Abstract
Previous studies have suggested that Moloney murine leukemia virus (MoMLV)-based vectors pseudotyped with the vesicular stomatitis virus G glycoprotein (VSV-G) have extensive ability to transduce nonmammalian cells. However, we have identified multiple cell lines from fish (FHM), mosquitoes (Mos-55), moths (Sf9 and High-5), flies (S2), and frogs (XPK2) that are not efficiently transduced by MoMLV-based vectors pseudotyped with many different viral envelope proteins, including VSV-G, while the same vectors are functional in these cells following transfection. A comparison of MoMLV-based vector transduction in mammalian and nonmammalian cells shows that the nonmammalian cells exhibit blocks at either entry, reverse transcription, or integration. Additionally, VSV-G-pseudotyped MoMLV-based vector transduction is attenuated in the zebrafish cell line ZF4 at entry and/or reverse transcription, whereas other transduction processes are unaffected. We show that the variation of transduction by MoMLV-based vectors in mammalian and nonmammalian cells is not due to differences in culture conditions or cell division rate but is likely the result of divergence in cellular factors required for retroviral transduction.
Several cellular factors are essential for infection by gammaretroviruses, a genus that includes murine leukemia viruses (MLVs) and other simple retroviruses. Infected cells express protein receptors which are needed for virus entry, as well as nucleotides and actin filaments that are required for reverse transcription (RT) (4, 10). Other proteins, such as barrier-to-autointegration factor (BAF) and high-mobility group I/Y (HMGI/Y) proteins, play a role in maintaining proviral DNA prior to integration (21, 40) and facilitate integration of the viral preintegration complex (PIC) (17, 23, 24), respectively. During mitosis the viral PIC gains access to the nucleus, where integration occurs (22), a process which is thought to be completed by cellular proteins (43). To better define the cellular factors which mediate retroviral infection, we examined transduction in cell types from organisms which are distantly related to mammals. A genetic approach to studying the processes of retroviral transduction may lead to the identification of cell types that can be used to discover additional host factors required for retrovirus replication.
Retroviral vectors based on gammaretroviruses, particularly those based on Moloney MLV (MoMLV), are useful for delivering heterologous DNA to many mammalian and avian cell types. In addition, previous investigations have indicated that MoMLV-based vectors pseudotyped with the G glycoprotein from vesicular stomatitis virus (VSV-G) can mediate gene transfer in cells from fish (5, 25), newts (6), mosquitoes (26), moths (13), and frogs (7). Since these initial studies, subsequent work has been done only with zebrafish, with which MoMLV-based vectors have been used for insertional mutagenesis in embryos (14). Aside from the work with fish, most studies of retroviral transduction in the other nonmammalian cells used nonquantitative PCR to detect integration events, and therefore gene transfer efficiency was not measured (6, 7, 13, 26). In addition, mainly VSV-G-pseudotyped MoMLV-based vectors have been evaluated for transduction of nonmammalian cells, whereas almost nothing is known about the expression and functionality of receptors for naturally occurring retrovirus envelope glycoproteins in these cell types.
As part of a study to investigate factors involved in receptor-mediated retrovirus entry into cells, we evaluated the susceptibilities of several nonmammalian cell lines to MoMLV-based vectors pseudotyped with various viral envelope proteins, including VSV-G. We were surprised to find that several cell types were not transduced by VSV-G-pseudotyped MoMLV-based vectors, which prompted us to further investigate the behavior of these vectors in nonmammalian cells. By comparing the early steps of retrovirus transduction in canine D17 cells to those in cells from lower-order organisms, we showed that the processes of virus entry, RT, and integration differ between the cell types tested. Most notably, we have observed that mammalian cells are permissive to VSV-G-pseudotyped MoMLV-based vectors, whereas several nonmammalian cells have one or more blocks to transduction by these viruses.
Phosphatidyl serine is thought to be the receptor for VSV-G-mediated virus entry (35, 36). Challenging cells with both VSV-G-pseudotyped retroviral vectors and a recombinant form of VSV that expresses gfp allowed us to evaluate the relative levels of receptor expression in the different cell types. Here we show that the VSV-G envelope poorly facilitates entry of MoMLV-based vectors or VSV cores into most nonmammalian cells in comparison to mammalian cells. Furthermore, several nonmammalian cells which were permissive to virus entry could not support MoMLV-based transduction. Variation in culture conditions did not explain the discrepancy of MoMLV-based transduction observed in the different cells, but rather the results of this study suggest that innate differences between the cell types dictate transduction efficiency. These findings have important implications for identifying cellular factors involved in retroviral transduction, as well as for using retroviral vectors to introduce heterologous DNA sequences into nonmammalian model organisms.
MATERIALS AND METHODS
Cell lines and cell culture.
Anopheles gambiae (Mos-55) (26), Brachydanio rerio (ZF4, ATCC CRL-2050), and Pimephales promelas (FHM, ATCC CCL-42) cells (kind gifts of Jane Burns, University of California at San Diego, La Jolla) were maintained at 26°C in Leibovitz's L-15 medium supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin. Xenopus laevis (XPK2) cells derived from adult tissue (kind gift of Ron Reeder, Fred Hutchinson Cancer Research Center, Seattle, Wash.) were maintained at 26°C in 42% L-15 medium–43% water–15% FBS with l-glutamine, penicillin, and streptomycin. Spodoptera frugiperda cells (Sf9; Invitrogen) were maintained at 26°C in TNM-FH medium (Sigma) supplemented with 10% FBS. Trichoplusia ni cells (High-5; Invitrogen) and Schneider's Drosophila melanogaster cells (S2; ATCC CRL-1963) (kind gift from Keith Fournier, Fred Hutchinson Cancer Research Center) were maintained at 26°C in Excell 400 with l-glutamine (JRH Biosciences). All nonmammalian cells were grown in air without supplemental CO2.
Canine D17 osteosarcoma cells (ATCC CCL-183), adenovirus 5-transformed human embryonal kidney 293 cells (ATCC CRL-1573.1), human HT-1080 cells (ATCC CCL-121), and all retrovirus packaging cells were maintained at 37°C in Dulbecco's minimal essential medium (DMEM) with a high concentration of glucose (4.5 g/liter) supplemented with 10% FBS, penicillin, and streptomycin. Cells were grown in a 5% CO2–air atmosphere.
FHM and D17 cells were adapted to grow at 33°C in a 3% CO2–air atmosphere. The cells were first grown to 60% confluence in 25-ml flasks in their normal culture environment (see above). Once the FHM cells reached 60% confluence, the medium was changed to 30% conditioned medium from FHM cells and 70% fresh DMEM supplemented with 10% FBS, penicillin, and streptomycin. The FHM cells were then passaged two times at 28°C in a 5% CO2–air atmosphere prior to being transferred to 33°C in a 3% CO2–air atmosphere. D17 cells which reached 60% confluence in a 25-ml flask were transferred to an incubator to grow at 33°C in a 3% CO2–air atmosphere. Both cell types were maintained in DMEM in the new culture environment for several weeks before they were used for infection assays.
