Abstract
A key factor involved in the processing of histone pre-mRNAs in the nucleus and translation of mature histone mRNAs in the cytoplasm is the stem–loop binding protein (SLBP). In this work, we have investigated SLBP nuclear transport and subcellular localization during the cell cycle. SLBP is predominantly nuclear under steady-state conditions and localizes to the cytoplasm during S phase when histone mRNAs accumulate. Consistently, SLBP mutants that are defective in histone mRNA binding remain nuclear. As assayed in heterokaryons, export of SLBP from the nucleus is dependent on histone mRNA binding, demonstrating that SLBP on its own does not possess any nuclear export signals. We find that SLBP interacts with the import receptors Impα/Impβ and Transportin-SR2. Moreover, complexes formed between SLBP and the two import receptors are disrupted by RanGTP. We have further shown that SLBP is imported by both receptors in vitro. Three sequences in SLBP required for Impα/Impβ binding were identified. Simultaneous mutation of all three sequences was necessary to abolish SLBP nuclear localization in vivo. In contrast, we were unable to identify an in vivo role for Transportin-SR2 in SLBP nuclear localization. Thus, only the Impα/Impβ pathway contributes to SLBP nuclear import in HeLa cells.
INTRODUCTION
The stem–loop binding protein (SLBP) is an evolutionarily conserved RNA binding protein involved in the metabolism of metazoan replication-dependent histone mRNAs (Wang et al., 1996; Martin et al., 1997; Marzluff and Duronio, 2002). Although the majority of mRNA 3′ ends result from a cleavage and polyadenylation reaction, those of replication-dependent histone mRNAs are formed by a unique mechanism characterized by endonucleolytic cleavage after a conserved stem–loop sequence (Marzluff and Duronio, 2002). SLBP is involved in the 3′ end cleavage reaction in the nucleus (Dominski and Marzluff, 1999) and in histone mRNA translation in the cytoplasm (Gallie et al., 1996; Sanchez and Marzluff, 2002).
SLBP is a cell cycle-regulated protein, which accumulates only during S phase, correlating with the production of histones during times of DNA synthesis (Whitfield et al., 2000). In late G1 phase, SLBP levels increase through activating the translation of SLBP mRNA (Whitfield et al., 2000). Subsequently, as cells approach G2 phase, SLBP levels are reduced by proteasome-mediated degradation (Whitfield et al., 2000; Zheng et al., 2003). Replication-dependent histone mRNAs similarly accumulate during S phase and are rapidly degraded upon completion of DNA synthesis (Marzluff and Duronio, 2002).
Many proteins and protein/RNA complexes, which are synthesized or assembled in the cytoplasm, have essential functions in the nucleus. Consequently, active transport mechanisms are in place to ensure their nuclear localization. Transport of cargoes between the nuclear and cytoplasmic compartments occurs through pores in the nuclear envelope termed nuclear pore complexes (NPCs) and is carried out by soluble transport receptors or karyopherins (Gorlich and Kutay, 1999). The majority of known transport receptors belong to a family of evolutionarily conserved proteins called Importin β-like transport receptors (Fried and Kutay, 2003; Mosammaparast and Pemberton, 2004). The similarities shared by these proteins include a Ran binding domain (see below) and the ability to bind NPCs.
The process of nuclear transport involves the small GTPase Ran, which is differentially bound to GTP and GDP in the nucleus and cytoplasm. Import receptors associate with their nuclear transport substrates in the cytoplasm (Fried and Kutay, 2003; Mosammaparast and Pemberton, 2004). After translocation to the nuclear side of the NPC, nuclear RanGTP binds to the receptor in the import complex, causing release of the cargo. Thus, import receptor–substrate interactions are favored in the cytoplasm where RanGTP levels are low.
A number of specific sequence motifs that confer binding to import receptors have been identified. Examples include the classical basic nuclear localization sequence (NLS) (Dingwall et al., 1982; Kalderon et al., 1984), the M9 domain (Siomi and Dreyfuss, 1995), and the phosphorylated arginine-serine-rich (RS) domain, which are recognized by the heterodimeric import receptor Importinα/Importinβ (Impα/Impβ) (Gorlich, 1997), Transportin (Pollard et al., 1996), and Transportin-SR2 (Lai et al., 2001), respectively. These signals are highly specific for their cognate importins, whereas other signals exist that can be recognized by multiple receptors, including receptors such as Transportin and Impβ. The best-defined signal of this second category is the β-like transport receptor binding domain of ribosomal protein L23a, a very basic region to which different importins can bind. Multiple import pathways exist not only for ribosomal proteins (Jakel and Gorlich, 1998; Muhlhausser et al., 2001) but also for core histones as shown in both mammalian cells and yeast (Rout et al., 1997; Schlenstedt et al., 1997; Mosammaparast et al., 2001; Muhlhausser et al., 2001).
The simplest mode of nuclear import, characteristic of most import receptors, involves the direct association of an import receptor with its cargo. However, there are examples of receptors that use adapter proteins to mediate substrate binding. Nuclear import of NLS-containing substrates by the Impα/Impβ heterodimer involves association of a cargo with the adapter Impα (Adam and Gerace, 1991; Gorlich et al., 1994; Moroianu et al., 1995a) and NPC binding by Impβ (Adam and Gerace, 1991; Chi et al., 1995; Moroianu et al., 1995a,b). In the nucleus, RanGTP binds to Impβ, causing the displacement of Impα from Impβ and the subsequent release of the NLS cargo from Impα (Rexach and Blobel, 1995; Gorlich et al., 1996). A feature specific for Impα/Impβ-mediated import (Ribbeck et al., 1999) is that RanGTP binding of Impβ also induces the dissociation of Impβ from nucleoporins (Rexach and Blobel, 1995; Gorlich et al., 1996). After import complex disassembly in the nucleus, Impα is exported to the cytoplasm by the export receptor CAS (Gorlich et al., 1997; Kutay et al., 1997a), and Impβ migrates back to the cytoplasm in an export receptor-independent manner (Gorlich et al., 1997; Izaurralde et al., 1997).
In this study, we have identified the nuclear import receptors that mediate SLBP import in vitro and have mapped the sequences in SLBP essential for its nuclear localization in vivo. We have also examined the subcellular localization of SLBP during the cell cycle.
MATERIALS AND METHODS
Molecular Cloning
Hemagglutinin (HA)-SLBP has been described previously (Zheng et al., 2003). The HA-SLBPSFTTP construct was generated by cloning the NcoI/XbaI fragment from His6-pcDNA3/SLBPSFTTP (Zheng et al., 2003) into HA-pcDNA3. HA-SLBPRR was generated by standard site-directed mutagenesis techniques. SLBP deletion and point mutants used for in vitro translation were constructed in the pXFRM/SLBP plasmid (Wang et al., 1999). pXFRM contains the SP6 promoter, and the 5′ and 3′ untranslated regions (UTRs) of β-globin as well as a 50-nt polyA stretch downstream of the β-globin 3′ UTR. Deletion and site-directed mutagenesis were performed using standard PCR and cloning methods.
The baculovirus His6-gluatathione S-transferase (GST)-SLBP expression plasmid (HTb/GST-SLBP) was made by amplifying the GST-SLBP sequence in pQE-60/GST-SLBP and cloning it into the KpnI and XbaI sites of pFastBac HTb.
The GFP-SLBP mammalian expression vector (GFP-SLBP) was constructed by insertion of the SLBP open reading frame into the BamHI and EcoRI sites of the green fluorescent protein (GFP) plasmid (pK7) kindly provided by I. Macara (Department of Cell Biology, University of Virginia School of Medicine, Charlottesville, VA) (McKiernan et al., 1996). GST-GFP-SLBP was made by cloning GST from pGEX-2T into the XbaI site upstream of GFP in GFP-SLBP. Site-directed mutagenesis and the combining of mutations were carried out using standard PCR and cloning methods. Annealed oligos containing the simian virus 40 (SV40) NLS (PKKKRKVED) or sequences required for SLBP nuclear localization (KRKL [RYKRKLLIN], RKRR [LGRKRRADG], and KVRH [PTKVRHMDS]) were cloned into the BamHI and EcoRI sites downstream of GFP in the GST-GFP vector to yield GST-GFPSV40, GST-GFPKRKL, GST-GFPRKRR and GST-GFPKVRH.
The coding region of human Transportin-SR2 was amplified by PCR by using HeLa cell cDNA as a template. The PCR fragment was cloned into the BamHI/XmaI sites of pQE30 to yield pQE30-TrnSR-2.
RNA Interference and Plasmid Transfections
HeLa cells were seeded at a density of 8.5 × 104 cells/ml in a total volume of 500 μl (DMEM supplemented with 10% fetal bovine serum) in a single well of a 24-well plate (Greiner Bio-One, Frickenhausen, Germany). The following day, the cells were transfected with small interfering RNA (siRNA) at a final concentration of 100 nM. The siRNA transfection protocol was performed essentially as described previously (Wagner and Garcia-Blanco, 2002). Briefly, 3 μl of LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) was added to 12 μl of Opti-MEM (Invitrogen) and allowed to incubate at room temperature for 7 min. After incubation, the mixture was added to a solution containing 47 μl of Opti-MEM and 3 μl of the 20 μM siRNA stock. The reaction was further incubated for 25 min at room temperature. Subsequently, 38 μl of Opti-MEM was added to the reaction, and 100 μl of the transfection mixture was pipetted onto the cells. The following day, the cells were split 1:2 from a 24-well plate to a six-well plate containing coverslips. On the next day, the cells were retransfected with siRNA in an identical manner as described above, except that the final concentration of siRNA was 50 nM. Two days later, the cells were either harvested and lysed for Western blot analysis or fixed for immunostaining. The siRNA sequence used to target SLBP is 5′-GAGAGAGAAAAUCAUCAUC-3′, whereas the C2 siRNA has been described previously (Wagner and Garcia-Blanco, 2002).
