Abstract
In animals, the nuclear envelope disassembles in mitosis, while budding and fission yeast form an intranuclear spindle. Ultrastructural data indicate that basidiomycetes, such as the pathogen Ustilago maydis, undergo an ‘open mitosis'. Here we describe the mechanism of nuclear envelope break-down in U. maydis. In interphase, the nucleus resides in the mother cell and the spindle pole body is inactive. Prior to mitosis, it becomes activated and nucleates microtubules that reach into the daughter cell. Dynein appears at microtubule tips and exerts force on the spindle pole body, which leads to the formation of a long nuclear extension that reaches into the bud. Chromosomes migrate through this extension and together with the spindle pole bodies leave the old envelope, which remains in the mother cell until late telophase. Inhibition of nuclear migration or deletion of a Tem1p-like GTPase leads to a ‘closed' mitosis, indicating that spindle pole bodies have to reach into the bud where MEN signalling participates in envelope removal. Our data indicate that dynein-mediated premitotic nuclear migration is essential for envelope removal in U. maydis.
Keywords: dynein, MEN signalling, nuclear envelope break-down, nuclear migration
Introduction
In eukaryotic cells, the chromosomes are surrounded by the nuclear envelope (NE), a membranous structure that is in continuum with the endoplasmic reticulum (ER). At the onset of mitosis, in animal cells a nucleus-associated microtubule organizing centre (MTOC), the centrosome, separates and subsequently forms the spindle apparatus. In order to establish contact between the centrosome-associated microtubules (MTs) and the chromosomes, the NE disappears in prometaphase (Lippincott-Schwartz, 2002). This is mediated by the activity of the MT-based motor dynein, which is recruited to the outer membrane of the NE (Salina et al, 2002) and interacts with MTs that surround the nucleus (Beaudouin et al, 2002). Dynein is a minus-end-directed motor, and consequently its transport activity along the astral MTs generates tension towards the centrosomes. This eventually leads to a breakage of the NE distal to the centrosome and a catastrophic disassembly of nuclear membranes (Beaudouin et al, 2002). In contrast to animal cells, the NE persists in mitosis of most fungi, including the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, and numerous filamentous fungi such as Neurospora crassa and Aspergillus nidulans (Gant and Wilson, 1997). While a closed mitosis is common among ascomycetes, ultrastructural data suggest that the NE of higher basidiomycete fungi disperses in mitosis (Heath, 1980). This includes important fungal pests such as the human pathogen Cryptococcus neoformans (Mochizuki et al, 1987) and the plant pathogens Ustilago maydis (O'Donnell and McLaughlin, 1984) and Ustilago violaceae (Heath, 1980). In contrast to ascomycete fungi, what these basidiomycetes have in common is that their nuclei migrate into the bud before they undergo mitosis (Holliday, 1965; Poon and Day, 1976; Mochizuki et al, 1987). Here we describe that premitotic nuclear migration and the removal of the NE are coupled in the plant pathogenic basidiomycete fungus U. maydis. This model organism shows remarkable similarities to animal cells (Kojic et al, 2002; Wedlich-Söldner et al, 2002b; Adamikova et al, 2004) and has numerous technical advantages, such as advanced molecular genetics, cytological tools and a published genome. In contrast to budding yeast, the interphase spindle pole body (SPB) of U. maydis is not embedded in the NE but is attached to the outer membrane of the nucleus (O'Donell, 1992). At this stage, the SPB is not active and interphase MTs are nucleated by cytoplasmic MTOCs, including a polar MTOC that focuses MT minus-ends at the bud neck (Steinberg et al, 2001; Straube et al, 2003). At prophase, the SPB becomes active and nucleates MTs, the NE disappears and subsequently a metaphase spindle is formed (O'Donell, 1992). We have previously reported that cytoplasmic dynein is responsible for premitotic migration of nuclei towards this MTOC and into the bud of U. maydis (Straube et al, 2001). Here we demonstrate that dynein at MT plus-ends pulls the SPBs into the bud, which stretches the nucleus and forms a long extension. Chromosomes and the SPBs migrate through this extension into the daughter cell, and the old envelope collapses back into the mother cell. NE breakage and removal involves MTs and requires dynein-dependent SPB migration into the bud. Interestingly, a Tem1p-like GTPase participates in envelope removal, suggesting that mitotic exit network (MEN) signalling is involved in this early mitotic event.
Results
Early in mitosis, chromosomes leave the nuclear envelope and migrate into the bud
In logarithmically growing cultures, most cells are unbudded, which corresponds to G1 or S phase (Snetselaar and McCann, 1997), and based on the doubling time of 2.1 h, this stage lasts about 73 min (Figure 1A). Polar budding cells that are in G2 phase contained long MTs that reached into the daughter cell (Figure 1B; overlay of histone4-CFP (cyan-shifted fluorescent protein), YFP-α-tubulin (YFP: yellow-shifted fluorescent protein) and DIC image). Mitosis lasted 8–9 min and was initiated by the migration of the nucleus into the daughter cell (Holliday, 1965), where a metaphase spindle was formed (Figure 1B). Chromosomes clustered around the spindle (Figure 1C) and were segregated by spindle elongation in anaphase, which took half of the chromosomes back into the mother cell (Figure 1B and C). Finally, chromosomes decondensed and two septae were formed, which was followed by cytokinesis (not shown). In order to gain first insight into the dynamics of the envelope during mitosis, we constructed nRFP, which is a fusion of three monomeric red fluorescent proteins (Campbell et al, 2002) targeted to the nucleus by a signal peptide (Straube et al, 2001). Because of its predicted molecular weight of ∼90 kDa, nRFP should not passively diffuse out of the nucleus (Görlich and Kutay, 1999). Consequently, nRFP was located in the interphase nucleus (Figure 1B, nRFP in red). Surprisingly, the fusion protein was released in large budded cells (Figure 1B), indicating that the mitotic NE becomes permeable or is removed in mitosis. This observation was in agreement with previous reports, which described that the NE is visible in growing interphase cells expressing ER-targeted fluorescent proteins (Wedlich-Söldner et al, 2002a; Figure 2A, arrow), but could not be detected during mitosis that occurred after migration of the nucleus into the daughter cell (Figure 2A, inset shows spindle). Furthermore, mitotic cells usually contained an accumulation of ER structures in the mother cell (Figure 2A, arrowhead). In order to investigate the relation between both phenomena, we then analysed NE dynamics at G2/M transition. Prior to mitosis, the nucleus was pulled towards the daughter cell and a nuclear membrane extension was formed that reached into the bud (Figure 2B, neck indicated by asterisk). Eventually, the NE opened at the leading tip of this extension (Figure 2B, arrowhead), started to collapse and finally remained as an accumulation of ER membranes in the mother cell (Figure 2B, arrow), while the spindle was formed within the daughter cell (Figure 2C). A 3D analysis demonstrated the disk-like shape of these NE remnants (Figure 2D, viewing angles are shown at upper left), which most likely coincide with the ‘nuclear parts' recognized in the mother cell during electron microscopic observation (O'Donnell and McLaughlin, 1984). Long-term observation of dividing cells revealed that the old envelope persists in the mother cell until telophase and is frequently contacted by one of the newly formed nuclei. A membranous bridge appeared to connect the new nucleus and the old envelope, which gradually disappeared while the new nucleus expanded, suggesting that material is recycled from the old envelope in telophase (Figure 2E; asterisk marks old envelope in mother cell; arrow indicates new nucleus; arrowhead points to membrane connection between both).
