Abstract
Dendritic cells (DCs) efficiently bind and transmit human immunodeficiency virus (HIV) to cocultured T cells and so may play an important role in HIV transmission. DC-SIGN, a novel C-type lectin that is expressed in DCs, has recently been shown to bind R5 HIV type 1 (HIV-1) strains and a laboratory-adapted X4 strain. To characterize the interaction of DC-SIGN with primate lentiviruses, we investigated the structural determinants of DC-SIGN required for virus binding and transmission to permissive cells. We constructed a panel of DC-SIGN mutants and established conditions which allowed comparable cell surface expression of all mutants. We found that R5, X4, and R5X4 HIV-1 isolates as well as simian immunodeficiency and HIV-2 strains bound to DC-SIGN and could be transmitted to CD4/coreceptor-positive cell types. DC-SIGN contains a single N-linked carbohydrate chain that is important for efficient cell surface expression but is not required for DC-SIGN-mediated virus binding and transmission. In contrast, C-terminal deletions removing either the lectin binding domain or the repeat region abrogated DC-SIGN function. Trypsin-EDTA treatment inhibited DC-SIGN mediated infection, indicating that virus was maintained at the surface of the DC-SIGN-expressing cells used in this study. Finally, quantitative fluorescence-activated cell sorting analysis of AU1-tagged DC-SIGN revealed that the efficiency of virus transmission was strongly affected by variations in DC-SIGN expression levels. Thus, variations in DC-SIGN expression levels on DCs could greatly affect the susceptibility of human individuals to HIV infection.
The entry of human immunodeficiency virus type 1 (HIV-1) into cells is a multistep process that requires, at the minimum, interactions between the viral Env protein and two cell surface receptors (1, 4, 8, 11, 12, 15). The CD4 molecule serves as a receptor for all primary HIV-1 strains studied to date and induces conformational changes in the gp120 subunit of Env that enable it to interact with a coreceptor (20, 31, 33), generally either the chemokine receptor CCR5 (R5 strains) or CXCR4 (X4 strains) (9). While binding to CD4 is required for efficient virus infection, attachment of virus to the cell surface can be mediated by interactions with a variety of molecules, only some of which have been well characterized (22, 32). Attachment to the cell surface per se can be a limiting step in the entry pathway. In vitro, infection of cell lines and peripheral blood mononuclear cells by HIV-1 can often be enhanced by inclusion of polycations in the virus inoculum or by centrifuging virus onto the cell surface (23). Infection of activated T cells can also be enhanced by first binding HIV-1 to dendritic cells (DCs) (3, 16). After removal of unbound virus, addition of activated T cells results in efficient transmission of virus to these cellular targets and a robust infection.
Recently, a type II integral membrane protein termed DC-SIGN has been shown to mediate binding of primary R5 and laboratory-adapted X4 strains of HIV-1 to DCs (16, 17). We have shown that a closely related homologue, termed DC-SIGNR (for DC-SIGN related [29]), also functions as an attachment factor for HIV-1, HIV-2, and simian immunodeficiency virus (SIV) (26). DC-SIGN contains a C-type (i.e., calcium-dependent) lectin-like domain that presumably mediates this process. Virus bound to DC-SIGN on DCs can remain infectious for several days, and virus-pulsed DCs efficiently transmit virus when they come into contact with CD4- and coreceptor-positive cell types (16). Transmission can be blocked by antibodies to DC-SIGN. Thus, DC-SIGN appears to be responsible for the ability of DCs to efficiently mediate infection of T cells in trans. Because DCs migrate from peripheral mucosal tissues to the lymph node upon encounter of antigen (2, 30), it has been proposed that HIV uses DCs as carriers allowing the virus to access lymphoid tissue, the major site of replication (16).
In this work, we confirm and extend the initial studies on this interesting virus binding factor. We demonstrate that SIV, HIV-2, and primary X4, R5, and R5X4 HIV-1 strains can all bind to DC-SIGN and be presented to susceptible cells. Mutagenesis studies indicated that DC-SIGN contains a single N-linked glycosylation site that is utilized, though glycosylation is not required for DC-SIGN function. The C-type lectin-like domain plays an important role in virus binding and transmission, since deletion of this domain abrogated these functions. Importantly, the ability of DC-SIGN to bind and transmit virus was strongly dependent on DC-SIGN surface expression levels. The threshold levels below which DC-SIGN concentrations became limiting for virus binding and transmission varied between different virus strains. Thus, DC-SIGN appears to be a universal binding factor for primate immunodeficiency viruses, at least under optimized conditions in vitro.
MATERIALS AND METHODS
Mutagenesis of DC-SIGN.
The DC-SIGN coding sequence was PCR amplified from cDNA obtained from PBMCs and DCs. Primers used for PCR amplification were p5-DC(5′-CCGGATCCAGAGTGGGGTGACATGAGTG-3′) and p3-DC (5′-CCGAATTCGGAAGTTCT-GCTACGCAGGAG-3′). The underlined BamHI and EcoRI restriction sites were used for cloning into pcDNA3 (Invitrogen, Carlsbad, Calif.). The amino acid sequence obtained was identical to GenBank sequence M98457. For detection of DC-SIGN expression via immunostaining, a C-terminal AU1 tag was added to the DC-SIGN sequence using primers p5-DC and p3-DC-AU1 (5′-CCGAATTCGTTATATGTATCTGTAGGTGTCCGCAGGAGGGGGGTTTGGGGTGGCAGG-3′). C-terminal deletions were introduced by PCR mutagenesis. Primers p5-DC and p3-D1 (5′-CC GAATTCGTTATATGTATCTGTAGGTGTCCAGGCGTTCCACTGCAGC CT-3′) were used for generation of variant ΔCter1, and primers p5-DC and p3-D2 (5′-CCGAATTCGTTATATGTATCTGTAGGTGTCCTCCTGCAGCTTAGATTTCT-3′)were used for generation of ΔCter2. The repeat region was deleted via overlap extension PCR (creating ΔRepeat). The 5′ PCR fragment was amplified using primers p5DC and p3-ΔRepeat (5′-CCATTCCCAGGGACAGGGGTGGCAGTTCTGGTAGATCGCGTCTTGCCTG-3′), and the 3′ PCR fragment was generated using primers p5N-lectin (5′-TGCCACCCCTGTCCCTGGGAATGG-3′) and p3DC-AU1. Both fragments were gel purified and used as the template in a second PCR with primers p5-DC and p3DC-AU1. Variants N80A and ΔInt + N80A were generated similarly but using primers which overlap the sequence encoding the glycosylation signal and the C terminus of the repeat region. All mutants were engineered to contain the C-terminal AU1 tag, and all PCR-amplified fragments were sequenced to ensure that only the intended changes were present.
