Abstract
Mutations in the gene encoding the zinc-finger transcription factor Ikaros (IKZF1) are found in patients with immunodeficiency, leukemia, and autoimmunity. Although Ikaros has a well-established function in modulating gene expression programs important for hematopoietic development, its role in other cell types is less well defined. Here, we uncover functions for Ikaros in thymic epithelial lineage development in mice and show that Ikzf1 expression in medullary thymic epithelial cells (mTECs) is required for both autoimmune regulator–positive (Aire+) mTEC development and tissue-specific antigen (TSA) gene expression. Accordingly, TEC-specific deletion of Ikzf1 in mice results in a profound decrease in Aire+ mTECs, a global loss of TSA gene expression, and the development of autoimmunity. Moreover, Ikaros shapes thymic mimetic cell diversity, and its deletion results in a marked expansion of thymic tuft cells and muscle-like mTECs and a loss of other Aire-dependent mimetic populations. Single-cell analysis reveals that Ikaros modulates core transcriptional programs in TECs that correlate with the observed cellular changes. Our findings highlight a previously undescribed role for Ikaros in regulating epithelial lineage development and function and suggest that failed thymic central tolerance could contribute to the autoimmunity seen in humans with IKZF1 mutations.
INTRODUCTION
The thymus is the anatomical site of T cell maturation and is required for generating a highly diverse yet self-tolerant T cell pool (1). These competing priorities are achieved, in part, through functional compartmentalization in which cortical epithelial cells (cTECs) select for functional T cell receptors (TCR) and medullary thymic epithelial cells (mTECs) screen for autoreactivity before release into the periphery. This screening is dependent on diverse expression and presentation of the protein-coding genome across the medullary compartment. The autoimmune regulator (Aire) and Fez family zinc finger 2 (Fezf2) proteins play critical and non-redundant roles in driving tissue-specific antigen (TSA) expression within a subset of major histocompatibility complex class II (MHC-II)high mTECs (2–4). Deletion of Aire or Fezf2 in mice results in failure to express subsets of TSA genes, the escape of autoreactive T cells, and organ-specific autoimmunity (5). Moreover, human mutations in AIRE result in autoimmune polyglandular syndrome type 1 (APS1), in which affected patients develop multiple-organ–specific autoimmune diseases (6). In addition to Aire-expressing MHC-IIhigh mTECs, single-cell transcriptomics has exposed considerable heterogeneity within MHC-IIlow mTECs (7). These MHC-IIlow mTECs include a transit-amplifying TEC population (TAC-TECs) (8), Ccl21a-expressing mTECs (9, 10), and a number of terminally differentiated epithelial lineages called “mimetic cells” because of their shared core transcriptional features with peripheral epithelial counterparts (11). The mimetic cell populations are diverse too and include highly differentiated thymic tuft cells, microfold (M) cells, muscle-like mTECs, and keratinocyte-like mTECs, among others (7, 8, 12). Moreover, mimetic cells express TSAs and are thought to contribute to the negative selection of T cells. Thymic tuft cells were the first mimetic cell population described and are functionally related to tuft cells located at mucosal epithelial barriers, which orchestrate interleukin-25 (IL-25)–mediated responses for the clearance of parasites and protozoa (13–16). Within the thymus, tuft cells have been shown to be important for the regulation of innate natural killer (NK) T cells (7, 17). Thus, the thymic epithelium is composed of both specialized cells broadly enriched for TSA expression and also highly orchestrated and terminally differentiated populations having distinct mimetic phenotypes and functions. Disordered development and regulation of these diverse populations may have consequences for T cell repertoire selection, immune tolerance, and adaptive immunity.
Recent work has shown that MHC-IIhigh mTECs express up to 80 to 90% of the protein-coding genome and that Aire and Fezf2 are important mediators of this process (18). Although experiments with deletion of either Aire or Fezf2 in mice reveal nonoverlapping TSA gene programs, they do not account for all TSA gene expression observed in mTECs, suggesting that other transcriptional regulators are needed to orchestrate the complete program of TSA gene expression (5, 18). Furthermore, although recent studies have cataloged putative transcription factor networks regulating mimetic cell diversity (11), there are likely other factors required to shape the extensive cellular heterogeneity within the thymic medulla. Prior studies on the transcriptional control of mTEC development have shown a central role for the noncanonical nuclear factor κB (NF-κB) pathway in MHC-IIhigh mTEC development and Aire expression (19–21), as well as for POU class 2 homeobox 3 (Pou2f3) in tuft cell development and SpiB/Sox8 for thymic M cell development (7, 11, 17).
To identify additional transcription factors relevant for mTEC function, we probed publicly available gene-expression databases (22) and observed substantial levels of Ikaros (Ikzf1) expression enriched in mTECs. Here, we show that Ikaros is expressed in a specific TEC population enriched for markers of cell proliferation termed TAC-TECs (8), as well as in Aire+ mTECs, and that the deletion of Ikzf1 in mTECs has a profound effect on mTEC cellular heterogeneity, TSA expression, and central tolerance. Specifically, TEC-specific deletion of Ikaros results in a loss of Aire+ mTECs and a marked expansion of thymic tuft and muscle-like mimetic cell populations. Single-cell RNA sequencing (scRNA-seq) and single-cell assay for transposase-accessible chromatin using sequencing on single cells (scATAC-seq) suggest that Ikaros mediates these effects through the modulation of key transcriptional programs in TECs. These findings highlight a function for Ikaros in thymic epithelial development and homeostasis and define Ikaros as a key mediator of central tolerance.
RESULTS
Ikaros is required for the expansion and maintenance of Aire+ mTECs
A broad survey of transcription factor expression in mTECs using publicly available datasets demonstrated relatively high levels of Ikzf1 transcript within mTECs (fig. S1A) (22). The transcription factor Ikzf1 has been extensively studied in hematopoietic lineages, and Ikaros is critical for B and T cell lineage development (23–36), but its expression in the thymic epithelium was unexpected. We confirmed Ikzf1 expression by flow-sorting mTECs and cTECs for gene expression analysis. In addition to the expected expression in hematopoietic T and B cells, we found that Ikzf1 transcript was detected in the thymic epithelium and was concentrated in the subset of mTECs expressing high levels of MHC-IIhigh relative to MHC-IIlow mTECs and cTECs (Fig. 1A and fig. S1B).
Fig. 1. Ikaros deletion in mTECs results in altered mTEC composition.
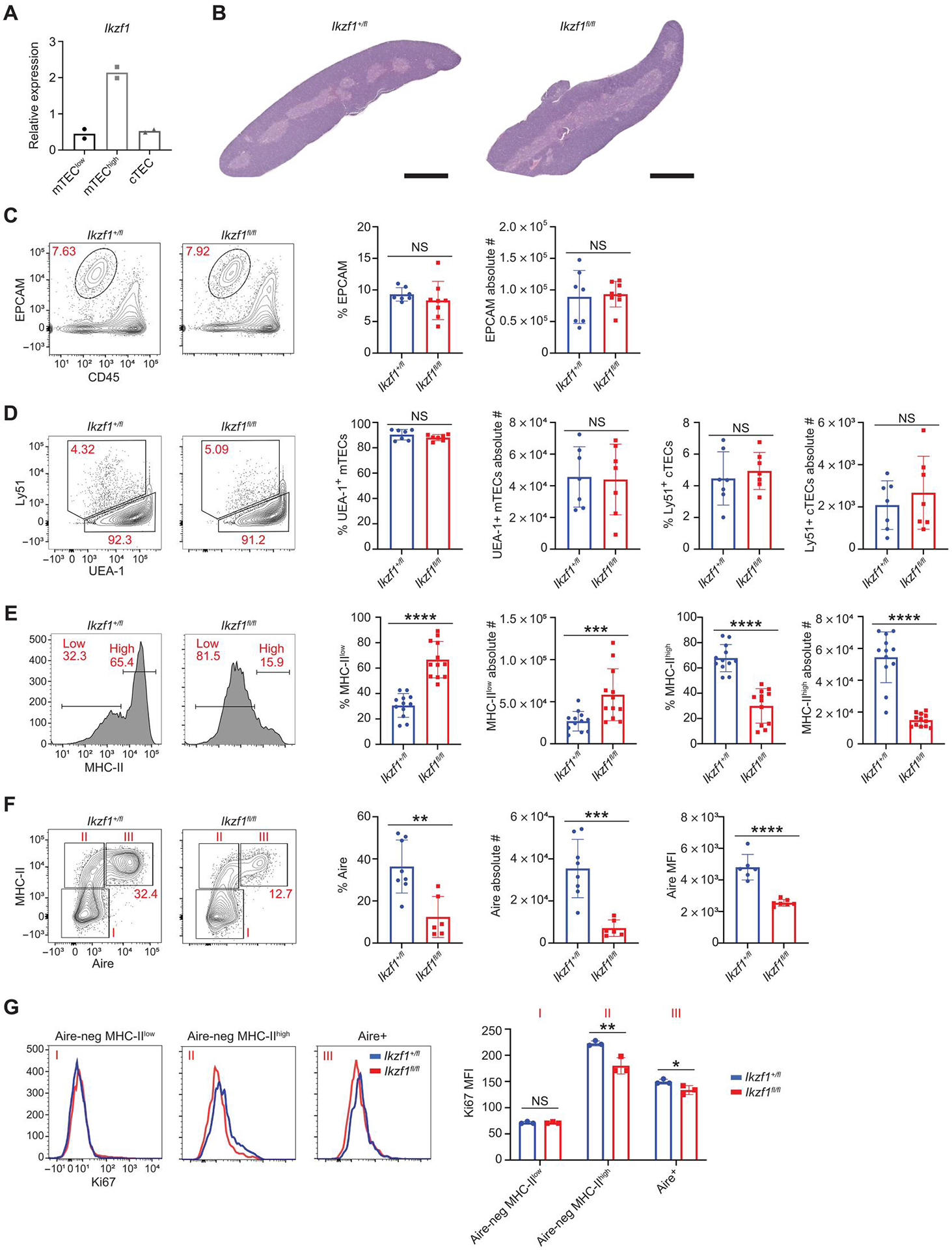
(A) qPCR of Ikzf1 gene expression in sorted MHC-IIhigh, MHC-IIlow mTECs, and cTECs from C57BL/6 mice. Sorted MHC-IIhigh and MHC-IIlow mTECs were gated on CD11c−CD45−EPCAM+Ly51−-IAb+ TECs. cTECs were gated on CD11c−CD45−EPCAM+Ly51+-UEA1− TECs. (B) Representative images of H&E staining of thymi from Foxn1-Cre/Ikzf1+/fl mice (Ikzf1+/fl) and Foxn1-Cre/Ikzf1fl/fl mice (Ikzf1fl/fl). Scale bars, 1 mm (1×). (C) Flow cytometry (left) and percentage and absolute number (right) of EPCAM+ TECs gated on CD11c−CD45−EPCAM+ thymic epithelial cells (n = 7 or 8 mice; three independent experiments). (D) Flow cytometry (left) and percentage and absolute number (right) of EPCAM+Ly51+UEA1− cTECs and EPCAM+Ly51−UEA1+ mTECs (n = 6 or 7 mice; two independent experiments). (E) Flow cytometry (left) and percentage and absolute number (right) of MHC-IIhigh and MHC-IIlow mTECs gated on CD11c−CD45−EPCAM+Ly51−IAb+ TECs (n = 12 mice; four independent experiments). (F) Flow cytometry (left) and percentage and absolute number (right) EPCAM+-MHC-IIhigh Aire+ mTECs. Aire MFI shown (n = 6 to 8 mice; three independent experiments). (G) Ki67 histograms corresponding to populations in (F): Aire-negative MHC-IIlow (I), Aire-negative MHC-IIhigh (II), and Aire+ (III). Bar graph shows Ki67 MFI (n = 3 mice per genotype; one representative experiment). (A and C to G) In graphs, the bar corresponds to the mean, with error bars showing ±SD of values shown, and each data point represents an individual mouse. (C to G) Statistical significance was determined using Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. NS, not significant.
Along with B and T cells, Ikaros has been reported to operate in other lineages including the central nervous system (37–39) and hypothalamic-pituitary axis (40), suggesting relevant roles beyond hematopoiesis. To investigate the potential function of Ikaros within the thymic epithelium, we conditionally deleted Ikzf1 in the thymic epithelium by crossing a floxed allele (Ikzf1fl/fl) (24) to the Foxn1-Cre line (41). We confirmed selective (although potentially incomplete) deletion of Ikaros transcripts containing the deleted exon in mTECs (fig. S1C) but found no appreciable change in Ikaros mRNA or protein levels in lymphocytes (fig. S1, D to F). Histologic analysis of Foxn1-Cre/Ikzf1fl/fl thymi by hematoxylin and eosin (H&E) staining and confocal microscopy did not indicate any substantial change to thymic architecture, with a separation of cortex and medulla (Fig. 1B and fig. S2A). Similarly, flow cytometric analysis of TECs from 30- to 40-day-old Foxn1-Cre/Ikzf1fl/fl mice revealed normal total numbers of epithelial cell adhesion molecule (EPCAM+) TECs, suggesting that deletion of Ikaros did not affect overall TEC cellularity (Fig. 1C and fig. S2B). Moreover, the percentage and absolute number of Ly51+/UEA1− cTECs were also unchanged (Fig. 1D). However, upon higher-resolution analysis of known mTEC subsets by flow cytometry, we noted substantial alterations in mTEC populations with a profound loss of MHC-IIhigh and Aire+ mTECs, including a reduction in Aire mean fluorescence intensity (MFI) (Fig. 1, E and F). Foxn1-Cre(+)/Ikzf1+/fl mice were used as controls because the percentage and absolute number of Aire+ mTECs were not distinguishable from those of Foxn1-Cre (−)/Ikzf1fl/fl littermates (figs. S1C and S3A), indicating that there was no haploinsufficient phenotype of heterozygote Ikaros expression for Aire+ mTEC development. Despite the strong reduction of Aire+ mTECs in Foxn1-Cre/Ikzf1fl/fl mice, confocal microscopy revealed normal subnuclear localization of Aire in characteristic nuclear speckles (fig. S3B).
To interrogate the mechanism underlying the loss of MHC-IIhigh Aire+ mTECs further, we measured cell proliferation by intracellular staining for Ki67. TEC-specific deletion of Ikzf1 resulted in an overall reduction of Ki67 staining in mTECs, with decreased expression most prominent in Aire-negative MHC-IIhigh mTECs (Fig. 1G and fig. S3C). Prior work has shown that Aire-negative MHC-IIhigh mTECs have increased proliferative capacity when compared with Aire+ mTECs (42) and that Aire-negative MHC-IIhigh mTECs are a developmental precursor to Aire+ mTECs. Thus, attenuated proliferation of Aire-negative MHC-IIhigh mTECs may explain the loss of Aire-negative and Aire+ MHC-IIhigh mTECs in Foxn1-Cre/Ikzf1fl/fl mice. Together, these results demonstrate that Ikaros has a role in modulating mTEC cellular homeostasis and proliferation.