Figure 1.
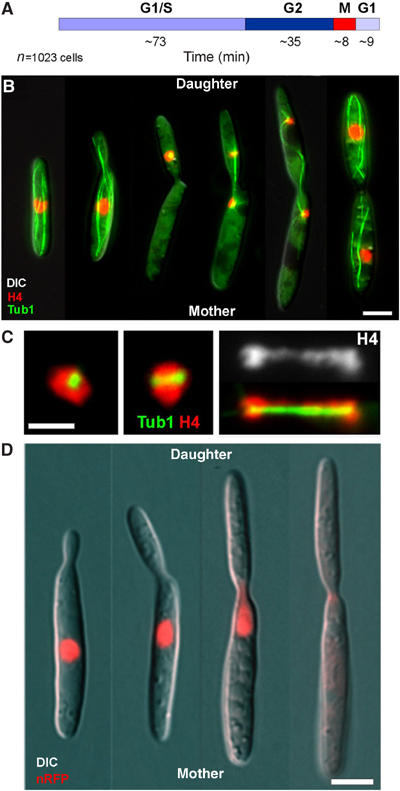
Premitotic nuclear migration in U. maydis. (A) Based on the abundance of unbudded cells (G1 or S phase), budded cells (G2) or mitotic cells in growing cultures that double every 2.1 h (CM-G, 28°C; our unpublished results), the duration of each cell cycle stage was estimated. (B) In interphase, the nucleus (DNA stained with histone4-CFP) is located in the centre of the mother cell, while a polar bud is formed. The SPB is inactive during these stages (O'Donell, 1992; Straube et al, 2003). Prior to mitosis, the nucleus migrates into the daughter cell. The initial phase of mitosis occurs inside the daughter, while the anaphase spindle takes half of the chromosomes back into the mother cell. Note that the late anaphase spindle is bent, suggesting that nuclear positioning is mediated by the astral MTs. Bar: 3 μm. (C) At metaphase, chromosomes gather around the newly formed spindle. Chromosome segregation is achieved by elongation of the anaphase spindle; no metaphase plate is formed. Note that some chromosomes trail, while the spindle elongates. Bar: 2 μm. (D) A fusion protein of 3 × RFP and an N-terminal nuclear localization signal (nRFP) locates to interphase nuclei. Due to its size of ∼90 kDa, the protein exceeds the exclusion limit of the nuclear pores and is therefore trapped in the nucleus. Note that the fusion protein is released from the mitotic nucleus and disperses in the cytoplasm. This is a first indication that the NE is permeable or is removed in mitosis. Bar: 3 μm.
Figure 2.
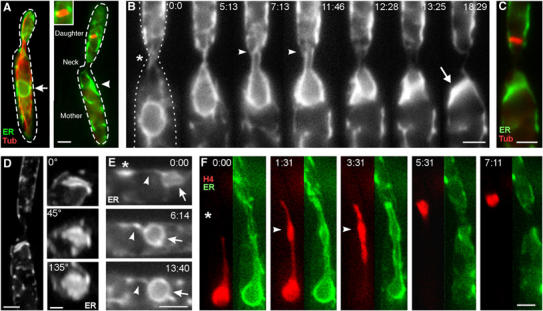
NEBD in U. maydis. (A) In interphase, the NE (arrow) surrounds the nucleus in the mother cell. Mitosis occurs in the daughter cell. The spindle is not surrounded by an NE (see also inset), but the mother cell contains an ER accumulation (arrowhead) that is always located close to the neck. Green, ER: ER-YFP; red, Tub: CFP-α-tubulin. Bar: 2 μm. (B) Prior to mitosis, the NE, marked with ER-YFP, elongates through the neck (asterisk) into the daughter cell. The NE breaks at its leading edge (arrowheads), and subsequently collapses and remains in the mother cell (arrow). ER: ER-YFP. Bar: 2 μm; elapsed time is given in minutes:seconds. (C) The mitotic spindle is formed in the bud. Green, ER: ER-YFP; red, Tub: CFP-α-tubulin. Bar: 2 μm. (D) 3D analysis reveals the disc-like shape of the collapsed NE. Viewing angles are given. ER: ER-GFP. Bars: 2and 1 μm. (E) In late anaphase, new NEs appear (arrow marks nucleus in mother cell), while the old envelope is still located in the mother cell (asterisk). The old and collapsed envelope is transiently contacted by the new nucleus and forms a membranous bridge (arrowhead). While the new nuclei enlarge, the old material is apparently absorbed. Bar: 2 μm. (F) Chromosomes within the nuclear extension condense while they migrate towards the daughter cell (arrowheads). Elapsed time is given in minutes, and neck is indicated by an asterisk. Green, ER: ER-YFP; red, H4, histone4-CFP. Bar: 2 μm. Supplementary movies for panels B, D and F are given on the EMBO website.
Chromosomes migrate through the nuclear extension while the envelope is still closed
In order to investigate the behaviour of chromosomes, we tagged them with histone4-CFP or histone4-GFP (green fluorescent protein) and observed the motility behaviour of chromosomes prior to mitosis at a stage where no spindle was formed. Double labelling of DNA (H4-CFP) and the NE (ERYFP) confirmed that chromosomes migrate up to 10 μm through the long extension of the NE into the bud (Figure 2F, neck indicted by asterisk). Chromosome condensation occurred while chromatin was actively migrating towards the tip of the extension (arrowheads in Figure 2F; see also Figure 3A). In animal cells, chromosome condensation is a characteristic of prophase (Alberts et al, 2002). Thus, we define the stage at which the nuclear extension is formed as prophase. However, in contrast to prophase in animals, no spindle is formed at this stage, but it appears shortly after the chromosomes have left the envelope (see below; Figure 5D). In order to gain further insight into the mechanism that underlies this chromosome motility, we made use of the nuclear RFP fusion protein (nRFP) and asked whether chromosomes migrate within the nuclear extension before or after NE opening. Surprisingly, chromosomes concentrated at the tip of the nuclear extension while nRFP was still trapped within the NE (Figure 3A; H4-GFP and nRFP; asterisk indicates tip of the nuclear extension), indicating that chromosome migration occurred within a closed envelope. The DNA usually migrated in bulky chromosome masses (not shown), but occasionally rapid motility of chromosomes at 0.26±0.03 μm/s was observed (Figure 3B, arrowheads mark H4-GFP-stained chromosomes; elapsed time is indicated in seconds). Again, this motility occurred while nRFP was enclosed by the intact NE (Figure 3C; nRFP image corresponds to Figure 3B; elapsed time is given in seconds).
Figure 3.