Retroviral vectors.
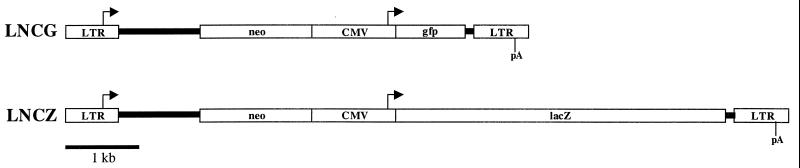
The retroviral vector LNCZ contains a neomycin phosphotransferase (neo) gene under the control of the MoMLV long terminal repeat (LTR) and lacZ under the control of a cytomegalovirus (CMV) immediate-early promoter. The retroviral vector LNCG is identical to LNCZ but contains egfp, which encodes the enhanced green fluorescent protein (GFP) (Clontech), under the control of the CMV promoter. The plasmid constructs pLNCZ and pLNCG were made by inserting either lacZ or egfp into the cloning site of pLNCX (30).
Transfection assay for gene transfer and expression from pLNCZ or pLNCG.
One day prior to transfection, the cells were plated in six-well dishes at 106 (Mos-55), 2 × 105 (S2, High-5, and Sf9), or 105 (ZF4, FHM, and XPK2) cells per well. Mos-55, S2, ZF4, and FHM cells were transfected with 10 μg of either pLNCZ or pLNCG by using calcium phosphate coprecipitation (9). XPK2 cells were transfected with 10 μg of either plasmid using Lipofectin by following the manufacturer's protocol (Life Technologies). High-5 and Sf9 cells were transfected with 4 μg of pLNCZ by using Insectin Plus according to the manufacturer's protocol (Invitrogen). Expression of β-galactosidase was determined 3 days after transfection by fixing cells with 0.5% glutaraldehyde, washing cells three times with phosphate-buffered saline (PBS), and staining for β-galactosidase activity with X-Gal staining solution [8 mM K3Fe(CN)6, 8 mM K4Fe(CN)6 · 3H2O, 2 mM MgCl2, 0.4 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml].
Virus production.
Different packaging lines that all contained the MoMLV gag and pol genes were used to produce the LNCZ vector with envelopes from amphotropic MLV or gibbon ape leukemia virus (GALV) or to produce the LNCG vector with envelopes from Mus dunni endogenous virus (MDEV), 10A1 virus, or RD114 virus. The LNCZ(amphotropic) and LNCZ(GALV) viruses were produced by transducing the PA317 (27) and PG13 (29) packaging lines, respectively, with LNCZ(ecotropic)- virus-containing medium, which was collected 1 day after the transfection of PE501 (30) packaging cells with pLNCZ. The LNCG(MDEV) and LNCG(10A1) viruses were produced by transducing the PD223 (42) and PT67 (28) packaging lines, respectively, with LNCG(ecotropic)-virus-containing medium, which was collected 1 day after the transfection of PE501 packaging cells with pLNCG. The LNCG(RD114) viruses were produced by transducing the FLYRD (11) packaging lines with LNCG(amphotropic)-virus-containing medium which was collected 1 day after the transfection of PA317 packaging cells with pLNCG. All transductions were done in the presence of 4 μg of Polybrene per ml. After 24 h, packaging lines containing LNCZ or LNCG were selected in 700 μg of G418 (active concentration) per ml for 7 to 10 days. Virus-containing media were collected from confluent dishes of vector-producing packaging cells and filtered through a 0.45-μm-pore-size filter. All virus stocks were stored at −70°C.
LNCZ was pseudotyped with the VSV-G glycoprotein by calcium phosphate cotransfection of 293 cells at 50% confluence in a 15-cm dish with 5 μg of pVSV-G, 10 μg of pJK3, 2 μg of pCMVtat, and 15 μg of pLNCZ per dish (2). pVSV-G encodes the G protein, and pJK3 contains MoMLV gag and pol under the control of the human immunodeficiency virus type 1 (HIV-1) LTR promoter (2). pCMVtat encodes the HIV-1 Tat protein, which is necessary for transactivation of the HIV-1 LTR promoter of pJK3 (2). Fifteen hours posttransfection, fresh medium was added to the cells, and virus-containing medium was collected every 12 h and filtered. Virus stocks were stored at −70°C. Virus titers were determined by exposing D17 cells to different amounts of virus, staining the cells for β-galactosidase, as described above, and scoring β-galactosidase-positive foci 2 days after virus exposure.
Infection of cells with different pseudotypes of LNCZ or LNCG.
Virus titers were determined by seeding cells in six-well plates at the following densities: 105 cells/well for D17 and HT-1080 cells; 5 × 105 cells/well for ZF4, FHM, XPK2, High-5, and Sf9 cells; and 106 cells/well for Mos-55 cells. After 24 h, the medium was changed to fresh medium containing 4 μg of Polybrene per ml and then the cells were inoculated with different dilutions of virus. Infected cells were incubated in their normal culture environment for 3 days. Afterwards, cells receiving MoMLV-based LNCZ vectors were washed once in PBS, fixed for 5 min in 0.5% glutaraldehyde, rinsed three times with PBS, and stained overnight at 37°C for β-galactosidase activity. Cells inoculated with virus having the LNCG vector were observed under a fluorescence microscope, and GFP+ foci were scored.
Production of LNCZ(VSV-G) for quantitative PCR.
LNCZ(VSV-G) was produced from a monoclonal population of 293 cells containing one copy of pLNCZ known to have the XbaI site only in the 3′ LTR. To generate these cells, 293 cells were transfected with 5 μg of pLNCZ using the calcium phosphate method and were subsequently selected in 700 μg of G418 (active concentration) per ml for 10 days. Clonal populations were first screened for β-galactosidase activity and then by Southern analysis for the presence of the XbaI site in the 3′ LTR of the vector. Clone 12 (293/LNCZ-c12) had the desired properties and was transfected with pVSV-G, pJK3, and pCMVtat for production of LNCZ(VSV-G) as described above. Virus titers were determined by exposing D17 cells to different amounts of virus and scoring positive foci of cells expressing the marker proteins.
Quantification of RT products.