Generation of replication-competent luciferase reporter viruses.
To generate HIV-1 luciferase reporter virus, a 2-kb BamHI/XhoI restriction fragment derived from pNL4-3.Luc.R−E− (6), containing the 3′ end of the viral genome and a luciferase reporter gene in place of nef, was inserted into a modified pBR322 vector containing the full-length HIV-1 NL4-3 provirus. This construct, named pBRNL4-3dnefluc, yields replication-competent HIV-1 luciferase reporter viruses after transient transfection of 293T cells. The env-defective pNL4-3.Luc.R−E− luciferase reporter construct was kindly provided by Nathaniel Landau (Salk Institute for Biological Studies, La Jolla, Calif.). For generation of the HIV-2 luciferase reporter virus, the luciferase gene was introduced into the proviral genome of HIV-2 Rod10. The full-length proviral genome of HIV-2 Rod10 (27), kindly provided by Klaus Strebel (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md.), was inserted into a modified pBR322 vector using standard cloning procedures to generate pBRod10. Site-directed mutagenesis was performed by spliced overlap extension PCR to replace the nef gene of pBRod10 with the luciferase reporter gene. Briefly, the env/nef region was amplified using primers K29-roddn1 (5′-GTGCGAGTGGATCCAAG-3′) and K30b-roddn2b (5′-CCCTTGTTTTTTATTAAA TACGCGTCGCGAGCGCGGCCGCTCACAGGAGGGCGATTTCTGC-3′), and the nef/long terminal repeat region was amplified using primers K76b-roduboxb (5′-CGACGCGTATTTAATAAAAAACAAGGGG-3′) and K8-rodltr3 (5′-CCGGAATTCCCGGGAATCTTGCTTCTAACTGGCAGC-3′). The 5′ and 3′ PCR products were gel purified, mixed at equimolar amounts, and subjected to a second PCR with primers K29 and K8. The PCR products were inserted into the pBRod10 vector by using the BamHI (bp 8569) and EcoRI (underlined) sites in the HIV-2 Rod envelope and the vector sequences flanking the 3′ end of the provirus. These modifications deleted bp 8725 to 8918 bp of the Rod10 nef gene and inserted unique NotI and MluI sites (bold) downstream of the Rod10 env gene. Subsequently, the luciferase gene was PCR amplified using primers K1-LUCATG (5′-ATAAGAATGCGGCCGCATGGAAGACGCCAAAAAC-3′) and K2-LUCTAA (5′-AACACGACGCGTTTACAATTTGGACTTTCCGC-3′). The PCR product was digested with NotI and MluI, purified, and cloned into the modified pBRod10 vector mentioned above. Numbers refer to the published HIV2 Rod sequence (Genbank accession number M15390) (5). Sequence analysis of the PCR-derived insert confirmed that only the intended changes were present in the pBRrod10Δnefluc luciferase reporter construct. The construction of replication-competent SIVmac239 harboring the luciferase gene in place of nef has been described previously (25).
Cell culture and production of virus stocks.
C8166 cells were maintained in RPMI 1640 with 10% fetal calf serum (FCS) and antibiotics. 293T cells were cultivated in Dulbecco modified Eagle medium (DMEM) with 10% FCS and antibiotics. All cells were grown at 37°C and 5% CO2. HIV-1 stocks were obtained from the Viral/Cell/Molecular Core of the Penn Center for AIDS Research. Replication-competent luciferase reporter viruses were produced by transfection of 293T cells using a calcium phosphate precipitation protocol as described previously (18).
p24 binding assay.
Binding of virus particles to DC-SIGN-expressing 293T cells was assessed by measuring cell-associated p24 levels. 293T cells were seeded in T25 flasks, incubated overnight, and transiently transfected with expression vectors encoding the DC-SIGN variants and a pcDNA3 control plasmid, using the calcium phosphate method as described above. At 24 or 48 h after transfection, cells were seeded in 48- or 96-well plates. Cells were grown for 24 h, and subsequently DC-SIGN expression and virus binding were analyzed in parallel. Expression was controlled in a fluorescence-activated cell sorting (FACS) assay as described below. For virus binding, 5 ng of p24 antigen was added in a total volume of 50 μl. After 3 to 5 h of incubation at 37°C, the supernatant was removed and cells were washed vigorously with fresh DMEM. Thereafter, cells were lysed in 100 μl of 0.5% Triton X-100 in H2O. The amount of bound virus was assessed using a commercially available p24 enzyme-linked immunosorbent assay (ELISA) (Coulter Beckman, Miami, Fla.).
FACS analysis of DC-SIGN expression.
To assess expression efficiency of the DC-SIGN variants, 293T cells were transfected with the indicated expression plasmids as described above and grown at 32°C. At 48 h after transfection, cells were harvested, washed with phosphate-buffered saline, (PBS) and recovered in ice-cold PBS containing 3% FCS and 0.05% sodium azide (FACS buffer). For staining of the AU1-tagged mutants, approximately 200,000 cells were incubated with 1 μg of anti-AU1 antibody (Covance, Richmond, Calif.) in a total volume of 100 μl. After a 30-min incubation at 4°C, cells were washed with FACS buffer and recovered in 100 μl of FACS buffer containing 1 μl of phycoerythrin-coupled anti-mouse antibody (Vector Laboratories, Burlingame, Calif.). Cells were incubated for 30 min at 4°C, washed with FACS buffer, and recovered in FACS buffer containing 2% paraformaldehyde. Staining of transfected cells was analyzed using a fluorescence-activated cell sorter (FACScan; Becton Dickinson).
Assessment of DC-SIGN-mediated infection in trans.
