Abstract
The susceptibility of sheep to scrapie is known to involve, as a major determinant, the nature of the prion protein (PrP) allele, with the VRQ allele conferring the highest susceptibility to the disease. Transgenic mice expressing in their brains three different ovine PrPVRQ-encoding transgenes under an endogenous PrP-deficient genetic background were established. Nine transgenic (tgOv) lines were selected and challenged with two scrapie field isolates derived from VRQ-homozygous affected sheep. All inoculated mice developed neurological signs associated with a transmissible spongiform encephalopathy (TSE) disease and accumulated a protease-resistant form of PrP (PrPres) in their brains. The incubation duration appeared to be inversely related to the PrP steady-state level in the brain, irrespective of the transgene construct. The survival time for animals from the line expressing the highest level of PrP was reduced by at least 1 year compared to those of two groups of conventional mice. With one isolate, the duration of incubation was as short as 2 months, which is comparable to that observed for the rodent TSE models with the briefest survival times. No survival time reduction was observed upon subpassaging of either isolate, suggesting no need for adaptation of the agent to its new host. Overexpression of the transgene was found not to be required for transmission to be accelerated compared to that observed with wild-type mice. Conversely, transgenic mice overexpressing murine PrP were found to be less susceptible than tgOv lines expressing ovine PrP at physiological levels. These data argue that ovine PrPVRQ provided a better substrate for sheep prion replication than did mouse PrP. Altogether, these tgOv mice could be an improved model for experimental studies on natural sheep scrapie.
Sheep scrapie is the most common transmissible spongiform encephalopathy (TSE) and naturally evolves following an infectious mode. TSEs are a group of neurodegenerative diseases which includes Creutzfeldt-Jakob disease (CJD) and Gerstmann-Sträussler-Scheinker syndrome (both affecting humans), as well as bovine spongifom encephalopathy (BSE). These diseases are characterized by the accumulation of an abnormal isoform of the host prion protein (PrP) called PrPsc, which is assumed to be responsible for the observed disorders (38). The transmissible nature of TSE is attributed to yet incompletely defined agents, designated prions, whose infectiveness is intimately associated with PrPsc (13, 37, 39).
Experimental transmission to laboratory rodents has been achieved with TSE agents from many naturally affected species. Such studies have set forth the notion of species barrier (16, 35, 38), which refers to the resistance to disease encountered following experimental inoculation with TSE agent derived from a foreign species. This resistance is manifested by either a lack of or an incomplete disease transmission associated with prolonged and heterogeneous incubation times prior to clinical disease. The adaptation of a TSE agent to its new host requires one or more passages, and a strain showing stable properties usually results upon further passaging in the same host. Multiple strains have been derived from TSE isolates (including isolates from sheep); these strains differ by their incubation times in various inbred mouse lines and by their neuropathological manifestations in affected brains (4, 5, 11, 17).
There is compelling genetic evidence that transmission of TSEs is tightly controlled by the PrP-encoding gene (Prnp) (38, 40, 52). Apart from modulating the individual susceptibility to the disease through natural polymorphism or mutation, PrP appears to play a critical role in determining the prion host range. Transgenetic studies have shown that the species barrier encountered during transmission from hamster to mice is overcome by introducing the hamster Prnp gene into recipient wild-type mice (36, 43). This observation has provided the rationale for the production of transgenic mice exhibiting an enhanced susceptibility to human or bovine prions (9, 15, 27, 44, 48).
Such investigations, however, have revealed that the expression of a donor-derived PrP transgene may not always be sufficient to erase the species barrier to TSE transmission. The susceptibility of mice to foreign prions has been found to be increased when a given PrP transgene is introduced into PrP knockout mice, implying that the resident murine gene may inhibit the propagation of the foreign prions (6, 48, 53). The so-called interfering effect of endogenous mouse PrP on transmission of most human prions is strongly reduced in mice expressing a chimeric mouse-human PrP transgene, suggesting the possible involvement of additional, species-specific factors in prion replication (47, 48). Though the transmission of classic CJD agent is greatly enhanced in transgenic mice expressing human PrP only, this is not the case for a variant CJD agent, despite its primary structure being identical to that of the human PrP, indicating that a strain-specific component may contribute to the transmission barrier (15, 27). Finally, introduction of a single-amino-acid change into the endogenous murine PrP gene was recently reported to have removed most of the human-to-mouse transmission barrier for a Gerstmann-Sträussler-Scheinker syndrome case (33).
Sheep scrapie strain characterization and infectivity measurement involve extensive use of mouse bioassays (5, 24). These studies, however, are hampered by the facts that the disease commonly requires between 1 and 2 years to develop and that a number of isolates do not transmit easily to mice (28). Recently, accelerated transmission of sheep scrapie to transgenic mice overexpressing bovine PrP has been mentioned (45). In an effort to develop an improved model for the experimental transmission of sheep scrapie, we have expanded transgenic lines of mice which express ovine PrP. In sheep, different Prnp alleles have been identified that closely modulate both the incidence and age of onset of natural or experimental scrapie. Three codons (at positions 136, 154, and 171) act as major determinants of this susceptibility. The 136V154R171Q allele (where V, R, and Q stand for valine, arginine, and glutamine, respectively) was found to confer the highest susceptibility, whereas the 136A154R171R allele (where A stands for alanine) was associated with an absolute clinical resistance (2, 14, 20, 21, 29, 56). We report here that the interspecies transmission of a TSE agent from sheep to mice expressing the PrPVRQ allelic variant was greatly facilitated compared to that from sheep to conventional mice.
(A portion of this research was presented as part of the Characterization and Diagnosis of Prion Diseases in Animal and Man Symposium, Tübingen, Germany, September 1999.)
MATERIALS AND METHODS
Constructs.
The tg1 construct was derived from the phgPrP half-genomic vector (18) by inserting the ovine PrPVRQ open reading frame (ORF) in place of the murine ORF. The ovine ORF was cloned by PCR using cDNA as a template and GAGCCGATACCCCGGGCAGGGCAGTC and TCATCCCACGATGAGAAAAATGAGG as the 5′ and 3′ primers. The 5′ primer is homologous to the region from positions 185 to 210 of the sheep Prnp cDNA (GenBank accession no. M31313) and introduced a SmaI restriction site. The 3′ primer is complementary to the 3′ end of the ovine ORF (starting at nucleotide [nt] 818) and includes 10 nt complementary to the murine Prnp sequence (nt 855 to 864, GenBank accession no. M13685). Another PCR was performed using CCTCATTTTTCTCATCGTGGATGA and GGCTGTTTTCCAGGGCGCCATCCCC as the 5′ and 3′ primers and phgPrP as a template, to amplify part of the 3′ murine Prnp untranslated region (UTR). This 5′ primer covers the same region of the murine cDNA as the 3′ primer did in the PCR described above, whereas this 3′ primer is complementary to the region from positions 1258 to 1282 of the murine cDNA and contains a NarI restriction site. The two PCR products, the ovine ORF and the murine 3′ UTR, were mixed in a third PCR with both the 5′ primer of the first PCR and the 3′ primer of the second PCR in order to obtain the ovine ORF linked to the murine 3′ UTR. This product was cloned into pUC19 and sequenced. The SmaI/NarI insert was released and inserted in place of the murine SmaI/NarI partial cDNA within the phgPrP vector, leading to the tg1 construct.
