Abstract
Purpose
The etiopathogenesis of coronal nonsyndromic craniosynostosis (cNCS), a congenital condition defined by premature fusion of 1 or both coronal sutures, remains largely unknown.
Methods
We conducted the largest genome-wide association study of cNCS followed by replication, fine mapping, and functional validation of the most significant region using zebrafish animal model.
Results
Genome-wide association study identified 6 independent genome-wide-significant risk alleles, 4 on chromosome 7q21.3 SEM1-DLX5-DLX6 locus, and their combination conferred over 7-fold increased risk of cNCS. The top variants were replicated in an independent cohort and showed pleiotropic effects on brain and facial morphology and bone mineral density. Fine mapping of 7q21.3 identified a craniofacial transcriptional enhancer (eDlx36) within the linkage region of the top variant (rs4727341; odds ratio [95% confidence interval], 0.48[0.39-0.59]; P = 1.2E−12) that was located in SEM1 intron and enriched in 4 rare risk variants. In zebrafish, the activity of the transfected human eDlx36 enhancer was observed in the frontonasal prominence and calvaria during skull development and was reduced when the 4 rare risk variants were introduced into the sequence.
Conclusion
Our findings support a polygenic nature of cNCS risk and functional role of craniofacial enhancers in cNCS susceptibility with potential broader implications for bone health.
Keywords: Coronal Nonsyndromic Craniosynostosis, DLX6 DLX5, GWAS, Regulatory elements, SEM1
Graphical abstract
Introduction
Craniosynostosis (CS), in which cranial vault sutures close prematurely, is the second most common congenital craniofacial abnormality after orofacial clefts.1 CS affects ∼1 in 2000 live births and leads to long-term complications for normal brain and skull growth and function.2 Currently, there are no pharmacological treatments available for CS, and affected infants typically require extensive, sometimes multiple, surgical treatments. Even with successful surgery, significant medical problems—such as increased intracranial pressure, vision and hearing impairments, breathing and dentition issues, and neurodevelopmental delays—may persist.2 As such, the human and financial impact of CS is considerable, which could be mitigated through screening and prevention.
The etiopathogenesis of CS is most likely multifactorial in its origins. Several modifiable and nonmodifiable environmental factors have been suggested as contributing to this major birth defect, but results remain inconclusive.2,3 In addition, genetic factors are considered to be significant predictors of CS risk.4 Although ∼200 syndromes manifest CS, 80% to 85% of affected infants present without additional major defects and are considered to have nonsyndromic CS (NCS).3, 4, 5
Fusion of the coronal suture is 1 of 3 major CS subtypes, along with sagittal and metopic synostoses, and accounts for 20% to 30% of all NCS.3,5 Unilateral coronal CS occurs ∼4 to 7 times as often as bilateral.6,7 The estimated prevalence of coronal NCS (cNCS) is ∼1 in 10,000 live births; 60% to 75% of those affected are female.1,3 Evidence of familial transmission suggests a genetic component for cNCS,3 but causative genetic variants have only been identified in a fraction of cases, suggesting a complex mode of inheritance, including low penetrance of causal variants and/or locus heterogeneity.
Candidate gene studies have identified several variants associated with cNCS.8, 9, 10 Our targeted sequencing study of genes previously implicated in syndromic and NCS identified several predicted to be damaging novel coding variants in EFNB1, BBS9, and TWIST1.11 Furthermore, an exome sequencing study reported variants in TCF12, a basic helix-loop-helix partner of TWIST1, that accounted for risk in 32% of bilateral and ∼10% of unilateral cNCS probands.12
Transcriptional regulation also plays a key role in craniofacial development,13, 14, 15, 16 suggesting that variation in gene regulatory elements may contribute to the development of CS. In humans, a number of noncoding variants have been identified: downstream of BMP2 and intronic BBS9 loci in a genome-wide association study (GWAS) of sagittal NCS17 and an intronic variant in BMP7 in a GWAS of metopic NCS.18 Furthermore, structural variants disrupting TWIST1 regulatory elements but not residing in its protein-coding sequence have been reportedly associated with a CS-like phenotype.16
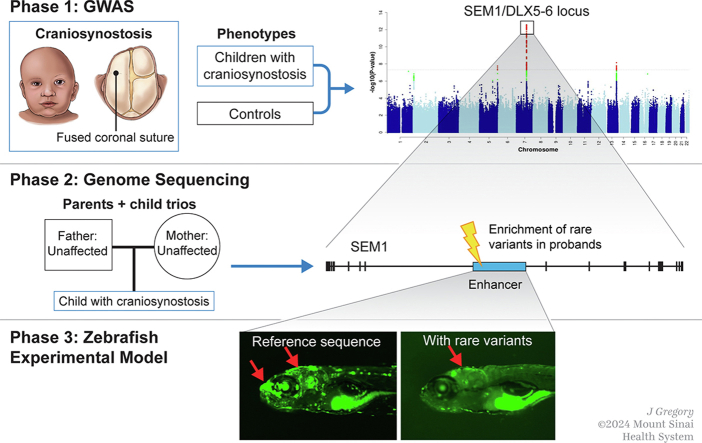
Systematic characterization of the complex landscape of cNCS etiopathogenesis, which would advance understanding of biological pathways required to develop therapeutics, is lacking. Herein, we performed a comprehensive, common variant analysis of cNCS using GWAS, followed by fine mapping of the top association signals using genome sequencing (GS) analyses. We then evaluated the functional impact of the most significant susceptibility locus in zebrafish (Danio rerio). Our findings advance the understanding of biological mechanisms involved in cNCS and identify potential directions for the development of early diagnostic strategies and therapeutic approaches.
Materials and Methods
Subjects
Discovery cohort
Our study was approved by the Institutional Review Boards of the participating institutions (Supplemental Table 1) and conducted in accordance with the institutional guidelines. Our initial, multiethnic discovery cohort comprised blood, saliva, or buccal cell specimens from 460 cNCS probands and their available parents (301 trios, 85 duos, and 74 singletons) recruited from several clinics and the population-based National Birth Defects Prevention Study19 (Supplemental Table 2). We excluded probands with synostosis of additional sutures and associated extra-cranial birth defects and those carrying variants in genes associated with syndromic forms of CS whenever this information was available. The ancestry of the study participants was categorized into European, Hispanic, and African American based on genotype data using the method described below (see Statistical Analysis). We applied ancestry analysis to the 460 probands, identifying 376 of European descent. Control DNA from 3376 unrelated individuals of European descent without major birth defects previously genotyped using a multiethnic global array for unrelated projects were selected from the Centers for AIDS Research Network of Integrated Clinical Systems cohort (n = 3318)20 and the Exploring Mechanisms of Disease Transmission In Utero Through the Microbiome study (n = 58).21
Replication cohort (European ancestry)
Our replication cohort included 59 cNCS cases and 289 controls of European ancestry. cNCS cases were identified from multiple clinics, the New York State Birth Defects Registry and the National Birth Defect Prevention Study19 (Supplemental Table 1). Controls were drawn from a random sample of live births without cNCS delivered during the same time period and recorded in the New York State Birth Defects Registry, the Iowa Pyloric Stenosis Study, and a cohort recruited for an unrelated study of osteogenic differentiation.22,23 The use of anonymous blood spots was approved by the Institutional Review Boards of the New York State Department of Health and University of Iowa. Participant exclusion criteria applied for our replication cohort were the same as those for our discovery cohort.
Replication cohort (Hispanic ancestry)
We conducted targeted validation of the top GWAS peaks by comparing allele frequencies of 77 cNCS cases of Hispanic ancestry (59 and 18 selected from discovery and replication collections, respectively) with those in the gnomAD v2.1 Hispanic population (n = 838). Individuals of African American ancestry were not analyzed because of a very small sample size.
GS cohort
We used the GS data of 89 cNCS trios (including 77 of European ancestry) from the Gabriella Miller Kids First Pediatric Research Program (dbGAP phs001806.v1.p1), 80 of which overlapped with our discovery GWAS cohort.
Genotyping and quality control
We extracted genomic DNA as described previously.20 Our discovery cohort was genotyped on the multiethnic global array at the Genomic Core Facility at Mount Sinai. Quality control procedures were carried out at both single-marker and subject levels before imputation. We excluded markers with a genotype call rate <95% or significant deviation from Hardy-Weinberg equilibrium (P < 1E−07) and probands or parents with >5% missing single-nucleotide polymorphism (SNP) genotypes. All probands were confirmed to be unrelated based on identity-by-descent (IBD) analysis implemented in PLINK.24 Because most of the probands in the discovery cohort were recruited with parents, pedigrees were reconstructed by pairwise IBD calculations performed using PLINK and Kinship-based inference for genome-wide association studies (KING)25 option available in PLINK2. The control specimens underwent the same quality control protocol.
Imputation
We used the Michigan Imputation Server26 to impute missing genotypes. Affected probands, parents, and unrelated controls from the discovery and replication cohorts were merged to predict ancestry and then imputed separately by ancestry. We used EAGLE 2.4 for phasing, the Haplotype Reference Consortium as the reference for European ancestry samples, and Trans-Omics for Precision Medicine as the reference for the other non-European ancestries.26, 27, 28, 29, 30 After the imputation of autosomal chromosomes, only the calls with imputation quality score r2 > 0.6 and genotype missingness rate <5% were included. Genotypes were dichotomized based on the posterior probability >0.9.31
Statistical analysis
We inferred genetic ancestry through principal components (PCs) analysis using EIGENSTRAT software32 and the 1000 Genomes Project as the reference database to evaluate population structure and predict the ancestry of each subject. Furthermore, we performed similar analyses within each ancestry group to derive PCs and used them as covariates in regression models.
We used PLINK to conduct the discovery case-control GWAS using logistic regression analysis under the additive genetic model in 376 cases of European descent and 3376 ancestry-matched controls.24 Each analysis included 10 ancestry-specific PCs as covariates. The genomic inflation factor was λ < 1.10 for the discovery cohort. Our discovery GWAS used a set of controls from our existing independent projects involving other health conditions (HIV or inflammatory bowel disease); therefore, we aimed to correct for potential batch effects that may have arisen because of project-specific protocols and procedures. Specifically, we compared minor allele frequencies (MAFs) of common variants in our control cohort with those in the gnomAD European population and excluded 3405 SNPs with statistically significant differences (P < 1E−06). We set the genome-wide significance P value threshold to 5E−08 for multiple testing correction.33 We applied conditional analysis to determine which variants were independent within the susceptibility locus by simultaneously including the SNPs most significantly associated with cNCS as covariates and testing the significance of other variants in the region.
