Abstract
SATB2-associated syndrome (SAS) is an autosomal dominant neurodevelopmental disorder caused by alterations in the SATB2 gene. Here we present a review of published pathogenic variants in the SATB2 gene to date and report 38 novel alterations found in 57 additional previously unreported individuals. Overall, we present a compilation of 120 unique variants identified in 155 unrelated families ranging from single nucleotide coding variants to genomic rearrangements distributed throughout the entire coding region of SATB2. Single nucleotide variants predicted to result in the occurrence of a premature stop codon were the most commonly seen (51/120 = 42.5%) followed by missense variants (31/120 = 25.8%). We review the rather limited functional characterization of pathogenic variants and discuss current understanding of the consequences of the different molecular alterations. We present an expansive phenotypic review along with novel genotype-phenotype correlations. Lastly, we discuss current knowledge of animal models and present future prospects. This review should help provide better guidance for the care of individuals diagnosed with SAS.
Keywords: genotype-phenotype correlation, pathogenic variants, SATB2, SATB2-associated syndrome, whole exome sequencing
1 ∣. BACKGROUND
SATB2-associated syndrome (SAS; MIM# 612313, Glass syndrome) is an autosomal dominant disorder first reported in 1989 in a 16-year-old male with severe intellectual disability and an interstitial deletion of 2q32.2-2q33.1 (Glass, Swindlehurst, Aitken, McCrea, & Boyd, 1989). Clinically, SAS is characterized by developmental delay/intellectual disability with absent or limited speech development, palatal and dental abnormalities, feeding difficulties, behavioral problems, and dysmorphic facial features (Docker et al., 2014; Zarate et al., 2015; Zarate, Smith-Hicks et al., 2018). Other supportive findings such as skeletal anomalies with low bone density and abnormal brain neuroimaging have been described (Zarate, Smith-Hicks et al., 2018; Zarate, Steinraths et al., 2018).
SAS is caused by the alterations of SATB2 that can include single nucleotide variants (loss-of-function as well as missense), intragenic deletions and duplications, contiguous deletions, and translocations with secondary gene disruption (Zarate & Fish, 2017; Zarate, Kaylor, & Fish, 1993). While haploinsufficiency of SATB2 seems the most likely mechanism of disease, a dominant negative effect has been suggested in at least one instance in an individual with a nonsense variant (Leoyklang et al., 2013). The SATB2 gene maps to 2q32-q33 and has three transcripts (NM_001172509, NM_001172517, and NM_015265), codes for SATB2, an 82.6 kDa protein of 733 amino acids.
SATB2 binds to nuclear matrix-attachment regions (MARs) where it organizes chromatin to regulate tissue-specific gene regulatory networks (GRNs), and thus has critical roles in multiple developmental processes (Britanova et al., 2006; Dobreva, Dambacher, & Grosschedl, 2003; Dobreva et al., 2006; Gyorgy, Szemes, de Juan Romero, Tarabykin, & Agoston, 2008). The SATB2 protein has two CUT domains and a homeodomain (FitzPatrick et al., 2003) that are highly conserved across vertebrate taxa (FitzPatrick et al., 2003; Sheehan-Rooney, Palinkasova, Eberhart, & Dixon, 2010). The CUT domains and homeodomains are both DNA-binding motifs, which may bind DNA independently or cooperatively.
Clinically, SAS has been characterized through two large cohort studies (Bengani et al., 2017; Zarate, Smith-Hicks et al., 2018). We recently presented the common clinical features and natural history of 72 individuals with SAS due to a variety of molecular mechanisms. In this study, we review the previously described individuals with SAS and present 57 additional individuals that expand the mutation spectrum seen in this condition and describe novel genotype-phenotype correlations. All families reported for the first time were enrolled under a research clinical registry protocol approved by the Institutional Review Board of the University of Arkansas for Medical Sciences.
2 ∣. VARIANTS
All SATB2 variants are described according to current HGVS mutation nomenclature guidelines based on Genbank accession number NM_015265 (den Dunnen et al., 2016). Novel variants are interpreted using ACMG classification recommendations (Richards et al., 2015). This report excludes larger deletions and duplications that encompass SATB2 along with adjacent genes.
Tables 1,2 detail all 101 previously published SATB2 intragenic alterations in the international peer-reviewed literature (PubMed database) and the Human Gene Mutation Database (HGMD professional 2018.3; Brewer et al., 1999; Leoyklang et al., 2007; Baptista et al., 2008; Rosenfeld et al., 2009; Tegay et al., 2009; Balasubramanian et al., 2011; Rauch et al., 2012; Talkowski et al., 2012; Asadollahi et al., 2014; Gilissen et al., 2014; Lieden, Kvarnung, Nilssson, Sahlin, & Lundberg, 2014; Rainger et al., 2014; Trakadis et al., 2014; Farwell et al., 2015; Kaiser et al., 2015; Zarate et al., 2015; Boone et al., 2016; Lee et al., 2016; Bengani et al., 2017; Bowling et al., 2017; Deciphering Developmental Disorders Study, 2017; Schwartz, Wilkens, Noon, Krantz, & Wu, 2017; Vissers et al., 2017; Zarate et al., 2017; Cherot et al., 2018; Kikuiri et al., 2018; Lv et al., 2018; Scott et al., 2018; Zarate, Smith-Hicks et al., 2018; Zarate, Steinraths et al., 2018). In this study, we also report 57 additional individuals with 47 SATB2 alterations (Tables 1,2) that have been submitted to the LOVD database: https://databases.lovd.nl/shared/genes/SATB2.
TABLE 1.
SATB2 variants found in individuals with SATB2-associated syndrome previously reported and from this study (numbering is according to the cDNA sequence (GenBank entry NM_015265.3)
| Variant type | Position | DNA variant | Predicted effect on protein |
Unrelated families in our cohort |
Unrelated families in the literature |
Variant reference (dbSNP, ClinVar) |
References |
|---|---|---|---|---|---|---|---|
| Nonsense | Exon 3 | c.124G>T | p.Gly42* | 1 | RCV000486598.1; rs1064793947 | Zarate et al. (1993); Zarate, Smith-Hicks et al. (2018) | |