The tg2 construct was derived from tg1 by substituting the human cytomegalovirus (CMV) promoter for the murine PrP promoter. The tg1 construct was first digested with SalI and BamHI, and the 8-kb internal fragment (encompassing the tg1 transcription unit, the 3′ flanking region, and part of the promoter) was gel purified. This fragment was digested by SmaI, and the resulting 4.5-kb SmaI/SalI and 3.5-kb BamHI/SmaI restriction fragments were isolated. The 3.5-kb fragment was further digested with EcoRI to derive a 1.9-kb EcoRI/SmaI subfragment, encompassing part of intron 1 and the start of exon II of tg1, that was in turn cloned into pPolyIII.I between the corresponding sites of the polylinker (31). The recombinant vector was digested with SmaI and SalI, and the above-mentioned 4.5-kb SmaI/SalI fragment, encompassing the end of exon II and the 3′ flanking region of tg1, was inserted into it. The resulting 6.4-kb BamHI/SalI insert was excised. To substitute a BamHI site at the EcoRI site located at intron 1 of the murine Prnp gene (nt 1695, GenBank accession no. U52821) and to introduce an EcoRI site within the 5′ UTR of exon I of this gene (nt 1993), this region was amplified by PCR using phgPrP as the template, as well as the primer 5′ GACGAATTCTGGGCGCTGCG and the primer 3′ GGAGGATCCTGCGCACCCGC. The PCR product was cloned into pUC19 and sequenced. This BamHI/EcoRI insert was subcloned into pPolyIII.I. The XhoI/EcoRI human CMV promoter (positions −675 to +75), isolated from the pUHG17-1 plasmid (23), was inserted into this recombinant plasmid between the SalI and EcoRI sites of the polylinker. The resulting 1.25-kb BamHI/NotI insert was excised. Finally, the two above-mentioned 6.4-kb BamHI/SalI and 1.25-kb BamHI/NotI inserts were ligated together within the SalI and NotI sites of pPolyII (31), leading to the tg2 construct. The tg1 (see above) and tg2 inserts were excised with NotI and SalI and gel purified for microinjection (51).
The tg3 construct was isolated from an ovine bacterial artificial chromosome (BAC) library constructed with PrnpVRQ/VRQ ram brain DNA (50). The library was screened by PCR using primers (5′ CAGAAGGTAGTGGAACAAAAG and 3′ GCTAAGGACAACACAGAAGAG) that amplified a 160-bp internal fragment of Prnp exon 3, as described previously (42). To evaluate possible chimerism, the DNA from the isolated BAC clone was analyzed by fluorescent in situ hybridization (FISH), as previously described (42). Location of the Prnp transcription unit within the BAC insert was assessed by restriction mapping analysis. Purification for microinjection of the BAC DNA insert following NotI digestion was performed as previously described (46).
Generation of transgenic mice.
Microinjection of the constructs directly into oocytes derived from Prnp0/0 mice (7) was unsuccessful, due to a very low rate of survival of these oocytes after injection. Subsequently, microinjection was performed on pronuclei of (C57BL/6 × CBA) × Prnp0/0 hybrid eggs. Tail biopsies were taken 2 weeks after birth, and the genomic DNA was purified as previously described (51). PstI-digested genomic DNA was size fractionated in 1% agarose gels, transferred to Hybond N filters and hybridized with an [α-32P]dCTP-oligonucleotide-labeled ovine and murine Prnp cDNA probe derived from tg1. This probe recognizes both the transgene and the endogenous Prnp loci. Twelve, six, and eight transgenic founder mice were obtained with transgenes tg1, tg2, and tg3, respectively. Transgenic mice were backcrossed with Prnp0/0 mice in order to establish the transgenes under a murine Prnp0/0 genetic background. This introgression was performed for at least three generations.
Expression analysis of ovine Prnp transgenes.
Northern blot analyses were performed as described previously (51), using 20 μg of total RNA per sample. Hybridization was carried out with an ovine and murine Prnp cDNA probe derived from tg1. Ten percent brain homogenates were obtained using an Ultra-Turrax Antrieb T25 in 0.32 M sucrose solution containing 0.5% Nonidet P-40 and 0.5% deoxycholic acid. Residual cell debris were removed by low-speed centrifugation, and the total protein contents of the homogenates were determined by the Lowry procedure. Homogenate samples were subjected to sodium dodecyl sulfate–13% polyacrylamide gel electrophoresis (SDS–13% PAGE) and transferred onto nitrocellulose membranes. PrP bands were visualized using the monoclonal antibody l42 (26) or 2D6 (41) and an enhanced chemiluminescence detection system (ECL; Amersham). PrP contents were determined using a Pharmacia LKB Imagemaster DTS scanning system.
Scrapie isolates.
Brain materials derived from two sheep affected with natural scrapie were used as a source of infectious agent: PG127/98 (referred to here as PG127) from a Cheviot-Welsh sheep breed (Veterinary Laboratory Agency, Addelstone, United Kingdom), and LA404 from a Romanov sheep (Institut National de la Recherche Agronomique [INRA], Langlade, France). Both animals were homozygous for the VRQ allele. Brain tissue was homogenized at 10% (wt/vol) in a sterile 5% glucose solution using a Ribolyser cell disrupter (Hybaid, Ashford, United Kingdom). Homogenates were kept at −80°C. Prior to inoculation, they were heated at 80°C for 20 min to inactivate possible viral or bacterial contaminants and sonicated for 1 to 2 min in a cup-horn apparatus.
Inoculation and determination of incubation period.
