Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) is a cAMP-dependent protein kinase- and ATP-regulated chloride channel, the activity of which determines the rate of electrolyte and fluid transport in a variety of epithelial tissues. Here we describe a mechanism that regulates CFTR channel activity, which is mediated by PDZ domains, a family of conserved protein-interaction modules. The Na+/H+ exchanger regulatory factor (NHERF) binds to the cytoplasmic tail of CFTR through either of its two PDZ (PDZ1 and PDZ2) domains. A recombinant fragment of NHERF (PDZ1–2) containing the two PDZ domains increases the open probability (Po) of single CFTR channels in excised membrane patches from a lung submucosal gland cell line. Both PDZ domains are required for this functional effect, because peptides containing mutations in either domain are unable to increase channel Po. The concentration dependence of the regulation by the bivalent PDZ1–2 domain is biphasic, i.e., activating at lower concentrations and inhibiting at higher concentrations. Furthermore, either PDZ domain alone or together is without effect on Po, but either domain can competitively inhibit the PDZ1–2-mediated stimulation of CFTR. Our results support a molecular model in which bivalent NHERF PDZ domains regulate channel gating by crosslinking the C-terminal tails in a single dimeric CFTR channel, and the magnitude of this regulation is coupled to the stoichiometry of these interactions.
The cystic fibrosis transmembrane conductance regulator (CFTR) is a member of the ATP-binding cassette family of membrane transporters, the structure of which is comprised of two homologous motifs. Each motif has six putative transmembrane helices followed by a cytoplasmic nucleotide-binding domain (NBD), with the two halves linked by a cytoplasmic regulatory domain with multiple consensus sites for phosphorylation by cAMP-dependent protein kinase and protein kinase C (1). CFTR is a Cl− ion channel, the activity (gating) of which depends on its state of phosphorylation, and ATP hydrolysis cycles at the two NBDs (2, 3). Dysfunctional CFTR activity is associated with the pathogenesis of diseases including cystic fibrosis, secretory diarrhea, and pancreatitis (2). CFTR is expressed in the apical membrane of epithelial cells in tissues affected in these diseases, where its activity contributes to the rate of transepithelial salt and fluid transport.
In addition, CFTR also may regulate other ion channels, including outwardly rectifying chloride channels, amiloride-sensitive epithelial sodium channels, and renal outer-medullary potassium channels (4). The mechanisms involved in these interactions are unknown but could possibly result from direct associations of CFTR with other channels or indirectly from CFTR interactions with other proteins. Recent studies suggest that CFTR may exist in macromolecular complexes, in which protein–protein interactions influence its Cl− channel activity. In Calu-3 lung submucosal gland cells, CFTR Cl− channel activity was shown to be coupled functionally to protein kinase A anchoring proteins (5, 6). The cytoplasmic N-terminal domain of CFTR, which is involved in CFTR gating (7), interacts with syntaxin 1A (8). This interaction inhibits CFTR-mediated whole cell Cl− currents (9) and in Xenopus oocytes inhibits the transport of CFTR to the cell surface (10). AMP-activated protein kinase, a metabolic sensor with kinase activity regulated by cellular AMP:ATP ratios, binds to the C terminus of CFTR and inhibits CFTR Cl− currents in Xenopus oocytes (11). Finally, the PDZ-domain-containing protein Na+/H+ exchanger regulatory factor (NHERF) binds to the C-terminal residues of CFTR (12–14).
NHERF was identified originally as a regulatory factor involved in the cAMP-dependent inhibition of Na+/H+ exchanger (type 3; refs. 15–17). It contains two PDZ (for PSD95, Discs-large and ZO-1) domains, which are conserved protein–protein interaction modules that mediate binding to specific C-terminal motifs on target proteins (18, 19). Based on in vitro binding studies, it was proposed that NHERF binds to CFTR through its PDZ1 domain (12–14), with the PDZ2 domain available to interact with other proteins such as Yes-associated protein 65 (20). Two human isoforms of NHERF have been identified and referred to as NHERF1 and NHERF2/E3KARP (14, 16). NHERF1 is also known as EBP50 [for ezrin/radixin/moesin (ERM)-binding phosphoprotein-50] and is linked to the actin cytoskeleton by binding to ERM proteins through its C-terminal ERM-binding domain (21). It has been proposed that NHERF might link CFTR with the actin cytoskeleton through association with ezrin, and that ezrin can serve also as an A kinase anchoring protein (22) to facilitate cAMP-dependent protein kinase-mediated phosphorylation of CFTR (6, 12, 13). Moreover, it was shown that the PDZ binding motif of CFTR is critical for its apical membrane localization in polarized epithelial cells (23). However, no function has been identified for the specific interaction of NHERF with CFTR, although it is conceivable that this interaction could influence CFTR function at multiple levels, including its channel activity.
