Summary
Background
A better understanding of the molecular events during liver normothermic machine perfusion (NMP) is warranted to develop a data-based approach for the identification of biomarkers representative of graft quality and posttransplant outcome. We analysed the dynamic transcriptional changes during NMP and linked them to clinical and biochemical parameters.
Methods
50 livers subjected to NMP for up to 24 h were enrolled. Bulk RNA sequencing was performed in serial biopsies collected pre and during NMP, and after reperfusion. Perfusate was sampled to monitor liver function. qPCR and immunohistochemistry were performed to validate findings. Molecular profiles were compared between transplanted and non-transplanted livers, and livers with and without early allograft dysfunction.
Findings
Pathways related to immune and cell stress responses, cell trafficking and cell regulation were activated during NMP, while cellular metabolism was downregulated over time. Anti-inflammatory responses and genes involved in tissue remodelling were induced at later time-points, suggesting a counter-response to the immediate damage. NMP strongly induced a gene signature associated with ischemia-reperfusion injury. A 7-gene signature corresponds with the benchmarking criteria for transplantation or discard at 6 h NMP (area under curve 0.99). CD274 gene expression (encoding programmed cell-death ligand-1) showed the highest predictive value. LEAP2 gene expression at 6 h NMP correlated with impaired graft function.
Interpretation
Assessment of gene expression markers could serve as a reliable tool to evaluate liver quality during NMP and predicts early graft function after transplantation.
Funding
The research was supported by “In Memoriam Dr. Gabriel Salzner Stiftung”, Tiroler Wissenschaftsfond, Jubiläumsfonds-Österreichische Nationalbank and MUI Start grant.
Keywords: Liver, Transplantation, Normothermic machine perfusion, Gene expression, Inflammation, Ischemia-reperfusion injury
Research in context.
Evidence before this study
Normothermic machine perfusion (NMP) allows for in-depth assessment of organ quality and function before transplantation. So far, only limited data is available on how NMP impacts human donor livers. Evidence suggests a quite unique and stringent immune activation during NMP, while there are also reports showing inhibition of hepatic inflammation and promotion of graft regeneration and cellular repair. A better understanding of the molecular events during liver NMP is warranted to develop a data-based approach for the identification of biomarkers representative for graft quality and posttransplant outcome. We herein analysed the dynamic transcriptional changes during liver NMP and linked them to clinical and biochemical parameters.
Added value of this study
NMP activates immune and cell stress response pathways early during NMP of human donor livers. In non-transplanted, pre-injured livers, a more pronounced inflammatory response was observed with prolonged perfusion time. Inflammatory responses were accompanied by the induction of regulatory and anti-inflammatory pathways and genes, supporting the idea of a counter-response to the immediate damage. We also provide evidence that ischemia-reperfusion injury is strongly induced at the transcriptomic level during NMP and post reperfusion. In the effort to more precisely assess graft quality early during NMP, we identified a 7-gene signature, which distinguishes between transplanted and non-transplanted livers at 6 h NMP. Notably, CD274 gene expression (encoding programmed cell-death ligand-1) was highly induced in livers which were not transplanted after NMP. Finally, we determined LEAP2 gene expression during NMP as a potential marker to predict early graft function after transplantation.
Implications of all the available evidence
We herein propose a 7-gene signature corresponding with graft quality early during liver NMP. This may help in the clinical decision-making process whether to transplant a liver or not. Assessment of gene expression markers may also help to predict early graft function and outcome after liver transplantation.
Introduction
Liver transplantation (LT) remains the sole treatment option for patients suffering from end-stage liver disease. However, organ shortage remains a major hurdle. Acceptance of livers from extended criteria donors (ECD) and donors after circulatory death (DCD) enable to increase the donor pool but bare the risk of graft dysfunction and failure after transplantation.1
Normothermic machine perfusion (NMP) allows for the assessment of organ function ex situ prior to transplantation.2 While intended to enhance organ preservation quality, NMP also helps to facilitate and ease logistics of transplantation. Further to the ability of in-depth evaluation of organ viability and performance, this technology enables organ treatment and reconditioning to improve organ quality and outcome after transplantation. Such interventions may help to expand the donor pool.3, 4, 5
Only limited data is available on how NMP impacts human donor livers. Evidence suggests, that NMP may inhibit hepatic inflammation, reduce ischemia-reperfusion injury (IRI), and promote graft regeneration and cellular repair mechanisms.6,7 On the other hand, our group and others have reported that a quite unique and stringent immune activation occurs during liver NMP.8, 9, 10 Building on these findings and the single-cell transcriptomic atlas depicting dynamic change of the hepatic immune cell repertoire during NMP,10 we aimed to establish a set of marker genes for the assessment of graft quality and posttransplant outcome during liver NMP. In a cohort of 50 livers subjected to NMP for up to 24 hours (h) serial biopsies and perfusate samples were collected during NMP and post reperfusion. Applying bulk RNA sequencing in tissue samples allowed to obtain a time-resolved global picture of transcriptomic changes during human liver NMP.
Methods
Study design and normothermic liver machine perfusion
The study was approved by the local ethics committee (Ethical committee of the Medical University Innsbruck, protocol #1175/2018) and all recipients signed an informed consent form prior to LT. Between April 2019 and November 2020, a total of 50 donor livers subjected to NMP consecutively, were enrolled. All livers were accepted by our institution with the intention for transplantation. A liver was defined as an ECD organ if one or more criteria according to Eurotransplant (https://www.eurotransplant.org/organ-match-characteristics/) and summarized by Resch et al.11 were met. The reason to apply NMP prior to transplantation included one or a combination of the following reasons: (D) suspected suboptimal organ quality (R) complex recipient, and/or (L) logistics. Livers were shipped on ice. Upon arrival at our institution (back-to-base concept), grafts were surgically prepared for NMP and connected to the OrganOx metra® device. NMP was performed and grafts monitored following the centre protocol.12 The decision to transplant or discard an organ after NMP was based on the following criteria previously reported by our group12: (i) Maintenance of physiological pH values (7.30–7.45) without need of repeated sodium bicarbonate addition, (ii) a functioning lactate metabolism with a prompt decline to lactate levels ≤2.5 mmol/l, (iii) lactate dehydrogenase (LDH), alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels <20,000 U/l. A sharp and constant incline of these parameters and excessively high IL-6 levels were considered warning signals urging for caution. Wedge liver biopsies were collected pre NMP, at 1 h, 6 h, 12 h, 20 h or at the end of NMP, and approximately 30 minutes (min) post reperfusion during transplantation. Samples were immediately snap frozen and stored at −80 °C until further processing. In addition to an observational analysis of the dynamics of the gene profile, gene expression was compared between transplanted and non-transplanted livers and between livers with initial function (IF) and those developing early allograft dysfunction (EAD) according to Olthoff et al.13 after LT (study design depicted in Fig. S1). qPCR and immunohistochemistry (IHC) were applied to validate gene expression data. Findings for EAD were correlated with risk scores Liver Graft Assessment Following Transplantation (L-GrAFT)14 and Model for Early Allograft Function (MEAF).15 Serial perfusate samples were taken to monitor liver function and the immune response. The sex of the donor (reported by Eurotransplant Liver Report) or the recipient (self-reported by study participant) was not taken into account in the design of this study.
RNA extraction and sequencing
Cryopreserved liver biopsies were homogenized using the ULTRA Turrax IKA T10 basic. Total RNA was extracted utilizing the RNeasy Micro Kit (Quiagen, #74004, Netherlands) according to manufacturer's guidelines. RNA integrity was determined using a 2100 Bioanalyzer and RNA 6000 Nano LabChip kits (Agilent Technologies, USA) and the RNA integrity number (RIN) was calculated. To allow for comparison of time-point samples in a paired fashion, livers were excluded from further analysis if one time-point sample showed a RIN below 6.
RNA-sequencing libraries were generated using the QuantSeq 3’ protocol (Lexogen, Austria) according to the manufacturer's recommendation at the Next Generation Sequencing (NGS) Core Facility at the Medical University of Innsbruck and paired-end sequencing (PE150) was performed at GENEWIZ (Azenta Life Science, Germany) on a NovaSeq 6000 sequencing device, resulting on average in 7.5 mio high-quality reads/sample. Sequencing reads in FastQ format were adapter-trimmed using BBDuk and quality checked using FastQC. Forward reads were mapped to the human reference genome (GRCh38/hg38) using the splice-aware aligner STAR (v2.7.1a).16 Read counts for NCBI Gene/Refseq annotation were determined using featureCounts (v2.0.0).17
Single-cell RNA sequencing analysis
For analysis of cell type-specific expression and to identify active crosstalk between cell types, previously published processed single-cell RNA sequencing (scRNAseq) data from eight livers before (T0) and at the end (T1) of NMP were used.10 Respective analyses (Jupiter notebook), visualization routines, and data (anndata object) were retrieved from https://doi.org/10.5281/zenodo.7249006. A pseudo-bulk matrix from this data under NMP at T1 from eight livers was extracted. DESeq2 was used to identify differentially expressed genes between scRNAseq and bulk RNA sequencing data at 20 h NMP from 18 livers. Since the gene expression in hepatocytes could not be assessed by scRNAseq at T1, upregulated genes could indicate hepatocyte-related gene expression. For further validation, pre-processed scRNAseq data from a human normal liver cell atlas18 and cell annotation were downloaded (https://www.livercellatlas.org) and analysed using the statistical software environment R and Seurat pipeline v4 (with filtering for cells ≥3, features ≥200, nFeature_RNA <6000, percent. mt <20, dimension reduction to 15 principal components, sctransform to perform normalization and variance stabilization, and cell type annotation as provided). DESeq2 was used to identify differentially expressed genes on pseudo-bulk data from hepatocytes vs other cell types. Commonly up-(log2 fold change >0) or commonly downregulated (log2 fold change <0) genes for both analyses were determined. To identify a potential crosstalk between different cell types we took advantage of 13 curated ligand-receptor databases as summarized in19 and https://github.com/LewisLabUCSD/Ligand-Receptor-Pairs (Table S1). To narrow down potential expression of ligands in hepatocytes and respective receptors in other cell types we identified an overlap of genes which were (i) significantly upregulated at 12 h and 20 h during NMP compared to baseline (pre) from our bulk RNA sequencing data, (ii) commonly up- or down regulated genes from bulk/pseudo bulk analysis in NMP single-cell analysis and normal liver cell atlas in hepatocytes vs other cell types, and (iii) annotation of ligand or receptors in at least two out of 13 curated ligand-receptor databases.
