Abstract
OBJECTIVE
Intracranial pressure (ICP) is an important therapeutic target in many critical neuropathologies. The current tools for ICP measurements are invasive; hence, these are only selectively applied in critical cases where the benefits surpass the risks. To address the need for low-risk ICP monitoring, the authors developed a noninvasive alternative.
METHODS
The authors recently demonstrated noninvasive quantification of ICP in an animal model by using morphological analysis of microvascular cerebral blood flow (CBF) measured with diffuse correlation spectroscopy (DCS). The current prospective observational study expanded on this preclinical study by translating the method to pediatric patients. Here, the CBF features, along with mean arterial pressure (MAP) and heart rate (HR) data, were used to build a random decision forest, machine learning model for estimation of ICP; the results of this model were compared with those of invasive monitoring.
RESULTS
Fifteen patients (mean age ± SD [range] 9.8 ± 5.1 [0.3–17.5] years; median age [interquartile range] 11 [7.4] years; 10 males and 5 females) who underwent invasive neuromonitoring for any purpose were enrolled. Estimated ICP (ICPest) very closely matched invasive ICP (ICPinv), with a root mean square error (RMSE) of 1.01 mm Hg and 95% limit of agreement of ≤ 1.99 mm Hg for ICPinv 0.01–41.25 mm Hg. When the ICP range (ICPinv 0.01–29.05 mm Hg) was narrowed on the basis of the sample population, both RMSE and limit of agreement improved to 0.81 mm Hg and ≤ 1.6 mm Hg, respectively. In addition, 0.3% of the test samples for ICPinv ≤ 20 mm Hg and 5.4% of the test samples for ICPinv > 20 mm Hg had a limit of agreement > 5 mm Hg, which may be considered the acceptable limit of agreement for clinical validity of ICP sensing. For the narrower case, 0.1% of test samples for ICPinv ≤ 20 mm Hg and 1.1% of the test samples for ICPinv > 20 mm Hg had a limit of agreement > 5 mm Hg. Although the CBF features were crucial, the best prediction accuracy was achieved when these features were combined with MAP and HR data. Lastly, preliminary leave-one-out analysis showed model accuracy with an RMSE of 6 mm Hg and limit of agreement of ≤ 7 mm Hg.
CONCLUSIONS
The authors have shown that DCS may enable ICP monitoring with additional clinical validation. The lower risk of such monitoring would allow ICP to be estimated for a wide spectrum of indications, thereby both reducing the use of invasive monitors and increasing the types of patients who may benefit from ICP-directed therapies.
Keywords: intracranial pressure, noninvasive sensing, light tissue interactions, diffuse correlation spectroscopy, machine learning, cerebral blood flow, trauma
Intracranial pressure (ICP) is defined as the pressure within the craniospinal compartment, a closed system that comprises a fixed total volume of neural tissue, cerebral blood, and cerebrospinal fluid (CSF).1 Within the rigid skull, any increase in one volume compartment needs to be matched by an equal decrease in another or ICP will rise, as illustrated by the Monro-Kellie doctrine.2,3 Although the CSF and the blood compartments provide some buffering of increasing volume, once the compensatory capacity is exhausted, further increases in volume lead to a rise in ICP, as routinely observed in patients with many neuropathologies.2,4–7 Thus, ICP is an important biomarker in the care of patients with these critical diseases.8–13
The techniques currently used in the clinic for ICP measurements are invasive, requiring pressure transducers to be placed into the parenchyma8 or the ventricular system.14 Due to the invasive nature of these procedures, there are associated risks that include infection and hemorrhagic complications.15 Hence, ICP monitoring is typically only recommended in critical cases where the benefits exceed the procedural risks.16 There is a need for a noninvasive alternative that minimizes the risk of complications and at the same time allows accurate ICP monitoring for a broader set of indications.
Over the past years, a variety of noninvasive approaches have been proposed for ICP measurement, including transcranial Doppler (TCD) ultrasonography and several diffuse optical techniques.17–23 Although TCD is considered promising and is being used in some regions of the world, factors such as operator dependence, motion sensitivity, and lack of ease for long-term bedside measurements limit its widespread clinical applicability.24,25 The measurement of optic nerve sheath diameter (ONSD), which increases with ICP, with standard intracranial imaging or more recently with ultrasound have also shown the promise of clinical utility.26,27 However, ONSD measurement is limited in that it is susceptible to inaccuracy due to differences in baseline ONSD between individuals, can only delineate between normal and elevated ICP, does not provide continuous monitoring, and presents additional shortcomings such as patient discomfort.26,27
In an attempt to derive absolute ICP values noninvasively, our prior work utilizing diffuse correlation spectroscopy (DCS) estimated ICP with an accuracy of < 4 mm Hg in a nonhuman primate model.28 DCS is an emerging diffuse optical technique that measures microvascular cerebral blood flow (CBF) noninvasively and continuously.29,30 DCS can resolve the cardiac pulsatility in CBF,31 and we have previously shown that the shape of the CBF cardiac pulse waveforms changes with baseline ICP; this association was previously explored and tested with TCD.32,33 Because DCS directly measures CBF instead of blood flow velocity, and also because the small cortical vessels probed with DCS are more susceptible to transmural pressure changes than the large vessels probed with TCD, DCS may be a superior option for noninvasive ICP measurement.34
This work specifically expanded on our preclinical study by translating the method to pediatric patients. First, we provide details on strategic and technical optimizations implemented in this work, including DCS data processing and machine learning tools. Then, we present data from 15 pediatric intensive care unit (PICU) patients and show estimation of ICP from the pulse morphological features of CBF with and without other physiological signals, e.g., mean arterial pressure (MAP) and heart rate (HR), by using a random forest regressor model.
Methods
To briefly summarize the methodology, pediatric patients were evaluated with DCS to extract CBF data. Next, morphological features were extracted from the CBF pulses, in addition to MAP and HR data, by using signal processing to train a machine learning model. Lastly, ICP estimated by the model was validated against invasively measured ICP.
Clinical Pediatric Population
The study was conducted at the PICU of the University of Pittsburgh Medical Center Children’s Hospital of Pittsburgh, Pittsburgh, Pennsylvania. The study protocol was approved by the University of Pittsburgh and Carnegie Mellon University Institutional Review Board.
Children between 1 month and under 18 years of age who underwent placement of an external ventricular drain (EVD) or a parenchymal pressure monitor for ICP monitoring as a part of their routine clinical care were eligible for enrollment. Written informed consent was provided by the legal guardian. Noninvasive monitoring was not performed in settings where it may have impeded patient care. For this study, a total of 18 patients were recorded and data from 3 patients were excluded. Two of these patients had a low signal-to-noise ratio (SNR), and 1 had a technical error resulting in incorrect synching of invasive and noninvasive data.