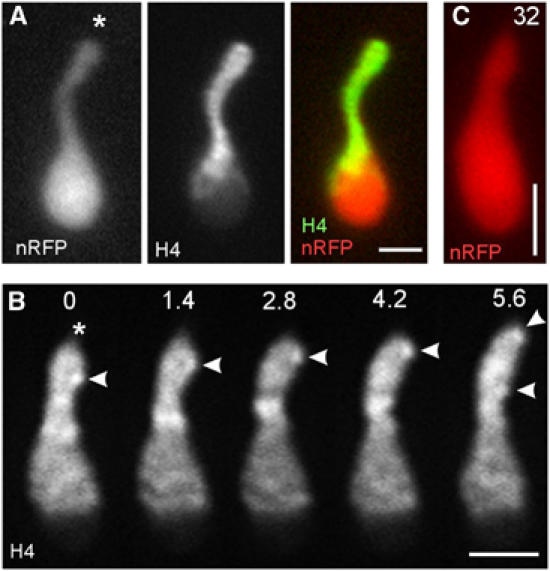
Motility of chromosomes within the nucleus. (A) Chromosomes concentrate at the tip of the nuclear extension, while nRFP is still trapped within the nucleus, indicating that chromosome migration occurs prior to breakage of the envelope. Asterisk indicates leading tip of the nucleus. nRFP: nRFP; H4: histone4-GFP. Bar: 2 μm. (B) Histone4-GFP stains patches within the nucleus that most likely correspond to chromosomes. These patches rapidly migrate towards the tip of the extension (asterisk) at ∼0.25 μm/s (arrowhead). Elapsed time is given in seconds. Bar: 2 μm. (C) nRFP is still present in the nucleus, shown in panel B, indicating that chromosome motility occurred within the closed nucleus. Compare elapsed time given in seconds with series shown in panel B. Bar: 2 μm. Supplementary movie for panel B is given on the EMBO website.
Figure 5.

MTs and formation of the nuclear extension. (A) In late G2 phase, nuclei are teardrop shaped. Fine extensions are formed that reach towards the bud. This motility does not result in nuclear displacement. Note that these extensions coalign with interphase MTs (red: nRFP; green: GFP-Tub1) and that their formation depends on dynein (Straube et al, 2001). Bars: 3 μm in series and 1 μm in merged image. (B) At the onset of mitosis, the SPB becomes active and nucleates MTs. Note that SPB-independent MTs are also found (see inset, lower MT has no contact with the SPB), indicating that two populations of MTs exist at the G2/M transition: Tub: GFP-Tub1, nRFP: nRFP. Bar: 5 μm. (C) In late G2 phase, the SPB (stained with γ-tubulin-GFP; Straube et al, 2003) is located at the side of the teardrop nucleus (nRFP). At the onset of prophase, the SPB migrates to the tip of the nucleus and pulls a long extension into the bud (bud neck indicated by asterisk). Elapsed time is given in minutes:seconds. Bar: 3 μm. (D) The SPB forms long astral MTs that reach into the daughter cell (indicated by ‘D') and the mother cell (indicated by ‘M'). After the envelope opens, indicated by the release of the nuclear RFP fusion protein, the SPB migrates into the bud and forms the metaphase spindle. Tub: GFP-Tub1; nRFP: nRFP. Bar: 3 μm. (E) At early prophase, MTs reach into the daughter cell (indicated by ‘D'), but are also tightly associated with the nucleus (indicated by ‘N') within the mother cell (indicated by ‘M', see arrows in overview). The plus-ends of astral MTs are in contact with the cortex (Tub, arrowheads), while the SPB (indicated by asterisk in upper image series) moves into the bud. Motility along the cortex coincides with the appearance of dynein at the tip of the MT (Dyn, arrowheads). Elapsed time is given in seconds. Bar: 3 μm. Supplementary movie for panels B and E is given on the EMBO website.
The envelope becomes permeable before it breaks at the tip of the nuclear extension
In early prophase, most nRFP was found in the prophase nuclei, while the nuclear membrane was apparently intact (Figure 4A, inset). However, at this stage, the RFP signal lost intensity (Figure 4A; nRFP, arrowheads indicate interphase nuclei; Figure 4C), indicating that the nucleus already released some of the fusion protein. In order to quantify this effect, we measured the total intensity of the RFP signal in prophase nuclei and compared these values with those of nuclei in growing interphase cells. These measurements confirmed that prophase nuclei lost about one-third of the RFP fusion protein (nRFP in G2 nuclei: 100±25%, n=13 cells; nRFP in prophase nuclei: 72±25%, n=11; different, P=0.0128). This indicates that the NE becomes partially permeable prior to nuclear envelope break-down (NEBD), an observation that confirms results from animal cells, where nuclear pore disassembly results in a partial permeability of the NE prior to NEBD (Terasaki et al, 2001; Lenart et al, 2003). A more detailed analysis revealed that small amounts of nRFP were continuously released from the nucleus until a more rapid decrease of the RFP signal occurred (Figure 4B, arrowhead marks the neck constriction; elapsed time is given in seconds; Figure 4C). This is most likely due to breakage of the envelope at the front of the extension. Indeed, false colouring of the nRFP signal visualized this release of the fusion protein into the daughter cell (Figure 4C, arrowhead at 54 s; daughter indicated by ‘D', mother indicated by ‘M'; elapsed time is given in seconds; Figure 4E, arrow). Shortly after, nRFP appeared in the mother cell (Figure 4D, 94 s; Figure 4D) and the NE collapsed (not shown). These data indicate that the envelope opens at the front of nuclear extension, a conclusion that supports our previous observations on ER-YFP-labelled envelopes (arrowheads in Figure 2B; see above).
Figure 4.
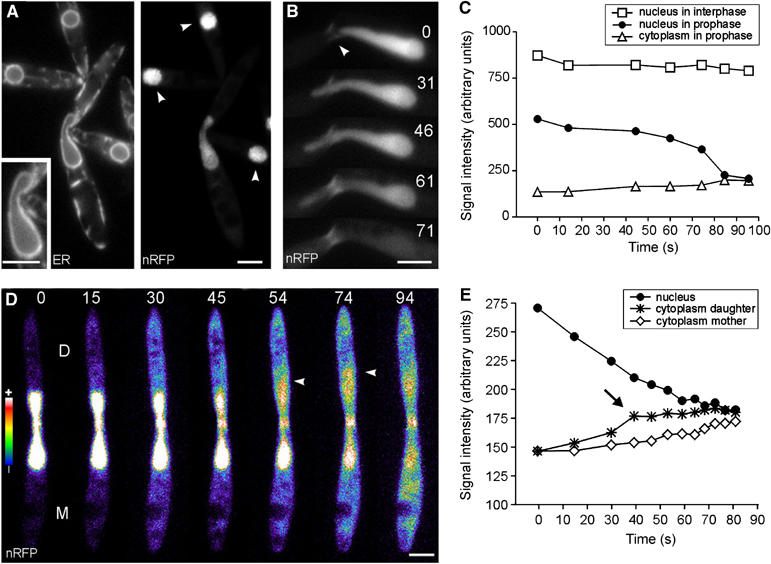
Release of a nucleus-targeted red fluorescent protein. (A) Elongated nuclei that reach into the daughter cell contained ∼30% less nRFP, although the NE appeared to be intact (inset). Arrowheads indicate neighbouring interphase nuclei. ER: ER-GFP; nRFP: nRFP. Bar: 3 μm. (B) A nuclear extension reaches into the daughter cell and rapidly releases the RFP fusion protein (nRFP). For unknown reasons, the neck also shows fluorescent staining (arrowhead). Bar: 3 μm. (C) Kinetic profile of nRFP fluorescence intensity in an interphase nucleus (square), in the basal region of the prophase nucleus (filled circles) and in the cytoplasm of the same cell (triangle). Note that the nucleus is permeable for nRFP at t=0 s (compare with panel A). While the nRFP signal in interphase remains fairly constant, the fusion protein continuously leaks out of the prophase nucleus until it is rapidly released at t=60 s. (D) Shortly before breakage, the envelope becomes permeable and releases some RFP fusion protein into the daughter cell (D; nRFP intensity is given in pseudocolours: blue indicates weak signal intensity, yellow to white is indicative of a strong signal). Rapid release of nRFP occurs with a first wave into the daughter cell (arrowhead at 54 and 74 s) and subsequent diffusion into the mother cell (M; arrowhead at 94 s). Elapsed time is given in seconds. Bar: 3 μm. (E) At t=0 s, the intensity of cytoplasmic nRFP in the daughter cell (star) is the same as in the mother cell (rhombus). However, in late stages, a peak of nRFP is found at the front of the extension (arrrow), indicating that the breakage of the NE occurs at the SPBs within the daughter cell. Supplementary movie for panel D is given on the EMBO website.
