Abstract
Genetically engineered mouse models (GEMM) have fundamentally changed how ovarian cancer etiology, early detection, and treatment are understood. MYC, an oncogene, is amongst the most amplified genes in high-grade serous ovarian cancer (HGSOC), but it has not previously been utilized to drive HGSOC GEMMs. We coupled Myc and dominant-negative mutant p53-R270H with a fallopian tube epithelium (FTE)-specific promoter Ovgp1 to generate a new GEMM of HGSOC. Female mice developed lethal cancer at an average of 14.5 months. Histopathologic examination of mice revealed HGSOC characteristics, including nuclear p53 and nuclear MYC in clusters of cells within the FTE and ovarian surface epithelium. Unexpectedly, nuclear p53 and MYC clustered cell expression was also identified in the uterine luminal epithelium, possibly from intraepithelial metastasis from the FTE. Extracted tumor cells exhibited strong loss of heterozygosity at the p53 locus, leaving the mutant allele. Copy-number alterations in these cancer cells were prevalent, disrupting a large fraction of genes. Transcriptome profiles most closely matched human HGSOC and serous endometrial cancer. Taken together, these results demonstrate that the Myc and Trp53-R270H transgenes were able to recapitulate many phenotypic hallmarks of HGSOC through the utilization of strictly human-mimetic genetic hallmarks of HGSOC. This new mouse model enables further exploration of ovarian cancer pathogenesis, particularly in the 50% of HGSOC which lack homology-directed repair mutations. Histologic and transcriptomic findings are consistent with the hypothesis that uterine serous cancer may originate from the FTE.
Significance:
Mouse models using transgenes which generate spontaneous cancers are essential tools to examine the etiology of human diseases. Here, the first Myc-driven spontaneous model is described as a valid HGSOC model. Surprisingly, aspects of uterine serous carcinoma were also observed in this model.
Introduction
Gynecologic oncology has only recently been able to take advantage of the targeted therapies generated over the last decade despite there being great need. Ovarian cancer is the fifth most common cause of cancer-related death in women, and uterine cancer is the sixth. Ovarian cancer deaths are dominated by the high-grade serous ovarian cancer (HGSOC) subtype. Uterine cancer, overall, is less lethal despite higher incidence rates, yet the serous carcinoma subtype accounts for 40% of endometrial cancer deaths (1). Uterine cancer incidence and mortality have increased due to a disproportionate increase in type 2 or nonendometrioid subtypes, including serous carcinoma (2).
Recent targeted therapies in HGSOC involve PARP inhibitors (PARPi), which best benefit patients with BRCA1 or BRCA2 tumor mutations or markers of homologous repair deficiency: half of patients in sum (3). Uterine cancers have shown promise in large phase III trials of immunotherapies, particularly in mismatch repair–deficient cases (4, 5), but only a small subset of patients with serous carcinoma is mismatch repair–deficient. Unfortunately, few targeted drugs have shown clinical benefit for HGSOC with normal homologous repair or in recurrent tumors which restore homologous repair after PARPi therapy. Similarly, serous endometrial cancer has few targetable alterations and thus few drugs in development (6). Serous endometrial cancer and HGSOC share similar disease etiology, such as postmenopausal disease presentation and peritoneal metastasis, and similar molecular characteristics, such as low mutation rates, high copy-number alterations (CNA), mutation in p53, and few other driver mutations.
Genetically engineered mouse models (GEMM) have fundamentally changed how ovarian cancer etiology and treatment is understood. Phylogenetic assessments of HGSOC compared primary tumors dissected from an ovary and metastases, which demonstrated that the earliest mutations often originated in serous tubal intraepithelial carcinoma (STIC; ref. 7). However, this study and other studies have shown that cells can arise first on the ovarian surface epithelium (OSE) or other origin sites, metastasize to the fallopian tube epithelium (FTE) and develop STIC, and finally develop into HGSOC (8–10). A recent pair of GEMMs indicated that OSE-originating cancer cells are phenotypically distinct from fallopian fimbriae–originating cancer cells despite identical drivers (9). Lgr5 was used to drive OSE gene deletion, whereas Pax8 was used to drive FTE gene deletion. Previous research established the Ovgp1 model of oviductal transgene expression, in which Ovgp1-mediated expression control was exclusive to the FTE (11, 12).
These previous mouse models revolutionized the field but have some important limitations. Most models utilized homozygous deletions of multiple tumor suppressors to generate tumors, which is not often, if ever, found in human disease. We previously investigated aneuploidy with or without a heterozygous deletion of the autophagy gene Becn1 in the Amhr2-SV40-T-antigen model (13, 14). These viral proteins are known to disrupt p53 function and RB1 function, amongst other proteins (15, 16). However, few aneuploid events were observed relative to human disease (14), similar to other GEMM findings (17). Previous HGSOC mouse models utilize BRCA deletions, PTEN deletions, or viral oncogenes. The field is left without a genetically representative model for HGSOC lacking BRCA1/2 or other homologous repair alterations (∼50% of human tumors) or HGSOC lacking a PTEN deletion (94% of human tumors). Here, we introduce a new mouse model of ovarian cancer driven by p53 mutation and MYC under the Ovgp1 promoter. Evidence of hallmarks of HGSOC and uterine serous cancer are described.
Materials and Methods
Transgenic OvTrpMyc mouse model creation
Transgenic mice were generated by the Taconic/Cyagen Transgenic Animal Center (Cyagen Biosciences Inc.). Briefly, Trp53 (NM_001127233.1) containing an R270H mutation and Myc (NM_001177352.1) were knocked into the endogenous Trp53 locus in a reverse orientation using CRISPR-Cas9–mediated gene editing. A construct with 985 bp of the murine Ovgp1 promoter, including its proximal epigenetic regulatory peak (Supplementary Fig. S1A; ref. 18), was cloned upstream of the murine Trp53-coding sequence containing a consensus Kozak sequence and an R270H mutation (CGT to CAT mutation in codon 270) without a stop codon. A P2A self-cleaving peptide sequence precedes the murine Myc-coding sequence which contains a 3× hemagglutinin (HA) C-terminal tag and is followed by a bovine growth hormone polyadenylation sequence. Homology arms were supplied by bacterial artificial chromosomes RP23-240I24 and RP23-243M15 (19). The complete transgenic sequence is supplied as Supplementary File 1.
Mouse husbandry, genotyping, and loss of heterozygosity
All mouse procedures were approved by the Medical University of South Carolina’s Institutional Animal Care and Use Committee, protocol #01305, prior to use. Animals were housed in individual vented cages in rooms with 12:12 light/dark cycle and were fed ad libitum (Purina Pro-Lab 5V75). Founder mice were generated in the C57BL/6Tc background and backcrossed to C57BL/6J (RRID: IMSR_JAX:000664) for experimentation. Litters were always formed by crossing a transgenic male with a wild-type (WT) C57BL/6 mother obtained externally (Jackson Laboratories, #000664). For genotyping, ∼2-mm sections of mice tails or ear punches were lysed at 55°C overnight in 350 μL of buffer containing 100 mmol/L Tris-HCl pH 8.8, 5 mmol/L ethylenediaminetetraacetic acid (EDTA) pH 8.0, 0.2% SDS, 200 mmol/L NaCl, and 100 μg/mL proteinase K (VWR, 97062-670). After briefly centrifuging to spin down debris, DNA in 300 μL of lysate was precipitated by adding 1 mL of 100% ethanol. DNA was then pelleted by centrifugation at 16,000 g for 30 minutes at 4°C. The supernatant was discarded, and the pellet was washed with 70% ethanol. After centrifugation at 16,000 g for 20 minutes at 4°C, the pellet was resuspended in 50 μL of tris-EDTA (TE: 10 mmol/L Tris-HCl, 0.2 mmol/L EDTA; pH 7.5). Finally, the tubes were placed in a heat block at 55°C for 2 hours with lids open to evaporate the remaining ethanol. Two PCRs per mouse were used to distinguish mutant and WT mice. Both PCRs used the same reverse primer (GTGAGATTTCATTGTAGGTGCCAG), and the forward primers were either specific to the knockin construct (CAGTTGAGATGGAGGAGACCTAGG) or the WT Trp53 allele (AGCCTGTTGAGCTTCACCCC). For loss of heterozygosity (LOH) studies, identical genotyping PCRs were performed, using genomic DNA extracted from ex vivo grown cells via Invitrogen PureLink Genomic DNA Mini Kit (#K182001).
Matings between male heterozygous OvTrpMyc and female C57BL/6J mice exhibited typical Mendelian inheritance. Of 585 offspring mice, 307 (52%) were male and 278 (48%) were female and 305 (52%) were WT and 280 (48%) were transgenic. Male mice were 47% transgenic and 53% WT; female mice were 49% transgenic and 51% WT. None of these differences were statistically different from the 1:1 ratios predicted by Mendelian segregation.
Longitudinal study of lethal tumor burden
All mice intended to be studied for end-of-life criteria were carefully monitored for signs of tumor burden starting at 2 months of age. The mice were monitored twice weekly for (i) indications of ascites by swollen abdomen, (ii) palpable tumors present externally on the mouse, and (iii) weight loss. Euthanasia was performed when the mice had a palpable tumor of >1 cm, any ascites was observed to increase over 2 days, gait was impaired, or weight loss surpassed 15% of the maximal observed weight. Mice that were found dead for unknown reasons were censored and not included in the Kaplan–Meier curve, and histology was never performed on these mice for quality control reasons. Comparison genotypes were previously published by other studies for p53+/− (20) and Pax8-driven models (17).