Female and male mice were kept in groups of 4 to 8 or 1 to 4 animals, respectively. Six- to 8-week-old mice were inoculated intracerebrally with 20 to 25 μl of undiluted 10% brain homogenate using a 27-gauge disposable hypodermic needle inserted into the right parietal lobe. In some experiments, dilutions of 10% homogenate in 5% glucose were used. Sonicated, unheated 10% brain homogenates were used for mouse-to-mouse passages. Beginning 1 month after inoculation, the mice were examined for neurologic dysfunction every 2 days, and once the first clinical signs were detected in a littermate, the mice were examined daily. Any animal whose death was clearly imminent was killed by cervical dislocation, and its brain was immediately removed and processed for histopathological examination and/or detection of a protease-resistant form of PrP (PrPres). Brains from animals found dead of scrapie (about 10% of animals) were also taken and processed for PrPres detection. Surgical instruments were decontaminated by immersion in 1 N NaOH overnight and then subjected to two cycles of autoclaving at 136°C for 20 min. The mean incubation period—actually, the survival time—was calculated as the interval between inoculation and death or the in extremis stage. Data for a few animals who died of intercurrent disease, as assessed by the lack of PrPres accumulation in Western blot analysis and/or of neurological symptoms, were excluded from the analysis.
PrPres detection in brain homogenate.
Brain homogenates (typically 200 μl of 10% brain homogenate) were digested for 1 h with 10 μg of proteinase K (PK) per ml, and the reactions were stopped with 4 mM Pefabloc. After addition of 10% sarcosyl and 10 mM Tris-HCl (pH 7.4), samples were incubated for 15 min at room temperature. They were centrifuged at 245,000 × g for 30 to 45 min at 20°C on 10% sucrose cushions. Pelleted material was resuspended in sample buffer, resolved by SDS–12% PAGE, and analyzed by immunoblotting. Visualization and comparative quantification of PrPres signal were performed as described for PrP.
Histopathological procedures.
Fixed, paraffin-embedded mouse brains were investigated by hematoxylin and eosin staining and immunohistochemistry as described previously (25). After formic acid and treatment with PK (4 μg/ml), followed by hydrated autoclaving, the slices were incubated with either monoclonal antibody (MAb) l42 for detection of ovine PrPres or an antibody to glial fibrillary acidic protein (DAKO, Roskilde, Denmark).
RESULTS
Generation and expression analyses of transgenic mice.
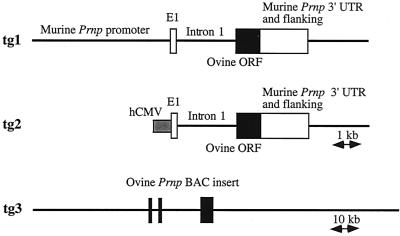
A schematic representation of the transgenes tg1, tg2, and tg3, designed for targeting expression of ovine PrPVRQ in mice, is given in Fig. 1. tg3 corresponds to a 125-kb BAC insert. Restriction fragment analysis revealed that it contains around 40 and 60 kb of the 5′ and 3′ Prnp-flanking sequences, respectively. FISH analysis showed that it maps to chromosome 13q17, as described previously (12), and that it is not chimeric. Preliminary data indicate that this BAC insert contains at least part of the Prnd (Doppel-encoding [34]) locus (data not shown).
FIG. 1.
Schematic representations of the ovine PrP-expressing constructs tg1, tg2, and tg3. The thin lines represent flanking and intronic regions, as indicated. Ovine and murine exonic sequences are represented by the black and the white boxes, respectively. The gray box indicates the human CMV (hCMV) promoter. E1, exon I.
The constructs were microinjected into (C57BL/6 × CBA) × Prnp0/0 hybrid eggs (see Material and Methods). Transgenic mice were backcrossed with Prnp0/0 mice (6) in order to obtain animals carrying the transgene on a murine Prnp knockout genotype (48, 53). Expression analyses were performed on ∼6-week-old mice heterozygous at the transgene locus. Detectable expression of the ovine PrP in the brain was revealed by Western blot analyses of 3 of 11 tg1 lines, 4 of 6 tg2 lines and 8 of 8 tg3 lines. Nonexpressing lines were discarded, as was one tg1-expressing line that carried the transgene close to the wild-type murine Prnp locus, as assessed by the consistent cosegregation of the two loci. The transgene of each of the 14 remaining lines was introgressed for at least three generations onto the murine Prnp0/0 genetic background, before reassessment of its expression in each line. The expression levels did not differ significantly from those observed in animals from the first generation. As often occurs with the use of large DNA fragments (19), expression of the tg3 construct was essentially site independent and copy number related (Table 1 and data not shown).
TABLE 1.
Susceptibility of transgenic and nontransgenic mice to intracerebral inoculation with two isolates of sheep scrapiea
| Line | Genotypeb | PrP-encoding gene
|
tg copy number | PrPc level (fold)c
|
Days to terminal disease (n/n0) following inoculation withd:
|
|||
|---|---|---|---|---|---|---|---|---|
| Sheep | Mouse | Sheep | Mouse | PG127 | LA404 | |||
| RIII | Prnp+/+ | Prnpa | 1 | 423 ± 6.1 (8/8) | >600 (2/5) | |||
| F1e | Prnp+/+ | Prnpa | 1 | 566 ± 8 (8/8) | 620 ± 22 (7/8) | |||
| tg301 | tg3+/−, Prnp0/0 | bacPrnpVRQ | 6 | 8 | 73 ± 3 (7/7)∗ | 205 ± 12.8 (10/10)∗ | ||
| tg328 | tg3+/−, Prnp0/0 | bacPrnpVRQ | 4 | 3.5 | 101 ± 5 (8/8)∗ | na | ||
| tg206 | tg2+/−, Prnp0/0 | cmv/phgPrnpVRQ | 9 | 2.2 | 113 ± 4.4 (7/7)∗ | na | ||
| tg211 | tg2+/−, Prnp0/0 | cmv/phgPrnpVRQ | 4 | 1.9 | 107 ± 2.7 (12/12)∗ | 341 ± 5 (8/8)∗ | ||
| tg223 | tg2+/−, Prnp0/0 | cmv/phgPrnpVRQ | 19 | 1.6 | 143 ± 5 (8/8)∗ | na | ||
| tg143 | tg1+/−, Prnp0/0 | phgPrnpVRQ | 30 | 1.5 | 149 ± 4.3 (8/8)∗ | 351 ± 5 (8/8)∗ | ||
| tg207 | tg2+/−, Prnp0/0 | cmv/phgPrnpVRQ | 8 | 1.2 | 196 ± 10 (8/8)∗ | na | ||
| tg335 | tg3+/−, Prnp0/0 | bacPrnpVRQ | 1 | 1.2 | 200 ± 9.3 (6/6)∗∗ | na | ||
| tg116 | tg1+/−, Prnp0/0 | phgPrnpVRQ | 4 | >0.4 | 399 ± 6 (7/7) | na | ||
| tga20f | tga20+/+, Prnp0/0 | phgPrnpa | 60f | 10f | 240 ± 3 (8/8) | 413 ± 16.8 (6/6) | ||
Heated 10% brain homogenate.
