Abstract
Thermal and chemical stresses induce the formation in human cells of novel and transient nuclear structures called nuclear stress bodies (nSBs). These contain heat shock factor 1 (HSF-1) and a specific subset of pre-mRNA processing factors. Nuclear stress bodies are assembled on specific pericentromeric heterochromatic domains containing satellite III (SatIII) DNA. In response to stress, these domains change their epigenetic status from heterochromatin to euchromatin and are transcribed in poly-adenylated RNAs that remain associated with nSBs. In this article, we describe the cloning, sequencing, and functional characterization of these transcripts. They are composed of SatIII repeats and originate from the transcription of multiple sites within the SatIII arrays. Interestingly, the level of SatIII RNAs can be down-regulated both by antisense oligonucleotides and small interfering RNAs (siRNA). Knockdown of SatIII RNA by siRNAs requires the activity of Argonaute 2, a component of the RNA-induced silencing complex. Down-regulation of satellite III RNAs significantly affects the recruitment of RNA processing factors to nSBs without altering the association of HSF-1 with these structures nor the presence of acetylated histones within nSBs. Thus, satellite III RNAs have a major role in the formation of nSBs.
INTRODUCTION
Traditionally, the genetic material has been divided into heterochromatin and euchromatin, the latter containing the active genes. The heterochromatin can be further divided in two. Whereas the so-called facultative heterochromatin contains silent genes, constitutive heterochromatin is virtually free of protein-coding genes. Constitutive heterochromatin is principally located at the pericentromeric regions and has been implicated in nuclear architecture and gene silencing. It is known to contain various repeated DNA sequences, such as satellites I–IV and α satellites (Grady et al., 1992). Heterochromatin has not been a major focus of genome sequencing programs and constitutes >200 Mb of unsequenced gaps of the human genome (She et al., 2004). To further complicate matters, these regions have proved extremely difficult to sequence and assemble not only because of their tandemly repetitive nature but also because of occurrences of segmental duplications, i.e., “fragments of genomic sequences with high sequence homology (>90% and 1 kb) that map to multiple regions” (Eichler et al., 2004).
In the past few years, it has become evident that constitutive heterochromatin may indeed have a function. Several studies describing transcription of satellite sequences in various organisms point toward a more active role of these regions (Diaz et al., 1981; Miyahara et al., 1985; Epstein et al., 1986; Schafer et al., 1986; Wu et al., 1986; Bonaccorsi et al., 1990; Gaubatz and Cutler, 1990; Belyaeva et al., 1992; Rudert et al., 1995; Rouleux-Bonnin et al., 1996; Renault et al., 1999). Work in yeast indicates the involvement of transcripts of repetitive DNA in silencing of heterochromatin regions (Volpe et al., 2002), whereas the large noncoding X-IST transcript is a key player in the process of X-chromosome inactivation in mammalian females (Boumil and Lee, 2001). Furthermore, the interaction of HP1 with heterochromatic domains has been shown to depend on as of yet unknown RNA molecules (Maison et al., 2002; Muchardt et al., 2002).
Recently, we and others have shown that in response to thermal and chemical stresses, specific pericentromeric heterochromatic domains of the human genome that colocalize with nuclear stress bodies (nSBs) change their epigenetic status and acquire euchromatic features (Jolly et al., 2004; Rizzi et al., 2004). This is accompanied by the transcriptional activation of these regions, with the production of RNA molecules recognized in Northern blotting by a probe specific for the satellite III (SatIII) repeats. The nature and function of these molecules are still a matter of investigation. In particular, it is unclear whether they derive from a single transcription unit or whether transcription occurs on various SatIII regions. The characterization performed so far indicates that SatIII RNAs are polyadenylated, they are of variable length, and they derive from only one strand of the SatIII region. Moreover, they seem to be stable components of nSBs, suggesting that they may have an important role in the recruitment of a number of RNA binding proteins to the bodies. This hypothesis is also supported by the fact that splicing factor SF2/ASF binds to a specific sequence element on the transcribed strand of the SatIII sequences (Chiodi et al., 2004). Interestingly, amino acid substitutions that abrogate binding of SF2/ASF to this sequence also prevent its recruitment to nSBs (Chiodi et al., 2004).
In this article, we set out to analyze further the satellite III transcripts, by cloning, sequencing, and specific knockdown. We show that these transcripts are composed entirely of SatIII repeats and seem to originate from transcription of multiple sites within the SatIII arrays. Disruption of these molecules driven by antisense oligonucleotides and by RNA interference (RNAi) technology drastically affects the recruitment of specific RNA processing factors to nSBs, supporting the idea that Sat III RNAs have a major role in the self-organization of these nuclear bodies.
MATERIALS AND METHODS
Cell Culture and Treatment
HeLa cells were grown in DMEM (Sigma-Aldrich, St. Louis, MO), 10% fetal calf serum (Sigma-Aldrich), 50 μg/ml gentamicin, and 2 mM l-glutamine. For heat shock experiments, cell monolayers were incubated for 1 h at 42°C in complete medium made with 40 mM HEPES, pH 7.0, and allowed to recover for different times at 37°C as indicated in the text.
Immunofluorescence
HeLa cells grown on coverslips were washed once with phosphate-buffered saline (PBS), fixed for 15 min in 4% formaldehyde, and subsequently permeabilized in 0.5% Triton X-100 on ice for 5 min. Primary antibodies were diluted to working concentration in PBS containing 5% skimmed milk (Difco, Detroit, MI) and added to the coverslips. Primary antibodies were as follows: affinity-purified rabbit anti-heterogeneous nuclear ribonucleoprotein (hnRNP) A1-interacting protein (HAP) polyclonal antibody (Weighardt et al., 1999), rat anti-heat shock factor (HSF)-1 monoclonal antibody (mAb) 10H8 (Neomarkers, Fremont, CA), mouse anti-SF2/ASF mAb-96 (Zymed Laboratories, South San Francisco, CA), and rabbit anti-hyperacetylated histone H4 (Upstate Biotechnology, Lake Placid, NY). After 1 h at room temperature (RT) in a humid chamber, coverslips were washed three times in PBS. Secondary antibodies used were rhodamine-conjugated anti-rabbit, anti-mouse, or anti-rat IgG goat antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit or anti-mouse IgG goat antibodies (Jackson ImmunoResearch Laboratories). These were diluted to the final concentration recommended by the supplier, in PBS with 5% skimmed milk, and added to the coverslips. After 1 h at RT in a humid chamber, the coverslips were washed three times in PBS and mounted with fluorescent mounting medium (DakoCytomation California, Carpinteria, CA). Confocal microscopy was performed with a TCS-NT digital scanning confocal microscope (Leica, Wetzlar, Germany) equipped with a 63×/1.32 numerical aperture oil immersion objective. A 488-nm laser line was used for excitation of FITC (detected at 500 nm < FITC < 540 nm) and the 543-nm laser line for the rhodamine fluorescence (detected at >590 nm). The pinhole diameter was kept at 1 μm. Images were exported to Adobe Photoshop (Adobe Systems, Mountain View, CA).