To understand the functional consequences of the interaction of NHERF with CFTR, we examined the interaction between CFTR and the PDZ domains of NHERF1. In this report, we have identified a previously unidentified role for NHERF1 in the regulation of CFTR Cl− channel activity. We demonstrate that NHERF1 binds to the cytoplasmic tail of CFTR through either of its two PDZ (PDZ1 and PDZ2) domains with affinities comparable to those observed for other PDZ-domain-mediated protein interactions. A recombinant fragment of NHERF containing the two PDZ domains (PDZ1–2) increases the Po of single CFTR channels in excised membrane patches from the Calu-3 cell line. This functional effect requires both PDZ domains because mutations in either domain abolish the PDZ1–2-mediated stimulation of channel Po. The regulation of CFTR channel gating by PDZ1–2 is biphasic, activating at lower concentrations and inhibiting at higher concentrations. Based on our results, we propose that a single functional CFTR Cl− channel is a dimer containing two PDZ binding motifs, which when bound and crosslinked by bivalent PDZ domains causes a conformational change in the channel that affects gating. Our results further imply that the magnitude of CFTR channel activity in epithelial cells can be modulated by the stoichiometry of NHERF–CFTR interactions.
Materials and Methods
Yeast Two-Hybrid Analysis.
Yeast two-hybrid screens using the C-terminal 70 aa of CFTR (1,411–1,480) as bait were performed as described (11). Wild-type and mutant CFTR C-terminal fragments were PCR-amplified from pcDNA–CFTR (24) by using primers containing desired mutations and subcloned into pLexA (CLONTECH). Hemagglutinin (HA)-tagged human NHERF1 was constructed as described (25) and various fragments of NHERF were PCR-amplified and cloned into pB42AD (CLONTECH). All plasmid constructs were verified by sequencing. Sequences of primers used for PCR amplification are available on request. Yeast “bait” and “prey” plasmids were cotransformed into the yeast reporter strain EGY48[p8op-lacZ] and plated on synthetic dropout (SD) plates [−his, −ura, −trp] to select for transformants. The transformants were restreaked onto both SD plates [−his, −ura, −trp, −leu] and 5-bromo-4-chloro-3-indolyl β-D-galactoside (X-gal)/SD plates. Growth on leucine-deficient plates and blue colonies on X-gal plates were used as indicators of protein interaction. The relative strengths of the interactions, designated as none (−) to high (+++) were based on growth rates and intensities of blue color of the colonies.
Cell Culture, Transfection, Immunoprecipitation, and Western Blotting.
Chinese hamster ovary (CHO) cells and a CHO cell line (CHO-BQ2) that stably expresses CFTR were maintained as described (26) and Calu-3 cells were cultured in DMEM/F12 medium (GIBCO/BRL) containing 10% (vol/vol) FBS. Subconfluent CHO cells were transfected by using lipofectamine (GIBCO/BRL) as per the manufacturer's instructions. Coimmunoprecipitation and Western blotting were performed as described (11).
Glutathione S-Transferase (GST) Affinity Binding Assays.
[35S]methionine-labeled full-length CFTR and CFTR-L1480A (Leu-1480 was replaced by Ala) was prepared by using an SP6 TnT (Promega) kit in the presence of canine pancreatic microsomes (Promega). In vitro-translated products were diluted 1:20 in binding buffer [(phosphate-buffered pH 7.5) 250 mM NaCl/2 mM EDTA/1% Triton X-100/BSA 2 mg/ml] and incubated overnight with 250 nM of GST–PDZ1–2 fusion protein (described below). GST fusion proteins were pulled down by using glutathione agarose beads preadsorbed with BSA. The bound beads were washed and eluted by using SDS sample buffer. The eluates were resolved by SDS/PAGE and processed for fluorography.
Preparation of GST Fusion Proteins.
Regions corresponding to residues of PDZ1–2 (), PDZ1 (), and PDZ2 () of NHERF were PCR-amplified and cloned into pGEX-6P-1 (Amersham Pharmacia) to create GST fusion constructs. Mutations in the PDZ domains (Lys to Ala; K19A, K158A, and K159A) were created in the GST fusion constructs by adopting the Quick-Change mutagenesis strategy (Stratagene), and all constructs were confirmed by sequence analysis. GST fusion proteins were expressed in Escherichia coli and purified on glutathione-Sepharose 4B (Amersham Pharmacia). The GST moiety was cleaved from the GST–PDZ fusion proteins by digesting with PreScission protease (Amersham Pharmacia). The cleaved PDZ domains were dialyzed by using Hepes-buffered saline, pH 7.5. The purified PDZ domains were quantified by using Bio-Rad protein assay reagent, and their purity was assessed to be >90% by Coomassie staining of SDS/PAGE gels.
Surface Plasmon Resonance (SPR) Measurements and Kinetic Analysis of Sensorgrams.
BIAcore 3000 (Biacore AB, Uppsala, Sweden) was used for amine coupling of streptavidin to the F1 chip. A biotinylated, HPLC-purified peptide corresponding to the terminal 18 residues of CFTR (biotinLC-QIAALKEETEEEVQDTRL-COOH) was captured at three different densities ranging from ≈15 to ≈60 response units. Analytes (GST–PDZ fusion proteins) at various concentrations in Hepes-buffered saline-EP [0.01 M Hepes/0.15 M NaCl/3 mM EDTA/0.005% (vol/vol) polysorbate 20, pH 7.4; Biacore AB] were perfused at a flow rate of 30 μl/min. The sensor chip was regenerated between successive injections with 10 mM NaOH and 1 M NaCl. At least two replicate experiments were performed for each fusion protein. Response curves were generated by subtraction of the background signal generated simultaneously on the control flow cell. The background-subtracted curves from replicates were averaged and prepared for fitting by subtracting the signal generated by buffer alone on experimental flow cells. In general, a short period after the injection start and stop were excluded from the fitting to avoid sample dispersion artifacts. Sensorgram curves were evaluated in BIAEVALUATION 3.0 software (Biacore AB) by using numerical integration algorithms. The response curves of various analyte concentrations were globally fitted to simple bimolecular binding (A + B = AB) or bivalent analyte (A + B = AB; AB + B = ABB) kinetic schemes.