The efficiency of DC-SIGN-mediated virus transfer was assessed in a cocultivation assay. 293T cells were transfected with the DC-SIGN variants and 24 h after transfection were seeded in 48-well dishes. The next day, expression was analyzed via FACS. To determine virus transmission, the transfected cells were incubated with 10 ng of luciferase reporter virus for 3 to 5 h at 37°C. Thereafter, cells were washed several times with fresh DMEM and cocultivated with C8166 cells. Two days after cocultivation, the medium was changed; 24 h thereafter, the cells were lysed with a commercially available lysis buffer (Promega, Madison, Ws.). Luciferase activity in 30 μl of cell lysate was determined using a commercially available kit (Promega).
Quantification of DC-SIGN copy numbers required for virus transmission.
To investigate the importance of DC-SIGN surface expression levels for virus transmission, a cell line which inducibly expresses DC-SIGN was generated. Commercially available 293 T-Rex cells (Invitrogen), which contain the gene for the tet repressor, were stably transfected with AU1-tagged DC-SIGN. DC-SIGN was expressed under the control of a cytomegalovirus promoter, which contains binding sites for the tet repressor. Rising concentrations of doxycycline in the culture medium lead to increased dissociation of the repressor from the promoter and subsequent activation of DC-SIGN expression. To assess the copy number of surface DC-SIGN required for efficient virus transmission, cells were seeded in duplicate wells and DC-SIGN expression was induced by addition of doxycycline. The following day, one panel of cells was used in the virus transmission assay as described above, whereas the other panel was used to quantify DC-SIGN surface expression by a quantitative FACS assay (QFACS) as described previously (21). Briefly, QFACS was performed by converting the geometrical mean channel fluorescence (GMCF) in antibody binding sites (ABS) by using a standardized microbeads kit (Sigma). Saturating amounts (10 μg/ml) of anti-AU1 (Covance) were added to about 100,000 beads, and the beads were processed like the samples being quantitated. An anti-mouse Fab fragment conjugated to phycoerythrin (Caltag, Burlingame, Calif.) was used as secondary antibody. The staining procedure was carried out according to the manufacturer's instructions. The binding capacities of the stained microbeads were regressed against the corresponding GMCF of each bead population, and the GMCF of the antigen analyzed on the sample cells was converted to ABS per cell by comparison with the regression curve generated. The GMCF of mouse immunoglobulins for each experiment was converted to ABS and subtracted from the ABS value obtained with the experimental sample. Since no anti-AU1 antibody coupled to an adequate fluorochrome was available, an indirect method of detection was used to quantify the ABS (primary anti-AU1 antibody; secondary phycoerythrin-conjugated anti-mouse Fab). Therefore, the degree of confidence on the numbers generated cannot be as accurate as with a directly conjugated anti-AU1 (up to threefold difference).
Western blot analysis.
293T cells were transfected in 12-well dishes, 12 to 16 h after transfection the medium was changed, and 48 h after transfection the cells were harvested. The cells were washed in PBS and lysed in 300 μl of sodium dodecyl sulfate sample buffer. Expression of DC-SIGN in cleared lysates was analyzed by immunoblotting. Proteins were detected with a 1:10,000 dilution of anti-AU1 antibody (Covance).
RESULTS
Generation of DC-SIGN mutants.
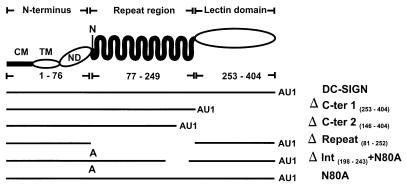
DC-SIGN is a 404-amino-acid-long type II transmembrane protein for which several distinct regions have been defined by amino acid homology (Fig. 1) (17). The N-terminal 40 amino acids are located in the cytoplasm, amino acids 41 to 61 constitute the transmembrane domain, and amino acids 62 to 404 form the ectodomain of the protein. The ectodomain consists of a short N-terminal region (amino acids 62 to 76), a domain containing seven complete copies and one incomplete copy of the sequence GELPEKSKMQEIYQELTRLKAAV, and a C-type lectin-like domain (amino acids 253 to 404). The N terminus of the repeat region harbors the protein's single N-linked glycosylation signal. The regions of DC-SIGN involved in HIV-1 binding and transmission have not been defined, nor is it known if DC-SIGN supports binding and transmission of HIV-2 and SIV.
FIG. 1.
Schematic representation of DC-SIGN and the mutants analyzed. DC-SIGN is a type II transmembrane protein for which several domains have been identified by sequence analysis: a cytoplasmic domain (CM), a transmembrane domain (TM), an N-terminal domain (ND), a repeat region, and a lectin-like domain. The asparagine at position 80, which is part of an N-linked glycosylation signal, is indicated by an N. The structures of the DC-SIGN mutants used in this study are indicated schematically; an A represents substitution of an alanine at position 80, and AU1 indicates the presence of an antigenic tag introduced to make detection of the proteins possible.
To confirm the role of DC-SIGN in HIV-1 binding and transmission, to determine if HIV-2 and SIV also interact with DC-SIGN, and to identify regions of DC-SIGN involved in these functions, we cloned and expressed DC-SIGN and generated a panel of DC-SIGN variants (Fig. 1). Employing PCR mutagenesis, we removed the lectin-like domain alone (ΔCter-1) or in combination with the 50 C-terminal amino acids of the repeat region (ΔC-ter2). Variant ΔRepeat was engineered to contain an internal deletion, which removed the repeat region but left the glycosylation signal, the N-terminal region, and the lectin-like domain intact. To determine if DC-SIGN is glycosylated and if carbohydrate addition affects its function, the potential N-linked glycosylation site was eliminated by changing the asparagine at position 80 to an alanine (N80A). In addition, this amino acid change was combined with a deletion of amino acids 198 to 243 located at the C terminus of the repeat region (ΔInt + N80A). Since no DC-SIGN-specific antibodies were available, a C-terminal AU1 tag was added to all DC-SIGN constructs. All variants were expressed under control of the cytomegalovirus promoter of pcDNA3.
Surface expression of the mutated DC-SIGN proteins.
