, 200 µL (50,000 cells) was seeded into the upper chamber of a 24-well plate. The lower chambers were filled with 600 µL full-serum media. Cells were incubated for 6–10 h (SUM159) or 14–16 h (MDA-MB-231). At the end of the experiment, cells that migrated into the bottom side of the transwell membrane were fixed with methanol, stained with 20% crystal violet solution, and then observed and photographed under a light microscope. Three random fields for each sample image (10 ×) were selected, and stained cells were counted with ImageJ software (NIH).
Matrigel invasion assay
Matrigel was purchased from Corning (Cat #354243) and was diluted in coating buffer (0.01 M Tris–HCl, pH 8.0, 0.7% NaCl in sterile water) at 1:8 ratio. Transwell chambers with 8 μm pores were precoated with 100 µL diluted Matrigel for 1 h at 37 °C. SUM159 cells were harvested and resuspended in serum-free media with 0.2% BSA at concentrations of 250,000 cells/mL and then seeded 200 µL (50,000 cells) into the upper chamber of a 24-well plate. The lower chambers were filled with 600 µL full-serum media. The cells were incubated for 6–14 h. At the end of the experiment, the cells that migrated into the bottom side of the transwell membrane were fixed with methanol, stained with 20% crystal violet solution, and then observed and photographed under a light microscope. Three random fields for each sample image (10 ×) were selected, and stained cells were counted with ImageJ software.
Wound healing assay
SUM159 cells were plated 20,000 cells per well in a 96-well ImageLock plate (Essen BioScience) for 24 h before treating with 0.5 µg/mL mitomycin C (Sigma, M4287). A wound maker was used to scratch, and the cells were removed from a discrete area of the monolayer to form a cell-free zone into which cells at the edges of the wound can migrate as previously described (64). The cells were imaged at 10 × magnification in an IncuCyte Zoom Live-content imaging system (Essen Bioscience) at 37 °C, 5% CO2. Relative wound densities were calculated as density in the wound area relative to that outside the wound area to account for confounding proliferation.
Cell proliferation assay
A total of 2,000 cells/well were plated into 96-well cell culture plates, and the plates were placed into the IncuCyte Zoom Live-content imaging system (Essen Bioscience). Images of cell movement were taken at regular intervals within a 48-h data collection period. Six to eight experimental replicates were used. Confluence level was calculated for data analysis.
qRT-PCR
Total RNA was extracted from each cell line using RNeasy Mini Kit (QIAGEN, #74106). RNA was reverse-transcribed into cDNA using iScript cDNA Synthesis Kit (Bio-Rad, #1708891) according to the manufacturer's instructions. qRT-PCR was conducted using equal amounts of cDNA incubated with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, #1725274). All samples were run in triplicate on a CFX96 thermal cycler (Bio-Rad). Detailed sequence information about qRT-PCR primers used is listed in Table S2.
Immunoblotting
Protein was extracted from cells using RIPA buffer supplemented with phosphatase and protease inhibitor (PhosSTOP and cOmplete mini, Roche, respectively). Protein concentration was determined by the Bradford assay (Bio-Rad). About 10–30 µg of total protein extract was run on a 4–12% SDS–PAGE gel (Invitrogen) and then transferred onto nitrocellulose membranes (GE Healthcare). The membranes were blocked with 5% nonfat milk (Cell Signaling Technology, #9999) dissolved in TBST buffer and then incubated with primary antibodies. The membranes were washed with TBST buffer, incubated with HRP-conjugated secondary antibodies (1:10,000), and visualized with the ChemiDoc Imaging System (Bio-Rad). Primary antibodies and secondary antibodies used are listed in Table S3.
RNA-sequencing analysis
RNA was extracted from SUM159 control and DAPK3 KO #3 cells using the RNeasy Mini Kit (QIAGEN, #74106) and treated with DNase (QIAGEN, #79254) to remove genomic DNA. The Genomic and RNA Profiling (GARP) Core at BCM measured concentration (using a NanoDrop spectrophotometer) and integrity (using an Agilent Bioanalyzer). The GARP core made mRNA libraries and performed sequencing using an Illumina NovaSeq 6000 sequencing instrument. RSEM (65) and Bowtie 2 (66) were used for the alignments against human genome version GRCh38. Transcripts per million) values calculated by RSEM were log2-transformed. R package edgeR (67) was used for differential expression analysis. Volcano plot was generated to visualize the differential expression between SUM159 control and DAPK3 KO #3 cells using ggplot2 (68).