Immunoblotting
Total cell extracts were separated on 10% SDS-PAGE in electrophoresis running buffer (25 mM Tris, 200 mM glycine, and 0.1% SDS) and electroblotted on Protran nitrocellulose transfer membrane (Schleicher & Schuell, Keene, NH) in blotting buffer [20% (vol/vol) methanol, 200 mM glycine, and 25 mM Tris]. After transfer the membranes were blocked for 1 h at RT in 3% skimmed milk in PBS containing 0.1% Tween 20 before incubation with primary antibody overnight at 4°C. In addition to antibodies described above, we used anti-α-tubulin (Sigma-Aldrich). The membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). The immunocomplexes were visualized using SuperSignal chemiluminescent substrate (Pierce Chemical, Rockford, IL) and detected with x-ray film.
In Situ Hybridization to RNA
The in situ hybridization to RNA was done as described previously (Rizzi et al., 2004). Briefly, cells were grown on coverslips and heat shocked as described above. Cells were fixed in 4% paraformaldehyde in PBS for 15 min, washed thoroughly, and permeabilized in 0.5% Triton X-100 on ice for 5 min. The cells were then hybridized overnight at 42°C with 5 ng/ml 5′ biotinylated oligonucleotide probe, subsequently washed in 2× SSC, and the biotinylated probe was detected with FITC-avidin (Vector Laboratories, Burlingame, CA). The signal was amplified by incubation with biotinylated anti-avidin D antibody (Vector Laboratories) followed by FITC-avidin. The oligonucleotide probes used were Direct and Reverse (Rizzi et al., 2004).
RNA Extraction and Northern Blotting
Total RNA was isolated from HeLa cells by using the RNeasy Mini kit (QIAGEN, Valencia, CA) and Tri Reagent (Sigma-Aldrich), following the manufacturer's protocol. Northern blotting was performed as described previously (Sambrook and Russel, 2001) on Z-probe GT membrane (Bio-Rad, Hercules, CA). Prehybridization and hybridization were done with Miracle-Hyb hybridization solution (Stratagene, La Jolla, CA) according to manufacturer's instructions. Probes were labeled with the Megaprime DNA labeling system (Amersham Biosciences, Piscataway, NJ). After hybridization membranes were washed twice in 2× SSC, 0.1% SDS at RT, and once in 1× SSC, 0.1% SDS at 65°C. Probes used were pHuR98 (American Type Culture Collection, Manassas, VA) and β-actin cDNA (BD Biosciences Clontech, Palo Alto, CA).
RNA was also purified from nuclear and cytoplasmic fractions prepared as described by Das et al. (2001). In brief, after washing in PBS, the cells were scraped off the flasks and centrifuged for 5 min at 800 × g. Cells were then lysed in 100 μl of lysis buffer (10 mM NaCl, 5 mM MgCl2, 50 mM Tris, pH 8.0, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, and 10 mM vanadyl ribonuclease complex). Cells were then vortexed for 15 s and centrifuged at full speed for 2 min. RNA was purified from the supernatant (cytoplasm) and the pellet (nucleus) by using 0.25 ml of Tri Reagent per sample, according to manufacturer's instructions.
Heterokaryon Assay
Interspecies heterokaryon assay was performed as described previously (Valgardsdottir et al., 2001). Briefly, equal number of human HeLa and mouse NIH/3T3 cells were grown on coverslips for 24 h before fusion for 2 min with PEG-4000 (Merck, Darmstadt, Germany). The cells were then washed thoroughly in PBS and incubated for 2 h in fresh medium. The cells were then stressed for 1 h as described above. After 2 h of recovery at 37°C, the cells were fixed and double stained with in situ hybridization to SatIII RNA (reverse oligonucleotide) and with anti-α-tubulin antibody (Sigma-Aldrich). The cells were counterstained with 4,6-diamidino-2-phenylindole (DAPI) to distinguish between the mouse and the human cells, mounted with fluorescent mounting medium (DakoCytomation California) and viewed with confocal microscopy as described above.
Selection of Satellite III Transcripts
Oligonucleotide (300 pmol) was bound on 10 μl of streptavidin-coated M-270 Dynabeads (Dynal Biotech, Lake Success, NY) that had been made RNase free following the manufacturer's instructions. After incubation for 45 min at RT, the beads were washed three times in bind and wash buffer (5 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, and 1 M NaCl). Then, 5 μg of total RNA in 2× SSC was added to the beads and left at RT for 1 h. The beads were washed thoroughly in 2× SSC, and the RNA was eluted in 2 mM EDTA at 65°C for 4 min. The RNA was blotted on Zeta Probe GT membrane (Bio-Rad) with a Slot blot minifold (Schleicher & Schuell) and hybridized as described above. The following 5′-biotinylated oligonucleotides were used: direct and reverse (as in in situ hybridization described above) and a control oligonucleotide recognizing the pBR322 vector sequence (Rizzi et al., 2004).
Cloning
The SatIII transcripts from heat-shocked cells, selected as described above, were cloned into pDNR-LIB vector with Creator SMART cDNA library construction kit (BD Biosciences, San Jose, CA) following the manufacturer's instructions. Positive clones containing satellite III sequences were selected by colony screening with pHuR98 as a probe and sequenced.
SatIII and Argonaute 2 (Ago-2) Knockdown
Phosphorothiorate-modified oligonucleotides were administered to cells by using Lipofectin reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions, by using 5 μg/ml Lipofectin and varying concentrations of oligonucleotide (see Results) in Opti-MEM reduced serum medium (Invitrogen). Twenty-four hours after transfection, the cells were heat shocked as described above and allowed to recover at 37°C for 3 h. The following phosphorothioate-modified antisense oligonucleotides (MWG Biotech, Ebsberg, Germany) were used: AS-REV, 5′-CCA TTC CAT TCC ATT CCA TT-3′, designed to bind and degrade the transcribed strand of the SatIII repeat; and its complimentary oligo, AS-Forw:5′-AAT GGA ATG GAA TGG AAT GG-3′, designed as a negative control.