qPCR
qPCR of liver sample RNA extracted for bulk RNA sequencing was performed with the New England Biolabs Lunascript #10151266 kit according to the manufacturer's protocols on a 96 Universal Gradient Peqstar. Approximately 1500 ng RNA was used as input per sample. qPCR was performed according to Thermo Fisher Scientific's TaqMan Universal PCR Master Mix #1907052 based on manufacturer's protocols on an Applied Biosystems 7500 Real Time PCR System (Thermo Fisher, USA). 50 ng of cDNA were used as input per well. TaqMan primers (Thermo Fisher, USA) used are presented in Table S2. Normalization was performed using the mean CT of the house keeping genes RAB2A and SNRPD3 as a reference. Correlation analysis of the library-size normalized counts and the 2ˆCT values of the qPCR were performed.
Immunohistochemistry
Liver biopsies were fixed in 4% paraformaldehyde and paraffin embedded. Immunohistochemical staining was performed on 4 μm sections for programmed cell death ligand-1 (PD-L1) in 6 h NMP samples using a commercially available assay (SP263, Cat No. 7419821001, Roche Diagnostics, USA). Overall staining was scored semiquantitatively by a blinded experienced pathologist as follows: 0 (<1% positive cells), 1 (1–10% positive cells), 2 (10–50% positive cells), 3 (>50% positive cells). Additionally, PD-L1 staining was scored separately for hepatocytes and immune cells.
Perfusate analysis
Perfusate was collected as part of a clinical routine and according to our centre protocol for blood gas analysis and liver function monitoring (Lactate, LDH, ALT, AST).12 Perfusate Interleukin-6 (IL-6) levels were measured to monitor the inflammatory and immunologic response. Analyses were performed at the Central Institute for Medical and Chemical Laboratory Diagnostics (ZIMCL) of the University Hospital Innsbruck.
Statistical analyses
Differentially expressed genes (DEG) between each time-point and pre NMP were calculated using the R package DESeq220 based on a negative binomial distribution in a paired fashion (including Liver IDs as covariate in the model) and at each time-point between non-transplanted vs transplanted and for the subgroup of transplanted livers between livers with IF vs EAD in an unpaired fashion. p-values were adjusted for multiple hypothesis testing based on the false discovery rate (FDR) according to the Benjamini-Hochberg method. Significantly differentially expressed genes were defined with more than two-fold change, adjusted p-value (FDR) <0.1 and mean of normalized counts >10. RNA sequencing data (raw counts) for human hepatic IRI (GSE151648)21 were re-analysed using DESeq220 and a signature including 64 genes were extracted from significantly upregulated genes in post transplantation IRI + livers. Enrichment of this IRI gene signature and hallmark gene sets (MSigDB) were analysed using gene set enrichment analysis (GSEA software v4.3.0)22 on pre-ranked log2 fold changes. For significantly overexpressed genes functional association (networks) were analysed with the Cytoscape app ClueGO,23 whereby overrepresented Reactome pathways (adjusted p < 0.05 using two-sided Fishers exact test) were represented as nodes and connected based on the number of shared genes (kappa score >0.4) and clustered into functional groups. The STRING database24 were used to construct gene interaction networks and MCL clustering (inflation parameter = 3) to divide the network into functional groups. Overrepresentation analysis (ORA) of significantly upregulated genes or selected genes were performed using the online tool ConsensusPathDB including various pathway databases.25 Enrichr26 and TRRUST database27 was used to identify potential transcription factor regulating overexpressed genes. Heatmaps and hierarchical cluster analyses were performed using Genesis (v1.8.0).28
To identify genes which are not only differentially expressed but are also able to classify non-transplanted from transplanted livers (or in the subgroup of transplanted livers with IF or EAD) at each time-point a logistic regression model with leave-one-out cross validation were performed using the R package ROCR. These analyses were performed for non-transplanted vs transplanted livers (EAD vs IF) as binary outcome and for the expression of each significantly DEG separately for each time-point as continuous dependent variable. The area under the receiver operating characteristics (ROC) curve (AUC) was used as a predictive value. To identify the set of genes out of the significantly DEGs corresponding with the classification of non-transplanted and transplanted livers (as binary outcome variable) at 6 h NMP, a regularized logistic regression (lasso)29 with 100 times 4-fold cross validation using the R package glmnet was performed. A final gene signature model was estimated based on genes, which are included in the optimal model (according selected λ with maximal AUC) more than 10 out of the 100 runs and 10-fold cross validation. Gene expression profiles of EAD livers were correlated with L-GrAFT or MEAF using Spearman's rank-based correlation analysis. Differential expression and logistic regression analyses were also performed with high-risk group based on L-GrAFT cutoff >−0.5 and a low-risk group based on L-GrAFT ≤−0.5 as described by Chen et al.30 as binary outcome.
Wilcoxon-Mann-Whitney test using R was applied to assess differences of LDH, lactate, ALT, AST and IL-6 perfusate levels between non-transplanted and transplanted livers at various time-points. Two-sided Student's t test was used to assess differences of PD-L1 staining between non-transplanted and transplanted livers at 6 h NMP. For this, p-value <0.05 was considered significant.
Role of the funding source
The funding sources had no role in study design, data collection, analysis, and interpretation, writing of the manuscript or the decision to submit the manuscript for publication.
Results
Donor liver characteristics and perfusion characteristics
In five out of 50 livers, at least one time-point sample revealed a RIN below 6, which indicates a low quality of the RNA. These livers were therefore excluded from NGS. Hence, serial biopsies of 45 livers subjected to NMP were analysed for their transcriptional landscape using NGS techniques (Fig. S1). An overview on these 45 donor livers and donor characteristics is presented in Table 1 (individual data are shown in Table S3). The indication criteria for NMP are depicted in Fig. S2. 12 livers were DCD organs, while 33 livers were from donors with brain death (DBD). 41 out of 45 livers met the ECD criteria.11 Median NMP time was 843 min (Interquartile range (IQR) 568–1267). Perfusate parameters LDH, lactate, AST and ALT revealed an overall stable perfusion over time (Fig. S3). Following pre-defined criteria (see methods),2,12,31 30 livers were transplanted after NMP, while 15 were declined.
Table 1.
Overview of donor liver characteristics.
| Characteristics | Median (IQR)| Categories | N (%) |
|---|---|---|
| Donor age | 59 (48–69) | 45 |
| Donor sex | female | 24 (53) |
| male | 21 (47) | |
| Donor type | DBD | 33 (73) |
| DCD | 12 (27) | |
| ECD | yes | 41 (91) |
| no | 4 (9) | |
| Cause of donor death | cerebrovascular | 27 (60) |
| trauma | 10 (22) | |
| circulatory | 6 (13) | |
| not otherwise specified | 2 (5) | |
| CIT [min] | 349 (298–415) | 45 |
| NMP time [min] | 843 (568–1267) | 45 |
| total preservation [min] | 1202 (967–1570) | 45 |
| Liver transplantation | transplanted | 30 (67) |
| not transplanted | 15 (33) | |
| Benchmark cases | no benchmark case | 15 (50) |
| benchmark case | 15 (50) |
n, number; DBD, donation after brain death; DCD, donation after circulatory death; ECD, extended criteria donor; CIT: cold ischemia time; min, minutes; NMP, normothermic machine perfusion; IQR, interquartile range.
Liver transplant recipients and post-operative outcome
The recipient median age was 59 years (IQR 52–67.75, n = 30). Recipient median MELD (model for end-stage liver disease) and BAR (balance of risk) score were 14 (IQR 10–18.75, n = 30) and 5.5 (IQR 4–8, n = 28), respectively. EAD occurred in 11 patients (36.7%). One-year patient survival was 100% for benchmarking cases according to Muller et al.,32 while six out of 15 patients died in the non-benchmarking group, mostly from sepsis-associated complications (Table 2). Death censored graft survival was 100%.
Table 2.