One day before infection, cells were plated in six-well dishes at 105 (D17, ZF4, XPK2, and FHM) or 106 (Mos-55) cells per well. Cells were exposed to LNCZ(VSV-G) at a multiplicity of infection (MOI) of 0.03. All infections were done in the presence of 4 μg of Polybrene per ml, whereas mock-infected cells received medium containing Polybrene only. Eighteen hours postinfection, the cells were trypsinized, collected, and rinsed in PBS. Fractions containing Hirt DNA and genomic DNA were obtained using the Hirt extraction method (18). Hirt DNA was resuspended in 25 μl of water, and 2 μl was used for PCR amplification in a total reaction volume of 100 μl. Hirt DNA was amplified for 35 rounds using primers U3a (5′ AGTTCAGATCAAGGTCAGG3′) and PSIa (5′TTAGGGTGTACAAAGGGC3′). For competitive PCR, Hirt DNA was amplified in the presence of 0, 0.1, 1, 10, 100, or 1,000 copies of a competitor (pLNCZ). Ten microliters of product from each PCR was digested with 5 U of XbaI at 37°C for 2 h. Samples were analyzed by gel electrophoresis on 1% agarose and were transferred to nylon membranes (Hybond-N; Amersham) for Southern analysis. Membranes were probed with a 677-bp Asp718-to-BstEII fragment of the LNCZ vector, which was purified with the QiaexII DNA purification kit (Qiagen). The 677-bp fragment hybridizes to R and U5 of the LTR, as well as to a portion of the packaging signal within the LNCZ vector.
Doubling times of cells.
All cell types were plated and cultured under the same conditions as were used for infection. Cells were seeded in six-well plates at the following densities: 105 cells/well for D17 and HT1080 cells; 5 × 105 cells/well for ZF4, FHM, XPK2, High-5, and Sf9 cells; and 106 cells/well for Mos-55 cells. Cells were trypsinized and counted every 24 h for 4 days. Doubling times were determined between days 2 and 4, the times during which the cells were exposed to virus in other experiments.
Exogenous RT assays.
Each reaction mixture contained 250 μl of RT cocktail {50 mM Tris (pH 7.8), 60 mM NaCl, 2 mM dithiothreitol, 0.6 mM MnCl2, 0.05% NP-40, 10 μM dTTP, 12.5 μg of poly(A)(dTTP)10 (Sigma), and 2 μl of 800-μCi/mol [32P]dTTP in a final volume of 1,000 μl}. Two hundred fifty microliters of cocktail was combined with 5 μl of LNCG(RD114), which had a titer of 105 focus-forming units (FFU)/ml in HT-1080 cells, or filtered conditioned medium from HT-1080 cells, the parental cell line used to make the LNCG(RD114) packaging cells. Reaction mixtures were incubated in water baths at 37 or 26°C. Three 10-μl samples from each reaction mixture were taken every 15 min, blotted onto DE-81 paper, and dried for 5 min at room temperature. Blots were washed four times at room temperature in saline sodium citrate for 5 min followed by two 5-min washes in 95% ethanol. Blots were dried and suspended in scintillation fluid for determining [32P]dTTP incorporation.
Endogenous RT assays.
Ten microliters of LNCG(RD114) or conditioned medium from the FLYRD packaging line was added to a 60-μl reaction mixture containing 50 mM Tris(pH 8.3), 15 mM dithiothreitol, 7 mM magnesium acetate, 0.01% NP-40, and 8 mM (each) dTTP, dATP, dGTP, and dCTP. Samples were incubated for 20 h, and aliquots were stored at −20°C until used for PCR amplification. Samples were amplified with primers PSIa and U3a (described above) in the presence of a pLNCZ competitor ranging from 102 to 107 molecules and pUC18 for a final DNA concentration of 1 pg/μl. Fifteen microliters of each sample was analyzed by 1% agarose gel electrophoresis and ethidium bromide staining.
RESULTS
The CMV immediate-early promoter and either β-galactosidase or GFP reporters work best to measure transduction in nonmammalian cells.
We assayed β-galactosidase, human placental alkaline phosphatase (AP), and GFP expression from several promoters to determine the best promoter and reporter proteins for use in the nonmammalian cells used in this study. Sf9, S2, and High-5 cells were transfected with expression constructs containing either LacZ or AP cDNA under the control of one of the following promoters: the CMV promoter, the Drosophila armadillo promoter (Arm), or the MoMLV LTR promoter. FHM, ZF4, Mos-55, and XPK2 cells were transfected with the same constructs except those containing the Arm promoter. Reporter expression was evaluated 3 days posttransfection by staining cells for β-galactosidase or AP activity. We found that the MoMLV LTR promoter functioned only in the FHM cells, whereas the CMV promoter drove the expression of β-galactosidase in all cell types tested. The Arm promoter also worked in a few cell lines (Sf9, S2, and High-5), but the marker gene activity in most cells was higher with the CMV promoter (data not shown).
Some cell lines (XPK2, ZF4, High-5, Sf9, and S2) had relatively high levels of endogenous, heat-resistant AP activity that masked detection of activity from the transferred AP gene. Since all cells transfected with the β-galactosidase marker were easy to score, we constructed a new retroviral vector (LNCZ) that contained lacZ under the control of the CMV promoter (Fig. 1). Expression of the reporter from both pLNCZ and a similar plasmid encoding GFP (pLNCG) was subsequently tested in most of the cell types used in this study (Fig. 1; Table 1). Bright foci were seen in all of the cell lines transfected with either plasmid, although some cell lines had fewer foci than others, suggesting poor transfection efficiency (Table 1). These results show that β-galactosidase and/or GFP reporters from these vectors are expressed in all cell types tested, making them good markers for the assay of retroviral transduction.
FIG. 1.
Diagram of MoMLV based retroviral vectors. Both LNCG and LNCZ contain retroviral LTRs, which drive the expression of the neomycin phosphotransferase (neo) gene and contain the polyadenylation signal (pA). The CMV promoter drives the expression of lacZ and gfp in LNCZ and LNCG, respectively.
TABLE 1.
Most nonmammalian cells tested were resistant to transduction by MoMLV-based vectors but susceptible to transfection by vector plasmids
| Cell line | Transfection by plasmida
|
Transduction by vectorb:
|
Infection by VSV-GFPb | ||||||
|---|---|---|---|---|---|---|---|---|---|
| pLNCZ | pLNCG | LNCG(10A1) | LNCZ(amphotropic) | LNCZ(GALV) | LNCG(MDEV) | LNCG(RD114) | LNCZ(VSV-G) | ||
| High-5 | ++ | ND | <2 | <1 | <1 | <2 | <2 | <10 | NA |
| SF9 | ++ | ND | <2 | <1 | <1 | <2 | <2 | <10 | NA |
| S2 | + | ND | <2 | <1 | <1 | <2 | <2 | <10 | ND |
| XPK2 | + | + | <2 | <1 | <1 | <2 | <2 | <10 | <200 |
| FHM | ++ | ++ | <2 | <1 | <1 | <1 | ND | <10 | 9 × 106 |
| ZF4 | + | + | <2 | <1 | 2 × 103 | <1 | 1 × 104 | 5 × 104 | 7 × 104 |
| Mos-55 | +++ | +++ | <2 | <1 | <1 | <1 | ND | <10 | 2 × 109 |
| D17 | ND | ND | 1 × 105 | 1 × 105 | 1 × 105 | ND | ND | 6 × 106 | 2 × 109 |
| HT-1080 | ND | ND | ND | ND | ND | 1 × 104 | 1 × 106 | ND | ND |
Percentages of GFP- or β-galactosidase-positive cells were determined 2 to 4 days after transfection. Results are means of two experiments. +, <1% but >0%; ++, 1 to 10%; +++, >10%; ND, not done.