Ikzf1 deficiency causes defects to the thymic epithelium
Given the major shifts in TEC populations in the medullas of Ikaros TEC-specific knockout (KO) mice, we profiled these changes at higher resolution by scRNA-seq of flow-sorted mTECs from 30-to 40-day-old Foxn1-Cre/Ikzf1+/fl and Foxn1-Cre/Ikzf1fl/fl mice. We sequenced 5245 CD45−/EPCAM+/Ly51− cells from Foxn1-Cre/Ikzf1+/fl mice and 2529 cells from Foxn1-Cre/Ikzf1fl/fl mice. Dimensional reduction and clustering of these data by uniform manifold approximation and projection (UMAP) revealed five mTEC subsets: TAC-TEC, Ccl21a+, Aire+, Late Aire, and tuft cell populations (Fig. 2, A to C, and fig. S4A) (7, 8). As described, TAC-TECs are a transit-amplifying mTEC population that express high levels of the high mobility group chromatin binding factors (Hmgb2, H2afz, Hmgn2, Hmgb1, and Hmgn1) and are predominantly in the G2-M phase of the cell cycle. We analyzed our single-cell data for fine mapping of Ikzf1 expression in TEC populations, which revealed the highest Ikzf1 expression within TAC-TECs and Aire+ mTECs (Fig. 2, D to F). Moreover, single-cell data showed substantial coexpression of Ikzf1 and Aire on a per-cell basis, with 90% of Ikzf1-expressing mTECs coexpressing Aire. Using previously published scRNA-seq data from human thymic epithelial cells (43), we also detected a similar pattern of expression of IKZF1 in the human thymus (fig. S4B). Because we observed alterations in Aire+ mTECs, as seen in flow cytometric analysis of Foxn1-Cre/Ikzf1fl/fl mice, we investigated whether we detected similar changes by scRNA-seq. In agreement with our flow cytometry findings (Fig. 1F), scRNA-seq showed altered mTEC cellular composition with a marked reduction of Aire+ mTECs. Moreover, there were substantial changes to other mTEC lineages, with a reduction in Late Aire mTECs and a profound expansion of tuft cells (Fig. 2, G to I). Last, there was an increase in the percentage of Ccl21a+ mTECs, and we confirmed an expansion in C-C motif chemokine ligand 21 (CCL21) protein–expressing mTECs by flow cytometry (Fig. 2J).
Fig. 2. Ikaros is expressed in TAC-TECs and Aire+ mTECs.
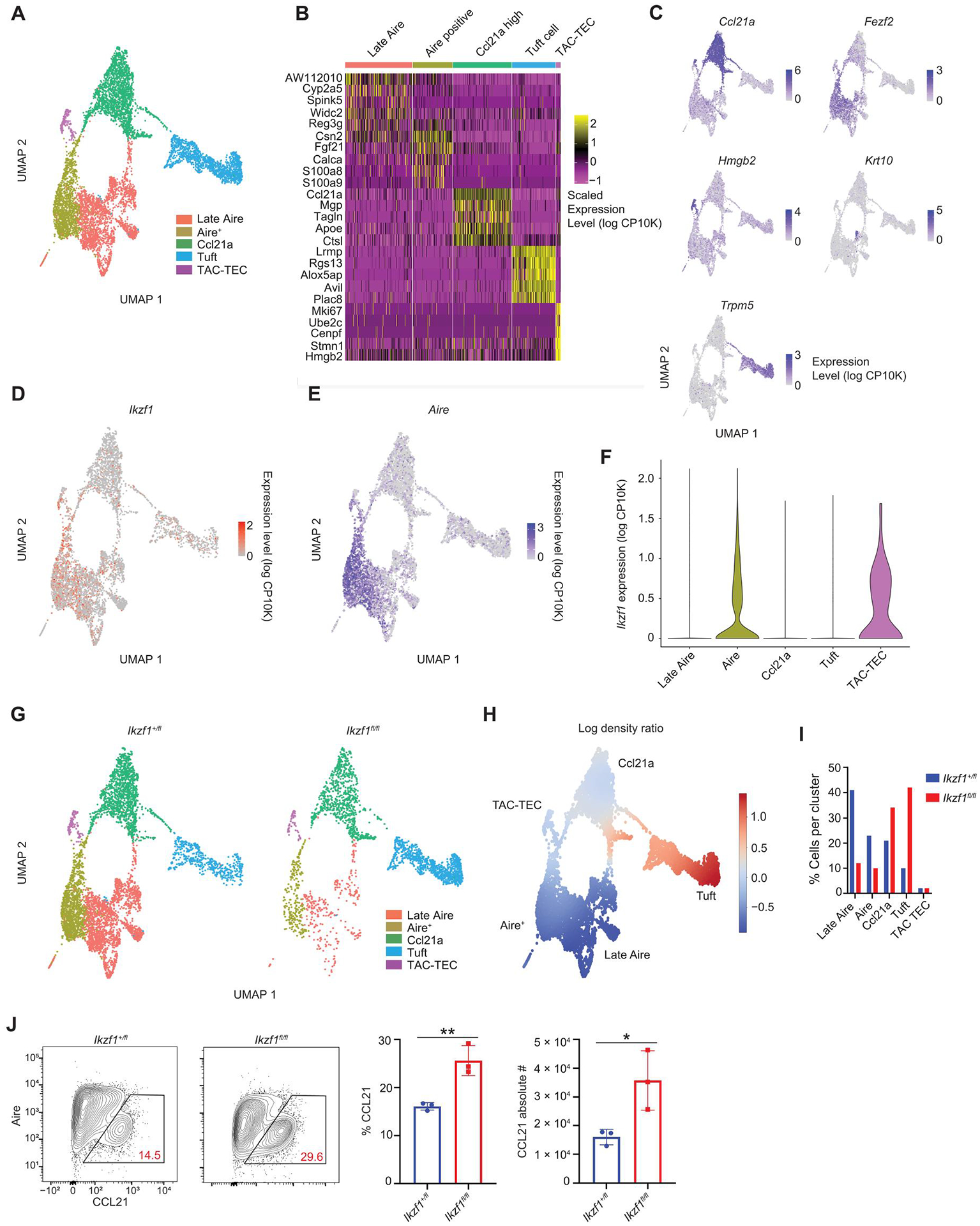
(A) Clustering of scRNA-seq data from sorted CD11c−EPCAM+Ly51− mTECs from Foxn1-Cre/Ikzf1+/fl mice (Ikzf1+/fl) and Foxn1-Cre/Ikzf1fl/fl mice (Ikzf1fl/fl). A total of 5245 mTECs from Ikzf1+/fl mice and 2529 mTECs from Ikzf1fl/fl mice were analyzed. Hematopoietic cells were removed from the UMAP. (B) Heatmap showing top five genes expressed in each mTEC cluster. (C) UMAPs of marker genes for each mTEC subset. (D) UMAP of Ikzf1 expression in Ikzf1+/fl mTECs. (E) UMAP of Aire expression in Ikzf1+/fl mTECs. (F) Violin plot showing Ikzf1 expression in Ikzf1+/fl mTECs. (G) UMAPs of cell distributions in Ikzf1+/fl mTECs and Ikzf1fl/fl mTECs. (H) Differential density plot showing distribution of increasing (red) or decreasing (blue) populations in Ikzf1fl/fl mTECs relative to Ikzf1+/ fl mTECs. (I) Bar graph of the percentage of cells in each cluster normalized to the total number cells sequenced. (J) Flow cytometry (left) and percentage and absolute number (right) of CCL21+ mTECs, gated on CD11c−CD45−EPCAM+Ly51− TECs (n = 3 mice per genotype; one representative experiment). (J) In graphs, the bars correspond to the mean, with error bars showing ±SD of values shown, and each data point represents an individual mouse. (J) Statistical significance was determined using Student’s t test. *P < 0.05 and **P < 0.01.
We next interrogated these data for changes in thymic mimetic cell populations (11, 17, 43). Most mimetic cells appear to pass through an Aire-expressing stage during their differentiation (11). Glycoprotein 2–positive (GP2+) microfold-like mTECs and cytokeratin 10–positive (KRT10+) keratinocyte-like mTECs are two such populations that have been well described (11). Other mimetic cells, such as tuft cells, are less dependent on the Aire-expressing lineage, with lineage-tracing experiments demonstrating that about 50% of tuft cells pass through an Aire-expressing stage (17). We reclustered our scRNA-seq data focusing on Late Aire mTECs and tuft cells and identified multiple previously described mimetic cell populations (Fig. 3A). This analysis revealed that Ikzf1 deletion in mTECs resulted in a substantial loss of mimetic subsets previously shown to go through an Aire-expressing stage, including goblet-like, neuroendocrine-like, microfold-like, and keratinocyte-like mTECs (Fig. 3B). In addition to confirming the expansion of thymic tuft cells, scRNA-seq revealed an expansion of muscle-like mTECs in the conditional KO. Consistent with these data, prior work has reported that muscle-like mTECs do not pass through an Aire-expressing stage in development (11). Flow cytometric analysis confirmed the loss of GP2+ microfold-like mTECs, KRT10+ keratinocyte-like mTECs, and the expansion of double cortin-like kinase 1 (DCLK1+) thymic tuft cells, which persisted in mice aged to 6 months (Fig. 3, C to E, and fig. S4C). Thus, Ikzf1 deletion in TECs results in a loss of Aire-dependent mimetic cell populations, which is balanced by an expansion of Aire-independent mimetic cells.
Fig. 3. Deletion of Ikaros alters mimetic cell diversity.
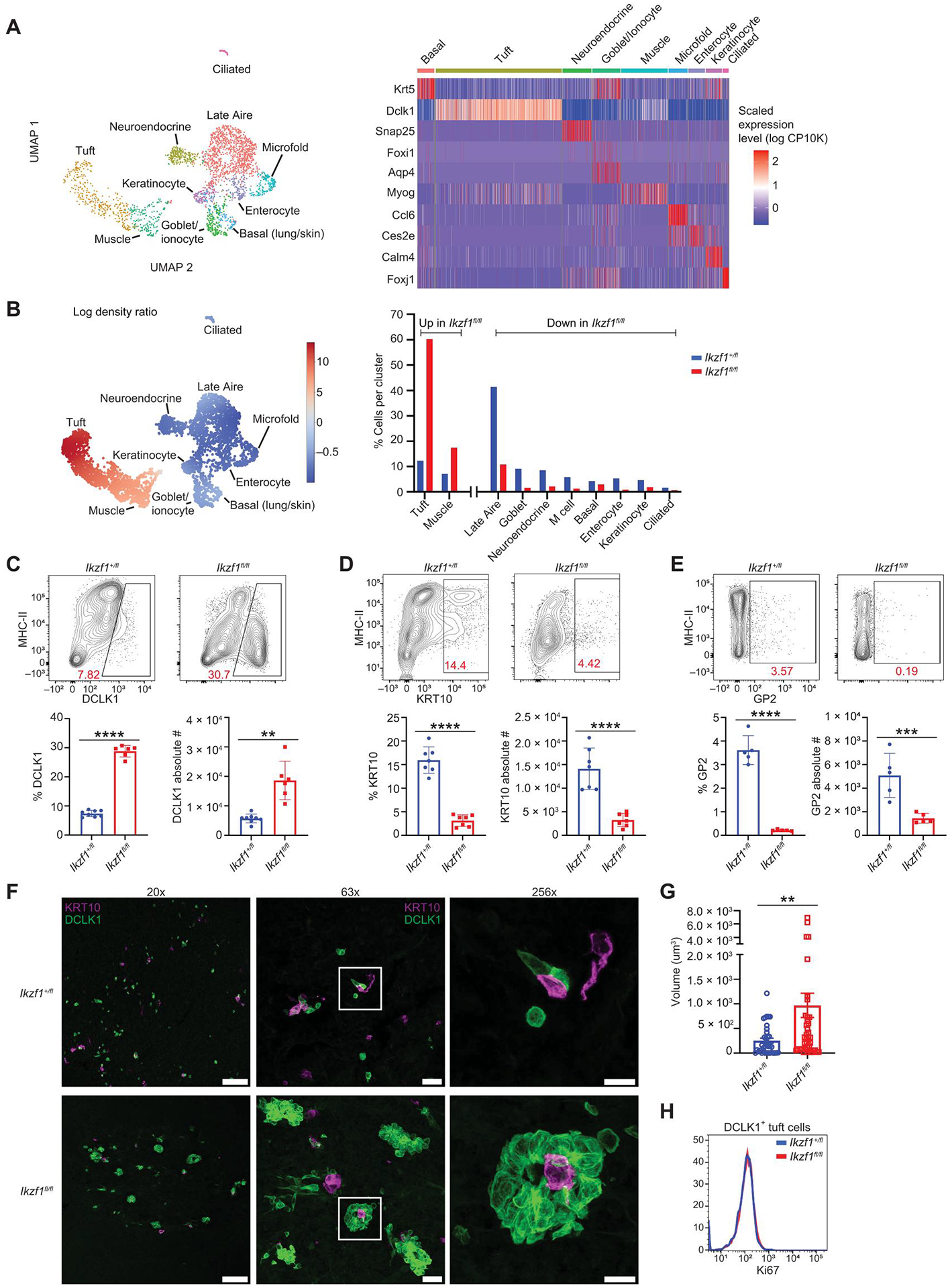
(A) UMAP of populations of mimetic cells found in Late Aire and tuft populations from Fig. 2A. Heatmap of marker genes in mimetic cell populations. A total of 2659 Late Aire and tuft cells from Ikf1+/fl mice and 1356 Late Aire and tuft cells from Ikzf1fl/fl mice were analyzed. (B) Differential density plot showing distribution of increasing (red) or decreasing (blue) populations in Ikzf1fl/fl mTECs relative to Ikzf1+/fl mTECs. Bar graph of the percentage of cells in each cluster normalized to the total number cells sequenced. (C) Flow cytometry (left) and percentage and absolute number (right) of DCLK1+ tuft cells, gated on CD11c−CD45−EPCAM+-Ly51− mTECs (n = 6 to 8 mice; two independent experiments). (D) Flow cytometry (left) and percentage and absolute number (right) of KRT10+ keratinocyte-like mTECs, gated on CD11c−CD45−EPCAM+-Ly51− mTECs (n = 6 or 7 mice; two independent experiments). (E) Flow cytometry (left) and percentage and absolute number (right) of GP2+ thymic M cells, gated on CD11c−CD45−EPCAM+Ly51− mTECs (n = 5 mice; two independent experiments). (F) Confocal microscopy of DCLK1+ tuft cells (green) and KRT10+ (magenta) keratinocyte mTECs. DAPI staining in blue. Scale bars, 100 μm (20×), 20 μm (63×), and 10 μm (256×). (G) Tuft cell volume quantification from thymic sections of two independent mice using Imaris software. Each data point is a DCLK1+ cluster (n = 36 to 43 DCLK1+ clusters). (H) Histogram of Ki67 in DCLK1+ tuft cells. (C to E and G) In graphs, the bars correspond to the mean, with error bars showing ±SD of values shown, and each data point represents an individual mouse (C to E) or an individual tuft cell cluster (G). (C to G) Statistical significance was determined using Student’s t test. **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Given the large increase in tuft cell numbers in the thymi of Foxn1-Cre/Ikzf1fl/fl mice, we further analyzed the organization and distribution of tuft cells with KRT10+ mTECs, which have previously been shown to colocalize with tuft cells in the medulla (17). Although there was a decrease in KRT10+ keratinocyte-like mTECs in the conditional KO, we did not observe altered colocalization of KRT10+ mTECs with DCLK1+ tuft cells in thymi from Foxn1-Cre/Ikzf1fl/fl mice and Foxn1-Cre/Ikzf1+/fl mice (Fig. 3F). Instead, we found that tuft cells in Foxn1-Cre/Ikzf1fl/fl thymi formed large, complex aggregates, which we confirmed by volumetric quantitative image analysis (Fig. 3, F and G, and fig. S4D). These large tuft aggregates often formed tubular structures with the appearance of a central lumen, suggesting a tendency toward ordered tertiary structure with tuft cells surrounding KRT10+ cells (Fig. 3F and fig. S4D). Although Ikzf1 deletion induced large tuft cell aggregates, tuft cells did not have increased proliferation, as measured by flow cytometric analysis of Ki67 (Fig. 3H). Together, our scRNA-seq data show that Ikzf1 is expressed in proliferating TECs and the Aire+ lineage and that deletion of Ikaros results in profound changes in mTEC cellular composition, with a loss of Aire+ mTECs and Aire-dependent mimetic cells counterbalanced by an expansion of tuft and muscle-like mTECs.