Astral microtubules and dynein pull the spindle pole body and the attached nuclear membrane into the bud
We next asked for the molecular mechanism by which the extension of the nucleus is pulled into the bud. It was previously reported that interphase MTs are nucleated at cytoplasmic MTOCs within the neck region, whereas the SPB is not active in interphase (O'Donell, 1992; Straube et al, 2003). At this stage, we observed the formation of long and thin extensions that coaligned with MTs and progressed into the daughter cell, but did not persist and were rapidly retracted (Figure 5A). This motility did not result in any nuclear displacement. In order to investigate whether these forces are applied directly on the nuclear membrane or on the nucleus-associated SBP, we visualized the SPB by fluorescent γ-tubulin fusion proteins (Straube et al, 2003) in a strain that expressed nRFP (Figure 5B). Surprisingly, the transient extensions were formed independently of the SPB (Figure 5B, inset; arrowhead indicates an extension), indicating that forces are indeed exerted on the envelope itself. At onset of mitosis, the SPB migrated from a lateral position to the tip of the teardrop-shaped nucleus (Figure 5B; elapsed time is given in minutes:seconds), and started to nucleate astral MTs (Figure 5C; note that SPB-independent MTs are also visible, inset). The long astral MTs made by the SPB reached far into the daughter cell, supporting the notion that they are connected to migration of the SPBs and attached chromosomes out of the envelope and into the bud (Figure 5D, nRFP and GFP-α-tubulin; elapsed time is given in minutes:seconds).
In other cell systems, it was shown that dynein locates at MT plus-ends (Han et al, 2001; Lee et al, 2003; Sheeman et al, 2003) and supports spindle motility by pulling on astral MTs (reviewed in Bloom, 2001). Indeed, premitotic nuclear migration into the bud depends on cytoplasmic dynein in U. maydis (Straube et al, 2001). In order to gain further proof for the role of dynein in spindle motility, we fused 2 × GFP to the N-terminus of the endogenous dyn1, which encodes the N-terminal part of the U. maydis dynein heavy chain (Straube et al, 2001), and expressed the fusion protein in a strain that contained RFP-labelled MTs. In prophase, astral MTs emanated from the nucleus (Figure 5E; ‘D' indicates daughter, ‘M' indicates mother cell, ‘N' marks the nucleus), and long MTs reached from the SPB into the bud and also the mother cell (indicated by arrows). MT tips often contacted the cortex of the bud (Tub1 series; MT tip indicated by arrowhead), and dynein localized at these plus-ends (Dyn, lower series). Dynein appeared at the end of the MT at the moment when the SPB began to move into the bud (Dyn; arrowheads; elapsed time is given in seconds). This localization and the previously demonstrated need for dynein activity for migration of DNA into the bud (Straube et al, 2001) is most consistent with the role of dynein in cortical sliding of astral MTs and the SPB into the daughter cell.
Nuclear envelope removal requires microtubules and dynein
Our data demonstrate that nuclear removal is coupled to the formation of a long nuclear extension that breaks within the bud, while the SPB and attached chromosomes migrate into the daughter cell. This suggests that the tip of the elongating nucleus has to reach into the daughter cell in order to trigger the opening of the envelope. To test this hypothesis, we impaired nuclear migration and analysed the efficiency of NEBD under these conditions. In untreated control cells, mitotic spindles were always found in the daughter cell and the NE was completely removed in all cases (Figure 6A, ‘Control', Figure 6B). We previously observed that cells inhibited in tip growth continued their cell cycle and formed spindles in the mother cell (our own unpublished results). In order to minimize the effect on the metabolism, we stopped polar cell growth by inhibition of cell wall synthesis with Calcofluor white, an inhibitor of chitin synthesis (Herth, 1980). Indeed, treatment with this inhibitor at 1–10 μg/ml impaired bud formation and often led to misplaced mitotic nuclei (Figure 6A, ‘growth inhibited'). In agreement with our model, all misplaced spindles were still surrounded by an envelope (Figure 6A, ‘growth inhibited'; Figure 6B, ‘Calc'), while all spindles that were correctly positioned in the bud had the envelope completely removed (Figure 6A, ‘growth inhibited'; Figure 6B, ‘Calc'). Similar results were obtained in other growth-defective mutants (not shown), indicating that the position relative to the neck region is crucial for envelope removal. In other words, in order to remove the envelope, the SPBs need to pass the neck region.
Figure 6.
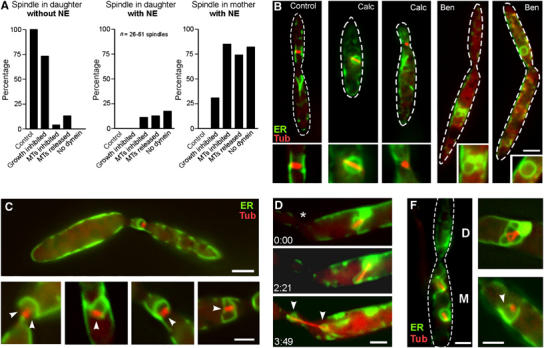
Nuclear migration and NE removal. (A) Quantitative analysis of the effect of nuclear position on NEBD. In control cells, spindles were located in the bud and were never surrounded by an envelope (‘spindle in daughter, without NE'). Treatment with Calcofluor, an inhibitor of chitin synthesis (Herth, 1980), impaired growth and resulted in the formation of spindles within the mother cell. All of the misplaced spindles were surrounded by an apparently intact NE (‘spindle in mother, with NE'). Disruption of MTs by benomyl (MTs inhibited) led to misplaced nuclei that almost always were surrounded by an envelope. Washout of benomyl (MTs released) initiated spindle formation within the closed and misplaced nuclei. No open mitosis was found in dynein mutants. Note that partially opened NEs were counted as ‘with NE'. For further details, see Table I. (B) At standard growth conditions, metaphase spindles were found in the daughter cell and the NE was removed (‘Control'). Treatment with Calcofluor, impaired growth and resulted in misplaced mitotic nuclei with an intact NE (‘Calc'). However, nuclei that had reached the bud did remove the NE. Treatment with the anti-MT drug benomyl (20 μM) impaired nuclear migration and led to condensed nuclei with closed envelopes (‘Ben'). In the absence of MTs, even correctly positioned nuclei were enclosed (‘Ben'). MTs are stained with CFP-Tub1, and NEs are marked with ER-YFP. Bar: 3 μm. (C) Washout of benomyl resulted in the immediate formation of spindles within the closed nuclei. Mitotic nuclei located at the border between mother and daughter cells, as well as in the daughter cell, often showed a partial opening of the NE (arrows indicate gaps in the envelope). Bars: 3 μm in overview and 1 μm in details. (D) After release from benomyl, nuclei that are positioned in the mother cell do not open their envelope. However, the elongating spindle finally disrupted the envelope while it was migrating towards the neck (indicated by asterisk). Note that the condensed chromosomes were still surrounded by small envelopes (arrowheads). Elapsed time is given in minutes:seconds. Spindles are labelled by CFP-Tub1, and envelope are labelled by ER-YFP. Bar: 2 μm. (F) In the absence of dynein, nuclear migration is impaired and nuclei remained in the mother cell (indicated by ‘M', daughter cell is indicated by ‘D'). An apparently intact NE surrounded these misplaced nuclei. Many, partially opened mitotic nuclei were found (see Table I). Red: Spindles labelled by CFP-Tub1, NE marked with ER-YFP. Bar: 2 μm.