Cell lines expressing HA-SLBPwt, HA-SLBPRR, and HA-SLBPSFTTP were produced by transfecting HeLa cells (in a six-well plate) with 1 μg of DNA (pcDNA3/HA-SLBPwt, pcDNA3/HA-SLBPRR, and pcDNA3/HA-SLBPSFTTP) by using the LipofectAMINE reagent (Invitrogen). One day after transfection, the cells were split into media containing 0.5 mg/ml G418 (Geneticin, Invitrogen). The cells were kept under selection until separate colonies could be observed by eye and were either pooled (HA-SLBPRR) or used to make clonal cell lines (HA-SLBPwt and HA-SLBPSFTTP) by using the limiting dilution method.
For the in vivo localization studies using either GFP- or GST-GFP–tagged SLBPs, 2 × 105 HeLa cells/well were plated onto coverslips in a six-well plate 1 d before transfection. The following day, cells were transfected with 100 ng of plasmid DNA by using the LipofectAMINE 2000 reagent (Invitrogen) according to the manufacturer's protocol.
Protein Purification and Labeling
Baculovirus expressed His6-GST-SLBP was purified by nickel-nitrilotriacetic acid (NTA) chromatography by using standard procedures. Reducing agents were excluded from all of the buffers used during the purification procedure. Proteins were labeled on cysteines with an equimolar amount of Alexa 488 (Molecular Probes, Eugene, OR) for 1 h on ice. Unincorporated Alexa 488 was removed by passage over a MicroSpin G-50 column (Amersham Biosciences, Piscataway, NJ) equilibrated in 50 mM Tris, pH 7.5, 200 mM potassium acetate, and 2 mM magnesium chloride. Escherichia coli expression and purification of RanQ69L, Impα, Impβ, Transportin, Imp5, and Imp7 were performed as described previously (Gorlich et al., 1996; Kutay et al., 1997b; Jakel and Gorlich, 1998). Trn-SR2 was expressed in E. coli XL1 (pBS161) at 22°C. Cells were lysed by sonication in 50 mM Tris-HCl, pH 7.5, 700 mM NaCl, 10 mM MgCl2, 5% glycerol, and 5 mM β-mercaptoethanol. Purification was performed by chromatography on nickel-NTA agarose, followed by MonoQ and gel filtration in 50 mM Tris-HCl, pH 7.5, 100 mM potassium acetate, and 2 mM dithiothreitol. Proteins tagged with two copies of the IgG binding domain or “z” domain of Protein A (termed “zz”) were prepared in a manner similar to their His-tagged counterparts.
In Vitro Binding Assays
[35S]Methionine-labeled SLBPs were produced by in vitro translation using the coupled transcription translation kit by Promega. Binding assays using purified recombinant zz-Trn-SR2 or zz-Impα/Impβ and [35S]SLBP were carried out as follows: zz-Impα and Impβ (70 pmol each) were first bound to one another for 20 min on ice. Depending on the relative amounts of each of the labeled proteins, between 10 and 12 μl of each in vitro translation reaction was mixed with 70 pmol of zz-Trn-SR2 or zz-Impα/Impβ heterodimers. The reactions were incubated for 30 min on ice and were subsequently diluted to 100 μl to achieve a final concentration of 225 mM NaCl, 20 mM HEPES, pH 7.0, 0.001% Triton X-100, and 2 mM Mg(OAc)2 (binding buffer). When included in the reaction, the final concentration of RanQ69L was 10 μM. Control reactions contained the [35S]SLBP and binding buffer. The binding reactions were then cleared of any aggregated material by centrifugation, and the supernatant was transferred to a fresh tube containing 10 μl of packed IgG-Sepharose beads (Amersham Biosciences) preequilibrated in the binding buffer. After a 30-min incubation, the beads were washed four times with the binding buffer and eluted with 50 μlof2× sample buffer. One-fifth of the eluted material was run on a 10% SDS-PAGE alongside 1/25 of the input. The gel was stained with Coomassie Blue to determine whether the same amount of receptor in each binding reaction was recovered. Subsequently, the gel was dried and exposed to a PhosphorImager screen.
In reactions using purified SLBP, 70 pmol of purified import receptor was incubated with 150 pmol of GST-SLBP in a final volume of 100 μl. The conditions for the binding of complexes to IgG-Sepharose beads and complex recovery were the same as for the binding assay using in vitro-translated SLBP.
In Vitro Import Assays
In vitro transport reactions into the nuclei of digitonin semipermeabilized cells were performed essentially as described previously (Jakel et al., 1999). Fluorescently labeled GST-SLBP (1 μM) was preincubated for 5 min on ice with equimolar amounts of purified import receptors. The import buffer contained 20 mM HEPES, pH 7.5, 100 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM EGTA, and 250 mM sucrose and was supplemented with an energy-regenerating system and Ran mix (Kutay et al., 1997b). Import was performed for 15 min at room temperature. The samples were then washed once with import buffer, fixed with 3.7% paraformaldehyde (PFA), and visualized by fluorescence microscopy.
Cell Synchronization and Drug Treatment
One hundred thousand cells per well of a six-well plate were seeded onto coverslips. On the next day, thymidine was added to the media at a final concentration of 2 mM. After 19 h, the cells were washed twice with 1× phosphate-buffered saline (PBS) and supplemented with media lacking thymidine. The cells were incubated for 9 h in the absence of thymidine and subsequently exposed to a second treatment with thymidine for 16 h. At this time, the cells were washed twice with 1× PBS and supplemented with fresh media lacking thymidine. Coverslips were fixed and cells were harvested every 2 h after release from the second thymidine block. Cells exposed to the proteasome inhibitor MG132 (50 μM) were treated 4 h after release from the second thymidine block and harvested 6 h later.
Lysate Preparation and Western Blotting
Cells were lysed in an NP-40 lysis buffer (0.1% NP-40, 20 mM Tris, pH 7.0, 150 mM NaCl, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride) for 10 min at 4°C. Lysates were cleared by centrifugation at 4°C for 10 min at maximum speed. Unless otherwise indicated in the figure legend, 40 μg of total protein was resolved on a 10% SDS-PAGE, transferred to nitrocellulose, and probed with the anti-SLBP or anti-GFP antibody (BD Biosciences Clontech, Palo Alto, CA) at a dilution of 1:1000. Detection was carried out using a horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA; diluted 1:10,000) and an enhanced chemiluminescence system (Amersham Biosciences).
Immunostaining and GFP Fusion Protein Localization
HeLa cells grown on coverslips were washed briefly with 1× PBS and fixed with 3.7% PFA. After permeabilization with 0.5% Triton X-100 and washing with 1× PBS, the coverslips were incubated for 1 h with either the anti-SLBP or anti-HA antibody diluted 1:100 in 1× PBS and 1% bovine serum albumin. The monoclonal anti-HA antibody was a kind gift of Dr. Yue Xiong (Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill, Chapel Hill, NC). Detection was carried out using either a rhodamine anti-rabbit (Jackson ImmunoResearch Laboratories) or Cy3-conjugated anti-mouse antibody (Jackson ImmunoResearch Laboratories). Cells were subsequently postfixed with 3.7% PFA and counterstained with 0.1 μg/ml 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO). Images were taken using a Zeiss confocal microscope and the LSM 510 software.
The localization of the GFP and GST-GFP–tagged proteins was assessed 2 d after transfection. Coverslips were washed briefly with 1× PBS, and the cells were fixed with 3.7% PFA. The cells were washed with PBS and permeabilized with 0.5% Triton X-100. Counterstaining with DAPI was carried out as described above.
Heterokaryon Assay
Heterokaryon assays were carried out essentially as described previously (Zhang and Xiong, 2001). Human U2-OS cells were transfected with plasmid DNA expressing the GFP-SLBP fusion proteins. After 12 h of incubation with the plasmid, cells were washed twice with prewarmed PBS to remove untransfected DNA. The cells were then trypsinized, mixed with wild-type mouse embryonic fibroblast (MEF) cells at a 1:1 ratio, and seeded onto a new dish. After another 12 h of incubation, the cells were rinsed once with PBS and covered with a solution of 50% (wt/vol) polyethylene glycol (PEG8000; Sigma-Aldrich) for 2 min at 37°C to induce cell fusion. After washing three times with PBS to remove the PEG, fused cells were continuously cultured in DMEM/10% fetal bovine serum media for 90 min. Cycloheximide (50 μg/ml; Sigma-Aldrich) was added to the culture 15 min before and immediately after cell fusion to block de novo protein synthesis. Subsequent cell fixation, permeabilization, and immunostaining followed standard immunofluorescence procedures. The cells were stained with a mouse antibody recognizing human but not murine Ku nuclear antigen to distinguish human and murine nuclei.
