Abstract
In budding yeast, Tup1 and Ssn6/Cyc8 form a corepressor that regulates a large number of genes. This Tup1-Ssn6 corepressor appears to be conserved from yeast to man. In the pathogenic fungus Candida albicans, Tup1 regulates cellular morphogenesis, phenotypic switching, and metabolism, but the role of Ssn6 remains unclear. We show that there are clear differences in the morphological and invasive phenotypes of C. albicans ssn6 and tup1 mutants. Unlike Tup1, Ssn6 depletion promoted morphological events reminiscent of phenotypic switching rather than filamentous growth. Transcript profiling revealed minimal overlap between the Ssn6 and Tup1 regulons. Hypha-specific genes, which are repressed by Tup1 and Nrg1, were not derepressed in ssn6 cells under the conditions studied. In contrast, the phase specific gene WH11 was derepressed in ssn6 cells, but not in tup1 or nrg1 cells. Hence Ssn6 and Tup1 play distinct roles in C. albicans. Nevertheless, both Ssn6 and Tup1 were required for the Nrg1-mediated repression of an artificial NRE promoter, and lexA-Nrg1 mediated repression in the C. albicans one-hybrid system. These observations are explained in models that are generally consistent with the Tup1-Ssn6 paradigm in budding yeast.
INTRODUCTION
Transcriptional repression plays a key role in controlling the growth and differentiation of eukaryotic cells. The fundamental importance of negative transcriptional regulators is reflected in their conservation during eukaryotic evolution. For example, orthologues of the Ssn6 (Cyc8) and Tup1 proteins, which act as global repressors in Saccharomyces cerevisiae, have been identified in humans, flies, worms, slime molds, and fungi (reviewed by Smith and Johnson, 2000).
Our understanding of the mechanisms of action of Ssn6 and Tup1 is based largely on studies in budding yeast. The S. cerevisiae paradigm suggests that Ssn6 and Tup1 interact physically to form a corepressor complex that actively represses the transcriptional machinery (Williams et al., 1991; Redd et al., 1997; Gounalaki et al., 2000). The relevance of this paradigm to eukaryotes in general is emphasized by the observation that yeast Ssn6 can interact with human Tup1-like proteins to mediate transcriptional repression in human cells (Grbavec et al., 1999).
The Ssn6-Tup1 corepressor is thought to repress transcription in yeast by two main mechanisms. First, the corepressor is thought to remodel chromatin on yeast promoters by recruiting histone deacetylases to these promoters and by positioning nucleosomes via direct interactions with histone tails (Edmondson et al., 1996; Davie et al., 2002, 2003; Zhang and Reese, 2004a). Second, Tup1 is thought to interact directly with the transcriptional machinery to attenuate its activity (Carlson, 1997; Redd et al., 1997; Gromoller and Lehming, 2000; Papamichos-Chronakis et al., 2000). Recent experiments suggest that both mechanisms operate in yeast, but that they are utilized differentially to regulate distinct sets of yeast genes (Green and Johnson, 2004). However, both mechanisms appear to operate in a redundant manner to repress some yeast genes (Green and Johnson, 2004; Zhang and Reese, 2004b). In addition, Ssn6-Tup1 presumably regulates the expression of other genes indirectly by controlling the levels of transcriptional activators or repressors that act directly on these genes.
The Ssn6-Tup1 corepressor complex does not interact with DNA directly. Instead the complex is targeted to specific promoters through interactions with sequence-specific DNA-binding proteins (Keleher et al., 1992; Smith and Johnson, 2000). For example, Mig1 and Nrg1 target Ssn6-Tup1 to glucose-repressed genes, whereas the a1/α2 heterodimer targets the corepressor complex to haploid-specific genes in S. cerevisiae. In contrast, Rox1 and Crt1 target Ssn6-Tup1 to hypoxia- and DNA damage-inducible genes in budding yeast. Therefore, Ssn6-Tup1 regulates the expression of functionally diverse sets of yeast genes via interactions with different DNA-binding proteins.
Ssn6-Tup1-mediated repression is generally controlled by regulating the levels or activities of these DNA-binding proteins (Smith and Johnson, 2000). For example, a1/α2 levels are controlled primarily at the transcriptional level in S. cerevisiae: a1 and α2 are synthesized by the MATa and MATα loci, respectively, and hence the a1/α2 heterodimer only forms in diploid cells. In contrast, the activity of Mig1 and Crt1 are regulated posttranscriptionally: the DNA-binding activity of Crt1 is inhibited by hyperphosphorylation, and the phosphorylation of Mig1 inhibits its interaction with Ssn6-Tup1 and leads to its export from the nucleus (Huang et al., 1998; Ostling and Ronne, 1998; Papamichos-Chroakis et al., 2004).
Ssn6 and Tup1 play different roles within the corepressor complex. Tzamarias and Struhl (1994) reported that the transcriptional repression mediated by a LexA-Ssn6 fusion in yeast was dependent on Tup1, whereas LexA-Tup1 repressed transcription even in the absence of Ssn6. Furthermore, the mating defect of yeast tup1 ssn6 cells can be suppressed by Tup1 overexpression, but not by Ssn6 overexpression (Komanchi et al., 1994). Hence, Tup1 is thought to mediate the transcriptional repression, whereas Ssn6 is thought to stabilize the interaction between Tup1 and the DNA-binding protein. As described above, a diverse set of Ssn6-Tup1-targeting proteins have been identified in S. cerevisiae. These diverse proteins appear to interact with different regions of the Ssn6-Tup1 corepressor complex (Komanchi et al., 1994; Tzamarias and Struhl, 1995; Smith and Johnson, 2000). For example, Rox1 and Mig1 contact different regions of the Ssn6 protein within the Ssn6-Tup1 corepressor, whereas α2 is thought to contact Tup1 directly (Komanchi et al., 1994; Tzamarias and Struhl, 1995).
Candida albicans is the major systemic fungal pathogen of humans (Odds, 1988; Calderone, 2002). This fungus causes oral and vaginal candidiasis and life-threatening blood-stream infections in immunocompromised patients. Braun and Johnson (1997) were the first to reveal the importance of transcriptional repression in the regulation of C. albicans virulence attributes by showing that Tup1 represses hyphal development in this fungus. Tup1 was subsequently shown to repress the transcription of hypha-specific genes (i.e., genes that are expressed specifically during the hyphal growth phase of C. albicans; Sharkey et al., 1999; Braun and Johnson, 2000; Braun et al., 2000, 2001; Murad et al., 2001a; Zheng et al., 2004). This was confirmed by a transcript profiling study that examined the effects of inactivating Tup1 or the Tup1-targeting protein, Nrg1, upon the expression of about one-third of C. albicans genes (Murad et al., 2001a, 2001b). This study also revealed that both Tup1 and Nrg1 play broader roles by coordinating yeast-hypha morphogenesis with the expression of other virulence attributes in this pathogen. Not surprisingly, a tup1 mutant displayed attenuated virulence in the mouse model of systemic candidiasis (Murad et al., 2001a). Hence transcriptional repression, and Tup1 in particular, is an important regulator of C. albicans virulence.
On the basis of the S. cerevisiae Ssn6-Tup1 paradigm, we and others predicted that C. albicans Ssn6 would play an important role in Tup1-mediated transcriptional repression in this fungus (Smith and Johnson, 2000; Garcia and Brown, 2001). We have tested this prediction by examining the cellular and molecular phenotypes of C. albicans ssn6 mutants. During the course of this study Hwang et al. (2003) reported in an elegant article that Ssn6 regulates yeast-hypha morphogenesis and virulence in C. albicans. They suggested that complex relationships exist between Ssn6 and the MAP kinase and cAMP-protein kinase A signaling pathways that activate morphogenesis in C. albicans. They also reported subtle differences in the morphological phenotypes of ssn6 and tup1 mutants, suggesting that Ssn6 might play distinct roles in C. albicans morphogenesis (Hwang et al., 2003). We show that, although ssn6 cells do display morphogenetic defects, these defects are distinct from those of tup1 or nrg1 cells and more reminiscent of phenotypic switching than yeast-hypha morphogenesis. We also show that although Ssn6 contributes to Tup1- and Nrg1-mediated repression at some promoters, Ssn6 is not essential for the repression of most Tup1- or Nrg1-regulated genes in C. albicans. The data suggest that Ssn6 executes roles that are independent of the Ssn6-Tup1 corepressor. These observations have important implications for the eukaryotic Ssn6-Tup1 paradigm.
MATERIALS AND METHODS
Strains and Growth Conditions
C. albicans strains (Table 1) were grown in YPD, SD, SC (Sherman, 1991, Kaiser et al., 1994), YPDA (YPD containing 0.01% adenine) or YPDAU (with 0.02% adenine and 0.008% uridine). Hyphal development was stimulated using YPD containing 10% serum at 37°C. Cell and colony morphology were analyzed using Olympus BX50 (Lake Success, NY) and Zeiss stereo microscopes (Thornwood, NY) as described previously (Murad et al., 2001a).
Table 1.