| Exon 4 | c.346G>T | p.Gly116* | 1 | Zarate, Smith-Hicks et al. (2018); Scott et al. (2018) | |||
| Exon 5 | c.390T>A | p.Tyr130* | 1 | Bengani et al. (2017); Zarate, Smith-Hicks et al. (2018) | |||
| Exon 6 | c.505C>T | p.Gln169* | 1 | Zarate, Smith-Hicks et al. (2018); Scott et al. (2018) | |||
| Exon 7 | c.688A>T | p.Lys230* | 1 | Vissers et al. (2017) | |||
| Exon 8 | c.715C>T | p.Arg239* | 3 | 3 | RCV000002627.6; RCV000256175.2; rs137853127 | Leoyklang et al. (2007); This study | |
| Exon 8 | c.748C>T | p.Gln250* | 1 | RCV000708552.1 | Zarate et al. (2017) | ||
| Exon 8 | c.847C>T | p.Arg283* | 2 | 4 | RCV000190685.3; RCV000254800.2; RCV000686152.1; rs797044874 | Zarate et al. (2017); Kikuiri et al. (2018); This study | |
| Exon 8 | c.868C>T | p.Gln290* | 2 | RCV000413829.1; RCV000624904.1; rs1057518496 | This study | ||
| Exon 8 | c.988C>T | p.Gln330* | 1 | This study | |||
| Exon 8 | c.997C>T | p.Gln333* | 1 | RCV000599413.1 | This study | ||
| Exon 8 | c.1135C>T | p.Gln379* | 1 | This study | |||
| Exon 8 | c.1171C>T | p.Gln391* | 1 | RCV000708554.1 | Zarate, Smith-Hicks et al. (2018); Scott et al. (2018) | ||
| Exon 9 | c.1255C>T | p.Gln419* | 1 | Zarate et al. (1993); Zarate, Smith-Hicks et al. (2018); Scott et al. (2018) | |||
| Exon 9 | c.1285C>T | p.Arg429* | 1 | 3 | RCV000307104.1; rs886041847 | Bengani et al. (2017); This study | |
| Exon 9 | c.1375C>T | p.Arg459* | 1 | 4 | RCV000625170, RCV000680089.2, RCV000719613.1 | Bengani et al. (2017); This study; This study | |
| Exon 10 | c.1495A>T | p.Lys499* | 1 | RCV000224980.1; rs878853163 | Zarate, Smith-Hicks et al. (2018); Bowling et al. (2017); Scott et al. (2018) | ||
| Exon 12 | c.1756C>T | p.Gln586* | 1 | This study | |||
| Exon 12 | c.2074G>T | p.Glu692* | 1 | RCV000708559.1 | Bengani et al. (2017) | ||
| Missense | Exon 4 | c.185T>A | p.Val62Asp | 1 | RCV000479021.1; rs1064796649 | Zarate, Smith-Hicks et al. (2018) | |
| Exon 4 | c.257T>G | p.Leu86Arg | 1 | This Study | |||
| Exon 4 | c.287T>G | p.Leu96Arg | 1 | RCV000485845.1; rs1064795247 | Zarate, Smith-Hicks et al. (2018) | ||
| Exon 4 | c.346G>C┼ | p.Gly116Arg | 1 | RCV000494383.1; rs1131691672 | Zarate et al. (1993); Zarate, Smith-Hicks et al. (2018) | ||
| Exon 5 | c.392T>A | p.Val131Glu | 1 | RCV000485195.1; rs1064794638 | Zarate, Smith-Hicks et al. (2018) | ||
| Exon 8 | c.760C>T | p.His254Tyr | 1 | This study | |||
| Exon 8 | c.983C>A | p.Ala328Asp | 1 | Farwell et al. (2015) | |||
| Exon 8 | c.1102G>T | p.Val368Phe | 1 | This study | |||
| Exon 8 | c.1136A>C | p.Gln379Pro | 1 | This study | |||
| Exon 8 | c.1142T>G | p.Val381Gly | 1 | RCV000708553.1 | Rauch et al. (2012) | ||
| Exon 8 | c.1157C>T | p.Ala386Val | 1 | Bengani et al. (2017); Zarate, Smith-Hicks et al. (2018) | |||
| Exon 8 | c.1165C>T | p.Arg389Cys | 5 | 5 | RCV000430827.1; RCV000623230.1; RCV000656508.2; rs1057521083 | Bengani et al. (2017); This study | |
| Exon 8 | c.1166G>T | p.Arg389Leu | 1 | 1 | Bengani et al. (2017); This study | ||
| Exon 8 | c.1169C>T | p.Thr390Ile | 1 | 1 | RCV000199456.1; rs863224917 | Zarate, Smith-Hicks et al. (2018); Zarate, Steinraths et al. (2018); This study | |
| Exon 9 | c.1174G>C | p.Gly392Arg | 1 | Zarate, Smith-Hicks et al. (2018); Zarate, Steinraths et al. (2018) | |||
| Exon 9 | c.1175G>A | p.Gly392Glu | 1 | RCV000489186.1; rs1085308028 | This study | ||
| Exon 9 | c.1181T>C | p.Leu394Ser | 1 | Bengani et al. (2017) | |||
| Exon 9 | c.1186G>C | p.Glu396Gln | 1 | RCV000708555.1 | Lee et al. (2016) | ||
| Exon 9 | c.1196G>A | p.Arg399His | 2 | 2 | RCV000413401.1; rs1057518190 | Bengani et al. (2017); Zarate, Smith-Hicks et al. (2018); Scott et al. (2018); This study | |
| Exon 9 | c.1196G>T | p.Arg399Leu | 2 | This study | |||
| Exon 9 | c.1196G>C | p.Arg399Pro | 1 | SCV000847591.1; rs1057518190 | Zarate, Smith-Hicks et al. (2018); Zarate, Steinraths et al. (2018) | ||
| Exon 9 | c.1204G>A | p.Glu402Lys | 1 | Bengani et al. (2017); Zarate, Smith-Hicks et al. (2018) | |||
| Exon 9 | c.1253T>G | p.Met418Arg | 1 | This study | |||
| Exon 9 | c.1286G>A | p.Arg429Gln | 1 | 3 | RCV000300452.2; RCV000708556.1; rs886041516 | Zarate et al. (1993); Zarate, Smith-Hicks et al. (2018); This study | |
| Exon 10 | c.1541A>G | p.Gln514Arg | 1 | This study | |||
| Exon 11 | c.1543G>A | p.Gly515Ser | 1 | SCV000778507.1 | Bengani et al. (2017) | ||
| Exon 11 | c.1554T>G | p.Cys518Trp | 1 | This study | |||
| Exon 11 | c.1564C>T | p.Arg522Cys | 1 | This study | |||
| Exon 11 | c.1696G>A | p.Glu566Lys | 1 | RCV000482629.1; RCV000656509.1; rs1064795530 | Bengani et al. (2017) | ||
| Exon 12 | c.1903G>T | p.Asp635Tyr | 1 | This study | |||
| Exon 12 | c.1964C>T | p.Pro655Leu | 1 | RCV000502290.1 | Zarate et al. (1993); Zarate, Smith-Hicks et al. (2018) | ||
| Frameshift | Exon 3 | c.9_16del | p.Arg4Glufs*68 | 1 | This study | ||
| Exon 3 | c.19dupA | p.Ser7Lysfs*68 | 1 | RCV000519084.1; RCV000709933.1; | This study | ||
| Exon 4 | c.245_263del | p.Ala82Glyfs*30 | 1 | This study | |||
| Exon 4 | c.334delG | p.Ala112Profs*6 | 1 | This study | |||
| Exon 5 | c.400delG | p.Ala134Hisfs*17 | 1 | Zarate et al. (1993); Zarate, Smith-Hicks et al. (2018) | |||
| Exon 6 | c.482delA | p.Lys161Serfs*19 | 1 | RCV000479959.1; rs1064793820 | Zarate et al. (1993); Zarate, Smith-Hicks et al. (2018) | ||
| Exon 6 | c.583dupT | p.Cys195Leufs*14 | 1 | Zarate et al. (1993); Bengani et al. (2017); Zarate, Smith-Hicks et al. (2018); Zarate, Steinraths et al. (2018) | |||
| Exon 6 | c.594_595delCC | p.Gln199Glufs*9 | 1 | Bengani et al. (2017) | |||
| Exon 8 | c.808_809delCA | p.Gln270Valfs*33 | 1 | RCV000623347.1 | Zarate, Smith-Hicks et al. (2018); Scott et al. (2018) | ||
| Exon 8 | c.816delT | p.His273Thrfs*21 | 1 | Zarate et al. (1993); Zarate, Smith-Hicks et al. (2018) | |||
| Exon 8 | c.832delC | p.His278Thrfs*16 | 1 | This study | |||
| Exon 8 | c.1131_1132delGT | p.Ser378Profs*18 | 1 | RCV000209866.1; rs875989830 | Zarate, Smith-Hicks et al. (2018) | ||
| Exon 8 | c.1132_1133insAC | p.Ser378Tyrfs*36 | 1 | This study | |||
| Exon 9 | c.1196delG | p.Arg399Leufs*14 | 1 | RCV000624362.1 | This study | ||
| Exon 9 | c.1203dupA | p.Glu402Argfs*35 | 1 | Bengani et al. (2017) | |||