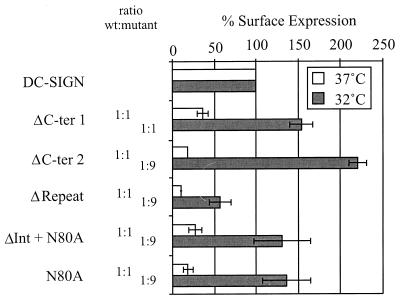
We investigated if the mutations introduced into DC-SIGN affected surface expression of the protein. The variants were transiently expressed in 293T cells, and surface expression levels were determined by FACS analysis using a monoclonal antibody directed against the AU1 antigenic tag. When equivalent amounts of DNA were used for the transfections and the cells were subsequently incubated at 37°C, expression of the various mutants ranged from 10 to 36% of wild-type (wt) DC-SIGN levels (Fig. 2). Thus, all mutations reduced but did not abrogate surface expression. Therefore, we sought conditions under which we could achieve comparable expression of all DC-SIGN constructs so that their abilities to support virus binding could be directly compared to that of wt DC-SIGN. To do this, we incubated the transfected cells at 32°C, since reduced temperature can facilitate protein folding and subsequent transport to the cell surface (10). Indeed, all mutants were expressed more efficiently at 32°C, but only ΔCter1 was expressed as efficiently as wt DC-SIGN (Fig. 2). Therefore, we titrated the amount of wt DC-SIGN plasmid, finding that ninefold less plasmid than mutant constructs gave conditions under which three DC-SIGN mutants were expressed at somewhat higher levels than wt DC-SIGN, while expression of the ΔRepeat variant reached 56% of the level for the parental construct (Fig. 2). Subsequent functional studies of the DC-SIGN variants were carried out using these conditions.
FIG. 2.
Surface expression of DC-SIGN variants. 293T cells were transfected with plasmid DNA encoding the indicated DC-SIGN mutants and incubated at 37 or 32°C. At 48 h after transfection, cells were stained with an anti-AU1 antibody and analyzed via FACS. Expression efficiency is indicated relative to that of wt DC-SIGN, and the amount of each mutant plasmid relative to that of wt plasmid is indicated. The data shown represent the average of three independent experiments.
Expression and glycosylation of the DC-SIGN mutants.
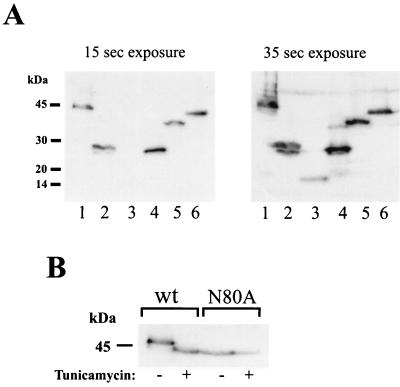
Expression of the DC-SIGN mutants in transfected cells was also investigated by Western blotting. 293T cells were transfected with the indicated mutants, and expression was analyzed 48 h after transfection. With the exception of ΔCter-2, which could be detected only after a prolonged exposure, all mutants were readily detectable (Fig. 3A). These data indicate that the mutations mainly interfere with protein folding and transport to the cell surface but have little impact on protein expression or stability. DC-SIGN harbors a glycosylation signal at the N terminus of the repeat region, with Asn 80 being potentially glycosylated. This glycosylation signal is disrupted in mutant N80A. To determine if DC-SIGN is glycosylated, cells were transfected with wt DC-SIGN or the N80A variant and incubated with or without tunicamycin, a compound that inhibits N-linked glycosylation. Tunicamycin treatment of wt DC-SIGN-transfected cells caused an increase in the gel mobility of the protein, causing it to comigrate with the N80A variant (Fig. 3B). Migration of the N80A variant was not affected by tunicamycin treatment. Therefore, the glycosylation site in the N-terminal domain of DC-SIGN is utilized.
FIG. 3.
Expression and glycosylation of DC-SIGN variants in transfected cells. (A) 293T cells were transfected with equal amounts of plasmid DNA encoding wt-DC-SIGN (lane 1) and mutants ΔCter1 (lane 2), ΔCter2 (lane 3),ΔRepeat (lane 4), ΔInt + N80A (lane 5), and N80A (lane 6). Two days after transfection, the cells were lysed and DC-SIGN expression was analyzed via immunoblotting using the anti-AU1 antibody as described in Materials and Methods. Two different exposure times are shown. (B) DC-SIGN is glycosylated. 293T cells were transfected with wt DC-SIGN or the N80A variant, incubated with tunicamycin, and analyzed via immunoblotting as described above. Tunicamycin treatment increased the gel mobility of wt DC-SIGN but not that of the N80A variant. Comparable results were obtained in an independent experiment.
HIV-1 isolates bind with different efficiency to DC-SIGN.
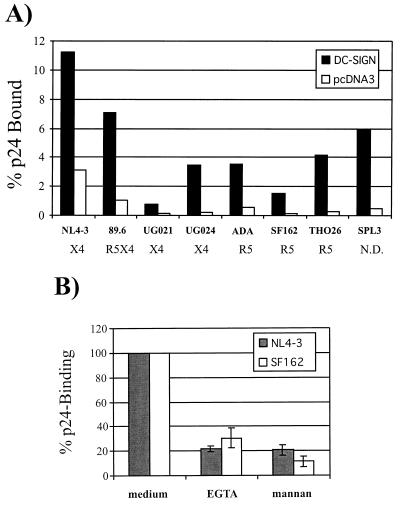
DC-SIGN has been shown to bind a number of R5 HIV-1 strains and a single laboratory-adapted X4 virus (16). To evaluate if different HIV isolates bind DC-SIGN with different efficiencies, we quantified binding of seven HIV-1 isolates including R5, R5X4, and primary X4 virus strains as well as the laboratory-adapted NL4-3 virus. Virus was added to 293T cells expressing DC-SIGN for 3 h. Thereafter, the cells were washed and lysed in detergent, and the amount of viral p24 antigen present in the lysate was determined by antigen capture ELISA. All virus strains tested bound to DC-SIGN-positive cells more efficiently than to cells transfected with empty vector (Fig. 4A). The increase in p24 association varied between 3.6- and 16.9-fold. We did not observe an obvious correlation between the binding efficiency and the viral phenotype. The laboratory-adapted X4 viral isolate NL4-3 and the primary R5X4 virus strain 89.6 bound to DC-SIGN with the highest efficiencies, with 11.3 and 7.1%, respectively, of the input virus binding. However, these virus strains also bound with the highest efficiency to pcDNA3-transfected control cells. These findings indicate that while all virus strains tested thus far bind to DC-SIGN, there may be differences in binding efficiencies.