Nuclear migration is a MT-dependent process in U. maydis (Steinberg et al, 2001), and MTs facilitate NEBD in animal cells (Beaudouin et al, 2002; Salina et al, 2002). We therefore analysed the role of MTs in NE removal in U. maydis. Disruption of MTs by the fungicide benomyl impaired nuclear migration and led to small and condensed nuclei that remained in the mother cell. In agreement with the results described above, an apparently intact NE always surrounded misplaced chromosomes, although no spindle was formed (Figure 6A, ‘MTs inhibited'; Figure 6B, ‘Ben'). Surprisingly, in the absence of MTs, apparently intact NEs were found even around correctly positioned nuclei (Figure 6A, ‘MTs inhibited'; Figure 6B, ‘Ben'). Only a small number successfully removed the envelope, indicating that these cells were arrested by benomyl in later mitotic stages, when envelope removal was already completed. This demonstrates that MTs are not only key players for the transport of the nucleus into the bud, but are also crucial for the opening process itself. The release of benomyl-treated cells from the mitotic block led to the immediate formation of spindles. In agreement with the position effect described above, nuclei that were correctly positioned in the neck region now partially opened the envelope (Figure 6A, ‘MTs released'; Figure 6C, arrowheads indicate gaps in the NE), but the spindle usually failed to migrate out of the old envelope. In contrast, nuclei within the mother cells did not open the envelope, but proceeded with spindle elongation until envelope rupture occurred (Figure 6D, neck indicated by asterisk). This led to two small envelopes around the spindle poles (Figure 6D, arrowheads). Interestingly, spindle migration during anaphase resulted in neck passage of one nucleus, indicating that astral MTs had contact with the cortex. These data suggest that MTs support NE removal in two ways. Firstly, they mediate migration of the SPBs into the daughter cell, which is, according to the proposed position effect, a prerequisite for successful NE removal. Secondly, MTs have additional functions in the breakage of the NE.
Next, we attempted to further elucidate the role of cytoplasmic dynein in NE removal in U. maydis. In agreement with its role in premitotic nuclear migration, depletion of dynein in a conditional dynein mutant (Straube et al, 2001) resulted in mitotic nuclei within the mother cell (Figure 6F; ‘D' indicates daughter, ‘M' indicates mother cell; Table I). Consistent with the results described above, an NE always surrounded misplaced mitotic spindles, although some envelopes were often not completely closed (Figure 6A, ‘no dynein', Figure 6F, arrowhead). In some cases, where the spindle was correctly positioned in the daughter cell, the envelope was opened (Table I). However, spindles never left the partially opened envelopes (Figure 6F). This indicates that dynein has a minor role in the initial opening of the membrane, but is crucial for the separation of the spindle and the NE. This again supports the notion that dynein-based SPB and chromosome motility into the bud is essential for envelope removal.
Table 1.
Envelope removal after growth inhibition, MT disruption and dynein inactivation
| Spindle in daughtera |
Spindle in mothera,b |
Number of spindles |
||||
|---|---|---|---|---|---|---|
| Without envelope | Partially opened envelope | Closed envelope | Partially opened envelope | Closed envelope | ||
| Control | 51/100 | 0/0 | 0/0 | 0/0 | 0/0 | 51 |
| Growth inhibitedc | 19/70.4 | 0/0 | 0/0 | 1/3.7 | 7/25.9 | 27 |
| MTs inhibitedd | 2/3.3 | 0/0 | 7/11.5 | 1/1.6 | 51/83.6 | 61 |
| MTs releasede | 4/12.9 | 3/9.7 | 1/3.2 | 6/19.4 | 17/54.8 | 31 |
| No dynein | 0/0 | 3/8.8 | 3/8.8 | 7/20.6 | 21/61.8 | 34 |
|
Δras3 |
21/26.6 |
31/39.2 |
17/21.5 |
4/5.1 |
6/7.6 |
79f |
| aValues are given as ‘absolute number/percentage'. | ||||||
| bIncludes spindles in the neck region. | ||||||
| cInhibition by 1–10 μM Calcofluor white for ∼2 h. | ||||||
| dInhibition by 20 μM benomyl for ∼4 h. | ||||||
| eImmediately after washout of the drug by fresh medium. | ||||||
| fFrom three independent transformants. | ||||||
MEN signalling participates in removal of the envelope
Our results demonstrated that SPBs have to migrate into the bud in order to remove the NE in U. maydis. This suggested that spatial cues within the bud might recognize the position of the SPB and trigger envelope removal. In budding yeast, mitotic exit is regulated by the activation of the Ras-like GTPase Tem1p, a central component of the MEN cascade that localizes at the leading SPB and becomes activated when the SPB reaches into the bud (reviewed in Pereira and Schiebel, 2001). U. maydis contains a Tem1p-like Ras-GTPase (UmRas3; Figure 7A; accession number: BN000729) that shows 43% identity with Tem1p. In order to check whether MEN signalling participates in envelope removal, we deleted ras3 (Figure 7B) in a strain that expressed CFP-α-tubulin and ER-YFP and analysed the effect on mitosis. Δras3 mutants neither showed defects in cell morphology or cytokinesis (Figure 7C), nor were they significantly affected in spindle positioning (Figure 7C and F). However, ∼60% of all correctly positioned spindles were still surrounded by an NE (Figure 7C, D and F) that was often partially opened (Figure 7E, arrowhead; Table I). In addition, many mother cells contained a membrane accumulation (asterisk in Figure 7C and D) that was in contact with the NE (Figure 7D, arrowhead), suggesting that parts of the envelope are still left behind in Δras3 mutants.
Figure 7.