Electrophysiology and Kinetic Analysis.
For patch-clamp recording, Calu-3 cells were plated at a low density on sterile cover slips and cultured in 35-mm tissue culture dishes for 1–4 days before use. The bath solution contained (in mM): 140 NaCl, 2 MgCl2, 0.1 CaCl2, 1 EGTA, 2 MgATP, and 10 Hepes (pH 7.5) with 200 units/ml cAMP-dependent protein kinase (PKA) catalytic subunit (Promega). Ca2+ activity was buffered to ≈40 nM (EGTA/CaCl2). The pipette solution was the same but without ATP and PKA. Forskolin (1 μM) was added to the bath solution to stimulate CFTR activity in intact cells and facilitate detection of channels in subsequently excised patches. All experiments were performed at room temperature and ± 60-mV transmembrane potential. Single-channel currents were filtered at 100 Hz, and digitized at 2 kHz and recorded by using PULSE + PULSEFIT software (HEKA Electronics, Lambrecht/Pfalz, Germany). Current records of at least 5-min duration were analyzed for each experimental condition by TAC software (Bruxton, Seattle, WA) for Po evaluation. The total number of channels in a patch was assumed to be the maximum number of open-channel current levels observed over the full duration of the experiment (15–180 min). The duration of each current record used for analysis was long enough to ensure a higher than 99% confidence level that the assumption is valid. The data were fitted and modeled by using IGOR PRO 3.4 (WaveMetrics, Lake Oswego, OR). Data are presented as mean ± SEM. Student's t test was used to determine the significance (P < 0.01).
Results
CFTR C Terminus Interacts with NHERF-PDZ Domains.
In a yeast two-hybrid screen performed to identify proteins that interact with the cytoplasmic C-terminal tail of CFTR, we isolated the C-terminal fragment (132–358, PDZ2–ERM) of NHERF1 from a human testis cDNA library. The interaction of CFTR with NHERF1 was verified in mammalian cells. HA-tagged NHERF was expressed in CFTR-expressing CHO-BQ2 cells. CFTR was specifically coimmunoprecipitated from cells expressing NHERFHA but not from cells that expressed CFTR alone (Fig. 1B), thus confirming their biochemical association in vivo in mammalian cells.
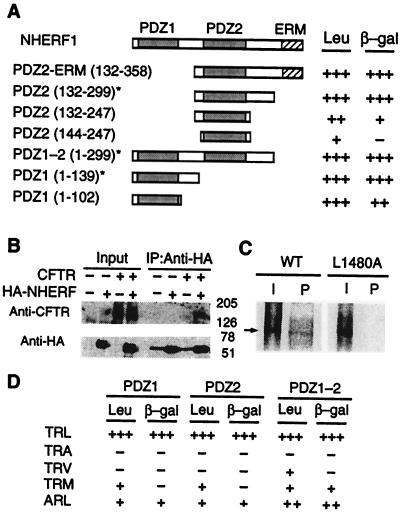
Figure 1.
Specific interaction of CFTR and NHERF. (A) Yeast two-hybrid analysis of CFTR–NHERF interaction. Full-length NHERF is shown at the top. NHERF cDNA isolated from the two-hybrid screen () and other deletion constructs are aligned beneath the full-length NHERF. Interaction strength of NHERF constructs with the C terminus (1,411–1,480) of CFTR are summarized based on induction of reporter genes β-galactosidase (β-gal) and Leu-2 (Leu). PDZ domains used in all subsequent experiments are marked (*). (B) Coimmunoprecipitation of CFTR and NHERF from CHO cells. Extracts (Input) from control cells and cells expressing HA-tagged NHERF and/or CFTR were immunoprecipitated (IP) with anti-HA antibodies, and the immunoprecipitates were blotted by using anti-CFTR or anti-HA antibodies, as indicated. Input lanes contain 5% of extract used for immunoprecipitation. (C) GST-affinity binding assays. [35S]methionine-labeled in vitro-translated products (I) and the GST-PDZ1–2 () “pull-down” products (P) of wild-type CFTR (WT) and mutant CFTR (L1480A) are indicated. (D) Specificity of PDZ1 and PDZ2 for CFTR C terminus. The terminal 3 aa of CFTR (1,411–1,480) and the NHERF PDZ domains are indicated.
Published results suggested that whereas the consensus binding sequence for the PDZ1 domain matches that of CFTR, the PDZ2 domain had a different binding specificity (12, 14). Surprisingly, the PDZ2 domain of NHERF interacted strongly with CFTR C terminus in our yeast two-hybrid assay. To resolve this discrepancy, we carried out a systematic deletion analysis to delineate the regions of NHERF that are critical for binding to CFTR. This analysis revealed that sequences adjacent to either PDZ domain (defined by sequence homology) are critical for optimal binding and confer to both domains comparable interactions with CFTR (Fig. 1A). Specificity of different PDZ domains for their binding partners is conferred by residues within the PDZ domain and at the (−2) and (0) residues in the C terminus of target proteins (27, 28) with the consensus sequence (T/S)-X-φ (in which φ is a hydrophobic amino acid; ref. 29). The specificity of CFTR–NHERF PDZ interactions was examined by mutating residues (0) and (−2) of CFTR and testing their ability to interact with PDZ1 and PDZ2 either individually or together. The mutation L1480A resulted in a complete loss of interaction, and mutations T1378A, L1480V, and L1480M resulted in markedly reduced interaction (Fig. 1D), with both PDZ1 and PDZ2 having similar specificities for their target ligands. Some PDZ domains, including NHERF PDZ domains, can interact with internal motifs (16, 30, 31). However, there are no additional PDZ binding sites in CFTR, because the PDZ1–2 domains bound to full-length in vitro-translated CFTR but not to mutant CFTR (L1480A) (Fig. 1C). Thus, whereas either PDZ domain can bind to CFTR, a CFTR molecule has only one PDZ binding site.