GSEA was performed using publicly available software (GSEA version 4.2) (69). The input of GSEA is the ranked gene list based on the signed (by direction of change) log10P-values from the differential expression analysis. The Hallmark gene sets within the Molecular Signatures Database (MSigDB) were used for GSEA (70). A FDR of 0.05 was utilized to define the statistical significance of GSEA in this study.
Mouse xenograft study
All animal procedures were approved by the Institutional Animal Care and Use Committee at BCM (protocol# AN-6934). For mouse xenograft experiments, luciferase-tagged SUM159 control or DAPK3 KO #3 cells were prepared in phosphate-buffered saline and mixed with Matrigel at 1:1 ratio (Corning Cat #354243). Five-week-old female NSG mice (Jackson Laboratory Strain #005557) were injected with 1 million cancer cells in 100 µL of solution in the mammary fat pad. Tumor growth was monitored by palpation, and tumor dimensions were measured using calipers. Tumor volume was calculated using the formula: Volume = 4/3 × 3.14 × (length/2)^2 × (width/2). Primary tumors were collected to measure the protein levels of DAPK3 (to confirm KO) and DSP, as well as proteomics profiling.
When tumors reached the size of ∼1,000 mm3, survival surgery was performed. Mice were anesthetized, and the primary tumors were excised aseptically. Care was taken to minimize trauma and bleeding during the procedure. After survival surgery, the in vivo bioluminescence imaging (BLI) was performed weekly to assay for sites of metastases (e.g. lung). For BLI, the anesthetized mice were imaged immediately after administration of 150 mg/kg D-luciferin via retro-orbital venous sinus. Imaging data were analyzed using Living Image software, and quantification was performed in regions of interest.
Label-free proteomics profiling
SUM159 DAPK3 KO #3 and control cells, as well as primary tumors from NSG mice orthotopically injected with SUM159 DAPK3 KO #3 or control cells, were subjected to proteomics profiling as described below.
The samples were dissolved in 20 µl of Evosep buffer A (0.1% formic acid) and loaded onto evotips. The samples were separated with an EV1109 column and the 44-min method on an evosep one. Data were acquired on a Thermo Exploris 480 with FAIMS set to −45 V. MS1 scans were acquired with a resolution of 30 K, a scan range of 500–740 m/z, 300% normalized AGC target, and 20 ms maximum injection time. Fifty-nine DIA scans were performed from 500 to 740 using 4 m/z isolation windows with optimized window placement. Stepped collision energy was used with normalized HCD collision energies of 22, 26, and 30. MS2 scans were acquired with 30 K resolving power with a scan range of 200–1,800, 300% normalized AGC target, and auto maximum injection time. All data were acquired with positive mode. Raw files were converted to mzml with MSConvert and analyzed with DIANN 1.8.1. FASTA files from uniprot for human and mouse were used individually and in combination. Library-free search was performed for all analysis with deep learning-based spectra, RTs, and ion mobility prediction. A precursor charge range of 2–6 was used with an m/z range of 500–740. Mass accuracy, MS1 accuracy, and scan window were set to auto and match bewteen runs was utilized.
Immunoprecipitation–mass spectrometry
For IP–MS, the pellets of SUM159 control or DAPK3 KO #3 cells were collected and lysed with lysis buffer (50 mM HEPES, pH 7.9, 0.5% Tween 20, 150 mM NaCl, 0.1 mM EDTA, and 10% glycerol). Five milligram of lysates was incubated with 5 μg DAPK3 antibody (Santa Cruz Biotechnology, sc-514223). Protein A Sepharose Beads (Cytiva) were then used to capture the antibody–antigen complex and then washed with lysis buffer four times. A mass spectrometric analysis was performed according to a previous study (71) with modifications. Immunoprecipitated proteins were eluted in Laemmli buffer and resolved on 4–12% SDS–PAGE gel (Invitrogen). The gels were stained with Coomassie blue. Gel lanes were sliced into six bands and in-gel digested with trypsin at 37 °C overnight. After digestion, peptides were extracted with acetonitrile and dried. Dried peptides were dissolved in a 5% methanol/0.1% formic acid solution. The samples were loaded through a 2-cm C18 trap, followed by 1 h 4–26% acetonitrile gradient on a 5-cm C18 column and were analyzed on a Orbitrap Fusion Lumos instrument (Thermo). The raw data were searched with Proteome Discoverer 2.0 Mascot (72) against a human RefSeq database, and the data were further grouped into gene products that were assigned homology and identification quality groups using an in-house developed algorithm.