C. albicans strains
| Strain | Genotype | Scource |
|---|---|---|
| SC5314 | Wild type | Gillum et al. (1984) |
| CAF2-1 | URA3/ura3::λ imm434 | Fonzi and Irwin (1993) |
| CAI4 | ura3::λ imm434/ura3::λ imm434, | Fonzi and Irwin (1993) |
| CAI8 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG | Fonzi and Irwin (1993) |
| BCA2-9 | ura3::λ imm434/ura3::λ imm434, tup1::hisG/tup1::hisG | Braun and Johnson (1997) |
| MMC3 | ura3::λ imm434/ura3::λ imm434, nrg1::hisG-URA3-hisG/nrg1::hisG | Murad et al. (2001a) |
| MMC4 | ura3::λ imm434/ura3::λ imm434, nrg1::hisG/nrg1::hisG | Murad et al. (2001a) |
| MMC9 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, nrg1::hisG/nrg1::hisG | Murad et al. (2001a) |
| HLC54 | ura3::1 imm434/ura3::1 imm434, cph1::hisG/cph1::hisG, efg1::hisG/efg1::hisG-URA3-hisG | Lo et al. (1997) |
| HLC54ura3 | ura3::1 imm434/ura3::1 imm434, cph1::hisG/cph1::hisG, efg1::hisG/efg1::hisG | This study |
| SGC121 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, SSN6/ssn6::hisG-URA3-hisG | This study |
| SGC122 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, SSN6/ssn6::hisG | This study |
| SGC123 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, ssn6::hisG-URA3-hisG/ssn6::hisG | This study |
| SGC124 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, ssn6::hisG/ssn6::hisG | This study |
| SGC131 | ura3::λ imm434/ura3::λ imm434, cph1::hisG/cph1::hisG, efg1::hisG/efg1::hisG, SSN6/ssn6::hisG-URA3-hisG | This study |
| SGC132 | ura3::λ imm434/ura3::λ imm434, cph1::hisG/cph1::hisG, efg1::hisG/efg1::hisG, SSN6/ssn6::hisG | This study |
| SGC133 | ura3::λ imm434/ura3::λ imm434, cph1::hisG/cph1::hisG, efg1::hisG/efg1::hisG, ssn6::hisG-URA3-hisG/ssn6::hisG | This study |
| SGC134 | ura3::λ imm434/ura3::λ imm434, cph1::hisG/cph1::hisG, efg1::hisG/efg1::hisG, ssn6::hisG/ssn6::hisG | This study |
| SGC179 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, ssn6::hisG/MET3-SSN6 | This study |
| CRC001 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, TUP1/tup1::hisG-URA3-hisG | This study |
| CRC002 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, TUP1/tup1::hisG | This study |
| CRC003 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, tup1::hisG-URA3-hisG/tup1::hisG | This study |
| CRC004 | ura3::λ imm434/ura3::λ imm434, ade2::hisG/ade2::hisG, tup1::hisG/tup1::hisG | This study |
Complementation tests were performed using the S. cerevisiae ssn6 strains MAP6, which was a generous gift from Markus Proft, and PHFT5 (Tzamarias and Struhl, 1994; Loubradou et al., 2001). C. albicans SSN6 was cloned as a 6-kb EcoRI fragment into YCp50, and the phenotypes of YCp50-SSN6 and YCp50-containing S. cerevisiae PHFT5 cells were compared.
Both alleles of the SSN6 locus were sequentially disrupted in C. albicans CAI8 by Ura-blasting (Fonzi and Irwin, 1993) using a ssn6::hisG-URA3-hisG cassette that deleted codons 385-955 of the 1081 codon ORF (Table 1). To generate this cassette the hisG-URA3-hisG was cloned between the KpnI and HindIII sites of a 6.2-kb PstI fragment from the SSN6 gene. The genotypes of homozygous ssn6/ssn6 mutants were confirmed by Southern blotting and PCR analysis (unpublished data). Eight independent ssn6/ssn6 mutants were generated, all with similar phenotypes (unpublished data). Uridine auxotrophic mutants were generated by selecting ura3 segregants on YPD containing 1 mg/ml 5-fluoroorotic acid (Fonzi and Irwin, 1993). SSN6 was also disrupted in C. albicans HLC54ura3 to generate the triple ssn6/ssn6 cph1/cph1 efg1/efg1 mutant, SGC134 (Table 1). A conditional ssn6 mutant, SGC179, was generated by inactivating one SSN6 allele and placing the remaining SSN6 allele under the control of the C. albicans MET3 promoter (Care et al., 1999). Again, the genotype of this ssn6/MET3-SSN6 strain was confirmed by Southern blotting and PCR analysis (unpublished data). The expression of MET3-SSN6 allele was repressed using 2.5 mM methionine and cysteine.
To create the NRE reporter, the sequence 5′-GTCGACGGATCCGCTAGCCCCCCTGACTGCTACCATCCCCCTAAATCGGATCCGCCCCCTTGCGAACAAGTCCCCCCTGCCTTGAACGAACTGCAG (SalI and PstI sites in italics; four C5T NRE elements underlined) was cloned between the SalI and PstI sites upstream in plac-basal. This plasmid contains the basal ADH1 promoter region (Tripathi et al., 2002) cloned upstream of the Streptococcus thermophilus lacZ reporter (Uhl and Johnson, 2001) in CIp10 (Murad et al., 2000). A second NRE reporter was generated with two A2C3T and two C5T elements. A single copy of each plasmid was integrated into the XbaI-plus allele of RPS1 (Murad et al., 2000). (RPS1 was previously known as RPS10 and RP10.)
To use the C. albicans one-hybrid system (Russell and Brown, 2005), an expression vector encoding a Staphylococcus aureus LexA-C. albicans Nrg1 protein fusion was constructed by PCR amplification of the NRG1 ORF and inserting it into CIp-LexA, to create CIp-LexA-Nrg1. The 5′ PCR primers used introduced a (Gly)3-Pro-(Gly)2 linker between the amino-terminal LexA domain and the carboxy terminal domains of Nrg1. CIp-LexA and CIp-LexA-Nrg1 were introduced into C. albicans reporter strains that carried pCR-lacZ (no lexA operator upstream of basic ADH1 promoter) or pCR-OplacZ (containing the lexA operator) integrated into the ade2::hisG locus (Doedt et al., 2004). The one-hybrid system requires both URA3 and ADE2 markers for the CIp- and pCR-based plasmids, respectively. Hence the analyses were performed in derivatives of CAI8 (Table 1). The nrg1 and ssn6 mutants were MMC9 and SGC124. The tup1 mutant, CRC004, was created in CAI8 by Ura-blasting (Fonzi and Irwin, 1993) using the same tup1::hisG-URA3-hisG cassette that was used to generate the original C. albicans tup1 mutants in CAI4 (Braun and Johnson, 1997).
DNA, RNA, and Protein Analyses
Southern, Northern (Murad et al., 2001a), and Western blotting (Cormack et al., 1997) were performed as described previously. RT-PCR was performed using standard methods using the intron-containing EFB1 product to control for loading and genomic DNA contamination (Schaller et al., 1998). To measure lacZ reporter activity, X-gal overlay assays and liquid β-galactosidase assays were performed as described previously and expressed in Miller units (Rupp, 2002).
Transcript Profiling
Transcript profiling was performed using congenic C. albicans strains: wild-type, CAI4 and CAI8; nrg1, MMC4; tup1, BCA2-9; ssn6, SGC124 (Table 1). For consistency, the nrg1, tup1, and ssn6 strains were compared directly to CAI4. Then, to control for possible differences between CAI4 and CAI8, these strains were compared directly. Cells were grown to midexponential phase (OD600 = 0.6-0.8) in YPDAU at 30°C. The cells were then harvested, frozen rapidly in liquid N2, sheared mechanically using a microdismembrator (Braun Melsungen, Germany) and RNA prepared by extraction with Trizol Reagent (Gibco-BRL), as described previously (Hauser et al., 1998). Cy3- and Cy5-labeled cDNAs were prepared from total RNA, and the probes were hybridized with (nearly) whole genome C. albicans microarrays (Eurogentec, Seraing, Belgium). Slides were scanned using a ScanArray Lite scanner (PerkinElmer Life Sciences, Beaconsfield, United Kingdom) and quantified using QuantArray software (version 2.0). Data normalization and analysis were performed using GeneSpring (Silicon Genetics, Redwood City, CA), and statistical analysis was performed using SAM (Significance Analysis of Microarrays: Tusher et al., 2001). Data from at least three independent biological replicates were used for each analysis, and the SAM False Discovery Rate was set at 10%. Expression ratios for each gene that displayed a reproducible and statistically significant change in expression in mutant cells relative to the wild-type control are available in the Supplementary Data and at the Galar Fungail website (http://www.pasteur.fr/recherche/unites/Galar_Fungail/; http://www.galarfungail.org/data.htm). C. albicans gene annotations were obtained from CandidaDB (http://genolist.pasteur.fr/CandidaDB: d'Enfert et al., 2005). Functional categories for C. albicans genes were assigned mainly on the basis of the MIPS functional assignments for S. cerevisiae homologues (http://mips.gsf.de/proj/yeast/CYGD/db/index.html: Yin et al., 2004). Genes were assigned to a new virulence category was based on the functional analysis of putative virulence attributes in C. albicans.
Chromatin Immunoprecipitation Assays
Polyclonal rabbit anti-peptide antibodies were raised against the carboxy-terminal region of C. albicans Ssn6 using the peptide CMRKIEEDENYDDE (Diagnostics Scotland, Carluke, United Kingdom). The specificity of the anti-Ssn6 antiserum was confirmed by Western blotting of protein extracts from C. albicans wild-type and ssn6 cells (unpublished data). Chromatin immunoprecipitation (ChIPs) was performed in triplicate (Strahl-Bolsinger et al., 1997) comparing preimmune and anti-Ssn6 antisera and comparing C. albicans wild-type and ssn6 cells.