Compared to level of sheep or wild-type (C57BL/6) mouse brain.
Values are means ± standard errors of the means. Each value in parentheses is the number of animals terminally ill/the number of animals inoculated. Statistical comparison to tga20 was performed with the unpaired Student's t test. ∗, P < 0.005; ∗∗, P ≤ 0.05. na, not available.
F1, (C57BL/6 × 129/Sv)F1.
As described in reference 18.
The tissue distribution of the expression of the transgenes analyzed by Northern blotting was found to be similar for mice bearing the same construct and was found to differ slightly for animals carrying different constructs (data not shown). In all lines, the highest level of ovine Prnp mRNA was observed in the brain. Lower levels could be detected in other tissues. The two Prnp mRNAs of 4.6 and 1.9 kb that occur in sheep tissues (22) were observed in the tg3 mice (data not shown).
A striking feature common to all tg2 lines (CMV promoter) was the occurrence of spontaneous neurological disorders. The predominant phenotype included kyphosis and paresis of the rear limbs, which led to complete paralysis as the animals aged. Such an abnormal phenotype is reminiscent of earlier observations of mice carrying a high copy number of cosmid constructs encoding mouse, hamster, or sheep PrP (54, 55). However, a simple gene dosage effect is unlikely to explain the tg2 phenotype, since no evidence of spontaneous disorders was found in any of the tg1 and tg3 lines, even at a higher PrP expression level. A more complete description of tg2 mice will be published elsewhere. Since the observed disorders did not notably affect life expectancy, the same mice were used for transmission experiments.
Susceptibility of tgOv(PrPVRQ) mice to natural sheep scrapie.
Two tg1, four tg2, and three tg3 lines (see Table 1) were selected for transmission experiments, which were all performed on Prnp0/0 mice heterozygous for the transgene. Two scrapie field isolates from distant geographical origins (isolate PG127, from the United Kingdom, and isolate LA404, from France) were used (both from a diseased sheep homozygous for the PrPVRQ allele). The mice were inoculated intracerebrally with heated brain homogenate from either one of these animals or from a healthy, presumably uninfected sheep. Conventional mice, mice from a RIII congenic line or (C57BL/6 × 129/Sv)F1 mice derived from congenic lines, as well as mouse PrP transgenic mice (tga20 line [18]), were challenged in the same way.
From the results summarized in Table 1, three main conclusions could be drawn. (i) Complete (100%) disease transmission was observed in all nine PG127-inoculated and all three LA404-inoculated tgOv lines. All mice showed acute neurological disorders typical of TSE before dying or being sacrificed at a terminal stage. The survival curves were essentially monophasic, and the latencies between inoculation and terminal clinical symptoms displayed standard deviations on the order of a few percentage points (Table 1 and data not shown). All mock-inoculated animals (two tg1 lines, four tg2 lines, and the tg328 line) remained free of scrapie. (ii) The brain PrP content, which ranged from <0.4- to 8-fold that in sheep brain among the tgOv lines, had a potent effect on the time of disease onset, i.e., mice with the highest steady-state level of ovine Prnp gene expression had the shortest survival time and vice versa. (iii) The observed survival times of tgOv mice were markedly reduced (by at least 40%) compared to those of wild-type RIII and (C57BL/6 × 129/Sv)F1 mice, and this was true for both isolates. The sole exception was the tg116 line, a result consistent with its very low level of transgene expression.
In the tg301 line, which expressed PrP at the highest level (eightfold that of the sheep brain), the mean survival time was dramatically shortened: 73 days versus 423 days in RIII mice after PG127 isolate infection, i.e., a reduction of more than 80% (Table 1). The survival time was even more reduced compared to that of the wild-type (C57BL/6 × 129/Sv)F1 mice, whose genetic background is closer to that of mice of the tgOv lines than to that of RIII mice (see Materials and Methods). The mean survival time of tg301 mice infected with the LA404 isolate (205 days) was significantly longer than that of mice infected with the PG127 isolate but, again, considerably shorter than that of (C57BL/6 × 129/Sv)F1 control mice. Incomplete transmission of LA404 to RIII mice was observed.
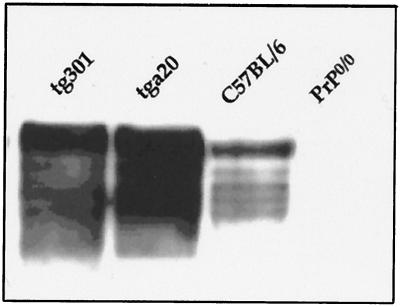
In order to examine whether overexpression of a murine PrP would also accelerate the transmission of sheep scrapie to mice, tga20 mice were infected with the same two isolates. Mice of the tga20 line (18) have the same genetic background as the tgOv mice and the same type of transgene as mice of the tg1 lines. All the inoculated mice died of scrapie, with mean survival times of 240 and 413 days for PG127- and LA404-infected animals, respectively (Table 1). Thus, although expressing PrP at an even higher level than tg301 mice (Fig. 2), tga20 mice appeared to be less susceptible than tgOv mice expressing ovine PrP at physiological levels (mice of the tg143, tg207, and tg335 lines).
FIG. 2.
Comparison of PrP levels of expression in ovine tg301 and murine tga20 transgenic mice. PrP levels in the brain of tg301 (tg3+/−, Prnp0/0), tga20 (tga20+/+, Prnp0/0 [18]), C56BL/6, and Prnp0/0 (6) mice were analyzed by Western blotting using 2D6 antibodies raised against the ovine PrP sequence. Fifty micrograms of total brain homogenates from 4- to 5-week-old female mice were analyzed as described in Materials and Methods.
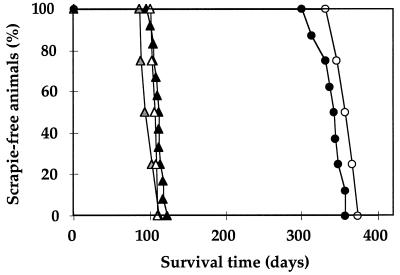
To determine whether a reduction of incubation time upon further passaging would occur in tgOv mice, serial transmissions were performed on mice of the tg211 line. As shown in Fig. 3, no significant (P > 0.01 by Student's t test) change was recorded on the second and third passages of the PG127 isolate or on the second passage of the LA404 isolate.
FIG. 3.