Histopathology
The mice had uterine horns and attached fallopian tubes, ovaries, and fat pads surgically removed. Tissues were fixed overnight in 10% normal buffered formalin (VWR 10790-714) and then transferred to 70% ethanol for 24 hours. The Shandon Citadel 2000 Processor (Thermo Fisher Scientific) was used to process tissues on a 4-hour cycle with the following reagent cycles: 70% ethanol followed by two changes of 95% ethanol, three changes of 100% ethanol, one cycle in a 1:1 solution of ethanol and xylene, three changes in xylene, and two changes in paraffin. During the embedding process, ovaries with a portion of the fallopian tube were laid down flat, and the remaining sections of the uterus were stood perpendicular for cross-sections, averaging 3 to 4 cross-sections of the uterus. After the paraffin hardened, the tissue block was cut in 20-μm sections until full sections of the ovaries and fallopian tubes were visible. Then, 5-μm sections were collected onto positively charged slides, air-dried at room temperature overnight, and heat-fixed at 60°C for 1 hour before hematoxylin and eosin (H&E) staining or IHC. IHC utilized Leica Bond III for automated processing. Antibodies used included rabbit anti-PAX8 antibody (Proteintech 10336-1-AP, RRID: AB_2236705) at a 1:2,000 dilution, rabbit anti-p53 antibody (Leica Biosystems NCL-p53-CM5p, RRID: AB_563933) at a 1:5,000 dilution, anti–c-MYC antibody (Abcam ab32072, RRID: AB_731658) at a 1:500 dilution, anti–pan keratin antibody (Roche 760-2595, RRID: AB_2941938) at its provided ready-to-use dilution, and rabbit anti-HA antibody (Cell Signaling Technology 3724, RRID: AB_1549585) at a 1:5,000 dilution. Sections were imaged on a BioTek Lionheart FX microscope, and images were processed using Gen5 and ImageJ software (RRID: SCR_003070). Mice without visible tumors were necropsied, and H&E slides were assessed by a veterinary pathologist for Supplementary Fig. S5 and by a gynecologic pathologist for Supplementary Fig. S6.
Ex vivo tumor cell culturing conditions
A cohort of mice was used to isolate cells from ovaries, uteruses, or visible tumors. The mice were euthanized and immediately dissected for cell extraction. Ovaries and fallopian tubes were excised from fat pads and uteruses. The mid-section of the uterus was used for uterine cell extraction. Visible tumors were excised from surrounding tissues. All tissues were first rinsed once in sterile PBS, and the PBS was then aspirated. Single, ethanol-sterilized razor blades were then used to mince 2-mm sections of tissue in a tissue culture hood. Thoroughly minced tissue sections were then pipetted using a P1000 pipette into a 6-well tissue culture dish, again dispersing cells by repeated pipetting. Cells were initially grown in RPMI 1640 containing glutamine (VWR, #95042-508), supplemented with 20% FBS (Thermo Fisher Scientific, #10437028), 5% penicillin–streptomycin solution (Sigma-Aldrich, P4333-100ML), 1% nonessential amino acids (Sigma-Aldrich, #TMS-001-C), and 1% sodium pyruvate solution (Sigma-Aldrich, #S8636-100ML). After 48 hours, media was aspirated and replaced with similar media containing only 2% penicillin–streptomycin solution. Media was then replaced every 3 to 4 days until the dish became 33% to 67% confluent. Cells were then trypsinized with Trypsin-EDTA (Sigma, #T3924-100ML) for 3 to 10 minutes until nonadherent, combined 1:1 with complete RPMI, and cell suspension was centrifuged at 1,000 g for 2 minutes. Cells were transferred to a new 6-well dish and allowed to confluence. Once confluency was reached, cells were transferred to larger plates for culturing and freezing aliquots in 10% DMSO. Once cell lines were established by growing in culture for a minimum of 2 weeks containing two trypsinization events, cells were grown as cell lines in complete RPMI. Complete RPMI consisted of 500 mL RPMI 1640 containing glutamine, 5 mL of sodium pyruvate solution, 5 mL of penicillin–streptomycin solution, and 50 mL FBS. Cells were passaged within a day of surpassing 80% confluency by 1:10 dilution. Cell lines from this study have been short tandem repeat–profiled by the ATCC Authentication Service, most recently in 3/13/24. Cells were tested for Mycoplasma and other pathogens prior to experimentation and freezing, by Charles River Research Animal Diagnostic Services Infectious Disease PCR, on 2/14/23. All cells were used at less than 10 passages from the original isolation or frozen tube.
Transcriptome sequencing and comparative analysis
RNA was extracted from cells using a Qiagen RNeasy Miniprep kit (#R1054). RNA quality was assessed using Agilent 4200 TapeStation, and RINe values ranged from 8.6 to 10. Polyadenylated RNA was captured from 500 ng of total RNA per sample, and libraries were prepared using NEBNext Poly(A) mRNA Magnetic Isolation Module (#7490L) and NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEBNext, #7760L). Paired-end sequencing was done at the Vanderbilt VANTAGE core laboratory (Vanderbilt University) to a depth of 25 million reads per library using Illumina NovaSeq 6000. Raw fastq.gz data were uploaded into Galaxy (21). GRCm38.86 (mm10) gtf.gz and toplevel.fa.gz reference files were obtained from Ensembl (RRID: SCR_002344; ref. 22). RNA STAR (23) was used to align paired-end 150 bp reads to mm10, followed by PCR duplicate removal by SAMtools RmDup (RRID:SCR_002105; ref. 24). Mapped reads were attributed to transcripts using featureCounts (25) and quantified for gene and transcript expression fragments per kilobase of transcript per million reads mapped (FPKM) using Cufflinks (26). Expression of CGT>CAT mutant p53 was manually verified in read data using mapped read visualization in the University of California Santa Cruz (UCSC) Genome Browser (RRID: SCR_005780) mm10 database around region chr11:69,589,594-69,589,622. One of eleven samples from an ex vivo cell line did not retain expression of the mutated copy of p53 (F361 tumor cells) despite having mapped RNA reads to this codon. FPKM values per sample were combined into transcripts per million reads tables using R to enable normalized sample-to-sample comparisons (27).
For human cancer cell lines to compare with, human Cancer Cell Line Encyclopedia (CCLE; 28) FPKM expression data were used and converted to TPM reads. To enable cross-species comparison, reciprocal ortholog data were utilized. Mousemine.org (29) and Humanmine.org (30) were used for ortholog identification, and the intersections of genes were used as reciprocal orthologous genes, resulting in 13,229 genes expressed and common orthologs between the two datasets. Human cell lines compared with mouse cancer cell lines included all lines annotated as from the cervix, endometrium, and ovary (N = 80 cell lines). Transcriptomes were pairwise compared using a Euclidean distance metric of all expressed genes. Nearest neighbor human cell lines comprise the cell lines shown. Gene expression in this figure was further normalized for visual comparison through mean transcription normalization, including the additional cell lines HELA, KURAMOCHI, SIHA, A2780, CAOV4, MDA-MB-231, MDA-MB-453, and KHYG (RRID: CVCL_0134, RRID: CVCL_0062, and RRID: CVCL_0418), chosen as representative cell lines which have mutational characteristics as meaningful to compare with the mouse cell lines (see “Results”). Shown signature genes were the top 25 genes with minimal expression differences within the mouse cell lines and nearest neighbor human cell line and maximal differences with non–nearest neighbor cell lines. Database for Annotation Visualization and Integrated Discovery (DAVID) (31) was used to generate the pathway annotations shown in the transcriptional signature.
CNA analysis
Raw counts from featureCounts were input into InferCNV to assess CNAs. Settings for InferCNV included i3 copy-number variant calling, cutoff value of 1, Hidden Markov Model transition probability of 1e−02, and a window length of 300. Samples F326 splenic tumor, F326 ROV, and F361 tumor were used as control inputs due to lower levels of initial copy number variant calls in these samples.
Syngeneic i.p. modeling
Cells, lentivirally labeled with GFP, were grown to 5 to 10 million per mouse and injected in 200 μL iced PBS via i.p. injection. Recipient mice were 3-month-old female OvTrpMyc mice. Mice were monitored for euthanasia criteria matching the longitudinal study. Tumors, uterine horns, attached fallopian tubes, ovaries, and fat pads were then immediately fixed and processed for IHC.
Protein collection and immunoblotting of cell lines
Lysis of cells was performed with modified RIPA buffer (50 mmol/L Tris, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% NP-40, and 1% sodium deoxycholate) with protease/phosphatase inhibitors (Thermo Fisher Scientific, #1861284) followed by sonication. SDS-PAGE was performed by standard methods, and antibodies were used for the detection of MYC (Abcam Cat# ab32072, RRID: AB_731658, 1:1,000 dilution) and GAPDH (Cell Signaling Technology Cat# 2118, RRID: AB_561053, 1:10,000 dilution). The secondary antibody was anti-rabbit IgG, horseradish peroxidase–linked (Cell Signaling Technology, Catalog #7074).