RESULTS
C. albicans Ssn6 Is Related to Ssn6-like Proteins in Other Eukaryotes
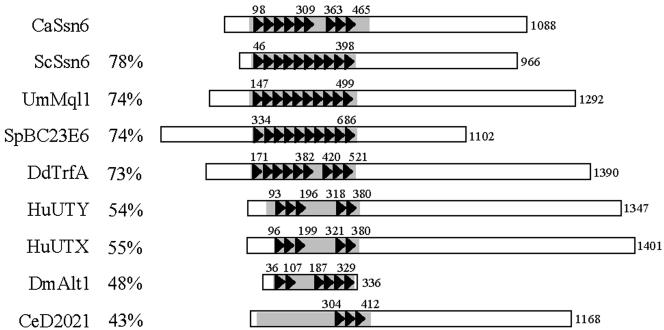
As reported by Hwang et al. (2003), C. albicans has a single locus with significant sequence similarity to S. cerevisiae SSN6 (orf19.6798; IPF5957.1). This gene encodes a 1081 amino acid protein, which contains nine tetratricopeptide (TPR) repeats between amino acids 98 and 430. An additional degenerate TPR-like sequence lies between TPR repeats six and seven (Figure 1). This TPR region of the C. albicans Ssn6 protein is closely related to the corresponding regions of Ssn6-like proteins from other fungi, slime molds, flies, worms, and mammals (Figure 1). However, the sequence similarity between these Ssn6-like proteins is relatively low outside the TPR region. The TPR repeats in S. cerevisiae Ssn6 mediate protein-protein interactions with Tup1 and with Ssn6-Tup1 targeting proteins (Tzamarias and Struhl, 1995).
Figure 1.
Ssn6-like proteins in eukaryotes. (A) S. cerevisiae paradigm of Ssn6-Tup1 function. (B) Arrangement of TPR repeats in Ssn6-like proteins from C. albicans, S. cerevisiae, Ustilago maydis, Schizosaccharomyces pombe, Dictyostelium discoideum, humans, Drosophila melanogaster, and Caenorhabditis elegans. Coordinates of TPR domains are shown, and percentage amino acid sequence similarities between the shaded regions of CaSsn6 and other Ssn6-like proteins are indicated.
We tested whether the C. albicans SSN6 gene encodes a functional homologue of S. cerevisiae Ssn6 (unpublished data). When transformed into S. cerevisiae using the single-copy vector YCp50, C. albicans SSN6 suppressed the flocculation defects of the ssn6 cells as well as their growth defects at 37°C and on glycerol. This was consistent with the observations of Hwang et al. (2003). We conclude that C. albicans Ssn6 is a functional homologue of S. cerevisiae Ssn6.
Phenotypes of C. albicans ssn6 Mutants
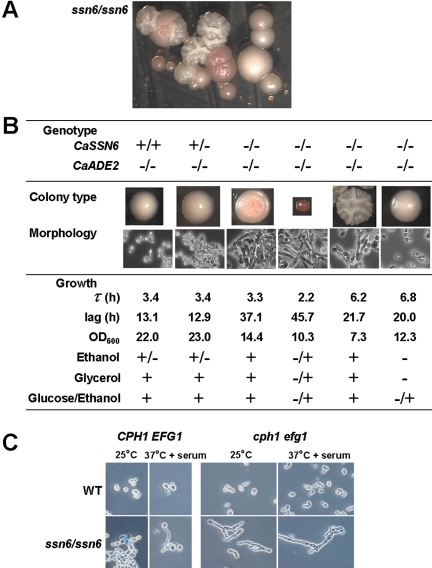
Tup1 inactivation derepresses morphogenesis in C. albicans, leading to the formation of characteristically wrinkly colonies (Braun and Johnson, 1997). If Ssn6 simply acts as a corepressor with Tup1 in C. albicans, one might predict that the inactivation of Ssn6 would generate a similar phenotype. However, Hwang et al. (2003) described subtle morphological differences between C. albicans ssn6 and tup1 mutants. To examine these differences in more detail, we generated independent homozygous ssn6/ssn6 mutants in C. albicans CAI8 (ura3, ade2) and carefully examined their morphologies.
As expected, wild-type SSN6/SSN6 cells and heterozygous ssn6/SSN6 cells generated large smooth white colonies when streaked onto YPDA plates (Figure 2). However, when freshly generated homozygous ssn6/ssn6 mutants were restreaked onto fresh YPDA, dramatic colony phenotypes were observed routinely. A range of colony phenotypes arose from a single ssn6/ssn6 colony: small versus large, red versus white, and wrinkled versus smooth colonies (Figure 2). Cells from all of these colonies were ade2/ade2 ssn6/ssn6 mutants as confirmed by diagnostic PCR. This colony variability was reproduced in eight independent mutants.
Figure 2.
Phenotypes of C. albicans ssn6 cells. (A) Mixed colony phenotypes of a freshly generated ssn6/ssn6 mutant (SGC123) restreaked on YPDA. (B) Characterization the various ssn6 colony types. Genotypes were rechecked by PCR diagnosis: +, wild-type allele; -, null allele (CAI8, SGC121 and SGC123: Table 1). For each colony type, cell morphology and growth in liquid medium in YPD at 30°C (length of lag phase, growth rate, and final OD600) were determined. Growth on nonfermentable carbon sources as sole carbon source was examined on YNB plates at 30°C: +, robust growth; +/-, weak growth; -/+, very weak growth; -, no growth. (C) Morphogenetic responses of ssn6 (SGC123) and ssn6, cph1, efg1 cells (SGC133) after 3 h in YPD containing 10% serum.
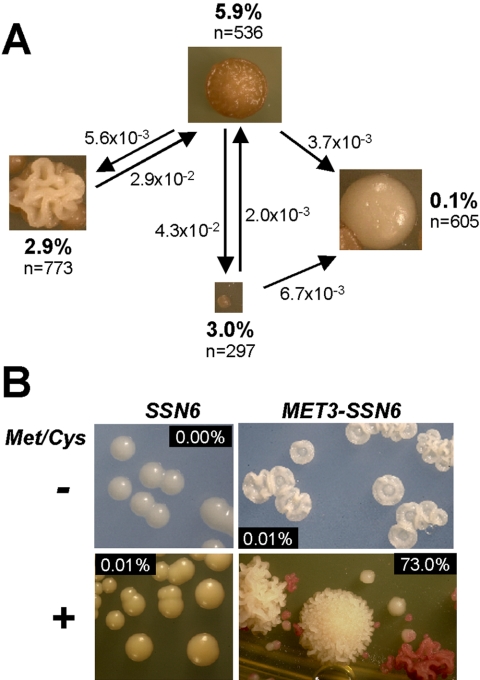
In addition to displaying colony variability, most ssn6 colony types displayed phenotypic instability. For example, 5.9% of cells from large pink wrinkly colonies switched to different colony morphologies when restreaked onto fresh YPDA (Figure 3). Ssn6 cells from large smooth white colonies were relatively stable, with only 0.1% of these cells switching colony morphologies. As a result, serial passaging of ssn6 strains generally led to an accumulation of the large smooth white growth form. Large wrinkly white colonies also arose relatively frequently after serial passaging.
Figure 3.
Phenotypic instability of C. albicans ssn6 cells. (A) The phenotypic stability of each colony type was assayed by replating cells on fresh YPD and measuring the proportion of colonies that had switched to an alternative form. (B) Phenotypic instability could be induced in a conditional ssn6/MET3-SSN6 strain following the addition of 2.5 mM methionine and cysteine to the medium: SSN6, CAI8; MET3-SSN6, SGC179 (Table 1).
A conditional ssn6/MET3-SSN6 mutant was constructed to confirm that these dramatic phenotypic effects were caused by Ssn6 depletion. In the absence of methionine and cysteine, when the MET3-SSN6 allele was expressed, this mutant displayed a poppy-like colony morphology (Figure 3), presumably because of abnormal Ssn6 expression patterns from the MET3 promoter. This poppy-like phenotype was stable. However, a range of different colony morphologies was observed when clonal ssn6/MET3-SSN6 cells were plated onto medium containing methionine and cysteine, and these phenotypes were unstable. Most cells in these colonies (73%) switched colony morphologies when replated onto medium containing methionine and cysteine (Figure 3). In contrast they reverted to the poppy-like morphology when replated onto medium lacking methionine and cysteine (unpublished data). These data confirmed that Ssn6 depletion dramatically effects C. albicans colony morphology and enhances switching between different colony phenotypes.
We examined the phenotypes of cells from the different colony types (Figure 2). During growth in liquid YPD, ssn6 cells from smaller colonies displayed longer lag periods and generated less biomass than ssn6 cells from larger colonies. With the exception of the large smooth white colonies, all cell types grew on media containing ethanol, glycerol as the sole carbon source.
Colonies formed by wild-type and SSN6/ssn6 strains on YPDA at 30°C contained the ovoid budding cells typical of the yeast growth form of C. albicans (Sudbery et al., 2004). In contrast, homozygous ssn6/ssn6 cells from some colonies resembled pseudohyphae with septa at mother-daughter junctions (Figure 2). However, there was significant population heterogeneity with respect to the morphology of these cells, and they did not divide synchronously like true C. albicans pseudohyphae. This suggested that these cells were displaying a pathological morphological phenotype caused by Ssn6 inactivation, rather than bone fide pseudohyphal development. Ssn6/ssn6 cells from other colonies were small and stumpy, reminiscent of C. albicans efg1 cells (Lo et al., 1997). There was no clear correlation between the degree of filamentation and the wrinkliness of ssn6 colonies.