| Exon 9 | c.1311_1314dupG-GAG | p.Arg439Glyfs*38 | 1 | SCV000845949.1 | Zarate, Smith-Hicks et al. (2018) | ||
| Exon 10 | c.1478_1479delAG | p.Gln493Argfs*19 | 1 | Kikuiri et al. (2018) | |||
| Exon 10 | c.1498delG Mosaic | p.Val500Cysfs*46 | 1 | This study | |||
| Exon 10 | c.1511_1512in-sCAAGCCT | p.Phe505Lysfs*10 | 1 | This study | |||
| Exon 10 | c.1515delT | p.Phe505Leufs*41 | 1 | RCV000255520.1; rs886039740 | Zarate, Smith-Hicks et al. (2018); Scott et al. (2018) | ||
| Exon 11 | c.1574delA | p.Glu525Glyfs*21 | 1 | Bengani et al. (2017) | |||
| Exon 11 | c.1592dupA | p.Asn531Lysfs*21 | 1 | RCV000533352.1 | Zarate, Smith-Hicks et al. (2018) | ||
| Exon 11 | c.1627delC | p.Arg543Alafs*3 | 1 | RCV000496200.1; rs1135401803 | Cherot et al. (2018) | ||
| Exon 11 | c.1639_1642de-linsTTT | p.Leu547Phefs*77 | 1 | This study | |||
| Exon 11 | c.1645delC | p.Gln549Serfs*75 | 1 | Bengani et al. (2017) | |||
| Exon 11 | c.1657delG | p.Asp553Metfs*71 | 1 | This study | |||
| Exon 11 | c.1728delT | p.Glu577Serfs*47 | 1 | Zarate et al. (1993); Zarate, Smith-Hicks et al. (2018) | |||
| Exon 12 | c.1945dupT | p.Ser649Phefs*40 | 1 | SCV000837674.1 | Zarate et al. (2017); Zarate, Smith-Hicks et al. (2018) | ||
| Exon 12 | c.1972_1973delAC | p.Thr658Hisfs*30 | 1 | Zarate, Smith-Hicks et al. (2018) | |||
| Exon 12 | c.2005dupC | p.His669Profs*20 | 1 | Zarate, Smith-Hicks et al. (2018); Zarate, Steinraths et al. (2018) | |||
| Exon 12 | c.2018dupA | p.His673Glnfs*16 | 1 | RCV000708558.1 | Boone et al. (2016) | ||
| Exon 12 | c.2028delG | p.Glu678Serfs*18 | 1 | Zarate, Smith-Hicks et al. (2018) | |||
| In-frame insertion | Exon 8 | c.929_930in-sTTGTAAGGCAAC | p.Q310delinsHis CysLysAlaThr | 1 | Gilissen et al. (2014) | ||
| Splice | Exon 4 | c.346+2T>G | p.? | 1 | SCV000837664.1 | Zarate et al. (2017) | |
| Exon 5 | c.473+1delG | p.? | 1 | RCV000598568.1 | This study | ||
| Exon 7 | c.598-2A>G | p.? | 1 | Bengani et al. (2017) | |||
| Exon 8 | c.1173+2T>C | p.? | 1 | Bengani et al. (2017) | |||
| Exon 9 | c.1174-2A>G | p.? | 1 | Zarate, Smith-Hicks et al. (2018); Scott et al. (2018) | |||
| Exon 12 | c.1741-1G>A | p.? | 1 | Kikuiri et al. (2018) | |||
| Translocation | t(2;11) cis regulatory | c.? | p.? | 1 | Rainger et al. (2014) | ||
| t(2;3) cis regulatory | c.? | p.? | 1 | Rainger et al. (2014) | |||
| t(2;7) intron 3 | c.? | p.? | 1 | Brewer et al. (1999) | |||
| t(2:14) intron 10 | c.? | p.? | 1 | Tegay et al. (2009) | |||
| t(2;10) Intron 8-to 11 | c.? | p.? | 1 | Baptista et al. (2008) | |||
| t(2;6) | c.? | p.? | 1 | Talkowski et al. (2012) |
This variant is also predicted to affect splicing if not resulting in a missense substitution.
TABLE 2.
Intragenic SATB2 chromosomal abnormalities previously reported and from this report.
| Abnormality | Position | Min sequence coordinates hg19 | Size (kb) | Methodology | Reference |
|---|---|---|---|---|---|
| Intragenic deletions | Exons 9–11 | chr2:200,168,993-200,203,730 | 35 | 105 K Oligo Agilent | Balasubramanian et al. (2011) |
| Exons 4–12 | chr2:200,128,960-200,312,555 | 183.6 | 105 K Oligo Agilent | Rosenfeld et al. (2009) | |
| Exons 3–11 | chr2:200,151,982-200,325,064 | 173.1 | 105 K Oligo Agilent | Rosenfeld et al. (2009) | |
| Exons 1–11 | chr2:200,151,782-200,336,956 | 185.2 | 105 K Oligo Agilent | Rosenfeld et al. (2009) | |
| Exons 1–8 | chr2: 200,199,481-200,340,178 | 141 | SNP Affymetrix | Zarate, Smith-Hicks et al. (2018) | |
| Exons 4–8 | chr2: 200,198,963-200,314,171 | 115 | SNP Affymetrix | This report | |
| Exons 9–10 | chr2: 200,180,939-200,200,560 | 19.6 | OGT 60k oligo | Zarate, Smith-Hicks et al. (2018) | |
| Exons 1–11 | chr2:200,151,782-200,336,956 | 185 | 105 K Oligo Agilent | Zarate, Smith-Hicks et al. (2018) | |
| Exons 5–12 | chr2: 200,018,395-200,246,466 | 228 | 60 K Oligo Agilent | Zarate, Smith-Hicks et al. (2018) | |
| Exon 9 | chr2:200,190,560-200,200,832 | 10 | Affymetrix 750k | Zarate, Smith-Hicks et al. (2018) | |
| Exons 2–4 | chr2:200,280,770-200,325,182 | 45 | OGT 8 × 60k | Zarate, Smith-Hicks et al. (2018) | |
| Exons 5–8 | chr2:200,212,737-200,256,585 | 44 | Affymetrix Cytoscan HD | Zarate, Smith-Hicks et al. (2018) | |
| Exon 4 | chr2:200,292,552-200,302,732 | 10.2 | Affymetrix Cytoscan HD | Zarate, Smith-Hicks et al. (2018) | |
| Exons 4–8 | chr2:200,195,604-200,315,398 | 120 | CombiSNP | This report | |
| Exons 8–9 | chr2:200,192,250-200,230,824 | 39 | Affymetrix Cytoscan HD | This report | |
| Exons 1–12 | chr2:200,133,428-200,450,474 | 317 | Agilent 8 × 60 K | This report | |
| Exons 7–8 | chr2:200,197,135-200,242,625 | 45.5 | Affymetrix Cytoscan HD | This report | |
| Exons 7–8 | chr2:200,194,727-200,240,771 | 46 | 180k Cytosure ISCA v2 | This report | |
| Exon 5 | chr2:200,246,407-200,296,329 | 50 | Agilent 4 × 180 K, PCR | This report | |
| Exons 1–4 | chr2:200,275,266-200,344,231 | 69 | Oligo array+SNP | This report | |
| Exon 7 | chr2:200,222,207-200,243,660 | 21.4 | Research WGS (MLPA confirmed) | This report | |
| Exon 9 | 1.chr2:200,193,421-200,193,634 | 0.213┼ | MLPA | This report | |
| Intragenic duplications | Exon 4 | chr2:200,278,502-200,310,272 | 32 | Affymetrix Cytoscan HD, MLPA | Asadollahi et al. (2014) |
| Exon 4 | chr2:200,256,546-200,310,885 | 54 | Affymetrix Cytoscan HD, PCR, MLPA | Kaiser et al. (2015) | |
| Exons 5–7 | chr2:200,233,354-200,255,458 | 35 | 180 K Oligo Agilent, MLPA | Lieden et al. (2014) |
Abbreviations: MLPA, multiplex ligation-dependent probe hybridization; PCR, polymerase chain reaction.
The exact location of the breakpoints of this deletion is unknown as the MLPA kit used only covered limited exons.
Overall, including our data and those of the literature, a total of 120 unique variants were found in 158 individuals from 155 unrelated families. While all types of pathogenic variants were found, single nucleotide variants that are predicted to result in the occurrence of a premature stop codon were the most commonly seen (51/120 = 42.5%). Missense variants were also frequently found (25.8%), followed by intragenic deletions (18.3%), translocations (5%), splice site alterations (5%), intragenic duplications (2.5%), and a single in-frame alteration (0.8%; Figure S1).