FIG. 4.
Binding of HIV-1 to DC-SIGN-transfected cells. (A) The DC-SIGN variants were transiently expressed in 293T cells. Cells were incubated with equal amounts of p24 antigen for each indicated virus, vigorously washed, and lysed in 0.5% Triton X-100, and p24 content quantified via ELISA. The data are shown as percentage of recovered antigen. The phenotype of each virus (e.g., X4, R5X4, or R5) is shown. Similar results were obtained in two independent experiments. (B) The binding assay was carried out as described above except that the cells were incubated in media containing EGTA (5 mM) and mannan (20 μg/ml), prior to the addition of virus.
It has been reported previously that HIV-1 Env protein binds to DC-SIGN-expressing cells and that binding can be inhibited by mannan and EGTA (16). Therefore, we tested if binding of virus to 293T cells expressing DC-SIGN can be inhibited by these reagents. The virus binding assay was carried out as described above except that the cells were incubated with EGTA or mannan prior to addition of virus (Fig. 4B). Preincubation with both reagents strongly reduced binding of the NL4-3 and the SF162 virus strains, indicating that HIV binding by DC-SIGN involves carbohydrate recognition.
The repeat and lectin-like regions in DC-SIGN are involved in HIV-1 binding.
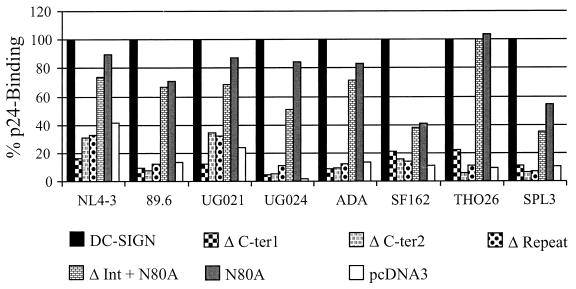
Next, we determined which regions of the DC-SIGN protein are required for virus binding. Expression of the mutants was performed under the conditions shown in Fig. 2, in which we showed by FACS that comparable surface expression levels were achieved. Deletion of the lectin-like domain (ΔCter1 and ΔCter2) as well as of the repeat region abrogated efficient p24 binding of all isolates tested (Fig. 5). In contrast, mutation of the glycosylation site did not affect efficient binding of most isolates to DC-SIGN-expressing cells. Binding of virus to cells expressing the N80A variant ranged from 71% (89.6) to 104% (TH026) of wt efficiency for most isolates, with the exception of the SF162 and SPL3 isolates, which bound with somewhat reduced efficiency to this variant. The deletion of amino acids 198 to 243 in addition to mutation of the glycosylation site did not result in a substantial loss of p24 binding compared to the N80A mutant, indicating that this part of the repeat region does not play a major role in p24 binding (Fig. 5).
FIG. 5.
Binding of HIV-1 to DC-SIGN mutants. The indicated DC-SIGN mutants were transfected into 293T cells, and equal expression was monitored as described in the legend to Fig. 2. The binding assays were performed as described in the legend to Fig. 4. Values were normalized to p24 binding to wt DC-SIGN-transfected cells for each virus type. Similar results were obtained in two independent experiments.
DC-SIGN mediates transmission of HIV-1, HIV-2 and SIV.
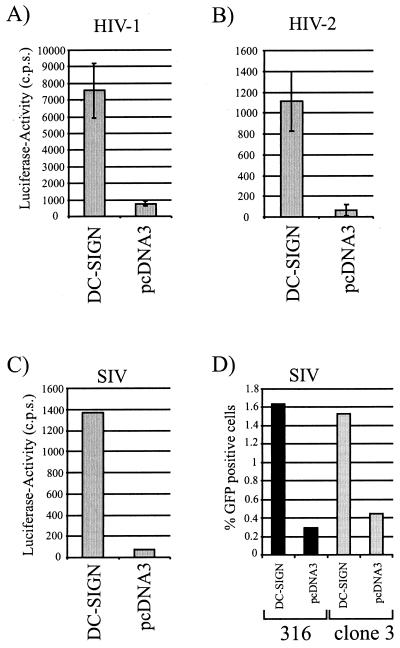
After binding to DC-SIGN, HIV-1 can be efficiently transmitted to susceptible cells (16). To determine if HIV-2 and SIV could also be transmitted by DC-SIGN, 293T cells expressing DC-SIGN or vector alone were incubated with replication-competent HIV-1 NL4-3, HIV-2 Rod10, and SIVmac239 reporter viruses harboring the luciferase gene in place of nef (Fig. 6A to C) as well as with replication defective green fluorescent protein (GFP) reporter viruses pseudotyped with the SIVmac316 and the SIVsmΔB670cl3 Envs (Fig. 6D). After virus binding, the cells were extensively washed and subsequently cocultured with T-cell lines, and the extent of infection was determined 3 days later. DC-SIGN-transfected cells transmitted HIV-1 NL4-3 about 5- to 10-fold more efficiently than pcDNA3-transfected control cells (Fig. 6). Similar results were obtained with HIV-2 Rod10, SIVmac239, SIVmac316, and SIVsmΔB670cl3. Thus, DC-SIGN can mediate transmission of SIV and HIV-2 strains as well as HIV-1.
FIG. 6.
DC-SIGN transmits HIV-1, HIV-2, and SIV. (A to C) 293T cells were transiently transfected with a DC-SIGN expression plasmid, incubated with replication-competent reporter virus (A, NL4-3; B, Rod10; C, SIVmac239), washed, and cocultured with C8166 T cells. Luciferase activity in cell lysates was determined 3 days after the start of the coculture. Data from one representative experiment out of three are shown. (D) SIVmac316- and SIVsmΔB670cl3 (clone 3)-pseudotyped GFP reporter virus was added to DC-SIGN-transfected 293T cells, and the coculture assay was performed as described above except that CEM cells stably expressing CCR5 (kindly provided by Michael Malim, University of Pennsylvania, Philadelphia) were used as target cells. The percentage of GFP-positive T cells was determined 3 days after cocultivation. Comparable results were obtained in an independent experiment.