RESULTS
Localization of SLBP during the Cell Cycle
Previous cell fractionation studies have indicated that SLBP is present in both the nuclear and cytoplasmic fractions (Hanson et al., 1996; Whitfield et al., 2004), consistent with its involvement in histone pre-mRNA processing in the nucleus and histone mRNA translation in the cytoplasm. To directly assess the localization of SLBP in intact cells, immunostaining was carried out on asynchronously growing HeLa cells using a polyclonal anti-SLBP antibody raised against the C-terminal 13 amino acids of the protein (Wang et al., 1996). SLBP was found to be predominantly nuclear, with some variability in the ratio of nuclear-to-cytoplasmic staining from cell to cell (Figure 1A, left). Consistent with the cell cycle-regulated expression of SLBP, the intensity of the signal generated by the SLBP antibody was not homogeneous in an asynchronous population of cells.
Figure 1.
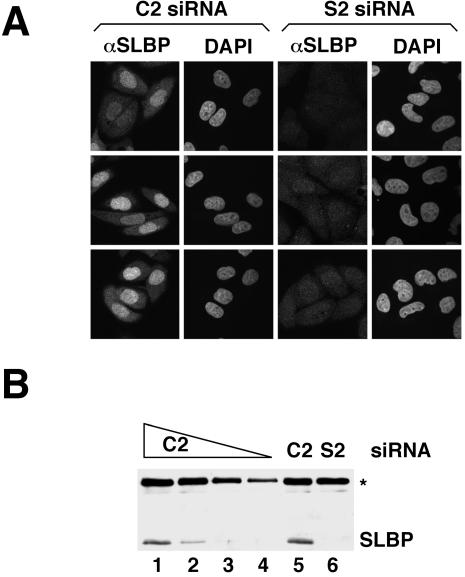
Specificity of the anti-SLBP antibody for SLBP in intact HeLa cells as assayed by immunofluorescence. (A) HeLa cells transfected with either the C2 control siRNA (left) or the S2 siRNA specific for SLBP (right) were subject to immunostaining with the SLBP antibody. (B) To evaluate the knockdown resulting from the S2 siRNA, Western blot analysis with the SLBP antibody was performed using lysates made from C2 (lanes 1–5) or SLBP (lane 6) siRNA-transfected cells. In lanes 1, 5, and 6, 10 μg of total protein was loaded, whereas in lanes 2, 3, and 4, 5, 2, and 1 μg of total protein was used, respectively. The asterisk marks the migration of the 75-kDa protein that cross-reacts with the SLBP antibody and serves as an internal loading control.
Western blotting of total HeLa cell extract resolved in denaturing SDS polyacrylamide gels shows that the SLBP antibody cross-reacts with two other proteins (75 and 90 kDa apparent molecular mass) in addition to SLBP (Figure 5B). To determine the specificity of the immunofluorescence signal generated with the SLBP antibody, we reduced the levels of SLBP by using RNA interference and performed immunostaining. Western analysis revealed that SLBP levels were reduced by at least 90% by an siRNA targeting the SLBP mRNA and not by a control siRNA (Figure 1B). The majority of the cells transfected with the SLBP siRNA exhibited only background staining with the SLBP antibody (Figure 1A, right). Thus, we conclude that, in intact HeLa cells assayed by immunofluorescence, the SLBP antibody recognizes only SLBP and that the cross-reacting proteins are only detected in Western blots.
Figure 5.
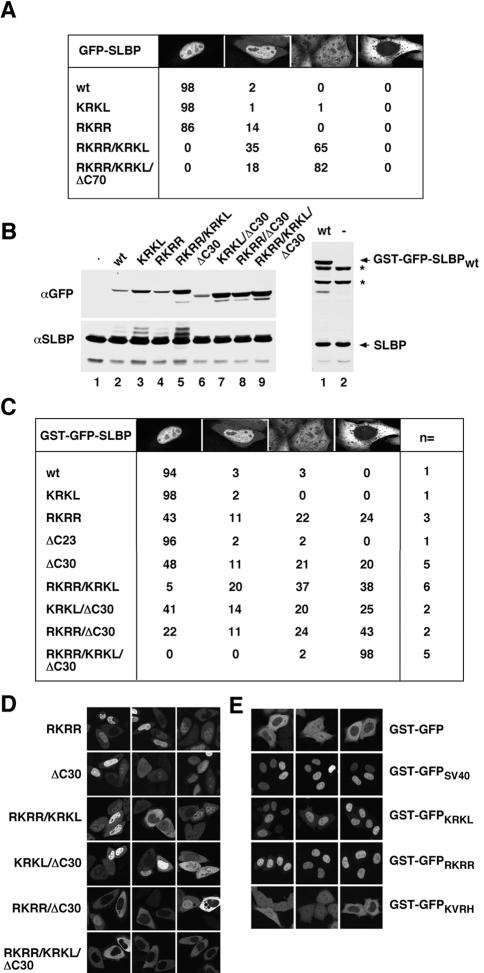
In vivo localization of SLBP mutants impaired for Impα binding. (A) Images of cells transiently transfected with the indicated GFP-SLBP construct were collected and at least 100 cells per construct were examined. Cells were scored for fusion protein localization and grouped into four categories: 1) exclusive nuclear localization; 2) greater nuclear than cytoplasmic localization; 3) even distribution between the nucleus and cytoplasm; or 4) exclusive cytoplasmic localization. (B) Western blot analysis with an anti-GFP antibody and anti-SLBP antibody, demonstrating the relative expression levels of the different GST-GFP–tagged SLBPs (top), and the levels of the endogenous SLBP as a loading control (bottom). The C-terminal GST-GFP-SLBP truncation mutants do not contain the epitope recognized by the anti-SLBP antibody necessitating their detection with the anti-GFP antibody (left, top). The expression of GST-GFP-SLBPwt (lane 1) relative to the endogenous SLBP is shown on the right. The asterisks mark the 75- and 90-kDa proteins that cross-react with the SLBP antibody. (C) Images of cells transiently transfected with the indicated GST-GFP-SLBP construct were collected and at least 300 cells per construct were examined. The number of independent transfections is indicated in the column labeled n=. Cells were scored as described in A. (D) Images of representative localization phenotypes for the indicated GST-GFP–tagged SLBP NLS mutants. (E) The predicted Imp α/β NLSs were fused to the C terminus of GST-GFP and examined for NLS activity by transient transfection. The construct transfected is indicated on the right side of each row of panels.
Because the staining pattern for SLBP in asynchronously growing cells (Figures 1A, left, and 2A, top left) was somewhat heterogeneous, we tested whether the variability could be explained by differences in cell cycle position. HeLa cells were synchronized using a double thymidine block protocol, which arrests cells at the beginning of S phase. These cells have accumulated SLBP, which is not present during G2, mitosis, or early G1 (Whitfield et al., 2000). After release from the double thymidine block, the cells progress synchronously through S phase and into G2 phase. Samples were collected at two-hour intervals after release of the cells from the second thymidine block. The cell cycle expression profile of the endogenous SLBP was analyzed by Western blotting by using the SLBP antibody (Figure 2B). Before and 2 h after release from the double thymidine block, SLBP was predominantly nuclear, with very little present in the cytoplasm (Figure 2A, 0 and 2 h). However, as cells progressed through S phase, there was an increase in the proportion of SLBP in the cytoplasm (Figure 2A, 4 and 6 h), indicating that during S phase, when histone mRNA levels are high, SLBP partially redistributes to the cytoplasm. A similar pattern of nucleocytoplasmic distribution of SLBP was observed in the following S phase (9 h later; our unpublished data), indicating that the staining pattern shown in Figure 2A accurately reflects the localization of SLBP during S phase. By 10 h after release from the double-thymidine block, the staining of SLBP is greatly reduced (Figure 2A, 10 h) as a result of the regulated degradation of SLBP 8 h after release (Figure 2B).
Figure 2.
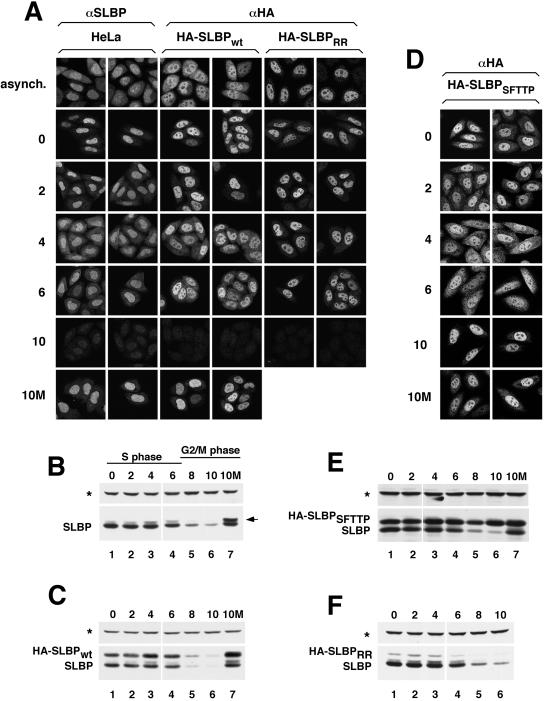
Subcellular distribution of SLBP changes during S phase and is dependent on RNA binding. (A) Untransfected HeLa cells (left two columns), a clonal cell line expressing HA-SLBPwt (center two columns) or a pool of stably transfected cells expressing HA-SLBPRR (right two columns) were synchronized by double-thymidine block. After release from the second thymidine block, the localization of the endogenous SLBP (left two columns) or HA-tagged SLBPs (remaining four columns) was monitored every 2 h by immunostaining with either the anti-SLBP or anti-HA antibody. The top row shows the localization of the various SLBPs in an asynchronous population of cells and the remaining rows show cells at different times (hours) after release from double thymidine block. The bottom row shows the localization of the endogenous SLBP and HA-SLBPwt in cells exposed to MG132 at 4 h (S phase) and harvested 10 h (G2 phase) after release (10M). (B and C) Western blot by using the anti-SLBP antibody to illustrate the expression profile of the endogenous SLBP (B) and HA-SLBPwt (C) in the same cells as those shown in A. The asterisk marks the 75-kDa protein that cross-reacts with the SLBP antibody on Western blots. The arrow indicates the phosphorylated SLBP, which accumulates when protein degradation is blocked at the end of S phase. (D) A clonal cell line expressing HA-SLBPSFTTP was synchronized, and the localization of HA-SLBPSFTTP was determined using the anti-HA antibody as in A. (E and F) Lysates prepared from HA-SLBPSFTTP (E) or HA-SLBPRR (panel F) expressing cells (at the indicated times postrelease from double-thymidine block) were tested for SLBP levels by Western blot analysis by using the SLBP antibody.