The mean ± SD (range) age of the population was 9.8 ± 5.1 (0.3–17.5) years, median age (interquartile range) was 11 (7.4) years, and 10 males and 5 females were included (Table 1). Optical measurement was taken on the left or right forehead, and the ICP sensor was surgically inserted at Kocher’s, Keen’s, and Frasier’s points as dictated by clinical necessity (Fig. 1). Each DCS measurement session spanned approximately 1 hour, with repeated measurements not exceeding a total of 6 hours/day for 7 consecutive days. During measurement, the patient was either supine or seated, as well as sedated or awake, depending on the clinical condition. Of 15 patients, 1 had a parenchymal pressure monitor for continuous ICP recording (Table 1). In patients with an EVD, a 3-way stopcock valve was turned to switch from draining CSF to recording ICP inside the closed catheter. Synched noninvasive recording was performed during the periods of time in which the EVD was clamped for clinical purposes.
TABLE 1.
Overview of the pediatric patient population, including measurement details
| Patient No. | Age (yrs)/Sex | Medical Diagnosis | Sensor Type | Location ICP | Location DCS | Value* |
|---|---|---|---|---|---|---|
| 1 | 11/M | Tumor | EVD | Lt | Not documented | 1 (1.9, 287) |
| 2 | 8.1/F | Possible hydrocephalus, unresponsiveness | EVD | Rt | Lt | 2 (6.7, 4311) |
| 3 | 4/M | TBI | ICP monitor | Rt | Lt | 1 (3, 1254) |
| 4 | 15/M | TBI | EVD | Rt | Lt | 1 (2.6, 863) |
| 5 | 14.4/F | Tumor | EVD | Rt | Rt | 1 (0.17, 60) |
| 6 | 13/F | Tumor | EVD | Rt | Rt | 3 (1.83, 556) |
| 7 | 14.3/M | Abscess/meningitis | EVD | Lt | Rt | 2 (1.6, 702) |
| 8 | 0.3/M | Shunt infection | EVD | Rt | Rt | 2 (7.5, 3023) |
| 9 | 12.1/M | TBI | EVD | Rt | Lt | 2 (8.1, 5089) |
| 10 | 5/M | Intraventricular hemorrhage | EVD | Lt | Rt | 1 (0.7, 188) |
| 11 | 13.2/F | Obstructive hydrocephalus | EVD | Lt | Not documented | 1 (1.9, 803) |
| 12 | 17.5/M | Abscess/meningitis | EVD | Rt | Not documented | 1 (0.8, 329) |
| 13 | 7.7/M | Abscess/meningitis | EVD | Rt | Lt | 2 (0.2, 79) |
| 14 | 2.3/M | Shunt infection | EVD | Rt | Lt/Rt | 3 (1.8, 1264) |
| 15 | 9/F | Tumor | EVD | Rt | Rt | 1 (0.6, 211) |
Location DCS = location of DCS probe on the forehead; Location ICP = location of invasive ICP sensor in the frontal lobe; TBI = traumatic brain injury.
Shown as days measured (hours analyzed, number of pulse averages).
FIG. 1.
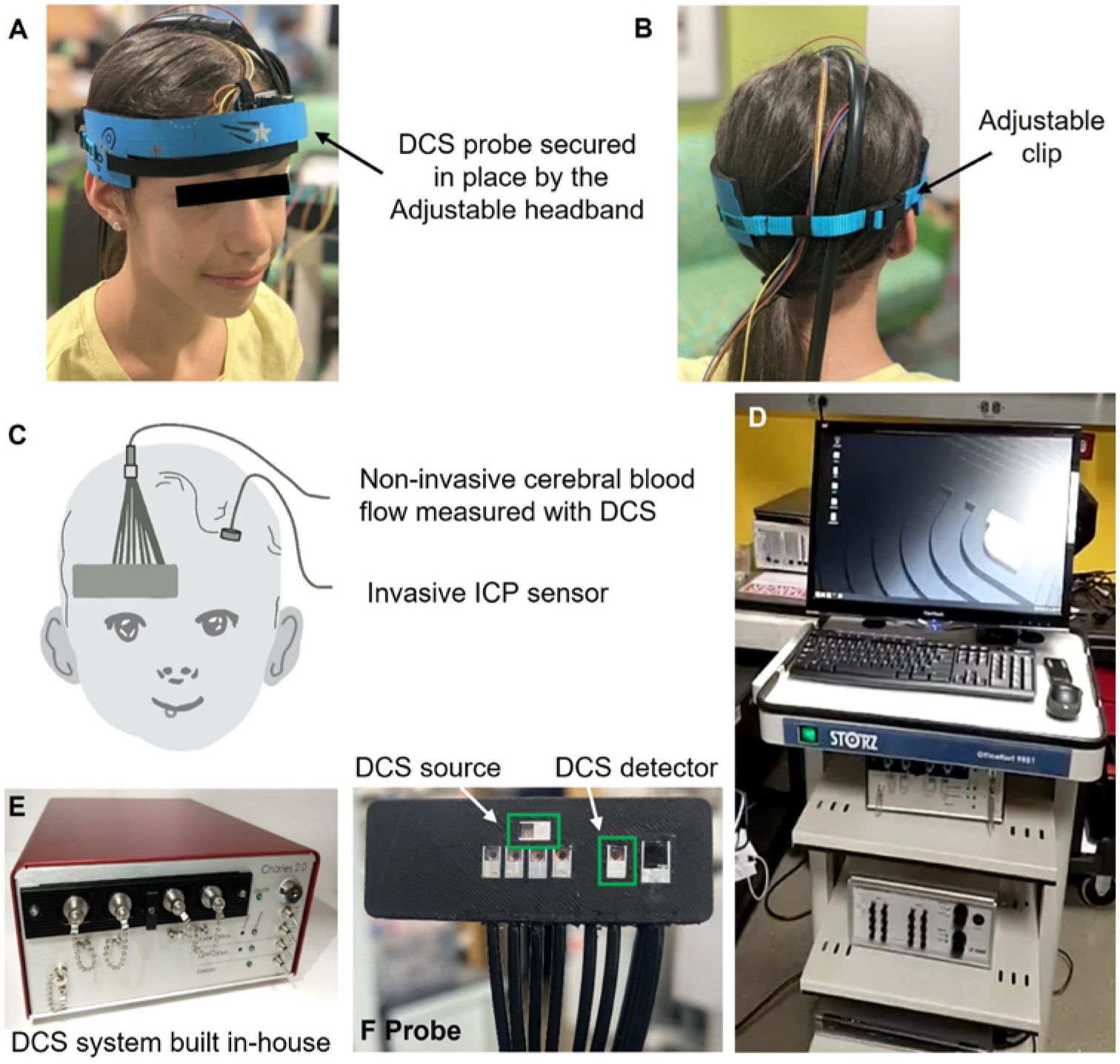
Details of DCS instrumentation and measurement setup. A–B: Probe secured on the left forehead with the adjustable headband. C: Probe placement on the right forehead of a patient, where the ICP monitor is surgically inserted in the left frontal lobe. D: Hospital cart with instruments placed. E: Custom-made DCS device. F: Custom-made DCS optical probe. The green regions refer to the DCS source and detector.