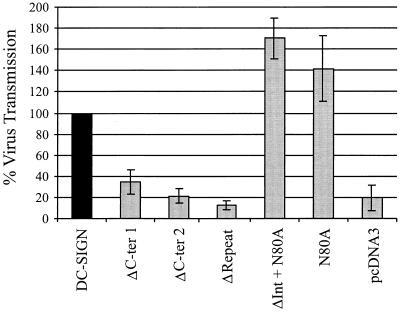
We next determined if virus binding correlated with efficient virus transmission to susceptible cells, once again using conditions that resulted in equivalent levels of surface expression for the DC-SIGN mutants (Fig. 2). Briefly, 293T cells expressing the DC-SIGN variants were incubated with the recombinant NL4-3 virus and washed, and C8166 T cells were added. Comparable surface expression of the variants was demonstrated by FACS analysis (data not shown). As observed for DC-SIGN-mediated p24 binding, deletion of the lectin-like domain as well as the repeat region abolished function (Fig. 7). In contrast, mutation of the glycosylation site alone and in combination with the deletion of amino acids 198 to 243 had no impact on viral transmission. These results indicate that the capacities of DC-SIGN to bind and transfer virus are linked and that virus binding and transmission require the same determinants of DC-SIGN.
FIG. 7.
DC-SIGN-mediated virus transmission. The DC-SIGN mutants were transiently expressed in 293T cells, and comparable surface expression was controlled as described in the legend to Fig. 2. The cells were incubated with equal amounts of p24 antigen of HIV-1 luciferase reporter virus, vigorously washed, and cocultivated with C8166 T cells. Three days after the start of the cocultivation, the cells were lysed and luciferase activity in the cell lysates was determined as described in Materials and Methods. Comparable results were obtained in two independent experiments.
Trypsin-EDTA inhibits DC-SIGN-mediated transmission.
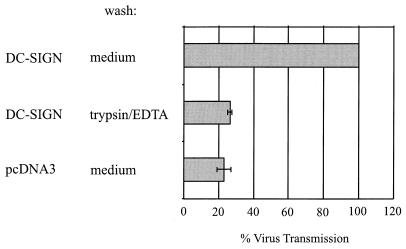
DC-SIGN contains motifs in its cytoplasmic domain that in some contexts mediate efficient endocytosis, raising the possibility that once bound to DC-SIGN, HIV may be internalized. To investigate this, we performed the virus transmission assay as described above but carried out one of the three wash steps with trypsin-EDTA. EGTA and EDTA both bind calcium ions, and EGTA has been shown to block HIV binding to DC-SIGN (7, 16), while trypsin nonspecifically digests proteins accessible on the cell surface. We found that washing virus-pulsed cells with trypsin-EDTA reduced viral transmission to values observed for control cells (Fig. 8). These data indicate that DC-SIGN-mediated transfer of HIV does not involve internalization of the virus in the cell system studied here.
FIG. 8.
Trypsin-EDTA inhibits virus transmission. The virus transmission assay was performed as described in the legend to Fig. 6 except that the first of three wash steps was carried out with trypsin-EDTA instead of medium. Cells were exposed to trypsin-EDTA for approximately 10 min at room temperature. Comparable results were obtained in an independent experiment.
DC-SIGN expression levels are important for the efficiency of virus transmission.
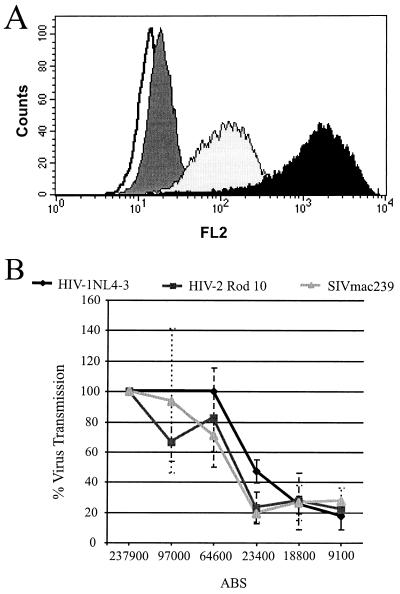
The efficiency with which HIV-1 infects cells is related to receptor density (19, 24, 28). To determine the relationship between DC-SIGN expression levels and the ability of DC-SIGN to bind and transmit virus, we generated a stable 293 T-Rex cell line expressing AU1-tagged DC-SIGN under the control of the tet repressor. Addition of increasing concentrations of doxycycline to the culture medium of these cells induced a corresponding increase in DC-SIGN surface expression (data not shown). Importantly, the cells responded to doxycycline homogeneously—there was little variability in the levels of DC-SIGN expression at any given drug concentration (Fig. 9A). This cell line was used to determine the number of surface DC-SIGN molecules required for efficient transmission of HIV-1, HIV-2, and SIVmac239. Cells were seeded in duplicate, with one panel being used in the virus transmission assay and the other used to quantify DC-SIGN surface expression by QFACS (21). Results from a single experiment in which each point was performed in triplicate (Fig. 9B) demonstrate that the efficiency of virus transmission was strongly related to DC-SIGN surface expression levels. Similar results were obtained with other experiments, though in each experiment the cells expressed somewhat different levels of DC-SIGN. A threshold level of about 60,000 copies was required for efficient transmission of all viruses tested. Below this threshold level, transmission efficiency decreased until a nadir of approximately 20% of maximum values was reached when DC-SIGN expression levels were below 20,000 copies per cell. These data suggest that efficient transmission of HIV and SIV requires high levels of DC-SIGN expression.
FIG. 9.