Binding Kinetics of CFTR C Terminus to NHERF PDZ Domains.
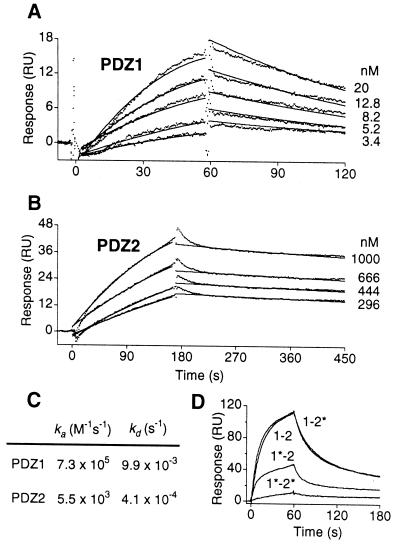
Quantitative binding kinetics of CFTR to NHERF PDZ domains were determined by SPR by using an immobilized peptide corresponding to the C terminus of CFTR. The binding affinities determined for the two PDZ binding domains (PDZ1, 14 nM and PDZ2, 74 nM) are within the range of affinities observed for other PDZ-domain–protein interactions (32, 33). The binding kinetics were distinct for the two domains, with the GST–PDZ1 (Fig. 2 A and C) peptide displaying faster association and dissociation kinetics compared with GST–PDZ2 (Fig. 2 B and C). Structural studies have revealed conserved positively charged amino acids in the GLGF loop of PDZ domains corresponding to Lys-19 (PDZ1) and Lys-158 and -159 (PDZ2) in NHERF that are essential for optimal binding to target peptides (27, 34). We mutated these lysines to alanine in either or both PDZ domains. In SPR experiments (Fig. 2D), PDZ1–2 peptide bound avidly to the CFTR C terminus. Whereas the binding of PDZ2 mutant (PDZ1–2*) was nearly indistinguishable from that of wild-type PDZ1–2, the binding of PDZ1 mutant (PDZ1*–2) was markedly reduced. The binding was significantly reduced further for the PDZ1*–2* double mutant. These results further confirm that both PDZ domains are involved in CFTR binding, with a larger contribution from PDZ1 than from PDZ2.
Figure 2.
Kinetics of CFTR–NHERF PDZ domain interactions. Experimental data (dots) represent average of repeated injections of analyte for each concentration, as indicated. Global fit of the data to a simple bimolecular reaction is shown by solid lines. (A) Kinetic-response data for GST–PDZ1 binding to CFTR. (B) Kinetics-response data for GST–PDZ2 binding to CFTR. (C) Association (ka) and the dissociation (kd) rate constants for binding of CFTR C terminus to PDZ1 and PDZ2 domains. (D) Sensorgram overlays for various GST–PDZ1–2 fusion proteins binding to CFTR C terminus at a concentration of 50 nM. Wild-type PDZ1–PDZ2 (1–2), PDZ1K19A–PDZ2 (1*–2), PDZ1–PDZ2 K158A, K159A (1–2*) and PDZ1K19A–PDZ2 K158A, K159A (1*–2*), are indicated.
Increase in CFTR Channel Po by Bivalent PDZ-Domain-Mediated Interaction.
NHERF can function as a scaffold to link CFTR with ezrin, a putative protein kinase A anchoring protein (22), by its C-terminal ERM-binding domain. To differentiate functional effects on CFTR channel activity caused by the binding of PDZ domains from those caused by the scaffolding function of NHERF, we used a truncated form of NHERF that lacks the ERM-binding domain but retains the PDZ domains. Various PDZ-domain-containing peptides were prepared by cleaving GST from the fusion proteins, and the functional consequences of PDZ-domain-mediated interactions with CFTR were studied by patch-clamp electrophysiology of excised inside-out membrane patches obtained from Calu-3 cells, which express CFTR endogenously.