Foci of GFP- or β-galactosidase-positive cells were counted 2 to 4 days after infection, and results are expressed as FFU per milliliter of virus added to the cells. Results are means of two to four experiments. ND, not done; NA, not applicable (gfp expression was difficult to distinguish due to the autofluorescence of the cells).
Of the nonmammalian cell lines tested, only one is transduced by MoMLV-based vectors.
To determine whether retroviral vectors could transduce nonmammalian cells, we challenged the cells with LNCZ or LNCG vectors pseudotyped with various viral envelopes that have relatively broad host ranges in mammalian cell lines. The MoMLV-based vector LNCZ was pseudotyped with either the amphotropic, GALV, or VSV-G envelope, and LNCG was pseudotyped with either the MLV 10A1, MDEV, or RD114 envelope. Nonmammalian cells were infected at MOIs ranging from 0.1 to 100, and titers were compared to those found with mammalian D17 and HT-1080 cells (Table 1).
All of the cells tested were resistant to transduction by the MoMLV-based vectors except for the ZF4 cells, which were permissive to transduction by vectors pseudotyped with either the GALV, RD114, or VSV-G envelope. However, these viruses consistently had lower titers in ZF4 cells than in D17 or HT-1080 cells (Table 1).
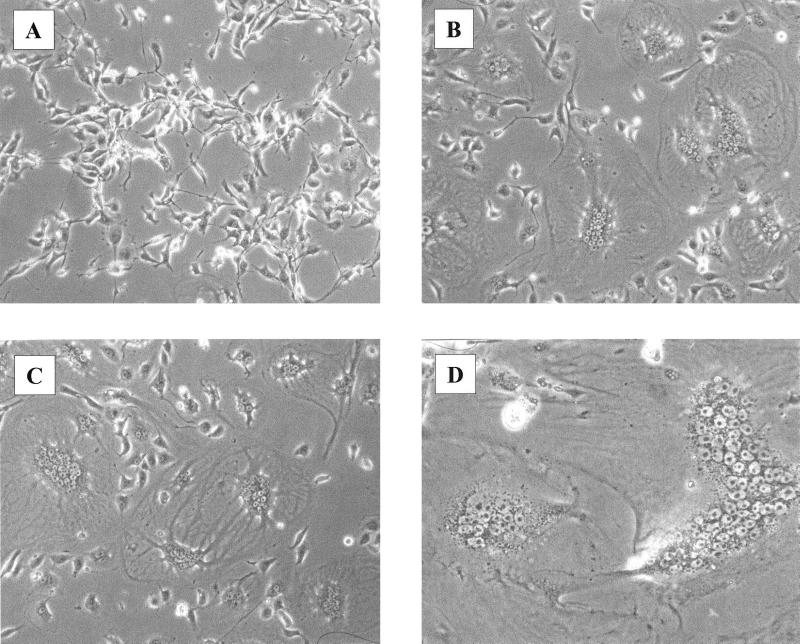
Unlike ZF4 cells, the other nonmammalian cells were not transduced by LNCZ or LNCG vectors pseudotyped with any envelope. However, exposure of the Xenopus cell type XPK2 to vectors having either the GALV or amphotropic pseudotypes produced remarkable syncytium formation in the cell monolayer (Fig. 2). Several other viral envelopes did not have this effect on XPK2 cells, suggesting that these cells specifically express high levels of the Pit1 (32) and Pit2 (31) receptors for GALV and the amphotropic virus, respectively. Despite the presence of virus-induced syncytia in XPK2 cells, the cells were not transduced by any vector pseudotyped with these two envelopes. The general lack of transduction by vectors having pseudotypes with a broad host range, including the pantropic VSV-G pseudotype, indicates that these cells may have blocks to MoMLV-based transduction that occur subsequent to virus entry.
FIG. 2.
Syncytium formation in frog cells exposed to MoMLV-based vectors pseudotyped with either GALV or amphotropic envelope. XPK2 cells were treated with 4 μg of Polybrene per ml with no additions (A) or with 104 FFU of LNCZ(PG13) (B) or LNCZ(PA317) (C). Panel D shows a syncytium with ∼75 nuclei. Photos were taken 24 h after virus exposure.
With the exception of XPK2 cells, all of the nonmammalian cell types tested are susceptible to infection by recombinant VSV.
The lack of transduction by VSV-G-pseudotyped vectors of so many nonmammalian cell types prompted us to investigate whether the VSV receptor was expressed in these cells. To address this we utilized a recombinant form of VSV which has the gfp gene inserted between the G (glycoprotein) and L (large subunit of RNA polymerase) genes of wild-type VSV. The normal gene order of VSV is 3′-N-P-M-G-L-5′, with expression controlled at the level of transcription by the viral RNA-dependent RNA polymerase (1, 21). VSV carries its RNA polymerase into the cytoplasm of cells, where it subsequently transcribes the single-stranded RNA genome in the absence of de novo protein synthesis. Because VSV replicates in most cell types, the expression of GFP by recombinant VSV (VSV-GFP) is a good marker for virus entry.
VSV-GFP could transduce all cells tested except XPK2 (Table 1). Equivalent titers of VSV-GFP were observed in Mos-55 and D17 cells, but the titers in other cells were reduced by as much as 7 orders of magnitude (Table 1). These data show that all but one of the nonmammalian cell lines express functional receptors for VSV-G and support the hypothesis that these cells have postentry blocks to transduction by VSV-G-pseudotyped MoMLV-based vectors.
Nonmammalian cells have less RT product than D17 cells after infection with LNCZ(VSV-G).
Although we observed entry of VSV-GFP in most of the nonmammalian cell types, it was still uncertain whether retroviral vectors pseudotyped with VSV-G could enter the cells. Detection of late products of RT within infected cells can be used as a marker for retrovirus entry (38). Strong-stop DNA is the first minus-strand DNA made during RT and is commonly found within incoming virions. However, late RT products such as full-length minus- and plus-strand DNAs are made only in newly infected cells. Therefore, we designed a PCR protocol that specifically identifies full-length RT products in cells infected with LNCZ(VSV-G) as follows (Fig. 3).
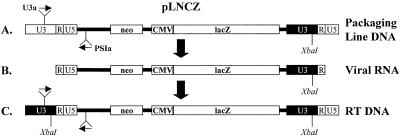
FIG. 3.