Ikaros modulates tuft cell gene expression and function
Given that deletion of Ikaros in TECs results in an expansion of DCLK1+ tuft cells with altered subcellular structure, we probed whether Ikzf1 deletion in mTECs altered tuft cell gene expression and function. We performed differential gene expression analysis on tuft cells using our scRNA-seq dataset and found altered expression of many key tuft cell genes upon Ikzf1 deletion, with increased expression of genes such as Dclkl and decreased expression of others such as Il25 and Gnat3 (Fig. 4A). We performed bulk RNA-seq on tuft cells to interrogate these changes using L1 cell adhesion molecule (L1CAM), which is coexpressed with DCLK1 as a tuft cell surface marker (fig. S5A and data file S1) as previously described (7). Bulk RNA-seq confirmed that Ikzf1 deletion in mTECs caused changes in gene expression in tuft cells, with 1054 genes down-regulated and 605 genes up-regulated [false discovery rate (FDR) < 0.05, log2 fold change (log2FC) > 1] (Fig. 4B and fig. S5B). Consistent with scRNA-seq showing increased cytokine expression in Foxn1-Cre/Ikzf1+/fl tuft cells, metascape pathway analysis (44) on the bulk RNA-seq data revealed that Ikaros-sufficient tuft cells were skewed toward an immune phenotype, whereas Ikzf1-deficient tuft cells had increased expression of vesicular trafficking and neuronal pathway transcriptional programs (Fig. 4C).
Fig. 4. Ikaros modulates tuft cell gene expression and function.
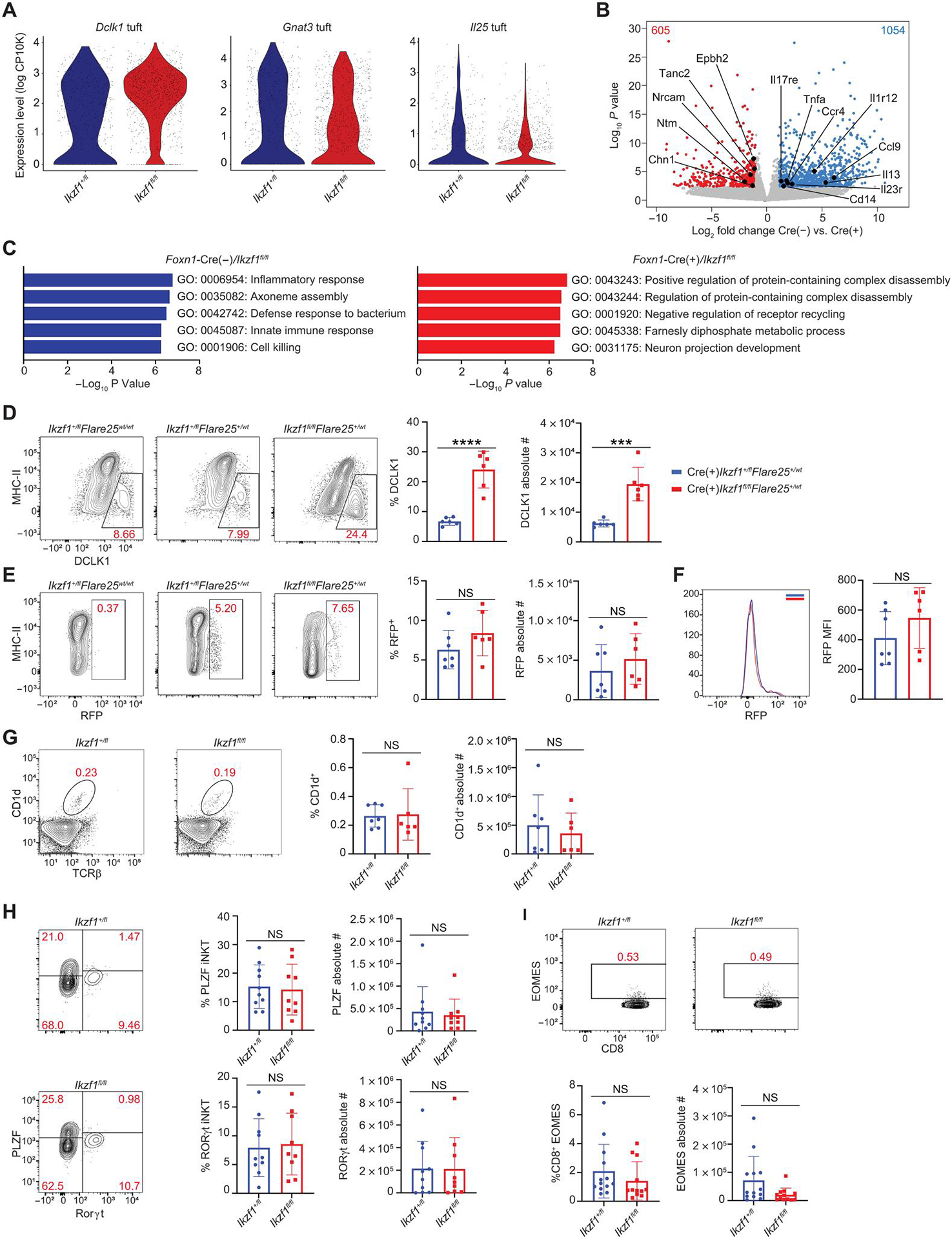
(A) Gene expression in Foxn1-Cre/Ikzf1+/fl (Ikzf1+/fl) versus Foxn1-Cre/Ikzf1fl/fl (Ikzf1fl/fl) tuft cells from scRNA-seq data. Violin plots of Il25, Gnat3, and Dclk1 expression. A total of 520 tuft cells from Ikzf1+/fl mice and 1056 tuft cells form Ikzf1fl/fl mice were analyzed. (B) Volcano plot of bulk RNA-seq of sorted CD11c− EPCAM+CD45−MHC-IIlowL1CAM+ tuft cells from Foxn1-Cre(−)/Ikzf1fl/fl mice and Foxn1-Cre(+)/Ikzf1fl/fl mice. Blue numbers indicate the number of genes significantly increased in Foxn1-Cre (−)/Ikzf1fl/fl tuft cells (FDR < 0.05, log2FC > 1). Red numbers indicate the number of genes significantly increased in Foxn1-Cre (+)/Ikzf1fl/fl tuft cells (FDR < 0.05, log2FC > 1) (n = 3 mice for Ikzf1fl/fl and n = 4 mice for Ikzf1+/fl). (C) Metascape pathway analysis of significant genes (FDR < 0.05, log2FC > 1) from bulk RNA-seq of L1CAM+ tuft cells. (D) Flow cytometry (left) and percentage and absolute number (right) of DCLK1+ thymic tuft cells from Foxn1-Cre/Ikzf1+/fl/Flare25+/ WT and Foxn1-Cre/Ikzf1fl/fl/Flare25+/WT mice. DCLK1+ tuft cells were gated on CD11c−-CD45−EPCAM+Ly51− mTECs (n = 6 mice; two independent experiments). (E) Flow cytometry (left) and percentage and absolute number (right) of RFP+ thymic tuft cells from Foxn1-Cre/Ikzf1+/fl/Flare25+/WT and Foxn1-Cre/Ikzf1fl/fl/Flare25+/WT mice. RFP+ tuft cells were gated on CD11c−CD45−-EPCAM+Ly51− mTECs (n = 6 or 7 mice; two independent experiments). (F) RFP MFI (n = 6 or 7 mice, two independent experiments). (G) Flow cytometry plots (left) and counts (right) of CD1d+TCRβ+ total iNKT from Foxn1-Cre/Ikzf1+/fl (Ikzf1+/fl) and Foxn1-Cre/Ikzf1fl/fl (Ikzf1fl/fl) mice (n = 6 or 7 mice; two independent experiments). (H) Flow cytometry plots (left) and counts (right) of PLZF+ iNKT2s and RAR-related orphan receptor gamma (RORγt)+ iNKT3s. Both iNKT2s and iNKT3s were gated on CD1d+-TCRβ+ cells (n = 9 mice; three independent experiments). (I) Flow cytometry plots (left) and counts (right) of CD8+EOMES+ innate T cells, gated on CD4−CD8+ thymocytes (n = 12 mice; four independent experiments). (D to I) In graphs, the bars correspond to the mean, with error bars showing ±SD of values shown, and each data point represents an individual mouse. (D to I) Statistical significance was determined using Student’s t test. ***P < 0.001 and ****P < 0.0001. NS, not significant.
We next sought to investigate IL-25 cytokine expression in tuft cells using the Flare25 reporter mouse in which tandem-dimer red fluorescent protein (RFP) is faithfully expressed from the endogenous Il25 locus by an internal ribosomal entry site sequence (16). We crossed Flare25 mice with Foxn1-Cre/Ikzf1fl/fl mice and analyzed cohorts of mice with heterozygous expression of the reporter allele (Foxn1-Cre/Ikzf1+/fl/Flare25+/WT and Foxn1-Cre/Ikzf1fl/fl/Flare25+/WT). As expected, immunofluorescent staining and confocal microscopy showed overlap of RFP in DCLK1+ tuft cells in Foxn1-Cre/Ikzf1+/fl/Flare25+/WT and Foxn1-Cre/Ikzf1fl/fl/Flare25+/WT mice (fig. S5C). Although TEC-specific deletion of Ikzf1 resulted in loss of MHC-IIhigh mTECs and an expansion of DCLK1+ tuft cells, there was not a difference in the percentage, absolute number, or MFI of IL-25–expressing cells in Foxn1-Cre/Ikzf1+/fl/Flare25+/WT and Foxn1-Cre/Ikzf1fl/fl/Flare25+/WT (Fig. 4, D to F). Thus, although Ikzf1 deletion results in a marked expansion of DCLK1+ tuft cells, the overall number of IL-25–expressing tuft cells remains unchanged—this could be consistent with an overall decrease in Il25 expression within the tuft compartment in Ikaros-deficient TECs.
Because TEC-specific deletion of Ikaros results in an expansion of DCLK1+ tuft cells, we next interrogated whether Ikzf1 deletion altered innate lymphocyte populations within the thymus. Prior work has shown that tuft cells are required for the development of innate lymphocytes including subsets of invariant natural killer cells (iNKT) and thymic CD8+ T cells expressing eomesodermin (EOMES+) (17, 45). In particular, development of promyelocytic leukemia zinc finger (PLZF+) iNKT2 cells has been shown to be dependent on IL-25 and thymic tuft cells (45). Given that there are more tuft cells but equivalent IL-25 expression in the absence of Ikzf1, we wanted to determine whether there was an impact on innate effector populations. Flow cytometric analysis revealed no differences in total CD1d+ iNKT cells, PLZF+ iNKT2, or CD8+-EOMES+ T cells in thymi from Foxn1-Cre/Ikzf1fl/fl mice and Foxn1-Cre/Ikzf1+/fl mice (Fig. 4, G to I, and fig. S6, A and B). This lack of alterations in innate lymphocyte populations would be consistent with there being similar total IL-25 levels despite an increase in DCLK1+ tuft cells. Thus, Ikzf1 deletion in TECs results in altered tuft cell gene expression and phenotypically altered tuft cell populations but does not change the frequency or absolute number of IL-25–dependent innate lymphocyte populations.
Ikaros regulates TSA expression in Aire+ mTECs
A fundamental property of Aire+ mTECs is their remarkable ability to express a diverse array of TSAs. Given the expression of Ikaros in Aire+ mTECs and their precursors (Fig. 2, D to F), we sought to determine whether Ikaros affects the ability of these cells to express TSAs. To determine the effect of Ikzf1 deletion on TSA gene expression, we performed differential gene expression analysis using our scRNA-seq dataset in Aire+ mTECs from Foxn1-Cre/Ikzf1+/fl mice versus Foxn1-Cre/Ikzf1fl/fl mice (fig. S7A). Pseudobulk analysis of Aire+ mTECs from Ikzf1-deleted TECs revealed a large number of differentially expressed genes, with 780 down-regulated and 590 up-regulated genes as compared with Foxn1-Cre/Ikzf1+/.fl Aire+ mTECs (P < 0.05, log2FC > 0.25). Analysis of the 780 down-regulated genes revealed the loss of many well-characterized Aire-dependent (Ins2, Pcp4, and S100a8) and Fezf2-dependent (Ttr, Timd2, and Fabp9) TSA genes, and Gene Ontology analysis (metascape) (44) showed enrichment of tissue-specific gene pathways (fig. S7B), suggesting that Ikaros is important for TSA gene expression. Moreover, the top 25 differentially expressed genes with Ikzf1 deletion in Aire+ mTECs were largely TSAs (fig. S7C). In addition to decreased Aire- and Fezf2-dependent TSAs, there was also decreased expression of many Aire- and Fezf2-independent TSA genes. Examples include Gad1 (central nervous system), Prg3 (bone), Serpinb12 (stomach), and Tkt (adipose and eye). Last, because TSA genes have stochastic expression in mTECs (46) and individual TSAs are often expressed in restricted numbers of mTECs (47, 48), we performed differential gene expression analysis between Ikaros wild-type (WT) and KO Aire+ mTECs, restricting the analysis to genes expressed in less than 25% of individual mTECs. Analysis of this TSA-enriched subset confirmed a marked decrease in TSAs expressed in the absence of Ikaros in mTECs (Fig. 5A).
Fig. 5. Ikaros is required for TSA gene expression.