Recipient data and post operative outcome.
| Characteristics | Median (IQR)|Categories | N (%) |
|---|---|---|
| Recipient sex | male | 22 (73) |
| female | 8 (27) | |
| Recipient age | 59 (52–67.75) | 30 |
| BAR score | 5.5 (4–8) | 28 |
| MELD | 14 (10–18.75) | |
| EAD | not observed | 19 (63) |
| observed | 11 (37) | |
| 1-year patient survival | benchmarking cases | 15 (100) |
| non-benchmarking cases | 9 (60) | |
| total | 24 (80) | |
| Time from transplantation to death in days | 39.5 (21.5–70.25) | 6 |
| COD recipient (non-benchmarking cases) | sepsis/necrotic fasciitis | 1 |
| septic shock/pancreatitis, MOF | 1 | |
| history of bleeding and perforation colon | 1 | |
| C.diff sepsis | 1 | |
| septic MOF/aspergillosis | 1 | |
| septic MOF/mycosis | 1 |
BAR, balance of risk; MELD, model for end-stage liver disease; EAD, extended criteria donor, COD, cause of death; MOF, Multiple organ failure; n, number.
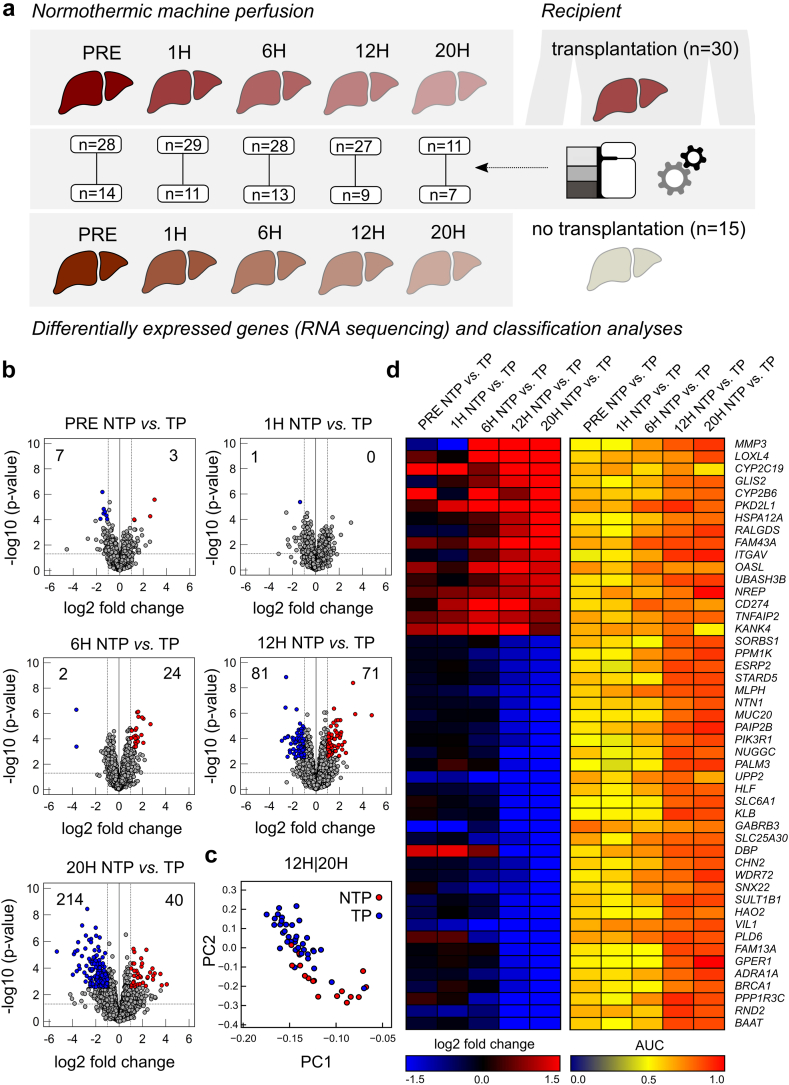
NMP significantly alters gene expression in donor livers
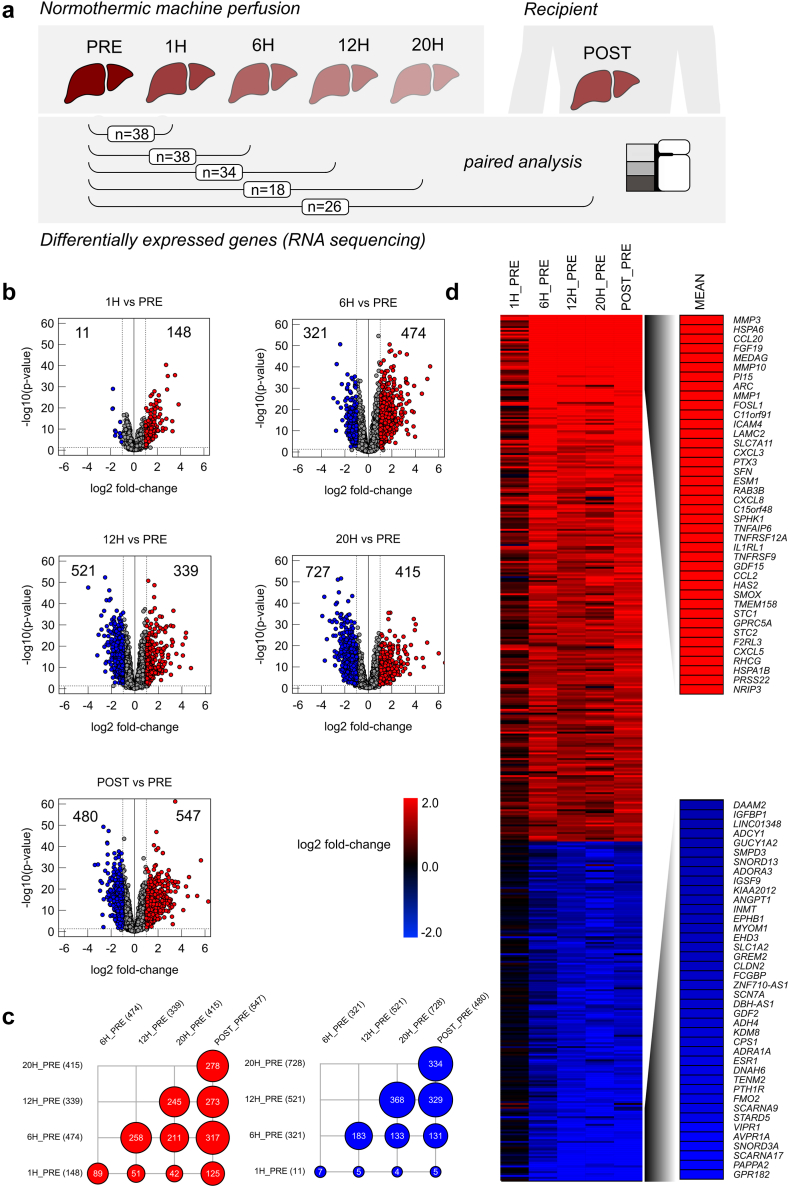
First, transcription levels were assessed in serial liver biopsies before, during and after NMP (Fig. 1a). Paired analysis of biopsies from the same donor livers (pre vs other time-points) guaranteed a high consistency of data from individual organs. Volcano plots in Fig. 1b depict up- and downregulation of gene expression pre NMP and at subsequent time-points. Gene expressions meeting the cutoff for up- or downregulation (FDR <0.1, more than two-fold change) are shown in red (upregulated) and blue (downregulated). Other genes are shown in grey. Gene upregulation during NMP peaked at 6 h (474), while gene downregulation was even more pronounced, especially at later time-points (12 h NMP: 521, 20 h NMP: 727). In liver samples collected post reperfusion, a high number of genes were up-as well downregulated, compared to pre NMP, indicating a second set or wave of gene regulations upon a second graft reperfusion. Numerous genes were found to be commonly up- and downregulated between the various NMP time-points—especially at 6 h, 12 h and 20 h NMP, and post reperfusion after LT (Fig. 1c). For upregulated genes, the highest conformity was observed between 6 h NMP and post reperfusion, suggesting that similar gene regulation mechanisms may occur upon both reperfusion processes in a time-dependent manner—at 6 h NMP and approximately 30 min after reperfusion over the course of LT. Obviously, the process of gene downregulation lags gene upregulation during NMP.
Fig. 1.
Results of differential gene expression analysis based on RNA bulk-sequencing during liver normothermic machine perfusion at different time-points compared to baseline (PRE). (a) Experimental design for paired analysis. (b) Volcano plots indicating significantly up- or downregulated genes (average normalized counts >10, more than two-fold change and false discovery rate (FDR) <0.1) at different time-points compared to baseline (PRE). (c) Dot plot indicating number of overlapping significantly up-regulated genes (red) or significantly down-regulated genes (blue). (d) Heatmap indicating log2 fold changes at different time points from significantly differentially expressed genes in at least on time point compared to baseline before NMP (PRE) and mean log2 fold change of top 40 up- and down regulated genes.
The top 40 up- and downregulated DEGs during liver NMP and post reperfusion after LT (Fig. 1d) include matrix metalloproteinases (MMP3, MMP10, MMP1), cytokines and chemokines involved in immune cell trafficking (CCL20, ICAM4, CXCL3, CXCL8), and factors regulating bile synthesis (FGF19) and metabolism (MEDAG). Also, immediate early genes (IEGs) and transcriptions factors (TFs) were among the top upregulated genes. Among the top downregulated genes were PAPPA2, AVPR1A, VIPR1A, STARD5, FMO2 and TENM2, which are involved in cell and oxidative metabolism, coagulation, lipid and bile synthesis, and cell signalling.