Immunoprecipitation–immunoblotting
For IP–immunoblotting, 1 mg of lysates was incubated with 1 µg DAPK3 antibody (Santa Cruz Biotechnology, sc-514223), or 1 µg LUZP1 antibody (Bethyl Laboratories, #A304–634A), or 1 µg HA antibody (Santa Cruz Biotechnology, sc-2367). Protein A Magnetic beads (Bio-Rad, #161-4013) were then used to capture the antibody–antigen complex and washed as above. Immunoprecipitated proteins and the whole-cell lysate as input control were analyzed by immunoblotting.
Bioinformatics analysis
The clinical information, RNA-Seq data, and iTRAQ global proteome data of WHIM PDXs were obtained from previous publications (9, 19). The sample information, RNA-Seq data, and TMT-based global proteome data of breast cancer cell lines were downloaded from the DepMap dataset (20). The proteome data of prospective CPTAC human breast cancer dataset were obtained from the previous publication (27). The clinical information and RNA-Seq data of the METABRIC (39) and TCGA (40) datasets were accessed online using cBioPortal. The data for the pan-cancer analysis were obtained by personal correspondence from the NCI-CPTAC Pan-Cancer working group (to be published soon). The protein-based EMT hallmark score was calculated using the newly developed inferred pathway activity score (IPAS) method (28).
The number of publications corresponding to each gene related to the field of TNBC was performed using R package RISmed (16). The peer-reviewed publications were extracted from the PubMed database, and each gene symbol was searched with the key word “triple-negative breast cancer” to find relevant publications. These tasks were performed in an in-house R script, where the final query for this publication was performed on 2024 June 8.
The total of 58 proteins of the DAPK3 interactome were subjected to GO overrepresentation analysis to identify enriched molecular function (MF) terms using the R package “WebGestaltR” (73). Pearson’s correlation analysis was performed between the levels of DAPK3 protein and all the proteins quantified by the iTRAQ data of the PDXs (19). R package ggplot2 (68) was applied to generate volcano plots, bar plots, and scatter plots. R package ComplexHeatmap (74) was applied to generate heatmaps.
Statistical analysis
Student's t test, one-way ANOVA test (with Dunnett's or Tukey's multiple comparisons test), and two-way ANOVA test (with Dunnett's or Sidak's multiple comparisons test) were performed with GraphPad Prism 9. Wilcoxon sum-rank tests were performed with R. P-values < 0.05 were considered statistically significant. For box and whiskers plots, the box shows inter-quartile range (IQR) with median marked in center and the whiskers indicate 1.5 × IQR.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Drs Eric C. Chang, Gloria V. Echeverria, Jeff M. Rosen, and Thomas Westbrook at BCM for their scientific input.
Contributor Information
Junkai Wang, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA.
Anh M Tran-Huynh, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Graduate Program in Cancer and Cell Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Beom-Jun Kim, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA.
Doug W Chan, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Dan L. Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA.
Matthew V Holt, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA.
Diana Fandino, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA.
Xin Yu, Verna and Marrs McLean Department of Biochemistry and Molecular Pharmacology, Baylor College of Medicine, Houston, TX 77030, USA.
Xiaoli Qi, Verna and Marrs McLean Department of Biochemistry and Molecular Pharmacology, Baylor College of Medicine, Houston, TX 77030, USA.
Jin Wang, Verna and Marrs McLean Department of Biochemistry and Molecular Pharmacology, Baylor College of Medicine, Houston, TX 77030, USA.
Weijie Zhang, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA.
Yi-Hsuan Wu, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA.
Meenakshi Anurag, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Dan L. Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA; Department of Medicine, Baylor College of Medicine, Houston, TX 77030, USA.
Xiang H F Zhang, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Dan L. Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA; Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Bing Zhang, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Dan L. Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA; Department of Human and Molecular Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Chonghui Cheng, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Dan L. Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA; Department of Human and Molecular Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Charles E Foulds, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Dan L. Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA; Department of Medicine, Baylor College of Medicine, Houston, TX 77030, USA.