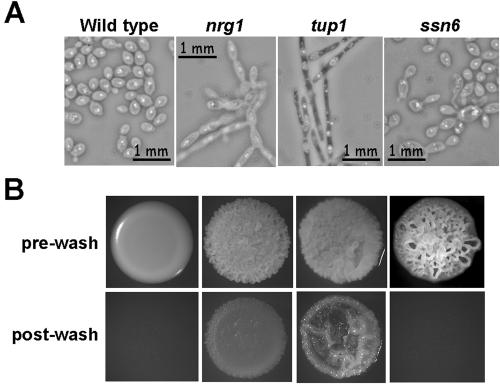
These ssn6 phenotypes were significantly different from those of tup1 and nrg1 cells, which display stable cellular and colonial phenotypes (Braun and Johnson, 1997; Braun et al., 2001; Murad et al., 2001a). A direct comparison of cells growing on YPDA at 30°C confirmed the contrasting cellular and colonial morphologies of wild-type, nrg1, tup1, and ssn6 cells (Figure 4). The ssn6 cells were more similar to the budding wild-type parent than the pseudohyphal nrg1 and tup1 cells. Furthermore, the wrinkly ssn6 colonies were not invasive, unlike the nrg1 and tup1 colonies (Figure 4).
Figure 4.
Phenotypic differences between wild-type, nrg1, tup1, and ssn6 cells. (A) Morphology of cells growing exponentially in YPD at 30°C: wild-type, CAI8 containing CIp10; nrg1, MMC3; tup1, BCA2-9; ssn6, SGC123 (Table 1). (B) Colony morphology (prewash) and invasiveness (postwash) of the same strains growing on YPD at 30°C. To measure invasiveness, cells were washed off the surface of the plate into H2O with a glass spreader.
Hwang et al. (2003) examined the role of Ssn6 in morphogenesis by examining the epistatic relationships between Ssn6, Tup1, Cph1, and Efg1. Cph1 and Efg1 define the MAP kinase and cAMP-protein kinase A pathways, respectively, that activate hyphal development in C. albicans (Liu et al., 1994; Bockmuhl and Ernst, 2001). Based on the complex phenotypes of the various mutants they examined, Hwang et al. (2003) suggested that Ssn6 inactivation induces pseudohyphal growth and that both Cph1 and Efg1 might activate this pseudohyphal growth. In our hands, Ssn6 inactivation did not lead to the formation of true pseudohyphae (Figures 2 and 4). We examined the effects of serum on ssn6 mutants because serum is a strong inducer of hyphal development in C. albicans. In response to serum stimulation, ssn6 cells appeared to retain the ability to form germ tubes, the progenitors of true hyphae (Figure 2C). Furthermore, unlike tup1 cells (Braun and Johnson, 1997; Murad et al., 2001), ssn6 cells were still able to respond to serum after Cph1 and Efg1 inactivation, although the filaments formed by the ssn6, efg1, cph1 triple mutants were not true hyphae. We suggest that although Cph1 and Efg1 can influence the morphology of ssn6 cells, the morphological phenotypes caused by Ssn6 inactivation are not related to bone fide morphogenetic developmental programs.
Global Molecular Phenotypes of Wrinkly and Smooth C. albicans ssn6 Cells
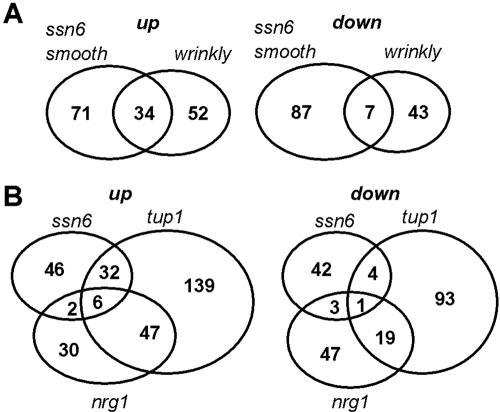
The global molecular phenotypes of C. albicans ssn6 cells from large white wrinkly and smooth colonies were compared by transcript profiling. The analysis of other growth forms was precluded by their phenotypic instability. About 1.5% of the ∼6000 C. albicans genes displayed reproducible and statistically significant increases in expression of twofold or more in the wrinkly or smooth growth forms relative to wild-type cells (Figure 5A). Similar numbers of C. albicans genes displayed decreased expression. A direct comparison of the transcript profiles of C. albicans CAI4 and CAI8 (Supplementary Data) confirmed that these differences were not due to differences between the parental strains use in this study (Table 1). The Ssn6 repressor might regulate these genes indirectly. Alternatively, Ssn6 might also be able to act as a transcriptional activator (Conlan et al., 1999).
Figure 5.
(A) Numbers of C. albicans genes that were up- or down-regulated twofold or more in smooth and wrinkly ssn6 cells (SGC123), relative to the wild-type control (CAF2-1). (B) Numbers of C. albicans genes that were significantly up- or down-regulated in wrinkly ssn6 (SGC123), tup1 (BCA2-9), and nrg1 cells (MMC3), relative to the wild-type control (CAF2-1). Significant changes of twofold or more were identified using SAM (FDR set at 10%) using data from three independent replicates for each cell type.
There was limited overlap between the transcript profiles of the wrinkly and smooth growth forms (Figure 5A), which might account for at least some of the phenotypic differences between these cells. A significant proportion (31%) of the genes that were up-regulated only in the wrinkly form encode functions involved in carbon metabolism (Table 2). In contrast, glycolytic genes were down-regulated in the smooth form. Ribosomal protein genes represented 28% of those that were up-regulated only in smooth cells. Iron assimilatory functions (FTR1, HEM1, RBT2) represented 9% of the genes that were up-regulated in both cell types. These observations were consistent with the differences in growth rates and carbon assimilation patterns of the various ssn6 growth forms (Figure 2).
Table 2.
C. albicans genes that are up-regulated in smooth and/or wrinkly ssn6 cells
| Smooth and wrinkly | Smooth only | Wrinkly only |
|---|---|---|
| ADH2, ALD5, CRD2, CTA1, FTR1, FRP6, GRP2, HEM1, HGT12, HSP12, INO1, MAM33, MET15, NTF2, OSM2, PCK1, PLB4.5, PLB4.3, PRY2, RBT2, RBT4, SAP7, IPF1580, IPF3277, IPF3282, IPF3282.3, IPF3964, IPF4181, IPF4182, IPF13493, IPF14109, IPF15839, IPF17186 | ATP7, ATP20, CAN2, CAR1, CAR1.3, CCC1, CIT1, CYB5, CYS3, DLD3, FRE32, FRP1, FTR2, GAP2, GAP5, HGT11, HHF21, HHF22, HNM1, IDP2, IFD7, LYS9, LYS21, MRPL10, MRPL37, OPT1, PLC3, QCR8, QCR9, RIM8, RPL11, RPL25, RPL29, RPL30, RPL34B, RPL35, RPL38, RPL39, RPL42, RPL43A, RPS18, RPS21B, RPS22A, RPS22, RPS24, RPS26A, RPS28B, RPS30, SOD1, STL1, TOM22, UBI4, IPF277, IPF643, IPF1617, IPF2277, IPF2690.5, IPF2690.3, IPF3485, IPF3912, IPF4553, IPF6629, IPF7109, IPF12942, IPF14452, IPF14850, IPF14895, IPF15679, IPF17190, IPF17237 | AAH1, ACS1, ADH1, ARC35, AYR2, CDR4, CRD1, CYB3, ECM41, ECM331, FBP1, GLC3, GLK1, GPD2, GSY1, HXK2, HXT5, HXT61, HXT62, MAE1, MLS1, PFK2, RHR2, RNR22, RPL23B, SNG3, SSA4, TFS1, TPS3, IPF1531, IPF4724, IPF5185, IPF6881, IPF7715, IPF8321, IPF8369, IPF10533, IPF10558, IPF12101, IPF13121.3, IPF13607, IPF13836, IPF15301, IPF15870, IPF15957, IPF16565, IPF17840, IPF19066, IPF19640, IPF19953, IPF20008 |
Genes that were up-regulated twofold or more in a reproducible and statistically significant manner are shown. Further details are provided in the Supplementary Data.
Global Comparison of the Ssn6, Tup1, and Nrg1 Regulons in C. albicans
Transcript profiling was also performed to compare the global roles of Ssn6, Tup1, and Nrg1 in C. albicans. Ssn6, Tup1, and Nrg1 regulons were defined operationally as those subsets of C. albicans genes whose expression was altered twofold or more in ssn6, tup1, and nrg1 cells, respectively. If Ssn6 acts simply as a corepressor with Tup1, one might expect their regulons to overlap. Nrg1 is thought to be one of a number of proteins that targets the Tup1-Ssn6 complex to specific C. albicans promoters. On this basis one might predict that the Nrg1 regulon would be a subset within the Ssn6-Tup1 regulon. However, neither of these simplistic predictions was supported by our data (Figure 5B), indicating that the roles of these factors are more complex.
C. albicans genes that are repressed by Tup1 or Ssn6 are expected to be overexpressed in tup1 and ssn6 cells, respectively. However, there was limited overlap between the Ssn6 and Tup1 regulons (Figure 5B). Most genes that were up-regulated in tup1 cells were not significantly up-regulated in ssn6 cells (186 of 224 genes). This suggests that Tup1 represses the expression of these genes, but that Ssn6 is not essential for their repression.