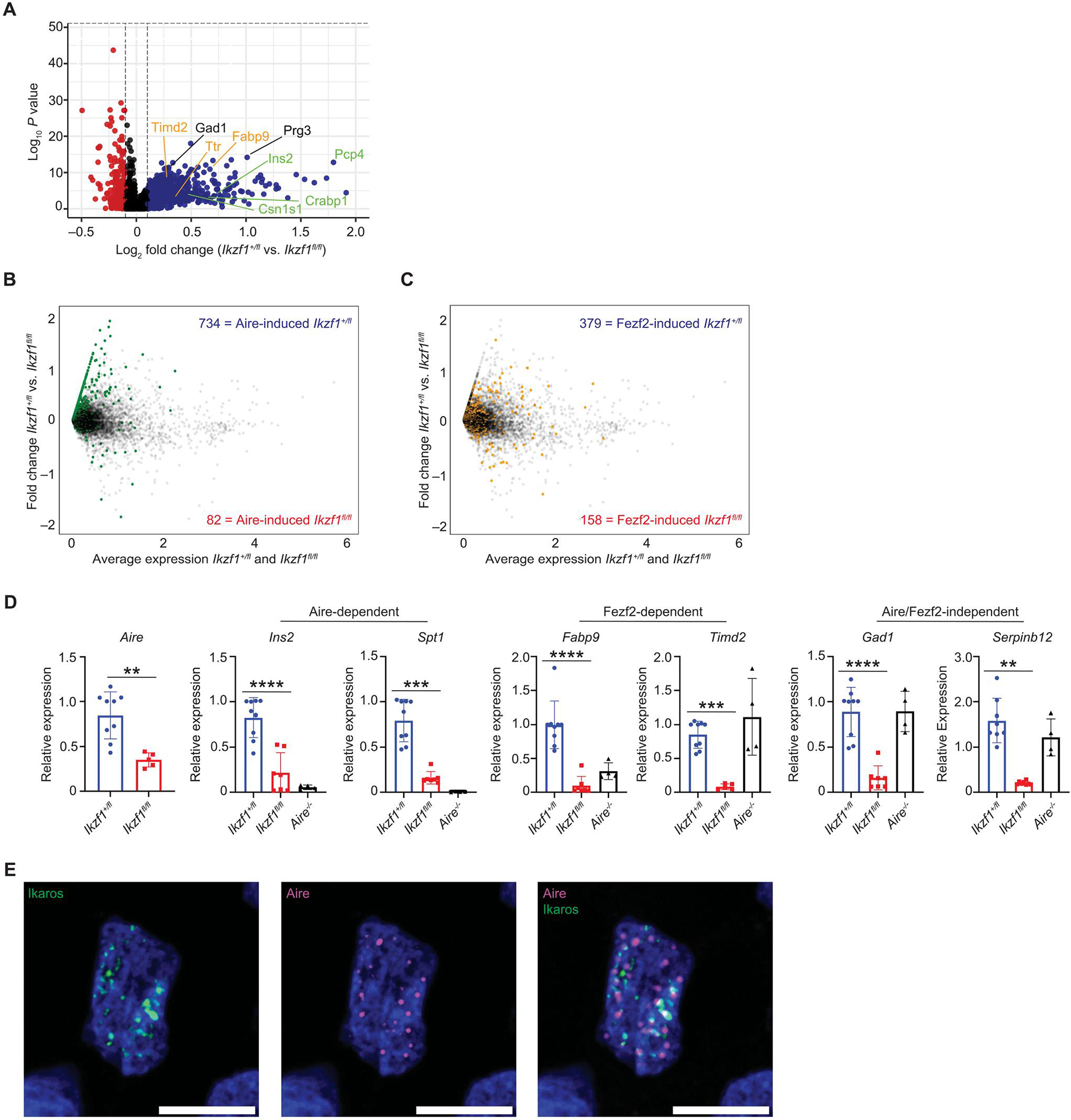
(A) Volcano plot of scRNA-seq from the Aire+ mTEC cluster. A total of 1185 Aire+ mTECs from Ikzf1+/fl mice and 262 Aire+ mTECs from Ikzf1fl/fl mice were analyzed. Plot shows genes expressed in less than 25% of mTECs. Blue dots show genes increased in Ikzf1+/fl mTECs, and red dots are increased Ikzf1fl/fl mTECs. Genes labeled in green are Aire-dependent TSA genes, genes in orange are Fezf2-dependent TSA genes, and genes in black are Aire/Fezf2-independent TSA genes. (B) Overlay of Aire-induced genes in the Aire+ mTEC cluster. Green dots represent genes that are induced by Aire (log2FC > 2, FDR < 0.05). Blue numbers enumerate the number of Aire-induced genes increased in Ikzf1+/fl mTECs, and red numbers equal the number of Aire-induced genes increased in Ikzf1fl/fl mTECs. (C) Overlay of Fezf2-induced genes in the Aire+ mTEC cluster. Yellow dots represent genes that are induced by Fezf2 (log2FC > 1, FDR < 0.05). Blue numbers enumerate the number of Fezf2-induced genes in Ikzf1+/fl mTECs, and red numbers equal the number of Fezf2-induced genes in Ikzf1fl/fl mTECs. (D) qPCR of sorted CD45−Ly51−EPCAM+MHC-IIhigh postnatal day 30 to 40 mTECs for Aire, Aire-dependent TSAs (Ins2 and Spt1), Fezf2-dependent TSAs (Fabp9 and Timd2), and Aire/Fezf2-independent TSAs (Gad1 and Serpinb12). Gene expression levels were normalized to ActB (n = 4 to 8 mice). (E) Immunofluorescence of epitope-tagged Aire (magenta) and Ikaros (green) in an individual 293T cell. One representative cell shown of >30 cells analyzed. Scale bars, 10 μm. (D) In graphs, the bars show the mean, with error bars showing ±SD of values shown, and each data point represents an individual mouse. (D) Statistical significance for Aire expression was calculated by a Student’s t test **P < 0.01. Statistical significance for Ins2, Spt1, Fabp9, Timd2, Gad1, and Serpinb12 expression was calculated using a one-way ANOVA, and grouped comparisons were corrected using Tukey’s multiple comparison test. **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To corroborate this loss of Aire-dependent and Fezf2-dependent TSA gene expression in the absence of Ikaros, we analyzed a curated list of Aire- and Fezf2-induced genes (5). Overlaying Aire-induced genes on the Foxn1-Cre/Ikzf1+/fl versus Foxn1-Cre/Ikzf1fl/fl differential expression plot showed that a portion of these genes decreased in Foxn1-Cre/Ikzf1fl/fl Aire+ mTECs (Fig. 5B). Likewise, overlay of Fezf2-induced genes showed a similar but less prominent decrease in expression in Foxn1-Cre/Ikzf1fl/fl Aire+ mTECs, suggesting that Ikaros may be more important in regulating Aire-dependent TSA genes (Fig. 5C). Using an orthogonal method to analyze decreased TSA gene expression, we calculated a TSA score as previously performed (43) by averaging expression of a list of TSA genes [compiled by Sansom et al. (18)] and subtracting the average expression of a reference set. The results as visualized by UMAP show loss of TSA gene expression with Ikzf1 deletion in Aire+ mTECs (fig. S8A), and aggregate expression of TSAs was decreased (fig. S8, B to D); this loss of TSA gene expression was also validated using quantitative polymerase chain reaction (qPCR) (Fig. 5D and fig. S9A). Decreased TSA gene expression persisted with proportional downsampling to match numbers of Aire+ mTECs between samples, suggesting that loss of TSA gene expression was not merely due to decreased numbers of Aire+ mTECs in Foxn1-Cre/Ikzf1fl/fl mice (fig. S9B). One potential mechanism of decreased Aire-dependent TSA gene expression is decreased Aire expression. Consistent with our flow cytometry results, we also observed reduced Aire expression by scRNA-seq, suggesting that reduced Aire expression itself may contribute to the observed reduction in TSA gene expression (Fig. 1F and fig. S9B). These data suggest that Ikzf1 deletion in mTECs results in a loss of TSA gene expression including Aire-dependent, Fezf2-dependent, and Aire/Fezf2-independent TSA genes.
Prior studies have shown that Ikaros binds to pericentromeric heterochromatin (49, 50) and forms a speckling pattern in cell nuclei, which appears similar to the well-described nuclear speckling pattern of Aire (51). Because we observed a pronounced effect of Ikaros loss on Aire-dependent TSA gene expression, we wanted to determine whether Ikaros colocalized with Aire in nuclei or interacted with Aire. We transfected Aire and Ikaros into human embryonic kidney (HEK) 293 cells and found that although both proteins formed characteristic speckles, there was no overlap of the nuclear distribution of the two proteins (Fig. 5E), suggesting that Ikaros and Aire localize to spatially distinct nuclear structures. Furthermore, transfection of epitope-tagged Aire and Ikaros in HEK293 cells showed that the two proteins could not be coimmunoprecipitated (fig. S9C). Thus, Ikaros and Aire do not appear to colocalize within nuclei or interact directly.
Loss of Pou2f3 fails to rescue TSA gene expression in TECs with Ikzf1 deletion
Our data show that Foxn1-Cre/Ikzf1fl/fl thymi are characterized by a marked expansion of thymic tuft cells and muscle-like mTECs with a loss of Aire+ mTECs and TSA gene expression. Prior literature suggests that Ikaros functions primarily as a transcriptional repressor through its interaction with multiple repressive chromatin complexes (24, 30, 36, 52, 53). Thus, we hypothesized that Ikaros may function, in part, by repressing tuft and muscle gene programs to allow proper Aire+ mTEC development and TSA gene expression. In support of this model, analysis of our scRNA-seq data showed that Ikzf1 deletion resulted in increased expression of the lineage-defining transcription factor for thymic tuft cells, Pou2f3, and the lineage-defining transcription factor for muscle-like mTECs, Myog, within Aire+ mTECs. Ikzf1 deletion resulted in a higher percentage of cells expressing Pou2f3, with 30% of Aire+ mTECs from Foxn1-Cre/Ikzf1fl/fl mice expressing Pou2f3 compared with 10% of Aire+ mTECs from Foxn1-Cre/Ikzf1+/fl mice (Fig. 6A). Similarly, Ikzf1-deletion in Aire+ mTECs resulted in a higher percentage of cells expressing Myog (fig. S9D), with 12% of Aire+ mTECs from Foxn1-Cre/Ikzf1fl/fl mice expressing Myog compared with 4% of Aire+ mTECs from Foxn1-Cre/Ikzf1+/fl mice.
Fig. 6. Deletion of Pou2f3 fails to rescue TSA gene expression.
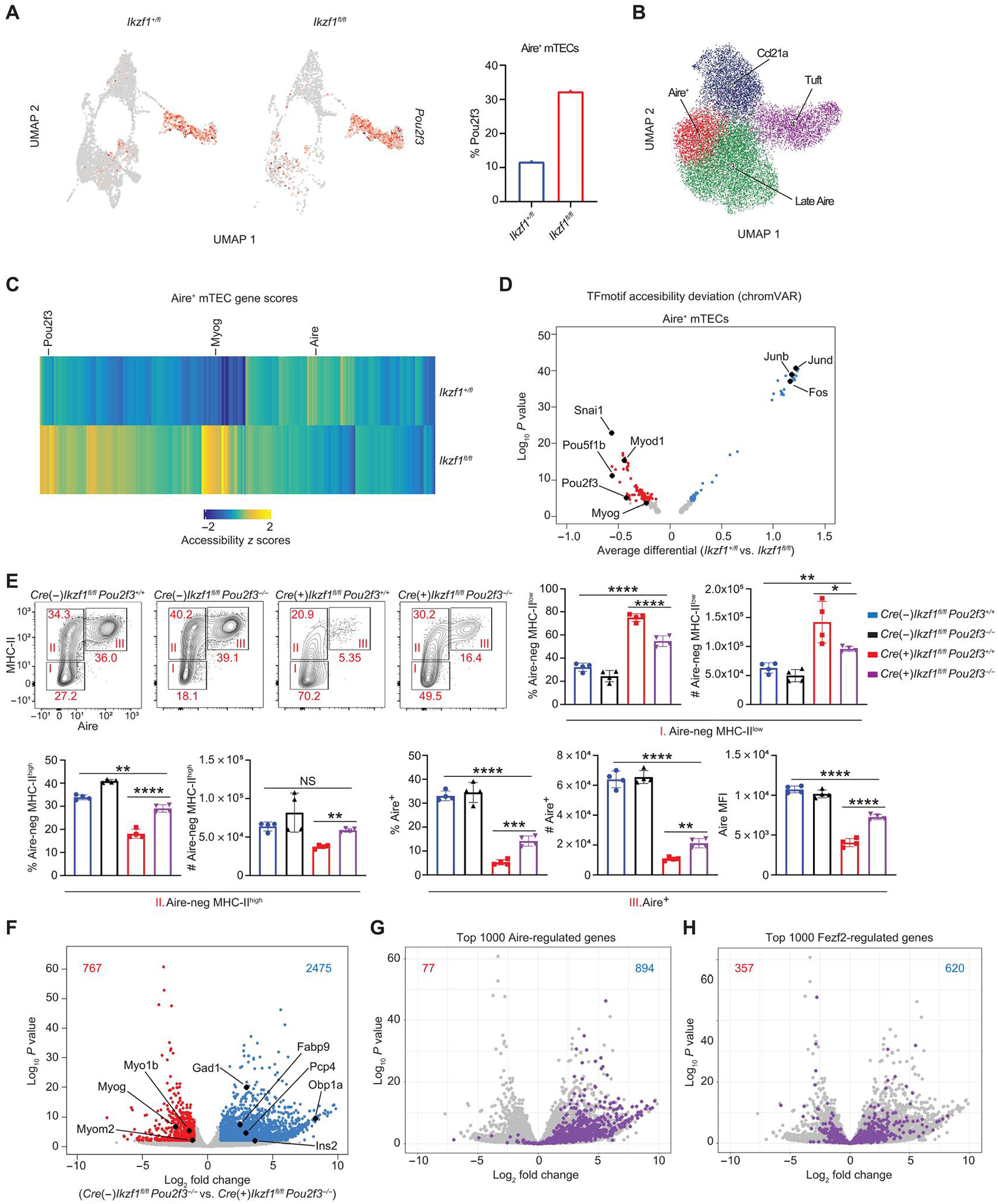
(A) UMAP of Pou2f3 expression in Aire+ mTECs from scRNA-seq data. A total of 1185 Aire+ mTECs from Ikzf1+/fl mice and 262 Aire+ mTECs from Ikzf1fl/fl mice were analyzed. Red color represents Pou2f3 expression. Bar graph represents the percentage of cells of each genotype expressing Pou2f3 in Aire+ mTECs. (B) scATAC-seq clustering with ArchR. (C) Heatmap of scATAC-seq gene score values (FDR < 0.05) comparing Foxn1-Cre/Ikzf1+/fl (Ikzf1+/fl) versus Foxn1-Cre/Ikzf1fl/fl (Ikzf1fl/fl) Aire+ mTECs. (D) chrom-VAR motif accessibility volcano plot in Aire+ mTECs of the indicated genotype. Blue dots show motifs significantly enriched (FDR < 0.05) in Foxn1-Cre/Ikzf1+/fl (Ikzf1+/fl) Aire+ mTECs. Red dots show motifs significantly enriched in Foxn1-Cre/Ikzf1fl/fl (Ikzf1fl/fl) Aire+ mTECs (FDR < 0.05). (E) Flow cytometry (left) and percentage and absolute number (right) of Aire-negative MHC-IIlow (I), Aire-negative MHC-IIhigh (II), and Aire+ mTECs (III), which were gated on CD11c−-CD45−EPCAM+Ly51− mTECs (n = 4 mice; one representative experiment of two independent experiments). (F) Volcano plot of bulk RNA-seq of sorted CD11c−-EpCAM+CD45−Ly51−MHC-IIhigh mTECs from Foxn1-Cre−/Ikzf1fl/fl/Pou2f3−/− mice and Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice. Blue numbers indicate the number of genes significantly increased in Foxn1-Cre−/Ikzf1fl/fl/Pou2f3−/− MHC-IIhigh mTECs (FDR < 0.05, log2FC > 1). Red numbers indicate the number of genes significantly increased in Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− MHC-IIhigh mTECs (FDR < 0.05, log2FC > 1). The top 1000 Aire-regulated genes are overlaid in (G), and the top 1000 Fezf2-regulated genes are overlaid in (H). In (G), the blue number represents number of Aire-induced genes in Foxn1-Cre−/Ikzf1fl/fl/Pou2f3−/− mice, and the red number represents number of Aire-induced genes in Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice. In (H), the blue number represents number of Fezf2-induced genes in Foxn1-Cre−/Ikzf1fl/fl/Pou2f3−/− mice, and the red number represents number of Fezf2-induced genes in Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice. (E) In graphs, the bars correspond to the mean, with error bars showing ±SD of values shown, and each data point represents an individual mouse. (E) Statistical significance was calculated using a one-way ANOVA, and grouped comparisons were corrected using Tukey’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We next used the scATAC-seq to define Ikaros-dependent changes in chromatin and transcription factor activity in TECs. Sorting was performed using an approach parallel to our prior TEC purification for single-cell transcriptomics, and high-quality scATAC-seq profiles were generated from 13,184 TECs from Foxn1-Cre/Ikzf1+/fl mice and 3305 TECs from Foxn1-Cre/Ikzf1fl/fl mice, with 102,944 peaks identified in our dataset (fig. S10, A to C). Clustering of the scATAC-seq data using ArchR (54) identified four mTEC subsets: Aire+, Late Aire, Ccl21a+, and tuft cells based on evaluating the cell type–defining transcription factor (TF) motif enrichments and associated locus-specific gene accessibility changes (Fig. 6B and fig. S10, D to H). For example, the cluster enriched with NF-κB motifs was consistent with Aire+ mTECs, and the cluster with an increase in Pou2f3 motifs was used to identify tuft cells. Analysis of the scATAC-seq data by genotype revealed the predicted changes in mTEC cell composition, with an increased percentage of tuft cells and Ccl21a+ mTECs and a loss of Aire+ mTECs and Late Aire mTECs in Foxn1-Cre/Ikzf1fl/fl mice (fig. S11A). Further analysis of the Late Aire cluster identified multiple mimetic cell clusters with distinct open chromatin regions (OCRs) (fig. S11B). We focused our scATAC-seq analysis on the Aire+ mTEC cluster and used ArchR to calculate gene activity scores—a method that predicts gene expression on the basis of accessibility of adjacent regulatory elements. We found increases in gene scores in Foxn1-Cre/Ikzf1fl/fl Aire+ mTECs for canonical tuft cell markers such as Pou2f3 and markers of muscle mTECs, including Myog (Fig. 6C), suggesting an increased tuft and muscle gene program in Aire+ mTECs that were deficient in Ikaros. Moreover, we found an increased gene score for the Aire locus in Foxn1-Cre/Ikzf1+/fl Aire+ mTECs, consistent with increased Aire protein and Aire gene expression in Ikaros-sufficient TECs (Fig. 6C).