Matthew J Ellis, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Dan L. Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
The authors acknowledge the following funding support: From the National Institutes of Health (NIH): U01CA214125 (M.J.E., M.A.), U24CA210954 (B.Z.), P50 CA186784-06 (M.J.E.), NCI-SPORE Career Enhancement Award to M.A. (part of P50 CA186784-06), and a NCI-SPORE Developmental Research Project grant (supported by P50CA186784-03) to J.W. From the Cancer Prevention & Research Institute of Texas (CPRIT): RR160027 (B.Z., Scholar in Cancer Research), RP220050 (B.Z.), RR140027 (M.J.E., CPRIT Scholar in Cancer Research), RP170691 (CPRIT Core Facilities Support Grants), and RP210027 (A.M.T.-H., BCM Comprehensive Cancer Training Program). M.J.E. was a Susan G. Komen Foundation Scholar. M.J.E. and B.Z. were/are McNair Scholars supported by the The Robert and Janice McNair Foundation. This project was also supported in part by the GARP Core at BCM with funding from the National Institutes of Health (NIH) grant (P30CA125123). This work was also supported by generous gifts from Lisa and Ralph Eads.
Author Contributions
M.J.E., C.E.F., J.W., C.C., and X.H.-F.Z. designed the research (conceptualization); J.W., B.-J.K., D.F., M.V.H., A.M.T.-H., X.Y., X.Q., W.Z., and Y.-H.W. performed the research (investigation); B.-J.K., D.W.C., and M.V.H. developed the methodology; M.A. and B.Z. analyzed the proteogenomic data; J.W., A.M.T.-H., B.-J.K., M.V.H., C.C., C.E.F., and M.J.E. analyzed the data; C.E.F. and M.J.E. provided project administration; J.W., C.E.F., and M.J.E. wrote the paper; all authors edited the paper.
Data Availability
Raw mass spectrometry proteomics data from SUM159 cells (DAPK3 WT/KO) and DAPK3 IP–MS experiments have been deposited to the ProteomeXchange Consortium via the PRIDE (75) partner repository with the dataset identifier PXD049187. Raw transcriptomics data from SUM159 cells (DAPK3 WT/KO) have been deposited in NCBI's Gene Expression Omnibus (GEO) (76) and are accessible through GEO Series accession number GSE273142 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE273142).
References
- 1. Dent R, et al. . 2007. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 13:4429–4434. [DOI] [PubMed] [Google Scholar]
- 2. Yin L, Duan J-J, Bian X-W, Yu S-C. 2020. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science. 298:1912–1934. [DOI] [PubMed] [Google Scholar]
- 4. Engh RA, Bossemeyer D. 2002. Structural aspects of protein kinase control-role of conformational flexibility. Pharmacol Ther. 93:99–111. [DOI] [PubMed] [Google Scholar]
- 5. Vogel CL, et al. . 2002. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 20:719–726. [DOI] [PubMed] [Google Scholar]
- 6. André F, et al. . 2019. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 380:1929–1940. [DOI] [PubMed] [Google Scholar]
- 7. Turner NC, et al. . 2023. Capivasertib in hormone receptor-positive advanced breast cancer. N Engl J Med. 388:2058–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saltzman AB, et al. . 2024. Kinase inhibitor pulldown assay (KiP) for clinical proteomics. Clin Proteomics. 21:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li S, et al. . 2013. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 4:1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shiloh R, Bialik S, Kimchi A. 2014. The DAPK family: a structure-function analysis. Apoptosis. 19:286–297. [DOI] [PubMed] [Google Scholar]
- 11. Kawai T, Matsumoto M, Takeda K, Sanjo H, Akira S. 1998. ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol Cell Biol. 18:1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawai T, Akira S, Reed JC. 2003. ZIP kinase triggers apoptosis from nuclear PML oncogenic domains. Mol Cell Biol. 23:6174–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Niiro N, Ikebe M. 2001. Zipper-interacting protein kinase induces Ca(2+)-free smooth muscle contraction via myosin light chain phosphorylation. J Biol Chem. 276:29567–29574. [DOI] [PubMed] [Google Scholar]
- 14. Morandi A, Plaza-Menacho I, Isacke CM. 2011. RET in breast cancer: functional and therapeutic implications. Trends Mol Med. 17:149–157. [DOI] [PubMed] [Google Scholar]
- 15. Farabaugh SM, Boone DN, Lee AV. 2015. Role of IGF1R in breast cancer subtypes, stemness, and lineage differentiation. Front Endocrinol (Lausanne). 6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kovalchik S. 2021. RISmed: download content from NCBI Databases. https://cran.r-project.org/web/packages/RISmed/index.html
- 17. Nakai K, Hung M-C, Yamaguchi H. 2016. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res. 6:1609–1623. [PMC free article] [PubMed] [Google Scholar]