Data availability
Next-generation sequencing raw and processed data are available via Gene Expression Omnibus (GEO) with accession number GSE238158.
Results
Generation of a human-mimetic genetic model of HGSOC and serous endometrial cancer
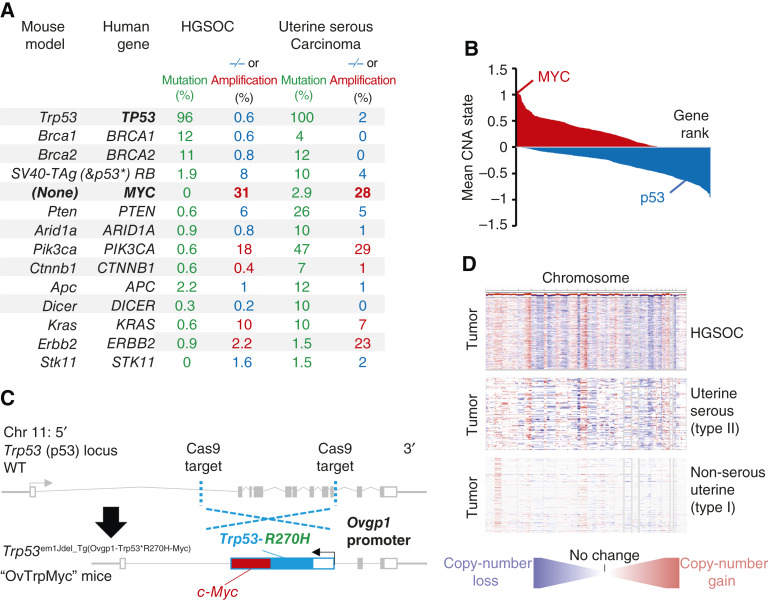
Previous models of epithelial ovarian cancer have been used to seek a better understanding of the disease using known mutations. Ovarian cancer most commonly presents as HGSOC, which contains p53 mutation in at least 94% of cases (32, 33) and sporadic BRCA1 or BRCA2 mutations in <15% of cases. Germline BRCA1 or BRCA2 mutations yield ovarian cancer, most often HGSOC, with a lifetime unmitigated risk of 30% to 70% (34). Other mutations are present in 15% or less of patient primary tumors, including APC, MUC16, ARID1A, RB1, NF1, CSMD3, CDK12, FAT3, and GABRA6 (32, 35). However, HGSOC exhibits unusually high levels of CNAs and aneuploidy, affecting two thirds of all genes. Low-grade serous carcinomas are driven by BRAF and KRAS and represent <5% of ovarian tumors (36). Mucinous tumors are driven by KRAS or ERBB2 amplification, representing about 3% of ovarian tumors. Endometrioid and clear-cell carcinoma each represent about 10% of ovarian tumors and have high rates of ARID1A mutations (30% and 50%, respectively) and PTEN alterations, including deletions and mutations. Clear-cell carcinomas mutate PIK3CA in 33% of tumors. Mouse models of ovarian cancer have utilized these known mutations, which are reviewed by Howell (37) and Zakarya and colleagues (38). A comparison of these mouse driver genes to known human genetic driver frequencies is shown in Fig. 1A. Notably, uterine serous carcinoma, a type II endometrial cancer, exhibits very similar mutation patterns to HGSOC.
Figure 1.
Rationale and design of the transgenic Ovgp1-Trp53-R270H-Myc mouse model. A, Comparison of currently available HGSOC mouse models genetics to genetic alterations in human HGSOC and serous endometrial cancers. −/− refers to homozygous deletion (blue, tumor suppressors), whereas Amp refers to copy-number amplification of at least two extra copies (red, oncogenes). B, Average copy-number state using TCGA HGSOC data at the gene level. Genes are plotted by ranking, with MYC and TP53 highlighted. C, Design of the transgenic OvTrpMyc mouse model. The endogenous Trp53 allele was targeted using a spCas9 and homology donor repair template strategy in the C57BL/6Tc background. Transgene genotyped pups were backcrossed with C57BL/6J mice and heterozygous mice used in all studies. The Ovgp1-driven transgene includes a dominant-negative murine p53-R270H mutant sequence and a murine c-Myc sequence separated by a P2A self-cleaving peptide. D, Comparison of CNAs across the genome for TCGA studied HGSOC and UCEC patient tumors. Red indicates copy-number gain, whereas blue indicates copy-number loss. TCGA, The Cancer Genome Atlas.
We sought to create a new genetic model, Trp53em1Jdel_Tg(Ovgp1-Trp53*R270H-Myc), nicknamed OvTrpMyc, which accurately depicts BRCA and PTEN normal HGSOC. We and other researchers have found that the most commonly and excessively amplified region of the HGSOC genome is 8q24 (Fig. 1B; refs. 14, 32). Expression of a strong oncogene located in 8q24, c-Myc, was coupled with the expression of mutant p53. MYC is a basic helix-loop-helix leucine zipper transcription factor that regulates cell stemness, metabolism, and apoptosis and cell-cycle progression (39). Ovarian cancers require MYC for proliferation (40). The most common mutation of p53 in HGSOC (32) is a dominant-negative, gain-of-function (41, 42) amino acid mutation R273H, which maps to R270H in mice. We engineered mice via CRISPR-Cas9–targeting to insert a transgene driven by Ovgp1 into the native Trp53 (p53) locus of chromosome 11 (Fig. 1C). Ovgp1 was previously demonstrated to express specifically in nonciliated fallopian fimbriae cells, not on the uterine epithelium and not on the OSE (11, 12). We confirmed FTE-selective staining patterns using the transgene’s HA peptide tag within the OvTrpMyc model (Supplementary Fig. S1B). However, the Ovgp1 promoter used in this study to drive the transgene differs from those used in previous studies, and it is possible that off-target expression also differs. The transgene was inserted at the p53 locus to allow for CNAs to evolve, including on the p53 resident chromosome, as CNAs are characteristic of HGSOC. All transgenic mice in this study were genotyped as heterozygous for the transgene. HGSOC CNAs are similar to uterine serous carcinoma but not type I uterine cancer (Fig. 1D).
Middle-age disease presentation
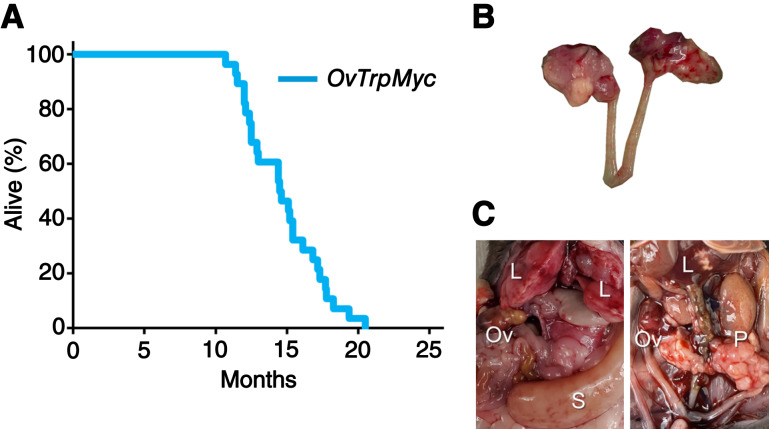
The median age of tumor burden–induced euthanasia for the OvTrpMyc mice was 14.5 months (Fig. 2A) or, given the limitations of human–mouse comparisons, about 40 to 60 years of human age [based on life expectancy of 80 years old, 896 days of female C567BL/6 mouse lifespan (43), with perimenopause in mice occurring around 9 months of age (44)].
Figure 2.
Middle-age presentation of ovarian and peritoneal tumors. A, Lifespan data from mice euthanized due to tumor burden criteria. B, Example of a dissected reproductive tract from a mouse with obvious macroscopic ovarian tumors. C, Examples of widespread intra-abdominal disease, including liver invasion (L), peritoneal tumors (P), splenic tumor (S), and ovarian tumors (Ov). L, liver invasion; Ov, ovarian tumors; P, peritoneal tumors; S, splenic tumor.
OvTrpMyc mice were designed with limited oncogenic stimuli with the intent of long latency, consistent with human disease. However, OvTrpMyc mice are effectively Trp53+/− in all cells. Heterozygous Trp53+/− mice develop lymphomas, osteosarcomas, fibrosarcomas, hemangiosarcomas, lung adenocarcinomas, and other tumors, which form lethal tumors in 25% of mice requiring euthanasia as early as 16.5 months in the C57BL/6 background (20). Complete knockout Trp53−/− mice develop primarily lymphomas and have a half-life of 4.5 months. We observed our OvTrpMyc mouse model to have a survival curve intermediate between these genotypes. Pax8-Cre–driven mixed-background models knocking out Pten and Trp53, in contrast, have a median age of euthanasia requirement between 3 and 6 months, depending on the full genotype (17).
Pathology of lethal ovarian and uterine cancers
Gross examination of mice meeting euthanasia criteria showed HGSOC and other cancer phenotypes. For HGSOC, tumors of the ovary were occasionally observed (Fig. 2B), but large tumors were rare. Widespread intra-abdominal disease was occasionally observed (Fig. 2C), with tumors throughout the mesentery as well as the peritoneal wall. Tumors involved with muscle external to the abdomen were observed, typically with invasion spreading from the peritoneal wall or from the ovary.