Serial transmission of sheep scrapie agent in ovine PrP transgenic mice. Survival curves of tg211 mice (tg2+/−, Prnp0/0) infected intracerebrally with 10% brain homogenate of a primary inoculum or a serially transmitted inoculum from two different scrapie isolates are shown. Black, gray, and white triangles, first passage (107 ± 2.7 days), second passage (99 ± 5 days), and third passage (106 ± 2 days), respectively, of the PG127 isolate; black and white circles, first passage (341 ± 5 days) and second passage (354 ± 6 days), respectively, of the LA404 isolate. (Values in parentheses are mean survival times ± standard errors of the means.)
Experiments aimed at analyzing the response of the tgOv lines to intracerebral injection of a serially diluted inoculum are still in progress. Preliminary data available for the PG127 isolate showed that the resulting survival time protraction differed strikingly between the tg301 and tg211 lines (with, respectively, approximately 1- and 3-week- increases per 10-fold dilution). This leads to an apparent multiplication rate of the infectious agent that is about threefold higher, for a fourfold increase in the PrP expression level (data not shown).
Clinical and neuropathological features.
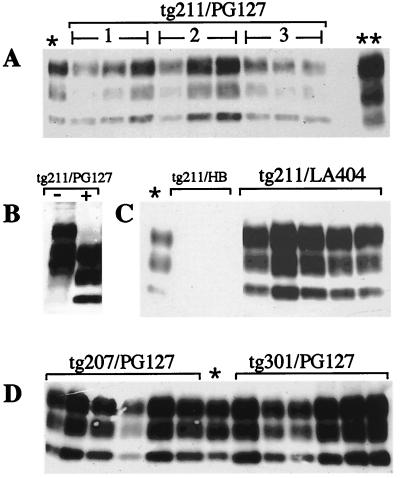
All scrapie-inoculated mice displayed typical symptoms, including at least two of the following signs: hindlimb paresis, waddling gait, plastic tail, and lethargy. In addition, obesity (11) was frequently observed in LA404-inoculated transgenic lines with incubation periods over 300 days. The duration of the clinical phase for transgenic animals was significantly decreased compared to that for nontransgenic animals, where it lasted from 1 month to several months. In tg301 mice, a rapid progression of the illness was observed after the appearance of the first symptoms, leading to death within 1 or 2 weeks after inoculation with PG127 or LA404, respectively. Western blot analysis revealed accumulation of PrPres in the brain of every inoculated mouse. The PrPres expression patterns of both isolates showed a predominance of a biglycosylated species (Fig. 4) similar to that observed with the sheep brain inocula. LA404-challenged mice accumulated PrPres at a slightly higher level than PG127-challenged mice (e.g., about fivefold relative increase in the tg211 line), consistent with immunohistochemistry analyses performed on the tg211 and tg301 lines (not shown). The PrPres levels did not greatly differ (at most three- to fivefold) among the different lines. Histopathological examination performed on the tg2 and tg3 lines revealed both spongiform changes and astrocytosis, mainly located in the midbrain and cerebellum. These effects were less pronounced in PG127- than in LA404-inoculated mice (to be reported elsewhere).
FIG. 4.
Results of immunoblot analysis of PK-resistant PrP in brains of scrapie-infected transgenic (tgOv+/−, Prnp0/0) mice. Brain homogenates (10%) were prepared from terminally ill animals. Samples (brain equivalent, 2 mg) were digested with PK as described in Material and Methods. After SDS–12% PAGE, Western blotting was performed using MAb 2D6. (A) tg211 mice inoculated with PG127 isolate (lanes 1) or with PG127 passaged once (lanes 2) or twice (lanes 3) in tg211 mice; (B) PG127-infected tg211 mice untreated (−) or treated with PK (+); (C) tg211 mice postinoculation with healthy brain (HB) homogenate (18 months postinoculation) or with LA404 isolate; (D) PG127-inoculated tg207 or tg301 mice. ∗, C506M3 (mouse-adapted) strain-infected C57BL/6 mice (brain equivalent, 0.1 mg); ∗∗, PG127-infected sheep.
DISCUSSION
Ovine PrP transgene expression may facilitate scrapie disease transmission from sheep to mice.
In this study, it was shown that expression of ovine PrPVRQ in transgenic mice lacking the mouse PrP gene (PrP0/0 mice) conferred an enhanced susceptibility to scrapie. The tgOv lines were generated from three different constructs (Fig. 1). Two of them were based on a half-genomic vector (18) in which the PrP coding sequence is driven by either mouse Prnp (tg1 lines) or CMV promoter (tg2 lines) sequences. The third construct introduced into the mouse genome a large piece of sheep DNA in which expression of Prnp is under the control of its natural regulatory sequences (tg3 lines). Following intracerebral inoculation with two sheep isolates, the tgOv mice showed a survival time that was dramatically reduced compared to those of two groups of conventional mice (Table 1). In the tg301 line, which expressed PrP at the highest level, the incubation duration was between 2 and 7 months, depending on the isolate. These are the shortest survival times after inoculation with sheep prion recorded to date.
The survival time in the different tgOv lines was overall inversely related to the transgene brain expression level, an observation which reiterates those from earlier studies on transgenic mice expressing hamster or mouse PrP (8, 32, 36, 49, 55). No influence (P ≥ 0.1 by Student's t test) of the type of construct was discernible (Table 1). All tgOv lines showed a preferential expression of the transgene in the brain. For tg2 mice, this corroborates the results of an earlier study in which the brain, including neuron-like cells, was a major target for CMV-driven expression in mice (1). To what extent the PrP regional distribution into the brain differs among the three types of lines, as well as whether extraneural routes of inoculation would reveal a differential effect of the construct, has to be examined.
Several lines of evidence argue that ovine PrP promoted a more favorable context than mouse PrP for sheep prion replication, at least for the two isolates studied. Firstly, overexpression of the ovine PrP transgene was not required for an accelerated transmission. For two lines expressing the transgene at a nearly physiological level (tg207 and tg335), the incubation duration was at least 200 days shorter than that for wild-type mice. Secondly, a transgenic line overexpressing mouse PrP, the tga20 line (18), was found to exhibit levels of susceptibility to both sheep isolates that were significantly higher than those exhibited by wild-type mice but not those exhibited by tgOv mice expressing lower levels of PrP (Table 1). The tga20 and tgOv mice have very close genetic backgrounds, since the lines were derived by introgression of the transgene onto the same PrP0/0 mice. Finally, another transgenic line, which is a murine counterpart of the tg2 lines and expresses mouse PrP at a level 1.2-fold greater than that of wild-type mice (tg440; data to be published elsewhere) was found to be poorly susceptible to scrapie. More than 370 days postinoculation with the fastest-acting sheep isolate, no disease has yet occurred in those mice.
