Abstract
P2Y1 purinergic receptors (P2Y1Rs) mediate rises in intracellular Ca2+ in response to ATP, but the duration and characteristics of this Ca2+ response are known to vary markedly in distinct cell types. We screened the P2Y1R carboxyl terminus against a recently created proteomic array of PDZ (PSD-95/Drosophila Discs large/ZO-1 homology) domains and identified a previously unrecognized, specific interaction with the second PDZ domain of the scaffold NHERF-2 (Na+/H+ exchanger regulatory factor type 2). Furthermore, we found that P2Y1R and NHERF-2 associate in cells, allowing NHERF-2-mediated tethering of P2Y1R to key downstream effectors such as phospholipase Cβ. Finally, we found that coexpression of P2Y1R with NHERF-2 in glial cells prolongs P2Y1R-mediated Ca2+ signaling, whereas disruption of the P2Y1R–NHERF-2 interaction by point mutations attenuates the duration of P2Y1R-mediated Ca2+ responses. These findings reveal that NHERF-2 is a key regulator of the cellular activity of P2Y1R and may therefore determine cell-specific differences in P2Y1R-mediated signaling.
Keywords: G protein-coupled receptor, purinergic, ATP, proteomic array
Adenine-based nucleotides such as ATP and ADP are prominent extracellular signaling molecules that mediate a wide variety of physiological actions in tissues throughout the body. Many of the physiological effects evoked by ATP and ADP are mediated by metabotropic P2Y receptors (P2YRs) (1), which are members of the G protein-coupled receptor (GPCR) superfamily. To date, seven distinct mammalian P2YR subtypes have been cloned: P2Y1, P2Y2, P2Y4, P2Y6, P2Y11, P2Y12, and P2Y13. Most P2YRs are coupled to Gαq proteins and thus to the activation of phospholipase Cβ (PLCβ) and generation of diacylglycerol and inositol-3,4,5-trisphosphate, ultimately leading to the activation of PKC and release of Ca2+ from internal stores (1). Some P2YRs, including P2Y1, P2Y2, P2Y12, and P2Y13, are also known to couple to Gαi and the inhibition of adenylyl cyclase (2–5). The purinergic P2YR type 1 (P2Y1R) subtype is abundantly expressed in a number of tissues, including the CNS (6, 7), where it plays a key role in the transmission of astrocytic Ca2+ waves (8), activation of mitogenic responses in astrocytes to brain trauma (9), inhibition of neuronal N-type voltage-activated Ca2+ channels (10), and embryonic brain development (11). P2Y1R also plays critical roles in the cardiovascular system, including the regulation of coronary vasodilation (12) and platelet aggregation (13).
Signaling by P2Y1Rs is known to be heavily dependent on cellular context. For example, stimulation of P2Y1Rs in some cell types is known to strongly promote cell proliferation (14), whereas P2Y1R stimulation in other cell types is known to induce apoptosis (15). Furthermore, P2Y1Rs can exert cellular effects that are quite different from those exerted by other P2YRs expressed in the same cell type and that couple to similar G proteins. For example, in astrocytes, both P2Y1Rs and P2Y2Rs can couple by means of Gq to the release of Ca2+ from a common intracellular pool (16). Yet, P2Y1Rs and P2Y2Rs exhibit striking temporal differences in their Ca2+ response patterns and susceptibility to regulation by PKC (17). At present, it is not clear why P2Y1R activity is so dependent on cellular context or why P2Y1Rs can exert physiological effects that are so different from related P2YR subtypes.
GPCR signaling can be regulated by various types of receptor-associated scaffold proteins (18). Many of the known interactions between GPCR scaffold proteins and their cognate GPCRs are mediated by the binding of PSD-95/Drosophila Discs large/ZO-1 homology (PDZ) domains within the scaffold proteins to specialized motifs at the carboxyl terminus (CT) of the GPCRs (18, 19). Among the >440 PDZ domains in the human genome, approximately one-quarter are believed to be class I PDZ domains, which are characterized by preferential binding to the carboxyl-terminal motif S/T-x-§ (where § represents a hydrophobic residue at the terminal position). Interestingly, P2Y1R-CT terminates in the motif D-T-S-L, suggesting that P2Y1R might associate with class I PDZ scaffold proteins. In this study, we used a recently created proteomic array of 96 distinct PDZ domains derived from a variety of cytosolic proteins to identify proteins that might selectively associate with the CT of P2Y1R. Using this technique, we found a previously unrecognized high-affinity association of P2Y1R-CT with the PDZ scaffold protein NHERF-2 (Na+/H+ exchanger regulatory factor type 2). We also observed that NHERF-2 and P2Y1R exhibit similar patterns of expression in native brain tissue and that NHERF-2 can exert substantial control over P2Y1R functional activity.
Materials and Methods
Construction of the PDZ Domain Proteomic Array. PDZ domains were expressed as His- and S-tagged fusion proteins by using the vector pET30A (Novagen) and were purified by using ProBond nickel resin (Invitrogen). We thank the large number of colleagues who sent us cDNA constructs encoding various PDZ proteins (listed in the supporting information, which is published on the PNAS web site). These cDNAs were used as templates to amplify by means of PCR the regions encoding various PDZ domains to subclone them into pET30A for fusion protein expression.
Overlays, Fusion Protein Pull-Downs, and Plate-Binding Assays. To assess the binding of receptor carboxyl-terminal GST fusion protein to the PDZ domain array, the purified PDZ domain fusion proteins were spotted at 1 μg per bin onto Nytran SuperCharge 96-grid nylon membranes (Schleicher & Schuell). The membranes were allowed to dry overnight and then blocked in “blot buffer” (2% nonfat dry milk/0.1% Tween 20/50 mM NaCl/10 mM Hepes, pH 7.4) for 1 h at room temperature. Overlays with P2Y1R-CT fusion protein (100 nM in blot buffer) were then performed by using an overlay technique that is described in refs. 20 and 21. Interactions were confirmed by means of fusion protein pull-downs, and affinity estimates were generated in saturation binding assays by using a plate-binding assay described in ref. 20.