A significant number of C. albicans genes were down-regulated in tup1 cells (Figure 5B). Tup1 might regulate these genes indirectly. Significantly, most genes that were down-regulated in tup1 cells were not down-regulated in ssn6 cells (112 of 117 genes). Again this suggests that Ssn6 is not essential for the Tup1-mediated regulation of these genes.
Of the 85 C. albicans genes that were up-regulated in nrg1 cells, 32 were not up-regulated in tup1 cells, and most were not up-regulated in ssn6 cells (Figure 5B). This suggested that Tup1 is not essential for the repression of some genes by Nrg1 and that Ssn6 is not essential for the repression of most Nrg1-repressed genes. Hence Nrg1 appears to act independently of Tup1 and Ssn6 to repress some C. albicans genes.
We examined the functions of the genes in the Nrg1, Tup1, and Ssn6 regulons in C. albicans. These genes and their functions are listed in the Supplementary Data. Previously, we reported that hypha-specific genes are regulated both by Nrg1 and Tup1 (Murad et al., 2001a). This observation, which was made using macroarrays containing about one-third of C. albicans genes, was confirmed in this study, which used (nearly) whole genome C. albicans microarrays. The hypha-specific genes HWP1, ECE1, RBT1, and RBT5 were among the most strongly regulated genes in the subset of genes regulated by Tup1 and Nrg1 (Table 3). These data are consistent with those of Kadosh and Johnson (2005) in the accompanying paper. The hypha-specific genes, ALS3 and ALS8, were absent from the subset of Tup1- and Nrg1-regulated genes because they were not on the microarray. However, we have shown previously that ALS3 is regulated both by Nrg1 and Tup1 (Murad et al., 2001a). (ALS8 is now known to be an allele of ALS3: Zhao et al., 2004.) The expression of HYR1, which encodes a hypha-specific cell wall glycoprotein (Bailey et al., 1996), was elevated 2.3-fold in tup1 cells, but was not elevated in nrg1 or ssn6 cells. The expression of HGC1/CLN21, which encodes a hypha-specific cyclin (Zheng et al., 2004), was elevated 4.0-fold in tup1 cells, 1.6-fold in nrg1 cells, and 1.0-fold in ssn6 cells. Therefore, whereas HGC1/CLN21 formally lies outside the Nrg1-Tup1 regulon, it does appear to be regulated by both of these factors.
Table 3.
C. albicans genes that are up-regulated in nrg1 and tup1 cells, but not in ssn6 cells
| Fold regulation
|
|||||
|---|---|---|---|---|---|
| Systematic name | Common name | nrg1 | tup1 | ssn6 | Functiona |
| CA2830 | RBT1 | 9.6 | 27.9 | 1.0 | Repressed by TUP1 protein 1 |
| CA4336 | DDR48 | 13.6 | 22.1 | 1.0 | Stress protein (by homology) |
| CA1402 | ECE1 | 8.7 | 20.0 | 1.0 | Cell elongation protein |
| CA2825 | HWP1 | 11.8 | 15.4 | 1.0 | Hyphal wall protein |
| CA2558 | RBT5 | 2.7 | 13.3 | 1.2 | Repressed by TUP1 protein 5 |
| CA5112 | IPF1341 | 5.9 | 10.0 | 1.7 | Similarity to mucin proteins (by homology) |
| CA2405 | IPF3844 | 3.4 | 7.4 | 1.1 | Unknown function |
| CA4381 | IPF20169 | 2.8 | 6.0 | 1.1 | Unknown function |
| CA0386 | IPF4065 | 2.1 | 5.4 | 1.1 | Unknown function |
| CA0448 | ALS10 | 8.5 | 4.7 | 1.1 | Agglutinin-like protein |
| CA4174 | IPF4119.5 | 2.4 | 4.6 | 1.1 | Unknown function, 5′ end |
| CA3154 | FUN34.5 | 2.1 | 4.4 | 1.8 | Unknown function, 5′ end |
| CA5641 | GAC1 | 2.7 | 4.3 | 1.0 | ser/thr phosphoprotein phosphatase 1, regulatory chain (by homology) |
| CA0074 | IFD7 | 2.5 | 4.3 | 1.3 | Putative aryl-alcohol dehydrogenase (by homology) |
| CA4857 | PHR1 | 3.2 | 4.2 | 1.1 | GPI-anchored pH responsive glycosyl transferase |
| CA3813 | FRP1 | 2.6 | 4.2 | 1.0 | Related to Y. lipolytica glyoxylate pathway regulator Gpr1p |
| CA4985 | IPF2050 | 3.6 | 3.9 | 1.1 | Similar to S. cerevisiae Kip1p kinesin-related protein (by homology) |
| CA1238 | IPF15781 | 3.9 | 3.8 | 1.1 | Unknown function |
| CA0722 | ERK1 | 2.3 | 3.8 | 1.2 | Mitogen-activated protein kinase (FUS3 homolog) |
| CA4120 | SOD1.3 | 3.9 | 3.7 | 1.0 | Cu,Zn-superoxide dismutase, 3-prime end |
| CA0671 | GRP4 | 2.2 | 3.6 | 1.0 | Putative reductase (by homology) |
| CA0316 | ALS1.3eoc | 3.4 | 3.5 | 0.8 | Agglutinin-like protein, 3-prime end |
| CA0840 | IFD1 | 2.1 | 3.5 | 1.1 | Putative aryl-alcohol dehydrogenase (by homology) |
| CA3827 | IPF10662 | 6.3 | 3.3 | 1.2 | Unknown function |
| CA3260 | IPF7968 | 2.3 | 3.3 | 1.0 | Unknown function |
| CA5650 | IPF7109 | 2.1 | 3.3 | 1.8 | Unknown function |
| CA3842 | YKE2.3 | 2.1 | 3.3 | 1.4 | gim complex component, 3-prime end (by homology) |
| CA4189 | IPF7527 | 3.0 | 3.0 | 1.0 | Unknown function |
| CA3173 | IPF14145 | 5.9 | 2.9 | 1.0 | Unknown function |
| CA4127 | IPF6629 | 2.6 | 2.9 | 1.6 | Unknown function |
| CA2302 | IPF6518 | 2.3 | 2.8 | 1.4 | Unknown function |
| CA2391 | ADH5 | 2.7 | 2.7 | 1.2 | Probable alcohol dehydrogenase (by homology) |
| CA5039 | GAP2 | 4.9 | 2.6 | 1.4 | General amino acid permease (by homology) |
| CA2291 | IPF9740 | 2.5 | 2.6 | 1.7 | Oligo-1,4 -1,4-glucantransferase/amylo-1,6-glucosidase (by homology) |
| CA1188 | RPL30.3 | 2.0 | 2.6 | 1.7 | RNA binding, 3′ end (by homology) |
| CA2937 | RPS21B.3 | 2.0 | 2.6 | 1.4 | Ribosomal protein S21, 3′ end |
| CA5339 | IPF885 | 2.8 | 2.5 | 1.2 | Glucan 1,3-beta-glucosidase (by homology) |
| CA5225 | ACB1.exon2 | 2.2 | 2.5 | 1.5 | Acyl-coenzyme-A-binding protein, exon 2 (by homology) |
| CA5953 | IPF3506 | 2.2 | 2.5 | 1.0 | Unknown function |
| CA0559 | GPX1 | 2.0 | 2.5 | 1.3 | Glutathione peroxidase (by homology) |
| CA2589 | KRE1 | 2.4 | 2.4 | 0.9 | Secretory pathway protein |
| CA3766 | IPF18298.3 | 2.6 | 2.3 | 1.1 | Unknown function, 3-prime end |
| CA4534 | RPS26A | 2.1 | 2.3 | 1.7 | Ribosomal protein S26.e.A, cytosolic (by homology) |
| CA3897 | PFY1 | 2.5 | 2.2 | 1.1 | Binds to actin |
| CA4113 | CHO1 | 2.3 | 2.2 | 1.1 | Phosphatidylserine synthase |
| CA0716 | DOG2 | 3.0 | 2.1 | 1.1 | 2-deoxyglucose-6-phosphate phosphatase (by homology) |
| CA2947 | IPF6298 | 2.4 | 2.0 | 1.2 | Unknown function |
Genes were considered to be up-regulated if their expression ratio in mutant vs. wild type cells was twofold or more in a reproducible and statistically significant manner. Further details are provided in the Supplementary Data.
Information on gene function was taken from CandidaDB (http://genolist.pasteur.fr/CandidaDB)
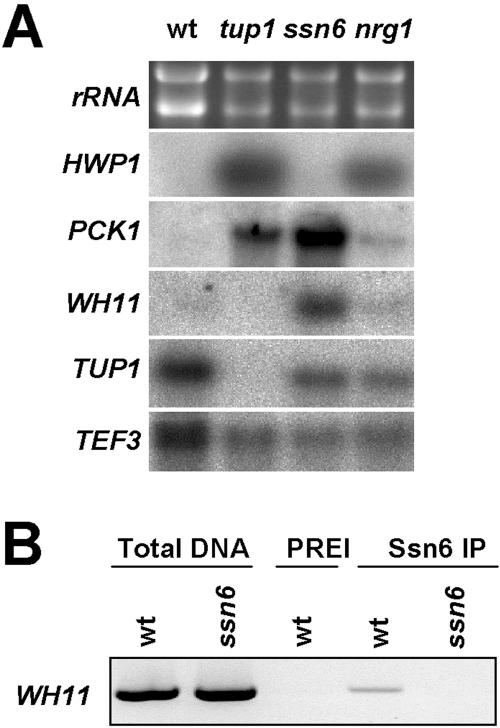
Interestingly, our transcript profiling indicated that hypha-specific genes were not derepressed in ssn6 cells grown at 30°C and was confirmed by northern blotting (Figure 6A). The HWP1 mRNA was derepressed in nrg1 and tup1 cells, but not in the ssn6 mutant. Hwang et al. (2003) reported that HWP1 and ECE1 were derepressed in ssn6 cells. However, this derepression was partial, only being observed at 37°C and not at 30°C. We conclude that Nrg1-Tup1 mediates the repression of most hypha-specific genes in C. albicans, but that Ssn6 is not essential for this repression, at least under the conditions examined in this study.