Signal Acquisition
DCS provides a measure of CBF in the microvasculature.29,35 A custom-built DCS device, which is not approved by the US Food and Drug Administration (FDA), was used (Fig. 1).28 DCS is similar to near-infrared spectroscopy, and some devices used for clinical purposes have been FDA approved and are often considered to have minimum risk. The device contained an auxiliary port to receive time markers used to time synchronize CBF with the signals obtained from the patient monitor. In addition to invasive ICP (ICPinv), electrocardiography (EKG) was performed and MAP was measured with the hospital bedside monitor (General Electric Solar 8000M, GE Healthcare). Although ICP and EKG data were measured continuously, MAP was measured sporadically, approximately every 30 minutes via a blood pressure cuff, or continuously measured with an arterial line. The optical probe was secured on the forehead with sports tape or a padded adjustable headband (Fig. 1A–B). More details on the DCS methodology can be found in the Supplemental Materials and Methods, Section 1.
Signal Processing
Signal processing was performed using MATLAB R2019b (The MathWorks Inc.). A total of 18 patients were recorded; however, 3 were excluded owing to poor SNRs or errors in patient monitoring data. Among the 15 included patients, the DCS sessions with poor SNRs (SNR < 4.8, empirically determined) were also removed. Movement, routine ICU procedures, room light, poor tissue contact, and poor probe placement were some of the probable reasons behind a poor SNR.
For each DCS measurement session, the CBF time series was preprocessed to remove noise such as respiration and time aligned with the signals from the patient monitor. Figure 2A shows example time traces of CBF and ICP from a representative patient, where the dashed vertical lines represent the R-peaks within the QRS complex of the EKG signal. The next step in data processing was to segment each individual CBF pulse from its time series, and an average of 100 consecutive pulses (150 for patients with an average SNR < 7) generated an average CBF cardiac pulse. The process was repeated, after the average window was shifted by 10% of the window length, to generate a series of average CBF pulses from the CBF time trace (Fig. 2B).
FIG. 2.
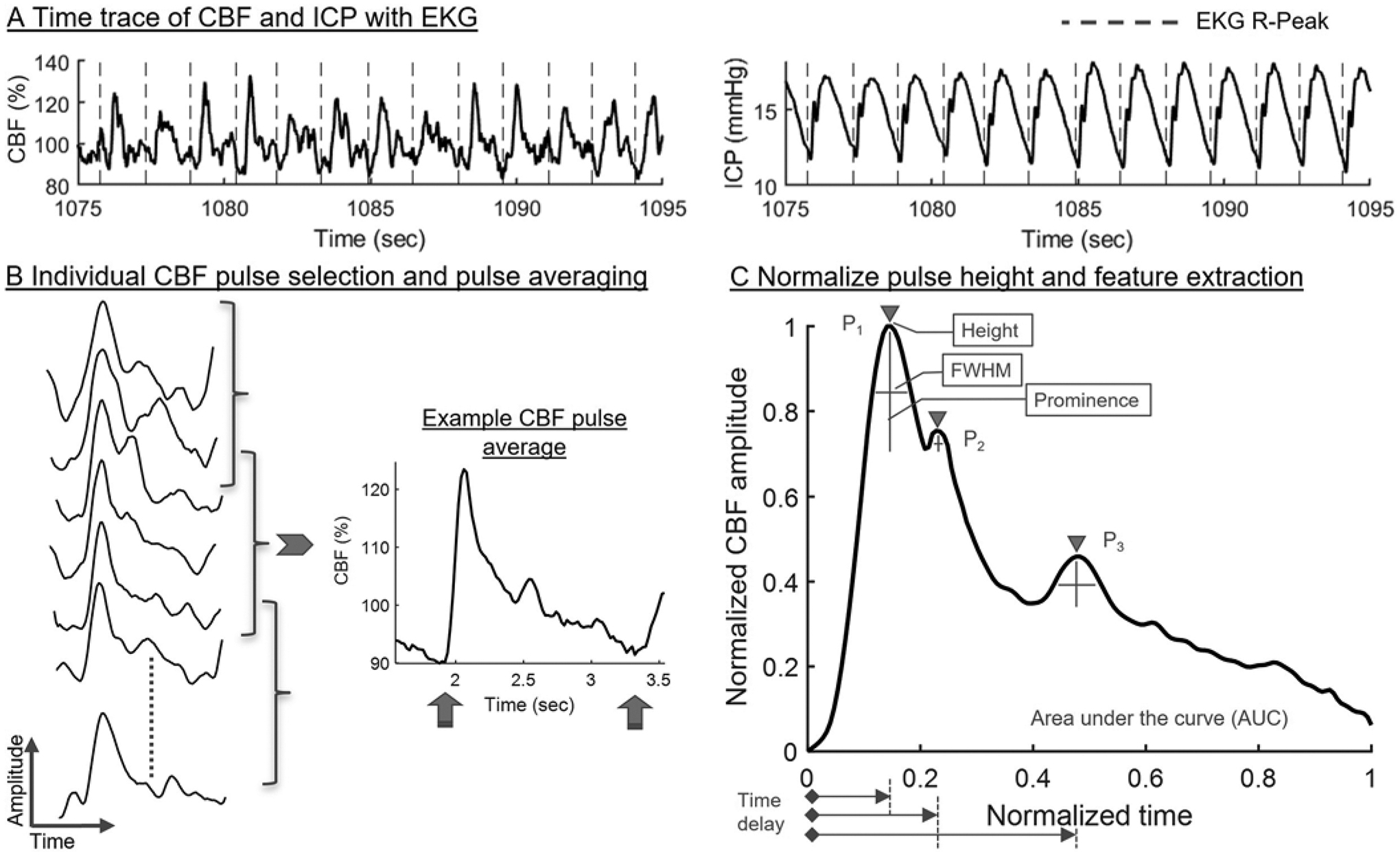
A: Example time traces of CBF and ICP from a representative patient, where the dashed vertical lines represent the R-peaks within the QRS complex of the EKG signal. B: Segmented CBF pulses on the left side, and an example CBF pulse average on the right side. The upward arrows indicate the two diastolic points utilized for pulse normalization in time. C: CBF pulse average normalized in amplitude and in time. The features extracted from the pulse average are described in the Regression Learner section of the Methods. FWHM = full width at half maximum.
Following that, the CBF pulse averages were normalized between 0 and 1 in time (x-axis) and amplitude (y-axis) (Fig. 2C) to achieve comparability across patients. At this step, ICPinv and MAP were averaged over the time span of CBF pulse averages to obtain average values. Additionally, the difference between the R-peaks within the QRS complex of the EKG signal was used to generate an HR value. Thus, at the end of signal processing, each CBF pulse average waveform was associated with a set of ICPinv, MAP, and HR values. The last column in Table 1 shows the number of days each patient was evaluated, the number of hours of the CBF time series that were analyzed, and the number of CBF pulse averages finally used for feature extraction. Altogether, a total of 19,019 pulse averages were extracted from 15 patients with an ICPinv range of 0.01–41.25 mm Hg.