Schematic representation of the assay used to detect and quantify RT products from infected cells. The packaging line contains plasmid DNA from the transfection of 293 cells with pLNCZ. After transcription of the pLNCZ vector, the viral RNA contains an XbaI restriction site in the U3 region of the 3′LTR. RT of the LNCZ vector copies the XbaI restriction site to the 5′LTR in the double-stranded DNA. Small arrows depict the location of primers used to amplify RT products and competitor plasmid DNA. The two products could be distinguished by restriction analysis with XbaI followed by gel electrophoresis.
During RT the U3 region of the 3′ LTR is copied to the 5′ LTR of the provirus DNA (15). The LNCZ vector contains an XbaI restriction site in the 3′ MoMLV LTR but not in the 5′ Moloney murine sarcoma virus LTR (Fig. 3B). Strand transfer steps which occur during RT of LNCZ produce a full-length RT product with an XbaI restriction site in the U3 region of the 5′ LTR (Fig. 3C). This site is distinguished from plasmid DNA which lacks the XbaI site in the same region of the provirus (Fig. 3A). Therefore, PCR amplification of this region followed by digestion with XbaI allowed us to detect and quantify the amount of RT product in cells infected with LNCZ(VSV-G) (Fig. 4A).
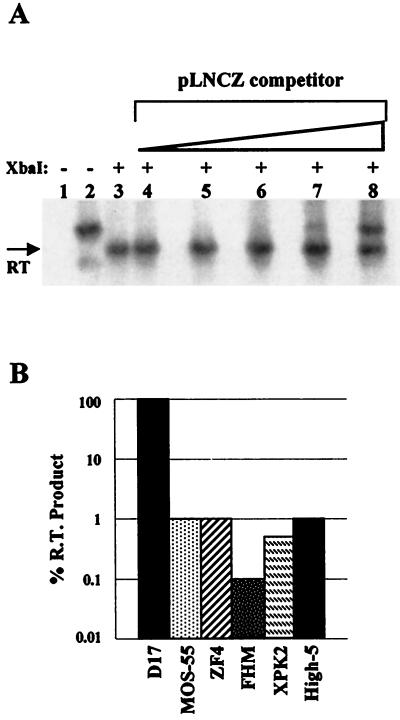
FIG. 4.
Quantification of RT products in cells infected with LNCZ(VSVG). (A) Analysis of D17 cells subjected to quantitative competitive PCR followed by Southern analysis. Lanes: 1 and 2, undigested PCR-amplified Hirt DNA from uninfected cells and cells infected with LNCZ(VSVG), respectively; 3 to 8, samples of Hirt DNA from infected cells which has been PCR amplified in the presence of 0, 0.1, 1, 10, 100, or 1,000 copies respectively, of competitor pLNCZ plasmid followed by digestion with XbaI. (B) Relative percentages of RT products found in nonmammalian cells normalized to D17 cells. Percentages are based on an average of two experiments. Variation between experiments was <5-fold.
Hirt DNA was obtained from cells infected with LNCZ(VSV-G) produced from 293 cells containing a single nonrearranged LNCZ provirus. Hirt DNA was PCR amplified, digested with XbaI, and subjected to Southern analysis. RT products were detected in all cells exposed to LNCZ(VSV-G) but in amounts which were 100- to 1,000-fold lower than those found in D17 cells (Fig. 4B). Using the same assay, viral stocks were analyzed for full-length RT products but none were detected (data not shown), indicating that all RT products were made after virion entry into cells. These data confirm that all nonmammalian cells tested can support both entry and RT of MoMLV-based vectors pseudotyped with VSV-G and that blocks to transduction in these cells occur after these processes. Furthermore, VSV-GFP infected Mos-55 and D17 cells equivalently, but the amount of RT product in Mos-55 cells infected with LNCZ(VSV-G) was 100-fold lower than the amount of RT product found in D17 cells infected with an equivalent MOI (Table 2; Fig. 4B). The VSV-G envelope promotes efficient virus entry in both D17 and Mos-55 cells; therefore, the difference between transduction with LNCZ(VSV-G) and infection with VSV-GFP in these two cell types is likely due to an inhibition which occurs before or during RT and prior to integration but is not a block at virus entry.
TABLE 2.
Nonmammalian cells divide at rates which are sufficient to promote nuclear entry of the MoMLV PIC
| Cell line | Doubling time (h)a | Efficiency of MoMLV-based vector transduction (% relative to that of D17 cells)b | RT product (% relative to that of D17 cells)c |
|---|---|---|---|
| D17 | 30 | 100 | 100 |
| ZF4 | 40 | 1 | 1 |
| FHM | 29 | <0.001 | 0.1 |
| High-5 | 36 | <0.001 | 1 |
| Mos-55 | 32 | <0.001 | 1 |
| XPK2 | 48 | <0.001 | 0.75 |
| Sf9 | 60 | <0.001 | ND |
Cells were plated into multiple dishes. Duplicate plates were trypsinized, and the cells were counted after treatment with trypsin every 24 h for 4 days. The doubling times were determined between days 2 and 4, the period in which the cells were exposed to virus for transduction assays.
Transduction by MoMLV-based vectors pseudotyped with either the GALV, RD114, or VSV-G envelope proteins.
Detected after exposure to MoMLV-based vectors. ND, not done.
The temperature at which nonmammalian cells are grown does not influence the RDDP activity of MoMLV reverse transcriptase in vitro.
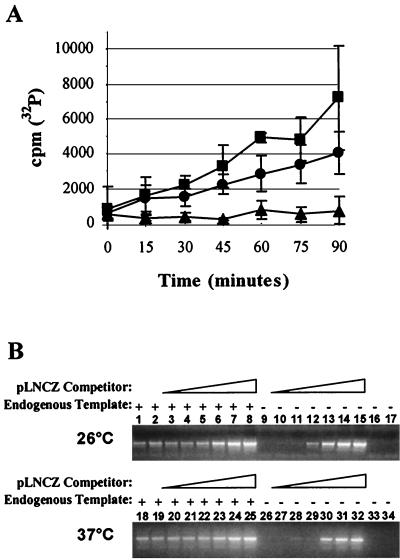
To investigate the postentry attenuation to RT in the nonmammalian cells, we determined whether culture temperatures could account for the low levels of RT products found in these cells. Therefore, we examined the RNA-dependent DNA polymerase (RDDP) activity of MoMLV reverse transcriptase at two different temperatures at which nonmammalian and D17 cells are grown, 26 and 37°C, respectively. Exogenous RDDP assays were performed using medium containing LNCG(RD114) as a source of MoMLV reverse transcriptase. Background activity was assessed with conditioned medium from HT-1080 cells, the parental cell type used to produce the FLYRD retrovirus packaging cells. We found that the RDDP activity of MoMLV reverse transcriptase was reduced by <2-fold at 26°C compared to the activity at 37°C (Fig. 5A). However, the small difference in RDDP activity at the two temperatures does not explain why nonmammalian and D17 cells infected with LNCZ(VSV-G) have 100- to 1,000-fold differences in the total amount of RT product.