Figure 6.
(A) Northern analysis of C. albicans mRNAs in wild-type (CAI8), tup1 (BCA2-9), ssn6 (SGC123), and nrg1 cells (MMC3). (B) Chromatin immunoprecipitation revealing association of Ssn6 with the WH11 promoter: wild-type cells (CAI8), ssn6 cells (SGC123); PCR with genomic DNA template, Total DNA; reaction with preimmune serum, PREI; reactions with anti-Ssn6 serum.
A comparison of the global cellular roles of Ssn6 and Tup1 was performed by examining whether specific functional categories were enriched in the subsets of genes whose expression levels were altered in ssn6 and tup1 cells (Table 4). This analysis reinforced the view that Ssn6 does not contribute to the regulation of hypha-specific genes under these growth conditions. The analysis also highlighted the important roles of Tup1 and Ssn6 in repressing genes involved in carbon metabolism. Most notably, the gluconeogenic genes PCK1 and FBP1 and the glyoxylate cycle gene MLS1 were derepressed in both ssn6 and tup1 cells (Table 4; Figure 6A). We note that phenotypic switching is associated with changes in the expression of genes involved in central carbon metabolism (Lan et al., 2002). Tup1 and Ssn6 also appear to play roles in the regulation of genes involved in amino acid metabolism. Further details are provided in the Supplementary Data.
Table 4.
Enrichment of functional categories in subsets of genes whose expression was significantly altered in ssn6 or tup1 cells
Relative enrichment of functional categorya
| Up
|
Down
|
|||||
|---|---|---|---|---|---|---|
| Functional category | tup1 | ssn6 | tup1 and ssn6 | tup1 | ssn6b | tup1 and ssn6 |
| Amino acid metabolism | 1.16 | 5.00 | 1.39 | 2.01 | 6.25 | 3.57 |
| Carbon metabolism | 2.88 | 1.67 | 5.56 | 0.43 | 0.00 | 2.38 |
| Cell rescue and defense | 0.63 | 2.50 | 1.39 | 0.46 | 2.08 | 0.00 |
| Hypha-specific genes | 16.88 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Unknown function | 1.76 | 0.45 | 1.52 | 0.88 | 0.00 | 0.16 |
The Relative Enrichment of Functional Category was calculated by determining the proportion of genes in the subset of Ssn6- or Tup1-regulated genes that belong to these functional categories and then dividing this by the proportion of all C. albicans genes that belong to the categories (Yin et al., 2004)
The number of genes in this subset was too low to generate significant data on the enrichment of Functional Category
Role of Tup1 and Ssn6 in NRE-mediated Repression
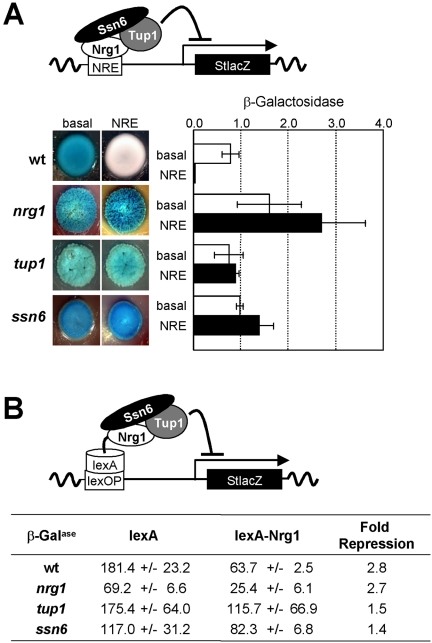
Nrg1 represses the hypha-specific gene expression via Nrg1 Response Elements (NREs: Murad et al., 2001a). Therefore, we tested whether Ssn6 and Tup1 are required for NRE-mediated repression in C. albicans. An oligonucleotide containing four NRE elements of the type C5T was cloned upstream of a basal lacZ reporter and transformed into C. albicans. The expression levels of the NRE-lacZ and basal-lacZ reporters were then compared in wild-type, nrg1, tup1, and ssn6 cells (Figure 7A). The introduction of the NRE elements repressed expression in wild-type cells, and this repression was released in nrg1 cells. This confirmed that Nrg1 was required for the repression imposed by the NRE element. Repression was also released in tup1 and ssn6 cells. This indicated that both Ssn6 and Tup1 were required for Nrg1-NRE-mediated repression in C. albicans. Similar findings were made using a second NRE reporter containing the A2C3T and C5T elements seen in the ALS3 promoter (unpublished data).
Figure 7.
(A) An NRE imposes repression on a lacZ reporter in C. albicans in an Nrg1-, Ssn6-, and Tup1-dependent manner: wild-type, CAI4; nrg1, MMC4; tup1, BCA2-9; ssn6, SGC124 (Table 1); basal reporter lacking NRE, basal; NRE [(C5T)4] containing reporter. (B) In a C. albicans one-hybrid assay, a lexA-Nrg1 fusion imposes repression on a Lex Operator-lacZ reporter in an Ssn6- and Tup1-dependent manner: wild-type, CAI8; nrg1, MMC9; tup1, CRC004; ssn6, SGC124. Fold repression was measured by comparing β-galactosidase levels (Miller Units) for C. albicans cells expressing LexA with those expressing the LexA-Nrg1 fusion. Means and standard deviations from triplicate assays on three independent transformants are shown.
The roles of Ssn6 and Tup1 in Nrg1-mediated repression were analyzed further using the C. albicans one-hybrid system (Doedt et al., 2004; Russell and Brown, 2005). Briefly, we tested the ability of a lexA-Nrg1 fusion to impose repression on a lacZ reporter that carries a lexA operator. The expression levels of the LexOP-lacZ reporter were analyzed in wild-type, nrg1, tup1, and ssn6 cells expressing either the lexA-Nrg1 fusion or the lexA protein alone (Figure 7B). The presence of Nrg1 in the lexA-fusion caused nearly threefold repression in wild-type and nrg1 cells. This repression was significantly attenuated in ssn6 and tup1 cells, indicating that both Ssn6 and Tup1 were required for repression by the lexA-Nrg1 fusion in C. albicans.
Regulation of WH11 by Ssn6
The phenotypic instability induced by Ssn6 depletion (Figure 3) was reminiscent of the phenotypic switching described by Soll and colleagues (Slutsky et al., 1985; Soll, 2002). They have reported that misexpression of TUP1 or the white-phase-specific gene WH11 can induce phenotypic switching in the C. albicans strain WO1 (Kvaal et al., 1997; Zhao et al., 2002). Therefore, we examined whether WH11 expression was affected in C. albicans ssn6 cells. WH11 mRNA levels were significantly elevated in ssn6 cells, but not in tup1 or nrg1 cells (Figure 6A). Then we tested whether Ssn6 is associated with the WH11 promoter by chromatin immunoprecipitation. WH11 promoter sequences were significantly enriched in wild-type cells, but not in ssn6 cells after chromatin immunoprecipitation with an anti-Ssn6 antibody (Figure 6B). This enrichment was not observed with preimmune sera. Therefore, Ssn6 appears to associate with the WH11 promoter in vivo.
These observations lend weight to the idea that Ssn6 can act independently of Tup1 and Nrg1 to regulate the expression of some C. albicans genes (Figure 5B). Furthermore, they provide a possible mechanism by which Ssn6 depletion can promote phenotypic instability. Ssn6 depletion appears to cause WH11 misexpression, which is known to promote phenotypic switching in C. albicans (Kvaal et al., 1997).
DISCUSSION
The prevailing view is that Ssn6 interacts with Tup1 to form a corepressor complex that regulates a large number of genes in budding yeast and that this corepressor complex is conserved from yeast to humans (reviewed by Smith and Johnson, 2000). Based on this S. cerevisiae paradigm, it was predicted that Ssn6 and Tup1 might execute equivalent functions in C. albicans. Consistent with this, Johnson and coworkers have shown that Tup1 plays a critical role in the repression of filamentous growth and the expression of hypha-specific genes in C. albicans (Braun and Johnson, 1997, 2000; Braun et al., 2000, 2001; Kadosh and Johnson, 2005). Others have added to this by confirming that Tup1 regulates hypha-specific genes and by identifying other putative Tup1 gene targets in C. albicans (Sharkey et al., 1999; Murad et al., 2001b).
The role of Ssn6 in C. albicans remained less clear. In their elegant article, Hwang et al. (2003) showed that C. albicans SSN6 is a functional homologue of S. cerevisiae SSN6. They also examined the morphological phenotypes of C. albicans ssn6 mutants and the epistatic relationships between Ssn6, Tup1, Cph1, and Efg1, which were complex. They suggested that Ssn6 plays an important role in regulating yeast-(pseudo)hypha morphogenesis, pointing out that Ssn6 might play some distinct roles in this process. However, the relationship between Ssn6 and Tup1 in C. albicans remained obscure. Hence we have examined this issue in some detail. As a result we are able to draw a number of important conclusions.