Replication analysis was conducted using the independent cohort of 59 cNCS cases and 289 unrelated controls of European ancestry using logistic regression model applying the same parameters as the discovery case-control analysis (additive genetic model, MAFs, statistical significance threshold) and including the top 10 PCs as covariates to correct for population structure. Inverse standard-error-weighted meta-analyses were performed using METAL to integrate the summary statistics from the discovery and replication GWASs of European cohorts, with test statistics corrected for inter-study heterogeneity.34 Transethnic meta-analysis of the top association signals was performed by integrating the association results from the European cohorts and the independent Hispanic cohort also by METAL.34 The transmission disequilibrium test (TDT) was used to compare the transmission of the top cNCS susceptibility alleles to the affected offspring vs the nontransmission in 301 proband-parent trios of various ancestries (probands from 241 trios were included as cases in the discovery case-control study) as applied in PLINK.24 All GWAS results were generated and visualized in R (version 3.0.2).35 Regional plots were created using LocusZoom.36 Lastly, a multi-marker association analysis, assessing the joint effect of carrying multiple risk alleles in the SEM1-DLX5-DLX6 locus, was performed using logistic regression in individuals of European ancestry from the combined discovery and replication cohorts. In this analysis, the number of risk alleles across the 4 most associated variants in this locus was counted in each individual, and then the odds of developing cNCS were compared between each combination of the risk alleles and a reference group consisting of no risk alleles. Multimarker test and multivariable logistic regression analyses applied to test the independence of the associated SNPs were carried out using STATA15.
Polygenic risk score (PRS) analysis
cNCS-specific PRS was derived by applying PRSice-2 R package.37 We used the summary statistics from our case-control discovery analysis (base data set) to derive the PRS, which was then tested using the European replication cohort (target data set). PRSice-2 implements clumping plus thresholding method with variant scores defined as the sum of the allele counts weighted by the effect sizes (beta estimates) from the discovery GWAS. To calculate the PRS, we retained variants with MAF >1% and genotype call rate >95% and clumped them based on the linkage disequilibrium (LD) pattern using the European-ancestry subcohort from the 1000 Genome Project as the reference (--clump-kb 250 kb; --climp-p 1 --clump-r2 0.10). We tested multiple P value thresholds (5E−08, 1E−05, 5E−05, 1E−04, 5E−04, 1E−03, .05, .1, .2, .3, .4, .5, 1) to select the best model. The best fit was achieved by a model that included 106 variants with association P < 5E−05.
Phenome-wide association analysis (PheWAS)
We conducted phenome-wide association analysis, PheWAS, by querying publicly available GWAS results using Open Targets Genetics38 to identify additional traits or diseases associated with our top susceptibility variants.
Functional annotation of significant SNPs
GWAS meta-analysis summary statistics were annotated using the Functional Mapping and Annotation (FUMA) platform.39 1000 Genome Project phase 3 was used as the reference panel for LD structure. All SNPs were annotated with databases of expression quantitative trait loci (eQTLs), chromatin states, and chromatin interaction information. We incorporated the genotype-tissue expression v8 data repository,40 the Brain eQTL Almanac repository,41 cis-eQTL and trans-eQTL data from CommonMind Consortium,42 xQTLServer,43 and cis-eQTL and trans-eQTL data from eQTLGen44 and the eQTL Catalog.45 We applied a false discovery rate46 of <5% to set significant eQTL associations. Additionally, we annotated the top regions with regulatory elements having craniofacial activity predicted by Wilderman et al.13 For chromatin interactions, we used Hi-C data of mesenchymal stem cell lines reported in the FUMA platform. We applied an false-discovery-rate-adjusted P value < 1E−06 to define significant interactions.
Pathway analysis
A gene-based association analysis was performed using Multimarker Analysis of GenoMic Annotation v1.08.47 Herein, variants were assigned to protein-coding genes (n = 19,294 genes) if they were within 10 kb of the gene, a standard distance as per FUMA instructions selected to best capture the LD block48 and cis-regulatory variants49 associated with each gene. The resulting SNP P values were combined into a gene-centered test statistic using the SNP-wise mean model. Bonferroni-corrected threshold was applied to evaluate statistical significance (P < 2.6E−06 [0.05/19,294]). To assess the joint effect of multiple genes on cNCS susceptibility, we performed pathway enrichment analysis with Multimarker Analysis of GenoMic Annotation against the canonical gene set libraries from the molecular signatures database50 and the database of gene-disease associations (DisGeNET).51 In addition, to determine potential biological connections between the top susceptibility loci, we performed gene set enrichment analysis with Enrichr52 across gene set libraries created from Orphanet,53 the Phenotype-Genotype Integrator,54 GWAS Catalog,55 DisGeNET,51 Rare Diseases Gene Reference Into Function,56 the Encyclopedia of DNA Elements,57 the Human Metabolome Database,58 and All RNA-seq and ChIP-seq sample and signature search transcription factors and All RNA-seq and ChIP-seq sample and signature search tissue expression.59 Enriched pathways that shared at least 2 genes from the queried gene set libraries were retained to construct a knowledge graph subnetwork that was visualized with Cytoscape.60
GS analysis
We used GS to fine map the top genome-wide significant regions. Standard sequencing and variant calling protocols were followed as previously described,61 and Annotate Variation (ANNOVAR) was used for variant annotation.62 Familial relationships were confirmed by pairwise IBD analysis performed using PLINK and KING option25 in PLINK2. We pulled out the sequence data of the flanking region spanning 100k from each genome-wide significant susceptibility variant identified in the European meta-analysis from the GS data set. We then performed gene-based rare-variant TDT analysis63 in 89 trios to evaluate the enrichment of rare damaging variants in each gene. Variants were defined as rare if their gnomAD MAF was <1% and damaging if they met all the following criteria: METASVM score > 0.80,64, 65, 66 CADD score > 20,67 and REVEL score > 0.50.68 We also divided each region into 6 kb sliding windows with an overlap of 2 kb and performed an aggregate analysis of rare variants for each interval using rare-variant TDT.63 Finally, we called copy-number variants (CNVs) by consensus calling between HMMcopy69 and CNVnator70 and identified de novo variants by applying the TrioDeNovo71 algorithm with stringent filtering quality controls, as previously described.64, 65, 66
Identification of craniofacial enhancer candidates
Publicly available chromatin immunoprecipitation assay combined with DNA sequencing (ChIP-seq) and assay for transposase-accessible chromatin using sequencing (ATAC-seq) data sets were obtained from mouse and human craniofacial tissues and analyzed to identify putative regulatory elements. Enhancer-associated marks (EP300; GSE49413, H3K27ac; GSE89435) from E10.5/E11.5 mouse embryos tissues (ie, maxilla [Mx], mandible [Mn], pharyngeal arch 2 [PA2], and frontonasal prominence [FNP]) were analyzed.13,72 Furthermore, human embryonic ChIP-seq data (H3K4me1, H3K4me2, H3K4me3, H3K36me3, H3K27ac, H3K27me3; GSE97752) of craniofacial tissues from Carnegie stages 13 through 20 were investigated.13 Overlapping peaks were considered when at least 1 base pair region intersected. Sequences were defined as enhancer candidates if they were marked in Carnegie stages 13 to 20 human data and in at least 1 of the craniofacial mouse data sets (H3K27ac and ATAC-seq).
Functional validation using transgenic zebrafish enhancer assay
We designed primers to amplify the candidate enhancer sequences from human genomic DNA (Supplemental Table 3). Polymerase chain reaction products were cloned into the E1b-GFP-Tol2 enhancer assay vector containing an E1b minimal promoter followed by green florescence protein (GFP) reporter gene.73 These constructs were injected into zebrafish embryos using standard procedures.74 For statistical power, at least 100 embryos were injected per construct in at least 2 different injection experiments along with Tol2 mRNA to facilitate genomic integration.75 The embryos were grown to sexual maturity to mate and generate stable lines for each enhancer candidate. GFP expression was observed and annotated during embryogenesis (24-72 hours after fertilization) and larvae maturation and adult stages (7, 14, 21, and 60 days after fertilization [dpf]). The 4 mutated eDlx36 sequences were synthesized and verified by Sanger sequencing and compared with the wild-type sequence.
Results
Genome-wide association analysis
Of the 460 probands in our discovery cohort, 385 (84%) presented with unilateral and 56 (12%) bilateral cNCS (Supplemental Table 2). The majority of the probands were females (65%). Similar clinical characteristics were identified in our replication cohort (Supplemental Table 2).
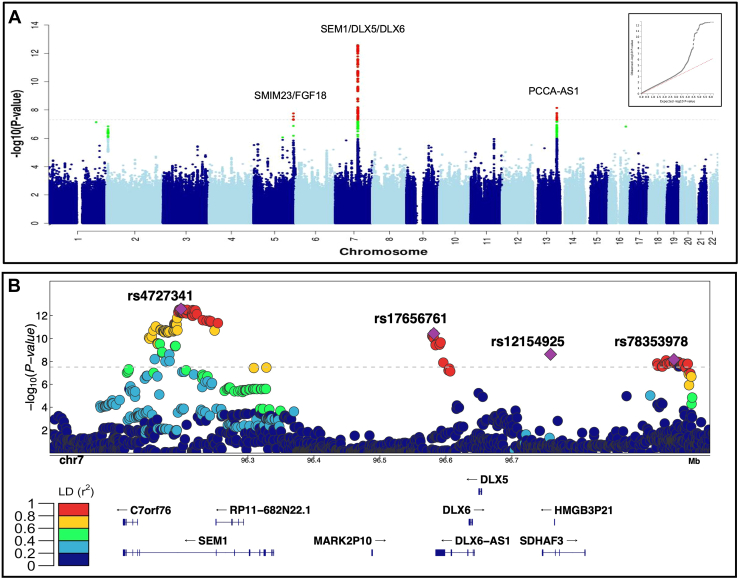
Our case-control analysis of 376 European ancestry cNCS cases and 3376 ancestry-matched controls found a genome-wide significant signal in the intronic region of SEM1 (26S proteasome subunit; rs4727341; OR [95% CI] = 0.48 [0.39-0.59], P = 1.2E−12). This conferred a reduction in risk for cNCS with a MAF of 18% vs 30% in case vs controls, with this latter estimate comparable to a MAF of 31% in the European gnomAD population (Table 1). We also identified 2 genome-wide significant variants that increased risk for cNCS: an intergenic variant 61 kb downstream of DLX6-AS1 (distal-less homeobox 6, antisense 1; rs17656761 (chr7:96581553:G>A), OR = 1.96 [1.61-2.38], P = 1.1E−11) and an intronic variant in PLEKHA6 (rs114264214, OR = 3.57 [2.26-5.65], P = 4.9E−8; Supplemental Figure 1A). rs4727341 and rs17656761 showed similar magnitude and direction of effect in an independent replication cohort of European ancestry (Table 1).