Nondegradable SLBP Is Nuclear in G2 Phase of the Cell Cycle
SLBP is degraded by the proteasome at the end of S phase in a phosphorylation-dependent manner (Whitfield et al., 2000; Zheng et al., 2003). Cells treated in late S phase with the proteasome inhibitor MG132 accumulate phosphorylated SLBP. To further characterize the location of SLBP present in G2-phase cells, we performed immunostaining on cells in G2 phase that had been exposed to MG132 during S phase. When cells were released from a double-thymidine block for 4 h and treated with MG132 for 6 h, we found that the majority of SLBP was nuclear (Figure 2A, 10M). This result is in contrast to our previous observations obtained by cellular fractionation, where the phosphorylated SLBP that accumulates after MG132 treatment was exclusively cytoplasmic (Zheng et al., 2003). It is likely that this difference results from leakage of phosphorylated SLBP from the nucleus during cell fractionation (see Discussion).
To rule out the possibility that our results might simply reflect that the SLBP antibody does not efficiently react with SLBP in the cytoplasm, possibly due to inaccessibility of the C-terminal end of the protein in this compartment, we examined the localization of an N-terminally HA-tagged SLBP (HA-SLBPwt), by using an anti-HA antibody. It has been previously shown that HA-SLBPwt is expressed at levels comparable with the endogenous SLBP and its expression profile mirrors that of the endogenous protein (Zheng et al., 2003; Figure 2C). A clonal HA-SLBPwt-expressing cell line stained with an HA antibody revealed that, like the endogenous SLBP, the HA-SLBPwt was predominantly nuclear in unsynchronized cells (Figure 2A, asynch.). In addition, HA-SLBPwt redistributed to the cytoplasm to a similar extent as the endogenous protein in mid- to late S phase and was predominantly nuclear in MG132-treated G2-phase cells (Figure 2A, 4 h, 6 h, and 10M). Thus, these results confirm the staining pattern observed using the SLBP antibody.
Because MG132 is a general inhibitor of the proteasome and likely perturbs many aspects of cellular regulation, we also used a mutant SLBP that is not degraded at the end of S phase to study the localization of SLBP in G2 phase. A critical sequence in SLBP responsible for its degradation at the end of S phase has been mapped to amino acids 59–63, containing the residues SFTTP. The two threonines in this sequence are phosphorylated and trigger the degradation of SLBP (Zheng et al., 2003). If this sequence is mutated to alanine, SLBP is stable at the S/G2 transition, and persists during G2 phase. HeLa cells stably expressing an HA-tagged SLBP construct containing the SFTTP/AAAAA mutation (HA-SLBPSFTTP) were synchronized (Figure 2E) and the localization of HA-SLBPSFTTP was observed with an anti-HA antibody (Figure 2D). HA-SLBPSFTTP accumulated in the nucleus of G2-phase cells (Figure 2D, 10 h), further demonstrating that SLBP present in G2 phase accumulates in the nucleus.
The RNA Binding Activity of SLBP Is Required for Cytoplasmic Localization
The data presented so far are consistent with the presence of SLBP in the cytoplasm being dependent on its association with histone mRNA. In agreement with this possibility, treatment of cells with hydroxyurea, which results in rapid degradation of histone mRNA, results in the relocalization of SLBP to the nucleus (Whitfield et al., 2004). To address this issue directly, we made a two-amino acid substitution in SLBP (R137K, R138K [HA-SLBPRR]), which is known to severely interfere with its RNA binding activity (Dominski et al., 2001), and subsequently examined the localization of HA-SLBPRR during S phase. A pool of cells stably transfected with HA-SLBPRR was synchronized (Figure 2F), and the localization of the protein was examined (Figure 2A). The HA-SLBPRR remained in the nucleus throughout the cell cycle (Figure 2A) and was degraded at the end of S phase (Figure 2F). These results thus demonstrate that the accumulation of SLBP in the cytoplasm requires that SLBP be able to bind to histone mRNA.
SLBP Is Imported by Transportin-SR2 and Impα/Impβ In Vitro
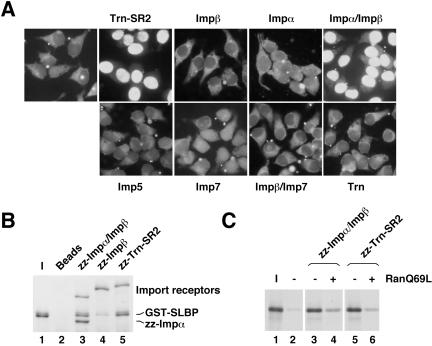
To identify potential SLBP import receptors, we used an in vitro nuclear import assay with semipermeabilized HeLa cells. A His6-tagged SLBP molecule (33 kDa) diffused into the nucleus of permeabilized HeLa cells without addition of any transport factors (our unpublished data). Therefore, a GST-tagged SLBP was constructed and expressed in insect cells. Incubation of permeabilized HeLa cells with purified, fluorescently labeled GST-SLBP and an energy-regenerating system did not allow for its nuclear import (Figure 3A). However, if the import reaction was supplemented with Trn-SR2 or Impα/Impβ, SLBP accumulated in the nucleus (Figure 3A). Significantly, Impβ on its own and four other import receptors were unable to support the accumulation of SLBP in the nucleus. All of these receptors were active in the import of their known cognate cargos (our unpublished data). These results therefore demonstrate that Trn-SR2 and Impα/Impβ can import SLBP in vitro.
Figure 3.
In vitro import of SLBP by Trn-SR2 and Impα/Impβ. (A) HeLa cells grown on coverslips were permeabilized with digitonin and incubated with fluorescently-labeled GST-SLBP in the presence of an energy regenerating system, buffer, and Ran without or with the purified import receptor indicated above or below each panel. After 15 min of incubation, the cells were washed to remove excess GST-SLBP. The cells were fixed, mounted onto microscope slides, and visualized using an epifluorescence microscope. (B) Purified GST-SLBP was tested for binding to import receptors zz-Impα/Impβ (lane 3), zz-Impβ (lane 4), and zz-Trn-SR2 (lane 5). The purified E. coli expressed import receptors tagged with two copies of the IgG binding domain or “z” domain of protein A were recovered on IgG-Sepharose beads. One-fifth of the eluted material was run alongside 1/20 of the input GST-SLBP. The background binding of GST-SLBP to the IgG-Sepharose beads is shown in lane 2. (C) In vitro-translated SLBP was incubated with zz-Impα/Impβ (lanes 3 and 4) or zz-Trn-SR2 (lanes 5 and 6). RanQ69L was added to the reactions in lanes 4 and 6. The proteins were bound to IgG-Sepharose beads, and 20% of the eluted material from each binding reaction was run on an 8% SDS-PAGE alongside 1/25 of the input SLBP (lane 1). The background binding of SLBP to the IgG-Sepharose beads is shown in lane 2.
SLBP Binds Trn-SR2 and Impα/Impβ Directly
To verify that SLBP can bind to Trn-SR2 and Impα/Impβ directly, a solution binding assay was performed. Complexes between purified baculovirus-expressed GST-SLBP and purified E. coli expressed import receptors were formed. IgG-Sepharose beads were used to recover the complexes by virtue of a tag containing two copies of the IgG binding domain of protein A (z domain) fused to Trn-SR2 and Impα (zz-Trn-SR2 and zz-Impα/Impβ). Bound proteins were eluted with SDS sample buffer and resolved on an SDS polyacrylamide gel. Coomassie staining of the gel revealed that both zz-Impα/Impβ (Figure 3B, lane 3) and zz-Trn-SR2 (lane 5) bound GST-SLBP, whereas the IgG-Sepharose beads alone (lane 2) did not. In addition, compared with the Impα/Impβ heterodimer, zz-Impβ without Impα interacted with SLBP only weakly (lane 4). Although the direct interaction between SLBP and zz-Impβ can be disrupted by RanQ69L (Supplemental Figure 1), it is insufficient to support nuclear localization of SLBP (Figure 3A). These results confirm that the interaction between SLBP and Trn-SR2 and Impα/Impβ is direct and that Impβ does not contribute substantially to the association of SLBP with the Impα/Impβ heterodimer.