Cell Culture and Transfection. HEK-293 and 1321N1 cells were regularly maintained in DMEM (GIBCO) supplemented with 10% FBS and 1% penicillin/streptomycin in a 37°C/5% CO2 incubator. C6 glioma cells were maintained in DMEM (GIBCO) supplemented with 10% heat-inactivated FBS and 1% penicillin/streptomycin. Cells were split every 3–4 days. For transfections, cells were split onto 100-mm plates, grown to 90–100% confluency, and transfected with the appropriate cDNA by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. cDNAs used in transfections were P2Y1R-pcDNA3, P2Y1R/L373A-pcDNA3, FLAG-tagged NHERF-2-pcDNA3, and hemagglutinin (HA)-tagged NHERF-2-pcDNA3.
Immunoprecipitation, Surface Expression Assay, and Western Blotting. Coimmunoprecipitation of full-length proteins from HEK-293 cells was performed by using appropriate primary antibodies and methods described in ref. 21. Surface expression of P2Y1R was verified by using a luminometer-based surface expression assay as described in ref. 21; no significant differences in surface expression between the WT P2Y1R and L373A mutant receptor were observed (data not shown). Purified proteins, cell extracts, and/or immunoprecipitated samples were separated by SDS/PAGE and blotted onto nitrocellulose as described in refs. 20 and 21. Monoclonal anti-HA 12CA5 antibody (Roche Applied Science, Indianapolis) and monoclonal anti-Flag M2 antibody (Sigma) were the primary antibodies used to detect and immunoprecipitate epitope-tagged proteins. To detect endogenous and/or recombinant P2Y1R, PLCβ1, and NHERF-2, we used polyclonal anti-P2Y1R antibody (Zymed), anti-PLCβ1 (Santa Cruz Biotechnology), and anti-NHERF-2 (22).
Immunocytochemistry and Electron Microscopy Analysis. To visualize immunostaining for P2Y1R and NHERF-2 in native brain tissue, C57BL/6 mice were killed, and brain sections were prepared and processed for the preembedding immunoperoxidase labeling as described in ref. 23 with the following modifications: 2% milk was used as a blocking agent, and rabbit anti-P2Y1R and anti-NHERF-2 antibodies were used at 1:1,000 and 1:7,000 dilutions, respectively. A random sampling of 33–62 micrographs of small capillaries were taken from ultrathin sections of the cerebral cortex with a charge-coupled device camera (DualView 300W, Gatan, Pleasanton, CA) controlled by digitalmicrograph 3.6.5 software (Gatan). Digitally acquired micrographs were adjusted for brightness and contrast; the image resolution was kept constant. All processes (147–218) in apposition with endothelial cells were analyzed for the presence or absence of amorphous diaminobenzidine deposit corresponding to immunoreactivity for P2Y1R or NHERF-2.
Calcium Imaging. The Ca2+-sensitive fluorophore fura-2 (Molecular Probes) was used for ratiometric Ca2+ imaging in C6 glioma and 132N1 human astrocytoma cells. All fluorescence measurements were made from subconfluent areas of the dishes so that individual cells could be readily identified. After transfection in 100-mm plates, cells were split onto coverslips immersed in 0.5 ml of media in 24-well plates and grown for 1–2 days. Before imaging, coverslips were incubated at room temperature for 30 min in extracellular recording solution (ECS) composed of 150 mM NaCl/10 mM Hepes/3mMKCl/2 mM CaCl2/2 mM MgCl2/5.5 mM glucose, pH 7.3, 325 milliosmolar. ECS was supplemented with pluronic acid (0.001%) and fura-2 AM (2 μM). Subsequently, coverslips were thoroughly rinsed with extracellular solution lacking fura-2 AM and BSA and mounted onto the microscope stage for imaging. Intensity images of 510-nm emission wavelengths were taken at 340- and 380-nm excitation wavelengths, and the two resulting images were taken from individual cells for ratio calculations. imaging work-bench 2.2.1 (Axon Instruments, Union City, CA) was used for acquisition of intensity images and conversion to ratios. The observed calcium responses were mainly due to liberation of calcium from intracellular stores (≈70%), with a minor component attributable to influx of extracellular calcium (≈30%), as assessed in experiments comparing responses in the presence versus absence of extracellular calcium (n = 95 cells; data not shown).
Results
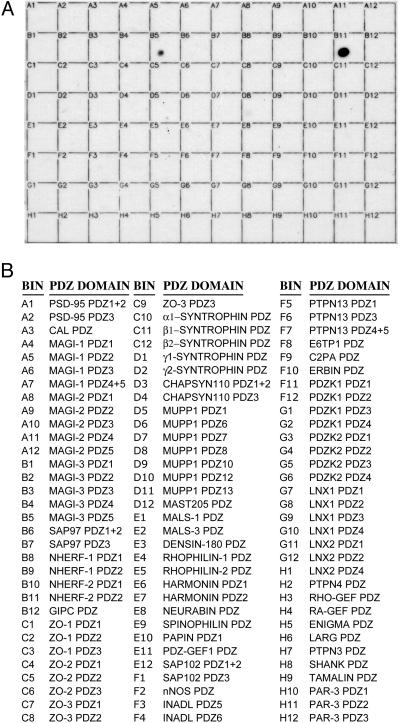
Screening of a PDZ Domain Proteomic Array Reveals Strong Association of P2Y1R-CT with the Second PDZ Domain (PDZ2) of NHERF-2. To identify PDZ domain-containing proteins that might associate with P2Y1R, we generated a GST fusion protein containing the last 50 aa of P2Y1R-CT, including the terminal D-T-S-L PDZ-domain binding motif. This P2Y1R-CT-GST fusion protein was used to screen a recently created proteomic array of various class I PDZ domains derived from multiple PDZ proteins. The PDZ domains were expressed as His- and S-tagged fusion proteins, purified, and spotted in equal amounts onto individual bins within gridded nylon membranes. P2Y1R-CT did not detectably associate with the vast majority of the 96 PDZ domains on the array. However, a robust association was found between P2Y1R-CT and PDZ2 of NHERF-2 (Fig. 1). A somewhat weaker interaction was also detected with the fifth PDZ domain of the multi-PDZ scaffold protein MAGI-3.