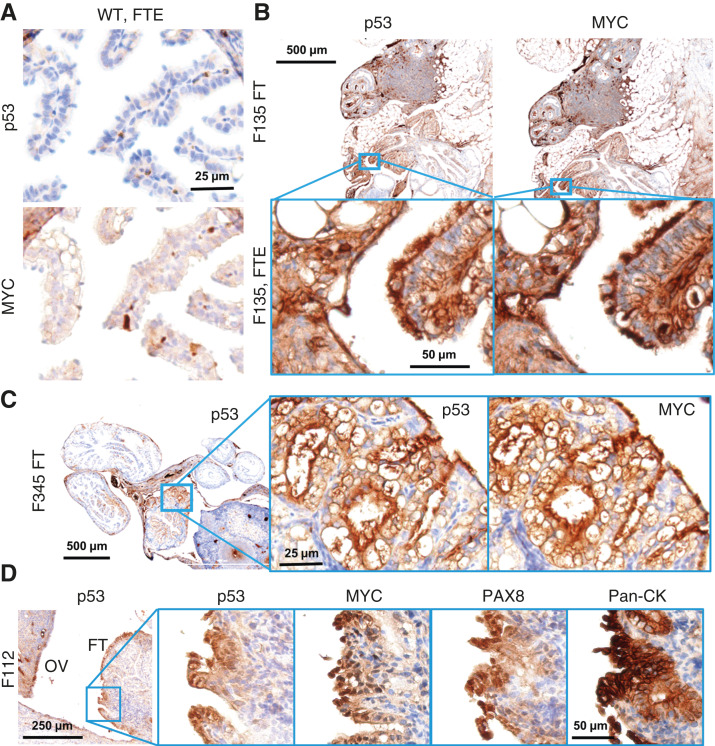
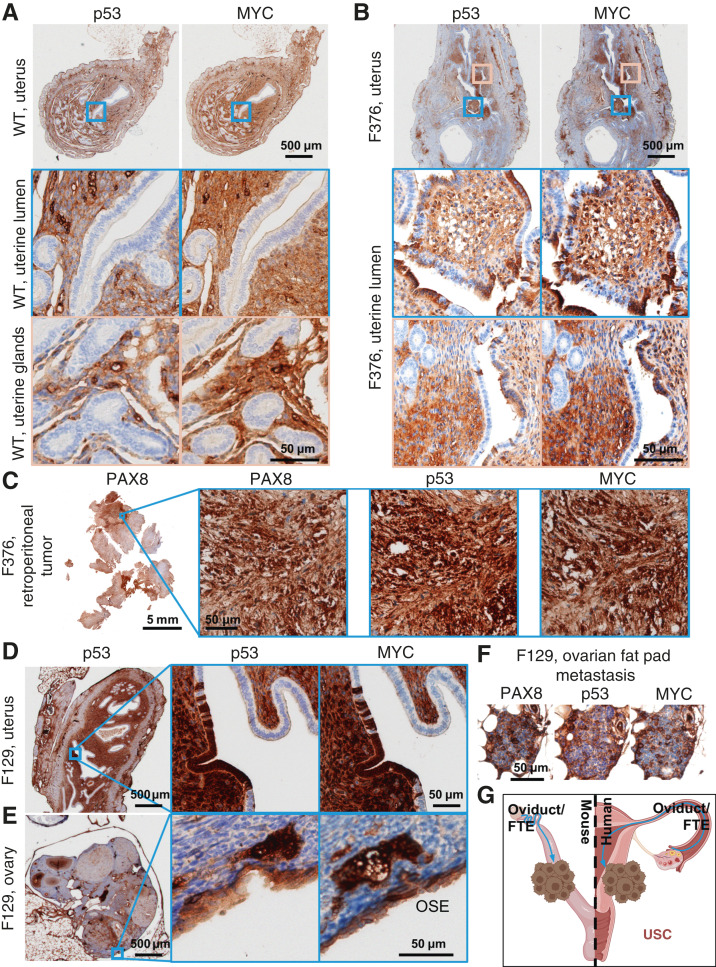
Previous, carefully controlled studies demonstrated Ovgp1 expresses on nonciliated epithelial cells within the FTE, not the OSE (11, 12). To investigate OvTrpMyc transgene protein products at end-stage disease, the gynecologic tract (including uteri, fallopian tubes, ovaries, and ovarian fat pads) was dissected from mice meeting euthanasia criteria at 10 to 16 months of age and processed for molecular pathology. Tissues were formalin-fixed and embedded in paraffin prior to sectioning and staining for p53 and MYC. Tumors, if observed, were additionally stained with paired-box transcription factor PAX8, a biomarker used to help define gynecologic tumors in humans (45–47) but is also expressed in embryogenesis of the thyroid, renal, and upper urinary tracts. Whereas background staining was observed, WT mice did not exhibit clusters of p53 or MYC positivity on the FTE, although scattered single cells were positive (Fig. 3A). In single sections, some FTEs from OvTrpMyc mice were also negative, like WT, perhaps because the entire fallopian tube was not examined due to limited material availability. However, 64% of mice requiring euthanasia exhibited positive FTE staining with p53 and MYC in the single section evaluated, which is likely an underestimate. This contrasts with young mice: in three of three 3-month-old OvTrpMyc mice, none showed positive staining despite clear transgene expression by 10 weeks of age (Supplementary Fig. S1B). Semiquantitative notes from all IHC-assessed mice are available in Table 1 for mice meeting humane euthanasia criteria and Supplementary Table S1 for healthy mice euthanized prior to lethal tumor burden.
Figure 3.
Molecular histology of the fallopian fimbriae. A, A WT mouse at 12 months of age showing little p53 or MYC staining in the FTE. B, OvTrpMyc fallopian tube at 15 months of age, F135, exhibiting p53 and MYC staining within the FTE. C, An OvTrpMyc mouse at 12 months of age, with vacuolated fallopian fimbriae–positive for p53 and MYC. D, An OvTrpMyc mouse at 15 months of age, with features resembling STIC.
Table 1.
Semiquantitative estimates of IHC staining in moribund mice
| Mouse ID | Genotype | Months | Euthanasia criteria met | Tissue | p53 | MYC | PAX8 |
|---|---|---|---|---|---|---|---|
| F129 | Het | 10.7 | Y | FTE | 0 | 0 | NE |
| F129 | Het | 10.7 | Y | OSE | 4 | 4 | 1 |
| F129 | Het | 10.7 | Y | ULE | 3 | 3 | 3 |
| F259 | Het | 11.4 | Y | FTE | 1 | 1 | NE |
| F259 | Het | 11.4 | Y | OSE | 0 | 0 | 0 |
| F259 | Het | 11.4 | Y | ULE | 4 | 3 | 3 |
| F345 | Het | 12 | Y | FTE | 3 | 3 | NE |
| F345 | Het | 12 | Y | OSE | 0 | 0 | 0 |
| F345 | Het | 12 | Y | ULE | 0 | 0 | 0 |
| F376 | Het | 12.1 | Y | FTE | 0 | 0 | NE |
| F376 | Het | 12.1 | Y | OSE | 0 | 0 | 0 |
| F376 | Het | 12.1 | Y | ULE | 3 | 3 | 2 |
| F127 | Het | 12.5 | Y | FTE | 1 | 1 | — |
| F127 | Het | 12.5 | Y | OSE | 4 | 4 | — |
| F127 | Het | 12.5 | Y | ULE | 3 | 3 | 4 |
| F340 | Het | 12.9 | Y | FTE | 0 | 3 | NE |
| F340 | Het | 12.9 | Y | OSE | 2 | 2 | 2 |
| F340 | Het | 12.9 | Y | ULE | 0 | 0 | 0 |
| F135 | Het | 14.6 | Y | FTE | 3 | 3 | NE |
| F135 | Het | 14.6 | Y | OSE | — | — | — |
| F135 | Het | 14.6 | Y | Tumor-RP | 4 | 4 | 4 |
| F135 | Het | 14.6 | Y | ULE | 0 | 0 | 0 |
| F2 | Het | 15.1 | Y | FTE | — | — | — |
| F2 | Het | 15.1 | Y | OSE | 0 | — | — |
| F2 | Het | 15.1 | Y | Tumor-RP | 4 | 4 | 4 |
| F2 | Het | 15.1 | Y | ULE | 3 | 4 | 3 |
| F283 | Het | 15.1 | Y | FTE | 0 | 3 | NE |
| F283 | Het | 15.1 | Y | OSE | — | — | — |
| F283 | Het | 15.1 | Y | ULE | 0 | 0 | 0 |
| F112 | Het | 15.4 | Y | FTE | 4 | 4 | NE |
| F112 | Het | 15.4 | Y | OSE | 4 | 4 | 4 |
| F112 | Het | 15.4 | Y | ULE | 4 | 4 | 4 |
| F278 | Het | 16.1 | Y | FTE | 1 | 1 | NE |
| F278 | Het | 16.1 | Y | OSE | 3 | 3 | 2 |
| F278 | Het | 16.1 | Y | Tumor-M | 4 | 4 | 4 |
| F278 | Het | 16.1 | Y | ULE | 1 | 1 | 1 |
| F12 | Het | 20.5 | Y | FTE | 1 | 2 | NE |
| F12 | Het | 20.5 | Y | OSE | 0 | 0 | 0 |
| F12 | Het | 20.5 | Y | ULE | 0 | 1 | 0 |
Abbreviations: M, mesenteric; RP, retroperitoneal.
p53, MYC, and PAX8 stains were enumerated: 0, no staining or scattered cells only; 1, clustered cells with predominantly cytoplasmic staining; 2, clustered cells with nuclear staining observed; 3, multiple clusters of cells with nuclear positivity; 4, majority of an area affected by positive staining.