Next, the morphological features of 3 different waveform peaks (P1–3) were extracted from the CBF pulse average (Fig. 2C). To describe the individual peaks, peak height (pk), prominence (p), full width at half maximum (w), and time point of the peak relative to the prior diastolic minimum (pos) were extracted. Area under the curve (AUC) was also acquired. Features were further obtained by quantifying the differences in pk, p, w, and pos between P1 and P2, as well as between P2 and P3. This resulted in a set of 21 features from each CBF pulse average for use in supervised machine learning. See the Supplemental Materials and Methods, Section 2, for further details on determining SNR, noise filtration, pulse averaging, normalization, and feature extraction.
Regression Learner
In this step, the entire data set of CBF pulses was randomly split, with 80% of the data used for training the model and 20% was held out from the training process for testing. The data points were randomly sampled in time. The model was optimized compared with our previous work in animals (Supplemental Materials and Methods, Section 3).28
Figure 3 describes the main aspects of the training and testing steps. The CBF features, MAP, HR, and ICPinv from the training set (total 15,215) were used to build and train a random decision forest with a bagged ensemble of 200 individual decision tree regressors. Each tree estimator was randomly assigned 60% of the training samples and 80% of the features (total 23) to reduce variance and limit overfitting.36 After training, the CBF features along with the corresponding MAP and HR data from the testing set were fed into each tree regressor to extract an estimated ICP. Lastly, the outputs of the 200 trees were averaged to obtain the estimated ICP value (ICPest). More details on the regression learner can be found in the Supplemental Materials and Methods, Section 3.
FIG. 3.
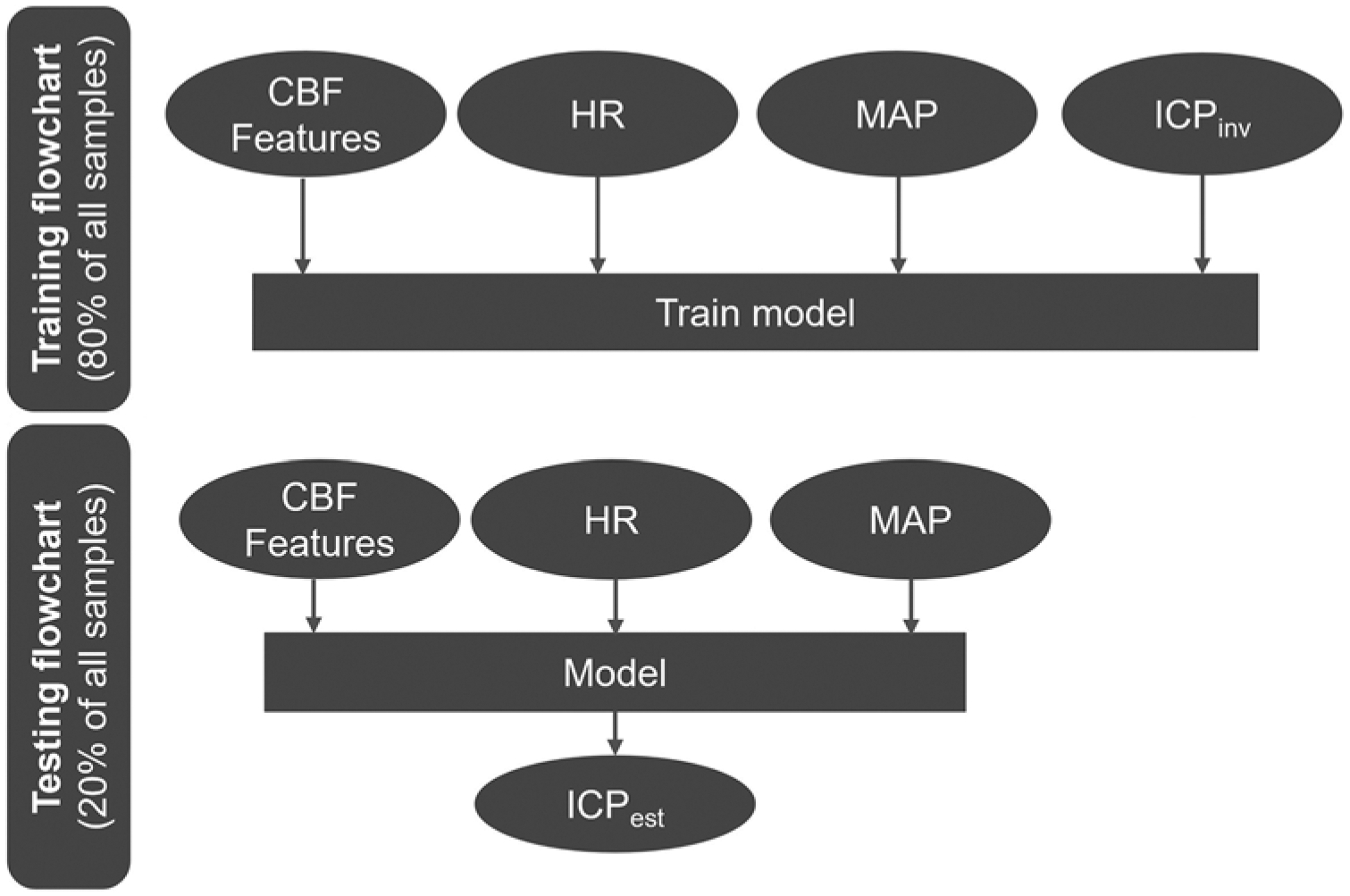
Algorithm flowchart of the machine learning model. A train/test split ratio of 80%/20% was used. Upper: The CBF morphological features and MAP, HR, and ICPinv values were used to train a random decision forest. Lower: During testing, the CBF features and MAP and HR values were fed into the model as input to obtain an estimated ICP.
Statistical Analysis
Statistical analyses were conducted using MATLAB R2019b (The MathWorks Inc.). For the statistical analyses, results from only the testing set were reported. The goodness of fit between ICPinv and ICPest was calculated using the coefficient of determination metric (R2) and root mean squared error (RMSE). Bland-Altman analysis was also conducted to determine the mean bias and 95% limit of agreement between the two methods. A discrepancy of 5 mm Hg between invasive and noninvasive monitoring was defined as the maximum difference that would justify use of DCS as, at minimum, a clinically useful screening adjunct.
Results
Random Decision Forest Regression Learner
The withheld testing data demonstrated that ICPest very closely matched ICPinv, with an R2 value of 0.97 and an RMSE error of 1.01 mm Hg (Fig. 4A); however, the sample population was not consistent across ICPinv values, as shown in the inset histogram. Noticeably, the size of the sample population was reduced drastically at high ICPinv values, which would subsequently reduce the sample size of the training set at these values. Because a small training set population is known to affect model performance,37 a new analysis was conducted with only ICPinv, for which the sample population matched or surpassed a certain threshold (as shown with the horizontal dotted line in the histogram shown Fig. 4A). This threshold represented 5% of the largest sample population. This masking based on the sample population narrowed down ICPinv to the range of 0.01–29.05 mm Hg. By using this narrower ICP range and by retraining the algorithm (Fig. 4B), we determined that the narrower ICPinv range improved both the R2 value and RMSE error to 0.98 and 0.81 mm Hg, respectively. The inset image shows a histogram of the sample population for the narrower ICPinv.