RanGTP Dissociates Trn-SR2 and Impα/Impβ from SLBP
Import receptor/cargo complexes are dissociated by RanGTP in the nucleus. Consequently, we assessed the ability of RanGTP to disrupt SLBP/Trn-SR2 and SLBP/Impα/Impβ complexes. RanQ69L-GTP, a Ran mutant lacking GTPase function that is constitutively GTP bound if loaded with GTP, was used for this purpose. zz-Trn-SR2 or zz-Impα/Impβ was incubated with in vitro-translated SLBP, and the complexes containing the radiolabeled SLBP were recovered on IgG-Sepharose beads (Figure 3C, lanes 3 and 5). When RanQ69L was included in the reaction, the amount of SLBP associated with zz-Impα/Impβ and zz-Trn-SR2 was reduced (Figure 3C, lanes 4 and 6). Dissociation of SLBP from Impα by RanQ69L was specific because SLBP and a second Impα substrate Kip (Sekimoto et al., 2004) were dissociated from Impα by RanQ69L to a similar extent (Supplemental Figure 2). These observations demonstrate that RanQ69L can dissociate SLBP from Trn-SR2 and Impα/Impβ and suggest that the complexes formed between SLBP and Impα/Impβ or Trn-SR2 are relevant to SLBP nuclear import.
Binding of Impα/Impβ to SLBP Is Mediated by Three Different Sequences
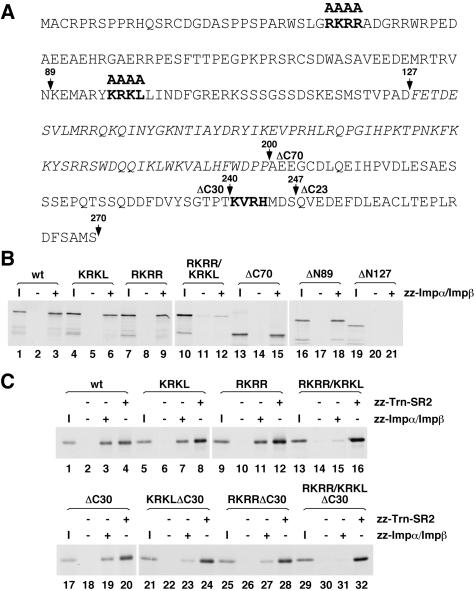
The classical, monopartite NLS recognized by Impα consists of a short stretch of basic residues exemplified by the SV40 large T antigen NLS (PKKKRKV) (Kalderon et al., 1984). Two stretches of basic amino acids in the N-terminal domain of SLBP (RKRR and KRKL) as well as one in the C-terminal domain of the protein (KVRH) are highlighted in bold in Figure 4A. As a first approach toward mapping the sequences required for binding to Impα/Impβ, SLBP constructs were designed which either lacked the C-terminal domain (ΔC70) or were truncated from the N-terminal end (ΔN89 and ΔN127), removing portions of the protein containing the predicted binding sites. The resulting proteins were then tested for the ability to interact with Impα/Impβ in a solution binding assay. SLBP lacking either the entire C-terminal domain (Figure 4B, lanes 13–15) or the first 89 amino acids (lanes 16–18) was able to interact with Impα/Impβ, albeit to a slightly reduced extent compared with intact SLBP, whereas a protein lacking the first 127 amino acids showed a significant impairment in binding Impα/Impβ (Figure 4B, lanes 19–21). Two of the predicted classical NLSs (RKRR and KRKL) reside within the first 127 amino acids, suggesting that these sequences may contribute to the binding of SLBP to Impα. To test this, the RKRR and KRKL sequences were mutated to alanines, yielding SLBPRKRR and SLBPKRKL. Mutation of either of these sequences on their own resulted in a reduction in binding by ∼50% (Figure 4B, lanes 4–9). Simultaneous mutation of both sequences (SLBPRKRR/KRKL, lanes 10–12) caused a binding defect nearly equivalent to that resulting from removing the entire N-terminal domain (Figure 4B, lanes 19–21). Thus, both the RKRR and KRKL sequences can mediate the association of Impα/Impβ through the N-terminal domain of SLBP.
Figure 4.
Mapping the sequences in SLBP required for Impα/Impβ binding. (A) The amino acid sequence of human SLBP is shown with the RNA binding domain in italics. The predicted Impα binding sites that were mutated to alanines are in bold. The arrows above the sequence mark the amino acids at which the protein was truncated either from the N-terminal end (ΔN89 and ΔN127) or the C-terminal end (ΔC70 and ΔC30). (B) The different in vitro-translated SLBP truncation or site-specific mutants labeled with [35S]methionine were incubated without import receptor (lanes 2, 5, 8, 11, 14, 17, and 20) or with zz-Impα/Impβ (lanes 3, 6, 9, 12, 15, 18, and 21). Complexes were recovered on IgG-Sepharose beads and eluted with sample buffer. Eluate (20%) was run on an 8% SDS-PAGE alongside 1/25 of the input SLBP for each binding reaction (lanes 1, 4, 7, 10, 13, 16, and 19). I, input. (C) The different in vitro-translated SLBP truncation or site-specific mutants labeled with [35S]methionine were incubated without import receptor (lanes 2, 6, 10, 14, 18, 22, 26, and 30), with zz-Impα/Impβ (lanes 3, 7, 11, 15, 19, 23, 27, and 31) or with zz-Trn-SR2 (lanes 4, 8, 12, 16, 20, 24, 28, and 32). The samples were subsequently processed as in B.
As noted above, the C-terminal deletion mutant SLBPΔC70 demonstrated reduced binding to Impα/Impβ (Figure 4B, lanes 13–15), suggesting that the C-terminal domain of SLBP may be involved in Impα/Impβ association. To test the involvement of the KVRH sequence in binding Impα/Impβ, we deleted the region of SLBP containing this sequence, yielding SLBPΔC30. To address potential cooperation with the other sites in SLBP, we also mutated the N-terminal sequences individually and together in the SLBPΔC30 construct. This series of SLBP mutants was then tested for the ability to bind Impα/Impβ and, as a negative control, Trn-SR2. Deleting the last 30 amino acids from SLBP resulted in an approximate 20% reduction in binding to Impα/Impβ relative to wild-type (Figure 4C, lanes 17–19), which is comparable with the reduction observed for the KRKL and RKRR alanine substitution mutants (Figure 4C, lanes 1–12). Introducing either the KRKL or RKRR mutation in SLBPΔC30 resulted in an 80 and 74% reduction in association with Impα/Impβ relative to wild-type (Figure 4C, lanes 21–28). The RKRR/KRKL mutant exhibited a 90% reduction in binding relative to wild-type SLBP (Figure 4C, lanes 13–15). Thus, combining the KRKL and RKRR mutations with one another or with the ΔC30 mutation resulted in a similar defect in association with Impα/Impβ. From these observations, we conclude that the C-terminal KVRH sequence in SLBP also contributes to Impα/Impβ binding, and that severe impairment of binding requires mutation of two of the three sites.
Sequences in SLBP Required for Impα Binding Are Critical for the Nuclear Accumulation of SLBP In Vivo
To assess whether the sequences required for binding Impα contributed to SLBP nuclear localization in vivo, constructs were generated containing GFP fused to the N terminus of either wild-type SLBP or SLBPs carrying mutations in the three sequences. HeLa cells were transiently transfected with each of the constructs, images were collected, and at least 100 GFP-expressing cells per construct were scored for fusion protein localization. Four categories were established to describe the different localization phenotypes: 1) completely nuclear; 2) partially cytoplasmic, with higher amounts in the nucleus; 3) evenly distributed between the nucleus and the cytoplasm; and 4) completely cytoplasmic. An example of each of these phenotypes is shown at the top of the table in Figure 5A.
GFP-SLBPwt as well as GFP-SLBPKRKL were almost exclusively nuclear (Figure 5A), whereas ∼15% of cells displayed partial cytoplasmic localization when the RKRR sequence was mutated (Figure 5A). Strikingly, GFP-SLBPRKRR/KRKL, which is mutated for both N-terminal sequences, was evenly distributed between the nucleus and cytoplasm in 65% of the cells. This result thus demonstrates that both N-terminal sequences can mediate the nuclear import of SLBP in vivo. We next examined the potential role of the C-terminal domain of SLBP in its nuclear localization. Indeed, GFP-SLBPRKRR/KRKL/ΔC70, which lacks the entire C-terminal domain, was evenly distributed in 80% of the cells, a 15% increase relative to GFP-SLBPRKRR/KRKL. From these data, we conclude that the N-terminal KRKL and RKRR sequences and perhaps the C-terminal domain of SLBP contribute to SLBP nuclear localization in vivo. An explicit analysis of the C-terminal region involved in import is given below.
Despite the fact that the KRKL and RKRR sequences and the C-terminal domain of SLBP seemed to be important for GFP-SLBP's nuclear localization, the mutant GFP-fusion proteins were never exclusively cytoplasmic. This observation could be explained either by Transportin-SR2–mediated import of SLBPs, which lack the putative Impα/Impβ NLSs, or diffusion of the GFP-SLBP. GFP-SLBPRKRR/KRKL and GFP-SLBPRKRR/KRKL/ΔC70 are 55 and 47 kDa, respectively, raising the possible question as to whether they might be small enough to diffuse in and out of the nucleus. To distinguish between these possibilities, a GST tag was added to the N terminus of GFP-SLBP, yielding GST-GFP-SLBP, which has a molecular mass of ∼90 kDa and is too large to cross NPCs by diffusion. Western blot analysis with the GFP antibody was carried out to compare the relative expression level of each of the GST-GFP fusion constructs (Figure 5B, left). Probing with the SLBP antibody revealed that the expression of GST-GFP-SLBPwt was similar to the endogenous SLBP (Figure 5B, right).