“—” indicates tissue was not available or otherwise not evaluable on this slide. “NE” indicates PAX8 is unevaluable by this scoring on the FTE due to positivity in the normal, WT FTE.
Morphology and cellular distribution of p53 and MYC varied between mice. Mouse F135 exhibited nuclear p53 and nuclear MYC within the FTE, indicative of HGSOC, with surrounding cells exhibiting a mix of nuclear and cytoplasmic staining (Fig. 3B). This mouse had a large retroperitoneal tumor extending from the ovary to the spinal cord. The tumor stained positive for PAX8 in addition to p53 and MYC, as did a small mass situated in the ovarian fat pad (Supplementary Fig. S2), consistent with known HGSOC homing to adipocytes (48). Other mice exhibited FTE staining with primarily cytoplasmic expression surrounded by vacuoles (Fig. 3C). STIC is a precursor to HGSOC found in as many as 8% of FTEs of patients with BRCA1/2 mutations from risk-reducing salpingo-oophorectomy and a higher proportion of patients with HGSOC (49). FTE from mouse F112 presented with features of STICs, including enlarged nuclei along the p53-positive epithelium, nuclear atypia, loss of polarity, and nearby morphologically normal regions (Fig. 3D). However, there were features which differ from human STICs, including occasional ciliated cell positivity and not strongly nuclear p53 in all affected cells, although MYC was strongly nuclear. Additional PAX8 and H&E images associated with p53 and MYC panels are provided as Supplementary Figs. S3 and S4.
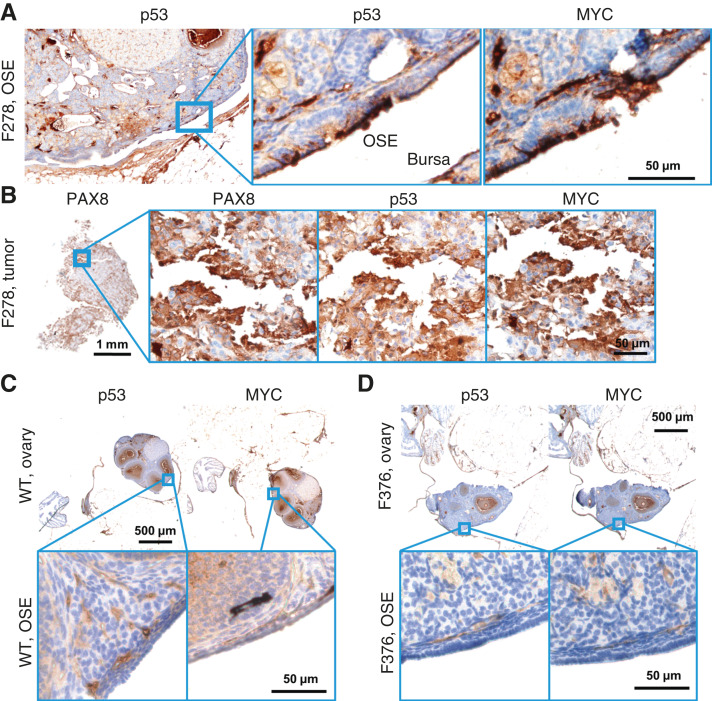
OSE staining of p53 and MYC was observed in 50% of OvTrpMyc mice studied. In OvTrpMyc mouse F278, nuclear positivity of p53 and MYC was observed in cell clusters on the OSE (Fig. 4A), as was PAX8 (Supplementary Fig. S3). Mouse F278 exhibited minimal cytoplasmic positivity in the FTE and uterine luminal epithelium (ULE). The intra-abdominal tumor found in mouse F278 displayed PAX8 staining in addition to p53 and MYC (Fig. 4B). Other transgenic mice, similar to WT mice, were not observed to have p53 or MYC positivity on the OSE. Like WT mice, other ovarian structures stained positive (Fig. 4C and D). The OSE observations positive for p53/MYC may be intraepithelial metastasis events.
Figure 4.
Molecular histology of the OSE. A, An OvTrpMyc mouse at 16 months of age with clear OSE staining of p53 and MYC. B, The intra-abdominal tumor extracted from the mouse shown in A exhibited PAX8, p53, and MYC staining. C, A WT mouse at 12 months of age is negative for p53 or MYC staining in OSE. D, Example of an OvTrpMyc mouse with a region of negative OSE staining of p53 and MYC, at 12 months of age.
Although the OvTrpMyc mice were originally designed to mimic HGSOC, strong staining of p53 and MYC on the ULE was noted in 58% of OvTrpMyc mice. Adjacent tissue exhibited p53 and MYC staining in normal, WT mice, whereas uterine epithelium was universally negative for staining (Fig. 5A). In OvTrpMyc mice, p53-, MYC-, and PAX8-positive epithelial clusters were observed, flanked by scattered cells evidently spreading into the adjacent epithelium, which was not observed in WT mice (Fig. 5A and B; Supplementary Fig. S3). This occurred in the absence of OSE (Fig. 4D) or FTE staining (Supplementary Fig. S3) in the single section from OvTrpMyc mouse F376 but coincided with macroscopic tumor staining (Fig. 5C). However, uterine lumen epithelial staining coincided with predominantly cytoplasmic p53 and MYC staining found in the OSE in one of the youngest mice euthanized, at 11 months (Fig. 5D and E). Ovarian fat pad metastatic cells were found and included p53, MYC, and PAX8 staining (Fig. 5F). These results are consistent with the model that the observed uterine p53/MYC-positive cells possibly originated on the FTE, as driven by Ovgp1, and migrated through the oviduct into the uterus (Fig. 5G).
Figure 5.
Uterine luminal epithelium staining of p53 and MYC. A, A WT mouse at 12 months of age showing negative p53 and MYC staining in the uterine epithelium and positive staining in adjacent tissues. B, Example of a 12 months old OvTrpMyc mouse with cytoplasmic p53 and whole-cell MYC staining along the ULE. C, A large retroperitoneal tumor, behind the kidney, stained for p53, MYC, and PAX8. D, An example of ULE expression in an OvTrpMyc mouse F129, with evidence of similar whole-cell staining of p53 and MYC on the (E) OSE and (F) ovarian fat pad metastasis. Mouse F129 was 11 months of age. G, Proposed model for observed dissemination of cells from the FTE to the ULE as an origin of uterine serous carcinoma. USC, uterine serous carcinoma.
Observation of non-gynecologic cancers
Macroscopic tumors were not always immediately observed upon dissection, despite a mouse reaching euthanasia criteria. Three such mice were subjected to full necropsies, revealing brain tumors and lung tumors (Supplementary Fig. S5). Tumors resembling sarcomas or carcinosarcomas were also identified from macroscopic neoplasms (Supplementary Fig. S6). It is unclear if these are metastatic events, a form of uterine carcinosarcoma, accelerated background strain tumor formation due to disruption of Trp53, or another event.
Aneuploidy and CNAs within OvTrpMyc tumor cells
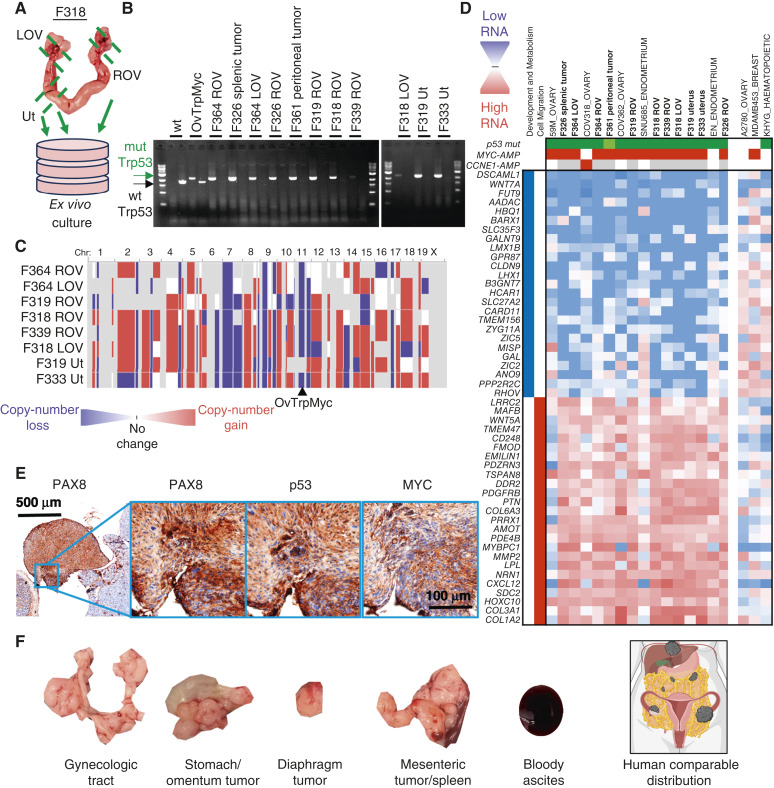
The primary rationale of integrating the OvTrpMyc transgene into the endogenous p53 locus was to allow for LOH of chromosome 11 (chromosome 17 in humans) during spontaneous tumor development. To test if LOH occurred in tumor cells from these mice, eight mice meeting euthanasia criteria were dissected (Fig. 6A) for small portions of the uterus, ovaries, and fallopian tubes and any macroscopic tumor presentation. These dissections were minced, washed, and allowed to grow ex vivo in tissue culture media (see “Materials and Methods”). Once a cell line was established with regular passaging, these cells were (i) genotyped for LOH and (ii) sequenced by bulk RNA sequencing for mRNA transcriptome analysis. Seven mice yielded a cell line from at least one of these origin sites. LOH was present in the ex vivo OvTrpMyc cell lines generated (Fig. 6B; mutant PCR bands were seen, but no WT bands were seen other than in controls). These results provide direct evidence that the WT allele of p53 was lost in these tumor cells. Each tumor cell line was shown to have ample chromosome alterations (Fig. 6C), consistent with patterns in human disease. Although synteny is partial for most chromosomes, loss of the p53-coding chromosome (11 in mice and 17 in humans) and gain of the mouse region syntenic to human amplicon 8q24, on murine 15qD1, was often found.