Table 1.
Summary statistics for the logistic model correcting for ancestry background of the genome-wide significant associated risk variants in the European discovery and European replication cohorts, and corresponding meta-analysis
| SNP | Location (Chr:Pos [A]) | Nearest Gene | Function | European Discovery (N = 376 vs 3376) |
European Replication (N = 59 vs 289) |
European Meta-Analysis |
gnomAD |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | AF CASE | AF CTRL |
OR (95% CI) | P | AF CASE | AF CTRL |
OR (95% CI) [Direction of Effect] | P | H | AF Eur | ||||
| rs4727341 | 7:96198615 [G] | SEM1 | Intronic | 0.48 (0.39-0.59) | 1.2 × 10−12 | 0.18 | 0.30 | 0.56 (0.34-0.93) | 0.02 | 0.21 | 0.34 | 0.49 (0.29-0.69) [−−] |
2.7 × 10−13 | 0.56 | 0.31 |
| rs17656761 | 7:96581553 [A] | DLX6-AS1 | Intergenic | 1.96 (1.61-2.38) | 1.1 × 10−11 | 0.22 | 0.13 | 1.31 (0.69-2.45) | 0.40 | 0.14 | 0.12 | 1.89 (1.69-2.09) [++] |
3.7 × 10−11 | 0.23 | 0.13 |
| rs12154925 | 7:96758550 [T] | SDHAF3 | Intronic | 1.87 (1.50-2.30) | 8.0 × 10−9 | 0.19 | 0.12 | 1.81 (0.97-3.35) | 0.06 | 0.15 | 0.09 | 1.86 (1.66-2.06) [++] |
2.4 × 10−9 | 0.92 | 0.13 |
| rs78353978 | 7:96945446 [A] | DLX5/TAC1 | Intergenic | 2.58 (1.88-3.55) | 5.9 × 10−9 | 0.07 | 0.03 | 1.82 (0.63-5.25) | 0.27 | 0.05 | 0.03 | 2.50 (2.19-2.81) [++] |
6.9 × 10−9 | 0.54 | 0.03 |
| rs7981517 | 13:101112917 [A] | PCCA/PCCA-AS1 | Intronic | 1.59 (1.34-1.90) | 1.7 × 10−7 | 0.31 | 0.22 | 1.91 (1.21-3.04) | 0.006 | 0.35 | 0.23 | 1.64 (1.47-1.79) [++] |
7.1 × 10−9 | 0.47 | 0.23 |
| rs33863 | 5:171166685 [A] | SM1M23/FGF18 | Intergenic | 1.53 (1.31-1.79) | 1.7 × 10−7 | 0.44 | 0.34 | 1.69 (1.09-2.62) | 0.02 | 0.51 | 0.35 | 1.55 (1.39 1.71) [++] |
1.7 × 10−8 | 0.69 | 0.35 |
| rs114264214 | 1:204310366 [A] | PLEKHA6 | Intronic | 3.57 (2.26-5.65) | 4.9 × 10−8 | 0.04 | 0.01 | 1.97 (0.47-8.20) | 0.35 | 0.03 | 0.01 | 3.37 (2.92-3.82) [++] |
7.3 × 10−8 | 0.44 | 0.01 |
We reported chromosome and genomic position in hg19 build (Chr:Pos), and the tested allele is reported in square brackets. P values in bold font are genome-wide statistically significant.
95% CI, 95% confidence intervals; AF CASE, minor allele frequency in cases; AF CTRL, minor allele frequency in controls; AF Eur, minor allele frequency in gnomAD for the European population; H, heterogeneity p-value; OR, odds ratio; P, P values are calculated by logistic regression analyses and corrected for population stratification with EIGENSTRAT axes within each cohort; SNP, single nucleotide polymorphism.
Our genome-wide meta-analysis (Figure 1A) that integrated summary statistics of the discovery and replication European GWASs revealed 6 independent genome-wide significant variants, with 4 located in the SEM1-DLX5-DLX6 region (Table 1). Within this region, we confirmed our findings for rs4727341 and rs17656761 and detected 2 novel independent loci, rs12154925 in an intron of SDHAF3 (human homolog for succinate dehydrogenase assembly factor 3), and rs78353978 (chr7:96945446:G>A), in an intergenic region (Figure 1B). Moreover, we identified rs7981517 (chr13:101112917:G>A), intronic to PCCA (propionyl-CoA carboxylase subunit alpha), near PCCA-AS1 (propionyl-CoA carboxylase subunit alpha antisense RNA 1) and rs33863 (chr5:171166685:G>A), an intergenic variant, located 282 kb away from FGF18 (fibroblast growth factor 18) and 46 kb away from SMIM23 (small integral membrane protein 23; Table 1). In our meta-analysis, rs114264214 in PLEKHA6 did not reach genome-wide significance (Table 1). None of the lead risk variants showed significant difference in frequency between unilateral and bilateral cases after adjustment for multiple testing (Supplemental Table 4) and an additive model best fit the underlying heritability compared with dominant or recessive models (Supplemental Table 5).
Figure 1.
Summary of the genome-wide analysis of coronal nonsyndromic craniosynostosis. A. Manhattan plot of the meta-analysis of discovery and replication genome-wide association analyses using common variants (minor allele frequency > 1%). The y-axis shows the −log10 transformed P value of each variant association found using a standard-error-weighted approach and controlling for population stratification, and the x-axis shows the chromosomal position. Variants crossing the genome-wide significance threshold of P < 5E−08 are color coded in red, and those with P < 5E−06 are in green. The top signals are annotated with the closest genes. Inset: quantile-quantile plot showing distribution of expected P values under the null model (red-dotted line) vs observed P values (black dots). B. Regional plot of the 4 top independent genomic association signals from the European meta-analyses. The y-axis shows −log10P values for individual variants annotated with the genes in the selected genomic interval. The top variants are marked as purple diamonds and other variants in pairwise linkage disequilibrium (r2) with the top variant, based on the 1000 Genomes Project Phase 3 European reference samples, are color coded as per the scale in legend.
Subsequent pathway analysis detected several DisGeNET gene-disease sets characterized by finger, limb, and facial developmental malformations that were significantly enriched among the common cNCS risk loci (Supplemental Table 6). No significant enrichment was detected using the molecular signatures database pathway libraries (such as Gene Ontology terms, Biocarta, and Reactome). Also, single-marker TDT analysis in 301 cNCS trios (241 cases overlapped with the case-control discovery) replicated the association of rs4727341, with the parents being less likely to transmit the reduced risk allele to the affected offspring (rs4727341, OR [CI] = 0.45 [0.33-0.60], P = 2.08E−08, Supplemental Figure 1B, Supplemental Table 7). No significant parent of origin effect was observed for any risk variant implying no differences in transmission rates between the father and the mother (Supplemental Table 7). Finally, a similar direction and magnitude of the associations were observed for rs4727341 (SEM1), rs17656761 (DLX6-AS1), and rs7981517 (PCCA/PCCA-AS1) in a small independent cohort of 77 cNCS Hispanic cases compared with the gnomAD Hispanic population (Supplemental Table 8). The 5 most significant variants in the European meta-analysis remained genome-wide statistically significant also in the trans-ethnic meta-analysis, with the exception of rs12154925 at SDHAF3 that was not present in the Hispanic cohort (Supplemental Table 8).
Multimarker analysis of top common variants
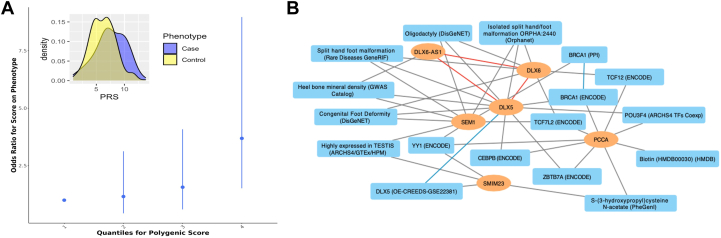
To evaluate the independence of associations of the 6 genome-wide significant signals, we included them simultaneously in a multivariable regression model and found that all of them remained significantly associated with cNCS risk (Supplemental Table 9). Moreover, our analysis showed that cNCS risk increased proportionally to the number of risk alleles across the 4 independent SNPs within the SEM1-DLX5-DLX6 locus (Table 2). Co-occurrence of the 3 alleles that increased cNCS risk and absence of the allele that reduced the risk conferred an over 7-fold increased risk of developing cNCS (OR [CI] = 7.15 [3.54-14.45]). Conversely, the absence of the 3 alleles that increased cNCS risk and the presence of the SEM1 allele that reduced cNCS risk was associated with a significantly lower risk of cNCS (OR [CI] = 0.54 [0.39-0.75], P = 2.3E−04). Lastly, we explored the predictive value of PRS for cNCS risk based on the summary statistics from the European discovery GWAS that included 106 independent markers with P < 5E−05 (Supplemental Table 10), which was selected from a series of PRS based on various P value thresholds (Supplemental Figure 2). PRS for cNCS was significantly associated with cNCS risk in the European replication cohort (P = 6E−04), explaining 5% of the trait variability. The risk increased with each quantile of PRS (Figure 2A), further suggesting the polygenic nature of cNCS.
Table 2.
