Abstract
The phloem macromolecular transport system plays a pivotal role in plant growth and development. However, little information is available regarding whether the long-distance trafficking of macromolecules is a controlled process or passive movement. Here, we demonstrate the destination-selective long-distance trafficking of phloem proteins. Direct introduction, into rice (Oryza sativa), of phloem proteins from pumpkin (Cucurbita maxima) was used to screen for the capacity of specific proteins to move long distance in rice sieve tubes. In our system, shoot-ward translocation appeared to be passively carried by bulk flow. By contrast, root-ward movement of the phloem RNA binding proteins 16-kD C. maxima phloem protein 1 (CmPP16-1) and CmPP16-2 was selectively controlled. When CmPP16 proteins were purified, the root-ward movement of CmPP16-1 became inefficient, suggesting the presence of pumpkin phloem factors that are responsible for determining protein destination. Gel-filtration chromatography and immunoprecipitation showed that CmPP16-1 formed a complex with other phloem sap proteins. These interacting proteins positively regulated the root-ward movement of CmPP16-1. The same proteins interacted with CmPP16-2 as well and did not positively regulate its root-ward movement. Our data demonstrate that, in addition to passive bulk flow transport, a destination-selective process is involved in long-distance movement control, and the selective movement is regulated by protein–protein interaction in the phloem sap.
INTRODUCTION
In vascular plants, phloem serves as a conduit for the delivery of photoassimilates and nutrients. It has been widely accepted that phloem translocation is driven by a pressure gradient from source to sink (Munch, 1930). In addition to the low molecular weight compounds, several recent findings have established that macromolecules, including peptides, proteins, and nucleic acids, also move long distance via the phloem (Golecki et al., 1999; Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Kim et al., 2001).
Long-distance movement of RNA through the phloem has been demonstrated for plant viral RNA (Carrington et al., 1996) and viroid RNA (Palukaitis, 1987). Moreover, plant endogenous mRNAs have been detected within functional sieve elements (Kuhn et al., 1997; Ruiz-Medrano et al., 1999; Kim et al., 2001; Doering-Saad et al., 2002), and long-distance movement of mRNA has been demonstrated (Ruiz-Medrano et al., 1999; Kim et al., 2001; Haywood et al., 2005). It has been shown that certain phloem-mobile RNAs play a pivotal role in regulating the development of distant tissues/organs (Kim et al., 2001; Haywood et al., 2005).
By contrast, the role of phloem sap proteins in long-distance signaling has yet to be defined. The presence of a wide variety of biochemically active proteins in phloem sap supports that they are involved in the coordination of the metabolism, development, and defense response at the whole plant level (Nakamura et al., 1993; Balachandran et al., 1997; Ishiwatari et al., 1998; Kehr et al., 1999; Schobert et al., 2000; Aoki et al., 2002; Yoo et al., 2002; Walz et al., 2004). Recently, increasing evidence has suggested that phloem proteins are involved in the trafficking of RNA. RNA binding proteins have been found from phloem of various plants (Xoconostle-Cázares et al., 1999, 2000; Owens et al., 2001; Yoo et al., 2004; Gomez et al., 2005). These findings provide insight into a novel function for phloem proteins as a component of an RNA-based systemic signaling mechanism.
Despite the recent progress in characterizing phloem-mobile macromolecules, our understanding of the control mechanisms for long-distance movement remains limited. It has been suggested that both plant viruses and phloem solutes are passively transported by bulk flow (Leisner and Turgeon, 1993; Roberts et al., 1997). An extensive analysis of green fluorescent protein (GFP) expressed in companion cells revealed that GFP moved nonselectively in sieve tubes (Imlau et al., 1999), indicating that GFP also moves by bulk flow. However, the occurrence of selective unloading, at specific cell boundaries, has been reported for viroid RNA (Zhu et al., 2002), viral RNA (Foster et al., 2002), posttranscriptional gene silencing signal (Voinnet et al., 1998), and viral movement protein (Itaya et al., 2002). These observations support the notion that the long-distance movement of macromolecules in the sieve tube system may not simply follow the stream of assimilates and that phloem exit in sink tissues is highly controlled. However, the control mechanism has not been elucidated.
In this study, we examined the long-distance movement of pumpkin (Cucurbita maxima) phloem proteins in the rice (Oryza sativa) sieve tube system using the tracer introduction method through cut stylets of insects. With this system, we demonstrate that certain pumpkin phloem proteins move long distance in rice. Under our experimental conditions, shoot-ward translocation of tracer appeared to be passively carried by bulk flow. On the other hand, detailed analysis of the movement of the pumpkin phloem RNA binding proteins 16-kD C. maxima phloem protein 1 (CmPP16-1) and CmPP16-2 revealed that they did not merely follow the direction of phloem bulk flow but rather moved preferentially to the root. Gel-filtration chromatography and coimmunoprecipitation experiments revealed that CmPP16-1 interacts with specific pumpkin phloem proteins, including eukaryotic initiation factor 5A, and a translationally controlled tumor protein. Cointroduction of these interacting proteins positively regulates the root-ward movement of CmPP16-1. It is also demonstrated that CmPP16-2 interacts with the same proteins, but the root-ward movement of CmPP16-2 was not positively regulated by the presence of these interacting proteins. Our results demonstrate that long-distance movement is a controlled process and that protein destination is regulated by protein–protein interaction within sieve tubes.
RESULTS
Tracer Protein Moves to Distant Organs via Phloem
To approach the question of whether the destination of phloem protein movement is controlled or not, we introduced pumpkin phloem proteins into a single rice sieve tube through a cut brown leafhopper stylet (Figures 1A to 1E). Application of tracer protein to the cut stylet allowed for protein diffusion into the sieve tube (Fujimaki et al., 2000). Typically, only a small fraction of applied tracer could diffuse into this sieve tube, and the amount of tracer successfully introduced was different from plant to plant. Once incorporated into the phloem translocation stream, tracer protein went down to the base of the leaf sheath, where branches of the rice vascular system are connected to one another, and then moved to the distant organs (Figure 1E). To estimate the sink–source status of tracer-applied leaf, we introduced radiolabeled nucleotide (see Supplemental Figure 1 online). Under our experimental conditions, most of the radioactivity was translocated to younger distant leaves and only trace amounts went to roots. This result indicated that the tracer-applied leaf functioned as a source for younger leaves and that shoot-ward bulk flow was predominant over root-ward flow.
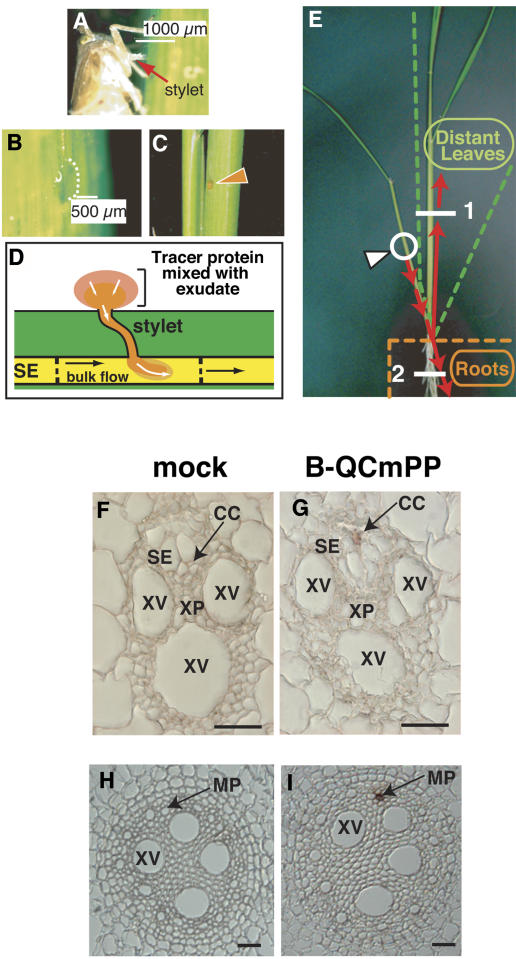
Figure 1.
Strategy to Introduce Tracer Proteins into a Single Sieve Tube.
(A) to (E) Outline of our experimental approach.
(A) Brown leafhopper feeding on the phloem sap of rice. Cut the stylet of the insect by laser.
(B) Phloem sap exudation through the cut stylet.
(C) Tracer proteins were mixed with phloem exudates.
(D) Tracer proteins entered the sieve element (SE) by diffusion. We frequently observed that the rate of exudation changed with time and could even go in the reverse (inward) direction.
(E) Translocation pathway of tracer proteins. Incorporated tracer moved toward the basal end of the leaf and then moved to another leaf or to roots. The white arrowhead indicates a tracer application point.
(F) to (I) Histochemical detection of tracer proteins in the distant organs. Leaf and root sections were obtained from positions 1 and 2, respectively, shown by the white lines in (E). No signal was detected in vascular tissues of leaf (H) or root (J) of plants treated with introduction buffer alone (mock). Tracer signal was detected in phloem tissues of distant leaves (I) and roots (K) of tracer-applied plants (B-QCmPP). The result was reproducible by sectioning three plants. CC, companion cell; MP, metaphloem tissue; SE, sieve element; XP, xylem parenchyma cell; XV, xylem vessel. Bars = 25 μm.