FIG. 5.
Temperature effect on MoMLV reverse transcriptase. (A) Exogenous RT assay to assess RDDP activity. Incorporation of [32P]dTTP into a poly(A)(dTTP)12 template-primer mixture by MoMLV reverse transcriptase at 26°C (●) or 37°C (■) was measured. Incorporation in HT-1080 conditioned medium at 37°C (▴) served as a control. (B) Endogenous RT assay to assess complete activity of MoMLV reverse transcriptase at 26 and 37°C. DNAs from the endogenous RT assays done at the two different temperatures were used as templates for PCR amplification of the LTR-ψ sequence without nonspecific (pUC18) DNA (lanes 1 and 18), with nonspecific DNA only (lanes 2 and 19), or in the presence of 102 to 107 copies of pLNCZ competitor and nonspecific DNA (lanes 3 to 8 and 20 to 25). Amplification of the competitor only (lanes 10 to 15 and 27 to 32), nonspecific DNA only (lanes 16 and 33), no template (lanes 9 and 26), and conditioned medium from mock producer cells (lanes 17 and 34) served as negative controls. The assay was repeated under the same conditions to confirm the results (data not shown).
DDDP activity and other processes of MoMLV RT are comparable at 26 and 37°C.
To investigate if MoMLV reverse transcriptase could efficiently complete strand transfer, strand displacement, and DNA-dependent DNA polymerase (DDDP) activities at lower temperatures, we performed endogenous RT assays at 26 and 37°C. Endogenous RT assays differ from exogenous RT assays in that the former are performed within permeabilized viruses and utilize the viral RNA as a template for RT (33, 39). Endogenous RT assays were done with LNCG(RD114) or conditioned medium from the FLYRD cell-based packaging line not containing a retroviral vector. A portion of each sample was subjected to PCR amplification in the presence of 102 to 107 molecules of plasmid DNA (pLNCZ) and nonspecific DNA (pUC18) to obtain an equal amount of DNA in each reaction mixture. By comparing amplified samples having no competitor (Fig. 5B, lanes 2 and 19) to those with competitor (Fig. 5B, lanes 3 to 8 and 20 to 25), we were able to determine the relative amounts of total RT products found in each endogenous reaction performed at either 26 or 37°C. We found that at both temperatures there were approximately 105 molecules of full-length RT product, thus indicating that RT does not significantly differ between 26 and 37°C.
Temperatures at which FHM cells grow do not influence MoMLV-based transduction in vivo.
To evaluate whether another step in transduction was affected by temperature, we adapted D17 cells and FHM cells to grow under identical culture conditions. Both cell types were adapted to grow at 33°C in a 3% CO2–air atmosphere and were later challenged with LNCZ(VSV-G). Titers were at least 5 orders of magnitude higher in D17 cells than in FHM cells. The lack of transduction by LNCZ(VSV-G) in FHM cells grown under the same culture conditions as D17 cells indicates that FHM cells lack cellular factors required for transduction by LNCZ(VSV-G) or that these cells contain an inhibitor of MoMLV-based vector transduction. These findings are supported by data which show that these vectors enter and undergo RT but fail to transduce the cells.
The lack of transduction by MoMLV-based vectors in nonmammalian cells is not due to a low rate of cell division at the time of infection.
Because the MoMLV PIC enters the nucleus only during mitosis, we wanted to determine whether the division rates of the nonmammalian cells might be lower than those of mammalian cells, thereby reducing the frequency of mitosis and thus the rate of transduction by MoMLV-based vectors. Therefore, we evaluated the doubling time of each cell line at the time of infection (Table 2). All cell types were plated and cultured under the same conditions as used for infection. We determined the doubling time for each cell type by counting cells every 24 h after plating. The FHM, High-5, and Mos-55 cells had shorter doubling times than the permissive ZF4 cells, yet none were permissive to transduction by MoMLV-based vectors. XPK2 cells divided at a rate which was slightly lower than that of ZF4 cells, whereas Sf9 cells divided at a markedly lower rate. Therefore, it is possible that the low division rate of Sf9 cells affects the transduction of these cells by MoMLV-based vectors. These data show that RT products found in most of the nonmammalian cell types 24 h posttransduction (Table 2; Fig. 4B) should have access to the nucleus and indicate that these cells likely have blocks to transduction by MoMLV-based vectors that are independent of nuclear entry.
DISCUSSION
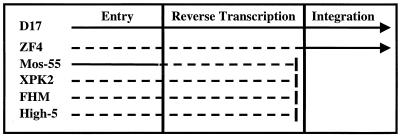
Here we report that several nonmammalian cell types exhibit blocks to MoMLV-based vector transduction. Many of the nonmammalian cell types tested were not transduced by MoMLV-based vectors pseudotyped with pantropic envelopes. The striking contrast between the transduction efficiency observed in mammalian and nonmammalian cells led us to investigate at what step MoMLV-based transduction was inhibited or blocked. Our data, summarized in Fig. 6, show that some nonmammalian cells have various postentry blocks or attenuations to MoMLV-based transduction during or prior to RT and at the step of integration. The differences in growth temperatures do not account for the relatively low levels of RT products found in the nonmammalian cells tested or for the failure of vector integration and lack of reporter expression in FHM cells. Interestingly, many nonmammalian cells that contain RT products after exposure to viruses containing MoMLV-based vectors are not permissive to transduction by these vectors even though they divide at rates which are comparable to those of permissive cells. In general, we found that in several nonmammalian cell types MoMLV-based transduction is blocked or attenuated at multiple early steps required for the expression of heterologous DNA from an MoMLV-based vector.
FIG. 6.
Schematic representation of blocks to MoMLV-based transduction in various cell types. Arrows show the procession of transduction through the three main steps. Solid lines represent uninterrupted events, and dashed lines represent disrupted events. Bars indicate a complete block to transduction after RT. D17 cells are the cells to which all of the other cell types were compared. ZF4 cells contain 100-fold fewer RT products than do D17 cells, and the titers of LNCZ(VSV-G) are also 100-fold lower than those of D17 cells. Mos-55 cells have 100-fold fewer RT products than do D17 cells, but the entry of LNCZ(VSV-G) is comparable in the two cell types as assessed by VSV-GFP titers. Furthermore, the RT products in Mos-55 cells do not lead to transduction events. XPK2, FHM, and High-5 cells contain 2 to 3 orders of magnitude fewer RT products than do D17 cells, and the RT products do not lead to transduction events.