2.1 ∣. Point pathogenic variants
Eighty-nine distinct point variants including single base substitutions and small deletions/insertions were found in 127 individuals from 125 families (Figure 1; Liu et al., 2015). Most molecular diagnostics were performed by whole exome sequencing (WES; 105/127 = 82.7%) with the remaining individuals obtaining the diagnosis through a different next-generation sequencing platform (epilepsy, intellectual disability or Angelman syndrome dedicated panels, 11.8%), SATB2 Sanger sequencing (3.1%), or whole genome sequencing (WGS; 2.4%). De novo status was confirmed in almost all instances when parental testing was performed (98.1%, 105/107 families), including a pair of monozygotic twins. The remaining two instances correspond to a pair of siblings found to have the same variant indicating germline mosaicism and a case of low-level blood mosaicism in a father of a single SAS-affected individual previously reported. Here, we present individual (SATB2-135) with a de novo mosaic pathogenic variant (c.1498delG) as determined by WES (32/143 reads) and presenting with the common phenotypic features.
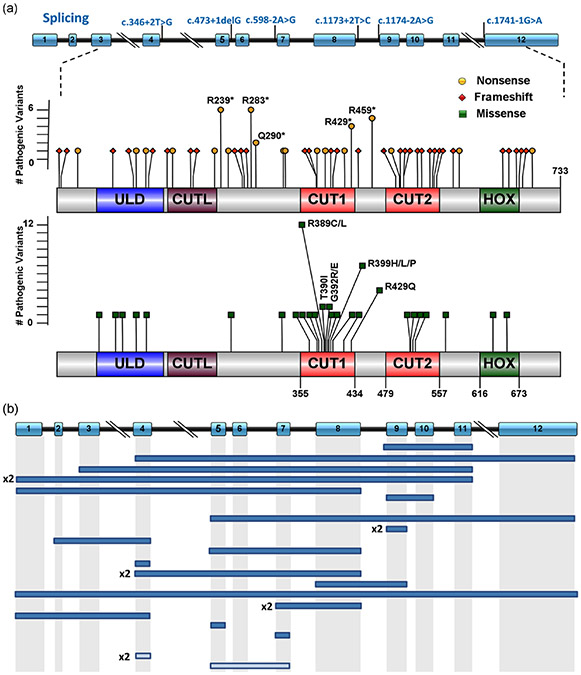
FIGURE 1.
Schematic representation of the spectrum of SATB2 variants previously described and from this study. (a) Splicing and coding exonic pathogenic variants. Splicing variants are represented along the genomic structure of the SATB2 gene according to NM_015265.3 including 12 exons (boxes) and introns (black horizontal lines). Exonic pathogenic variants are illustrated according to changes at the protein level (p.) by corresponding mutation types as follows: green squares for missense variants, red diamonds for frameshift variants, and yellow circles for nonsense variants. Codons 239 and 283 for nonsense (6 each), and 389 (12 individuals) and 399 (7 individuals) for missense variants are affected by the highest number of pathogenic variants. Diagrams were constructed using Illustrator for Biosequence (IBS1.0.1). (b) SATB2 intragenic rearrangements. Full boxes correspond to deletions while lighter rectangles to duplications. In case of recurrence of the same exon being involved, the number of occurrences is indicated next to the rearrangement
The 89 pathogenic variants were distributed along the entire coding sequence of SATB2, and while variants were present in every coding exon (exon 3-12), the distribution was not uniform (Figure S2A). Unique pathogenic variants found in single individuals were most common (Figure 1). However, 7.9% of the pathogenic variants (7/89) were seen in two to four different families and 4.5% (4/89) were present in five or more families. Nearly half (41/89 = 46.1%) of the pathogenic variants were found in exons 8 and 9. There was a clear overrepresentation of exon 9 in particular when adjusting by size of each individual exon. This suggests a hotspot of pathogenic variants (Supp. Figure S2B).
2.1.1 ∣. Missense variants
Thirty-one unique missense variants were found in 49 individuals. Most missense variants were located within exons 8 and 9 (19/31 = 61.3%; Figure S2A) and located within the CUT1 domain (17/31 = 54.8%) of the SATB2 protein. Missense variants were often shared by multiple individuals with alterations in codons 389 (12 individuals) and 399 (7 individuals) being particularly common, suggesting hotspots. Most missense variants were confirmed to be de novo (44/45 = 97.8%). To assess the predicted pathogenicity of the 11 novel variants reported in this study, Polyphen2, SIFT, Provean, Mutation Taster, and CADD prediction programs were used (Table S1). Ten variants were interpreted as deleterious by all five programs, the remaining (p.Gln514Arg) with 4/5 programs predicting damaging effects.
Functional studies have been performed for three missense variants: c.1165C>T, p.(Arg389Cys), c.1543G>A, p.(Gly515Ser), and c.1696G>A p.(Gln566Lys; Bengani et al., 2017). The p.Arg389Cys change located in the CUT1 domain led to a marked increase in the proportion of soluble fraction of the protein whereas the p.Gly515-Ser and p.Gln566Lys variants located within the CUT2 domain and the region between CUT2 and the HOX domains respectively had the opposite effect. These experiments suggested a role of the CUT1 domain in initiating interaction with chromatin and a requirement for the CUT2 domain to facilitate dissociation of SATB2 from bound chromatin. These alterations in the kinetics of chromatin association resulting from missense variants have been postulated to functionally result in alterations that resemble complete loss-of-protein function (Bengani et al., 2017).
2.1.2 ∣. In-frame insertion
A single de novo in-frame insertion of 12 nucleotides not predicted to alter the reading frame has been reported in SATB2: c.929_930insTTGTAAGGCAAC, p.(Q310delinsHCKAT). The individual had a history of moderate to severe intellectual disability, autism, macrocephaly, frontal bossing, and deep-set eyes (Gilissen et al., 2014).
2.1.3 ∣. Predicted truncating variants
Fifty-one SATB2 variants reported were predicted to be disruptive to protein production resulting in loss-of-function, including 5 nonsense and 11 frameshift novel variants from this study. The variants were distributed throughout the reading frame but were frequently found within exons 8 to 12 (38/51 = 74.5%). Stop-gain pathogenic variants located within the last two exons might be expected to escape nonsense-mediated decay (NMD) and result in a shorter protein. In keeping with this hypothesis, a single variant located within the last exon of SATB2 (c.2074G>T; p.Glu692*) was shown to result in a shorter protein consistent with the predicted 692aa protein product of the mutant cDNA (Bengani et al., 2017). Two additional predicted loss-of-function variants have been studied in greater detail: c.715C>T (p.R239*) and c.847C>T (p.R283*) both found in six individuals each. The c.715C>T variant located in exon 8, was documented at the RNA level in two previously described unrelated individuals, indicating that the RNA was stable enough to escape NMD (Docker et al., 2014; Leoyklang et al., 2007). Further, the translated truncated protein retained the SATB2 dimerization domain. Through luciferase assays using a MAR sequence binding domain, it was documented to interfere with the repressive MAR-regulated transcriptional activity of the wild-type SATB2, suggesting a dominant negative effect for this mutation (Leoyklang et al., 2013). Conversely, the c.847C>T variant, also located in exon 8, was studied from tooth mesenchymal cells from an affected individual. Diminished SATB2 expression by Sanger sequencing and reduced SATB2 mRNA compared to control was demonstrated for this variant, suggesting NMD of the mutant RNA transcript (Kikuiri et al., 2018).
2.1.4 ∣. Splice site variants
Six variants (1 novel from this study) disrupted an essential splice site consensus sequence: three affecting the donor (5′) site at the end of exons 4, 5, and 8, and three affecting the acceptor (3′) site at the start of exons 7, 9, and 12. Splice-prediction programs (MaxEntScan and Human Splicing Finder) predicted abnormal splicing for all variants and potential activation of cryptic donor site or acceptor site for c.473+1delG and c.1741-1G>A, respectively. No RNA analysis has been performed to confirm the impact on splicing for any of these variants.