Small interfering RNAs (siRNA) duplexes were administered to cells by using Oligofectamine reagent (Invitrogen) following the manufacturer's instructions, by using various concentration of siRNA (see Results). Twenty-four hours after transfection, the cells were heat shocked and allowed to recover at 37°C for 3 h. The following siRNAs (Dharmacon, Boulder, CO) were used: siSatIII (5′-UGG AAU GGA AUG GAA UGG AdTdT-3′ and 5′-UCC AUU CCA UUC CAU UCC AdTdT-3′) designed to bind and degrade the SatIII transcripts. This molecule targets the same sequence as AS-REV/AS-Forw. As a negative control, we used siLUC directed against the luciferase gene (5′-CGU ACG CGG AAU ACU UCG AdTdT-3′ and 5′-UCG AAG UAU UCC GCG UAC GdTdT-3′). The siGenome SMARTpool siRNA (Dharmacon) was used to knock down Argonaute-2.
To study the effect of the siRNA and antisense treatments on protein recruitment to the nSBs, HeLa cells were grown on coverslips, two for each antibody. The cells were stressed and allowed to recover for 0 h (anti-HSF-1 and anti-hyperacetylated histone H4) or 1 h (anti-HAP and anti-ASF/SF2) before fixing and staining with the respective antibodies. Pictures were taken from random fields on each coverslip with the confocal microscopy as described above, and 471–869 (average 607) cells were scored for each protein for the association with nSBs.
RESULTS
Subcellular Distribution of SatIII RNA
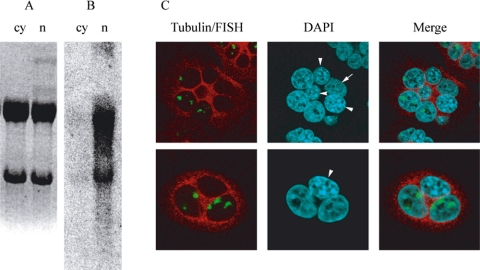
nSBs are assembled on arrays of satellite III DNA that are mainly found at pericentromeric heterochromatic domains of several human chromosomes. They correspond to RNA polymerase II transcription factories for heterogeneously sized polyadenylated SatIII RNA (Rizzi et al., 2004). These transcripts sojourn for long time intervals in nSBs where they are detectable even when the activity of RNA polymerase II is inhibited with actinomycin D (Jolly et al., 2004). It is presently unknown, however, whether they are exclusively nuclear or are exported to the cytoplasm. To clarify this point, we have purified nuclear and cytoplasmic RNA from HeLa cells heat-shocked 1 h at 42°C and then allowed to recover for 3 h at 37°C before being harvested. The RNA was then analyzed in Northern blotting with a probe recognizing SatIII sequences (pHuR98). As expected, a smear of SatIII transcripts is detectable in the fraction of nuclear RNA (Figure 1), whereas only a faint signal corresponding to the rRNA is detectable in the cytoplasmic fraction. This result strongly indicates that the SatIII RNAs are mainly nuclear; however, it does not rule out the possibility that nucleocytoplasmic shuttling can occur and that SatIII RNAs are first exported to the cytoplasm and then reaccumulate in proximity of the chromatin domains where they were transcribed. Therefore, we performed an interspecies heterokaryon assay to assess whether SatIII transcripts could exit from the human nuclei to enter murine NIH/3T3 nuclei that are not proficient for the formation of nSBs (Denegri et al., 2002). Three independent experiments were performed, with at least two coverslips examined for each experiment. As exemplified in Figure 1C, no murine nucleus in the heterokaryons was stained by the reverse SatIII oligonucleotide in RNA in situ hybridization, not even with a diffused hybridization signal. Moreover, we frequently noticed heterokaryons in which at least one human nucleus did not contain SatIII transcripts (Figure 1C). Collectively, these results support the idea that SatIII RNAs do not shuttle between nuclei but remain associated with their sites of transcription.
Figure 1.
Subcellular localization of SatIII transcripts. For Northern analysis, HeLa cells were heat shocked 1 h at 42°C and allowed to recover at 37°C for 3 h before harvesting. Nuclear (n) and cytoplasmic (cy) RNA was purified as described in Materials and Methods. The RNA was electrophoresed on an agarose gel and stained with ethidium bromide (A). The RNA was then transferred to a nylon membrane and analyzed in Northern blotting with a probe recognizing SatIII sequences (pHuR98) (B). To assess for nucleocytoplasmic shuttling, an interspecies heterokaryon assay was performed. HeLa and NIH/3T3 cells were grown together on coverslips and fused for 2 min with PEG-4000. Two hours later, the cells were incubated at 42°C for 1 h and then allowed to recover for 2 h at 37°C before fixing. Cells were then stained with RNA-FISH by using the reverse oligo, and with anti-tubulin antibody, and counterstained with DAPI to distinguish between the HeLa cells that stain diffusely and NIH/3T3 that stain with a distinct speckled pattern. Arrow, HeLa cell without nSBs. Arrowheads, mouse nuclei.
Selection and Cloning of Satellite III Transcripts
The isolation of SatIII transcripts is an important step toward the characterization of nSBs. To this aim, we developed a method to select these transcripts based on a biotinylated oligonucleotide complementary to the transcribed strand of the SatIII sequence (reverse oligo). After annealing to total RNA purified from heat-shocked HeLa cells, the oligonucleotide was captured on streptavidin-coated magnetic beads. As a control, we repeated the same procedure with 1) an oligonucleotide, complementary to the reverse oligo (forward oligo) and 2) with an oligonucleotide specific for a vector sequence (pBR322). The reverse oligo was also hybridized to RNA purified from unstressed cells. Bound and unbound fractions were then analyzed in slot blot by using the pHuR98 probe. As shown in Figure 2, ∼50% of the SatIII RNA in heat-shocked cells was selected by the reverse oligo, whereas no signal was observed in the material selected by the forward and the pBR322 oligos or in total RNA prepared from unstressed cells.
Figure 2.