Summary statistics for the combinations of the 4 top variants in the 7q21.3 locus from the multimarker logistic regression model in individuals from combined European discovery and replication cohorts
| rs4727341 | rs17656761 | rs12154925 | rs78353978 | AF (N) Cases | AF (N) Controls | OR (95%CI) | P |
|---|---|---|---|---|---|---|---|
| − | − | − | + | 1% (6) | 1% (18) | 3.02 (1.17-7.75) | .02 |
| − | − | + | − | 8% (31) | 7% (248) | 1.13 (0.74-1.72) | .56 |
| − | − | + | + | 3% (13) | 1% (37) | 3.18 (1.64-6.16) | 6.0 × 10−4 |
| − | + | − | − | 14% (57) | 7% (249) | 2.07 (1.47-2.93) | 3.70 × 10−5 |
| − | + | − | + | 0.2% (1) | 0.2% (6) | 1.51 (0.18-12.64) | .7 |
| − | + | + | − | 8% (31) | 2% (65) | 4.32 (2.70-6.90) | 1.0 × 10−9 |
| − | + | + | + | 4% (15) | 1% (19) | 7.15 (3.54-14.45) | 4.3 × 10−8 |
| + | − | − | − | 15% (60) | 29% (999) | 0.54 (0.39-0.75) | 2.3 × 10−4 |
| + | − | − | + | 1% (3) | 1% (28) | 0.97 (0.29-3.24) | .96 |
| + | − | + | − | 5% (20) | 7% (240) | 0.75 (0.46-1.24) | .26 |
| + | + | − | − | 7% (29) | 11% (377) | 0.7 (0.46-1.06) | .09 |
| + | − | + | + | 2% (10) | 2% (58) | 1.56 (0.78-3.14) | .21 |
| + | + | − | + | 0.05% (2) | 0.02% (8) | 2.26 (0.47-10.78) | .31 |
| + | + | + | − | 3% (11) | 2% (82) | 1.21 (0.63-2.35) | .56 |
| + | + | + | + | 1% (3) | 1% (20) | 1.36 (0.40-4.64) | .63 |
| − | − | − | − | 28% (115) | 30% (1041) | Reference | - |
Results in bold are nominally statistically significant. “+” indicates presence of the minor allele. “−” indicates absence of the minor allele.
95% CI, 95% confidence intervals; AF, allele frequency; N, number of samples in the group; OR, odds ratio; P, P values when compared with noncarriers of the 4 variants as the reference group.
Figure 2.
Multilocus analysis of the top susceptibility loci. A. Polygenic risk score calculated using genome-wide association study summary statistics from our discovery cohort was used to predict the risk for coronal nonsyndromic craniosynostosis in the replication cohort. Best fit model was achieved by PRsice with 106 variants with association P < 5E−05. Inset: difference in means of Polygenic risk score values in the craniosynostosis cases (blue) and the control group (yellow) are shown. B. Knowledge graph connecting the identified genes with shared enriched functional terms from Enrichr. In the network, identified genes are represented as orange ovals, whereas shared enriched annotations from Enrichr are shown as blue rectangles. Known physical interactions between the protein products of the identified genes are depicted by red lines, connections to functional terms are depicted by gray lines, and related terms are connected by blue lines.
Candidate gene enrichment and PheWAS analysis of top susceptibility loci
We performed enrichment analysis using the candidate genes within the susceptibility loci (SEM1, PCCA, PCCA-AS1, DLX5, DLX6, DLX6-AS1, and SMIM23) as input for Enrichr.52 The genes shared many known functional annotations, especially within the SEM1-DLX5-DLX6 locus in which these 3 genes were previously reported to physically interact. Moreover, PCCA and DLX5 were functionally related to several other genes. These include identified targets for the transcription factor TCF12, a gene with known variants linked to cNCS,12 and BRCA1, an established breast cancer gene that potentially regulates expression of PCCA and DLX5, and also shows protein-protein interactions with SEM176,77 (Figure 2B, Supplemental Table 11).
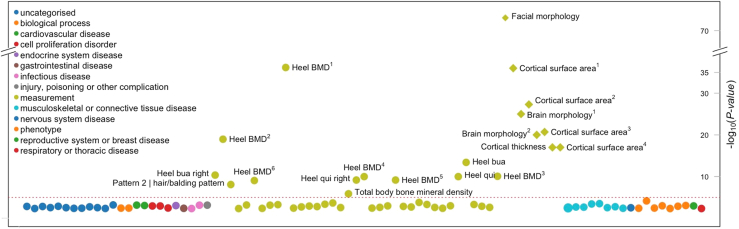
We also explored if our 6 most significant cNCS variants were previously associated with other morphological and skeletal phenotypes. Using PheWAS analysis, we queried public GWAS results using Open Targets Genetics38 to identify additional traits or diseases associated with these variants. The variant-level PheWAS analysis showed that SEM1 rs4727341 was implicated in several bone density traits, facial morphology, and brain cortical surface area measurements (Figure 3). Bone density traits were also significantly associated with DLX6-AS1 rs17656761 but not with rs12154925 or rs78353978, whereas rs33863 (chr5:171166685:G>A) was linked to height, facial morphology, and cortical surface, supporting the role of our observed cNCS variants in bone-related phenotypes (Supplemental Figure 3).
Figure 3.
Phenome-wide association analysis of rs4727341, the top risk variant. Phenotype-wide association analysis of complex traits associated with rs4727341. Summary statistics from the UK Biobank, FinnGen, and genome-wide association study catalog repositories were downloaded from Open Target (https://genetics.opentargets.org/). Only traits with P value < .005 are shown in the diagram. x axis shows traits and y axis shows the variant’s P value of association to each trait. The circles are color coded by the trait category (see legend) as reported in Open Target website. The red dashed line shows the significance threshold corrected for the number of traits shown. In the figure, heel bone mineral density and other traits appear multiple times since the association was reported in many independent studies/publications as follows: heel bone mineral density (Heel BMD): 1GCST006979, 2GCST006288, 3NEALE2_3148_raw, 4NEALE2_78_raw (t score automated), 5NEALE2_4125_raw (t score automated right), 6NEALE2_4124_raw (right); cortical surface area: 1GCST010282_20: pars triangularis, 2GCST010701: MOSTest, 3GCST010697: min P and 4GCST90091060; other heel measurements are also shown—heel broadband ultrasound attenuation (heel bua): NEALE2_3144_raw: direct entry and NEALE2_4120_raw: right); and heel quantitative ultrasound index (heel qui): NEALE2_3147_raw: direct entry and NEALE2_4123_raw: right.
Fine mapping of rare variants across the candidate regions
Next, to identify causal variants in our top cNCS susceptibility locus, we performed fine mapping using rare variant enrichment analysis of GS data from 89 cNCS trios (267 individuals) in selected candidate genes and open reading frames spanning the SEM1-DLX5-DLX6 region: C7orf76, SEM1, RP11-682N22/1, MARK2P10, DLX5, DLX6, DLX6-AS1, SDHAF3, and HMGB3P21. We did not detect any de novo SNP, de novo CNVs, or enrichment in damaging protein-coding variants in these genes regardless of the MAF (Supplemental Table 12). Furthermore, none of the exonic variants identified in some candidate genes were “predicted damaging” or carried by more than 1 proband (Supplemental Table 13). Similarly, no enrichment of deleterious coding variants or de novo variants were identified within the PCCA-AS1, PCCA, SM1M23, and FGF18 genes associated with the other 2 (rs78353978 and rs33863) genome-wide significant hits (Supplemental Table 14). Additionally, because of the important role in bicoronal cNCS, we evaluated the damaging variants in TCF12.12 We detected 3 predicted damaging SNPs, and 2 de novo frameshift deletions. Overall, the damaging variants were carried by 20% of bicoronal and 3% of unicoronal cNCS cases. However, the gene was not significantly enriched in damaging variants (P = .07) likely because of a limited number of bicoronal cases. We also detected 3 additional missense variants, but they were predicted to be tolerated (Supplemental Table 15).
Given that no protein-coding variants in the SEM1-DLX5-DLX6 locus were associated with cNCS, we examined regulatory elements in this region. Although this locus encompasses several tissue-specific developmental enhancers that regulate DLX5/DLX6 expression,15 craniofacial enhancers around SEM1 have not been fully elucidated. Therefore, we focused on identifying and characterizing enhancers with craniofacial activity across the SEM1-DLX5-DLX6 region (chr7:96,070,205-96,696,725). We initially analyzed the chromatin organization of this locus and found that SEM1, DLX5, and DLX6 were positioned in the same topological-associated domain in human-cultured cranial neural crest cells and Carnegie stage 17, when the skull has a membranous roof before ossification78 (Supplemental Figure 4). We next analyzed ATAC-seq and histone modification ChIP-seq of mouse E10.5 maxilla, mandible, pharyngeal arch 2, and frontonasal process,14 searching for sequences marked as active enhancers in mouse craniofacial developmental tissues. These analyses identified 16 candidate regulatory sequences that may play a role during craniofacial development (Supplemental Table 16). The lead intronic variant (rs4727341) belongs to a long haplotype (r2 > 0.9, Figure 4A-C) encompassing 3 candidate regulatory sequences, eDlx34, eDlx35, and eDlx36 (Figure 4D). Although publicly available eQTL data sets did not have tissues relevant to the cNCS phenotype, we found that rs4727341 and several LD variants belonging to the long haplotype showed an eQTL effect modulating DLX5 expression conserved in various tissues (P = 9.7E−05) and SLC25A13 expression (P = 2.5E−06, Supplemental Figure 5 and Supplemental Table 17). These observations support the involvement of regulatory elements that control the expression of craniofacial genes, rather than protein-coding variants, in cNCS risk.
Figure 4.