Figure 6.
CNAs, transcriptomics, and tumorigenicity of extracted OvTrpMyc tumor cells. A, Dissection diagram for ex vivo expansion. Gynecologic tract for mouse F318 is shown. B, LOH was observed by PCR of the Trp53 transgene (mutation) in isolated cells. C, Whole-genome CNA calls of tumor cells. Blue indicates copy-number loss, and red indicates copy-number gain. D, Transcriptomic profiling of OvTrpMyc cancer cells compared with human gynecologic cancer within the human CCLE. Header genes refer to mutation in p53 (green), MYC amplification (red), or CCNE1 amplification (red), shaded white if absent or gray if unknown. Nearest neighbor human cell lines comprise the cell lines shown. A2780, MDA-MB-453, and KHYG cell lines are shown as representative cell lines which have driver gene characteristics comparable with the mouse cell lines but clearly different transcriptional profiles. E, 8 × 106 F339 ROV OvTrpMyc cell line cells were injected i.p. into a young OvTrpMyc female recipient mouse. The mouse lost weight, meeting euthanasia criteria after 54 days. IHC staining of a dissected tumor attached to the intestines is shown. F, Distribution of tumors found in F318 LOV i.p. syngeneic injected mice (5 × 106 cells, 132 days) relative to a human image depiction of HGSOC metastatic spread. LOV, left ovary; ROV, right ovary; Ut, uterus.
OvTrpMyc tumor cells transcriptionally pair with HGSOC and uterine serous cancer
Transcriptomes from OvTrpMyc tumor cells were next analyzed to characterize which human gynecologic cancers they best mimic. Because cell lines have clear differences from endogenous disease (50), transcriptomes, we compared between mouse and human cell lines. Clustering of transcriptomes revealed that tumor cells from OvTrpMyc mice most closely mimicked well-characterized HGSOC and, unexpectedly, p53-mutant endometrial cancer cell lines but not cervical nor other subtypes of ovarian cancer (Fig. 6D). A singular exception in cell identity clustering was the EN cell line, which in the CCLE dataset, utilized for transcriptomes in Fig. 6D, was not annotated as p53-mutant nor MYC- or CCNE1-amplified. However, the founders of the EN line note a p53-R273H mutation (51), and the transcriptomic data from the CCLE (52) also show MYC, CCNE1, and p53 expressions as comparable with HGSOC lines. Cell lines with features that are only a portion of the transgene are shown for comparison. A2780 is a non-HGSOC ovarian cancer cell line but has neither a p53 mutation nor a MYC amplification. Triple-negative breast cancers have many similar characteristics from a genetic standpoint to HGSOC, including frequent BRCA mutation, p53 mutation, and high aneuploidy, but a representative cell line MDA-MB-453 displayed a clearly different transcriptomic profile. Lastly, the leukemia cell line KHYG, which has a p53 mutation, also displayed a clearly different transcriptomic profile. OvTrpMyc clusters revealed a downregulation of developmental and metabolic genes and an upregulation of cell migration genes. These transcriptome-wide data provide molecular evidence that complements histopathology to confirm HGSOC and serous endometrial cancers arise in the OvTrpMyc GEMM.
Cell lines were amplified and injected intraperitoneally into recipient young female OvTrpMyc mice. The mice were monitored for disease meeting the same euthanasia criteria as the spontaneous disease model. Mice required euthanasia due to ascites or distorted abdomen at days 40 (F319 uterus), 54 (F339 ROV), 65 (F326 ROV), and 132 (F318 LOV). Upon euthanasia, gross pathology observations were noted, the gynecologic tract was dissected, and tissues were processed for histopathologic analysis. Histopathologic examination of intra-abdominal tumors showed evidence of HGSOC, including nuclear atypia and pleomorphism, cellular budding, PAX8 staining, nuclear p53 staining, and nuclear MYC staining (Fig. 6E). OvTrpMyc cell line protein levels of MYC were intermediate between high-MYC and low-MYC human cell lines (Supplementary Fig. S7). Macroscopic dissemination was similar to human HGSOC for F318 LOV cells (Fig. 6F). These F339 ROV and F318 LOV cells are evidently useful for syngeneic injections for controlled experimentation about HGSOC tumor formation and development. The F319 uterus cell line exhibited rapid ascites and may be considered for uterine serous carcinoma syngeneic experiments.
Discussion
Our study demonstrates that MYC expression and a p53-R270H mutation drives HGSOC in a mouse model of Ovgp1 promoter–driven transgene expression. No BRCA or homologous repair gene was altered in the GEMM design. The single transgene is sufficient for experimentation without extensive breeding protocols. Latency is long, 14.5 months on average, allowing for the development of HGSOC characteristics such as peritoneal metastasis and extensive CNAs. The FTE was aberrant in most moribund mice, with features consistent with STICs observed. Histopathologic observations were made, which were consistent with intraepithelial metastases to the OSE and ULE. Cells grown ex vivo from the gynecologic tract from transgenic mice formed tumors following i.p. injections. This mouse model may be used to study aspects of spontaneous cancer development and for syngeneic injection experimentation.
At morbidity, 100% of the studied mice exhibited evidence of p53 and MYC positivity along epithelial clusters in at least one site within the FTE, OSE, and ULE. More than half of the mice exhibited positivity at multiple sites. However, the largest tumor observed in these mice tended to be metastases, not the ovary itself. Indeed, when tumor cells were isolated for ex vivo growth, the ovary and oviduct seemed within the reference range of macroscopic size, albeit somewhat morphologically different. We speculate that this may be due to high metastatic potential of these cells which have both p53 mutation and MYC overexpression, consistent with the finding that syngeneic mouse injections yielded lethal tumor burden. Given that Myc is a Yamanaka factor enabling dedifferentiation and redifferentiation into many tissue sites (53), it may be unsurprising that metastases predominate in this GEMM.
The OvTrpMyc mice have a longer latency than other models with strong drivers. SV40 T-antigen MISIIR mice develop ovarian tumors with complete penetrance, usually within 4 to 6 months of age (54). With Ovgp1, SV40-driven mice develop histologic lesions in the epithelium of the fallopian tube as early as 6 weeks of age (55). Mice with Pax8-Cre–driven PTEN knockouts were lethal at 2.2 to 5.7 months, from disease originating in fallopian STICs (17). Similar to Brca1−/−p53−/−Rb−/−Nrf1+/− mice (12), OvTrpMyc mice often require a year or more to develop observable disease. We predicted latency is required to accumulate aneuploidy, which we previously observed was somewhat rare in SV40 T-antigen–driven disease (14). Aneuploidy is characteristic of HGSOC and uterine serous cancer (32, 56, 57). A handful of aneuploid events were observed previously in Brca2−/−Pten−/−p53R270H/−–driven tumors (17). Here, OvTrpMyc mice clearly utilized aneuploid mechanisms for tumor development processes, with strong positive selection for the mutant allele of p53 on chromosome 11. CNAs were ubiquitous in cell lines derived from OvTrpMyc.
Fallopian fimbriae as a likely origin site of ovarian cancer was first suggested from histopathology of prophylactic oophorectomy from women with germline BRCA mutations (58). GEMMs have now provided ample support for epithelium within the fallopian fimbriae as a site with characteristics of disease initiation (37, 38). However, Ovgp1-driven mouse models all included a Brca1+/− or Brca1−/− genotype along with other driver genes. There remains clinical debate about whether salpingectomies should be performed in patients of average risk, particularly opportunistic procedures for contraception (10, 59). The OvTrpMyc model presented here is the first HGSOC GEMM showing evidence of precursor lesions in the fallopian fimbriae epithelium using an Ovgp1 promoter without Brca deletions. This is supportive of the thesis that patients without germline BRCA mutation may nonetheless benefit from salpingectomies to reduce ovarian cancer risk.