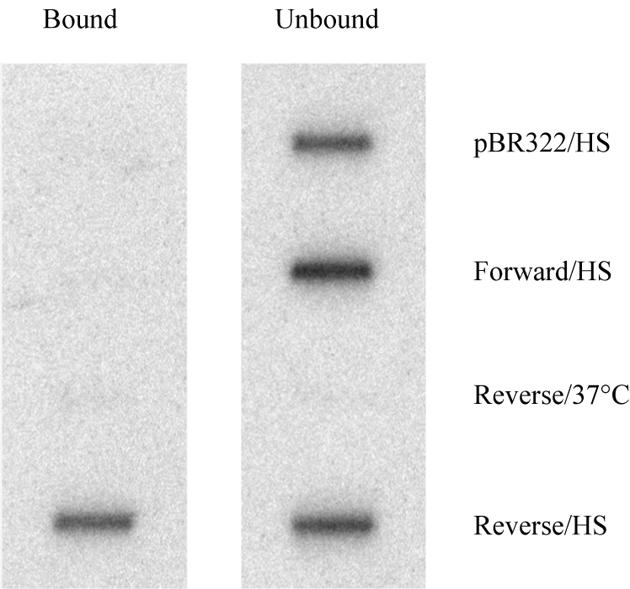
Selection of SatIII transcripts. Total RNA from heat-shocked HeLa cells was annealed to a biotinylated oligonucleotide complementary to the transcribed strand of the SatIII sequences (Reverse/HS). The oligonucleotide was then captured on streptavidin-coated magnetic beads and unbound RNA washed away. As a control, we repeated the same procedure with 1) an oligonucleotide complementary to the reverse oligo (Forward/HS) and 2) with an oligonucleotide specific for a vector sequence (pBR322/HS). The reverse oligo was also hybridized to RNA purified from unstressed cells (Reverse/37°C). Bound and unbound fractions were then analyzed in slot blot by using the pHuR98 probe.
On the basis of this result, we decided to apply this approach to produce a library of SatIII cDNAs. Because the SatIII RNAs are polyadenylated (Rizzi et al., 2004), we used poly(A)+ RNA as a source. From 35 μg of poly(A)+ RNA, we obtained ∼150 ng of selected RNA that was reverse transcribed with an oligo dT primer and cloned into pDNR-LIB vector (see Materials and Methods). This resulted in a library consisting of ∼250,000 independent clones that was enriched for SatIII cDNA, but not free from other cDNAs. Twenty clones were randomly selected by colony hybridization by using pHuR98 as a probe (Supplemental Figure 1). These clones were then sequenced and analyzed (Table 1). The size of the inserts ranged from 19 to >1400 nucleotides (not counting the polyA tail). Of these, 18 were fully sequenced, one was sequenced only from the 5′ end (clone 14), and one was too large to be completely sequenced with primers annealing to the cloning site (clone 20). All the cDNAs had a polyA tail, which ranged in size between 23 and 87 nt. In only few cases was this preceded by a canonical PolyA signal site. The cDNAs had the typical signatures of SatIII sequences, made of two types of repeats, a pentanucleotide (GGATT) and a decanucleotide (CAACCCGAGT). The ∼1500 copies of the pentanucleotide in our clones occur in tandem arrays composed of a variable number of repeats (from 1 to 39). Of these, 60% are identical to the consensus and most of the remaining 40% are one-nucleotide variations of this sequence. The most frequent variations were GCAAT (60 times), GGAAA (54 times), and GGATT, GTAAT, GGAAC, and AGAAT (43–45 times each). The pentanucleotide arrays are terminated by a decanucleotide monomer. The consensus decanucleotide was found a total of 29 times and accounted for ∼25% of all decanucleotides in the clones. Several variations of the termination signal were found, usually differing from the consensus sequence for one to two nucleotides. The most frequent ones (CAACCAGAGT, CAACCCGAAT, and CGTTCCGAGT) were found four times each. Because of the variations in the penta- and decanucleotides sequences and in the number of repeats in each array, each SatIII transcript has a unique structure.
Table 1.
Cloned SatIII transcripts
| Clone | Size | PolyA | GeneBank accession no. |
|---|---|---|---|
| 1 | 19 | 31 | AY845709 |
| 2 | 22 | 26 | NA |
| 3 | 55 | 33 | AY845690 |
| 4 | 139 | 28 | AY845691 |
| 5 | 185 | 33 | AY845692 |
| 6 | 196 | 87 | AY845693 |
| 7 | 256 | 29 | AY845694 |
| 8 | 270 | 31 | AY845695 |
| 9 | 326 | 29 | AY845696 |
| 10 | 414 | 26 | AY845697 |
| 11 | 403 | 29 | AY845698 |
| 12 | 449 | 29 | AY845699 |
| 13 | 471 | 26 | AY845700 |
| 14 | >491 | — | AY845701 |
| 15 | 507 | 30 | AY845702 |
| 16 | 551 | 23 | AY845703 |
| 17 | 666 | 30 | AY845704 |
| 18 | 711 | 30 | AY845705 |
| 19 | 1157 | 49 | AY845706 |
| 20 | >1421 | 30 | AY845707 |
| AY845708 |
Cloned SatIII transcripts. The number and the GeneBank accession number of each clone are listed together with the length of each cDNA and of their respective polyA tails. —, not known; NA, not available.
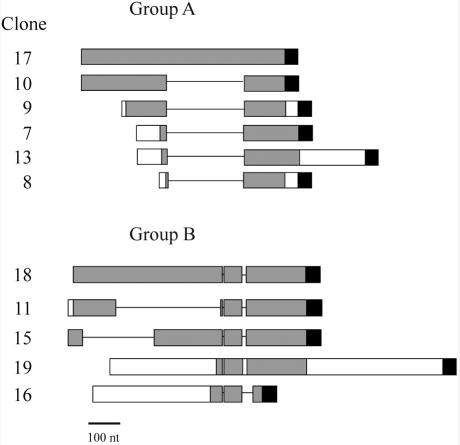
Sequence analysis, however, revealed blocks of 100%, or near 100%, identity between different clones. Two groups of cDNAs were identified. Group A is made of six cDNAs (Figure 3 and Supplemental Figure 2), with the strongest similarity between clones 10 and 17 that contain two blocks of complete identity. Block 1 comprises the first 278 nt from the 5′ end, whereas block 2 is composed of the last 135 nt before the polyA tail. The two blocks are separated by 256 nt that are only present in clone 17. The remaining clones in this group differ from clone 10 for the presence of specific tails at both ends and for the fraction of block 1 they contain. Interestingly, the junctions between the two identity blocks and the intervening sequence specific for clone 17 are perfectly conserved in all clones of group A (Figure 3 and Supplemental Figure 2; see Discussion). Blocks of identity separated by clone specific sequences are clearly detectable also in group B, which contains five clones (Figure 3 and Supplemental Figure 2). However, the pattern is more complicated than in group A. The remaining transcripts that are not included in these two groups contain regions of complete identity <50 nt, mainly formed by tandem arrays of the consensus GGAAT repeat. Because of the high overall similarity between cDNAs, it was not possible to assess the frequency of each of them within the library.