Fig. 1.
P2Y1R-CT binds selectively to NHERF-2 PDZ2. The CT of P2Y1R was prepared as a GST fusion protein and overlaid onto a proteomic array containing 96 putative class I PDZ domains. (A) The results of the overlay. (B) A list of the PDZ domains that were spotted in each bin of the array. P2Y1R-CT exhibited strong binding to bin B11, the PDZ2 of NHERF-2, and weaker binding to bin B5, the fifth PDZ domain of MAGI-3. Much longer exposures (not shown) revealed faint binding of P2Y1R-CT to bin B8 (NHERF-1 PDZ1) and bin A12 (MAGI-2 PDZ5). These results are representative of five independent experiments.
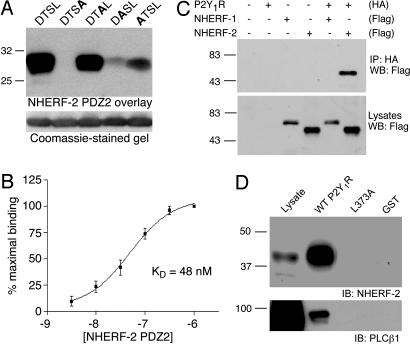
Structural Determinants of the P2Y1R–NHERF-2 Interaction. Because the D-T-S-L motif at the P2Y1R-CT is a canonical class I PDZ-domain binding motif, we generated a series of mutant P2Y1R-CT fusion proteins with sequential substitutions of the various residues in this motif. Overlay studies revealed a robust association of the NHERF-2 PDZ2 fusion protein with the WT P2Y1R-CT as well as with the mutant P2Y1R-CT in which the serine at the –1 position was replaced by alanine, suggesting that this residue is not obligatory for the interaction (Fig. 2A). Mutations of the –2 and –3 positions of P2Y1R-CT, however, resulted in markedly reduced binding of NHERF-2 PDZ2, suggesting that the threonine and aspartic acid residues at these positions are involved in the P2Y1R–NHERF-2 association. Most striking, however, was the lack of any detectable association of NHERF-2 PDZ2 with the P2Y1R-CT fusion protein in which the terminal leucine residue was mutated to alanine. Taken together, these results reveal that three of the last four residues of P2Y1R are key determinants of the interaction with NHERF-2 PDZ2.
Fig. 2.
Structural determinants of the P2Y1R–NHERF-2 PDZ2 interaction in vitro and in cells. (A) The last four amino acids of P2Y1R-CT (D-T-S-L) were sequentially mutated to alanine, and WT P2Y1R-CT as well as the four mutants were expressed as GST fusion proteins. These fusion proteins were loaded equally (2 μg) onto SDS/PAGE gels (Lower), transferred to nitrocellulose, and overlaid with 100 nM His-tagged NHERF-2 PDZ2 fusion protein (Upper). The data shown are representative of three independent experiments. (B) Affinity of the P2Y1R-CT–NHERF-2 PDZ2 interaction. The wells of a 96-well plate were coated with P2Y1R-CT-GST and overlaid with six different concentrations of His-tagged NHERF-2 PDZ2 (3 nM to 1 μM). Binding was expressed as a percentage of the binding observed at 1 μM NHERF-2 PDZ2. The binding was saturable and yielded an estimated dissociation constant of 48 nM (n = 3). Points and error bars represent mean ± SEM. (C) Coimmunoprecipitation of P2Y1R and NHERF-2 from cells. HA-tagged P2Y1R was expressed in HEK-293 cells in the absence and presence of Flag-tagged NHERF-1 and NHERF-2. P2Y1R was immunoprecipitated by using anti-HA antibodies, and the immunoprecipitates were probed with anti-Flag antibodies (Upper). The total expression of Flag-tagged NHERF-1 and NHERF-2 in the cell lysates is shown by anti-Flag Western blot (Lower). The data shown are representative of four independent experiments. (D) NHERF-2 physically links P2Y1R-CT to PLCβ1. Lysates from untransfected HEK-293 cells were incubated with fusion proteins corresponding to GST alone, GST-P2Y1R-CT, or L373A P2Y1R-CT. Pull-down samples were probed with antibodies against NHERF-2 (Upper) or PLCβ1(Lower). These data are representative of four independent experiments.
To more quantitatively assess the association of NHERF-2 PDZ2 with P2Y1R-CT, we performed in vitro binding assays overlaying NHERF-2 PDZ2 fusion proteins at various concentrations onto 96-well plates coated with P2Y1R-CT fusion protein. As shown in Fig. 2B, binding of NHERF-2 PDZ2 was saturable within the range of concentrations tested, yielding an estimated dissociation constant (Kd) of 48 nM. This estimated affinity is within the same range of other PDZ–receptor interactions that are known to occur in cells and have physiological relevance for receptor function (19).