2.2 ∣. Large intragenic rearrangements
Twenty-five SATB2 exonic 1–12 inclusive) rearrangements including 22 deletions (10 from this report) and three duplications have been described (Table 2). For the 10 individuals that underwent parental testing, de novo status was confirmed. A pair of siblings (SATB2-02 and SATB2-128) with an intragenic deletion from phenotypically unaffected parents is also reported here for the first time suggesting germline mosaicism. Except for one individual (SATB2-92) identified through WGS and another as part of subtelomere multiplex ligation-dependent probe hybridization (MLPA) analysis (SATB2-140), all other copy number variations were initially identified by chromosomal microarray performed as part of the clinical evaluation with sizes ranging from 10 to 317 kb for deletions and between 32 and 54 kb for duplications (Table 2). For all individuals, SATB2 was the only gene affected with no involvement of adjacent genes. Multi-exonic rearrangements were more common (19/25 = 76.0%) than single exon involvement (Figure 1). Abnormalities involving at least exon 7 were present in half of these individuals (14 total from 13 unrelated families). No deletions have been documented at the cDNA level and only 2 refined by MLPA. A single duplication was studied at the cDNA level (Kaiser et al., 2015). In this female individual, an initial chromosomal microarray (Cytoscan HD; Affymetrix, Santa Clara, CA) revealed an 84-kb duplication within chromosomal region 2q33.1 (200,256,583–200,340,204) encompassing a part of the SATB2 gene. Reverse transcription PCR analysis with cDNA primers flanking the duplicated exon and subsequent Sanger sequencing of the extracted cDNA fragments showed the tandem in-frame duplication consistent with coexpression of transcripts with a duplicated exon and wild-type transcripts.
2.3 ∣. Large chromosomal rearrangements including 2q33.1
Thirty-five individuals with larger chromosomal alterations that include 2q33.1 (33 deletions, 2 duplications) have been reported and reviewed in the past (Zarate & Fish, 2017; Zarate, Smith-Hicks et al. (2018)). For deletions, some of these include dozens of adjacent genes and are as large as 26.3 Mb (Rifai et al., 2010). Although overlapping features with SAS are present in most individuals with larger deletions, other less common abnormalities, such as genitourinary anomalies, cardiac defects, and ectodermal changes (other than dental), appear to be more common (or exclusively present) when compared with intragenic molecular alterations (Zarate & Fish, 2017). For this group of individuals with larger rearrangements of 2q33.1, it remains difficult to establish genotype/phenotype correlations given the potential phenotype contribution of other genes besides SATB2.
2.4 ∣. Translocations
The original report of two de novo apparently balanced autosomal translocations t(2;7)(q33;p21) and t(2;11)(q32;p14) allowed the recognition of SATB2 as the causative gene for this syndrome (Brewer et al., 1999). High resolution mapping of the t(2;7) translocation showed disruption of the coding region of the SATB2 gene between exons 2 and 3 (FitzPatrick et al., 2003), whereas t(2;11) disrupted the long-range cis regulatory elements located in the centromeric gene desert 3′ of SATB2 (Rainger et al., 2014). A few additional individuals with SATB2 disruption secondary to de novo chromosomal translocations were subsequently described to result in SAS providing further supporting evidence for this molecular mechanism of disease (Baptista et al., 2008; Rainger et al., 2014; Talkowski et al., 2012; Tegay et al., 2009).
3 ∣. BIOLOGICAL RELEVANCE
SATB2 pathogenic alterations are distributed throughout the coding regions of the gene but exons 8, 9, and 11 are most commonly involved. For missense variants, most (23/31 = 74.2%) are located within the CUT1, CUT2, or HOX DNA protein domains. While the joint function of these DNA-binding domains is not fully clear, it is suggested that missense variants within the core of the CUT domain are likely to result in loss of DNA-binding activity; CUT1 being required to initiate interaction with chromatin and CUT2 (and the region between CUT2 and HOX) required to facilitate dissociation of SATB2 from bound chromatin (Bengani et al., 2017). The vast majority of pathogenic alterations of SATB2 are null variants (frameshift, nonsense, canonical splice site, and single or multiexon deletions) and while predicted to result in a loss-of-function and haploinsufficiency, limited functional studies have suggested a potential dominant negative effect for some. Of note, an increasing number of missense pathogenic variants have been described with functional alterations that resemble the complete loss-of-protein function.
4 ∣. CLINICAL AND DIAGNOSTIC RELEVANCE
A summary of clinical features of all 158 individuals previously reported and from this report, excluding those with larger deletions and duplications that include SATB2 and surrounding genes, is presented in Table 3. With rare exceptions, SAS diagnosis was not clinically recognized a priori. However, although the diagnosis of SAS still relies on molecular confirmation of a pathogenic variant in SATB2, a distinctive phenotype can often be identified. Speech delay is present in all individuals older than 2 years of age.
TABLE 3.
Demographic and phenotypic features of 158 individuals with SATB2-associated syndrome.
| Characteristic (N with data) | All patients (n = 158) |
|---|---|
| Demographics | |
| Male (152) | 90 (59.2%) |
| Mean age, years (152) | 9.5 ± 7.5 |
| Molecular mechanism | |
| Missense | 49 (31.0%) |
| Nonsense | 38 (24.1%) |
| Frameshift | 32 (20.3%) |
| Splicing | 7 (4.4%) |
| In-frame insertion | 1 (0.6%) |
| Intragenic deletion | 22 (13.9%) |
| Translocations | 6 (3.8%) |
| Intragenic duplication | 3 (1.9%) |
| Phenotype | |
| Neurodevelopmental abnormalities | |
| Developmental delay (157) | 157 (100%) |
| No words for speech (152) | 66 (43.4%) |
| Autistic behavior (145) | 29 (20.0%) |
| Dental anomalies (137) | 135 (98.5%) |
| Sialorrhea (109) | 96 (88.1%) |
| Facial dysmorphism (127) | 107 (84%) |
| Low BMD (46) | 33 (71.7%) |
| Feeding difficulties (130) | 89 (68.5%) |
| Hypotonia (114) | 67 (58.8%) |
| Sleeping difficulties (109) | 55 (50.5%) |
| Abnormal neuroimaging (108) | 50 (46.3%) |
| Sleeping difficulties (145) | 66 (45.5%) |
| Cleft palate (154) | 70 (45.5%) |
| Strabismus (143) | 51 (35.7%) |
| Agitation/aggressive (145) | 45 (31.0%) |
| Growth retardation (144) | 44 (30.6%) |
| Clinical seizures (143) | 29 (20.3%) |
| Hyperactivity (145) | 29 (20.0%) |
Severe expressive language delay is common with 84.3% (102/121) of individuals older than 4 years of age having 10 or fewer words in their expressive vocabulary, with 42.1% (51/121) demonstrating completely absent verbal communication (Figure S3). Other areas of neurodevelopment can also be compromised as evidenced by an average age at the first steps of 25.5 months. Intellectual disability has been reported in several individuals old enough to undergo cognitive evaluations and often in the moderate to severe range (Zarate, Smith-Hicks et al. (2018)). Dental abnormalities are present in all individuals and include delayed development of the mandibular second bicuspids or the roots of the permanent teeth, severely rotated or malformed teeth, taurodontism, and multiple odontomas (Kikuiri et al., 2018; Scott et al., 2018). Behavioral difficulties, feeding issues, abnormal brain neuroimaging, low bone density, suggestive facial features, and cleft palate complete the characteristic phenotype of SAS (Table 3). Through previous reports and the evaluation of dozens of patients by at least a single examiner (Y.A.Z.), broad thumbs and/or halluces appear to be another distinctive feature, present in a third of individuals evaluated (16/47 = 34%), that could raise the clinical suspicion of this diagnosis (Figure 2 a).
FIGURE 2.