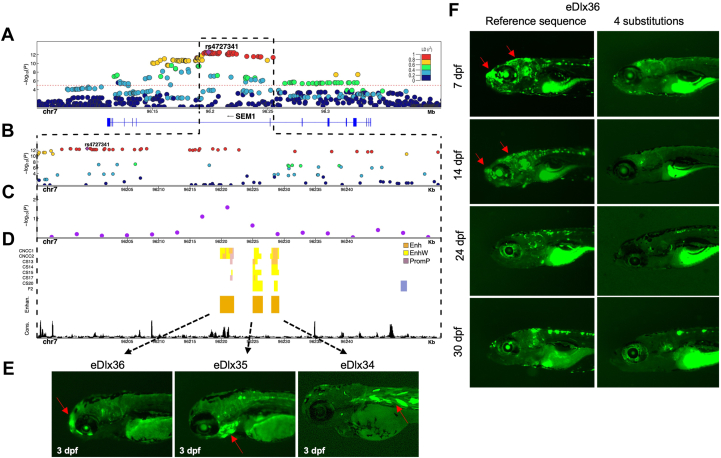
Enhancer analysis of the SEM1 locus. A. A regional association plot surrounding the top risk variant, rs4727341 (shown as a purple diamond). The x axis represents a 0.2 Mb region, 100 kb upstream and downstream of the lead variant; the y axis shows −log10P values for individual variant associations from the European meta-analysis annotated with the genes in the selected genomic interval. Pairwise linkage disequilibrium (LD) (r2) with the lead variant color coded based on the 1000 Genomes Project Phase 3 European reference samples. B. Zoom-in of the top signal region to highlight the rs4727341-LD region (r2 > 0.9) (dashed black box). The y axis shows −log10P values for individual variant associations from the European meta-analysis as above. Pairwise LD (r2) with the lead variant color coded based on the 1000 Genomes Project Phase 3 European reference samples. C. Zoom-in genomic region annotated with −log10P values for individual 6 kb sliding windows from rare variant TDT aggregate analysis of rare variants in family-based study (purple dots). The dot is represented at the start of each window. D. Predicted craniofacial regulatory elements near rs4727341. Histone modification marks are associated with active craniofacial predicted enhancers (CNCC1 through F2).13 Highlighted are 3 predicted enhancer candidates (Enhan.), eDlx34, eDlx35, and eDlx36, which were tested in this study (orange bar). Phylop conservation track (Cons.) from University of California Santa Cruz (UCSC) Genome Browser is shown in black color. E. Zebrafish enhancer assays at 3 days after fertilization (dpf). eDlx34 drove green fluorescent protein (GFP) expression in the heart and somitic muscles. eDlx35 drove specific GFP expression in the mandibular arch and branchial arches (basibranchials, hypobranchials, and ceratobranchial 1-5) and notochord. eDlx36 drove specific GFP expression in the premaxillary, maxillary, FNP, and apical region of the skull. F. The effect of 4 rare variants on the in vivo eDlx36 enhancer activity. GFP-positive cells drove by eDlx36 enhancer in the FNP and apical region of the skull at 7 and 14 dpf, whereas the mutated eDlx36 embryos showed low GFP expression and fewer GFP-positive embryos in the craniofacial tissues at 7,14, 24, and 30 dpf.
Functional validation of candidate transcriptional enhancers in the SEM1 intron
To determine the in vivo activity of the candidate enhancers at the SEM1 locus, we used the transgenic zebrafish enhancer assay. The candidate enhancers, eDlx34-eDlx36, were each cloned into a vector upstream of the E1b minimal promoter and GFP as a reporter gene and then individually injected into 1-cell-stage zebrafish embryos to generate a transgenic zebrafish. Using this enhancer assay, we showed that eDlx34 drove specific GFP expression in the heart and somitic muscles at 3 dpf, while, eDlx35 drove specific GFP expression in the mandibular and branchial arches (basibranchials, hypobranchials, and ceratobranchial 1-5) and notochord at 3 dpf. Moreover, eDlx36 drove specific GFP expression in the premaxillary, maxillary, and FNP at 3 dpf (Figure 4E). In addition, eDlx35 and eDlx36 enhancers drove GFP expression in the head of larval zebrafish, indicating their role in craniofacial and skull development (Figure 4E). Interestingly, the activity of these 2 enhancers resembles the expression patterns of dlx5a/6a in zebrafish (Supplemental Figure 6). Importantly, each enhancer had a discrete activity pattern and, along with additional enhancers,15 may comprise a potential spatiotemporal regulatory network that controls the expression of genes, such as DLX5/DLX6, during craniofacial development.
To test which enhancer is likely affecting the abnormal cranial phenotype, we performed TDT-based aggregate analysis of rare variants (MAF < 1%) by dividing the rs4727341-associated high LD region (r2 > 0.90) into 6 kb ± 2 kb sliding genomic segments (Figure 4A). The interval chr7:96,220,956-96,226,956 showed the most significant enrichment (unadjusted min P = .02) and fully overlapped with eDlx36 (Figure 4B).
Importantly, eDlx36 contained 4 rare intronic variants carried by 6 independent cNCS probands, all heterozygotes, in our GS study sample (Supplemental Table 18). To test whether these variants alter eDlx36 enhancer activity, we generated an enhancer sequence with all 4 variants and compared its activity with the reference sequence in our zebrafish model (Figure 4F). We annotated the enhancer activity at 7, 14, 24, and 30 dpf and noticed that eDlx36 is active in the FNP and the apical region of the head at 7 and 14 dpf. The number of GFP-positive cells in the FNP and the apical region of the head decreased at 24 and 30 dpf. (Figure 4F). Moreover, we found that the mutated eDlx36 embryos showed low GFP expression and fewer GFP-positive embryos already at 7 dpf (Supplemental Figure 7), suggesting that these variants affect the enhancer activity during early craniofacial development. Taken together, our results suggest that rare variants clustered in the SEM1 intron affect the activity of eDlx36 enhancer, which may potentially modulate the expression of candidate cNCS genes, such as DLX5 and DLX6.
Functional annotation of the other genome-wide significant cNCS susceptibility loci
To also determine the relevance in cNCS risk of rs17656761, rs12154925, and rs78353978 on chromosome 7q21, rs7981517 (PCCA-AS1) on chromosome 13, and rs33863 (SM1M23/FGF18) on chromosome 5, we performed FUMA functional annotation and fine-mapping analysis of each of these regions. Because we did not identify any enrichment of deleterious coding variants or de novo variants, we hypothesize that these cNCS risk variants could also affect gene regulation. We found the regions of LD around each of the 5 variants spanned, in various ways, known craniofacial predicted enhancers or heterochromatin elements (Supplemental Figure 8). However, the most important evidence is that the LD regions of 2 of the 5 lead variants contained long noncoding RNA. In fact, rs17656761 and its LD region on chromosome 7 spanned DLX6-AS1, and the LD block of rs7981517 on chromosome 13 spanned another antisense gene, PCCA-AS1. Although the LD region of rs17656761 was not associated with any known eQTL signals (Supplemental Figure 8), rs7981517 and other variants in LD with it contained established eQTL signals that strongly modulated the expression of TMTC4 (P = 3.75E−73), GGACT (P = 5.49E−73), and PCCA (P = 8.73E−135) in different tissues (eg, fibroblasts and peripheral blood mononuclear cells) (Supplemental Table 17). Of note, rs33863, the intergenic variant on chromosome 5, resided in a region with predicted craniofacial enhancers and a heterochromatin region (Supplemental Figure 8). This region showed evidence of potential chromatin interaction with FGF18 in mesenchymal stem cell culture (Supplemental Figure 9). Lastly, we did not find significant enrichment of transmitted noncoding rare variants for any sliding segments encompassing these genomic regions, possibly due to limited statistical power (Supplemental Figure 8). These results suggest that there might be additional regulatory elements, outside of the extended SEM1 locus, which may affect the cranial suture development, even in the absence of nearby rare causative variants.
Discussion
In this comprehensive meta-analysis of common genetic variation in cNCS across 2 independent European cohorts, we identified 6 genome-wide significant signals, 4 of which were located in the 7q21.3 locus. Individuals with all 4 risk alleles showed an over 7-fold increased risk of cNCS compared with those who had none. The 7q21.3 locus has previously been linked to split hand/split foot malformation type 1, also known as ectrodactyly, autosomal dominant syndrome with a deep median cleft of the hand and/or foot and aplasia/hypoplasia of the phalanges, metacarpals, and metatarsals.79 Our top associated risk variant, rs4727341, resides in an intronic region of SEM1, a gene known to play a role in cell cycle progression, apoptosis, and DNA damage repair. Some individuals with genetic alterations in SEM1 (also named SHFM1) present with intellectual disability, craniofacial findings, orofacial clefting,80 and hearing loss.79 Three other independent lead SNPs that occupy the same locus reside within the proximity of 2 transcription factors, DLX5 and DLX6, known to influence craniofacial development: rs17656761 near DLX6-AS1, rs12154925, and rs78353978.81
A previous study has reported craniofacial malformations in dlx5a/6a morpholino-based knockdown zebrafish, although no details on a particular phenotype were provided.82 In mice, Dlx5 and Dlx6 are highly expressed in embryonic coronal suture osteogenic cell subsets.83 Dlx5 is activated in proliferating osteoblast precursors that are upregulated by bone morphogenetic proteins (BMPs) and inhibited by the BMP-antagonist Noggin.81 Dlx5 also drives expression of the master regulator of osteogenesis and the transcription factor Runx2 and induces osteogenic differentiation in developing cranial suture mesenchyme.84 Although no obvious neurocranial abnormalities were reported, inactivation of Dlx5 and Dlx6 in mice resulted in severe craniofacial, axial, and appendicular skeletal abnormalities, leading to perinatal lethality.81
In our human study, the premature suture closure could not be explained by protein-coding variants in SEM1 or DLX5/DLX6 because none of these or other genes within the locus were enriched in damaging missense variants in cNCS cases. Because transcriptional regulation plays a key role in craniofacial development,13,14 it is plausible that variants in regulatory elements alter the expression of the target gene(s) that lead(s) to this craniofacial condition. We have already shown that deletions of enhancers that reside in the HDAC9 protein-coding sequence alter the expression of the neighboring gene, TWIST1, which plays a role in cranial suture closure during skull development and leads to CS-like phenotypes.16 Here, we showed a marginally significant enrichment in rare noncoding variants of a segment located within the rs4727341-high LD intronic region in SEM1, in which 4 rare variants (carried by 6 independent cNCS cases) overlapped with a novel craniofacial enhancer, eDlx36. Introduction of these rare variants in zebrafish enhancer assays altered activity in the FNP and apical region of the skull during development. Taken together, these findings suggest that the eDlx36 enhancer that resides in the SEM1 intron regulates the expression of target genes that are important during craniofacial development, potentially contributing to CS. We could hypothesize that eDlx36 variants disrupting enhancer activity may affect DLX5 and/or DLX6 expression given their known pivotal role in craniofacial development. Interestingly, we found that the eDlx36 activity resembles the expression patterns of dlx5a and dlx6a in zebrafish. Moreover, rs4727341 and its LD variants have a marginal eQTLs function modulating DLX5 expression conserved in various tissues, although not directly relevant to cNCS pathogenesis. We further speculate that DLX5 and/or DLX6 expression could be independently regulated by DLX6-AS1, contributing to the risk of premature suture closing. Future in vivo functional studies could elucidate the target genes and transcription regulation mechanism of action for eDlx36.