Association of Full-Length P2Y1R with NHERF-2 in Cells. Next, we determined whether association of NHERF-2 with P2Y1R occurs between full-length forms of the proteins in living cells. Full-length HA-tagged P2Y1R was expressed in HEK-293 cells either alone or in combination with Flag-tagged NHERF-2 or a FLAG-tagged version of a closely related protein, NHERF-1. When HA-P2Y1R was immunoprecipitated from the lysates of cells cotransfected with HA-P2Y1R and FLAG-NHERF-2, robust coimmunoprecipitation of FLAG-NHERF-2 was observed (Fig. 2C). No detectable coimmunoprecipitation, however, was observed from lysates of cells in which HA-P2Y1R was cotransfected with FLAG-NHERF-1. Similarly, we failed to detect coimmunoprecipitation of NHERF-2 from lysates of HEK-293 cells cotransfected with FLAG-NHERF-2 and a full-length mutant P2Y1R in which the terminal leucine was changed to alanine (P2Y1R L373A). These results demonstrate that NHERF-2 strongly associates with P2Y1R in a cellular context and that this interaction depends on the same motif identified in the fusion protein studies.
P2Y1R, PLCβ, and NHERF-2 Form a Multiprotein Complex. P2Y1R exerts effects on cellular physiology mainly through coupling to Gαq. Interestingly, NHERF family proteins are known to interact with a number of proteins implicated in Gαq-mediated signal transduction, including Gαq itself (24), PKCα (25), and subtypes of PLCβ (26). NHERF-2 might therefore be able to strongly influence P2Y1R-mediated signaling efficacy by clustering these key signaling components in the vicinity of the receptor. To determine whether NHERF-2 might serve as a scaffold to physically link P2Y1R and PLCβ in cells, we pulled down protein complexes from HEK-293 cell lysates by using the P2Y1R-CT-GST fusion protein. Interestingly, both endogenous PLCβ1 and endogenous NHERF-2 were robustly pulled down by WT P2Y1R-CT but not by L373A P2Y1R-CT or control GST fusion (Fig. 2D). Given that L373A P2Y1R is unable to bind NHERF-2, these data suggest that P2Y1R-CT associates with PLCβ in a NHERF-2-dependent manner.
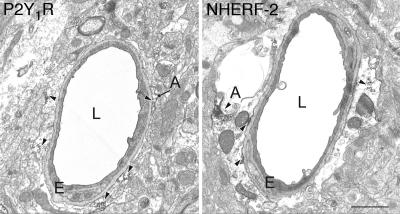
P2Y1R and NHERF-2 Are both Expressed in Perivascular Glial Cells in the CNS. Our finding that NHERF-2 and P2Y1R are able to physically interact with one another would only be relevant for in vivo function if the two proteins are coexpressed in native tissues. It has been shown that P2Y1R is expressed in both neurons and glia throughout the CNS (6), but the localization of NHERF-2 in the CNS remains to be established. Therefore, we performed an electron microscopic analysis of NHERF-2 and P2Y1R expression at the cellular and subcellular level to determine whether the expression patterns of these two proteins might overlap. At the electron microscopic level, immunostaining for both P2Y1R and NHERF-2 was abundantly found in astrocytic processes (Fig. 3). Within astrocytes, the labeling for both proteins was associated with the plasma membrane or with vesicle-like structures. However, because of technical limitations inherent in the fact that the P2Y1R and NHERF-2 antibodies used were raised in the same species, we could not perform a strict colocalization study to assess the degree of coexistence of these two proteins in individual elements. Instead, we determined the proportion of glial processes that displayed P2Y1R and NHERF-2 immunoreactivity. To increase the likelihood of examining a common population of glial processes in both sets of immunostained sections, we analyzed the population of glial processes surrounding small capillaries (27). To make sure that the lack of immunoreactivity in some of these processes was not due to limited antibody penetration, we selected only ultrathin sections from the top of tissue blocks for this analysis. Furthermore, nonimmunoreactive elements were quantified only if they were found to be in the close vicinity of labeled structures. Using this approach, we found that 69% (102 of 147 elements) of perivascular glial processes displayed immunoreactivity for P2Y1R, whereas 65% (142 of 218 elements) were immunoreactive for NHERF-2, suggesting that P2Y1R and NHERF-2 expression is likely to overlap in this population of cells.
Fig. 3.
P2Y1R and NHERF-2 exhibit similar patterns of expression in native brain tissue. Examples of P2Y1R (Left) and NHERF-2 (Right) immunopositive astrocytic processes in mouse cerebral cortex. Several of the most prominent immunoperoxidase deposits in each frame are indicated with arrowheads. A, astrocytic process; E, endothelial cell; L, lumen of capillary. (Scale bar, which applies to Left and Right:1 μm.)
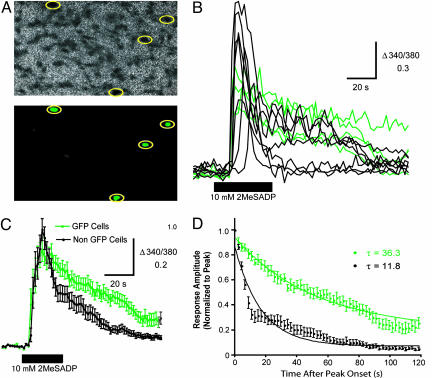
NHERF-2 Controls the Duration of P2Y1R-Mediated Ca2+ Signaling. To determine whether NHERF-2 is able to regulate P2Y1R function, we investigated the role of NHERF-2 in the Gαq- and PLCβ-dependent mobilization of Ca2+ by P2Y1R by performing fura-2-based Ca2+ imaging in C6 glioma cells. These glial cells express endogenous P2Y1R (28) but do not express detectable levels of endogenous NHERF-2 (data not shown). The setup used in these experiments allowed for switching between nonratiometric imaging of GFP fluorescence and ratiometric imaging of fura-2 fluorescence. Therefore, by making use a GFP-NHERF-2 construct that allowed for identification on a cell-by-cell basis of the cells that expressed transfected NHERF-2 versus those that did not (Fig. 4A), we could directly measure the effect of transfected NHERF-2 on the signaling efficacy of endogenous P2Y1Rs.