FIG. 4.
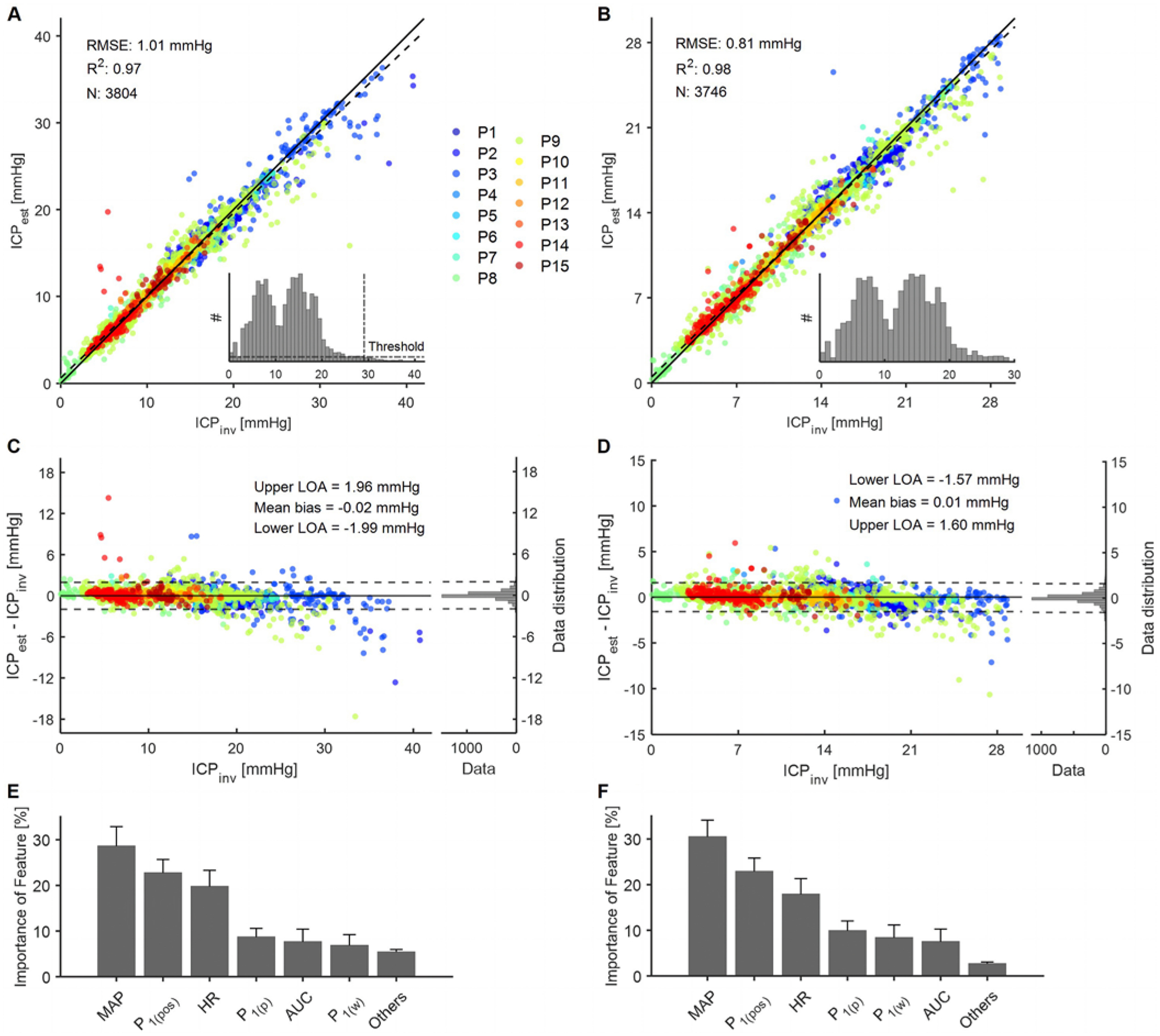
Results of the regression forest machine learning model for the entire range of ICPinv (0.01–41.25 mm Hg) and a narrower range of ICPinv (0.01–29.05 mm Hg). The data points are color coded according to patient identification. A–B: Performance of the regression forest. The solid line shows the ideal fit between ICPinv and ICPest, whereas the dashed line shows the linear fit. R2 represents goodness of fit, and N represents the sample size of the testing set. The inset images show histograms of ICPinv of the entire sample population before splitting into the train/test sets. A bin width of 0.83 mm Hg was used in the histogram. C–D: The Bland-Altman plot shows the difference between the two methods with respect to ICPinv. The solid line represents mean bias and the dashed lines represent the upper and lower 95% LOA. The histogram plots on the right side of each panel display the distributions of the differences. E–F: Distribution of features used in the regression forest model as a percentage of all chosen features of all decision criteria generated. The standard deviation across 200 individual trees is shown as error bars. Others refers to all other features apart from the ones shown in the plot. For nomenclature, please see the Signal Processing section of the Methods and Fig. 2C. LOA = limit of agreement.
Further evidence of goodness of fit for both ICPinv ranges was provided by the Bland-Altman plots shown in Fig. 4C and D, where the difference between ICPest and ICPinv was plotted against ICPinv. Negligible bias was observed for both the entire and narrower ICPinv ranges. The narrower ICPinv range also slightly improved the upper and lower limits of the agreement values and demonstrated that 95% of the testing data points were within ± 1.6 mm Hg of the corresponding ICPinv. Because 5 mm Hg was considered the acceptable limit of agreement for clinical validity of ICP sensing, we explored what percentage of the testing samples, less than and greater than 20 mm Hg, was outside the limit of agreement of 5 mm Hg. To summarize, for the entire ICP range, 0.3% of the test samples for ICPinv ≤ 20 mm Hg and 5.4% of the test samples for ICPinv > 20 mm Hg were outside the limit of agreement of 5 mm Hg. For an ICP range of 0.01–29.05 mm Hg, only 0.1% of data points for ICPinv ≤ 20 mm Hg and 1.1% of data points for ICPinv > 20 mm Hg were outside the limit of agreement of 5 mm Hg.
To further delve into the model, the importance of the features was analyzed for both ICPinv ranges (Fig. 4E and F). The order of features in terms of importance was consistent between the entire and narrower ICPinv ranges. MAP was the most widely used, as observed in our previous work.28 HR was also promising and turned out to be the third strongest predictor. Among the CBF features, the features related to the P1 peak were found to be the most useful compared with the other two peaks. For example, P1(pos) was the second strongest feature and P1(p) was the fourth strongest feature. P1(w) and AUC were interchangeably the fifth and sixth best features.
Leave-one-out analysis was also conducted, during which data points from 1 patient were used in the testing set and data points from the other 14 patients were used in the training set. This analysis mimics how well the algorithm would work for a new patient who was not a part of the training set. The process was repeated for each patient. When averaged over all 15 patients, an RMSE of 6.1 mm Hg and limit of agreement ≤ 7.4 mm Hg was observed for the entire ICP range, and an RMSE of 6 mm Hg and limit of agreement ≤ 7 mm Hg were observed for the narrower ICP range.