Along with the SEM1-DLX5-DLX6 locus, we identified 2 additional statistically significant signals associated with cNCS risk: rs78353978 in PCCA/PCCA-AS1 and intergenic rs33863 near SMIM23. PCCA encodes the alpha subunit of the heterodimeric mitochondrial enzyme propionyl-CoA carboxylase, and variants in this gene have been linked to propionic acidemia, an autosomal recessive organic acid disorder, which can also be accompanied by osteoporosis.85 The variants are located in the proximity of PCCA-AS1 that could potentially possess regulatory effects on nearby target genes, including ZIC2 and ZIC5 that are located nearby PCCA. Members of the Zic family of zinc-finger transcription factors have been previously implicated in early development.86 ZIC1 variants have been linked to coronal CS,87 whereas ZIC2 has been implicated in neural crest and craniofacial development.88,89 In our study, ZIC1 did not show damaging variants in the GS data sets. Finally, we found that the risk variant rs33863 showed evidence of chromatin interaction with FGF18 in a mesenchymal stem cell line. FGF18 is expressed in both osteogenic mesenchymal cells and differentiating osteoblasts during calvarial bone development90 and has been shown to be enriched in 2 of the ectocranial clusters identified in the coronal suture datasets.83 Interestingly, the progress of suture closure is delayed in Fgf18-deficient mice.90 Functional studies are warranted to determine if regulatory elements in this region are involved in craniofacial development and contribute to CS.
Our multivariable regression analysis including all top 6 genome-wide significant signals confirmed the independent effects of these variants. Assuming a polygenic nature of cNCS, we calculated PRS that evaluates genetic burden across multiple susceptibility loci and has been previously shown to have a greater predictive value for complex diseases than individual variants.91 We detected that the PRS consisting of the 106 SNPs associated with cNCS at P < 1E−05 was predictive of cNCS diagnosis in a European population. Finally, our network analysis indicated extensive connections across the top loci and with TCF12, a previously reported cNCS gene, and with BRCA1, 1 of the 2 most common causes of hereditary breast cancer.
Our top variants associated with cNCS, especially the 4 in 7q21.3, have also been implicated in other skeletal traits, such as femur, spine, and heel bone mineral density, cortical brain measurements, and dysmorphic facial features. This is consistent with a recent GWAS of skull bone mineral density that identified loci common to both osteoporosis and CS, suggesting a shared pathophysiology between craniofacial defects and bone diseases.92 In addition, our top variants were found among those associated with skull bone mineral density in a recent GWAS meta-analysis92 and overlapped with the 76 genomic loci influencing both brain and face shape from a large GWAS of cortical surface morphology.93 A number of earlier studies have reported opposite effects of variants in the same gene resulting in accelerated ossification (ie, CS) and deficient ossification (eg, parietal foramina, large fontanelles, or osteoporosis), including MSX2,94, 95, 96 TWIST1,97,98 RUNX2,99,100 Nell1,101,102 ALX4,103,104 and BMP2.17,105 These observations are consistent with other reports implying extensive genetic pleiotropy across the genome. Of note, pleiotropic variants have been shown more likely to be functional compared with nonpleiotropic variants,106 suggesting that the identification of shared genetic risks may provide a better insight into biological mechanisms underlying various conditions. Taken together and coupled with a recent report in the literature,93 our findings demonstrate a shared genetic control of skull, brain, and face shape development and suggest that variants regulating accelerated ossification and CS early in life may also affect whole body bone density with potential implications on bone health, fracture healing, and ultimately, aging. These observations can help inform further investigations into risk stratification and drug repurposing, especially when both cNCS and osteoporosis have substantial female predominance.
Our top cNCS susceptibility loci did not overlap with the signals within the BMP2 and BBS9 loci identified in a GWAS of sagittal NCS17 or BMP7 detected in a metopic NCS GWAS.18 Differences in phenotypic manifestation, extra-cranial complications, population incidence, heritability, and male/female ratios indicate that each type of sutural synostosis might represent a separate birth defect with a distinct set of etiologic factors107 and suggest that future screening and treatment strategies should be suture specific. Importantly, the top loci associated with NCS of other sutures were located in noncoding regulatory regions containing functional enhancers,17,18 suggesting an overarching regulatory control of cranial suture development and patency.
The strengths of our study include the largest cohort of cNCS cases to date supporting the discovery and replication analyses, fine mapping using GS data, and validation in an in vivo model system. However, our study has limitations. First, population controls in the case-control cohort were not screened for CS, which could result in phenotype misclassification. However, CS is relatively rare, and an inadvertent inclusion of cNCS cases in the control group would bias the results toward the null. Moreover, the possibility of syndromic CS among some of our cNCS cases cannot be ruled out. There are recorded instances of reduced penetrance and more subtle syndromic clinical manifestations, which may become more obvious later in life.108 However, nonsyndromic status was confirmed by clinicians and clinical geneticists through a review of clinical and imaging data at most of the participating sites, and the majority of cNCS cases were also screened using various craniofacial clinical genetic panels and were excluded if variants in previously reported CS genes were detected. Even if some syndromic CS cases were inadvertently included, it is unlikely that they systematically affected our results. Importantly, we had a limited number of cases of non-European ancestry preventing us from generating a multiethnic PRS and limiting the applicability of the PRS to other populations. A larger multiethnic study is warranted to validate the predictive value of our PRS for cNSC. Additionally, our sample sizes for GWAS and GS studies, although the largest to date for this condition, were still modest and our analyses only focused on autosomal chromosomes, excluding sex chromosomes and mitochondria. For functional studies, we used the well-established zebrafish models. However, zebrafish may not accurately reflect the genetic regulation of mammalian or human skull development. Nevertheless, previous work has shown that human enhancer sequences can function as active enhancers in zebrafish, even without homologous sequences.109 Future studies aimed at the elucidation of a broader role of the SEM1-DLX5-DLX6 locus in other more common skeletal phenotypes, such as osteoporosis, have the potential to be impactful.
In summary, our findings indicate that variation in predicted regulatory elements residing in the SEM1-DLX5-DLX6 locus plays a role in craniofacial and suture development and may contribute to the pathogenesis of cNCS by most likely deregulating the DLX5/DLX6 pathways. Moreover, the top cNCS susceptibility variants possess pleiotropic effects on bone mineral density and brain and facial morphology traits, opening potential avenues into shared diagnostic and therapeutic strategies.
Data Availability
GWAS summary level data that support the findings of this study are available in Supplemental Table 10 and upon request. Requests should be addressed to Dr Inga Peter (inga.peter@mssm.edu). The GS data that support the findings of this study are available in dbGAP with the identifier phs001806.v1.p1. Row genotyping data are in the process of being deposited to dbGAP.
Conflict of Interest
The authors declare no competing interests in relation to the work described.
Acknowledgments
The authors thank the Centers for AIDS Research Network of Integrated Clinical Systems (CNICS) cohort study for providing the control genotype frequencies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Funding
This work was supported in part by the National Institutes of Health, United States (NIH) R01 DE16886 (S.A.B. and P.A.R.), R03 DE031061 (S.A.B. and P.A.R.), X01 HL140535 Gabriela Miller KidsFirst program (S.A.B.), U01 DE024448 (E.W.J.), P01 HD078233 (E.W.J., J.T.R., I.P., and P.A.R.), and R01 DE030596 (G.H.), the NIH Intramural Research Program (HHSN01DK73431, N275201100001I, HHSN275201100001C, and HHSN275201100001G to J.M.), Centers for Disease Control and Prevention (CDC) R01 DD000350 (E.W.J.), cooperative agreements PA #96043, PA #02081, FOA #DD09-001, FOA #DD13-003, and NOFO #DD18-001 to the Centers for Birth Defects Research and Prevention participating in the National Birth Defects Prevention Study and/or the Birth Defects Study To Evaluate Pregnancy exposureS (BD-STEPS), grants (U01 DD001035 and U01 DD001223) awarded to the Iowa Center for Birth Defects Research and Prevention (P.A.R. and K.M.C.), US-Israel Binational Science Foundation BSF #2021102 (I.P. and R.B.), and Wellcome Investigator Award 102731 (A.O.M.W.). This work was supported in part through the computational and data resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai supported by the Office of Research Infrastructure of the National Institutes of Health under award S10OD026880 and S10OD030463 and by the Clinical and Translational Science Award grant UL1TR004419 from the NIH National Center for Advancing Translational Sciences.
Author Information
Conceptualization: E.W.J., J.T.R., R.Y.B., P.A.R., P.Nicoletti, D.Karasik, I.P.; Methodology: E.W.J., J.T.R., R.Y.B., P.A.R., P.Nicoletti, D.K, A.M.; Supervision: E.W.J., J.T.R., R.Y.B., P.A.R., P.Nicoletti, D.Karasik, A.M.; Data Curation: A.K.G., R.F., P.Noons, J.E.V.M., J.S., M.M.J., J.R., J.T., J.L.M., L.C.W., S.C., M.E., C.F., G.G.-F., E.W.J., P.J.T., R.R., E.S., A.E.L., J.F., L.C.B., C.M.J., A.F.W., E.A., S.F., M.H., D.S., A.T.T., S.A.W., D.J., J.C.B., D.C., J.T.R., T.D.S., E.R., N.S., C.H., G.M.S., Y.H., F.D.R., L.L.-M., S.A.B., A.C., K.Conway, R.H., P.A.R., A.O., A.O.M.W., J.M.P., K.Crawford, P.L., L.B., M.L.F., R.J.H., A.K., M.C., D.Kay, R.S., J.P.; Project Administration: S.R.B.; Formal Analysis: S.Z., W.O.L., A.S., L.M. M.P., R.J.H., I.I., J.E.E., G.B.M., G.H., Y.R., M.W., P.Nicoletti; Writing-original draft: E.W.J., J.T.R., R.Y.B., P.A.R., P.Nicoletti, D.Karasik; Writing-review & editing: A.K.G., R.F., P.Noons, J.E.V.M., J.S., M.M.J., J.R., J.T., J.L.M., L.C.W., M.E., C.F., G.G.-F., E.W.J., P.J.T., R.R., E.S., A.E.L., J.F., L.C.B., C.M.J., A.F.W., E.A., S.F., M.H., D.S., A.T.T., S.A.W., D.J., J.C.B., D.C., J.T.R., T.D.S., E.R., N.S., C.H., G.M.S., Y.H., F.D.R., L.L.-M., S.A.B., S.C., A.C., K.Conway, R.H., P.A.R., A.O., A.O.M.W., J.M.P., K.Crawford, P.L, L.B., M.L.F., R.J.H., A.K., M.C., D.Kay, D.Karasik, R.S., J.P., A.M., S.Z., W.O.L., A.S., L.M., M.P., R.J.H., I.I., J.E.E., G.B.M., G.H., Y.R., M.W., S.R.B.