For eight highly expressed and biologically interesting genes out of the top 40 regulated DEGs (ATF3, CCL20, CD83, CD274, CXCL3, CXCL8, FGF19 and MMP1), qPCR analysis was conducted in 10 out of 45 livers to verify NGS results. Correlation analysis of the library-size normalized counts and the 2ˆCT values of the qPCR revealed a robust correlation (Pearson) for all genes analysed (Fig. S4). Taken together, NMP induces up-as well as downregulation of a high number of genes in human donor livers, especially after 6 h perfusion. Regulated genes are associated with a broad variety of cellular functions.
NMP activates inflammatory, cell regulation and tissue remodelling pathways
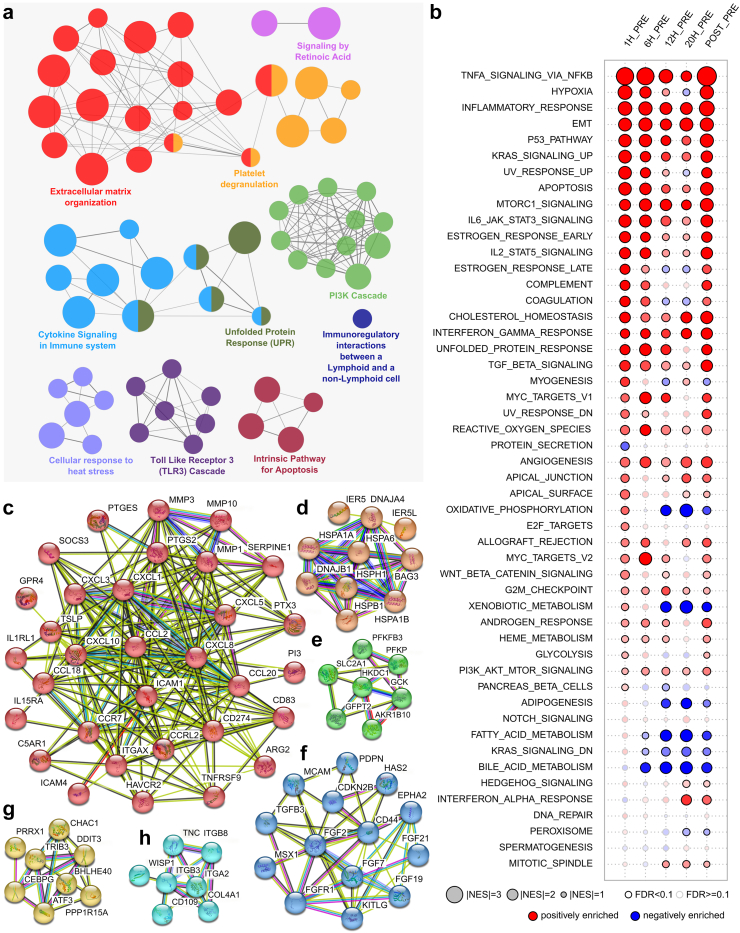
Network analysis based on the top upregulated DEGs revealed that biological processes such as extracellular matrix organization, immunologic processes, cell migration and regulation, and stress responses are overrepresented (Fig. 2a).
Fig. 2.
Functional analyses of differentially expressed genes. (a) ClueGO analyses indicating biological processed upregulated during liver NMP. Most relevant terms are highlighted. Connection between nodes (biological processes) indicate shared genes based on kappa statistics. (b) Gene set enrichment analyses for 50 hallmark gene sets with normalized enrichment score is indicated by size and significance (false discovery rate (FDR) <0.1) by strong border. Gene sets enriched in upregulated genes (positive enrichment) is indicated in red, gene sets enriched in downregulated genes (negative enrichment) is indicated in blue. (c–h) Different clusters of up-regulated genes and their related protein interactions using the STRING database and MCL clustering; connections indicate (different) evidences for interactions.
Using gene set enrichment analysis (GSEA), we assessed changes in the expression of groups of genes within an established biological pathway. Fig. 2b shows the positively (red) and negatively (blue) enriched pathways at all time-points compared to the pre sample against the hallmarks data set. Numerous pathways involved in inflammation were enriched during liver NMP, especially during the first 6 h NMP, and post reperfusion after LT, supporting the assumption of a second wave of gene regulation. TNF-α signalling via NFκB was most pronounced. In line with this, members of the tumour necrosis factor (TNF) superfamily such as TNFAIP6, TNFRSF12A and TNFRSF9 were among the top upregulated DEGs (Fig. 1d). Further immune response-associated pathways included inflammatory response, IL6 JAK STAT3 signalling, IL2 STAT5 signalling, and interferon gamma response. Concordantly, numerous leucocyte-recruiting chemokines such as CCL20, ICAM4, CXCL3, CXCL8, CCL2 and CXCL5 (Fig. 1d) were among the top upregulated genes. Complement was induced early during NMP (at 1 h and 6 h) and again post reperfusion. Also, the induction of cell stress response and apoptosis pathways showed a similar pattern. Further, gene signatures representing biologic processes such as cell growth and differentiation, protein synthesis and maintenance of proper cell function were enhanced during liver NMP. A pronounced downregulation was observed for fatty acid and bile metabolism during prolonged NMP. Concordantly, FGF19 gene expression, encoding for fibroblast growth factor (FGF)-19, which is involved in the suppression of bile acid synthesis, was found among the top upregulated DEGs. Interestingly, genes involved in oxidative phosphorylation (OXPHOS), a key pathway required to generate adenosine triphosphate (ATP), were induced at 1 h NMP compared to pre NMP, but markedly downregulated after 12 h NMP.
Protein interaction analysis using STRING of the 40 top upregulated genes identified six major clusters related to: Inflammatory processes and immune cell trafficking (Fig. 2c), cells stress responses (Fig. 2d), glucose metabolism (Fig. 2e), tissue repair and angiogenesis (Fig. 2f), TFs and IEGs (Fig. 2g), and cell adhesion (Fig. 2h). Taken together, DEGs upregulated during liver NMP and post reperfusion display a profound activation of biologic processes involved in immunologic and cell stress responses, but also facilitate cellular homeostasis, regeneration, wound healing, and cell repair.
Upregulated genes during NMP are involved in the crosstalk between hepatocytes and immune cells
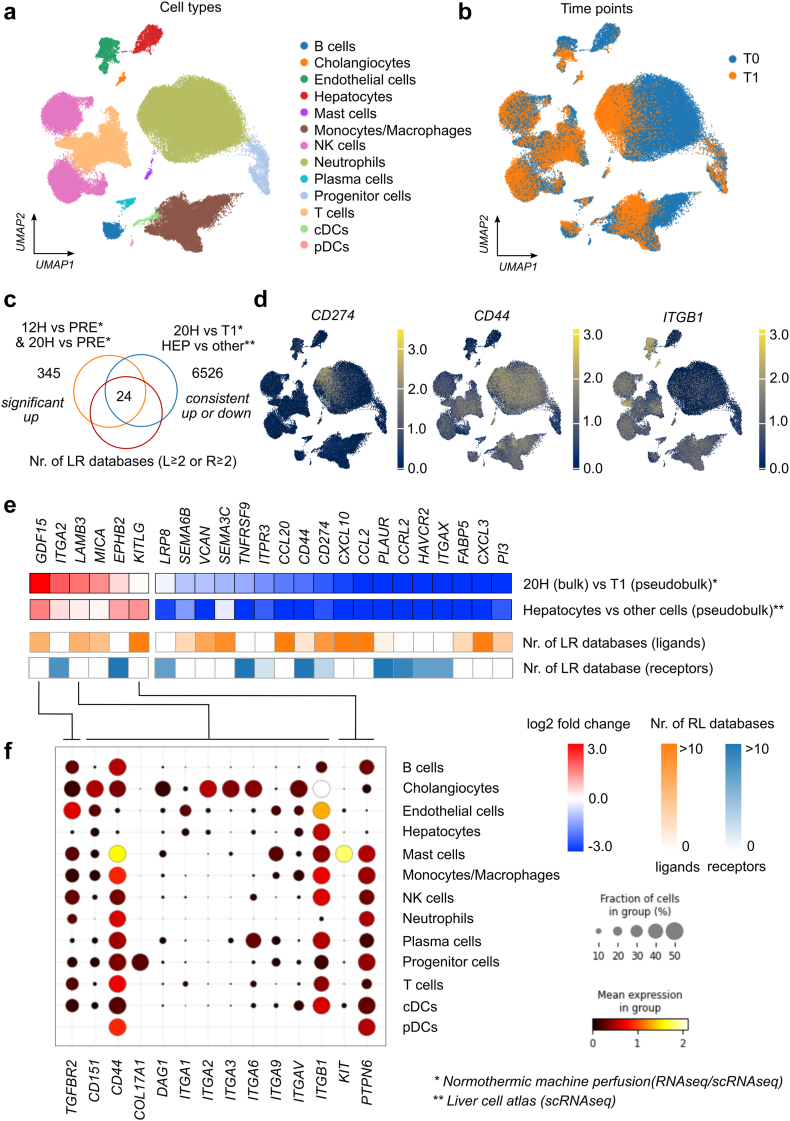
To identify upregulated genes, which were predominantly expressed in hepatocytes, we correlated our findings from this study with the scRNAseq data of livers during NMP10 and data from a liver cell atlas.18 Thus, we performed differential gene expression analyses (i) between our bulk RNA sequencing expression data after 20 h NMP with the pseudo-bulk data from scRNAseq data at T1 (at the end of NMP), and (ii) between hepatocytes and all other cell types using the pseudo-bulk data from single-cell analyses of healthy liver tissue (Fig. 3a and b). From the 6526 genes consistently up- (log2 fold change>0) or down-regulated genes (log2 fold change <0) in these two comparisons (Fig. 3c) and from 245 commonly significantly upregulated genes at 12 h and 20 h compared to pre (Fig. 1, Fig. 3c), 24 genes were identified as ligands or receptors in at least two out of 13 curated ligand-receptor databases.19 Four out of these 24 genes (GDF15, LAMB3, MICA, KITLG) were consistently upregulated and encode ligands. While the expression of the corresponding natural killer cell receptor NKG2D for MICA could not be detected based on scRNAseq data during NMP, we found specific expression patterns on various immune cells for TGFBR2, CD44 and several integrins such as ITGB1, as well as PTPN6 for the upregulated ligands GDF15, LAMB3, and KITLG, respectively, which are specifically expressed in hepatocytes. A high expression of CD44 was found in T cells, macrophages, and neutrophils (Fig. 3d–f). ITGB1 was strongly expressed in endothelial cells (Fig. 3d–f). Interestingly, using the scRNAseq data, we could observe a high expression of CD274, encoding for the immune checkpoint PD-L1, particularly in neutrophils at the end of NMP (Fig. 3d). On the contrary, CD274 expression was found downregulated in hepatocytes vs other cell types (Fig. 3e). In summary, our analyses prove an active crosstalk between hepatocytes and immune cells at a transcriptional level during NMP.