Fig. 4.
GFP-NHERF-2 prolongs the duration of P2Y1R-mediated Ca2+ responses in C6 glioma cells. (A) (Upper) The ratiometric image of fura-2 fluorescence for a field of C6 glioma cells acquired before the start of the experiment. (Lower) The nonratiometric image of GFP fluorescence acquired from the same field. GFP-positive cells are identified by the yellow circles. (B) Individual traces show records of the ratio of fluorescence intensities of fura-2 at 340 and 380 nm for GFP-positive (green) and GFP-negative (black) cells from A. 2-MeSADP (10 μM) was applied as indicated by the black bar. (C) Each trace represents the mean ± SEM Δ340/380 measured from all GFP-positive (green; n = 9) and GFP-negative (black; n = 30) cells before, during, and after application of 2-MeSADP (10 μM) as indicated by the black bar. (D) The mean ± SEM amplitudes of responses evoked in GFP-positive (green; n = 9) and GFP-negative (black; n = 30) cells by applying 2-MeSADP (10 μM) plotted as a function of time elapsed since the onset of peak amplitude. Response amplitudes are normalized to the peak amplitude of each response. The lines represent the best single exponential fit approximated for the decay phase of each data set and are defined by the equations y = 0.06197e–0.01280x (GFP cells) and y = 0.059e–0.03606x (non-GFP cells).
Application of the P2Y1R agonist 2-methylthio-ADP (2-MeSADP) (10 μM for 40 sec) evoked transient increases in intracellular Ca2+ concentration in the majority of C6 glioma cells, whether they expressed GFP-NHERF-2 or not (Fig. 4B). We observed that responses in GFP-NHERF-2-positive cells were slightly smaller in amplitude, on average, but persisted for much longer than responses evoked in GFP-NHERF-2-negative cells. The mean ± SEM Δ340/380 evoked by 2-MeSADP in GFP-positive and GFP-negative cells was 0.67 ± 0.06 (n = 8) and 0.96 ± 0.05 (n = 30), respectively (P < 0.05, Student's t test). However, Ca2+ responses evoked in GFP-positive cells persisted, on average, 3-fold longer than those evoked in GFP-negative cells (Fig. 4 C and D). Meanwhile, no difference in the latency of Ca2+ responses was observed between GFP-NHERF-2-positive and GFP-NHERF-2-negative cells (14.8 ± 1.8 s, n = 8; 16.3 ± 0.8 s, n = 30; P = 0.41, Student's t test). Put together, these results reveal that expression of NHERF-2 in C6 glioma cells greatly prolongs the duration of P2Y1R-mediated Ca2+ signaling while having a slight effect on the response magnitude.
Because NHERF-2 can interact with and regulate the activity of various Gq signaling components (24–26), the effects of NHERF-2 on P2Y1R-mediated Ca2+ signaling observed in the experiments with the C6 glioma cells might reflect general effects of NHERF-2 on the efficiency of Gq signaling (or on Ca2+ signaling in general) rather than specific effects on P2Y1R-mediated signaling due to the NHERF-2 interaction with P2Y1R. Thus, we performed parallel experiments in human 1321N1 astrocytoma cells that lack endogenous P2YRs (8) but express significant levels of endogenous NHERF-2 (data not shown). These glial cells lacking endogenous P2Y1R allowed for direct comparisons between WT P2Y1Rs and L373A mutant receptors that are unable to bind to the endogenous NHERF-2 expressed in these cells. As expected, 1321N1 cells transfected with either WT P2Y1R or L373A mutant P2Y1R exhibited transient Ca2+ responses after application of 2-MeSADP (Fig. 5 A and B). Comparisons of the Ca2+ responses evoked in WT versus L373A-expressing cells revealed no significant differences in either the response magnitude (0.56 ± 0.10, n = 5 versus 0.92 ± 0.12, n = 7; P = 0.068, Student's t test) or the latency (11.4 ± 1.1 s, n = 8 versus 16.0 ± 2.6 s, n = 5; P = 0.0876, Student's t test). However, there was a dramatic difference in the duration of Ca2+ responses evoked by WT versus L373A mutant P2Y1Rs, with the WT P2Y1R-mediated responses persisting >3-fold longer than those mediated by activation of L373A mutant receptors (Fig. 5C). This difference between the WT and L373A mutant P2Y1Rs observed in 132N1 cells expressing endogenous NHERF-2, considered together with a similar difference observed for endogenous P2Y1Rs in C6 glioma cells in the presence versus absence of transfected NHERF-2, provides strong evidence that NHERF-2 association with P2Y1R controls receptor signaling by prolonging the duration of P2Y1R-mediated responses.
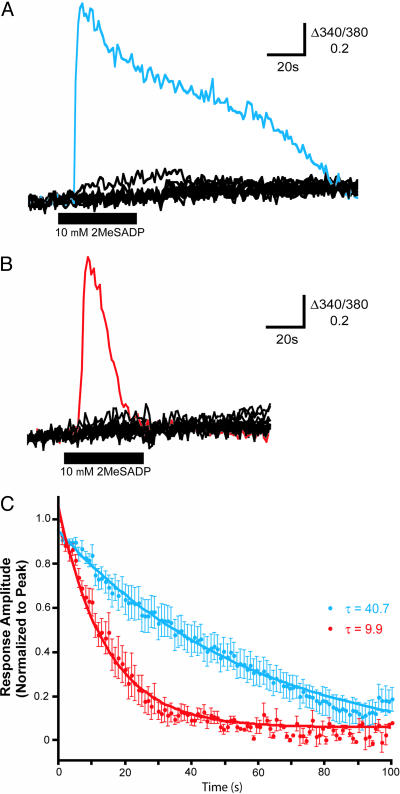
Fig. 5.