Limitations of this model are varied. Some of the nongynecologic tumors found in late-life euthanized mice were analogous to those found in Trp53+/− mice. Although we also found that gynecologic cancer cells could be isolated in 88% of cases of a dissected mouse at end-stage disease, metastatic modeling of HGSOC in this spontaneous model is difficult. PAX8 and other biomarker staining can aid in interpretation of the site of origin. Mice envelop the ovary with a layer of tissue forming the bursa, which humans do not have, and this may affect metastatic properties. The bursa is additionally known to develop leiomyosarcomas in response to intrabursal viral inactivation of Trp53 and Brca1 (60). The IHC staining presented here contained significant background, which, if removed through optimization, would improve the understanding of tissue expression specificity and nuclear/cytoplasmic ratios. The mice did not always exhibit obvious macroscopic tumors, including on the ovaries, even in mice which clearly had viable cancer cells once dissected from the ovary (Fig. 6A), and ultimately formed tumors in a syngeneic setting. Lastly, although the Ovgp1-driven transgene product HA expressed primarily in the FTE (Supplementary Fig. S1), it is possible that some off-target expression occurs during the lifespan of a mouse.
The unexpected, yet exciting, finding that hallmarks of uterine serous carcinomas, such as cells resembling endometrial intraepithelial carcinoma, are found in the OvTrpMyc model provides further support that uterine serous cancer is similar genetically and biologically to HGSOC. Like HGSOC, the most common p53 alteration in uterine serous carcinomas is a mutation in R273 (murine R270). For CNAs, 8q24, containing MYC, is the second most common amplification just behind the unstable MECOM locus near PIK3CA (56). Previous serous endometrial GEMMs required loss of PTEN in a p53 null background or a mutation in a telomere shelterin complex gene to yield disease within the lifetime of a mouse (61, 62). Uterine serous carcinoma is aggressive and treatment-resistant like HGSOC; it only accounts for 10% of endometrial cancer cases, yet it is responsible for 39% of endometrial cancer–related deaths (63). Retrospective studies have revealed a link between tubal ligation, which may prevent intraepithelial metastasis from the fallopian tube, and reduced rates of endometrial cancer (64). In a carefully controlled cohort study of tubal ligation, the hazard ratio for type I endometrioid uterine cancer was 0.78, but the HR was strikingly 0.25 for type II, typically p53-mutant uterine serous carcinoma, indicating surprisingly low incidence of type II tumors following tubal ligation (65). Fallopian STICs and other fallopian epithelium irregularities were observed in patients with uterine serous carcinoma and included the same clonal p53 mutation (66, 67). Taken together, these human retrospective studies, the current OvTrpMyc GEMM study, and human genetic data (68, 69) are all consistent with the emerging hypothesis that uterine serous carcinoma can originate from the same site as HGSOC: the FTE.
Supplementary Material
Ovgp1 transgene expression
Tumor immunohistochemistry from FTE affected mice
PAX8 staining corresponding to other immunohistochemical figures
H&E staining corresponding to immunohistochemical figures
Necropsy of mice lacking macroscopic disease
Other malignancies
MYC levels in syngeneic OvTrpMyc cell lines relative to human lines
Semi-quantitative estimates of immunohistochemical staining in healthy mice
Acknowledgments
Assistance in the preparation and specimen staining were provided by the Histology and Immunohistochemistry Laboratory, Department of Pathology and Laboratory Medicine, Medical University of South Carolina (MUSC). We thank Audreanna Miserendino and Sydney Oesch for helpful comments on manuscript preparation. We thank all private donors, particularly Matt Prisby for organizing the Sheryl Prisby gynecologic oncology fund, for their individually perhaps small yet cumulatively enormous contributions to advancing cancer research and health. We thank the Division of Laboratory Animal Resources staff, Jackson Laboratories staff, and Cyagen/Taconic staff for technical aspects of mouse care and transport. This study was supported in part by a grant from the Rivkin Center for Ovarian Cancer. Research reported in this publication was supported by the NIH under award numbers DP2CA280626 (J.R. Delaney), CA207729 (J.R. Delaney), GM132055 (C.M. Jones), K22CA207575 (R.L. Carpenter), and GM119512 (D.T. Long). This study was supported in part by the Translational Science Laboratory Shared Resource, Hollings Cancer Center, MUSC (P30 CA138313, J.R. Delaney). The MUSC Molecular Analytics Core was used and is supported by GM103499 (J.R. Delaney) and the MUSC’s Office of the Vice President for Research. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NI H or other funders.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
G. Fullbright reports grants from the NIH/National Institute of General Medical Sciences during the conduct of the study. R.L. Carpenter reports a patent to 18/418,110 pending. D.T. Long reports grants from the NIH/National Institute of General Medical Sciences during the conduct of the study. J.R. Delaney reports grants from the NIH and Rivkin Foundation during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
A. Blackman: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. A.C. Rees: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. R.R. Bowers: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. C.M. Jones: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. S.G. Vaena: Data curation, formal analysis, investigation, methodology, writing–review and editing. M.A. Clark: Data curation, formal analysis, investigation, writing–review and editing. S. Carter: Investigation, methodology, writing–review and editing. E.D. Villamor: Investigation, methodology, writing–review and editing. D. Evans: Investigation, methodology, writing–review and editing. A.J. Emanuel: Investigation, methodology, writing–review and editing. G. Fullbright: Investigation, methodology, writing–review and editing. M.S. O’Malley: Formal analysis, investigation, methodology, writing–review and editing. R.L. Carpenter: Resources, supervision, investigation, visualization, project administration, writing–review and editing. D.T. Long: Conceptualization, formal analysis, supervision, investigation, methodology, project administration, writing–review and editing. L.S. Spruill: Formal analysis, investigation, visualization, methodology, writing–review and editing. M.J. Romeo: Data curation, formal analysis, supervision, validation, investigation, project administration, writing–review and editing. B.C. Orr: Conceptualization, formal analysis, supervision, validation, investigation, visualization, writing–review and editing. K.L. Helke: Conceptualization, formal analysis, supervision, validation, investigation, visualization, methodology, project administration, writing–review and editing. J.R. Delaney: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Ferriss JS, Erickson BK, Shih I-M, Fader AN. Uterine serous carcinoma: key advances and novel treatment approaches. Int J Gynecol Cancer 2021;31:1165–74. [DOI] [PubMed] [Google Scholar]
- 2. Clarke MA, Devesa SS, Hammer A, Wentzensen N. Racial and ethnic differences in hysterectomy-corrected uterine corpus cancer mortality by stage and histologic subtype. JAMA Oncol 2022;8:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. González-Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2019;381:2391–402. [DOI] [PubMed] [Google Scholar]
- 4. Mirza MR, Chase DM, Slomovitz BM, dePont Christensen R, Novák Z, Black D, et al. Dostarlimab for primary advanced or recurrent endometrial cancer. N Engl J Med 2023;388:2145–58. [DOI] [PubMed] [Google Scholar]
- 5. Eskander RN, Sill MW, Beffa L, Moore RG, Hope JM, Musa FB, et al. Pembrolizumab plus chemotherapy in advanced endometrial cancer. N Engl J Med 2023;388:2159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu KH, Broaddus RR. Endometrial cancer. N Engl J Med 2020;383:2053–64. [DOI] [PubMed] [Google Scholar]
- 7. Eckert MA, Pan S, Hernandez KM, Loth RM, Andrade J, Volchenboum SL, et al. Genomics of ovarian cancer progression reveals diverse metastatic trajectories including intraepithelial metastasis to the fallopian tube. Cancer Discov 2016;6:1342–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh R, Cho KR. Serous tubal intraepithelial carcinoma or not? Metastases to fallopian tube mucosa can masquerade as in situ lesions. Arch Pathol Lab Med 2017;141:1313–5. [DOI] [PubMed] [Google Scholar]
- 9. Zhang S, Dolgalev I, Zhang T, Ran H, Levine DA, Neel BG. Both fallopian tube and ovarian surface epithelium are cells-of-origin for high-grade serous ovarian carcinoma. Nat Commun 2019;10:5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim J, Park EY, Kim O, Schilder JM, Coffey DM, Cho C-H, et al. Cell origins of high-grade serous ovarian cancer. Cancers (Basel) 2018;10:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu R, Zhai Y, Kuick R, Karnezis AN, Garcia P, Naseem A, et al. Impact of oviductal versus ovarian epithelial cell of origin on ovarian endometrioid carcinoma phenotype in the mouse. J Pathol 2016;240:341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhai Y, Wu R, Kuick R, Sessine MS, Schulman S, Green M, et al. High-grade serous carcinomas arise in the mouse oviduct via defects linked to the human disease. J Pathol 2017;243:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hensley H, Quinn BA, Wolf RL, Litwin SL, Mabuchi S, Williams SJ, et al. Magnetic resonance imaging for detection and determination of tumor volume in a genetically engineered mouse model of ovarian cancer. Cancer Biol Ther 2007;6:1717–25. [DOI] [PubMed] [Google Scholar]