It was unexpected that most of the nonmammalian cells could not be transduced by MoMLV-based vectors pseudotyped with envelopes from viruses which have broad host ranges. Earlier work has shown that MoMLV-based vectors pseudotyped with the MDEV envelope can efficiently transduce species such as the rat, hamster, quail, cat, dog, human, and mouse (3). Previously, there were no reports of a cell line which MDEV vectors could not transduce. Likewise, VSV-G-pseudotyped viruses have been referred to as pantropic because the receptor is thought to be a phospholipid, a component of most cellular membranes. Therefore, we were surprised to find that several nonmammalian cells were not transduced by MoMLV-based vectors pseudotyped with MDEV or VSV-G viral envelopes, even though these vectors are expressed when transfected into the same nonmammalian cells. Similarly, we expected to observe transduction in XPK2 cells infected with either LNCZ(GALV) or LNCZ(amphotropic) because these viruses caused dramatic syncytium formation shortly after infection, and yet they were negative for β-galactosidase expression. It remains unclear if High-5, Sf9, S2, FHM, and Mos-55 cells express the receptors for MDEV, 10A1 virus, amphotropic virus, and GALV.
The identification of de novo RT products in the nonmammalian cells infected with LNCZ(VSV-G) confirmed that the pseudotyped viruses entered the cells. However, the efficiency of virus entry in some of the cell types is not known because some of the cells were poorly infected by both LNCZ(VSV-G) and VSV-GFP. For example, the XPK2 cells did not support the replication of VSV-GFP, but entry was established by the detection of RT products after infection with LNCZ(VSV-G). Therefore, it is possible that the relatively low levels of RT products in some of the cell types can be ascribed to poor virus entry. On the other hand, infection with VSV-GFP showed that a few of the nonmammalian cell types should be very permissive to vectors pseudotyped with VSV-G envelopes. In agreement with previous studies that have demonstrated productive infection of wild-type VSV in mosquitoes (37), we found that Mos-55 cells were extremely permissive to VSV-GFP. Remarkably, the same cells were not transduced by MoMLV-based vectors pseudotyped with VSV-G, thus demonstrating that transduction is blocked for reasons other than inefficient virus entry. In contrast to our results, one study has demonstrated integration of MoMLV-based vectors pseudotyped with VSV-G in Mos-55 cells (26). However, in that study nested PCR identified only two integrated events from a pool of cells which were infected at an MOI of 0.3. These findings suggest that integration of MoMLV-based vectors in Mos-55 cells is extremely rare at best. In our study, we used colorimetric titering assays to measure transduction in Mos-55 cells and did not find any such events even when infections were done at an MOI of >5 (data not shown). Regardless, we were able to detect entry of either LNCZ(VSV-G) or VSV-GFP in the nonmammalian cell types, and we hypothesize that the lack of transduction by LNCZ(VSV-G) or LNCG(VSV-G) must be attributable to postentry blocks.
The dramatic variation between MoMLV-based transduction in mammalian and nonmammalian cells is not due to differences in culture conditions. Previous reports indicated that the optimal activity of MoMLV reverse transcriptase occurs at temperatures between 39 and 41°C (34). We found the RDDP activity of MoMLV reverse transcriptase to be only slightly greater at 37°C than at 26°C. Consistent with another report by Whiting and Champoux (41), we found the processes of strand transfer and strand displacement and the DDDP activity of reverse transcriptase to be comparable at 37 and 26°C. Therefore, the relatively low levels of RT products found in the nonmammalian cells were not due to differences in culture temperatures. Along these lines, MoMLV-based transduction in FHM cells could not be rescued by adapting the cells to grow at temperatures which are sufficient for transduction in mammalian cells. Therefore, other reasons, such as a deficiency in specific cellular factors or the presence of an inhibitory factor, account for the lack of transduction in the nonmammalian cells.
Cellular factors which are required for early events in MoMLV replication may be absent in the nonmammalian cells tested in this study. MoMLV enzymatic proteins are sufficient to produce full-length double-stranded DNA products in vitro, but it is unclear if other factors are required in vivo (12, 15). Previous studies of HIV-1 show a role for casein kinase II (16), the HIV-1 coreceptor (8), actin filaments, and possibly actin-binding proteins in the early steps of RT (4). Since little is known about the intracellular composition of the MoMLV RT machinery, it is unclear if host factors other than nucleotides are needed to produce a full-length cDNA from incoming viral RNA. The MoMLV PIC includes BAF, a cellular factor which binds retroviral cDNA at a relatively late time during RT (17). Although a function for BAF in RT has not been identified, it is known that BAF's association with the MoMLV PIC prevents autointegration (21) and may play an indirect role in creating a proper intasome structure necessary for integration of viral DNA into the host cell genome (40). Interestingly, a mutant of BAF that cannot bind DNA restores the integration activity of salt-stripped HIV-1 PICs in vitro, suggesting that BAF interacts with other components of the viral PIC in addition to DNA (17). BAF's interaction with HIV-1 PICs may be through association with other cellular factors which might also be present in MoMLV PICs. Another group has used reconstituted salt-stripped MoMLV PICs for in vitro intermolecular integration assays and established the need for the host factor HMGI/Y in retroviral integration (23). However, in that study extracts from NIH 3T3 cells were able to complement salt-stripped PICs better than purified HMGI/Y, suggesting that additional cellular factors may play a role in MoMLV integration.
With so many cellular factors involved in MoMLV-based transduction, it would perhaps be unlikely that nonmammalian cells contain all of the mammalian homologs which are required for the process or that they would contain them in sufficient quantities. For example, poor MoMLV-based transduction efficiencies in several zebrafish cell types (data not shown) could be the result of insufficient quantities of cellular factors. In agreement with our results, other groups found that titers of VSV-G-pseudotyped MoMLV-based vectors are 100-fold lower in zebrafish cells than in hamster cells (5) and proposed that the difference may be due to the dissimilarity of host factors within the two cell types (20). Likewise, species-specific postentry blocks have been reported for human and primate immunodeficiency viruses in various mammalian cell types (19). Alternatively, we cannot rule out an inhibitory mechanism which would prevent MoMLV-based transduction in many of the nonmammalian cells which displayed a complete block to transduction. In either case, the identification of postentry blocks to transduction in the nonmammalian cells provides a method for identifying cellular factors involved in the early steps of MoMLV replication. Currently, we are attempting to rescue MoMLV-based transduction in FHM cells by providing these cells with chromosomes from zebrafish cells which are transduced by these vectors. The many cell lines displaying different postentry blocks to transduction by MoMLV-based vectors create new opportunities for studying retroviral infection.
ACKNOWLEDGMENTS
We thank Marie Vodicka for her suggestions and comments on the manuscript; Jane Burns for the Mos-55, FHM, and ZF4 cells; Ron Reeder for the XPK2 cells; Keith Fournier for the S2 cells; Jack Rose for recombinant VSV; Michael Emerman for the Sf9 and High-5 cells; and Becca Gottschalk and Neal Van Hoeven for LNCG.