WT P2Y1R exhibits Ca2+ responses of longer duration than mutant L373A P2Y1R. (A and B) Individual traces show records of Δ340/380 fura-2 fluorescence in a field of 1321N1 cells transfected with WT (A) or mutant L373A (B) P2Y1R. 2-MeSADP (10 μM) was applied as indicated by the black bar. Representative traces of responsive cells in each field are shown in blue (WT) and red (L373A), respectively. (C) The mean ± SEM amplitudes of Ca2+ responses in 1321N1 cells expressing WT (blue; n = 8) or mutant L373A (red; n = 5) P2Y1R evoked by applying 2-MeSADP (10 μM) plotted as a function of time elapsed since the onset of peak amplitude. Response amplitudes are normalized to the peak amplitude of each response. Comparable differences between WT P2Y1R and the L373A mutant receptor were also observed at lower doses of agonist (1 nM and 20 nM 2-MeSADP; data not shown). The lines represent the best single exponential fit approximated for the decay phase of each data set and are defined by the equations y = 1.002e–0.017030x (WT P2Y1R cells) and y = 1.001e–0.02822x (L373A P2Y1R cells).
Discussion
The interaction between P2Y1R and NHERF-2 described in this study was found by screening the P2Y1R-CT against a recently created proteomic array of PDZ domains. Potential PDZ interaction motifs are found at the carboxyl termini of many receptors, ion channels, enzymes, and other signaling proteins (19). For such proteins with predicted PDZ interaction motifs, the focused screening of a specific proteomic array such as the PDZ domain array described here can be a useful alternative to yeast two-hybrid library screening and other unbiased approaches for finding protein–protein interactions. The P2Y1R–NHERF-2 interaction identified in the screens of the PDZ domain array was found to be quite specific; the vast majority of PDZ domains on the array did not detectably associate with the P2Y1R-CT.
The key structural determinants of the P2Y1R–NHERF-2 interaction are PDZ2 of NHERF-2 and the D-T-S-L motif at the CT of P2Y1R. Interestingly, it has previously been shown that there is a very weak interaction between the P2Y1R-CT and NHERF-1 (29). This interaction, however, is too weak to be detected in cells and is therefore probably not of physiological significance (29). Indeed, in the present study, extended overexposures of the P2Y1R-CT-GST overlays of the PDZ domain array revealed weak binding of P2Y1R-CT to NHERF-1 PDZ1 and a handful of other PDZ domains, but these interactions were all determined to be at least 10- to 50-fold lower in affinity than the P2Y1R–NHERF-2 interaction (data not shown). The strong and specific binding observed between P2Y1R and NHERF-2, as well as the similar patterns of expression of these two proteins in native tissues and the striking functional effects of NHERF-2 on P2Y1R signaling, suggest that NHERF-2 is a primary cellular partner of P2Y1R.
We observed that NHERF-2 dramatically prolongs the time course of P2Y1R-mediated Ca2+ signaling. Overexpression of NHERF-2 in C6 glioma cells, which do not detectably express endogenous NHERF-2 according to our Western blots, resulted in a 3-fold increase in the duration of Ca2+ responses evoked by activation of endogenous P2Y1R. Similarly, in 1321N1 cells, which express substantial levels of endogenous NHERF-2 according to our Western blots, activation of WT P2Y1R evoked Ca2+ responses that persisted for >3-fold longer than those evoked by activation of L373A P2Y1R. Thus, both sets of experiments revealed that association of P2Y1R with NHERF-2 allows the receptor to couple to sustained increases in intracellular Ca2+. Based on these observations, one might predict that cells expressing endogenous NHERF-2 should exhibit much more sustained P2Y1R-mediated Ca2+ responses than cells lacking endogenous NHERF-2. Indeed, we observed that the duration of Ca2+ signaling of WT P2Y1Rs in 1321N1 versus C6 cells was, in fact, profoundly different, with the Ca2+ responses lasting markedly longer in cells expressing endogenous NHERF-2 (the 1321N1 cells) than in cells lacking endogenous NHERF-2 (the C6 cells). It is well known that the activity levels of various calcium-dependent signaling cascades depend heavily on the localization, magnitude, frequency, and duration of the Ca2+ response (30, 31). By prolonging the ability of P2Y1R to mobilize intracellular Ca2+, NHERF-2 may determine which signaling pathways are ultimately engaged after P2Y1R stimulation in different cell types.
NHERF family proteins are known to associate with many of the intermediates involved in Gαq-mediated signaling, including Gαq itself (24), PKCα (25), and PLCβ (26). Thus, NHERF-2 may control P2Y1R activity by acting as a scaffold to link these key signaling proteins in the vicinity of the receptor, allowing for a more sustained signaling response. This model is analogous to the model proposed for the action of the PDZ domain-containing protein InaD in regulating rhodopsin-mediated visual signaling in Drosophila (32) and the action of the non-PDZ protein Homer in regulating metabotropic glutamate receptor-mediated calcium signaling in mammalian neurons (33). However, InaD, Homer, and NHERF-2 have quite distinct effects on the dynamics of the calcium responses induced by their associated receptors, presumably tailored to meet the unique requirements of the cell types in which these receptor/scaffold combinations are expressed. Beyond the interaction with P2Y1R described here, NHERF-2 is also known to associate with and regulate the activity of at least one other GPCR, the parathyroid hormone receptor 1 (PTH1R), with the primary demonstrated action of NHERF-2 on PTH1R being an effect on the receptor's G protein-coupling specificity (34). Interestingly, PTH1R and P2Y1R are known to exhibit substantial cross-talk and synergistic signaling (35), which might be related to the fact that both of these receptors can associate with NHERF-2 in a functionally relevant manner.