Earlier experimental transmissions of hamster, human, and bovine prions to transgenic mice expressing a cognate Prnp gene has led to the proposition of a complete or nearly complete removal of the species barrier (15, 36, 44). In this study, no substantial change of the incubation period occurred upon subsequent passage of either of the two sheep isolates in tg211 mice, tg211 being the only tgOv line examined so far in this regard (Fig. 3). This is in sharp contrast with the common observation that the incubation duration of natural scrapie isolates is markedly reduced from the initial to the second passage in conventional mice (3). Collectively, these data may indicate that transmission of the sheep TSE agent to tgOv mice requires little or no adaptation. This issue, however, has to be further documented through transmission studies including a larger number of isolates, additional tgOv lines, and a comparison of the infectivity detection limit with that in the natural host.
Transgenic lines expressing ovine Prnp were described several years ago, (54) but, as was mentioned in a subsequent review (53), their susceptibility did not exceed that of nontransgenic controls. Although some relevant information, such as the PrP protein expression levels and the number and genotype of the donor animal(s), is lacking, differences in both the transgene construct and the mouse genetic background might account for the inefficient transmission previously observed. Firstly, a sheep cosmid clone harboring an ARQ allele was used. While the nature of the vector is unlikely to be of concern (see above), the divergence at codon 136 (i.e., arginine instead of valine [56]) may explain, at least in part, these discrepant results. Secondly, the transgene was not expressed onto a Prnp0/0 knockout background. The presence of endogenous PrP may thus have impaired the propagation of scrapie agent, as documented for other transgenic mice (6, 48, 53). Experiments are under way in an attempt to determine the extent to which the above-mentioned parameters modulate the susceptibility of tgOv mice to sheep TSE agent.
TgOv mice: an improved model for experimental studies on natural sheep scrapie?
Despite intensive studies performed during the last few decades on sheep scrapie, a number of crucially important questions, such as the natural mode of sheep to sheep transmission and the diversity and possible evolution of the strains which circulate in the field, remain unanswered. The existence of sheep-derived scrapie strains with clearly distinct biological properties though propagated in the same inbred mouse line is a well established fact (3, 4, 11, 17). Such strains, however, result from the passage of a TSE agent into a new host species, and there is ample evidence that both agent and host contribute to strain properties (4, 21, 38). As a consequence, a precise idea of the natural scrapie strain diversity has yet to be gained. Addressing this point has become even more important since the emergence of the BSE epidemics, because of the consequences that an accidental transmission of cattle TSE to sheep would entail for the public health (10). The availability of a laboratory animal endowed with increased susceptibility to sheep agent, such as these tgOv mice, may bring new opportunities with regard to field strains characterization and infectivity measurement.
The term susceptibility was dealt with in this study solely in reference to transmission pace. Whether tgOv mice would allow not only for the faster but also for the more sensitive detection of infectivity is not yet known, and parallel endpoint titrations on conventional and tgOv mice have been undertaken to answer this question. With regard to strain characterization, it came to light that the two isolates tested, issuing from one British and one French natural scrapie case, while showing indistinguishable PrPres banding patterns, exhibited fairly distinct properties in vivo. Their respective incubation periods in each of the three tgOv lines analyzed differed strikingly and remained essentially unchanged after subpassaging (Fig. 3). Moreover, the PrP deposition and lesion patterns in the brains of infected mice (for both primary and secondary infection) clearly differed between the two isolates (data to be published elsewhere). Transmission to tgOv mice thus allowed these two isolates to be differentiated within a much shorter period of time than would have been the case with wild-type mice. A further advantage of transmission from sheep to tgOv mice could be a lower selection pressure on the incoming agent, possibly resulting in a more accurate assessment of the natural scrapie strain diversity. Evaluating tgOv mice as an alternative tool for strain typing will require the analysis of an enlarged panel of field isolates. In particular, it would be worth learning whether and to what extent the donor sheep PrP genotype influences the efficiency of transmission in tgOv, as only transmissions in which PrP alleles from donors and recipients were homologous are reported here. It would also be informative to test isolates whose incubation times in conventional mice appear to exceed the natural life span of the host (28).
Finally, an unanticipated outcome of this work was the development of a model of TSE with one of the shortest incubation times of any such model. Indeed, one combination, tg301 mice inoculated with the PG127 isolate, led to a survival time of around 10 weeks postinoculation, with the clinical phase lasting less than 1 week. This time is remarkably brief compared to that reported for TSE human and bovine isolates in relevant tg mice (from ∼25 to ∼35 weeks [15, 27, 44, 48]). Furthermore, while resulting from a sheep-to-mouse primary transmission, such an incubation time is comparable to that reported for the the available rodent TSE models with the shortest incubation times (18, 30, 43). Therefore, the new model introduced here might be of benefit for various aspects of TSE research.
ACKNOWLEDGMENTS
We are grateful to J. P. Deslys (CEA, Fontenay-aux-Roses, France) for teaching us basic TSE-specific skills, O. Andreoletti (ENVT-INRA, Toulouse, France) for histopathological analysis on tg301 mice, and J. M. Elsen (INRA, Toulouse, France) for providing scrapie-infected material. We also thank the following persons from INRA, Jouy, France: A. Vaiman for performing the FISH analysis, J. Grosclaude for providing the antibody 2D6, B. Cayron and colleagues for animal care, and E. Cribiu and C. La Bonnardière for support and helpful discussions. We are particularly indebted to C. Weissmann for making the PrP0/0 mice, the tga20 mice, and the phgPrP vector available to us. The RIII mice originated from the Neuropathogenesis Unit, Edinburgh, United Kingdom, and were kindly provided by P. Sarradin (INRA, Tours, France).
This work was supported by grants from the European Community (FAIR 5CT-973304), from the CCI-EST (Cellule de Coordination Interorganismes sur les EST), and from INRA.