First, C. albicans Ssn6 is member of a conserved family of eukaryotic Ssn6-like proteins that display a relatively high level of sequence similarity in their TPR domains (Figure 1). This is significant because these domains mediate interactions with Tup1-like proteins (Tzamarias and Struhl, 1995). On this basis, C. albicans Ssn6 would be expected to form a corepressor complex with Tup1.
Second, Ssn6 and Tup1 do not execute equivalent cellular roles in C. albicans. This conclusion is based on several complementary observations. C. albicans ssn6 and tup1 mutants display dramatically different phenotypes. C. albicans tup1 cells display stable phenotypes, growing constitutively as pseudohyphae that form highly wrinkled and invasive colonies (Braun and Johnson, 1997). In contrast, independent C. albicans ssn6 mutants and conditional MET3-ssn6 mutants displayed unstable phenotypes, switching at high frequencies between different growth forms (strains SGC123 and SGC179: Table 1, Figures 2 and 3). Some ssn6 growth forms generated pseudohyphal-like cells. However, these cells did not divide synchronously, they showed a high degree of population heterogeneity, and they were not invasive (Figure 4). We also observed that ssn6 cells were able to form filaments in response to serum, even in the absence of Cph1 and Efg1 (Figure 2C). This indicated that the filamentous structures generated by ssn6 cells were not dependent on the MAP kinase and cAMP-protein kinase A signaling pathways that trigger hyphal development in response to serum (Lo et al., 1997; Ernst, 2000; Brown 2002). Furthermore, the inactivation of Ssn6 led to the derepression of the phase-specific gene WH11, but did not affect the expression of hypha-specific genes (Figures 5 and 6). These data are consistent with the idea that Ssn6 depletion does affect cell morphology (Hwang et al., 2003). However, we conclude that, under the conditions analyzed in this study, Ssn6 depletion promotes phenotypic switching rather than morphogenesis.
Third, Ssn6 is not essential for Tup1-mediated repression of many genes in C. albicans. If Ssn6 were essential for Tup1-mediated repression, transcript profiling would have revealed a high degree of concordance between the Ssn6 and Tup1 regulons, as was the case in S. cerevisiae (Hughes et al., 2000). In general, S. cerevisiae ssn6 and tup1 cells display only quantitative differences in expression levels. However, this was not the case in C. albicans. The repression of most Tup1-regulated genes was not dependent on Ssn6 (Figure 5). Hence Ssn6 does not simply act as a corepressor with Tup1 in C. albicans.
Fourth, Ssn6 can act independently of Tup1 to repress some C. albicans genes. The repression of a significant number of Ssn6-regulated genes was not dependent on Tup1 (Figure 5). Furthermore, although the repression of the HWP1 gene was dependent on Tup1 but not Ssn6, the repression of WH11 was dependent on Ssn6 but not Tup1 (Figure 6).
Fifth, Ssn6 and Tup1 are required to differing extents for Nrg1-mediated repression at specific C. albicans promoters. Nrg1 represses the transcription of some C. albicans promoters in a Tup1-dependent manner via the NRE element (Braun et al., 2001; Murad et al., 2001a). This knowledge facilitated the functional analysis of Ssn6 in C. albicans by allowing us to investigate the role of this protein at specific promoters. Under the conditions analyzed in this study, it is clear that Ssn6 is not essential for the Nrg1-mediated repression of C. albicans genes, including hypha-specific genes (Figures 5 and 6). However, both Ssn6 and Tup1 are essential for Nrg1-mediated repression at two artificial promoters (Figure 7).
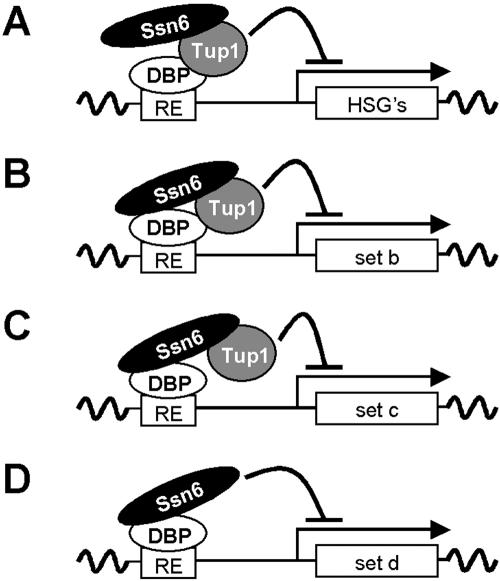
Many of these observations can be explained in the context of the S. cerevisiae Ssn6-Tup1 paradigm (Tzamarias and Struhl, 1995; Smith and Johnson, 2000). According to this paradigm, Ssn6 stabilizes the interaction between Tup1 and the Tup1 targeting protein. We suggest that the extent to which Ssn6 is required to stabilize this interaction depends on the C. albicans promoter (Figure 8). Ssn6 might essentially be redundant at some C. albicans promoters, but this does not exclude the possibility that Ssn6 remains bound to Tup1 at these promoters (Figure 8A). At other promoters, Ssn6 might promote the interaction between Tup1 and the Tup1-targeting protein. Under some conditions the role of Ssn6 might be minimal, but this does not exclude the possibility that under other conditions, Ssn6 might become more important to maintain the stability of the complex (Figure 8B). Finally at other promoters, Ssn6 might be essential for the integrity of the complex (Figure 8C). In reality, Tup1-regulated C. albicans promoters probably represent a continuum between the extremes shown in Figure 8.
Figure 8.
Models illustrating Ssn6 function in C. albicans. (A) Ssn6 is not required for Tup1-mediated repression at some promoters, suggesting that Tup1 might interact directly with the DNA-binding protein (DBP). (B) Ssn6 is partially required for Tup1-mediated repression at some C. albicans promoters, suggesting that Ssn6 stabilizes Tup1 interactions with the DNA-binding protein. (C) Ssn6 is essential for Tup1-mediated repression at other promoters, suggesting that Tup1 depends on Ssn6 for its interaction with the DNA-binding protein. The DNA-binding protein might differ between these promoters, or alternatively differences in the sequence context of the Response Element (RE) might affect the Ssn6 dependency. (D) The repression of some C. albicans promoters by Ssn6 is not dependent on Tup1 (see text).
These models predict that different Tup1 targeting proteins will depend to differing extents on Ssn6 for their interaction with Tup1 (Tzamarias and Struhl, 1995; Smith and Johnson, 2000). However, if one assumes that the sequence context of the NRE can influence Nrg1 conformation, then these models can also explain why the Nrg1-mediated repression of some promoters, but not others, depends on Ssn6 (Figures 5 and 7). Furthermore, these models also account for the observation that Ssn6 is not required for the repression of hypha-specific promoters at 30°C (Figures 5 and 6), but is apparently essential at 37°C (Hwang et al., 2003). However, an additional model is required to account for the observation that the repression of some promoters, such as WH11, is dependent on Ssn6, but not Tup1 (Figures 5 and 6). Clearly, Ssn6 can repress some C. albicans genes independently of Tup1 (Figure 8D). It remains to be established whether Ssn6 can act in concert with an alternative corepressor at some C. albicans promoters.
Supplementary Material
Acknowledgments
We are grateful to David Kadosh and Sandy Johnson for communicating data before publication. We also thank Sandy Johnson for helpful discussions and providing strains. We thank Markus Proft for providing strains and Susan Budge for excellent technical assistance. S.G. and S.A. were supported by fellowships from the Basque Government and the Argentinean Antorchas Foundation, respectively. A.M. and B.E. were supported by the European Commission (QLRT-1999-30795; MRTN-CT-2003-504148). P.D. and C.R. were supported by the BBSRC. A.B. is supported by the BBSRC (1/P17124, BBS/B06679) and the Wellcome Trust (063204, 068143).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-01-0071) on April 6, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bailey, D. A., Feldmann, P.J.F., Bovey, M., Gow, N.A.R., and Brown, A.J.P. (1996). The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178, 5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B. R., and Johnson, A. D. (1997). Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277, 105-109. [DOI] [PubMed] [Google Scholar]
- Braun, B. R. and Johnson, A. D. (2000). TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155, 57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B. R., Head, W. S., Wang, M. X., and Johnson, A. D. (2000). Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156, 31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B. R., Kadosh, D., and Johnson, A. D. (2001). NRG1, a TUP1-dependent repressor of filamentous growth in Candida albicans, is down-regulated during filament induction. 20, 4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmuhl, D. P., and Ernst, J. F. (2001). A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157, 1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A.J.P. (2002). Morphogenetic signalling pathways in Candida albicans. In: Candida and Candidiasis, ed. R. Calderone, Washington, DC: ASM Press, 95-106.
- Calderone, R. A. (2002). Candida and Candidiasis, Washington, DC: ASM Press.