Ikaros regulates gene expression by recruiting multiple chromatin remodeling complexes, including the nucleosome remodeling and deacetylase complex SWItch/sucrose nonfermentable and polycomb repressive complex 2 (52, 53, 55–57). Ikaros is thought to act primarily as a transcriptional repressor, although in specific developmental contexts, it can also activate transcription by modulating the enhancer/superenhancer landscape (24, 55, 58–61). We, therefore, assessed whether Ikzf1 deletion in TECs altered global chromatin accessibility. Ikzf1 deletion resulted in an increased number of OCRs within Aire+ mTECs, tuft cells, and Ccl21a mTECs, in agreement with the well-described function of Ikaros as a transcriptional repressor (fig. S11C).
Given that there was increased expression of Myog and Pou2f3 in Aire+ mTECs where Ikzf1 had been deleted, we investigated whether there was increased enrichment of Myog and Pou2f3 binding sites in OCRs from Aire+ mTECs. We analyzed chromatin accessibility at TF binding sites using chromVAR (Fig. 6D and fig. S11D). From this, we found that tuft cell and muscle mTEC-associated TF motifs, including Myod1, Myog, and Pou2f3 motifs, were more accessible in Aire+ mTECs and Late Aire mTECs from Foxn1-Cre/Ikzf1fl/fl mice. Conversely, activator protein 1 (AP-1) TF motifs including Jun and Fos were enriched in Foxn1-Cre/Ikzf1+/fl Aire+ mTECs and Late Aire mTECs. Prior work has shown that NF-κB is critical for the maturation of Aire+ mTECs (19–21). We also performed TF binding motif analysis in Foxn1-Cre/Ikzf1+/fl and Foxn1-Cre/Ikzf1fl/fl Aire+ mTECs and Late Aire mTECs for NF-κB motifs (fig. S12A), but we did not find any changes in OCR induced by Ikaros deficiency. Moreover, we completed TF binding motif analysis for murine Foxn1, a transcription factor critical for TEC development (62), and did not see a difference in chromatin accessibility in Foxn1-Cre/Ikzf1+/fl and Foxn1-Cre/Ikzf1fl/fl Aire+ mTECs and Late Aire mTECs (fig. S12B). Together, these results suggest that the Ikzf1-deletion results in enhanced expression and increased chromatin accessibility of lineage-defining tuft and muscle mTEC transcription factors Myod1, Myog, and Pou2f3, resulting in altered mTEC development.
Because of the marked expansion of thymic tuft cell populations in the absence of Ikzf1, we tested whether loss of Pou2f3, the lineage-defining transcription factor of tuft cells, would rescue the cellular and transcriptomic changes seen with Ikzf1 deletion. We crossed Pou2f3−/− mice with Foxn1-Cre/Ikzf1fl/fl mice, and, as expected, deletion of Pou2f3 resulted in a complete loss of DCLK1+ thymic tuft cells in both Foxn1-Cre−/Ikzf1fl/fl/Pou2f3−/− mice and Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice (fig. S12C). Deletion of Pou2f3 resulted in a near-complete rescue in the percentage and absolute number of Aire-negative MHC-IIhigh mTECs in Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice (Fig. 6E), with the percentage and absolute number of Aire-negative MHC-IIhigh mTECs comparable in Foxn1-Cre−/Ikzf1fl/fl/Pou2f3+/+ and Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice. Moreover, deletion of Pou2f3 restored the proliferative capacity of mTECs with Ikzf1 deletion (fig. S12D).
Prior work has shown that Aire-negative MHC-IIhigh mTECs are the developmental precursor to Aire+ mTECs (42). Unexpectedly, although Pou2f3 deficiency restored the percentages and absolute number of Aire-negative MHC-IIhigh mTECs associated with deletion of Ikaros, the rescue of numbers of Aire+ MHC-IIhigh mTECs in Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice compared with Foxn1-Cre+/Ikzf1fl/fl/Pou2f3+/+ mice was less pronounced (Fig. 6E). Consistent with a defect in the maturation of Aire+ mTECs in Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice, these mice also showed persistent loss of Aire-dependent mimetic cells, including GP2+ microfold-like mTECs and KRT10+ keratinocyte-like mTECs (fig. S12, E and F).
We next assessed the effect of combined Ikzf1/Pou2f3 deletion on TSA gene expression. We sorted MHC-IIhigh mTECs from Foxn1-Cre−/Ikzf1fl/fl/Pou2f3−/− mice and Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice and performed bulk RNA-seq (fig. S13A and data file S2). Differential gene expression analysis revealed 2475 genes down-regulated and 767 genes up-regulated (FDR < 0.05, log2FC > 1) in MHC-IIhigh mTECs from Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mice relative to their Cre– littermates (Fig. 6F). Even in the absence of thymic tuft cells, the most highly Ikaros-dependent genes were again skewed toward TSAs (fig. S13B). Moreover, consistent with our scRNA-seq data analysis of Aire+ mTECs, overlay of Aire-induced genes and Fezf1-induced genes on this differential gene list showed a portion of Aire-induced and Fezf2-induced genes decreased in Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mTECs (Fig. 6, G and H). This loss of TSA gene expression in MHC-IIhigh Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mTECs was confirmed with qPCR (fig. S13C). A large number of transcripts up-regulated in MHC-IIhigh Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− mTECs are also found in muscle, suggesting failure to suppress a muscle gene program and muscle-like mTECs. In summary, lack of Pou2f3 in the setting of an Ikaros-deficient TEC compartment results in a partial rescue of Aire-negative MHC-IIhigh mTECs but is unable to rescue either Aire or TSA gene expression.
Ikaros expression in thymic epithelial cells is essential for immune tolerance
Given that Ikaros in thymic epithelium plays a key role in both the composition of TEC populations and thymic TSA expression, we next wanted to determine its role in T cell selection and immune tolerance. Foxn1-Cre/Ikzf1fl/fl mice did not display altered frequencies or absolute numbers of thymic single-positive (SP) CD4+/CD8+ T cells or double-positive T cells (fig. S14A). Moreover, TEC-specific deletion of Ikzf1 did not affect late-stage thymocyte frequencies (fig. S14, B and C) (63). The deletion of Ikaros resulted in a decreased frequency and absolute number of CD73−CD25+Foxp3+ regulatory T cells (Tregs) (Fig. 7A and fig. S6B), a phenotype also observed with thymic deletion of Aire (4, 64), but thymocyte development and maturation was otherwise intact in Foxn1-Cre/Ikzf1fl/fl mice.
Fig. 7. Deletion of Ikaros in mTECs results in defects in negative selection and organ specific autoimmunity.
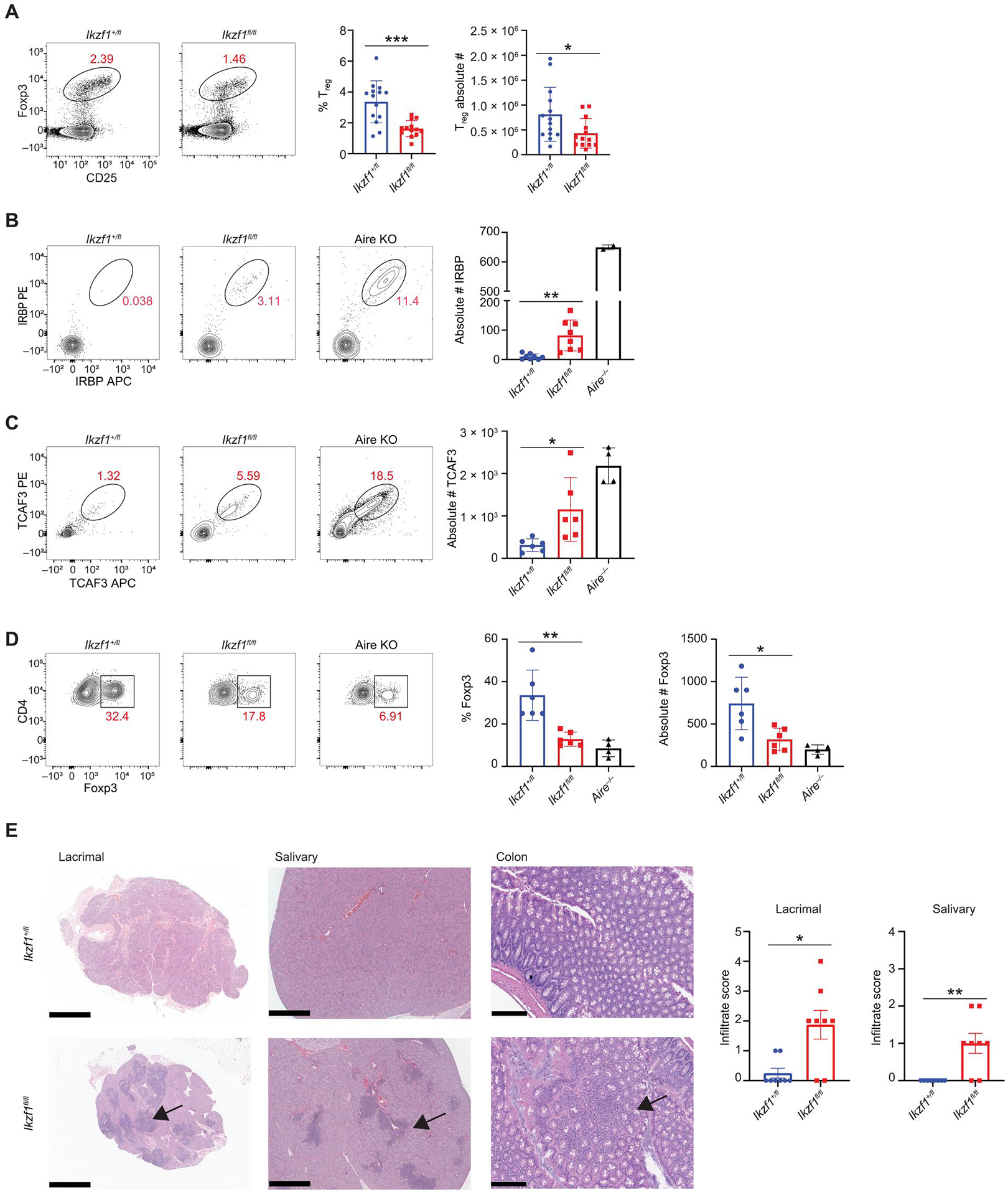
(A) Flow cytometry (left) and percentage and absolute number (right) of thymic CD4+CD73−Foxp3+CD25+ regulatory T cells from Foxn1-Cre/Ikzf1+/fl (Ikzf1+/fl) mice and Foxn1-Cre/Ikzf1fl/fl (Ikzf1fl/fl) mice (n = 13 or 14 mice; five independent experiments). (B) Flow cytometry (left) and absolute number (right) of IRBP+ CD4+ T cells from Ikzf1+/fl, Ikzf1fl/fl, and Aire−/− mice 11 days after immunization with IRBP peptide [n = 8 mice per genotype (Ikzf1+/fl and Ikzf1fl/fl) and n = 2 for Aire−/− mice]. (C) Flow cytometry (left) and absolute number (right) of TCAF3+ CD4+ T cells from male Ikzf1+/fl, Ikzf1fl/fl, and Aire−/− mice 11 days after immunization with TCAF3 peptide [n = 6 mice per genotype (Ikzf1+/fl and Ikzf1fl/fl), n = 4 for Aire−/− mice]. (D) Representative flow plots of CD4+TCAF3+Foxp3+ T cells and percentage of TCAF3+ Tregs [n = 6 mice per genotype (Ikzf1+/fl and Ikzf1fl/fl) and n = 4 for Aire−/− mice]. (E) H&E staining of the lacrimal gland, salivary gland, and colon. Scale bars, 1 mm (lacrimal/salivary) and 200 μm (colon). Bar graphs quantitate lymphocytic infiltrate (n = 8 mice per genotype). (A to E) In graphs, the bar corresponds to the mean, with error bars showing ±SD of values shown, and each data point represents an individual mouse. (A and E) Statistical signify+cance was determined using Student’s t test (A) and Mann-Whitney test (E). *P < 0.05, **P < 0.01, and ***P < 0.001. (B to D) Statistical significance was calculated using a one-way ANOVA, and grouped comparisons were corrected using Tukey’s multiple comparison test. *P < 0.05 and **P < 0.01.