Dependence of Learner Performance on Model Parameters
Because learner performance can depend on the model parameters, several model parameters were tested against learner outputs; the results are shown in Table 2. All analyses were conducted for the narrow ICPinv range of 0.01–29.05 mm Hg. We investigated train/test split ratios of 80%/20%, 70%/30%, 60%/40%, and 50%/50%. The results showed that R2 changed slightly; however, both RMSE and limit of agreement values increased by as much as approximately 32% because the training population size was reduced from 80% to 50% of the train/test ratio.
TABLE 2.
Dependence of learner performance on algorithm parameters
| Analysis of Model Variables | Stationary Model Variables | Accuracy | Bland-Altman Analysis | ||||
|---|---|---|---|---|---|---|---|
| RMSE (mm Hg) | R2 | Mean (mm Hg) | Upper LOA (mm Hg) Lower LOA (mm Hg) | ||||
| Train/test split ratio | 80%/20% | Random forest, CBF, MAP, & HR | 0.81 | 0.98 | 0.01 | 1.6 | −1.57 |
| 70%/30% | 0.86 | 0.98 | 0.03 | 1.71 | −1.66 | ||
| 60%/40% | 0.98 | 0.97 | −0.02 | 1.9 | −1.93 | ||
| 50%/50% | 1.06 | 0.97 | −0.0 | 2.07 | −2.08 | ||
| Regressor type | Random forest | 80%/20% split, CBF, MAP, & HR | 0.81 | 0.98 | 0.01 | 1.6 | −1.57 |
| Bagging | 0.94 | 0.97 | 0.01 | 1.85 | −1.84 | ||
| Feature performance | CBF, MAP, & HR | Random forest & 80%/20% split | 0.81 | 0.98 | 0.01 | 1.6 | −1.57 |
| CBF & MAP | 1.03 | 0.97 | −0.03 | 1.99 | −2.05 | ||
| CBF & HR | 1.77 | 0.91 | 0.02 | 3.49 | −3.44 | ||
| CBF features only | 2.38 | 0.82 | −0.03 | 4.64 | −4.69 | ||
| MAP & HR only | 2.79 | 0.76 | 0.04 | 5.51 | −5.43 | ||
LOA = limit of agreement.
Because our previous work in animals used a different type of decision forest classifier called the bagging regressor, we also compared the bagging and random forest regressors.38 The parameters for the bagging regressor were kept as previously used,28 except the number of trees was kept at 200 for an even comparison. To summarize, although R2 showed only a slight improvement, the bagging regressor underperformed because it increased both the RMSE and limit of agreement values by approximately 17% (Table 2).
Lastly, features were tested to find the optimum feature set for the most accurate learner prediction. Because MAP and HR were among the top three important features in this analysis (Fig. 4E and F), either one or both features were excluded from the feature set to investigate how that could affect model performance. The results showed that truncating one or both features increased both RMSE and limit of agreement values by as much as approximately 200% compared with CBF, MAP, and HR (Table 2). However, when the same analysis was conducted with only MAP and HR as features (i.e., no CBF features), RMSE and the limit of agreement increased by approximately 246% compared with CBF, MAP, and HR. This confirmed that although MAP and HR were important features, the CBF features played a very important role in learner performance, and the best performance was achieved when MAP and HR were added to the model with CBF features.
Discussion
In this work, we have demonstrated the clinical translation of our previously developed, noninvasive DCS-based ICP estimation method to pediatric patients admitted to the PICU. We found that the waveform features in CBF measured in the cerebral microvasculature are influenced by ICP. These CBF features, along with HR and MAP data, were used to train a random forest regressor for mapping CBF onto ICP.28 To summarize the results, strong similarity between estimates of ICP and the invasive measurements (R2 = 0.97, RMSE = 1.01 mm Hg) were obtained with a limit of agreement ≤ 1.99 mm Hg. Model performance was improved for a narrower ICPinv range of 0.01–29.05 mm Hg, with RMSE reduced to 0.81 mm Hg and the limit of agreement reduced to ≤ 1.6 mm Hg. For this ICP range, only 0.1% of data points for ICPinv ≤ 20 mm Hg and 1.1% for ICPinv > 20 mm Hg were found with the limit of agreement > 5 mm Hg. These results show the potential of DCS as a valid screening tool for ICP.
More accurate ICP extraction was possible when evaluating a narrower ICPinv range of 0.01–29.05 mm Hg compared with the data set that included ICP > 30 mm Hg. The increase in estimation error likely stems from the lack of data at higher ICP and not the model inaccuracy. We believe that, with the addition of more data at higher ICP values, the model could be trained across a wider ICP range and thus improve overall model accuracy even at ICP > 30 mm Hg. Our results suggest that absolute ICP estimation based on noninvasive measurements of CBF cardiac waveforms is possible, with further enhanced performance with concurrent MAP and HR data. Although we used EKG signals to obtain HR, other means such as a wrist-worn wearable device should suffice.39 The invasively measured ICP used as the reference/gold standard was certainly not free from measurement error. The combined mean (95% CI) difference between invasive ICP probes, reportedly 1.6 (1.3–1.9) mm Hg in the literature,40 was comparable to the noninvasive limit of agreement values calculated in this study.
Machine learning approaches such as regression forests provide information about feature importance in terms of how often a feature was used for ICP splits at split nodes.28 This information could be used to improve algorithm performance in the future by optimizing good features and rejecting bad ones. In this work, MAP and HR were among the topmost features, as both signals were strongly related to the cardiac cycle. Among the CBF features, P1 peak was the most useful compared with the P2 and P3 features, which is consistent with our previous preclinical work. AUC was found to be the fifth or sixth best feature, which implies that changes in P1 peak features (pos, p, and w) were more significant than the overall broadening of the CBF pulse in contributing to change in AUC. Most notably, in contrast to the preclinical results, none of the differences in peak features between P1 and P2, or between P2 and P3, were found to be useful.
Because MAP and HR turned out to be among the top three features, analyses were conducted to explore how the model performed without these features. Truncating MAP and HR data from the feature set increased both RMSE and limit of agreement values by approximately 200%, but using only these two features in the feature set affected the algorithm outcome even more by raising the percentage error to approximately 246% compared with that of the feature set that also included CBF. This indicated that although MAP and HR were valuable components to the model, the CBF features were crucial, thus offering the best prediction accuracy when combined. In the comparison of regressor types, the random forest regressor performed slightly better than the bagging regressor, and we assume this was a result of the additional subsampling of the features (80%) and samples (60%) assigned to each tree during training; this an important attribute of the random forest regressor known to curb overfitting.36 The leave-one-out analysis showed an RMSE ≤ 6.1 mm Hg and a limit of agreement ≤ 7.4 mm Hg. Although this suggests that the model loses accuracy when validated against unknown data, we believe this is avoidable by increasing the number of patients and thus the training set population.37 With additional refinement, a noninvasive model capable of consistently predicting a limit of agreement < 5 mm Hg will have clinically meaningful applications. Invasive monitors could be then applied in the setting of equipoise, such as a result of ≥ 20 mm Hg. In our current model, we used 100–150 consecutive pulses to estimate 1 ICP value, corresponding to a readout of 1–2 per minute. Although the data were downsampled by averaging multiple waveforms, a readout of every pulsation should be feasible when SNR is high, producing real-time or near real-time ICP estimation.