Figure 3.
Homologies among the cloned transcripts. Two groups of sequences with high homology were found (A and B). Blocks with 100% or near 100% homology are shown in gray, white rectangles represent sequences with <95% homology, lines represent gaps and black boxes stand for the polyA tail. Bar, 100 nt.
A RepeatMasker analysis of the cDNAs showed that several of them contained short fragments that were not identified as SatIII repeats (our unpublished data). In most cases, however, these are sequences that are very similar to the SatIII arrays and may thus represent more diverged regions. In one case (23 nt in clone 8 of group A), the nonmasked sequence was found to be homologous to rRNA. Incidentally, this sequence forms the 5′ tail upstream of block 1 in clone 8. Interestingly, ribosomal genes on acrocentric chromosomes have been mapped in proximity of SatIII arrays (Bandyopadhyay et al., 2001), suggesting that the transcriptional activation at the SatIII regions may extend to the nearby sequences.
Sequence Analysis
A BLAST search identified genomic sequences similar to the SatIII transcripts on most of the human chromosomes, consistently with the spreading of the SatIII elements throughout the genome. None of these sequences, however, showed a homology >80% with any of the cloned cDNAs. One of the largest blocks of SatIII repeats in the human genome is located on the heterochromatic pericentromeric band q12 of chromosome 9 and forms one of the platforms for the assembly of nSBs. We have identified >50 sequences from this region in GenBank, most of which were annotated as “unfinished sequence.” The level of homology with the SatIII clones described here ranged between 60 and 85%, and blocks of complete homology were short (20–25 base pairs). Together, these results indicate that our pool of RNA derives from transcription of unsequenced portions of the genome.
An analogous search in human expressed sequence tag (EST) databanks identified >50 partially sequenced ESTs with a similarity to the cloned SatIII transcripts ranging between 70 and 90%. These ESTs are 400–600 nt in length and derive from various normal tissues and tumors. Again, no large blocks of complete homology were found. Interestingly, these ESTs are representative of both the strands of the SatIII sequence, suggesting that, contrary to what occurs in heat-shocked cells, in normal human tissues and cell lines, transcription of SatIII repeats may take place on both strands. The fact that we failed to detect SatIII RNA in Northern blot analysis of unstressed HeLa cells (Rizzi et al., 2004) may imply that the SatIII regions are transcribed at a very low level under physiological conditions.
Targeting of SatIII Transcripts by Antisense Oligonucleotides and siRNA Molecules
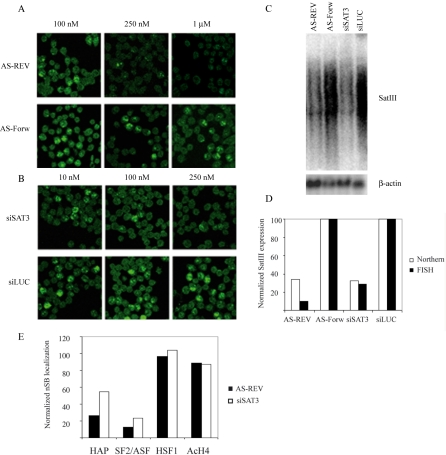
We have previously proposed a major role for the SatIII transcripts in the recruitment of specific RNA binding proteins to nSBs (Biamonti, 2004; Rizzi et al., 2004). Indeed, RNA integrity and transcription are necessary for the association of these factors with nSBs (Weighardt et al., 1999; Chiodi et al., 2004; Metz et al., 2004). Consistently, with this hypothesis, we and others have found that the recruitment of splicing factor SF2/ASF is mediated by the second RNA recognition motif (RRM2) of the protein (Chiodi et al., 2004; Metz et al., 2004). SatIII RNAs contain several putative binding sites for SF2/ASF, and amino acid substitutions that abrogate RRM2 binding to the SatIII sequence also prevent the recruitment of SF2/ASF to the nSBs (Chiodi et al., 2004). This, however, does not rule out the possibility that other RNA molecules are present in these nuclear compartments and mediate the recruitment of RNA processing factors. To elucidate this, we investigated whether the integrity of the nSBs could be affected by antisense oligonucleotide. These are known to trigger the degradation of RNAs in the cell nucleus (Vickers et al., 2003). We designed a 20-mer complementary to the repeated pentanucleotide of the SatIII sequence. The targeted site for this oligo is present at least once in each of the larger cDNAs described above. Transfection of this 20-mer reduces the abundance of SatIII transcripts in heat-shocked cells as estimated by both in situ hybridization analysis and Northern blotting (Figure 4, A and C). This effect can be detected for oligonucleotide concentrations higher than 100 nM and peaks at 1 μM when the level of SatIII transcripts is ∼30% of that detectable in cells treated with a control 20-mer. This resulted in a 90% reduction of the number of cells in which nSBs are detectable by RNA in situ hybridization (Figure 4A). Incidentally, these results show that antisense oligos are also effective at the site of transcription.
Figure 4.
Knockdown of SatIII transcripts. The SatIII transcripts were specifically targeted with different concentrations of antisense oligonucleotide complementary to the transcribed strand of the SatIII sequence (AS-REV) and with a siRNA targeting the same sequence (siSAT3). As negative controls, the same concentrations of an oligonucleotide complementary to the antisense oligo (AS-Forw) and a siRNA directed against the luciferase gene (siLUC) were used. Cells were analyzed by RNA in situ hybridization (A and B) and the RNA by Northern blotting (C). The effect of both treatments on the formation of nSBs (FISH, black column) and the SatIII RNA level (Northern, white column) was quantified and normalized to control values (D). The effect of both treatments on the localization of hnRNP HAP (HAP), SF2/ASF, HSF-1, and hyperacetylated histone H4 (Ac-H4) to the nSBs in cells transfected with AS-REV (black column) or siSAT3 (white column) was quantified and normalized to values in untreated cells for each protein (E).