REFERENCES
- 1.Baskar J F, Smith P P, Nilaver G, Jupp R A, Hoffmann S, Peffer N J, Tenney D J, Colberg-Poley A M, Ghazal P, Nelson J A. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J Virol. 1996;70:3207–3214. doi: 10.1128/jvi.70.5.3207-3214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossers A, Schreuder B E C, Muileman I H, Belt P B G M, Smits M A. PrP genotype contributes to determining survival times of sheep with natural scrapie. J Gen Virol. 1996;77:2669–2673. doi: 10.1099/0022-1317-77-10-2669. [DOI] [PubMed] [Google Scholar]
- 3.Bruce M E, Dickinson A G. Biological evidence that scrapie agent has an independent genome. J Gen Virol. 1987;68:78–89. doi: 10.1099/0022-1317-68-1-79. [DOI] [PubMed] [Google Scholar]
- 4.Bruce M E, McConnell I, Fraser H, Dickinson A G. The disease characteristics of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol. 1991;72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 5.Bruce M E, Fraser H, McBride P A, Scott J R, Dickinson A G. The basis of strain variation in scrapie. In: Prusiner S B, Collinge J, Powell J, Anderton B, editors. Prion diseases of humans and animals. New York, N.Y: Ellis Horwood; 1992. pp. 497–508. [Google Scholar]
- 6.Büeler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 7.Büeler H, Fisher M, Lang Y, Bluethmann H, Lipp H P, DeArmond S J, Prusiner S B, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 8.Büeler H, Raeber A, Sailer A, Fischer M, Aguzzi A, Weissmann C. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol Med. 1994;1:19–30. [PMC free article] [PubMed] [Google Scholar]
- 9.Buschmann A, Pfaff E, Reifenberg K, Müller H M, Groschup M H. Detection of cattle-derived BSE prions using transgenic mice overexpressing bovine PrPc. Arch Virol Suppl. 2000;16:75–86. doi: 10.1007/978-3-7091-6308-5_6. [DOI] [PubMed] [Google Scholar]
- 10.Butler D. Doubts over ability to monitor risks of BSE spread to sheep. Nature. 1998;395:6–7. doi: 10.1038/25573. [DOI] [PubMed] [Google Scholar]
- 11.Carp R I, Rubenstein R. Diversity and significance of scrapie strains. Semin Virol. 1991;2:203–213. [Google Scholar]
- 12.Castiglioni B, Comincini S, Drisaldi B, Motta T, Ferretti L. Comparative mapping of the prion gene (PRNP) locus in cattle, sheep and human with PCR-generated probes. Mamm Genome. 1998;9:853–855. doi: 10.1007/s003359900882. [DOI] [PubMed] [Google Scholar]
- 13.Caughey B, Chesebro B. Prion protein and the transmissible spongiform encephalopathies. Trends Cell Biol. 1997;7:56–63. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 14.Clouscard C, Beaudry P, Elsen J M, Milan D, Dussaucy M, Bounneau C, Schelcher F, Chatelain J, Launay J M, Laplanche J L. Different allelic effects of the codons 136 and 171 of the prion protein gene in sheep with natural scrapie. J Gen Virol. 1995;76:2097–2101. doi: 10.1099/0022-1317-76-8-2097. [DOI] [PubMed] [Google Scholar]
- 15.Collinge J, Palmer M S, Sidle K C L, Hill A F, Gowland I, Meads J, Asante E, Bradley R, Doey L J, Lantos P L. Unaltered susceptibility to BSE in transgenic mice expressing human prion protein. Nature. 1995;378:21–28. doi: 10.1038/378779a0. [DOI] [PubMed] [Google Scholar]
- 16.Dickinson A G. Scrapie in sheep and goats, p 209–241. In: Kimrelin R H, editor. Slow virus diseases of animals and man. Amsterdam, The Netherlands: North Holland; 1976. [Google Scholar]
- 17.Dickinson A G, Meikle V M. A comparison of some biological characteristics of the mouse-passaged scrapie agents, 22A and ME7. Genet Res. 1969;13:213–225. doi: 10.1017/s0016672300002895. [DOI] [PubMed] [Google Scholar]
- 18.Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 19.Forget B G. YAC transgenes: bigger is probably better. Proc Natl Acad Sci USA. 1993;90:7909–7911. doi: 10.1073/pnas.90.17.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldmann W, Hunter N, Foster J D, Salbaum J M, Beyreuther K, Hope J. Two alleles of a neural protein gene linked to scrapie in sheep. Biochemistry. 1990;87:2476–2480. doi: 10.1073/pnas.87.7.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldmann W, Hunter N, Smith G, Foster J, Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J Gen Virol. 1994;75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 22.Goldmann W, O'Neill G, Cheung F, Charleson F, Ford P, Hunter N. PrP (prion) gene expression in sheep may be modulated by alternative polyadenylation of its messenger RNA. J Gen Virol. 1999;80:2275–2283. doi: 10.1099/0022-1317-80-8-2275. [DOI] [PubMed] [Google Scholar]
- 23.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 24.Hadlow W J, Kennedy R C, Race R E. Natural infection of Suffolk sheep with scrapie virus. J Infect Dis. 1982;146:657–664. doi: 10.1093/infdis/146.5.657. [DOI] [PubMed] [Google Scholar]
- 25.Hardt M, Baron T, Groschup M H. A comparative study of immunochemical studies for detecting abnormal prion protein with monoclonal and polyclonal antibodies. J Comp Pathol. 2000;1:43–53. doi: 10.1053/jcpa.1999.0343. [DOI] [PubMed] [Google Scholar]
- 26.Harmeyer S, Pfaff E, Groschup M H. Synthetic peptide vaccines yield monoclonal antibodies to cellular and pathological prion proteins of ruminants. J Gen Virol. 1998;79:937–945. doi: 10.1099/0022-1317-79-4-937. [DOI] [PubMed] [Google Scholar]
- 27.Hill A F, Desbruslais M, Joiner S, Sidle K C L, Gowland I, Collinge J, Doey L J, Lantos P. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 28.Hunter N. PrP genetics in sheep and the applications for scrapie and BSE. Trends Microbiol. 1997;8:331–334. doi: 10.1016/s0966-842x(97)01081-0. [DOI] [PubMed] [Google Scholar]
- 29.Hunter N, Foster J D, Goldmann W, Stear M J, Hope J, Bostock C. Natural scrapie in a closed flock of Cheviot sheep occurs only in specific PrP genotypes. Arch Virol. 1996;141:809–824. doi: 10.1007/BF01718157. [DOI] [PubMed] [Google Scholar]
- 30.Kimberlin R H, Walker C A. Characteristics of a short incubation model of scrapie in the golden hamster. J Gen Virol. 1977;34:295–304. doi: 10.1099/0022-1317-34-2-295. [DOI] [PubMed] [Google Scholar]
- 31.Lathe R, Vilotte J L, Clark A J. Plasmid and bacteriophage vectors for excision of intact inserts. Gene. 1987;57:193–201. doi: 10.1016/0378-1119(87)90122-3. [DOI] [PubMed] [Google Scholar]