It has been shown that incorporated fluorescent tracer could be detected in the vicinity of the injection point (Fujimaki et al., 2000). However, we could not detect fluorescent tracer in distant organs (data not shown). To overcome this technical limitation, covalently biotinylated proteins were used as tracer to increase detection sensitivity. We first introduced biotinylated Q-Sepharose–bound C. maxima phloem sap proteins (B-QCmPP) as a tracer. In this fraction, P-protein 1 and P-protein 2, which readily form water-insoluble polymers (Beyenbach et al., 1974), were removed. It was earlier shown that the insect stylet–assisted delivery method enabled the introduction of sample into a single sieve element (Fujimaki et al., 2000). We checked whether the tracer moved long distance through the phloem by histochemical staining of distant tissues. In distant leaves, signal from tracer proteins was detected in companion cells of the phloem tissue (Figure 1G), suggesting that tracer accumulated in companion cells after long-distance translocation. In the root, signal was detected in the metaphloem tissue (Figure 1I). In mock-treated plants, background staining was detected in epidermal cells of leaves and roots (data not shown), but not in the vascular tissue (Figures 1F and 1H). Reproducible results were obtained after analysis of another two plants. These results collectively demonstrated that exogenously applied tracer proteins moved long distance via the phloem.
Profile of Tracer Proteins Detected in Distant Organs
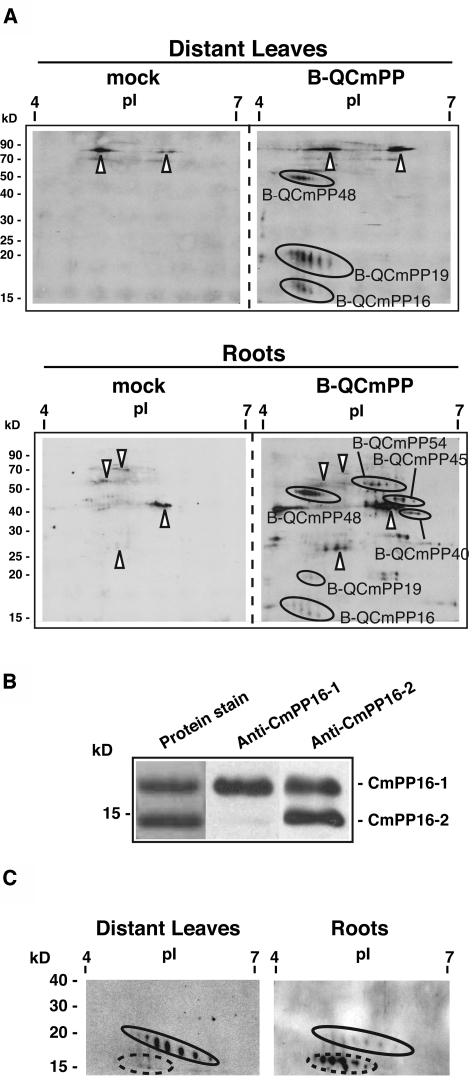
B-QCmPP tracer proteins were detected by two-dimensional electrophoresis followed by the avidin-biotin complex (ABC) detection method (Figure 2A). In distant leaves, three spot series were detected and designated B-QCmPP48, B-QCmPP19, and B-QCmPP16. In roots, in addition to spot series detected in distant leaves, three spot series were detected and designated B-QCmPP54, B-QCmPP45, and B-QCmPP40. This result implied that proteins detected in distant leaves and roots were not the same. Furthermore, the signal intensity ratio of B-QCmPP48, B-QCmPP19, and B-QCmPP16 seemed to be different in distant leaves and roots.
Figure 2.
Profile of Tracer Proteins Detected in Rice Distant Organs.
(A) Tracer proteins were detected by the combination of two-dimensional electrophoresis, on a 12% acrylamide gel, followed by ABC detection (2DE-ABC). Signals in the mock plant represent background signals from rice endogenous proteins (arrowheads). Several spot series were detected specifically in B-QCmPP-applied plants (B-QCmPP). Indicated spot series were subjected to internal peptide sequencing. BQCmPP48 was identified as SLW1, BQCmPP19 was identified as CmPP16-1, and BQCmPP16 was identified as CmPP16-2. BQCmPP54, BQCmPP45, and BQCmPP40 were left unidentified. Reproducible results were obtained from analysis of five plants. Forty micrograms of soluble protein was loaded per gel.
(B) To confirm the presence of CmPP16 proteins in rice distant organs, anti-CmPP16-1 antibody and anti-CmPP16-2 antibody were prepared. The specificity of these antibodies was examined against phloem-purified CmPP16-1 and CmPP16-2. Anti-CmPP16-1 antibody reacted with CmPP16-1 50-fold more specifically than with CmPP16-2. Anti-CmPP16-2 antibody reacted with both CmPP16-1 and CmPP16-2 to the same extent.
(C) Immunoblotting using anti-CmPP16-2 antibody revealed that B-CmPP19 (oval) and B-CmPP16 (broken oval) cross-reacted with the antibody, indicating that they were CmPP16-1 and CmPP16-2, respectively. The signal intensity ratio of the immunoreaction was similar to that of ABC detection. Proteins were run on 12% acrylamide gels. Note that immunodetection of tracer CmPP16 proteins was possible only when the biotin-derived signal in distant organs was very strong.
CmPP16-1, CmPP16-2, and SLW1 Are Identified as Long-Distance Tracers
The protein spots of B-QCmPP tracer detected in distant organs were subjected to internal peptide sequence analysis (see Supplemental Table 1 online). The spot series B-QCmPP19 was CmPP16-1, a characterized pumpkin phloem RNA binding protein (Xoconostle-Cázares et al., 1999). The spot series B-QCmPP16 was CmPP16-2 (Xoconostle-Cázares et al., 1999). The spot series B-QCmPP48 was an ortholog of silver leaf whitefly-inducible protein 1 (SLW1) (van de Ven et al., 2000). Three other spot series detected only in roots, B-QCmPP54, B-QCmPP45, and B-QCmPP40, were left unidentified because of their low abundance.
Internal peptide sequence analysis (see Supplemental Table 1 online) revealed that CmPP16-1 and CmPP16-2 identified from our sample had amino acid substitutions of Tyr-146→Asp and His-19→Leu, respectively. Additionally, it was revealed that the N-terminal Met of CmPP16-1 and CmPP16-2 was cleaved off and the exposed α-amino group of the second Gly residue was biotinylated.
The presence of the introduced CmPP16-1 and CmPP16-2 in the rice distant organs was further confirmed by immunoblot analysis. Using anti-CmPP16-2 antibody that recognized both CmPP16-1 and CmPP16-2 to the same extent (Figure 2B), biotinylated CmPP16-1 and CmPP16-2 were detected in distant leaves and roots of B-QCmPP–applied plants (Figure 2C). No immunoreaction was detected in mock-treated plants (data not shown).
Phloem Protein Moves Long Distance in a Destination-Selective Manner
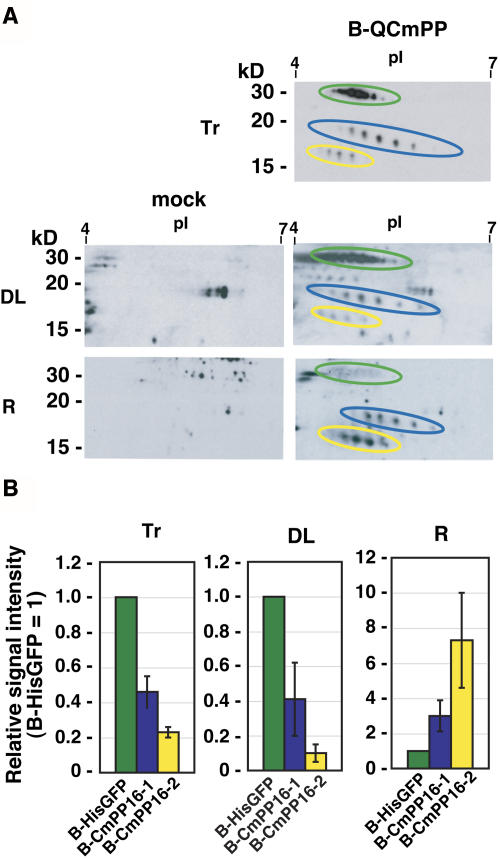
To further investigate the long-distance movement of tracer, we focused on the comparative analysis of CmPP16-1 and CmPP16-2. We included biotinylated N-terminally His-tagged GFP (B-HisGFP) in tracer as a phloem-mobile marker (Figure 3A). Hereafter, we use the term tracer to represent a mixture of biotinylated pumpkin phloem proteins and B-HisGFP. If CmPP16-1 and CmPP16-2 were just passively transported by bulk flow, the signal ratio of the B-HisGFP, biotinylated CmPP16-1 (B-CmPP16-1), and biotinylated CmPP16-2 (B-CmPP16-2) would be the same in distant leaves and roots. We first analyzed the relative signal intensity of B-QCmPP tracer (Figure 3A). The [B-HisGFP/B-CmPP16-1/B-CmPP16-2] ratio in distant leaves (1/0.41 ± 0.21/0.1 ± 0.05) was similar to the ratio in introduced tracer (1/0.46 ± 0.09/0.23 ± 0.03). By contrast, the ratio in roots was (1/3.0 ± 0.88/8.4 ± 2.7), which was significantly different from that in tracer (Figure 3B).