Given the global loss of TSA gene expression in Foxn1-Cre/Ikzf1fl/fl mice, we tested for failed negative selection of T cells to TSAs. We used tetramer reagents to monitor the expansion of T cells with defined specificity after immunization. Prior studies have shown that immunizing Aire-deficient mice with the eye-specific protein interphotoreceptor retinoid-binding protein (IRBP) or the prostate-specific protein TRPM8 channel-associated factor 3 (TCAF3) results in an expansion of IRBP- or TCAF3-specific T cells (65–68). Both Rbp3 (gene encoding IRBP) and Tcaf3 are well-characterized Aire-dependent genes in mTECs. Consistent with the loss of Aire+ mTECs and TSA gene expression in Foxn1-Cre/Ikzf1fl/fl mice, there was an expansion of IRBP- and TCAF3-specific T cells in the lymph nodes after immunization (Fig. 7, B and C, and fig. S15A). Thymic expression of TCAF3 is also critical for the positive selection of Tregs, and Aire-deficient mice have decreased TCAF3+Foxp3+ T cells after immunization with TCAF3. Similarly, Foxn1-Cre/Ikzf1fl/fl mice also had decreased TCAF3-specific Tregs (Fig. 7D). Thus, Foxn1-Cre/Ikzf1fl/fl mice have defects in the medullary selection of autoreactive T cells. To determine whether failed thymic selection of autoreactive T cells resulted in systemic autoimmunity, we collected tissues from a cohort of 6-month-old Foxn1-Cre/Ikzf1+/fl and Foxn1-Cre/Ikzf1fl/fl mice. Foxn1-Cre/Ikzf1fl/fl mice showed lymphocytic infiltrates in multiple organs, including the salivary gland, lacrimal gland, and colon, indicative of systemic autoimmunity and failed central tolerance (Fig. 7E). The increased salivary and lacrimal infiltrates observed in Foxn1-Cre/Ikzf1fl/fl mice are similar in degree and distribution to those in mutant mice with an autosomal-dominant Aire mutation on the C57BL/6 background, which have reduced but not abolished TSA gene expression (69). Thus, the conditional loss of Ikaros in the thymic epithelium leads to defects in thymic selection and decreases in both Aire and TSA gene expression, resulting in systemic autoimmunity.
DISCUSSION
Our findings reveal a previously uncharacterized role for the transcription factor Ikaros in thymic epithelial biology, where it is required for proper mTEC development and TSA gene expression. The function of Ikaros in mTEC lineage commitment is reminiscent of its role in hematopoietic development and underscores a similar function for Ikaros in diverse tissues involved in immune cell regulation. Ikaros is expressed beginning early in hematopoietic development and is detected in hematopoietic stem cells (29), and Ikaros-null mice have a complete block in both fetal and postnatal B cell development (28). Although postnatal T cells do emerge, Ikaros-null mice also have severe defects in T cell development (28). Moreover, similar to AIRE, defects in IKZF1 are associated with autoimmunity in humans. Coding mutations in IKZF1 have been linked to a familial autoimmune syndrome (70–72), and small-nucleotide polymorphisms in Ikaros have been linked to multiple autoimmune diseases, including systemic lupus erythematosus and Sjogren’s syndrome (73, 74).
Studies on Ikaros have largely been confined to the hematopoietic compartment, but there are reports of its functional roles in other tissues, including the brain (37–39) and hypothalamic-pituitary axis (40). Our study describes the role of Ikaros within the thymic epithelial lineage. In thymic epithelial cells, scRNA-seq shows that Ikzf1 is expressed in TAC-TECs and Aire+ mTECs specifically and is required for the expansion and maintenance of the Aire+ mTEC compartment and downstream mimetic cell populations. Ikzf1 expression in TAC-TECs suggests a function for Ikaros in proliferating mTECs, and Ikaros may act at multiple steps of mTEC development, similar to its function in hematopoietic development. Because Foxn1 is expressed early in mTEC development (embryonic day 11.5), the defects we observe with TEC-specific deletion of Ikzf1 could be due to Ikaros deletion at multiple stages of thymic organogenesis. Further work will be required to map its roles within various stages of mTEC development more precisely.
Recent studies have also highlighted the substantial diversity found within the mTEC compartment in both mice and humans (8, 11, 17, 43). Mimetic cells are thought to be critical for the presentation of TSA genes and central tolerance. Each of these mimetic cell types appears to use similar lineage-defining transcription factors as their peripheral counterparts. Here, we show that deletion of Ikaros in TECs results in profound changes in mimetic cell distribution, with an expansion of thymic tuft cells and muscle-like mTECs and a loss of the Aire-dependent mimetic populations. Thus, Ikaros plays a previously unappreciated role in the maintenance of mTEC homeostasis and normal mimetic cell development. In addition to increased numbers of tuft cells with Ikzf1 deletion, these tuft cells are marked by changes in gene expression and cytokine production. Future studies should more deeply interrogate the impact of TEC-specific deletion of Ikzf1 on the biology of mimetic cell subsets.
In addition to a function for Ikaros in mTEC development, our data show that Ikaros is required for the expression of Aire-dependent, Fezf2-dependent, and Aire/Fezf2-independent TSA genes. Therefore, Ikaros is a transcriptional regulator required for the proper induction of central tolerance. One possible mechanism for decreased TSA gene expression with Ikzf1 deletion is failure of mTECs to reach a developmental stage permissive for TSA gene expression; data supporting this model include the loss of both Aire-negative MHC-IIhigh mTECs and Aire+ MHC-IIhigh mTECs with Ikzf1 deletion, suggesting that Ikaros may block the development of the MHC-IIhigh mTEC population and, consequently, these TSA-enriched cells. However, the finding that combined Ikzf1/Pou2f3 deletion rescues MHC-II expression but fails to rescue TSA gene expression may indicate a more complex function for Ikaros in orchestrating TSA gene expression, including modulation of Aire gene expression or Aire protein function, or direct regulation of TSA gene expression. Arguing against direct regulation of Aire protein function is that Ikaros does not appear to colocalize or interact with Aire, at least in vitro; however, future studies performed in TECs are needed to completely rule out this possibility. Prior studies have shown that Ikaros binds to pericentromeric heterochromatin; Aire has also been shown to bind to repressive DNA through interaction with hypomethylated histones (H3K4me0) (75) or repressive complexes (76). Thus, Ikaros may interact with repressive chromatin, and this interaction could be required to allow for stochastic accessibility of linear DNA regions encoding TSA genes through its well-described interaction with multiple chromatin-modifying complexes (52, 53, 55–57). Last, Ikaros may be required to repress the aberrant expression of lineage-defining transcription factors to allow proper TSA gene expression. Prior work in B and T cells supports a function for Ikaros in gene repression (24, 30, 36, 52, 53), and in pre-B cells, Ikaros has been shown to induce a repressive chromatin landscape and to silence extralineage-specific transcription factors that are aberrantly activated with Ikzf1 deletion in transformed cells. Our data showing increased Myog and Pou2f3 expression with Ikzf1 deletion in Aire+ mTECs support this model, and future studies investigating the phenotype of triple depletion mice (Ikzf1/Pou2f3/Myog) may be illuminating.
The loss of TSA gene expression with Ikzf1 deletion in TECs results in defects in T cell selection and organ-specific autoimmunity in mice. scRNA-seq data from human thymus suggest that Ikaros and Aire are coexpressed, similar to our observations in mouse, suggesting that Ikaros may have a similar function in humans. Given that Ikaros mutations are found in patients with autoimmunity, an intriguing possibility is that failed thymic central tolerance due to loss of Ikaros expression in mTECs contributes to patient autoimmune syndromes. A cohort of patients with IKZF1 mutations have undergone hematopoietic stem cell transplant because of severe mutations (77, 78). It will be interesting to investigate whether these patients continue to develop autoimmunity despite correction of IKZF1 mutations in their hematopoietic compartment. If so, this would suggest a parallel and critical function for Ikaros in human mTECs. Last, there is growing interest to create artificial thymi from stem cells to induce tolerance in humans (79); because of the essential function of Ikaros in mTEC development, it may be critical to monitor and perhaps modulate its expression during the development of human thymic organoids.
MATERIALS AND METHODS
Study design
This study was designed to determine transcription factors involved in thymic epithelial development and thymic tissue–specific antigen expression. We focused our investigation on the function of the transcription factor Ikaros. We examined the effect of thymic epithelium–specific Ikaros deletion on thymocyte development and innate lymphocytes. To delete Ikaros in the thymic epithelium, we crossed an Ikaros-floxed mouse line to the Foxn1-Cre mouse line that expresses Cre recombinase in both the thymic epithelium and skin. To determine how loss of tuft cells affects the phenotype observed with Ikzf1 deficiency, we used mice that had combined deletion of Ikzf1/Pou2f3 in the thymic epithelium. We used a variety of techniques to probe the effect of Ikzf1 deletion in the thymic epithelium, including flow cytometry of thymic epithelial cells and genomic studies including scRNA-seq and scATAC-seq. Both male and female mice were included in the study. Investigators were not blinded before data analysis. All experiments were repeated at least three times unless noted in the figure legend.
Mice
Ikzf1fl/fl mice, Foxn1-Cre mice, Flare25 mice, Pou2f3−/−, mice, and Aire−/− mice were previously published (3, 16, 17, 24, 41). All mice were maintained on the C57BL/6 genetic background, and both male and female mice were used. In most experiments, mice were between 30 to 40 days old except when mice were aged to 6 months to assay for immune infiltrates. Both Foxn1-Cre+/Ikzf1+/fl and Foxn1-Cre−/Ikzf1fl/fl were used as controls. Mice were maintained in the University of California San Francisco (UCSF) specific pathogen–free animal facility in accordance with the guidelines established by the Institutional Animal Care and Use Committee and Laboratory Animal Resource Center, and all experimental procedures were approved by the Laboratory Animal Resource Center at UCSF.
Histology
Organs from mice were harvested and fixed overnight in 10% formalin and dehydrated with 30% EtOH followed by 70% EtOH. Dehydrated tissues were then embedded, sectioned, and stained with H&E. Scoring of the infiltrates of the lacrimal gland and salivary gland was performed blinded as previously described (69): scores of 0, 1, 2, 3, or 4 indicating no, <25% infiltrate, 25 to 50% infiltrate, 50 to 75% infiltrate, or > 75% infiltrate, respectively. Histology for colitis was scored blinded using the following scoring system: (0) no immune cell infiltration, (1) minimal inflammation with <10% infiltration of the mucosa, (2) mild inflammation 10 to 25% of the mucosa infiltrated, (3) moderate inflammation 26 to 50% infiltration, (4) extensive infiltration 51 to 75% affected, (5) diffuse infiltration >75% of the mucosa infiltrated.
Cell culture and transfection
HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), l-glutamate, and penicillin-streptomycin in a humidified incubator supplemented with 5% CO2. The day before transfection, cells were counted and then seeded in six-well plates. Cells were transfected with appropriate plasmid(s) using Mirus Trans-IT HEK293 transfection reagent according to the manufacturer’s protocol.
Immunofluorescence
Whole thymi were harvested and gently washed with phosphate-buffered saline (PBS) and fixed in 2% paraformaldehyde (PFA) for 2 hours. Fixed tissues were dehydrated overnight with 30% sucrose and embedded in optimal cutting temperature compound. Twenty-five-micrometer sections were cut and air-dried for 1 hour or stored at −80°C. Dried sections were blocked for an hour using BlockAid (Life Technologies) and stained with KRT10, KRT5, KRT8, Aire, rabbit polyclonal RFP, or DCLK1 primary antibodies (table S1) diluted in BlockAid overnight in a humidified chamber at 4°C. Sections were washed with PBS and stained with 1:10,000 4′,6-diamidino-2-phenylindole (DAPI; 5 mg/ml stock) in PBS. Secondary antibody staining for unconjugated Krt8 was done with goat anti-rabbit A488 (2 mg/ml stock) at 1:1000 in BlockAid for 2 hours in a humidified chamber at room temperature. Secondary antibody staining for RFP was done with anti-rabbit A647 (2 mg/ml stock) at 1:1000 in BlockAid for 2 hours in a humidified chamber at room temperature. Stained thymus sections were then mounted using ProLong Diamond antifade mount (Thermo Fisher Scientific) and imaged on the SP5 Leica confocal microscope. Images were analyzed using Fiji (based on ImageJ2). Transfected HEK293 cells were grown on circular glass coverslips at the bottom of six-well tissue culture–treated plates at a concentration of 2 million to 4 million cells per well. Immunofluorescence of transfected HEK293 cells was performed by carefully removing coverslips with fine-tipped forceps to six-well tissue culture–treated plates containing PBS. The cover slips were then removed and blocked for 1 hour with BlockAid and stained with primary antibodies for 2 hours in a humidified chamber. After staining with secondary antibodies and DAPI, the cover slips were mounted on slides with ProLong Diamond antifade mount. The slides were then imaged on the SP5 Leica confocal microscope and analyzed using Fiji.
Imaris quantitative image analysis
To measure the volumes of tuft cell aggregates, images from thymic sections were taken on the SP5 Leica confocal microscope and imported to be analyzed in Bitplane’s Imaris 9.6 with the Filament Module software package. Thresholds of intensities and surface volumes of the three-dimensional segments were used to filter the images’ voxels, and volumes of tuft cells were subsequently measured. Surface volumes > 5 (1 × 104 μm3) were measured to be recognized as a tuft cell aggregate.
Immunohistochemistry
Whole thymi were dehydrated overnight in 70% EtOH and were embedded in paraffin. The sections were then faced and chilled overnight at 4°C. Subsequent sections (5 μm) were cut on the microtome (Leica Biosystems) and dried for 2 hours. After drying, the sections were baked for an hour to remove excess paraffin and were deparaffinized and rehydrated. Antigen retrieval was done using a citrate-based antigen retrieval solution (Biogenex Laboratories), and endogenous peroxidase activity was quenched with 3% H2O2. The sections were then blocked with 10% FBS in 0.2% Tween 20 in PBS and incubated with primary antibody overnight (table S1). Next, the sections were incubated with biotinylated anti-rabbit secondary in a humidified chamber at room temperature. VECTAS-TAIN Elite ABC system (Vector Laboratories) was used to introduce the peroxidase enzyme and 3,3’-diaminobenzidine (DAB) reagent (Vector Laboratories) to brown our antigen of interest. Sections were counterstained with hematoxylin (MilliporeSigma), dehydrated, and mounted using Cytoseal mounting media (Thermo Fisher Scientific).