In addition to linking P2Y1R to downstream effectors and enhancing P2Y1R signaling, the P2Y1R–NHERF-2 interaction may have other physiologically important consequences. First, NHERF-2 is known to interact with actin-associated ezrin-radixinmoesin proteins (36) and thus might serve as a link between P2Y1R and the actin cytoskeleton. Second, the ability of NHERF proteins to interact with PKC (25) might facilitate the tethering of PKC in the vicinity of P2Y1R and thereby modulate the ability of PKC to phosphorylate and regulate the activity of P2Y1R (17). Third, NHERF-2 is known to regulate the activity of Na+/H+ exchangers (37) and therefore may play a role in linking P2Y1R to modulation of Na+/H+ exchange (38). Finally, it is interesting to note that NHERF-2 has been shown to form a high-affinity complex with the cystic fibrosis transmembrane regulator (CFTR) in A6 cells (39). It has also recently been shown that CFTR activity in A6 cells is regulated by P2Y1R in a manner that is blocked by overexpression of a dominant-negative NHERF-2 construct (40). These unusual data, considered together with our description of a direct interaction between P2Y1R and NHERF-2, suggest the possibility that P2Y1R, NHERF-2, and CFTR form a cellular triple complex that is critical for purinergic regulation of CFTR channel activity in certain cell types.
P2YRs are major targets for commonly prescribed antithrombotic drugs (41) and are also targets for pharmaceuticals that might be useful in the future treatment of epilepsy, chronic pain, brain tumors, heart disease, atherosclerosis, cystic fibrosis, and other diseases (42). To fully exploit the enormous therapeutic potential of the P2YR family, however, it is essential to understand how the various P2YR subtypes are different from one another and how their activity might be differentially regulated in distinct tissues. We have found that the PDZ scaffold protein NHERF-2 associates with a specific motif on the P2Y1R CT and profoundly regulates P2Y1R functional activity in a cell-specific manner. These data provide a striking example of GPCR regulation by a receptor-associated scaffold protein and also represent an important step toward understanding the molecular mechanisms underlying subtype-specific and cell-specific regulation of P2YRs.
Supplementary Material
Acknowledgments
We thank Anna Goldschmidt for technical assistance in the Ca2+ imaging experiments and Michael Salter (Univerisity of Toronto, Toronto) for kindly providing 1321N1 human astrocytoma cells. This work was supported by National Institutes of Health (NIH) grants (to R.A.H., Y.S., S.F.T., and C.C.Y.), a Distinguished Young Scholar in Medical Research award from the W. M. Keck Foundation (to R.A.H.), a Canadian Institutes of Health Research Postdoctoral Fellowship (to S.R.F.), and a Postdoctoral National Research Service Award grant from the NIH (to C.J.L.). Equipment used in Ca2+ imaging experiments was purchased through National Alliance for Research on Schizophrenia and Depression funds.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: P2YR, purinergic P2Y receptor; P2Y1R, P2YR subtype 1; GPCR, G protein-coupled receptor; NHERF-2, Na+/H+ exchanger regulatory factor type 2; PDZ, PSD-95/Drosophila Discs large/ZO-1 homology; PDZ2, second PDZ domain; PLCβ, phospholipase Cβ; CT, carboxyl terminus; HA, hemagglutinin; 2-MeSADP, 2-methylthio-ADP.
References
- 1.Ralevic, V. & Burnstock, G. (1998) Pharmacol. Rev. 50, 413–492. [PubMed] [Google Scholar]
- 2.Webb, T. E., Feolde, E., Vigne, P., Neary, J. T., Runberg, A., Frelin, C. & Barnard, E. A. (1996) Br. J. Pharmacol. 119, 1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, W. C. & Chen, C. C. (1998) Glia 22, 360–370. [DOI] [PubMed] [Google Scholar]
- 4.Czajkowski, R., Lei, L., Sabala, P. & Baranska, J. (2002) FEBS Lett. 513, 179–183. [DOI] [PubMed] [Google Scholar]
- 5.Marteau, F., Le Poul, E., Communi, D., Communi, D., Labouret, C., Savi, P., Boeynaems, J. M. & Gonzalez, N. S. (2003) Mol. Pharmacol. 64, 104–112. [DOI] [PubMed] [Google Scholar]
- 6.Moran-Jimenez, M. J. & Matute, C. (2000) Brain Res. Mol. Brain Res. 78, 50–58. [DOI] [PubMed] [Google Scholar]
- 7.Janssens, R., Communi, D., Pirotton, S., Samson, M., Parmentier, M. & Boeynaems, J. M. (1996) Biochem. Biophys. Res. Commun. 221, 588–593. [DOI] [PubMed] [Google Scholar]
- 8.Fam, S. R., Gallagher, C. J. & Salter, M. W. (2000) J. Neurosci. 20, 2800–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neary, J. T., Kang, Y., Willoughby, K. A. & Ellis, E. F. (2003) J. Neurosci. 23, 2348–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippov, A. K., Brown, D. A. & Barnard, E. A. (2000) Br. J. Pharmacol. 129, 1063–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weissman, T. A., Riquelme, P. A., Ivic, L., Flint, A. C. & Kriegstein, A. R. (2004) Neuron 43, 647–661. [DOI] [PubMed] [Google Scholar]