GST-GFP-SLBPwt and GST-GFP-SLBPKRKL demonstrated similar localization to GFP-SLBPwt and GFP-SLBPKRKL (Figure 5, A and C), indicating that mutation of the KRKL sequence alone had no effect on SLBP localization. In contrast, when the RKRR sequence was mutated in GST-GFP-SLBP, the resulting protein (GST-GFP-SLBPRKRR) distributed evenly between the nucleus and cytoplasm in 22% of cells and was completely cytoplasmic in 24% of cells (Figure 5, C and D). If the KRKL mutation was introduced simultaneously with the RKRR mutation in GST-GFP-SLBP (GST-GFP-SLBPRKRR/KRKL), more of the cells showed cytoplasmic localization than with the RKRR mutation alone, indicating that the KRKL sequence does contribute to the import of SLBP (Figure 5, C and D). Deleting the last 23 amino acids from the C-terminal end of SLBP (GST-GFP-SLBPΔC23), just beyond the KVRH sequence, did not affect SLBP's nuclear localization (Figure 5C). However, removing the next seven amino acids containing the KVRH sequence (GST-GFP-SLBPΔC30) resulted in a pattern of localization similar to that observed for the RKRR mutant (Figure 5, C and D), indicating that the KVRH sequence is important for SLBP nuclear localization. Strikingly, when the two N-terminal sequences were mutated and the C-terminal KVRH sequence was removed (GST-GFP-SLBPRKRR/KRKLΔC30), SLBP was excluded from the nucleus of all cells (Figure 5, C and D).
Although we had identified which sequences were required for SLBP nuclear localization, we did not know whether these sequences were in fact NLSs. The proof for NLS activity is the ability to direct a cytoplasmic protein into the nucleus. Therefore, we fused each of the sequences to GST-GFP, which is normally cytoplasmic (Figure 5E, top row), and analyzed the localization of the fusion proteins. The well-characterized SV40 NLS also was fused to GST-GFP as a positive control (Figure 5E, second row). The results of our analysis show that the KRKL and RKRR sequences indeed have NLS activity (Figure 5E, third and fourth rows), whereas the KVRH sequence does not (bottom row). Thus, we conclude that the RKRR and KRKL sequences are bona fide NLSs and that the KVRH sequence is not. Perhaps the KVRH sequence helps stabilize the interaction between SLBP and Impα/β and for this reason is needed for optimal import (see Discussion).
To determine which of the N-terminal NLSs contributed most significantly to the nuclear localization defect of GST-GFP-SLBPRKRR/KRKL/ΔC30, the KRKL and RKRR sequences were mutated individually in the context of the GST-GFP-SLBPΔC30 construct (yielding GST-GFP-SLBPKRKL/ΔC30 and GST-GFP-SLBPRKRR/ΔC30). GST-GFP-SLBPRKRR/ΔC30 and GST-GFP-SLBPKRKL/ΔC30 were exclusively cytoplasmic in 43 and 25% of the cells examined (Figure 5, C and D), a 23 and 5% increase relative to GST-GFP-SLBPΔC30. These observations are consistent with those showing that GST-GFP-SLBPRKRR was more severely impaired in nuclear localization than GST-GFP-SLBPKRKL. From these observations, we conclude that the C-terminal KVRH sequence and the RKRR sequence make the largest contribution to the nuclear localization of SLBP (see Discussion).
Trn-SR2 Does Not Import SLBP in HeLa Cells
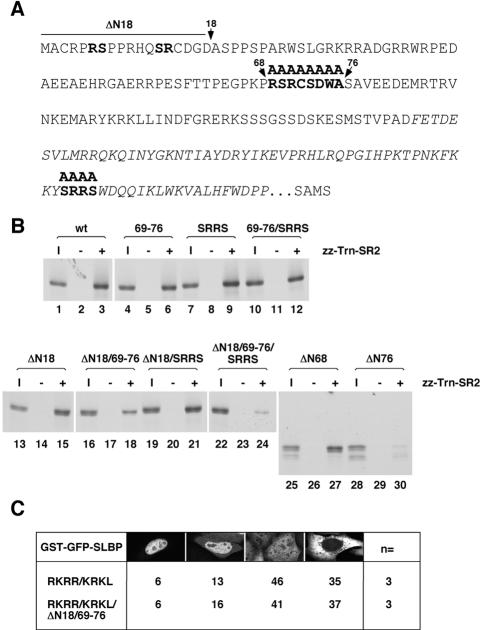
We also determined which part of SLBP is required for association with Trn-SR2. SLBP does not contain a conventional RS domain as found in other import substrates of Trn-SR2, but it contains only a few dispersed SR dipeptides. Candidate Trn-SR2 binding motifs in SLBP are shown in bold in Figure 6A.
Figure 6.
Characterization of the sequences in SLBP needed for Trn-SR2 binding. (A) The amino acid sequence of human SLBP is shown with the RNA binding domain in italics. The sequence after the RNA binding domain and before the last four amino acids of the protein (SAMS) is not shown. The predicted Trn-SR2 binding sites that were mutated to alanines are in bold. The arrows above the sequence mark the amino acids at which the protein was truncated from the N-terminal end (ΔN18, ΔN68, and ΔN76). (B) The different in vitro-translated SLBP truncation or site-specific mutants labeled with [35S]methionine were incubated without import receptor (lanes 2, 5, 8, 11, 14, 17, 20, 23, 26, and 29) or with zz-Trn-SR2 (lanes 3, 6, 9, 12, 15, 18, 21, 24, 27, and 30). Samples were subsequently processed as in Figure 4B. I, 1/25 of the input (lanes 1, 4, 7, 10, 13, 16, 19, 22, 25, and 28). (C) Images of cells transiently transfected with the indicated GST-GFP-SLBP construct were collected, and at least 300 cells per construct were examined. The number of independent transfections analyzed is indicated in the column labeled n=. Cells were scored as described in Figure 5A.
Importantly, we found that impairing the two N-terminal Impα binding sites either independently or at the same time had no effect on Trn-SR2 binding (Figure 4C, lanes 4, 8, 12, and 16). As a first approach toward mapping the Trn-SR2 binding site(s), SLBP was serially truncated from the N terminus (ΔN68 and ΔN76), removing portions of the protein containing the predicted binding sites. The resulting proteins were then tested for the ability to interact with Trn-SR2 in a solution binding assay. Through this analysis, we found that the first 68 amino acids are dispensable for Trn-SR2 binding (Figure 6B, lanes 1–3 and 25–27). However, if the first 76 amino acids were deleted, there was a significant reduction in the association with Trn-SR2 (Figure 6B, lanes 28–30). These data suggest that the region between amino acids 68 and 76, which contains one of the predicted Trn-SR2 binding sites, might be involved in recruiting Trn-SR2 to SLBP. To test this hypothesis, the amino acids between residues 68 and 76 were substituted with alanines (69–76). We also deleted the first 18 amino acids of SLBP (ΔN18 SLBP) and mutated the third potential Trn-SR2 binding site in the RNA binding domain to alanines (SRRS). SLBP 69–76 demonstrated a 40% reduction in binding to Trn-SR2 relative to wild-type SLBP (Figure 6B, lanes 1–6). In contrast, the ΔN18 mutation did not significantly interfere with Trn-SR2 binding (Figure 6B, lanes 13–15). However, because both of these sequences reside within the first 76 amino acids of SLBP, we tested whether combining these two mutations (ΔN18/69-76) would result in a Trn-SR2 binding defect similar to ΔN76 SLBP. The association of ΔN18/69-76 SLBP with Trn-SR2 was reduced by ∼70% relative to wild-type (Figure 6B, lanes 16–18), but this mutant was not as defective as ΔN76 SLBP, which exhibited a 90% reduction in binding to Trn-SR2 under the same conditions (Figure 6B, lanes 28–30).
A final possible Trn-SR2 binding site is the SRRS sequence, which lies in the RNA binding domain of SLBP. Mutation of this sequence did not interfere with Trn-SR2 binding (Figure 6B, lanes 7–9). However, when all three predicted Trn-SR2 binding sites were mutated, the binding of Trn-SR2 was reduced by ∼90% relative to wild-type (Figure 6B, lanes 22–24). From these results, we conclude that the binding of Trn-SR2 to SLBP occurs through an extended region rather than a precise set of residues in the first 76 amino acids of SLBP and also may involve the SRRS sequence in the RNA binding domain.
Mutation of the three sequences in SLBP required for binding to Impα/Impβ, which was sufficient to completely impair SLBP nuclear accumulation in vivo, did not affect Trn-SR2 binding in vitro (Figure 4C, lane 32). These observations suggest that Trn-SR2 does not contribute to SLBP import in vivo. Indeed, simultaneous mutation of all three Trn-SR2 binding sites had no effect on SLBP nuclear localization (our unpublished data). We reasoned that if Impα/Impβ represents the dominant import pathway for SLBP, perhaps reducing the ability of SLBP to be imported by this pathway may reveal any potential import activity of Trn-SR2 toward SLBP in this system. To test this, we determined whether mutating the N-terminal sequences that contribute to Trn-SR2 binding added to the defect in nuclear localization resulting from mutation of the two N-terminal Impα/Impβ NLSs. HeLa cells were transfected with GST-GFP-SLBPRKRR/KRKL or GST-GFP-SLBPRKRR/KRKL/ΔN18/69–76 and scored for fusion protein localization. There was no significant difference in the localization of these two SLBPs (Figure 6C). Thus, we conclude that Trn-SR2 is not involved in SLBP nuclear import in HeLa cells.