- 32.Manson J C, Clarke A R, McBride P A, McConnell I, Hope J. PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegeneration. 1994;3:331–340. [PubMed] [Google Scholar]
- 33.Manson J C, Jamieson E, Baybutt H, Tuzi N L, Barron R, McConnell I, Somerville R, Ironside J, Will R, Sy M S, Melton D W, Hope J, Bostock C. A single amino acid alteration (101L) introduced into murine PrP dramatically alters incubation time of transmissible spongiform encephalopathy. EMBO J. 1999;18:6855–6864. doi: 10.1093/emboj/18.23.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore R C, Lee I Y, Silverman G L, Harrison P M, Strome R, Heinrich C, Karunaratne A, Pasternak S H, Chishti M A, Liang Y, Mastrangelo P, Wang K, Smit A F A, Katamine S, Carlson G A, Cohen F E, Prusiner S B, Melton D W, Tremblay P, Hood L E, Westaway D. Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein Doppel. J Mol Biol. 1999;292:797–817. doi: 10.1006/jmbi.1999.3108. [DOI] [PubMed] [Google Scholar]
- 35.Pattison I H. Experiments with scrapie with special reference to the nature of the agent and the pathology of the disease. In: Gajdusek D C, Gibbs C J Jr, Alpers M P, editors. Slow, latent and temparate virus infections. NINDB monograph 2. U.S. Washington, D.C.: Government Printing Office; 1965. pp. 249–257. [Google Scholar]
- 36.Prusiner S B, Scott M, Foster D, Pan K M, Groth D, Mirenda C, Torchia M, Yand S L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 37.Prusiner S B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 38.Prusiner S B. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prusiner S B, Scott M R, DeArmond S J, Cohen F E. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 40.Raeber A J, Brandner S, Klein M A, Benninger Y, Musahl C, Frigg R, Roeckl C, Fischer M B, Weissmann C, Aguzzi A. Transgenic and knockout mice in research on prion diseases. Brain Pathol. 1998;8:715–733. doi: 10.1111/j.1750-3639.1998.tb00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rezaeï H, Marc D, Choiset Y, Takahashi M, Hui Bon Hoa G, Heartlé T, Grosclaude J, Debey P. High yield purification and physico-chemical properties of full-length recombinant allelic variants of sheep prion protein linked to susceptibility. Eur J Biochem. 2000;267:2833–2839. doi: 10.1046/j.1432-1033.2000.01347.x. [DOI] [PubMed] [Google Scholar]
- 42.Schibler L, Vaiman D, Oustry A, Guinec N, Dangy-Caye A L, Billault A, Cribiu E P. Construction and extensive characterization of a goat bacterial artificial library with threefold genome coverage. Mamm Genome. 1998;9:119–124. doi: 10.1007/s003359900701. [DOI] [PubMed] [Google Scholar]
- 43.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, Torchia M, Groth D, Carlson G, DeArmond S J, Westaway D, Prusiner S B. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 44.Scott M R, Safar J, Telling G, Nguyen O, Groth D, Torchia M, Koehler R, Tremblay P, Walther D, Cohen F E, DeArmond S J, Prusiner S B. Identification of a prion protein epitope modulating transmission of bovine spongiform encephalopathy prions to transgenic mice. Proc Natl Acad Sci USA. 1997;94:14279–14284. doi: 10.1073/pnas.94.26.14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott M R, Will R, Ironside J, Nguyen H O B, Tremblay P, DeArmond S J, Prusiner S B. Compelling transgenic evidence for transmission of bovine spongiform encephalopathy prions to humans. Proc Natl Acad Sci USA. 1999;96:15137–15142. doi: 10.1073/pnas.96.26.15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stinnakre M G, Soulier S, Schibler L, Lepourry L, Mercier J C, Vilotte J L. Position-independent and copy-number-related expression of a goat bacterial artificial chromosome α-lactalbumin gene in transgenic mice. Biochem J. 1999;339:33–36. [PMC free article] [PubMed] [Google Scholar]
- 47.Telling G C, Scott M, Hsiao K K, Foster D, Yang S L, Torchia M, Sidle K C L, Collinge J, DeArmond S J, Prusiner S B. Transmission of Creutzfeldt-Jakob disease from humans to transgenic mice expressing chimeric human-mouse prion protein. Proc Natl Acad Sci USA. 1994;91:9936–9940. doi: 10.1073/pnas.91.21.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 49.Tremblay P, Meiner Z, Galou M, Heinrich C, Petromilli C, Lisse T, Cayetano J, Torchia M, Mobley W, Bujard H, DeArmond S J, Prusiner S B. Doxycycline control of prion protein transgene expression modulates prion disease in mice. Proc Natl Acad Sci USA. 1998;95:12580–12585. doi: 10.1073/pnas.95.21.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaiman D, Billault A, Tabet-Aoul K, Schibler L, Vilette D, Oustry-Vaiman A, Soravito C, Cribiu E P. Construction and characterization of a sheep BAC library of three genome equivalents. Mamm Genome. 1999;10:585–587. doi: 10.1007/s003359901049. [DOI] [PubMed] [Google Scholar]
- 51.Vilotte J L, Soulier S, Stinnakre M G, Massoud M, Mercier J C. Efficient tissue-specific expression of bovine α-lactalbumin in transgenic mice. Eur J Biochem. 1989;186:43–48. doi: 10.1111/j.1432-1033.1989.tb15175.x. [DOI] [PubMed] [Google Scholar]
- 52.Weissmann C. Molecular biology of transmissible spongiform encephalopathies. FEBS Lett. 1996;389:3–11. doi: 10.1016/0014-5793(96)00610-2. [DOI] [PubMed] [Google Scholar]
- 53.Westaway D. Transgenic approaches to prion “species-barrier” effects. In: Baker H, Ridley R M, editors. Methods in molecular medicine: prion diseases. Totowa, N.J: Humana Press, Inc.; 1996. pp. 251–263. [Google Scholar]
- 54.Westaway D, DeArmond S J, Cayetano-Canlas J, Groth D, Foster D, Yang S L, Torchia M, Carlson G A, Prusiner S B. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell. 1994;76:117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 55.Westaway D, Mirenda C A, Foster D, Zerbajadian Y, Scott M, Torchia M, Yang S-L, Serban H, DeArmond S J, Ebeling C, Prusiner S B, Carlson G A. Paradoxical shortening of incubation times by expression of prion protein transgenes derived from long incubation period mice. Neuron. 1991;7:56–68. doi: 10.1016/0896-6273(91)90074-a. [DOI] [PubMed] [Google Scholar]
- 56.Westaway D, Zuliani V, Cooper C M, Da Costa M, Neuman S, Jenny A L, Detwiler L, Prusiner S B. Homozygosity for prion protein alleles encoding glutamine-171 renders sheep susceptibility to natural scrapie. Genes Dev. 1994;8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]