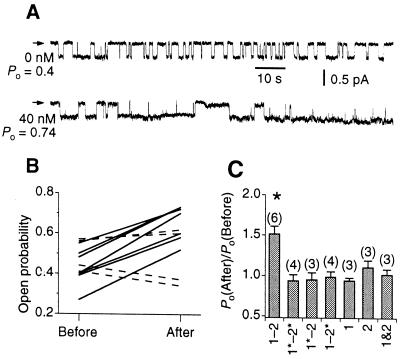
Addition of PDZ1–2 peptide to the bath solution markedly increased the single-channel Po of CFTR with no observable change in the single-channel conductance or the number of channels (Fig. 3A). In contrast, a peptide containing mutations in both PDZ domains (PDZ1*–2*) was without effect on Po (Fig. 3B), although channel activities in these patches were stimulated by addition of wild-type PDZ1–2 peptide to the bath containing the mutated peptide (data not shown). Surprisingly, mutations in either PDZ1 (PDZ1*–2) or PDZ2 (PDZ1–2*) also abolished the ability of PDZ1–2 peptide to increase channel activity (Fig. 3C). Furthermore, neither individual PDZ domain added separately or mixed together up to a concentration of 120 nM was able to stimulate CFTR (Fig. 3C). These results indicate that although CFTR can bind to either PDZ domain, an interaction with a bivalent PDZ1–2 domain is necessary for stimulation of CFTR channel activity. The PDZ1–2-domain-mediated increases in Po were observed in all our experiments and in membrane patches containing single as well as multiple CFTR channels. The Po distribution of CFTR channels in multichannel patches (number of channels ≤8, total of five multiple-channel experiments) conformed to a binomial distribution (i.e., the probability of k channels being open in an n-channel patch is (1 − Po)n − kPokn!/[k!(n − k)!]) both before and after stimulation by the peptide, suggesting that the channels were equivalent and they opened and closed independently of each other after stimulation by the bivalent PDZ1–2 peptide (data not shown). Together, these observations indicate that a functional CFTR channel contains at least two PDZ binding sites. Binding and crosslinking of these sites by a bivalent PDZ1–2 domain results in stimulation of channel activity.
Figure 3.
Effect of NHERF PDZ-domain-mediated interactions on CFTR channel activity. (A) Single-channel current traces of CFTR before and after addition of 40 nM PDZ1–2. The closed level is indicated by arrowheads. (B) Po of CFTR before and after addition of 40 nM PDZ1–2 (solid lines) or PDZ1*–2* (dashed line). Each trace represents individual experiments. (C) Mean ratio of CFTR Po after and before addition of 40 nM of various PDZ peptides. * Indicates significant difference of Po (P < 0.01) after addition of peptide. PDZ1–2 (1–2), PDZ1*–PDZ2* (1*–2*), PDZ1*–PDZ2 (1*–2), PDZ1–PDZ2* (1–2*), PDZ1 (1), PDZ2 (2), and PDZ1 and PDZ2 mixed together (1&2) are indicated. Number of experiments indicated in parentheses.
Biphasic Regulation of CFTR Channel Po by PDZ1–2.
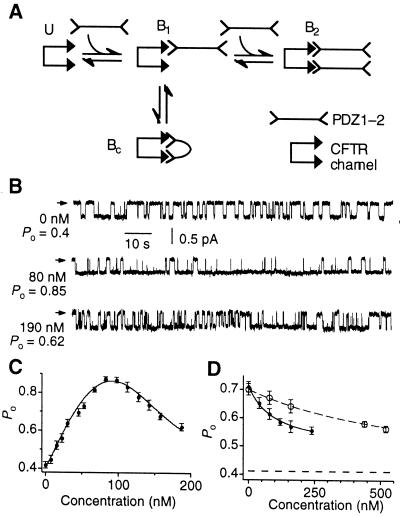
A simple equilibrium model describing the binding of a bivalent ligand to a bivalent molecule that can account for our results is shown in Fig. 4A. In this scheme, the kinetically faster PDZ1 domain of the PDZ1–2 peptide binds initially to one of the PDZ binding sites of an unbound CFTR channel (U). The resulting complex B1 facilitates the interaction of the slower-binding PDZ2 domain with the second PDZ binding site to create a crosslinked channel (Bc), which has a higher Po than either the U or the B1 states. The slower dissociation rate of PDZ2 further assists in stabilizing the crosslinked channel. With increasing concentrations of PDZ1–2 peptides, the PDZ1 domain of another PDZ1–2 peptide will compete with either PDZ domain of the bound PDZ1–2 peptide to generate state B2, in which the CFTR channel is associated with two different PDZ1–2 peptides. Because the channel in this state is not crosslinked, it is expected to have a lower Po. We therefore examined the concentration dependence of PDZ1–2 on membrane patches containing only one active CFTR channel. Increasing the concentration of PDZ1–2 up to 100 nM stimulated the Po of CFTR. At optimal concentrations of PDZ1–2 (80–100 nM), the maximal Po attained was as high as 0.85–0.9 (Fig. 4 B and C). Further increases in PDZ1–2 concentrations decreased channel activity (Fig. 4 B and C). The dependence of CFTR Po on PDZ1–2 concentration could be fitted by a biphasic Hill equation with half-maximal activating concentration (Kact) of ≈80 nM and half-maximal inhibiting concentration (Kinh) of ≈145 nM. The similar magnitudes of Kact and Kinh suggest that the activating and inhibitory PDZ binding sites have similar affinities, consistent with the hypothesis that inhibition is caused by self-competition of a bivalent molecule with itself. These results therefore support a model in which the bivalent PDZ1–2 peptide stimulates CFTR-channel activity by crosslinking two PDZ binding sites associated with a CFTR channel. Higher concentrations of PDZ1–2 reverse this effect by competing for the PDZ binding sites.
Figure 4.