Figure 3.
Semiquantitative Estimation of Long-Distance Movement of CmPP16 Proteins.
(A) B-QCmPP tracer (Tr) was introduced, and then CmPP16 proteins were detected in rice distant leaves (DL) and roots (R) by 2DE-ABC detection. 2DE-ABC detection of mock-treated plants (mock) showed the background signals from rice endogenous proteins. B-HisGFP, B-CmPP16-1, and B-CmPP16-2 are indicated by green, blue, and yellow ovals, respectively.
(B) Comparison of B-HisGFP (green bars), B-CmPP16-1 (blue bars), and B-CmPP16-2 (yellow bars) signals in tracer, distant leaves, and roots. The graph shows the signal intensity ratio of B-CmPP16-1 or B-CmPP16-2 to B-HisGFP. Values represent means ± se of three independent plants.
This result strongly suggested the presence of two translocation modes. First, tracer could be passively carried by bulk flow. In particular, movement to distant leaves appeared largely attributable to bulk flow transport, because the B-HisGFP:B-CmPP16-1:B-CmPP16-2 ratio was nearly identical to that of the tracer. Second, root-ward movement of CmPP16-1 and CmPP16-2 could be selectively regulated by a mechanism distinct from B-HisGFP translocation or bulk flow transport. Furthermore, CmPP16-1 and CmPP16-2 appeared to move differentially to the root.
To test the possibility that the accumulation pattern of B-CmPP16-1 and B-CmPP16-2 was a result of differential degradation, we measured their in vitro degradation rate in distant organs (see Supplemental Figure 2 online). In leaf extracts, B-CmPP16-1 and B-CmPP16-2 degraded at nearly the same rate. In root extracts, B-CmPP16-2 degraded more rapidly than did B-CmPP16-1. This result did not explain the results described above that showed that B-CmPP16-2 accumulated to higher levels in roots than did B-CmPP16-1 (Figure 3B). Collectively, these results excluded the possibility that the differential accumulation is a consequence of differential degradation of CmPP16 proteins. The differential accumulation appeared to be the result of some form of selective movement.
Root-Ward Movement of CmPP16-1 and CmPP16-2 Is Differentially Regulated by the Presence of Other Pumpkin Phloem Proteins
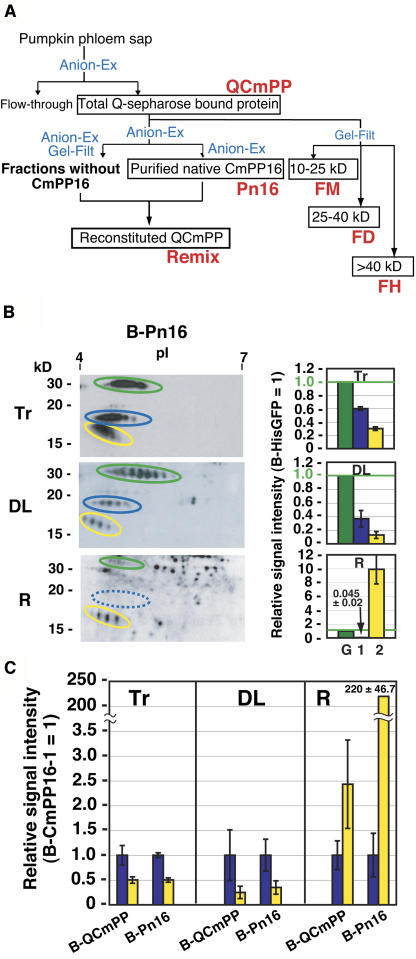
The question of how the destination-selective long-distance movement is controlled was explored next. To approach this question, pumpkin phloem sap proteins were fractionated by liquid chromatography (Figure 4A), and these fractions were subjected to movement analysis.
Figure 4.
Long-Distance Movement of Purified CmPP16 Proteins.
(A) Fractionation of pumpkin phloem sap protein by anion-exchange chromatography (Anion-Ex) and gel-filtration chromatography (Gel-Filt). Crude phloem sap was first loaded onto an anion-exchange column, and then total bound proteins were designated QCmPP. QCmPP was further fractionated by either Anion-Ex or Gel-Filt. Anion-Ex–purified CmPP16 was designated purified native CmPP16 (Pn16). All other Anion-Ex–eluted fractions free from CmPP16 protein were pooled and designated fraction without CmPP16. In reconstitution (shown in Figure 5B), Pn16 and fraction without CmPP16 were mixed to give reconstituted phloem proteins (Remix). Gel-Filt elution was subdivided into FM, FD, and FH according to molecular mass.
(B) Biotinylated Pn16 (B-Pn16; Tr) was introduced into a rice sieve tube, and then the movement of B-HisGFP (green ovals), B-CmPP16-1 (blue ovals), and B-CmPP16-2 (yellow ovals) was detected in distant leaves (DL) and roots (R) by 2DE-ABC. In the root, B-CmPP16-1 signal was under the detection limit (blue broken oval). At right, relative signal intensities of B-CmPP16-1 (bar 1) and B-CmPP16-2 (bar 2) to B-HisGFP (bar G) are shown.
(C) Comparison of relative signal intensities of B-CmPP16-2 (yellow bars) to B-CmPP16-1 (blue bars) in B-QCmPP and B-Pn16 introductions. Values represent means ± se of the signal intensity ratios from six independent plants. In the root, the [B-CmPP16-2/B-CmPP16-1] ratio increased significantly when B-Pn16 tracer was introduced. Values for B-QCmPP introduction were calculated from the relative signal intensity shown in Figure 3B.
We first compared long-distance movement patterns of CmPP16 proteins in QCmPP (Figure 3B) and in purified native CmPP16 fraction (Pn16) (Figure 4B). We compared the movement of B-CmPP16-1 and B-CmPP16-2 on the basis of the [B-CmPP16-2/B-CmPP16-1] signal ratio (Figure 4C). The ratios in distant leaves were similar to that of tracer in both B-QCmPP and B-Pn16 introduction (Figure 4C). By contrast, when B-Pn16 was introduced, the [B-CmPP16-2/B-CmPP16-1] signal ratio increased significantly in roots (Figure 4C). This increase is attributable to a remarkable decrease of B-CmPP16-1 signal in roots (Figure 4B, R), suggesting that root-ward movement of B-CmPP16-1 became inefficient by purification.
First, these results demonstrated that shoot-ward movement of B-CmPP16-1 or B-CmPP16-2 was not influenced by CmPP16 purification. This finding supported the hypothesis that, in our experimental setting, the contribution of bulk flow transport was predominant in shoot-ward translocation. Second, the root-ward movement was affected by CmPP16 purification. The decrease of apparent signal intensity of B-CmPP16-1 was conspicuous, and consequently, the [B-CmPP16-2/B-CmPP16-1] ratio increased significantly. This result demonstrated that root-ward movement of B-CmPP16-1 and B-CmPP16-2 was differentially regulated by the presence of other phloem sap proteins.
CmPP16-1 Interacts with Other Pumpkin Phloem Sap Proteins
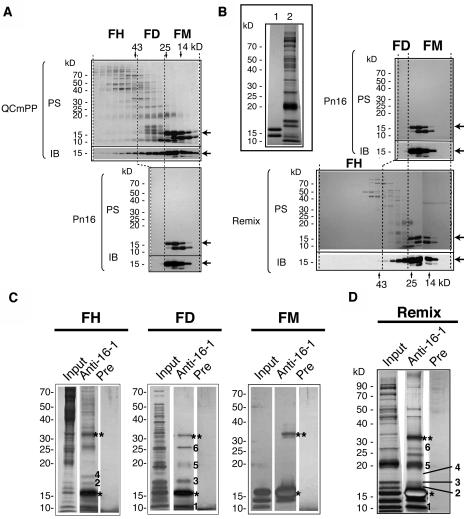
We hypothesized that a putative effector of root-ward movement was present in QCmPP but absent in Pn16 and was most likely a phloem protein. To identify the effector, we first performed gel-filtration chromatography of QCmPP to test whether CmPP16-1 interacted with other proteins (Figure 5A). Gel-filtration elution of QCmPP was fractionated into three groups: molecular mass fraction 10 to 25 kD (FM), 25 to 40 kD (FD), and >40 kD (FH) (Figures 4A and 5A). Eluted fractions were immunoblotted using anti-CmPP16-1 antibody. Anti-CmPP16-1 antibody reacted with CmPP16-1 50-fold more specifically than with CmPP16-2 (Figure 2B), although it cross-reacted with enriched CmPP16-2 in FM (Figure 5A). Immunoblotting showed that CmPP16-1 was eluted in FM, FD, and FH. Because the deduced molecular mass of CmPP16-1 is 16.5 kD, CmPP16-1 in FM was a monomer and CmPP16-1 in FD and FH should form either homocomplexes or heterocomplexes. By contrast, CmPP16-1 in Pn16 was eluted predominantly in FM and absent in FD and FH (Figure 5A). The comparison of gel-filtration profiles suggested that Pn16 did not contain the CmPP16-1 complex. Gel-filtration profiles of biotinylated QCmPP and biotinylated Pn16 were qualitatively comparable to those of the nonbiotinylated counterparts (data not shown). This result indicated that biotinylation did not disturb the complex formation of CmPP16-1, a finding confirmed by reconstitution experiments (Figure 5B). Pn16 and fraction without CmPP16 proteins were prepared separately and then mixed (Figure 4A, Remix; for protein profile, see the top left panel of Figure 5B). By gel-filtration chromatography, CmPP16-1 in Remix was eluted over a broad range, covering FM, FD, and the 40- to 50-kD range of FH (Figure 5B). This result demonstrated that putative CmPP16-1-containing complexes were reconstituted in Remix.