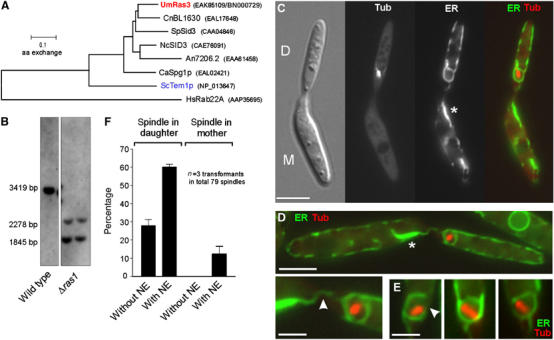
The role of UmRas3, a Temp1-like GTPase in ‘open mitosis'. (A) The genome of U. maydis contains a Ras3-like GTPase that has remarkable similarity to a predicted gene in the pathogenic basidiomycete C. neoformans. The tree is based on a comparison of ∼190 amino acids of the core region of all predicted proteins. Um: Ustilago maydis; Cn: Cryptococcus neoformans; Sp: Schizosaccharomyces pombe; Nc: Neurospora crassa; An: Aspergillus nidulans; Ca: Candida albicans; Sc: Saccharomyces cerevisiae; Hs: Homo sapiens. Accession numbers are given in parentheses. (B) Southern blot confirming that ras3 is deleted. After digestion with BsiWI, wild-type genomic DNA contains a fragment of 3419 bp that is split into smaller fragments in ras3 null mutants. (C) Δras3 mutants show defects neither in morphology nor in cytokinesis. Spindles are positioned correctly within the bud (Tub: CFP-α-tubulin), but are often surrounded by ER (ER-YFP), suggesting that they fail to remove the envelope. Membrane accumulations are found in the mother cell (asterisk), indicating that parts of the envelope are left behind. Bar: 5 μm. (D) The NE often has contact with the membrane accumulation in the mother cell (arrowhead in detail). Bars: 5 and 2 μm. (E) Correctly positioned spindles are often surrounded by a partially opened envelope (arrowhead; see also Table I). (F) Quantitative analysis reveals that Δras3 mutants have only minor defects in spindle positioning (‘spindle in mother'), but ∼60% of the spindle in the daughter bud are surrounded by ER (‘Spindle in daughter, with NE'). Note that all spindles in control strains are in the first category (‘Spindle in daughter, without NE'; see Figure 6A).
Discussion
The mechanism of NEBD differs between animals and fungi
It was suggested that the removal of the NE is required to allow contact between the chromosomal kinetochores and the MTs emanating from the extranuclear centrosomes (Lippincott-Schwartz, 2002). A similar situation is found in the basidiomycete U. maydis. In this organism, the SPB is attached to the outer membrane of the NE (O'Donell, 1992). Therefore, astral MTs that are nucleated in prophase (O'Donell, 1992; Straube et al, 2003) have to overcome the NE barrier to reach the kinetochores. In analogy to animal cells, removal of the fungal NE might therefore be a necessary consequence of the SPB organization. However, in contrast to animal cells, fungal envelope removal does not involve the fragmentation of the nuclear membranes. Instead, the envelope is stripped off and deposited within the mother cell until it gets recycled in telophase. This fundamental difference suggests that the mechanism of removal of the fungal and animal NE evolved independently. This notion is further supported by the role of the cytoskeleton in this process. In animal cells, astral MTs embrace the nucleus and dynein pulls the envelope towards the minus-ends of the MTs at the centrosomes. This activity generates a tension, which results in envelope rupture at the opposite side of the centrosomes, but also removes fragments of the disassembling envelope (Beaudouin et al, 2002; Salina et al, 2002). In U. maydis, MTs and dynein also participate in the breakage of the nuclear membranes. However, the nucleus breaks directly at the SPB, where chromosomes exit the envelope. Dynein appears to pull on the SPB and stretches the nucleus into the bud. Thus, breakage at the SPB might be a consequence of overstretching the envelope. Such a model is supported by the fact that lamins that stabilize the NE in animal cells (overview in Moir and Spann, 2001) are not found in fungi. Consequently, fungal NEs might be more flexible, which could favour a tension-based mechanism of envelope breakage. However, in dynein mutants, the envelopes often open partially but are not removed from the spindle. This indicates that breakage of the envelope might also happen independent of dynein-mediated pulling forces. Therefore, we consider it likely that MTs in U. maydis participate in tension-independent mechanism to break up the NE, while the removal might involve dynein-mediated motility of the SPB and attached chromosomes.
Dynein-dependent nuclear migration is essential for open mitosis in U. maydis
In interphase, thin and transient NE extensions are pulled towards the growing bud (see Figure 5A). In dynein mutants, these thin extensions were not observed (Straube et al, 2001; Wedlich-Söldner et al, 2002a). These extensions were formed at the front of nuclei in G2, but are independent of the SPB that is inactive and located at the lateral region of the nucleus at this stage (Figure 8). This indicates that dynein directly pulls on the NE, a mechanism that was also postulated for animal cells at the onset of NEBD (Salina et al, 2002). However, dynein-dependent formation of the thin extensions does not result in obvious net movement of the nucleus. In contrast, mitosis is initiated by the activation of the SPB that migrates to the front of the nucleus (Figure 8). Our data argue that dynein pulls on these SPB-nucleated cytoplasmic MTs and thereby drags a long nuclear extension into the daughter cell. After chromosomes migrate within the closed nucleus towards the SPB, the envelope eventually breaks and dynein pulls the SPB and attached chromosomes into the bud (Figure 8). The empty envelope collapses and is left behind in the mother cell until its recycling during telophase. In our model, dynein plays a key role in NE removal. Two major lines of evidence support this notion. Firstly, we show here that SPB migration into the bud is a prerequisite for envelope removal, and this premitotic nuclear migration is based on dynein (Straube et al, 2001). In addition, we report that the NE is never removed in dynein mutants, although the envelope was often opened. Secondly, astral MTs that emanate from the SPBs reach into the bud and apparently slide along the cortex. Dynein appeared at the plus-ends of MTs, which confirms previous findings in other fungi (Han et al, 2001; Lee et al, 2003; Sheeman et al, 2003). Interestingly, dynein appeared during the time when SPB movement occurred, indicating that dynein participates in cortical MT sliding and thereby mediates SPB and chromosome migration out of the envelope. This notion is very much supported by recent findings in S. cerevisiae, where it was shown that astral MTs deliver plus-end-located dynein to cortical anchors (Lee et al, 2003; Sheeman et al, 2003). This results in cortical MT sliding and correct positioning of the spindle (Adames and Cooper, 2000; Yeh et al, 2000; overview in Bloom, 2001).
Figure 8.
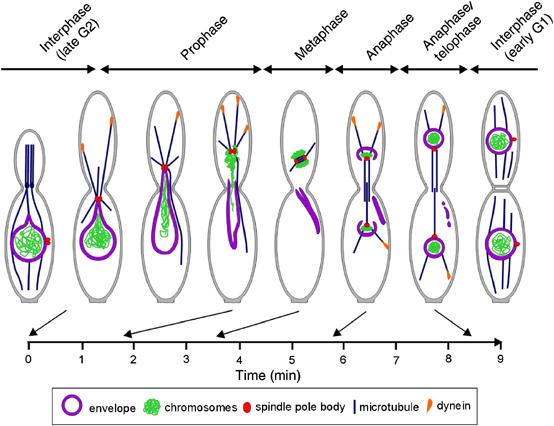
Model of the role of the cytoskeleton in NEBD in U. maydis. Interphase: Nuclei are located in the mother cell while a polar bud is formed (G2). The SPB is inactive and MTs are nucleated at polar MTOCs in the bud neck. At this stage, forces are exerted on the nucleus that pull membrane extensions towards the bud, without displacing the nucleus. Prophase: The SPB becomes actived, migrates to the front of the nucleus and nucleates MTs that reach into the bud. Dynein at the plus-end of these MTs appears to pull a nuclear extension into the bud. At this time, chromosomes condense and migrate through the extension into the daughter. The NE becomes permeable and finally breaks at the SPB. Chromosomes leave the old envelope and gather around the SPB, while the old envelope is left behind. Metaphase: A short spindle is formed within the daughter cell, which is surrounded by chromosomes. Anaphase: The spindle elongates and pushes chromosomes back into the mother. New envelopes appear at the distal poles of the spindle. Telophase: The NE closes up and the DNA decondenses. The old envelope is resorbed. Interphase (G1): Finally, the SPB becomes inactivated, new MT arrays appear and septa are formed. After cell separation and DNA duplication, a new bud is formed at the opposite cell pole (Wedlich-Söldner et al, 2002a). An estimated time scale for the mitotic steps is given below.