(a) Broad halluces from four different individuals. (b) Composite images of individuals with nonsense, missense, and frameshift variants. Across all three images a flat philtrum with thin vermillion of the upper lip can be recognized
Because the identification of SATB2 as the gene responsible for SAS, the number of described individuals has continued to grow over the last few years. Most current molecular cytogenetic platforms should be able to detect exon level intragenic deletions involving SATB2. Likewise, SATB2 is part of several commercially available panels targeting broad phenotypes such as developmental delay, autism, or seizures. As families receive counseling, it is important to discuss the potential recurrence risk considering a few instances of suspected germline mosaicism documented. With a previous report of coding variants in siblings and the sibling pair with intragenic deletions reported here, we estimate a 1 to 2% germline mosaicism risk (2/155 families = 1.3%). Of note, paternal blood mosaicism has also been documented on one occasion. Lastly, management and surveillance guidelines for SAS have been proposed and typically need the participation of a multidisciplinary team with a heavy emphasis on pediatric dentistry and speech therapy (Zarate & Fish, 2017; Zarate et al., 1993).
5 ∣. GENOTYPE/PHENOTYPE CORRELATIONS
Table S2 presents detailed clinical features present in each of the 158 individuals (114 individuals enrolled in the SAS registry and 44 additional reported in the literature). We identified differences in clinical characteristics by a molecular mechanism. Specific changes to the gene were compared to all other changes, using either Chi-square or Fisher's exact tests (when at least one cell had an expected count of less than 5) for categorical variables and t tests for continuous variables. While there were no differences in the average age at walking or talking, other clinically relevant distinctions were identified (Table S3). The proportion of individuals with no verbal words to communicate older than 4 years of age was lowest for nonsense variants (8/29 = 27.6%) and highest for missense pathogenic variants mutations (20/39 = 51.3%; p = 0.0496). Individuals with missense pathogenic variants were less likely to have cleft palate (11/49 = 22.5% vs. 59/105 = 56.2% for other groups; p < 0.0001) but more likely to have clinical seizures (14/46 = 30.4% vs. 15/97 = 15.5% for other groups; p = 0.0375). Conversely, individuals with nonsense variants had significantly fewer clinical seizures (3/35 = 8.6% vs. 26/108 = 24.1% for other groups; p = 0.0474). Lastly, individuals with frameshift variants were more likely to have feeding difficulties (25/26 = 96.2% vs. 64/104 = 61.5% for other groups; p = 0.0007). If this difference in the prevalence of feeding difficulties is the result of a true biologically different mechanism for frameshift variants compared to nonsense variants or merely the result of a statistical anomaly, is unclear.
To determine if facial dysmorphisms were different enough among the most common molecular mechanisms, 102 2D photographs from 69 individuals (19 nonsense, 31 missense, and 19 frameshift variants) were analyzed using Face2Gene (FDNA Inc., Boston, MA) analytic tool vs.18.2.0. No statistically significant differences were found among the three composite images (Figure 2 b). However, binary comparisons for each of the three groups against a respective age and gender-matched cohort of typical individuals revealed statistically significant differences for all three groups (nonsense, p = 0.04; missense, p = 0.018; frameshift, p = 0.019).
6 ∣. ANIMAL MODELS
Much of what we know about SATB2 function in human development has come from studies in animal models. In mice, Satb2 is expressed in tissues that are affected in patients with SAS. During development, Satb2 is expressed in upper layer neurons, neural crest progenitors of the jaw, osteoblasts, odontoblasts, and other dental progenitor cells (Britanova et al., 2006; Dobreva et al., 2006; He et al., 2017). In adults, Satb2 continues to contribute to bone and brain function through its expression in osteoblasts and pyramidal neurons of the cerebral cortex and hippocampus (Huang et al., 2013; Jaitner et al., 2016; Wei et al., 2012).
In general, Satb2 functions as a transcriptional regulator that is important for tissue-specific functions. In mice bone progenitors, Satb2 regulates osteogenic differentiation genes (Dobreva et al., 2006), while in postmitotic neurons Satb2 regulates expression of genes involved in upper layer identity, synaptic transmission, and axon guidance (Jaitner et al., 2016; Li et al., 2017; Whitton et al., 2018). The consequence of loss of Satb2 in bone progenitors is reduced bone size and density due to increased cell death and reduced differentiation potential (Britanova et al., 2006; Dobreva et al., 2006).
In murine brain development, Satb2 is required to specify upper layer cortical neurons that project axons across the corpus callosum to the contralateral hemisphere (Alcamo et al., 2008; Britanova et al., 2008). Loss of Satb2 also results in reduced branches and spine density in basal dendrites of hippocampal neurons (Li et al., 2017). Similar to patients with SAS, Satb2+/− heterozygous mice have no reported corpus callosum defects (Alcamo et al., 2008; Zarate, Smith-Hicks et al. (2018)). However, Satb2+/− heterozygous mice suffer from impaired working and spatial memory. This deficit is exacerbated in mice where Satb2 has been deleted postnatally in hippocampal neurons (Li et al., 2017). These mice also have difficulties in locomotion, short-term novel object recognition memory, and long-term contextual fear memory (Jaitner et al., 2016; Li et al., 2017).
Taken together, research in animal systems has provided molecular and cellular mechanisms underlying SAS pathogenesis. These data help outline expectations of long-term care and provide medical practitioners with guidance about the potential spectrum of defects to manage in patients with SAS. However, current treatments are symptom-guided and do not specifically target pathogenic mechanisms. Future research is needed to further explore potential therapies that directly target SAS pathology.
7 ∣. FUTURE PROSPECTS
A key avenue of future research is the use of human-induced pluripotent stem cells (hiPSCs) to model disease and evaluate potential treatments. hiPSCs can be generated from patients with defined clinical phenotypes, thus enabling in vitro cellular phenotypes to be linked to individual clinical presentation. Use of hiPSCs allows the pathogenic mechanisms of different types of mutations to be evaluated. The potential to have hiPSCs from an unaffected parent provides a control for genetic background, allowing molecular and cellular outcomes, including the effect of different mutations on SATB2 mRNA and protein levels to be efficiently compared. Importantly, this in vitro system allows for reasonably high throughput testing of pharmacological agents.
To facilitate patient treatment, it will be important to elucidate how variability in individual disease pathogenesis contributes to SAS phenotypes. This variability may derive from the type of molecular alteration to the SATB2 locus, differences in genetic background, or even differences in lifestyle. Although therapies attempting to exogenously supplement reduced protein levels have achieved little success (Dietz, 2010), it may be possible to increase the endogenous amount of active protein in several ways. For example, in individuals with nonsense pathogenic variants, low levels of SATB2 are thought to result from NMD of prematurely terminated mRNA transcripts. Several pharmacological agents are now being used to promote “read-through” of stop codons, thus increasing levels of full-length mRNA (Baradaran-Heravi et al., 2017; Landfeldt, Sejersen, & Tulinius, 2018; Roy et al., 2016). This is an especially attractive avenue for treatment; however, not all nonsense mutations may be equally amenable to these treatments as sequences around the mutation may affect read-through activity (Bolze, Mocek, Zimmermann, & Klingenspor, 2017). The power of testing multiple different nonsense mutations in an hiPSC in vitro system has great potential to elucidate the details of this potential therapeutic mechanism. Of note, loss-of-function mutations may be amenable to some treatment types that would not alleviate the effects of dominant negative mutations. Also important to consider, knowing that SATB2 has an important role in early neurodevelopment as it has been demonstrated in mice models, the degree of correction or reversal of cognitive and speech deficits even if achieving normalization of SATB2 protein levels in potential human patients treated at later ages could be limited.
SATB2 function is affected by both posttranscriptional regulation by microRNAs (miRNAs) and posttranslational modification via sumoylation (Deng et al., 2013; Dobreva et al., 2003; Wei et al., 2012). While miRNAs regulate amounts of SATB2 protein, sumoylation affects SATB2 activation potential and association with endogenous MARs in vivo (Dobreva et al., 2003). Sumoylation targets SATB2 to the nuclear periphery (Dobreva et al., 2003). Therefore, inhibiting sumoylation may increase the amount of active SATB2 in association with MARs.