Figure 5.
CmPP16-1 Interacts with Specific Pumpkin Phloem Sap Proteins.
(A) and (B) Comparison of gel-filtration profiles of QCmPP (top gel) and Pn16 (bottom gel) (A) and Pn16 (top gel) and Remix (bottom gel) (B). The inset in (B) shows protein profiles of Pn16 (lane 1) and a fraction without CmPP16 proteins (lane 2). Protein samples were run on a 12.5% acrylamide gel. In each gel, the top images show protein stain by Coomassie Brilliant Blue (PS) and the bottom images show immunoblotting using anti-CmPP16-1 antibody (IB). Arrows indicate CmPP16-1. Gel-filtration elution fractions were grouped into FM (10 to 25 kD), FD (25 to 40 kD), and FH (>40 kD). In QCmPP, CmPP16-1 was eluted in FM, FD, and FH. In Pn16, most of the CmPP16-1 was eluted in FM.
(C) and (D) Immunoprecipitation assay using anti-CmPP16-1 antibody against QCmPP-derived FH, FD, and FM (C) and Remix fraction (D). Input proteins were immunoprecipitated using either anti-CmPP16-1 IgG (Anti-16-1) or preimmune IgG (Pre). Protein samples were run on a 12.5% acrylamide gel and silver-stained. The numbered bands represent coimmunoprecipitated proteins. Asterisks and double asterisks indicate CmPP16-1 monomer and CmPP16-1 dimer, respectively.
CmPP16-1 Forms a Complex with Other Pumpkin Phloem Proteins
To examine whether CmPP16-1 formed a complex with other phloem sap proteins, FM, FD, and FH were separately subjected to immunoprecipitation using anti-CmPP16-1 antibody (Figure 5C). From FM, CmPP16-2 was immunoprecipitated by inevitable cross-reaction, but no other proteins were coprecipitated. In FH and FD, several proteins were coprecipitated with CmPP16-1, suggesting that CmPP16-1 was present in heterocomplexes with other phloem sap proteins (Figure 5C). We should mention that CmPP16-2 was not immunoprecipitated from FD or FH. The Remix fraction was also subjected to immunoprecipitation. Bands 3, 5, and 6 seen in FD and FH (Figure 5C) were coimmunoprecipitated (Figure 5D), confirming that complexes were reconstituted. Obviously, Pn16 did not contain these coimmunoprecipitated proteins, suggesting that the presence of interacting proteins should be necessary for root-ward movement of CmPP16-1.
CmPP16-2 interacts with the Same Phloem Sap Proteins as CmPP16-1
We performed the same analysis for CmPP16-2. To investigate whether CmPP16-2 interacts with other phloem sap proteins, gel-filtration elution of QCmPP was probed with anti-CmPP16-2 antibody (Figure 6A). Immunoblotting revealed that CmPP16-2 was present in FD as well as in FM, although the amount of CmPP16-2 protein in FD was much less than that of CmPP16-1 (Figure 6A). This result suggested that CmPP16-2 could interact with proteins in FD.
Figure 6.
CmPP16-2 Interacts with the Same Pumpkin Phloem Sap Proteins as CmPP16-1.
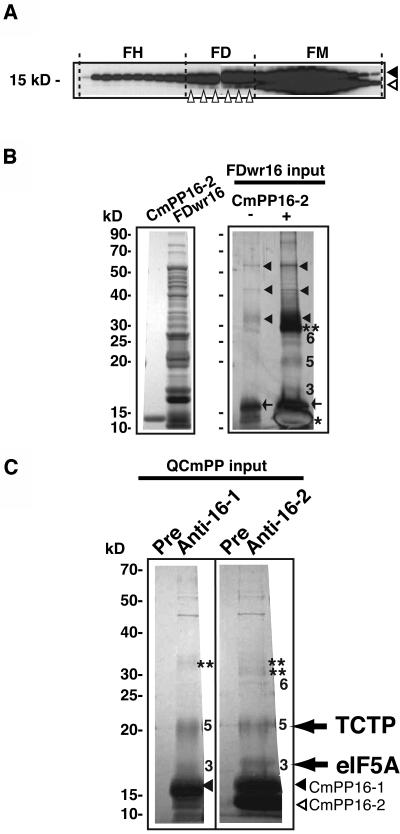
(A) Gel-filtration fractions of QCmPP were immunoblotted using anti-CmPP16-2 antibody (which cross-reacts with CmPP16-1 and CmPP16-2 equally well). Black and white arrowheads indicate CmPP16-1 and CmPP16-2, respectively. Small white arrowheads in FD indicate CmPP16-2 present in FD. CmPP16-2 was present in FM (monomer) and FD but not in FH.
(B) Phloem-purified CmPP16-2 and FD with reduced amount of CmPP16 (FDwr16) were prepared separately (left). Coimmunoprecipitation was performed to isolate CmPP16-2–interacting protein from FD using anti-CmPP16-2 antibody (right). The same amount of FDwr16 (300 μg) was applied to the anti-CmPP16-2 antibody column with (lane +, right) or without (lane −, right) the addition of CmPP16-2 (30 μg). Samples were run on a 2.5% acrylamide gel and silver-stained. Asterisks and double asterisks indicate CmPP16-2 monomer and CmPP16-2 dimer, respectively. Arrowheads indicate the nonspecifically precipitated FDwr16 proteins. Arrows indicate residual CmPP16-1 in FDwr16 captured by anti-CmPP16-2 antibody. Bands 3, 5, and 6 were identical to those in Figure 5C.
(C) Immunoprecipitation of QCmPP input using anti-CmPP16-1 IgG (Anti-16-1), anti-CmPP16-2 IgG (Anti-16-2), and preimmune IgG (Pre). Bands 3 and 5 were coimmunoprecipitated from unfractionated QCmPP. A faint signal of band 6 was detected in the Anti-16-2 lane but not in the Anti-16-1 lane. Double asterisks indicate dimers of CmPP16-1 (top) and CmPP16-2 (bottom). Internal peptide analysis revealed that bands 3 and 5 represent orthologs of eIF5A and TCTP, respectively.
This hypothesis was tested by coimmunoprecipitation. CmPP16 proteins in FD were first removed by gel-filtration chromatography in the presence of β-mercaptoethanol (Figure 6B). The resulting FD with reduced amount of CmPP16 (FDwr16) was supplemented with phloem-purified CmPP16-2 and then applied to an anti-CmPP16-2 antibody column (Figure 6B). The 17- and 21-kD proteins were coimmunoprecipitated with CmPP16-2. Interestingly, these proteins were identical to bands 3 and 5 that were coimmunoprecipitated with CmPP16-1 (Figure 5C and 5D). A 27-kD protein (band 6) was also coimmunoprecipitated with CmPP16-2. This band also seemed to be identical to band 6 that coimmunoprecipitated with CmPP16-1; however, its identity has not yet been confirmed. In control immunoprecipitation without CmPP16-2, residual CmPP16-1 in FD was immunoprecipitated. However, 17- and 21-kD proteins were not detectable, probably because CmPP16-1 was insufficient to coimmunoprecipitate a detectable amount of these proteins. Thus, we conclude that these proteins were indeed coimmunoprecipitated with CmPP16-2.
To confirm the interaction of CmPP16-1 and CmPP16-2 with pumpkin phloem sap proteins under natural conditions, a coimmunoprecipitation assay was performed using QCmPP, before any fractionation, as an input (Figure 6C). Under this condition, 17-kD (band 3) and 21-kD (band 5) proteins were coimmunoprecipitated with CmPP16-1 and CmPP16-2. This result demonstrated that CmPP16-1 and CmPP16-2 interacted with common phloem sap proteins under natural conditions.
Identification of Coimmunoprecipitated Proteins
Protein bands 3 and 5, which coimmunoprecipitated with both CmPP16-1 and CmPP16-2, were subjected to internal peptide analysis (Figure 6C; see Supplemental Table 2 online). Three peptide sequences obtained from band 3 (17 kD) matched the internal sequence of eukaryotic translation initiation factor 5A (eIF5A). The identity of band 3 to eIF5A was further confirmed by immunoblotting using anti-eIF5A antibody (see Supplemental Figure 3 online). Band 5 (21 kD) was identified as translationally controlled tumor-associated protein (TCTP). TCTP has also been characterized as IgE-dependent histamine-releasing factor (MacDonald et al., 1995), and this protein has previously been found in the phloem sap of Ricinus communis (Barnes et al., 2004). The identity of band 5 to TCTP was further confirmed by immunoblotting using anti-human histamine-releasing factor antibody (see Supplemental Figure 3 online).