Having obtained positive results with the antisense nucleotide, we decided to use nSBs as a model system to assess whether the RNA interference (RNAi) mechanisms could operate on noncoding RNA molecules in the cell nucleus. Thus, cells were transfected with siRNA (siSAT3, targeting the same sequence as the antisense oligonucleotide), or with a control siRNA directed against the luciferase transcript (siLUC). After 24 h, cells were heat shocked and analyzed with in situ hybridization and Northern blotting (Figure 4, B and C) to determine the level of SatIII transcripts and the fraction of cells in which nSBs were detectable. As shown in Figure 4B, the level of SatIII transcripts and the formation of nSBs were drastically affected by siSAT3 but not by the control siRNA. The effect was already detectable at low concentrations of siRNA (10 nM) and reached a plateau at 250 nM. Neither the treatment with the antisense oligonucleotide nor the siRNA directed against the SatIII transcripts had effects on the level of transcription of the β-actin (Figure 4C), indicating that the effect was specific for the SatIII RNA.
Involvement of Argonaute-2 in the siRNA-mediated Down-Regulation of SatIII RNA
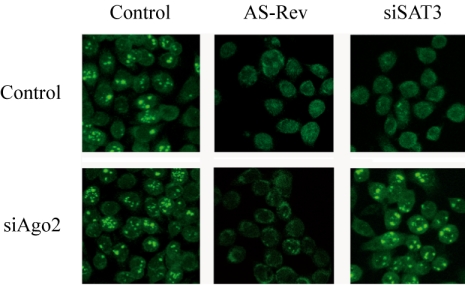
With few exceptions, previous studies have considered RNAi as exclusively cytoplasmic. To our knowledge, only one study has so far reported the use of siRNA duplex directed against an intron to obtain specific gene silencing (Bosher et al., 1999), and a recent study showed that RNAi could specifically target small nuclear RNA 7SK and U6 snRNA in the nucleus (Robb et al., 2005). Furthermore, Meister et al. (2004) have shown that a mature micro-RNA and its cleavage products can be detected both in nucleus and cytoplasm, implying the ability of RNA-induced silencing complex (RISC) to translocate to the nucleus (Meister et al., 2004). As the SatIII transcripts are exclusively nuclear, it was of interest to study whether the siSAT3 worked through the conventional RNAi mechanism. Recently Ago-2 has been shown to account for the slicer-activity in RISC (Liu et al., 2004; Meister et al., 2004; Song et al., 2004). We decided to assess whether Ago-2 is involved in the down-regulation of SatIII RNA by siSAT3. HeLa cells were transfected twice at 3-d intervals with small interfering Ago-2 (siAgo2). Five days after the first siAgo2 transfection, cells were treated either with the antisense oligonucleotide against the SatIII sequence or with the siSAT3. The following day, the cells were stressed and fixed 2 h later. In mock-transfected cells, both the antisense oligonucleotide and the siSAT3 siRNA were effective in triggering degradation of the SatIII transcripts (Figure 5). As expected, siAgo2 did not affect the function of the antisense oligonucleotide. On the contrary, the activity of siSAT3 was greatly diminished in siAgo2-transfected cells and nSBs were detected by in situ hybridization in most of the cells (Figure 5). The same results were obtained when cells were stained with anti-HAP antibody (our unpublished data). Thus, siRNA-induced knockdown of the SatIII RNAs is mediated by a RISC complex that includes Ago-2 and that is active in the cell nucleus.
Figure 5.
Ago-2 is required for down-regulation of SatIII RNAs with siSAT3. HeLa cells were transfected twice with siAgo2 at 3-d intervals. Five days after the first transfection, cells were treated with the antisense oligonucleotide against the SatIII sequence (AS-REV), with siSAT3 or with mock. The following day, the cells were stressed and fixed 2 h later. The cells were then analyzed by RNA in situ hybridization with the biotinylated oligo complementary to the SatIII RNAs to visualize the formation of nSBs.
Role of SatIII Transcripts in the Formation of nSBs
The results in the previous sections opened the possibility to assess the role of SatIII transcripts in the recruitment of different proteins to nSBs. As expected, down-regulation of these transcripts neither affected the distribution of HSF-1 in stressed cells nor the presence of hyperacetylated histones in the nSBs (Figure 4E and Supplemental Figure 3A), suggesting that neither of these phenomena requires the presence of the SatIII RNAs. On the contrary, the recruitment of RNA binding proteins such as hnRNP HAP and SF2/ASF to the nSBs was greatly reduced (Figure 4 and Supplemental Figure 3, C and D). Indeed, the extent of this effect matches the reduction of SatIII RNA levels as measured by both Northern blotting and RNA-fluorescence in situ hybridization (FISH) analysis. This was further supported by the findings that the localization of both proteins to the nSBs was detectable only in cells containing SatIII RNAs (Supplemental Figure 3, C and D). Moreover, the expression levels of the proteins were not altered by the treatment with RNAi or antisense oligos, as evaluated by immunoblotting (Supplemental Figure 3B), indicating that the treatment affects the recruitment of these proteins to the nSBs rather than their expression levels. On the basis of these results, we conclude that the synthesis of SatIII RNAs is critical for the recruitment of RNA processing factors to nSBs.
DISCUSSION
In this article, we describe the cloning and analysis of polyadenylated RNAs deriving from the transcription of chromosomal domains composed of SatIII repeats. These transcripts are produced in response to thermal and chemical stresses, are exclusively nuclear, and remain associated with the site of transcription, the recruitment centers of nSBs, as was evidenced by cellular fractionation of the RNA, and by interspecies heterokaryon assay, which excluded the possibility of nuclear-cytoplasmic shuttling of the SatIII transcripts. The analyzed cDNAs range in size from 19 to >1400 nt, not counting the polyA tail, and are composed of a variable number of tandem SatIII repeats ((GGAAT)n.CAACCCGAGT)n,. We were unable to identify the genomic origin of any of these transcripts, indicating that they derive from unsequenced parts of the genome. Indeed, it has been estimated that ∼200 Mb of the unsequenced gaps of the human genome arise from heterochromatic regions. This hampers the possibility to determine the identity of the transcription units and the nature of the promoters involved. However, our results strongly support the idea that different arrays of the SatIII regions are transcribed. This is also in agreement with the finding that different human chromosomes in human hamster cell hybrid can direct the production of SatIII transcripts in human>hamster cell hybrids after heat shock (Denegri et al., 2002).