Ethics Declaration
The study is approved by Institutional Review Boards (IRB) at each participating institution with the principal site at the Icahn School of Medicine IRB docket #14-00822. Informed consent was obtained from all participants and all data were deidentified.
Footnotes
The Article Publishing Charge (APC) for this article was paid by Inga Peter and Ramon Birnbaum.
Paola Nicoletti and Samreen Zafer contributed equally to this work.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gimo.2024.101851) contains supplemental material, which is available to authorized users.
Contributor Information
Ramon Y. Birnbaum, Email: ramonb@bgu.ac.il.
Inga Peter, Email: inga.peter@mssm.edu.
Supplementary Material
(SM) collect eighteen supplementary tables (1-18) and 9 supplementary figures (1-9) supporting the results described in the current article. The supplementary tables and figures are all mentioned in the text within the appropriate scientific contents. SM is linked to the online version of the paper at https://www.gimopen.org/
References
- 1.Cornelissen M., Ottelander B., Rizopoulos D., et al. Increase of prevalence of craniosynostosis. J Craniomaxillofac Surg. 2016;44(9):1273–1279. doi: 10.1016/j.jcms.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Shlobin N.A., Baticulon R.E., Ortega C.A., et al. Global epidemiology of craniosynostosis: a systematic review and meta-analysis. World Neurosurg. 2022;164:413–423.e3. doi: 10.1016/j.wneu.2022.05.093. [DOI] [PubMed] [Google Scholar]
- 3.Blessing M., Gallagher E.R. Epidemiology, genetics, and pathophysiology of craniosynostosis. Oral Maxillofac Surg Clin North Am. 2022;34(3):341–352. doi: 10.1016/j.coms.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Heuzé Y., Holmes G., Peter I., Richtsmeier J.T., Jabs E.W. Closing the gap: genetic and genomic continuum from syndromic to nonsyndromic craniosynostoses. Curr Genet Med Rep. 2014;2(3):135–145. doi: 10.1007/s40142-014-0042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkie A.O.M., Johnson D., Wall S.A. Clinical genetics of craniosynostosis. Curr Opin Pediatr. 2017;29(6):622–628. doi: 10.1097/MOP.0000000000000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulet S.L., Rasmussen S.A., Honein M.A. A population-based study of craniosynostosis in metropolitan Atlanta, 1989-2003. Am J Med Genet A. 2008;146A(8):984–991. doi: 10.1002/ajmg.a.32208. [DOI] [PubMed] [Google Scholar]
- 7.Wilkie A.O.M., Byren J.C., Hurst J.A., et al. Prevalence and complications of single-gene and chromosomal disorders in craniosynostosis. Pediatrics. 2010;126(2):e391–e400. doi: 10.1542/peds.2009-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson D., Wall S.A., Mann S., Wilkie A.O. A novel mutation, Ala315Ser, in FGFR2: a gene-environment interaction leading to craniosynostosis? Eur J Hum Genet. 2000;8(8):571–577. doi: 10.1038/sj.ejhg.5200499. [DOI] [PubMed] [Google Scholar]
- 9.Merrill A.E., Bochukova E.G., Brugger S.M., et al. Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum Mol Genet. 2006;15(8):1319–1328. doi: 10.1093/hmg/ddl052. [DOI] [PubMed] [Google Scholar]
- 10.Seto M.L., Hing A.V., Chang J., et al. Isolated sagittal and coronal craniosynostosis associated with TWIST box mutations. Am J Med Genet A. 2007;143A(7):678–686. doi: 10.1002/ajmg.a.31630. [DOI] [PubMed] [Google Scholar]
- 11.Sewda A., White S.R., Erazo M., et al. Nonsyndromic craniosynostosis: novel coding variants. Pediatr Res. 2019;85(4):463–468. doi: 10.1038/s41390-019-0274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma V.P., Fenwick A.L., Brockop M.S., et al. Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nat Genet. 2013;45(3):304–307. doi: 10.1038/ng.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilderman A., VanOudenhove J., Kron J., Noonan J.P., Cotney J. High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Rep. 2018;23(5):1581–1597. doi: 10.1016/j.celrep.2018.03.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minoux M., Holwerda S., Vitobello A., et al. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science. 2017;355(6332) doi: 10.1126/science.aal2913. [DOI] [PubMed] [Google Scholar]
- 15.Birnbaum R.Y., Everman D.B., Murphy K.K., Gurrieri F., Schwartz C.E., Ahituv N. Functional characterization of tissue-specific enhancers in the DLX5/6 locus. Hum Mol Genet. 2012;21(22):4930–4938. doi: 10.1093/hmg/dds336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirsch N., Dahan I., D‘Haene E., et al. HDAC9 structural variants disrupting TWIST1 transcriptional regulation lead to craniofacial and limb malformations. Genome Res. 2022;32(7):1242–1253. doi: 10.1101/gr.276196.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Justice C.M., Yagnik G., Kim Y., et al. A genome-wide association study identifies susceptibility loci for nonsyndromic sagittal craniosynostosis near BMP2 and within BBS9. Nat Genet. 2012;44(12):1360–1364. doi: 10.1038/ng.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Justice C.M., Cuellar A., Bala K., et al. A genome-wide association study implicates the BMP7 locus as a risk factor for nonsyndromic metopic craniosynostosis. Hum Genet. 2020;139(8):1077–1090. doi: 10.1007/s00439-020-02157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reefhuis J., Gilboa S.M., Anderka M., et al. The National Birth Defects Prevention Study: a review of the methods. Birth Defects Res A Clin Mol Teratol. 2015;103(8):656–669. doi: 10.1002/bdra.23384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng H., Sewda A., Marquez-Luna C., et al. Genetic architecture of cardiometabolic risks in people living with HIV. BMC Med. 2020;18(1):288. doi: 10.1186/s12916-020-01762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres J., Hu J., Seki A., et al. Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ-free mice. Gut. 2020;69(1):42–51. doi: 10.1136/gutjnl-2018-317855. [DOI] [PubMed] [Google Scholar]
- 22.Carcamo-Orive I., Hoffman G.E., Cundiff P., et al. Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell. 2022;29(10):1505. doi: 10.1016/j.stem.2022.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Schaniel C., Dhanan P., Hu B., et al. A library of induced pluripotent stem cells from clinically well-characterized, diverse healthy human individuals. Stem Cell Rep. 2021;16(12):3036–3049. doi: 10.1016/j.stemcr.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S., Neale B., Todd-Brown K., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das S., Forer L., Schönherr S., et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy S., Das S., Kretzschmar W., et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaneau O., Marchini J., Zagury J.F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 29.Delaneau O., Marchini J., 1000 Genomes Project Consortium C, Genomes Project, 1000 Genomes Project Consortium Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.1000 Genomes Project Consortium. Auton A., Brooks L.D., et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicoletti P., Aithal G.P., Bjornsson E.S., et al. Association of liver injury from specific drugs, or groups of drugs, with polymorphisms in HLA and other genes in a genome-wide association study. Gastroenterology. 2017;152(5):1078–1089. doi: 10.1053/j.gastro.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 33.McCarthy M.I. Casting a wider net for diabetes susceptibility genes. Nat Genet. 2008;40(9):1039–1040. doi: 10.1038/ng0908-1039. [DOI] [PubMed] [Google Scholar]
- 34.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The R. Project for Statistical Computing. Version R 3.0.2. http://wwwr-projectorg
- 36.Pruim R.J., Welch R.P., Sanna S., et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi S.W., O‘Reilly P.F. PRSice-2: polygenic Risk Score software for biobank-scale data. GigaScience. 2019;8(7) doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghoussaini M., Mountjoy E., Carmona M., et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021;49(D1):D1311–D1320. doi: 10.1093/nar/gkaa840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sng L.M.F., Thomson P.C., Trabzuni D. Genome-wide human brain eQTLs: in-depth analysis and insights using the UKBEC dataset. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-55590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fromer M., Roussos P., Sieberts S.K., et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19(11):1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Võsa U., Claringbould A., Westra H.J., et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–1310. doi: 10.1038/s41588-021-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerimov N., Hayhurst J.D., Peikova K., et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat Genet. 2021;53(9):1290–1299. doi: 10.1038/s41588-021-00924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 47.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4) doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petersen A., Alvarez C., DeClaire S., Tintle N.L. Assessing methods for assigning SNPs to genes in gene-based tests of association using common variants. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0062161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tehranchi A., Hie B., Dacre M., et al. Fine-mapping cis-regulatory variants in diverse human populations. Elife. 2019;8 doi: 10.7554/eLife.39595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen E.Y., Tan C.M., Kou Y., et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinreich S.S., Mangon R., Sikkens J.J., Teeuw M.E., Cornel M.C. [Orphanet: a European database for rare diseases] Ned Tijdschr Geneeskd. 2008;152(9):518–519. [PubMed] [Google Scholar]
- 54.Ramos E.M., Hoffman D., Junkins H.A., et al. Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur J Hum Genet. 2014;22(1):144–147. doi: 10.1038/ejhg.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welter D., MacArthur J., Morales J., et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(database issue):D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell J.A., Aronson A.R., Mork J.G., Folk L.C., Humphrey S.M., Ward J.M. Gene indexing: characterization and analysis of NLM‘s GeneRIFs. AMIA Annu Symp Proc. 2003;2003:460–464. [PMC free article] [PubMed] [Google Scholar]