Biphasic regulation of CFTR by bivalent PDZ-domain-mediated interactions. (A) An equilibrium model illustrating various physical states (U, B1, Bc, and B2) of a CFTR channel bound to bivalent PDZ1–2 is described. The CFTR channel is represented by two PDZ binding motifs. See text for details. (B) Single-channel current traces of CFTR at various concentrations of PDZ1–2. Arrowheads indicate closed level. (C) A representative single-channel experiment showing the biphasic dependence of CFTR Po on PDZ1–2 concentration. (D) Representative single-channel experiments showing inhibition of PDZ1–2-mediated effect on CFTR Po by monovalent PDZ domains (PDZ1, filled circles; PDZ2, open circles). The Po of CFTR before addition of 40 nM PDZ1–2 is indicated (lower dashed line).
Accordingly, this model predicts that monovalent single PDZ domains, which are capable of binding to CFTR (Figs. 1 and 2) but are without effect on Po (Fig. 3C), should nevertheless competitively inhibit the increase in CFTR Po mediated by the bivalent PDZ1–2 domain. This prediction was confirmed experimentally. Membrane patches containing a single active CFTR Cl− channel stimulated by the addition of 40 nM PDZ1–2 were exposed to increasing concentrations of single PDZ domains in the bath (Fig. 4D). The increase in Po caused by the bivalent PDZ1–2 peptide was competitively inhibited by either monovalent PDZ domain, with Kinh ≈100 nM and ≈667 nM for PDZ1 and PDZ2, respectively (Fig. 4D). Competitive inhibition by individual PDZ domains provides further support for a model involving crosslinking of CFTR PDZ binding sites by bivalent PDZ domains.
Discussion
In the present study, we have examined the interactions of the PDZ domains of NHERF with CFTR and determined the consequences of these interactions for the ion channel activity of CFTR. Our results demonstrate that (i) both PDZ domains of NHERF are capable of binding to CFTR, (ii) a bivalent PDZ-domain-mediated interaction with CFTR leads to an increase in its Po, (iii) the regulation of CFTR channel activity by NHERF is biphasic, and (iv) addition of monovalent PDZ domains either alone or together, which by themselves have no effect on Po, inhibits this increase in activity. These results have implications for the regulation of CFTR channel activity and for the oligomeric structure of the channel.
The associations of ion channels and membrane receptors with distinct sets of intracellular proteins to form macromolecular complexes are often mediated by scaffolding proteins containing PDZ domains (18, 19). Spatial grouping of key transduction components into multiprotein complexes is functionally important for achieving speed and efficacy of signaling by reducing diffusion distances. Like other PDZ-domain-containing proteins, NHERF has been characterized mainly as a cytoskeletal-associated scaffold protein involved in the assembly of protein complexes. By demonstrating that the binding of NHERF to CFTR has a direct effect on CFTR channel gating, our data provide evidence that PDZ-domain-mediated interactions may have catalytic functions, in addition to the passive scaffold function of providing docking sites for multiprotein complexes. Thus, in addition to colocalizing key components of signal transduction pathways, PDZ-domain-containing proteins may play an active role by directly modulating the gating of ion channels or the activities of other proteins.
Previous studies have demonstrated that the PDZ1 domain of NHERF binds to CFTR C terminus but suggested that PDZ2 had a different specificity (12, 14, 17). We suggest that the discrepancy between those results and our data here is likely attributable to two factors. First, our results from yeast two-hybrid analysis revealed that sequences adjacent to either PDZ domain (defined by sequence homology) are necessary for optimal binding and confer to both domains comparable interactions with CFTR. Second, steady-state binding equilibria might not have been established by the end of the binding assays in previous studies because of the slower association rate of PDZ2. Our SPR experiments revealed a striking difference between the two PDZ domains in that the PDZ1–CFTR complex is formed much faster than PDZ2–CFTR, whereas the PDZ2–CFTR complex is much more stable than the PDZ1–CFTR complex (Fig. 2C). Importantly, our functional analyses suggest that both PDZ domains are critical for PDZ-domain-mediated stimulation of CFTR channel activity.
We observed an increase in the Po of CFTR only when both PDZ domains were functional and linked in a bivalent construct. Neither of the individual PDZ domains (PDZ1 and PDZ2) increased the channel Po, whether alone or when mixed together. Furthermore, mutation of either PDZ domain in the bivalent construct abolished the stimulatory activity. These results demonstrate that, although either PDZ domain is sufficient for binding to CFTR, a bivalent molecule containing two PDZ domains is required to increase CFTR channel activity. Several observations indicate that the two required sites for binding to both PDZ domains in a bivalent molecule reside within a single CFTR channel. First, and most important, the PDZ1–2-domain-mediated increase in CFTR Po was observed even in membrane patches containing only one CFTR channel. Of note, stimulation of gating in patches containing only one or several CFTR channels was never associated with an increase in the number of active channels after the addition of PDZ1–2. Second, in membrane patches that contained more than one CFTR channel, gating of the individual channels remained independent of each other after stimulation by the bivalent construct. This result suggests that the channels are not coordinately regulated after the addition of PDZ1–2 peptide, as might occur if the bivalent PDZ domain was stimulating gating by crosslinking two CFTR channels. Our data indicate that, although a CFTR molecule has only one PDZ-domain-binding site (Fig. 1 C and D), a single functional channel is a dimer containing two PDZ-domain-binding sites.
Implications of Bivalent PDZ-Domain-Mediated Regulation for CFTR Channel Structure.