Fig. 3.
Analyses of crosstalk between hepatocytes and other cells during normothermic machine perfusion (NMP) using single-cell RNA sequencing data. (a) UMAP showing clusters of single-cell data with cell type annotation. (b) Cells from livers during NMP (T1 refers to end of NMP and T0 refers to baseline before NMP also named PRE) (c) Venn diagram indicating 24 genes which are consistently up- (log2FC >0) or down regulated (log2FC <0) 1) between bulk RNA sequencing expression data after 20 h NMP with pseudo-bulk data from scRNAseq at T1, 2) between hepatocytes and all other cell types using the pseudo-bulk data from single-cell analyses of normal liver tissue, and 3) are annotated as a ligand or receptor in at least two out of 13 curated ligand-receptor databases. (d) UMAP plot indicating expression of selected genes (receptors). (e) Heatmap of the 24 genes indicating differential expression (log2 fold change) for the two comparisons and number of annotation as ligand or receptor in ligand-receptor databases. (f) Expression of respective receptors in various cell types during liver NMP based on scRNAseq data according to the legend at the right.
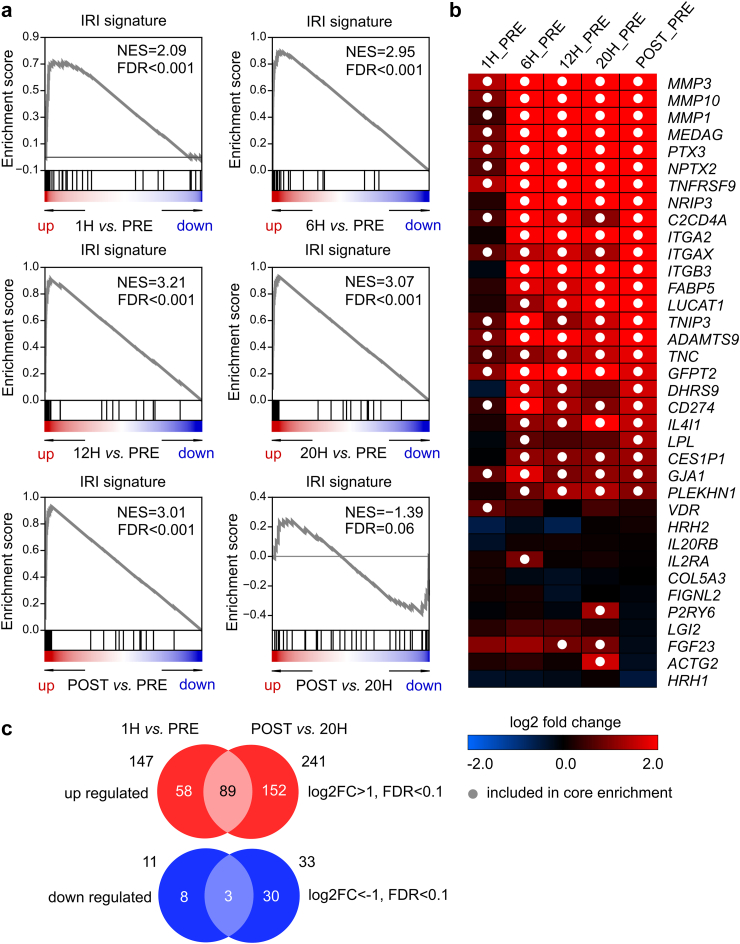
An in vivo signature of ischemia-reperfusion injury is strongly induced during liver NMP
Next, gene expression during NMP was compared with a previously established liver IRI signature based on 64 genes.21 Compared to pre NMP, a strong enrichment of this IRI gene signature and hallmark gene sets were observed at all time-points of NMP and post reperfusion after LT. Enrichment was most significant at 12 h NMP (Fig. 4a). Leading-edge genes which contribute most to the maximum enrichment are shown in Fig. 4b: MMPs were the top three candidates. Compared to the sample taken at the end of NMP (20 h), the post reperfusion sample did not reveal the activation of the IRI pattern. This may indicate that transcriptomic changes associated with IRI in the post reperfusion sample were already induced previously upon NMP. When comparing overall DEGs of the two different reperfusion processes, either due to NMP (1 h_pre) or reperfusion in the recipient post transplantation (post_24 h), 89 genes with a log2 fold change >1, FDR <0.1 were upregulated in common, while still a numerous genes (152) were solely upregulated due to reperfusion in the recipient (Fig. 4c, DEGs shown in Supplementary data file). Pathway analysis revealed that selectively upregulated genes during the first reperfusion event at start of NMP were attributed to apoptotic events, while during LT, they were mainly involved in broad regulatory events, including cell proliferation and differentiation (Pathways shown in Supplementary data file).
Fig. 4.
Gene set enrichment analyses indicate a signature of ischemia-reperfusion injury (IRI) is highly enriched in upregulated genes during liver normothermic machine perfusion (NMP). (a) Enrichment plots for different time points. (b) Heatmap indicating log2 fold change of genes which are also included in the IRI signature, dots indicate that genes are included in core enrichment (this refers to genes before the maximum enrichment is reached). (c) Venn diagrams indicating the number of overlapping significantly upregulated (downregulated) genes between 1 h and before NMP and upregulated (downregulated) genes between transplanted livers (POST) and the last time point (20 h) of NMP.
Enhanced inflammatory, but also counter responses are observed in non-transplanted livers during NMP
Next, the transcriptomic landscape and its dynamic changes during NMP were compared between transplanted (n = 30) and non-transplanted (n = 15) livers (Fig. 5a). Perfusate parameters LDH, lactate, AST and ALT were significantly higher in non-transplanted vs transplanted livers throughout NMP (Wilcoxon-Mann-Whitney test, Fig. S5). Volcano plots in Fig. 5b depict the number of significantly up- (red) and downregulated (blue) genes in non-transplanted vs transplanted livers at various time-points. Most apparent differences were observed with prolonged NMP at 12 h and 20 h. Along the same line, principal component analysis (PCA) revealed that both groups can be clearly distinguished at 12 and 20 h NMP based on their transcriptome (Fig. 5c), but not earlier (Fig. S6). Also, the heatmap and AUC reveal clear differences in gene regulation between both groups at later time-points (Fig. 5d). Thereby, the following genes were most strikingly upregulated in non-transplanted livers: MMP3, LOXL4, PKP2L1, ITGAV (involved in tissue remodelling and collagen synthesis), NREP (regeneration), CYP2C19, CYP2B6, SORBS1, STARD5 (metabolism) and UBASH3B, CD274 (immunosuppressive responses).
Fig. 5.
Comparison of gene expression between non-transplanted livers (NTP) and transplanted livers (TP) at different time-points. (a) Experimental design for differential expression analyses based on RNA sequencing and logistic regression classification at different time points. (b) Volcano plots indicating number of significantly up- or down-regulated genes. (c) Heatmap visualizing log2 fold change and area under curve (AUC) from receiver operating characteristics (ROC) from logistic regression analyses predicting the binary outcome (NTP vs TP). (d) Principal component analyses for genes from (c) showing separation between NTP and TP livers.