In summary, we consider it most likely that dynein-mediated SBP motility first extends the nucleus through the bud and later pulls chromosomes out of the old envelope. However, the interesting question remains of which counterforce holds the old envelope back in the mother cell. Occasionally, we observed the formation of transient and fine nuclear extensions at the rear end of the interphase nucleus (our unpublished observations). MT plus-ends are located at distal ends of the mother cell (Steinberg et al, 2001; Straube et al, 2003), thus indicating that kinesin motors pull on the NE. It remains to be seen whether kinesins counteract dynein in the removal of the envelope during the ‘open' mitosis of U. maydis.
MEN signalling participates in envelope removal
The results presented here indicate that the SPBs have to reach the bud in order to remove the envelope, suggesting that spatial cues in the bud could trigger envelope removal. In budding yeast, it was shown that Tem1p, a Ras-like GTPase that is a central component of the MEN (Shirayama et al, 1994), preferentially localizes at the leading SPB (Bardin et al, 2000; Pereira et al, 2000; Molk et al, 2004). In anaphase, the SPB reaches the bud and Tem1p becomes activated, which triggers the MEN signalling cascade and finally leads to mitotic exit (summarized in Pereira and Schiebel, 2001). The genome of U. maydis contains compounds of this signalling cascade, including an Lte1p-like putative GDP/GTP exchange factor (accession number: EAK85923) and a highly conserved Tem1p-like Ras-GTPase (termed Ras3; accession number: EAK85109/BN000729; see Figure 7A). Thus, it was tempting to speculate that MEN signalling could participate in removing the NE in U. maydis. Indeed we found that Δras3 mutants often undergo a ‘closed' mitosis even when the spindle is correctly positioned within the bud. However, most of the envelopes were partially open (see Table I), indicating that Ras3 is not required for envelope breakage. This concides with the observation that 50–60% of the misplaced spindles in dynein mutants or benomyl-treated cells were surrounded by a partially opened envelope (see Table I). Thus, breakage of the envelope is apparently independent of spindle localization, while envelope removal is based on dynein-dependent nuclear migration and Ras3. It is presently unknown how MEN signalling regulates NEBD in U. maydis. Spindle positioning was not impaired in ras3 null mutants, which argues against a regulation of dynein activity by Ras3. Further studies are needed to elucidate the role of MEN signalling in these early steps of mitosis in U. maydis.
Conclusions
We show here that dynein-dependent migration of the SPB and attached chromosomes participates in the removal of the NE in U. maydis. Premitotic nuclear migration into the bud is typical for basidiomycete yeasts, and these organisms are also reported to undergo an ‘open mitosis'. Therefore, we consider it likely that nuclear migration and NE removal are generally linked in basidiomycete fungi. Both dynein-dependent nuclear migration and SPB-based MEN signalling exist in ascomycete fungi such as budding yeast. Thus, it appears that basidiomycetes made use of already existing regulatory and structural compounds in order to remove the envelope. Therefore, it is not surprising that the mechanism of NE removal in animals shows some fundamental differences. However, the interesting question remains of why an ‘open mitosis' evolved in different kingdoms. It was suggested that for mammalian cells NEBD allowed contact between chromosomes and the MTs formed at extranuclear centrosomes (Lippincott-Schwartz, 2002). This explanation might also be valid for U. maydis cells, where the SPB was observed to be associated with the outside of meiotic nuclei in prophase (O'Donnell and McLaughlin, 1984). Thus, the structure of the perinuclear MTOC may dictate the need for the removal of the NE. However, we report that misplaced mitotic nuclei underwent a ‘closed' mitosis, which questions this simple model. Further studies on the model fungus U. maydis will help to further elucidate the importance and molecular mechanisms of nuclear dynamics in higher eukaryotic cells.
Materials and methods
Sequence analyses
UmH4 was identified in the genomic sequence of U. maydis (www.genome.wi.mit.edu/annotation/fungi/ustilago_maydis/"). The accession number is AJ646896. Dendrograms were generated using ClustalX (Thompson et al, 1997) and MEGA 2.1 (Kumar et al, 2001). UmRas3 was identified as a hypothetical protein (UM04012.1; EAK85109); the new accession number is BN000729.
Strains and plasmids
The ER was labelled by plasmids carrying eGFP, CFP or YFP (Clontech, Palo Alto, USA) fused to the N-terminal signal peptide from rat calreticulin and the retention signal HDEL at the C-terminus (Wedlich-Söldner et al, 2002a). Chromosomes were stained with UmH4 C-terminally fused to CFP or GFP. nRFP consists of three copies of mRFP (Campbell et al, 2002) fused to a nuclear localization signal (Straube et al, 2001). α-Tubulin and γ-tubulin fusion constructs were described elsewhere (Steinberg et al, 2001; Straube et al, 2003). In Δras1 strains, the complete open reading frame was deleted by homologous recombination. For details, see Table II. Duration of cell cycle stages was estimated by counting cells in all stages in strain FB1GT grown to OD600=0.3–0.4 in glucose-containing complete medium at 28°C. Duration of stages was calculated by dividing the relative abundance by the doubling time of 2.1 h (our own unpublished results).
Table 2.