This work was supported by NIH training grant GM07270, distributed through the Molecular and Cellular Biology Program at the University of Washington, and NIH grant HL54881.
REFERENCES
- 1.Ball L A, Pringle C R, Flanagan B, Perepelitsa V P, Wertz G W. Phenotypic consequences of rearranging the P, M, and G genes of vesicular stomatitis virus. J Virol. 1999;73:4705–4712. doi: 10.1128/jvi.73.6.4705-4712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartz S R, Vodicka M A. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 3.Bonham L, Wolgamot G, Miller A D. Molecular cloning of Mus dunni endogenous virus: an unusual retrovirus in a new murine viral interference group with a wide host range. J Virol. 1997;71:4663–4670. doi: 10.1128/jvi.71.6.4663-4670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinskaya A, Brichacek B, Mann A, Stevenson M. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J Exp Med. 1998;188:2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns J C, Matsubara T, Lozinski G, Yee J K, Friedmann T, Washabaugh C H, Tsonis P A. Pantropic retroviral vector-mediated gene transfer, integration, and expression in cultured newt limb cells. Dev Biol. 1994;165:285–289. doi: 10.1006/dbio.1994.1253. [DOI] [PubMed] [Google Scholar]
- 7.Burns J C, McNeill L, Shimizu C, Matsubara T, Yee J K, Friedmann T, Kurdi-Haidar B, Maliwat E, Holt C E. Retroviral gene transfer in Xenopus cell lines and embryos. In Vitro Cell Dev Biol Anim. 1996;32:78–84. doi: 10.1007/BF02723038. [DOI] [PubMed] [Google Scholar]
- 8.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 11.Cosset F-L, Takeuchi Y, Battini J-L, Weiss R A, Collins M K L. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fassati A, Goff S P. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J Virol. 1999;73:8919–8925. doi: 10.1128/jvi.73.11.8919-8925.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franco M, Rogers M E, Shimizu C, Shike H, Vogt R G, Burns J C. Infection of lepidoptera with a pseudotyped retroviral vector. Insect Biochem Mol Biol. 1998;28:819–825. doi: 10.1016/s0965-1748(98)00056-3. [DOI] [PubMed] [Google Scholar]
- 14.Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature. 1996;383:829–832. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 15.Gilboa E, Mitra S W, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 16.Harada S, Maekawa T, Haneda E, Morikawa Y, Nagata N, Ohtsuki K. Biochemical characterization of recombinant HIV-1 reverse transcriptase (rRT) as a glycyrrhizin-binding protein and the CK-II-mediated stimulation of rRT activity potently inhibited by glycyrrhetinic acid derivative. Biol Pharm Bull. 1998;21:1282–1285. doi: 10.1248/bpb.21.1282. [DOI] [PubMed] [Google Scholar]
- 17.Harris D, Engelman A. Both the structure and DNA binding function of the barrier-to-autointegration factor contribute to reconstitution of HIV type 1 integration in vitro. J Biol Chem. 2000;275:39671–39677. doi: 10.1074/jbc.M002626200. [DOI] [PubMed] [Google Scholar]
- 18.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins N. High titers of retrovirus (vesicular stomatitis virus) pseudotypes, at last. Proc Natl Acad Sci USA. 1993;90:8759–8760. doi: 10.1073/pnas.90.19.8759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M S, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis P F, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Farnet C M, Anderson W F, Bushman F D. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J Virol. 1998;72:2125–2131. doi: 10.1128/jvi.72.3.2125-2131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Yoder K, Hansen M S T, Olvera J, Miller M D, Bushman F D. Retroviral cDNA integration: stimulation by HMG I family proteins. J Virol. 2000;74:10965–10974. doi: 10.1128/jvi.74.23.10965-10974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin S, Gaiano N, Culp P, Burns J C, Friedmann T, Yee J K, Hopkins N. Integration and germ-line transmission of a pseudotyped retroviral vector in zebrafish. Science. 1994;265:666–669. doi: 10.1126/science.8036514. [DOI] [PubMed] [Google Scholar]
- 26.Matsubara T, Beeman R W, Shike H, Besansky N J, Mukabayire O, Higgs S, James A A, Burns J C. Pantropic retroviral vectors integrate and express in cells of the malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci USA. 1996;93:6181–6185. doi: 10.1073/pnas.93.12.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller A D, Chen F. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J Virol. 1996;70:5564–5571. doi: 10.1128/jvi.70.8.5564-5571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–982. , 984–986, 989–990. [PMC free article] [PubMed] [Google Scholar]
- 31.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Sass P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 33.Rothenberg E, Baltimore D. Increased length of DNA made by virions of murine leukemia virus at limiting magnesium ion concentration. J Virol. 1977;21:168–178. doi: 10.1128/jvi.21.1.168-178.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothenberg E, Smotkin D, Baltimore D, Weinberg R A. In vitro synthesis of infectious DNA of murine leukaemia virus. Nature. 1977;269:122–126. doi: 10.1038/269122a0. [DOI] [PubMed] [Google Scholar]
- 35.Schlegel R, Tralka T S, Willingham M C, Pastan I. Inhibition of VSV binding and infectivity by phosphatidylserine: is phosphatidylserine a VSV-binding site? Cell. 1983;32:639–646. doi: 10.1016/0092-8674(83)90483-x. [DOI] [PubMed] [Google Scholar]
- 36.Schlegel R, Willingham M C, Pastan I H. Saturable binding sites for vesicular stomatitis virus on the surface of Vero cells. J Virol. 1982;43:871–875. doi: 10.1128/jvi.43.3.871-875.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schloemer R H, Wagner R R. Mosquito cells infected with vesicular stomatitis virus yield unsialylated virions of low infectivity. J Virol. 1975;15:1029–1032. doi: 10.1128/jvi.15.4.1029-1032.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma I M. Studies on reverse transcriptase of RNA tumor viruses. III. Properties of purified Moloney murine leukemia virus DNA polymerase and associated RNase H. J Virol. 1975;15:843–854. doi: 10.1128/jvi.15.4.843-854.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei S Q, Mizuuchi K, Craigie R. Footprints on the viral DNA ends in moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whiting S H, Champoux J J. Properties of strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase: mechanistic implications. J Mol Biol. 1998;278:559–577. doi: 10.1006/jmbi.1998.1720. [DOI] [PubMed] [Google Scholar]
- 42.Wolgamot G, Rasko J E J, Miller A D. Retrovirus packaging cells expressing the Mus dunni endogenous virus envelope facilitate transduction of CHO and primary hematopoietic cells. J Virol. 1998;72:10242–10245. doi: 10.1128/jvi.72.12.10242-10245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoder K E, Bushman F D. Repair of gaps in retroviral DNA integration intermediates. J Virol. 2000;74:11191–11200. doi: 10.1128/jvi.74.23.11191-11200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]