Depth of measurement inside the brain varied due to differences in patient age, scalp thickness, and head size.41 Although different brain volumes were measured between patients, SNR was not age dependent (data not shown). Our previous animal study was conducted directly on the skull to avoid the effect of the scalp on DCS measurements. Although the clinical measurements in this work were taken with the scalp in place, we did not see an effect on data quality or model performance. This implies that the implementation of this technology is feasible across a wide span of pediatric and adult patients.
Our work compares favorably to other noninvasive ICP monitors that use TCD, which have shown an overall accuracy of ± 12 mm Hg,25 and acoustic signals propagating through the cranium within an accuracy of ≤ 7.38 mm Hg.42 In terms of methodology, a close similarity can be drawn to Fischer et al., who applied a neural network model with solely CBF time traces from DCS without using MAP or HR data.43 Although the accuracy of ICP estimation was comparable to the results shown here, we believe the added benefit of our approach is the interpretability of the features found. Given that the same importance of peak features was found between our animal studies and human data, we believe that the presented model can be generalized to different diseases, ages, and patient populations.
The current study was limited to its demonstration of potential efficacy in pediatric populations, although older patients in the cohort may be physiologically comparable to some adult populations. To show generalization, the next steps will involve recruiting a larger cohort of patients with various ages and conditions. We anticipate several challenges in translation to an adult population, e.g., increased scalp thickness and vascular stiffness. The increase in central arterial stiffening due to aging may alter MAP.44 However, we believe that the proposed method will remain highly accurate in the adult population because it performed well as MAP changed in children and adolescents.45
With decreasing risk of harm to patients, the possibility of new applications for ICP measurement may arise for traditional pathologies such as hydrocephalus and shunt malfunction, but also for novel ones such as concussion, intraoperative monitoring, and Chiari malformation. It is our hope, with time, that ICP will transition to becoming the next commonly used vital sign for patients with neurological disease and allow for a greater understanding of normal and abnormal ICP across a wide spectrum of patients.
Conclusions
DCS-based sensing of ICP showed a high level of similarity with invasive measurements. CBF waveforms were used for ICP estimation, but inclusion of MAP and HR data improved accuracy. Our preliminary leave-one-out analysis showed that the algorithm could be applied to a new patient not included in the training set, but accuracy of prediction requires a larger data set, particularly at high ICP. Further work is needed to streamline and automate the entire process of DCS measurement and ICP estimation. We believe that our method made promising advancements in the translation of DCS-based ICP estimation to humans. The wide availability of noninvasive ICP measurement technology would minimize procedural risk and at the same time expand ICP monitoring to a wide array of clinical circumstances.
Supplementary Material
Acknowledgments
We thank the University of Pittsburgh Medical Center Children’s Hospital of Pittsburgh, specifically the nurses who made sure that the measurements were smooth and complete. A special thanks goes to all the patients and their parents who voluntarily participated in the study. We acknowledge financial support through the Center for Machine Learning and Health, the Pennsylvania Infrastructure Technology Alliance, the American Heart Association (grant no. 17SDG33700047), and the National Institutes of Health (grant nos. R21-EB024675, T32 HD40686, and R21 NS115174).
ABBREVIATIONS
- AUC
area under the curve
- CBF
cerebral blood flow
- CSF
cerebrospinal fluid
- DCS
diffuse correlation spectroscopy
- EKG
electrocardiography
- EVD
external ventricular drain
- FDA
US Food and Drug Administration
- HR
heart rate
- ICP
intracranial pressure
- ICPest
estimated ICP
- ICPinv
invasive ICP
- MAP
mean arterial pressure
- ONSD
optic nerve sheath diameter
- p
prominence
- PICU
pediatric intensive care unit
- pk
peak height
- pos
time point of the peak relative to the prior diastolic minimum
- P1
waveform peak
- RMSE
root mean square error
- SNR
signal-to-noise ratio
- TCD
transcranial Doppler
- w
full width at half maximum
Footnotes
Disclosures
The authors report no conflict of interest concerning the materials or methods used in this study or the findings specified in this paper.
Supplemental Information
Online-Only Content
Supplemental material is available with the online version of the article.
Supplemental Materials and Methods. https://thejns.org/doi/suppl/10.3171/2022.9.JNS221203.
References
- 1.Irani D Cerebrospinal Fluid in Clinical Practice. Saunders Elsevier; 2009. [Google Scholar]
- 2.Gupta AK, Gelb A. Essentials of Neuroanesthesia and Neurointensive Care. Saunders; 2008. [Google Scholar]
- 3.Mokri B The Monro-Kellie hypothesis: applications in CSF volume depletion. Neurology. 2001; 56(12): 1746–1748. [DOI] [PubMed] [Google Scholar]
- 4.Mckee AC, Daneshvar DH. The neuropathology of traumatic brain injury. Handb Clin Neurol. 2015; 127: 45–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prabhakaran S, Naidech AM. Ischemic brain injury after intracerebral hemorrhage: a critical review. Stroke. 2012; 43(8): 2258–2263. [DOI] [PubMed] [Google Scholar]
- 6.Johanson CE, Duncan JA III, Klinge PM, Brinker T, Stopa EG, Silverberg GD. Multiplicity of cerebrospinal fluid functions: new challenges in health and disease. Cerebrospinal Fluid Res. 2008; 5: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rekate HL. The definition and classification of hydrocephalus: a personal recommendation to stimulate debate. Cerebrospinal Fluid Res. 2008; 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harary M, Dolmans RGF, Gormley WB. Intracranial pressure monitoring—review and avenues for development. Sensors (Basel). 2018; 18(2): 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawoos U, McCarron RM, Auker CR, Chavko M. Advances in intracranial pressure monitoring and its significance in managing traumatic brain injury. Int J Mol Sci. 2015; 16(12): 28979–28997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czosnyka Z, Czosnyka M. Long-term monitoring of intracranial pressure in normal pressure hydrocephalus and other CSF disorders. Acta Neurochir (Wien). 2017; 159(10): 1979–1980. [DOI] [PubMed] [Google Scholar]
- 11.Chen CJ, Ding D, Ironside N, et al. Intracranial pressure monitoring in patients with spontaneous intracerebral hemorrhage. J Neurosurg. 2019; 132(6): 1854–1864. [DOI] [PubMed] [Google Scholar]