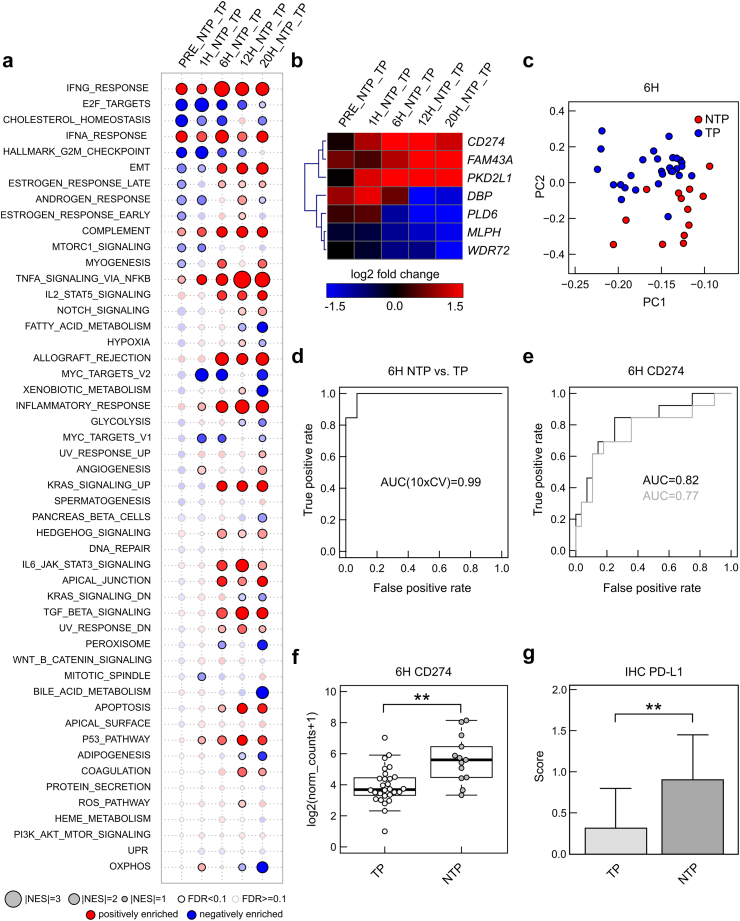
GSEA showed, that several pathways involved in inflammatory and immune processes were overrepresented in non-transplanted vs transplanted livers over perfusion time (Fig. 6a). Overall, the interferon gamma response was most pronounced, while TNF-α signalling via NFκB gradually increased over time and inflammatory response, allograft rejection, complement, but also TGF beta signalling were overrepresented especially after 6 h perfusion in non-transplanted organs. Concordantly, IL-6 perfusate levels were significantly increased in non-transplanted vs transplanted livers as from 6 h perfusion (Wilcoxon-Mann-Whitney test, Fig. S5e). In contrast, gene sets related to cell cycle/proliferation, metabolism and OXPHOS were negatively enriched over perfusion time in non-transplanted vs transplanted livers. In summary, gene expression patterns differ between non-transplanted and transplanted organs, especially with prolonged NMP. While numerous inflammatory pathways are specifically activated after 6 h NMP in non-transplanted livers, genes involved in repair mechanisms and immunosuppression are concomitantly upregulated, suggesting for a counter-response to mitigate inflammation and damage.
Fig. 6.
Differential genes (signatures) between non-transplanted livers (NTP) and transplanted livers (TP). (a) Gene set enrichment analyses indicating positively (red) or negatively (blue) enrichment of hallmark gene sets in differential expression of genes in NTP vs TP at different time points (size indicate normalized enrichment score, solid border indicates significant enrichment with false discovery rate (FDR) <0.1). (b) Heatmap indicating log2 fold changes of 7 genes as identified using regularized logistic regression (LASSO). (c) Principal component analysis showing separation of NTP vs TP at 6 h. (d) Receiver operating characteristics of this 7-gene model and area under curve (AUC) (mean of 10-fold cross validation). (e) Receiver operating characteristics (ROC) curve and AUC for the CD274 gene (grey line: leave-one-out cross validated logistic regression model). (f) Boxplot indicating differences in gene expression of CD274 (log2 from normalized counts adding pseudocount of 1) between NTP (n = 13) and TP (n = 28) at 6 h. ∗∗ FDR<0.01 (test based on a negative binomial distribution using DESeq2 and correction based on the FDR according to the Benjamini-Hochberg method) (g) PD-L1 protein expression (encoded by CD274 gene) in immune cells using immunohistochemical stains in liver biopsies at 6 h in NTP (n = 13) and TP (n = 28) livers. ∗∗p < 0.05 (two-sided Student's t test, error bars indicate standard deviation).
A 7-gene signature differentiates between non-transplanted and transplanted livers at 6 h NMP
To identify a gene signature, which allows a distinction between both groups at an early time-point, we performed logistic regression analyses using the least absolute shrinkage and selection operator (lasso). Through this, a 7-gene signature could be identified (Table 3, heatmap Fig. 6b) and a clear separation of non-transplanted and transplanted livers at 6 h NMP using PCA (Fig. 6c) with an area under curve (AUC) of 0.99 (Fig. 6d) was found. CD274 gene expression had the highest predictive value (Fig. 6e) and was found significantly upregulated in non-transplanted livers at 6 h NMP, compared to transplanted livers (log2 from normalized counts adding pseudocount of 1, Fig. 6f). In line, PD-L1 staining was found significantly increased in immune cells by then (two-sided Student's t test, Fig. 6g). No significant differences were observed for overall PD-L1 staining and PD-L1 expression in hepatocytes (two-sided Student's t test, Fig. S7).
Table 3.
7-gene signature for prediction of non-transplanted livers.
| Variable | Weights |
|---|---|
| INTERCEPT | −0.385 |
| FAM43A | 0.593 |
| PKD2L1 | 0.917 |
| CD274 | 0.081 |
| PLD6 | −0.619 |
| MLPH | −0.437 |
| DBP | 0.415 |
| WDR72 | 0.592 |
log(p/(1-p)) = −0.385 + 0.593xFAM43A + 0.917xPKD2L1 + 0.081xCD274-0.619xPLD6-0.437xMLPH + 0.415xDBP + 0.592xWDR72 using log2(normalized counts + 1) and p indicating probability of non-transplanted (dysfunctional) livers.
Weights are from a logistic regression model.
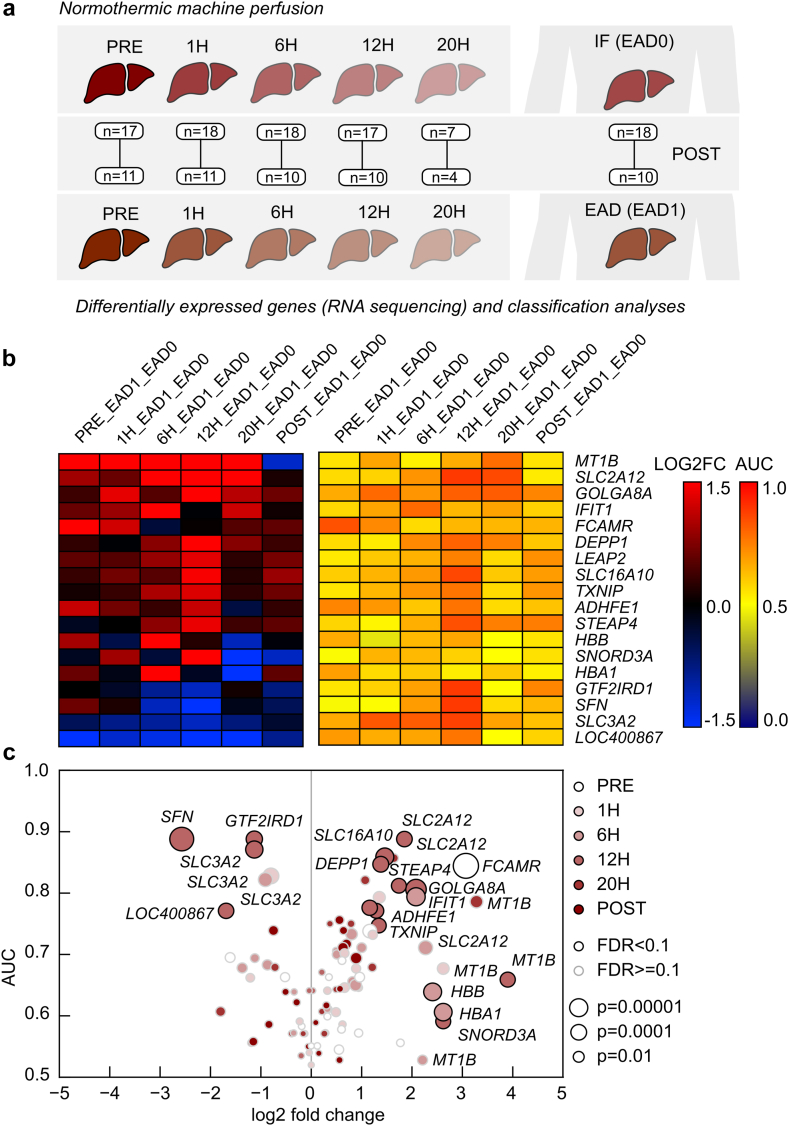
Marker genes may indicate early graft function and outcome
Finally, DEGs were compared between grafts developing EAD (n = 11) and grafts with good initial function (IF, n = 19) posttransplant (Fig. 7a). The heatmap reveals the top up- and downregulated DEGs in EAD vs IF livers at the various time-points (Fig. 7b). Most pronounced differences were observed at 12 h NMP. Significantly upregulated genes such as MT1B, TXNIP, ADHFE1 and STEAP4 are involved in cellular stress responses and oxidative/reductive pathways. MT1B is continuously and most significantly upregulated in EAD livers pre and during NMP. It encodes for metallothionein 1 B, which exerts a protective role upon oxidative stress and IRI, and regulates immune homeostasis. SLC2A12, SLC16A10 and LEAP2 encode for transporter proteins, which a play a role in cell and glucose metabolism. Significantly regulated DEGs (Fig. 7c) were correlated with L-GrAFT and MEAF as further risk scores evaluating early graft function after LT. While quite a high number of DEGs correlate well between EAD and MEAF (eg. LEAP2, STEAP4, IFIT1, TXNIP), this tendency was not confirmed in the comparison between L-GrAFT and EAD (Fig. S8). The most significant correlation (Spearman) between the three risk scores was found for LEAP2 expression at 6 h and IFIT1 expression at 20 h.