What are the molecular identities of the two PDZ binding sites? Two hypotheses suggest themselves. First, NHERF could crosslink CFTR with a nearby, stably associated, as-yet-unidentified protein containing a NHERF PDZ binding motif. In this model, NHERF acts as an adapter, facilitating a heterodimeric interaction that modulates the activity of CFTR. However, enhancement of CFTR-channel activity by the NHERF PDZ1–2 domain also has been observed in heterologous cell types (CHO, NIH 3T3, and HEK293) that do not express CFTR endogenously (data not shown), which suggests that the unidentified molecule must be ubiquitously expressed. An alternate hypothesis is that a CFTR channel is a homodimer composed of two CFTR molecules, so that even in patches with only one CFTR channel, NHERF can crosslink two CFTR tails, thereby inducing conformational changes in the channel structure that affect its gating. This conclusion that a CFTR channel is a homodimer of CFTR molecules is at variance with biochemical studies that have suggested that CFTR is a monomer (35). However, recent electron microscopy and electrophysiology studies also have suggested that the CFTR chloride channel is a homodimer (36, 37). Further studies will be required to determine the oligomeric state of a CFTR channel.
Functional Implications for CFTR Gating.
We and others have observed CFTR channels with high Po (0.8–0.9) in cell-attached patches from Calu-3 cells (38) and in excised giant patches from cardiac myocytes (39), whereas the maximal Po rarely exceeds 0.5 in excised inside-out patches in numerous studies. Various kinetic models of CFTR gating fail to explain the high Po of CFTR in cell-attached patches (40). One possibility that might account for this discrepancy is that a cytosolic factor, which affects CFTR gating, is rapidly lost after membrane-patch excision. Our experiments have shown that a Po of 0.85 can be achieved in excised inside-out patches in the presence of optimal concentrations (80–100 nM) of NHERF PDZ1–2. An intriguing hypothesis is that NHERF, or another bivalent PDZ-domain-containing protein, is the cytosolic factor that is lost after excision. By adding the NHERF PDZ domains to excised membrane patches, we have functionally reconstituted the physiological conditions required for generating high Po CFTR channels.
The biphasic concentration dependence of the modulation of CFTR Po by NHERF suggests that the stoichiometries of these proteins will be critical in determining the magnitude of this regulation. Depending on the native concentration of CFTR and the relative concentration of NHERF, both positive and negative regulatory roles could be ascribed to NHERF. Long-term and dynamic regulation of CFTR- and NHERF-associated protein complexes may influence the activity of CFTR by affecting these stoichiometries, with implications for the rates of transepithelial salt and water transport in tissues affected in cystic fibrosis, pancreatitis, and secretory diarrhea. Our results raise the possibility that PDZ-domain-mediated interactions could affect gating of other ion channels, which may have important functional consequences.
Acknowledgments
We thank W. Skach for A2 antibodies; V. Ramesh for NHERF cDNA; S. Tanaka, K. Hallows, S. McBride, and C. Shi for technical assistance; and M. Ostap and G. Chandry for comments on the manuscript. We also thank G. Canziani and the University of Pennsylvania Biosensor and Structural Biology Cores Group for assistance with the SPR experimental protocols and support services. V.R. is supported by a Cystic Fibrosis Foundation Fellowship. These studies were supported by the Cystic Fibrosis Foundation.
Abbreviations
- CFTR
cystic fibrosis transmembrane conductance regulator
- NHERF
Na+/H+ exchanger regulatory factor
- ERM
ezrin/radixin/moesin
- CHO,Chinese hamster ovary
HA, hemagglutinin
- GST
glutathione S-transferase
- SPR
surface plasmon resonance
Footnotes
See commentary on page 787.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031538898.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031538898
References
- 1.Riordan J R, Rommens J M, Kerem B-S, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J-L, et al. Science. 1989;245:1066–1072. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard D N, Welsh M J. Physiol Rev. 1999;79:S23–S45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- 3.Gadsby D C, Nairn A C. Physiol Rev. 1999;79:S77–S107. doi: 10.1152/physrev.1999.79.1.S77. [DOI] [PubMed] [Google Scholar]
- 4.Schwiebert E M, Benos D J, Egan M E, Stutts M J, Guggino W B. Physiol Rev. 1999;79:S145–S166. doi: 10.1152/physrev.1999.79.1.S145. [DOI] [PubMed] [Google Scholar]
- 5.Huang P, Trotter K, Boucher R C, Milgram S L, Stutts M J. Am J Physiol. 2000;278:C417–C422. doi: 10.1152/ajpcell.2000.278.2.C417. [DOI] [PubMed] [Google Scholar]
- 6.Sun F, Hug M J, Bradbury N A, Frizzel R A. J Biol Chem. 2000;275:14360–14366. doi: 10.1074/jbc.275.19.14360. [DOI] [PubMed] [Google Scholar]