- 14. Delaney JR, Patel CB, Bapat J, Jones CM, Ramos-Zapatero M, Ortell KK, et al. Autophagy gene haploinsufficiency drives chromosome instability, increases migration, and promotes early ovarian tumors. PLoS Genet 2020;16:e1008558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ali SH, DeCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol 2001;11:15–23. [DOI] [PubMed] [Google Scholar]
- 16. Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005;24:7729–45. [DOI] [PubMed] [Google Scholar]
- 17. Perets R, Wyant GA, Muto KW, Bijron JG, Poole BB, Chin KT, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell 2013;24:751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hammal F, de Langen P, Bergon A, Lopez F, Ballester B. ReMap 2022: a database of human, mouse, drosophila and arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res 2022;50:D316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Osoegawa K, Tateno M, Woon PY, Frengen E, Mammoser AG, Catanese JJ, et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res 2000;10:116–28. [PMC free article] [PubMed] [Google Scholar]
- 20. Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol 1994;4:1–7. [DOI] [PubMed] [Google Scholar]
- 21. Jalili V, Afgan E, Gu Q, Clements D, Blankenberg D, Goecks J, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res 2020;48:W395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, et al. Ensembl 2022. Nucleic Acids Res 2022;50:D988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 26. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao Y, Li MC, Konaté MM, Chen L, Das B, Karlovich C, et al. TPM, FPKM, or normalized counts? A comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J Transl Med 2021;19:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER III, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Motenko H, Neuhauser SB, O’Keefe M, Richardson JE. MouseMine: a new data warehouse for MGI. Mamm Genome 2015;26:325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lyne R, Bazaga A, Butano D, Contrino S, Heimbach J, Hu F, et al. HumanMine: advanced data searching, analysis and cross-species comparison. Database (Oxford) 2022;2022:baac054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022;50:W216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cancer Genome Atlas Research Network . Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vang R, Levine DA, Soslow RA, Zaloudek C, Shih I-M, Kurman RJ. Molecular alterations of TP53 are a defining feature of ovarian high-grade serous carcinoma: a rereview of cases lacking TP53 mutations in the cancer genome atlas ovarian study. Int J Gynecol Pathol 2016;35:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Fan I, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst 2006;98:1694–706. [DOI] [PubMed] [Google Scholar]
- 35. Chowdhury S, Kennedy JJ, Ivey RG, Murillo OD, Hosseini N, Song X, et al. Proteogenomic analysis of chemo-refractory high-grade serous ovarian cancer. Cell 2023;186:3476–98.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prat J, D’Angelo E, Espinosa I. Ovarian carcinomas: at least five different diseases with distinct histological features and molecular genetics. Hum Pathol 2018;80:11–27. [DOI] [PubMed] [Google Scholar]
- 37. Howell VM. Genetically engineered mouse models for epithelial ovarian cancer: are we there yet? Semin Cell Dev Biol 2014;27:106–17. [DOI] [PubMed] [Google Scholar]
- 38. Zakarya R, Howell VM, Colvin EK. Modelling epithelial ovarian cancer in mice: classical and emerging approaches. Int J Mol Sci 2020;21:4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reyes-González JM, Vivas-Mejía PE. c-MYC and epithelial ovarian cancer. Front Oncol 2021;11:601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeng M, Kwiatkowski NP, Zhang T, Nabet B, Xu M, Liang Y, et al. Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. Elife 2018;7:e39030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiao G, Lundine D, Annor GK, Canar J, Ellison V, Polotskaia A, et al. Gain-of-function mutant p53 R273H interacts with replicating DNA and PARP1 in breast cancer. Cancer Res 2020;80:394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang C, Liu J, Xu D, Zhang T, Hu W, Feng Z. Gain-of-function mutant p53 in cancer progression and therapy. J Mol Cell Biol 2020;12:674–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller RA, Harrison DE, Astle CM, Fernandez E, Flurkey K, Han M, et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell 2014;13:468–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Diaz Brinton R. Minireview: translational animal models of human menopause: challenges and emerging opportunities. Endocrinology 2012;153:3571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tacha D, Zhou D, Cheng L. Expression of PAX8 in normal and neoplastic tissues: a comprehensive immunohistochemical study. Appl Immunohistochem Mol Morphol 2011;19:293–9. [DOI] [PubMed] [Google Scholar]
- 46. Laury AR, Perets R, Piao H, Krane JF, Barletta JA, French C, et al. A comprehensive analysis of PAX8 expression in human epithelial tumors. Am J Surg Pathol 2011;35:816–26. [DOI] [PubMed] [Google Scholar]
- 47. Xiang L, Kong B. PAX8 is a novel marker for differentiating between various types of tumor, particularly ovarian epithelial carcinomas. Oncol Lett 2013;5:735–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 2011;17:1498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bachert SE, McDowell A Jr., Piecoro D, Baldwin Branch L. Serous tubal intraepithelial carcinoma: a concise review for the practicing pathologist and clinician. Diagnostics (Basel) 2020;10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Isaka K, Nishi H, Sagawa Y, Nakada T, Osakabe Y, Serizawa H, et al. Establishment of a new human cell line (EN) with TP53 mutation derived from endometrial carcinoma. Cancer Genet Cytogenet 2003;141:20–5. [DOI] [PubMed] [Google Scholar]
- 52. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–76. [DOI] [PubMed] [Google Scholar]
- 54. Connolly DC, Bao R, Nikitin AY, Stephens KC, Poole TW, Hua X, et al. Female mice chimeric for expression of the simian virus 40 TAg under control of the MISIIR promoter develop epithelial ovarian cancer. Cancer Res 2003;63:1389–97. [PubMed] [Google Scholar]
- 55. Sherman-Baust CA, Kuhn E, Valle BL, Shih IM, Kurman RJ, Wang TL, et al. A genetically engineered ovarian cancer mouse model based on fallopian tube transformation mimics human high-grade serous carcinoma development. J Pathol 2014;233:228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cancer Genome Atlas Research Network; Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bowers RR, Jones CM, Paz EA, Barrows JK, Armeson KE, Long DT, et al. SWAN pathway-network identification of common aneuploidy-based oncogenic drivers. Nucleic Acids Res 2022;50:3673–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leeper K, Garcia R, Swisher E, Goff B, Greer B, Paley P. Pathologic findings in prophylactic oophorectomy specimens in high-risk women. Gynecol Oncol 2002;87:52–6. [DOI] [PubMed] [Google Scholar]
- 59. Gockley AA, Elias KM. Fallopian tube tumorigenesis and clinical implications for ovarian cancer risk-reduction. Cancer Treat Rev 2018;69:66–71. [DOI] [PubMed] [Google Scholar]
- 60. Clark-Knowles KV, Senterman MK, Collins O, Vanderhyden BC. Conditional inactivation of Brca1, p53 and Rb in mouse ovaries results in the development of leiomyosarcomas. PLoS One 2009;4:e8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Daikoku T, Hirota Y, Tranguch S, Joshi AR, DeMayo FJ, Lydon JP, et al. Conditional loss of uterine Pten unfailingly and rapidly induces endometrial cancer in mice. Cancer Res 2008;68:5619–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Akbay EA, Peña CG, Ruder D, Michel JA, Nakada Y, Pathak S, et al. Cooperation between p53 and the telomere-protecting shelterin component Pot1a in endometrial carcinogenesis. Oncogene 2013;32:2211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hamilton CA, Cheung MK, Osann K, Chen L, Teng NN, Longacre TA, et al. Uterine papillary serous and clear cell carcinomas predict for poorer survival compared to grade 3 endometrioid corpus cancers. Br J Cancer 2006;94:642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Loghmani L, Saedi N, Omani-Samani R, Safiri S, Sepidarkish M, Maroufizadeh S, et al. Tubal ligation and endometrial Cancer risk: a global systematic review and meta-analysis. BMC Cancer 2019;19:942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Falconer H, Yin L, Altman D. Association between tubal ligation and endometrial cancer risk: a Swedish population-based cohort study. Int J Cancer 2018;143:16–21. [DOI] [PubMed] [Google Scholar]
- 66. Tang S, Onuma K, Deb P, Wang E, Lytwyn A, Sur M, et al. Frequency of serous tubal intraepithelial carcinoma in various gynecologic malignancies: a study of 300 consecutive cases. Int J Gynecol Pathol 2012;31:103–10. [DOI] [PubMed] [Google Scholar]
- 67. Steenbeek MP, Bulten J, Snijders M, Lombaers M, Hendriks J, van den Brand M, et al. Fallopian tube abnormalities in uterine serous carcinoma. Gynecol Oncol 2020;158:339–46. [DOI] [PubMed] [Google Scholar]
- 68. Jia L, Yuan Z, Wang Y, Cragun JM, Kong B, Zheng W. Primary sources of pelvic serous cancer in patients with endometrial intraepithelial carcinoma. Mod Pathol 2015;28:118–27. [DOI] [PubMed] [Google Scholar]
- 69. Jarboe EA, Miron A, Carlson JW, Hirsch MS, Kindelberger D, Mutter GL, et al. Coexisting intraepithelial serous carcinomas of the endometrium and fallopian tube: frequency and potential significance. Int J Gynecol Pathol 2009;28:308–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ovgp1 transgene expression
Tumor immunohistochemistry from FTE affected mice
PAX8 staining corresponding to other immunohistochemical figures
H&E staining corresponding to immunohistochemical figures
Necropsy of mice lacking macroscopic disease
Other malignancies
MYC levels in syngeneic OvTrpMyc cell lines relative to human lines
Semi-quantitative estimates of immunohistochemical staining in healthy mice
Data Availability Statement
Next-generation sequencing raw and processed data are available via Gene Expression Omnibus (GEO) with accession number GSE238158.