- Care, R. S., Trevethick, J., Binley, K. M., and Sudbery, P. E. (1999). The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34, 792-798. [DOI] [PubMed] [Google Scholar]
- Carlson. M. (1997). Genetics of transcriptional regulation in yeast: connections to the RNA polymerase. Annu. Rev. Cell Dev. Biol. 13, 1-23. [DOI] [PubMed] [Google Scholar]
- Conlan, R. S., Gounalaki, N., Hatzis, P., and Tzamarias, D. (1999). The Tup1-Cyc8 protein complex can shift from a transcriptional co-repressor to a transcriptional co-activator. J. Biol. Chem. 274, 205-210. [DOI] [PubMed] [Google Scholar]
- Cormack, B., Bertram, G., Egerton, M., Gow, N.A.R., Falkow, S., and Brown, A.J.P. (1997). Yeast enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143, 303-311. [DOI] [PubMed] [Google Scholar]
- d'Enfert, C. et al.. (2005). CandidaDB: a genome database for Candida albicans pathogenomics. Nucleic Acids Res. 33, D353-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie, J. K., Trumbly, R. J., and Dent, S.Y.R. (2002). Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22, 693-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie, J. K., Edmondson, D. G., Coco, C. B., and Dent, S.Y.R. (2003). Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J. Biol. Chem. 278, 50158-50162. [DOI] [PubMed] [Google Scholar]
- Doedt, T., Krishnamurthy, S., Bockmühl, D. P., Tebarth, B., Stempel, C., Russell, C. L., Brown, A.J.P., and Ernst, J. F. (2004). APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15, 3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson, D. G., Smith, M. M., and Roth, S. Y. (1996). Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10, 1247-1259. [DOI] [PubMed] [Google Scholar]
- Ernst, J. F. (2000). Transcription factors in Candida albicans-environmental control of morphogenesis. Microbiology 146, 1763-1774. [DOI] [PubMed] [Google Scholar]
- Fonzi, W. A., and Irwin, M. Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, S., and Brown, A.J.P. (2001). Characterisation of Candida albicans Ssn6 and its negative role in the regulation of morphogenesis. Yeast 18, S143-S143. [Google Scholar]
- Gillum, A. M., Tsay, E. Y., and Kirsch, D. R. (1984). Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198, 179-182. [DOI] [PubMed] [Google Scholar]
- Gounalaki, N., Tzamarias, D., and Vlassi, M. (2000). Identification of residues in the TPR domain of ssn6 responsible for interaction with the Tup1 protein. FEBS Lett. 473, 37-41. [DOI] [PubMed] [Google Scholar]
- Grbavec, D., Lo, R., Liu, Y., Greenfield, A., and Stifani, S. (1999). Groucho/transducin-like Enhancer of split (TLE) family members interact with the yeast transcriptional co-repressor SSN6 and mammalian SSN6-related proteins: implications for evolutionary conservation of transcription repression mechanisms. Biochem. J. 337, 13-17. [PMC free article] [PubMed] [Google Scholar]
- Green, S. R., and Johnson, A. D. (2004). Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 4191-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromoller, A., and Lehming, N. (2000). Srb7 is a physical and physiological target of Tup1p. EMBO J. 19, 6845-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, N. C., Vingron, M., Scheideler, M., Krems, B., Hellmuth, K., Entian, K.-D., and Hoheisel, J. D. (1998). Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast 14, 1209-1221. [DOI] [PubMed] [Google Scholar]
- Huang, M., Zhou, Z., and Elledge, S. J. (1998). The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94, 595-605. [DOI] [PubMed] [Google Scholar]
- Hughes, T. R. et al. (2000). Functional discovery via a compendium of expression profiles. Cell 102, 109-126. [DOI] [PubMed] [Google Scholar]
- Hwang, C.-S., Oh, J.-H., Huh, W.-K., Yim, H.-S., and Kang, S.-O. (2003). Ssn6, an important factor of morphological conversion and virulence in Candida albicans. Mol. Microbiol. 47, 1029-1043. [DOI] [PubMed] [Google Scholar]
- Kadosh, D., and Johnson, A. D. (2005). Induction of the Candida albicans filamentous growth programme by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16, 2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C., Michaelis, S., and Mitchell, A. (1994). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Keleher, C. A., Redd, M. J., Schultz, J., Carlson, M., and Johnson, A. D. (1992). Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68, 709-719. [DOI] [PubMed] [Google Scholar]
- Komanchi, K., Redd, M. J., and Johnson, A. D. (1994). The WD repeats of Tup1 interact with the homeo domain protein α2. Genes Dev. 8, 2857-2867. [DOI] [PubMed] [Google Scholar]
- Kvaal, C. A., Srikantha, T., and Soll, D. R. (1997). Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect. Immun. 65, 4468-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, C. Y., Newport, G., Murillo, L. A., Jones, T., Scherer, S., Davis, R. W., and Agabian, N. (2002). Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 99, 14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Kohler, J. R., and Fink, G. R. (1994). Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266, 1723-1726. [DOI] [PubMed] [Google Scholar]
- Lo, H. J., Kohler, J. R., DiDomenico, B., Loebenberg, D., Cacciapuoti, A., and Fink, G. R. (1997). Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939-949. [DOI] [PubMed] [Google Scholar]
- Loubradou, G., Brachmann, A., Feldbrugge, M., and Kahmann, R. (2001). A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signaling in Ustilago maydis. Mol. Microbiol. 40, 719-730. [DOI] [PubMed] [Google Scholar]
- Murad, A.M.A., Lee, P. R., Broadbent, I. D., Barelle, C. J., and Brown, A.J.P. (2000). CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16, 325-327. [DOI] [PubMed] [Google Scholar]
- Murad, A.M.A. et al.. (2001a). NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20, 4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad, A.M.A., d'Enfert, C., Gaillardin, C., Tournu, H., Tekaia, F., Talibi, D., Marechal, D., Marchais, V., Cottin, J., and Brown, A.J.P. (2001b). Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors, CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42, 981-993. [DOI] [PubMed] [Google Scholar]
- Odds, F. C. (1988). Candida and Candidosis, 2nd ed. London: Bailliere Tindall.
- Ostling, J., and Ronne, H. (1998). Negative control of the Mig1 repressor by Snf1-dependent phosphorylation in the absence of glucose. Eur. J. Biochem. 252, 162-168. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis, M., Conlan, R. S., Gounalaki, N., Copf, T., and Tzamarias, D. (2000). Hrs1/Med3 is a Cyc8-Tup1 corepressor target in the RNA polymerase II holoenzyme. J. Biol. Chem. 275, 8397-8403. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis, M., Gligoris, T., and Tzamarias, D. (2004). The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 5, 368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd, M. J., Arnaud, M. B., and Johnson, A. D. (1997). A complex composed of Tup1 and Ssn6 represses transcription in vitro. J. Biol. Chem. 272, 11193-11197. [DOI] [PubMed] [Google Scholar]
- Rupp, S. (2002). LacZ assays in yeast. Methods Enzymol. 350, 112-131. [DOI] [PubMed] [Google Scholar]
- Russell, C. L., and Brown, A.J.P. (2005). Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gen4 is a transcriptional activator. Fungal Genet. Biol. (in press). [DOI] [PubMed]
- Schaller, M., Schafer, W., Korting, H. C. and Hube, B. (1998). Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol. Microbiol. 29, 605-615. [DOI] [PubMed] [Google Scholar]
- Sharkey, L. L., McNemar, M. D., Saporito-Irwin, S. M., Sypherd, P. S., and Fonzi, W. A. (1999). HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1 and RBF1. J. Bacteriol. 181, 5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- Slutsky, B., Buffo, J., and Soll, D. R. (1985). High frequency “switching” of colony morphology in Candida albicans. Science 230, 666-669. [DOI] [PubMed] [Google Scholar]
- Smith, R. L., and Johnson, A. D. (2000). Turning off genes by Ssn6-Tup1, a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25, 325-330. [DOI] [PubMed] [Google Scholar]
- Soll, D. R. (2002). Phenotypic switching. In: Candida and Candidiasis, ed. R. Calderone, ed. Washington, DC: ASM Press, 123-142.
- Strahl-Bolsinger, S., Hecht, A., Luo, K., and Grunstein, M. (1997). SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11, 83-93. [DOI] [PubMed] [Google Scholar]
- Sudbery, P. E., Gow, N.A.R., and Berman, J. (2004). The distinct morphological states of Candida albicans. Trends Microbiol. 12, 317-324. [DOI] [PubMed] [Google Scholar]
- Tripathi, G., Wiltshire, C., Macaskill, S., Tournu, H., Budge, S., and Brown, A.J.P. (2002). CaGcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21, 5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V. G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias, D., and Struhl, K. (1994). Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature 369, 758-761. [DOI] [PubMed] [Google Scholar]
- Tzamarias, D., and Struhl, K. (1995). Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 co-repressor complex to differentially regulated promoters. Genes Dev. 9, 821-831. [DOI] [PubMed] [Google Scholar]
- Uhl, M. A., and Johnson, A. D. (2001). Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology 147, 1189-1195. [DOI] [PubMed] [Google Scholar]
- Williams, F. E., Varanasi, U., and Trumbly, R. J. (1991). The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol. Cell. Biol. 11, 3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Z., Stead, D., Selway, L., Walker, J., Riba-Garcia, I., Mclnerney, T., Gaskell, S., Oliver, S. G., Cash, P., and Brown, A.J.P. (2004). Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics 4, 2425-2436. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and Reese, J. C. (2004a). Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J. 23, 2246-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., and Reese, J. C. (2004b). Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279, 39240-39250. [DOI] [PubMed] [Google Scholar]
- Zheng, X., Wang, Y., and Wang, Y. (2004). Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23, 1845-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R., Lockhart, S. R., Daniels, K., and Soll, D. R. (2002). Roles of TUP1 in switching, phase maintenance, and phase-specific gene expression in Candida albicans. Eukar. Cell 1, 353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Oh, S. H., Cheng, G., Green, C. B., Nuessen, J. A., Yeater, K., Leng, R. P., Brown, A.J.P., and Hoyer, L. L. (2004). ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150, 2415-2428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.