Immunizations (TCAF3 and IRBP)/tetramer staining
Immunization cocktail was prepared by emulsifying 200 μg of P2-IRBP peptide (PLGGG GQTWE GSGVL, Genemed Synthesis) or TCAF3 peptide (THYKAPWGELATD, Genemed Synthesis) in 100 μl of incomplete Freund’s adjuvant with Mycobacterium tuber-culosis (BD Difco). Immunization was performed by injecting mice subcutaneously (200 μl) into both sides of the chest (100 μl per side). Eleven days after immunization, lymphocytes from the cervical, brachial, and axillary lymph nodes were isolated through a 40-μm filter and transferred to a 5-ml filter-topped fluorescence-activated cell sorting (FACS) tube. The cells were stained with tetramer for 1 hour at room temperature and incubated for 15 min with anti-phycoerythrin (anti-PE) and anti-allophycocyanin (anti-APC) magnetic-activated cell sorting (MACS) beads at a 1:10 dilution in MACS buffer. To positively isolate tetramer-positive cells, magnetic bead separation was done using LS MACS columns (Miltenyi Biotec). The columns were prewetted with MACS buffer, and the cell solution was loaded into the LS columns. The columns were eluted with 5 ml of MACS buffer, washed with PBS, and stained with the appropriate antibodies/tetramers for flow cytometry analysis (table S1).
Single-cell tissue preparation
Thymic epithelial cells were isolated by harvesting mouse thymi into DMEM (DME H-21, UCSF Cell Culture Facility) containing 2% FBS (HyClone). Thymi were minced by hand with razor blades and were moved to 15-ml conical tubes using glass Pasteur pipettes. Minced thymi were vortexed for 45 s, and tissue remains were allowed to settle on ice before the medium was removed and replaced with 4 ml of digestion medium [deoxyribonuclease I (100 μg/ml; Roche), Liberase (100 μg/ml; MilliporeSigma), and 2% FBS in DMEM]. To aid in digestion, the thymi were placed in a 37°C water bath and mixed vigorously with the glass Pasteur pipettes at intervals of every 4 min for 24 min. At every 8 min, the thymi pieces were briefly spun down at 340g for 10 s, and the supernatant fractions were collected into 20 ml of chilled MACS buffer [5 g of bovine serum albumin (Sigma-Aldrich) and 4 ml of 0.5 M EDTA (Gentrox) in 1 liter of 1× PBS]. The combined fractions were then spun down and incubated in 20 ml of MACS buffer on ice to quench enzymatic activity. To isolate stromal cells, the cells were centrifuged for 30 min at 1500g (acceleration 5, deceleration 0) using a three-layer Percoll density gradient (MilliporeSigma). The three layers had 1.115, 1.065, and 1.0 relative densities, and the stromal cells were isolated from the interphase of 1.065 and 1.0 Percoll layers. Isolated cells were then resuspended in MACS buffer and stained for FACS analysis (table S1). To isolate lymphocytes for flow cytometry, the thymus, lymph nodes, or spleen were isolated by dissection from mice and then mashed through a 70-μm filter. Spleen cells were lysed in ammonium-chloride-potassium (ACK) lysis buffer to remove red blood cells. Isolated cells were counted, and between 1 × 106 and 5 × 106 cells were prepared for flow cytometry.
Flow cytometry and intracellular staining
Cells were isolated into suspension and were first stained with LIVE/DEAD Fixable Blue Stain Kit (Thermo Fisher Scientific) or Ghost Dye (Tonbo), followed by blocking in 2.4G2 (anti-mouse CD16/CD32, Bio X Cell) before staining with the appropriate surface antibodies for flow cytometry (table S1). For intracellular and transcription factor staining, cells were fixed for 20 min with 2% PFA or with the eBioscience Foxp3/transcription factor staining buffer set (Thermo Fisher Scientific). After fixation, cells were permeabilized with 1× permeabilization buffer (eBioscience) and stained with the appropriate intracellular antibodies (table S1).
RNA isolation and qPCR
RNA was isolated using the RNeasy Micro Kit (QIAGEN). Isolated RNA was reverse-transcribed using SuperScript IV reverse transcription (Invitrogen) with OligoDT20 primers (Invitrogen). qPCR was performed on the Applied Biosystems QuantStudio3 machine using Quantabio Perfecta qPCR ToughMix Low Rox. All reactions were normalized to Actin (ActB), and fold induction was calculated using the Delta-Delta CT (ddCT) method. All primer probes for TaqMan assays were purchased from Applied Biosystems (ABI, under Thermo Fisher Scientific): Ikzf1 Mm01187882, Aire Mm00477461, Ins2 Mm00731595, Gad1 Mm04207422, Pcp4 Mm00500973, Spt1 Mm00839568, Fabp9 Mm07297047, Timd2 Mm00506693, Serbinp12 Mm00510925, Gpr50 Mm00439147, Fgg Mm00513575, Crabp1 Mm00442776, Gal Mm00439056, Pyy Mm00520716, Crhbp Mm01283832, Prg3 Mm00450410, and ActB (Life Technologies).
scRNA-seq and data analysis
Sorted EPCAM+Ly51− mTECs were pooled from three mice of the appropriate genotype and were resuspended at approximately 20,000 cells per 100 μl and sent to the UCSF Genomics core for processing. Single cells were captured using the 10X Chromium micro-fluidics system (10x Genomics). The cells were encapsulated, and barcoded complementary DNA libraries were prepared using the single-cell 3′mRNA Kit (10x Genomics). Single-cell libraries were sequenced using a NovaSeq 6000 (Illumina). FASTQ files were converted into single-cell matrices using 10x Cell Ranger. Matrices were transformed into a Seurat object using the Seurat package in R. Initial exclusion criteria included features that were present in fewer than 100 cells and cells where fewer than 1000 features were detected. Ikzf1+/fl and Ikzf1fl/fl samples were merged and integrated using 2000 variable features and dimensions 1:20. Cells with greater than 50% ribosomal RNA, greater than 10% mitochondrial RNA, and greater than 5000 features were excluded. Principal components analysis was performed using 30 principal components, and UMAPs were generated using dimensions 1:20. Cluster identities were assigned cell types, and dendritic cells, T cells, fibroblasts, and endothelial cells were excluded from further analysis. Differential gene expression analysis was performed in Seurat.
scATAC-seq and data analysis
Sorted EPCAM+Ly51− mTECs were pooled from three mice of the appropriate genotype and were lysed, and nuclei were counted and resuspended at approximately 20,000 cells/100 μl in 10x nuclei buffer and sent to the UCSF Genomics core for processing. Using chromium scATAC workflow (10x Genomics), nuclei were transposed and then encapsulated and barcoded. Libraries were sequenced using a NovaSeq 6000 (Illumina). Fragment files were created from FASTQ files using 10x Cell Ranger. Fragment files were transformed into ArrowFiles in R using the ArchR package (54), with an initial exclusion criterion of cells with a transcription start site enrichment score of less than four or cells with fewer than 1000 mapped ATAC-seq fragments. Gene annotation was performed with the mm10 genome. Doublets were removed using the filterDoublets function in ArchR. Iterative latent semantic indexing (LSI)–based dimensional reduction was performed with four iterations, a resolution of 0.8, 250,000 variable features, and dimensions 2:30. After clustering and marker features analysis, T cell and dendritic cell clusters and clusters containing a low number of variable features were removed from further analysis. Further analysis was performed with the ArchR pipeline. Additional analysis was also performed via the Signac package. FASTQ files were converted into single-cell matrices using 10x Cell Ranger, and then Ikzf1+/fl and Ikzf1fl/fl matrices were integrated using Cell Ranger Aggr and were transformed into a Seurat object. Features that were present in fewer than 10 cells, and cells where fewer than 500 fragments were detected or more than 8000 fragments were detected, cells with less than 15% of reads in peaks, cells with a blacklist ratio greater than 0.1, cells with a nucleosome signal greater than 1.5, or cells that had a transcription start site enrichment score of less than 2 were excluded. Iterative LSI-based dimensional reduction and clustering were done with dims 2:30. After clustering and marker features analysis, T cell and dendritic cell clusters and clusters containing a low number of variable features were removed from further analysis. Differential peak expression analysis was performed in Signac, and motifs were analyzed with JASPAR2020 (species = 9606) and chromVAR (80). Additional TF footprinting motif analysis was performed in ArchR and Signac (Foxn1 motifs). Sequence data were visualized on UCSC genome browser (31) after creation of aggregated ATAC-seq reads or downloaded bed-files for peak annotation from published datasets (82).
RNA-seq and data analysis
About 500 L1 cell adhesion molecule (L1CAM+) Tuft cells or 500 MHC-IIhigh mTECs were sorted from individual mice directly into lysis buffer from the SMART-seq v4 (Takara Bio) kit. Either three or four mice were used for each genotype. Lysed cells were given to the UCSF Genomics COLAB for Library Preparation. Libraries prepared by the core were submitted to the UCSF Center for Advanced Technology for single-end sequencing (30 million reads). Aire-dependent genes and Fezf2-dependent genes were identified using previously published RNA-seq data (5). Sequence alignment was performed using STAR (version 2.7.9a) (83). Mappings were restricted to those that were uniquely assigned to the mouse genome, and unique read alignments were used to quantify expression and aggregated on a per-gene basis using the Ensembl (GRCm39) annotation. Differentially expressed genes between experimental groups were then determined using DESeq2 (v4.0.3) (84).
Coimmunoprecipitation
293T cells were grown in six-well plates and then transfected with appropriate plasmids: Aire-FLAG (85) (a gift from J. Abramson) or hemagglutinin (HA)–Ikaros (contains HA-tagged mouse Ikzf1 Ik1 isoform cloned into pcDNA) (50). Forty-eight hours after transfection, cells were lysed with cytoplasmic buffer containing 10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, EDTA-free protease inhibitor cocktail, and 0.1% octylphenoxypo-lyethoxyethanol (IGEPAL CA-630). The nuclear pellet was isolated by centrifugation at 500g for 5 min. Total nuclear extract was isolated by incubation of the nuclear pellet with 1% SDS and 1:100 benzonase. Nuclear lysates were then used for coimmunoprecipitation with the Dynabead Protein G Immunoprecipitation Kit (Life Technologies). Eluted proteins were separated by electrophoresis through Novex Life Technologies minigels and then were transferred to polyvinylidene difluoride membranes for immunoblot analysis with the appropriate antibodies (table S1), followed by enhanced chemiluminescence.
Statistical analysis
All experiments were performed using randomly assigned mice without investigator blinding. No data were excluded. Statistical significance between two groups was calculated using an unpaired, parametric, two-tailed Student’s t test or the nonparametric Mann-Whitney test. For experiments with three groups, statistical significance was calculated using a one-way analysis of variance (ANOVA), and grouped comparisons were corrected using Tukey’s multiple comparison test. Experimental groups included a minimum of three biological replicates. Intragroup variation was not assessed. All statistical analysis was performed using Prism 9 (GraphPad Software). A P value of < 0.05 was considered statistically significant. No statistical methods were used to predetermine sample size.
Supplementary Material
Acknowledgments:
We thank W. Howell for the technical assistance. Flow cytometry was performed at the UCSF Parnassus Flow Core, and high-throughput sequencing was performed at the UCSF Functional Genomics Core Facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Funding:
This work was supported by NIH grants R01AI165829 (to M.R.W.), R01AI127709 (to H.S.), and R25GM056847-23 (to J.S.); the UCSF Institute for Rheumatic Disease (to M.R.W. and H.S.); the UCSF Sandler PSSP (to J.M.G.); the UCSF IMSD Program (to J.S.); and the UCSF Parnassus Flow Core (RRID:SCR_018206, supported by NIH P30DK063720)
Footnotes
Supplementary Materials
This PDF file includes:
Other Supplementary Material for this manuscript includes the following:
Competing interests:
The authors declare that they have no competing interests.