- 7.Naren A P, Cormet-Boyaka E, Fu J, Villain M, Blalock J E, Quick M W, Kirk K L. Science. 1999;286:544–548. doi: 10.1126/science.286.5439.544. [DOI] [PubMed] [Google Scholar]
- 8.Naren A P, Nelson D J, Xie W W, Jovov B, Pevsner J, Bennett M K, Benos D J, Quick M W, Kirk K L. Nature (London) 1997;390:302–305. doi: 10.1038/36882. [DOI] [PubMed] [Google Scholar]
- 9.Naren A P, Quick M W, Collawn J F, Nelson D J, Kirk K L. Proc Natl Acad Sci USA. 1998;95:10972–10977. doi: 10.1073/pnas.95.18.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters K W, Qi J, Watkins S C, Frizzell R A. Am J Physiol. 1999;277:C174–C180. doi: 10.1152/ajpcell.1999.277.1.C174. [DOI] [PubMed] [Google Scholar]
- 11.Hallows K R, Raghuram V, Kemp B E, Witters L A, Foskett J K. J Clin Invest. 2000;105:1711–1721. doi: 10.1172/JCI9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S S, Raab R W, Schatz P J, Guggino W B, Li M. FEBS Lett. 1998;427:103–108. doi: 10.1016/s0014-5793(98)00402-5. [DOI] [PubMed] [Google Scholar]
- 13.Short D B, Trotter K W, Reczek D, Kreda S M, Bretscher A, Boucher R C, Stutts M J, Milgram S L. J Biol Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- 14.Hall R A, Ostedgaard L S, Premont R T, Blitzer J T, Rahman N, Welsh M J, Lefkowitz R J. Proc Natl Acad Sci USA. 1998;95:8496–8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinman E J, Steplock D, Wang Y, Shenolikar S. J Clin Invest. 1995;95:2143–2149. doi: 10.1172/JCI117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun C H C, Oh S, Zizak M, Steplock D, Tsao S, Tse C M, Weinman E J, Donowitz M. Proc Natl Acad Sci USA. 1997;94:3010–3015. doi: 10.1073/pnas.94.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall R A, Premont R T, Chow C W, Blitzer J T, Pitcher J A, Claing A, Stoffel R H, Barak L S, Shenolikar S, Weinman E J, et al. Nature (London) 1998;392:626–630. doi: 10.1038/33458. [DOI] [PubMed] [Google Scholar]
- 18.Craven S E, Bredt D S. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 19.Sheng M, Pak D T S. Annu Rev Physiol. 2000;62:755–778. doi: 10.1146/annurev.physiol.62.1.755. [DOI] [PubMed] [Google Scholar]
- 20.Mohler P J, Kreda S M, Boucher R C, Sudol M, Stutts M J, Milgram S L. J Cell Biol. 1999;147:879–890. doi: 10.1083/jcb.147.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reczek D, Berryman M, Bretscher A. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dransfield D T, Bradford A J, Smith J, Martin M, Roy C, Mangeat P H, Goldenring J R. EMBO J. 1998;16:35–43. doi: 10.1093/emboj/16.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyer B D, Denton J, Karlson K H, Reynolds D, Wang S S, Mickle J E, Milewski H, Cutting G R, Guggino W B, Li M, Stanton B A. J Clin Invest. 1999;104:1353–1361. doi: 10.1172/JCI7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugita M, Yue Y, Foskett J K. EMBO J. 1998;17:898–908. doi: 10.1093/emboj/17.4.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murthy A, Gonzalez-Agosti C, Cordero E, Pinney D, Candia C, Solomon F, Gusella J, Ramesh V. J Biol Chem. 1998;273:1273–1276. doi: 10.1074/jbc.273.3.1273. [DOI] [PubMed] [Google Scholar]
- 26.Tabcharani J A, Chang X-B, Riordan J R, Hanrahan J W. Nature (London) 1991;352:628–631. doi: 10.1038/352628a0. [DOI] [PubMed] [Google Scholar]
- 27.Doyle D A, Lee A, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 28.Daniels D L, Cohen A R, Anderson J M, Bruger A T. Nat Struct Biol. 1998;5:317–325. doi: 10.1038/nsb0498-317. [DOI] [PubMed] [Google Scholar]
- 29.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 30.Shieh B H, Zhu M Y. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 31.Hillier B J, Christopherson K S, Prehoda K E, Bredt D S, Lim W A. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 32.Saras J, Heldin C H. Trends Biochem Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- 33.Garner C C, Nash J, Huganir R L. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- 34.Cabral J H M, Petosa C, Sutcliffe M J, Raza S, Byron O, Poy F, Marfatia S M, Chishti A H, Liddington R C. Nature (London) 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- 35.Marshall J, Fang S, Ostedgaard L S, O'Riordan C R, Ferrara D, Amara J F, Hoppe H, Scheule R K, Welsh M J, Smith A E, Cheng S H. J Biol Chem. 1994;269:2987–2995. [PubMed] [Google Scholar]
- 36.Eskandari S, Wright E M, Kreman M, Starace D M, Zampighi G A. Proc Natl Acad Sci USA. 1998;95:11235–11240. doi: 10.1073/pnas.95.19.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerhusen B, Zhao J Y, Xie J X, Davis P B, Ma J J. J Biol Chem. 1999;274:7627–7630. doi: 10.1074/jbc.274.12.7627. [DOI] [PubMed] [Google Scholar]
- 38.Haws C, Finkbeiner W E, Widdicombe J H, Wine J J. Am J Physiol. 1994;266:L502–L512. doi: 10.1152/ajplung.1994.266.5.L502. [DOI] [PubMed] [Google Scholar]
- 39.Hwang T-C, Nagel G, Nairn A C, Gadsby D C. Proc Natl Acad Sci USA. 1994;91:4698–4702. doi: 10.1073/pnas.91.11.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeltwanger S, Wang F, Wang G T, Gillis K D, Hwang T C. J Gen Physiol. 1999;113:541–554. doi: 10.1085/jgp.113.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]