SLBP Does Not Contain Nuclear Export Signals
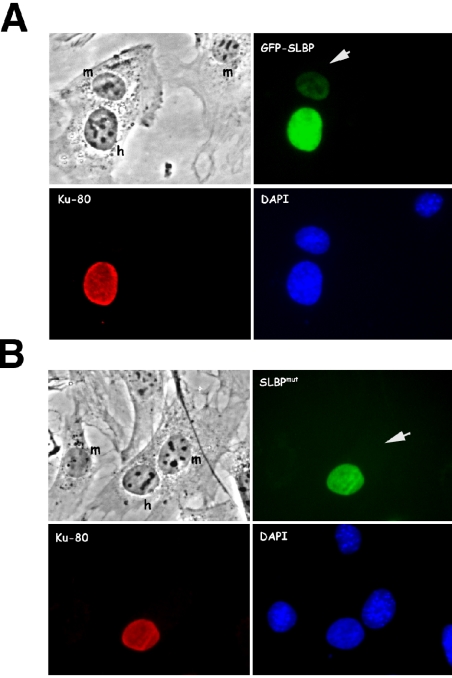
SLBP present in the cytoplasm of S-phase cells is associated with the histone messenger ribonucleoprotein particle (mRNP) (Whitfield et al., 2004). Because SLBP is not required for histone mRNA export (Erkmann et al., 2005), we considered it possible that it accompanies the histone mRNP to the cytoplasm but is not actively exported. To test this hypothesis, we used an interspecies heterokaryon assay to examine the shuttling capabilities of wild-type SLBP (wt) and an SLBP mutant (mut) that cannot bind histone mRNA (Dominski et al., 2001). Human U2-OS cells were transfected with either wt or mut GFP-SLBP and were subsequently treated with protein synthesis inhibitors and fused to MEF cells. Accumulation of the GFP-tagged protein in the mouse nucleus of the heterokaryon indicates that the protein has been exported out of the human nucleus into the shared cytoplasm and imported into the mouse nucleus. A small amount of GFP-SLBP wt relocalized to the mouse nucleus after ninety minutes of incubation (Figure 7A), but GFP-SLBP mut, which cannot bind histone mRNA, remained in the human nucleus in the heterokaryon (Figure 7B). Thus, SLBP does not contain any nuclear export signals and is likely exported from the nucleus as a passive cargo of the histone mRNP.
Figure 7.
SLBP does not contain nuclear export signals. U2-OS cells were transfected with plasmids expressing GFP-SLBP wt (A) or the RNA binding mutant GFP-SLBP mut (B), which contains two tyrosines in the RBD replaced with alanines (Dominski et al., 2001). Twenty-four hours after transfection, the cells were washed and fused to wild-type MEF cells, and de novo protein synthesis was blocked by cycloheximide. Ninety minutes after fusion, the cells were fixed and stained with an antibody to human Ku antigen followed by staining with a Texas Red-conjugated secondary antibody. The heterokaryons were photographed. Nuclei of human (h) and mouse (m) origin are identified by DAPI staining. Mouse nuclei are indicated with white arrows.
DISCUSSION
The processing-specific hSLBP orthologues that have been cloned, Xenopus SLBP1 (xSLBP1), mouse SLBP, and Drosophila SLBP, are found in both the nuclear and cytoplasmic compartments in oocytes and embryos (Wang et al., 1999; Allard et al., 2002; Lanzotti et al., 2004). In addition, in Xenopus oocytes a small fraction of xSLBP1 localizes to subnuclear structures termed Cajal bodies, which are implicated in histone gene transcription and mRNA processing (Abbott et al., 1999). In this report, we describe the identification of two import receptors that are able to import human SLBP into HeLa nuclei in vitro and have determined the sequences in SLBP necessary for its nuclear localization in vivo. Additionally, we have examined the changes in the localization of SLBP during the cell cycle.
SLBP Localization during S Phase
During S phase, SLBP is localized predominantly in the nucleus and does not seem to concentrate in Cajal bodies. In cells arrested at the beginning of S phase, SLBP is nuclear. As cells progress through S phase and the levels of histone mRNA increase, SLBP partially redistributes to the cytoplasm. Previously, we have shown that the treatment of cells with inhibitors of DNA replication, which results in the degradation of histone mRNA, also results in SLBP localizing to the nucleus (Whitfield et al., 2004). These observations therefore suggest that the cytoplasmic localization of SLBP may result from its association with histone mRNAs in the cytoplasm. In agreement with this interpretation, we find that SLBP with a two-amino acid mutation that prevents RNA binding is restricted to the nucleus (Figure 2A). Also, using a heterokaryon assay, we show that SLBP incapable of histone mRNA binding does not shuttle out of the nucleus (Figure 7). Previous observations made in Drosophila embryos also support the notion that the cytoplasmic localization of SLBP requires histone mRNA production (Lanzotti et al., 2004). During the early cell cycles of Drosophila development, dSLBP persists throughout the cell cycle but its localization varies with the phase of the cell cycle (Lanzotti et al., 2002; Lanzotti et al., 2004). During S phase, when histone mRNAs are made, dSLBP is present in both the cytoplasm and nucleus, whereas in G1 and G2 phase, SLBP is exclusively nuclear (Lanzotti et al., 2004).
At the end of S phase, human SLBP is degraded in a phosphorylation-dependent manner (Whitfield et al., 2000; Zheng et al., 2003). Treatment of cells with proteasome inhibitors prevents this degradation, resulting in the accumulation of phosphorylated SLBP in G2 phase. SLBP containing mutations in the phosphorylation sites responsible for triggering its degradation is also stable in G2 phase. Interestingly, we have found that various nondegraded forms of SLBP are nuclear in G2 phase. These results, however, are in contrast to what we previously observed by cellular fractionation, which showed that phosphorylated SLBP from G2-phase cells was exclusively cytoplasmic (Zheng et al., 2003). We believe that our immunofluorescence studies accurately reflect the localization of SLBP in G2-phase cells because we obtained identical staining patterns in these cells using antibodies targeting both the N- and C-terminal ends of the protein. Therefore, epitope masking can be ruled out as the reason for not detecting SLBP in the cytoplasm of G2-phase cells. SLBP is only 28 kDa, which is below the size threshold for diffusion through NPCs. Therefore, the presence of SLBP in cytoplasmic extracts from G2 cells is most likely due to its diffusion out of the nucleus during the fractionation procedure. We note that in the cellular fractionation experiments (Zheng et al., 2003), only the phosphorylated form of SLBP was entirely cytoplasmic, whereas the unphosphorylated form was present in both the nuclear and cytoplasmic fractions. Most likely, unphosphorylated SLBP was retained in the nuclear fraction by associating with either a subnuclear structure or macromolecular complex that is incapable of diffusing through nuclear pores.
Multiple Sequences in SLBP Mediate Impα/Impβ Association
We have determined that Impα/Impβ interacts with SLBP and can import it in vitro. Consistent with this observation, Impα/Impβ was dissociated from SLBP by RanGTP. We mapped three sequences in SLBP (RKRR, KRKL, and KVRH) that mediate the interaction with Impα/Impβ. One of these sequences (KVRH) is more distant than the other two from the SV40 NLS, because it contains only two basic residues. We have determined that, although this sequence is important for binding to Impα and the nuclear localization of SLBP in vivo, it does not possess NLS activity when fused to a heterologous protein. We interpret these data to mean that the KVRH sequence is necessary for optimal Impα/β association but is probably not a binding site for the receptor. Perhaps it helps stabilize the interaction between SLBP and Impα/β and for this reason is needed for optimal import. Single mutation of the N-terminal (RKRR) and the C-terminal (KVRH) sequences resulted in similar reductions in SLBP nuclear localization in vivo. In contrast, mutating the N-terminal KRKL sequence had no effect on the localization of SLBP, despite our demonstration that this sequence is a functional NLS. The activity of the KRKL sequence as an NLS was revealed only if the RKRR NLS also was mutated, whether it be in the GST-GFP-SLBPRKRR/KRKL or GST-GFP-SLBPRKRR/KRKL/ΔC30 construct. The KRKL sequence is also a cyclin A/cdk2 binding site, required for SLBP phosphorylation and degradation (Zheng et al., 2003; Koseoglu, unpublished data). These observations therefore suggest that perhaps the KRKL sequence is not the preferred NLS in the wild-type protein, but only is used in the absence of the nearby RKRR Impα/Impβ binding site. In sum, we have identified two NLSs in SLBP (KRKL and RKRR) and a third sequence (KVRH) that contributes to SLBP nuclear localization.
SLBP Binding and Import by Trn-SR2
We have demonstrated that complexes containing Trn-SR2 and SLBP form efficiently in vitro and are dissociated by Ran-GTP. In an in vitro import assay, in which other import receptors were unable to import SLBP, Trn-SR2 efficiently imported SLBP. Despite this in vitro activity, we find that Trn-SR2 does not play a role in the import of SLBP in HeLa cells. The best evidence for this is that SLBP containing mutations in the three sequences required for Impα/Impβ binding is exclusively present in the cytoplasm, although it retains its full ability to bind to Trn-SR2. Although we were not able to precisely map the Trn-SR2 binding site, the ΔN18/69-76 mutant, which had reduced Trn-SR2 binding in vitro, was not defective in nuclear localization. Moreover, combining this mutation with mutations in the two Impα NLSs resulted in no further decrease in the nuclear localization of SLBP compared with SLBP lacking only the two Impα NLSs. It remains possible that Trn-SR2 has a role in SLBP nuclear localization perhaps during a specific stage in development or in a particular cell type.