Fig. 7.
Differential gene expression analyses between livers with early allograft dysfunction (EAD, EAD1) and no early allograft dysfunction (=initial function (IF), EAD0). (a) Experimental design and number of livers for differential expression analyses and classification of EAD vs IF. (b) Heatmap of log2 fold changes (LOG2FC) and area under curve from receiver operating characteristics (AUC) at different time-points. (c) Visualization of AUC vs log2 fold changes (size of dots indicate p-values according to the legends and different time-points are indicated by colour intensity). (Test based on the negative binomial distribution using DESeq2).
To enlighten the potential different signalling pathways related to EAD and L-GrAFT, GSEA was performed for DEGs of both scores. Unfolded protein response and Myc targets V1 and V2 were most significantly decreased pre NMP and early during NMP in EAD, compared to IF livers, while inflammatory pathways were overrepresented in EAD livers, especially post reperfusion (TNF-α signalling via NFκB, allograft rejection, inflammatory response, IL2 STAT5 signalling, IL6 JAK STAT3 signalling, Fig. S9). In livers with a high L-GrAFT inflammatory pathways (inflammatory response, TNF-α signalling via NFκB, interferon α response, interferon γ response) were strikingly enhanced at 12 h and 20 h NMP and after reperfusion (Fig. S10).
As six out of 15 patients died within the first year posttransplant in the non-benchmarking case group, we finally compared gene expression profiles in benchmarking vs non-benchmarking cases at 6 h NMP. While PCA was not able to clearly distinguish between both groups based on a bunch of significantly regulated DEGs (Fig. S11a), these genes showed a significant differential expression (log2 fold change) between the two groups, as visualized in the heatmap (Fig. S11b) and AUC (Fig. S11c) at 6 h NMP.
Discussion
NMP has advanced to a clinical routine in LT in a growing number of centres. In addition to improved preservation compared to static cold storage (SCS), it offers the unique possibility to assess organs during perfusion. While the rich datasets generated during NMP offer a comprehensive description of the graft, only lactate clearing capacity has been validated in a multicentre trial as a biomarker for the risk of liver allograft dysfunction.33 A better understanding of the molecular events occurring during liver NMP and a function and pathway-oriented read of the data is warranted to develop a more fact-based approach to biomarker identification. Applying bulk RNA sequencing in a series of 45 human donor livers for up to 24 h NMP and post reperfusion, we observed upregulation of inflammatory pathways and genes involved in immune cell trafficking, but also tissue remodelling and repair. In addition, a robust IRI signature was observed on transcriptomic level throughout NMP and post reperfusion. In search of markers indicative of graft quality, a 7-gene signature was identified which discriminates between livers performing well and less adequate after 6 h NMP. Transcriptomic profiling further suggests markers predicting early graft function and outcome after LT.
To date, there is little evidence on how NMP impacts liver grafts during perfusion. Such a knowledge and understanding are required for optimization of perfusion protocols, early readout and endpoints in modification studies. We found that NMP activates immune and cell stress response pathways in human livers as early as 1 h NMP. In non-transplanted livers, a more pronounced inflammatory response was observed after a longer period of NMP on both, gene and protein levels. Immune activation and inflammation on a transcriptomic level was also reported in rat livers perfused for a short period of 4 h.8,9 Our team recently demonstrated immune cell activation along with massive mobilization of neutrophils into the perfusate during human liver NMP.10 This is in line with this more in-depth analysis presented here. Inflammatory responses were accompanied by the induction of regulatory and anti-inflammatory pathways and genes such as CD274, which inhibits T-cell proliferation and induces IL-10 producing T-cell subsets in the liver,34 and SOCS3, a suppressor of cytokine signalling.35 Interestingly, these counter-regulatory mechanisms were even more prominent in non-transplanted livers, suggesting the ability of injured livers to concomitantly induce compensating processes to mitigate further damage during NMP. Also, Jassem et al6 and Ohman et al7 found induction of regenerative processes on a transcriptomic level in a limited number of human livers during NMP. Here, cellular metabolism and OXPHOS were found downregulated after prolonged NMP and during reperfusion after LT. We have previously investigated mitochondrial function using high-resolution respirometry and did not observe any significant decline of bioenergetic performance during liver NMP.5 In fact, we have analysed this aspect in an experimental trial and found a relatively robust correlation of bioenergetic performance and outcome.36 The complexity of the electron transport chain (ETC) makes it difficult to make predictions regarding its overall function based on alterations in the expression of a small subset of genes, or whether there is a real discrepancy in these findings.
Our previously published single-cell transcriptomic data from liver NMP, generated using the microwell-based BD Rhapsody scRNAseq platform, allowed us to assess the immune cell repertoire including neutrophils, but was less focused on the gene expression analysis in hepatocytes.10 Combining our datasets with a single-cell atlas from healthy livers,18 we observed an active crosstalk between hepatocytes and immune cells on transcriptomic level during liver NMP. The identified ligands and their corresponding receptors are primarily involved in cell–cell interactions, cell adhesion and migration. Such molecules could be targeted for modulation of cell migration during liver NMP.
IRI upon organ reperfusion causes major damage, particularly in pre-injured and ECD organs. This has been investigated comprehensively at the transcriptional level in LT.37,38 To date, there is no clear evidence that NMP positively impacts the condition of a liver, but rather limits the exposure towards SCS and allows to expand the time of preservation.2,39,40 Our data suggest that genes associated with IRI21 are strongly upregulated at 1 h NMP and that this gene profile persists during NMP and after the second reperfusion event in the recipient (after approx. 30 min). Further to that, various pathways are induced upon both reperfusion events. This needs investigation in more detail, considering also graft quality and recipient-related factors. NMP could serve as a platform for targeted amelioration of molecular events related to IRI before LT.41
In search of reliable markers for graft quality and viability assessment during NMP, we compared the transcriptome and its dynamic changes of non-transplanted (pre-injured) and transplanted livers. We were able to distinguish between both groups based on the transcriptome as early as 6 h NMP and identified a 7-gene signature with a high predictive value corresponding with graft quality early during NMP. This is of particular importance as the decision whether to transplant a liver or not is often made on the dynamic changes in the perfusate, requiring NMP durations of at least several hours. Notably, CD274 gene expression was highly induced in livers which were not transplanted even at 6 h NMP. To increase feasibility in the clinical setting, its encoded protein PD-L1 could be measured in the perfusate and may therefore serve as a valuable biomarker for organ quality assessment during liver NMP. As a next step, correlation/comparison of gene expression and serum makers such as ALT, AST, LDH and lactate should be performed. Furthermore, the relevance of gene expression in predicting early graft function and outcome (survival) needs to be addressed in future studies in more detail. Since numerous scores are available for the assessment of early graft function and outcome after LT, our analysis was based on the development of EAD as a rather simple and easy score, and findings correlated to MEAF and L-GrAFT. While significantly regulated DEGs for EAD correlate well with high-risk MEAF, this was not confirmed for high-risk L-GrAFT. This could in part be explained by the various parameters and different weighting between these scores. LEAP2 expression showing the highest correlation between all three risk scores at 6 h NMP may serve as a novel marker to predict impaired early graft function after LT. This observation needs to be evaluated on protein level.
We herein provide evidence that gene expression differs between benchmarking and non-benchmarking cases. If this translates into an inferior outcome or even death, remains to be elucidated.
This study has several limitations. The decision whether to transplant a liver or not has been made primarily on perfusate markers which have not yet been formally established and approved in multicentre studies, except for lactate.33 Livers subjected to NMP reveal quite a substantial heterogeneity, especially the non-transplanted livers, as they are not standard quality organs. A comparison to optimal quality donor livers is missing. Moreover, potential outcome data of the non-transplanted livers is lacking. Finally, changes on transcriptomic level do not necessarily fully translate into proteomic changes.
NMP offers the unique possibility to expand the preservation time, allowing for a more comprehensive assessment of organs prior to transplantation. The data obtained during NMP could be of help in the decision-making process whether to transplant an organ or not, to choose the “optimal” recipient for an organ, and to predict complications and outcome in the future.
Contributors
Conceptualization and study design (TH, JT, SS), data collection (TH, HG, JH, SSa, BZ, RO, BC, AW, TR, SS), data analysis (HH, HG, RG, JH), data verification (JT, SS), visualization (TH, HH, HG, RG, JH), writing–original draft (TH, HH), writing–review & editing (all). All authors read and approved the final manuscript. TH was responsible for the decision to submit the manuscript.
Data sharing statement
Sequencing data that support the findings of this study is available through GEO (GSE263614). All additional data are available at zenodo (https://doi.org/10.5281/zenodo.10939822).