- 12.Korchazhkina, O., Wright, G. & Exley, C. (1999) Br. J. Pharmacol. 127, 701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabre, J. E., Nguyen, M., Latour, A., Keifer, J. A., Audoly, L. P., Coffman, T. M. & Koller, B. H. (1999) Nat. Med. 10, 1199–1202. [DOI] [PubMed] [Google Scholar]
- 14.Greig, A. V., Linge, C., Terenghi, G., McGrouther, D. A. & Burnstock, G. (2003) J. Invest. Dermatol. 120, 1007–1015. [DOI] [PubMed] [Google Scholar]
- 15.Sellers, L. A., Simon, J., Lundahl, T. S., Cousens, D. J., Humphrey, P. P. & Barnard, E. A. (2001) J. Biol. Chem. 276, 16379–16390. [DOI] [PubMed] [Google Scholar]
- 16.Idestrup, C. P. & Salter, M. W. (1998) Neuroscience 86, 913–923. [DOI] [PubMed] [Google Scholar]
- 17.Fam, S. R., Gallagher, C. J., Kalia, L. V. & Salter, M. W. (2003) J. Neurosci. 23, 4437–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, R. A. & Lefkowitz, R. J. (2002) Circ. Res. 91, 672–680. [DOI] [PubMed] [Google Scholar]
- 19.Sheng, M. & Sala, C. (2001) Annu. Rev. Neurosci. 24, 1–29. [DOI] [PubMed] [Google Scholar]
- 20.Lau, A. G. & Hall, R. A. (2001) Biochemistry 30, 8572–8580. [DOI] [PubMed] [Google Scholar]
- 21.Balasubramanian, S., Teissere, J. A., Raju, D. V. & Hall, R. A. (2004) J. Biol. Chem. 279, 18840–18850. [DOI] [PubMed] [Google Scholar]
- 22.Yun, C. H., Lamprecht, G., Forster, D. V. & Sidor, A. (1998) J. Biol. Chem. 273, 25856–25863. [DOI] [PubMed] [Google Scholar]
- 23.Paquet, M. & Smith, Y. (2003) J. Neurosci. 23, 7659–7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rochdi, M. D., Watier, V., La Madeleine, C., Nakata, H., Kozasa, T. & Parent, J. L. (2002) J. Biol. Chem. 277, 40751–40759. [DOI] [PubMed] [Google Scholar]
- 25.Lee-Kwon, W., Kim, J. H., Choi, J. W., Kawano, K., Cha, B., Dartt, D. A., Zoukhri, D. & Donowitz, M. (2004) Am. J. Physiol. Cell Physiol. 285, 1527–1536. [DOI] [PubMed] [Google Scholar]
- 26.Hwang, J. I., Heo, K., Shin, K. J., Kim, E., Yun, C., Ryu, S. H., Shin, H. S. & Suh, P. G. (2000) J. Biol. Chem. 275, 16632–16637. [DOI] [PubMed] [Google Scholar]
- 27.Peters, A., Palay, S. L. & Webster, H. (1991) The Fine Structure of the Nervous System: Neurons and Their Supporting Cells (Oxford Univ. Press, New York).
- 28.Sabala, P., Czajkowski, R., Przybylek, K., Kalita, K., Kaczmarek, L. & Baranska, J. (2001) Br. J. Pharmacol. 132, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall, R. A., Ostedgaard, L. S., Premont, R. T., Blitzer, J. T., Rahman, N., Welsh, M. J., Lefkowitz & R. J. (1998) Proc. Natl. Acad. Sci. USA 95, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berridge, M. J., Lipp, P. & Bootman, M. D. (2000) Nat. Rev. Mol. Cell Biol. 1, 11–21. [DOI] [PubMed] [Google Scholar]
- 31.Dolmetsch, R. E., Lewis, R. S., Goodnow, C. C. & Healy, J. I. (1997) Nature 386, 855–858. [DOI] [PubMed] [Google Scholar]
- 32.Tsunoda, S., Sierralta, J., Sun, Y., Bodner, R., Suzuki, E., Becker, A., Socolich, M. & Zuker, C. S. (1997) Nature 388, 243–249. [DOI] [PubMed] [Google Scholar]
- 33.Tu, J. C., Xiao, B., Yuan, J. P., Lanahan, A. A., Leoffert, K., Li, M., Linden, D. J. & Worley, P. F. (1998) Neuron 21, 717–726. [DOI] [PubMed] [Google Scholar]
- 34.Mahon, M. J., Donowitz, M., Yun, C. C. & Segre, G. V. (2002) Nature 417, 858–861. [DOI] [PubMed] [Google Scholar]
- 35.Buckley, K. A., Wagstaff, S. C., McKay, G., Gaw, A., Hipskind, R. A., Bilbe, G., Gallagher, J. A. & Bowler, W. B. (2001) J. Biol. Chem. 276, 9565–9571. [DOI] [PubMed] [Google Scholar]
- 36.Bretscher, A., Chambers, D., Nguyen, R. & Reczek, D. (2000) Annu. Rev. Cell Dev. Biol. 16, 113–143. [DOI] [PubMed] [Google Scholar]
- 37.Yun, C. H., Oh, S., Zizak, M., Steplock, D., Tsao, S., Tse, C. M., Weinman, E. J. & Donowitz, M. (1997) Proc. Natl. Acad. Sci. USA. 94, 3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixon, S. J., Yu, R., Panupinthu, N. & Wilson, J. X. (2004) Glia 47, 367–376. [DOI] [PubMed] [Google Scholar]
- 39.Raghuram, V., Mak, D. D. & Foskett, J. K. (2001) Proc. Natl. Acad. Sci. USA. 98, 1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guerra, L., Favia, M., Fanelli, T., Calamita, G., Svelto, M., Bagorda, A., Jacobson, K. A., Reshkin, S. J. & Casavola, V. (2004) Pflugers Arch. 449, 66–75. [DOI] [PubMed] [Google Scholar]
- 41.Hollopeter, G., Jantzen, H. M., Vincent, D., Li, G., England, L., Ramakrishnan, V., Yang, R. B., Nurden, P., Nurden, A., Julius, D. & Conley, P. B. (2001) Nature 409, 202–207. [DOI] [PubMed] [Google Scholar]
- 42.Burnstock, G. & Williams, M. (2000) J. Pharmacol. Exp. Ther. 295, 862–869. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.