Model for SLBP Localization
Together, our data suggest a model for SLBP localization where SLBP is imported into the nucleus by Impα/Impβ, and the cytoplasmic accumulation of SLBP is dependent on the presence of histone mRNA. We have recently shown that neither SLBP nor the 3′ end of histone mRNA is essential for histone mRNA export (Erkmann et al., 2005). Thus, it is most probable that after histone pre-mRNA processing in the nucleus SLBP is exported to the cytoplasm as a passive cargo of the histone mRNA. SLBP present in the cytoplasm then participates in histone mRNA translation (Sanchez and Marzluff, 2002). As histone mRNAs turnover in the cytoplasm, SLBP reenters the nucleus (Whitfield et al., 2004). At the end of S phase, SLBP is degraded by a phosphorylation-dependent mechanism requiring cyclin A/cdk 2 (Zheng et al., 2003). Because histone mRNA decay does not require the prior degradation of SLBP (Whitfield et al., 2004), it is possible that SLBP is degraded in the nucleus after dissociation from the mRNA.
Acknowledgments
We thank the members of the Kutay and Marzluff laboratories for support during the course of this project. In addition, we thank Dr. Yue Xiong for providing us with the anti-HA antibody and Dr. Ian Macara for the GFP plasmid. This work was supported by the Swiss National Science Foundation and an intramural Swiss Federal Institute of Technology grant to U. K. and National Institutes of Health Grants GM-58921 and GM-29832 to W.F.M. Y. Z. was supported by National Institutes of Health Grants CA-100302 and CA-87580. E.J.W. acknowledges funding from a National Research Service Award Postdoctoral Fellowship (GM-070101-01) and the Cottrell Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-11-1023) on April 13, 2005.
References
- Abbott, J., Marzluff, W. F., and Gall, J. G. (1999). The stem–loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol. Biol. Cell 10, 487-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, S. A., and Gerace, L. (1991). Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66, 837-847. [DOI] [PubMed] [Google Scholar]
- Allard, P., Champigny, M. J., Skoggard, S., Erkmann, J. A., Whitfield, M. L., Marzluff, W. F., and Clarke, H. J. (2002). Stem-loop binding protein accumulates during oocyte maturation and is not cell-cycle-regulated in the early mouse embryo. J. Cell Sci. 115, 4577-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, N. C., Adam, E. J., and Adam, S. A. (1995). Sequence and characterization of cytoplasmic nuclear protein import factor p97. J. Cell Biol. 130, 265-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall, C., Sharnick, S. V., and Laskey, R. A. (1982). A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 30, 449-458. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Erkmann, J. A., Greenland, J. A., and Marzluff, W. F. (2001). Mutations in the RNA binding domain of stem–loop binding protein define separable requirements for RNA binding and for histone pre-mRNA processing. Mol. Cell. Biol. 21, 2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., and Marzluff, W. F. (1999). Formation of the 3′ end of histone mRNA. Gene 239, 1-14. [DOI] [PubMed] [Google Scholar]
- Erkmann, J. A., Sanchez, R., Treichel, N., Marzluff, W. F., and Kutay, U. (2005). Nuclear export of metazoan replication-dependent histone mRNAs is dependent on RNA length and is mediated by TAP. RNA 11, 45-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried, H., and Kutay, U. (2003). Nucleocytoplasmic transport: taking an inventory. Cell Mol. Life Sci. 60, 1659-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie, D. R., Lewis, N. J., and Marzluff, W. F. (1996). The histone 3′-terminal stem–loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 24, 1954-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D. (1997). Nuclear protein import. Curr. Opin. Cell Biol. 9, 412-419. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., Dabrowski, M., Bischoff, F. R., Kutay, U., Bork, P., Hartmann, E., Prehn, S., and Izaurralde, E. (1997). A novel class of RanGTP binding proteins. J. Cell Biol. 138, 65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607-660. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., Pante, N., Kutay, U., Aebi, U., and Bischoff, F. R. (1996). Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15, 5584-5594. [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., Prehn, S., Laskey, R. A., and Hartmann, E. (1994). Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79, 767-778. [DOI] [PubMed] [Google Scholar]
- Hanson, R. J., Sun, J., Willis, D. G., and Marzluff, W. F. (1996). Efficient extraction and partial purification of the polyribosome-associated stem–loop binding protein bound to the 3′ end of histone mRNA. Biochemistry 35, 2146-2156. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I. W., and Gorlich, D. (1997). The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16, 6535-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel, S., Albig, W., Kutay, U., Bischoff, F. R., Schwamborn, K., Doenecke, D., and Gorlich, D. (1999). The importin β/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 18, 2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel, S., and Gorlich, D. (1998). Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, D., Richardson, W. D., Markham, A. F., and Smith, A. E. (1984). Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33-38. [DOI] [PubMed] [Google Scholar]
- Kutay, U., Bischoff, F. R., Kostka, S., Kraft, R., and Gorlich, D. (1997a). Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90, 1061-1071. [DOI] [PubMed] [Google Scholar]
- Kutay, U., Izaurralde, E., Bischoff, F. R., Mattaj, I. W., and Gorlich, D. (1997b). Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16, 1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M. C., Lin, R. I., and Tarn, W. Y. (2001). Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. USA 98, 10154-10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti, D. J., Kaygun, H., Yang, X., Duronio, R. J., and Marzluff, W. F. (2002). Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol. Cell. Biol. 22, 2267-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti, D. J., Kupsco, J. M., Yang, X. C., Dominski, Z., Marzluff, W. F., and Duronio, R. J. (2004). Drosophila stem–loop binding protein intracellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Mol. Biol. Cell 15, 1112-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, F., Schaller, A., Eglite, S., Schumperli, D., and Muller, B. (1997). The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16, 769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff, W. F., and Duronio, R. J. (2002). Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14, 692-699. [DOI] [PubMed] [Google Scholar]
- McKiernan, C. J., Stabila, P. F., and Macara, I. G. (1996). Role of the Rab3A-binding domain in targeting of rabphilin-3A to vesicle membranes of PC12 cells. Mol. Cell. Biol. 16, 4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu, J., Blobel, G., and Radu, A. (1995a). Previously identified protein of uncertain function is karyopherin α and together with karyopherin β docks import substrate at nuclear pore complexes. Proc. Natl. Acad. Sci. USA 92, 2008-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu, J., Hijikata, M., Blobel, G., and Radu, A. (1995b). Mammalian karyopherin α1β and α2β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. USA 92, 6532-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast, N., Jackson, K. R., Guo, Y., Brame, C. J., Shabanowitz, J., Hunt, D. F., and Pemberton, L. F. (2001). Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol. 153, 251-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast, N., and Pemberton, L. F. (2004). Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547-556. [DOI] [PubMed] [Google Scholar]
- Muhlhausser, P., Muller, E. C., Otto, A., and Kutay, U. (2001). Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2, 690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, V. W., Michael, W. M., Nakielny, S., Siomi, M. C., Wang, F., and Dreyfuss, G. (1996). A novel receptor-mediated nuclear protein import pathway. Cell 86, 985-994. [DOI] [PubMed] [Google Scholar]
- Rexach, M., and Blobel, G. (1995). Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83, 683-692. [DOI] [PubMed] [Google Scholar]
- Ribbeck, K., Kutay, U., Paraskeva, E., and Gorlich, D. (1999). The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr. Biol. 9, 47-50. [DOI] [PubMed] [Google Scholar]
- Rout, M. P., Blobel, G., and Aitchison, J. D. (1997). A distinct nuclear import pathway used by ribosomal proteins. Cell 89, 715-725. [DOI] [PubMed] [Google Scholar]
- Sanchez, R., and Marzluff, W. F. (2002). The stem–loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 22, 7093-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt, G., Smirnova, E., Deane, R., Solsbacher, J., Kutay, U., Gorlich, D., Ponstingl, H., and Bischoff, F. R. (1997). Yrb4p, a yeast ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J. 16, 6237-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto, T., Fukumoto, M., and Yoneda, Y. (2004). 14–3-3 suppresses the nuclear localization of threonine 157-phosphorylated p27(Kip1). EMBO J. 23, 1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, H., and Dreyfuss, G. (1995). A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129, 551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E. J., and Garcia-Blanco, M. A. (2002). RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10, 943-949. [DOI] [PubMed] [Google Scholar]
- Wang, Z. F., Ingledue, T. C., Dominski, Z., Sanchez, R., and Marzluff, W. F. (1999). Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol. 19, 835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. F., Whitfield, M. L., Ingledue, T. C., 3rd, Dominski, Z., and Marzluff, W. F. (1996). The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10, 3028-3040. [DOI] [PubMed] [Google Scholar]
- Whitfield, M. L., Kaygun, H., Erkmann, J. A., Townley-Tilson, W. H., Dominski, Z., and Marzluff, W. F. (2004). SLBP is associated with histone mRNA on polyribosomes as a component of the histone mRNP. Nucleic Acids Res. 32, 4833-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, M. L., Zheng, L. X., Baldwin, A., Ohta, T., Hurt, M. M., and Marzluff, W. F. (2000). Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20, 4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Xiong, Y. (2001). A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292, 1910-1915. [DOI] [PubMed] [Google Scholar]
- Zheng, L., Dominski, Z., Yang, X. C., Elms, P., Raska, C. S., Borchers, C. H., and Marzluff, W. F. (2003). Phosphorylation of stem–loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell. Biol. 23, 1590-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]