Strains and plasmids used in this study
| Nomenclature | Genotype | Reference |
|---|---|---|
| FB1 | a1b1 | Banuett and Herskowitz (1989) |
| FB2 | a2b2 | Banuett and Herskowitz (1989) |
| FB1GT | a1b2/pGFPTub1 | Steinberg et al (2001) |
| FB1YT1-H4C | a1b1/pYFPTub1/pH4CFP | This study |
| FB2nRFP3 | a2b2/pC_NLS3xRFP | This study |
| FB1ERY-CT1 | a1b1/pERYFP/pCFPTub1 | This study |
| FB2EG | a2b2/pERGFP | Adamikova et al (2004) |
| FB1ERY-H4C | a1b1/pERYFP/pH4CFP | This study |
| FB2nRFP-H4G | a2b2/pC_NLS3xRFP/pH4GFP | This study |
| FB2nRFP-ERG | a2b2/pC_NLS3xRFP/pN_ERGFP | This study |
| FB2nRFP-GT1 | a2b2/pH_NLS3xRFP/pGFPTub1 | This study |
| FB2nRFP-T2G | a2b2/pH_NLS3xRFP/pTub2GFP | This study |
| FB2G2Dyn1-RT1 | a2b2Pdyn1-2xgfp-dyn1, hygR/pRFPTub1 | This study |
| FB1rDyn2 | a1b1Pcrg-dyn2, bleR | Wedlich-Söldner et al (2002a) |
| FB1rDyn2ERY-CT1 | a1b1Pcrg-dyn2, bleR/pERYFP/pCFPTub1 | This study |
| FB1ΔRas1-ERY-CT1 | a1b1Δras3R/pERYFP/pCFPTub1 | This study |
| pYFPTub1 | Potef-yfp-tub1, cbxR | This study |
| pH4CFP | Potef-h4-cfp, hygR | This study |
| pH_NLS3xRFP | Potef-gal4s-mrfp-mrfp-mrfp, hygR | This study |
| pC_NLS3xRFP | Potef-gal4s-mrfp-mrfp-mrfp, cbxR | This study |
| PERYFP | Potef-cals-yfp-HDEL, cbxR | This study |
| pCFPTub1 | Potef-cfp-tub1, hygR | Wedlich-Söldner et al (2002b) |
| PERGFP | Potef-cals-gfp-HDEL, cbxR | Wedlich-Söldner et al (2002a) |
| pN_ERGFP | Potef-cals-gfp-HDEL, natR | This study |
| pH4GFP | Potef-h4-gfp, hygR | This study |
| pGFPTub1 | Potef-gfp-tub1, cbxR | Steinberg et al (2001) |
| pTub2GFP | Potef-tub2-gfp, cbxR | Straube et al (2003) |
| pRFPTub1 |
Potef-mrfp-tub1, cbxR |
This study |
| a, b, mating type genes; P, promoter; -, fusion; hygR, hygromycin resistance; bleR, phleomycin resistance; cbxR, carboxin resistance; /, ectopically integrated; tub1, α-tubulin; tub2, γ-tubulin; h4, histone 4; Dyn1/Dyn2, N- and C-terminal half of the dynein heavy chain; gfp, enhanced green flourescent protein; yfp/cfp, yellow-shifted/cyan-shifted fluorescent protein; mrfp, monomeric red fluorecent protein; calS, signal sequence of calreticulin from rabbit (nt 1–51); HDEL, ER retention signal; gal4s, nuclear localization signal of the GAL-4 DNA binding domain from pC-ACT1 (Clontech). | ||
Light microscopy and image analysis
Logarithmically growing cells were embedded in low melt agarose and observed using a Zeiss Axiophot microscope (Oberkochen, Germany) and filter sets for YFP (BP500/20, FT515, BP535/30) and CFP (BP436, FT455, BP480–500), and standard filters for FITC and rhodamine. Image aquisition were carried out using a CoolSNAP-HQ CCD camera (Photometrics, Tucson, AZ, USA) controlled by the imaging software MetaMorph (Universal Imaging, Downing Town, USA). 3D analysis was performed as described (Straube et al, 2003). Image scaling and processing was carried out with Metamorph and Adobe Photoshop.
Supplementary Material
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Movie 7
Acknowledgments
We thank Daniela Assmann, Petra Happel and Isabel Manns for technical support and the Bayer CropScience AG for providing the genomic sequence of UmH4. Regine Kahmann and I Brent Heath are acknowledged for helpful comments on the manuscript. We also wish to acknowledge Robert Bauer, Johannes Wöstenmeyer, Kerstin Voigt and Klaus Lengeler for helpful discussion. Finally, we wish to thank the anonymous referees for excellent suggestions. This work was supported by a grant of the Deutsche Forschungsgemeinschaft to GS (STE 799/4).
References
- Adames NR, Cooper JA (2000) Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol 149: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamikova L, Straube A, Schulz I, Steinberg G (2004) Calcium signaling is involved in dynein-dependent microtubule organization. Mol Biol Cell 15: 1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) Molecular Biology of the Cell. 4th edn. New York: Garland Science [Google Scholar]
- Banuett F, Herskowitz I (1989) Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci USA 86: 5878–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A (2000) A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102: 21–31 [DOI] [PubMed] [Google Scholar]
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J (2002) Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108: 83–96 [DOI] [PubMed] [Google Scholar]
- Bloom K (2001) Nuclear migration: cortical anchors for cytoplasmic dynein. Curr Biol 11: R326–R329 [DOI] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gant TM, Wilson KL (1997) Nuclear assembly. Annu Rev Cell Dev Biol 13: 669–695 [DOI] [PubMed] [Google Scholar]
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X (2001) The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr Biol 11: 719–724 [DOI] [PubMed] [Google Scholar]
- Heath IB (1980) Variant mitosis in lower eukaryotes: indicators of the evolution of mitosis? Int Rev Cytol 64: 1–80 [DOI] [PubMed] [Google Scholar]
- Herth W (1980) Calcofluor white and Congo red inhibit chitin microfibril assembly of Poterioochromonas: evidence for a gap between polymerization and microfibril formation. J Cell Biol 887: 442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R (1965) Induced mitotic crossing-over in relation to genetic replication in asynchronously dividing cells of Ustilago maydis. Genet Res 6: 104–120 [DOI] [PubMed] [Google Scholar]
- Kojic M, Kostrub CF, Buchman AR, Holloman WK (2002) BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell 10: 683–691 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245 [DOI] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA (2003) The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol 160: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P, Rabut G, Daigle N, Hand AR, Terasaki M, Ellenberg J (2003) Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J Cell Biol 160: 1055–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J (2002) Cell biology: ripping up the nuclear envelope. Nature 416: 31–32 [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Tanaka S, Watanabe S (1987) Ultrastructure of the mitotic apparatus in Cryptococcus neoformans. J Med Vet Mycol 25: 223–233 [PubMed] [Google Scholar]
- Moir RD, Spann TP (2001) The structure and function of nuclear lamins: implication for disease. Cell Mol Life Sci 58: 1748–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk JN, Schulyler SC, Liu JY, Evans JG, Salmon ED, Pellman D, Bloom K (2004) The differential roles of budding yeast Temp1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol Biol Cell 15: 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donell K (1992) Ultrastructure of meiosis and the spindle pole body in freeze-substituted basidia of the smut fungi Ustilago maydis and Ustilago avenae. Can J Bot 70: 629–638 [Google Scholar]
- O'Donnell KL, McLaughlin DJ (1984) Postmeiotic mitosis, basidiospore development, and septation in Ustilago maydis. Mycologia 76: 486–502 [Google Scholar]
- Pereira G, Schiebel E (2001) The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr Opin Cell Biol 13: 762–769 [DOI] [PubMed] [Google Scholar]
- Pereira G, Höfken T, Grindlay J, Manson C, Schiebel E (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell 6: 1–10 [PubMed] [Google Scholar]
- Poon NH, Day AW (1976) Somatic nuclear division in the sporidia of Ustilago violacea. III. Ultrastructural observations. Can J Microbiol 22: 495–506 [DOI] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B (2002) Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108: 97–107 [DOI] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D (2003) Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol 13: 364–372 [DOI] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Toh EA (1994) The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Biol Cell 14: 7476–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snetselaar KM, McCann MP (1997) Using microdensitometry to correlate cell morphology with the nuclear cycle in Ustilago maydis. Mycologia 89: 689–697 [Google Scholar]
- Steinberg G, Wedlich-Söldner R, Brill M, Schulz I (2001) Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J Cell Sci 114: 609–622 [DOI] [PubMed] [Google Scholar]
- Straube A, Brill M, Oakley BR, Horio T, Steinberg G (2003) Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol Biol Cell 14: 642–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A, Enard W, Berner A, Wedlich-Soldner R, Kahmann R, Steinberg G (2001) A split motor domain in a cytoplasmic dynein. EMBO J 20: 5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M, Campagnola P, Rolls MM, Stein PA, Ellenberg J, Hinkle B, Slepchenko B (2001) A new model for nuclear envelope breakdown. Mol Biol Cell 12: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R, Schulz I, Straube A, Steinberg G (2002a) Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol Biol Cell 13: 965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R, Straube A, Friedrich MW, Steinberg G (2002b) A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J 21: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K (2000) Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell 11: 3949–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Movie 7