- 18. Toy KA, et al. . 2015. Tyrosine kinase discoidin domain receptors DDR1 and DDR2 are coordinately deregulated in triple-negative breast cancer. Breast Cancer Res Treat. 150:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang K-L, et al. . 2017. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat Commun. 8:14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghandi M, et al. . 2019. Next-generation characterization of the cancer cell line encyclopedia. Nature. 569:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tewari M, et al. . 1995. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 81:801–809. [DOI] [PubMed] [Google Scholar]
- 22. Li X, He S, Ma B. 2020. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 19:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haystead TAJ. 2005. ZIP kinase, a key regulator of myosin protein phosphatase 1. Cell Signal. 17:1313–1322. [DOI] [PubMed] [Google Scholar]
- 24. Deng J-T, Bhaidani S, Sutherland C, MacDonald JA, Walsh MP. 2019. Rho-associated kinase and zipper-interacting protein kinase, but not myosin light chain kinase, are involved in the regulation of myosin phosphorylation in serum-stimulated human arterial smooth muscle cells. PLoS One. 14:e0226406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moffat LD, et al. . 2011. Chemical genetics of zipper-interacting protein kinase reveal myosin light chain as a bona fide substrate in permeabilized arterial smooth muscle. J Biol Chem. 286:36978–36991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brognard J, Zhang Y-W, Puto LA, Hunter T. 2011. Cancer-associated loss-of-function mutations implicate DAPK3 as a tumor-suppressing kinase. Cancer Res. 71:3152–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krug K, et al. . 2020. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell. 183:1436–1456.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reva B, et al. . 2020. Prioritization of prostate cancer to immune checkpoint therapy by ranking tumors along IFN-γ axis and identification of immune resistance mechanisms. bioRxiv 2020.10.19.345629, : not peer reviewed. 10.1101/2020.10.19.345629 [DOI] [Google Scholar]
- 29. Dou Y, et al. . 2023. Proteogenomic insights suggest druggable pathways in endometrial carcinoma. Cancer Cell. 41:1586–1605.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dusek RL, Attardi LD. 2011. Desmosomes: new perpetrators in tumour suppression. Nat Rev Cancer. 11:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aberle H, et al. . 1995. The human plakoglobin gene localizes on chromosome 17q21 and is subjected to loss of heterozygosity in breast and ovarian cancers. Proc Natl Acad Sci U S A. 92:6384–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chidgey M, Dawson C. 2007. Desmosomes: a role in cancer? Br J Cancer. 96:1783–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davies EL, et al. . 1999. The immunohistochemical expression of desmoplakin and its role in vivo in the progression and metastasis of breast cancer. Eur J Cancer. 35:902–907. [DOI] [PubMed] [Google Scholar]
- 34. Yang L, et al. . 2012. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/β-catenin signaling pathway in human lung cancer. Carcinogenesis. 33:1863–1870. [DOI] [PubMed] [Google Scholar]
- 35. Winn RA, et al. . 2002. gamma-Catenin expression is reduced or absent in a subset of human lung cancers and re-expression inhibits transformed cell growth. Oncogene. 21:7497–7506. [DOI] [PubMed] [Google Scholar]
- 36. Chun MGH, Hanahan D. 2010. Genetic deletion of the desmosomal component desmoplakin promotes tumor microinvasion in a mouse model of pancreatic neuroendocrine carcinogenesis. PLoS Genet. 6:e1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Landschulz WH, Johnson PF, McKnight SL. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 240:1759–1764. [DOI] [PubMed] [Google Scholar]
- 38. Mellacheruvu D, et al. . 2013. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 10:730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Curtis C, et al. . 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 486:346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoadley KA, et al. . 2018. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 173:291–304.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bi J, et al. . 2009. Downregulation of ZIP kinase is associated with tumor invasion, metastasis and poor prognosis in gastric cancer. Int J Cancer. 124:1587–1593. [DOI] [PubMed] [Google Scholar]
- 42. Das TP, et al. . 2016. Activation of AKT negatively regulates the pro-apoptotic function of death-associated protein kinase 3 (DAPK3) in prostate cancer. Cancer Lett. 377:134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kocher BA, White LS, Piwnica-Worms D. 2015. DAPK3 suppresses acini morphogenesis and is required for mouse development. Mol Cancer Res. 13:358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan J, et al. . 2017. Anacardic acid induces cell apoptosis of prostatic cancer through autophagy by ER stress/DAPK3/Akt signaling pathway. Oncol Rep. 38:1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kake S, Usui T, Ohama T, Yamawaki H, Sato K. 2017. Death-associated protein kinase 3 controls the tumor progression of A549 cells through ERK MAPK/c-Myc signaling. Oncol Rep. 37:1100–1106. [DOI] [PubMed] [Google Scholar]