The Presence of Interacting Proteins Has Different Effects on the Root-Ward Movement of CmPP16-1 and CmPP16-2
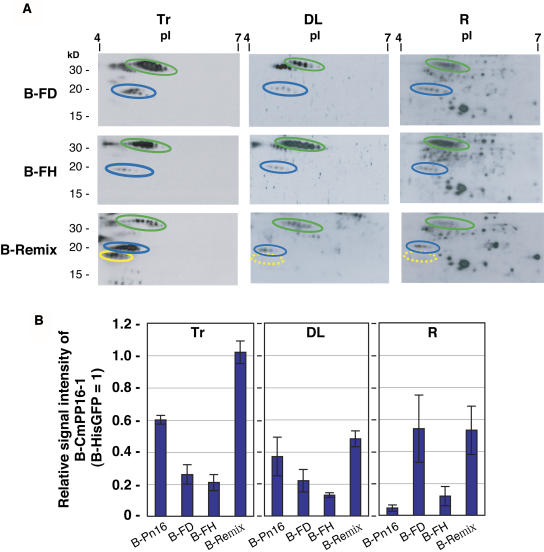
We investigated the effect of CmPP16-interacting proteins on the long-distance movement of CmPP16-1 by introducing B-Pn16, biotinylated FD (B-FD), biotinylated FH (B-FH), and biotinylated Remix (B-Remix) (Figure 7A). In vitro degradation assay demonstrated that neither FD nor FH affected the degradation of B-HisGFP (see Supplemental Figure 4 online). Unlike the faint signal of B-CmPP16-1 in roots of B-Pn16 introduction (Figure 4B), B-CmPP16-1 in B-FD was clearly detected in roots and in distant leaves (Figure 7A). B-CmPP16-1 in B-FH and B-Remix was also clearly detected in roots. These results demonstrated that the apparent signal intensity of B-CmPP16-1 in roots became higher in the presence of CmPP16-interacting proteins. Next, we estimated the movement efficiency based on the [B-CmPP16-1/B-HisGFP] signal ratio, instead of the [B-CmPP16-2/B-CmPP16-1] signal ratio, because B-FH did not contain CmPP16-2 (Figure 7B). Relative signal intensities in distant leaves were similar to those of tracers. The relative signal intensity in roots of B-FD was 10 times higher than that of B-Pn16 and even higher than that of B-FD tracer, demonstrating that FD restored the root-ward movement of B-CmPP16-1. Relative signal intensities of B-FH and B-remix were also higher than that of B-Pn16, demonstrating that FH and Remix also restored the root-ward movement of B-CmPP16-1, although less effectively than FD. These results demonstrated that fractions containing interacting proteins, particularly FD, had a positive effect on the root-ward movement of CmPP16-1.
Figure 7.
Restoration of Root-Ward Movement of CmPP16-1 by Introducing Interacting Protein–Containing Fractions.
(A) Biotinylated FD, FH, and Remix (B-FD, B-FH, and B-Remix, respectively; tracer profiles are shown in column Tr) were introduced, and then B-HisGFP (green ovals) and B-CmPP16-1 (blue ovals) movement was estimated by 2DE-ABC in distant leaves (DL) and roots (R). B-CmPP16-1 was clearly detected in roots. B-CmPP16-2 content of B-FD tracer was very low. B-FH tracer did not contain B-CmPP16-2. Although Remix tracer contained B-CmPP16-2 (∼20% the level of B-CmPP16-1), B-CmPP16-2 was not detected in distant leaves or roots (yellow broken ovals).
(B) Comparison of the relative signal intensity of B-CmPP16-1 (blue bars) to B-HisGFP. Values represent means ± se of three independent plants. Values of B-Pn16 introduction were adapted from Figure 4C. In B-FD, the relative signal intensity of B-CmPP16-1 in roots was greater than that in B-Pn16 and greater than that of B-FD tracer, demonstrating that the root-ward movement of B-CmPP16-1 was indeed restored. In FH and Remix, the relative signal intensity of B-CmPP16-1 in roots was greater than that in B-Pn16 but smaller than that of each tracer, suggesting that the extent of restoration was smaller than in B-FD.
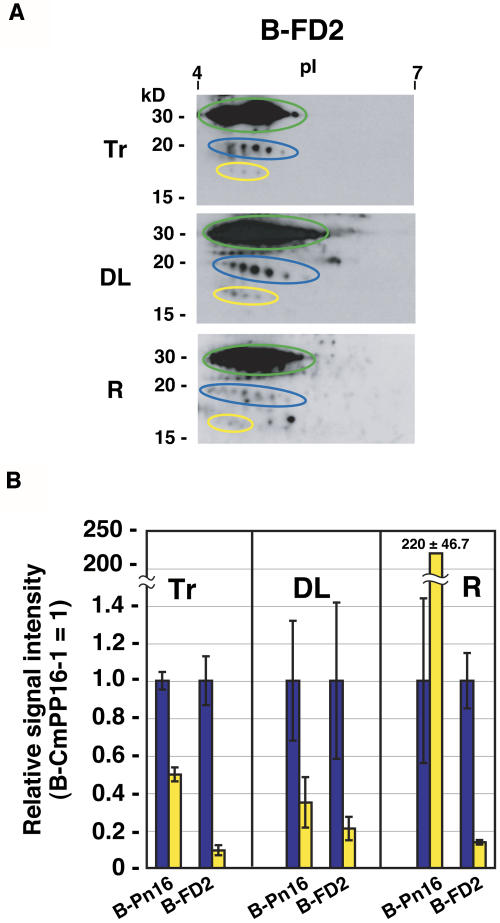
B-CmPP16-2 movement was also analyzed. We found that B-CmPP16-2 was undetectable in distant organs in B-Remix introduction, even though B-Remix tracer contained B-CmPP16-2 (Figure 7A, dotted yellow ovals). This result led us to test the possibility that the presence of interacting proteins affects the root-ward movement of CmPP16-2. Because CmPP16-2 interacted with proteins in FD (Figures 6A and 6B), we tested the effect of FD on CmPP16-2 movement. Phloem-purified CmPP16-2 was supplemented with native FD, and then a biotinylated mixture of CmPP16-2 and FD (B-FD2) was introduced (Figure 8A). The result was compared with that of B-Pn16 on the basis of the [B-CmPP16-2/B-CmPP16-1] signal ratio (Figure 8B). By B-FD2 introduction, the [B-CmPP16-2/B-CmPP16-1] signal ratio in roots was similar to that of B-FD2 tracer and was much lower than that of B-Pn16. This result demonstrated that B-CmPP16-2 in B-FD2 moved to the root less efficiently than did purified CmPP16-2, suggesting that the addition of FD did not positively regulate the root-ward movement of CmPP16-2. CmPP16-interacting proteins had a differential effect on the root-ward movement of CmPP16-1 and CmPP16-2.
Figure 8.
The Presence of Interacting Proteins Did Not Positively Regulate the Root-Ward Movement of CmPP16-2.
(A) Effect of FD addition on CmPP16-2 movement. Phloem-purified CmPP16-2 was mixed with native FD and biotinylated to obtain B-FD2. B-FD2 tracer (Tr) was introduced, and then B-HisGFP (green ovals), B-CmPP16-1 (blue ovals), and B-CmPP16-2 (yellow ovals) movement was estimated by 2DE-ABC in distant leaves (DL) and roots (R).
(B) Comparison of the relative signal intensity of B-CmPP16-2 (yellow bars) and B-CmPP16-1 (blue bars). Values represent means ± se from three independent plants. Values of B-Pn16 introduction were adapted from Figure 4C. In B-FD2 introduction, the relative signal intensity of B-CmPP16-2 in roots was much smaller than that in B-Pn16 and similar to that of B-FD2 tracer, indicating that the root-ward movement of B-CmPP16-2 was not positively regulated in the presence of FD proteins. B-CmPP16-1 was clearly detected in roots, indicating that the root-ward movement was restored, as seen in Figure 6B.
DISCUSSION
Our results demonstrate that phloem proteins move long distance by a combination of two modes: a nonselective transport by bulk flow, and a selectively regulated movement. Analysis of the root-ward movement of CmPP16-1 and CmPP16-2 revealed that their transport was under selective control. Protein–protein interaction within sieve tubes appeared to be involved in the differential regulation of the root-ward movement of CmPP16-1 and CmPP16-2.
The signal intensity of tracer in distant organs depends on three factors: the amount of tracer that was successfully diffused into the sieve tube, movement efficiency, and tracer stability. We routinely cointroduced B-HisGFP as a phloem-mobile marker. Although it had been expected that B-HisGFP moved nonselectively, as has been implied by transgenic plants expressing GFP in phloem (Imlau et al., 1999), we frequently observed that B-HisGFP signal was weaker in roots than in distant leaves (Figures 3A, 4B, and 7A). Thus, it was suspected that the movement of B-HisGFP, or the degradation of B-HisGFP (see Supplemental Figure 2 online), was selectively regulated. Because of these possibilities, B-HisGFP was not used as a reference to compare signal intensities. We basically compared the [B-CmPP16-2/B-CmPP16-1] signal ratio, so that we could demonstrate the difference between CmPP16-1 and CmPP16-2 movement.