Quantification of surface DC-SIGN required for virus transmission. 293 T-Rex DC-SIGN cells were seeded in duplicate and incubated overnight with increasing concentrations of doxycycline. One panel was used in the virus transmission assay as described in Materials and Methods; the other panel was used for quantification of surface DC-SIGN expression via QFACS, employing the anti-AU1 antibody. (A) DC-SIGN expression upon induction with doxycycline. Overlay histogram shows DC-SIGN staining with the anti-AU1 antibody upon induction with, doxycycline at 0.01 (black) and 0.0001 (light grey) μg/ml compared to control staining with mouse (dark grey) immunoglobulin G and no antibody (white). (B) Virus transmission to C8166 T cells. Virus transmission efficiency was normalized to optimal transmission and is shown relative to the number of ABS. Data from one representative experiment carried out in triplicate are shown. Comparable results were obtained in two independent experiments.
DISCUSSION
DC-SIGN mediates interactions between DCs and T cells by binding to intracellular adhesion molecule 3 and also serves as an attachment factor for HIV-1 (7, 16, 17). Geijtenbeek and colleagues (16) have shown that DC-SIGN is largely responsible for the ability of DCs to bind HIV-1 and to present virus particles to cells expressing CD4 and an appropriate coreceptor, resulting in efficient virus infection. DC-SIGN is a type II membrane protein that contains a C-type lectin-like domain, and interactions between DC-SIGN and HIV can be prevented by EGTA and by mannan (7, 16). This makes it likely that DC-SIGN interacts with one or more carbohydrate structures on the HIV-1 Env protein. If this is so, it could explain the ability of DC-SIGN to bind disparate primate immunodeficiency viruses. We found little difference in the ability of DC-SIGN to bind to HIV-1, HIV-2, and SIV strains, suggesting that DC-SIGN may serve as a universal binding factor for primate lentiviruses.
C-type lectin-like domains have been found in numerous proteins with a wide array of functions (13). Proteins that contain C-type lectin-like domains are often type II membrane proteins, are typically oligomeric, and contain a neck region and a short cytoplasmic domain (13). While it is not known if DC-SIGN is oligomeric, its lectin domain is situated at the C terminus of the protein, being separated from the membrane by the neck region. Elimination of the lectin-like domain abrogated the ability of DC-SIGN to bind and transmit HIV, consistent with the ability of mannan and EGTA to inhibit these functions (16). It is not known if recognition of Env by DC-SIGN is due solely to carbohydrate recognition or if direct protein-protein interactions are also involved.
Deletion of the repeat, or neck, region also prevented HIV binding and transmission. However, surface expression of this protein was reduced under all conditions tested. Therefore, it is unlikely that this protein has an entirely native conformation, and the loss of DC-SIGN function resulting from deletion of the repeat region could be due to altered folding of the lectin-like domain. At present we have no way to determine if this is the case, but this point can be addressed when conformation-dependent monoclonal antibodies become available. The repeat region could also contribute to DC-SIGN function in several other ways. It could mediate oligomerization of DC-SIGN, though proteins that fail to oligomerize are rarely transported from the endoplasmic reticulum, suggesting that if DC-SIGN is oligomeric, other regions of the molecule are likely to be involved in subunit-subunit interactions (14). Elimination of the repeat region would also be expected to bring the lectin-like domain closer to the cellular membrane. It will be interesting to determine if distance from the membrane is an important parameter for DC-SIGN function.
The mechanism by which DC-SIGN mediates virus transmission is not clear. In general, it appears that virus attachment to the surface is a rate-limiting step for infection of many cell types, regardless of the presence (or expression levels) of CD4 and coreceptor. Attachment can be enhanced by the use of polycations or by spinoculation, in which virus is centrifuged onto the cell surface (23). By virtue of its ability to interact with HIV-1, DC-SIGN appears to greatly increase the efficiency of virus attachment. Does attachment to DC-SIGN necessarily mean that HIV-1 can be transmitted efficiently to all receptor-positive cell types? While information on this point is limited, we note that the efficiency of virus transfer in our study was less than that reported for transfer between a THP-1 cell line expressing DC-SIGN and primary T cells (16). The efficiency of virus transmission could be dependent in part on interactions between DC-SIGN and molecules such as intracellular adhesion molecule 3 on the surface of receptor-positive cells (17). In the presence of strong cell-to-cell interactions, as occurs between DCs and T cells, virus that is bound to DC-SIGN on the surface of one cell could be brought into close proximity to the surface of another. As a result, interactions with CD4 and coreceptors could be enhanced. If this speculation is correct, then one might expect the efficiency with which DC-SIGN binds virus to be relatively independent of the cell type in which it is expressed, while the ability of DC-SIGN to transmit virus could be much more dependent on the cell types involved and the interactions that occur between them. Another possibility by which DC-SIGN could enhance virus infection would be to alter Env structure in a manner that improves the efficiency of receptor interactions or perhaps makes Env easier to trigger, thus enhancing fusogenicity. These issues will have to be addressed to more fully understand the mechanisms by which DC-SIGN binds and transmits virus.
Once bound to DC-SIGN, virus could potentially remain stably associated with the cell surface or could be endocytosed. The cytoplasmic domain of DC-SIGN contains two motifs that have been shown to mediate endocytosis and recycling in multiple contexts. However, we found that virus bound to DC-SIGN was susceptible to trypsin, indicating that it remained associated with the cell surface. However, this should not be taken to mean that DC-SIGN–virus complexes are never internalized. Endocytosis of cell surface proteins can be highly context dependent, and it is possible the DC-SIGN transiently expressed in 293T cells is simply not endocytosed efficiently. It will be important to determine if DC-SIGN is internalized in DCs and, if it is, whether this process is influenced by virus binding.
The efficiency of HIV-1 infection is related in part to receptor density due to the cooperative nature of the fusion reaction. Several HIV trimers are needed to form a fusion pore, and several CD4 and coreceptor binding events are needed to activate individual Env trimers (19). On most primary CD4-positive cell types, coreceptor levels are more limiting than CD4 (21). We found that DC-SIGN expression levels can also be limiting for virus binding and transmission. With the cell system that we used and the three virus strains examined, transmission efficiency was strongly affected by differences in DC-SIGN expression between approximately 30,000 and 100,000 copies per cell. Transmission efficiency did not increase at higher levels of DC-SIGN expression, while a small amount of virus transmission was observed when DC-SIGN levels fell below 30,000 copies per cell. This finding indicates that it will be important to measure DC-SIGN expression levels on primary cell types and to determine if there are significant differences in DC-SIGN expression between individuals, as there is for coreceptor expression. If this is so, it could greatly affect the efficiency with which DCs capture HIV and could impact virus transmission. Our results also suggest that in order to determine if diverse virus strains vary in the ability to interact with DC-SIGN, it will be important to titrate DC-SIGN levels so that they reflect levels attained in vivo. Finally, the discovery of DC-SIGN underscores the importance of investigating Env interactions with other cell surface molecules in a variety of cell types to determine if changes in attachment efficiency strongly impact viral infectivity and perhaps viral tropism and pathogenicity. Our recent finding that DC-SIGNR, a DC-SIGN homologue (29) that is expressed on endothelial cells in placenta, liver, and lymph node sinuses, also supports binding and transmission of HIV-1, HIV-2, and SIV strains illustrates this point (26).