Origin of the SatIII RNAs
Despite the lack of perfect homology, several SatIII transcripts, as those in groups A and B, share blocks with 100% homology flanked by transcript specific sequences (Figure 3 and Supplemental Figure 2). Two hypotheses can be raised to explain this occurrence. They can either reflect the arrangement of the genomic sequence or originate from alternative splicing of a common primary transcript. According to the splicing hypothesis, the shared and unique sequences would correspond to different exons brought together by alternative splicing. The occurrence of splicing was already suggested by Metz and collaborators (Metz et al., 2004) who found that the SatIII RNAs interact in vivo with components of the splicing machinery such as small nuclear ribonucleoproteins (Metz et al., 2004). Alternative splicing of these transcripts, mediated by the presence of high-affinity binding sites for specific splicing factors such as SF2/ASF (Chiodi et al., 2004) could account, at least in part, for the smear of hybridization signals observed in Northern blotting. From this perspective, nSBs could be viewed as sites of transcription and processing of SatIII RNAs.
Alternatively, blocks of homology could reflect the presence of duplicated sequences within the arrays of SatIII DNA. This is consistent with the notion that intra- and interchromosomal duplications are a characteristic feature of the pericentromeric regions (Humphray et al., 2004). Furthermore, sequence analysis of a 60,000-bp genomic region consisting of an array of SatIII repeats (GenBank accession no. AC118282) identifies several blocks of 100–300 base pairs repeated throughout the sequence (our unpublished data). These homology blocks are different in length and, similarly to what observed in our pool of SatIII cDNAs, they are flanked by unique sequences. Moreover, we have identified a genomic fragment (GenBank accession no. Z79305) that contains blocks 1 and 2 present in group A without any intervening sequence in between. This speaks against the possibility that the difference between clones 17 and 10 can arise from alternative splicing and favors the idea that it reflects the genomic organization of the transcribed units. Because the transcripts described in this work derive from unsequenced portions of the genome, it is not possible at present to discriminate between the two, not mutually exclusive, hypotheses.
Interestingly, SatIII sequences are present in EST databanks from various normal and tumor tissues, indicating that transcription of SatIII regions occurs even at 37°C. These molecules are probably present at very low levels, as suggested by our failure to detect them in Northern blot analysis of unstressed HeLa cells (Rizzi et al., 2004). In accordance with this interpretation, Jolly et al. (2004) have recently observed SatIII RNAs in unstressed cells by applying highly sensitive radioactive reverse transcription. The possibility that SatIII RNAs are generated even in unstressed cells is consistent also with the recent observation that SatIII-containing regions on chromosome 9 have a heterogeneous chromatin fiber structure, being partially heterochromatic and partially euchromatic (Gilbert et al., 2004). We are presently investigating whether these SatIII RNAs might have a role in establishing heterochromatic domains, possibly through RNAi mechanisms.
Role of SatIII in the Assembly of nSBs
An important aspect of our analysis is the observation that SatIII transcripts are substrates for RNase H and RNAi-based degradation. To our knowledge, this is the first time that transcripts formed by repeated sequences have been targeted to degradation with these methods. Notably, our results convey the idea that targeting occurs in the nucleus when the RNA is still associated with sites of transcription. Until recently, siRNA has been thought to function only in the cytoplasm (Zeng and Cullen, 2002; Vickers et al., 2003). Our results, however, support recent results (Meister et al., 2004; Robb et al., 2005) that suggest that the RISC complex is present in the nucleus and capable of degrading nuclear residing transcripts. The SatIII sequences may thus provide an important tool for further analysis of RNAi in the nucleus.
Down-regulation of the SatIII RNAs is paralleled by the decrease in the recruitment of RNA processing factors to nSBs, indicating a direct role of these RNAs in the recruitment of pre-mRNA processing factors. This is consistent with the fact that the recruitment of SF2/ASF is mediated by the same protein domain, i.e., the second RNA recognition motif, that is responsible for the interaction with SatIII sequences (Chiodi et al., 2004; Metz et al., 2004). Down-regulation of the SatIII RNAs, however, does not affect the association of transcription factor HSF-1 to the SatIII regions, which is due to DNA binding, nor the acetylation of histones associated with nSBs. This finding unequivocally confirms the central role that SatIII transcripts have in the recruitment of RNA processing factors to nSBs and raises the intriguing possibility that these RNAs, by perturbing the ratio between RNA binding proteins in the nucleoplasm, may represent novel splicing regulators.
Supplementary Material
Acknowledgments
This work was supported by grants of the “Associazione Italiana per la Ricerca sul Cancro” (to G.B.), the program Ministero dell'Istruzione, dell'Università e della Ricerca/Fondo per gli Investimenti della Ricerca di Base “PostGenoma” contract nos. RBNEOKXC9_001 and RBNE015MPB_003 (to S.R. and G.B. respectively), and from Progetto Consiglio Nazionale delle Ricerche-Ministero dell'Istruzione, dell'Università e della Ricerca “Genomica Funzionale” L.449/97 (to G.B.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1078) on March 23, 2005.