Another promising approach for the treatment of SAS is to focus on modifiers that buffer or compensate for reductions in protein function (Chen et al., 2016). In mice, loss of Satb2 dysregulates the expression of multiple miRNAs involved in memory and synaptic plasticity (Jaitner et al., 2016). The effects of dietary supplementation with phospholipidic concentrates of krill oil and buttermilk on the expression of miRNAs in hippocampal neurons have been studied in murine models (Crespo et al., 2018). Finally, neurological defects in patients with SAS have been reported to share molecular and cellular mechanisms with other neurodevelopmental or neurodegenerative diseases, such as schizophrenia and Alzheimer's disease (Whitton et al., 2018). Therefore, similar treatments could be explored for these diseases where shared molecular mechanisms are identified.
8 ∣. CONCLUSION
In this mutation update, we present data from 158 SAS individuals and review the current state of knowledge and future prospects on human SATB2 alterations. The 120 unique variants from 155 unrelated families range from single nucleotide variations to complex genomic rearrangements involving the entire coding region of SATB2. While germline mosaicism has been found in some instances, most pathogenic variants have been confirmed to be de novo. Missense pathogenic variants are often found, and for those studied functionally, are predicted to act as loss-of-protein function pathogenic variants. Almost invariably, the diagnosis of SAS is made after molecular investigations are performed and for individuals with genomic coding variants, next-generation sequencing technologies are most often used. Through our extensive review of individuals with SAS, we present an emerging phenotype that appears more recognizable with age. Our broad molecular and clinical descriptions of individuals with SATB2 should help clinicians and families establish the diagnosis of SAS and develop future therapies.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to all participating families. B.C is a senior clinical investigator of the Research Foundation-Flanders.
Footnotes
CONFLICT OF INTERESTS
The authors declare that there is no conflict of interests.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, & McConnell SK (2008). Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron, 57(3), 364–377. [DOI] [PubMed] [Google Scholar]
- Asadollahi R, Oneda B, Joset P, Azzarello-Burri S, Bartholdi D, Steindl K, & Rauch A (2014). The clinical significance of small copy number variants in neurodevelopmental disorders. Journal of Medical Genetics, 51(10), 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M, Smith K, Basel-Vanagaite L, Feingold MF, Brock P, Gowans GC, & Parker MJ (2011). Case series: 2q33.1 microdeletion syndrome--further delineation of the phenotype. Journal of Medical Genetics, 48(5), 290–298. [DOI] [PubMed] [Google Scholar]
- Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, Maloney V, & Crolla JA (2008). Breakpoint mapping and array CGH in translocations: Comparison of a phenotypically normal and an abnormal cohort. American Journal of Human Genetics, 82(4), 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran-Heravi A, Niesser J, Balgi AD, Choi K, Zimmerman C, South AP, & Roberge M (2017). Gentamicin B1 is a minor gentamicin component with major nonsense mutation suppression activity. Proceedings of the National Academy of Sciences of the USA, 114(13), 3479–3484. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bengani H, Handley M, Alvi M, Ibitoye R, Lees M, Lynch SA, & FitzPatrick DR (2017). Clinical and molecular consequences of disease-associated de novo mutations in SATB2. Genetics in Medicine, 19(8), 900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolze F, Mocek S, Zimmermann A, & Klingenspor M (2017). Aminoglycosides, but not PTC124 (Ataluren), rescue nonsense mutations in the leptin receptor and in luciferase reporter genes. Scientific Reports, 7(1), 1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Chan YM, Hunter JV, Pottkotter LE, Davino NA, Yang Y, & Bacino CA (2016). Increased bone turnover, osteoporosis, progressive tibial bowing, fractures, and scoliosis in a patient with a final-exon SATB2 frameshift mutation. American Journal of Medical Genetics. Part A, 170(11), 3028–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling KM, Thompson ML, Amaral MD, Finnila CR, Hiatt SM, Engel KL, & Cooper GM (2017). Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Medicine, 9(1), 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer CM, Leek JP, Green AJ, Holloway S, Bonthron DT, Markham AF, & FitzPatrick DR (1999). A locus for isolated cleft palate, located on human chromosome 2q32. American Journal of Human Genetics, 65(2), 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, Depew MJ, Schwark M, Thomas BL, Miletich I, Sharpe P, & Tarabykin V (2006). Satb2 haploinsufficiency pheno-copies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. American Journal of Human Genetics, 79(4), 668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, & Tarabykin V (2008). Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron, 57(3), 378–392. [DOI] [PubMed] [Google Scholar]
- Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, & Friend SH (2016). Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nature Biotechnology, 34(5), 531–538. [DOI] [PubMed] [Google Scholar]
- Cherot E, Keren B, Dubourg C, Carre W, Fradin M, Lavillaureix A, & Mignot C (2018). Using medical exome sequencing to identify the causes of neurodevelopmental disorders: Experience of 2 clinical units and 216 patients. Clinical Genetics, 93(3), 567–576. [DOI] [PubMed] [Google Scholar]
- Crespo MC, Tome-Carneiro J, Gomez-Coronado D, Burgos-Ramos E, Garcia-Serrano A, Martin-Hernandez R, & Visioli F (2018). Modulation of miRNA expression in aged rat hippocampus by buttermilk and krill oil. Scientific Reports, 8(1), 3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deciphering Developmental Disorders Study (2017). Prevalence and architecture of de novo mutations in developmental disorders. Nature, 542(7642), 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wu S, Zhou H, Bi X, Wang Y, Hu Y, & Fan X (2013). Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells. Stem Cells and Development, 22(16), 2278–2286. [DOI] [PubMed] [Google Scholar]
- Dietz HC (2010). New therapeutic approaches to mendelian disorders. New England Journal of Medicine, 363(9), 852–863. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Dambacher J, & Grosschedl R (2003). SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes and Development, 17(24), 3048–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, & Grosschedl R (2006). SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell, 125(5), 971–986. [DOI] [PubMed] [Google Scholar]
- Docker D, Schubach M, Menzel M, Munz M, Spaich C, Biskup S, & Bartholdi D (2014). Further delineation of the SATB2 phenotype. European Journal of Human Genetics, 22(8), 1034–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, & Taschner PE (2016). HGVS recommendations for the description of sequence variants: 2016 Update. Human Mutation, 37(6), 564–569. [DOI] [PubMed] [Google Scholar]
- Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Tippin Davis B, & Tang S (2015). Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: Results from 500 unselected families with undiagnosed genetic conditions. Genetics in Medicine, 17(7), 578–586. [DOI] [PubMed] [Google Scholar]
- FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, & Bonthron DT (2003). Identification of SATB2 as the cleft palate gene on 2q32-q33. Human Molecular Genetics, 12(19), 2491–2501. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, & Veltman JA (2014). Genome sequencing identifies major causes of severe intellectual disability. Nature, 511(7509), 344–347. [DOI] [PubMed] [Google Scholar]
- Glass IA, Swindlehurst CA, Aitken DA, McCrea W, & Boyd E (1989). Interstitial deletion of the long arm of chromosome 2 with normal levels of isocitrate dehydrogenase. Journal of Medical Genetics, 26(2), 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy AB, Szemes M, de Juan Romero C, Tarabykin V, & Agoston DV (2008). SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. European Journal of Neuroscience, 27(4), 865–873. [DOI] [PubMed] [Google Scholar]