In the analysis shown in Figure 7B, we had to use B-HisGFP as a reference, because one of the tracers of interest (B-FH) did not contain B-CmPP16-2. It was confirmed that neither FD nor FH affected the degradation of B-HisGFP in vitro; however, the 45-kD protein in FH interacted with HisGFP (data not shown). Thus, it is possible that we overestimated or underestimated the effect of FH as it might affect His-GFP movement rather than CmPP16-1 movement. On the other hand, FD did not interact with HisGFP, suggesting that FD should not affect HisGFP movement by protein–protein interaction. Therefore, we conclude that FD indeed positively regulated the root-ward movement of CmPP16-1.
In distant leaves, the [B-CmPP16-2/B-CmPP16-1] signal ratio was nearly identical to that of tracer, irrespective of the tracer species (Figures 3B, 4B, 7B, and 8B). This result strongly suggested that shoot-ward translocation was passively carried by bulk flow transport under our experimental conditions. We could roughly estimate the shoot:root allocation ratio of total incorporated tracer, and in many cases, ∼90% of incorporated B-HisGFP accumulated in distant leaves (data not shown). This allocation pattern was consistent with that of radiolabeled nucleotide. This may also explain why tracer-component ratio was constant in the shoot even when the root-ward movement pattern was affected. It is likely that relatively small perturbations in root-ward movement did not affect the bulk allocation to the shoot.
By contrast, the [B-CmPP16-2/B-CmPP16-1] signal ratio in roots was significantly different from that of tracer (Figures 3B and 4B), indicating that root-ward movement was selectively regulated by a mechanism(s) distinct from bulk flow transport. The comparison of CmPP16-1 and CmPP16-2 movement indicated that the root-ward movement of CmPP16-2 was more efficient than that of CmPP16-1. The difference was conspicuous when CmPP16 proteins were purified (Figures 4B and 4C). Furthermore, CmPP16-1 and CmPP16-2 were differentially regulated in the presence of interacting proteins of CmPP16 (IP16) (Figures 6B and 7C). An interacting protein of CmPP16-1, NtNCAPP1, has been characterized (Lee et al., 2003). In this study, we identified phloem sap-derived IP16s. Interestingly, eIF5A and TCTP interacted with both CmPP16-1 and CmPP16-2 and had different effects on their root-ward movement (Figures 6B and 7C). This differential regulation on CmPP16-1 and CmPP16-2 is consistent with the different movement patterns seen in B-QCmPP and B-Pn16 introductions. In the absence of IP16s, the root-ward movement efficiency of CmPP16-1 and CmPP16-2 was quite different, as indicated by the very high [B-CmPP16-2/B-CmPP16-1] ratio in B-Pn16 introduction (Figure 4C). When IP16s were present, they interacted with CmPP16-1 and CmPP16-2 (Figure 6C) and then positively regulated the root-ward movement of CmPP16-1 but did not upregulate CmPP16-2. Consequently, the difference between the root-ward movement of CmPP16-1 and CmPP16-2 became smaller in B-QCmPP introduction, as indicated by the smaller [B-CmPP16-2/B-CmPP16-1] ratio.
With respect to the effect of IP16s on the root-ward movement of CmPP16-2, B-FD2 introduction demonstrated that the presence of IP16s did not have a positive effect (Figure 8B). Do IP16s have a negative effect on the root-ward movement of CmPP16-2? If so, purification of CmPP16 proteins could have upregulated the root-ward movement of CmPP16-2. Comparing B-QCmPP and B-Pn16 introductions, the [B-CmPP16-2/B-CmPP16-1] ratio increased by the purification (Figure 4C). However, the increase in the ratio seemed to result from a decrease in the CmPP16-1 signal, because CmPP16-2 appeared to move to the root in B-QCmPP as efficiently as in B-Pn16. Therefore, it is still unclear whether or not IP16s have a negative effect on CmPP16-2 movement. Nevertheless, this argument does not undermine our finding that the root-ward movement of CmPP16-1 and CmPP16-2 was differentially regulated. Considering the high structural similarity between CmPP16-1 and CmPP16-2, it seems likely that they could interact with eIF5A and TCTP competitively. A study is currently under way to characterize the CmPP16–IP16 interaction and to elucidate how IP16s control CmPP16 protein movement.
In the coimmunoprecipitation experiments (Figures 5C and 5D), the amount of each IP16 was much less than that of CmPP16-1 or CmPP16-2. This result implies that the interaction between CmPP16 and IP16 is not stable. This raises the question of whether or not CmPP16 moved long distance with IP16. In the root-ward movement analysis of the IP16-containing fractions (Figures 6A and 7B), we could not confirm the presence of signals from additional proteins. Thus, it remains to be proven whether CmPP16 and IP16 move as a complex.
With respect to the functions of IP16s, it has been demonstrated that eIF5A is capable of binding to RNA (Xu and Chen, 2001). As for TCTP, it has been reported that TCTP interacts with the ribosome-associated proteins eEF1A and eEF1B and functions as a guanine nucleotide dissociation inhibitor (Cans et al., 2003). However, these known functions of eIF5A and TCTP have yet to be demonstrated for their phloem counterparts.
We propose two models for how the destination of CmPP16 movement is determined. First, a lateral transfer of CmPP16 proteins between sieve tubes could be controlled selectively. It has been reported that the distribution of photosynthate could be controlled by vascular anatomy and resistance to lateral transfer at the nodal region where vascular bundles are cross-connected (Patrick and Wardlaw, 1984). Lateral transfer of CmPP16-1 and CmPP16-2 might be controlled selectively at the node. Our second model is that destination is determined by selective exit from the sieve tube system. The selective exit from sieve elements has been reported for RNA (Ruiz-Medrano et al., 1999; Foster et al., 2002; Zhu et al., 2002), specific phloem proteins (Fisher et al., 1992), viral movement protein (Itaya et al., 2002), and small solutes (Ayre et al., 2003). The exit of CmPP16 proteins from sieve tubes might be controlled by this discrimination mechanism.
Combining the capacity for RNA binding (Xoconostle-Cázares et al., 1999), destination-selective long-distance movement of CmPP16 proteins will provide new insights into the mechanism of long-distance signaling. Our results provide an experimental basis with which to examine the possibility that CmPP16-1 and CmPP16-2 mediate the targeting of different populations of RNA selectively to the shoot or root.
METHODS
Plant Materials
Pumpkin (Cucurbita maxima cv Guiness Ponkin) plants were grown in soil culture with Arnon-Hoagland solution (Arnon and Hoagland, 1940) in a greenhouse under natural sunlight. Rice (Oryza sativa cv Kanto) plants were grown in hydroponic culture as described previously (Nakamura et al., 1993).
Collecting Pumpkin Phloem Sap Protein
Phloem exudates from cut stems and cut petioles of a 6-week-old pumpkin plant were collected in 0.1 M sodium phosphate buffer, pH 8.0, 2% (v/v) β-mercaptoethanol, and 10% (v/v) glycerol and dialyzed against EQ buffer (50 mM sodium phosphate buffer, pH 8.0, 1% [v/v] β-mercaptoethanol, and 10% [v/v] glycerol).
Fractionation of Phloem Sap Protein
Phloem sap protein was fractionated as shown in Figure 4A. All fractionation procedures were performed using an AKTA fast protein liquid chromatography system (Amersham Biosciences, Buckinghamshire, UK) at 4°C. Crude, dialyzed phloem sap was loaded onto HiTrap Q-Sepharose (Amersham Biosciences) preequilibrated with EQ buffer, and bound proteins were eluted with EQ buffer containing 1 M NaCl. At this step, most of P-protein 1 and P-protein 2 were removed. The eluate was designated QCmPP. QCmPP was loaded onto a HiTrap Q-Sepharose column after dialysis against EQ buffer, and proteins were eluted with a 0 to 500 mM NaCl linear gradient. Fractions containing CmPP16 proteins were pooled and subjected to a second round of HiTrap Q-Sepharose purification. Fractions containing CmPP16 proteins, free from contaminating proteins, were pooled, dialyzed against GF buffer (20 mM sodium phosphate buffer, pH 8.0, and 200 mM NaCl), and designated Pn16. All other fractions of first and second Q-Sepharose elution that did not contain CmPP16 proteins were pooled, and residual CmPP16 proteins were removed by gel-filtration chromatography using a Superose 12 HR 10/30 (Amersham Biosciences) preequilibrated with EQ buffer. CmPP16-free fractions were pooled and dialyzed against GF buffer. This fraction was designated fraction without CmPP16. A Remix fraction was prepared by mixing Pn16 and fraction without CmPP16 at a 1:10 weight:weight ratio followed by incubation for 3 h at 4°C with gentle agitation. To size fractionate QCmPP, QCmPP was dialyzed against GF buffer and then applied onto a Superose 12 HR 10/30 (Amersham Biosciences) preequilibrated with GF buffer at the flow rate 0.4 mL/min. Elution was subgrouped according to molecular mass to obtain FM, FD, and FH. Phloem-purified CmPP16-2, free from CmPP16-1, was obtained by passing Pn16 through a nickel-nitrilotriacetic acid (Ni-NTA) agarose Superflow column (Qiagen, Hilden, Germany) preequilibrated with GF buffer. The flow-through was collected, and then the column was washed with 5 column volumes of GF buffer. The flow-through and wash fractions were pooled and concentrated to obtain phloem-purified CmPP16-2.
cDNA Cloning
cDNA clones of CmPP16-1 and CmPP16-2 were obtained from a pumpkin stem cDNA library. Poly(A)+ RNA was prepared from young stems of a 6-week-old pumpkin plant. cDNAs were synthesized with the ZAP cDNA synthesis kit (Stratagene, La Jolla, CA). They were ligated into the λZAP vector (Stratagene) and then packaged in λ phage particles. The library was used for screening by the nucleic hybridization method (Sambrook et al., 1989). The CmPP16-1 coding sequence was used as a screening probe.