- 12.Nabeta HW, Bahr NC, Rhein J, et al. Accuracy of noninvasive intraocular pressure or optic nerve sheath diameter measurements for predicting elevated intracranial pressure in cryptococcal meningitis. Open Forum Infect Dis. 2014; 1(3): ofu093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajajee V, Williamson CA, Fontana RJ, Courey AJ, Patil PG. Noninvasive intracranial pressure assessment in acute liver failure. Neurocrit Care. 2018; 29(2): 280–290. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Griffith M, Jang HJ, et al. Intracranial pressure monitoring via external ventricular drain: are we waiting long enough before recording the real value? J Neurosci Nurs. 2020; 52(1): 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raboel PH, Bartek J Jr, Andresen M, Bellander BM, Romner B. Intracranial pressure monitoring: Invasive versus non-invasive methods—a review. Crit Care Res Pract. 2012; 2012: 950393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall SA, Riechers RG II. Diagnosis and management of moderate and severe traumatic brain injury sustained in combat. Mil Med. 2012; 177(8 suppl): 76–85. [DOI] [PubMed] [Google Scholar]
- 17.Robba C, Bacigaluppi S, Cardim D, Donnelly J, Bertuccio A, Czosnyka M. Non-invasive assessment of intracranial pressure. Acta Neurol Scand. 2016; 134(1): 4–21. [DOI] [PubMed] [Google Scholar]
- 18.Reid A, Marchbanks RJ, Burge DM, et al. The relationship between intracranial pressure and tympanic membrane displacement. Br J Audiol. 1990; 24(2): 123–129. [DOI] [PubMed] [Google Scholar]
- 19.Amini A, Kariman H, Arhami Dolatabadi A, et al. Use of the sonographic diameter of optic nerve sheath to estimate intracranial pressure. Am J Emerg Med. 2013; 31(1): 236–239. [DOI] [PubMed] [Google Scholar]
- 20.Varsos GV, Kolias AG, Smielewski P, et al. A noninvasive estimation of cerebral perfusion pressure using critical closing pressure. J Neurosurg. 2015; 123(3): 638–648. [DOI] [PubMed] [Google Scholar]
- 21.Khan MN, Shallwani H, Khan MU, Shamim MS. Noninvasive monitoring intracranial pressure—a review of available modalities. Surg Neurol Int. 2017; 8: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg JB, Shiloh AL, Savel RH, Eisen LA. Non-invasive methods of estimating intracranial pressure. Neurocrit Care. 2011; 15(3): 599–608. [DOI] [PubMed] [Google Scholar]
- 23.Ruesch A, Schmitt S, Yang J, Smith MA, Kainerstorfer JM. Fluctuations in intracranial pressure can be estimated noninvasively using near-infrared spectroscopy in non-human primates. J Cereb Blood Flow Metab. 2020; 40(11): 2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purkayastha S, Sorond F. Transcranial Doppler ultrasound: technique and application. Semin Neurol. 2012; 32(4): 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cardim D, Robba C, Bohdanowicz M, et al. Non-invasive monitoring of intracranial pressure using transcranial Doppler ultrasonography: is it possible? Neurocrit Care. 2016; 25(3): 473–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robba C, Santori G, Czosnyka M, et al. Optic nerve sheath diameter measured sonographically as non-invasive estimator of intracranial pressure: a systematic review and meta-analysis. Intensive Care Med. 2018; 44(8): 1284–1294. [DOI] [PubMed] [Google Scholar]
- 27.Dong J, Li Q, Wang X, Fan Y. A review of the methods of non-invasive assessment of intracranial pressure through ocular measurement. Bioengineering (Basel). 2022; 9(7): 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruesch A, Yang J, Schmitt S, Acharya D, Smith MA, Kainerstorfer JM. Estimating intracranial pressure using pulsatile cerebral blood flow measured with diffuse correlation spectroscopy. Biomed Opt Express. 2020; 11(3): 1462–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durduran T, Yodh AG. Diffuse correlation spectroscopy for non-invasive, micro-vascular cerebral blood flow measurement. Neuroimage. 2014; 85(Pt 1): 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutin J, Zimmerman B, Tyulmankov D, et al. Time-domain diffuse correlation spectroscopy. Optica. 2016; 3(9): 1006–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Parthasarathy AB, Baker WB, et al. Fast blood flow monitoring in deep tissues with real-time software correlators. Biomed Opt Express. 2016; 7(3): 776–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, Hamilton R, Pineles S, Bergsneider M, Hu X. Noninvasive intracranial hypertension detection utilizing semisupervised learning. IEEE Trans Biomed Eng. 2013; 60(4): 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quachtran B, Hamilton R, Scalzo F. Detection of intracranial hypertension using deep learning. In: Proceedings of the 23rd International Conference on Pattern Recognition (ICPR). Vol 2016. IEEE; 2016: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farzam P, Sutin J, Wu KC, et al. Fast diffuse correlation spectroscopy (DCS) for non-invasive measurement of intracranial pressure (ICP) (Conference Presentation). In: Proceedings Volume 10050: SPIE BIOS: Clinical and Translational Neurophotonics. SPIE; 2017. [Google Scholar]
- 35.Boas DA, Yodh AG. Spatially varying dynamical properties of turbid media probed with diffusing temporal light correlation. J Opt Soc Am A Opt Image Sci Vis. 1997; 14(1): 192–215. [Google Scholar]
- 36.Fawagreh K, Gaber MM, Elyan E. Random forests: from early developments to recent advancements. Syst Sci Control Eng. 2014; 2(1): 602–609. [Google Scholar]
- 37.Domingo J, Galal G, Huang J, et al. Preventing delayed and missed care by applying artificial intelligence to trigger radiology imaging follow-up. NEJM Catal Innov Care Deliv. 2022; 3(4). [Google Scholar]
- 38.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011; 12(85): 2825–2830. [Google Scholar]
- 39.Nelson BW, Low CA, Jacobson N, et al. Guidelines for wrist-worn consumer wearable assessment of heart rate in biobehavioral research. NPJ Digit Med. 2020; 3(1): 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zacchetti L, Magnoni S, Di Corte F, Zanier ER, Stocchetti N. Accuracy of intracranial pressure monitoring: systematic review and meta-analysis. Crit Care. 2015; 19: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Sullivan TD, Cerussi AE, Cuccia DJ, Tromberg BJ. Diffuse optical imaging using spatially and temporally modulated light. J Biomed Opt. 2012; 17(7): 071311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganslandt O, Mourtzoukos S, Stadlbauer A, Sommer B, Rammensee R. Evaluation of a novel noninvasive ICP monitoring device in patients undergoing invasive ICP monitoring: preliminary results. J Neurosurg. 2018; 128(6): 1653–1660. [DOI] [PubMed] [Google Scholar]
- 43.Fischer JB, Ghouse A, Tagliabue S, et al. Non-invasive estimation of intracranial pressure by diffuse optics: a proof-of-concept study. J Neurotrauma. 2020; 37(23): 2569–2579. [DOI] [PubMed] [Google Scholar]
- 44.Zieman SJ, Melenovsky V, Kass DA. Mechanisms, pathophysiology, and therapy of arterial stiffness. Arterioscler Thromb Vasc Biol. 2005; 25(5): 932–943. [DOI] [PubMed] [Google Scholar]
- 45.Ahmadi N, Mahdieh Namayandeh S, Bafghi SMS, et al. Age-, sex-, and height-based blood pressure reference charts, Yazd children 6–18 years, Iran. Clin Exp Pediatr. 2020; 63(8): 321–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