View the article online
Data and materials availability:
Ikzf1-floxed mice were made available from S.C. and P.K. under a material transfer agreement with the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC) in France. Sequencing data for Foxn1-Cre/Ikzf1+/fl and Foxn1-Cre/Ikzf1fl/fl murine mTEC scATAC-seq, Foxn1-Cre−/Ikzf1fl/fl and Foxn1-Cre+/Ikzf1fl/fl murine tuft cell bulk RNA-seq, and Foxn1-Cre−/Ikzf1fl/fl/Pou2f3−/− and Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− murine mTEC bulk RNA-seq are available under accession code GSE238150. Sequencing data for Foxn1-Cre/Ikzf1+/fl and Foxn1-Cre/Ikzf1fl/fl murine mTEC scRNA-seq are available via GSE241742. Aire KO and Fezf2 KO murine mTEC RNA-seq is available via GSE144877 (5); human mTEC scRNA-seq is available via GSE147520 (43). All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Abramson J, Anderson G, Thymic epithelial cells. Annu. Rev. Immunol 35, 85–118 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D, The cellular mechanism of Aire control of T cell tolerance. Immunity 23, 227–239 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D, Projection of an immunological self shadow within the thymus by the Aire protein. Science 298, 1395–1401 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Takaba H, Morishita Y, Tomofuji Y, Danks L, Nitta T, Komatsu N, Kodama T, Takayanagi H, Fezf2 orchestrates a thymic program of self-antigen expression for immune tolerance. Cell 163, 975–987 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Tomofuji Y, Takaba H, Suzuki HI, Benlaribi R, Martinez CDP, Abe Y, Morishita Y, Okamura T, Taguchi A, Kodama T, Takayanagi H, Chd4 choreographs self-antigen expression for central immune tolerance. Nat. Immunol 21, 892–901 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJE, Lalioti MD, Mullis PE, Antonarakis SE, Kawasaki K, Asakawa S, Ito F, Shimizu N, Positional cloning of the APECED gene. Nat. Genet 17, 393–398 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Bornstein C, Nevo S, Giladi A, Kadouri N, Pouzolles M, Gerbe F, David E, Machado A, Chuprin A, Tóth B, Goldberg O, Itzkovitz S, Taylor N, Jay P, Zimmermann VS, Abramson J, Amit I, Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559, 622–626 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Wells KL, Miller CN, Gschwind AR, Wei W, Phipps JD, Anderson MS, Steinmetz LM, Combined transient ablation and single-cell RNA-sequencing reveals the development of medullary thymic epithelial cells. eLife 9, e60188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozai M, Kubo Y, Katakai T, Kondo H, Kiyonari H, Schaeuble K, Luther SA, Ishimaru N, Ohigashi I, Takahama Y, Essential role of CCL21 in establishment of central self-tolerance in T cells. J. Exp. Med 214, 1925–1935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lkhagvasuren E, Sakata M, Ohigashi I, Takahama Y, Lymphotoxin β receptor regulates the development of CCL21-expressing subset of postnatal medullary thymic epithelial cells. J. Immunol 190, 5110–5117 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Michelson DA, Hase K, Kaisho T, Benoist C, Mathis D, Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell 185, 2542–2558.e18 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadouri N, Nevo S, Goldfarb Y, Abramson J, Thymic epithelial cell heterogeneity: TEC by TEC. Nat. Rev. Immunol 20, 239–253 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, Bruschi M, Harcus Y, Zimmermann VS, Taylor N, Maizels RM, Jay P, Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, Artis D, Garrett WS, Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Leary CE, Schneider C, Locksley RM, Tuft cells-systemically dispersed sensory epithelia integrating immune and neural circuitry. Annu. Rev. Immunol 37, 47–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Moltke J, Ji M, Liang HE, Locksley RM, Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller CN, Proekt I, von Moltke J, Wells KL, Rajpurkar AR, Wang H, Rattay K, Khan IS, Metzger TC, Pollack JL, Fries AC, Lwin WW, Wigton EJ, Parent AV, Kyewski B, Erle DJ, Hogquist KA, Steinmetz LM, Locksley RM, Anderson MS, Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature 559, 627–631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, Heger A, Ponting CP, Holländer GA, Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24, 1918–1931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burkly L, Hession C, Ogata L, Reilly C, Marconl LA, Olson D, Tizard R, Gate R, Lo D, Expression of relB is required for the development of thymic medulla and dendritic cells. Nature 373, 531–536 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Riemann M, Andreas N, Fedoseeva M, Meier E, Weih D, Freytag H, Schmidt-Ullrich R, Klein U, Wang ZQ, Weih F, Central immune tolerance depends on cross-talk between the classical and alternative NF-κB pathways in medullary thymic epithelial cells. J. Autoimmun 81, 56–67 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck RP, Lira SA, Bravo R, Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κB /Rel family. Cell 80, 331–340 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Immunological Genome Project, Immunological Genome, ImmGen at 15. Nat. Immunol 21, 700–703 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Ferreiros-Vidal I, Carroll T, Taylor B, Terry A, Liang Z, Bruno L, Dharmalingam G, Khadayate S, Cobb BS, Smale ST, Spivakov M, Srivastava P, Petretto E, Fisher AG, Merkenschlager M, Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood 121, 1769–1782 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Heizmann B, Kastner P, Chan S, Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J. Exp. Med 210, 2823–2832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyon de Ana C, Arakcheeva K, Agnihotri P, Derosia N, Winandy S, Lack of Ikaros deregulates inflammatory gene programs in T cells. J. Immunol 202, 1112–1123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwickert TA, Tagoh H, Gültekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, Dickins RA, Zuber J, Jaritz M, Busslinger M, Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol 15, 283–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwickert TA, Tagoh H, Schindler K, Fischer M, Jaritz M, Busslinger M, Ikaros prevents autoimmunity by controlling anergy and Toll-like receptor signaling in B cells. Nat. Immunol 20, 1517–1529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K, Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5, 537–549 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K, Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol 7, 382–391 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schjerven H, Ayongaba EF, Aghajanirefah A, McLaughlin J, Cheng D, Geng H, Boyd JR, Eggesbø LM, Lindeman I, Heath JL, Park E, Witte ON, Smale ST, Frietze S, Müschen M, Genetic analysis of Ikaros target genes and tumor suppressor function in BCR-ABL1+ pre-B ALL. J. Exp. Med 214, 793–814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schjerven H, McLaughlin J, Arenzana TL, Frietze S, Cheng D, Wadsworth SE, Lawson GW, Bensinger SJ, Farnham PJ, Witte ON, Smale ST, Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat. Immunol 14, 1073–1083 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arenzana TL, Schjerven H, Smale ST, Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor. Genes Dev. 29, 1801–1816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heller JJ, Schjerven H, Li S, Lee A, Qiu J, Chen ZME, Smale ST, Zhou L, Restriction of IL-22-producing T cell responses and differential regulation of regulatory T cell compartments by zinc finger transcription factor Ikaros. J. Immunol 193, 3934–3946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynaud D, A Demarco I, L Reddy K, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H, Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol 9, 927–936 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernardi C, Maurer G, Ye T, Marchal P, Jost B, Wissler M, Maurer U, Kastner P, Chan S, Charvet C, CD4+ T cells require Ikaros to inhibit their differentiation toward a pathogenic cell fate. Proc. Natl. Acad. Sci. U.S.A 118, e2023172118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleinmann E, Geimer Le Lay AS, Sellars M, Kastner P, Chan S, Ikaros represses the transcriptional response to Notch signaling in T-cell development. Mol. Cell. Biol 28, 7465–7475 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agoston DV, Szemes M, Dobi A, Palkovits M, Georgopoulos K, Gyorgy A, Ring MA, Ikaros is expressed in developing striatal neurons and involved in enkephalinergic differentiation. J. Neurochem 102, 1805–1816 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Alsio JM, Tarchini B, Cayouette M, Livesey FJ, Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A 110, E716–E725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Ibanez R, Crespo E, Urbán N, Sergent-Tanguy S, Herranz C, Jaumot M, Valiente M, Long JE, Pineda JR, Andreu C, Rubenstein JLR, Marín O, Georgopoulos K, Mengod G, Fariñas I, Bachs O, Alberch J, Canals JM, Ikaros-1 couples cell cycle arrest of late striatal precursors with neurogenesis of enkephalinergic neurons. J. Comp. Neurol 518, 329–351 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Ezzat S, Mader R, Fischer S, Yu SJ, Ackerley C, Asa SL, An essential role for the hematopoietic transcription factor Ikaros in hypothalamic-pituitary-mediated somatic growth. Proc. Natl. Acad. Sci. U.S.A 103, 2214–2219 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon J, Xiao S, Hughes B III, Su DM, Navarre SP, Condie BG, Manley NR, Specific expression of lacZ and cre recombinase in fetal thymic epithelial cells by multiplex gene targeting at the Foxn1 locus. BMC Dev. Biol 7, 69 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray D, Abramson J, Benoist C, Mathis D, Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J. Exp. Med 204, 2521–2528 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bautista JL, Cramer NT, Miller CN, Chavez J, Berrios DI, Byrnes LE, Germino J, Ntranos V, Sneddon JB, Burt TD, Gardner JM, Ye CJ, Anderson MS, Parent AV, Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun 12, 1096 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK, Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas B, White AJ, Cosway EJ, Parnell SM, James KD, Jones ND, Ohigashi I, Takahama Y, Jenkinson WE, Anderson G, Diversity in medullary thymic epithelial cells controls the activity and availability of iNKT cells. Nat. Commun 11, 2198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derbinski J, Pinto S, Rosch S, Hexel K, Kyewski B, Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc. Natl. Acad. Sci. U.S.A 105, 657–662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennecke P, Reyes A, Pinto S, Rattay K, Nguyen M, Küchler R, Huber W, Kyewski B, Steinmetz LM, Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat. Immunol 16, 933–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meredith M, Zemmour D, Mathis D, Benoist C, Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat. Immunol 16, 942–949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG, Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91, 845–854 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST, Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 14, 2146–2160 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinderle C, Christensen HM, Schweiger S, Lehrach H, Yaspo ML, AIRE encodes a nuclear protein co-localizing with cytoskeletal filaments: Altered sub-cellular distribution of mutants lacking the PHD zinc fingers. Hum. Mol. Genet 8, 277–290 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Koipally J, Georgopoulos K, A molecular dissection of the repression circuitry of Ikaros. J. Biol. Chem 277, 27697–27705 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Oravecz A, Apostolov A, Polak K, Jost B, le Gras S, Chan S, Kastner P, Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2. Nat. Commun 6, 8823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang HY, Greenleaf WJ, ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet 53, 403–411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K, Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10, 345–355 (1999). [DOI] [PubMed] [Google Scholar]
- 56.O’Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, Han S, Seong RH, Park SD, Agalioti T, Munshi N, Thanos D, Erdjument-Bromage H, Tempst P, Bank A, An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell. Biol 20, 7572–7582 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sridharan R, Smale ST, Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J. Biol. Chem 282, 30227–30238 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Georgopoulos K, The making of a lymphocyte: The choice among disparate cell fates and the IKAROS enigma. Genes Dev. 31, 439–450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heizmann B, Kastner P, Chan S, The Ikaros family in lymphocyte development. Curr. Opin. Immunol 51, 14–23 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Katerndahl CDS, Heltemes-Harris LM, Willette MJL, Henzler CM, Frietze S, Yang R, Schjerven H, Silverstein KAT, Ramsey LB, Hubbard G, Wells AD, Kuiper RP, Scheijen B, van Leeuwen FN, Müschen M, Kornblau SM, Farrar MA, Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival. Nat. Immunol 18, 694–704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu Y, Zhang Z, Kashiwagi M, Yoshida T, Joshi I, Jena N, Somasundaram R, Emmanuel AO, Sigvardsson M, Fitamant J, el-Bardeesy N, Gounari F, van Etten RA, Georgopoulos K, Superenhancer reprogramming drives a B-cell-epithelial transition and high-risk leukemia. Genes Dev. 30, 1971–1990 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nehls M, Kyewski B, Messerle M, Waldschütz R, Schüddekopf K, Smith AJH, Boehm T, Two genetically separable steps in the differentiation of thymic epithelium. Science 272, 886–889 (1996). [DOI] [PubMed] [Google Scholar]
- 63.Hogquist KA, Xing Y, Hsu FC, Shapiro VS, T cell adolescence: Maturation events beyond positive selection. J. Immunol 195, 1351–1357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D, Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science 348, 589–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malchow S, Leventhal DS, Lee V, Nishi S, Socci ND, Savage PA, Aire enforces immune tolerance by directing autoreactive T cells into the regulatory T cell lineage. Immunity 44, 1102–1113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malchow S, Leventhal DS, Nishi S, Fischer BI, Shen L, Paner GP, Amit AS, Kang C, Geddes JE, Allison JP, Socci ND, Savage PA, Aire-dependent thymic development of tumor-associated regulatory T cells. Science 339, 1219–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Proekt I, Miller CN, Jeanne M, Fasano KJ, Moon JJ, Lowell CA, Gould DB, Anderson MS, DeFranco AL, LYN- and AIRE-mediated tolerance checkpoint defects synergize to trigger organ-specific autoimmunity. J. Clin. Invest 126, 3758–3771 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taniguchi RT, DeVoss JJ, Moon JJ, Sidney J, Sette A, Jenkins MK, Anderson MS, Detection of an autoreactive T-cell population within the polyclonal repertoire that undergoes distinct autoimmune regulator (Aire)-mediated selection. Proc. Natl. Acad. Sci. U.S. A 109, 7847–7852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su MA, Giang K, }umer K, Jiang H, Oven I, Rinn JL, DeVoss JJ, Johannes KPA, Lu W, Gardner J, Chang A, Bubulya P, Chang HY, Peterlin BM, Anderson MS, Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J. Clin. Invest 118, 1712–1726 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Nieuwenhove E, Garcia-Perez JE, Helsen C, Rodriguez PD, van Schouwenburg PA, Dooley J, Schlenner S, van der Burg M, Verhoeyen E, Gijsbers R, Frietze S, Schjerven H, Meyts I, Claessens F, Humblet-Baron S, Wouters C, Liston A, A kindred with mutant IKAROS and autoimmunity. J. Allergy Clin. Immunol 142, 699–702.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoshino A, Okada S, Yoshida K, Nishida N, Okuno Y, Ueno H, Yamashita M, Okano T, Tsumura M, Nishimura S, Sakata S, Kobayashi M, Nakamura H, Kamizono J, Mitsui-Sekinaka K, Ichimura T, Ohga S, Nakazawa Y, Takagi M, Imai K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Kojima S, Nonoyama S, Morio T, Kanegane H, Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J. Allergy Clin. Immunol 140, 223–231 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Eskandarian Z, Fliegauf M, Bulashevska A, Proietti M, Hague R, Smulski CR, Schubert D, Warnatz K, Grimbacher B, Assessing the functional relevance of variants in the IKAROS family zinc finger protein 1 (IKZF1) in a cohort of patients with primary immunodeficiency. Front. Immunol 10, 568 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, Xie HF, Fang H, Lu QJ, Xu JH, Li XP, Pan YF, Deng DQ, Zeng FQ, Ye ZZ, Zhang XY, Wang QW, Hao F, Ma L, Zuo XB, Zhou FS, du WH, Cheng YL, Yang JQ, Shen SK, Li J, Sheng YJ, Zuo XX, Zhu WF, Gao F, Zhang PL, Guo Q, Li B, Gao M, Xiao FL, Quan C, Zhang C, Zhang Z, Zhu KJ, Li Y, Hu DY, Lu WS, Huang JL, Liu SX, Li H, Ren YQ, Wang ZX, Yang CJ, Wang PG, Zhou WM, Lv YM, Zhang AP, Zhang SQ, Lin D, Li Y, Low HQ, Shen M, Zhai ZF, Wang Y, Zhang FY, Yang S, Liu JJ, Zhang XJ, Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet 41, 1234–1237 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Qu S, du Y, Chang S, Guo L, Fang K, Li Y, Zhang F, Zhang K, Wang J, Common variants near IKZF1 are associated with primary Sjögren’s syndrome in Han Chinese. PLOS ONE 12, e0177320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koh AS, Kingston RE, Benoist C, Mathis D, Global relevance of Aire binding to hypomethylated lysine-4 of histone-3. Proc. Natl. Acad. Sci. U.S.A 107, 13016–13021 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waterfield M, Khan IS, Cortez JT, Fan U, Metzger T, Greer A, Fasano K, Martinez-Llordella M, Pollack JL, Erle DJ, Su M, Anderson MS, The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat. Immunol 15, 258–265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boutboul D, Kuehn HS, van de Wyngaert Z, Niemela JE, Callebaut I, Stoddard J, Lenoir C, Barlogis V, Farnarier C, Vely F, Yoshida N, Kojima S, Kanegane H, Hoshino A, Hauck F, Lhermitte L, Asnafi V, Roehrs P, Chen S, Verbsky JW, Calvo KR, Husami A, Zhang K, Roberts J, Amrol D, Sleaseman J, Hsu AP, Holland SM, Marsh R, Fischer A, Fleisher TA, Picard C, Latour S, Rosenzweig SD, Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency. J. Clin. Invest 128, 3071–3087 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kellner ES, Krupski C, Kuehn HS, Rosenzweig SD, Yoshida N, Kojima S, Boutboul D, Latour S, Barlogis V, Galambrun C, Stray-Pedersen A, Erichsen HC, Marsh RA, Allogeneic hematopoietic stem cell transplant outcomes for patients with dominant negative IKZF1/IKAROS mutations. J. Allergy Clin. Immunol 144, 339–342 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Parent AV, Russ HA, Khan IS, LaFlam TN, Metzger TC, Anderson MS, Hebrok M, Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development. Cell Stem Cell 13, 219–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ, chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D, The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boyd J, Rodriguez P, Schjerven H, Frietze S, ssvQC: An integrated CUT&RUN quality control workflow for histone modifications and transcription factors. BMC. Res. Notes 14, 366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abramson J, Giraud M, Benoist C, Mathis D, Aire’s partners in the molecular control of immunological tolerance. Cell 140, 123–158 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Ikzf1-floxed mice were made available from S.C. and P.K. under a material transfer agreement with the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC) in France. Sequencing data for Foxn1-Cre/Ikzf1+/fl and Foxn1-Cre/Ikzf1fl/fl murine mTEC scATAC-seq, Foxn1-Cre−/Ikzf1fl/fl and Foxn1-Cre+/Ikzf1fl/fl murine tuft cell bulk RNA-seq, and Foxn1-Cre−/Ikzf1fl/fl/Pou2f3−/− and Foxn1-Cre+/Ikzf1fl/fl/Pou2f3−/− murine mTEC bulk RNA-seq are available under accession code GSE238150. Sequencing data for Foxn1-Cre/Ikzf1+/fl and Foxn1-Cre/Ikzf1fl/fl murine mTEC scRNA-seq are available via GSE241742. Aire KO and Fezf2 KO murine mTEC RNA-seq is available via GSE144877 (5); human mTEC scRNA-seq is available via GSE147520 (43). All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.