Declaration of interests
SS received consulting fees, honoraria for lectures or travel support from the following entities, not related to the present study: Merck, Atara, Nefro Health, ITB, iCoat Medical, Dialectica, Johnson & Johnson, Sanofi, OrganOx, Chiesi, Astra Zenica, XVIVO, Intuitive, Takeda. SS received an EP-Patent not related to this study (23150338.4). SS is member of the DSMB of the following study: Statins for Improving orGaN outcomE in Transplantation (SIGNET) Trial, and in the following Trial Boards: Trial Steering Committee Chair: Perfused, Liver, Utilization Study (PLUS Study), NHS, UK. The other authors have no potential conflict of interest to report.
Acknowledgements
The research was supported by the “In Memoriam Dr. Gabriel Salzner Stiftung” granted to SS, Tiroler Wissenschaftsfond granted to TH, Jubiläumsfonds—Österreichische Nationalbank granted to RO, and MUI Start grant granted to AW.
The authors would like to thank Dr. Anne Krogsdam, Astrid Drasche and Anh-Vu Nguyen for technical support.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105330.
Appendix A. Supplementary data
References
- 1.Nemes B., Gaman G., Polak W.G., et al. Extended criteria donors in liver transplantation Part I: reviewing the impact of determining factors. Expert Rev Gastroenterol Hepatol. 2016;10(7):827–839. doi: 10.1586/17474124.2016.1149061. [DOI] [PubMed] [Google Scholar]
- 2.Nasralla D., Coussios C.C., Mergental H., et al. A randomized trial of normothermic preservation in liver transplantation. Nature. 2018;557(7703):50–56. doi: 10.1038/s41586-018-0047-9. [DOI] [PubMed] [Google Scholar]
- 3.Watson C.J.E., Kosmoliaptsis V., Pley C., et al. Observations on the ex situ perfusion of livers for transplantation. Am J Transplant. 2018;18(8):2005–2020. doi: 10.1111/ajt.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mergental H., Laing R.W., Kirkham A.J., et al. Transplantation of discarded livers following viability testing with normothermic machine perfusion. Nat Commun. 2020;11(1):2939. doi: 10.1038/s41467-020-16251-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann J., Meszaros A.T., Buch M.L., et al. Bioenergetic and cytokine profiling may help to rescue more DCD livers for transplantation. Int J Mol Sci. 2023;24(11):9536. doi: 10.3390/ijms24119536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jassem W., Xystrakis E., Ghnewa Y.G., et al. Normothermic machine perfusion (NMP) inhibits proinflammatory responses in the liver and promotes regeneration. Hepatology. 2019;70(2):682–695. doi: 10.1002/hep.30475. [DOI] [PubMed] [Google Scholar]
- 7.Ohman A., Raigani S., Santiago J.C., et al. Activation of autophagy during normothermic machine perfusion of discarded livers is associated with improved hepatocellular function. Am J Physiol Gastrointest Liver Physiol. 2022;322(1):G21–G33. doi: 10.1152/ajpgi.00266.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson K.N., Pavan-Guimaraes J., Verhagen J.C., et al. Interleukin-10 and transforming growth factor-beta cytokines decrease immune activation during normothermic ex vivo machine perfusion of the rat liver. Liver Transpl. 2021;27(11):1577–1591. doi: 10.1002/lt.26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheuermann U., Zhu M., Song M., et al. Damage-associated molecular patterns induce inflammatory injury during machine preservation of the liver: potential targets to enhance a promising technology. Liver Transpl. 2019;25(4):610–626. doi: 10.1002/lt.25429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hautz T., Salcher S., Fodor M., et al. Immune cell dynamics deconvoluted by single-cell RNA sequencing in normothermic machine perfusion of the liver. Nat Commun. 2023;14(1):2285. doi: 10.1038/s41467-023-37674-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Resch T., Cardini B., Oberhuber R., et al. Transplanting marginal organs in the era of modern machine perfusion and advanced organ monitoring. Front Immunol. 2020;11:631. doi: 10.3389/fimmu.2020.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardini B., Oberhuber R., Fodor M., et al. Clinical implementation of prolonged liver preservation and monitoring through normothermic machine perfusion in liver transplantation. Transplantation. 2020;104(9):1917–1928. doi: 10.1097/TP.0000000000003296. [DOI] [PubMed] [Google Scholar]
- 13.Olthoff K.M., Kulik L., Samstein B., et al. Validation of a current definition of early allograft dysfunction in liver transplant recipients and analysis of risk factors. Liver Transpl. 2010;16(8):943–949. doi: 10.1002/lt.22091. [DOI] [PubMed] [Google Scholar]
- 14.Agopian V.G., Harlander-Locke M.P., Markovic D., et al. Evaluation of early allograft function using the liver graft assessment following transplantation risk score model. JAMA Surg. 2018;153(5):436–444. doi: 10.1001/jamasurg.2017.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pareja E., Cortes M., Hervas D., et al. A score model for the continuous grading of early allograft dysfunction severity. Liver Transpl. 2015;21(1):38–46. doi: 10.1002/lt.23990. [DOI] [PubMed] [Google Scholar]
- 16.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 18.Guilliams M., Bonnardel J., Haest B., et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185(2):379–396.e38. doi: 10.1016/j.cell.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armingol E., Officer A., Harismendy O., Lewis N.E. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet. 2021;22(2):71–88. doi: 10.1038/s41576-020-00292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sosa R.A., Terry A.Q., Kaldas F.M., et al. Disulfide high-mobility group box 1 Drives ischemia-reperfusion injury in human liver transplantation. Hepatology. 2021;73(3):1158–1175. doi: 10.1002/hep.31324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bindea G., Mlecnik B., Hackl H., et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D., Gable A.L., Nastou K.C., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamburov A., Wierling C., Lehrach H., Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37(Database issue):D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Z., Bailey A., Kuleshov M.V., et al. Gene set knowledge discovery with Enrichr. Curr Protoc. 2021;1(3) doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han H., Cho J.W., Lee S., et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sturn A., Quackenbush J., Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18(1):207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 29.Xie Y., Shi H., Han B. Bioinformatic analysis of underlying mechanisms of Kawasaki disease via weighted gene correlation network analysis (WGCNA) and the least absolute shrinkage and selection operator method (LASSO) regression model. BMC Pediatr. 2023;23(1):90. doi: 10.1186/s12887-023-03896-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S., Wang T., Luo T., et al. Prediction of graft survival post-liver transplantation by L-GrAFT risk score model, EASE score, MEAF scoring, and EAD. Front Surg. 2021;8 doi: 10.3389/fsurg.2021.753056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bral M., Gala-Lopez B., Bigam D., et al. Preliminary single-center Canadian experience of human Normothermic ex vivo liver perfusion: results of a clinical trial. Am J Transplant. 2017;17(4):1071–1080. doi: 10.1111/ajt.14049. [DOI] [PubMed] [Google Scholar]
- 32.Muller X., Marcon F., Sapisochin G., et al. Defining benchmarks in liver transplantation: a multicenter outcome analysis determining best achievable results. Ann Surg. 2018;267(3):419–425. doi: 10.1097/SLA.0000000000002477. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann J., Meszaros A.T., Butler A., et al. Predictive value of early postoperative lactate (<6 h) during normothermic machine perfusion and outcome after liver transplantation: results from a multicentre study. Br J Surg. 2024;111(6) doi: 10.1093/bjs/znae084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiegs G., Lohse A.W. Immune tolerance: what is unique about the liver. J Autoimmun. 2010;34(1):1–6. doi: 10.1016/j.jaut.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Carow B., Rottenberg M.E. SOCS3, a major regulator of infection and inflammation. Front Immunol. 2014;5:58. doi: 10.3389/fimmu.2014.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meszaros A.T., Hofmann J., Buch M.L., et al. Mitochondrial respiration during normothermic liver machine perfusion predicts clinical outcome. eBioMedicine. 2022;85 doi: 10.1016/j.ebiom.2022.104311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borozan I., Chen L., Sun J., et al. Gene expression profiling of acute liver stress during living donor liver transplantation. Am J Transplant. 2006;6(4):806–824. doi: 10.1111/j.1600-6143.2006.01254.x. [DOI] [PubMed] [Google Scholar]
- 38.Conti A., Scala S., D'Agostino P., et al. Wide gene expression profiling of ischemia-reperfusion injury in human liver transplantation. Liver Transpl. 2007;13(1):99–113. doi: 10.1002/lt.20960. [DOI] [PubMed] [Google Scholar]
- 39.Riveros S., Marino C., Ochoa G., et al. Customized normothermic machine perfusion decreases ischemia-reperfusion injury compared with static cold storage in a porcine model of liver transplantation. Artif Organs. 2023;47(1):148–159. doi: 10.1111/aor.14390. [DOI] [PubMed] [Google Scholar]
- 40.Fodor M., Cardini B., Peter W., et al. Static cold storage compared with normothermic machine perfusion of the liver and effect on ischaemic-type biliary lesions after transplantation: a propensity score-matched study. Br J Surg. 2021;108(9):1082–1089. doi: 10.1093/bjs/znab118. [DOI] [PubMed] [Google Scholar]
- 41.Liu W., Fan Y., Ding H., et al. Normothermic machine perfusion attenuates hepatic ischaemia-reperfusion injury by inhibiting CIRP-mediated oxidative stress and mitochondrial fission. J Cell Mol Med. 2021;25(24):11310–11321. doi: 10.1111/jcmm.17062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.