- 46. Takahashi M, et al. . 2021. The tumor suppressor kinase DAPK3 drives tumor-intrinsic immunity through the STING-IFN-β pathway. Nat Immunol. 22:485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bouhaddou M, et al. . 2019. Mapping the protein-protein and genetic interactions of cancer to guide precision medicine. Curr Opin Genet Dev. 54:110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Del-Toro N, et al. . 2019. Capturing variation impact on molecular interactions in the IMEx consortium mutations data set. Nat Commun. 10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim M, et al. . 2021. A protein interaction landscape of breast cancer. Science. 374:eabf3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paczkowska M, et al. . 2020. Integrative pathway enrichment analysis of multivariate omics data. Nat Commun. 11:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reyna MA, et al. . 2020. Pathway and network analysis of more than 2500 whole cancer genomes. Nat Commun. 11:729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hyodo T, et al. . 2023. Leucine zipper protein 1 (LUZP1) regulates the constriction velocity of the contractile ring during cytokinesis. FEBS J. 291:927–944. 10.1111/febs.17017 [DOI] [PubMed] [Google Scholar]
- 53. Sun DS, et al. . 1996. Identification, molecular characterization, and chromosomal localization of the cDNA encoding a novel leucine zipper motif-containing protein. Genomics. 36:54–62. [DOI] [PubMed] [Google Scholar]
- 54. Hsu C-Y, et al. . 2008. LUZP deficiency affects neural tube closure during brain development. Biochem Biophys Res Commun. 376:466–471. [DOI] [PubMed] [Google Scholar]
- 55. Lee MW, Chang AC, Sun DS, Hsu CY, Chang NC. 2001. Restricted expression of LUZP in neural lineage cells: a study in embryonic stem cells. J Biomed Sci. 8:504–511. [DOI] [PubMed] [Google Scholar]
- 56. Krebs AR, et al. . 2010. ATAC and mediator coactivators form a stable complex and regulate a set of non-coding RNA genes. EMBO Rep. 11:541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang J, Nakamura F. 2019. Identification of filamin A mechanobinding partner II: fimbacin is a novel actin cross-linking and filamin A binding protein. Biochemistry. 58:4737–4743. [DOI] [PubMed] [Google Scholar]
- 58. Juric D, et al. . 2019. Alpelisib plus fulvestrant in PIK3CA-altered and PIK3CA-wild-type estrogen receptor-positive advanced breast cancer: a phase 1b clinical trial. JAMA Oncol. 5:e184475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mertins P, et al. . 2016. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 534:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Satpathy S, et al. . 2020. Microscaled proteogenomic methods for precision oncology. Nat Commun. 11:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cho ES, et al. . 2020. Breast cancer subtypes underlying EMT-mediated catabolic metabolism. Cells. 9:2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kapałczyńska M, et al. . 2018. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci. 14:910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vinci M, et al. . 2012. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Matsunuma R, et al. . 2018. DPYSL3 modulates mitosis, migration, and epithelial-to-mesenchymal transition in claudin-low breast cancer. Proc Natl Acad Sci U S A. 115:E11978–E11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wickham H. 2016. Ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- 69. Subramanian A, et al. . 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liberzon A, et al. . 2015. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Malovannaya A, et al. . 2011. Analysis of the human endogenous coregulator complexome. Cell. 145:787–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 20:3551–3567. [DOI] [PubMed] [Google Scholar]
- 73. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47:W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32:2847–2849. [DOI] [PubMed] [Google Scholar]
- 75. Perez-Riverol Y, et al. . 2022. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50:D543–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Edgar R, Domrachev M, Lash AE. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry proteomics data from SUM159 cells (DAPK3 WT/KO) and DAPK3 IP–MS experiments have been deposited to the ProteomeXchange Consortium via the PRIDE (75) partner repository with the dataset identifier PXD049187. Raw transcriptomics data from SUM159 cells (DAPK3 WT/KO) have been deposited in NCBI's Gene Expression Omnibus (GEO) (76) and are accessible through GEO Series accession number GSE273142 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE273142).