Abbreviations used: Ago-2, Argonaute 2; HSF-1, heat shock factor 1; nSB, nuclear stress body; RISC, RNA-induced silencing complex; SatIII, satellite III; siRNA, small interfering RNA.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bandyopadhyay, R., McQuillan, C., Page, S. L., Choo, K.H.A., and Shaffer, L. G. (2001). Identification and characterization of satellite III subfamilies to the acrocentric chromosomes. Chrom. Res. 9, 223–233. [DOI] [PubMed] [Google Scholar]
- Belyaeva, T. A., Vishnivetsky, P. N., Potapov, V. A., Zhelezova, A. I., and Romashchenko, A. G. (1992). Species- and tissue-specific transcription of complex, highly repeated satellite-like Bsp elements in the fox genome. Mamm. Genome 3, 233–236. [DOI] [PubMed] [Google Scholar]
- Biamonti, G. (2004). Nuclear stress bodies: a heterochromatin affair? Nat. Rev. Mol. Cell. Biol. 5, 493–498. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi, S., Gatti, M., Pisano, C., and Lohe, A. (1990). Transcription of a satellite DNA on two Y chromosome loops of Drosophila melanogaster. Chromosoma 99, 260–266. [DOI] [PubMed] [Google Scholar]
- Bosher, J. M., Dufourcq, P., Sookhareea, S., and Labouesse, M. (1999). RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics 153, 1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumil, R. M., and Lee, J. T. (2001). Forty years of decoding the silence in X-chromosome inactivation. Hum. Mol. Genet. 10, 2225–32. [DOI] [PubMed] [Google Scholar]
- Chiodi, I., Corioni, M., Giordano, M., Valgardsdottir, R., Ghigna, C., Cobianchi, F., Xu, R. M., Riva, S., and Biamonti, G. (2004). RNA recognition motif 2 directs the recruitment of SF2/ASF to nuclear stress bodies. Nucleic Acids Res. 32, 4127–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M., Harvey, I., Chu, L. L., Sinha, M., and Pelletier, J. (2001). Full-length cDNAs: more than just reaching the ends. J. Physiol. Genomics 6, 57–80. [DOI] [PubMed] [Google Scholar]
- Denegri, M., Moralli, D., Rocchi, M., Biggiogera, M., Raimondi, E., Cobianchi, F., De Carli, L., Riva, S., and Biamonti, G. (2002). Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol. Biol. Cell 13, 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, M. O., Barsacchi-Pilone, G., Mahon, K. A., and Gall, J. G. (1981). Transcripts from both strands of a satellite DNA occur on lampbrush chromosome loops of the newt Notophthalmus. Cell, 24, 649–659. [DOI] [PubMed] [Google Scholar]
- Eichler, E. E., Clark, R. A., and She, X. (2004). An assessment of the sequence gaps: unfinished business in a finished human genome. Nat. Rev. Genet. 5, 345–354. [DOI] [PubMed] [Google Scholar]
- Epstein, L. M., Mahon, K. A., and Gall, J. G. (1986). Transcription of a satellite DNA in the newt. J. Cell Biol. 103, 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz, J. W., and Cutler, R. G. (1990). Mouse satellite DNA is transcribed in senescent cardiac muscle. J. Biol. Chem. 265, 17753–17758. [PubMed] [Google Scholar]
- Gilbert, N., Boyle, S., Fiegler, H., Woodfine, K., Carter, N. P., and Bickmore, W. A. (2004). Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118, 555–566. [DOI] [PubMed] [Google Scholar]
- Grady, D. L., Ratliff, R. L., Robinson, D. L., McCanlies, E. C., Meyne, J., and Moyzis, R. K. (1992). Highly conserved repetitive DNA sequences are present at human centromeres. Proc. Natl. Acad. Sci. USA 89, 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphray, S. J., et al. (2004). DNA sequence and analysis of human chromosome 9. Nature 429, 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly, C., Metz, A., Govin, J., Vigneron, M., Turner, B. M., Khochbin, S., and Vourc'h, C. (2004). Stress-induced transcription of satellite III repeats. J. Cell Biol. 164, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Carmell, M. A., Rivas, F. V., Marsden, C. G., Thomson, J. M., Song, J. J., Hammond, S. M., Joshua-Tor, L., and Hannon, G. J. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–14341. [DOI] [PubMed] [Google Scholar]
- Maison, C., Bailly, D., Peters, A. H., Quivy, J. P., Roche, D., Taddei, A., Lachner, M., Jenuwein, T., and Almouzni, G. (2002). Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329–334. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197. [DOI] [PubMed] [Google Scholar]
- Metz, A., Soret, J., Vourc'h, C., Tazi, J., and Jolly, C. (2004). A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J. Cell Sci. 117, 4551–8. [DOI] [PubMed] [Google Scholar]
- Miyahara, M., Sumiyoshi, H., Yamamoto, M., and Endo, H. (1985). Strand specific transcription of satellite DNA I in rat ascites hepatoma cells. Biochem. Biophys. Res. Commun. 130, 897–903. [DOI] [PubMed] [Google Scholar]
- Muchardt, C., Guilleme, M., Seeler, J. S., Trouche, D., Dejean, A., and Yaniv, M. (2002). Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 3, 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault, S., Rouleux-Bonnin, F., Periquet, G., and Bigot, Y. (1999). Satellite DNA transcription in Diadromus pulchellus (Hymenoptera). Insect Biochem. Mol. Biol. 29, 103–111. [DOI] [PubMed] [Google Scholar]
- Rizzi, N., Denegri, M., Chiodi, I., Corioni, M., Valgardsdottir, R., Cobianchi, F., Riva, S., and Biamonti, G. (2004). Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol. Biol. Cell 15, 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb, G. B., Brown, K. M., Khurana, J., and Rana, T. M. (2005). Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 12, 133–137. [DOI] [PubMed] [Google Scholar]
- Rouleux-Bonnin, F., Renault, S., Bigot, Y., and Periquet, G. (1996). Transcription of four satellite DNA subfamilies in Diprion pini (Hymenoptera, Symphyta, Diprionidae). Eur. J. Biochem. 238, 752–759. [DOI] [PubMed] [Google Scholar]
- Rudert, F., Bronner, S., Garnier, J. M., and Dolle, P. (1995). Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm. Genome 6, 76–83. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russel, D. W. (2001). Molecular Cloning, 3rd ed., Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schafer, R., Boltz, E., Becker, A., Bartels, F., and Epplen, J. T. (1986). The expression of the evolutionarily conserved GATA/GACA repeats in mouse tissues. Chromosoma 93, 496–501. [DOI] [PubMed] [Google Scholar]
- She, X., et al. (2004). The structure and evolution of centromeric transition regions within the human genome. Nature 430, 857–864. [DOI] [PubMed] [Google Scholar]
- Song, J. J., Smith, S. K., Hannon, G. J., and Joshua-Tor, L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. [DOI] [PubMed] [Google Scholar]
- Valgardsdottir, R., Brede, G., Eide, L. G., Frengen, E., and Prydz, H. (2001). Cloning and Characterization of MDDX28, a putative DEAD-box helicase with mitochondrial and nuclear localization. J. Biol. Chem., 276, 32056–32063. [DOI] [PubMed] [Google Scholar]
- Vickers, T. A., Koo, S., Bennett, C. F., Crooke, S. T., Dean, N. M., and Baker, B. F. (2003). Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 278, 7108–7118. [DOI] [PubMed] [Google Scholar]
- Volpe, T. A., Kidner, C., Hall, I. M., Teng, G., Grewal, S. I., and Martienssen, R. A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- Weighardt, F., Cobianchi, F., Cartegni, L., Chiodi, I., Villa, A., Riva, S., and Biamonti, G. (1999). A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell Sci. 112, 1465–1476. [DOI] [PubMed] [Google Scholar]
- Wu, Z. G., Murphy, C., and Gall, J. G. (1986). A transcribed satellite DNA from the bullfrog Rana catesbeiana. Chromosoma 93, 291–7. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., and Cullen, B. R. (2002). RNA interference in human cells is restricted to the cytoplasm RNA. RNA 8, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.