ACKNOWLEDGMENTS
We thank Nathaly Finze and Mandy Krumbiegel for excellent technical assistance. We also thank Victor Holubowsky and Farida Shaheen for generation and quantification of virus stocks.
R.W.D. was supported by NIH grants AI 35383 and 40880, a Burroughs Wellcome Fund Translational Research Award, and an Elizabeth Glaser Scientist Award from the Pediatric AIDS Foundation. S.P. was supported by a fellowship from the Deutsche Forschungsgemeinschaft. F.B. was supported by a fellowship from the Swiss National Science Foundation, grant 823A-611772. This work was supported by grant P30-AI45008 of the Viral/Cell/Molecular Core of the Penn Center for AIDS Research, the Wilhelm-Sander Foundation, and grant SFB466.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: A RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Barratt-Boyes S M, Watkins S C, Finn O J. In vivo migration of dendritic cells differentiated in vitro: a chimpanzee model. J Immunol. 1997;158:4543–4547. [PubMed] [Google Scholar]
- 3.Cameron P U, Freudenthal P S, Barker J M, Gezelter S, Inaba K, Steinman R M. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science. 1992;257:383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 4.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 5.Clavel F, Mansinho K, Chamaret S, Guetard D, Favier V, Nina J, Santos-Ferreira M O, Champalimaud J L, Montagnier L. Human immunodeficiency virus type 2 infection associated with AIDS in West Africa. N Engl J Med. 1987;316:1180–1185. doi: 10.1056/NEJM198705073161903. [DOI] [PubMed] [Google Scholar]
- 6.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 7.Curtis B M, Scharnowske S, Watson A J. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc Natl Acad Sci USA. 1992;89:8356–8360. doi: 10.1073/pnas.89.17.8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Marzio P D, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 9.Doms R, Edinger A, Moore J. Coreceptor use by primate lentiviruses, p. III-1–III-28. In: Korber B, Foley B, Leitner T, Myers G, Hahn B, McCutchan F, Mellors J, Kuiken C, editors. Human retroviruses and AIDS. Los Alamos, N.M: Los Alamos National Laboratory, Theoretical Biology and Biophysics; 1999. [Google Scholar]
- 10.Doms R W, Lamb R, Rose J K, Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 11.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 12.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 13.Drickamer K. C-type lectin-like domains. Curr Opin Struct Biol. 1999;9:585–590. doi: 10.1016/s0959-440x(99)00009-3. [DOI] [PubMed] [Google Scholar]
- 14.Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane domain, G-protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Geijtenbeek T B H, Kwon D S, Torensma R, van Vliet S J, van Duijnhoven G C F, Middel J, Cornelissen I L M H A, Nottet H S L M, Kewalramani V N, Littman D R, Figdor C G, van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 17.Geijtenbeek T B H, Torensma R, van Vliet S J, van Duijnhoven G C F, Adema G J, van Kooyk Y, Figdor C G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 18.Kirchhoff F, Pöhlmann S, Hamacher M, Means R E, Kraus T, Überla K, Marzio P D. Simian immunodeficiency virus variants with differential t-cell and macrophage tropism use CCR5 and an unidentified cofactor expressed in CEMX174 cells for efficient entry. J Virol. 1997;71:6509–6516. doi: 10.1128/jvi.71.9.6509-6516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between Fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 21.Lee B, Sharron M, Montaner L, Weissman D, Doms R. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mondor I, Ugolini S, Sattentau Q J. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Doherty U, Swiggard W J, Malim M H. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations by macrophage-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pöhlmann S, Lee B, Meister S, Krumbiegel M, Leslie G, Doms R W, Kirchoff F. Human immunodeficiency virus utilizes human but not rhesus macaque STRL33 for efficient entry. J Virol. 2000;74:5075–5082. doi: 10.1128/jvi.74.11.5075-5082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pöhlmann S, Soilleux E J, Baribaud F, Leslie G, Morris L S, Trowsdale J, Lee B, Coleman N, Doms R W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and immunodeficiency viruses and activates infection in trans. Proc Natl Acad Sci USA. 2001;98:2670–2675. doi: 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan-Graham M A, Peden K W. Both virus and host components are important for the manifestation of a Nef-phenotype in HIV-1 and HIV-2. Virology. 1995;213:158–168. doi: 10.1006/viro.1995.1556. [DOI] [PubMed] [Google Scholar]
- 28.Sharron M P, Pöhlmann S, Price K, Tsang M, Kirchoff F, Doms R W, Lee B. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood. 2000;96:41–49. [PubMed] [Google Scholar]
- 29.Soilleux E J, Barten R, Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 30.Stahl-Hennig C, Steinman R M, Tenner-Racz K, ope M, Stolte N, Matz-Rensing K, Grobschupff G, Raschdorff B, Hunsmann G, Racz P. Rapid infection of oral mucosal-associated lymphoid tissue with simian immunodeficiency virus. Science. 1999;285:1261–1265. doi: 10.1126/science.285.5431.1261. [DOI] [PubMed] [Google Scholar]
- 31.Trkola A, Drajic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its coreceptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 32.Ugolini S, Moulard M, Mondor I, Barois N, Demandolx D, Hoxie J, Brelot A, Alizon M, Davoust J, Sattentau Q J. HIV-1 gp120 induces an association between CD4 and the chemokine receptor CXCR4. J Immunol. 1997;159:3000–3008. [PubMed] [Google Scholar]
- 33.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]