- 57.ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 58.Wishart D.S., Guo A., Oler E., et al. HMDB 5.0: the human metabolome database for 2022. Nucleic Acids Res. 2022;50(D1):D622–D631. doi: 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lachmann A., Torre D., Keenan A.B., et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat Commun. 2018;9(1):1366. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Byrska-Bishop M., Evani U.S., Zhao X., et al. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell. 2022;185(18):3426–3440.e19. doi: 10.1016/j.cell.2022.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang H., Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10(10):1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He Z., Zhang D., Renton A.E., et al. The rare-variant generalized disequilibrium test for association analysis of nuclear and extended pedigrees with application to Alzheimer disease WGS data. Am J Hum Genet. 2017;100(2):193–204. doi: 10.1016/j.ajhg.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Homsy J., Zaidi S., Shen Y., et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350(6265):1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samocha K.E., Robinson E.B., Sanders S.J., et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46(9):944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Timberlake A.T., Furey C.G., Choi J., et al. De novo mutations in inhibitors of Wnt, BMP, and Ras/ERK signaling pathways in non-syndromic midline craniosynostosis. Proc Natl Acad Sci U S A. 2017;114(35):E7341–E7347. doi: 10.1073/pnas.1709255114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schubach M., Maass T., Nazaretyan L., Röner S., Kircher M. CADD v1.7: using protein language models, regulatory CNNs and other nucleotide-level scores to improve genome-wide variant predictions. Nucleic Acids Res. 2024;52(D1):D1143–D1154. doi: 10.1093/nar/gkad989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ioannidis N.M., Rothstein J.H., Pejaver V., et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shah S.P., Roth A., Goya R., et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21(6):974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei Q., Zhan X., Zhong X., et al. A Bayesian framework for de novo mutation calling in parents-offspring trios. Bioinformatics. 2015;31(9):1375–1381. doi: 10.1093/bioinformatics/btu839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Attanasio C., Nord A.S., Zhu Y., et al. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342(6157) doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Q., Ritter D., Yang N., et al. A systematic approach to identify functional motifs within vertebrate developmental enhancers. Dev Biol. 2010;337(2):484–495. doi: 10.1016/j.ydbio.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shochat C., Wang Z., Mo C., et al. Deletion of SREBF1, a functional bone-muscle pleiotropic gene, alters bone density and lipid signaling in zebrafish. Endocrinology. 2021;162(1) doi: 10.1210/endocr/bqaa189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fisher S., Grice E.A., Vinton R.M., et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1(3):1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 76.Jeyasekharan A.D., Liu Y., Hattori H., et al. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat Struct Mol Biol. 2013;20(10):1191–1198. doi: 10.1038/nsmb.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gudmundsdottir K., Lord C.J., Ashworth A. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene. 2007;26(54):7601–7606. doi: 10.1038/sj.onc.1210579. [DOI] [PubMed] [Google Scholar]
- 78.Wilderman A., D’haene E., Baetens M., et al. A distant global control region is essential for normal expression of anterior HOXA genes during mouse and human craniofacial development. Nat Commun. 2024;15(1):136. doi: 10.1038/s41467-023-44506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tackels-Horne D., Toburen A., Sangiorgi E., et al. Split hand/split foot malformation with hearing loss: first report of families linked to the SHFM1 locus in 7q21. Clin Genet. 2001;59(1):28–36. doi: 10.1034/j.1399-0004.2001.590105.x. [DOI] [PubMed] [Google Scholar]
- 80.Elliott A.M., Evans J.A. Genotype-phenotype correlations in mapped split hand foot malformation (SHFM) patients. Am J Med Genet A. 2006;140(13):1419–1427. doi: 10.1002/ajmg.a.31244. [DOI] [PubMed] [Google Scholar]
- 81.Robledo R.F., Rajan L., Li X., Lufkin T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002;16(9):1089–1101. doi: 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heude E., Shaikho S., Ekker M. The dlx5a/dlx6a genes play essential roles in the early development of zebrafish median fin and pectoral structures. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0098505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farmer D.T., Mlcochova H., Zhou Y., et al. The developing mouse coronal suture at single-cell resolution. Nat Commun. 2021;12(1):4797. doi: 10.1038/s41467-021-24917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holleville N., Matéos S., Bontoux M., Bollerot K., Monsoro-Burq A.H. Dlx5 drives Runx2 expression and osteogenic differentiation in developing cranial suture mesenchyme. Dev Biol. 2007;304(2):860–874. doi: 10.1016/j.ydbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 85.Valdés-Flores M., Casas-Avila L., Ponce de León-Suárez V. In: Topics in Osteoporosis. IntechOpen. Valdés-Flores M., editor. 2013. Genetic diseases related with osteoporosis; pp. 29–65. [Google Scholar]
- 86.Merzdorf C.S. Emerging roles for zic genes in early development. Dev Dyn. 2007;236(4):922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- 87.Twigg S.R.F., Forecki J., Goos J.A.C., et al. Gain-of-function mutations in ZIC1 are associated with coronal craniosynostosis and learning disability. Am J Hum Genet. 2015;97(3):378–388. doi: 10.1016/j.ajhg.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elms P., Siggers P., Napper D., Greenfield A., Arkell R. Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev Biol. 2003;264(2):391–406. doi: 10.1016/j.ydbio.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 89.Teslaa J.J., Keller A.N., Nyholm M.K., Grinblat Y. Zebrafish Zic2a and Zic2b regulate neural crest and craniofacial development. Dev Biol. 2013;380(1):73–86. doi: 10.1016/j.ydbio.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohbayashi N., Shibayama M., Kurotaki Y., et al. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16(7):870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khera A.V., Chaffin M., Aragam K.G., et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Medina-Gomez C., Mullin B.H., Chesi A., et al. Bone mineral density loci specific to the skull portray potential pleiotropic effects on craniosynostosis. Commun Biol. 2023;6(1):691. doi: 10.1038/s42003-023-04869-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Naqvi S., Sleyp Y., Hoskens H., et al. Shared heritability of human face and brain shape. Nat Genet. 2021;53(6):830–839. doi: 10.1038/s41588-021-00827-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jabs E.W., Müller U., Li X., et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75(3):443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- 95.Spruijt L., Verdyck P., Van Hul W., Wuyts W., de Die-Smulders C. A novel mutation in the MSX2 gene in a family with foramina parietalia permagna (FPP) Am J Med Genet A. 2005;139(1):45–47. doi: 10.1002/ajmg.a.30923. [DOI] [PubMed] [Google Scholar]
- 96.Wilkie A.O., Tang Z., Elanko N., et al. Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat Genet. 2000;24(4):387–390. doi: 10.1038/74224. [DOI] [PubMed] [Google Scholar]
- 97.Howard T.D., Paznekas W.A., Green E.D., et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15(1):36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 98.Stankiewicz P., Thiele H., Baldermann C., et al. Phenotypic findings due to trisomy 7p15.3-pter including the TWIST locus. Am J Med Genet. 2001;103(1):56–62. doi: 10.1002/ajmg.1512. [DOI] [PubMed] [Google Scholar]
- 99.Maruyama Z., Yoshida C.A., Furuichi T., et al. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev Dyn. 2007;236(7):1876–1890. doi: 10.1002/dvdy.21187. [DOI] [PubMed] [Google Scholar]
- 100.Mefford H.C., Shafer N., Antonacci F., et al. Copy number variation analysis in single-suture craniosynostosis: multiple rare variants including RUNX2 duplication in two cousins with metopic craniosynostosis. Am J Med Genet A. 2010;152A(9):2203–2210. doi: 10.1002/ajmg.a.33557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Desai J., Shannon M.E., Johnson M.D., et al. Nell1-deficient mice have reduced expression of extracellular matrix proteins causing cranial and vertebral defects. Hum Mol Genet. 2006;15(8):1329–1341. doi: 10.1093/hmg/ddl053. [DOI] [PubMed] [Google Scholar]
- 102.Zhang X., Zara J., Siu R.K., Ting K., Soo C. The role of NELL-1, a growth factor associated with craniosynostosis, in promoting bone regeneration. J Dent Res. 2010;89(9):865–878. doi: 10.1177/0022034510376401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu Y.Q., Badano J.L., McCaskill C., Vogel H., Potocki L., Shaffer L.G. Haploinsufficiency of ALX4 as a potential cause of parietal foramina in the 11p11.2 contiguous gene-deletion syndrome. Am J Hum Genet. 2000;67(5):1327–1332. doi: 10.1016/S0002-9297(07)62963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yagnik G., Ghuman A., Kim S., et al. ALX4 gain-of-function mutations in nonsyndromic craniosynostosis. Hum Mutat. 2012;33(12):1626–1629. doi: 10.1002/humu.22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Styrkarsdottir U., Cazier J.B., Kong A., et al. Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLOS Biol. 2003;1(3) doi: 10.1371/journal.pbio.0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sivakumaran S., Agakov F., Theodoratou E., et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89(5):607–618. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Greenwood J., Flodman P., Osann K., Boyadjiev S.A., Kimonis V. Familial incidence and associated symptoms in a population of individuals with nonsyndromic craniosynostosis. Genet Med. 2014;16(4):302–310. doi: 10.1038/gim.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Robin N.H., Scott J.A., Cohen A.R., Goldstein J.A. Nonpenetrance in FGFR3-associated coronal synostosis syndrome. Am J Med Genet. 1998;80(3):296–297. doi: 10.1002/(SICI)1096-8628(19981116)80:3<296::AID-AJMG25>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 109.Birnbaum R.Y., Clowney E.J., Agamy O., et al. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 2012;22(6):1059–1068. doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(SM) collect eighteen supplementary tables (1-18) and 9 supplementary figures (1-9) supporting the results described in the current article. The supplementary tables and figures are all mentioned in the text within the appropriate scientific contents. SM is linked to the online version of the paper at https://www.gimopen.org/
Data Availability Statement
GWAS summary level data that support the findings of this study are available in Supplemental Table 10 and upon request. Requests should be addressed to Dr Inga Peter (inga.peter@mssm.edu). The GS data that support the findings of this study are available in dbGAP with the identifier phs001806.v1.p1. Row genotyping data are in the process of being deposited to dbGAP.