Recombinant Proteins
Recombinant proteins were obtained using the QIAexpress protein purification system (Qiagen). Standard PCR cloning techniques were used to introduce sGFP (S65T) (Chiu et al., 1996), CmPP16-1, and CmPP16-2 coding sequences into the BamHI-KpnI site of the pQE30 expression vector. Proteins were expressed in Escherichia coli M15[pREP4] strain. Expressed His-tagged proteins were then purified using a Ni-NTA Superflow column (Qiagen) and subsequently using a HiLoad 16/60 Superdex 75pg (Amersham Biosciences) gel-filtration column in combination with an AKTA fast protein liquid chromatography system.
Preparation of Biotinylated Tracer Proteins
Proteins were mixed with a 50-fold molar excess of sulfo-NHS-LC-biotin (Pierce Biotechnology, Rockford, IL) in 20 mM sodium phosphate buffer, pH 8.0, and 150 mM NaCl, and the mixture was incubated on ice in the dark for 8 h. To get rid of free sulfo-NHS-LC-biotin, the reaction mixture was passed through a Micro Bio-Spin 6 chromatography column (Bio-Rad, Hercules, CA) preequilibrated with 50 mM Tris-HCl, pH 8.0, and 150 mM NaCl. Finally, biotinylated protein was dialyzed against introduction buffer (0.9 M sucrose, 30 mM Asp, 50 mM Asn, 20 mM Glu, and 40 mM Gln).
Insect Stylet–Assisted Introduction of Tracer Proteins into a Rice Sieve Tube
A modified micro-introduction using stylet of insects method (Fujimaki et al., 2000) was used to introduce biotinylated pumpkin protein into a rice sieve tube. For controls, buffer without tracer protein was mixed with exudates, which is referred to as mock-treated plant (mock). The plastic cage was carefully put back to prevent the wicking of protein solution along the leaf surface and the accidental contamination by insects. After 16 h, tracer protein was thoroughly rinsed, and rice tissues were subjected to protein extraction. The tracer protein mixture was prepared by mixing biotinylated pumpkin phloem proteins (2.5 mg/mL B-QCmPP, 1 mg/mL B-Pn16, 3 mg/mL B-FD, 5 mg/mL B-FH, or 5 mg/mL B-Remix) with 2 mg/mL B-HisGFP. Protein concentrations indicated here represent final concentrations.
Detection of Tracer Proteins
Distant leaves included tillers. Roots included both seminal roots and crown roots. Rice tissues were ground in liquid nitrogen and homogenized in extraction buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, 1 μg/mL pepstatin A, and 1 μg/mL leupeptin) using a Polytron homogenizer (PT 3100; Kinematica, Littau, Switzerland). The homogenates were subsequently filtered through Miracloth (Calbiochem, La Jolla, CA) and then centrifuged at 38,000g for 30 min. Supernatant was concentrated using an ultrafiltration concentrator (VIVASPIN 20 mL, 5000 MWCO; Vivascience, Binbrook, UK). After determining protein concentration using Protein Assay (Bio-Rad), protein solution was dissolved in SDS-PAGE sample buffer immediately. For two-dimensional gel electrophoresis, 250 μg of distant leaf protein or 100 μg of root protein was treated with 50 units of DNase I (Roche Diagnostics, Mannheim, Germany) at 25°C for 2 h. After concentrating the DNase I reaction mixture to <100 μL, protein was precipitated using the 2-D Clean-Up kit (Amersham Biosciences). Protein precipitation was dissolved in immobilized pH gradient rehydration solution and then subjected to isoelectric focusing using an IPG Dry Strip (pH 4 to 7, 7 cm; Amersham Biosciences), followed by SDS-PAGE separation on a 12.5 or 15% acrylamide gel. Proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P transfer membrane; Millipore, Bedford, MA). The following detection procedure was performed at an ambient temperature. The membrane was blocked using 4% (w/v) Block Ace (Dainihon Seiyaku, Suita, Japan). Avidin-biotin complex was prepared 30 min before use by mixing 0.4 μg/mL Neutravidin (Pierce Biotechnology) and 0.2 μg/mL biotinylated peroxidase (Zymed, South San Francisco, CA) in TBST (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% [v/v] Tween 20). The membrane was incubated in avidin-biotin complex solution for 1 h, followed by rinsing with TBST. Signal from peroxidase was detected using the Western Lightning Chemiluminescence Reagent Plus kit (Perkin-Elmer Life Sciences, Boston, MA). Chemiluminescence signal was captured using Typhoon 8600 (Amersham Biosciences), or BioMax MS film (Eastman Kodak, Rochester, NY) when long exposure time was required, and then analyzed by ImageMaster (Amersham Biosciences) and ImageQuant (Molecular Dynamics, Sunnyvale, CA) software.
Peptide Analysis
Protein spots or bands were excised, and then gel pieces were incubated with 0.2 μg of Achromobacter protease I (a gift from Takeharu Masaki, University of Ibaraki, Ibaraki, Japan) (Masaki et al., 1981) in 0.1 M Tris-HCl, pH 9.0, 1 mM EDTA, and 0.1% (w/v) SDS at 37°C. Digested peptides were extracted and then purified as described previously (Takeda et al., 2001). Peptides were analyzed using a Procise 494 c LC peptide sequencer (Applied Biosystems) and a Reflex mass spectrometer (Bruker Daltonics, Billerica, MA).
Histochemical Detection of Tracer Proteins
Rice tissues were fixed, embedded, sectioned, and rehydrated by a conventional procedure (Hayakawa et al., 1994). After blocking with 4% Block Ace (Dainihon Seiyaku), sections were incubated at ambient temperature for 1 h with avidin-biotin complex solution in PBS containing 2 μg/mL biotinylated alkaline phosphatase (Vector Laboratories, Burlingame, CA) and 4 μg/mL Neutravidin (Pierce Biotechnology), which was preincubated at ambient temperature for 30 min before use. Finally, alkaline phosphatase activity was visualized using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium.
Antibodies and Immunoprecipitation
Anti-eIF5A antiserum was a gift from Chris Kuhlenmeier (University of Bern, Bern, Switzerland). Anti-human histamine-releasing factor antibody (HRF [L-20]: sc-20427) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-CmPP16-1 and anti-CmPP16-2 antibodies were raised against recombinant His-tagged CmPP16 proteins in rabbit. Anti-CmPP16-1/CmPP16-2 antiserum was affinity purified using a MAbTrap column (Amersham Biosciences) and subsequently using a HiTrap NHS-activated HP column (Amersham Biosciences) to which recombinant His-tagged CmPP16-1/CmPP16-2 was immobilized. Affinity-purified anti-CmPP16-1/CmPP16-2 IgG was then immobilized to ImmunoPure Plus Immobilized Protein G (Pierce Biotechnology). Immunoprecipitation assay was performed using the Seize X Immunoprecipitation kit (Pierce Biotechnology). First, to preimmune the IgG-immobilized spin column, input proteins (0.1 mg of FM, 1.1 mg of FD, 1.8 mg of FH, 1.1 mg of Remix, or 0.3 mg of FD-16 mixed with or without 30 μg of CmPP16-2) were applied and incubated at 4°C for 16 h. Unbound flow-through was collected. The flow-through was then applied to the anti-CmPP16-1/CmPP16-2 IgG spin column, and the column was incubated at 4°C for 16 h. Binding buffer and washing buffer consisted of 20 mM sodium phosphate buffer, pH 8.0, 200 mM NaCl, 1% (v/v) Triton X-100, 1 mM EDTA, and 1 μg/mL aprotinin. Protein was eluted with elution buffer (supplied with the kit), and elution was immediately neutralized by a 1/20 volume of 1 M Tris-HCl, pH 9.5. QCmPP (0.67 mg per experiment) was also subjected to immunoprecipitation as described above. Silver staining was performed as described previously (Gharahdaghi et al., 1999).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers Q9ZT47 and Q9ZT46 for CmPP16-1 and CmPP16-2, respectively.