- He L, Liu H, Shi L, Pan S, Yang X, Zhang L, & Niu Y (2017). Expression and localization of special AT-rich sequence binding protein 2 in murine molar development and the pulp-dentin complex of human healthy teeth and teeth with pulpitis. Experimental and Therapeutic Medicine, 14(4), 3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Song NN, Lan W, Hu L, Su CJ, Ding YQ, & Zhang L (2013). Expression of transcription factor Satb2 in adult mouse brain. Anatomical Record (Hoboken), 296(3), 452–461. [DOI] [PubMed] [Google Scholar]
- Jaitner C, Reddy C, Abentung A, Whittle N, Rieder D, Delekate A, & Apostolova G (2016). Satb2 determines miRNA expression and long-term memory in the adult central nervous system. eLife, 5, e17361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser AS, Maas B, Wolff A, Sutter C, Janssen JW, Hinderhofer K, & Moog U (2015). Characterization of the first intragenic SATB2 duplication in a girl with intellectual disability, nearly absent speech and suspected hypodontia. European Journal of Human Genetics, 23(5), 704–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuiri T, Mishima H, Imura H, Suzuki S, Matsuzawa Y, Nakamura T, & Yoshiura KI (2018). Patients with SATB2-associated syndrome exhibiting multiple odontomas. American Journal of Medical Genetics. Part A, 176, 2614–2622. [DOI] [PubMed] [Google Scholar]
- Landfeldt E, Sejersen T, & Tulinius M (2018). A mini-review and implementation model for using ataluren to treat nonsense mutation Duchenne muscular dystrophy. Acta Paediatrica, 108, 224–230. [DOI] [PubMed] [Google Scholar]
- Lee JS, Yoo Y, Lim BC, Kim KJ, Choi M, & Chae JH (2016). SATB2-associated syndrome presenting with Rett-like phenotypes. Clinical Genetics, 89(6), 728–732. [DOI] [PubMed] [Google Scholar]
- Leoyklang P, Suphapeetiporn K, Siriwan P, Desudchit T, Chaowana-panja P, Gahl WA, & Shotelersuk V (2007). Heterozygous nonsense mutation SATB2 associated with cleft palate, osteoporosis, and cognitive defects. Human Mutation, 28(7), 732–738. [DOI] [PubMed] [Google Scholar]
- Leoyklang P, Suphapeetiporn K, Srichomthong C, Tongkobpetch S, Fietze S, Dorward H, & Shotelersuk V (2013). Disorders with similar clinical phenotypes reveal underlying genetic interaction: SATB2 acts as an activator of the UPF3B gene. Human Genetics, 132(12), 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, You QL, Zhang SR, Huang WY, Zou WJ, Jie W, & Li JM (2017). Satb2 ablation impairs hippocampus-based long-term spatial memory and short-term working memory and immediate early genes (IEGs)-mediated hippocampal synaptic plasticity. Molecular Neurobiology[Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Lieden A, Kvarnung M, Nilssson D, Sahlin E, & Lundberg ES (2014). Intragenic duplication—A novel causative mechanism for SATB2-associated syndrome. American Journal of Medical Genetics. Part A, 164A(12), 3083–3087. [DOI] [PubMed] [Google Scholar]
- Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, & Ren J (2015). IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics, 31(20), 3359–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv HY, Zheng RJ, Wang QL, Ren PS, Jin LH, Gu XL, & Li LX (2018). SATB2-associated syndrome: A case report of a de novo nonsense mutation in SATB2 from China and review of literature. Clinica y Laboratorio, 64(4), 627–637. [DOI] [PubMed] [Google Scholar]
- Rainger JK, Bhatia S, Bengani H, Gautier P, Rainger J, Pearson M, & Fitzpatrick DR (2014). Disruption of SATB2 or its long-range cis-regulation by SOX9 causes a syndromic form of Pierre Robin sequence. Human Molecular Genetics, 23(10), 2569–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, & Strom TM (2012). Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: An exome sequencing study. Lancet, 380(9854), 1674–1682. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, & Committee ALQA (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17(5), 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifai L, Port-Lis M, Tabet AC, Bailleul-Forestier I, Benzacken B, Drunat S, & Aboura A (2010). Ectodermal dysplasia-like syndrome with mental retardation due to contiguous gene deletion: Further clinical and molecular delineation of del(2q32) syndrome. American Journal of Medical Genetics. Part A, 152A(1), 111–117. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Ballif BC, Lucas A, Spence EJ, Powell C, Aylsworth AS, & Shaffer LG (2009). Small deletions of SATB2 cause some of the clinical features of the 2q33.1 microdeletion syndrome. PLoS One, 4(8), e6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Friesen WJ, Tomizawa Y, Leszyk JD, Zhuo J, Johnson B, & Jacobson A (2016). Ataluren stimulates ribosomal selection of near-cognate tRNAs to promote nonsense suppression. Proceedings of the National Academy of Sciences of the USA, 113(44), 12508–12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E, Wilkens A, Noon SE, Krantz ID, & Wu Y (2017). A de novo SATB2 mutation in monozygotic twins with cleft palate, dental anomalies, and developmental delay. American Journal of Medical Genetics. Part A, 173(3), 809–812. [DOI] [PubMed] [Google Scholar]
- Scott J, Adams C, Simmons K, Feather A, Jones J, Hartzell L, & Zarate YA (2018). Dental radiographic findings in 18 individuals with SATB2-associated syndrome. Clinical Oral Investigations, 22(8), 2947–2951. [DOI] [PubMed] [Google Scholar]
- Sheehan-Rooney K, Palinkasova B, Eberhart JK, & Dixon MJ (2010). A cross-species analysis of Satb2 expression suggests deep conservation across vertebrate lineages. Developmental Dynamics, 239(12), 3481–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, & Gusella JF (2012). Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell, 149(3), 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegay DH, Chan KK, Leung L, Wang C, Burkett S, Stone G, & Hatchwell E (2009). Toriello-Carey syndrome in a patient with a de novo balanced translocation [46,XY,t(2;14)(q33;q22)] interrupting SATB2. Clinical Genetics, 75(3), 259–264. [DOI] [PubMed] [Google Scholar]
- Trakadis YJ, Buote C, Therriault JF, Jacques PE, Larochelle H, & Levesque S (2014). PhenoVar: A phenotype-driven approach in clinical genomics for the diagnosis of polymalformative syndromes. BMC Medical Genomics, 7, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers L, van Nimwegen KJM, Schieving JH, Kamsteeg EJ, Kleefstra T, Yntema HG, & Willemsen M (2017). A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genetics in Medicine, 19(9), 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, & Karsenty G (2012). miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. Journal of Cell Biology, 197(4), 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton L, Apostolova G, Rieder D, Dechant G, Rea S, Donohoe G, & Morris DW (2018). Genes regulated by SATB2 during neurodevelopment contribute to schizophrenia and educational attainment. PLoS Genetics, 14(7), e1007515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate YA, & Fish JL (2017). SATB2-associated syndrome: Mechanisms, phenotype, and practical recommendations. American Journal of Medical Genetics. Part A, 173(2), 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate YA, Kaylor J, & Fish J (1993). Adam MP, Ardinger HH, Pagon HH, Wallace SE, Bean LJH, Mefford HC, Stephens K, Amemiya A, & Ledbetter N (Eds.), SATB2-Associated Syndrome. Seattle (WA): GeneReviews(R). [PubMed] [Google Scholar]
- Zarate YA, Perry H, Ben-Omran T, Sellars EA, Stein Q, Almureikhi M, & Slavotinek A (2015). Further supporting evidence for the SATB2-associated syndrome found through whole exome sequencing. American Journal of Medical Genetics. Part A, 167A(5), 1026–1032. [DOI] [PubMed] [Google Scholar]
- Zarate YA, Kalsner L, Basinger A, Jones JR, Li C, Szybowska M, & Everman DB (2017). Genotype and phenotype in 12 additional individuals with SATB2-associated syndrome. Clinical Genetics, 92(4), 423–429. [DOI] [PubMed] [Google Scholar]
- Zarate YA, Smith-Hicks CL, Greene C, Abbott MA, Siu VM, Calhoun A, & Chung WK (2018). Natural history and genotype-phenotype correlations in 72 individuals with SATB2-associated syndrome. American Journal of Medical Genetics. Part A, 176(4), 925–935. [DOI] [PubMed] [Google Scholar]
- Zarate YA, Steinraths M, Matthews A, Smith WE, Sun A, Wilson LC, & Ma NS (2018). Bone health and SATB2-associated syndrome. Clinical Genetics, 93(3), 588–594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.