Supplementary Material
Acknowledgments
We thank William J. Lucas (University of California, Davis) for providing CmPP16-1 cDNA. Anti-eIF5A antiserum was a gift from Chris Kuhlenmeier (University of Berne). Achromobacter protease I was a gift from Takeharu Masaki (University of Ibaraki). We thank Tomohiro Tanaka (University of Tokyo) for providing brown leafhopper and Amy Ngom and Yuichiro Miyata for technical assistance. We also thank Taku Demura (Plant Science Center, RIKEN, Institute of Physical and Chemical Research) for the two-dimensional electrophoresis equipment. This work was partly supported by a Grant-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant 15031227) to K.A.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Koh Aoki (kaoki@kazusa.or.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031419.
References
- Aoki, K., Kragler, F., Xoconostle-Cazares, B., and Lucas, W.J. (2002). A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc. Natl. Acad. Sci. USA 99, 16342–16347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon, D.I., and Hoagland, D.R. (1940). Crop production in artificial solutions and soils with special reference to factors influencing yield and absorption of inorganic nutrients. Soil Sci. 50, 463–471. [Google Scholar]
- Ayre, B.G., Keller, F., and Turgeon, R. (2003). Symplastic continuity between companion cells and the translocation stream: Long-distance transport is controlled by retention and retrieval mechanisms in the phloem. Plant Physiol. 131, 1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran, S., Xiang, Y., Schobert, C., Thompson, G.A., and Lucas, W.J. (1997). Phloem sap proteins from Cucurbita maxima and Ricinus communis have the capacity to traffic cell to cell through plasmodesmata. Proc. Natl. Acad. Sci. USA 94, 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, A., Bale, J., Constantinidou, C., Ashton, P., Jones, A., and Pritchard, J. (2004). Determining protein identity from sieve element sap in Ricinus communis L. by quadrupole time of flight (Q-TOF) mass spectrometry. J. Exp. Bot. 55, 1473–1481. [DOI] [PubMed] [Google Scholar]
- Beyenbach, J., Weber, C., and Kleinig, H. (1974). Sieve-tube proteins from Cucurbita maxima. Planta 119, 113–124. [DOI] [PubMed] [Google Scholar]
- Cans, C., et al. (2003). Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc. Natl. Acad. Sci. USA 100, 13892–13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., Kasschau, K.D., Mahajan, S.K., and Schaad, M.C. (1996). Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Doering-Saad, C., Newbury, H.J., Bale, J.S., and Pritchard, J. (2002). Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. J. Exp. Bot. 53, 631–637. [DOI] [PubMed] [Google Scholar]
- Fisher, D.B., Wu, Y., and Ku, M.S.B. (1992). Turnover of soluble proteins in the wheat sieve tube. Plant Physiol. 100, 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, T.M., Lough, T.J., Emerson, S.J., Lee, R.H., Bowman, J.L., Forster, R.L., and Lucas, W.J. (2002). A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14, 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimaki, S., Fujiwara, T., and Hayashi, H. (2000). A new method for direct introduction of chemicals into a single sieve tube of intact rice plants. Plant Cell Physiol. 41, 124–128. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi, F., Weinberg, C.R., Meagher, D.A., Imai, B.S., and Mische, S.M. (1999). Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis 20, 601–605. [DOI] [PubMed] [Google Scholar]
- Golecki, B., Schulz, A., and Thompson, G.A. (1999). Translocation of structural P proteins in the phloem. Plant Cell 11, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, G., Torres, H., and Pallas, V. (2005). Identification of translocatable RNA-binding phloem proteins from melon, potential components of the long-distance RNA transport system. Plant J. 41, 107–116. [DOI] [PubMed] [Google Scholar]
- Hayakawa, T., Nakamura, T., Hattori, F., Mae, T., Ojima, K., and Yamaya, T. (1994). Cellular localization of NADH-dependent glutamate-synthase protein in vascular bundles of unexpanded leaf blades and young grains of rice plants. Planta 193, 455–460. [Google Scholar]
- Haywood, V., Yu, T.S., Huang, N.C., and Lucas, W.J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42, 49–68. [DOI] [PubMed] [Google Scholar]
- Imlau, A., Truernit, E., and Sauer, N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwatari, Y., Fujiwara, T., McFarland, K.C., Nemoto, K., Hayashi, H., Chino, M., and Lucas, W.J. (1998). Rice phloem thioredoxin h has the capacity to mediate its own cell-to-cell transport through plasmodesmata. Planta 205, 12–22. [DOI] [PubMed] [Google Scholar]
- Itaya, A., Ma, F., Qi, Y., Matsuda, Y., Zhu, Y., Liang, G., and Ding, B. (2002). Plasmodesma-mediated selective protein traffic between “symplasmically isolated” cells probed by a viral movement protein. Plant Cell 14, 2071–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr, J., Haebel, S., Blechschmidt-Schneider, S., Willmitzer, L., Steup, M., and Fisahn, J. (1999). Analysis of phloem protein patterns from different organs of Cucurbita maxima Duch. by matrix-assisted laser desorption/ionization time of flight mass spectroscopy combined with sodium dodecyl sulfate polyacrylamide gel electrophoresis. Planta 207, 612–619. [DOI] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289. [DOI] [PubMed] [Google Scholar]
- Kuhn, C., Franceschi, V.R., Schulz, A., Lemoine, R., and Frommer, W.B. (1997). Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275, 1298–1300. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Yoo, B.C., Rojas, M.R., Gomez-Ospina, N., Staehelin, L.A., and Lucas, W.J. (2003). Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299, 392–396. [DOI] [PubMed] [Google Scholar]
- Leisner, S.M., and Turgeon, R. (1993). Movement of virus and photoassimilate in the phloem: A comparative analysis. Bioessays 15, 741–748. [DOI] [PubMed] [Google Scholar]
- MacDonald, S.M., Rafnar, T., Langdon, J., and Lichtenstein, L.M. (1995). Molecular identification of an IgE-dependent histamine-releasing factor. Science 269, 688–690. [DOI] [PubMed] [Google Scholar]
- Masaki, T., Tanabe, M., Nakamura, K., and Soejima, M. (1981). Studies on a new proteolytic enzyme from Achromobacter lyticus M497–1. I. Purification and some enzymatic properties. Biochim. Biophys. Acta 660, 44–50. [DOI] [PubMed] [Google Scholar]
- Munch, E. (1930). Die Stoffbewegungen in der Pflanze. (Jena, Germany: Fischer).
- Nakamura, S., Hayashi, H., Mori, S., and Chino, M. (1993). Protein phosphorylation in the sieve tubes of rice plants. Plant Cell Physiol. 34, 927–933. [Google Scholar]
- Owens, R.A., Blackburn, M., and Ding, B. (2001). Possible involvement of the phloem lectin in long-distance viroid movement. Mol. Plant-Microbe Interact. 14, 905–909. [DOI] [PubMed] [Google Scholar]
- Palukaitis, P. (1987). Potato spindle tuber viroid: Investigation of the long-distance, intra-plant transport route. Virology 158, 239–241. [DOI] [PubMed] [Google Scholar]
- Patrick, J.W., and Wardlaw, I.F. (1984). Vascular control of photosynthate transfer from the flag leaf to the ear of wheat. Aust. J. Plant Physiol. 11, 235–241. [Google Scholar]
- Roberts, A.G., Cruz, S.S., Roberts, I.M., Prior, D., Turgeon, R., and Oparka, K.J. (1997). Phloem unloading in sink leaves of Nicotiana benthamiana: Comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126, 4405–4419. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schobert, C., Gottschalk, M., Kovar, D.R., Staiger, C.J., Yoo, B.C., and Lucas, W.J. (2000). Characterization of Ricinus communis phloem profilin, RcPRO1. Plant Mol. Biol. 42, 719–730. [DOI] [PubMed] [Google Scholar]
- Takeda, M., Dohmae, N., Takio, K., Arai, K., and Watanabe, S. (2001). Cell cycle-dependent interaction of Mad2 with conserved Box1/2 region of human granulocyte-macrophage colony-stimulating factor receptor common betac. J. Biol. Chem. 276, 41803–41809. [DOI] [PubMed] [Google Scholar]
- van de Ven, W.T., LeVesque, C.S., Perring, T.M., and Walling, L.L. (2000). Local and systemic changes in squash gene expression in response to silverleaf whitefly feeding. Plant Cell 12, 1409–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Walz, C., Giavalisco, P., Schad, M., Juenger, M., Klose, J., and Kehr, J. (2004). Proteomics of curcurbit phloem exudate reveals a network of defence proteins. Phytochemistry 65, 1795–1804. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares, B., Ruiz-Medrano, R., and Lucas, W.J. (2000). Proteolytic processing of CmPP36, a protein from the cytochrome b(5) reductase family, is required for entry into the phloem translocation pathway. Plant J. 24, 735–747. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283, 94–98. [DOI] [PubMed] [Google Scholar]
- Xu, A., and Chen, K.Y. (2001). Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with postsystematic evolution of ligands by exponential enrichment RNA. J. Biol. Chem. 276, 2555–2561. [DOI] [PubMed] [Google Scholar]
- Yoo, B.C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y.M., Lough, T.J., and Lucas, W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16, 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, B.C., Lee, J.Y., and Lucas, W.J. (2002). Analysis of the complexity of protein kinases within the phloem sieve tube system: Characterization of Cucurbita maxima calmodulin-like domain protein kinase 1. J. Biol. Chem. 277, 15325–15332. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Qi, Y., Xun, Y., Owens, R., and Ding, B. (2002). Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.